Abstract
This article presents maplet, an open-source R package for the creation of highly customizable, fully reproducible statistical pipelines for metabolomics data analysis. It builds on the SummarizedExperiment data structure to create a centralized pipeline framework for storing data, analysis steps, results and visualizations. maplet’s key design feature is its modularity, which offers several advantages, such as ensuring code quality through the maintenance of individual functions and promoting collaborative development by removing technical barriers to code contribution. With over 90 functions, the package includes a wide range of functionalities, covering many widely used statistical approaches and data visualization techniques.
Availability and implementation
The maplet package is implemented in R and freely available at https://github.com/krumsieklab/maplet
1 Introduction
A major shift in the biomedical community in recent years has been a push to promote reproducibility in research (Baker, 2016a; Brito et al., 2020; Sandve et al., 2013; Winchester, 2018). This has led to substantial changes in scientific publishing, including new rules for the mandatory sharing of source code and accompanying data for publication in peer-reviewed journals (Baker, 2016b). Adapting data analysis workflows to these new guidelines requires the development of code that is easily readable and maintainable, which often demands substantial time and effort.
An effective way to mitigate this burden is to utilize computational toolboxes that include the following features. (i) To allow users to quickly start developing their own customized workflows, the toolbox must be straightforward to use with easily readable code pipelines. (ii) The source code should be modular to enable rapid development and efficient maintenance of its functionality. (iii) In order to provide full transparency of the workflow, the code should allow users to retrace and inspect the intermediate results generated at every step of the pipeline; this can be achieved using a container object that records all functions, parameters and results in a central location. (iv) The toolbox should be extensible—adding new functionalities should be straightforward for core developers and members of the open-source community alike.
In recent years, several packages for the generation of metabolomics analysis pipelines have been published (Stanstrup et al., 2019). To the best of our knowledge, only two of these packages, MetaboAnalystR (Chong and Xia, 2018) and structToolbox (Lloyd and Weber, 2021), partially address the requirements we listed above; however, they are missing a few key features from our list which results in several limitations. structToolbox does not store results and parameters in a single location, forcing the user to keep track of numerous result variables. Neither structToolbox nor MetaboAnalystR records intermediate steps, making it difficult to determine how results were generated subsequently. On the developer’s side, neither package appears to be designed with community development in mind, as structToolbox depends on a complex system of interconnected classes and MetaboAnalystR must be integrated with the MetaboAnalyst web service.
Here, we present maplet, an open-source R package that combines modular design with a container to automatically record all steps, parameters and results to create a framework for flexible and reproducible metabolomics data analysis (Fig. 1). maplet contains a diverse collection of functions, covering preprocessing, differential analysis, pathway analysis, visualization and various other functionalities. The toolbox is under active development by an international team and its simple template-based design makes contributing new functionalities intuitive for external users. maplet code is readable and easy-to-use making it convenient for new users to integrate into their workflows.
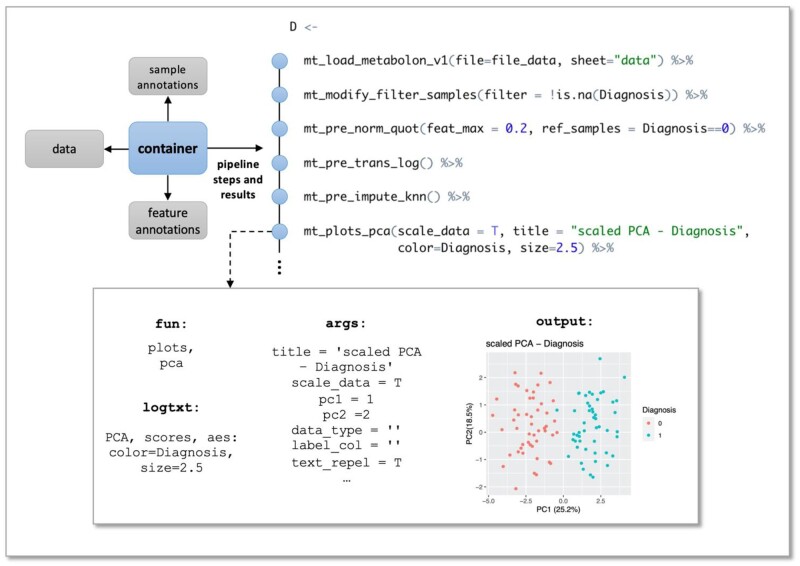
Fig. 1.
maplet pipeline. All data and annotations are stored in a central Summarized Experiment container, which is passed between functions. Each function generates a result entry, containing all function-specific information as well as the results the function generated (such as statistics tables and plots)
2 Toolbox
2.1 Pipeline design
The maplet package allows for the creation of fully transparent analytical pipelines in which each intermediate result can be retraced and inspected by the user. This is achieved by using a container that is passed between each function and records all data, results, plots, R and package versions, analysis steps and their parameters. The pipeline container builds on SummarizedExperiment (Morgan et al., 2021), a container class provided by Bioconductor (Huber et al., 2015) which stores datasets and all corresponding annotations in a single object.
maplet is designed to be used with a pipe operator—either the popular %>% operator from the magrittr package (Bache and Wickham, 2020) or the recently introduced |> operator from base R. Pipe operators enable the smooth connection of processing steps in a maplet pipeline—seamlessly passing the container from function to function. This makes code more readable and eliminates the need for intermediate result variables. Figure 1 presents a subsection of a pipeline and a diagram representing how each step in the pipeline is stored in the container.
2.2 Modularity
maplet follows a modular ‘one function, one operation’ design. Each task is encapsulated in a single function, which enables the rapid development of pipelines where any step can be flexibly inserted, removed or rearranged. Another key advantage of this modular design is the ability to maintain high-quality code. Since functions have no interdependencies, they can be rigorously evaluated and maintained separately. Finally, the modular structure promotes a culture of open-source development by removing the technical barriers to code contribution for unfamiliar developers. Any interested user can add a desired functionality based on a simple function template and only minimal knowledge of the inner workings of the package.
2.3 Functionality
Currently, maplet contains a growing set of over 90 functions organized into various groups, such as data loading, annotation, data modification, preprocessing, statistical analysis, visualization, reporting results, exporting data and pipeline maintenance. This covers many commonly used analytical methods necessary for standard data analysis encountered in everyday research projects. There are specialized loading functions for working with data from various popular metabolomics platforms, including loaders for the online metabolomics data repositories ‘Metabolomics Workbench’ (Sud et al., 2016) and ‘MetaboLights’ (Haug et al., 2020). The package provides a wide variety of functionalities commonly used by bioinformatics researchers, including linear models, missing-value imputation, principal component analysis (PCA), heatmaps, as well as more advanced functionalities such as pathway analysis and network inference. Notably, maplet can be used for other types of omics data and already contains loaders for the Olink proteomics platform. The toolbox comes with several extensive example pipelines and documentation to aid new users in the design of new workflows and a specialized testing framework to ensure stable functionality as the package is further developed.
2.4 Report generation and result access
Once a maplet pipeline has been executed, results can be visualized through comprehensive reports automatically assembled by maplet using R markdown/knitr. These reports lay out all functions in the pipeline in the order they were executed, including the name of the function, arguments and any plots or statistics tables produced by the function. The report is compiled into a single HTML, PDF or Word document, which stores all results in a single location and can be easily shared. Moreover, maplet comes with a series of accessor functions, which allow the user to extract processed data, statistical results or plots from the pipeline container and further analyze them using their own R code.
3 Conclusion
The maplet R package facilitates the fast development of reproducible analysis pipelines for metabolomics data. Its modular design allows for highly customizable, fully reproducible metabolomics pipelines, while also improving readability, ensuring code quality and promoting open-source development.
Funding
This work was supported by the ‘Biomedical Research Program’ funds at Weill Cornell Medical College in Qatar, a program funded by the Qatar Foundation and multiple grants from the Qatar National Research Fund (QNRF) to K.S.; the National Institute of Aging of the National Institutes of Health [U19AG063744 and 1R01AG069901-01A1 to J.K. and R.B.]; and the National Institute on Aging [U19AG063744, U01AG061359, RF1AG058942 and RF1AG059093 to M.A.].
Conflict of Interest: none declared.
Contributor Information
Kelsey Chetnik, Department of Physiology and Biophysics, Institute for Computational Biomedicine, Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10021, USA.
Elisa Benedetti, Department of Physiology and Biophysics, Institute for Computational Biomedicine, Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10021, USA.
Daniel P Gomari, Institute of Computational Biology, Helmholtz Zentrum München—German Research Center for Environmental Health, Neuherberg, Germany.
Annalise Schweickart, Department of Physiology and Biophysics, Institute for Computational Biomedicine, Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10021, USA.
Richa Batra, Department of Physiology and Biophysics, Institute for Computational Biomedicine, Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10021, USA.
Mustafa Buyukozkan, Department of Physiology and Biophysics, Institute for Computational Biomedicine, Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10021, USA.
Zeyu Wang, Department of Physiology and Biophysics, Institute for Computational Biomedicine, Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10021, USA.
Matthias Arnold, Institute of Computational Biology, Helmholtz Zentrum München—German Research Center for Environmental Health, Neuherberg, Germany.
Jonas Zierer, Department of Physiology and Biophysics, Institute for Computational Biomedicine, Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10021, USA.
Karsten Suhre, Department of Physiology and Biophysics, Weill Cornell Medical College—Qatar Education City, Doha, Qatar.
Jan Krumsiek, Department of Physiology and Biophysics, Institute for Computational Biomedicine, Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10021, USA.
References
- Bache S.M., Wickham H. (2020) magrittr: a forward-pipe operator for R. https://CRAN.R-project.org/package=magrittr.
- Baker M. (2016a) 1,500 scientists lift the lid on reproducibility. Nat. News, 533, 452–454. [DOI] [PubMed] [Google Scholar]
- Baker M. (2016b) Why scientists must share their research code. Nat. News, [Google Scholar]
- Brito J.J. et al. (2020) Recommendations to enhance rigor and reproducibility in biomedical research. GigaScience, 9, giaa056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Xia J. (2018) MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics, 34, 4313–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug K. et al. (2020) MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res., 48, D440–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W. et al. (2015) Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods, 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd G.R., Weber R.J.M. (2021) structToolbox: data processing & analysis tools for metabolomics and other omics Bioconductor version: release (3.13). https://bioconductor.org/packages/release/bioc/html/SummarizedExperiment.html.
- Morgan M. et al. (2021) SummarizedExperiment: SummarizedExperiment container Bioconductor version: release (3.12). https://bioconductor.org/packages/release/bioc/html/structToolbox.html.
- Sandve G.K. et al. (2013) Ten simple rules for reproducible computational research. PLoS Comput. Biol., 9, e1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanstrup J. et al. (2019) The metaRbolomics Toolbox in Bioconductor and beyond. Metabolites, 9, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud M. et al. (2016) Metabolomics workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res., 44, D463–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester C. (2018) Give every paper a read for reproducibility. Nature, 557, 281–281. [DOI] [PubMed] [Google Scholar]