Abstract
Purpose:
Gastrointestinal stromal tumor (GIST) commonly arise in different regions of the stomach and are driven by various mutations (most often in KIT, PDGFRA and SDHx). We hypothesized that the anatomic location of gastric GIST is associated with unique genomic profiles and distinct driver mutations.
Experimental Design:
We compared KIT versus non-KIT status with tumor location within the National Cancer Database (NCDB) for 2,418 patients with primary gastric GIST. Additionally, we compiled an international cohort (TAGC) of 236 patients and reviewed sequencing results, cross-sectional imaging, and operative reports. Subgroup analyses were performed for tumors located proximally versus distally. Risk factors for KIT versus non-KIT tumors were identified using multivariate regression analysis. A random forest machine learning model was then developed to determine feature importance.
Results:
Within the NCDB cohort, non-KIT mutants dominated distal tumor locations (p<0.03). Proximal GIST were almost exclusively KIT mutant (96%) in the TAGC cohort, while 100% of PDGFRA and SDH-mutant GIST occurred in the distal stomach. On multivariate regression analysis, tumor location was associated with KIT versus non-KIT mutations. Using random forest machine learning analysis, stomach location was the most important feature for predicting mutation status.
Conclusions:
We provide the first evidence that the mutational landscape of gastric GIST is related to tumor location. Proximal gastric GIST are overwhelmingly KIT mutant, irrespective of morphology or age, while distal tumors display non-KIT genomic diversity. Anatomic location of gastric GIST may therefore provide immediate guidance for clinical treatment decisions and selective confirmatory genomic testing when resources are limited.
Keywords: Next generation sequencing (NGS), Genomic landscape, Mutant, KIT, PDGFRA, SDH, BRAF, NF1, KRAS
Introduction
Gastrointestinal stromal tumor (GIST) is the most common mesenchymal tumor of the GI tract, with the majority tending to arise within the stomach (1). GIST has been found to be genetically diverse with KIT, PDGFRA, and SDHx mutations being the most common primary drivers (2), and mutations in the RAS pathway (e.g., KRAS, BRAF, NF1) being less frequent drivers (3). These various mutations confer differing tumor biologies and varying sensitivities to tyrosine kinase inhibitor (TKI) therapies, with KIT-mutant GIST being responsive and non-KIT-mutant GIST being resistant to TKIs. Next Generation Sequencing (NGS) of GIST helps tailor medical management according to KIT- versus non-KIT-mutant genotype. Moreover, we have recently shown that matching genotype to treatment is a more cost-effective approach compared with empirical imatinib therapy for all patients with newly diagnosed metastatic GIST (4). Thus, NGS is becoming an essential tool for guiding medical management of GIST in many developed countries, including the United States, The Netherlands, and Germany (5-9). However, frequent barriers to routine genetic profiling remain, including inadequate tissue availability on biopsies and high testing costs. The latter is a barrier to testing in single-payer healthcare systems of developed countries (e.g., United Kingdom, Israel, and Japan) and in developing countries, despite there being coverage for TKIs through universal health care systems or through programs like the Glivec International Patient Assistance Program (GIPAP) (4,10).
Previous genomic analyses have demonstrated that the anatomic location of malignancies, including gastric adenocarcinoma, colon cancer, and biliary tract cancers correlate with underlying driving genomic profiles (7,11,12). GIST mutation profiles have also previously been associated with different anatomic segments of the GI tract (i.e., stomach versus small bowel versus rectum). The most common driver mutations in gastric GIST are KIT exon 11, PDGFRA and SDHx; the latter two mutations are almost exclusively found in tumors of gastric origin. In contrast, the small intestine has a greater incidence of KIT exon 9, BRAF and NF1 mutations, as well as gene fusions (13). These mutations are not evenly distributed along the small intestine. For example, our group first reported that the hepatoduodenal ligament is a “hot spot” for BRAF V600E and somatic NF1 mutant tumors (i.e., non-germline NF1 mutations without associated Neurofibromatosis type-1)(14). However, it remains unknown whether all gastric GIST mutant subtypes are evenly distributed within the stomach.
To date, there have been no studies that have examined whether GIST mutation profiles vary based on location within the stomach. We hypothesized that GIST arising from distinct regions of the stomach are associated with unique genomic profiles. We show that anatomical location of gastric GIST may predict the probability of KIT versus non-KIT mutations, which would help guide treatment decisions when there is a lack of access to genomic testing, lack of adequate tissue for genomic testing, or a need for immediate clinical decision-making that cannot wait for molecular testing results.
Materials and Methods
Patient Selection and Data
NCDB Cohort
A cohort study was conducted utilizing data from the American College of Surgeons National Cancer Database (NCDB). Patients with primary gastric GIST diagnosed between 2010-2016 were included (n=2,418). Exclusion criteria included unknown mutation status, multi-focal GIST, and missing data on tumor location. Data on patient age, mutation status classification for KIT and PDGFRA, tumor size, mitotic index, and tumor location were retrospectively collected. Only deidentified data was utilized for this analysis, therefore the need for Institutional Review Board and informed consent was waived. Based on these preliminary findings, further studies were conducted utilizing data from a collaborative GIST database containing more granular clinical data.
TAGC Cohort
An international, retrospective cohort study was conducted with 236 patients from the TransAtlantic GIST Collaborative (TAGC), which includes three specialized sarcoma centers [University of California San Diego (UCSD), Netherlands Cancer Institute (NKI), Knight Cancer Institute at Oregon Health & Science University (OHSU)] following institutional review board approval (UCSD HRPP #141555/190358, NKI PTC14.0057, and OHSU 00020791, respectively). Patients diagnosed with gastric GIST between the years of 2001-2019 were included in the study. Patients were required to have available NGS results, and accessible preoperative and/or postoperative cross-sectional imaging and/or operative reports. Exclusion criteria included patients with multi-focal GIST, or patients with very large GISTs replacing the majority of the stomach for which the exact location of the tumor could not be determined. An additional 15 patients were included as a run-on cohort (for a total of n=251) and included in some subsequent analyses.
Data on patient demographics (e.g., age at diagnosis, gender) and tumor characteristics (e.g., baseline tumor size, mitotic rate, cell morphology and mutation status) were retrospectively collected. Risk at diagnosis was classified according to the modified NIH risk assessment scale. Mutation status classification was performed for known genomic drivers, including KIT, PDGRFA, SDHx, KRAS, BRAF, and NF1, as well as for additional cancer-associated genes, depending on the NGS panel used at each institution [UCSD: Foundation One, Tempus; OHSU: Genetrails Solid Tumor Panel, GIST Mutational Profile; NKI: KIT/PDGFRA Sanger sequencing 2009-2014, NGS (Illumina MiSeq) 2015-2019 with additional SDHB immunohistochemistry if indicated. Mutations that were deemed nonsynonomous were not included in further analyses. Patients without mutations in any of the above genes were labeled as Unclassified (15). No patients had gene fusions.
Assessment of Tumor Location
NCDB Cohort
Tumor location was classified into the following regions: cardia or fundus, anterior/posterior body, greater curvature or lesser curvature, and antrum or pylorus. This was determined by the ICD-O-3 Primary Site Code within the NCDB.
TAGC Cohort
Tumor locations were classified into six distinct regions: cardia, fundus, lesser curvature, greater curvature, anterior/posterior body, and antrum (including pre-pyloric region) in order to standardize assessments of tumor location. The primary differentiating factor between tumors of the gastric body was origination from the anterior or posterior gastric wall, rather than from the lesser/greater curvature. Tumor location and growth pattern (endophytic or bulging into the gastric lumen versus exophytic or bulging into the peritoneal cavity) were determined by review of cross-sectional imaging (UCSD: JdlT and JKS, NKI: WJH and NSIJ, OHSU: TLS and SCM) and operative notes (when available). Reviewers of imaging were blinded to patient mutational status.
Statistical Analysis
Baseline characteristics were tabulated for all 2,418 patients within the NCDB dataset and 236 patients within the TAGC cohort. The association between KIT or non-KIT mutation status and tumor location was evaluated using Fisher exact tests then corrected for multiple comparisons using the Holm’s correction. Within the NCDB cohort, a subgroup analysis was conducted for patients with documented results for both KIT and PDGFRA. This cohort was meant to approximate patients within the TAGC cohort who had all undergone testing for multiple genomic drivers through NGS.
Within the TAGC cohort, a subgroup analysis was conducted for patients with proximal versus distal tumors. Proximal tumors were defined as those within the cardia or fundus, while distal tumors involved those within the anterior/posterior body, lesser curvature, greater curvature, or antrum. These analyses were performed using Fisher’s exact and Mann-Whitney U tests.
In order to identify independent predictors of KIT mutation status, a multivariable regression analysis was performed. First, variables were selected based on clinical significance. Then, a univariable logistic regression was performed for each variable. Variables with a p-value <0.20 (a level set a priori) from the univariable analysis were then included in the multivariable logistic regression model. All analyses were performed by SPSS Statistics (IBM Corp, Version 25, Armonk, NY). Level of statistical significance was set as p<0.05 and all statistical tests were two-sided.
Predictive Machine Learning Model for GIST
We performed random forest analysis, a machine learning algorithm commonly utilized for developing classification models, to identify the feature importance of the risk factors identified during the logistic regression analysis. Feature importance identifies the most important variables based on their contribution to the predictive accuracy of the model. The model was developed in Python (version 3.7.8) using the scikit-learn (version 0.23.1) package. The model was created with 1000 estimators and non-KIT mutants were assigned a class weight of 5, while KIT mutants were assigned a weight of 1 in order to train the model preferentially for non-KIT mutants. Age, tumor cell morphology (i.e., spindle, epithelioid, mixed), and location in the stomach were selected as features for the model to utilize. For this model, the original 236 gastric GIST patients were included, as well as 15 additional patients were included as a run-on cohort (n=251).The dataset was randomly split into a training and testing subset, with 80% of patients in the training set and 20% in the testing set. The model was bootstrapped on multiple random samples obtained with replacement 50 times. Relative importance of the features were computed.
Results
NCDB Cohort
We performed a retrospective cohort study using the National Cancer Database (NCDB) to correlate gastric GIST location and tumor mutational profile. The NCDB is a clinical oncology database sourced from hospital registry data that includes more than 70 percent of newly diagnosed cancer cases in the United States.
Patient and Tumor Characteristics
A total of 2,418 eligible patients were identified and included in the NCDB analysis. The median age at diagnosis was 64 years (interquartile range 56-74 years) (Table 1). Most patients had tumors located in the anterior/posterior body, greater curvature (GC), or lesser curvature (LC) (n=1,379, 57%) while a minority of patients had tumors located in the cardia or fundus (n=785, 32%) or antrum or pylorus (n=254, 11%).
Table 1.
Baseline characteristics of the National Cancer Database (NCDB) and TransAtlantic GIST Collaborative (TAGC) Cohort with sub-analysis.
| National Cancer Database (NCDB) Cohort | TransAtlantic GIST Collaborative (TAGC) Cohort | ||||||
|---|---|---|---|---|---|---|---|
| Characteristic | Number (%) | Characteristic | All Tumors n=236 |
Characteristic | Proximal Tumors n=71 |
Distal Tumors n=165 |
p-value |
|
Age at diagnosis Median [IQR] |
64.5 [56-74] |
Age at Diagnosis Median [IQR] |
62 [52-71] |
Age at Diagnosis Median [IQR] |
57 [49-69] | 63 [53-72] | 0.061 |
| Gender | Gender | 0.573 | |||||
| Male | 116 (49.2) | Male | 37 (52.1) | 79 (47.9) | |||
| Female | 120 (50.8) | Female | 34 (47.9) | 86 (52.1) | |||
| Tumor Size |
Tumor Size (mm) Median [IQR] |
70 [43-112] |
Tumor Size (mm) Median [IQR] |
60 [40-87] | 70 [45-120] | 0.054 | |
| < 2 cm | 222 (9.2) | Location within Stomach | |||||
| 2.1 – 5.0 cm | 828 (34.3) | Cardia | 22 (9.3) | ||||
| > 5 cm | 934 (38.7) | Fundus | 49 (20.8) | ||||
| Unknown | 429 (17.8) | Lesser Curvature | 65 (27.5) | ||||
| Location within Stomach | Greater Curvature | 57 (24.2) | |||||
| Cardia/Fundus | 785 (32.5) | Anterior/Posterior Body | 3 (1.3) | ||||
| Body/GC/LC | 1379 (57.0) | Antrum | 40 (16.9) | ||||
| Antrum/Pylorus | 254 (10.5) | Location Subgroups ‡ | |||||
| Gene Testing | Proximal | 71 (30) | |||||
| KIT only | 2162 (89.4) | Distal | 165 (70) | ||||
| PDGFRA only | 22 (0.9) | Baseline Mitotic Rate | Baseline Mitotic Rate | 0.09* | |||
| Multi-gene** | 234 (9.7) | Low (≤5/5 mm2) | 130 (55.1) | Low (≤5/5 mm2) | 33 (46.5) | 97 (58.8) | |
| Mutation Status | High (>5/5 mm2) | 58 (24.6) | High (>5/5 mm2) | 22 (31.0) | 36 (21.8) | ||
| KIT | 2270 (93.9) | Unknown | 48 (20.3) | Unknown | 16 (22.5) | 32 (19.4) | |
| PDGFRA | 81 (3.3) | Tumor Cell Morphology | Tumor Cell Morphology | <0.001* | |||
| KIT/PDGFRA Wild-type† | 67 (2.8) | Spindle | 168 (71.2) | Spindle | 61 (85.9) | 107 (64.9) | |
| Baseline Mitotic Rate | Epithelioid | 24 (10.2) | Epithelioid | 1 (1.4) | 23 (13.9) | ||
| Low (≤5/5 mm2) | 1770 (73.4) | Mixed | 40 (16.9) | Mixed | 7 (9.9) | 33 (20.0) | |
| High (>5/5 mm2) | 440 (0.182) | Unknown | 4 (1.7) | Unknown | 2 (2.8) | 2 (1.2) | |
| Unknown | 203 (8.41) | Growth Pattern | Growth Pattern | <0.001* | |||
| Endophytic | 93 (39.4) | Endophytic | 40 (56.3) | 53 (32.1) | |||
| Exophytic | 142 (60.2) | Exophytic | 31 (43.7) | 111 (67.3) | |||
| Unknown | 1 (0.4) | Unknown | - | 1 (0.6) | |||
| Mutation Status | Mutation Status | ||||||
| KIT | 170 (72.0) | KIT | 68 (95.8) | 102 (61.8) | <0.001* | ||
| PDGFRA | 46 (19.5) | PDGFRA | - | 46 (27.9) | |||
| SDHx (A/B/C/D) | 15 (5.9) | SDHx (A/B/C/D) | - | 15 (8.5) | |||
| NF1 | 1 (0.4) | Other§ | 3 (4.2) | 2 (1.8) | |||
| KRAS | 1 (0.4) | ||||||
| Unclassified§ | 3 (1.6) | ||||||
Multi-gene testing refers to those tested for both KIT and PDGFRA mutations.
No mutation was found, while patients were at least tested for KIT and PDGFRA mutations.
Proximal: Cardia and Fundus; Distal: Lesser curvature, greater curvature, anterior/posterior body, antrum
Includes NF1, KRAS, or unclassified mutations.
Mutational and Histopathologic Profiles within the Stomach
A majority of the tumors in this cohort were found to be less than 5 cm in size, and 73% of these tumors had a low mitotic rate (Table 1). Most notably, 89% (n=2,162) had only single gene KIT testing while 9.7% (n=234) had both KIT and PDGFRA mutation testing. A majority of gastric tumors had KIT mutations (n=2,270, 94%) while a minority had PDGFRA mutations (n=81, 3%), which are mutually exclusive with KIT mutations. A small subset were classified as KIT/PDGFRA wild-type (WT) GIST, lacking both KIT and PDGFRA mutations (n=67, 3%).
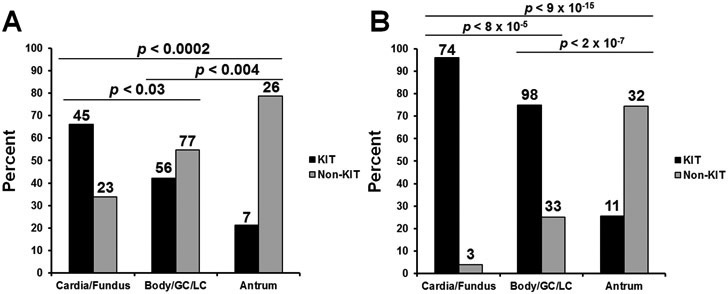
In the overall NCDB cohort (n=2,418), the percentage of non-KIT mutant tumors increased with more distal tumor locations (p < 0.005 for all comparisons) (Supplemental Figure 1). Since over 90% had only one gene tested, we performed a sub-analysis of 234 patients (10%) with both KIT and PDGFRA testing results. Forty-six percent had KIT mutations (n=108), twenty-five percent had PDGFRA mutations (n=59), and a subset were KIT/PDGFRA wild-type (n=67, 29%). In patients with both KIT and PDGFRA testing, those with GISTs of the antrum/pylorus and body/lesser curvature/greater curvature were more likely to have non-KIT mutations (n=26, 79%; n=77, 58%, respectively) than GISTs of the cardia/fundus (n=23, 34%). The percentage of non-KIT mutants increased as tumor location became more distal and this was significantly different between all locations after performing pairwise comparisons (p < 0.03 for all comparisons) (Figure 1A).
Figure 1. KIT vs non-KIT mutation rates within the stomach.
A. Bar graph representing analysis of 234 gastric GIST cases in the National Cancer Database (NCDB) database with multi-gene genetic testing. Cases are stratified by stomach location and KIT mutation (black) versus non-KIT (grey) status. B. Bar graph representing analysis of 236 gastric GIST cases in the in the Collaborative Cohort (UCSD/NKI/OHSU) with genetic testing. Cases are stratified by stomach location and KIT mutation status.
TAGC Cohort
Based on results from the NCDB cohort, we next sought to validate these findings from the TransAtlantic GIST Collaborative (TAGC), a cohort pooled from three high-volume GIST centers: University of California-San Diego (UCSD), USA, Oregon Health & Science University (OHSU), USA, and The Netherlands Cancer Institute (NKI), the Netherlands that routinely perform tumor NGS.
Patient and Tumor Characteristics
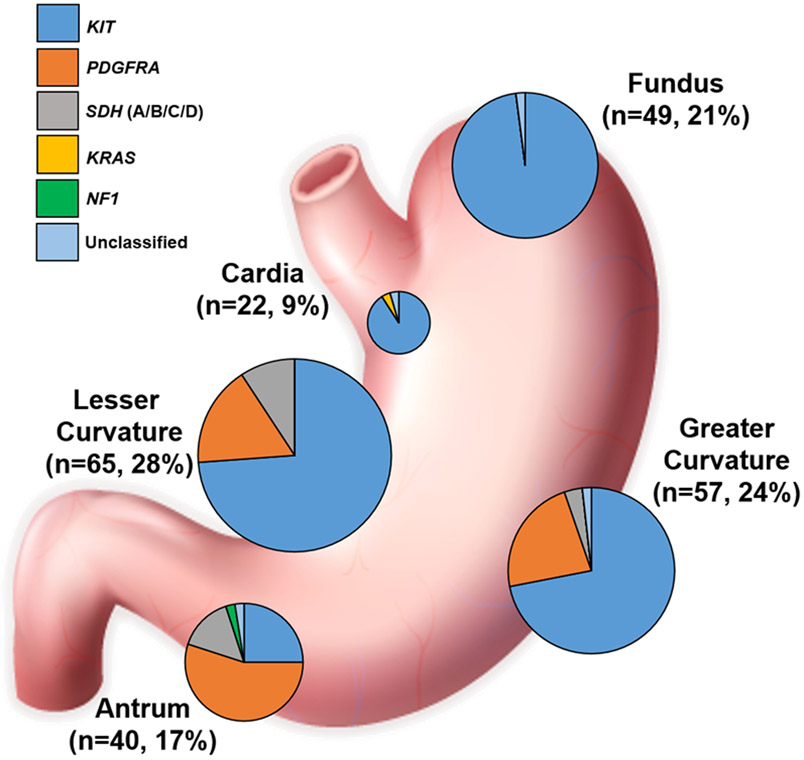
In total, 236 eligible patients were identified and included in our analysis (NKI: 54%, UCSD: 25% and OHSU: 21%). The median age at diagnosis was 62 years, and 51% of patients were female (Table 1). The median baseline tumor size was 7.0 cm. At the time of diagnosis, 3% of patients were considered very low risk and 17% were considered low risk according to the modified NIH classification (16). Another 32% of patients were classified as high risk and 16% had metastatic disease at the time of diagnosis. The majority of GIST were localized to the distal portion (i.e., non-cardia/fundus) of the stomach (70%). In descending order, gastric GIST were located in the lesser curvature (28%), greater curvature (24%), fundus (21%) and antrum (17%). In regard to tumor pathology, the majority demonstrated spindle cell morphology (72%), an exophytic growth pattern (60%), and a low baseline mitotic index (<5 mitoses per 5 mm2; 55%).
Mutational and Histopathologic Profiles within the Stomach
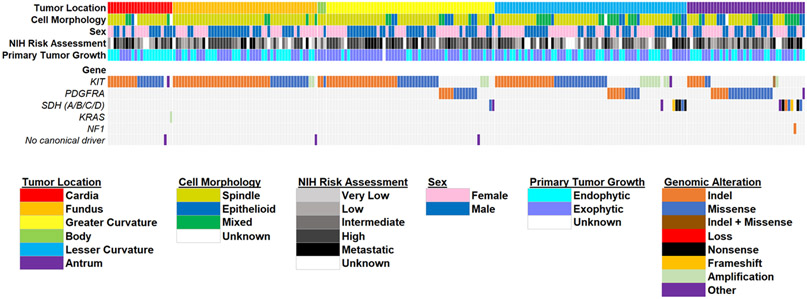
Patients with cardia/fundus tumors were more likely to have KIT mutant GIST (n=68, 96%). The percentage of non-KIT mutants were significantly different between all locations and increased as tumor location became more distal (p<0.001 for all pairwise comparisons) (Figure 1B). Figure 2 represents a co-mutation plot of all patients in the TAGC and represents a location-stratified overview of patient demographics, tumor clinicopathologic features, and driver mutations. Of the 236 GIST, most harbored KIT mutations (n=170, 72%). As expected, 98% of these patients harbored KIT exon 11 genomic alterations (Table 2). Similar to the findings in NCDB, patients were more likely to have non-KIT mutant GIST within the antrum/pylorus (n=30, 75%) and the body/lesser curvature/greater curvature (n=33, 26%) (Figure 3). Patients with cardia/fundus tumors were more likely to have KIT mutant GIST (n=68, 96%). As expected, most non-KIT mutations were in PDGFRA (n=46, 20%; with 76% occurring in exon 18 [Table 2]) or SDHx (n=15, 6%) genes.
Figure 2. Somatic mutations in gastric GIST.
Co-mutation plot from next generation sequencing of 236 gastric GISTs in the TAGC Cohort (UCSD/NKI/OHSU). Data from three international institutions were pooled for analysis. Plot including patient demographics and tumor clinicopathologic features. Tumors are sorted according to gastric location.
Table 2:
Exon data for TAGC cohort by driver mutation.
| Driver mutations | ||||
|---|---|---|---|---|
| Cohort | UCSD N (% for each gene) |
NKI N (% for each gene) |
OHSU N (% for each gene) |
Total N (% for each gene) |
| KIT | 38 (63.3)* | 99 (78.6) | 33 (66.0)** | 170 (72.0) |
| Exon 11 | 37 | 97 | 32 | 166 |
| Exon 9 | 0 | 2 | 2 | 4 |
| Exon 17 | 5 | 0 | 0 | 5 |
| Exon 13 | 0 | 0 | 2 | 2 |
| PDGFRA | 11 (18.3) | 22 (17.5) | 13 (26.0) | 46 (19.5) |
| Exon 18 | 7 | 19 | 9 | 35 |
| Exon 12 | 4 | 3 | 3 | 10 |
| Exon 14 | 0 | 0 | 1 | 1 |
| SDH (A/B/C/D) | 8 (13.3) | 4 (2.4) | 3 (6.0) | 15 (6.4) |
| KRAS | 1 (1.7) | 0 | 0 | 1 (0.4) |
| NF1 | 1 (1.7) | 0 | 0 | 1 (0.4) |
| Unclassified | 0 | 2 (1.5) | 1 (2.0) | 3 (1.3) |
Four KIT exon 11 have exon 17 mutations; 1 KIT exon 17 only; total of 38.
One KIT exon 11 also had exon 13 mutation; one KIT Exon 13 only; total of 33.
Figure 3. Distribution of Mutations in gastric GIST.
Schematic of the stomach including the percent of tumors in each location and pie graphs representing the relative incidence of mutated genes in each location. Pie graph sizes correlate with the relative number of cases in each site.
Tumor location sub-analysis revealed that proximal tumors were more likely to harbor spindle cell morphology compared to distal tumors (86% vs. 65%, p<0.001) (Supplemental Figure 2A). Distal tumors tended to occur in both patients ≤40 years of age and >40 years of age (79% vs. 69%, p=0.444) (Supplemental Figure 2B). Moreover, distal tumors were more likely to exhibit an exophytic growth pattern compared to proximal tumors (68% vs 44%, respectively, p<0.001), which more often had an endophytic growth pattern.
On univariate and subsequent multivariate logistic regression analyses (Table 3), tumors were found to be significantly more likely to be KIT mutant among patients aged >65 years [odds ratio (OR) 2.3, 95% confidence interval (CI) 1.07-5.16, p<0.05] and within proximal tumors [OR 17.7, 95% CI 3.91-80.55, p<0.001]. Furthermore, tumors were significantly less likely to be KIT mutant with an epithelioid morphology (OR 0.04, 95% CI 0.01-0.13, p<0.001) or mixed cell morphology (OR 0.11, 95% CI 0.05-0.26, p<0.001).
Table 3.
Model to identify independent predictors for oncogenic KIT versus non-KIT mutations.
| Characteristic | Univariable regression |
95% CI | p-value | Multivariable regression |
95% CI | p-value |
|---|---|---|---|---|---|---|
| Age at diagnosis | ||||||
| ≤65 years | Reference | |||||
| >65 years | 1.500 | 0.822-2.737 | 0.186* | 2.343 | 1.065-5.157 | 0.034** |
| Tumor size | ||||||
| ≤7 cm | Reference | |||||
| >7 cm | 1.029 | 0.581-1.821 | 0.923 | |||
| Gender | ||||||
| Male | Reference | |||||
| Female | 1.048 | 0.594-1.851 | 0.871 | |||
| Location | ||||||
| Proximal | 14.000 | 4.244-46.397 | <0.001* | 17.735 | 3.905-80.550 | <0.001** |
| Distal | Reference | |||||
| Growth pattern | ||||||
| Endophytic | Reference | |||||
| Exophytic | 0.756 | 0.419-1.367 | 0.355 | |||
| Mitotic rate | ||||||
| Low (≤5/5 mm2) | Reference | |||||
| High (>5/5 mm2) | 1.320 | 0.657-2.649 | 0.435 | |||
| Cell morphology | ||||||
| Spindle | Reference | <0.001* | <0.001** | |||
| Epithelioid | 0.029 | 0.009-0.092 | 0.038 | 0.011-0.134 | ||
| Mixed | 0.106 | 0.049-0.229 | 0.111 | 0.047-0.261 |
Variables with a p-value <0.2 were included in the multivariable regression analysis.
Variables with a p-value <0.05 were considered significant in multivariable analysis
Machine Learning Model for GIST Mutation Profiling
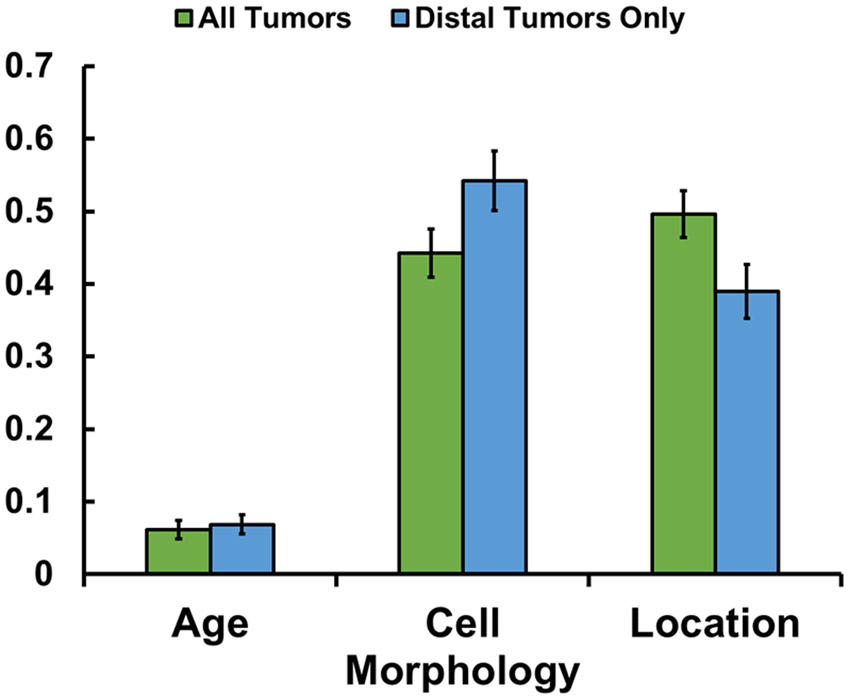
The relative importance of all factors based on our previous multivariate regression analysis was determined using an artificial intelligence (AI) random forest machine learning algorithm (Figure 4). The most important independent variable was tumor location (0.496). Age was found to have a minor level of importance (0.061). The model achieved an average accuracy of 83.3% for prediction of driver mutation using a testing cohort. In addition, since 72% of patients had KIT mutations, we also tested the model using only distal tumors (greater/lesser curvature and antrum), where only 62.5% of tumors were KIT positive. This yielded an average accuracy of 79.0% for prediction of driver mutation. Once again, location (0.389) and cell morphology (0.542) were considered important features (Figure 4).
Figure 4. Feature Importance for GIST.
Feature importance based on random forest machine learning algorithm using all tumors in all locations (green), and based on only distal tumors (blue; includes lesser curvature, greater curvature, and antrum).
Discussion
To our knowledge, this is the first study describing the correlation between gastric GIST location and the genomic profile of tumors, as well as the largest genomic analysis of resected gastric GIST. We initially evaluated a large, national cancer registry to test this hypothesis, and noted that there is an increasing proportion of non-KIT mutant tumors more distally in the stomach. However, while extensive, this cancer registry lacked granularity. The dataset missed covariates needed for robust statistical modeling, including non-KIT and non-PDGFRA mutational status, as well as tumor cell morphology. To address these limitations, we created an international cohort, the TransAtlantic GIST Consortium, of 236 patients with resected gastric GIST from three high-volume centers. We classified the tumors based on re-assessment of the exact location in the stomach, multi-gene mutational testing using NGS, and tumor histopathology. Our findings were consistent with those from the NCDB, and suggest mutation profiles of GIST are not evenly distributed. Tumors located in the proximal stomach were almost uniformly found to have a KIT mutation. Distal tumors were found to exhibit a more diverse and heterogenous mutational profile, with greater incidence of PDGFRA and SDHx subunit mutations.
Classically, tumor morphology and patient age have been used to predict non-KIT mutations. Based on the findings mentioned above, we sought to predict KIT mutational status using an artificial intelligence machine learning algorithm. Using tumor location in conjunction with patient age and tumor cell morphology under a random forest algorithm, we were able to predict tumors as either KIT or non-KIT on testing datasets with an accuracy of approximately 83%. Interestingly, the model demonstrated that tumor location was the most important variable used to classify tumors as KIT vs non-KIT mutant status compared to tumor morphology or age, with age having almost no importance in predicting mutational status. These findings therefore suggest that tumor location is a better predictor compared to tumor morphology or age. But, morphology still has important value in prediction. If used in conjunction with tumor location, they may lead to more accurate classification of GIST when genomic testing is unavailable. Overall, this model has potential to be expanded to allow for prediction of mutational profiles and aid in clinical decision making for guiding administration of targeted therapies, especially when mutation profiling may not be readily available. Thus, our findings now provide definitive evidence that there is a previously unappreciated correlation between location in the stomach and the likelihood of having a KIT mutant versus non-KIT mutant GIST, and can be easily utilized by clinicians to stratify patients into those who may require confirmatory molecular testing versus those who likely will respond well to imatinib or other adjuvant therapy.
There is mounting evidence that proper molecular matching of TKIs to GIST is associated with improved outcomes. In addition, for many non-KIT GIST, there are no current established adjuvant therapy regimens, and molecular matching of TKIs with genotyping becomes even more important to avoid overtreatment for patients who cannot benefit from imatinib or other TKIs. However, barriers to tumor genomic profiling remain a significant issue. A recent study by Florindez and Trent, in which the authors assessed KIT mutation testing in 3,866 patients using the Surveillance Epidemiology and End Results (SEER) database (2010-2015) (17). While targeted therapy in the metastatic setting was associated with improved overall survival, only 26.7% of patients underwent testing. Moreover, in many resource-limited developed countries with single-payer healthcare systems where there is a lack of national health insurance coverage for molecular testing, this may not be possible at all (18). This disparity is further exaggerated in developing countries. The unfortunate reality is that despite the benefits of molecular testing, the vast majority of patients with GIST do not undergo testing, and many will undergo therapy with imatinib or other un-matched TKIs, which may lead to unnecessary risk to the patient without any perceivable benefits.
Recently, Sicklick’s group reported development of a Markov model to compare the cost-effectiveness of genomic testing and tailored first-line therapy compared with empirical imatinib therapy among patients with a new diagnosis of metastatic GIST (4). Based on this modeling, therapy directed by genomic testing remained cost-effective for testing costs up to $3,730. While ideally every patient diagnosed with GIST would be able to undergo molecular testing, these barriers mentioned above make this difficult to achieve, and therefore, there is an important need to stratify which patients require confirmatory testing. The findings reported in the current study have potential for immediate application in clinical practice since age, location, and tumor cell morphology will be available for any core needle biopsy-proven GIST or any resected gastric GIST to help guide appropriate neoadjuvant and adjuvant therapy. Other clinical applications include cases where the diagnosis of GIST is secured through a fine needle aspiration, which may produce inadequate tissue for genomic testing and require an invasive and expensive repeat core-needle biopsy, or cases when there is a need for timely clinical decision-making that cannot wait for several weeks to obtain molecular testing results. Thus, in many cases, a cheap and easy surrogate for tumor genomic sequencing would be crical for making educated decisions about prescribing imatinib only to patients with a high likelihood of a KIT mutant GIST.
For example, a 50 year old with a mixed epithelioid-spindeloid GIST in the antrum that expresses SDHB is highly likely (i.e., 75%) to have a non-KIT mutant (i.e., PDGFRA D842V) GIST that is completely imatinib resistant. This patient could be identified and undergo tumor genomic testing on a selective basis before initiating any systemic treatment. In fact, not only are KIT mutations rare in the antrum and distal stomach, but our data also demonstrate a more diverse and heterogenous mutational landscape for these tumors, many of which are likely resistant to standard empiric imatinib therapy. On the other hand, a 50 year old with any morphology GIST in the cardia or fundus that expresses SDHB is almost guaranteed (i.e., 98%) to be KIT exon 11 mutant and be imatinib sensitive. In the case of aggressive disease biology, we would readily start this patient on imatinib therapy while waiting 3-6 weeks for their tumor sequencing. For the other former patient with the antral GIST, we would not immediately start treatment but rather wait for molecular testing, because they are more likely to have toxicity (i.e., harm) than response (i.e., benefit). In other words, our findings have important implications for not just administering the “right” drug, but also for not administering the “wrong” drug to patients. Especially outside the U.S., this straightforward and cost-effective clinical findings, which strongly correlates to genomic status, can help guide treatment decisions and the pursuit of confirmatory genomic testing for PDGFRA and SDH mutations in perhaps a more cost-conscious manner. While this clinical decision support has potential to be effective, ultimately, with larger patient numbers, machine learning algorithms, such as the one developed in this study, offer the potential to significantly aid with clinical decision support for GIST patients.
In addition, our findings raise biological questions regarding the origins of GIST. GIST is thought to originate from the interstitial cells of Cajal (ICC). These cells function as intestinal pacemaker cells, which regulate motility throughout the entire GI tract. The regional distribution of ICCs in the stomach has previously been investigated, noting that at least two subtypes are present, namely an ICC-MY subtype in the myenteric plexus region, and a second population, which is distributed outside the myenteric region and is abundant in deep muscular plexus through the muscle layers (ICC-IM)(19). These ICCs are found ubiquitously in human stomach, though the density of ICC was significantly lower in the muscularis mucosa of both fundus and body, while higher in the submucosa of gastric fundus (19). But, there is new evidence that there may be alternative cells-of-origin for GIST. Ricci et al. demonstrated that a interstitial cell-type, the telocyte, also thought to be involved in neurotransmission, may be implicated in PDGFRA mutant GIST (20). In addition, a more recent study has demonstrated that smooth muscle cells are capable of resulting in BRAF mutated GIST in mouse models, suggesting a third potential cell-of-origin for GIST (21). Given the emerging evidence of several potential GIST cells of origin, we hypothesized that it may be possible that different gastric locations have unique predispositions to driver mutations. Thus, GIST arising from distinct regions of the stomach may possess unique genomic profiles, which predict drug sensitivities. Our findings give possible credence to this hypothesis given the non-homogenous distribution of driver mutations throughout the stomach.
There are a few limitations of our current study. Both the NCDB and TAGC cohort were retrospective studies that were subject to missing data for a small subset of patients. There were some limitations in mutational testing in the NCDB cohort. First, mutational testing was only recently incorporated into the dataset in 2010, and thus only patients since 2010 have been included. Second, a vast majority of patients only underwent KIT without PDGFRA testing, likely contributing to the very low rate of PDGFRA mutations in the overall dataset. When looking at patients who underwent PDGFRA testing, the rate (~31%) more closely resembles the TAGC cohort and other previously established rates. In addition, despite having a standardized system for identifying tumor location within the NCDB cohort, there is a strong possibility of inter-observer variability. Finally, the NCDB dataset only collects data on “malignant” tumors, and thus there may be concern that this dataset could be biased by excluding GIST that have very low or low risk of recurrence and therefore would not be representative of gastric GIST. However, in our analysis, more than 40% of GIST within the NCDB were < 5 cm in size, and nearly 75% of these tumors had low mitotic indices, consistent with GIST with lower risk of recurrence, which suggests that the dataset is representative of the spectrum of disease. For our TAGC dataset, we developed a standard approach to determine location. One important limitation is that the NCDB dataset is a collection of nationwide cases throughout the country, and represents approximately 70% of all newly diagnosed cancers in the United States. Therefore, it is possible that a proportion of the patients in the TAGC cohort may be included in the NCDB dataset as well. However, it is impossible to ascertain which cases, if any, were in both the NCDB and the TAGC cohort. Regardless, the majority of institutional cases were comprised of patients from the Netherlands and would therefore not be included in the NCDB. Additionally, the cohort in this study has a fairly high proportion of patients with SDHx mutations and a relatively low number of low-risk GIST. This can be attributed to referral bias as many patients with SDHx mutations are ultimately referred to specialized, tertiary care centers with multi-disciplinary expertise in GIST/sarcoma, such as the ones included in this study. Finally, while our machine learning model works well, it is limited by the relatively small sample size of our cohort.
To our knowledge, this is the first study describing the anatomic-genomic landscape of gastric GIST, as well as characterizing the largest cohort of gastric GIST evaluated with NGS. We discovered that proximal gastric GIST were overwhelmingly KIT mutant irrespective of tumor cell morphology and patient age, while tumors arising in the distal stomach displayed much more genomic diversity that is associated with tumor cell morphology and patient age. These findings suggest that gastric GIST is not a homogenous disease and that distinct regions of the stomach carry varying risk of developing certain GIST subtypes with unique drug sensitivity/resistance profiles. Based on these findings, tumor location, tumor cell morphology, and patient age may help to: 1) guide NGS in resource-limited practice settings; 2) prompt NGS prior to initiating neoadjuvant or adjuvant imatinib therapy (especially in cases where there is a high likelihood of a non-KIT mutation); and 3) to alert physicians to cases with a higher potential of non-KIT mutant GIST (including germline SDHx mutant GIST with implications for familial genetic counseling and development of paraganglioma). In the future, it will be important to validate our findings in a larger cohort, which will allow for more sophisticated machine learning models and development of more precise prediction tools.
Supplementary Material
Translational Relevance.
The genetic profile of GIST can determine the treatment course, but only one in four GIST patients in the United States undergo tumor genomic testing. In other developed countries with universal health care systems and in developing countries, even fewer GIST patients have routine access to genomic testing despite having health care coverage or international patient assistance programs. Therefore, predictors of GIST genetic mutations may help inform early clinical decisions when genomic testing is not accesible. We now provide the first evidence that the mutational landscape of gastric GIST correlates with tumor location. Based on these results, gastric tumor location may predict the presence of KIT versus non-KIT mutations, which are associated with first line therapy (imatinib) sensitivity versus resistance, respectively. Thus, our findings present a potential rapid and cost-effective approach for guiding initial GIST treatment or selective genomic testing when time, resources, and/or tissue samples are limited.
Acknowledgements:
We appreciate funding support from the Surgical Society of the Alimentary Tract (SSAT) Mentored Research Award (S. Banerjee) and NIH T32 CA121938 Cancer Therapeutics (CT2) Training Fellowship (S.Banerjee, A.K. Sharma). NIH U24 CA248457 (J.P. Mesirov), NIH U01 CA184898 (J.P. Mesirov), Lighting the Path Forward for GIST Cancer Research (J.K. Sicklick), GIST Cancer Research Fund (M.C. Heinrich and C.L. Corless), The David Foundation (J.K. Sicklick, M.C.H), VA Merit Award Grant (M.C. Heinrich, 2I01BX000338-05), NIH R01 CA226803 (J.K. Sicklick), and FDA R01 FD006334 (J.K.Sicklick, A.M. Burgoyne).
We appreciate our insightful discussions with GIST patient advocates, Sarah Rothschild and Peter Knox of the GIST Life Raft Group, regarding inequities in GIST care globally.
This data has been presented in part at the Connective Tissue Oncology Society (CTOS) Meeting in Tokyo, Japan on November 15, 2019.
Footnotes
Conflict of Interest:
A.M. Burgoyne serves as consultant fees from Genentech, Deciphera, Exelixis, and Eisai.
P. Snaebjornsson. has participated in an Expert Input Forum for MSD B.V.
M.C. Heinrich has received honoraria from Novartis and Deciphera Pharmaceuticals; has served in advisory or consultancy roles for Novartis, Blueprint Medicines, and Deciphera Pharmaceuticals; and has provided expert testimony for Novartis. He has a patent “Activating Mutations of PDGFRA”. His institution receives royalties for a patent “Treatment of Gastrointestinal Stromal Tumors” licensed by Novartis.
N. Steeghs provided consultation or attended advisory boards for AIMM Therapeutics, Boehringer Ingelheim, Ellipses Pharma. She received research grants for the institute from AB Science, Abbvie, Actuate Therapeutics, Amgen, Array, AstraZeneca/MedImmune, Bayer, Blueprint Medicines, Boehringer Ingelheim, Bristol-Myers Squibb, Cantargia, Cytovation, Deciphera, Genentech/Roche, GlaxoSmithKline, Incyte, InteRNA, Lilly, Merck Sharp & Dohme, Merus, Novartis, Pfizer, Pierre Fabre, Roche, Sanofi, Taiho, Takeda (outside the submitted work).
J.K. Sicklick receives research funding from Novartis Pharmaceuticals, Amgen Pharmaceuticals, and Foundation Medicine, consultant fees from Grand Rounds, Loxo, and Deciphera, and speaker’s fees from Roche.
All other authors have no disclosures to claim.
References
- 1.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer 2011;11(12):865–78 doi 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 2.Boikos SA, Pappo AS, Killian JK, LaQuaglia MP, Weldon CB, George S, et al. Molecular Subtypes of KIT/PDGFRA Wild-Type Gastrointestinal Stromal Tumors: A Report From the National Institutes of Health Gastrointestinal Stromal Tumor Clinic. JAMA Oncol 2016;2(7):922–8 doi 10.1001/jamaoncol.2016.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniels M, Lurkin I, Pauli R, Erbstosser E, Hildebrandt U, Hellwig K, et al. Spectrum of KIT/PDGFRA/BRAF mutations and Phosphatidylinositol-3-Kinase pathway gene alterations in gastrointestinal stromal tumors (GIST). Cancer Lett 2011;312(1):43–54 doi 10.1016/j.canlet.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee S, Kumar A, Lopez N, Zhao B, Tang CM, Yebra M, et al. Cost-effectiveness Analysis of Genetic Testing and Tailored First-Line Therapy for Patients With Metastatic Gastrointestinal Stromal Tumors. JAMA Netw Open 2020;3(9):e2013565 doi 10.1001/jamanetworkopen.2020.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Network CGAR. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513(7517):202–9 doi 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sicklick JK, Fanta PT, Shimabukuro K, Kurzrock R. Genomics of gallbladder cancer: the case for biomarker-driven clinical trial design. Cancer Metastasis Rev 2016;35(2):263–75 doi 10.1007/s10555-016-9602-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MS, Menter DG, Kopetz S. Right Versus Left Colon Cancer Biology: Integrating the Consensus Molecular Subtypes. J Natl Compr Canc Netw 2017;15(3):411–9 doi 10.6004/jnccn.2017.0038. [DOI] [PubMed] [Google Scholar]
- 8.Sicklick JK, Kato S, Okamura R, Schwaederle M, Hahn ME, Williams CB, et al. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med 2019;25(5):744–50 doi 10.1038/s41591-019-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waseem D, Tushar P. Intrahepatic, perihilar and distal cholangiocarcinoma: Management and outcomes. Ann Hepatol 2017;16(1):133–9 doi 10.5604/16652681.1226927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tekinturhan E, Audureau E, Tavolacci MP, Garcia-Gonzalez P, Ladner J, Saba J. Improving access to care in low and middle-income countries: institutional factors related to enrollment and patient outcome in a cancer drug access program. BMC Health Serv Res 2013;13:304 doi 10.1186/1472-6963-13-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513(7517):202–9 doi 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47(9):1003–10 doi 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 13.Szucs Z, Thway K, Fisher C, Bulusu R, Constantinidou A, Benson C, et al. Molecular subtypes of gastrointestinal stromal tumors and their prognostic and therapeutic implications. Future Oncol 2017;13(1):93–107 doi 10.2217/fon-2016-0192. [DOI] [PubMed] [Google Scholar]
- 14.Burgoyne AM, De Siena M, Alkhuziem M, Tang CM, Medina B, Fanta PT, et al. Duodenal-Jejunal Flexure GI Stromal Tumor Frequently Heralds Somatic. JCO Precis Oncol 2017;2017 doi 10.1200/PO.17.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alkhuziem M, Burgoyne AM, Fanta PT, Tang CM, Sicklick JK. The Call of "The Wild"-Type GIST: It's Time for Domestication. J Natl Compr Canc Netw 2017;15(5):551–4 doi 10.6004/jnccn.2017.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joensuu H. Risk stratification of patients diagnosed with gastrointestinal stromal tumor. Hum Pathol 2008;39(10):1411–9 doi 10.1016/j.humpath.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Florindez J, Trent J. Low Frequency of Mutation Testing in the United States: An Analysis of 3866 GIST Patients. Am J Clin Oncol 2020;43(4):270–8 doi 10.1097/COC.0000000000000659. [DOI] [PubMed] [Google Scholar]
- 18.Schoffski P, Wozniak A, Schoffski O, van Eycken L, Debiec-Rychter M. Overcoming Cost Implications of Mutational Analysis in Patients with Gastrointestinal Stromal Tumors: A Pragmatic Approach. Oncol Res Treat 2016;39(12):811–6 doi 10.1159/000453057. [DOI] [PubMed] [Google Scholar]
- 19.Yun HY, Sung R, Kim YC, Choi W, Kim HS, Kim H, et al. Regional Distribution of Interstitial Cells of Cajal (ICC) in Human Stomach. Korean J Physiol Pharmacol 2010;14(5):317–24 doi 10.4196/kjpp.2010.14.5.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ricci R, Giustiniani MC, Gessi M, Lanza P, Castri F, Biondi A, et al. Telocytes are the physiological counterpart of inflammatory fibroid polyps and PDGFRA-mutant GISTs. J Cell Mol Med 2018;22(10):4856–62 doi 10.1111/jcmm.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo J, Huh WJ, Franklin JL, Heinrich MC, Rubin BP, Coffey RJ. A smooth muscle-derived, Braf-driven mouse model of gastrointestinal stromal tumor (GIST): evidence for an alternative GIST cell-of-origin. J Pathol 2020. doi 10.1002/path.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.