Abstract
Summary
Somatic mutations and gene fusions can produce immunogenic neoantigens mediating anticancer immune responses. However, their computational prediction from sequencing data requires complex computational workflows to identify tumor-specific aberrations, derive the resulting peptides, infer patients’ Human Leukocyte Antigen types and predict neoepitopes binding to them, together with a set of features underlying their immunogenicity. Here, we present nextNEOpi (nextflow NEOantigen prediction pipeline) a comprehensive and fully automated bioinformatic pipeline to predict tumor neoantigens from raw DNA and RNA sequencing data. In addition, nextNEOpi quantifies neoepitope- and patient-specific features associated with tumor immunogenicity and response to immunotherapy.
Availability and implementation
nextNEOpi source code and documentation are available at https://github.com/icbi-lab/nextNEOpi
Contact
dietmar.rieder@i-med.ac.at or francesca.finotello@uibk.ac.at
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
T-cell mediated recognition of tumor neoantigens is pivotal for the success of anticancer immunotherapies (Schumacher et al., 2019). Thus, in silico prediction of patient-specific neoepitopes from whole-exome (WES), whole-genome (WGS), and RNA sequencing (RNA-seq) data is a fundamental task in immuno-oncology. To this end, complex computational pipelines must be assembled to predict tumor-specific, mutated peptides and their likelihood of binding the patients’ Human Leukocyte Antigen (HLA) molecules and being recognized by T cells (Finotello et al., 2019; Wells et al., 2020). In addition to neoantigens derived from single-nucleotide variants (SNVs) and insertions or deletions (indels), gene fusions can be a source of noncanonical neoantigens (Yang et al., 2019).
In recent years, several pipelines for the prediction of neoantigens have been developed (see recent review, Finotello et al., 2019), but most of them require cumbersome software installation and extensive data preprocessing with third-party tools to predict somatic mutations and HLA types. Moreover, to the best of our knowledge, most of the available pipelines are not able to predict class-II and noncanonical neoantigens, or to extract features associated to anticancer immune responses like mutation clonality and immune-cell receptor repertoires (Supplementary Table S1). Here, we present nextNEOpi (nextflow NEOantigen prediction pipeline), a fully automated and comprehensive computational workflow that overcomes these shortcomings. nextNEOpi predicts class-I and -II neoantigens originating from SNVs, indels and gene fusions through the analysis of raw sequencing data and derives a set of features associated with tumor immunogenicity and response to immunotherapy.
2 The nextNEOpi pipeline
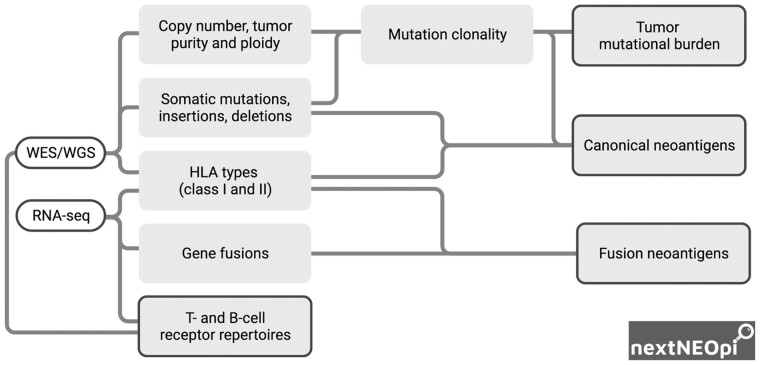
nextNEOpi takes as input raw WES or WGS data from matched tumor-normal samples and, optionally, bulk-tumor RNA-seq data (Fig. 1 and Supplementary Fig. S1). After data preprocessing, nextNEOpi derives germline and phased somatic mutations, copy number variants, tumor purity and ploidy, and selects high-confidence variants through majority voting (Supplementary Methods).
Fig. 1.
Schematization of nextNEOpi pipeline with main input data (white boxes), data flow (lines), intermediate (grey boxes) and final (grey boxes with borders) outputs.
nextNEOpi infers class-I and -II HLA types from WES/WGS (DNA-seq) and RNA-seq data using OptiType (Szolek et al., 2014) and HLA-HD (Kawaguchi et al., 2017), respectively, and can employ an RNA-seq-informed strategy to correct DNA-seq calls for missing HLA genes or alleles (Supplementary Methods). HLA typing benchmarking using data from the 1000 Genomes Project confirmed the high performance of OptiType and HLA-HD, especially on RNA-seq data (Supplementary Figs S2–S4). DNA-seq calls showed a lower accuracy, a systematic underestimation of zygosity and a higher number of missing calls likely due to the low sequencing depth of WGS data (∼4–29 million reads per sample), which were improved using the RNA-seq-informed approach.
nextNEOpi uses pVACseq (Hundal et al., 2020) to predict class-I and -II neoepitopes and derive features associated with neoantigen presentation, including peptide-HLA-binding affinity quantified as half-maximal inhibitory concentration (IC50) and percentile rank quantified by NetMHCpan (Jurtz et al., 2017) and MHCflurry (O’Donnell et al., 2020). Class-I and -II fusion neoepitopes are predicted with NeoFuse (Fotakis et al., 2020).
nextNEOpi exploits tumor purity information to derive the cancer cell fraction (CCF) and clonality of mutations and resulting neoantigens. Tumor mutational burden (TMB) is computed as the number of somatic mutations over the entire read-covered genome or exome. In addition, clonal TMB is computed by considering only clonal somatic mutations (Litchfield et al., 2021). MiXCR is used to predict B-cell receptor and T-cell receptor (BCR and TCR) repertoires (Bolotin et al., 2015). An overview of nextNEOpi results is provided in Supplementary Tables S2–S4.
We implemented nextNEOpi in the Nextflow workflow language (Di Tommaso et al., 2017) to assure portability, scalability, and reproducibility. Parallelization is implicitly defined by inputs and outputs of the individual pipeline tasks, enabling transparent scale-up without requiring adaptation to a specific platform architecture. By leveraging conda environment and singularity container capabilities, the installation demands for nextNEOpi are kept on a minimal level facilitating its usage by users with limited bioinformatics expertise.
3 Analysis of TESLA data with nextNEOpi
To benchmark nextNEOpi, we considered WES and RNA-seq data from two cohorts of melanoma and non-small cell lung cancer patients (n = 8) generated by the Tumor Neoantigen Selection Alliance (TESLA) initiative (Wells et al., 2020). nextNEOpi predicted 30 912–364 532 putative HLA-binding peptides (pMHC) per patient, spanning 5152–90 819 unique peptides (Supplementary Table S5). The identified pMHC represented 76.40–92.59% of those assessed by TESLA and covered 75.00–100% of the total immunogenic pMHC. In total, 36 over 38 immunogenic pMHC were identified. Prioritization of candidate neoepitopes based on relaxed filtering (Supplementary Methods) resulted in the identification of 32 over 38 immunogenic pMHC (Supplementary Table S6).
We considered all pMHC experimentally assessed by TESLA to investigate the features associated with immunogenicity. All scores related to HLA-binding affinity of the mutated peptides were strongly associated with immunogenicity (Supplementary Fig. S5): immunogenic peptides showed a lower IC50 (P = 5.8e−11) and percentile rank (P = 1.6e−10). The IC50 fold-change was not discriminative (P = 0.45), whereas the expression of the mutated gene was higher for immunogenic peptides (P = 9.9e−5). Clonality features showed different distributions for immunogenic and non-immunogenic peptides, with the former having higher CCF 5% confidence interval (P = 0.017) and probability of being clonal (P = 0.024). Clonal neoantigens were enriched in patients responding to immune checkpoint blockers, whereas subclonal mutations were associated with a single patient (Pat_8) with progressive disease (PD).
The investigation of TMB and diversity of immune receptor repertoires can provide further insights into the antigenicity of the tumors and immune-cell infiltration and expansion in the single patients (Supplementary Fig. S6).
4 Conclusions
nextNEOpi is a comprehensive and fully automated pipeline that predicts tumor neoepitopes from raw sequencing data. It is implemented in Nextflow to ensure easy installation and usage, as well as high portability, scalability (see example computational times in Supplementary Table S7), and reproducibility. nextNEOpi quantifies neoepitope- and patient-specific features associated with tumor immunogenicity and response to immunotherapy, and uses multi-method consensus approaches to guarantee robust results in case of suboptimal data. In the near future, we plan to extend nextNEOpi to other classes of noncanonical neoantigens and to introduce DSL 2 Nextflow syntax to facilitate the integration of additional features relevant for neoantigen prioritization.
Data availability
References and accession numbers to the published data analyzed in this work are reported in the Supplementary Material.
Funding
This work was supported by the Austrian Science Fund (FWF) [T 974-B30 to F.F.], by the Oesterreichische Nationalbank (OeNB) [18496 to F.F.] and by the European Research Council (ERC) [786295 to Z.T.].
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Dietmar Rieder, Biocenter, Institute of Bioinformatics, Medical University of Innsbruck, Innsbruck 6020, Austria.
Georgios Fotakis, Biocenter, Institute of Bioinformatics, Medical University of Innsbruck, Innsbruck 6020, Austria.
Markus Ausserhofer, Biocenter, Institute of Bioinformatics, Medical University of Innsbruck, Innsbruck 6020, Austria.
Geyeregger René, St. Anna Children’s Cancer Research Institute, Vienna 1090, Austria.
Wolfgang Paster, St. Anna Children’s Cancer Research Institute, Vienna 1090, Austria.
Zlatko Trajanoski, Biocenter, Institute of Bioinformatics, Medical University of Innsbruck, Innsbruck 6020, Austria.
Francesca Finotello, Biocenter, Institute of Bioinformatics, Medical University of Innsbruck, Innsbruck 6020, Austria; Institute of Molecular Biology, University Innsbruck, Innsbruck 6020, Austria; Digital Science Center (DiSC), University Innsbruck, Innsbruck 6020, Austria.
References
- Bolotin D.A. et al. (2015) MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods, 12, 380–381. [DOI] [PubMed] [Google Scholar]
- Di Tommaso P. et al. (2017) Nextflow enables reproducible computational workflows. Nat. Biotechnol., 35, 316–319. [DOI] [PubMed] [Google Scholar]
- Finotello F. et al. (2019) Next-generation computational tools for interrogating cancer immunity. Nat. Rev. Genet., 20, 724–746. [DOI] [PubMed] [Google Scholar]
- Fotakis G. et al. (2020) NeoFuse: predicting fusion neoantigens from RNA sequencing data. Bioinformatics, 36, 2260–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundal J. et al. (2020) pVACtools: a computational toolkit to identify and visualize cancer neoantigens. Cancer Immunol Res., 8, 409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurtz V. et al. (2017) NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol., 199, 3360–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi S. et al. (2017) HLA-HD: an accurate HLA typing algorithm for next-generation sequencing data. Hum. Mutat., 38, 788–797. [DOI] [PubMed] [Google Scholar]
- Litchfield K. et al. (2021) Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell, 184, 596–614.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell T.J. et al. (2020) MHCflurry 2.0: improved pan-allele prediction of MHC class I-presented peptides by incorporating antigen processing. Cell Syst., 11, 42–48.e7. [DOI] [PubMed] [Google Scholar]
- Schumacher T.N. et al. (2019) Cancer neoantigens. Annu. Rev. Immunol., 37, 173–200. [DOI] [PubMed] [Google Scholar]
- Szolek A. et al. (2014) OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics, 30, 3310–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D.K. et al. ; Tumor Neoantigen Selection Alliance. (2020) Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell, 183, 818–834.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. et al. (2019) Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat. Med., 25, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
References and accession numbers to the published data analyzed in this work are reported in the Supplementary Material.