Abstract
Background
Acute myeloid leukemia (AML) is one of the most common hematopoietic malignancies. The cure rate of currently intensive chemotherapy in AML was only 40% or less, and there is an urgent need to develop novel effective therapeutic targets or drugs. The TATA-box binding protein associated factor 1 (TAF1) plays important roles in transcriptional regulation and leukemogenesis. However, the potential of TAF1 as a therapeutic target for AML remains unclear. The present study examined the effects of the TAF1 inhibitor Bay-299 on AML cells and the underlying molecular mechanisms.
Methods
The expression of TAF1 in various types of tumors was analyzed using The Cancer Genome Atlas (TCGA) and the UALCAN database. The effects of Bay-299 on cell proliferation were evaluated using the Cell Counting Kit-8 (CCK-8) assay. Cell death, EdU incorporation, and cell differentiation were detected using flow cytometry. Western blot analysis was utilized to confirm the activation of the apoptotic pathway. Expression of cell cycle and cell death-related genes was analyzed by quantitative real-time polymerase chain reaction (qRT-PCR).
Results
Analysis of the public databases showed that TAF1 expression was elevated in multiple types of tumors. Treatment of AML cells with the TAF1 inhibitor Bay-299 resulted in a remarkable inhibition of cell growth, increased cell death, reduced Edu incorporation, and increased cell differentiation. The apoptosis inhibitor Z-VAD and the receptor-interacting protein kinase 1 (RIPK1) inhibitor Nec-2 could rescue cell death induced by Bay-299. Bay-299 treatment increased the cleavage of key pro-apoptotic proteins, and this effect was ameliorated by administration of Z-VAD and Nec-2. Moreover, Bay-299 treatment was associated with increased expression of cell cycle inhibitor genes and multiple pyroptosis-promoting genes, contributing to the phenotypes observed in AML cell lines.
Conclusions
The TAF1 inhibitor Bay-299 induced AML cell death through multiple mechanisms and may be a promising candidate for the treatment of patients with AML.
Keywords: Acute myeloid leukemia (AML), Bay-299, cell death, epigenetics, TATA-box binding protein associated factor 1 (TAF1)
Introduction
Acute myeloid leukemia (AML) is one of the most common hematopoietic malignancies characterized by clonal expansion of abnormally differentiated and occasionally poorly differentiated cells of the hematopoietic system (1). Currently, intensive chemotherapy is the main form of treatment with a cure rate of approximately 40% in patients aged 60 years or younger, and 5% to 15% for those older than 60 years (1,2). With recent advancements in the understanding of the leukemogenesis process, many novel targets have been identified including epigenetic factors such as DNA methyltransferase 1 (DNMT1), Bromodomain-containing protein 4 (BRD4), DOT1 like histone lysine methyltransferase (Dot1l), Lysine-specific demethylase 1 (LSD1), Histone deacetylases (HDACs), Type I protein arginine methyltransferase (PRMT), and protein arginine methyltransferase 5 (PRMT5) (3-9). Furthermore, inhibitors targeting these factors have shown potent AML cytotoxicity both in vitro and in vivo (10-12), suggesting that epigenetic factors are important players in leukemogenesis and AML development and may be suitable targets for treating AML patients. Indeed, some of these inhibitors are currently undergoing clinical trials (10-12).
Bromodomain-containing proteins are a large family of proteins that function as epigenetic reader proteins (13). Bromodomains are evolutionarily conserved motifs on certain proteins that recognize and facilitate its binding to acetylated lysines on genomes, especially histone tails, thereby regulating transcription (13). Recently the importance of bromodomain-containing proteins in myeloid leukemogenesis and AML development has been recognized (10). Bromodomain-containing protein 4 (BRD4) is one of the most studied among these proteins, and it was identified in an RNA interference (RNAi) screen as a therapeutic target in AML (9). Subsequent studies showed that many inhibitors targeting BRD4 or BRD2/3/4 exert strong cytotoxicity in AML cells in vitro and in vivo (10,11,14). Importantly, some of these inhibitors such as I-BET762 and OTX015 are being tested in clinical trials for the treatment of AML as well as other cancers (15). In addition to BRD2/3/4, many other bromodomain-containing proteins are also known to play important roles in leukemogenesis or AML development. These include but are not limited to Bromodomain adjacent to zinc finger domain 2A/B (BAZ2A/B), Bromodomain-containing protein 9 (BRD9), SWI/SNF-related, matrix-associated, actin- dependent regulator of chromatin, subfamily a, member 2/4 (SMARCA2/4), CREB binding protein (CREBBP), and TATA-box binding protein associated factor 1 (TAF1) (16). Inhibitors targeting these proteins, such asN-(1,1-Dioxidotetrahydro-2H-thiopyran-4-yl)-5-ethyl-4-oxo-7-(3-(trifluoromethyl) phenyl)-4,5-dihydrothieno [3,2-c]pyridine-2-carboximidamide(I-BRD9), (E)-1-(2-Hydroxyphenyl)-3-((1R,4R)-5-(pyridin-2-yl)-2,5-diazabicyclo[2.2.1]heptan-2-yl)prop-2-en-1-one (PFI-3), 1-[7-(3,4-Dimethoxyphenyl)-9-[[(3S)-1-methylpiperidin-3-yl]methoxy]-2,3,4,5-tetrahydro-1,4-benzoxazepin-4-yl]propan-1-one (I-CBP112), and SGC-CBP30, have shown efficacy in inhibiting cancer cell growth (16,17). All these studies strongly suggested that targeting bromodomains may be an effective method to treat cancers, including AML.
The bromodomain-containing protein TAF1 is a subunit in the polymerase II preinitiation complex (PIC) and functions as a bridge to bring other TAF proteins to the promoter regions (18). TAF1 has a critical role in regulating genes involved in the cell cycle and cell death in cancer cells (19,20). Recently, TAF1 was reported to play a crucial role in AML1-ETO driven leukemogenesis (21). Knockdown of TAF1 impaired leukemic cell self-renewal and induced differentiation and apoptosis, therefore hampering leukemia cell growth (21). Moreover, the association of AML1-ETO fusion protein with chromatin and the capability of AML1-ETO to regulate the expression of related genes, but not the normal HSCs, were decreased by TAF1 depletion (21). These data suggested that TAF1 may be a potential therapeutic target in AML. However, to the best of our knowledge, there have not been any reports investigating the effects of targeting TAF1 in AML cells. BAY-299 is an efficient TAF1 inhibitor, which is currently the most potent compound to inhibit TAF1, and has been used to investigate the role of TAF1 (22). This current study used Bay-299 to explore the therapeutic potential of targeting TAF1 in AML cells. Indeed, Bay-299 efficiently inhibited the growth of AML cells, causing massive cell death and cell cycle inhibition, as well as cell differentiation. The expression of cell cycle inhibitor genes and genes involved in pyroptosis were upregulated. Furthermore, the pan-caspase inhibitor and the receptor-interacting protein kinase 1 (RIPK1) inhibitor partially rescued the AML cell death induced by Bay-299. Together, these data suggested that Bay-299 induced cell death through multiple mechanisms, and it is a promising candidate for AML treatment. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-2295).
Methods
Public dataset analysis
The expression profiles of TAF1 were downloaded from The Cancer Genome Atlas (TCGA) official website. The RNA-seq transcriptomic data of TAF1 was acquired from the UCSC Xena browser (https://xenabrowser.net/datapages/). Transcripts per million (TPM) format data was log2 transformed before further analysis. Finally, a total of 18 enrolled tumor types involving 4,335 patient cases and 5,577 normal cases were analyzed. Additionally, the expression of TAF1 was analyzed based on different French-American-British (FAB) classifications using the UALCAN database (http://ualcan.path.uab.edu). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
The human AML cell lines MV4-11 and NB4 were obtained from Cellcook (China) and cultured in Iscove’s Modified Dulbecco’s Medium (IMDM, Gibco, #12440053) or Roswell Park Memorial Institute (RPMI)1640 (Gibco, #11875093), respectively, supplemented with 10% fetal bovine serum (FBS, Hyclone, #SH30396) and 1% Penicillin-Streptomycin Liquid (Hyclone, #SV30010) at 37 °C in a humidified atmosphere of 5% CO2.
Cell viability assay
Cells were seeded in 96-well plates at a density of 1×104 cells per well and incubated with 2 or 4 µM Bay-299 (MCE, # HY-107424) for 24, 48, 72, or 96 hours (The concentration of Bay-299 was determined according to literature methods (21) and our pre-experiments). Cell viability was determined using the Cell Counting Kit-8 assay (CCK-8; Bioos, #BA00208). Briefly, 10 µL CCK-8 was added to each well and incubated for 2 hours at 37 °C. The absorbance at 450 nm was measured using a microplate reader (Thermo Fisher Scientific, Varioskan Flash). Before and after Bay-299 treatment, cell images were obtained using 20X objective of a microscope (Leica DMi8).
To determine the ability of inhibitors to rescue cell viability, 1×104 cells were seeded into 96-well plates and pre-treated with 20 µM Z-VAD-FMK (Z-VAD, MCE, #HY16658B), 10 µM necrostatin-2 (Nec-2, MCE, #HY-14622), 1 µM (E)-necrosulfonamide (MLKLi, MCE, #HY-100573), 5 µM chloroquine (CQ, Sigma, #C6628), 5 nM bafilomycin A1 (Baf, Solarbio, #A8510), 1 µM ferrostatin-1 (Fer-1, MCE, #HY-100579), or 100 µM α-tocopherol (α-Toco, MCE, #HY-N068) for 2 hours. Following the pre-treatment, 4 µM Bay-299 was added and cells were co-incubated with the inhibitors and Bay-299 for 72 hours before the CCK-8 assay was performed.
Flow cytometry
Apoptosis assay
The flow cytometry Annexin V/Propidium Iodide (PI) Kit (Vazyme, #A211-01) was used to detect cell death. Briefly, 2×105cells were plated in 12-well plates and treated with 4 µM Bay-299. After incubation for 48 hours, cells were collected, washed with phosphate buffered saline (PBS) and stained with Annexin V and PI in binding buffer for 20 minutes at room temperature in the dark. The samples were then analyzed using the BD FACSCANTO II system (BD Biosciences) to quantify the percentage of dead cells.
For the inhibitor rescue assay, cells were pre-treated with 20 µM Z-VAD-FMK (Z-VAD, MCE, #HY-16658B) or 10 µM necrostatin-2 (Nec-2, MCE, #HY-14622) for 2 hours prior to co-incubation with 4 µM Bay-299 for 48 hours. Cells were then assessed by flow cytometry using the Annexin V/PI kit.
Edu assay
Cells were seeded into 12-well plates at a density of 2×105 cells/well and treated with 4 µM Bay-299 for 48 hours. The Edu incorporation assay was performed according to the manufacturer’s instructions. Briefly, Edu (Thermo Fisher Scientific, Click-iT™ Plus Edu Alexa Fluor™ 647 Flow Cytometry Assay Kit, #C10635) was added to the cells at a final concentration of 10 µM. After incubation for 2 hours, cells were harvested, washed twice with PBS, and stained using the Click-iT™ Plus Edu Alexa Fluor™ 647 Flow Cytometry Assay Kit. The samples were analyzed with the BD FACSCANTO II system (BD Biosciences) to determine the percentage of Edu incorporation.
Cell differentiation
Cells were seeded into 12-well plates at a density of 2×105 cells/well and treated with 4 µM Bay-299 for 48 hours. Cells were then collected, washed with PBS, and stained with CD11b (Biolegend, #101217, 1:200 dilution), CD14 (Biolegend, #301812, 1:200 dilution), or CD38 (Biolegend, #303506, 1:400 dilution) for 20 minutes in the dark at room temperature. The samples were detected using the BD FACSCANTO II system (BD Biosciences). The FlowJo V10 software was used to analyze all acquired flow cytometry data.
Western blot analysis
A total of 1×106 cells were seeded in a 100-mm dish and treated with 4 µM Bay-299 for 48 hours. After treatment, cells were harvested and washed with PBS. The cell pellets were lysed in loading buffer (Thermo Fisher Scientific, #NP0008) containing DTT (Thermo Fisher Scientific, #R0861) and the proteins samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to PVDF membranes. Membranes were then probed using primary antibodies against caspase 9 (Cell Signaling Technology, #52873, 1:1,000 dilution), caspase 3 (Cell Signaling Technology, #14220,1:1,000 dilution), or poly adenosine diphosphate-ribose polymerase (PARP; Cell Signaling Technology, #9542,1:1,000 dilution), tubulin (Beyotime, #AF0001, 1:1,000 dilution) was used as the internal reference. The next day, after incubation with peroxidase (HRP) goat anti-Rabbit IgG secondary antibody (Jackson, #111-035-003), the blots were detected by ECL HRP Substrate (Millipore, WBULS0100) and scanned using a Gel Imager machine (Bio-Rad, CHEMIDOCTMXRS+ System) and the ImageJ software was utilized to quantify the intensity of the bands.
Real-time quantitative polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cells using TRIzol solution (Thermo Fisher Scientific, #15596018) and the RNA concentrations were measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific). The RNA was reverse transcribed using the PrimeScript RT Reagent Kit with the gDNA Eraser Kit (Takara, #RR047A). qRT-PCR was performed using SYBR Green Master Mix (Takara, #RR820A) according to the manufacturer’s protocol. The samples were analyzed using the Bio-Rad Realtime PCR System (CFX96 Touch). The primers used are list in Table 1.
Table 1. List of the primers (5' to 3') used in this study for qRT-PCR.
| Gene (human) | Forward | Reverse |
|---|---|---|
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
| CDKN1A | TGTCCGTCAGAACCCATGC | AAAGTCGA AGTTCCATCGCTC |
| CDKN1B | TAATTGGGGCTCCGGCTAACT | TGCAGGTCGCTTCCTTATTCC |
| CDKN2B | TGTCCGTCAGAACCCATGC | AAAGTCGAAGTTCCATCGCTC |
| Caspase 1 | TTTCCGCAAGGTTCGATTTTCA | GGCATCTGCGCTCTACCATC |
| Caspase 4 | CAAGAGAAGCAACGTATGGCA | AGGCAGATGGTCAAACTCTGTA |
| GSDMA | AGACAAGGGACATGAAGTGACC | CTCCAACGAAATAGAGGATGGC |
| GSDMB | ATGTAGACTCAACGGGAGAGTT | GTAGCCAGATACTGCTGGGATA |
| GSDMC | TCCATGTTGGAACGCATTAGC | CAAACTGACGTAATTTGGTGGC |
| GSDMD | GAGTGTGGCCTAGAGCTGG | GGCTCAGTCCTGATAGCAGTG |
| GSDME | ACATGCAGGTCGAGGAGAAGT | TCAATGACACCGTAGGCAATG |
qRT-PCR, quantitative real-time polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; CDKN1A, cyclin dependent kinase inhibitor 1A; CDKN1B, cyclin dependent kinase inhibitor 1B; CDKN2B, cyclin dependent kinase inhibitor 2B; GSDMA, gasdermin A; GSDMB, gasdermin B; GSDMC, gasdermin C; GSDMD, gasdermin D; GSDME, gasdermin E.
Statistical analysis and data presentation
All experiments were performed in triplicates or more and the data are displayed as mean ± standard deviation (SD). Statistical comparisons were determined by the two-tailed Student’s t-test unless stated otherwise. The GraphPad Prism 8.0 software was used to analyze data. Significant differences are shown by asterisks (*P<0.05; **P<0.01; ***P<0.001; ****P<0.0001; ns, no significance).
Results
TAF1 is highly expressed in multiple types of tumors
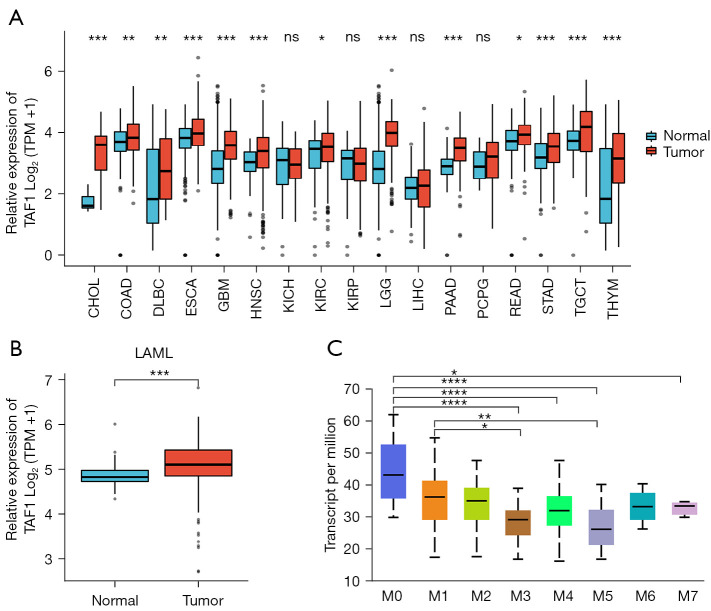
The TCGA database was used to explore the expression of TAF1 in various tumors compared to control healthy tissues. The results revealed that TAF1 RNA expression was upregulated in multiple types of tumors, including cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ), skin cutaneous melanoma (STAD), testicular germ cell tumor (TGCT), and thymoma (THYM) (Figure 1A,1B; Table 2). Furthermore, the expression profiles of TAF1 according to the different French-American-British (FAB) classifications were investigated using the UALCAN database. In AML, higher TAF1 expression was found in AML-M0 and AML-M1, which suggested that TAF1 expression levels may be negatively correlated with the degree of differentiation of AML cells (Figure 1C). In summary, TAF1 expression was elevated in multiple types of tumors compared to normal healthy tissues and may be positively associated with poorly differentiated AML.
Figure 1.
Analysis of TAF1 expression in multiple types of tumors using samples from the TCGA and the UALCAN database. (A) The expression of TAF1 in various types of tumors compared to normal tissues; (B) the expression of TAF1 is elevated in acute myeloid leukemia samples (AML) compared to healthy samples; (C) the expression of TAF1 according to the French-American-British (FAB) classifications of AML patients. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. ns, no significance; TCGA, The Cancer Genome Atlas; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; READ, rectum adenocarcinoma; STAD, skin cutaneous melanoma; TGCT, testicular germ cell tumor; THYM, thymoma.
Table 2. TAF1 expression in multiple types of tumors.
| Type of cancer | Number of normal subjects | Number of patients | Significance (patients/normal) |
|---|---|---|---|
| CHOL | 9 | 36 | *** |
| COAD | 349 | 290 | ** |
| DLBC | 444 | 47 | ** |
| ESCA | 666 | 182 | *** |
| GBM | 1157 | 166 | *** |
| HNSC | 44 | 520 | *** |
| KICH | 53 | 66 | ns |
| KIRC | 100 | 531 | * |
| KIRP | 60 | 289 | ns |
| LAML | 70 | 173 | *** |
| LGG | 1152 | 523 | *** |
| LIHC | 160 | 371 | ns |
| PAAD | 171 | 179 | *** |
| PCPG | 3 | 182 | ns |
| READ | 318 | 93 | * |
| STAD | 210 | 414 | *** |
| TGCT | 165 | 154 | *** |
| THYM | 446 | 119 | *** |
*, P<0.05; **, P<0.01; ***, P<0.001; ns, no significance. CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; READ, rectum adenocarcinoma; STAD, skin cutaneous melanoma; TGCT, testicular germ cell tumor; THYM, thymoma.
Bay-299 efficiently inhibits AML cell growth and induces cell differentiation
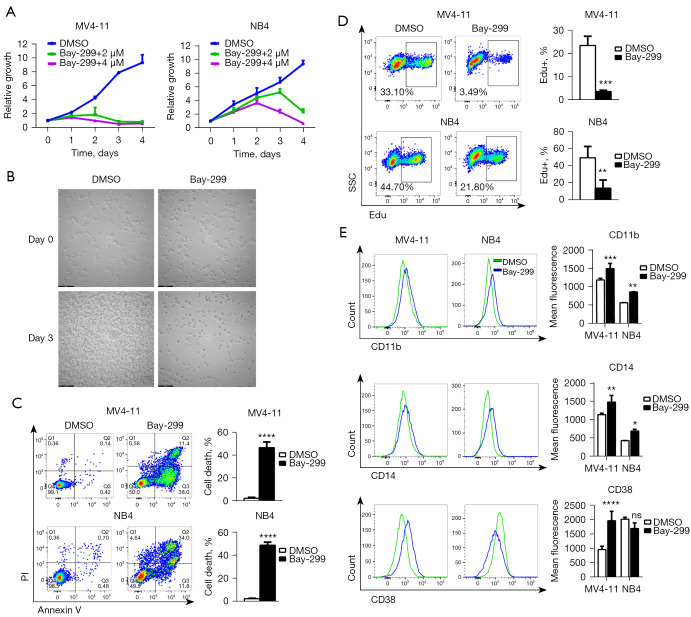
Treatment of the human AML cell lines MV4-11 and NB4 with Bay-299 resulted in a dramatic inhibition of cell growth in a dose and time-dependent manner, with MV4-11 cells being more sensitive to Bay-299 than NB4 cells (Figure 2A). The strong proliferation inhibition and disruption of cells after Bay-299 treatment were confirmed by microscopy observation (Figure 2B). At the higher concentration of 4 µM Bay-299, flow cytometry analysis showed total cell death in both cell lines after 48 hours (Figure 2C). Moreover, Edu incorporation was also significantly decreased after Bay-299 treatment (Figure 2D). These data suggested that Bay-299 inhibited cell proliferation and caused cell death in both AML cell lines. Since differentiation of AML cells is an important mechanism involved in the regulation of cell death and cell proliferation, the cell differentiation status of AML cells was examined. Indeed, treatment with Bay-299 resulted in elevated expression of the myeloid differentiation markers CD11b and CD14 in both cell lines, and increased CD38 expression in MV4-11 cells but not in NB4 cells (Figure 2E). Overall, these results suggested that Bay-299 could promote AML cell differentiation, cell death, and cell cycle inhibition.
Figure 2.
Bay-299 inhibits the growth of MV4-11 and NB4 cells. (A) CCK-8 viability assay for MV4-11 and NB4 cells treated with 2 and 4 µM Bay-299 for 96 hours; (B) representative microscopic images in NB4 cells before and after treatment with DMSO or 4 µM Bay-299 for 72 hours. Scale bar =100 µm; (C) flow cytometry analysis of cell death induced by 4 µM Bay-299 treatment for 48 hours in MV4-11 and NB4 cells; (D) flow cytometry Edu assay analysis of cell proliferation in MV4-11 and NB4 cells treated with 4 µM Bay-299 for 48 hours; (E) flow cytometry analysis of CD11b, CD14, and CD38 expression in MV4-11 and NB4 cells treated with 4 µM Bay-299 for 48 hours. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. ns, no significance; CCK-8, Cell-Counting Kit-8; DMSO, dimethyl sulfoxide.
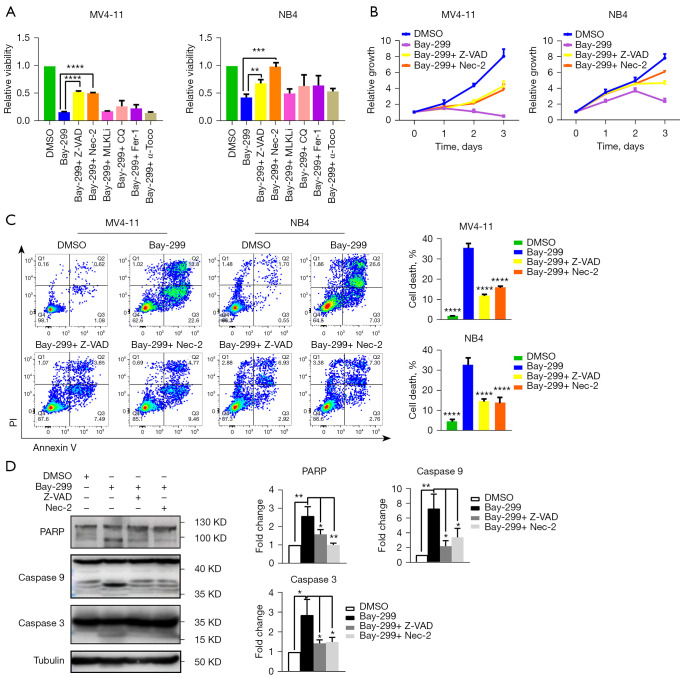
Bay-299 activates the apoptosis of AML cells via RIKP1 signaling
To investigate the mechanism of Bay-299-induced cell death, MV4-11 and NB4 cells were pretreated with the cell death pathway inhibitors Z-VAD-FMK (Z-VAD), necrostatin-2 (Nec-2), chloroquine (CQ), (E)-necrosulfonamide (MLKLi), bafilomycin A1 (Baf), ferrostatin-1 (Fer-1), or α-tocopherol prior to Bay-299 treatment. The CCK8 assays demonstrated that the apoptosis inhibitor Z-VAD and the RIPK1 inhibitor Nec-2 significantly ameliorated the reduction in cell viability induced by Bay-299 in both cell lines (Figure 3A,3B). However, none of the other cell death pathway inhibitors were effective at preventing the decrease in cell viability caused by Bay-299. Furthermore, flow cytometry analysis revealed that pre-incubation with Z-VAD and Nec-2 could partially recue cell death induced by Bay-299 (Figure 3C). These data suggested that Bay-299-induced cell death is partially mediated through the apoptosis pathway.
Figure 3.
Bay-299 induces apoptosis in MV4-11 and NB4 cells and this can be rescued by Z-VAD-FMK and necrostatin-2. (A) CCK-8 viability assay in MV4-11 and NB4 cells treated with the indicated inhibitors and Bay-299 (4 µM) for 72 hours; (B) CCK-8 viability assay in MV4-11 and NB4 cells pre-treated with Z-VAD (20 µM) or Nec-2 (10 µM) followed by incubation with Bay-299 (4 µM) for 72 hours; (C) flow cytometry analysis of cell death in MV4-11 and NB4 cells pre-treated with Z-VAD (20 µM) or Nec-2 (10 µM), followed by Bay-299 (4 µM) treatment for 48 hours; (D) Western blot analysis and quantification of PARP, caspase 9, and caspase 3 protein expression in MV4-11 cells pre-treated with Z-VAD (20 µM) or Nec-2 (10 µM), followed by incubation with Bay-299 (4 µM) for 48 hours. *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001. DMSO, dimethyl sulfoxide; PARP, poly adenosine diphosphate-ribose polymerase.
To further confirm the involvement of the apoptotic pathway, the hallmarks of apoptosis such as protein expression of cleaved PARP, caspase 9, and caspase3 was assessed. Western blot analysis revealed that the cleavage of all 3 key apoptotic promoting proteins was increased after Bay-299 treatment, and this was partially rescued by pre-treatment with Z-VAD and Nec-2 (Figure 3D). These results further support the notion that Bay-299 induces apoptosis via activating RIKP1 signaling.
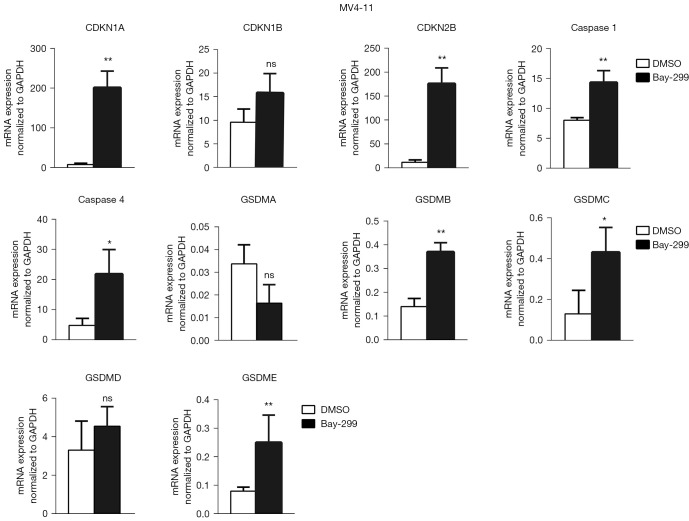
Bay-299 regulates cell cycle and cell death-related genes
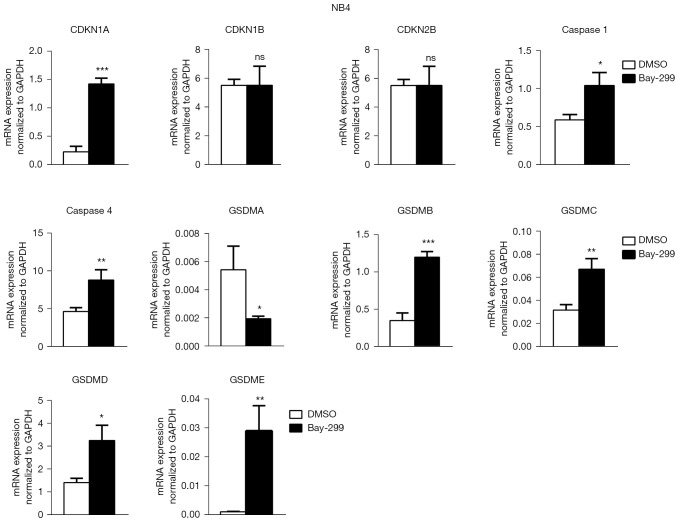
To further understand how Bay-299 regulates cell proliferation and cell death, the expression of genes involved in cell death and cell cycle progression were analyzed. Both AML cell lines showed similar changes in the expression of these genes after Bay-299 treatment. Bay-299 increased the expression of cell cycle inhibitor genes CDKN1A and CDKN2B, but not the expression of CDKN1B. The expression of genes that promote pyroptosis, including caspase 1, caspase 4, GSDMB, GSDMC, and GSDME, were all significantly elevated in both cell lines after Bay-299 treatment. However, while GSDMD expression was increased in NB4 cells after Bay-299 treatment, only a slight increase was observed in MV4-11 cells. Surprisingly, GSDMA expression was significantly decreased in NB4 cells, and a slight decreasing trend was observed in MV4-11 cells (Figures 4,5). Aberrant expression of these genes may contribute to the phenotypes observed in AML cell lines treated with Bay-299.
Figure 4.
Bay-299 regulates cell cycle and cell death-related genes in MV4-11 cells. qRT-PCR analysis of CDKN1A, CDKN1B, CDKN2B, caspase 1, caspase 4, GSDMA, GSDMB, GSDMC, GSDMD, and GSDME expression in MV4-11 cells treated with 4 µM Bay-299 for 24 hours. *, P<0.05; **, P<0.01. ns, no significance; qRT-PCR, quantitative real-time polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; CDKN, cyclin dependent kinase inhibitor; GSDM, gasdermin; DMSO, dimethyl sulfoxide.
Figure 5.
Bay-299 regulates cell cycle and cell death-related genes in NB4 cells. qRT-PCR analysis of CDKN1A, CDKN1B, CDKN2B, caspase 1, caspase 4, GSDMA, GSDMB, GSDMC, GSDMD, and GSDME expression in NB4 cells treated with 4 µM Bay-299 for 24 hours. *, P<0.05; **, P<0.01; ***, P<0.001. ns, no significance; qRT-PCR, quantitative real-time polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; CDKN, cyclin dependent kinase inhibitor; GSDM, gasdermin; DMSO, dimethyl sulfoxide.
Discussion
TAF1 is the largest subunit of the polymerase II PIC. It is also a bromodomain-containing protein that can recognize acetylated lysines (23). Despite its important function in the regulation of gene expression, there is a paucity of data relating to its role in tumor development. Overexpression of TAF1 is involved in prostate cancer progression and castration resistance (24). Knockdown of TAF1 in leukemic initiation cells strongly impaired their self-renewal ability and promoted differentiation and apoptosis (21). Silencing TAF1 has also been shown to inhibit leukemogenesis induced by AML1-ETO in the mouse model. Importantly, TAF1 knockdown showed little effect on normal hematopoietic stem cells (21). These data suggests that TAF1 may be an ideal target for treating AML.
This current study demonstrated that targeting TAF1 with the small molecular inhibitor Bay-299 could effectively inhibit the growth of AML cells. Bay-299 induced cell death through multiple mechanisms. Experiments revealed that the pan-caspase inhibitor Z-VAD could partially rescue cell death induced by Bay-299. Moreover, increased cleavage of PARP, caspase 9, and caspase 3 was observed after Bay-299 treatment. These results demonstrated that Bay-299 can induce apoptosis in AML cells. Furthermore, inhibition of RIPK1 by Nec-2 also rescued AML cell death induced by Bay-299, suggesting that Bay-299 induced apoptosis via activation of RIKP1 signaling. The upregulation of cell cycle inhibitor genes CDKN1A and CDKN2B may explain the decreased Edu incorporation after Bay-299 treatment. Thus, Bay-299 can induce both cell death and cell cycle inhibition.
Because apoptosis is only one form of cell death, and our results showed that Z-VAD could not ultimately rescue cell death induced by Bay-299. We wondered whether Bay-299 treatment might cause some other form of cell death in AML cells. Interestingly, gene expression analysis indeed revealed that Bay-299 upregulates genes involved in promoting pyroptosis. Upon induction, activated caspase 1 or caspase 4/5/11 can cleave GSDMD to induce pyroptosis (25). Alternatively, activated caspase 3 cleaves GSDME to induce pyroptosis independent from caspase 1 and GSDMD (26). In addition to these two important pathways, GSDMA, GSDMB, and GSDMC also contain a pore-formation domain that can induce pyroptosis (27). Recently, it was shown that granzyme A from cytotoxic lymphocytes could cleave GSDMB to induce pyroptosis in cancer cells (28). Another study suggested that GSDMB can promote pyroptosis by enhancing caspase 4 activity (29). However, the precise mechanisms by which GSDMA and GSDMC are activated and their roles in regulating pyroptosis remain unclear. The present analysis demonstrated that the expression of caspase 1, caspase 4, GSDMB, GSDMC, GSDMD, and GSDME was elevated after Bay-299 treatment, suggesting Bay-299 treatment may induce apoptosis and may trigger pyroptosis. Furthermore, Bay-299 treatment may sensitize AML cells to pyroptosis induction. Notably, a study using dipeptidyl peptidase (DPP)8/DPP9 inhibitors suggested that the induction of pyroptosis may be an effective method for the treatment of AML (30). Thus, it is possible that combined Bay-299 administration and pyroptosis induction may be a more potent treatment strategy against AML compared to a single treatment regimen.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (81972564) and the Military Logistics Project (AWS17J007).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-2295
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-2295
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-2295). The authors have no conflicts of interest to declare.
(English Language Editor: J. Teoh)
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute Myeloid Leukemia. N Engl J Med 2015;373:1136-52. 10.1056/NEJMra1406184 [DOI] [PubMed] [Google Scholar]
- 2.Estey EH. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am J Hematol 2018;93:1267-91. 10.1002/ajh.25214 [DOI] [PubMed] [Google Scholar]
- 3.Cheung N, Fung TK, Zeisig BB, et al. Targeting Aberrant Epigenetic Networks Mediated by PRMT1 and KDM4C in Acute Myeloid Leukemia. Cancer Cell 2016;29:32-48. 10.1016/j.ccell.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Wei J, Huang X, et al. Comprehensively analyze the expression and prognostic role for ten-eleven translocations (TETs) in acute myeloid leukemia. Transl Cancer Res 2020;9:7259-83. 10.21037/tcr-20-3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen AT, Taranova O, He J, et al. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 2011;117:6912-22. 10.1182/blood-2011-02-334359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi L, Huang Y, Huang X, et al. Analyzing the key gene expression and prognostics values for acute myeloid leukemia. Transl Cancer Res 2020;9:7284-98. 10.21037/tcr-20-3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarighat SS, Santhanam R, Frankhouser D, et al. The dual epigenetic role of PRMT5 in acute myeloid leukemia: gene activation and repression via histone arginine methylation. Leukemia 2016;30:789-99. 10.1038/leu.2015.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong KK, Lawrie CH, Green TM. Oncogenic Roles and Inhibitors of DNMT1, DNMT3A, and DNMT3B in Acute Myeloid Leukaemia. Biomark Insights 2019;14:1177271919846454. 10.1177/1177271919846454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478:524-8. 10.1038/nature10334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alqahtani A, Choucair K, Ashraf M, et al. Bromodomain and extra-terminal motif inhibitors: a review of preclinical and clinical advances in cancer therapy. Future Sci OA 2019;5:FSO372. 10.4155/fsoa-2018-0115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bewersdorf JP, Shallis R, Stahl M, et al. Epigenetic therapy combinations in acute myeloid leukemia: what are the options? Ther Adv Hematol 2019;10:2040620718816698. 10.1177/2040620718816698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wingelhofer B, Somervaille TCP. Emerging Epigenetic Therapeutic Targets in Acute Myeloid Leukemia. Front Oncol 2019;9:850. 10.3389/fonc.2019.00850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng L, Zhou MM. Bromodomain: an acetyl-lysine binding domain. FEBS Lett 2002;513:124-8. 10.1016/S0014-5793(01)03309-9 [DOI] [PubMed] [Google Scholar]
- 14.Gambacorta V, Gnani D, Vago L, et al. Epigenetic Therapies for Acute Myeloid Leukemia and Their Immune-Related Effects. Front Cell Dev Biol 2019;7:207. 10.3389/fcell.2019.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyes-Garau D, Ribeiro ML, Roué G. Pharmacological Targeting of BET Bromodomain Proteins in Acute Myeloid Leukemia and Malignant Lymphomas: From Molecular Characterization to Clinical Applications. Cancers (Basel) 2019;11:1483. 10.3390/cancers11101483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics 2017;12:323-39. 10.1080/15592294.2016.1265710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Q, Heidenreich D, Zhou S, et al. A chemical toolbox for the study of bromodomains and epigenetic signaling. Nat Commun 2019;10:1915. 10.1038/s41467-019-09672-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Louder RK, He Y, López-Blanco JR, et al. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 2016;531:604-9. 10.1038/nature17394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Brien T, Tjian R. Different functional domains of TAFII250 modulate expression of distinct subsets of mammalian genes. Proc Natl Acad Sci U S A 2000;97:2456-61. 10.1073/pnas.97.6.2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki-Yagawa Y, Guermah M, Roeder RG. The ts13 mutation in the TAF(II)250 subunit (CCG1) of TFIID directly affects transcription of D-type cyclin genes in cells arrested in G1 at the nonpermissive temperature. Mol Cell Biol 1997;17:3284-94. 10.1128/MCB.17.6.3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Man N, Karl D, et al. TAF1 plays a critical role in AML1-ETO driven leukemogenesis. Nat Commun 2019;10:4925. 10.1038/s41467-019-12735-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouché L, Christ CD, Siegel S, et al. Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains. J Med Chem 2017;60:4002-22. 10.1021/acs.jmedchem.7b00306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Gural A, Sun XJ, et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 2011;333:765-9. 10.1126/science.1201662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavassoli P, Wafa LA, Cheng H, et al. TAF1 differentially enhances androgen receptor transcriptional activity via its N-terminal kinase and ubiquitin-activating and -conjugating domains. Mol Endocrinol 2010;24:696-708. 10.1210/me.2009-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J, Gao W, Shao F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem Sci 2017;42:245-54. 10.1016/j.tibs.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Gao W, Shi X, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 2017;547:99-103. 10.1038/nature22393 [DOI] [PubMed] [Google Scholar]
- 27.Tang R, Xu J, Zhang B, et al. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J Hematol Oncol 2020;13:110. 10.1186/s13045-020-00946-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, He H, Wang K, et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science 2020;368:eaaz7548. 10.1126/science.aaz7548 [DOI] [PubMed] [Google Scholar]
- 29.Chen Q, Shi P, Wang Y, et al. GSDMB promotes non-canonical pyroptosis by enhancing caspase-4 activity. J Mol Cell Biol 2019;11:496-508. 10.1093/jmcb/mjy056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson DC, Taabazuing CY, Okondo MC, et al. DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat Med 2018;24:1151-6. 10.1038/s41591-018-0082-y [DOI] [PMC free article] [PubMed] [Google Scholar]