Abstract
Biotechnological approaches have always sought to utilize novel and efficient methods in the prevention, diagnosis, and treatment of diseases. This science has consistently tried to revolutionize medical science by employing state-of-the-art technologies in genomic and proteomic engineering. CRISPR–Cas system is one of the emerging techniques in the field of biotechnology. To date, the CRISPR–Cas system has been extensively applied in gene editing, targeting genomic sequences for diagnosis, treatment of diseases through genomic manipulation, and in creating animal models for preclinical researches. With the emergence of the COVID-19 pandemic in 2019, there is need for the development and modification of novel tools such as the CRISPR–Cas system for use in diagnostic emergencies. This system can compete with other existing biotechnological methods in accuracy, precision, and wide performance that could guarantee its future in these conditions. In this article, we review the various platforms of the CRISPR–Cas system meant for SARS-CoV-2 diagnosis, anti-viral therapeutic procedures, producing animal models for preclinical studies, and genome-wide screening studies toward drug and vaccine development.
Keywords: CRISPR–Cas systems, SARS-CoV-2, Diagnosis, Anti-viral approach, Genome-wide association study, Animal model
Introduction
The SARS-CoV-2 virus, a member of the Coronaviridae family, Betacoronavirus genus, and the causative agent of Coronavirus Disease 2019 (COVID-19), is responsible for the third large-scale human outbreak of its family in the last two decades. This zoonotic virus is a non-segmented enveloped positive-sense single-stranded RNA virus with a large genomic size that has high animal-to-human and human-to-human transmission compared to its family members [1]. The genomic sequence of this new emerging virus has a high rate of mutation and recombination, due to its unique self-replication phenomenon [2]. At the beginning of the pandemic, the complete sequence of the viral genome was shared through metagenomic approaches and the three-dimensional structure of its key proteins was determined [3]. The fourteen open reading frames (ORF) of the SARS-CoV-2 genome encode sixteen nonstructural proteins to form replicase complex (such as 3-chymotrypsin-like protease (3CLpro), papain-like protease (PLpro), helicase, and RNA-dependent RNA polymerase (RdRp), nine accessory proteins, and four structural proteins (spike (S), membrane (M), envelope (E), and nucleocapsid (N)). Spike protein has two functional segments (S1 and S2) that are activated by host cell proteases (cathepsin L and transmembrane protease serine 2 (TMPRSS2) [4]. In SARS-CoV-2 the receptor-binding domain (RBD) of spike protein binds to the human cell receptors, Angiotensin-Converting Enzyme 2 (ACE2), and defines the tropism and the pathogenicity of the virus, While the spike protein of other human coronaviruses can bind to the other cellular entry receptors such as aminopeptidase N (APN) and dipeptidyl peptidase 4 (DPP4) too. Mutation in RBD enhances the binding affinity of the virus to human ACE2 and consequently higher transmissibility compared to SARS-CoV and MERS-CoV [1, 5].
The creation of new and fast platforms at three compasses of prevention, diagnosis, and treatment of virus infections are remarkable biotechnological approaches in the COVID-19 pandemic. Therefore, knowing more about the biology and pathogenesis of the virus will help in diagnosis, treatment regimens, and vaccine design. Regions of RdRP gene sequences in the ORF1ab, envelope protein gene (E), and nucleocapsid protein gene (N) are hotspot regions used for molecular detection of the SARS-CoV-2 virus due to their conserved sequences [2]. The use of intervention strategies, such as targeting the virus cell entry pathway, including RBD, as well as the virus replication complex, including RdRp, are critical targets in the design of anti-viral molecular therapeutic platforms [6, 7].
CRISPR–Cas System
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and its associated proteins (Cas) are encoded by bacteria and archaea as defensive mechanisms against invasive genetic agents such as viruses and plasmids through a three-stage process of adaptation, maturation, and interference [8, 9]. This two-component system (CRISPR array and Cas protein) is classified into class I and class II, including different types and subtypes based on computational sequence and protein analysis of their effector subunits. The discovery of the CRISPR system has revolutionized biotechnology approaches because of its genomic sequence recognition and enzymatic cleavage ability to genetic engineering of the genome and beyond [10]. Types II, V, and VI from class II of the CRISPR/Cas system have enzymatic activity with a similar nuclease domain in their effector protein through the detection of target DNA or RNA sequences. In CRISPR/Cas complex, the CRISPR RNA (crRNA) is responsible for target sequence (RNA or DNA) identification and can depend on protospacer-associated motif (PAM) or protospacer flanking site (PFS) based on CRISPR types. In addition to crRNA and Cas complex, type II and type V-B have additional trans-activating RNA (tracrRNA) which mediates the binding between them. The single-guide RNA (sgRNA) sequence is chimerically designed with about 20 nucleotides complementary of the target sequence, instead of the crRNA-tracrRNA complex, in the laboratory to use as a biotechnological tool. In these complexes, the nuclease domains of the Cas protein have cleavage activity, resulting in blunt or overhang on-target sequences [11, 12]. Table 1 summarizes the three most applied types of class II CRISPR–Cas systems [13–16].
Table 1.
Applied types of class II CRISPR–Cas system
| Type | Subtypes | Nuclease domain | PAM/PFS | Cleavage activity | Applications |
|---|---|---|---|---|---|
| II (Cas9) |
II-A II-B II-C |
HNH and RuvC domains | 3′ G-rich motif | dsDNA blunt cleavage activity |
Diagnostic platforms Genome-wide screening Animal model designing |
| V (Cas12) | V-A (Cas12a/Cpf1) | RuvC and Nuc domains | 5′ T-rich motif | dsDNA overhang cleavage activity and ssDNA collateral cleavage activity | Diagnostic platforms |
| V-B (Cas12b/C2c1) | RuvC domain | ||||
| VI (Cas13) | VI-A (Cas13a/C2c2) | 2 HEPN domains | 3′ none G PFS | ssRNA overhang cleavage activity and ssRNA collateral cleavage activity |
Diagnostic platforms Anti-viral therapy |
| VI-D (Cas13d) | None |
PAM protospacer adjacent motif, PFS protospacer flanking site
Applications of CRISPR–Cas Systems in COVID-19 Disease
Some aspects of the CRISPR system beyond genome editing have been considered by researchers during this course of the COVID-19 pandemic. In this review, we screened research articles from PubMed and LitCovid for CRISPR–Cas system application in SARS-CoV-2 using ‘‘COVID-19 and CRISPR’’ as keywords for the period 2020 and 2021. We returned 290 articles and after deleting articles that were unrelated, review, and preprint articles, we were left with 86 articles focusing on diagnosis, anti-viral therapeutics, preclinical models, and genome-wide screening. The percent distribution of the diagnosis researches and the other categorized research articles is presented via a pie diagram in Fig. 1.
Fig. 1.
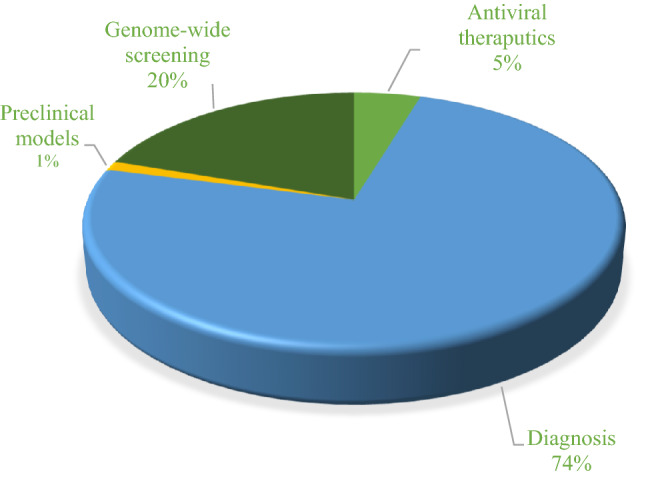
Diagram of published research articles related to CRISPR-based studies for COVID-19
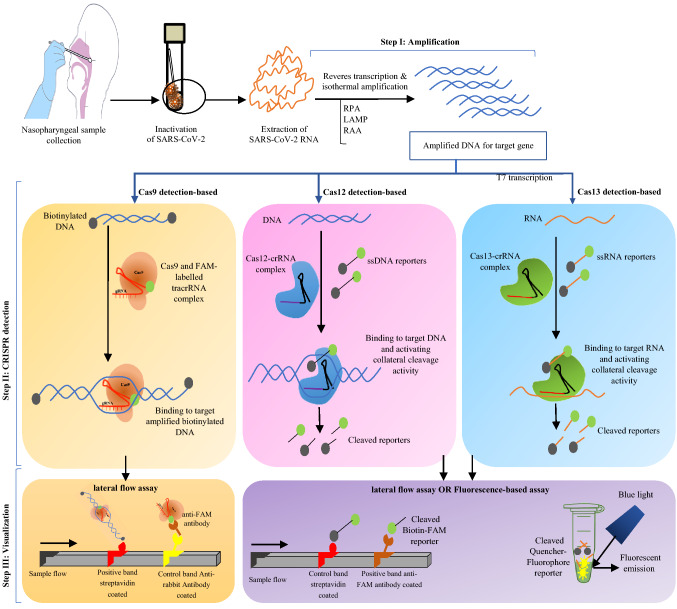
CRISPR-Based Diagnosis Assays
For most in vitro molecular diagnostic assays of infectious agents, such as viruses, the target nucleic acid must first be amplified. Thermocycling-based amplifications, namely, polymerase chain reaction (PCR) and real-time-PCR methods, are the gold standard techniques. Also, isothermal-based amplification methods, namely, loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA), are other available rapid nucleic acid amplification tests. Parameters such as sensitivity, specificity, efficiency, accuracy, simplicity, cost, and especially in this case, the speed of reaction are significant criteria for creating a suitable molecular diagnostic platform for identification of agents [17, 18]. The goal of designing diagnostic platforms is receiving point-of-care (POC) methods, considering detection with the least tools and even outdoors away from the laboratory. Therefore, simultaneous detection of multiple agents in the shortest time is changing approach from routine methods to novel methods [19, 20].
One of the new applications of the CRISPR–Cas system, beyond genome editing, is the diagnosis of infectious and non-infectious diseases. Since 2017 CRISPR-based diagnosis (CRISPR-DX) platforms based on nucleic acid detection have been developed. The design of CRISPR-DX platforms has been able to consider most of the necessary parameters by incorporating other amplification and detection methods to develop optimal systems with high performance [21]. To date, there is an urgent need to develop new POC detection methods in the emerging condition of the COVID-19 pandemic. In 2020 the CRISPR-based detection systems for SARS-CoV-2 have been able to receive the US food and drug administration (FDA) emergency use authorization (EUA) approval [22].
Due to the structure of the CRISPR–Cas system, designing different and innovative CRISPR-based diagnostic tools often considers the diversity in classes and types of Cas nucleases. Cas subunit with their enzymatic RNase activity properties along with crRNA can identify the target DNA or RNA and make cleavage [23]. The most important classes of this enzyme, applicable in detection, are class II (types II/Cas9, V/Cas12a/b, and VI/Cas13), with attomolar sensitivity for detecting RNA or DNA viruses. Usually, using each Cas protein type for virus genome detection should combine with amplification methods, especially isothermal methods, to improve the target sample sequence. Then, through the crRNA and Cas complex, the target sequence can be identified more accurately with visual fluorescent-based readout or lateral flow assay (LFA) in final detection step [10, 24]. In cases with low viral RNA copy numbers, it is difficult to interpret by visual detection. On the other hand, there is a correlation between LFA signals and Ct values in Real-Time PCR results. So, the visual results can be semi-quantified through machine learning tools to enhance the analytical sensitivity. There have been efforts to use smartphone applications for detection and quantification of POC SARS-CoV-2 CRISPR-DX platforms by imaging [25]. In fluorescent-based readouts using diode laser [26], capturing of image by companion smart-phone application [27] and 3D printing instrument for helping smart-phones detection [28] are used to develop the POC purposes.
For detecting the SARS-CoV-2 virus, different CRISPR-based platforms are using the diverse Cas nuclease enzymes listed in Table 2. The Cas9 endonuclease protein in combination with gRNA targets the dsDNA sequence. One of the advantages of using the Cas9 type is the ability to identify single-nucleotide polymorphisms that are useful for virus genotyping as well as low-frequency gene mutations [29, 30]. This type of capability has enabled the Cas9 from Campylobacter jejuni NCTC11168 (CjeCas9) to be used to distinguish the SARS-CoV-2 and its D614G (Asp614 → Gly) variant with single-base resolution in patient samples [31]. Amplification-free electrochemical detection methods are based on binding affinity between the dCas9 (dead activity-Cas9) and the viral genome on graphene-based FET chips, which also have the potential for SARS-CoV-2 detection [32].
Table 2.
Notable CRISPR–Cas-based diagnostic platforms for SARS-CoV-2 designed in 2020 and 2021
| CRISPR–Cas type | Technique name | Attendant methods | Target genes | Limit of detection (LOD) | Duration time | References | ||
|---|---|---|---|---|---|---|---|---|
| Amplification method | Detection method | |||||||
| Type II (Cas9) | Cas9/D10A with nickase activity |
CRISPR array-mediated primer exchange reaction-based biochemical circuit cascades |
PER | Electrochemical biosensing | Nucleocapsid (N) | 5 nM for synthesized SARS-CoV-2 genome in the human cell lysate | Three steps (CRISPR reactant, PER reactant, and detection) within 110 min | [33] |
| Francisella novicidae Cas9 | CRISPR/Cas9-mediated triple-line lateral flow assay (TL-LFA) | RT-RPA | Lateral flow visual readout | Simultaneous dual genes; envelope (E) and Orf1ab | 100 RNA copies per reaction (25 μl) | Two steps at 37 °C within 40 min | [34] | |
| Campylobacter jejuni NCTC11168 (CjeCas9) | Leveraging engineered tracrRNAs and on-target DNAs for parallel RNA detection (LEOPARD) | RT-PCR or free-amplification | Polyacrylamide gel electrophoresis or Bioanalyzer | Spike (S) | One copy, or 1.7 aM in the original dilution, of RNA compared with 3 × 108 copies, or 0.6 nM in the original dilution, without preamplification | unspecified | [31] | |
| Type V (Cas12) | Cas12a (Cpf1) | DNA endonuclease-targeted CRISPR trans reporter (DETECTR) | RT–LAMP | Lateral flow visual readout | Nucleocapsid (N) and envelope (E) | 10 copies per µl of nasopharyngeal swab samples | Three steps within 30–40 min | [35] |
| CRISPR-based fluorescent diagnosis system for COVID-19 (COVID-19 CRISPR-FDS) | RT-RPA | Fluorescence-based detection in 96-well microtiter plate and adaptable to smartphone-read chip format | Nucleocapsid (N) and envelope (E) | 2 copies per µl of nasal swab samples | Three steps within 50 min | [36, 37] | ||
| In vitro Specific CRISPR-based Assay for Nucleic acid detection (iSCAN) | RT-LAMP | Lateral flow and fluorescence-based detection | Nucleocapsid (N) and envelope (E) | 10 copies per µl of Nasopharyngeal swab samples | One and two steps at 62 °C within 60 min | [38] | ||
| CRISPR–Cas12a naked eye readout (CRISPR–Cas12a-NER) | RT-RAA | Fluorescence-based detection | orf1a, orf1b nucleocapsid (N) and envelope (E) | 10 copies per µl input | Three steps within 60 min | [39] | ||
| Specific enhancer for PCR-amplified Nucleic Acid (SENA) | rRT-PCR | Fluorescence-based detection | Orf1ab (O) and nucleocapsid (N) | 1.6 copies per µl of pharyngeal and nasopharyngeal swabs with 95% confidence | Three steps within rRT-PCR time and CRISPR detection time | [40] | ||
| Manganese-enhanced Cas12a detection (MeCas12a) | RT-RAA | Fluorescence-based detection | Envelope (E) | 5 copies with Mn2+ compared to 10 copies with Mg2+ | Three steps within 45 min | [41] | ||
| ENHANCE system | RT-LAMP | Fluorescence-based assay and lateral flow assay | Nucleocapsid (N) | 3–300 copies per µl | Three steps within 40–60 min | [42] | ||
| One-pot visual SARS-CoV-2 detection system named (opvCRISPR) | RT-LAMP | Fluorescence-based assay | Spike (S) | 5 copies per µl | Three steps within 45 min | [43] | ||
| All-In-One Dual CRISPR–Cas12a (AIOD-CRISPR) | RT-RPA | Fluorescence-based assay | Nucleocapsid (N) | 5 copies per µl | One-step amplification and detection at room temperature (37 °C) within 20 min | [44] | ||
| Microfluidic Isotachophoresis (ITP)-CRISPR-based | RT-LAMP | Fluorescence-based assay | Nucleocapsid (N) and envelope (E) | 10 copies per µl of Nasopharyngeal swab samples | Within 30 min | [45] | ||
|
Digital warm-start CRISPR (DWS-CRISPR) |
RT-DAMP | Fluorescence-based assay | Nucleocapsid (N) | 5 copies/μl RNA in the chip tenfold higher sensitivity than tube-based bulk assay format | One step in QuantStudio 3D digital chip initiating at above 50 °C | [46] | ||
| Cas12b (C2c1) | CRISPR-assisted detection (CASdetec) | RT-RAA | Fluorescence-based assay | RdRp | 10 copies per µl | one step at 42 °C within 60 min | [47] | |
| SHERLOCK Testing in One Pot (STOP) | RT-LAMP | Fluorescence-based assay and lateral flow assay | Nucleocapsid (N) | 100 copies/reaction | One-step amplification and detection at 60 °C within 45 min | [48] | ||
| Type VI (Cas13) | Cas13a (C2c2) | Specific High-sensitivity Enzymatic Reporter unlocking (SHERLOCK) | RT–RPA and T7 transcription | Fluorescence-based assay and lateral flow assay | Spike (S), nucleoprotein (N) replicase polyprotein 1ab (Orf1ab) | 42 RNA copies per reaction of Nasopharyngeal swab | Three steps within 60 min | [26, 49] |
| Combinatorial arrayed reactions for multiplexed evaluation of nucleic acids (CARMEN-Cas13) | PCR or RT-RPA | Fluorescence-based assay using mixes of color-coded amplified sequences | spike (S), nucleoprotein (N) replicase polyprotein 1ab (Orf1ab) | 104 copies per µl for synthetic targets | Three steps | [50, 51] | ||
| Streamlined highlighting of infections to navigate epidemics (SHINE) | RT–RPA and T7 transcription | Fluorescence-based assay and lateral flow assay | ORF1a | 10 copies per µl with 100% specificity | One step | [27] | ||
| Cas13 assisted saliva-based & smartphone integrated testing (CASSPIT) | RT-RPA and T7 transcription | Lateral flow assay integrated with a smartphone application | S and Orf1ab | 200 copies/reaction for the S gene with a corresponding Ct value of 35.4 | Two steps dual-heat inactivation (37 °C for 10 min and 95 °C for 5 min) | [25] | ||
PER primer exchange reaction, LAMP loop-mediated isothermal amplification, rLAMP RNA transcription following LAMP, RAA recombinase-aided amplification, RPA recombinase polymerase amplification, DAMP dual-priming isothermal amplifications
The first diagnostic platform to be designed based on Cas12 was a DNA endonuclease-targeted CRISPR trans reporter (DETECTR) for the detection of HPV in human patient samples. In this platform, the crRNA-Cas12a complex was used to bind and cleave the amplified virus genome sequence. Collateral cleavage of a non-specific short ssDNA sequence, which is a fluorophore quencher (FQ) labeled as a reporter was used to visualize this binding by light emission [52]. The DETECTR platform has been redesigned for SARS-CoV-2 detection from the extracted RNA of a respiratory swab, by lateral flow assay. The collaboration of this method with reverse transcriptase-loop-mediated isothermal amplification (RT-LAMP) to convert and amplify RNA virus to DNA effectively increased the detection efficiency [35]. The advantages of this method are laboratory instrument independence and rapidness, along with acceptable accuracy and specificity [28, 53]. Other DETECTR-like platforms for COVID-19 are listed in Table 2.
The pioneer of the CRISPR-DX system is Specific High-sensitivity Enzymatic Reporter unlocking (SHERLOCK) that uses type VI/Cas13 for detection [54]. This platform was designed to detect pathogens, including viruses, in combination with reverse transcriptase-recombinase polymerase amplification (RT-RPA) and T7 transcription to increase detection accuracy using a quenched fluorescent ssRNA reporter suitable for visual or lateral flow readout [55]. This platform was successful in the diagnosis of the SARS-CoV-2 virus during its pandemic. It is the first CRISPR-based platform to be used on clinical samples with the US FDA-EUA approval and has acceptable results compared with other diagnostic methods, such as next-generation sequencing (NGS) and RT-PCR (q-PCR) [22, 49, 56]. Discrimination of the wild and mutated SARS-CoV-2, such as D614G mutation with Cas13a-gRNA and Cas12a-gRNA CRISPR-based platforms, is valuable for monitoring of epidemiological analyses and can improve the ability of POC tests [57, 58]. Other notable CRISPR–Cas13-based platforms for SARS-CoV-2 virus detection are listed in Table 2. Figure 2 shows the steps of the available three types of CRISPR-DX platforms.
Fig. 2.
An overview of CRISPR-based diagnostic assays with three types of Cas enzymes; Cas9, Cas12, and Cas13 detections with names of FELUDA, DETECTR, and SHERLOCK, respectively, after extraction of RNA of SARS-CoV-2, pass through three steps and can be visualized through the lateral flow assay or fluorescence-based assay
CRISPR-Based Therapeutics
Conventional therapies for treating viral diseases are usually based on preventing the virus infections by targeting biomolecules in the entrance pathway up to proliferation. In the emergence of viruses, the first step in anti-viral therapy is drug repurposing to use existing drugs for the benefit of treatment in the shortest possible time. The second approach could be target-based drug designing by focusing on the genomics and proteomics of the desired virus [59, 60]. Based on these two approaches, drug candidates for the treatment of SARS-CoV-2 virus infectious usually focus on disruption of the enzymatic function of the virus (especially RNA-dependent RNA polymerase), prevention of virus endocytosis, blocking viral proteins (envelope, membrane, nucleocapsid, and accessory proteins), helping innate immunity or suppressing the excessive inflammatory response, and prevention of SARS-CoV-2 replication [61]. On the other hand, viruses that have new mutations and cause pandemics usually cannot respond to old drugs and vaccines. These limitations in the treatment and prevention of the COVID-19 could be applied to other viruses of the same family, such as SARS and MERS. Therefore, under these conditions, there is an urgent need for new therapies based on targeting the genomic sequences of the virus. The disruption of gene function in anti-viral therapy incorporates biotechnological tools, such as siRNA or CRISPR–Cas system. One of the well-known applications of the CRISPR–Cas system is the possibility of identifying the DNA or RNA sequence of the virus to cut them and destroy their function [62].
Related studies have suggested CRISPR an acceptable candidate for targeting human pathogens such as HIV, hepatitis B virus (HBV), herpesviruses, human papillomavirus (HPV), and JC virus (JCV) [63]. The Cas9 and Cas13 as endonuclease enzyme subunits in the CRISPR system can identify the DNA and RNA of viruses in infected mammalian cells. Studies have shown that Cas9 has a lower cleavage efficacy for ssRNA and also the ability to create off-targets in the DNA sequence of virus-infected host cells [15, 64].The CRISPR–Cas13 system is not dependent on the detection of PAM and the crRNA targets ssRNA coding sequences of the virus without interfering with the human transcriptome, causing sequence degradation, interrupting gene expression, and eventually blocking viral function [65]. Therefore, using CRISPR–Cas13 is superior to CRISPR–Cas9 as an anti-viral programmable inhibition system.
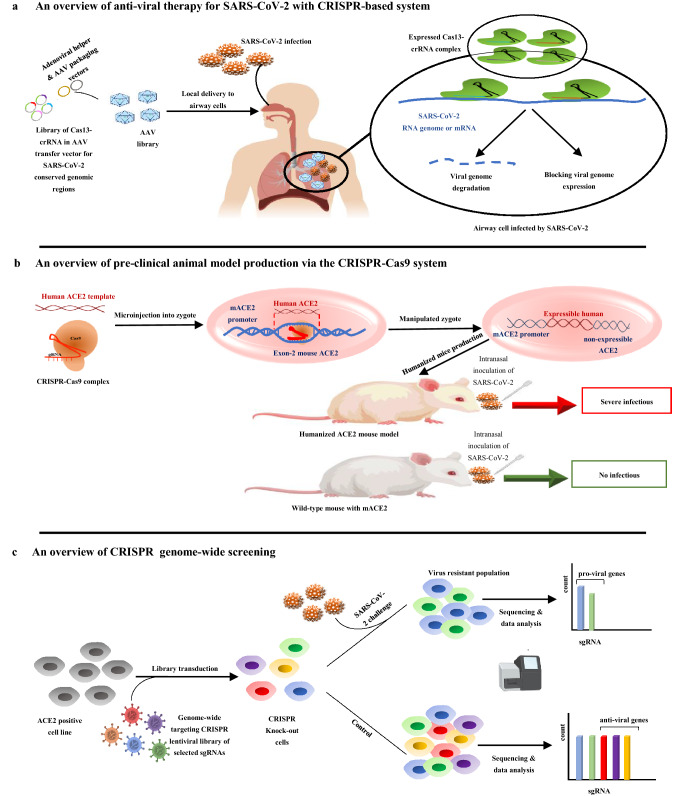
Prophylactic anti-viral CRISPR in human cells (PAC-MAN), effectively can identify and degrade the virus sequence and its mutants in the human lung epithelial cells. The PAC-MAN system uses the class 2, type VI-D, CRISPR–Cas13d system derived from Ruminococcus flavefaciens XPD3002. PAC-MAN simultaneously identifies more than 90% of all coronaviruses using a combination of 6 crRNAs by identifying protected regions (such as ORF1ab, RdRp, and N genes) as well as detecting virus ssRNA in their replication and transcription phases [66]. The comprehensive set of bioinformatics methods for receiving in silico optimal crRNA candidates can improve the efficiency of laboratory tests for combating SARS-CoV-2 in the least time [67]. Besides in silico designing and human cell line tests, using animal models will improve preclinical studies with the simulation of respiratory infections in vivo and will help to screen the safety and effectiveness of CRISPR-based platforms for anti-viral approaches [68]. The advantages of the CRISPR-based anti-viral systems are high flexibility in identifying viral sequences, speed of detection, and directness against the virus [65, 66]. The Cas13d expression vector could be transmitted via the adeno-associated virus-packaging system (AAV delivery method) with high tropism for respiratory tissues and expressed under the induction of expression promoters in specific tissues, especially airway cells [65]. The basis of the anti-viral CRISPR-based system for SARS-CoV-2 is depicted in Fig. 3a.
Fig. 3.
a Cas13-crRNA complex through variable delivery systems (such as AAV delivery method) can target virus genes (such as ORF1ab, RdRp, S, and N genes) to degrade viral genome and block genome expression of virus. b Production of humanized ACE2 mouse model through the microinjection of CRISPR–Cas9 complex and human ACE2 template sequence in mouse zygote to substitute mouse ACE2 gene and express in the lung, intestine, and brain of a mouse under its promoter. This humanized model is compared with a wild-type mouse in SARS-CoV-2 infectious conditions. c The pooled or arrayed CRISPR genome-wide screening is done for analysis of top-ranked gene clusters in the pathway of virus infection in host cells. In this direction, designing of sgRNA library for targeting candidate host cell gene to production of knocked out cells and challenging with SARS-CoV-2 virus are done for determining of anti-viral and pro-viral genes from sensitive and resistant cells
CRISPR in preclinical Researches
Preclinical researches related to COVID-19 require suitable animal model that can be infected with the SARS-CoV-2 virus and subsequently access mild-to-severe disease. As in human cells, the target cells in the selected animal should possess ACE2 as the SARS-CoV-2-binding receptor to let virus cell entrance. Animals such as hamsters, ferrets, African green monkeys, cynomolgus, and rhesus macaques have similar human ACE2 receptors. For extensive researches the best candidate is mice because of reducing costs and increasing the number of animals but mouse ACE2 has much lower binding affinity for the viral spike protein, compared to its human counterpart. This low binding affinity will develop a mild disease in mice models that is not suitable for multi-aspect studies [69, 70]. The application of transgenic mice can solve this problem by inserting the human ACE2 receptor gene into the mouse embryo genome in an engineered manner that will pass on to the next generation. In this case, humanized ACE2-incorporated mice will be more sensitive to SARS-CoV-2 infection via intranasal inoculation. Notably, the pathological outcomes and subsequent lung damages will be similar to human infection [71]. In addition to the biotechnology-based techniques used to produce transgene mice, CRISPR technology could also insert the target gene into the mouse genome by the knock-in process. For COVID-19 preclinical studies, the CRISPR–Cas9 knock-in technology can help the expression of the human ACE2 receptor in various mouse tissues. Humanized mice sensitize to SARS-CoV-2 intranasal inoculation and intragastric inoculation [72], which is depicted in Fig. 3b. The production of humanized mice has facilitated anti-viral therapeutic research, the development of vaccines, studying virus transmission and pathogenesis, and screening severe symptoms in COVID-19.
Genome-Wide Screening by CRISPR
Functional studies of genes via conditional mutagenesis in mammalian cells have been considered for decades. Genome engineering tools such as Cre-recombinase, RNAi, and designer nucleases such as ZFN and TALENs have been used for forwarding genetic screening until 2012. The discovery of the CRISPR–Cas system as a novel biotechnological method of designer nucleases generation is an alternative technique to overcome the limitations. One of the advantages of genome-wide CRISPR screening is the designing of sgRNA libraries to target thousands of genes simultaneously for pooled or arrayed screening and directly identify the candidates of desired phenotype [73, 74]. In addition to genome-wide mutagenesis through CRISPR knock-out approach, this system is a suitable tool for insertion and deletion of the target genes. Also, site-directed base editing and up/down gene regulation via gene-specific transcription factors can be done with the new modifications of CRISPR system. CRISPRing has provided an opportunity to understand the biology of mammalian cells related to diseases, such as cancers and pathogen infectious, including viruses, and designing the pharmacological studies through the creation of targeted mutant cell libraries [75, 76].
Usually the host genes that coordinate the virus entrance and pathogenesis processes are unknown. The identification of principal host genes in the viral infection, regulation, or suppression pathway can provide a better understanding of the virus function in the host cell and can help to find novel potential therapeutic marks such as antagonist drugs and design an appropriate vaccine. All of these studies in the host cell lines can be done by CRISPR–Cas9 genome-wide mutagenesis screening. Recently screening of the Noroviruses, Zika virus, West Nile virus, and HIV in host cell lines has been performed by CRISPRing [77–79].
The necessity to identify the pathogenesis of the SARS-CoV-2 virus pandemic, new screening studies are extended for revealing of viral entry and pathogenesis of host genes through genome-wide pooled or array CRISPR libraries in the appropriate cell lines. In these studies, the candidate host genes are determined by systems biology approaches and host–viral protein interactome [61]. The targeted sgRNA libraries are designed for candidate host genes to produce knock-out cell lines. These mutant cells will be challenged by virus and finally, the sequencing of the target region of the sgRNAs on mutant cell genome will determine the causative genes of resistance and sensitivity [80–82]. The process of the CRISPR genome-wide screening of SARS-CoV-2 host cell is depicted in Fig. 3c. After knocking-out steps the genes that make the virus-resistant cells are pro-viral, and the genes that make the virus-sensitive cells are anti-viral. By propagating resistant knock-out cells and sequencing data analysis, anti-viral drugs would be designable based on these gene pathways [83]. In Table 3, the CRISPR-based genome-wide screening of important cellular pathways and processes to determine top-ranked resistant and sensitive genes involved in SARS-CoV-2 infection is reported. In some cases, the candidate small molecules that mimic anti-viral gene pathways or blocking pro-viral gene pathways have been specified. The studies show that due to the lack of adequate knowledge about specific genetic pathways involved in cytotoxicity and metabolism associated with drug repurposing in SARS-CoV-2, research in this field has begun by genome-wide CRISPR.
Table 3.
Genome-wide CRISPR screens of pathways and processes involved in SARS-CoV-2 infection
| Screened pathways & process | Screened cell line | Top-ranked gene clusters in pathway | Highlights | Similar small molecule | References | |
|---|---|---|---|---|---|---|
| Resistant genes | Sensitize genes | |||||
| Cholesterol biosynthesis pathway (part of the endosomal entry pathway from the vacuolar ATPase proton pump, Retromer, and Commander complexes) | A549 |
ATP6AP1 ATP6V1A NPC1 RAB7A CCDC22 PIK3C3 |
Loss of resistant genes such as RAB7A reduces viral entry by sequestering the ACE2 receptor inside cell | RNA-sequencing of cells treated with amlodipine small molecule shows a similar differential gene expression profile as seen in CRISPR knock-out of genes in cholesterol biosynthesis pathway | [84] | |
|
Rab-GTPase requirements Glycosylphosphatidylinositol-anchored biosynthesis Cholesterol biosynthesis |
HAP1 Huh-7.5-Cas9 |
SREBP/SCAP HS2ST1 EIF4E2 RAB2A RAB10 RAB14 |
MRPS2 MRPS5 MRPS25 MRPS27 |
SCAP regulates lipid and cholesterol homeostasis by sequestering SREBPs in the ER in the presence of sterols |
Fatostatin molecule as SCAP inhibitors shows anti-viral properties 27-Hydroxycholesterol (27OHC) and 25OHC have SARS-CoV-2 anti-viral activity in VeroE6 cells |
[83] |
| Replication cycle | A549 | TMEM41B | SARS-CoV-2 require TMEM41B for replication cycles in cell | None | [85] | |
|
Chromatin remodeling Histone modification Cellular signaling RNA regulation |
Huh7.5 VeroE6 |
ARID1A DYRK1A KDM6A CTSL ACE2 SMARCA4 DYRK1A KDM6A HMGB1 HIRA CABIN1 TRIP12 BPTF PIAS2 |
TARDBP | Viral RNA-RNA and RNA–protein interactions reveal specific SARS-CoV-2-mediated mitochondrial dysfunction during infection pro-viral genes and pathways, including HMGB1, and the SWI/SNF chromatin remodeling complex are SARS-CoV-2 lineage HMGB1 is critical for SARS lineage viral entry with critical role in ACE2 expression |
PFI-3, which targets the bromodomains of the SWI/SNF proteins SMARCA4 and SMARCA2 SIS3, which targets the pro-viral gene SMAD3 identified in the screen |
[80, 81] |
|
Glycosaminoglycan biosynthesis SREBP signaling Glycosylphosphatidylinositol biosynthesis Cholesterol biosynthesis |
Huh-7.5 hepatoma cells (Huh-7.5-Cas9) |
SCAP TMEM106B TMEM41B VAC14 ACE2 HMGCS1 MVK PMVK RAB6A RAB10 |
Absolute requirement for the VTT-domain containing protein TMEM41B for infection by SARS-CoV-2 | None | [86] | |
|
Phosphatidylinositol phosphate biosynthesis Cholesterol homeostasis Heparan sulfate biosynthetic genes |
Huh7.5.1 hepatoma cells |
TMEM106B B3GALT6 B3GAT3 B4GALT7 EXT1 EXT2 EXTL3 FAM20B NDST1 SLC35B2 UGDH XYLT2 SREBP/SCAP |
Deletions in TMEM106B caused defects in lysosome trafficking, impaired acidification, and reduced levels of lysosomal enzymes but its precise molecular function remains enigmatic | None | [87, 88] | |
|
O-glycan biosynthesis N-glycan biosynthesis |
HEK293T |
C1GalT1 MGAT1 |
Knocking-out N-glycan biosynthesis on Spike-abrogated viral entry | Kifunensine small molecule inhibits N-linked glycosylation to reduce viral entry | [89] | |
| Cholesterol biosynthesis pathway (part of the endosomal entry pathway from the vacuolar ATPase proton pump, Retromer, and Commander complexes) | A549 |
ATP6AP1 ATP6V1A NPC1 RAB7A CCDC22 PIK3C3 |
Loss of resistant genes such as RAB7A reduces viral entry by sequestering the ACE2 receptor inside cell | RNA-sequencing of cells treated with amlodipine small molecule shows a similar differential gene expression profile as seen in CRISPR knock-out of genes in cholesterol biosynthesis pathway | [84] | |
|
Rab-GTPase requirements Glycosylphosphatidylinositol-anchored biosynthesis Cholesterol biosynthesis |
HAP1 Huh-7.5-Cas9 |
SREBP/SCAP HS2ST1 EIF4E2 RAB2A RAB10 RAB14 |
MRPS2 MRPS5 MRPS25 MRPS27 |
SCAP regulates lipid and cholesterol homeostasis by sequestering SREBPs in the ER in the presence of sterols |
Fatostatin molecule as SCAP inhibitors shows anti-viral properties 27-hydroxycholesterol (27OHC) and 25OHC have SARS-CoV-2 anti-viral activity in VeroE6 cells |
[83] |
| Replication cycle | A549 | TMEM41B | SARS-CoV-2 require TMEM41B for replication cycles in cell | None | [85] | |
|
Chromatin remodeling Histone modification Cellular signaling RNA regulation |
Huh7.5 VeroE6 |
ARID1A DYRK1A KDM6A CTSL ACE2 SMARCA4 DYRK1A KDM6A HMGB1 HIRA CABIN1 TRIP12 BPTF PIAS2 |
TARDBP | Viral RNA-RNA and RNA–protein interactions reveal specific SARS-CoV-2-mediated mitochondrial dysfunction during infection pro-viral genes and pathways, including HMGB1, and the SWI/SNF chromatin remodeling complex are SARS-CoV-2 lineage HMGB1 is critical for SARS lineage viral entry with critical role in ACE2 expression |
PFI-3, which targets the bromodomains of the SWI/SNF proteins SMARCA4 and SMARCA2 SIS3, which targets the pro-viral gene SMAD3 identified in the screen |
[80, 81] |
|
Glycosaminoglycan biosynthesis SREBP signaling Glycosylphosphatidylinositol biosynthesis Cholesterol biosynthesis |
Huh-7.5 hepatoma cells (Huh-7.5-Cas9) |
SCAP TMEM106B TMEM41B VAC14 ACE2 HMGCS1 MVK PMVK RAB6A RAB10 |
Absolute requirement for the VTT-domain containing protein TMEM41B for infection by SARS-CoV-2 | None | [86] | |
|
Phosphatidylinositol phosphate biosynthesis Cholesterol homeostasis Heparan sulfate biosynthetic genes |
Huh7.5.1 hepatoma cells |
TMEM106B B3GALT6 B3GAT3 B4GALT7 EXT1 EXT2 EXTL3 FAM20B NDST1 SLC35B2 UGDH XYLT2 SREBP/SCAP |
Deletions in TMEM106B caused defects in lysosome trafficking, impaired acidification, and reduced levels of lysosomal enzymes but its precise molecular function remains enigmatic | None | [87, 88] | |
|
O-glycan biosynthesis N-glycan biosynthesis |
HEK293T |
C1GalT1 MGAT1 |
Knocking-out N-glycan biosynthesis on Spike-abrogated viral entry | Kifunensine small molecule inhibits N-linked glycosylation to reduce viral entry | [89] | |
Conclusion
After less than two years of the COVID-19 pandemic, CRISPR technology has shown some potential in all applications, from diagnosis to treatment of this disease. All of these capabilities are as a result of CRISPR’s high sensitivity, flexibility, adaptability, and developable platform. In addition, based on new SARS-CoV-2 mutations and the need for newly designed vaccines, we have to find other treatment strategies like CRISPR technology for COVID-19 and other future viral infections. In addition to all advantages of using the CRISPR system in the detection of the virus variants, producing a preclinical animal model for drug and vaccine researches, its application in anti-viral therapeutics based on eliminating the virus in the infectious cells, and studies on genes involved in the disease; this system certainly has shortcomings.
One of the disadvantages of this system in diagnosis is difficult to access results for clinicians in clinical studies, so diagnostic protocols based on CRISPR-DX platforms should be toward automation as well as better reading using the mobile application with provision for offline or online mode.
In studies related to anti-viral applications of the CRISPR system, the biggest limitation is the development of effective and safe in vivo delivery methods. Therefore, more studies on the transfer of structures are needed. Also, sufficient expression of CRISPR/Cas in the target cells has a direct effect on achieving efficient viral inhibition through viral genome cleavage.
Off-target effects and limited on-target activity as well as low knock-out efficiency for designed sgRNAs are other problems facing CRISPR genome-wide screening researches, which require optimizing CRISPR systems through structural engineering and more precise design of sgRNAs for the target genome.
Author Contributions
Conceptualization, ZY and SF; supervision, GK; writing—original draft preparation, ZY; reviewing and editing of the manuscript, GK, RF, and SK; all authors read and agreed to the final version of the manuscript.
Funding
This research was not funded.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zeinab Yousefi Najafabadi, Email: z-yousefin@razi.tums.ac.ir.
Gholam Ali Kardar, Email: gakardar@tums.ac.ir.
References
- 1.Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunology. 2020 doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sood S, et al. COVID-19 pandemic: From molecular biology, pathogenesis, detection, and treatment to global societal impact. Current Pharmacology Reports. 2020 doi: 10.1007/s40495-020-00229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dömling A, Gao L. Chemistry and biology of SARS-CoV-2. Chem. 2020;6(6):1283–1295. doi: 10.1016/j.chempr.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nature Reviews Microbiology. 2020 doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang J, et al. Cell entry mechanisms of SARS-CoV-2. PNAS. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. ChemBioChem. 2020;21(5):730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elfiky AA. Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Science. 2020 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova WY, Koonim E. Classification and nomenclature of CRISPR-Cas systems: Where from here? The CRISPR Journal. 2018;1(5):325–336. doi: 10.1089/crispr.2018.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nature Reviews Microbiology. 2011;9(6):467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y, Fu Y. Class 2 CRISPR/Cas: An expanding biotechnology toolbox for and beyond genome editing. Cell & Bioscience. 2018;8:59. doi: 10.1186/s13578-018-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmakov S, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Molecular Cell. 2015;60(3):385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shmakov S, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nature reviews. Microbiology. 2017;15(3):169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burmistrz M, Krakowski K, Krawczyk-Balska A. RNA-targeting CRISPR-Cas systems and their applications. International Journal of Molecular Sciences. 2020 doi: 10.3390/ijms21031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable dual-RNA—Guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freije CA, et al. Programmable inhibition and detection of RNA viruses using Cas13. Molecular Cell. 2019;76(5):826–837.e11. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zetsche B, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esbin MN, Whitney ON, Chong S, Maurer A, Darzacq X, Tjian R. Overcoming the bottleneck to widespread testing: A rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26(7):771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen M, et al. Recent advances and perspectives of nucleic acid detection for coronavirus. Journal of Pharmaceutical Analysis. 2020 doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng W, et al. Molecular diagnosis of COVID-19: Challenges and research needs. Analytical Chemistry. 2020;92(15):10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 20.R. Jalandra et al., Strategies and perspectives to develop SARS-CoV-2 detection methods and diagnostics. Biomed Pharmacother, 2020. [DOI] [PMC free article] [PubMed]
- 21.Murugan K, Babu K, Sundaresan R, Rajan R, Sashital DG. The Revolution continues: Newly discovered systems expand the CRISPR-Cas toolkit. Molecular Cell. 2017;68(1):15–25. doi: 10.1016/j.molcel.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guglielmi G. First CRISPR test for the coronavirus approved in the United States. Nature. 2020 doi: 10.1038/d41586-020-01402-9. [DOI] [PubMed] [Google Scholar]
- 23.Sashital DG. Pathogen detection in the CRISPR-Cas era. Genome Medicine. 2018;10(1):32. doi: 10.1186/s13073-018-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolany Vangah S, Katalani C, Booneh HA, Hajizade A, Sijercic A, Ahmadian G. CRISPR-based diagnosis of infectious and noninfectious diseases. Biological Procedures Online. 2020 doi: 10.1186/s12575-020-00135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azmi I, et al. A saliva-based RNA extraction-free workflow integrated with Cas13a for SARS-CoV-2 detection. Frontiers in Cellular and Infection Microbiology. 2021 doi: 10.3389/fcimb.2021.632646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fozouni P, et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2020 doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arizti-Sanz J, et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nature Communications. 2020;11(1):5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosensors and Bioelectronics. 2020 doi: 10.1016/j.bios.2020.112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia C, et al. New applications of CRISPR/Cas9 system on mutant DNA detection. Gene. 2018;641:55–62. doi: 10.1016/j.gene.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Zhang B, Xu X, Long F, Wang J. CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method. Scientific Reports. 2018;8(1):14126. doi: 10.1038/s41598-018-32329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiao C, et al. Noncanonical crRNAs derived from host transcripts enable multiplexable RNA detection by Cas9. Science. 2021;372(6545):941–948. doi: 10.1126/science.abe7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hajian R, et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nature Biomedical Engineering. 2019;3(6):427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai Y, Xu W, Somoza RA, Welter JF, Caplan AI, Liu CC. Integrated multi-function heterogeneous biochemical circuit for high-resolution electrochemistry based genetic analysis. Angewandte Chemie International Edition. 2020 doi: 10.1002/anie.202010648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong E, et al. Simultaneous dual-gene diagnosis of SARS-CoV-2 based on CRISPR/Cas9-mediated lateral flow assay. Angewandte Chemie International Edition. 2020 doi: 10.1002/anie.202014506. [DOI] [PubMed] [Google Scholar]
- 35.Broughton JP, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nature Biotechnology. 2020 doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Z, et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosensors and. 2020 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ning B, et al. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Science Advances. 2020 doi: 10.1126/sciadv.abe3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ali Z, et al. iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Research. 2020;9:221. doi: 10.1016/j.virusres.2020.198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo L, et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang W, et al. A CRISPR-Cas12a-based specific enhancer for more sensitive detection of SARS-CoV-2 infection. EBioMedicine. 2020;61:103036. doi: 10.1016/j.ebiom.2020.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma P, et al. MeCas12a, a highly sensitive and specific system for COVID-19 detection. Advanced Science (Weinheim) 2020;7(20):2001300. doi: 10.1002/advs.202001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen LT, Smith BM, Jain PK. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nature Communications. 2020;11(1):4906. doi: 10.1038/s41467-020-18615-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang R, et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens Bioelectron. 2020;172:112766. doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding X, et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nature Communications. 2020;11(1):4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramachandran A, et al. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. PNAS. 2020 doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding X, Yin K, Li Z, Sfeir MM, Liu C. Sensitive quantitative detection of SARS-CoV-2 in clinical samples using digital warm-start CRISPR assay. Biosens Bioelectron. 2021;184:113218. doi: 10.1016/j.bios.2021.113218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teng F, et al. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biology. 2019;20(1):132. doi: 10.1186/s13059-019-1742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joung J, et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. New England Journal of Medicine. 2020;383(15):1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patchsung M, et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nature Biomedical Engineering. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 50.Ackerman CM, et al. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582(7811):277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storch GA. CRISPR tool scales up to interrogate a huge line-up of viral suspects. Nature. 2020;582(7811):188–189. doi: 10.1038/d41586-020-01447-w. [DOI] [PubMed] [Google Scholar]
- 52.Chen JS, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saha A, Arantes PR, Hsu RV, Narkhede YB, Jinek M, Palermo G. Molecular dynamics reveals a DNA-induced dynamic switch triggering activation of CRISPR-Cas12a. Journal of Chemical Information and Modeling. 2020 doi: 10.1021/acs.jcim.0c00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gootenberg JS, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science (New York, NY) 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hou T, et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathogens. 2020 doi: 10.1371/journal.ppat.1008705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, et al. Detection of SARS-CoV-2 and its mutated variants via CRISPR-Cas13-based transcription amplification. Analytical Chemistry. 2021;93(7):3393–3402. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- 58.Meng Q, et al. Detection of the SARS-CoV-2 D614G mutation using engineered Cas12a guide RNA. Biotechnology Journal. 2021 doi: 10.1002/biot.202100040. [DOI] [PubMed] [Google Scholar]
- 59.Saxena A. Drug targets for COVID-19 therapeutics: Ongoing global efforts. Journal of Biosciences. 2020;45(1):87. doi: 10.1007/s12038-020-00067-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chellapandi P, Saranya S. Genomics insights of SARS-CoV-2 (COVID-19) into target-based drug discovery. Medicinal Chemistry Research. 2020 doi: 10.1007/s00044-020-02610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Atzrodt CL, et al. A Guide to COVID-19: a global pandemic caused by the novel coronavirus SARS-CoV-2. FEBS Journal. 2020 doi: 10.1111/febs.15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soppe JA, Lebbink RJ. Antiviral goes viral: Harnessing CRISPR/Cas9 to combat viruses in humans. Trends in Microbiology. 2017;25(10):833–850. doi: 10.1016/j.tim.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 64.Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proceedings of the National Academy of Sciences USA. 2015;112(19):6164–6169. doi: 10.1073/pnas.1422340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nguyen TM, Zhang Y, Pandolfi PP. Virus against virus: A potential treatment for 2019-nCov (SARS-CoV-2) and other RNA viruses. Cell Research. 2020;30(3):189–190. doi: 10.1038/s41422-020-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.T. R. Abbott et al., Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell, 2020. [DOI] [PMC free article] [PubMed]
- 67.Wang L, Zhou J, Wang Q, Wang Y, Kang C. Rapid design and development of CRISPR-Cas13a targeting SARS-CoV-2 spike protein. Theranostics. 2021;11(2):649–664. doi: 10.7150/thno.51479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blanchard EL, et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nature Biotechnology. 2021;64:4893. doi: 10.1038/s41587-021-00822-w. [DOI] [PubMed] [Google Scholar]
- 69.Subbarao K, Roberts A. Is there an ideal animal model for SARS? Trends in Microbiology. 2006;14(7):299–303. doi: 10.1016/j.tim.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ni W, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Critical Care. 2020;24(1):422. doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lutz C, Maher L, Lee C, Kang W. COVID-19 preclinical models: Human angiotensin-converting enzyme 2 transgenic mice. Human Genomics. 2020;14(1):20. doi: 10.1186/s40246-020-00272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun SH, et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host & Microbe. 2020;28(1):124–133.e4. doi: 10.1016/j.chom.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Agrotis A, Ketteler R. A new age in functional genomics using CRISPR/Cas9 in arrayed library screening. Frontiers in Genetics. 2015;6:300. doi: 10.3389/fgene.2015.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Francis Stewart A, von Melchner H. New methods for extracting function from the mammalian genome. Methods. 2019;164–165:1–2. doi: 10.1016/j.ymeth.2019.06.026. [DOI] [PubMed] [Google Scholar]
- 75.Ludwig ML, et al. Generation and utilization of CRISPR/Cas9 screening libraries in mammalian cells. In: Appasani K, et al., editors. Genome editing and engineering: From TALENs, ZFNs and CRISPRs to molecular surgery. Cambridge University Press; 2018. pp. 223–234. [Google Scholar]
- 76.Yu JSL, Yusa K. Genome-wide CRISPR-Cas9 screening in mammalian cells. Methods. 2019;164–165:29–35. doi: 10.1016/j.ymeth.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 77.Ma H, et al. A CRISPR-based screen identifies genes essential for west-nile-virus-induced cell death. Cell Reports. 2015;12(4):673–683. doi: 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Savidis G, et al. Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Reports. 2016;16(1):232–246. doi: 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 79.Park RJ, et al. A genome-wide CRISPR screen identifies a restricted set of HIV host dependency factors. Nature Genetics. 2017;49(2):193–203. doi: 10.1038/ng.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei J, et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2020 doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flynn RA, et al. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell. 2021;184(9):2394–2411.e16. doi: 10.1016/j.cell.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu F, Tellier M. CRISPRing for host genes regulating SARS-CoV-2. Nature Reviews Immunology. 2020 doi: 10.1038/s41577-020-0400-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hoffmann HH, et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host & Microbes. 2020;5:4921. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Daniloski Z, et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2020 doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoffmann HH, et al. TMEM41B is a pan-flavivirus host factor. Cell. 2021;184(1):133–148.e20. doi: 10.1016/j.cell.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneider WM, et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184(1):120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang R, et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184(1):106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baggen J, et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nature Genetics. 2021;53(4):435–444. doi: 10.1038/s41588-021-00805-2. [DOI] [PubMed] [Google Scholar]
- 89.Yang Q, et al. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration,". Elife. 2020 doi: 10.7554/eLife.61552. [DOI] [PMC free article] [PubMed] [Google Scholar]