Abstract
SARS-CoV-2 variants of concern (VOCs) display enhanced transmissibility and resistance to antibody neutralization. Comparing the early 2020 isolate EU-1 to the VOCs Alpha, Beta, and Gamma in mice transgenic for human ACE2 reveals that VOCs induce a broadened scope of symptoms, expand systemic infection to the gastrointestinal tract, elicit the depletion of natural killer cells, and trigger variant-specific cytokine production patterns. Gamma infections result in accelerated disease progression associated with increased immune activation and inflammation. All four SARS-CoV-2 variants induce pDC depletion in the lungs, paralleled by reduced interferon responses. Remarkably, VOCs also use the murine ACE2 receptor for infection to replicate in the lungs of wild-type animals, which induce cellular and innate immune responses that apparently curtail the spread of overt disease. VOCs thus display distinct intrinsic pathogenic properties with broadened tissue and host range. The enhanced pathogenicity of VOCs and their potential for reverse zoonotic transmission pose challenges to clinical and pandemic management.
Keywords: SARS-CoV-2, variants of concern, Alpha variant, Beta variant, Gamma variant, K18-hACE2, cytokine production, ISG expression, cellular immunity
Graphical abstract
Stolp et al. show that the infection of transgenic mice with early SARS-CoV-2 variant EU-1 or variants of concern (VOCs) Alpha, Beta, and Gamma results in lethal infection, with strain-specific patterns of immune cell recruitment, cytokine production, and organ tropism. Gamma displays enhanced pathogenicity. VOCs replicate in the lungs of wild-type mice.
Introduction
The coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) rapidly evolved to a pandemic in early 2020, with more than 363 million confirmed cases and 5.63 million deaths by January 27, 2022 (https://coronavirus.jhu.edu/). While effective and broadly acting therapeutics against SARS-CoV-2 infection have only recently become available, a series of effective vaccines has been implemented on a global scale at an unprecedented speed (Kyriakidis et al., 2021). The availability of vaccines remains, however, limited in many countries, and the emergence of SARS-CoV-2 variants that are increasingly resistant to neutralization by antibodies triggered by the current vaccination strategies raises concerns about the long-term efficacy of the current vaccination campaigns. Efficient proofreading by the RNA-dependent RNA polymerase ensures relatively low mutation rates of the SARS-CoV-2 genome (V’kovski et al., 2021). However, high case incidences and long-lasting viral replication, especially in immunocompromised and elderly patients, result in the generation of new variants with selective advantages during infection, transmission, and adaptation to host immune responses (Peacock et al., 2021). Particular attention is given to mutations in the gene encoding for the receptor-binding Spike protein since modifications therein have the potential to alter the affinity and/or specificity to entry receptors on target cells and thus potentially affect virulence and viral tropism (Harvey et al., 2021). Early in the pandemic, emerging SARS-CoV-2 variants carrying Spike proteins with a D614G mutation rapidly became dominant and are now considered the prototype of early pandemic SARS-CoV-2 (e.g., EU-1, B.1.177 used in the present study) (Hou et al., 2020; Korber et al., 2020; Zhou et al., 2021). Subsequently, additional virus variants with variations in Spike emerged and, due to their ability to spread rapidly and displace local SARS-CoV-2 variants, were classified as variants of concern (VOCs). These VOCs include the variants B.1.1.7 (referred to as Alpha variant), B.1.351 (referred to as Beta variant), and P.1/B.1.1.28.1 (referred to as Gamma variant). These VOCs display enhanced infectivity and transmissibility in human populations, but also in cell culture and animal models (Boehm et al., 2021; Cai et al., 2021; Gómez et al., 2021; Harvey et al., 2021; Wang et al., 2021). Moreover, their sensitivity to neutralization by antibodies induced by natural infection or vaccination is in part reduced (Bates et al., 2021; Garcia-Beltran et al., 2021; Harvey et al., 2021). While these observations suggest that VOCs are able to more efficiently spread between hosts and evade humoral immunity, information on their intrinsic pathogenic potential is lacking. In this study, we therefore compared pathogenesis, organ tropism, and immune responses of infections of transgenic (Tg) mice expressing the human ortholog of the SARS-CoV-2 entry receptor angiotensin-converting enzyme 2 (ACE2) under the control of the epithelial cell cytokeratin-18 promoter (K18-hACE2 mice, henceforth: hACE2 Tg mice) as well as non-transgenic wild-type (WT) mice following intranasal challenge with SARS-CoV-2 variant EU-1 or VOCs Alpha, Beta, or Gamma. The results reveal that (1) disease progression and outcome, induction of cellular and innate immune responses, and organ tropism differ between EU-1 and VOCs but also among VOCs, and (2) VOCs display expanded host tropism, enabling them to replicate in WT mice.
Results
Enhanced virulence and pathogenicity of SARS-CoV-2 VOC Gamma in hACE2 Tg mice
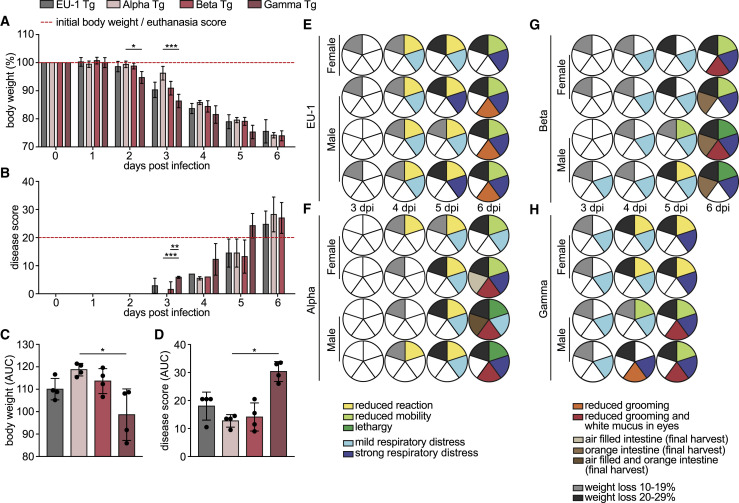
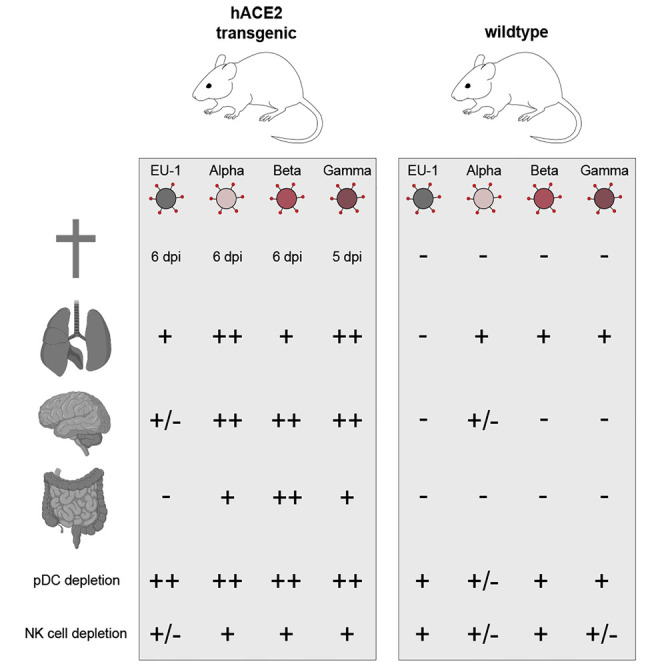
To compare intrinsic pathogenic properties of SARS-CoV-2 VOCs, we intranasally (i.n.) challenged 8-week-old heterozygous hACE2 Tg mice of both sexes with 104 plaque forming units (pfu) of the early SARS-CoV-2 isolate EU-1 or VOCs Alpha, Beta, or Gamma. Consistent with previous reports (Golden et al., 2020; Oladunni et al., 2020; Winkler et al., 2020; Yinda et al., 2021; Zheng et al., 2021), infection with EU-1 caused a steady reduction in body weight, first notable on day 3 post-infection (p.i.) (Figure 1A, see Figure 1C for overall body weight development). At day 6 p.i., animals lost more than 20% of their initial body weight, paralleled by the manifestation of additional symptoms of pathogenic SARS-CoV-2 infection, including reduced mobility, signs of respiratory distress, and reduced grooming activity (Figures 1B and 1E). At this stage, these animals reached an overall disease score of above 20 that required euthanasia (red dashed line in Figure 1B; see Figure 1D for overall disease score development, Figure 1E for symptom profiles, and STAR Methods and Video S1 for disease score criteria and examples). Infections with Alpha or Beta VOCs resulted in disease progression comparable to that observed with EU-1, although the loss of body weight and onset of disease were slightly delayed for infections with Alpha (Figures 1A–1D, 1F, and 1G). In contrast, infections with Gamma resulted in significantly accelerated and more pronounced body weight loss, already notable at day 2 p.i., and accelerated disease progression necessitating euthanasia already at day 5 p.i. (Figures 1A–1D, 1H, S1A, and S1B). Key disease symptoms, including lethargy and respiratory distress, associated with macroscopic signs of lung inflammation such as the deep-red appearance (Figure S1C), were common to infections by all four SARS-CoV-2 variants. In addition, infections with the three VOCs caused the generation of white mucus in the eyes, and infections with Alpha and Beta affected the intestine, which appeared macroscopically orange and bloated (Figures 1E–1H and S1D). SARS-CoV-2 VOCs can thus trigger variant-specific symptoms, and Gamma displays significantly enhanced pathogenicity in hACE2 Tg mice.
Figure 1.
Disease progression and range of symptoms differ in hACE2 Tg mice upon infection with SARS-CoV-2 VOCs
hACE2 Tg mice were infected i.n. with 104 pfu of indicated SARS-CoV-2 variants on day 0 and monitored for 5–6 days p.i. Body weight and symptoms were monitored daily according to the score sheet as described in STAR Methods.
(A) Body weight relative to 0 days p.i. Dashed red line indicates initial body weight (100%).
(B) Disease score according to the score sheet. Dashed red line indicates score at which animals become eligible for euthanasia. Shown are mean values ± SDs from 7 (0–3 days p.i.) and 4 (4–6 days p.i.) mice per time point.
(C and D) Area under the curve (AUC) of body weight (C) and disease score (D) of the animals analyzed in (A) and (B) calculated for 0–5 days p.i., as shown in Figures S1A and S1B. Shown are mean values ± SDs. Black dots represent individual animals. p values were calculated performing a Kruskal-Wallis test with a Dunn’s multiple comparison test (∗p ≤ 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005).
(E–H) Breakdown of symptoms for individual mice 3 to 6 days p.i. with (E) EU-1, (F) Alpha, (G) Beta, or (H) Gamma. Each row of pie charts indicates 1 animal over the course of the experiment. Sex of mice is indicated. Individual slices indicate reaction (green), breathing (blue), grooming (red), appearance of intestine upon harvest (brown), and body weight (gray), starting clockwise from 12:00. See Figure S1 and Video S1 for further information on the observed symptoms.
K18-hACE2 transgenic mice, infected with SARS-CoV-2 that reached the maximum disease score. The first part displays lethargic behavior. Lethargic mice hardly move in the cage and display limited motivation to return to an upright position, when gently moved to the side. The second part displays a mouse with mild respiratory distress, defined as clearly visible and enhanced breathing motion. The third part displays a mouse with strong respiratory distress, defined as dramatically strained breathing motion.
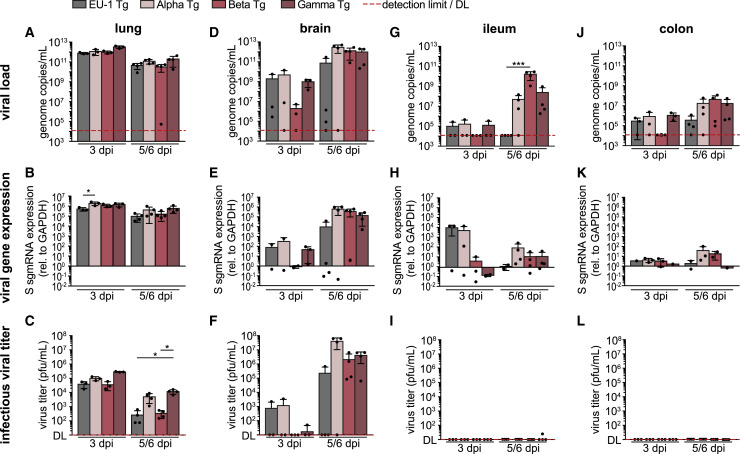
SARS-CoV-2 VOCs differ in their organ tropism and organ viral load in hACE2 Tg mice
We next analyzed the viral load by qPCR for the SARS-CoV-2 N1 gene, viral gene expression by qPCR for viral S gene subgenomic (sgm) RNA, and the production of infectious viral progeny by plaque assay on Vero E6 cells in various organs at day 3 p.i. and day 5 (Gamma) or 6 (EU-1, Alpha, and Beta) p.i. (Figures 2 and S2A–S2N). Infection with 104 pfu corresponded to 6.24 × 106, 2.21 × 106, 4.27 × 105, and 3.11 × 107 copies of virion-associated N1 gene for EU-1, Alpha, Beta, and Gamma, respectively, and viral loads of up to 6 orders of magnitude higher were detected by qPCR for N1 gene on day 3 p.i., revealing local viral replication. During the viral replication cycle, viral genes such as the S gene are detectable only in infected cells as they are expressed from sgmRNAs generated by splicing of full-length viral RNA and not packaged into virions. Using qPCR primer sets binding to distant positions in the viral genome restricts amplification to sgmRNAs, thus representing markers of viral gene expression. The stability of sgmRNAs allows their detection over extended periods of time in tissue (Alexandersen et al., 2020), thus not only reporting current but also previous viral gene expression, enabling us to detect the transient replication of SARS-CoV-2 variants in tissues even in the absence of detectable infectious virus.
Figure 2.
SARS-CoV-2 VOCs differ in organ tropism and organ viral load upon infection of hACE2 Tg mice
hACE2 Tg mice were infected i.n. with 104 pfu of indicated SARS-CoV-2 variants 0 days p.i. Lung (A–C), brain (D–F), ileum (G–I), and colon (J–L) were harvested 3 and 5 (Gamma) or 6 (EU-1, Alpha, Beta) days p.i. and analyzed for viral load by qPCR for SARS-CoV-2 N1 gene (A, D, G, and J), viral gene expression by qPCR for SARS-CoV-2 S gene sgmRNA (B, E, H, and K), and infectious viral titer by plaque assay on Vero E6 cells (C, F, I, and L). Shown are mean values ± SDs from 3 (3 days p.i.) to 4 (5/6 days p.i.) mice. Each black dot represents an individual animal. Dashed red line indicates detection limit (DL), which is 12,042 genome copies/mL for the N1 gene qPCR and 0 plaques/mL for the plaque assay. S gene sgmRNA is presented as 2−ΔΔCT with 100 representing mock; down error bars are omitted in case of negative values, due to the logarithmic scale. p values were calculated performing a Kruskal-Wallis test with a Dunn’s multiple comparison test (∗p ≤ 0.05).
See Figure S2 for the analysis of further organs and immunohistology of lung and brain tissue.
In line with previous reports (Golden et al., 2020; Oladunni et al., 2020; Winkler et al., 2020; Yinda et al., 2021; Zheng et al., 2021), lung and brain, together with trachea (at lower levels), were primary target organs infected by EU-1, with high viral load paralleled by efficient viral gene expression and production of infectious progeny (Figures 2A–2F, S2A, S2H, and S2N). Virus replication in the lung reached its maximum already by day 3 p.i., while infection of the brain was delayed but increased over the course of the experiment (Figures 2A and 2D). Kidney and liver tissue displayed viral loads barely above the detection limit (Figures S2B and S2C), while the spleen, submandibular salivary gland (SMG), and heart contained low amounts of viral RNA that remained stable or decreased during the course of infection (Figures S2D–S2F) and did not result in the production of infectious progeny (Figures S2J–S2L). Infections with the three VOCs in general resembled EU-1, with predominant replication in lung, brain, and trachea and lack thereof in most other organs analyzed, with the exception of Alpha supporting negligible production of infectious virus in liver (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 72I). However, we also observed several notable differences from infections with EU-1: Viral gene expression of VOCs in lung tended to be higher than for EU-1, reaching statistical significance for Alpha on day 3 p.i. (Figure 2B). Infectious virus production by Alpha and Gamma was significantly increased in lung at day 5/6 p.i. relative to infection with the other variants (Figure 2C). In the brain, the replication of Beta started with some delay, but at day 5/6 p.i., at least 50% of mice infected with VOCs reached viral loads and produced levels of infectious virus that were at least one order of magnitude higher than the highest levels observed in EU-1 (Figures 2D–2F). This tropism was in line with immunohistochemical staining of lungs and brains for nucleocapsid protein at day 5/6 p.i. that revealed pronounced nucleocapsid expression in the lungs of animals infected by all of the variants. Notably, infection with Gamma resulted in a significantly higher amount of nucleocapsid-positive cells than infection with the other variants (Figures S2O and S2P). In turn and in line with the detected amounts of infectious virus, nucleocapsid expression in the brains of infected animals was markedly increased for all VOCs (at half of the animals with more than 20% positive cells) relative to animals infected with EU-1 (only one in four animals positive for nucleocapsid expression [5% of cells]) (Figures S2O and S2Q).
Figure 3.
Infection of hACE2 Tg mice with SARS-CoV-2 variants induced changes in immune cell composition of the lung, most pronounced for VOC Gamma
(A–J) Flow cytometric analysis of lung single-cell suspensions of hACE2 Tg mice infected i.n. with 104 pfu of indicated SARS-CoV-2 variants on 0 days p.i., 3 and 5 days p.i. (Gamma), or 6 days p.i. (EU-1, Alpha, Beta), analyzed as shown in Figure S3. Shown are mean fold changes in frequency ±SD of (A) hematopoietic cells, (B) CD8+ T cells, (C) PD-1pos CD8+ T cells, (D) CD4+ T cells, (E) inflammatory monocytes, (F) neutrophils, (G) CD11b+ DCs, (H) inflammatory DCs, (I) pDCs, and (J) NK cells, relative to the mean of mock infected hACE2 Tg mice (n = 4). Percentages of immune cells and single mock measurements are shown in Figure S4.
(K–M) Relative amount of (K) IFN-α4, (L) IFN-β, and (M) IFN-λ mRNA quantified by qRT-PCR of lung homogenates relative to the mean of mock infected mice (n = 4, 100) and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as housekeeper. Each black dot represents an individual animal. Red dashed line or 100 indicates the mean of mock, arbitrarily set to 1. p values were calculated performing a Kruskal-Wallis test with a Dunn’s multiple comparison test (∗p ≤ 0.05).
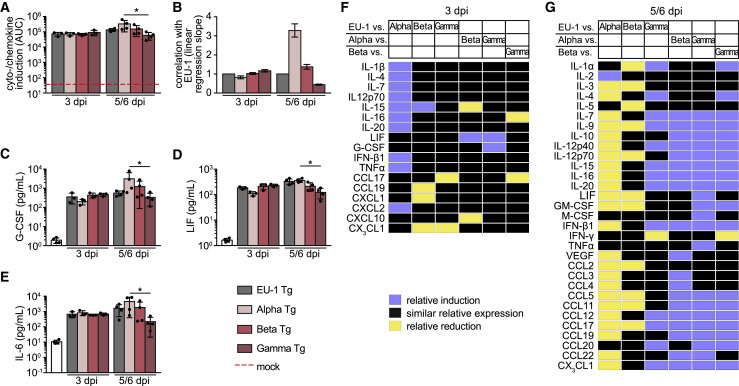
Figure 4.
Differential cytokines and chemokines are induced by infection of hACE2 Tg mice with SARS-CoV-2 variants
Lungs of hACE2 Tg mice infected i.n. with 104 pfu of indicated SARS-CoV-2 variants were harvested 0, 3 (n = 4), and 5 (Gamma, n = 4)) or 6 (EU-1, Alpha, Beta; n = 4) days p.i., homogenized and analyzed for cytokines/chemokines. For a complete set of cytokines/chemokines, see Figures S5 and S6.
(A) AUC of the fold induction of cytokines/chemokines relative to mock, as shown in Figure S5. Dashed red line indicates AUC of mock samples, which were arbitrarily set to 1.
(B) Slopes with 95% confidence intervals of the linear regression curves, as plotted in the correlation graphs of the fold cytokine induction of the different SARS-CoV-2 variants relative to EU-1 shown in Figure S7. Slopes were forced to go through the center of origin.
(C–E) Absolute amounts of (C) G-CSF, (D) LIF, and (E) IL-6 measured in lung homogenates. Shown are mean values ± SDs. Each black dot represents an individual animal. p values were calculated performing a Kruskal-Wallis test with a Dunn’s multiple comparison test (∗p ≤ 0.05).
(F and G) Heatmaps depicting the relative induction (blue) or reduction (yellow) of specific cytokines and chemokines in lung homogenates of SARS-CoV-2-infected hACE2 Tg mice at day 3 (F) or day 5/6 (G) p.i. Shown are differences in the relative amounts of cytokines/chemokines between indicated variants that are significantly outside of the slope and 95% confidence interval of the correlation plots expecting similar levels of induction (see Figure S7).
Figure 5.
SARS-CoV-2 VOCs exhibit broader organ tropism and replication capacity in WT mice
WT mice were infected i.n. with 104 pfu of the indicated SARS-CoV-2 variants on 0 days p.i.
(A) Body weight was monitored daily and is shown relative to 0 days p.i. Red dashed line indicates initial body weight (100%). Mean values of hACE2 Tg mice infected with the respective VOCs, as shown in Figure 1A, are indicated by transparent bars.
(B) AUC of the body weight, calculated for 0–5 days p.i., as shown in Figure S8A. Lung (C–E), trachea (F–H), and brain (I–K) were harvested 3 (n = 3) and 5 (Gamma) or 6 (EU-1, Alpha, Beta) (n = 4) days p.i. and analyzed for viral load by qPCR for SARS-CoV-2 N1 gene (C, F, and I), viral gene expression by qPCR for SARS-CoV-2 S gene sgmRNA (D, G, and J), and infectious viral titer by plaque assay on Vero E6 cells (E, H, and K). Shown are mean values ± SDs. Each black dot represents an individual animal. Mean values of hACE2 Tg mice infected with the respective VOCs, as shown in Figures 1C, 2A–2F, S2A, S2H, and S2N, are indicated by colored round symbols. Dashed red line indicates DL, which is 12,042 genome copies/mL for the N1 qPCR and 0 plaques/mL for the plaque assay. S gene sgmRNA is presented as 2−ΔΔCT, with 100 representing mock; down error bars are omitted in case of negative values, due to the logarithmic scale. p values were calculated performing a Kruskal-Wallis test with a Dunn’s multiple comparison test (∗p ≤ 0.05). Analysis of additional organs is shown in Figure S8. Mutations in the different SARS-CoV-2 variants that may account for the observed differences are summarized in Table S1.
Figure 6.
Infection of WT mice with SARS-CoV-2 variants induced pronounced changes in immune cell composition of the lung, even upon limited local replication
(A–J) Flow cytometric analysis of lung single-cell suspensions of the WT mice analyzed in Figure 5: infected i.n. with 104 pfu of indicated SARS-CoV-2 variants 0 days p.i., 3 (n = 3) and 5 (Gamma), or 6 (EU-1, Alpha, Beta) days p.i. (n = 4), analyzed as shown in Figure S3. Shown are mean fold changes in frequency ±SDs of (A) hematopoietic cells, (B) CD8+ T cells, (C) PD-1pos CD8+ T cells, (D) CD4+ T cells, (E) inflammatory monocytes, (F) neutrophils, (G) CD11b+ DCs, (H) inflammatory DCs, (I) pDCs, and (J) NK cells relative to the mean of mock infected WT mice (n = 4, 1). Percentages of immune cells and single mock measurements are shown in Figure S9.
(K–M) Relative amount of (K) IFN-α4, (L) IFN-β, and (M) IFN-λ mRNA quantified by qRT-PCR of lung homogenates relative to the mean of mock infected mice (n = 4) and normalized to GAPDH as housekeeper. Each black dot represents 1 individual animal. Red dashed line or 100 indicates the mean of mock, arbitrarily set to 1. Mean values of K18-hACE2 transgenic (Tg) mice infected with the respective VOCs, as shown in Figure 3, are indicated by colored round symbols. p values were calculated performing a Kruskal-Wallis test with a Dunn’s multiple comparison test (∗p ≤ 0.05).
Figure 7.
Differential cytokines and chemokines are induced by infection of WT mice with variants of SARS-CoV-2
Lungs of the WT mice analyzed in Figure 5: infected i.n. with 104 pfu of indicated SARS-CoV-2 variants were harvested 0, 3 (n = 3), and 5 (Gamma, n = 4), or 6 (EU-1, Alpha, Beta; n = 4) days p.i., homogenized, and analyzed for cytokines/chemokines. A complete set of analyzed cytokines/chemokines are presented in Figures S10 and S11.
(A) AUC of the fold induction of cytokines/chemokines relative to mock, as shown in Figure S10. Dashed red line indicated AUC of mock samples that were arbitrarily set to 1.
(B) Slopes with 95% confidence intervals of the linear regression curves, as plotted in the correlation graphs of the fold cytokine induction of the different SARS-CoV-2 variants relative to EU-1 shown in Figure S12. Slopes were forced to go through the center of origin.
(C–G) Absolute amounts of (B) CCL2, (C) LIF, (D) IL-6, (E) IL-16, and (F) CX3CL1 measured in lung homogenates. Shown are mean values ± SDs. Each black dot represents an individual animal. Mean values of hACE2 Tg mice infected with the respective VOCs, as shown in Figures 4A, 4B, 4D, 4E, S6A, S6G, and S6L, are indicated by colored round symbols. p values were calculated performing a Kruskal-Wallis test with a Dunn’s multiple comparison test (∗p ≤ 0.05). Heatmaps depicting the relative induction (green) or reduction (red) of specific cytokines and chemokines in lung homogenates of SARS-CoV-2-infected WT mice at days 3 (F) or 5/6 (G) p.i. Shown are differences in the relative amounts of cytokines and chemokines between the indicated SARS-CoV-2 variants that are significantly outside of the slope and 95% confidence interval of the correlation plots expecting similar levels of induction (see Figure S12).
Another important difference was the detection of robust viral loads in the colon and particularly the ileum for all VOCs but not EU-1 (Figures 2G–2L). However, these robust amounts of VOC RNA were not associated with a detectable production of infectious progeny. Consistently, in feces, viral RNA was mainly detected at days 5 and 6 p.i., but no infectious virus was detectable (Figures S2G and S2M). This suggests that VOCs have the ability to infect the intestine, but either do not actively replicate and spread in this tissue or are readily inactivated by components in the gastrointestinal (GI) tract. These results reveal that compared to EU-1, VOCs have a broadened tissue tropism and different replication kinetics in hACE2 Tg mice.
Cellular immune activation and immune cell recruitment into lungs of hACE2 Tg mice upon VOC infection
We next assessed immune cell activation and composition in the lungs of hACE2 Tg mice in response to infection by flow cytometry of single-cell suspensions (Figures 3 and S4; see Figure S3 for gating strategy). CD45+ hematopoietic cells (Figures 3A and S4A) were reduced by roughly 50% on day 3 p.i. by all of the variants in comparison to mock-infected animals; however, they remained reduced only in infections with Alpha and Gamma, while numbers recovered in EU-1- and Beta-infected animals. SARS-CoV-2 infection did not affect the overall abundance of CD8+ T cells (Figures 3B and S4B), but especially on day 3 p.i., the fraction of PD-1pos CD8+ T cells (Figures 3C and S4C), indicative of CD8+ T cell activation, was increased upon infection with Beta and Gamma relative to uninfected mice. Infection with all of the variants increased the amounts of lung CD4+ T cells (Figures 3D and S4D), inflammatory monocytes (Figures 3E and S4E), neutrophils (Figures 3F and S4F), CD11b+ dendritic cells (DCs) (Figures 3G and S4G), and inflammatory DCs (Figures 3H and S4H), and this effect was most pronounced for Gamma at day 5 p.i. Strikingly, the relative amount of pDCs (Figures 3I and S4I) in lung tissue gradually decreased over time in infected animals irrespective of the virus variant, resulting in the apparent depletion of pDCs at days 5 or 6 p.i. Since pDCs are one of the main producers of all types of interferons (IFNs) in response to infection (Ali et al., 2019; Gary-Gouy et al., 2002; Hagberg et al., 2011; Swiecki and Colonna, 2015), we quantified IFN mRNA levels (i.e., IFN-α4, IFN-β, and IFN-λ mRNA) in lung homogenates and found that their expression reduced markedly upon SARS-CoV-2 infection compared to uninfected mice (Figures 3K–3M). Finally, the abundance of natural killer (NK) cells was also reduced upon infection and this effect was more pronounced for all VOCs compared to EU-1 at the final harvest (Figures 3J and S4J). Infection with all of the SARS-CoV-2 variants triggered marked immune activation and inflammation, and this effect was most pronounced for Gamma. Depletion of lung pDCs was observed upon infection with all of the variants, and this interference with innate immune cell abundance was further emphasized by an additional reduction of NK cell numbers.
Infections with SARS-CoV-2 VOCs induce distinct cytokine and chemokine profiles in hACE2 Tg mice
To complement the characterization of cellular immune responses, we determined the concentrations of 42 cytokines and chemokines in lung homogenates of infected animals on days 3 and 5 or 6 p.i. (Figures S5 and S6). In line with previous reports from infected hACE2 Tg mice and COVID-19 patients (Olbei et al., 2021; Pum et al., 2021; Winkler et al., 2020; Yinda et al., 2021), infection with all SARS-CoV-2 variants triggered the production of a wide array of proinflammatory cytokines (interleukin-6 [IL-6], IL-1-α and -β, and tumor necrosis factor α [TNF-α]), T cell-derived cytokines (IL-17, IFN-γ, IL-2, and IL-4), and myeloid cell-derived chemokines (C-C motif chemokine ligand 2 [CCL2], CCL3, CCL4, and C-X-C motif chemokine ligand 1 [CXCL1]) (Figure S5). Across all cytokines examined, this induction was more pronounced for Alpha relative to EU-1 in comparison to the other VOCs at the later time point (Figures 4A and 4B), and Gamma tended to trigger less potent cytokine responses, with specific cytokines significantly less induced (e.g., granulocyte-colony-stimulating factor [G-CSF], leukemia inhibitory factor [LIF], IL-6 (Figures 4C–4E), CCL3, IFN-γ, CCL5, IL-17, IL-2, IL-11 [Figures S6B, S6I–S6K, S6M, and S6U]). The relative overall induction of the cytokine response differed between infections with the SARS-CoV-2 variants, as made apparent by the slope of the linear regression of curves in the correlation plots (Figures 4B and S7). In addition, correlation plots highlighted variant-specific, significant reductions or inductions of individual cytokines (Figures 4F and 4G). These results suggest that VOCs induce different sets of cytokines in the lungs of hACE2 Tg mice and indicate that the accelerated disease progression induced by Gamma is associated by an attenuated cytokine response.
SARS-CoV-2 VOCs replicate in WT mice but virus spread is restricted
Earlier SARS-CoV-2 variants such as EU-1 are unable to use murine ACE2 for cell entry (Hoffmann et al., 2020). Pathogenic mouse models of SARS-CoV-2 infection therefore are either based on Tg animals that express human ACE2 or use virus variants that have been experimentally adapted to the use of murine ACE2 (Dinnon et al., 2020; Jia et al., 2020; Lutz et al., 2020). Since the above analyses had revealed that Alpha, Beta, and Gamma display different and variant-specific pathogenic properties and tissue tropism in hACE2 Tg mice, we wondered whether their species tropism may have expanded. Following i.n. challenge with EU-1 or each of the VOCs, none of the WT mice showed symptoms of SARS-CoV-2-induced disease. Due to the high-calorie diet used, animals even gained weight during the course of the experiment in the absence of disease, an effect that was less pronounced for Alpha-infected WT mice (Figures 5A, 5B, and S8A). As expected, infection with EU-1 resulted in the detection of viral RNA amounts close to the detection limit, even in primary target organs, and viral gene expression or production of infectious progeny was not observed (Figures 5C–5K). In sharp contrast, infection with all VOCs resulted in the detection of up to 109 viral genome copies per milliliter of lung homogenate 3 days p.i. that decreased to approximately 107 viral genome copies per milliliter at day 5/6 p.i. (Figure 5C). These amounts of viral genomes in the lung were several orders of magnitude lower than those observed in hACE2 Tg mice, but 103- to 104-fold above those contained in the inoculum and thus clearly resulting from the production of new virus particles. Similarly, at day 3 p.i., VOC infection of WT mice resulted in robust viral gene expression and production of infectious progeny, albeit at lower levels relative to infections in ACE2 Tg mice, and these levels decreased or became undetectable in the case of Beta at day 6 p.i. (Figures 5D and 5E). Viral RNA was also detected in the tracheas and brains of WT mice infected with VOCs, but to significantly lower levels than in hACE2 Tg mice and without production of infectious progeny (Figures 5F–5K). However, in the case of Alpha, viral RNA levels in brains increased from day 3 to 5/6 p.i. (Figure 5I). Viral RNA was essentially undetectable in kidney, liver, spleen, SMG, heart, ileum, colon, and feces, with the exception of negligible amounts of viral RNA in the hearts of animals infected with VOCs and spleens of Gamma-infected animals at day 3 p.i. (Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6, Figure 78B–S8I). Similar to hACE2 Tg mice, infection of WT mice with all of the SARS-CoV-2 variants resulted in a reduction of CD45+ hematopoietic cells in lungs, which also persisted at late time points (Figures 6A and S9A). Levels of PD-1pos CD8+ T cells, CD4+ T cells, inflammatory monocytes, neutrophils, CD11b+ DCs, and inflammatory DCs were increased, but to a lesser extent than in hACE2 Tg mice. Amounts of overall CD8+ T cells and NK cells were stable or reduced, respectively, to similar extents as in hACE2 Tg mice (Figures 6B–6J and S9B–S9J). Of note, although less pronounced than in hACE2 Tg mice, all of the variants triggered a reduction in pDC frequency in lung tissue compared to mock infected mice (Figures 6I and S9I), and this phenotype was again paralleled by a significant reduction in the levels of IFN-α4, IFN-β, and IFN-λ mRNA (Figures 6K–6M). Moreover, challenge of WT animals resulted in a broad cytokine response for all virus variants that was overall less pronounced than in hACE2 Tg mice and did not increase from early to late time points (Figures 7A and S10). Interestingly, infections with Gamma, which showed the highest virus titers in the lungs of WT mice, tended to trigger the strongest overall cytokine response at day 3 p.i. (Figures 7A, 7B, and S10D). This overall induction resulted from the deregulation of a specific set of cytokines that were partly significantly induced in comparison to the other variants (e.g., CCL2, LIF, IL-6 [Figures 7C–7E], CXCL1 [Figure S11F]). Another set of cytokines remained unaffected by Gamma infection but reached similar levels of induction as in hACE2 Tg mice by the other variants (e.g., IL-16, CX3CL1 [Figures 7F and 7G], IL-20, CCL22 [Figures S11L and S11N]). Correlation plots highlighted variant-specific, significant reductions or inductions of individual cytokines, revealing variant-specific induction patterns also in SARS-CoV-2-infected WT mice (Figures 7H, 7I, and S12). These results revealed that SARS-CoV-2 VOCs have gained an expanded species tropism following evolution in the human species that allows them to carry out at least initial rounds of replication in the lungs of WT mice. These infections triggered cellular immune and cytokine responses that likely contributed to the self-limiting nature of the infection. Indicating the immunogenic potency of the virus inoculum, robust immune cell activation and cytokine responses were also induced by EU-1 in the apparent absence of virus replication.
Discussion
The emergence of SARS-CoV-2 VOCs has been recognized as a significant complication for combating the pandemic, as they often display an increased capacity for transmission, higher virulence, and immune escape (Boehm et al., 2021; Garcia-Beltran et al., 2021; Harvey et al., 2021). Here, we sought to assess the intrinsic pathogenic properties of VOCs Alpha, Beta, and Gamma relative to the early SARS-CoV-2 isolate EU-1. Using identical amounts of infectious virus for the intranasal challenge of mice, we found that all of the variants induced rapid weight loss in hACE2 Tg mice associated with increasing lethargy and respiratory distress. These disease symptoms coincided with productive infection of the lung, trachea, and brain and were associated with immune activation and recruitment of inflammatory myeloid cells to the lung as well as the marked production of pro-inflammatory and T cell- and myeloid cell-produced cytokines and chemokines. However, VOCs displayed variant-specific characteristics that include differences in disease progression, symptoms, tissue tropism, induction of cellular immune activation, and cytokine responses. These differences were particularly pronounced for Gamma, which replicated to higher titers in the lung and led to significantly accelerated disease progression. This increased virulence was paralleled by a more pronounced induction of inflammatory monocytes and DCs as well as neutrophil recruitment to infected lungs. Possibly reflecting this accelerated replication kinetics, cytokine responses of Gamma-infected animals tended to be more pronounced early p.i., but showed reduced magnitude at the late stage relative to that of animals infected with the other variants. Gamma thus represents the prototype of a variant with significantly enhanced pathogenic potential in an in vivo model of pathogenic infection. These results reveal that SARS-CoV-2 VOCs bear a different intrinsic pathogenic potential and establish hACE2 Tg mice as a readily accessible disease model for the characterization of newly emerging variants of this pandemic respiratory pathogen.
Interestingly, we also noted that infection of the lung with all of the SARS-CoV-2 variants tested resulted in a drastic reduction in pDCs numbers, one of the main producer cell types of IFNs in this tissue. Consistently, lungs from infected animals showed a marked reduction in IFN-α4, IFN-β, and IFN-λ expression, and both parameters were correlated (pDC depletion versus IFN-β protein p = 0.0014; pDC depletion versus IFN-λ mRNA p = 0.0022). This phenotype was absent in tissues not supporting robust virus replication (data not shown), and no correlation was observed between pDC depletion and the production of IFN-γ, which is predominantly produced by lymphocytes. Interestingly, pDCs are not productively infected ex vivo, but are activated upon SARS-CoV-2 exposure (Onodi et al., 2021). Our in vivo study demonstrates that SARS-CoV-2 replication in lung tissue results in the depletion of this cell type that is critical for immunosurveillance and early innate responses. Since pDC depletion was most pronounced in the context of active virus replication but still triggered with reduced efficacy by the replication-incompetent EU-1 inoculum in WT mice, the extent of pDC depletion is likely a function of local virus load. In particular, in the case of VOCs, the reduction of NK cell numbers, which was positively correlated with the depletion of pDC (p = 0.0058), may synergize toward a potent blockade of the first line of innate defense by uninfected bystander cells (Ali et al., 2019). Depletion of pDCs and reduction of NK cells may thus constitute an effective addition to the suppression of innate immune signaling that SARS-CoV-2 exerts in productively infected cells (Kasuga et al., 2021; Taefehshokr et al., 2020). Of note, reductions in pDC and NK cell levels have also been observed in the peripheral blood of COVID-19 patients (Krämer et al., 2021; Zingaropoli et al., 2021) or patients with H1N1 influenza virus-induced pneumonia (Lichtner et al., 2011). The molecular mode of action and the functional consequences of this SARS-CoV-2-induced pDC-centered pathology warrant further investigation.
SARS-CoV-2 infection of hACE2 Tg mice reflects important aspects of COVID-19 in humans, but results in rapid and—relative to the human disease—overemphasized brain infection, associated with severe encephalitis within 1 week after challenge. Of note, the use of identical infectious titers for all of the variants analyzed and the lack of neutralizing antibody production during the first days of fulminant SARS-CoV-2 infection in this model (Yinda et al., 2021) allowed us to specifically assess the intrinsic potential of VOCs to spread and cause disease in infected animals. Despite the focus of this infection model on acute and fulminant disease, our study revealed significant differences in the pathogenic profiles of VOCs, including development of an expanded spectrum of symptoms, accelerated disease course, and variant-specific cytokine induction profiles. A particularly interesting feature of all VOCs was the ability to infect cells in the ileum and colon of hACE2 Tg mice, in addition to the respiratory tract. For animals harvested on day 6 p.i. (infection with Alpha and Beta), this coincided with macroscopically detectable pathology in the GI tract. Such variant-specific alterations in tissue tropism may explain why the GI tract has been suggested to harbor neutralization-resistant viral reservoirs in some COVID-19 patients (Gaebler et al., 2021). Although robust amounts of VOCs were observed in the GI tract, viral transcriptional activity was moderate and did not result in the measurable production of infectious progeny. This may reflect a loss in infectivity upon exposure to intestinal fluids. Moreover, and in contrast to productively infected tissues such as the lung, the infection of the GI tract resulted in the induction rather than the suppression of IFN responses that are likely to limit virus spread (data not shown). VOCs may have just not yet optimized their replication and immune evasion strategies with these additional tissues.
A surprising and important observation was that all of the VOCs, but not EU-1, were able to replicate in WT mice that only express murine ACE2. This could reflect that the Spike proteins of Alpha, Beta, and Gamma have a 27- to 108-fold increased affinity for mouse ACE2 over that of early SARS-CoV-2 variants enabling cell entry (Wang et al., 2021). However, infections of WT mice were generally asymptomatic and limited to the lung, where new infectious progeny was produced to markedly lower levels than in hACE2 Tg mice (up to 1,600-fold less viral genome copies and 22-fold less infectious viral progeny). The magnitude of affinity increase for the replication capacity of mouse ACE2 and VOC in WT mice was not correlated with another, suggesting that in addition to the Spike protein, other viral determinants may contribute to the ability of VOCs to replicate in WT mice. In comparison to EU-1, Alpha, Beta, and Gamma display 24, 29, and 25 mutations throughout the genome and 11, 10, and 13 within Spike, respectively (Table S1). Most striking was that a three-amino acid deletion in the non-structural protein Nsp6 (Δ106–108) is shared by all VOCs and may contribute to the observed replication capacity in WT mice (e.g., by its ability to promote evasion from type I IFN and its potential role in autophagosome expansion) (Peacock et al., 2021; Sargazi et al., 2021; Xia et al., 2020). In addition, several candidate mutations present only in Gamma may account for its enhanced pathogenicity in Tg mice and the ability to mount the highest viral load in the lungs of WT animals at day 3 p.i. These include the L3829F mutation in Nsp6; the A105V mutation in ORF7a, a protein that modulates the immune function of monocytes, can inhibit the host cell restriction factor tetherin, and interferes with the translation of host proteins (Martin-Sancho et al., 2021); the E92K mutation in ORF8, a viral protein that promotes evasion from CD8 T cell recognition (Zhang et al., 2021); as well as polymorphisms in ORF9b, a suppressor of antiviral innate signaling (Wu et al., 2021) (see Table S1). Future studies will assess the impact of these mutations on SARS-CoV-2 replication and pathogenesis. Similarly, pDC depletion, immune cell recruitment, suppression of IFN expression, and induction of cytokine production were observed, but were less pronounced than in Tg mice. Our results suggest that VOCs gained the ability to infect the lungs of WT mice and that initial triggering of immune responses allowed the animals to systemically control these infections. Importantly, such rapid basal immune responses were triggered by allof the variants analyzed, including EU-1. Since EU-1 failed to replicate in WT mice, these responses were likely elicited by the virus inoculum without the need for active virus replication. These results thus emphasize the protective potential of initial immune responses to transmitted virus before the onset of robust virus replication and spread.
The identification of distinct pathogenic properties of VOCs in these animal models illustrates that the continuous evolution of this pandemic virus can result in accelerated disease progression, broadened tissue tropism, and host range. The latter may develop into the ability of SARS-CoV-2 for reverse zoonosis (i.e. the transfer of variants from humans to rodents or other animal species). These properties of VOCs must be considered in public health decisions aimed at limiting virus spread and monitoring viral reservoirs, including those in vaccinated human populations.
Limitations of the study
We recognize that the relatively small animal group sizes in combination with the observed variations in the magnitude of parameter differences between individual animals limit the statistical power of our analysis. This study thus served to document, for example, the induction of overall distinct cytokine and chemokine profiles between infections of mice by different SARS-CoV-2 variants, and to identify candidate cytokines for reduced and increased expression levels as a subject for further studies. The size of the study also did not allow us to test whether different minimal infectious doses of SARS-CoV-2 variants are required to cause pathogenic infection, but our results predict that VOCs such as Gamma may require significantly less input virus to cause acute lethal disease than, for example, EU-1. For the same reason, we were unable to conduct studies with sublethal doses to study the induction of humoral immune responses. Finally, the more recently emerged VOCs Delta and Omicron could not be included in the study. Testing the intrinsic pathogenic properties in mice should be included in the routine characterization of new SARS-CoV-2 VOCs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-mouse CD45-PerCP (clone 30-F11) | Biolegend | Cat#103130; RRID:AB_893339 |
| Hamster anti-mouse CD3ε-brilliant violet 711 (clone 145-2C11) | Biolegend | Cat#100349; RRID:AB_2565841 |
| Rat anti-mouse CD4-AlexaFluor488 (clone RM4-5) | Biolegend | Cat#100529; RRID:AB_389303 |
| Rat anti-mouse CD8a-PE-Cy5 (clone 53-6.7) | Biolegend | Cat#100710; RRID:AB_312749 |
| Rat anti-mouse PD-1-PE (clone 29F.1A12) | Biolegend | Cat#135206; RRID:AB_1877231 |
| Hamster anti-mouse CD11c-brilliant violet 605 (clone N418) | Biolegend | Cat#117334; RRID:AB_2562415 |
| Rat anti-mouse CD11b-PE (clone M1/70) | Biolegend | Cat#101208; RRID:AB_312791 |
| Rat anti-mouse Ly-6C-PE/Cy7 (clone HK1.4) | Biolegend | Cat#128018; RRID:AB_1732082 |
| Rat anti-mouse Ly-6G-AlexaFluor488 (clone 1A8) | Biolegend | Cat#127626; RRID:AB_2561340 |
| Mouse anti-mouse NK1.1/CD161-brilliant violet 711 (clone PK136) | Biolegend | Cat#108745; RRID: AB_2563186 |
| rabbit anti-SARS-CoV-2 nucleocapsid protein (clone HL344) | Cell Signaling Technology | Cat#26369 |
| Bacterial and virus strains | ||
| SARS-CoV-2 (lineage B.1.1.7, Alpha) | European Virus Archive global | Ref-SKU: 004V-04032 |
| EU-1 (D614G) | Patient swab / diagnostic MVP | GISAID EPI ISL: 2967222 |
| B1.351 (Beta) | LGL | GISAID EPI ISL: 1752394 |
| P.1 (Gamma) | LGL | GISAID EPI ISL: 2095177 |
| Chemicals, peptides, and recombinant proteins | ||
| Antibiotic/Antimycotic (100x) | Gibco, ThermoFisher Scientific | Cat#15240062 |
| Carboxymethylcellulose | Sigma-Aldrich | Cat#21902-250G |
| Collagenase/Dispase | Merck | Cat#10269638001 |
| Crystal violet solution | Sigma-Aldrich | Cat#HT90132 |
| DMEM, high glucose, GlutaMAX™ Supplement | Gibco, ThermoFisher Scientific | Cat#10566016 |
| Dako Target Retrieval Solution pH6 | Dako | Cat#S236984-2 |
| Fetal Bovine Serum | Capricorn Scientific | Cat#FBS-11A |
| Formaldehyde solution 35% | Carl Roth | Cat#4980.4 |
| Isoflurane | Fresenius Cabi | Cat#M60303 |
| MagNA Pure LC Total Nucleic Acid Lysis Buffer | Roche | Cat#03246779001 |
| MEM | Gibco, ThermoFisher Scientific | Cat#31095052 |
| 16% Paraformaldehyde | Fisher Scientific | Cat#50-980-487 |
| ImmPress Horse anti rabbit IgG Kit | Linearis | Cat#MP-7401-50 |
| PBS | Sigma-Aldrich | Cat#P38135-10PAK |
| Penicillin/Streptomycin Solution 100x | Capricorn Scientific | Cat#PS-B |
| Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#P8340-5ml |
| Critical commercial assays | ||
| Mouse Cytokine Array / Chemokine Array 44-Plex (MD44) | Eve technologies | Cat#MD44 |
| Deposited data | ||
| Video S1 | This paper | |
| Experimental models: Cell lines | ||
| Vero clone E6 | ATCC | Cat#CRL-1586 |
| Experimental models: Organisms/strains | ||
| Mouse: C57Bl/6 wildtype | Charles River | Cat#027 |
| Mouse: K18-hACE2 mice (B6.Cg-Tg(K18-ACE2)2Prlmn/J | The Jackson Laboratory | Stock No. 034860 |
| Oligonucleotides | ||
| 53437: 5`-GACCCCTGAGGGTTTCATATAG-3` | Sigma-Aldrich | The Jackson Laboratory, Stock No. 034860 |
| 53438: 5`-CACCAACACAGTTTCCCAAC-3` | Sigma-Aldrich | The Jackson Laboratory, Stock No. 034860 |
| 53439: 5’-AAGTTGGAGAAGATGCTGAAAGA-3’ | Sigma-Aldrich | The Jackson Laboratory, Stock No. 034860 |
| HPRT Fwd: 5’-GCGTCGTGATTAGCGATGATG-3` | Sigma-Aldrich | Li et al. (2016) |
| HPRT Rev: 5’-CTCGAGCAAGTCTTTCAGTCC-3` | Sigma-Aldrich | Li et al. (2016) |
| IFNα4 Fwd: 5`-TCCATCAGCAGCTCAATGAC-3` | Sigma-Aldrich | Li et al. (2016) |
| IFNα4 Rev: 5`-AGGAAGAGAGGGCTCTCCAG-3` | Sigma-Aldrich | Li et al. (2016) |
| IFNβ Fwd: 5`-TCAGAATGAGTGGTGGTTGC-3’ | Sigma-Aldrich | Li et al. (2016) |
| IFNβ Rev: 5`-GACCTTTCAAATGCAGTAGATTCA-3’ | Sigma-Aldrich | Li et al. (2016) |
| IFNλ Fwd: 5`-AGCTGCAGGTCCAAGAGCG-3’ | Sigma-Aldrich | Li et al. (2016) |
| IFNλ Rev: 5`-GGTGGTCAGGGCTGAGTCATT-3’ | Sigma-Aldrich | Li et al. (2016) |
| GAPDH Fwd: 5’-CTGACGTGCCGCCTGGAGAAA-3` | Sigma-Aldrich | This Paper |
| GAPDH Rev: 5’-AGCCCCGGCATCGAAGGTGG-3` | Sigma-Aldrich | This Paper |
| 2019-nCoV_N1 Fwd: 5’-GACCCCAAAATCAGCGAA AT-3’ | IDT Technologies | Lu et al. (2020) |
| 2019-nCoV_N1: 5’-TCTGGTTACTGCCAGTTGAATCTG-3’ | IDT Technologies | Lu et al. (2020) |
| 2019-nCoV_N1: 5’-ACCCCGCATTACGTTTGGTGGACC-3’ | IDT Technologies | Lu et al. (2020) |
| Leader sgmRNA Fwd: 5’-CCTTCCCAGGTAACAAACCAACC-3’ | IDT Technologies | This paper |
| S gene sgm RNA Rev: 5‘-ACACACTGACTAGAGACTAGTGGC-3’ | IDT Technologies | This paper |
| Recombinant DNA | ||
| 2019-nCoV_N_Positive Control | IDT Technologies | Cat#10006625 |
| Software and algorithms | ||
| GraphPad Prism 7.0a | GraphPad | www.graphpad.com/scientific-software/prism/ |
| BD FACSDiva | BD Biosciences | www.bdbiosciences.com |
| FlowJo 10.7 | BD Biosciences | www.flowjo.com |
| Microsoft Office 365 | Microsoft | www.microsoft.com |
| Adobe Illustrator CS6 | Adobe | www.adobe.de |
| SnapGene 5.2 | Snapgene | www.snapgene.com |
| Other | ||
| Breeding diet mice | Altromin | Cat#1314 – 10mm Pellets |
| Food mice | Altromin | Cat#1324 – 10mm Pellets |
| 5 mL Round bottom polystyrene test tube, with cell strainer snap cap | Falcon | Cat#352235 |
| Bead Ruptor 12A | Omni lab | Cat#21-19-050A |
| Cage bedding | ABBEDD | Cat.#LT-E-001 |
| pluriStrainer mini 70μm | Pluriselect | Cat#SKU 43-10070-70 |
| Diet Gel® Boost | ClearH2O | Cat#72-04-5022 |
| Homogenization tubes with 1.4mm ceramic beads | Omni lab | Cat#21-19-627D |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Dr. Oliver T. Fackler (oliver.fackler@med.uni-heidelberg.de).
Materials availability
There are no new materials generated in this paper. All material has to be requested from authors of cited references.
Experimental model and subject details
Cells and viruses
Vero E6 cell were grown in complete Dulbecco’s modified Eagle medium (DMEM) with high glucose and GlutaMAX™ containing 10% fetal bovine serum (FBS), 100U/mL penicillin and 100μg/mL streptomycin. Isolation and expansion of EU-1: (EU-1; GISAID EPI ISL: 2967222). Caco-2 cells, cultured in virus isolation medium (DMEM containing 2% fetal bovine serum, 100U/mL penicillin-streptomycin, 1x non-essential amino acids, 0.25μg/mL amphotericin B and 0.5μg/mL gentamicin) were challenged for 2h with a clinical isolate previously obtained from a nasopharyngeal swab of a COVID-19 patient. Subsequently, virus isolation medium was replaced with culture medium, and three days post infection supernatant was collected and passaged onto Vero E6 cells. After three additional days, cell culture supernatants were harvested and stored at −80°C. Further propagation of virus was performed in expansion medium (DMEM containing 5% fetal bovine serum, 100U/mL penicillin-streptomycin, 1x non-essential amino acids. Expansion of VOCs. VOCs Beta (B.1.351; (GISAID EPI ISL: 1752394)) and Gamma (P.1; (GISAID EPI ISL: 2095177)) were kindly provided by the Bavarian Landesamt für Gesundheit und Lebensmittelsicherheit (LGL) and expanded in expansion medium on Vero E6 cells. Virus stocks were characterized by RT-qPCR, as reported previously (Muenchhoff et al., 2020). In parallel, for expanded stocks of SARS-CoV-2 near full-length genome sequences were generated following the ARTIC network nCoV-2019 sequencing protocol v2 (Tyson et al., 2020) as described previously (Weinberger et al., 2021). The human nCoV19 isolate/England/MIG457/2020 (20I/501Y.V1, lineage B.1.1.7., Alpha) was kindly provided by Dr. Babak Afrough (Public Health England, London) through the European Virus Archive Global (EVAg, Ref-SKU: 004V-04032) at passage 3. For alignment of viral protein sequences, ORFs were identified using SnapGene and amino acid sequences were aligned using MUSCLE algorithm. Amino acid changes of the used SARS-CoV-2 variants in comparison to the official GISAID reference sequence (hCoV-19/Wuhan/WIV04/2019, EPI_ISL_402124) are highlighted in Table S1.
Biosafety
All aspects of the study were approved by the local authorities (Regierungspräsidien Karlsruhe and Tübingen) before initiation of this study. Work with SARS-CoV-2 was performed in a biosafety level 3 laboratory by personnel equipped with powered air-purifying respirators.
Mice
Animal experiments were carried out in accordance with the standards approved by the central animal facility of the University of Heidelberg (T-39/19, G-96/20). Male heterozygous K18-hACE2 (B6.Cg-Tg(K18-ACE2)2Prlmn/J) in the C57Bl/6 background were obtained from The Jackson Laboratory, imported via embryo transfer into the general animal facility at the University of Heidelberg and bred heterozygous with wildtype C57Bl/6 females. Genotype of mice was assessed by PCR using primers that distinguish between homozygous and heterozygous K18-hACE2 transgenic and wildtype mice using the following primers: 53437: 5`-GAC CCC TGA GGG TTT CAT ATA G-3`, 53438: 5`-CAC CAA CAC AGT TTC CCA AC-3` and 53439: 5’-AAG TTG GAG AAG ATG CTG AAA GA-3’ on total genomic DNA from ear punches. Wildytpe C57Bl/6 mice were purchased from Charles River. Mice were kept at 22 ± 2°C, 45–65% relative humidity with a 12-h light/dark cycle, fed ad libitum with Altromin Rod 16 or Rod 18. Cages were supplemented with ABBEDD LT-E−001 bedding and Crincklets Nest-Pads. At the general animal facility mice were kept in conventional type II cages with filter tops. At the age of 9 weeks, 2 days before initiation of the experiment, mice were transferred into the BSL3 area, kept in individually ventilated cages (Tecniplast green line) under negative pressure and were provided with DietGel® Boost to reduce severity of weight loss upon infection. Male and female mice were used in the study. For the 3 day post infection time point 2 males and one female were used per experimental group. Sex of mice that were harvested at the final time point is indicated in Figures 1E–1H. No obvious sex differences were observed.
Method details
Infection and harvesting of mice
K18-hACE2 transgenic mice, wildtype littermates and wildtype mice were infected intranasally (i.n.) under short term anesthesia with 5% isoflurane, applying 104 plaque forming units of the indicated SARS-CoV-2 variants in a total volume of 50μL, diluted in DMEM without supplements, to both nostrils. Male and female mice were used. Mice were visually inspected every day to assess disease score and body weight. 3 mice per group were harvested on day 3 post infection and 4 mice on day 5 or 6 post infection. Wildtype animals infected with the same VOC were always harvested in parallel to the K18-hACE2 transgenic animals. Gamma infected transgenic mice reached the maximum score one day earlier and were harvested already 5 days’ post infection. Mock animals were transferred into the BSL3 area and directly harvested.
Score sheet
| Symptom | Score |
|---|---|
| Body weight | |
| Body weight loss 10–19% | 5 |
| Body weight loss 20–29% | 15 |
| Body weight loss ≥30% | 20 |
| Skin and Fur | |
| Ruffled fur, reduced hygiene | 3 |
| Dirty and ruffled fur, clotted orifices | 8 |
| Spontaneous reaction | |
| Reduced reaction | 1 |
| Reduced mobility, reluctance to move, enhanced muscle tone, coordination disorders | 5 |
| Self isolation, lethargy, apathy, hunched back, seizures | 10 |
| Breathing | |
| Mild respiratory distress | 1 |
| Strongly respiratory distress, abdominal, accelerated breathing | 5 |
For organ harvest, mice were deeply anesthetized with 5% isoflurane, sacrificed by cervical dislocation and organs were harvested in the following order.
| Organ | Histology | Homogenization | Flow cytometry |
|---|---|---|---|
| spleen | 50% | 50% | – |
| kidney | one kidney | one kidney | – |
| liver | ca. 5 × 10 mm piece | ca. 5 × 10 mm piece | – |
| lung | Right lobe | Left lung and one small right lobe | rest |
| heart | 50% | 50% | – |
| Submandibular salivary gland | one lobe | one lobe | – |
| trachea | upper 3mm | Rest | – |
| brain | one lobe | one lobe | – |
| ileum | ca. 5 cm | ca. 5 cm | – |
| feces | – | taken directly from colon if applicable | – |
| colon | ca. 3 cm | ca. 3 cm |
Organ homogenization
Organs were harvested and directly transferred into precooled 2mL homogenization tubes, filled with 1.4mm ceramic beads, 500μL DMEM without supplements and 1:1000 protease inhibitor cocktail. Tissue was homogenized for one minute at 4m/s using a Bead Ruptor 12A, centrifuged at 4°C for one minute at maximum speed and supernatant was transferred into a fresh tube. Samples for qPCR, cytokine analysis and plaque assay were generated from cleared organ homogenates.
Cytokine/chemokine analysis
For cytokine/chemokine analysis 75μL of lung homogenate mixed with a final concentration of 0.5% Tx-100, incubated for 30min at room temperature and stored at −80°C until shipping on dry ice to Eve technologies (Calgary, Canada). Undiluted, filtered samples were measured using a Mouse Cytokine Array/Chemokine Array 44-Plex (MD44). Only 38 cytokines and chemokines are presented in Figures S5–S7 and S10–S12 as some cytokines were frequently saturated (TIMP-1, CCL21, CXCL9), not or not reproducibly detected in mock samples (IL-11, IL-13), or not detectable in our samples at all (Erythropoietin). Mean values ± SD of secreted cytokine amounts of animals from the same group and harvesting time point were normalized to mean values of wildtype and hACE2 transgenic mock control animals (n = 4), respectively. These mean values ± SD were plotted in correlation graphs (see Figures S7 and S12) and linear regression curves with 95% confidence intervals were fitted to the data with slopes forced to go through the center of origin expecting similar levels of induction. Cytokines/chemokines, for which error bars were not overlapping with the 95% confidence interval of the linear regression curves were considered differentially induced or reduced by the respective SARS-CoV-2 variants and presented in heat maps (Figures 4F, 4G and 7H, 7I) as variant-specific cytokines and chemokines.
qPCR analysis
100μL of cleared organ homogenate was mixed with 100μL MagNA Pure LC Total Nucleic Acid Lysis buffer and stored at −80°C until RNA extraction. Viral RNA extraction and cDNA synthesis: Viral nucleic acid extraction of inactivated cell culture supernatants was done using the Beckmann Biomek NX robotics platform and the RNAdvance Viral kit according to manufacturer’s instructions. Subsequently, cDNA synthesis was performed using the High-Capacity RNA-to-cDNA kit according to manufacturer’s instructions. cDNA synthesis was performed for 60min at 37°C, 5min at 95°C on a PCR cycler. Detection of SARS-CoV-2 N1 gene (Taqman). Real-time RT-PCR (genomic) was performed using SARS-CoV-2 N1 gene primers (500nM) and probe (125nM) in a standard Taqman PCR in a QuantStudio 3 Real-Time PCR System. Dilution series of the N positive control reference standard plasmid were prepared in nuclease-free water. Slope of the curve was calculated by performing a linear regression. Detection of Interferons and housekeeping genes (SYBR Green): RT-qPCR was performed using either indicated IFNs or GAPDH forward and reverse primers (600nM each). Quantification of ISGs and GAPDH was done in a standard PowerUp SYBR Green PCR master mix on a QuantStudio 3 Real-Time PCR System. Detection of SARS-CoV-2 sgmRNAs. RT-qPCR was performed using either SARS-CoV-2 forward primer leader universal (600nM) in combination with a gene-specific sgmRNA reverse primer (600nM). Quantification of sgmRNAs was done in a standard PowerUp SYBR Green PCR on a QuantStudio 3 Real-Time PCR System.
Plaque assay
Plaque assays were essentially performed as previously described (Klein et al., 2020). Briefly, Vero E6 cells were seeded into 24-well plates at 2.5 × 105 cells/well. Next day, six times ten-fold serial dilutions of infectious supernatants or cleared organ homogenates were prepared in complete DMEM, medium was removed from the plates and 200μL were added to the cells in duplicates. Infection was performed at 37°C for 1 to 3hrs, followed by removal of supernatant and overlaying with 1mL of minimum essential medium (MEM) containing 0.8% Carboxymethylcellulose and incubation for 72hrs at 37°C. For testing of ileum, colon or feces homogenates 1:100 antibiotic/antimycotic solutions was added at any step. Cells were then fixed by overlay with 1mL 10% PBS-buffered formaldehyde for at least 30min at room temperature, followed by immersion of the whole plates in 6% H2O-diluted formaldehyde for at least 30min at room temperature before they were taken out of the BSL3 area. Water-rinsed cell monolayers were stained for 15 to 30min with 2.3% crystal violet solution, extensively washed with tap water and plaques were manually counted after drying. Virus stocks used to infect mice, were plaqued in quadruplicates.
Flow cytometry
Lung pieces were minced, incubated in collagenase/dispase (0.1U/mL collagenase, 0.8U/mL dispase) for 30min at 37°C and single cell suspensions were generated by squeezing digested organ pieces through a 70μm pores size cell strainer using a metal stick and an excess of FACS buffer (1x PBS with 0.5% BSA, 2% FBS, 2mM EDTA). After washing with FACS buffer, single cell suspensions were stained for 20min at 4°C, with two separate staining mixtures: a) anti-CD45-PerCP (clone 30-F11), anti-CD3ε-brilliant violet 711 (clone 145-2C11), anti-CD4-AlexaFluor488 (clone RM4-5), anti-CD8a-PE-Cy5 (clone 53–6.7) and anti-PD-1-PE (clone 29F.1A12) b) anti-CD45-PerCP (clone 30-F11), anti-CD11c-brilliant violet 605 (clone N418), anti-CD11b-PE (clone M1/70), anti-Ly-6C-PE/Cy7 (clone HK1.4), anti-Ly-6G-AlexaFluor488 (clone 1A8) and anti-NK1.1/CD161-brilliant violet 711 (clone PK136). After washing in FACS buffer, cells were fixed in 4% paraformaldehyde for 90min at room temperature before taking samples out of the BSL3 area. Cells were again washed, resuspended in FACS buffer, filtered through a cell strainer snap cap 5mL FACS tube and acquired using FACSCelesta with BD FACSDiva Software. Cell populations were gated and quantified as shown in Figure S3 using FlowJo 10.7 Software.
Histology
Organs were fixed in 4mL 4% PBS-buffered formaldehyde for 48hrs at room temperature, then transferred into 70% ethanol and stored at room temperature until paraffin embedding. After deparaffinization of 2μm slides, antigen retrieval was performed using DAKO ph6 antigen retrieval buffer. Immunohistochemistry was performed using a rabbit anti-SARS-CoV-2 nucleocapsid protein antibody (1:200) and immunodetection was completed with the ImmPress horse anti rabbit IgG kit. Per section three high power fields were evaluated by an experienced pathologist and mean percentages with standard deviation of positive cells were calculated.
Quantification and statistical analysis
Statistical analysis of datasets was carried out using GraphPad Prism version 7.0a. Statistical significance was calculated using the Kruskal-Wallis test with a Dunn’s multiple comparison test. Comparisons were always performed among all 4 SARS-CoV-2 variants separately for each time point. For correlation plots linear regression curves with 95% confidence intervals were calculated using GraphPad Prism and r- and p values were calculated using nonparametric Spearman correlation. Cytokines and chemokines were considered as statistically significantly induced or reduced upon infection between the different SARS-CoV-2 variants if SD does not overlap with the 95% confidence interval of the linear regression curve. N.s., not significant; ∗, p < 0.05; ∗∗, p < 0.005; ∗∗∗, p < 0.0005. See figure legends for details.
Acknowledgments
This study was supported by the DFG (German Research Foundation), Projektnummer 240245660–SFB 1129, to O.T.F. (TP8), R.B. (TP11), and A.R. (TP13) and Projektnummer 272983813–TRR 179, to R.B. O.T.F. acknowledges support from the Sonderfördermaßnahmen Covid19 des Landes Baden-Württemberg. O.T.K. acknowledges support from the Bavarian Ministry of State (Bayerische Forschungsstiftung, FORCOVID BayVOC) and thanks Helga Mairhofer, Christopher Dächert, and Natascha Grzimek-Koschewa for excellent technical support and discussion; Ulrike Protzer for providing access to the P3 laboratory of the Institute of Virology, TUM; Jochen Rech and Veit Hornung for help with the qPCR analysis and laboratory access, respectively; Andreas Sing for providing SARS-CoV-2 clinical isolates; and Maximilian Münchhoff, Helmut Blum, Stefan Krebs, and Alexander Graf for SARS-CoV-2 sequencing and analysis. F.G. is supported by the Chica and Heinz Schaller (CHS) Foundation. We are extremely grateful to Roland Kehm and Cordula Ackermann for their generous help and support with setting up the BSL3 animal facility for this project, to Kathrin Bajak for support during the preparation of this manuscript, and to Steeve Boulant for helpful discussions.
Author contributions
Conceptualization, O.T.F., B.S., and O.T.K.; methodology, B.S., I.A., A.R., M.C., and R.B.; investigation, B.S., M.S., I.A., K.H., L.G., and K.M.; data analysis, B.S. and F.G.; writing – original draft, O.T.F. and B.S.; writing – review & editing, O.T.F., B.S., and O.T.K.; funding acquisition, O.T.F. and O.T.K.; resources, M.C., R.B., and A.R.; supervision, O.T.F. and O.T.K.
Declaration of interests
The authors declare no competing interests.
Published: January 28, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110387.
Supplemental information
Amino acid sequences of known SARS-CoV-2 open reading frames (ORFs) of the used SARS-CoV-2 variants were extracted from the full-length viral genome, aligned using the MUSCLE alignment algorithm in SnapGene 5.2. Observed alterations in the amino acid sequences are summarized here. The left column specifies processed proteins in case of polyproteins (ORF1ab) or domains of the protein (Spike). Features of viral proteins that may be of interest for the present study are seen in column 7 (features). The measured changes in affinity of Spike to murine ACE2, as published by Wang et al. (2021) are provided in the last column.
Data and code availability
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alexandersen S., Chamings A., Bhatta T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020;11:6059. doi: 10.1038/s41467-020-19883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Mann-Nüttel R., Schulze A., Richter L., Alferink J., Scheu S. Sources of type I interferons in infectious immunity: plasmacytoid dendritic cells not always in the driver's seat. Front. Immunol. 2019;10:778. doi: 10.3389/fimmu.2019.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates T.A., Leier H.C., Lyski Z.L., McBride S.K., Coulter F.J., Weinstein J.B., Goodman J.R., Lu Z., Siegel S.A.R., Sullivan P., et al. Neutralization of SARS-CoV-2 variants by convalescent and BNT162b2 vaccinated serum. Nat. Commun. 2021;12:5135. doi: 10.1038/s41467-021-25479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm E., Kronig I., Neher R.A., Eckerle I., Vetter P., Kaiser L. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin. Microbiol. Infec. 2021;27:1109–1117. doi: 10.1016/j.cmi.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Lavine C.L., Rawson S., Peng H., Zhu H., Anand K., Tong P., Gautam A., et al. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science. 2021;373:642–648. doi: 10.1126/science.abi9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., 3rd, Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Jr., Hou Y.J., Adams L.E., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St. Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e2379. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary-Gouy H., Lebon P., Dalloul A.H. Type I interferon production by plasmacytoid dendritic cells and monocytes is triggered by viruses, but the level of production is controlled by distinct cytokines. J. Interferon Cytokine Res. 2002;22:653–659. doi: 10.1089/10799900260100132. [DOI] [PubMed] [Google Scholar]
- Golden J.W., Cline C.R., Zeng X., Garrison A.R., Carey B.D., Mucker E.M., White L.E., Shamblin J.D., Brocato R.L., Liu J., et al. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI insight. 2020;5:e142032. doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez C.E., Perdiguero B., Esteban M. Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccines. 2021;9:243. doi: 10.3390/vaccines9030243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg N., Berggren O., Leonard D., Weber G., Bryceson Y.T., Alm G.V., Eloranta M.L., Rönnblom L. IFN-α production by plasmacytoid dendritic cells stimulated with RNA-containing immune complexes is promoted by NK cells via MIP-1β and LFA-1. J. Immunol. 2011;186:5085–5094. doi: 10.4049/jimmunol.1003349. [DOI] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Yue X., Lazartigues E. ACE2 mouse models: a toolbox for cardiovascular and pulmonary research. Nat. Commun. 2020;11:5165. doi: 10.1038/s41467-020-18880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga Y., Zhu B., Jang K.-J., Yoo J.-S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021;53:723–736. doi: 10.1038/s12276-021-00602-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e819. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer B., Knoll R., Bonaguro L., ToVinh M., Raabe J., Astaburuaga-García R., Schulte-Schrepping J., Kaiser K.M., Rieke G.J., Bischoff J., et al. Early IFN-α signatures and persistent dysfunction are distinguishing features of NK cells in severe COVID-19. Immunity. 2021;54:2650–2669.e14. doi: 10.1016/j.immuni.2021.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakidis N.C., López-Cortés A., González E.V., Grimaldos A.B., Prado E.O. SARS-CoV-2 vaccines strategies: a comprehensive review of phase 3 candidates. NPJ Vaccin. 2021;6:28. doi: 10.1038/s41541-021-00292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wohlford-Lenane C., Perlman S., Zhao J., Jewell A.K., Reznikov L.R., Gibson-Corley K.N., Meyerholz D.K., McCray P.B., Jr. Middle East Respiratory Syndrome Coronavirus Causes Multiple Organ Damage and Lethal Disease in Mice Transgenic for Human Dipeptidyl Peptidase 4. The Journal of Infectious Diseases. 2016;213:712–722. doi: 10.1093/infdis/jiv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtner M., Mastroianni C.M., Rossi R., Russo G., Belvisi V., Marocco R., Mascia C., Del Borgo C., Mengoni F., Sauzullo I., et al. Severe and persistent depletion of circulating plasmacytoid dendritic cells in patients with 2009 pandemic H1N1 infection. PloS one. 2011;6:e19872. doi: 10.1371/journal.pone.0019872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S.K., Whitaker B., Murray J., Kamili S., Lynch B., Malapati L., Burke S.A., Harcourt J., et al. US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 2020;26:1654–1655. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C., Maher L., Lee C., Kang W. COVID-19 preclinical models: human angiotensin-converting enzyme 2 transgenic mice. Hum. Genomics. 2020;14:20. doi: 10.1186/s40246-020-00272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Sancho L., Lewinski M.K., Pache L., Stoneham C.A., Yin X., Becker M.E., Pratt D., Churas C., Rosenthal S.B., Liu S., et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol. Cell. 2021;81:2656–2668.e2658. doi: 10.1016/j.molcel.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenchhoff M., Mairhofer H., Nitschko H., Grzimek-Koschewa N., Hoffmann D., Berger A., Rabenau H., Widera M., Ackermann N., Konrad R., et al. Multicentre comparison of quantitative PCR-based assays to detect SARS-CoV-2, Germany, March 2020. Euro Surveill. 2020;25:2001057. doi: 10.2807/1560-7917.ES.2020.25.24.2001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oladunni F.S., Park J.-G., Pino P.A., Gonzalez O., Akhter A., Allué-Guardia A., Olmo-Fontánez A., Gautam S., Garcia-Vilanova A., Ye C., et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbei M., Hautefort I., Modos D., Treveil A., Poletti M., Gul L., Shannon-Lowe C.D., Korcsmaros T. SARS-CoV-2 causes a different cytokine response compared to other cytokine storm-causing respiratory viruses in severely ill patients. Front. Immunol. 2021;12:629193. doi: 10.3389/fimmu.2021.629193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodi F., Bonnet-Madin L., Meertens L., Karpf L., Poirot J., Zhang S.Y., Picard C., Puel A., Jouanguy E., Zhang Q., et al. SARS-CoV-2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4. J. Exp. Med. 2021;218:e20201387. doi: 10.1084/jem.20201387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock T.P., Penrice-Randal R., Hiscox J.A., Barclay W.S. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J. Gen. Virol. 2021;102:001584. doi: 10.1099/jgv.0.001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pum A., Ennemoser M., Adage T., Kungl A.J. Cytokines and chemokines in SARS-CoV-2 infections-therapeutic strategies targeting cytokine storm. Biomolecules. 2021;11:91. doi: 10.3390/biom11010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargazi S., Sheervalilou R., Rokni M., Shirvaliloo M., Shahraki O., Rezaei N. The role of autophagy in controlling SARS-CoV-2 infection: an overview on virophagy-mediated molecular drug targets. Cell Biol. Int. 2021;45:1599–1612. doi: 10.1002/cbin.11609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015;15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taefehshokr N., Taefehshokr S., Hemmat N., Heit B. Covid-19: perspectives on innate immune evasion. Front. Immunol. 2020;11:580641. doi: 10.3389/fimmu.2020.580641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J.R., James P., Stoddart D., Sparks N., Wickenhagen A., Hall G., Choi J.H., Lapointe H., Kamelian K., Smith A.D., et al. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv. 2020 doi: 10.1101/2020.09.04.283077. [DOI] [Google Scholar]
- V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54:1611–1621.e1615. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger T., Steffen J., Osterman A., Mueller T.T., Muenchhoff M., Wratil P.R., Graf A., Krebs S., Quartucci C., Spaeth P.M., et al. Prospective longitudinal serosurvey of health care workers in the first wave of the SARS-CoV-2 pandemic in a quaternary care hospital in munich, Germany. Clin. Infect. Dis. 2021;73:e3055–e3065. doi: 10.1093/cid/ciaa1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Shi Y., Pan X., Wu S., Hou R., Zhang Y., Zhong T., Tang H., Du W., Wang L., et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021;34:108761. doi: 10.1016/j.celrep.2021.108761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y., Wang H., Menachery V.D., Rajsbaum R., Shi P.Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33:108234. doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinda C.K., Port J.R., Bushmaker T., Offei Owusu I., Purushotham J.N., Avanzato V.A., Fischer R.J., Schulz J.E., Holbrook M.G., Hebner M.J., et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. Plos Pathog. 2021;17:e1009195. doi: 10.1371/journal.ppat.1009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen Y., Li Y., Huang F., Luo B., Yuan Y., Xia B., Ma X., Yang T., Yu F., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wong L.R., Li K., Verma A.K., Ortiz M.E., Wohlford-Lenane C., Leidinger M.R., Knudson C.M., Meyerholz D.K., McCray P.B., Jr., et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Thao T.T.N., Hoffmann D., Taddeo A., Ebert N., Labroussaa F., Pohlmann A., King J., Steiner S., Kelly J.N., et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. 2021;592:122–127. doi: 10.1038/s41586-021-03361-1. [DOI] [PubMed] [Google Scholar]
- Zingaropoli M.A., Nijhawan P., Carraro A., Pasculli P., Zuccalà P., Perri V., Marocco R., Kertusha B., Siccardi G., Del Borgo C., et al. Increased sCD163 and sCD14 plasmatic levels and depletion of peripheral blood pro-inflammatory monocytes, myeloid and plasmacytoid dendritic cells in patients with severe COVID-19 pneumonia. Front. Immunol. 2021;12:627548. doi: 10.3389/fimmu.2021.627548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
K18-hACE2 transgenic mice, infected with SARS-CoV-2 that reached the maximum disease score. The first part displays lethargic behavior. Lethargic mice hardly move in the cage and display limited motivation to return to an upright position, when gently moved to the side. The second part displays a mouse with mild respiratory distress, defined as clearly visible and enhanced breathing motion. The third part displays a mouse with strong respiratory distress, defined as dramatically strained breathing motion.
Amino acid sequences of known SARS-CoV-2 open reading frames (ORFs) of the used SARS-CoV-2 variants were extracted from the full-length viral genome, aligned using the MUSCLE alignment algorithm in SnapGene 5.2. Observed alterations in the amino acid sequences are summarized here. The left column specifies processed proteins in case of polyproteins (ORF1ab) or domains of the protein (Spike). Features of viral proteins that may be of interest for the present study are seen in column 7 (features). The measured changes in affinity of Spike to murine ACE2, as published by Wang et al. (2021) are provided in the last column.
Data Availability Statement
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.