Abstract
Lung cancer leads to the most cancer-related death in the world. It was shown from the increasing evidences that long non-coding RNAs (lncRNAs) are emerging as molecules for diagnosis, prognosis and even therapy of lung cancer and other malignancies. The biological functions or involved signaling pathways of lncRNAs are always found to be inconsistent among different types of malignancies. However, no available literature has systemically summarized differences in the functions and underlying molecular mechanisms of lncRNAs between lung cancer and other cancers. In this review, the biological functions and molecular mechanisms of lncRNAs in lung cancer were introduced. Furthermore, their functional differences between lung cancer and other malignancies were discussed. Finally, their potential clinical applications in future lung cancer therapy were focused on.
Keywords: Lung cancer, long non-coding RNAs (lncRNAs), biomarkers, targeted therapy
Introduction
In the long history of medical science, the cancer has always been troubling us as the most insurmountable disease. Lung cancer is one of the most lethal and prevalent cancers in the world, which is reported to cause 26% of all cancer-related deaths (1). However, there are still some limitations in current therapy approaches (surgery, chemotherapy and radiotherapy), such as high recurrence rates and serious side effects. Furthermore, effective clinical approaches are still lacked for early-stage diagnosis. In order to improve therapeutic effect and decrease high lethality of lung malignancy, it is urgently needed to discover new biomarkers and therapeutic targets.
According to the classical theory of the central dogma, the RNA molecule is considered as a template for protein synthesis. Although all of the genome is expected to be transcribed into RNA, only 5–10% of the genome can be transcribed. During the transcription, the protein coding RNAs only account for lower than 10% due to evolutionary conservation (2). Distinct from protein-coding RNAs, the large group of RNAs is lack of protein-coding function, and therefore, they are designated as non-coding RNAs (ncRNA). Accumulated evidences have demonstrated that ncRNAs have effects on the regulation of transcription or pre-mRNA processing, even some ncRNAs can interact with proteins (3). During the last decade, the emerging of ncRNAs has opened a new field for both basic and clinical oncologists.
NcRNAs are classified into small non-coding RNAs and long non-coding RNAs (lncRNAs). In general, the former is featured with a length of shorter than 200 nt, which consists of microRNA (miRNA), small nuclear RNA (snRNA), small interfering RNA (siRNA), Piwi interacting RNA (piRNA), transfer RNA (tRNA) and ribosomal RNA (rRNA). Whereas, lncRNAs comprise a group of transcripts and they are longer than 200 nucleotides. According to their genomic locations, lncRNAs can be subdivided into intergenic and intragenic lncRNAs (lincRNA). LincRNAs are further classified as exonic, intronic and overlapping lncRNAs (4). Interestingly, several lncRNAs have been reported to be actually the precursor of other small RNAs (5). Unlike extensively studied miRNAs in Cancer Res, the association between lncRNAs and malignancies is relatively less characterized.
Thousands of lncRNAs are screened in human’s transcriptome, and however, only dozens of them have been well studied, such as X-inactive specific transcript (XIST), HOX transcript antisense RNA (HOTAIR), nuclear enriched abundant transcript 1 (NEAT1), and metastasis associated lung adenocarcinoma transcript 1 (MALAT1). LncRNAs have shared characteristics with mRNA. For examples, they are both transcribed by RNA polymerase II and usually followed by splicing. Their mature molecules have both capped 5' termini and polyadenylated 3' termini. The only difference between lncRNAs and mRNAs is that lncRNAs are lack of a protein-coding potential (6).
It was shown from many studies that lncRNAs can modulate specific gene expression, and they are thus involved in various biological processes of cancer cells. Moreover, the increasing evidences show that the lncRNA play a key role in cancer initiation, progression, and metastasis (7). On the basis of previous studies, we found that one lncRNA always relates to different signaling pathways or biological functions in different types of malignancies. Therefore, researching the functional differences of lncRNAs in multiple malignancies including lung cancer is an important work for further understanding the etiology and pathology of cancer.
In this review, dozens of known lung-cancer-associated lncRNAs were summarized, and their roles in lung cancer biology were described. Firstly, the biological functions and molecular mechanism of these lncRNAs in lung cancer were presented. Furthermore, their functional differences between lung cancer and other malignancies were discussed. Finally, their potential clinical applications in future lung cancer therapy were focused on.
Action mechanisms of lncRNAs in cancers
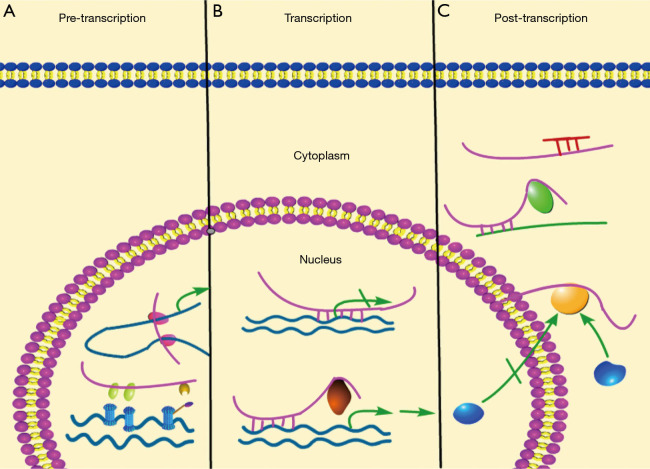
So far, the known molecular mechanisms by which lncRNAs affect cell functions involve interfering of transcription, initiation of chromatin remodeling, and silencing of gene clusters (8,9). In this part, the molecular mechanisms of lncRNA functions (Figure 1) were described.
Figure 1.
Biological mechanisms of lncRNAs are shown in this figure. Pink, blue, green lines stand for lncRNA, mRNA and DNA, respectively. lncRNA mechanisms can be divided into three parts. (A) At pre-transcription level, the regulation of lncRNAs associated with epigenetic regulation, involves histone modification, DNA methylation, gene imprinting, and chromosome dosage compensation. (B) At transcription level, lncRNAs can affect transcription of neighboring genes and genes on different chromosomes. Sometimes, lncRNAs may also cooperate with transcription factors to regulate the transcription of target genes. (C) At post-transcriptional level, lncRNAs always act as a ceRNA, or is engaged to modulate mRNA stability and translation, and influence cellular signaling cascades. LncRNAs’ functions lie on their subcellular localization. Nuclear lncRNAs mainly functions in pre-transcription level and transcription level regulation, while cytoplasmic lncRNAs are involved in post-transcription regulation.
Roughly, lncRNAs exert their functions on affecting transcription via pre-transcriptional, transcriptional and post-transcriptional levels. At pre-transcription level, the regulation of lncRNAs on genome involves histone modification, DNA methylation, X-chromosome inactivation, gene imprinting, and chromosome dosage compensation. For instance, XIST, a well-studied lncRNA, regulates dosage compensation in female mammals. After localized to the X chromosome, XIST can recruit multiple factors indirectly to execute X chromosome inactivation (XCI) (10-13). At transcription level, lncRNAs can affect transcription of neighboring genes and genes on different chromosomes (11,14). According to the study, NEAT1 binds to the promoter region of oncogenic genes to increases their transcription (15). LncRNA may also cooperate with transcription factors to regulate the transcription of target genes. For example, p53-inducible lincRNA-p21 directly binds to hnRNP-K, and in turn, mediates gene repression during cellular response to DNA damage (13). At post-transcriptional level, lncRNAs always act as a competing endogenous RNAs (ceRNA) by competitively binding miRNAs (small non-coding RNAs located in cytoplasm, which always target 3' UTR of mRNAs to degrade them or prevent their translation), and consequently, they indirectly protect their predestinated mRNAs from being silencing or degraded (16,17). In addition, lncRNAs also affect translation by influencing the alternative splicing of pre-mRNAs (5). Notably, some lncRNAs can improve the stability of mRNA via their complementary sequences with target molecules, without involving miRNAs (18).
Furthermore, depending on their subcellular localizations, lncRNAs can be classified into nuclear and cytoplasmic lncRNAs. Nuclear lncRNAs are enriched for functionality involving transcriptional regulation, chromatin interactions and RNA processing. Unlike RNA located in nucleus, cytoplasmic lncRNAs are primarily engaged to improve mRNA stability and influence cellular signaling cascades (19-21). The recent study showed that some nuclear lncRNAs could also function as a ceRNA, such as NEAT1 (nuclear enriched abundant transcript 1) and MALAT1 etc. (22-24), thereby suggesting that ceRNA-associated molecular mechanisms may be much more complicated than that we thought.
LncRNAs on different malignant characters of lung cancer
Dozens of lncRNAs are implicated in different types of lung cancer (Table 1). From the table we can see that most of these lncRNAs function as cancer promoters. Furthermore, all the oncogenic lncRNAs are upregulated in lung cancer, while tumor-inhibiting ones are downregulated. Several lncRNAs have been well characterized to be involved in cancer initiation and metastasis, insensitivity of patients to chemotherapeutic compounds, radiations and targeted therapy, and the proliferation, apoptosis and invasion in lung cancer cells, while the biological functions of several lncRNAs remain to be further explored in lung cancer, including LINC00115, MIAT, HMlincRNA717, GAS6-as1 and DLX6-AS1 (39,46,55,84). Additionally, one lncRNA can exert multiple functions in different cancers. These lung-cancer-associated lncRNAs were subsequently summarized according to their biological functions.
Table 1. Lung cancer-associated lncRNAs.
| LncRNAs | Alterations | Lung cancer phenotypes | Function in lung cancer | References |
|---|---|---|---|---|
| AFAP1-AS1 | Upregulated | Invasion and metastasis | Promoting | (25) |
| AC006050.3-003 | Upregulated | Drug-resistance | Promoting | (26) |
| AK126698 | Downregulated | Drug-resistance | Inhibiting | (27) |
| BANCR | Downregulated | Invasion and metastasis | Inhibiting | (28,29) |
| ANRIL | Upregulated | Proliferation and apoptosis, invasion and metastasis | Promoting | (30,31) |
| BCYRN1 | Upregulated | Invasion and metastasis | Promoting | (32) |
| CAR10 | Upregulated | Proliferation and apoptosis | Promoting | (33) |
| CCAT1 | Upregulated | Cancer initiation, proliferation and apoptosis, drug-resistance | Promoting | (34,35) |
| CCAT2 | Upregulated | Proliferation and apoptosis, invasion and metastasis | Promoting | (36-38) |
| DLX6-AS1 | Upregulated | Unexplored | Promoting | (39) |
| DQ786227 | Upregulated | Cancer initiation | Promoting | (40) |
| GAS5 | Downregulated | Proliferation and apoptosis, drug-resistance | Inhibiting | (41-45) |
| GAS6-as1 | Downregulated | Unexplored | Inhibiting | (46) |
| GHSROS | Upregulated | Invasion and metastasis | Promoting | (47) |
| H19 | Upregulated | Cancer initiation, proliferation and apoptosis | Promoting | (48-54) |
| HMlincRNA717 | Downregulated | Unexplored | Inhibiting | (55) |
| HNF1A-AS1 | Upregulated | Proliferation and apoptosis, invasion and metastasis | Promoting | (56) |
| HOTAIR | Upregulated | Cancer initiation, drug-resistance, proliferation and apoptosis, invasion and metastasis | Promoting | (57-64) |
| LINC01133 | Upregulated | Proliferation and apoptosis, invasion and metastasis | Promoting | (65-67) |
| LINCRNA-p21 | Downregulated | Proliferation and apoptosis | Inhibiting | (13,68) |
| lnc-bc060912 | Upregulated | Proliferation and apoptosis | Promoting | (69) |
| loc728228 | Upregulated | Proliferation and apoptosis, invasion and metastasis | Promoting | (70) |
| LUADT1 | Upregulated | Proliferation and apoptosis | Promoting | (71) |
| MALAT1 | Upregulated | Invasion and metastasis | Promoting | (72-80) |
| MEG3 | Downregulated | Cancer initiation, proliferation and apoptosis, drug-resistance | Inhibiting | (81-83) |
| LINC00115 | Upregulated | Unexplored | Unexplored | (84) |
| MIAT1 | Upregulated | Unexplored | Unexplored | (84) |
| MVIH | Upregulated | Proliferation and apoptosis, invasion and metastasis | Promoting | (85) |
| NEAT1 | Upregulated | Proliferation and apoptosis, drug-resistance | Promoting | (86,87) |
| NKX2-1-AS1 | Upregulated | Proliferation and apoptosis | Promoting | (88) |
| PANDAR | Downregulated | Proliferation and apoptosis | Inhibiting | (89) |
| DRAIC | Downregulated | Invasion and metastasis | Inhibiting | (90) |
| PVT1 | Upregulated | Proliferation and apoptosis, invasion and metastasis | Promoting | (91,92) |
| RGMB-AS1 | Upregulated | Proliferation and apoptosis, invasion and metastasis | Promoting | (93) |
| SCAL1 | Upregulated | Proliferation and apoptosis, anti-cytotoxic effect | Promoting | (94,95) |
| SOX2-OT | Upregulated | Proliferation and apoptosis | Promoting | (96) |
| SPRY4-IT1 | Upregulated | Invasion and metastasis | Promoting | (97) |
| TUG1 | Downregulated | Proliferation and apoptosis | Inhibiting | (98) |
| ZXF1 | Upregulated | Invasion and metastasis | Promoting | (99) |
| lncRNA-LET | Downregulated | Invasion and metastasis | Inhibiting | (100) |
| UCA1 | Upregulated | Drug-resistance | Promoting | (101) |
| XIST | Upregulated | Invasion and metastasis | Promoting | (102) |
LncRNAs associated with lung cancer initiation
The mechanism of cancer initiation is always of great importance for the study on lung cancer, but is still far from complete understanding. Identification of the relevant players in the process of malignant transformation is a key step to Cancer Res. Previous studies show that 5 lncRNAs can induce malignant transforming of lung and human bronchial epithelial (HBE) cells under specific condition.
H19 is the first lncRNA discovered in human cells and highly expressed during the development of the embryo and fetus, while it is shut down in most tissues shortly after birth (103). Interestingly, this molecule was initially described as an anti-tumor molecule, but the most recent studies have demonstrated that H19 exerts an oncogenic function in various malignancies including lung, breast, cervix, esophageal, ovarian, bone and bladder cancers (54,104-110). Some studies have suggested a distinctive function of H19 in the initiation of lung cancer. Kaplan et al. reported that loss of imprinting (LOI)-caused H19 overexpression acted as one of early markers in the progression of airway epithelium malignant transformation. They found that H19 presented a high expression level in airway epithelium of smokers without alterations of other imprinted genes. Then, they treated HBE cells with cigarette smoke exposure (CSE) and found that a significant increase in H19 level was followed by malignant transformation (48). Another study by Hu et al. reported that HBE cells induced by cigarette smoke condensate showed a significant alteration in transcription profile of cancer-associated genes, including H19, IGF2, and MEG3. This study further demonstrated that long-term treatment with cigarette smoke condensate led to malignant transformation of HEB cells (111). Expression of H19 has been revealed to be improved by mineral dust-induced gene (MDIG), and then, it leads to shortened survival of lung cancer patients and increased incidence of smokers to suffer of lung cancer. MDIG is involved in the demethylation of H3K9me3 in the promoter region of H19, and therefore, it can promote its expression (112,113). Besides the cancer-initiated function mentioned above in lung cancer, H19 has also been reported to be associated with cancer cell proliferation. As a direct transcriptional target of c-Myc, H19 knockdown significantly inhibits NSCLC cell proliferation both in vitro and in vivo (50-52). LncRNA maternally expressed gene 3 (MEG3), a tumor-inhibiting molecule, has also been reported to be associated with various types of tumor tissues and cell lines. Different from H19, MEG3 exerts a distinctively anti-malignant-transformed function in lung carcinoma. MEG3 downregulation is attributed to environmental carcinogennickel-induced DNA methyltransferase 3b (DNMT3b) upregulation in HBE cells. Downregulation of MEG3 contributes to malignant transformation via reducing its binding to transcription factor c-Jun, an inhibitor of PHLPP1. The further study showed that decreased PHLPP1 can lead to Akt/p70S6K/S6 pathway activation, thereby increased the expression of HIF-1α and malignant transformation in HBE cells (81). This study indicates a unique anti-malignant-transformed function of MEG3 in lung cancer. CCAT1 (Colon cancer associated transcript 1, CARLo-5) was first found to be upregulated in colon carcinoma, and it has been reported to be downregulated in various cancers, including lung carcinoma (114-118). CCAT1 is mainly associated with malignant transformation of lung cancer. Lu et al. reported that CCAT1 functioned as a ceRNA to inhibit miRNA let-7, and then, it induced a malignant transformation through indirectly promoting activity of c-Myc in CSE-induced HBE cells (34). In a lncRNA expression microarray analysis, Gao et al. reported a lncRNADQ786227, which was found to be overexpressed in benzo(a)pyrene-induced HBE cells and it induced malignant transforming. According to the study, DQ786227 is upregulated in transformed HBE cells. Further studies on lncRNA DQ786227 is required (40).
LncRNAs on tumor cell proliferation and apoptosis
The uncontrollable cell proliferation and apoptosis is one of the most common malignant characters of cancer. There are 23 known lncRNAs which are associated with lung cancer cells proliferation and apoptosis, such as ANRIL, HOTAIR and lincRNA-p21 etc. In this part, several important proliferation-and-apoptosis-associated lncRNAs and their underlying mechanisms in lung cancer are summarized, and the others can be seen in Table 1.
ANRIL (antisense non-coding RNA in the INK4 locus) was first uncovered in a genetic analysis of patients (119). It is reported that ANRIL functions as a promoter of cell proliferation in lung cancer. With the loss-of-function study, Nie et al. showed that ANRIL (mostly located in the nucleus) affected lung cancer cell proliferation and apoptosis partly via enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2)-dependent silencing of Kruppel like factor 2 (KLF2) and p21 transcription (30). Another study revealed that ANRIL is transactivated by c-Myc (31). Apoptosis-regulated lncRNA, lincRNA-p21, acts as a repressor in p53-dependenttranscriptional responses. The inhibition of this lncRNA affects expressions of hundreds of gene targets enriched for genes normally repressed by p53 in lung cancer cells. It was reported that this effect was mediated via a physical association with hnRNP-K (13). Up to now, the function of lincRNA-p21 as a promoter of cancer cell apoptosis has been reported in several malignancies, including prostate, hepatocellular and colorectal cancer (68,120-123). Recently, Thai et al. identified a novel cancer-inhibiting lncRNA, SCAL1 (the smoke and cancer–associated lncRNA-1), which is upregulated in the airways of smokers and various lung cancer cell lines, and transcriptionally regulated by NRF2 (nuclear factor erythroid 2-related factor). In vitro assays demonstrated that upregulated SCAL1 exerted an anti-cytotoxic function either in CSE-treated HBE cells or in lung cancer cells (95). Another study reported that LUCAT1 (SCAL1) was correlated to poor prognosis and promotes cell proliferation via repressing the expression of p21 and p57 in lung cancer (94). A novel lncRNA, LUADT1 (LUAD transcript 1), was reported to be highly expressed in lung cancer tissues and correlated with T staging. It functionally promotes cell proliferation in lung cancer. This lncRNA exerts cancer-promoting function via binding to proteasome component 2 (PRC2), and therefore, suppresses oncogene p21 expression (71).
LncRNAs associated with cancer invasion and metastasis
Metastasis is regarded as one of the most important lethal factors for cancer, and therefore, it is significant to explore the mechanisms of lung cancer cell invasion and metastasis for improving patients’ survival. Seventeen cancer-associated lncRNAs have been revealed to be implicated in invasion and metastases in lung carcinoma (Table 1). In this part, several lncRNAs which exert important functions in lung cancer invasion and metastasis are introduced
Since its discovery in non-small cell lung cancer (NSCLC) (75), MALAT1 has been linked to various human malignancies, including lung, bladder, breast, colorectal, endometrial, esophageal, gastric, hepatocellular, ovarian, prostate, and renal cell carcinoma (124). MALAT1 is significantly overexpressed and can improve cancer invasion and metastasis in lung cancer (125). In lung carcinoma, it was reported that MALAT1 was regulated by transactive response DNA-binding protein-43 (TDP43) (72), Specificity protein 1 (SP1) (72), and DNA methylation (73), which played as a ceRNA through binding multiple miRNAs, such as miR-204 (74). Thus, MALAT1 regulates several EMT-associated proteins, including SLUG, TWIST and E-cadherin (126). It has also been reported that MALAT1 is associated with some other clinical parameters and malignant phenotypes in multiple malignancies. In cervical cancer cells, MALAT1 impacts cell viability, proliferation, metastasis, cell cycle progression, and tumor growth (127). In colorectal cancer, MALAT1 is correlated with the sensitivity to chemotherapy (128). These two studies indicate that there may be some uncovered associations between this lncRNA and lung cancer except for metastasis. LncRNA HOX transcript antisense RNA (HOTAIR) is a dual-located lncRNA which is located both in nucleus and cytoplasm (72). It has been reported that the elevated expression of HOTAIR is associated with cell proliferation, migration and invasion in pancreatic, prostate, gastric, colorectal, cancer and melanoma (73,74,126-128). While the main function of HOTAIR in lung carcinoma refers to invasion and metastasis. In lung carcinoma, HOTAIR level is always accompanied by advanced stages, metastases, and shortened patient survivals in lung malignant disease (29,129). By means of supporting a role for HOTAIR in lung cancer metastasis, siRNA-mediated downregulation of this lncRNA decreased the migration and invasion of lung cancer cells in vitro and their metastatic potential in vivo (28). The further study showed that HOTAIR regulated the ratio of FoxA1 to FoxA2 via interacting with chromatin remodeling factor LSH, thereby affecting invasive and metastatic phenotypes of lung cancer cells (130). It has also been indicated that HOTAIR is associated with lung malignant transformation. HOTAIR promotes malignant transformation process via activating autocrine IL-6/STAT3 signaling in cigarette smoke extract (CSE)-treated HBE cells (131). Another metastasis-associated lncRNA BANCR (BRAF-regulated lncRNA1), which is induced by oncogene BRAFV600E (129), functions via suppressing MAPK signaling and then inhibiting the transcription of EMT-related proteins in lung carcinoma (28,29). It has also been revealed that BANCR exerted an anti-tumor function to suppress cell invasion and metastasis in colorectal, thyroid, bladder cancer and melanoma (130-133). LncRNA-LET has been shown to be a tumor suppressor in various malignancies. In nasopharyngeal carcinoma, it has been revealed that a significant downregulated level of lncRNA-LET is regulated by EZH2-mediated H3K27 histone methylation (134). The tumor-suppressive function of LET has been reported in gallbladder and gastric cancer (135,136). The hypoxia-induced histone deacetylase 3 suppresses the expression of lncRNA-LET by affecting its promoter, and it eventually facilitates NF90 accumulation and hypoxia-induced cell invasion in lung cancer (100). This finding suggests a lncRNA-mediated connection between metastasis and hypoxia. Recently, Sakurai et al. reported a novel lncRNA downregulated RNA in androgen independent cells (DRAIC), which is downregulated in lung cancer, acts as an anti-tumor player. Overexpression of DRAIC significantly represses cell migration and invasion in lung cancer (90).
LncRNAs involved in therapy-resistant effects
Chemotherapy and radiotherapy are both important modalities against malignancies. However, resistance of cancer cells remains an impediment to the therapeutics. In this part, 8 lncRNAs which increase or decrease therapy-resistant of lung cancer patients are summarized.
Growth arrest-specific transcript 5 (GAS5) is a well-known anti-tumor lncRNA associated with cancer cell apoptosis. It mainly affects sensitivity of chemotherapy in lung carcinoma. Zhang et al. reported that GAS5 can enhance cisplatin sensitivity in lung cancer via autophagy inhibition (137). Another study demonstrated that GAS5 can promote gefitinib-induced lung cancer cell death through inhibiting IGF-1R expression (45). It is revealed by Xue et al. that GAS5 enhanced radiosensitivity through interacting with miR-135b in lung cancer cells (42). Moreover, Xia et al. showed that MEG3 functioned as increasing cisplatin sensitivity of lung cancer cells through regulation of p53, β-catenin and surviving (83). One study on NEAT1 in lung cancer revealed that the interaction between NEAT1 and CTR1 (Copper transporter 1) promoted the internalization of a significant fraction of cDDP (Platinum-based chemotherapy, such as cisplatin) in tumor cells, thereby enhancing cisplatin sensitivity (87). LncRNA HOTAIR has been well demonstrated to be implicated in multiple aspects of malignant characters, including drug-resistance. Fang et al. found that downregulation of HOTAIR can promote cell sensitivity to anti-cancer drugs, thus suppressing cell viability, cell cycle arrest, and tumor growth (63). Except for its role in drug-resistance, a study on HOTAIR reported that over-expression of this lncRNA decreased radio-sensitivity via inactivating β-catenin (64). Additionally, Hou et al. found that a lncRNA, AC006050.3-003, can potentially play a key role in chemo-resistance (26). Yang et al. reported that AK126698 appeared to induce cisplatin resistance by targeting the Wnt pathway in lung cancer cells (27). Urothelial cancer-associated 1 (UCA1) was previously reported to downregulate and exert its oncogenic function in bladder and hepatocellular carcinoma (138,139). Cheng et al. revealed that this molecule was upregulated to induce cancer cell acquired resistances to EGFR-TKIs (epidermal growth factor receptor tyrosine kinase inhibitors) through activating AKT/mTOR pathway (101).
Lung cancer lncRNAs functionally distinct from other malignancies
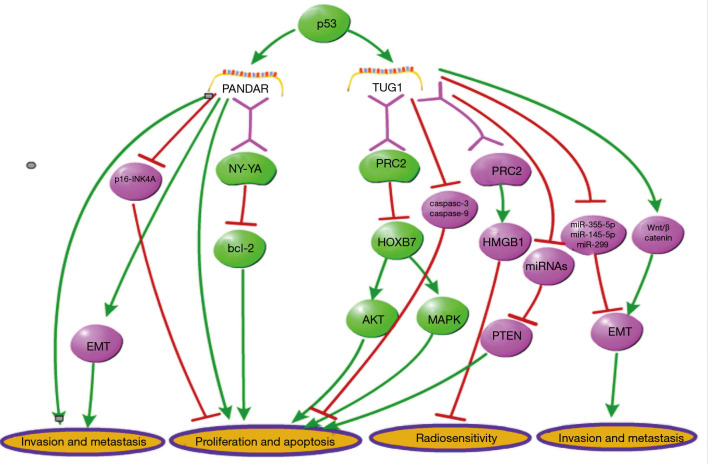
According to previous researches, the biological functions of several specific lncRNAs in lung cancer are always distinguished from other human malignant tumors, through affecting different signaling pathways. In this part, two unique lncRNAs in lung cancer (PANDAR and TUG1) were mainly discussed with comparing to multiple malignancies (Figure 2).
Figure 2.
The known functions and mechanisms of two special lung cancer lncRNAs, TUG1 and PANDAR, are demonstrated in this figure. Green lines stand for the molecular interaction specific for lung carcinoma, while red lines stand for the common pathways that mediate the action of TUG1 and PANDAR in other cancers. These two lncRNAs function as cancer inhibitors in lung carcinoma. The unique cancer-inhibiting functions of TUG1 and PANDAR are both mediated by RNA binding proteins, and regulated by p53 in lung cancer, while their p53-dependent anti-tumor mechanism has not been reported to be involved in other malignancies.
It was reported that the lncRNA promoter of CDKN1Aantisense DNA damage-activated RNA (PANDAR) was induced by DNA damage, and then, it functionally regulated the proliferation and apoptosis of cancer cells (140). According to previous studies, this lncRNA is overexpressed in tumor tissue, and then, boost cell proliferation or metastasis in most malignancies, except for lung carcinoma. In liver cancer, it was reported that PANDAR was overexpressed in cancer cells and associated with poor prognosis. Silencing of PANDAR represses cell proliferation, colony formation and cycle progression (141). In colorectal cancer, PANDAR functions as an independent prognostic factor which correlated to tumor size and prognosis of patients, and its overexpression promotes EMT of cancer cells (142). Besides, it has been reported that PANDAR expression is positively correlated with advanced stages and poor prognosis in bladder cancer. Suppression of this lncRNA decreased invasion and metastasis, and increased apoptosis in bladder cancer cells (143). Additionally, it has been reported that PANDAR is upregulated and it facilitates cell proliferation in breast cancer. Knockdown of PANDAR inhibits G1/S transition of breast cancer cells via suppressing p16INK4A (144). In gastric cancer, it has been demonstrated that PANDAR was highly expressed in cancerous tissues, which is positively correlated to tumor size, lymph node burden and lower survival rates (145). Moreover, Wu et al. has showed an early-stage diagnosis value of circulating PANDAR in clear cell renal cell carcinoma (146). The expression profile and function of PANDAR is completely different in lung carcinoma from other cancers mentioned above, although it is consistently upregulated in cancer and functions as a cancer promoter in most malignancies. Han et al. reported that PANDER exhibited a downregulated expression level in lung cancer and was negatively associated with tumor size and TNM stages. Furthermore, PANDAR was found to be regulated by p53. Upregulation of PANDAR inhibits Bcl-2 expression via interacting with RNA binding protein NY-YA, thus, functionally inhibiting cell growth in vitro and in vivo (89).
LncRNA TUG1 (taurine upregulated gene1) was firstly reported to be upregulated in taurine-treated mouse retinal cells and associated with retinal development (147). Since Khalil et al. revealed its function of recruiting PRC2 via genome-wide RNA immune precipitation (148), TUG1 has been demonstrated to be associated with several malignant diseases. The same as PANDAR, the expression and function of TUG1 in lung carcinoma is also opposite to other cancers. According to a study on colorectal cancer, an elevated expression level of TUG1 was detected in tumor tissues and associated with metastasis phenotype of tumor cells (149). Wang et al. demonstrated that TUG1 functions as a ceRNA to regulate miRNA-355-5p, thereby promoting migration and invasion in osteosarcoma cells (150). Moreover, Jiang et al. revealed that TUG1 decreases radiosensitivity in bladder carcinoma via regulating the expression of HMGB1 (high mobility group box 1 protein), a chromosome-binding protein associated with DNA repair, duplication, transcription, and nucleosome packaging (151). Additionally, the function of promoting cell metastasis in TUG1 has also been reported in breast cancer, gastric cancer, endometrial cancer, small cell lung cancer and other malignancies (152-157). Unlike the tumorigenic role in other the above-mentioned cancers, TUG1 exerts a distinctively tumor-inhibiting function in NSCLC. Recently, Zhang et al. demonstrated that TUG1 is downregulated and functions as a cancer inhibitor in lung carcinoma. Through univariate and multivariate analyses in a cohort of 192 lung cancer patients, it was revealed that TUG1expression was negatively associated with TNM stage, tumor size, and shortened overall survival in lung cancer. Furthermore, this study showed that TUG1 is regulated by p53 via directly regulating its transcription. They also found that TUG1 inhibits cell proliferation via PRC2-mediated inhibiting of homeobox B7 (HOXB7), thus affecting AKT and AMPK pathways. The p53/TUG1/PRC2/HOXB7 axis reveals a distinctive anti-tumor function of TUG1 in lung carcinoma. The anti-tumor function of TUG1 in lung cancer has also been certified in another study (98,158).
These above studies showed that the role of lncRNA may be not consistent in different types of malignancies. The unique cancer-inhibiting functions of TUG1 and PANDAR are both mediated by RNA binding proteins, and regulated by p53 in lung cancer. The p53-dependent mechanisms have not been revealed in other malignant diseases yet. This unique mechanism of the two lncRNAs’ function may be associated with some molecular features specific for lung carcinoma. Thus, it is instrumental in improving lung cancer therapy to further elucidate the molecular mechanism of lung cancer specific lncRNAs.
Potential clinical applications of lung cancer-related lncRNAs
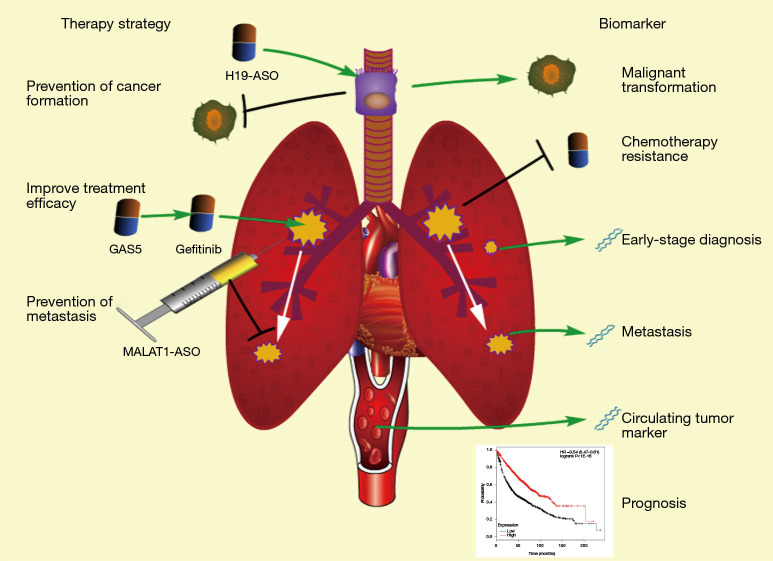
In addition to their roles in diseases, accumulated references suggest that lncRNAs obtained from either blood, tissue, or exhaled breath may provide reliable biomarkers for the diagnosis and prognosis of lung cancer. Moreover, the potential of lncRNAs on cancer therapy remains to be developed (Figure 3).
Figure 3.
The potential clinical applications of lncRNAs are demonstrated in this figure. The items in the left stand for lncRNAs’ potential in lung cancer prevention and treatment; right ones stand for the potential applications in prediction, diagnosis and prognosis of lung cancer.
Biomarkers for prediction, diagnosis and prognosis
It has been demonstrated that smoking is correlated to the incidence of lung cancer (159,160). As mentioned above, the expression levels of H19 and MEG3 are significantly increased and decreased in CSE-treated bronchial epithelial cells respectively, and these two lncRNAs have critical functions in malignant transformation. Besides, it has been also revealed that HOTAIR and DQ786227 are associated with lung cancer initiation. Thus, it is significant for long-term smokers to screen the expression level of these lncRNAs in their bronchial epithelial cells to evaluate the incidence of lung cancer. This screening can be accomplished with their bronchial washing fluids or bronchial epithelial cells during bronchoscopy. Additionally, for better clinical application, it is necessary to define the threshold concentrations of these lncRNA biomarkers for lung cancer prediction.
The intraoperative pathological diagnosis is a main approach to differentiate benign and malignant tumors (161). Since it is difficult to detect these cancer cells in some early-stage cases, more biomarkers are required for the diagnosis of lung cancer. Some lncRNAs (H19, MALAT1, HOTAIR, etc.) always significantly are overexpressed in early stage of lung cancer (50,60,78). Thus, the detection of these lncRNAs can provide us with an approach to timely detect pulmonary malignant tumors. Furthermore, circulating lncRNAs can be used in auxiliary lung cancer diagnosis. It has been reported that both GAS5 and MALAT1 can exhibit elevated expression levels in whole blood of early-stage lung cancer patients (162,163). Screening of MALAT1 and GAS5 in blood is significant for diagnosis of early-stage lung cancer.
Chemotherapy is one of primary therapeutic strategies against malignancies. According to previous studies, the overexpression of these chemotherapy-resistance-associated lncRNAs (GAS5, NEAT1, UCA1, etc.) can affect therapeutic effect of chemotherapy. In order to improve the efficacy, it is highly required to screen on chemotherapy-resistance-associated lncRNAs in lung tumor. In order to obtain necessary biological information for postoperative chemotherapy, it is of important clinical significance to detect these lncRNAs in intraoperative pathological tissue. Unlike the patients who are undergoing surgery, the detection of predictive lncRNA can be performed for these preoperative or non-operative patients during CT-guided percutaneous biopsy.
The invasion and metastasis are closely related to prognosis of lung cancer patients (164). MALAT1 is overexpressed in lung cancer tissues, as well as circulating system, and plays a critical role in lung cancer metastasis (80,163,165). High MALAT1 level in intraoperative pathological tissues or in whole blood indicated a metastatic potential for patients with lung carcinoma.
In addition, it has been reported that several lncRNAs are associated with prognosis of lung cancer patients (60,67,78,162,165-168). With the help of available big data, such as The Cancer Genome Atlas (TCGA), we can identify multifactor prognostic markers through analyzing the correlation between lncRNA expression and survival rate. It is more likely to achieve a more precise prognosis through a combined application of these lncRNA biomarkers for lung cancer patients.
Potentially therapeutic applications
The molecule-targeted therapy is a novel and potential approach against malignancies featured with better outcomes and fewer side effects. Several lung-cancer-associated lncRNAs are critical for initiation, progression, and metastasis of lung cancer, thus having potential to act as therapeutic targets. Theoretically, the regulation of expression of these target lncRNAs can prevent the initiation of lung carcinoma and decrease the malignancy of tumor cells. Antisense oligonucleotides (ASOs) are short single-stranded deoxyribonucleotide analogs, which can downregulate RNAs via binding their complementary sequence. One study has demonstrated that injection of MALAT1-specific ASO into lung tumor can significantly inhibit lung cancer metastasis (79). Maybe, similar approaches can also be used to silence H19, which is elevated in SCE-treated bronchial epithelial cells and induces lung cancer initiation as mentioned above, to prevent the formation of lung carcinoma induced by smoking. Regarding that MEG3 can prevent malignant transformation in bronchial epithelial cells (81), it is possible to prevent lung cancer through the restoration of MEG3 in human lung and bronchial epithelial cells with effective transfer vectors. In addition, lncRNAs involved in chemotherapy sensitivity can improve therapeutic effect of drugs on lung cancer. As noted above, HOTAIR can reduce the sensitivity of lung carcinoma cells to chemotherapeutic drugs, such as cisplatin (63). Theoretically, the cisplatin effect can be improved by treatment of lung cancer patients with HOTAIR-specific ASO. Moreover, several lncRNAs can also be used for the enhancement of chemotherapy, including UCA1, NEAT1 and GAS5.
Discussion
Although numerous studies have reported that many lncRNAs are closely associated with lung cancer, the functions and clinical potential of these lncRNAs are far from being well-understood. Tumor-derived lncRNAs are not suitable to widely and clinically predict the progression of lung cancer due to its relatively low accessibility, while lncRNAs in blood are more valuable in lung cancer diagnosis and prognosis prediction. Therefore, it is urgently needed to study circulating lncRNAs more intensely for future clinical application. The uncontrollable recurrence and metastasis are the highest lethality factors for lung cancer patients, as well as other malignancies, and however, it is yet to report which lncRNAs are correlated to cancer recurrence. Additionally, it has been revealed that several cancer-inhibiting lncRNAs are implicated in lung cancer, such as MEG3, PANDAR, DRAIC. It is promising to further develop anti-tumor therapeutic strategies based on utilizing these tumor suppressor lncRNAs.
Up to now, several miRNAs, like miR-34, have been well-studied, and ready to be used for diagnosis and therapy of malignancies (169), while lncRNA-based therapeutics are still far from clinical applications. Some problems are needed to be solved to facilitate the clinical application of lncRNAs, such as safety of lncRNA-based drugs, and delivery of therapeutic agent into target cells.
Circular RNAs represent a novel class of endogenous RNAs with diversity and universality (170). Recent studies have well demonstrated the relationship between circular and linear RNAs and the possible mechanisms of cyclization (171,172), thus implying that lncRNAs may transform from linear form into circular one. For instance, the cyclization of ANRIL is induced by Alu repeats flanking its exons, and exerts different functions from linear ANRIL (173). Therefore, the emerging of circular RNA provides a new direction for lncRNAs’ study.
Conclusions
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Acknowledgments
Funding: This study was funded by Key R & D programs in Shandong Province (grant number: 2018GSF118119).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.22). The authors have no conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629-41. 10.1016/j.cell.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 3.Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 2016;17:47-62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu Rev Biochem 2015;84:405-33. 10.1146/annurev-biochem-060614-034258 [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Zhu JK. Emerging roles of RNA processing factors in regulating long non-coding RNAs. RNA Biol 2014;11:793-7. 10.4161/rna.29731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wierzbicki AT, Ream TS, Haag JR, et al. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 2009;41:630-4. 10.1038/ng.365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doerr A. Unraveling the lncRNA mystery. Nat Methods 2014;11:890. 10.1038/nmeth.3089 [DOI] [PubMed] [Google Scholar]
- 8.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet 2014;48:433-55. 10.1146/annurev-genet-120213-092323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y, Cheung BB, Atmadibrata B, et al. The regulatory role of long noncoding RNAs in cancer. Cancer Lett 2017;391:12-9. 10.1016/j.canlet.2017.01.010 [DOI] [PubMed] [Google Scholar]
- 10.Morlando M, Ballarino M, Fatica A, et al. The role of long noncoding RNAs in the epigenetic control of gene expression. ChemMedChem 2014;9:505-10. 10.1002/cmdc.201300569 [DOI] [PubMed] [Google Scholar]
- 11.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet 2014;30:348-55. 10.1016/j.tig.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plath K, Mlynarczyk-Evans S, Nusinow DA, et al. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet 2002;36:233-78. 10.1146/annurev.genet.36.042902.092433 [DOI] [PubMed] [Google Scholar]
- 13.Bao X, Wu H, Zhu X, et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res 2015;25:80-92. 10.1038/cr.2014.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vennin C, Spruyt N, Robin YM, et al. The long non-coding RNA 91H increases aggressive phenotype of breast cancer cells and up-regulates H19/IGF2 expression through epigenetic modifications. Cancer Lett 2017;385:198-206. 10.1016/j.canlet.2016.10.023 [DOI] [PubMed] [Google Scholar]
- 15.West JA, Davis CP, Sunwoo H, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 2014;55:791-802. 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X, Sun M, Wu Y, et al. Post-transcriptional regulation of long noncoding RNAs in cancer. Tumour Biol 2015;36:503-13. 10.1007/s13277-015-3106-y [DOI] [PubMed] [Google Scholar]
- 17.Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunten S, Kaller M, Drepper F, et al. p53-Regulated Networks of Protein, mRNA, miRNA, and lncRNA Expression Revealed by Integrated Pulsed Stable Isotope Labeling With Amino Acids in Cell Culture (pSILAC) and Next Generation Sequencing (NGS) Analyses. Mol Cell Proteomics 2015;14:2609-29. 10.1074/mcp.M115.050237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rashid F, Shah A, Shan G. Long Non-coding RNAs in the Cytoplasm. Genomics Proteomics Bioinformatics 2016;14:73-80. 10.1016/j.gpb.2016.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang K, Shi ZM, Chang YN, et al. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene 2014;547:1-9. 10.1016/j.gene.2014.06.043 [DOI] [PubMed] [Google Scholar]
- 21.Khanduja JS, Calvo IA, Joh RI, et al. Nuclear Noncoding RNAs and Genome Stability. Mol Cell 2016;63:7-20. 10.1016/j.molcel.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 2016;44:863-77. 10.1093/nar/gkv1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirata H, Hinoda Y, Shahryari V, et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res 2015;75:1322-31. 10.1158/0008-5472.CAN-14-2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao J, Zhang Y, Yang J, et al. NEAT1 regulates pancreatic cancer cell growth, invasion and migration though mircroRNA-335-5p/c-met axis. Am J Cancer Res 2016;6:2361-74. [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng Z, Bo H, Gong Z, et al. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol 2016;37:729-37. 10.1007/s13277-015-3860-x [DOI] [PubMed] [Google Scholar]
- 26.Hou Z, Xu C, Xie H, et al. Long noncoding RNAs expression patterns associated with chemo response to cisplatin based chemotherapy in lung squamous cell carcinoma patients. PLoS One 2014;9:e108133. 10.1371/journal.pone.0108133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y, Li H, Hou S, et al. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One 2013;8:e65309. 10.1371/journal.pone.0065309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang W, Zhang D, Xu B, et al. Long non-coding RNA BANCR promotes proliferation and migration of lung carcinoma via MAPK pathways. Biomed Pharmacother 2015;69:90-5. 10.1016/j.biopha.2014.11.027 [DOI] [PubMed] [Google Scholar]
- 29.Sun M, Liu XH, Wang KM, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer 2014;13:68. 10.1186/1476-4598-13-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nie FQ, Sun M, Yang JS, et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther 2015;14:268-77. 10.1158/1535-7163.MCT-14-0492 [DOI] [PubMed] [Google Scholar]
- 31.Lu Y, Zhou X, Xu L, et al. Long noncoding RNA ANRIL could be transactivated by c-Myc and promote tumor progression of non-small-cell lung cancer. Onco Targets Ther 2016;9:3077-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu T, Lu YR. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int 2015;15:36. 10.1186/s12935-015-0183-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei MM, Zhou YC, Wen ZS, et al. Long non-coding RNA stabilizes the Y-box-binding protein 1 and regulates the epidermal growth factor receptor to promote lung carcinogenesis. Oncotarget 2016;7:59556-71. 10.18632/oncotarget.10006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu L, Qi H, Luo F, et al. Feedback circuitry via let-7c between lncRNA CCAT1 and c-Myc is involved in cigarette smoke extract-induced malignant transformation of HBE cells. Oncotarget 2017;8:19285-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Zhang K, Song H, et al. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget 2016;7:62474-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Z, Wang J, Wang S, et al. LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomed Pharmacother 2017;87:692-7. 10.1016/j.biopha.2016.12.122 [DOI] [PubMed] [Google Scholar]
- 37.Qiu M, Xu Y, Yang X, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol 2014;35:5375-80. 10.1007/s13277-014-1700-z [DOI] [PubMed] [Google Scholar]
- 38.Chen S, Wu H, Lv N, et al. LncRNA CCAT2 predicts poor prognosis and regulates growth and metastasis in small cell lung cancer. Biomed Pharmacother 2016;82:583-8. 10.1016/j.biopha.2016.05.017 [DOI] [PubMed] [Google Scholar]
- 39.Li J, Li P, Zhao W, et al. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int 2015;15:48. 10.1186/s12935-015-0201-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao L, Mai A, Li X, et al. LncRNA-DQ786227-mediated cell malignant transformation induced by benzo(a)pyrene. Toxicol Lett 2013;223:205-10. 10.1016/j.toxlet.2013.09.015 [DOI] [PubMed] [Google Scholar]
- 41.Mei Y, Si J, Wang Y, et al. Long noncoding RNA GAS5 suppresses tumorigenesis by inhibiting miR-23a 5 expression in non-small cell lung cancer. Oncol Res 2017;25:1027-37. 10.3727/096504016X14822800040451 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Xue Y, Ni T, Jiang Y, et al. LncRNA GAS5 Inhibits Tumorigenesis and Enhances Radiosensitivity By Suppressing miR-135b Expression in Non-Small Cell Lung Cancer. Oncol Res 2017;25:1305-16. 10.3727/096504017X14850182723737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y, Lyu H, Liu H, et al. Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep 2016;6:31093. 10.1038/srep31093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi X, Sun M, Liu H, et al. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non-small-cell lung cancer. Mol Carcinog 2015;54 Suppl 1:E1-E12. 10.1002/mc.22120 [DOI] [PubMed] [Google Scholar]
- 45.Dong S, Qu X, Li W, et al. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J Hematol Oncol 2015;8:43. 10.1186/s13045-015-0140-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han L, Kong R, Yin DD, et al. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med Oncol 2013;30:694. 10.1007/s12032-013-0694-5 [DOI] [PubMed] [Google Scholar]
- 47.Whiteside EJ, Seim I, Pauli JP, et al. Identification of a long non-coding RNA gene, growth hormone secretagogue receptor opposite strand, which stimulates cell migration in non-small cell lung cancer cell lines. Int J Oncol 2013;43:566-74. 10.3892/ijo.2013.1969 [DOI] [PubMed] [Google Scholar]
- 48.Kaplan R, Luettich K, Heguy A, et al. Monoallelic up-regulation of the imprinted H19 gene in airway epithelium of phenotypically normal cigarette smokers. Cancer Res 2003;63:1475-82. [PubMed] [Google Scholar]
- 49.Luo X, Li Z, Yan Q, et al. The human WW45 protein enhances MST1-mediated apoptosis in vivo. Int J Mol Med 2009;23:357-62. [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang E, Li W, Yin D, et al. c-Myc-regulated long non-coding RNA H19 indicates a poor prognosis and affects cell proliferation in non-small-cell lung cancer. Tumour Biol 2016;37:4007-15. 10.1007/s13277-015-4185-5 [DOI] [PubMed] [Google Scholar]
- 51.Zhang E, Li W, Yin D, et al. Erratum to: c-Myc-regulated long non-coding RNA H19 indicates a poor prognosis and affects cell proliferation in non-small-cell lung cancer. Tumour Biol 2016;37:5653. 10.1007/s13277-015-4347-5 [DOI] [PubMed] [Google Scholar]
- 52.Cui J, Mo J, Luo M, et al. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int J Clin Exp Pathol 2015;8:12400-9. [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Q, Cheng N, Li X, et al. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget 2017;8:2558-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matouk IJ, Halle D, Gilon M, et al. The non-coding RNAs of the H19-IGF2 imprinted loci: a focus on biological roles and therapeutic potential in Lung Cancer. J Transl Med 2015;13:113. 10.1186/s12967-015-0467-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie X, Liu HT, Mei J, et al. LncRNA HMlincRNA717 is down-regulated in non-small cell lung cancer and associated with poor prognosis. Int J Clin Exp Pathol 2014;7:8881-6. [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Y, Liu H, Shi X, et al. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget 2015;6:9160-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Luo F, Xu Y, et al. Epithelial-mesenchymal transition and cancer stem cells, mediated by a long non-coding RNA, HOTAIR, are involved in cell malignant transformation induced by cigarette smoke extract. Toxicol Appl Pharmacol 2015;282:9-19. 10.1016/j.taap.2014.10.022 [DOI] [PubMed] [Google Scholar]
- 58.Zhou C, Ye L, Jiang C, et al. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1alpha activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol 2015;36:9179-88. 10.1007/s13277-015-3453-8 [DOI] [PubMed] [Google Scholar]
- 59.Ono H, Motoi N, Nagano H, et al. Long noncoding RNA HOTAIR is relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancer. Cancer Med 2014;3:632-42. 10.1002/cam4.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakagawa T, Endo H, Yokoyama M, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun 2013;436:319-24. 10.1016/j.bbrc.2013.05.101 [DOI] [PubMed] [Google Scholar]
- 61.Liu XH, Liu ZL, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 2013;13:464. 10.1186/1471-2407-13-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang R, Shi Y, Chen L, et al. The ratio of FoxA1 to FoxA2 in lung adenocarcinoma is regulated by LncRNA HOTAIR and chromatin remodeling factor LSH. Sci Rep 2015;5:17826. 10.1038/srep17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang S, Gao H, Tong Y, et al. Long noncoding RNA-HOTAIR affects chemoresistance by regulating HOXA1 methylation in small cell lung cancer cells. Lab Invest 2016;96:60-8. 10.1038/labinvest.2015.123 [DOI] [PubMed] [Google Scholar]
- 64.Chen J, Shen Z, Zheng Y, et al. Radiotherapy induced Lewis lung cancer cell apoptosis via inactivating beta-catenin mediated by upregulated HOTAIR. Int J Clin Exp Pathol 2015;8:7878-86. [PMC free article] [PubMed] [Google Scholar]
- 65.Kong J, Sun W, Li C, et al. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett 2016;380:476-84. 10.1016/j.canlet.2016.07.015 [DOI] [PubMed] [Google Scholar]
- 66.Zang C, Nie FQ, Wang Q, et al. Long non-coding RNA LINC01133 represses KLF2, P21 and E-cadherin transcription through binding with EZH2, LSD1 in non small cell lung cancer. Oncotarget 2016;7:11696-707. 10.18632/oncotarget.7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang J, Zhu N, Chen X. A novel long noncoding RNA LINC01133 is upregulated in lung squamous cell cancer and predicts survival. Tumour Biol 2015;36:7465-71. 10.1007/s13277-015-3460-9 [DOI] [PubMed] [Google Scholar]
- 68.Castellano JJ, Navarro A, Vinolas N, et al. LincRNA-p21 Impacts Prognosis in Resected Non-Small Cell Lung Cancer Patients through Angiogenesis Regulation. J Thorac Oncol 2016;11:2173-82. 10.1016/j.jtho.2016.07.015 [DOI] [PubMed] [Google Scholar]
- 69.Luo H, Sun Y, Wei G, et al. Functional Characterization of Long Noncoding RNA Lnc_bc060912 in Human Lung Carcinoma Cells. Biochemistry 2015;54:2895-902. 10.1021/acs.biochem.5b00259 [DOI] [PubMed] [Google Scholar]
- 70.Hu G, Yang T, Zheng J, et al. Functional role and mechanism of lncRNA LOC728228 in malignant 16HBE cells transformed by anti-benzopyrene-trans-7,8-dihydrodiol-9,10-epoxide. Mol Carcinog 2015;54 Suppl 1:E192-204. 10.1002/mc.22314 [DOI] [PubMed] [Google Scholar]
- 71.Qiu M, Xu Y, Wang J, et al. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis 2015;6:e1858. 10.1038/cddis.2015.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo F, Jiao F, Song Z, et al. Regulation of MALAT1 expression by TDP43 controls the migration and invasion of non-small cell lung cancer cells in vitro. Biochem Biophys Res Commun 2015;465:293-8. 10.1016/j.bbrc.2015.08.027 [DOI] [PubMed] [Google Scholar]
- 73.Guo F, Guo L, Li Y, et al. MALAT1 is an oncogenic long non-coding RNA associated with tumor invasion in non-small cell lung cancer regulated by DNA methylation. Int J Clin Exp Pathol 2015;8:15903-10. [PMC free article] [PubMed] [Google Scholar]
- 74.Li J, Wang J, Chen Y, et al. LncRNA MALAT1 exerts oncogenic functions in lung adenocarcinoma by targeting miR-204. Am J Cancer Res 2016;6:1099-107. [PMC free article] [PubMed] [Google Scholar]
- 75.Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031-41. 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- 76.Huang NS, Chi YY, Xue JY, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget 2016;7:37957-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu M, Sun W, Liu Y, et al. The role of lncRNA MALAT1 in bone metastasis in patients with non-small cell lung cancer. Oncol Rep 2016;36:1679-85. 10.3892/or.2016.4909 [DOI] [PubMed] [Google Scholar]
- 78.Weber DG, Johnen G, Casjens S, et al. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes 2013;6:518. 10.1186/1756-0500-6-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013;73:1180-9. 10.1158/0008-5472.CAN-12-2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmidt LH, Gorlich D, Spieker T, et al. Prognostic impact of Bcl-2 depends on tumor histology and expression of MALAT-1 lncRNA in non-small-cell lung cancer. J Thorac Oncol 2014;9:1294-304. 10.1097/JTO.0000000000000243 [DOI] [PubMed] [Google Scholar]
- 81.Zhou C, Huang C, Wang J, et al. LncRNA MEG3 downregulation mediated by DNMT3b contributes to nickel malignant transformation of human bronchial epithelial cells via modulating PHLPP1 transcription and HIF-1alpha translation. Oncogene 2017;36:3878-89. 10.1038/onc.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kruer TL, Dougherty SM, Reynolds L, et al. Expression of the lncRNA Maternally Expressed Gene 3 (MEG3) Contributes to the Control of Lung Cancer Cell Proliferation by the Rb Pathway. PLoS One 2016;11:e0166363. 10.1371/journal.pone.0166363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia Y, He Z, Liu B, et al. Downregulation of Meg3 enhances cisplatin resistance of lung cancer cells through activation of the WNT/beta-catenin signaling pathway. Mol Med Rep 2015;12:4530-7. 10.3892/mmr.2015.3897 [DOI] [PubMed] [Google Scholar]
- 84.Li DS, Ainiwaer JL, Sheyhiding I, et al. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci 2016;20:2285-95. [PubMed] [Google Scholar]
- 85.Nie FQ, Zhu Q, Xu TP, et al. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol 2014;35:7587-94. 10.1007/s13277-014-2009-7 [DOI] [PubMed] [Google Scholar]
- 86.Sun C, Li S, Zhang F, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget 2016;7:51784-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang P, Wu X, Wang X, et al. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget 2016;7:43337-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Balbin OA, Malik R, Dhanasekaran SM, et al. The landscape of antisense gene expression in human cancers. Genome Res 2015;25:1068-79. 10.1101/gr.180596.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Han L, Zhang EB, Yin DD, et al. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis 2015;6:e1665. 10.1038/cddis.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sakurai K, Reon BJ, Anaya J, et al. The lncRNA DRAIC/PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Mol Cancer Res 2015;13:828-38. 10.1158/1541-7786.MCR-15-0016-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wan L, Sun M, Liu GJ, et al. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation through Epigenetically Regulating LATS2 Expression. Mol Cancer Ther 2016;15:1082-94. 10.1158/1535-7163.MCT-15-0707 [DOI] [PubMed] [Google Scholar]
- 92.Huang C, Liu S, Wang H, et al. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am J Transl Res 2016;8:5025-34. [PMC free article] [PubMed] [Google Scholar]
- 93.Li P, Li J, Yang R, et al. Study on expression of lncRNA RGMB-AS1 and repulsive guidance molecule b in non-small cell lung cancer. Diagn Pathol 2015;10:63. 10.1186/s13000-015-0297-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Renhua G, Yue S, Shidai J, et al. 165P: Long noncoding RNA LUCAT1 is associated with poor prognosis in human non-small cell lung cancer and affects cell proliferation via regulating p21 and p57 expression. J Thorac Oncol 2016;11:S129. 10.1016/S1556-0864(16)30275-1 [DOI] [Google Scholar]
- 95.Thai P, Statt S, Chen CH, et al. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol 2013;49:204-11. 10.1165/rcmb.2013-0159RC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saghaeian Jazi M, Samaei NM, Ghanei M, et al. Overexpression of the non-coding SOX2OT variants 4 and 7 in lung tumors suggests an oncogenic role in lung cancer. Tumour Biol 2016;37:10329-38. 10.1007/s13277-016-4901-9 [DOI] [PubMed] [Google Scholar]
- 97.Sun M, Liu XH, Lu KH, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis 2014;5:e1298. 10.1038/cddis.2014.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang EB, Yin DD, Sun M, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis 2014;5:e1243. 10.1038/cddis.2014.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang L, Zhou XF, Pan GF, et al. Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinoma. Biomed Pharmacother 2014;68:401-7. 10.1016/j.biopha.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 100.Liu B, Pan CF, He ZC, et al. Long Noncoding RNA-LET Suppresses Tumor Growth and EMT in Lung Adenocarcinoma. Biomed Res Int 2016;2016:4693471. 10.1155/2016/4693471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cheng N, Cai W, Ren S, et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 2015;6:23582-93. 10.18632/oncotarget.4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun 2016;478:811-7. 10.1016/j.bbrc.2016.08.030 [DOI] [PubMed] [Google Scholar]
- 103.Barsyte-Lovejoy D, Lau SK, Boutros PC, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 2006;66:5330-7. 10.1158/0008-5472.CAN-06-0037 [DOI] [PubMed] [Google Scholar]
- 104.Yang W, Ning N, Jin X. The lncRNA H19 Promotes Cell Proliferation by Competitively Binding to miR-200a and Derepressing beta-Catenin Expression in Colorectal Cancer. Biomed Res Int 2017;2017:2767484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peng F, Li TT, Wang KL, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis 2017;8:e2569. 10.1038/cddis.2016.438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Conigliaro A, Costa V, Lo Dico A, et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer 2015;14:155. 10.1186/s12943-015-0426-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014;5:2318-29. 10.18632/oncotarget.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Murrell A, Ito Y, Verde G, et al. Distinct methylation changes at the IGF2-H19 locus in congenital growth disorders and cancer. PLoS One 2008;3:e1849. 10.1371/journal.pone.0001849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Byun HM, Wong HL, Birnstein EA, et al. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res 2007;67:10753-8. 10.1158/0008-5472.CAN-07-0329 [DOI] [PubMed] [Google Scholar]
- 110.Chen CL, Ip SM, Cheng D, et al. Loss of imprinting of the IGF-II and H19 genes in epithelial ovarian cancer. Clin Cancer Res 2000;6:474-9. [PubMed] [Google Scholar]
- 111.Hu YC, Yang ZH, Zhong KJ, et al. Alteration of transcriptional profile in human bronchial epithelial cells induced by cigarette smoke condensate. Toxicol Lett 2009;190:23-31. 10.1016/j.toxlet.2009.06.860 [DOI] [PubMed] [Google Scholar]
- 112.Chen B, Yu M, Chang Q, et al. Mdig de-represses H19 large intergenic non-coding RNA (lincRNA) by down-regulating H3K9me3 and heterochromatin. Oncotarget 2013;4:1427-37. 10.18632/oncotarget.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Y, Lu Y, Yuan BZ, et al. The Human mineral dust-induced gene, mdig, is a cell growth regulating gene associated with lung cancer. Oncogene 2005;24:4873-82. 10.1038/sj.onc.1208668 [DOI] [PubMed] [Google Scholar]
- 114.Guo X, Hua Y. CCAT1: an oncogenic long noncoding RNA in human cancers. J Cancer Res Clin Oncol 2017;143:555-62. 10.1007/s00432-016-2268-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhao X, Wei X, Zhao L, et al. The rs6983267 SNP and long non-coding RNA CARLo-5 are associated with endometrial carcinoma. Environ Mol Mutagen 2016;57:508-15. 10.1002/em.22031 [DOI] [PubMed] [Google Scholar]
- 116.Zhang Y, Ma M, Liu W, et al. Enhanced expression of long noncoding RNA CARLo-5 is associated with the development of gastric cancer. Int J Clin Exp Pathol 2014;7:8471-9. [PMC free article] [PubMed] [Google Scholar]
- 117.Wang F, Xie C, Zhao W, et al. Long non-coding RNA CARLo-5 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Clin Exp Med 2017;17:33-43. 10.1007/s10238-015-0395-9 [DOI] [PubMed] [Google Scholar]
- 118.Liu JN, Shangguan YM. Long non-coding RNA CARLo-5 upregulation associates with poor prognosis in patients suffering gastric cancer. Eur Rev Med Pharmacol Sci 2017;21:530-4. 10.1016/j.ejps.2016.10.024 [DOI] [PubMed] [Google Scholar]
- 119.Pasmant E, Laurendeau I, Heron D, et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 2007;67:3963-9. 10.1158/0008-5472.CAN-06-2004 [DOI] [PubMed] [Google Scholar]
- 120.Yang N, Fu Y, Zhang H, et al. LincRNA-p21 activates endoplasmic reticulum stress and inhibits hepatocellular carcinoma. Oncotarget 2015;6:28151-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang X, Ruan Y, Wang X, et al. Long intragenic non-coding RNA lincRNA-p21 suppresses development of human prostate cancer. Cell Prolif 2017;50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tran UM, Rajarajacholan U, Soh J, et al. LincRNA-p21 acts as a mediator of ING1b-induced apoptosis. Cell Death Dis 2015;6:e1668. 10.1038/cddis.2015.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tang SS, Zheng BY, Xiong XD. LincRNA-p21: Implications in Human Diseases. Int J Mol Sci 2015;16:18732-40. 10.3390/ijms160818732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gutschner T, Hammerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791-801. 10.1007/s00109-013-1028-y [DOI] [PubMed] [Google Scholar]
- 125.Wu Y, Huang C, Meng X, et al. Long Noncoding RNA MALAT1: Insights into its Biogenesis and Implications in Human Disease. Curr Pharm Des 2015;21:5017-28. 10.2174/1381612821666150724115625 [DOI] [PubMed] [Google Scholar]
- 126.Shen L, Chen L, Wang Y, et al. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol 2015;121:101-8. 10.1007/s11060-014-1613-0 [DOI] [PubMed] [Google Scholar]
- 127.Sun R, Qin C, Jiang B, et al. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial-mesenchymal transition. Mol Biosyst 2016;12:952-62. 10.1039/C5MB00685F [DOI] [PubMed] [Google Scholar]
- 128.Li P, Zhang X, Wang H, et al. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol Cancer Ther 2017;16:739-51. 10.1158/1535-7163.MCT-16-0591 [DOI] [PubMed] [Google Scholar]
- 129.Flockhart RJ, Webster DE, Qu K, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res 2012;22:1006-14. 10.1101/gr.140061.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Staff PO. Correction: Long non-coding RNA BANCR promotes proliferation in malignant melanoma by regulating MAPK pathway activation. PLoS One 2015;10:e0118728. 10.1371/journal.pone.0118728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shi Y, Liu Y, Wang J, et al. Downregulated Long Noncoding RNA BANCR Promotes the Proliferation of Colorectal Cancer Cells via Downregualtion of p21 Expression. PLoS One 2015;10:e0122679. 10.1371/journal.pone.0122679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.He A, Liu Y, Chen Z, et al. Over-expression of long noncoding RNA BANCR inhibits malignant phenotypes of human bladder cancer. J Exp Clin Cancer Res 2016;35:125. 10.1186/s13046-016-0397-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liao T, Qu N, Shi RL, et al. BRAF-activated LncRNA functions as a tumor suppressor in papillary thyroid cancer. Oncotarget 2017;8:238-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sun Q, Liu H, Li L, et al. Long noncoding RNA-LET, which is repressed by EZH2, inhibits cell proliferation and induces apoptosis of nasopharyngeal carcinoma cell. Med Oncol 2015;32:226. 10.1007/s12032-015-0673-0 [DOI] [PubMed] [Google Scholar]
- 135.Tian J, Hu X, Gao W, et al. Identification of the long noncoding RNA LET as a novel tumor suppressor in gastric cancer. Mol Med Rep 2017;15:2229-34. 10.3892/mmr.2017.6263 [DOI] [PubMed] [Google Scholar]
- 136.Ma MZ, Kong X, Weng MZ, et al. Long non-coding RNA-LET is a positive prognostic factor and exhibits tumor-suppressive activity in gallbladder cancer. Mol Carcinog 2015;54:1397-406. 10.1002/mc.22215 [DOI] [PubMed] [Google Scholar]
- 137.Zhang N, Yang GQ, Shao XM, et al. GAS5 modulated autophagy is a mechanism modulating cisplatin sensitivity in NSCLC cells. Eur Rev Med Pharmacol Sci 2016;20:2271-7. [PubMed] [Google Scholar]
- 138.Zhen S, Hua L, Liu YH, et al. Inhibition of long non-coding RNA UCA1 by CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer. Oncotarget 2017;8:9634-46. 10.18632/oncotarget.14176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang F, Ying HQ, He BS, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget 2015;6:7899-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011;43:621-9. 10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Peng W, Fan H. Long non-coding RNA PANDAR correlates with poor prognosis and promotes tumorigenesis in hepatocellular carcinoma. Biomed Pharmacother 2015;72:113-8. 10.1016/j.biopha.2015.04.014 [DOI] [PubMed] [Google Scholar]
- 142.Li X, Wang F, Sun Y, et al. Expression of long non-coding RNA PANDAR and its prognostic value in colorectal cancer patients. Int J Biol Markers 2017;32:e218-e23. 10.5301/jbm.5000249 [DOI] [PubMed] [Google Scholar]
- 143.Zhan Y, Lin J, Liu Y, et al. Up-regulation of long non-coding RNA PANDAR is associated with poor prognosis and promotes tumorigenesis in bladder cancer. J Exp Clin Cancer Res 2016;35:83. 10.1186/s13046-016-0354-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sang Y, Tang J, Li S, et al. LncRNA PANDAR regulates the G1/S transition of breast cancer cells by suppressing p16(INK4A) expression. Sci Rep 2016;6:22366. 10.1038/srep22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ma P, Xu T, Huang M, et al. Increased expression of LncRNA PANDAR predicts a poor prognosis in gastric cancer. Biomed Pharmacother 2016;78:172-6. 10.1016/j.biopha.2016.01.025 [DOI] [PubMed] [Google Scholar]
- 146.Wu Y, Wang YQ, Weng WW, et al. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis 2016;5:e192. 10.1038/oncsis.2015.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 2005;15:501-12. 10.1016/j.cub.2005.02.027 [DOI] [PubMed] [Google Scholar]
- 148.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009;106:11667-72. 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang L, Zhao Z, Feng W, et al. Long non-coding RNA TUG1 promotes colorectal cancer metastasis via EMT pathway. Oncotarget 2016;7:51713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang Y, Yang T, Zhang Z, et al. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci 2017;108:859-67. 10.1111/cas.13201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jiang H, Hu X, Zhang H, et al. Down-regulation of LncRNA TUG1 enhances radiosensitivity in bladder cancer via suppressing HMGB1 expression. Radiat Oncol 2017;12:65. 10.1186/s13014-017-0802-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu L, Chen X, Zhang Y, et al. Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget 2017;8:31386-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hu Y, Sun X, Mao C, et al. Upregulation of long noncoding RNA TUG1 promotes cervical cancer cell proliferation and migration. Cancer Med 2017;6:471-82. 10.1002/cam4.994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Niu Y, Ma F, Huang W, et al. Long non-coding RNA TUG1 is involved in cell growth and chemoresistance of small cell lung cancer by regulating LIMK2b via EZH2. Mol Cancer 2017;16:5. 10.1186/s12943-016-0575-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li T, Liu Y, Xiao H, et al. Long non-coding RNA TUG1 promotes cell proliferation and metastasis in human breast cancer. Breast Cancer 2017;24:535-43. 10.1007/s12282-016-0736-x [DOI] [PubMed] [Google Scholar]
- 156.Kuang D, Zhang X, Hua S, et al. Long non-coding RNA TUG1 regulates ovarian cancer proliferation and metastasis via affecting epithelial-mesenchymal transition. Exp Mol Pathol 2016;101:267-73. 10.1016/j.yexmp.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 157.Ji TT, Huang X, Jin J, et al. Inhibition of long non-coding RNA TUG1 on gastric cancer cell transference and invasion through regulating and controlling the expression of miR-144/c-Met axis. Asian Pac J Trop Med 2016;9:508-12. 10.1016/j.apjtm.2016.03.026 [DOI] [PubMed] [Google Scholar]
- 158.Lin PC, Huang HD, Chang CC, et al. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer 2016;16:583. 10.1186/s12885-016-2569-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Stone EC, Zhou C, International Association for the Study of Lung Cancer Tobacco Control C. Slowing the Titanic: China's Epic Struggle with Tobacco. J Thorac Oncol 2016;11:2053-65. 10.1016/j.jtho.2016.07.020 [DOI] [PubMed] [Google Scholar]
- 160.Filosso PL, Oliaro A, Ruffini E, et al. Outcome and prognostic factors in bronchial carcinoids: a single-center experience. J Thorac Oncol 2013;8:1282-8. 10.1097/JTO.0b013e31829f097a [DOI] [PubMed] [Google Scholar]
- 161.Xu X, Chung JH, Jheon S, et al. The accuracy of frozen section diagnosis of pulmonary nodules: evaluation of inflation method during intraoperative pathology consultation with cryosection. J Thorac Oncol 2010;5:39-44. 10.1097/JTO.0b013e3181c09f9c [DOI] [PubMed] [Google Scholar]
- 162.Tan Q, Zuo J, Qiu S, et al. Identification of circulating long non-coding RNA GAS5 as a potential biomarker for non-small cell lung cancer diagnosisnon-small cell lung cancer, long non-coding RNA, plasma, GAS5, biomarker. Int J Oncol 2017;50:1729-38. 10.3892/ijo.2017.3925 [DOI] [PubMed] [Google Scholar]
- 163.Guo F, Yu F, Wang J, et al. Expression of MALAT1 in the peripheral whole blood of patients with lung cancer. Biomed Rep 2015;3:309-12. 10.3892/br.2015.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Detterbeck F, Youssef S, Ruffini E, et al. A review of prognostic factors in thymic malignancies. J Thorac Oncol 2011;6:S1698-704. 10.1097/JTO.0b013e31821e7b12 [DOI] [PubMed] [Google Scholar]
- 165.Schmidt LH, Spieker T, Koschmieder S, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 2011;6:1984-92. 10.1097/JTO.0b013e3182307eac [DOI] [PubMed] [Google Scholar]
- 166.Hou Z, Zhao W, Zhou J, et al. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol 2014;53:380-8. 10.1016/j.biocel.2014.06.004 [DOI] [PubMed] [Google Scholar]
- 167.Deng J, Liang Y, Liu C, et al. The up-regulation of long non-coding RNA AFAP1-AS1 is associated with the poor prognosis of NSCLC patients. Biomed Pharmacother 2015;75:8-11. 10.1016/j.biopha.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 168.Cui D, Yu CH, Liu M, et al. Long non-coding RNA PVT1 as a novel biomarker for diagnosis and prognosis of non-small cell lung cancer. Tumour Biol 2016;37:4127-34. 10.1007/s13277-015-4261-x [DOI] [PubMed] [Google Scholar]
- 169.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16:203-22. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 170.Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet 2016;32:309-16. 10.1016/j.tig.2016.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014;28:2233-47. 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kramer MC, Liang D, Tatomer DC, et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 2015;29:2168-82. 10.1101/gad.270421.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010;6:e1001233. 10.1371/journal.pgen.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]