Summary
COVID-19-associated acute kidney injury (COVID-AKI) is a common complication of SARS-CoV-2 infection in hospitalized patients. The susceptibility of human kidneys to direct SARS-CoV-2 infection and modulation of the renin-angiotensin II signaling (RAS) pathway by viral infection remain poorly characterized. Using induced pluripotent stem cell-derived kidney organoids, SARS-CoV-1, SARS-CoV-2, and MERS-CoV tropism, defined by the paired expression of a host receptor (ACE2, NRP1 or DPP4) and protease (TMPRSS2, TMPRSS4, FURIN, CTSB or CTSL), was identified primarily among proximal tubule cells. Losartan, an angiotensin II receptor blocker being tested in patients with COVID-19, inhibited angiotensin II-mediated internalization of ACE2, upregulated interferon-stimulated genes (IFITM1 and BST2) known to restrict viral entry, and attenuated the infection of proximal tubule cells by SARS-CoV-2. Our work highlights the susceptibility of proximal tubule cells to SARS-CoV-2 and reveals a putative protective role for RAS inhibitors during SARS-CoV-2 infection.
Subject areas: Nephrology, Virology, Cell biology
Graphical abstract

Highlights
-
•
SARS-CoV-2 kidney organoid tropism is primarily among proximal tubule cells
-
•
Losartan attenuates angiotensin II-mediated ACE2 internalization
-
•
Losartan upregulates viral restrictive genes IFITM1 and BST2
-
•
SARS-CoV-2 infection is enhanced by angiotensin II and attenuated by losartan
Nephrology; Virology; Cell biology
Introduction
COVID-19-associated acute kidney injuries (COVID-AKI) are frequent complications of SARS-CoV-2 infection. Throughout the COVID-19 pandemic, the incidence of COVID-AKI ranged from one-third to one-half of hospitalized patients varying across geographical regions with a declining incidence as the pandemic progressed (Chan et al., 2021; Dellepiane et al., 2021; Hirsch et al., 2020). This complication portends a poor prognosis as approximately 35–50% die compared to 8% among those without AKI (Chan et al., 2021; Hirsch et al., 2020). The pathophysiology of COVID-AKI is thought to involve a wide range of local and systemic inflammatory responses typical of the multiorgan failure seen in critically ill patients (Legrand et al., 2021). Interestingly, the predominant kidney biopsy finding in ICU cohorts was acute tubular injury whereas glomerular pathologies such as collapsing glomerulopathy and focal segmental glomerulosclerosis were more prevalent in non-critically ill patients (Batlle et al., 2020; Ferlicot et al., 2021; Hanley et al., 2020; Santoriello et al., 2020). The diverse histopathological patterns of kidney injury suggest that a multitude of pathologic processes may be responsible for the development of COVID-AKI in SARS-CoV-2 infected individuals (Legrand et al., 2021).
Whether or not kidney injury occurs from direct viral toxicity remains unresolved (Batlle et al., 2020). Angiotensin-converting enzyme 2 (ACE2) was the first SARS-CoV-2 host receptor identified; a carboxypeptidase found in human kidney tubular epithelial cells responsible for counter-regulating the renin-angiotensin signaling (RAS) pathway by enzymatically deactivating angiotensin II (Ang II). Mounting evidence suggests that ACE2 facilitates SARS-CoV-1 and SARS-CoV-2 entry and disease (Donoghue et al., 2000; Forrester et al., 2018; Hoffmann et al., 2020b; Li et al., 2003). Indeed, postmortem analyses have identified coronavirus-like particles, SARS-CoV-2 protein, and SARS-CoV-2 RNA in kidney proximal tubular and glomerular epithelial cells (Braun et al., 2020; Puelles et al., 2020; Su et al., 2020; Varga et al., 2020; Werion et al., 2020). Moreover, SARS-CoV-2 RNA was more frequently observed in biopsy specimens obtained from patients with COVID-19 with AKIs compared with those without (72 vs 43%) (Braun et al., 2020). Nonetheless, some reports have questioned the validity of these findings, particularly the non-specific appearance findings in electron micrographs and the paucity of viral RNA in multiple postmortem analyses and urine specimens (Miller and Goldsmith, 2020; Kudose et al., 2020; Rossi et al., 2020; Santoriello et al., 2020; Wölfel et al., 2020). Whether SARS-CoV-2 can infect and replicate within human kidney cells and contribute to the pathogenesis of COVID-AKI remains unclear.
Early in the pandemic, the discovery that ACE2 was SARS-CoV-2's primary entry receptor raised significant concerns around the continued use of ACEI (ACE inhibitors) and angiotensin II receptor blockers (ARBs) such as losartan (South et al., 2020). There were concerns that ACEI and ARBs could increase the pathogenicity of SARS-CoV-2 (Vaduganathan et al., 2020). These two classes of drugs are the mainstay of therapy for many cardiovascular and kidney disorders characterized by chronic and maladaptive RAS activation (Brenner et al., 2001; Carey, 2015; Griffin and Bidani, 2006; Kobori et al., 2007; Lewis et al., 1993, 2001). Their therapeutic effects are derived from the downregulation of Ang II-mediated AT1R signaling and upregulation of ACE2 expression and activity (Ferrario et al., 2005; Furuhashi et al., 2015; Ishiyama et al., 2004; Klimas et al., 2015; Soler et al., 2009). Fortunately, multiple retrospective studies have shown that ACEI and ARBs are not associated with higher rates of COVID-19 diagnosis, disease severity, or mortality (Fosbøl et al., 2020; Jung et al., 2020; Li et al., 2020; Lopes et al., 2021; Mehta et al., 2020; Reynolds et al., 2020). In fact, several studies have reported a lower risk of infection and all-cause mortality associated with RAS inhibitors (Baral et al., 2021; Guo et al., 2020; Hippisley-Cox et al., 2020; Zhang et al., 2020), although the mechanism for the decreased risk remains unclear. Therefore, several randomized control trials are pending to evaluate the RAS inhibitors on outcomes of COVID-19 infections (Cohen et al., 2021; Lopes et al., 2021; Puskarich et al., 2021; Russell et al., 2020).
Human-induced pluripotent stem cell (iPSC)-derived organoids have gained significant traction for modeling COVID-19 pathogenesis (Clevers, 2020). Multiple groups have demonstrated that SARS-CoV-2 can infect iPSC-derived kidney organoids (Monteil et al., 2020; Wysocki et al., 2021). To better understand the susceptibility of kidney cell types to coronavirus infection and the impact of RAS inhibition on this susceptibility, we performed an in-depth receptor expression analysis for SARS-CoV-1, SARS-CoV-2, and MERS-CoV in human iPSC-derived kidney organoids using single-cell RNA sequencing (scRNA-seq). We examined the impact of Ang II and losartan on the infectivity of SARS-CoV-2 to present evidence supporting the potential use of losartan as adjunctive therapy for SARS-CoV-2.
Results
Human-induced pluripotent stem cell -derived kidney organoids express coronavirus entry receptors
To investigate SARS-CoV-2's kidney tropism, we generated iPSC-derived human kidney organoids with minor modifications from the protocol used by Takasato et al. (Takasato et al., 2016). Immunofluorescence staining of our kidney organoids was positive for LTL (Lotus tetragonolobuslectin; proximal tubular cells), ECAD (E-cadherin; distal tubular cells), NPHS1 (nephrin; podocytes), and PDGFR-β (platelet-derived growth factor-beta; mesangial and stromal cells) (Figure 1). Unsupervised clustering of pooled scRNA-seq data containing 21,592 cells that met quality control gating revealed 21 transcriptionally distinct cell clusters and annotated as described in the literature (Figure S1) (Combes et al., 2019; Tran et al., 2019). We regrouped clusters as proximal tubular cells (PTCs), distal tubule cells (DTCs), podocytes, stromal cells, endothelial cells, and five off-target cells including muscle cells, neuronal cells, melanocyte and two unknown populations (UK1, and UK2) (Figure 1B). Cluster-specific violin plots show the five most enriched genes across the five kidney-specific cell types (Figure 1C; Table S1). We compared our scRNA-seq dataset with one generated from SARS-CoV-2 infectable kidney organoids and observed significant overlap between organoid preparations after the normalization and integration of datasets (Figure 1D; Pearson correlation coefficient [PCC] = 0.926) (Monteil et al., 2020).
Figure 1.
Cellular diversity of human iPSC-derived kidney organoids
(A) Immunofluorescence staining showing the major nephron structures generated in kidney tubules (LTL, Lotus tetragonolobusLectin, fluorescein – proximal tubules; ECAD, E-cadherin – distal tubules; NPHS1, nephrin – podocytes and PDGFR-β, platelet-derived growth factor-beta – stromal and mesangial cells). Scale bars, 50 µm.
(B) UMAP (Uniform Manifold Approximation and Projection) plot grouping 10 identified clusters annotated by querying canonical marker genes.
(C) Stacked violin plot of the top 5 marker genes (S, stromal; E, endothelial cells; DTC, distal tubular cells; PTC, proximal tubular cells and Pod, podocytes (Wilcoxon rank-sum test, adjusted p value < 0.001).
(D) UMAP projection of 4,222 randomly sampled cells from infectable human kidney organoids integrated with 4,222 cells from our control kidney organoids using a mutual nearest neighbor-based approach. UK1 and UK2 denote unknown one and 2 clusters, respectively.
We focused our analysis on experimentally established host receptors and proteases for three highly pathogenic human beta-coronaviruses SARS-CoV-1 (Glowacka et al., 2011; Li et al., 2003; Simmons et al., 2005), SARS-CoV-2 (Cantuti-Castelvetri et al., 2020; Daly et al., 2020; Hoffmann et al., 2020a, 2020b; Shang et al., 2020; Zang et al., 2020) and MERS-CoV (Gierer et al., 2013; Millet and Whittaker, 2014; Raj et al., 2013). Previously described host receptors: ACE2, NRP1 (neuropilin 1), DPP4 (dipeptidyl-peptidase-4), and protease activators: FURIN, TMPRSS2 (transmembrane serine protease 2), TMPRSS4 (transmembrane serine protease 4), CTSB (cathepsin B), and CTSL (cathepsin L) are shown in Figure 2A. Coronavirus receptors or activating proteases were expressed in either a tubular restricted fashion (ACE2, TMPRSS2, and TMPRSS4) or across all kidney-specific cell types (NRP1, DDP4, FURIN, CTSB, and CTSL) in our organoids and SARS-CoV-2 infectable organoids (Figure 2B). Taken together, iPSC-derived kidney organoids generate multicellular three-dimensional nephron-like structures that reproducibly express coronavirus entry receptors.
Figure 2.
Coronavirus tropism analysis in kidney organoids reveal PTC susceptibility to infection
(A) Graphical illustration of the host receptor and proteases required by SARS-CoV-1, SARS-CoV-2, and MERS-CoV for cell host entry.
(B) Violin plots for the relative expression of coronavirus entry receptors compared across the major kidney cell-types from scRNA-seq analysis for control organoids used in this study and from Monteil et al. S, stromal; E, endothelial cells; DTC, distal tubular cells; PTC, proximal tubular cells; and GEC, glomerular epithelial cells.
(C) Graphical representation of coronavirus (CoV) Spike protein (S protein) i) receptor binding to the host receptor, ii) proteolytic activation, and iii) cell entry after activation by a protease.
(D) Feature plots of cells expressing the eight coronavirus entry receptors across the five major kidney cell types. Red dots = positive cell. CoV-1, SARS-CoV-1; CoV-2, SARS-CoV-2 and MERS, MERS-CoV.
(E) Percentage of cells predicted to be susceptible to infection determined by co-expression of at least one compatible host receptor/protease pair. SARS-CoV-1: ACE2 + TMPRSS2 or CTSB or CTSL. SARS-CoV-2: NRP1 + FURIN or ACE2 + TMPRSS2 or TMPRSS4 or CTSB or CTSL. MERS-CoV: DPP4 and FURIN or TMPRSS2 or CTSB or CTSL. S, stromal; E, endothelial cells; DTC, distal tubular cells; PTC, proximal tubular cells; Pod – podocytes.
(F) Immunoperoxidase staining for the eight coronavirus entry receptors in human kidneys from the Human Protein Atlas (http://www.proteinatlas.org).
Analysis of coronavirus kidney tropism in kidney organoids
Tissue tropism for SARS-CoV-2 can be determined by ACE2 and TMPRSS2 expression but a broader repertoire of receptors or proteases may facilitate viral entry into kidney cells. A complex cascade of molecular triggers orchestrates virus-cell fusion and entry (Figure 2C). First, virion attachment to the host receptor is mediated by the Spike (S) protein. The S protein is then cleaved at two sites by a host protease before or during endocytosis to facilitate viral-cell membrane fusion (Hulswit et al., 2016). The SARS-CoV-1 S protein can bind to ACE2 or NRP1 and is proteolytically cleaved by TMPRSS2, CTSB, or CTSL for ACE2 or FURIN for NRP1 (Glowacka et al., 2011; Li et al., 2003; Simmons et al., 2005) (Hoffmann et al., 2020b). MERS-CoV S protein binds to DPP4 after proteolytic activation by TMPRSS2, FURIN, CTSB, or CTSL (Gierer et al., 2013; Millet and Whittaker, 2014; Raj et al., 2013). To map the potential susceptibility to coronavirus infection, we identified kidney organoid cells with a receptor-protease permutation conducive for viral entry (Figures 2D and 2E). PTCs expressed ACE2, NRP1, and DPP4 in greatest proportion: 9.7%, 48 and 73%, respectively (Figure 2D). Very few DTCs expressed ACE2 (0.3%) relative to NRP1 (15.2%) and DPP4 (48.0%) (Figure 2D). ACE2 expression was negligible in glomerular and interstitial cells. NRP1 was the predominant host receptor among glomerular and interstitial cells: podocytes (34.0%), stromal cells (23.0%), and endothelial cells (66.6%). DPP4 was expressed in 15.0% of podocytes and 8.9% of endothelial cells (Figure 2D). The greatest proportion of cells with a receptor-protease pair conducive for SARS-CoV-2 entry was within the PTC and endothelial cell clusters (18.7 vs 36.6% expressed NRP1 + FURIN or ACE2 + TMPRSS2 or TMPRSS4 or CTSB or CTSL) (Figure 2E). With 6.5-fold fewer endothelial cells, PTCs represented 76.9% of SARS-CoV-2 potentially susceptible cells. Predicted SARS-CoV-1 susceptibility was also found among PTCs (9.3% expressed ACE2 + TMPRSS2 or CTSB or CTSL). Interestingly, MERS-CoV potential susceptibility was the most common cell type among PTC and DTC clusters (64.9 vs 38.3% expressed DPP4 + FURIN or TMPRSS2 or CTSB or CTSL) owing to diffuse DPP4 and CTSB/L co-expression (Figure 2E). MERS virions have only been reported in tubular epithelial cells (Alsaad et al., 2018). Protein analysis of coronavirus receptors as reported by the Human Protein Atlas generally corroborated our scRNA-seq data (Figure 2F) (Uhlén et al., 2015).
Intracellular host factors involved in RNA processing, protein translation, and innate immune signaling also contribute to viral infection and replication. Therefore, we expanded our analysis of SARS-CoV-2 kidney tropism by generating twenty-six gene sets containing human proteins known to interact with SARS-CoV-2 proteins, the viral-host interactome (Gordon et al., 2020) and scored them across the major kidney cell clusters (Figure S2 and Table S2). Relative to other cells, PTCs were enriched for mRNA whose protein products interact with SARS-CoV-2 proteins involved cell entry (S), viral packaging (M), suppressing the interferon response (Orf9b and Orf9c) (Jiang et al., 2020), and viral replication (Nsp1, two and 4) (Schubert et al., 2020). Endothelial cells were also enriched with mRNA whose protein products interact with SARS-CoV-2 proteins involved in apoptosis (Orf3a) (Ren et al., 2020), suppressing interferon signaling (Orf6) (Miorin et al., 2020) and viral RNA processing (Nsp7 and 10) (Krafcikova et al., 2020). Overall, our tropism analysis reaffirms the PTC’s susceptibility to coronavirus infection.
Renin-angiotensin II signaling modulation of coronavirus susceptibility factors
To determine the impact of RAS modulation on viral entry, we evaluated coronavirus receptor and activating protease expression in the presence of Ang II, losartan, or Ang II with losartan. Losartan had no measurable impact on ACE2, NRP1, DPP4, TMPRSS2/4, FURIN, and CTSB/L in the organoid cell clusters when compared to vehicle treated (Figure S3A). This finding corroborates transcriptional analyses of human airway cells in patients with SARS-CoV-2 treated with ACE inhibitors or ARBs (Trump et al., 2021). In DTCs, Ang II downregulated NRP1 and FURIN in endothelial cells, DPP4 in PTCs, and TMPRSS4. Co-treatment with losartan normalized NRP1, FURIN, DPP4, and TMPRSS4 expression in respective cells (Wilcoxon signed-rank test; p = 0.01; Figure S3A). The viral-host interactome genes were unperturbed by RAS modulation (Figure S3B).
In murine models, RAS inhibitors upregulate surface ACE2 expression by inhibiting Ang II-mediated ACE2 internalization (Deshotels et al., 2014). To the best of our knowledge, this has not been documented in human kidney tubular cells. ACE2 was primarily found on the cell membrane of PTCs colocalizing with LRP2 (megalin), a tubular-specific receptor expressed exclusively on the apical membrane (Figure 3A). We observed a robust downregulation of luminal ACE2 in Ang II-treated organoids (Figure 3B). The fraction of LRP2 signal overlapping with ACE2 was one-third that of vehicle control (p < 0.001; Figures 3B and 3C). There was greater ACE2 signal within the cytoplasm after Ang II treatment consistent with ACE2 internalization (Figure 3B). Ang II treatment with losartan (A + L) showed a trend toward a higher colocalization ACE2 with LRP2 compared to Ang II alone but did not reach statistical significance. Losartan alone had no statistically significant impact on ACE2 and LRP2 colocalization (ANOVA Tukey post hoc test, vehicle vs losartan, p = 1.0, Figures 3B and 3C). Total ACE2 levels were unchanged in human kidney organoids and human tubular epithelial cells (hTECs) in response to Ang II or losartan (Figure 3D). This corroborates observations made in human sinonasal tracts; ciliary ACE2 levels were unchanged in patients taking ACE inhibitors or ARBs (Lee et al., 2020). Taken together, acute modulation of RAS had little effect on the transcription of coronavirus receptors in human PTCs. The most dramatic effect was Ang II-mediated intracellular trafficking of ACE2 in an AT1R dependent fashion.
Figure 3.
Losartan ameliorates Ang II-mediated internalization of ACE2 in PTCs
(A) Immunofluorescence probing for ACE2 (red) in kidney organoids (n = 2) co-stained with the proximal tubular maker LRP2 (green) and Hoechst 33342 (white) for nuclei. White arrowheads indicate ACE2 localized to LTL-labeled cell membrane. Scale bars, 25 μm.
(B) Immunofluorescence of ACE2 (red) and LRP2 (green) colocalization in kidney organoids (n =2 per group) treated with DMSO vehicle, Ang II (1 μM), Ang II + losartan (100 μM), and losartan alone for 24 h. Ang II induces cell surface ACE2 receptor internalization, which is reduced by losartan. Hoechst 33342 (white) was used to stain nuclei. Dashed line indicates the outline of tubules. Scale bars, 25 μm.
(C) Quantification of ACE2 and LRP2 colocalization in treated organoids using Mander's coefficient method on ImageJ with JACoP plugin (mean ± SD, ∗p < 0.05, ∗∗∗p < 0.001, ns - non-significant, n = 14–26 areas per treatment group). Data were analyzed using ANOVA with Tukey post hoc test.
(D) Representative immunoblots for ACE1 and ACE2 proteins in kidney organoid and tubular epithelial cells treated with DMSO vehicle, Ang II, Ang II + losartan, and losartan for 24 hr. n = two independent experiments. Abbreviations: V, vehicle; A, Ang II; L, losartan; A + L, angiotensin II + losartan.
Losartan upregulated viral restrictive genes in proximal tubular cells
Viral-specific RNA sequences engage host antiviral mechanisms by activating hundreds of IFN-stimulated genes (ISGs) that promote a state of viral resistance (Lazear et al., 2019). Severe COVID-19 presentations are believed to be a result of blunted or delayed IFN responses (Blanco-Melo et al., 2020; Hadjadj et al., 2020). We wondered whether losartan modulated ISG expression. 354 human ISGs were curated and measured across the main kidney organoid cells (Schoggins et al., 2011). Under control conditions, each cell type had a distinct ISG module (Figure 4A). Endothelial cells, podocytes, and PTCs were the highest expressors of ISGs. Interestingly, losartan upregulated two ISGs, in podocytes and PTCs, known to restrict coronavirus infection: BST2 (CD317/tethrin) and IFITM1 (Figure 4B). BST2 inhibits the cellular release of hCoV-229E and SARS-CoV-1 by tethering the budding virions to the host cell membrane (Taylor et al., 2015). IFITM1 inhibits SARS-CoV-2 cell entry and is downregulated in critically ill patients with COVID-19 (Buchrieser et al., 2020; Hadjadj et al., 2020). In our kidney organoids, IFITM1 and BST2 were expressed in ACE2+ PTCs and upregulated by losartan (Figures 4B and 4C). In human kidney tissue, IFITM1 and BST2 were expressed in glomeruli whilst BST2 was also expressed in tubular cells (Figure 4D). Taken together, the upregulation of antiviral ISGs such as BST2 and IFITM1 by losartan may contribute to reduced susceptibility of SARS-CoV-2 infection in PTCs.
Figure 4.
Losartan upregulates viral restrictive genes IFITM1 and BST2 in PTCs
(A) Violin plots ranking the average module scores for interferon-stimulated genes (ISG) across five organoid cell clusters.
(B) Volcano plots of Wilcox rank-sum determined differentially expressed genes in podocytes, PTCs, and DTCs from kidney organoids treated with losartan. ISGs are highlighted in red and non-ISGs in gray.
(C) Feature plots of IFITM1 and BST2 (green) co-expression with ACE2 (red) in PTCs.
(D) Immunoperoxidase staining of human kidneys for IFITM1 and BST2 obtained from the Human Protein Atlas.
Losartan diminished the number of ACE2+ tubular cells infected by SARS-CoV-2
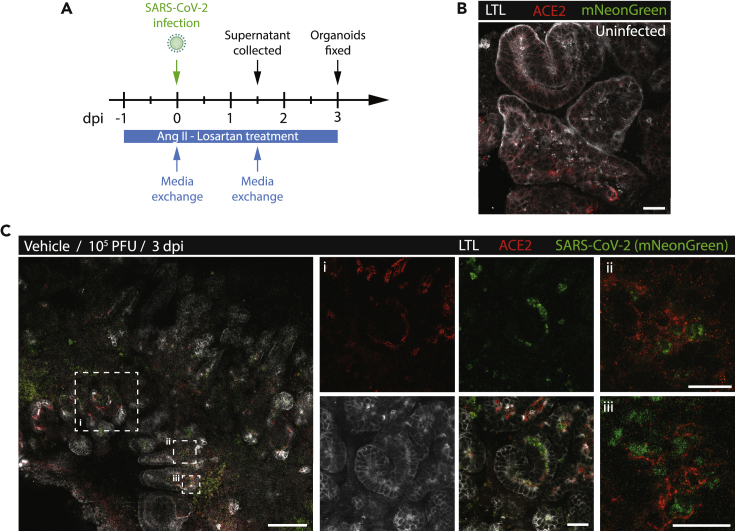
Next, we performed live virus experiments in kidney organoids to examine the impact of Ang II and losartan on PTC susceptibility to SARS-CoV-2 infection. The SARS-CoV-2 infectious clone that expressed the reporter gene mNeonGreen was used for these experiments to help visualize infected cells (Xie et al., 2020). Organoids were pretreated with vehicle, Ang II, losartan, or Ang II with losartan and then exposed to 103 or 105 plaque-forming units (PFU)/mL of virus after 24 h (Figure 5A). Supernatant collected 1.5 days postinfection (dpi) was reserved to assess viral titers and organoids were fixed at three dpi for confocal imaging (Figure 5A). At three dpi we observed robust mNeonGreen signal in LTL+ PTCs (Figures 5C and S4) which was absent in uninfected organoids (Figures 5B andS4). As expected, a significant proportion of mNeonGreen was observed within the cytoplasm of ACE2+ PTCs (Figure 5C, insets ii and iii). Remarkably, PTC susceptibility to SARS-CoV-2 infection was sensitive to RAS modulation (Figures 6A–6C). By three dpi for kidney organoids exposed to 103 PFU/mL SARS-CoV-2, Ang II significantly increased the number mNeonGreen+ LTL+ cells compared to vehicle controls (6.1 for Ang II vs 3.2 for control mNeonGreen+/total LTL+ cells, p = 0.006) (Figures 6A and 6B). Losartan alone did not diminish the number mNeonGreen+ LTL+ cells from control (2.5 vs 3.2, p = 0.844) but in combination with Ang II treatment, it significantly blunted Ang II promotion of SARS-CoV-2 infectivity (6.1 for Ang II vs 3.8 for Ang II + losartan, p = 0.045) (Figure 6B). Kidney organoids exposed to 105 PFU/mL three dpi showed similar results to 103 PFU/mL infected kidney organoids with a trend toward higher number of infected cells with Ang II (Figures 6C and S4). We performed tissue culture infectious dose (TCID50) assays using reserved culture supernatant. Kidney organoids produced infectious SARS-CoV-2 when treated with vehicle in comparison to uninfected controls confirming susceptibility of kidney cells to infection (Figure 6D). The median TCID50 of supernatant collected from losartan-treated organoids showed fewer SARS-CoV-2 infected cells with higher concentrations of losartan in a dose-dependent manner (Figure 6D). Taken together, our findings suggest that PTCs are susceptible and permissive to SARS-CoV-2 infection and this susceptibility depends on AT1R signaling. Our data indicate that treatment with losartan may provide a protective advantage in COVID-AKI.
Figure 5.
SARS-CoV-2 infection of ACE2+ PTCs in kidney organoids
(A) Schematic of the experimental plan for treating and infecting kidney organoids. CoV2, SARS-CoV-2.
(B) Immunofluorescence of control uninfected kidney organoids stained with LTL (white) and ACE2. Uninfected kidney organoid shows the absence of SARS-CoV-2+ signal (green). Scale bar, 25μm.
(C) Immunofluorescence of ACE2 (red), LTL (white) vehicle control organoids 3 days postinfection (dpi) with 105 PFU (plaque-forming units)/mL of SARS-CoV-2 expressing mNeonGreen. Scale bars, 100 μm (left panel) and 25 μm (insets). Panels i, ii, and iii are magnified views of hatched boxes from left panel showing that robust cytoplasmic expressions of SARS-CoV-2 (green) within ACE2+ (red) cells.
Figure 6.
SARS-CoV-2 infection enhanced by Ang II and attenuated by losartan in PTCs
(A) Immunofluorescence staining of LTL (white, proximal tubular cells), NPHS1 (red, glomerulus), and endogenous SARS-CoV-2 (mNeonGreen) in the infected kidney organoids (103 PFU/mL – three dpi) treated with vehicle, Ang II, Ang II with losartan and losartan alone. Images are representative of two independent infection experiments. Scale bar, 50 μm.
(B and C) Quantification of 103 PFU/mL and (C) 105 PFU/mL of SARS-CoV-2 infected proximal tubular cells (LTL+mNeonGreen+) per total proximal tubular cells (LTL+), expressed as a percentage, per field of view across four treatment groups (mean ± SE, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, n = 10 field of views per treatment group). Data were analyzed using one-way ANOVA with Tukey post hoc test.
(D) Tissue Culture Infectious Dose (TCID50) assay for viral titer analysis using supernatant from kidney organoids after treatment with losartan at the indicated concentrations collected at 1.5 dpi. (mean ± SE, ∗p < 0.05, ∗∗p < 0.01). Kidney organoids plated at 500,000 cells (500K; open circles) and 250,000 cells (250K, closed circles). Data are analyzed using one-way ANOVA with Fisher's LSD post hoc test.
Discussion
In this study, we use human kidney organoids to understand the kidney tropism and susceptibility to SARS-CoV-2 in response to the modulation of the RAS pathway. We use scRNA-sequencing to perform a comprehensive expression analysis of coronavirus receptors and protease which revealed that PTCs have high co-expression of several potential host receptor and protease pairs. Using immunofluorescence staining of kidney organoids, we provide evidence for altered ACE2 expression and localization in response treatment with Ang II and losartan. We show a reduction of apical membrane expression of ACE2 with Ang II treatment in PTCs that was blunted by losartan. Using live virus experiments, we show SARS-CoV-2 enrichment at the apical membrane of PTCs providing evidence for potential susceptibility to SARS-CoV-2 infection. Overall, our results demonstrate enhanced susceptibility to SARS-CoV-2 infection by Ang II in PTCs which is attenuated by losartan supporting a potential therapeutic use for losartan to inhibit SARS-CoV-2 infection.
Before our study, kidney organoids were shown to be susceptible to SARS-CoV-2 infection but the kidney cells targeted for viral infection were not clearly established (Monteil et al., 2020; Wysocki et al., 2021). In this study, we used scRNA-sequencing of iPSC-derived kidney organoids to perform in silico analysis of SARS-CoV-2 receptor and protease expression to identify kidney cells susceptible to SARS-CoV-1, SARS-CoV-2, and MERS-CoV. Our scRNA-sequencing experiments show that PTCs are most susceptible to infection by beta-coronaviruses based on the expression of surface and intracellular host factors. Our results show that PTCs of kidney organoids express ACE2 and TMPRSS2 supporting the possibility of SARS-CoV-2 entry in PTCs via this host receptor-protease pair. However, in the adult human kidney, the co-expression is restricted to a very small population of PTCs (0.5–5.0%) (Qi et al., 2020; Singh et al., 2020; Sungnak et al., 2020). Moreover, histopathological analyses of patients with COVID-19 identified viral inclusion bodies and SARS-CoV-2 RNA in tubules, endothelial cells, and podocytes (Puelles et al., 2020; Varga et al., 2020). This discrepancy may be explained by recent observations that SARS-CoV-2 can gain entry using different host receptors or proteases particularly in the presence of the FURIN-cleavage site (Cantuti-Castelvetri et al., 2020; Daly et al., 2020; Hoffmann et al., 2020b). Our data also suggest that SARS-CoV-2 has an expanded kidney tropism for endothelial cells analogous to the expanded tropism witnessed in the upper respiratory tract and nasal epithelium (Chu et al., 2020; Hulswit et al., 2016; Sungnak et al., 2020). The detection of LTL-negative SARS-CoV-2 infected cells suggests the presence of host receptors/proteases other cell types within the kidney organoids (Figure 6A). Therefore, the expanded tropism of host receptors and proteases in glomerular epithelial cells and endothelial cells may contribute to glomerular lesions in patients with COVID-19 and requires further investigation. Future immunofluorescence studies of kidney organoids and tissue co-stained with antibodies directed against SARS-CoV-2, receptors/proteases, and kidney cells would help to validate the potential involvement of cell types other than the PTCs.
At the beginning of the pandemic, there were concerns that patients with COVID-19 taking ACEIs and ARBs had upregulated expression of ACE2 which would lead to worse clinical outcomes (Ferrario et al., 2005). However, a recent meta-analysis of 52 studies evaluating the clinical outcomes of over 26,000 patients with COVD-19 receiving ACEIs and ARBs suggested a protective effect of these medications, especially among patients with hypertension (Baral et al., 2021). Consistent with the recent clinical studies, our results provide evidence for a beneficial effect of losartan. We demonstrate that losartan prevents the internalization of ACE2 and attenuates SARS-CoV-2 infection in PTCs of kidney organoids. Our data suggest that AT1R inhibition may diminish SARS-CoV-2 infections by preventing the internalization of ACE2-virus complex. Losartan represents a potential adjunctive therapy for SARS-CoV-2 infection and our findings support the rationale for losartan in patients with COVID-19 currently being investigated in pending clinical trials (e.g., NCT04312009, NCT04311177, and NCT04510623).
Our work potentially establishes a transcriptional relationship between RAS inhibition and IFN signaling in podocytes tubular cells (Figure 4). BST2 and IFITM1, two ISGs involved in coronavirus restriction, were significantly upregulated by losartan. Giant multinucleated syncytial cells were found in the alveoli of patients with COVID-19 (Schaller et al., 2020). Syncytia formation (cell-cell fusion) is virus mediated, promotes the spread of infection, cell death, and is enhanced by SARS-CoV-2's S-protein in cultured kidney cells (Hoffmann et al., 2020a). Interestingly, IFITM1 inhibits syncytia formation in placental trophoblasts (Buchrieser et al., 2019) and human osteosarcoma epithelial cells infected with SARS-CoV-2 (Buchrieser et al., 2021). Losartan may ameliorate viral tubular cytotoxicity through IFITM1 and the inhibition of syncytia formation. Of note, a recent study using human lung cells, cardiomyocytes, and gut organoids suggests that IFITM proteins promote SARS-CoV-2 infection by acting as cofactors for efficient SARS-CoV-2 infection (Prelli Bozzo et al., 2021). It is unclear if IFITM proteins would promote SARS-CoV-2 infection in the context of viral infection and interferon stimulation of kidney organoids. Measuring the cytotoxic effects of SARS-COV-2 and secretion of cytokines will help to establish a causal relationship between COVID-associated AKIs and direct viral infections.
Induced PSC-derived kidney organoids represent an immature model of the human kidney but express the appropriate genes and proteins for each type of kidney cell (Combes et al., 2019; Takasato et al., 2015; Tran et al., 2019). Our study highlights the strengths and reproducibility of data generated using the kidney organoid model system to recapitulate SARS-CoV-2 infection and RAS signaling in the kidney. At the transcriptomic level, scRNA-sequencing data generated from our kidney organoids had significant overlap with data reported by Monteil et al. (Monteil et al., 2020). At the protein level, our immunofluorescence staining results showed that the 3D arrangement of cells in the organoids are necessary for ACE2 protein localization and function which would not be observed in cultured tubular epithelial cells as they lack expression of important transporters and proteins including ACE2 (Xia et al., 2020). Our results corroborate prior reports of apical membrane-bound ACE2 in kidney organoids and a 3D renal epithelial tabloid system (Ichimura et al., 2020; Wysocki et al., 2021). Therefore, kidney organoids can represent a more physiological experimental setting compared to cell lines thus should be considered to study SARS-CoV-2 infection.
In summary, our work supports the use of kidney organoids as a model system to study the impact of the RAS signaling pathways on kidney cell susceptibility to SARS-CoV-2 infection. Our work highlights the susceptibility of kidney cells to SARS-CoV-2 infection relative to related human beta-coronaviruses, the plausibility of direct infection leading to COVID-AKI, and the potential utility of RAS inhibitors to prevent SARS-CoV-2 infection.
Limitations of the study
Human kidney organoids model the developing human fetal kidney and extrapolation of organoid data to humans must be conducted with caution and clinically validated. To minimize the physiologic off-target effects of Ang II and losartan, we limited exposure of organoids to short drug treatment as our research question was to determine the acute ability of losartan to inhibit viral infection.
The SARS-CoV-2 infection of kidney organoids in untreated and treated organoids was reproducible but exhibited heterogeneity between batches thus requiring numerous replicates to achieve statistical significance. The limited number of replicate experiments was impacted by the availability of the biosafety level 3 facility required for SARS-CoV-2 research.
As only losartan at a standard in vitro dose was used for the majority of this study, extrapolating our findings to in vivo dosing and to other ARBs and ACE inhibitors will need to be validated in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sheep polyclonal anti-NPHS1 | R&D Systems | Cat. #AF4269; RRID:AB_2154851 |
| Rabbit monoclonal anti-PDGFRβ (clone C82A3) | Cell Signaling Technology | Cat. #4564; RRID:AB_2236927 |
| Mouse monoclonal anti-E-Cadherin (clone 36) | BD Transduction laboratories | Cat. #610181; RRID:AB_397580 |
| Goat polyclonal anti-ACE2 | R&D Systems | Cat. #AF933; RRID:AB_355722 |
| Rabbit polyclonal anti-ACE2 | ProteinTech | Cat. #21115-1-AP; RRID:AB_10732845 |
| Rabbit monoclonal anti-ACE1 (clone EPR22291-247) | Abcam | Cat. #254222 |
| Mouse monoclonal anti-LRP2 (clone H-10) | Santa Cruz Biotechnology | Cat. #sc-515772; RRID:AB_2783023 |
| Rabbit monoclonal anti-GAPDH (clone 14C10) | Cell Signaling Technology | Cat. #2118; RRID:AB_561053 |
| Alexa Fluor 405 conjugated anti-mouse IgG | Abcam | Cat. # ab175658; RRID:AB_2687445 |
| Alexa Fluor 488 conjugated anti-mouse IgG | Thermo Fisher Scientific | Cat. #A-21202; RRID:AB_141607 |
| Alexa Fluor 555 conjugated anti-goat IgG | Thermo Fisher Scientific | Cat. #A-21432; RRID: AB_2535853 |
| Alexa Fluor 568 conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat. #A-10042; RRID:AB_2757564 |
| Alexa Fluor 568 conjugated anti-sheep IgG | Thermo Fisher Scientific | Cat. #A-21099; RRID:AB_2535753 |
| Alexa Fluor 647 conjugated anti-mouse IgG | Thermo Fisher Scientific | Cat. #A-31571; RRID:AB_162542 |
| Alexa Fluor 647 conjugated anti-sheep IgG | Thermo Fisher Scientific | Cat. #A-21448; RRID:AB_1500712 |
| Lotus Tetragonolobus Lectin (LTL), Fluorescein labelled | Vector Laboratories | Cat. #FL-1321-2; RRID:AB_2336559 |
| Lotus Tetragonolobus Lectin (LTL), Cy5 | GlycoMatrix | Cat. #21761117-1 |
| Biological samples | ||
| Human kidney tissue | Non-diseased segments of human kidney nephrectomy samples obtained from deidentified patients approved by the Conjoint Health Research Ethics Board (CHREB) at the University of Calgary | n/a |
| Chemicals, peptides, and recombinant proteins | ||
| mTeSR™1 | Stem Cell Technologies | Cat. #85850 |
| Gentle Cell Dissociation Reagent | Stem Cell Technologies | Cat. #7174 |
| STEMdiff™ APEL2™ medium | Stem Cell Technologies | Cat. #05275 |
| Protein-Free Hybridoma Medium | Thermo Fisher Scientific | Cat. #12040077 |
| 0.05% Trypsin-EDTA | Thermo Fisher Scientific | Cat. #25300-062 |
| Dulbecco's modified Eagle's medium | Thermo Fisher Scientific | Cat. #11965-092 |
| Dulbecco's modified Eagle's medium/F12 | Thermo Fisher Scientific | Cat. #11330032 |
| Advanced RPMI 1640 medium | Thermo Fisher Scientific | Cat. #12633012 |
| Embryonic stem cell-qualified fetal bovine serum | Thermo Fisher Scientific | Cat. #10439016 |
| Matrigel hESC-Qualified Matrix LDEV-free | Corning | Cat. #354277 |
| Donkey serum | Millipore Sigma | Cat. #S30 |
| Fetal Bovine Serum | Millipore Sigma | Cat. #F1051 |
| Angiotensin II | Millipore Sigma | Cat. #A9525 |
| Heparin sodium salt | Millipore Sigma | Cat. #4784 |
| CHIR99021 | R&D Systems | Cat. #4423 |
| InSolution™ Y-27632 | Millipore Sigma | Cat. #688001 |
| 16% aqueous Paraformaldehyde (PFA) | EMS | Cat. #15710 |
| Recombinant human FGF-9 | R&D Systems | Cat. #273-F9-025 |
| Losartan | Tocris | Cat. #3798 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat. #H3570 |
| Penicillin/streptomycin (100x) | Thermo Fisher Scientific | Cat. #15070-063 |
| Penicillin-Streptomycin-Glutamine (100X) | Thermo Fisher Scientific | Cat. #10378016 |
| Collagenase IV | Worthington | Cat. #LS004188 |
| Prostaglandin E1 | Millipore Sigma | Cat. #P8908 |
| Human EGF | Millipore Sigma | Cat. #SRP3027 |
| T3 (3,3’,5-triiodo-L-thyronine) | Millipore Sigma | Cat. #T2877 |
| Hydrocortisone | Millipore Sigma | Cat. #H4001 |
| ITSS | Millipore Sigma | Cat. #I1884-1VL |
| TrypLE Select | Thermo Fisher Scientific | Cat. #12563011 |
| Critical commercial assays | ||
| 10X Chromium™ Controller | 10X Genomics | N/A |
| Chromium™ Single Cell A Chip kit, 48 rxns | 10X Genomics | Cat. #120236 |
| Chromium™ Single cell 3′Library & Gel beaded kit V3 and 3.1 Next GEM, 16 rxns | 10X Genomics | Cat. #1000075; Cat. #1000128 |
| Chromium i7 multiplex Kit 96 rxns | 10X Genomics | Cat. #120262 |
| Illumina NovaSeq S2 Flowcell | Illumina, Centre for Health Genomics and Informatics, University of Calgary | N/A |
| Scepter™ 2.0 Handheld Automated Cell Counter | Millipore | Cat. #PHCC20040 |
| Agilent D1000 ScreenTape System | Agilent Technologies | Part # 5067-5582 |
| Deposited data | ||
| Kidney organoid scRNA-seq | This study | GEO:GSE149687 |
| SARS-CoV-2 infectable kidney organoids scRNA-seq | Monteil et al. (2020) | GEO:GSNE147863 |
| Experimental models: cell lines | ||
| Human iPSC – Gibco episomal iPSC line | Thermo Fisher Scientific | Cat. #A18945 |
| Human iPSC line (derived from human dermal fibroblast, Cell Applications, Inc. Cat#106-05n, Lot# 1481) | Gift from Dr. Martin Pollak, Harvard Medical School | n/a |
| Vero E6 | ATCC | Cat. #C1008 |
| Software and algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/; RRID: SCR_003070 |
| GraphPad Prism 8.3.0 | GraphPad | https://www.graphpad.com/;SCR_015807 |
| R Studio | https://www.rstudio.com/products/rstudio/ | Version 3.6.2-4.1.0 |
| R | https://www.r-project.org/ | Version 3.6.2-4.0.5 |
| Cell Ranger | 10X Genomics | Version 3.0.1 |
| Seurat | https://github.com/satijalab/seurat | Version 3.2.3 - 4.0.3 |
| Leica software | Leica | N/A |
| BioRender | BioRender.com | |
| Other | ||
| μ-slide 8 well glass-bottom chamber | ibidi | Cat. #80826 |
| Transwell | Corning | Cat. #07-200-170 |
| Human Protein Atlas | https://www.proteinatlas.org/ | Version 19.3 |
Resource availability
Lead contact
Information and reagents for resources and reagents should be directed to and will be fulfilled by the lead contact, Justin Chun (chuj@ucalgary.ca)
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell lines
Human iPSC lines were purchased from Gibco (human episomal iPSC line, Cat. #A18944-derived from neonate female umbilical cord blood) or were derived from cryopreserved human dermal fibroblast (Cell Applications, Inc., Cat. #106-05n, lot #1481) using CytoTune™-iPS 2.0 Sendai reprogramming kit (Life Technologies, Cat. #A16517) at the Harvard Stem Cell Institute iPSC Core Facility, as a gift from Dr. Martin Pollak at Harvard Medical School. iPSCs were confirmed to be of normal karyotype and maintained at 37°C with 5% CO2 with daily medium changes of mTeSR1 medium in 6 well plates coated with Matrigel® (Corning, Cat. #354277). iPSC colonies were passaged using GCDR (Stem Cell Technologies, Cat. #05275) and transferred to T25 flasks coated with Matrigel® prior to differentiation. iPSCs were routinely tested and confirmed negative for mycoplasma. Vero E6 cells (ATCC, Cat. #C1008) were maintained in a humidified 37 °C incubator with 5% CO2 and cultured in DMEM (Thermo Fisher) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine (Thermo Fisher, Cat. #10378-016; herein referred to as 1% PSQ) and 10% heat-inactivated FBS (Thermo Fisher; herein referred to as complete DMEM).
Primary culture of tubular epithelial cells
Primary human tubular epithelial cells (hTEC) were isolated and cultured from non-diseased segment of nephrectomy samples from de-identified patients with renal cell carcinoma. The use of human kidney samples was approved by the Conjoint Health Research Ethics Board (CHREB) at the University of Calgary. Renal cortex segments of the nephrectomy tissues were finely dissected, minced and digested with collagenase IV (Worthington, Cat. #LS004188, 1.5 mg/mL) for 30 min at 37°C. Cells were passed through a 75-μm nylon sieve and the filtrate were washed in HBSS twice and resuspended in Dulbecco's modified Eagle's medium/F-12 (Thermo Fisher Scientific, Cat. #11330032) containing 10% heat-inactivated FBS (Millipore Sigma, Cat. #F1051), 1% penicillin-streptomycin (Thermo Fisher, Cat. #15070-063), 125 ng/mL of prostaglandin E1 (Millipore Sigma, Cat. #P8908), 25 ng/mL of human EGF (Millipore Sigma, Cat. #SRP3027), 1.8 μg/mL of 3,3′,5-triiodo-L-thyronine (Millipore Sigma, Cat. #T2877), 3.38 ng/mL of hydrocortisone (Millipore Sigma, Cat. #50-23-7) and 1% ITSS hormone mix (Millipore Sigma, Cat. #I1884-1VL). hTEC culture was established on a collagen IV-coated plates at 37°C with 5% CO2 and treated at 90% confluence at passage 5.
Methods details
Generation of kidney organoids
Kidney organoids were differentiated using the Takasato protocol (Takasato et al., 2016)with minor modifications that included APEL2 medium (Stem Cell Technologies, Cat. #05275) supplemented with 5% PFHM-II (Thermo Fisher, Cat. #12040077) (APEL2 + PFHM) in replacement of APEL. Induced PSCs were grown on a feeder-free system using Matrigel matrix coated plates. Cells were treated with 8 μM CHIR99021 (R&D Systems, Cat. #4423) for 4 days followed by recombinant human FGF-9 (200 ng/mL) and heparin (1 μg/mL) for an additional 3 days. At day 7, cells were dissociated into single cell suspension using trypsin-EDTA (0.05%) for 2 min and 250,000 or 500,000 cells were centrifuged to make a pellet at 400 × g for 2 min. The pellet was transferred onto a six-well transwell plate (Corning, Cat. # 07-200-170) with four or nine pellets per well, respectively. Pellets were incubated with a pulse of 5 μM CHIR99021 in APEL2 + PFHM medium for 1 hr at 37 °C. After 1 hr, the medium was changed to APEL2 + PFHM supplemented with FGF-9 (200 ng/mL, R&D Systems, Cat. #273-F9-025) and heparin (1 μg/mL, Millipore Sigma, Cat. #4784) for an additional 5 days, and then were maintained in APEL2 + PFHM medium for 14 days with medium change every other day.
Treatment with RAS modulators
Kidney organoids on day 28 or hTEC at passage 5 were pretreated with or without losartan (100 μM, Tocris, Cat. #3798) for 30 min prior to Ang II (1 μM, Millipore Sigma, Cat. #A9525) treatment for 24 hr. DMSO was used as a vehicle control.
SARS-CoV-2 Propagation
The mNeon-Green infectious clone of SARS-CoV-2 (icSARS-CoV-2-mNG) was generously provided by Dr. Pei Yong Shi of the World Reference Center for Emerging Viruses and Arboviruses through the University of Texas Medical Branch at Galveston (Xie et al., 2020). All experiments with SARS-CoV-2 were conducted in a containment level 3 (CL3) facility, and all standard operating procedures were approved by the CL3 Oversight Committee and Biosafety Office at the University of Calgary. The icSARS-CoV-2-mNG was propagated in Vero E6 cells by infecting cells in a T-175 cm2 flask at a multiplicity of infection of 0.01 for 1 h in 2.5 mL of serum-free DMEM at 37°C. Following the incubation, 17.5 mL of DMEM supplemented with 2% heat-inactivated FBS and 1% PSQ (herein referred to as low-serum DMEM) was added directly to the viral inoculum. The supernatant was harvested 40–48 hpi and centrifuged at 500 × g for 10 min to remove cellular debris and particulates. Virus stocks were aliquoted and stored at −80°C. Stock titers were determined via plaque assay using equal parts 2.4% w/v semi-solid colloidal cellulose overlay (Sigma; prepared in ddH20) and 2 × DMEM (Wisent), all supplemented with 1% heat-inactivated FBS and 1% PSQ (Mendoza et al., 2020).
SARS-CoV-2 infection of kidney organoids
On day 28, kidney organoids from 250,000 or 500,000 cells were pre-treated in duplicate with vehicle, Ang II (1 μM) in presence or absence of losartan (100 μM) for 24 hr. After 24 hr, organoids were infected with 103 or 105 PFU of icSARS-CoV-2-mNeonGreen (mNG) in 0.5 mL or 0.25 mL of advanced RPMI medium (Thermo fisher, Cat. #12633012) for 1h. After 1h incubation, organoids were washed in 0.5 mL of PBS for three times and then replenished with 0.5 mL of fresh RPMI medium containing Ang II (1 μM) and/or losartan (100 μM) as per previous treatment. At 1.5 dpi, supernatants were collected and new fresh medium with Ang II and/or losartan were added. The collected supernatant was centrifuged at 500 × g for 10 min to remove cellular debris and particulates. Resulting supernatant was transferred to a new Eppendorf tube and stored at −80°C for downstream viral titer analysis. At 3 dpi, organoids were washed once with 0.5 mL of PBS and then fixed in 1 mL of 4% PFA for 1 h at room temperature. Organoids were washed 3 times with 1 mL of PBS prior to further analysis.
Tissue culture infectious dose assay
Vero E6 cells were seeded in a 96-well plate at a seeding cell density of 30,000 cells/well in complete DMEM. The next day, a 1:10 dilution series using 1.5 dpi supernatants was prepared in low-serum DMEM, up to 7 times (10−7). Medium from the 96-well plate was removed and Vero E6 monolayers were exposed to 200 μL of diluted supernatant in duplicate for 5 d at 37°C with 5% CO2. At 5 dpi, Vero E6 cells were monitored and scored for cytopathic effects. Viral titer was calculated from the plate scoring using the Kärber method (Kärber, 1931) to determine the tissue culture infectious dose wherein 50% of cells are infected (TCID50/mL).
Immunofluorescence
Kidney organoids treated with Ang II and/or losartan for 24 hr were washed in PBS and fixed with 2% PFA for 20 min at 4°C. Next, the organoids were washed in PBS twice and blocked in a blocking buffer containing 10% donkey serum, 0.05% Triton-X in PBS for 3 hr at room temperature in μ-slide 8 well glass-bottom chamber (iBidi, Cat. #80826). Primary antibodies to NPHS1 (R&D Systems, Cat. #AF4269, 1:100) PDGFR-β (Cell Signaling, Cat. #4564, 1:100), E-Cadherin (BD Transduction laboratories, Cat. #610181, 1:200), ACE2 (R&D, Cat. #AF-933, 1:100), LRP2 (Santa Cruz, Cat. #SC-515772, 1:100) were incubated at 4°C overnight. After 6 washes with wash buffer (0.3% Triton X-100 in PBS) for 10 min each wash, samples were incubated in wash buffer with fluorescent conjugated secondary antibodies that include Alexa Fluor donkey anti-mouse 405 (Abcam, 1:250), donkey anti-mouse 488 (Thermo Fisher Scientific, 1:250), donkey anti-goat 555 (Thermo Fisher Scientific, 1:250), donkey anti-rabbit 568 (Thermo Fisher Scientific, 1:250), donkey anti-sheep 568 (Thermo Fisher Scientific, 1:250), donkey anti-sheep 647 (Thermo Fisher Scientific, 1:250) and/or donkey anti-mouse 647 (Thermo Fisher Scientific, 1:250) for 2–3 hr at room temperature. LTL-Cy5 (GlycoMatrix, Cat. #21761117-1, 1:100) or FITC-conjugated LTL (Vector laboratories, FL1321-2, 1:200) was incubated with the secondary antibodies. After 3 washes with PBS, confocal images were acquired with Zeiss LSM 880 confocal microscope or Leica TCS SP8.
Immunoblotting
Protein samples from kidney organoids and hTEC were prepared using RIPA buffer (50 mM Tris, 150 mM NaCl, 1mM EDTA, 1% NP40, 1% sodium deoxycholate, 0.1% SDS). Proteins were separated on SDS-PAGE gels under reducing conditions and transferred onto nitrocellulose membrane. After blocking membranes in 5% milk PBS-0.1% Tween (PBST) for 1 hr at room temperature, primary antibodies to ACE1(Abcam, Cat. #254222, 1:1,000), ACE2 (ProteinTech, Cat. #21115-1-AP, 1:1,000) and GAPDH (Cell signaling Technology, Cat. #2118, 1:2,000) were incubated in blocking buffer overnight at 4°C. Membranes were washed in PBST for 4 times and followed by HRP-conjugated secondary antibodies for 1 hr. After additional washes, proteins were visualized with ECL western blotting detection system (GE Healthcare) and digitally captured using a Chemidoc MP device (Bio-Rad).
scRNA-Seq library construction
Single (n = 1) technical replicates of each treatment were kidney organoid group derived from the 1481 iPSC line were prepared for sequencing. Organoids were dissociated to single cells using TrypLE Select (Thermo Fisher Scientific) at 37°C with mechanical trituration. Single cell suspensions were visually inspected under a microscope, counted using an automated hand-held cell counter (Millipore Scepter™ 2.0 Cell Counter), and resuspended at a concentration of 15,000 single-cells in 50 microliters of FACS buffer (0.1% BSA–PBS). All four samples were processed according to 10X Genomics ChromiumTM Next GEM Single Cell 3' Reagent Kits v3.1 as per manufacturer’s protocol. Briefly, single cells were partitioned together with barcoded Gel Bead-In-EMulsions (GEMs) using 10x GemCodeTM Technology. This process lysed cells and enabled barcoded reverse transcription of RNA, generating cDNA from poly-adenylated mRNA. DynaBeads® MyOneTM Silane magnetic beads were used to remove leftover biochemical reagents, then cDNA was amplified by PCR. Quality control size gating was used to select cDNA amplicon size prior to library construction. Read 1 primer sequences were added to cDNA during GEM incubation. P5 primers, P7 primers, i7 sample index, and Read 2 primer sequences were added during library construction. Quality control and cDNA quantification was performed using Agilent D1000 ScreenTape System. Libraries were pooled and first sequenced on NovaSeq 100 (plus 25 additional) Cycle SP dual lane flow cell to approximate the number of recovered cells in each sample. We recovered 4,235 cells in Control, 5,702 cells in losartan-alone, 5,800 cells in angiotensin II alone, and 5,855 cells in angiotensin II plus losartan treated groups with an estimated doublet rate of ≈4%. Based on these, we determined proportions for Illumina NovaSeq 100 (plus 25 additional) Cycle S2 dual lane flow cell with a targeted sequencing depth of ∼30,000 reads per cell. Illumina sequencer's BCL files were demultiplexed using cellranger mkfastq (a wrapper around Illumina's bcl2fastq). Downstream barcode processing, alignment to pre-built GRCh38, and single-cell 3′ gene counting were performed using CellRanger software version 3.0.1 (10X Genomics). All four libraries were aggregated using cellranger aggr (with mapped normalization). The resulting filtered feature-barcode matrix was imported into Seurat v3 for quality control, dimensionality reduction, cell clustering, and differential expression analysis using default or recommended parameters (Stuart et al., 2019).
Bioinformatic analyses of integrated organoid datasets
Low quality cells (<200 or >7,500 genes detected, or >20% mitochondrial genes) were filtered out from downstream analysis as they may have arisen from broken cells, multiplets, or other technical artifacts. To identify anchor genes for integrating the four aggregated scRNA-Seq samples, FindIntegrationAnchors and IntegrateData functions were used with 20 principal components. Percentage of cells expressing a gene was calculated using the PrctCellExpringGene function found at github.com/satijalab/seurat/issues/371. Gene expression signatures were visualized using DoHeatmap, Vlnplot, RidgePlot, and FeaturePlot functions in Seurat. Supervised gene set enrichment scores were calculated using the AddModuleScore function which calculates the average expression of a custom gene set subtracted by the aggregated expression of a randomly selected control feature set. The curated lists of genes scored are provided below.
Corroboration with previously published scRNA-Seq datasets
Integration of our control organoid (GSM4509001) with previously infectable kidney organoids (GSNE147863) (Monteil et al., 2020) was performed using a mutual nearest neighbor (MNN)-based anchoring strategy implemented in Seurat v3. Log-normalized and scaled gene expression was jointly queried across both datasets.
Gene set analysis
Interferon-stimulated genes (Schoggins et al., 2011).
IFIH1, CXCL9, CREB3L3, GTPBP1, DDX58, G6PC, VAMP5, CTCFL, IRF1, IGFBP2, B2M, SPTLC2, IRF7, OASL, CD69, IFI44L, CCL2, PDGFRL, IRF9, MT1X, MKX, CCL19, SOCS1, CCL4, HSH2D, GEM, SOCS2, IL28RA, SLC1A1, XAF1, DEFB1, CD9, LRG1, HLA-G, ZNF313, SLFN5, C5orf39, IFI35, FAM46C, APOL2, USP18, APOL1, RPL22, FBXO6, JUNB, SLC25A28, HLA-E, MAB21L2, C4orf33, WARS, NDC80, IRF2, SAT3, CCL5, RNASE4, C22orf28, TNFSF10, C10orf10, LINCR, GZMB, VEGFC, IFITM2, TRIM21, TNFAIP6, CD80, IFIT1, ADAMDEC1, PI4K2B, CCDC92, FLJ39739, HK2, PBEF1, C5orf27, NUP50, IFITM3, CCDC109B, THBD, MYD88, GAK, GK, CCL8, DYNLT1, SP110, IFIT5, ETV7, DTX3L, ANKRD22, ARG2, MT1H, NAPA, SAA1, MAFB, NRN1, CSDA, CXCL10, SIRPA, C15orf48, NOD2, MT1G, UPP2, ADFP, NT5C3, C2orf31, EIF2AK2, DDIT4, PMM2, HESX1, LAP3, LY6E, MT1F, ABTB2, GTPBP2, TLR3, S100A8, GMPR, BATF2, P2RY6, ISG15, TNFRSF10A, GCH1, GBP2, CES1, EIF3L, SERPINE1, AGPAT9, IL17RB, DCP1A, PUS1, NCF1, FNDC4, SMAD3, PFKFB3, EPSTI1, MT1M, BUB1, MAP3K14, AIM2, NFIL3, BTN3A3, BCL2L14, PNRC1, PXK, KIAA0040, ANGPTL1, STEAP4, ENPP1, PML, PDK1, LGALS9, CASP7, MAP3K5, LGALS3, PARP12, HES4, OAS2, IFNGR1, IFI44, IFI30, TRIM5, SERPING1, CMAH, EPAS1, MAFF, CRP, PRAME, LGMN, UBE2L6, IMPA2, CEBPD, SSBP3, RASSF4, CYP1B1, SECTM1, PADI2, TAP2, ALDH1A1, COMMD3, PMAIP1, CXCL11, TMEM49, TMEM51, FLJ23556, CDKN1A, TNFSF13B, PCTK2, EXT1, ZBP1, HPSE, FAM125B, KIAA1618, CCDC75, TYMP, BLVRA, MAX, TREX1, TRIM14, FUT4, SCARB2, CCND3, FAM70A, TRIM38, MTHFD2L, CPT1A, CCR1, TBX3, SLC15A3, CD74, ISG20, MX1, DHX58, CHMP5, BCL3, MSR1, LAMP3, EHD4, TDRD7, BAG1, DUSP5, HLA-C, SNN, AQP9, THOC4, IL6ST, GBP4, GLRX, PHF11, MCOLN2, TLK2, IFI6, PPM1K, STAP1, UNC84B, NCOA3, ATP10D, GBP5, WHDC1, SERPINB9, PTMA, BST2, AMPH, RARRES3, CX3CL1, SCO2, MARCKS, STARD5, ADM, ETV6, TRIM34, PHF15, IL1RN, TAGAP, GPX2, PPM1K, ARNTL, MX1, FKBP5, UNC93B1, IFI27, GBP1, CLEC4D, AXUD1, MCL1, C6orf150, ANKFY1, FFAR2, SAMD4A, CLEC2B, IL1R1, SLC16A1, IFIT3, AKT3, FAM134B, IFI16, AHNAK2, HLA-F, FNDC3B, NPAS2, FER1L3, LMO2, RNF19B, IFITM3, Gluc, HEG1, PRKD2, PSCD1, CCNA1, PSMB9, LIPA, C1S, OPTN, PRIC285, NMI, ADAR, IDO1, DDX3X, TNFAIP3, TIMP1, CLEC4E, IFIT2, ODC1, ABLIM3, RGS1, JAK2, STAT2, ATF3, PSMB8, ARHGEF3, HERC6, ERLIN1, MICB, GCA, GJA4, RBCK1, PIM3, GBP3, OGFR, SAMHD1, IL15, ELF1, PNPT1, RIPK2, CRY1, SLC25A30, ULK4, DNAPTP6, IFI6, IL15RA, TRAFD1, CD38, CD274, FAM46A, SPSB1, PLSCR1, TRIM25, STAT1, MASTL, GALNT2, TMEM140, C9orf91, CD163, ZNF385B, TXNIP, LEPR, APOBEC3A, RTP4, B4GALT5, TCF7L2, ACSL1, TAP1, KIAA0082, FCGR1A, C4orf32, IFITM1, CFB.
SARS-CoV-2 protein-protein interactome (Gordon et al., 2020).
Nsp1: PRIM1, PRIM2, POLA1, POLA2, COLGALT1, PKP2.
Nsp2: EIF4E2, GIGYF2, POR, WASHC4, FKBP15, RAP1GDS1, SLC27A2
Nsp4: TIMM3, TIMM10, TIMM10B, TIMM29, TIMM9, IDE, ALG11, NUP210, DNAJC11
Nsp5: HDAC3
nsp5-C857A: TRMT1, GPX1
Nsp6: SIGMAR1, ATP6AP1, ATP5M6, ATP13A3
Nsp7: RAB18, RAB14, RAB8A, RAB7A, RAB10, RAB5C, RAB2A, RAB1A, LMAN2, GNG5, GNB1, NDUFAF2, COMT, HS2ST1, QSOX2, RALA, RHOA, MTARC1, SCCPDH, SELENOS, AGPS, TOR1AIP1, DNAJC19, MOGS, ACSL3, FAM162A, DCAKD, NAT14, SCARB1, CYB5B, PTGES2, CYB5R3
Nsp8: EXOSC2, EXOSC5, EXOSC3, EXOSC8, LARP7, MEPCE, MRPS27, MRPS5, MRPS25, MRPS2, SEPSECS, ATE1, MPHOSPH10, NSD2, DDX10, HECTD1, NGDN, NOL10, AATF, SRP54, SRP19, SRP72, CCDC86, NARS2
Nsp9: ZNF503, FBN2, FBN1, FBLN5, SPART, MIB1, NEK9, GTF2F2, DCAF7, EIF4H, NUP62, NUP214, NUP58, NUP88, NUP54, MAT2B, ZNF503
Nsp10: AP2M1, AP2A2, ERGIC1, GFER, GRPEL1
Nsp11: TBCA
Nsp12: RIPK1, RBM41, PRRC2B, PLEKHA5, PDZD11, MYCBP2, LARP4B, CRTC3, AKAP8, UBAP2L, AKAP8, UBAP2L, UBAP2L, ZNF318, ZC3H7A, BCKDK, USP54, TYSND1, TCF12, SBNO1, PPIL3, SLU7
Nsp13: GOLGA2, GOLGA3, GOLGB1, GORASP1, GCC2, GCC1, FYCO1, HSBP1, USP13, MIPOL1, TLE1, TLE3, TLE5, CIT, TBKBP1, TBK1, GRIPAP1, JAKMIP1, CLIP4, RDX, C1orf50, ERC1, CENPF, CDK5RAP2, PCNT, NINL, NINL, CNTRL, CEP68, CEP43, CEP350, CEP250, CEP135, CEP112, AKAP9, PRKACA, PRKAR2A, PRKAR2B, PDE4DIP, HOOK1
Nsp14: SIRT5, GLA, IMPDH2
Nsp15: RNF41, NUTF2, ARF6
S: GOLGA7, ZDHHC5
E: SLC44A2, ZC3H18, AP3B1, CWC27, BRD2, BRD4
N: FAM98A, SNIP1, CSNK2B, CSNK2A2, G3BP2, G3BP1, DDX21, UPF1, RPL36, RBM28, MOV10, PABPC1, LARP1, PABPC4, RRP9, RPL36
M: AASS, ACADM, AKAP8L, ANO6, TUBGCP3, TUBGCP2, RTN4, YIF1A, REEP5, REEP6, SLC30A9, SLC30A7, SLC25A21, SAAL1, AAR2, TARS2, STOM, PSMD8, PMPCB, PMPCA, PITRM1, INTS4, GGCX, FASTKD5, FAM8A1, ETFA, COQ8B, BZW2, ATP6V1A, ATP1B1
Orf3a: HMOX1, TRIM59, ALG5, ARL6IP6, CLCC1, SUN2, VP239, VPS11
Orf3b: STOML2
Orf6: MTCH1, NUP98, RAE1
Orf7a: MDN1, HEATR3
Orf8: EDEM3, ERLEC1, OS9, UGGT2, ERO1B, SIL1, HYOU1, NGLY1, TOR1A, FOXRED2, SDF2, ADMTS1, TM2D3, NPC2, CHPF, LOX, PVR, COL6A1, PLOD2, FKBP10, FKBP7, GGH, IL17RA, PUSL1, PLD3, PCSK6, ADM9, ITGB1, DNMT1, EMC1, CHPF2, ERP44, MFGE8, HS6ST2, POFUT1, NPTX1, STC2, POGLUT2, PLAT, NEU1, SMOC1, GDF15, INHBE, CISD3, PLEKHF2, POGLUT3, FBXL12
Orf9b: DCTPP1, TOMM70, SLC9A3R1, CHMP2A, DPH5, CSDE1, PTBP2, BAG5, MARK2, MARK1, MARK3
Orf9c: TMEM97, ERMP1, TAPT1, PIGS, GPAA1, SLC30A6, TMED5, SCAP, BCS1L, NDFIP2, DPY19L1, F2RL1, GHITM, ABCC1, TMEM39B, ALG8, ECSIT, NDUFB9, ACAD9, NDUFAF1, FAR2, WFS1, PIGO, RETREG3, UBXN8, NLRX1
Orf10: MAP7D1, THTPA, TIMM8B, PPT1, ZYG11B, ELOC, ELOB, RBX1, CUL2
Activator Protein-1 (AP-1): ATF2, ATF3, ATF4, ATF5, ATF1, ATF6B, ATF7, ATF6, BATF2, BATF3, JDP2, FOS, FOSB, FOSL1, FOSL2, JUN, JUNB, JUND, MAF, MAFA, MAFB, MAFF, MAFG, MAFK, NRL
Quantification and statistical analysis
ACE2 and LRP2 co-localization analysis
To measure ACE2 receptor localization change upon Ang II and losartan treatment, Mander’s coefficient was used on ImageJ with JACoP (just another colocalization plugin). LRP2 was used as a marker for proximal tubular cells that do not change localization in kidney organoids. First, 153 x 153 μm2 area was selected where LRP2 is expressed in the kidney organoids. Auto-thresholding was applied to each channel for ACE2 and LRP2 using RenyiEntropy. 50 × 50 μm2 area with relatively highest ACE2 and LRP2 signal intensities was cropped, and JACoP plugin was run to measure Pearson’s M1 and M2 coefficients. M2, a fraction of LRP2 overlapping with ACE2 was used to indicator of localization change for ACE2 from the plasma membrane to intracellular region. Two regions of interest were selected per image. In total, 7 to 13 field of view were used per each treatment.
Quantification of SARS-CoV-2 infected cells in proximal tubules
To examine the impact of losartan on susceptibility of proximal tubular cells to SARS-CoV-2 infection, quantifying of SARS-CoV-2 infected cells were confined to proximal tubules. In each field of view, total number of proximal tubular cells (LTL+) and SARS-CoV-2 infected cells with a robust cytoplasmic mNeonGreen (mNG) signal expression (LTL+/mNG+) within proximal tubular cells were quantified. SARS-CoV-2 positive proximal tubular cells (LTL+/mNG+) per total proximal tubular cells (LTL+) in each field of view was expressed as a percentage. 10 field of view were acquired for each treatment condition.
Statistical analysis
All statistical analyses and graphical representations were performed using Microsoft Excel v16, R version 3.6.3 and GraphPad Prism v8.3.0. Significance between two parametric groups was determined using student’s t-test. Significance between two non-parametric groups was determined using Wilcoxon signed-rank test. For comparisons between multiple groups, one-way ANOVA comparisons and Tukey corrections were employed. For all tests, significance was defined as p value <0.05. Single (n = 1) technical replicates of each treatment were kidney organoid group derived from the 1481 iPSC line were prepared for sequencing.
Acknowledgments
We thank Dr. Nicole Rosin for sequencing coordination, Dr. Adrienne Kline for support on statistical analyses, Carolyn-Ann Robinson for helpful discussions, and the sequencing staff at Center for Health Genomics and Informatics (CHGI). The mNeon-Green infectious clone of SARS-CoV-2 was generously provided by Dr. Pei-Yong Shi of the World Reference Center for Emerging Viruses and Arboviruses through the University of Texas Medical Branch at Galveston. HC was supported the Roy and Vi Baay Chair in Kidney Research. This research was supported by the Department of Medicine, Division of Nephrology at the University of Calgary and the Snyder Institute for Chronic Diseases (startup funds to JC). JC is supported by a new investigator award from the Kidney Research Scientist Core Education and National Training Program.
Author contributions
WR and JC wrote the article and drafted the figures. HC and SS co-wrote the article and contributed figures. JC and HC cultured and performed experiments using human tubular epithelial cells and kidney organoids. HC, JC, and WR stained and imaged the kidney organoids. SS and JB performed single-cell RNA sequencing preparation and analysis. SS and WR analyzed the scRNA-seq dataset. MB performed the SARS-CoV-2 infection experiments and JAC designed and analyzed data. RA and AJ assisted with statistical analyses. JC conceived of the experiments and supervised the study with JB.
Declaration of interests
The authors have no competing interests to declare.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103818.
Contributor Information
Jeff Biernaskie, Email: jeff.biernaskie@ucalgary.ca.
Justin Chun, Email: chuj@ucalgary.ca.
Supplemental information
Data and code availability
Single cell RNA-Seq data have been deposited in NCBI GEO (which automatically makes SRA deposit) and are publicly available as of the date of publication with the following accession number: GSE149687. Single-cell RNA-Seq datasets were integrated with previously infectable kidney organoids which can be accessed with the following accession number: GSNE147863. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alsaad K.O., Hajeer A.H., Al Balwi M., Al Moaiqel M., Al Oudah N., Al Ajlan A., AlJohani S., Alsolamy S., Gmati G.E., Balkhy H., et al. Histopathology of Middle East respiratory syndrome coronovirus (MERS-CoV) infection - clinicopathological and ultrastructural study. Histopathology. 2018;72:516–524. doi: 10.1111/his.13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baral R., Tsampasian V., Debski M., Moran B., Garg P., Clark A., Vassiliou V.S. Association between renin-angiotensin-aldosterone system inhibitors and clinical outcomes in patients with COVID-19: a systematic review and meta-analysis. JAMA Netw. Open. 2021;4:e213594. doi: 10.1001/jamanetworkopen.2021.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle D., Soler M.J., Sparks M.A., Hiremath S., South A.M., Welling P.A., Swaminathan S. Acute kidney injury in COVID-19: emerging evidence of a distinct pathophysiology. J. Am. Soc. Nephrol. 2020;31:1380–1383. doi: 10.1681/asn.2020040419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun F., Lütgehetmann M., Pfefferle S., Wong M.N., Carsten A., Lindenmeyer M.T., Nörz D., Heinrich F., Meißner K., Wichmann D., et al. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet. 2020;396:597–598. doi: 10.1016/s0140-6736(20)31759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner B.M., Cooper M.E., de Zeeuw D., Keane W.F., Mitch W.E., Parving H.H., Remuzzi G., Snapinn S.M., Zhang Z., Shahinfar S. Effects of losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N. Engl. J. Med. 2001;345:861–869. doi: 10.1056/NEJMoa011161. [DOI] [PubMed] [Google Scholar]
- Buchrieser J., Degrelle S.A., Couderc T., Nevers Q., Disson O., Manet C., Donahue D.A., Porrot F., Hillion K.H., Perthame E., et al. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science. 2019;365:176–180. doi: 10.1126/science.aaw7733. [DOI] [PubMed] [Google Scholar]
- Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel-Benhassine F., Van der Werf S., et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39:e106267. doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel-Benhassine F., Van der Werf S., et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2021;40:e107405. doi: 10.15252/embj.2020107405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey R.M. The intrarenal renin-angiotensin system in hypertension. Adv. Chron. Kidney Dis. 2015;22:204–210. doi: 10.1053/j.ackd.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Chan L., Chaudhary K., Saha A., Chauhan K., Vaid A., Zhao S., Paranjpe I., Somani S., Richter F., Miotto R., et al. AKI in hospitalized patients with COVID-19. J. Am. Soc. Nephrol. 2021;32:151–160. doi: 10.1681/asn.2020050615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F., Yuen T.T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.C., Tsang J.O., Huang X., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/s2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. COVID-19: organoids go viral. Nat. Rev. Mol. Cell Biol. 2020;21:355–356. doi: 10.1038/s41580-020-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J.B., Hanff T.C., William P., Sweitzer N., Rosado-Santander N.R., Medina C., Rodriguez-Mori J.E., Renna N., Chang T.I., Corrales-Medina V., et al. Continuation versus discontinuation of renin-angiotensin system inhibitors in patients admitted to hospital with COVID-19: a prospective, randomised, open-label trial. Lancet. 2021;9:275–284. doi: 10.1016/s2213-2600(20)30558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes A.N., Zappia L., Er P.X., Oshlack A., Little M.H. Single-cell analysis reveals congruence between kidney organoids and human fetal kidney. Genome Med. 2019;11:3. doi: 10.1186/s13073-019-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellepiane S., Vaid A., Jaladanki S.K., Coca S., Fayad Z.A., Charney A.W., Bottinger E.P., He J.C., Glicksberg B.S., Chan L., Nadkarni G. Acute kidney injury in patients hospitalized with COVID-19 in New York city: temporal trends from march 2020 to april 2021. Kidney Med. 2021;3:877–879. doi: 10.1016/j.xkme.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshotels M.R., Xia H., Sriramula S., Lazartigues E., Filipeanu C.M. Angiotensin II mediates angiotensin converting enzyme type 2 internalization and degradation through an angiotensin II type I receptor-dependent mechanism. Hypertension. 2014;64:1368–1375. doi: 10.1161/HYPERTENSIONAHA.114.03743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Ferlicot S., Jamme M., Gaillard F., Oniszczuk J., Couturier A., May O., Grünenwald A., Sannier A., Moktefi A., Le Monnier O., et al. The spectrum of kidney biopsies in hospitalized patients with COVID-19, acute kidney injury, and/or proteinuria. Nephrol. Dial. Transplant. 2021 doi: 10.1093/ndt/gfab042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario C.M., Jessup J., Chappell M.C., Averill D.B., Brosnihan K.B., Tallant E.A., Diz D.I., Gallagher P.E. Effect of angiotensin-converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin-converting enzyme 2. Circulation. 2005;111:2605–2610. doi: 10.1161/CIRCULATIONAHA.104.510461. [DOI] [PubMed] [Google Scholar]
- Forrester S.J., Booz G.W., Sigmund C.D., Coffman T.M., Kawai T., Rizzo V., Scalia R., Eguchi S. Angiotensin II signal transduction: an update on mechanisms of physiology and pathophysiology. Physiol. Rev. 2018;98:1627–1738. doi: 10.1152/physrev.00038.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosbøl E.L., Butt J.H., Østergaard L., Andersson C., Selmer C., Kragholm K., Schou M., Phelps M., Gislason G.H., Gerds T.A., et al. Association of Angiotensin-Converting Enzyme Inhibitor or Angiotensin Receptor Blocker Use With COVID-19 Diagnosis and Mortality. JAMA. 2020;324:168–177. doi: 10.1001/jama.2020.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M., Moniwa N., Mita T., Fuseya T., Ishimura S., Ohno K., Shibata S., Tanaka M., Watanabe Y., Akasaka H., et al. Urinary angiotensin-converting enzyme 2 in hypertensive patients may be increased by olmesartan, an angiotensin II receptor blocker. Am. J. Hypertens. 2015;28:15–21. doi: 10.1093/ajh/hpu086. [DOI] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Krämer-Kühl A., Welsch K., Winkler M., Meyer B., Drosten C., et al. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/jvi.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/jvi.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin K.A., Bidani A.K. Progression of renal disease: renoprotective specificity of renin-angiotensin system blockade. Clin. J. Am. Soc. Nephrol. 2006;1:1054–1065. doi: 10.2215/cjn.02231205. [DOI] [PubMed] [Google Scholar]
- Guo X., Zhu Y., Hong Y. Decreased mortality of COVID-19 with renin-angiotensin-aldosterone system inhibitors therapy in patients with hypertension: a meta-analysis. Hypertension. 2020;76:e13–e14. doi: 10.1161/hypertensionaha.120.15572. [DOI] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley B., Naresh K.N., Roufosse C., Nicholson A.G., Weir J., Cooke G.S., Thursz M., Manousou P., Corbett R., Goldin R., et al. Histopathological findings and viral tropism in UK patients with severe fatal COVID-19: a post-mortem study. Lancet Microbe. 2020;1:e245–e253. doi: 10.1016/s2666-5247(20)30115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippisley-Cox J., Tan P.S., Coupland C. Risk of severe COVID-19 disease with ACE inhibitors and angiotensin receptor blockers: cohort study including 8.3 million people. Heart. 2020 doi: 10.1136/heartjnl-2020-318312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J.S., Ng J.H., Ross D.W., Sharma P., Shah H.H., Barnett R.L., Hazzan A.D., Fishbane S., Jhaveri K.D. Acute kidney injury in patients hospitalized with COVID-19. Kidney Int. 2020;98:209–218. doi: 10.1016/j.kint.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e775. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T., Mori Y., Aschauer P., Padmanabha Das K.M., Padera R.F., Weins A., Nasr M.L., Bonventre J.V. KIM-1/TIM-1 is a receptor for SARS-CoV-2 in lung and kidney. medRxiv. 2020 doi: 10.1101/2020.09.16.20190694. [DOI] [Google Scholar]
- Ishiyama Y., Gallagher P.E., Averill D.B., Tallant E.A., Brosnihan K.B., Ferrario C.M. Upregulation of angiotensin-converting enzyme 2 after myocardial infarction by blockade of angiotensin II receptors. Hypertension. 2004;43:970–976. doi: 10.1161/01.HYP.0000124667.34652.1a. [DOI] [PubMed] [Google Scholar]
- Jiang H.W., Zhang H.N., Meng Q.F., Xie J., Li Y., Chen H., Zheng Y.X., Wang X.N., Qi H., Zhang J., et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.Y., Choi J.C., You S.H., Kim W.Y. Association of Renin-angiotensin-aldosterone System Inhibitors With Coronavirus Disease 2019 (COVID-19)- Related Outcomes in Korea: A Nationwide Population-based Cohort Study. Clinical Infectious Diseases. 2020;71:2121–2128. doi: 10.1093/cid/ciaa624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärber G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Schmiedeb. Arch. für Exp. Pathol. Pharmakol. 1931;162:480–483. doi: 10.1007/BF01863914. [DOI] [Google Scholar]
- Klimas J., Olvedy M., Ochodnicka-Mackovicova K., Kruzliak P., Cacanyiova S., Kristek F., Krenek P., Ochodnicky P. Perinatally administered losartan augments renal ACE2 expression but not cardiac or renal Mas receptor in spontaneously hypertensive rats. J. Cell Mol. Med. 2015;19:1965–1974. doi: 10.1111/jcmm.12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori H., Nangaku M., Navar L.G., Nishiyama A. The intrarenal renin-angiotensin system: from physiology to the pathobiology of hypertension and kidney disease. Pharmacol. Rev. 2007;59:251–287. doi: 10.1124/pr.59.3.3. [DOI] [PubMed] [Google Scholar]
- Krafcikova P., Silhan J., Nencka R., Boura E. Structural analysis of the SARS-CoV-2 methyltransferase complex involved in RNA cap creation bound to sinefungin. Nat. Commun. 2020;11:3717. doi: 10.1038/s41467-020-17495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudose S., Batal I., Santoriello D., Xu K., Barasch J., Peleg Y., Canetta P., Ratner L.E., Marasa M., Gharavi A.G., et al. Kidney biopsy findings in patients with COVID-19. J. Am. Soc. Nephrol. 2020;31:1959–1968. doi: 10.1681/asn.2020060802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Schoggins J.W., Diamond M.S. Shared and distinct functions of type I and type III interferons. Immunity. 2019;50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.T., Nakayama T., Wu C.T., Goltsev Y., Jiang S., Gall P.A., Liao C.K., Shih L.C., Schürch C.M., McIlwain D.R., et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat. Commun. 2020;11:5453. doi: 10.1038/s41467-020-19145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand M., Bell S., Forni L., Joannidis M., Koyner J.L., Liu K., Cantaluppi V. Pathophysiology of COVID-19-associated acute kidney injury. Nat. Rev. Nephrol. 2021;17:751–764. doi: 10.1038/s41581-021-00452-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.J., Hunsicker L.G., Bain R.P., Rohde R.D. The effect of angiotensin-converting-enzyme inhibition on diabetic nephropathy. The Collaborative Study Group. N. Engl. J. Med. 1993;329:1456–1462. doi: 10.1056/NEJM199311113292004. [DOI] [PubMed] [Google Scholar]
- Lewis E.J., Hunsicker L.G., Clarke W.R., Berl T., Pohl M.A., Lewis J.B., Ritz E., Atkins R.C., Rohde R., Raz I. Renoprotective effect of the angiotensin-receptor antagonist irbesartan in patients with nephropathy due to type 2 diabetes. N. Engl. J. Med. 2001;345:851–860. doi: 10.1056/NEJMoa011303. [DOI] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang X., Chen J., Zhang H., Deng A. Association of Renin-Angiotensin System Inhibitors With Severity or Risk of Death in Patients With Hypertension Hospitalized for Coronavirus Disease 2019 (COVID-19) Infection in Wuhan, China. JAMA Cardiol. 2020;5:825–830. doi: 10.1001/jamacardio.2020.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes R.D., Macedo A.V.S., de Barros E.S.P.G.M., Moll-Bernardes R.J., Dos Santos T.M., Mazza L., Feldman A., D'Andréa Saba Arruda G., de Albuquerque D.C., Camiletti A.S., et al. Effect of discontinuing vs continuing angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers on days alive and out of the hospital in patients admitted with COVID-19: a randomized clinical trial. JAMA. 2021;325:254–264. doi: 10.1001/jama.2020.25864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta N., Kalra A., Nowacki A.S., Anjewierden S., Han Z., Bhat P., Carmona-Rubio A.E., Jacob M., Procop G.W., Harrington S., et al. Association of Use of Angiotensin-Converting Enzyme Inhibitors and Angiotensin II Receptor Blockers With Testing Positive for Coronavirus Disease 2019 (COVID-19) JAMA Cardiol. 2020;5:1020–1026. doi: 10.1001/jamacardio.2020.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza E.J., Manguiat K., Wood H., Drebot M. Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2. Curr. Protoc. Microbiol. 2020;57:ecpmc105. doi: 10.1002/cpmc.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S.E., Goldsmith C.S. Caution in identifying coronaviruses by electron microscopy. J. Am. Soc. Nephrol. 2020;31:2223–2224. doi: 10.1681/asn.2020050755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. U S A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. U S A. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913 e907. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelli Bozzo C., Nchioua R., Volcic M., Koepke L., Krüger J., Schütz D., Heller S., Stürzel C.M., Kmiec D., Conzelmann C., et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 2021;12:4584. doi: 10.1038/s41467-021-24817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles V.G., Lütgehetmann M., Lindenmeyer M.T., Sperhake J.P., Wong M.N., Allweiss L., Chilla S., Heinemann A., Wanner N., Liu S., et al. Multiorgan and renal tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puskarich M.A., Cummins N.W., Ingraham N.E., Wacker D.A., Reilkoff R.A., Driver B.E., Biros M.H., Bellolio F., Chipman J.G., Nelson A.C., et al. A multi-center phase II randomized clinical trial of losartan on symptomatic outpatients with COVID-19. EClinicalMedicine. 2021;37:100957. doi: 10.1016/j.eclinm.2021.100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., Han Y., Zhang X.Y., Zhou W., Qiu Y., Zhou X. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol. Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds H.R., Adhikari S., Pulgarin C., Troxel A.B., Iturrate E., Johnson S.B., Hausvater A., Newman J.D., Berger J.S., Bangalore S., et al. Renin-Angiotensin-Aldosterone System Inhibitors and Risk of Covid-19. N. Engl. J. Med. 2020;382:2441–2448. doi: 10.1056/NEJMoa2008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G.M., Delsante M., Pilato F.P., Gnetti L., Gabrielli L., Rossini G., Re M.C., Cenacchi G., Affanni P., Colucci M.E., et al. Kidney biopsy findings in a critically ill COVID-19 patient with dialysis-dependent acute kidney injury: a case against “SARS-CoV-2 nephropathy”. Kidney Int. Rep. 2020;5:1100–1105. doi: 10.1016/j.ekir.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J.A., Marshall J.C., Slutsky A., Murthy S., Sweet D., Lee T., Singer J., Patrick D.M., Du B., Peng Z., et al. Study protocol for a multicentre, prospective cohort study of the association of angiotensin II type 1 receptor blockers on outcomes of coronavirus infection. BMJ Open. 2020;10:e040768. doi: 10.1136/bmjopen-2020-040768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoriello D., Khairallah P., Bomback A.S., Xu K., Kudose S., Batal I., Barasch J., Radhakrishnan J., D'Agati V., Markowitz G. Postmortem kidney pathology findings in patients with COVID-19. J. Am. Soc. Nephrol. 2020;31:2158–2167. doi: 10.1681/asn.2020050744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller T., Hirschbühl K., Burkhardt K., Braun G., Trepel M., Märkl B., Claus R. Postmortem examination of patients with COVID-19. JAMA. 2020;323:2518–2520. doi: 10.1001/jama.2020.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.A., Leibundgut M., Thiel V., Mühlemann O., Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U S A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Bansal V., Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 2020;32:108175. doi: 10.1016/j.celrep.2020.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M.J., Ye M., Wysocki J., William J., Lloveras J., Batlle D. Localization of ACE2 in the renal vasculature: amplification by angiotensin II type 1 receptor blockade using telmisartan. Am. J. Physiol. Ren. Physiol. 2009;296:F398–F405. doi: 10.1152/ajprenal.90488.2008. [DOI] [PubMed] [Google Scholar]
- South A.M., Tomlinson L., Edmonston D., Hiremath S., Sparks M.A. Controversies of renin-angiotensin system inhibition during the COVID-19 pandemic. Nature Reviews Nephrology. 2020;16:305–307. doi: 10.1038/s41581-020-0279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Yang M., Wan C., Yi L.X., Tang F., Zhu H.Y., Yi F., Yang H.C., Fogo A.B., Nie X., Zhang C. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Chiu H.S., Little M.H. Generation of kidney organoids from human pluripotent stem cells. Nat. Protoc. 2016;11:1681–1692. doi: 10.1038/nprot.2016.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasato M., Er P.X., Chiu H.S., Maier B., Baillie G.J., Ferguson C., Parton R.G., Wolvetang E.J., Roost M.S., Chuva de Sousa Lopes S.M., Little M.H. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526:564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- Taylor J.K., Coleman C.M., Postel S., Sisk J.M., Bernbaum J.G., Venkataraman T., Sundberg E.J., Frieman M.B. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J. Virol. 2015;89:11820–11833. doi: 10.1128/jvi.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T., Lindström N.O., Ransick A., De Sena Brandine G., Guo Q., Kim A.D., Der B., Peti-Peterdi J., Smith A.D., Thornton M., et al. In vivo developmental trajectories of human podocyte inform in vitro differentiation of pluripotent stem cell-derived podocytes. Dev. Cell. 2019;50:102–116.e106. doi: 10.1016/j.devcel.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trump S., Lukassen S., Anker M.S., Chua R.L., Liebig J., Thürmann L., Corman V.M., Binder M., Loske J., Klasa C., et al. Hypertension delays viral clearance and exacerbates airway hyperinflammation in patients with COVID-19. Nat. Biotechnol. 2021;39:705–716. doi: 10.1038/s41587-020-00796-1. [DOI] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Vaduganathan M., Vardeny O., Michel T., McMurray J.J.V., Pfeffer M.A., Solomon S.D. Renin-angiotensin-aldosterone system inhibitors in patients with covid-19. N. Engl. J. Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/s0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werion A., Belkhir L., Perrot M., Schmit G., Aydin S., Chen Z., Penaloza A., De Greef J., Yildiz H., Pothen L., et al. SARS-CoV-2 causes a specific dysfunction of the kidney proximal tubule. Kidney Int. 2020;98:1296–1307. doi: 10.1016/j.kint.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wysocki J., Ye M., Hassler L., Gupta A.K., Wang Y., Nicoleascu V., Randall G., Wertheim J.A., Batlle D. A novel soluble ACE2 variant with prolonged duration of action neutralizes SARS-CoV-2 infection in human kidney organoids. J. Am. Soc. Nephrol. 2021;32:795–803. doi: 10.1681/asn.2020101537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Wu M., Chen S., Zhang T., Ye L., Liu J., Li H. Long term culture of human kidney proximal tubule epithelial cells maintains lineage functions and serves as an ex vivo model for coronavirus associated kidney injury. Virol. Sin. 2020;35:311–320. doi: 10.1007/s12250-020-00253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Castro Maria Florencia G., McCune Broc T., Zeng Q., Rothlauf Paul W., Sonnek Naomi M., Liu Z., Brulois Kevin F., Wang X., Greenberg Harry B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Zhu L., Cai J., Lei F., Qin J.J., Xie J., Liu Y.M., Zhao Y.C., Huang X., Lin L., et al. Association of inpatient use of angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers with mortality among patients with hypertension hospitalized with COVID-19. Circ. Res. 2020;126:1671–1681. doi: 10.1161/circresaha.120.317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single cell RNA-Seq data have been deposited in NCBI GEO (which automatically makes SRA deposit) and are publicly available as of the date of publication with the following accession number: GSE149687. Single-cell RNA-Seq datasets were integrated with previously infectable kidney organoids which can be accessed with the following accession number: GSNE147863. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.