Abstract
Mutation of the CDC73 gene, which encodes parafibromin, has been linked with parathyroid cancer. However, no correlation between genotypes of germline CDC73 mutations and the risk of parathyroid cancer has been known. In this study, subjects with germline CDC73 mutations were identified from the participants of two clinical protocols at National Institutes of Health (Discovery Cohort) and from the literature (Validation Cohort). The relative risk of developing parathyroid cancer was analyzed as a function of CDC73 genotype, and the impact of representative mutations on structure of parafibromin was compared between genotype groups. A total of 419 subjects, 68 in Discovery Cohort and 351 in Validation Cohort, were included. In both cohorts, percentages of CDC73 germline mutations that predicted significant conformational disruption or loss of expression of parafibromin (referred as ‘high-impact mutations’) were significantly higher among the subjects with parathyroid cancers compared to all other subjects. The Kaplan–Meier analysis showed that high-impact mutations were associated with a 6.6-fold higher risk of parathyroid carcinoma compared to low-impact mutations, despite a similar risk of developing primary hyperparathyroidism between two groups. Disruption of the C-terminal domain (CTD) of parafibromin is directly involved in predisposition to parathyroid carcinoma, since only the mutations impacting this domain were associated with an increased risk of parathyroid carcinoma. Structural analysis revealed that a conserved surface structure in the CTD is universally disrupted by the mutations affecting this domain. In conclusion, high-impact germline CDC73 mutations were found to increase risk of parathyroid carcinoma by disrupting the CTD of parafibromin.
Keywords: atypical parathyroid adenoma, hyperparathyroidism-jaw tumor syndrome (HPT-JT), genotype-phenotype correlation, HRPT2, jaw tumor
Introduction
Parathyroid cancer is an uncommon disease with an estimated annual incidence of 5.73 per 10,000,000 population in the United States (Lee et al. 2007). Most parathyroid cancers are functional and secrete excessive parathyroid hormone (PTH), which causes primary hyperparathyroidism (PHPT) and typically severe complications secondary to hypercalcemia. Because it is insensitive to chemotherapy or radiotherapy, surgery is the only effective treatment of parathyroid cancer (Rodrigo et al. 2020). However, about 50% of patients had persistent or recurrent disease and PHPT is the main cause of death (Wei & Harari 2012). Identifying those at high risk of developing parathyroid cancer could potentially lead to earlier-stage diagnosis and increased chance of cure.
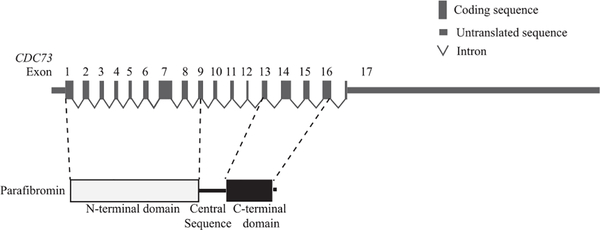
CDC73, formerly known as HRPT2, is a key gene implicated in development of parathyroid cancer. It is a putative tumor suppressor gene with 17 exons (Fig. 1), and multiple germline and somatic mutations have been identified and are presumed to cause partial or complete inactivation of this gene (Newey et al. 2010). It is estimated that at least two thirds of seemingly sporadic parathyroid carcinomas harbor somatic or germline mutations in the CDC73 gene (Shattuck et al. 2003, Yu et al. 2015, Pandya et al. 2017, Clarke et al. 2019, Cui et al. 2019), while 10–15% of subjects with germline CDC73 mutations develop parathyroid carcinoma (Jackson et al. 1993). In atypical parathyroid adenomas, germline CDC73 mutations have so far been the most commonly identified genomic abnormalities (Cetani et al. 2019). In contrast, somatic CDC73 mutation occurs at a much lower frequency in sporadic typical parathyroid adenomas (Krebs et al. 2005, Wei et al. 2018). Besides parathyroid tumors, germline CDC73 mutation could lead to fibro-osseous jaw tumors and tumors in kidney or uterus, all of which are manifestations of a familial cancer syndrome named the hyperparathyroidism-jaw tumor syndrome (HPT-JT syndrome; OMIM #145001) (Li & Simonds 2016).
Figure 1.
A schematic presentation of CDC73 gene and parafibromin protein domains.
The CDC73 gene encodes an evolutionally conserved protein, parafibromin (Carpten et al. 2002). Parafibromin functions as a component of the polymerase-associated factor-1 (PAF1) complex that regulates gene transcription through its interaction with RNA polymerase II (Yart et al. 2005). Parafibromin contains two recognized protein domains, an N-terminal domain (NTD; Pfam PF16050) and a C-terminal domain (CTD; Pfam PF05179; Fig. 1). The NTD contains one nuclear localization sequence (Hahn & Marsh 2005) and two nucleolar localization signals (Hahn & Marsh 2007, Panicker et al. 2010, Pazienza et al. 2013). The crystal structure of the N-terminal 111 amino acids of parafibromin revealed a highly-conserved hydrophobic groove with unknown function (Sun et al. 2017). In contrast, the structure of the CTD of parafibromin has not been solved. Yeast Cdc73, a homolog of the CTD of human parafibromin, is similar in structure to the Ras family of small GTPase but lacks GTPase activity (Amrich et al. 2012, Chen et al. 2012). The central sequence of parafibromin is important for binding to multiple proteins, such as β-catenin, GLI (glioma-associated oncogene transcription factor) and the PAF1 (polymerase-associated factor 1) complex (Yart et al. 2005, Mosimann et al. 2006, 2009, Iwata et al. 2007).
Unlike the multiple endocrine neoplasia type 2 syndrome in which specific germline RET proto-oncogene mutations predict a higher risk of medullary thyroid carcinoma (Eng et al. 1996, Frank-Raue et al. 1996, Machens et al. 2003), any correlation between the risk of parathyroid cancer and genotypes of germline CDC73 mutations has not been known. In this study, we explored the connection(s) between genotypes of CDC73 germline mutations and parathyroid cancer/HPT-JT syndrome and investigated the structural basis of such connection(s).
Materials and methods
Subjects
The subjects in this study were identified from the participants of two natural history protocols, NCT00001345 for inheritable metabolic disease and NCT00001277 for hyperparathyroidism, at the National Institutes of Health (NIH), as well as from the published cases and case series included in Human Genetic Mutation Database (HGMD; http://www.hgmd.cf.ac.uk/ac/index.php) from 2002 to 2016. The clinical protocols were approved by the Institutional Review Board of National Institute of Diabetes and Digestive and Kidney Diseases and consents were obtained for all subjects enrolled in the NIH protocols. Genomic CDC73 alterations were detected by targeted gene sequencing or by copy number analysis if the former analysis was negative.
Data collection
Information about subject demographics, CDC73 germline mutation, parathyroid disease status, age of onset of parathyroid disease and other clinical manifestations of HPT-JT syndrome, and biochemical test results were collected, if available. In instances when CDC73 mutation-positive subjects had no evidence of parathyroid disease, the ages at the last follow-up were recorded. Parathyroid disease status was recorded according to the surgical pathological diagnosis. Among the NIH cases, diagnosis of parathyroid tumors was made according to the WHO criteria (Bondeson et al. 2004, DeLellis et al. 2017): parathyroid carcinoma must have histological evidence of vascular, capsular, or gross adjacent tissue invasion or obvious metastases, while atypical parathyroid adenoma should have aggressive histological features (e.g., pleomorphism and atypia, mitoses, necrosis, and microscopic capsular infiltration) but must not have extensive invasion into adjacent tissues or metastasis (two diagnostic criteria for carcinoma). Immunohistochemical (IHC) staining for parafibromin was not performed for diagnosis of parathyroid disease at the NIH. The levels of serum total calcium and PTH were recorded at initial diagnosis or at recurrence(s) of PHPT if the former values were not available, and units were converted to the same unit (e.g., mmol/L for calcium and pg/mL for PTH). The germline mutations of the CDC73 gene in the general population were queried in the ExAC database (https://gnomad.broadinstitute.org/).
Structure modeling
The crystal structure of the N-terminal 1–111 amino acid fragment of human parafibromin (PDB ID: 5YDE) was visualized using PyMOL (Sun et al. 2017). The highly conserved residues in or at both ends of a previously reported hydrophobic groove (Sun et al. 2017) were highlighted based on their hydrophobicity scores (Wimley & White 1996). The missense mutations were highlighted to show their locations in or near the hydrophobic groove.
The structure of the CTD of human CDC73 was modeled based on the structure of yeast Cdc73 (PDB ID: 3V46) (Amrich et al. 2012). The human parafibromin (aa 350–531) sequence, which corresponds to the sequence of yeast Cdc73, was analyzed by I-TASSER (Yang et al. 2015). The resulted five models were aligned with the yeast Cdc73 to determine the best fit human parafibromin CTD model. Next, patient-specific mutations were introduced into the sequence, and models for each mutant were generated by I-TASSER and visualized by PyMOL.
Statistical analysis
Continuous variables were compared with the use of the Student-t test (for two-group comparison) or ANOVA and Dunnett test with a single pooled variance (multiple comparisons) under the presumption of same S.D, and normal distribution. Log transformation was applied to the PTH results to convert the data from a highly skewed distribution into a normal distribution before quantitative statistical analysis. Categorical variables were compared using a chi-square test or Fisher’s exact test, as appropriate. The Kaplan–Meier analysis and log-rank test were used to estimate time-to-event curves, hazard ratio and 95% CI. All tests were performed with a two-sided significance level of 5%.
Results
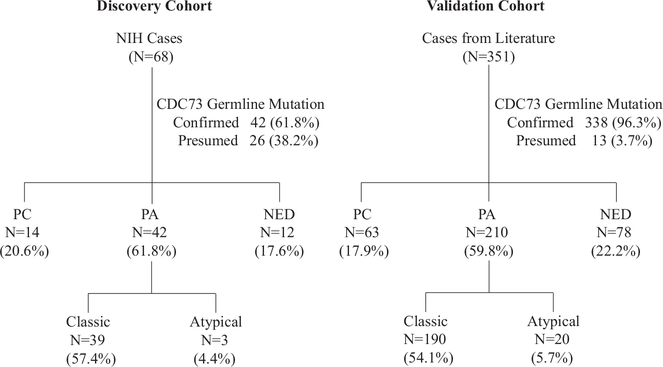
We identified 68 and 351 subjects with germline CDC73 mutations from the participants of two clinical protocols in the NIH and from the published case reports/series ranging from 2002 to 2016, respectively (Supplementary Table 1, see section on supplementary materials given at the end of this article). A total of 86 distinct germline CDC73 mutations were found among these 419 subjects (Supplementary Table 2). There were 42 (61.8%) and 338 (96.3%) subjects whose germline CDC73 mutations were confirmed by DNA analysis in the NIH cases and the cases reported in literature, respectively; and the remaining subjects in each group were presumed to possess CDC73 mutations, because they were either obligate carriers or clinically affected first- or second-degree relatives of an index case with a confirmed CDC73 mutation (Fig. 2). Among the NIH cases and the cases reported in literature, 14 (20.6%) and 63 (17.9%) subjects had a history of parathyroid cancer, and 3 (4.4%) and 20 (5.7%) subjects were diagnosed with atypical parathyroid adenomas, respectively (Fig. 2). Among the subjects who had detailed surgical pathology information (n = 128), 59.1% (13 out of 22) in the Parathyroid Carcinoma group and 42.5% (45 out of 106) in the Parathyroid Adenoma group were found to have multi-glandular parathyroid involvement. Among the NIH parathyroid carcinoma cases with detailed pathology report (n = 8), 62.5% (n = 5) had evidence of metastasis and the remaining three cases (37.5%) had features of local invasion on histopathology. Compared to those with classic and atypical parathyroid adenoma, the subjects who developed parathyroid carcinoma had slightly older age as well as higher serum PTH and calcium levels, all of which were statistically significant (Supplementary Fig. 1a, c, and d). The median tumor size of parathyroid carcinomas was marginally bigger than those of atypical adenomas and classic adenomas (P = 0.05) (Supplementary Fig. 1b).
Figure 2.
CONSORT diagram of the cases with CDC73 germline mutations in this study. ‘Confirmed’ referred to the cases with CDC73 germline mutation verified by DNA sequencing, and ‘Presumed’ were those considered to have CDC73 germline mutation but without DNA sequencing verification. NIH, National Institutes of Health; PC, parathyroid carcinoma; PA, parathyroid adenoma; NED, no evidence of parathyroid disease.
Correlation between CDC73 genotype and parathyroid cancer
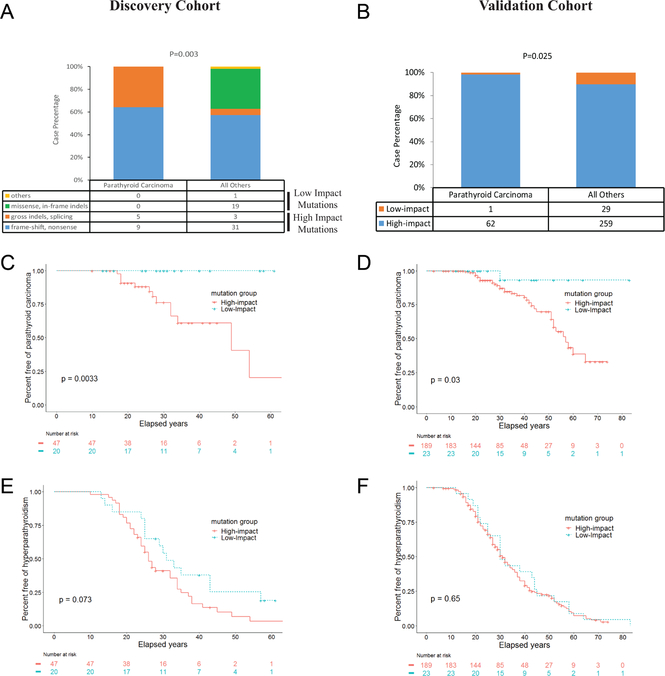
To assess what CDC73 mutation genotype is more commonly present in parathyroid cancer, we grouped the mutations based on their effects on the coding region of the CDC73 gene. We used the NIH cases as the Discovery cohort and the cases reported in literature as the Validation cohort. In the Discovery cohort, composition of CDC73 mutation types in the parathyroid carcinoma cases was strikingly different (P = 0.003) from that of all other cases (Fig. 3a). The four types of mutations (gross indels, splicing, frame-shift and nonsense), which were all predicted to cause a significant impact on parafibromin function or a loss of parafibromin expression and were thus considered as ‘high-impact’ mutations, were more common in the Parathyroid Carcinoma group. In contrast, other types of mutations (missense, in-frame indels and others), which were predicted to have a more limited impact on parafibromin function and were thus referred as ‘low-impact’ mutations, were absent in the Parathyroid Carcinoma group (Fig. 3a). Similarly, in the Validation cohort, there was only one case (1.6%) with low-impact mutation in the Parathyroid Carcinoma group (n = 63) compared to 29 cases (37%) in all other subjects (n = 288; P = 0.0251; Fig. 3b).
Figure 3.
High-impact germline mutations of the CDC73 gene, but not low-impact ones, were associated with an increased risk of parathyroid cancer. (A, B) Comparison of mutation composition between the subjects with parathyroid carcinoma and all other subjects in the Discovery Cohort (A) and Validation cohort (B). (C, D) Kaplan-Meier plots of event-free survival of parathyroid carcinoma in the subjects of the Discovery cohort (c) and the Validation cohort (D) grouped by predicted impact of the CDC73 germline mutations. (E, F) Kaplan-Meier plots of event-free survival of hyperparathyroidism in the subjects of the Discovery cohort (E) and the Validation cohort (F) grouped by predicted impact of the CDC73 germline mutations.
Kaplan–Meier analysis was next performed to estimate risks of parathyroid carcinoma in the high-impact and low-impact CDC73 mutation groups. In the Discovery cohort, the subjects with high-impact mutations had a significantly higher risk of developing parathyroid carcinoma compared to those with low-impact mutations (hazard ratio: not applicable; P = 0.0033; Fig. 3c). Similar results were found in the Validation cohort (hazard ratio = 6.60; P = 0.03; 95% CI, 2.7–16.1; Fig. 3d). In contrast, in both Discovery and Validation cohorts, the subjects in the high-impact mutation groups had similar risks of developing PHPT as those in the low-impact mutation groups (Fig. 3e and f). These analyses were also performed in the subsets of the subjects who had sequencing-confirmed germline CDC73 mutations, in those who were the index cases, and in those after atypical parathyroid adenoma was excluded. Significantly higher risks of parathyroid carcinoma, but similar risks of developing PHPT, were consistently found in the subjects harboring high-impact mutations compared to those with low-impact mutations in all subsets (Supplementary Fig. 2). We also compared among various types (namely gross indels, splicing, frame-shift or nonsense) of high-impact mutations, and found all types of high-impact mutations increased risk for parathyroid carcinomas without changing risk for PHPT in comparison to low-impact mutations (Supplementary Fig. 3). Taken together, these findings showed that high-impact germline mutations, but not low-impact mutations, of the CDC73 gene are associated with an increased risk of parathyroid cancer.
Differential effect of high-impact and low-impact mutations on parafibromin protein domains
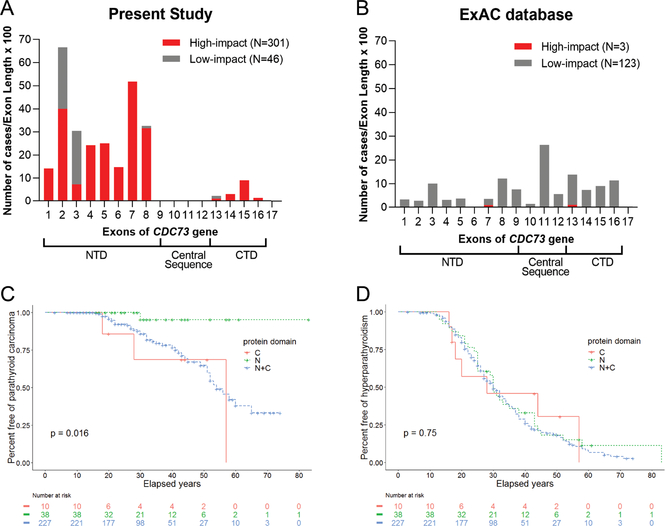
To investigate how the high-impact mutations differ from the low-impact ones, we first examined mapping locations of all nonsynonymous mutations included in this study in the coding sequence of the CDC73 gene. Most of the nonsynonymous mutations mapped to exons 1–8 that constitute the NTD of parafibromin and the remaining mutations mapped to exons 13–16 that constitute the CTD, with no mutations present in exons 9–12 that encode the central sequence of parafibromin or in exon 17 that encodes the last 12 amino acids at the carboxy terminus of parafibromin (Fig. 4a). The mapping locations of low-impact mutations differed from those of high-impact ones in that the former mutations predominantly affect the NTD of parafibromin and are rarely present in the exons encoding the CTD (Fig. 4a). Although high-impact mutations were also more commonly mapped to the coding region of the NTD, they were all predicted to disrupt the CTD (either the CTD alone or both the CTD and the NTD) because the coding sequence of CTD is altered by these mutations. Next, we selected 19 mutations from both genotype groups that were predicted not to cause a complete loss of parafibromin. Irrespective of mutation genotypes, the mutant forms of parafibromin could all be expressed in a transient-transfection system but commonly at much lower levels than WT parafibromin except for two low-impact mutations (c.1135G > A and c.815 A > G; Supplementary Fig. 4a). In addition, mutations from both genotype groups did not typically affect binding of parafibromin to PAF1 complex when the central sequence of parafibromin remains intact (Supplementary Fig. 4b). These results indicate that mutant forms of parafibromin could retain certain properties of the native protein and mutations of various genotype groups may cause loss of separate function of parafibromin by disrupting different protein domains.
Figure 4.
Germline CDC73 mutations that impact the CTD of parafibromin are associated with an increased risk of parathyroid carcinoma. (A, B) Mapping of high-impact and low-impact CDC73 germline mutations from the present study (A) or in the ExAC database (B) in the coding regions of the CDC73 gene. The gross-indel mutations were not included in this analysis because these genomic alterations affect multiple exons of the CDC73 gene. The Y axis represents the case number in each exon of the CDC73 gene divided by length of the corresponding exon and then multiplied by 100. (C, D) Kaplan–Meier plots of event-free survival of parathyroid carcinoma (c) and hyperparathyroidism (d) in the subjects with germline mutations that are predicted to impact the NTD only (N), the CTD only (C) or both the NTD and CTD (N + C) of parafibromin.
We next compared the germline CDC73 mutations in our dataset with those identified in the general population. To this end, we queried the CDC73 gene mutations collected in the Exome Aggregation Consortium (ExAC) database. In contrast to the findings in our dataset, most of the nonsynonymous variants in the ExAC database were low-impact mutations and were mapped throughout the CDC73 coding sequence, with the highest number of variants in exon 11 that encodes part of the central sequence of parafibromin (Fig. 4b). These findings suggest that not all germline CDC73 mutations are equally damaging and mutations that disrupt either the CTD or the NTD of parafibromin are more pathogenic.
To determine disruption of which protein domain of parafibromin increases risk of parathyroid cancer, we grouped the subjects based on the predicted effects of the mutations on two protein domains of parafibromin (Supplementary Table 2), namely impacting the NTD-only, CTD-only and NTD + CTD groups. The cases from both Discovery and Validation cohorts were combined to increase power of the analysis. The Kaplan–Meier analysis revealed a significant difference in the risk of developing parathyroid cancer among the three groups (P = 0.016; Fig. 4c). Compared to the NTD-only group, the hazard ratios for parathyroid cancer were 14.64 (95% CI, 1.078–198.8; P = 0.002) and 9.625 (95% CI, 4.52–20.49; P = 0.006) in the CTD-only and NTD + CTD groups, respectively. In contrast, the subjects in all three groups had a similar risk of PHPT (Fig. 4d). The analyses performed on the subsets of the subjects produced very similar results (Supplementary Fig. 5).
In summary, high-impact mutations differ from low-impact ones with respect to the protein domains of parafibromin that are affected. The mutations involving the CTD (either CTD only or both NTD and CTD), but not those involving the NTD only, are associated with an increased risk of parathyroid carcinoma.
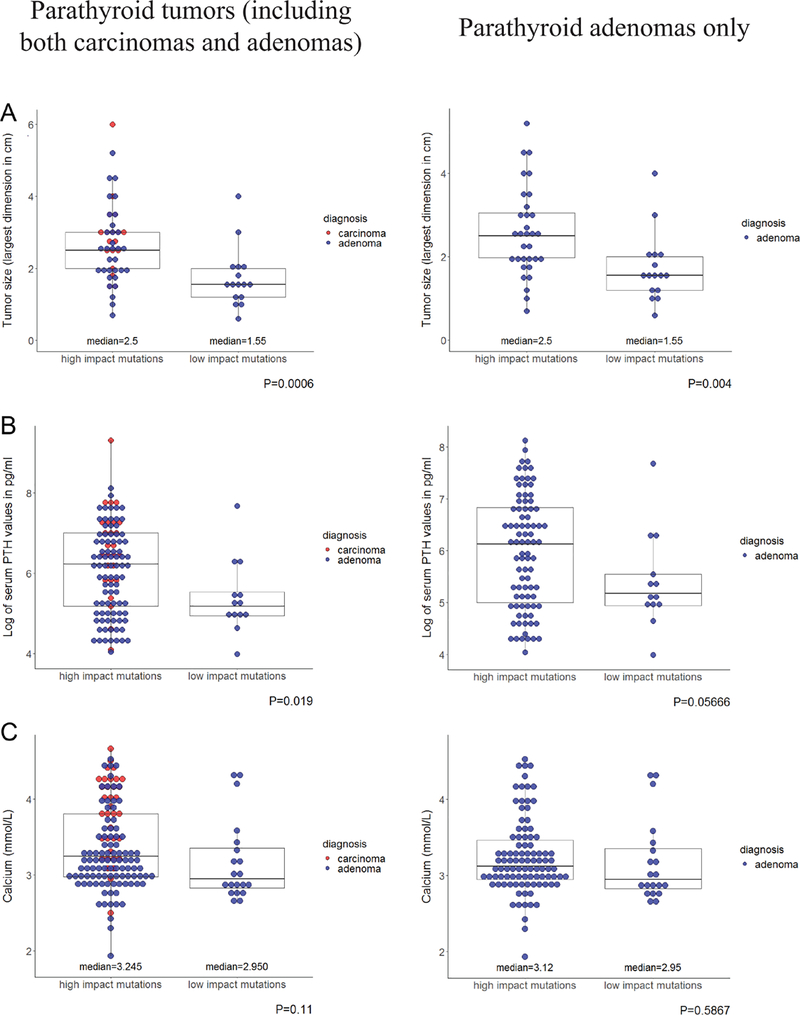
Correlation of CDC73 mutation genotype and severity of PHPT
We next examined if the subjects with high-impact CDC73 mutations could have a more advanced clinical presentation of parathyroid disease. As shown in Fig. 5a, b, and c left panel, the average size of the parathyroid tumors (including both carcinomas and adenomas), serum PTH and calcium levels were all greater in the high-impact mutation group than those in the low-impact mutation group, although the difference in the serum calcium levels was not statistically significant. To eliminate the potential bias of disproportionally higher numbers of parathyroid carcinomas in the high-impact mutation group, we repeated this analysis by including only parathyroid adenomas. As shown in Fig. 5a, b, and c right panel, the high-impact mutation group still had a greater size of parathyroid adenoma as well as higher serum PTH and calcium levels, though the latter differences were not statistically significant. Because clinical information was available only in a few subjects within the CTD-only group, a comparison of PHPT severity was not performed between the NTD, CTD and NTD + CTD groups. Together, these results demonstrate that the high-impact CDC73 germline mutations are associated with a more advanced clinical presentation of parathyroid disease.
Figure 5.
The subjects harboring high-impact CDC73 germline mutations had more advanced clinical presentation of PHPT than those with low-impact mutations. Comparisons of parathyroid tumor size (A), serum PTH levels (B), and serum calcium levels (C) between the high-impact mutation group and the low-impact mutation group among the subjects who were diagnosed of parathyroid tumors (including both adenomas and carcinomas, left panel) or in the subjects who were diagnosed of parathyroid adenoma only (right panel).
CDC73 mutation genotype in atypical parathyroid adenoma
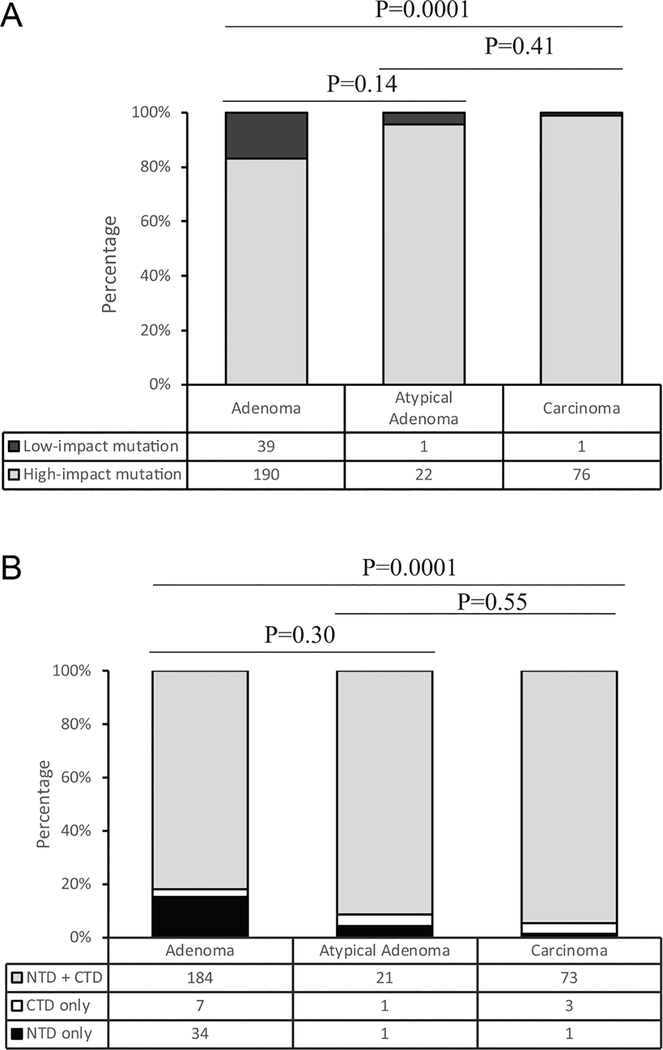
Atypical parathyroid adenoma represents an intermediate form of parathyroid neoplasms of uncertain malignant potential (Cetani et al. 2019). We asked how genotype of CDC73 mutation in this tumor group differs from those in typical parathyroid adenomas and parathyroid cancers. As shown in Fig. 6a and b, the percentage of high-impact mutations or mutations involving the CTD in the subjects with atypical parathyroid adenomas resembled that in the subjects with parathyroid carcinomas, indicating the genotypes of CDC73 mutations in these two groups are more similar.
Figure 6.
The genotypes of CDC73 germline mutations in atypical parathyroid carcinomas resemble those in parathyroid carcinomas. (A) Composition of the low-impact and the high-impact mutations in the subjects with atypical parathyroid adenomas, parathyroid typical adenomas or parathyroid carcinomas; (B) Composition of the NTD-only, the CTD-only, or both NTD + CTD mutations in the subjects with atypical parathyroid adenomas, parathyroid typical adenomas or parathyroid carcinomas.
Genotype-phenotype correlation between CDC73 germline mutations and other manifestations of the HPT-JT syndrome
The HPT-JT syndrome, which has been linked to CDC73 germline mutations, is featured with PHPT resulting from parathyroid tumors (typical adenoma, atypical adenoma, or carcinoma), fibro-osseous jaw tumors, and/or tumors in kidney and uterus (Li & Simonds 2016). We asked if genotypes of CDC73 germline mutations are also correlated with the manifestations of the HPT-JT syndrome other than parathyroid carcinoma. Because specific pathology information was often not available for the lesions found in jaw, kidney and uterus, we included all diseases in each organ for this analysis. As shown in Supplementary Table 3, the percentage of high-impact mutations was significantly higher in the subjects who developed jaw diseases but not in those with kidney and uterine diseases. Similarly, the mutations affecting the CTD only or NTD + CTD were significantly more common in the subjects who developed jaw diseases but not in the subjects with kidney diseases and uterine diseases (Supplementary Table 4). Because information on age of onset of jaw disease was unavailable in most of the subjects, Kaplan-Meier analysis could not be performed. In summary, these results demonstrate that, besides parathyroid carcinoma, the high-impact CDC73 germline mutations were associated with an increased risk of jaw disease.
Structural impact of different genotype mutations on parafibromin
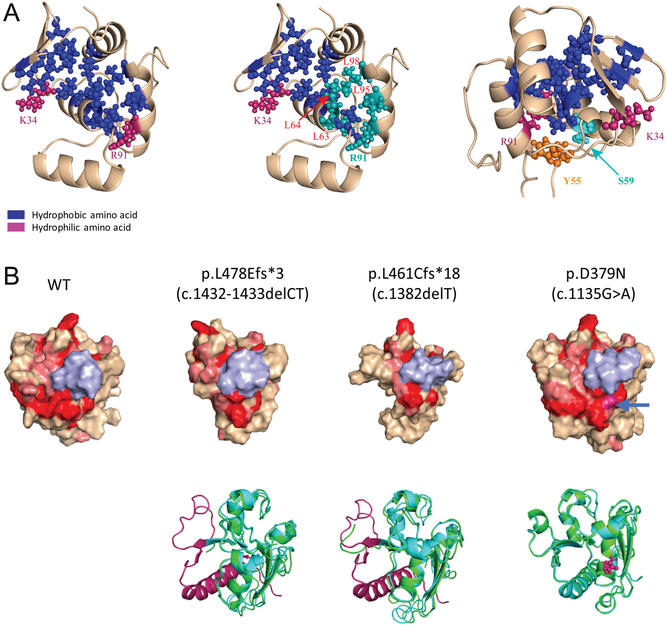
To gain further insight into how the NTD-only and CTD-only mutations affect the function of parafibromin, we assessed impact of mutations within each genotype group on the structure of parafibromin. Since the crystal structure of part of the NTD of human parafibromin has been solved, we examined structural impact of the NTD-only mutations by modeling on the crystal structure of amino acids 1–111 of parafibromin (Sun et al. 2017). Strikingly, many of the N-terminal missense mutations affect the residues in a highly conserved hydrophobic groove in the NTD of parafibromin (Fig. 7a middle panel). The c.176c > T (S59F) mutation, the only missense germline CDC73 mutation associated with parathyroid carcinoma, affects a residue adjacent to the highly-conserved hydrophobic groove, while the Y55 residue, which is affected by two distinct germline missense mutations (c.163T > A [Y55N] and c.164A > C [Y55S]), is at some distance away from the groove (Fig. 7a right panel).
Figure 7.
Structural impact of the germline mutations that affect the CTD only or the NTD only. (A) The germline mutations that impact the NTD only commonly affect the residues in or near the conserved hydrophobic groove of the NTD. Left: The conserved residues that form the hydrophobic groove in the NTD were highlighted in blue color; K34 and R91, two hydrophilic residues at the bilateral ends of the groove, were highlighted in dark red color. Middle: The residues L63, L64, L95, L98 and R91 (highlighted in light blue), which are affected by the germline mutations, are in or at one end of the hydrophobic groove. Right: The two residues that are affected by the germline mutations, S59 and Y55, are located proximal to or away from the hydrophobic groove, respectively. (B) Mutations that impact the CTD only disrupt a conserved ‘flat’ surface structure. Far left: The residues 350–531 in the CTD were modeled based on the yeast Cdc73 structure. Red and pink colors highlight identical and highly conserved residues between human and yeast CDC73, respectively. The region from 387–394 is colored in light blue to display the similar orientation of each model. Top 3 on the right: the surface view of the mutant structures as specified. The blue arrow shows the location of the residue D379; Bottom: Superimposition of the modeled mutant structure (light blue) over the WT structure (green), with the missing/affected structure in the mutants highlighted in magenta color.
We next assessed the structural impact of the CTD-only mutations. Because the structure of the CTD of parafibromin is still unsolved, we modeled its structure based on the structure of yeast Cdc73, which is a homolog of the CTD of human CDC73 but without an NTD-like sequence. All of the seven high-impact mutations affecting the CTD only cause disruption of a previously-reported ‘flat’ surface (highlighted in red in Fig. 7b left panel), which is formed by the residues that are evolutionarily-conserved across different species (Amrich et al. 2012); and the only missense germline mutation present in the CTD (c.1135G > A [p.D379N]) also affected one of the residues present on this surface (Fig. 7b right panel). Together, these data suggest that the conserved ‘flat’ surface structure in the CTD is critical for tumor suppressor activity of parafibromin and is disrupted by ‘high-impact’ mutations or those affecting the CTD.
Discussion
In this study, we discovered that high-impact germline CDC73 mutation is associated with an increased risk of parathyroid carcinoma and jaw disease. Consistent with our findings, in three published studies that performed targeted gene sequencing or whole exome sequencing on parathyroid carcinomas, all of the subjects harboring germline CDC73 mutations had high-impact mutations (either frame-shift or nonsense mutations) in the CDC73 gene (Shattuck et al. 2003, Yu et al. 2015, Pandya et al. 2017). In contrast, we found that the subjects with low-and high-impact germline mutations had a similar risk of developing PHPT. In addition, the clinical presentation of PHPT was more advanced in the subjects with high-impact germline CDC73 mutations. These results suggest that impairment of parafibromin function by the high-impact mutations have pathophysiologic ramifications that overlap with, but are distinct from, those resulting from the low-impact mutations.
One common feature we found between the low- and high-impact mutations of the CDC73 gene is that most of the mutations in both genotype groups diminished expression of mutant parafibromin. Despite being unstable, the parafibromin mutants resulting from these germline mutations were still able to bind to CTR9 and thus likely retained certain biological function(s) of native parafibromin. A notable distinction between the low-and high-impact mutations is that most of the former mutations only affect the NTD while all of the latter mutations disrupt the CTD of parafibromin. Structure modeling revealed that the mutations within the NTD-only group or the CTD-only group are predicted to disrupt a conserved structure within each protein domain. We speculate that each domain of parafibromin has distinct functions (e.g., the CTD is more critical for the tumor suppressor function of parafibromin), and disruption of separate domains of parafibromin by different genotype groups of germline mutations could be the mechanistic basis for their differential risk of parathyroid cancer. A previous study in yeast Cdc73 has suggested that the CTD of parafibromin may play an important role in transcription elongation (Amrich et al. 2012), but evidence is still lacking in human parafibromin. Future study should investigate how disruption of the conserved ‘flat’ surface structure in the CTD could compromise the tumor suppressor function of parafibromin.
It has been suggested that IHC staining of parafibromin could be used as a screening test for not-so-widely-available genomic testing of CDC73 germline mutation (Gill et al. 2019). However, IHC test of parafibromin has not been standardized and could be difficult to perform with variable results (Gill et al. 2019). In addition, there is a concern that this approach may favor the detection of germline mutations that cause a complete/significant loss of parafibromin expression over those that do not affect parafibromin expression. Our data in Supplementary Fig. 4 showed that, among the mutations we tested, all the high-impact germline mutations and most of the low-impact germline mutations of the CDC73 gene (which account for 88% of all the low-impact mutation cases in our study) caused a significantly diminished expression of parafibromin mutants compared to WT parafibromin. Two low-impact mutations (c.1135G > A and c.815 A > G, which account for 4% of the low-impact mutation cases in our study) did not affect expression of parafibromin mutants, while the effect of remaining three low-impact mutations (g.95151T > C, *12C > A and c.-4dupG, which account for 8% of the low-impact mutation cases) were unknown. These results suggest that the approach of using parafibromin IHC as a screening test could lead to missed diagnosis of a small percentage of low-impact germline mutations that do not affect or have unknown effect on expression of parafibromin mutants. Because this approach was not adopted in our institution and may only affect a small percentage of the cases reported in the literature, it is unlikely to be a significant confounding factor in our study.
The findings in this study have several clinical implications. First, subjects who are known to harbor the high-impact germline CDC73 mutations that damage or eliminate the CTD of parafibromin should be educated and counseled for an increased risk of parathyroid carcinoma. These subjects may also benefit from closer monitoring, particularly for parathyroid cancer. On the contrary, subjects with the low-impact mutations or mutations not affecting CTD of parafibromin, have a much lower risk of parathyroid carcinoma and may be suitable for less frequent surveillance. Secondly, our findings are consistent with the view that atypical parathyroid adenoma may represent a precursor of parathyroid carcinoma and has the potential to transform into cancer after accumulation of addition genomic alteration(s). Our findings, therefore, support the recommendation to closely follow patients with atypical parathyroid adenoma after parathyroid surgery (Cetani et al. 2019).
This study has several limitations. First, most of the case reports and case series were cross-sectional observations without long-term follow up. Therefore, the rates of parathyroid tumor are likely to be underestimated, and we could not assess effect of genotypes of germline CDC73 mutation on long-term outcome (e.g., parathyroid tumor recurrence or all-cause survival). Secondly, the results of laboratory tests (e.g., calcium and PTH levels) came from many different centers over a span of 14 years. These results may be obtained by employing different assays and with different normal ranges and it is not possible to normalize these values. Therefore, the analysis results on these data need be interpreted with this limitation in mind. Lastly, diagnosis of parathyroid carcinomas and atypical parathyroid adenomas could be challenging if only based on histological findings, and pathological diagnosis is not necessary to predict the outcome of parathyroid disease (Gill 2014). It would be ideal to have all parathyroid pathologies reviewed by an expert pathologist in a blinded fashion to ensure the same set of criteria being applied to all cases. However, because most of the parathyroid specimens were unavailable for blinded pathology review in both cohorts, we could only rely on the diagnoses reported in the original reports for our analysis. Nevertheless, because of retrospective nature of our study, the diagnoses of parathyroid diseases were made by pathologists without knowing specific genotype of CDC73 mutations for the purposes of this study.
In conclusion, this is the first study to show a strong correlation between CDC73 genotype and the development of parathyroid cancer and jaw disease. Appropriate counseling and surveillance for parathyroid carcinoma and jaw disease should be considered for those harboring high-risk germline mutations of the CDC73 gene, which could lead to a better clinical outcome of these subjects.
Supplementary Material
Acknowledgements
The authors thank all the subjects and clinical staff who participated in this study at the National Institutes of Health. The authors also thank Drs Sunita K Agarwal and Stephen J Marx for helpful discussions, and Dr Rashad M Riazuddin for his assistance with the initial data collection.
Funding
This research was supported by the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases (DK043012-18; to W F S) and the National Cancer Institute (BC011787-18; to H C) as well as the Dr Richard J Santen Research Fund (to Y L). These funding sources had no direct role in any aspect of the research or article.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Supplementary materials
This is linked to the online version of the paper at https://doi.org/10.1530/erc-20-0149.
References
- Amrich CG, Davis CP, Rogal WP, Shirra MK, Heroux A, Gardner RG, Arndt KM & VanDemark AP 2012. Cdc73 subunit of Paf1 complex contains C-terminal Ras-like domain that promotes association of Paf1 complex with chromatin. Journal of Biological Chemistry 287 10863–10875. ( 10.1074/jbc.M111.325647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson L, Grimelius L, DeLellis RA, Lloyd R, Akerstrom G, Larsson C, Arnold A, Eng C, Shane E & Bilezikian JP 2004. Parathyroid carcinoma. In World Health Organization Classification of Tumors: Pathology & Genetics: Tumors of Endocrine Organs, pp 124–127. Eds DeLellis RA, Lloyd RV, Heitz PU & Eng C. Lyon, France: IARC Press. [Google Scholar]
- Carpten JD, Robbins CM, Villablanca A, Forsberg L, Presciuttini S, Bailey-Wilson J, Simonds WF, Gillanders EM, Kennedy AM, Chen JD, et al. 2002. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nature Genetics 32 676–680. ( 10.1038/ng1048) [DOI] [PubMed] [Google Scholar]
- Cetani F, Marcocci C, Torregrossa L & Pardi E 2019. Atypical parathyroid adenomas: challenging lesions in the differential diagnosis of endocrine tumors. Endocrine-Related Cancer 26 R441–R464. ( 10.1530/ERC-19-0135) [DOI] [PubMed] [Google Scholar]
- Chen H, Shi N, Gao Y, Li X, Teng M & Niu L 2012. Crystallographic analysis of the conserved C-terminal domain of transcription factor Cdc73 from Saccharomyces cerevisiae reveals a GTPase-like fold. Acta Crystallographica: Section D, Biological Crystallography 68 953–959. ( 10.1107/S0907444912017325) [DOI] [PubMed] [Google Scholar]
- Clarke CN, Katsonis P, Hsu TK, Koire AM, Silva-Figueroa A, Christakis I, Williams MD, Kutahyalioglu M, Kwatampora L, Xi Y, et al. 2019. Comprehensive genomic characterization of parathyroid cancer identifies novel candidate driver mutations and core pathways. Journal of the Endocrine Society 3 544–559. ( 10.1210/js.2018-00043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M, Hu Y, Bi Y, Wang W, Wang M, Zhang X, Zhang R, Wang P, Su Z, Gao X, et al. 2019. Preliminary exploration of potential molecular therapeutic targets in recurrent and metastatic parathyroid carcinomas. International Journal of Cancer 144 525–532. ( 10.1002/ijc.31948) [DOI] [PubMed] [Google Scholar]
- DeLellis R, Larsson C, Arnold A, Lloy R, Bilezikian J, Mete O & Eng C 2017. Tumors of the parathyroid glands. In WHO Classification of Tumors of Endocrine Organs, pp 145–159. Eds Lloyd R, Osamura R, Kloppel G & Rosai J. Lyon, France: IARC Press. [Google Scholar]
- Eng C, Clayton D, Schuffenecker I, Lenoir G, Cote G, Gagel RF, van Amstel HK, Lips CJ, Nishisho I, Takai SI, et al. 1996. The relationship between specific RET proto-oncogene mutations and disease phenotype in multiple endocrine neoplasia type 2. International RET mutation consortium analysis. JAMA 276 1575–1579. ( 10.1001/jama.1996.03540190047028) [DOI] [PubMed] [Google Scholar]
- Frank-Raue K, Hoppner W, Frilling A, Kotzerke J, Dralle H, Haase R, Mann K, Seif F, Kirchner R, Rendl J, et al. 1996. Mutations of the RET protooncogene in German multiple endocrine neoplasia families: relation between genotype and phenotype. German Medullary Thyroid Carcinoma Study Group. Journal of Clinical Endocrinology & Metabolism 81 1780–1783. ( 10.1210/jcem.81.5.8626834) [DOI] [PubMed] [Google Scholar]
- Gill AJ 2014. Understanding the genetic basis of parathyroid carcinoma. Endocrine Pathology 25 30–34. ( 10.1007/s12022-013-9294-3) [DOI] [PubMed] [Google Scholar]
- Gill AJ, Lim G, Cheung VKY, Andrici J, Perry-Keene JL, Paik J, Sioson L, Clarkson A, Sheen A, Luxford C, et al. 2019. Parafibromin-deficient (HPT-JT type, CDC73 mutated) parathyroid tumors demonstrate distinctive morphologic features. American Journal of Surgical Pathology 43 35–46. ( 10.1097/PAS.0000000000001017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA & Marsh DJ 2005. Identification of a functional bipartite nuclear localization signal in the tumor suppressor parafibromin. Oncogene 24 6241–6248. ( 10.1038/sj.onc.1208778) [DOI] [PubMed] [Google Scholar]
- Hahn MA & Marsh DJ 2007. Nucleolar localization of parafibromin is mediated by three nucleolar localization signals. FEBS Letters 581 5070–5074. ( 10.1016/j.febslet.2007.09.050) [DOI] [PubMed] [Google Scholar]
- Iwata T, Mizusawa N, Taketani Y, Itakura M & Yoshimoto K 2007. Parafibromin tumor suppressor enhances cell growth in the cells expressing SV40 large T antigen. Oncogene 26 6176–6183. ( 10.1038/sj.onc.1210445) [DOI] [PubMed] [Google Scholar]
- Jackson MA, Rich TA, Hu MI, Perrier ND & Waguespack SG 1993. CDC73-related disorders. In Gene Reviews (R). Eds Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Ledbetter N, Mefford HC, Smith RJH, et al. Seattle, WA, USA: University of Washington. (available at: https://www.ncbi.nlm.nih.gov/books/NBK3789/) [Google Scholar]
- Krebs LJ, Shattuck TM & Arnold A 2005. HRPT2 mutational analysis of typical sporadic parathyroid adenomas. Journal of Clinical Endocrinology & Metabolism 90 5015–5017. ( 10.1210/jc.2005-0717) [DOI] [PubMed] [Google Scholar]
- Lee PK, Jarosek SL, Virnig BA, Evasovich M & Tuttle TM 2007. Trends in the incidence and treatment of parathyroid cancer in the United States. Cancer 109 1736–1741. ( 10.1002/cncr.22599) [DOI] [PubMed] [Google Scholar]
- Li Y & Simonds WF 2016. Endocrine neoplasms in familial syndromes of hyperparathyroidism. Endocrine-Related Cancer 23 R229–R247. ( 10.1530/ERC-16-0059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machens A, Niccoli-Sire P, Hoegel J, Frank-Raue K, van Vroonhoven TJ, Roeher HD, Wahl RA, Lamesch P, Raue F, Conte-Devolx B, et al. 2003. Early malignant progression of hereditary medullary thyroid cancer. New England Journal of Medicine 349 1517–1525. ( 10.1056/NEJMoa012915) [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G & Basler K 2006. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell 125 327–341. ( 10.1016/j.cell.2006.01.053) [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G & Basler K 2009. The role of Parafibromin/Hyrax as a nuclear Gli/Ci-interacting protein in Hedgehog target gene control. Mechanisms of Development 126 394–405. ( 10.1016/j.mod.2009.02.002) [DOI] [PubMed] [Google Scholar]
- Newey PJ, Bowl MR, Cranston T & Thakker RV 2010. 2010 cell division cycle protein 73 homolog (CDC73) mutations in the hyperparathyroidism-jaw tumor syndrome (HPT-JT) and parathyroid tumors. Human Mutation 31 295–307. ( 10.1002/humu.21188) [DOI] [PubMed] [Google Scholar]
- Pandya C, Uzilov AV, Bellizzi J, Lau CY, Moe AS, Strahl M, Hamou W, Newman LC, Fink MY, Antipin Y, et al. 2017. Genomic profiling reveals mutational landscape in parathyroid carcinomas. JCI Insight 2 e92061. ( 10.1172/jci.insight.92061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker LM, Zhang JH, Dagur PK, Gastinger MJ & Simonds WF 2010. Defective nucleolar localization and dominant interfering properties of a parafibromin L95P missense mutant causing the hyperparathyroidism-jaw tumor syndrome. Endocrine-Related Cancer 17 513–524. ( 10.1677/ERC-09-0272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazienza V, la Torre A, Baorda F, Alfarano M, Chetta M, Muscarella LA, Battista C, Copetti M, Kotzot D, Kapelari K, et al. 2013. Identification and functional characterization of three NoLS (nucleolar localisation signals) mutations of the CDC73 gene. PLoS One 8 e82292. ( 10.1371/journal.pone.0082292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo JP, Hernandez-Prera JC, Randolph GW, Zafereo ME, Hartl DM, Silver CE, Suarez C, Owen RP, Bradford CR, Makitie AA, et al. 2020. Parathyroid cancer: an update. Cancer Treatment Reviews 86 102012. ( 10.1016/j.ctrv.2020.102012) [DOI] [PubMed] [Google Scholar]
- Shattuck TM, Valimaki S, Obara T, Gaz RD, Clark OH, Shoback D, Wierman ME, Tojo K, Robbins CM, Carpten JD, et al. 2003. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. New England Journal of Medicine 349 1722–1729. ( 10.1056/NEJMoa031237) [DOI] [PubMed] [Google Scholar]
- Sun W, Kuang XL, Liu YP, Tian LF, Yan XX & Xu W 2017. Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome. Scientific Reports 7 15638. ( 10.1038/s41598-017-15715-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CH & Harari A 2012. Parathyroid carcinoma: update and guidelines for management. Current Treatment Options in Oncology 13 11–23. ( 10.1007/s11864-011-0171-3) [DOI] [PubMed] [Google Scholar]
- Wei Z, Sun B, Wang ZP, He JW, Fu WZ, Fan YB & Zhang ZL 2018. Whole-exome sequencing identifies novel recurrent somatic mutations in sporadic parathyroid adenomas. Endocrinology 159 3061–3068. ( 10.1210/en.2018-00246) [DOI] [PubMed] [Google Scholar]
- Wimley WC & White SH 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nature Structural Biology 3 842–848. ( 10.1038/nsb1096-842) [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J & Zhang Y 2015. The I-TASSER Suite: protein structure and function prediction. Nature Methods 12 7–8. ( 10.1038/nmeth.3213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D & Krek W 2005. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Molecular & Cellular Biology 25 5052–5060. ( 10.1128/MCB.25.12.5052-5060.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, McPherson JR, Stevenson M, van Eijk R, Heng HL, Newey P, Gan A, Ruano D, Huang D, Poon SL, et al. 2015. Whole-exome sequencing studies of parathyroid carcinomas reveal novel PRUNE2 mutations, distinctive mutational spectra related to APOBEC-catalyzed DNA mutagenesis and mutational enrichment in kinases associated with cell migration and invasion. Journal of Clinical Endocrinology & Metabolism 100 E360–E364. (doi: 10.1210/jc.2014-3238) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.