Abstract
Multiple myeloma (MM) patients with suboptimal response to induction therapy or early relapse, classified as the functional high-risk (FHR) patients, have been shown to have poor outcomes. We evaluated newly-diagnosed MM patients in the CoMMpass dataset and divided them into three groups: genomic high-risk (GHR) group for patients with t(4;14) or t(14;16) or complete loss of functional TP53 (bi-allelic deletion of TP53 or mono-allelic deletion of 17p13 (del17p13) and TP53 mutation) or 1q21 gain and International Staging System (ISS) stage 3; FHR group for patients who had no markers of GHR group but were refractory to induction therapy or had early relapse within 12 months; and standard-risk (SR) group for patients who did not fulfill any of the criteria for GHR or FHR. FHR patients had the worst survival. FHR patients are characterized by increased mutations affecting the IL-6/JAK/STAT3 pathway, and a gene expression profile associated with aberrant mitosis and DNA damage response. This is also corroborated by the association with the mutational signature associated with abnormal DNA damage response. We have also developed a machine learning based classifier that can identify most of these patients at diagnosis.
Subject terms: Cancer genomics, Myeloma
Introduction
There is an increasing appreciation that risk stratification is important in the management of multiple myeloma (MM) [1]. Despite advancement of MM treatment in the past decades following the introduction of proteasome inhibitors and immunomodulatory drugs, which, together with autologous stem cell transplantation (ASCT), have improved the median overall survival from 3 to 8 years [2–4], there are still ~20% of patients who survive for only ~3 years. Improving the outcome of these high-risk patients is one of the most important current therapeutic challenge in MM.
The current approach to risk stratification uses genetic information from FISH, with t(4;14) or t(14;16) or del17p13 by FISH identified as high-risk genetic abnormalities, as well as clinical information such as serum albumin and beta-2 microglobulin and lactate dehydrogenase (LDH), in the R-ISS staging system [5]. Recently, a specific entity named the double-hit MM with del17p13 and TP53 mutation or 1q amplification (four or more copies) and International Staging System (ISS) stage 3 has been shown to have poor survival with median progression-free survival (PFS) of 15.4 months and median overall survival (OS) of 20.7 months [6]. Many published and ongoing studies have also evaluated MM gene expression signatures such as EMC92 [7], GEP70 [8] to identify high-risk patients, although this has not been routinely used in clinical practice. Some clinical characteristics such as extramedullary plasmacytomas [9], presence of circulating tumor cells [10] and renal failure [11, 12] have also been associated with high-risk disease, but they have not been consistently included as criteria for clinical trials.
Recently, a number of studies have highlighted that multiple myeloma patients with suboptimal response to induction therapy or early relapse have been shown to have poor outcomes [13, 14].
A real-world outcome study of 1320 newly diagnosed patients by Australian and New Zealand Myeloma and Related Diseases Registry (MRDR) showed that 40% of patients with suboptimal response to induction therapy died within 3 years of diagnosis, and patients who had early disease progression within 12 months of starting induction therapy had median OS of only 20.2 months [13]. These patients are categorized as the functional high-risk (FHR) MM patients. However, it is not clear how many of these patients also have high-risk genetic features.
In this study, we assess the criteria of FHR patients with a more refined definition including only those who either have suboptimal response to therapy or progress within 12 months of starting induction therapy but yet do not have any of the clinically applicable high-risk genetic features. In addition, we comprehensively compare their genomic profiles (DNA mutations, mutational signatures (MS), transcriptional signatures, copy number abnormalities) with other MM patients to gain insights into what may drive this phenotype of extremely poor outcome.
Methods
We evaluated genomic sequencing and high-throughput molecular assay data of newly diagnosed MM patients in the CoMMpass dataset (IA13a version), a publicly available dataset from the Multiple Myeloma Research Foundation (MMRF).
We divided the patients into three groups: genomic high-risk (GHR) group for patients with t(4;14) or t(14;16) or complete loss of functional TP53 (bi-allelic deletion of TP53 or mono-allelic deletion of 17p13 (del17p13) and TP53 mutation) or 1q21 gain and International Staging System (ISS) stage 3; FHR group for patients who had no markers of GHR group but were refractory to induction therapy or had early relapse within 12 months; and standard-risk (SR) group for patients who did not fulfill any of the criteria for GHR or FHR.
To categorize the patients into their respective risk groups, we used the following CoMMpass data: translocations of IGH locus and their partners using RNA-seq data for canonical Ig translocation, copy number aberrations (CNAs) using CNA segmentation data, response to the first line therapy using per patient treatment response data, ISS staging information using per patient aggregate of clinical information, and disease progression information using per patient survival data.
We analyzed the transcriptome data to obtain clues of underlying molecular mechanisms driving FHR or GHR. We first identified differentially expressed genes (DEGs) using SAM [15] for multiple groups and then queried top DEGs to DAVID [16] to infer their functional consequences. To account for the distortion introduced during the selection of top DEGs, we also employed gene set enrichment analysis (GSEA; v4.1) [17] and captured transcriptomic changes in the genome-wide scale. We also used the following gene expression signatures to investigate the utility of these signature indices in identifying FHR patients based on transcriptomic data: centrosome index (CI) [18], chromosomal instability (CIN) index by Carter et al. (CIN70) [19], CIN index from sarcoma study (CINSARC) [20], CIN index of our own (CINGEC) [21], 92-gene survival index from HOVON-65/GMMG-HD4 study (EMC92) [7], 7-gene survival index from an MM cell line study (HMCL7) [22], a signature of cell death genes affected by homozygous deletion (HZDCD) [23], 15-gene survival index from Intergroupe Francophone du Myeloma study (IFM15) [24], proliferation index (PI) [25], a gene signature index proliferation associated genes from HOVON-65/GMMG-HD4 study (PR) [7], 70-gene and 80-gene survival index from University of Arkansas Medical School (UAMS70 [8], UAMS80 [26]).
We evaluated the copy number aberrations (CNAs) to uncover CNA features associated with FHR or GHR. Starting from the segmentation data provided in the CoMMpass data portal, we determined optimal threshold values for different CNA status as well as minimal segment size to retain and applied them to the segmentation data to determine CNA status for each segment. Subsequently, CNA status was transformed into a matrix format (Supplementary Fig. 1). The CIN index [21] we had developed before was also employed to see if there could be any difference in CIN and hence the biological mechanism to ensure chromosomal integrity among different risk groups. Mutation status of each patient was also analyzed to identify genes and pathways that were preferentially mutated among risk groups using non-synonymous (NS) mutation data compiled in CoMMpass. MS were also assessed and examined for difference in propensity among risk groups using tools and data compiled in COSMIC (v3.2) [27].
To build a predictive model to identify FHR patients using machine learning, we generated six sets of data for each patient based on various features: the number of mutations for each of 15,633 genes that harbored at least one NS mutation (mutation_matrix; 15,633 features), the number of all NS mutations for each of 44 chromosomal arms (mutation_count_by_arm; 44 features), the CNA status for each of 13,155 genes that harbored at least one CNA (cna_by_gene_reduced; 13,155 features), the CNA status for each of 44 chromosomal arms (cna_by_arm; 44 features), transcriptome data of 25,554 genes from RNA-seq where each gene’s expression profile was normalized against its median level (gep_normed; 25,554 features), and six parameters including age, gender, creatinine level, ECOG status, ISS staging, and proliferation index (clinical_parameters; 6 features). In determining cna_by_gene_reduced and cna_by_arm, if multiple CNA statuses appear in a gene or a chromosomal arm, we selected the CNA status of dominant span in a gene or a chromosomal arm. The details of these features are listed in the Supplementary Table 1 and Supplementary File 1.
We undertook the following pre-processing of the six datasets. We first removed highly correlated features from each dataset to reduce the overall feature space and to concentrate on the features that had more meaningful information. We used an absolute correlation threshold of 0.5, and, for some datasets, we performed a significance test to further limit the feature space. In training and testing machine learning models, each dataset was divided into two subsets using a 70–30 rule where 70% of the data were used for training models whereas the remaining 30% were used for testing. During data division, we took a special effort to maintain the distribution of positive and negative cases in both testing and training datasets.
As CoMMpass data is greatly affected by severe class imbalance, we employed a widely used oversampling technique called Synthetic Minority Oversampling Technique (SMOTE) [28] to reduce the bias towards the majority class during the modeling stage. SMOTE creates synthetic minority class samples using KNN technique and potentially performs better than simple oversampling [29]. It has been used in several studies such as breast cancer detection [30], miRNA gene prediction [31, 32], and for the identification of the binding specificity of the regulatory proteins [33].
We used the random forest algorithm for predicting FHR cases in this study. Random forest is an ensemble decision tree-based technique where each tree registers a vote for the most prevalent class and the final decision is made on consensus majority votes [34]. Despite its algorithmic simplicity, it is known to perform fairly well and has been widely used in bioinformatics e.g. for the classification of mRNA microarray data [35], to detect biomarkers for prostate cancer progression [36] and more. The performance of prediction results on test datasets were evaluated for all six individual models on multiple measures such as specificity, sensitivity, false-negative rate (FNR), False positive rate (FPR), accuracy, F1-Score, and Matthews correlation coefficient (MCC) (Supplementary Fig. 2).
Results
FHR patients had the worst survival
Of the 512 evaluable patients, there were 345 patients in the SR group, 106 patients in the GHR group, and 61 patients in the FHR group. The available baseline clinical characteristics of these patients were listed in Table 1. As expected, there are more ISS III and Revised ISS III patients in the GHR group. Interestingly, there are no unique clinical characteristics to FHR patients, in particular, very few are R-ISS III. Most patients had proteasome inhibitor (PI) and immunomodulatory (IMiD) drug combination as first line treatment, while a smaller proportion of patients had PI-based or IMiD-based treatment. The treatment received across the three groups were similar.
Table 1.
Baseline characteristics.
| SR (N = 345) (%) | GHR (N = 106) (%) | FHR (N = 61) (%) | ||
|---|---|---|---|---|
| Gender | Male | 186 (53.9) | 67 (63.2) | 39 (63.9) |
| Female | 159 (46.1) | 39 (36.8) | 22 (36.1) | |
| Age | Median (year) | 62 | 61 | 65 |
| ISS | Stage I | 142 (41.2) | 31 (29.2) | 17 (27.9) |
| Stage II | 115 (33.3) | 42 (39.6) | 20 (32.8) | |
| Stage III | 88 (25.5) | 33 (31.1) | 24 (39.3) | |
| Revised-ISS | Stage I | 111 (32.2) | 1 (0.9) | 13 (21.3) |
| Stage II | 211 (61.2) | 74 (69.8) | 43 (70.5) | |
| Stage III | 23 (6.7) | 31 (29.2) | 5 (8.2) | |
| Treatment class | PI/IMiD combination-based | 367 (48.1) | 99 (46) | 50 (42.7) |
| PI-based | 233 (30.5) | 68 (31.6) | 43 (36.8) | |
| IMiD-based | 126 (16.5) | 39 (18.1) | 18 (15.4) | |
| Others | 37 (4.8) | 9 (4.2) | 6 (5.1) |
SR Standard Risk group, GHR Genomic High-Risk group, FHR Functional High-Risk group, ISS International Staging System, PI Proteasome Inhibitor, IMiD Immunomodulatory Drug.
Data are number (%).
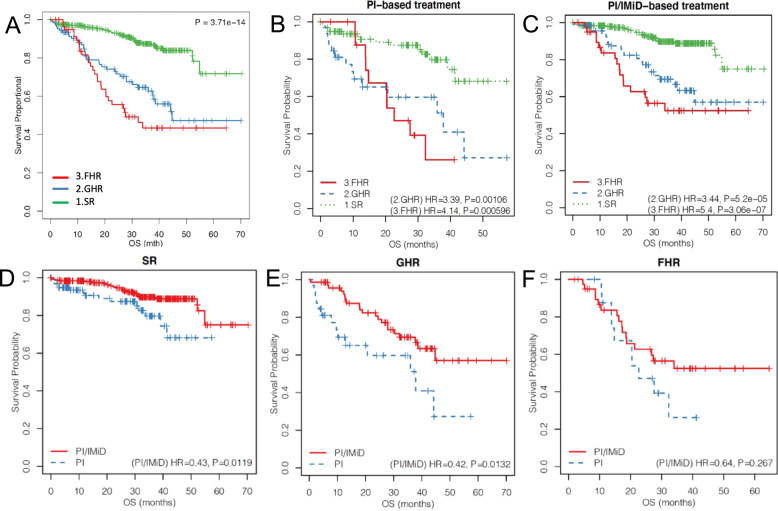
On the survival analysis, both FHR and GHR groups had significantly poorer outcomes compared to the SR group, with FHR group being the worst. The median OS for the FHR group was 27.6 months, while the median OS was 44.7 months for the GHR group, and not reached for the SR group (FHR: HR = 5.19, p = 3.42 × 10−11; GHR: HR = 3.55, p = 3.5 × 10−8) (Fig. 1A). Similar patterns were seen for these group of patients when treated with PI-based and PI/IMiD combination induction therapies (Fig. 1B–F).
Fig. 1. Survival curves for FHR, GHR, and SR MM patients in CoMMpass dataset.
A Overall (B) FPI-based induction treatment and (C) PI/IMiD-based induction treatment. Survival curves for patients treated with PI and PI/IMiD in (D) SR, (E) GHR, (F) FHR groups, respectively. P-values indicate Cox regression test of PI/IMiD treatment against PI treatment.
FHR patients cannot be easily identified using known high-risk gene expression signatures or combination of high-risk genetic features
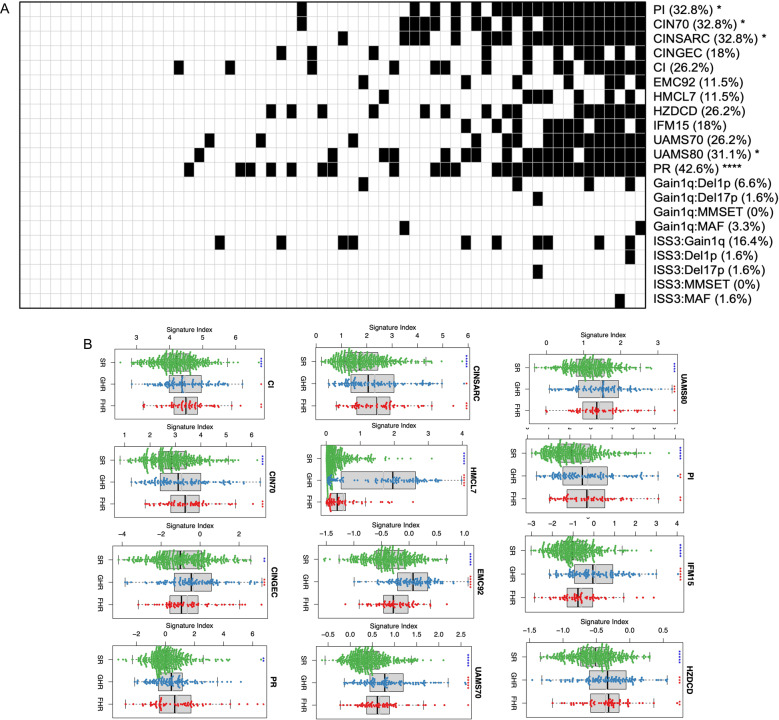
We applied established gene expression signatures of high-risk disease, including proliferation (PI), chromosomal instability (CIN70, CINSARC, CINGEC), centrosome (CI), cell death (HZDCD), and others (EMC92, HMCL7, IFM15, UAMS70, and UAMS80), to see if these FHR cases are characterized by them. Interestingly, most of the FHR patients do not have these high-risk gene expression signature (Fig. 2A). In fact, when compared across the three groups (SR, GHR, and FHR), these signatures are not significantly different between SR and FHR (HMCL7, UAMS80, UAMS70, EMC92, IFM15, and CINGEC) or between GHR and FHR (PI, HZDCD, CINSARC, CI, CIN70, and PR), although the indices associated with chromosomal instability (CINSARC, CI, and CIN70) or tumor aggression (PI and PR) appear to be generally higher in the FHR patients (Fig. 2B). We also used several different combinations of high-risk features including the PR index, gain1q21 + del1p, gain1q21 + del17p13, gain1q21+MMSET, gain1q21+MAF, ISS3 + gain1q21, ISS3 + del1p, ISS3 + del17p13, ISS3 + MMSET, and ISS3 + MAF, to evaluate if they could identify these FHR patients. These combinations of markers are rarely present in FHR patients (Fig. 2A).
Fig. 2. FHR MM patients and known high-risk signatures.
A Each column indicates individual FHR patient. For gene expression signatures—PI, CIN70, CINSARC, CINGEC, CI, EMC92, HMCL7, HZDCD, IFM15, UAMS70, UAMS80, PR—we arbitrarily set patients with top 20% signature indices as high risk (black square). For other combination high-risk genetic markers—Gain1q:Del1p, Gain1q:Del17p, Gain1q:MMSET, Gain1q:MAF, ISS3:Gain1q, ISS3:Del1p, ISS3:Del17p, ISS3:MMSET, ISS3:MAF—the presence of such combination markers was indicated (black square). Individual genetic markers are as follow: gain1q (gain of 1q), del1p (deletion of 1p), del17p (deletion of 17p), MMSET (dysregulation of MMSET), MAF (dysregulation of MAF/MAFB/MAFC), and ISS3 (ISS stage 3). B Gene expression signature box plots. Symbols above each risk group indicate statistical significance (*: 0.01 < p ≤ 0.05; **: 10−3 < p ≤ 10−2; ***: 10−4 < p ≤ 10−3; ****: 10−5 < p ≤ 10−4; *****: p ≤ 10−5) of comparison between a specific group and all the rest. Symbol colors indicate whether the mean level of a specific group is above (red) or below (blue) that of all the rest.
This suggests that the FHR patients are generally not characterized by known high-risk signatures that have been described.
What are the genomic features of FHR MM patients?
We next explored the mutational, transcriptional and copy number landscape of these patients to see if unique molecular and genomic abnormalities can be identified.
Mutation analysis
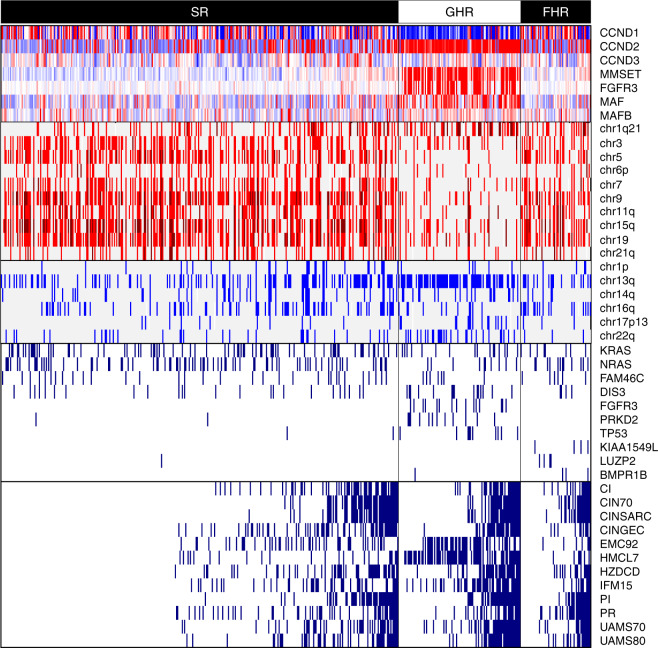
We analyzed the NS mutations in the CoMMpass data and evaluated the prevalence of mutation for genes known to be frequently mutated in MM (KRAS, NRAS, and FAM46C) between different risk groups using the Fisher’s exact test. There was no obvious concentration of mutations in any risk group. However, we uncovered that the GHR group had higher mutational load (p = 0.00331), and genes such as FGFR3 (p = 1.63 × 10−11), PRKD2 (p = 2.82 × 10−7), and TP53 (p = 8.7 × 10−6) were predominantly mutated in GHR group as compared with others. On the other hand, KIAA1549L, LUZP2, and BMPR1B were predominantly mutated in FHR (Fig. 3).
Fig. 3. Composite heat map combining gene expression, copy number aberration, mutation, and gene expression signatures.
Top panel (CCND1, CCND2, CCND3, MMSET, FGFR3, MAF, MAFB) shows the median-normalized gene expression profiles of important TC class marker genes. For each gene, expression above/below the median level is indicated as red/blue, and the median level is indicated as white. The second panel comprises chr1q21, chr3, chr5, chr6p, chr7, chr9, chr11q, chr15q, chr19, chr21q which displays gain of the respective chromosomal regions. Single copy gain is indicated as red and two or more copy gain is indicated as dark red. The GHR group clearly shows the dominance of non-hyperdiploid cases while the SR and FHR groups show prevalence of hyperdiploid cases. The third panel comprises chr1p, chr13q, chr14q, chr16q, chr17p13, chr22q which exhibits loss of respective chromosomal regions. Single copy loss is indicated as blue and two-copy loss is indicated as dark blue. The GHR group clearly shows the dominance of chr13q deletion, possibly indicating the involvement of RB1. The fourth panel comprises KRAS, NRAS, FAM46C, DIS3, FGFR3, PRKD2, TP53, KIAA1549L, LUZP2, and BMPR1B which shows presence of NS mutations for the respective genes. KRAS, NRAS, FAM46C, and DIS3 are known to be frequently mutated in MM. However, FGFR3, PRKD2, and TP53 genes are found to be mutated specifically in the GHR group, while KIAA1549L, LUZP2, and BMPR1B genes are found to be mutated specifically in the FHR group in this study. The bottom panel comprises CI, CIN70, CINSARC, CINGEC, EMC92, HMCL7, HZDCD, IFM15, PI, PR, UAMS70, and UAMS80 which represents gene expression signatures. Here, patients with top 20% respective indices are marked as high-risk.
We then assessed whether the mutated genes were enriched for any gene sets in each risk group, and analyzed the differences with Wilcoxon’s rank-sum test. In the FHR group, the IL6-JAK-STAT3 pathway was found to be significantly enriched (p = 0.00924), while estrogen response (p = 0.000369), KRAS (p = 0.000417), and WNT β catenin (p = 0.000447) signaling pathways were found to be enriched in the GHR group.
Copy number aberrations
Of the 471 evaluable patients, 224 (47.6%) were non-hyperdiploid and 247 (52.4%) were hyperdiploid. Interestingly, FHR group was predominantly hyperdiploid (57.9% vs 42.1% non-hyperdiploid) while GHR group was mostly non-hyperdiploid (90.8% vs 9.2% hyperdiploid). GHR group also had more pronounced 13q deletion, and increased 1q21 gain compared to FHR (p = 1.45 × 10−10) and SR groups (p < 2.2 × 10−16). The SR group had more hyperdiploid (64.9% vs 35.1% non-hyperdiploid) patients, which was statistically insignificant compared to the FHR group (p = 0.369) (Fig. 3 and Supplementary Fig. 3). Therefore, the copy number profile of FHR patients were similar to that of SR patients.
We also estimated CIN using CNAs over autosomal chromosomes. There were no statistically significant differences between FHR and SR groups (p = 0.194) or between GHR and SR groups (p = 0.516), or between FHR and GHR groups (p =0.427) (Supplementary Fig. 4).
Differentially expressed genes and enriched pathways
Using the RNA-seq data, we evaluated the DEGs. The DEGs in FHR and GHR groups were distinct. The list of DEGs from SAM are listed in Supplementary File 2.
In order to understand the biological processes implicated by the list of DEGs, we queried the functional annotations using DAVID for top 200 genes from the comparison between FHR and SR groups but excluding those turned out to be significant from the comparison between GHR and SR groups (Supplementary File 3). FHR patients were found to be enriched for genes linked to mitotic cell cycle and DNA replication, C2H2 zinc finger, and DNA repair. For instance, the first, second, and several of the top sixth and eighth annotation clusters were composed of highly significant terms such as centromere, mitotic cell cycle, and DNA replication, displaying the significant association of mitotic cell cycle processes to FHR. The third annotation cluster was composed of mostly C2H2 zinc finger related terms that are rather too generic to infer further biological context directly. However, a recent publication [37] links the recruitment of C2H2 zinc finger domain to cereblon (CRBN) and the induction of the ubiquitination and proteasomal degradation of genes targeted by small molecules thalidomide and its analogs, lenalidomide and pomalidomide, thereby suggesting potential relevance of this cluster. The fourth, fifth, and top of seventh annotation clusters were associated with DNA repair.
We also queried the functional annotations using the top 499 genes from the comparison of GHR and SR but excluding those significant between FHR and SR groups (Supplementary File 4) to DAVID. GHR patients were enriched for genes linked to ribosomal RNA/protein and protein translation initiation and Ig subtype clusters.
To mitigate the limitation of functional annotations due to artificial selection of top DEGs, we additionally performed GSEA. Compared to the SR group, FHR group showed enrichment in a number of gene sets of the hallmark group known to be involved in MM (P < 0.05 & FDR < 0.25), including E2F Targets, G2M Checkpoint, MTORC1 signaling, Glycolysis, Unfolded protein response, Myc targets, DNA repair, while no significant gene set enrichment were found for the SR group (Table 2). The full list of GSEA result is shown on the Supplementary File 5. For the GHR group, five gene sets were found enriched (Androgen response, Estrogen response, Glycolysis, UV response, and IL2-STAT5 signaling) as compared to the SR group (Table 3). The full list is shown on the Supplementary File 6.
Table 2.
Top hallmark gene sets (P < 0.05 & FDR < 0.25) enriched in FHR group.
| Term | ES | NES | P-value | FDR | Size |
|---|---|---|---|---|---|
| Hallmark_E2F_Targets | 0.72 | 1.77 | 0.00412 | 0.165 | 197 |
| Hallmark_G2M_Checkpoint | 0.65 | 1.74 | 0.01030 | 0.118 | 195 |
| Hallmark_MTORC1_Signaling | 0.54 | 1.70 | 0.00795 | 0.110 | 196 |
| Hallmark_Glycolysis | 0.45 | 1.66 | 0.00877 | 0.124 | 198 |
| Hallmark_Unfolded_Protein_Response | 0.57 | 1.66 | 0.01170 | 0.100 | 109 |
| Hallmark_MYC_Targets_V1 | 0.62 | 1.59 | 0.02760 | 0.141 | 194 |
| Hallmark_Bile_Acid_Metabolism | 0.42 | 1.54 | 0.00952 | 0.171 | 110 |
| Hallmark_Fatty_Acid_Metabolism | 0.45 | 1.54 | 0.02650 | 0.158 | 156 |
| Hallmark_DNA_Repair | 0.54 | 1.51 | 0.02330 | 0.171 | 146 |
| Hallmark_Peroxisome | 0.44 | 1.47 | 0.03650 | 0.203 | 104 |
| Hallmark_Estrogen_Response_Late | 0.33 | 1.40 | 0.01760 | 0.205 | 198 |
ES Enrichment Score, NES Normalized Enrichment Score, FDR False Discovery Rate.
Table 3.
Top hallmark gene sets (P < 0.05 and FDR < 0.25) enriched in GHR group.
| Term | ES | NES | P-value | FDR | Size |
|---|---|---|---|---|---|
| Hallmark_Androgen_Response | 0.46 | 1.64 | 0.01300 | 0.233 | 98 |
| Hallmark_Estrogen_Response_Early | 0.39 | 1.64 | 0.00515 | 0.160 | 198 |
| Hallmark_Glycolysis | 0.41 | 1.49 | 0.04490 | 0.218 | 198 |
| Hallmark_UV_Response_DN | 0.37 | 1.43 | 0.04280 | 0.226 | 142 |
| Hallmark_IL2_STAT5_Signaling | 0.35 | 1.43 | 0.04950 | 0.207 | 198 |
ES Enrichment Score, NES Normalized Enrichment Score, FDR False Discovery Rate.
Mutational signatures
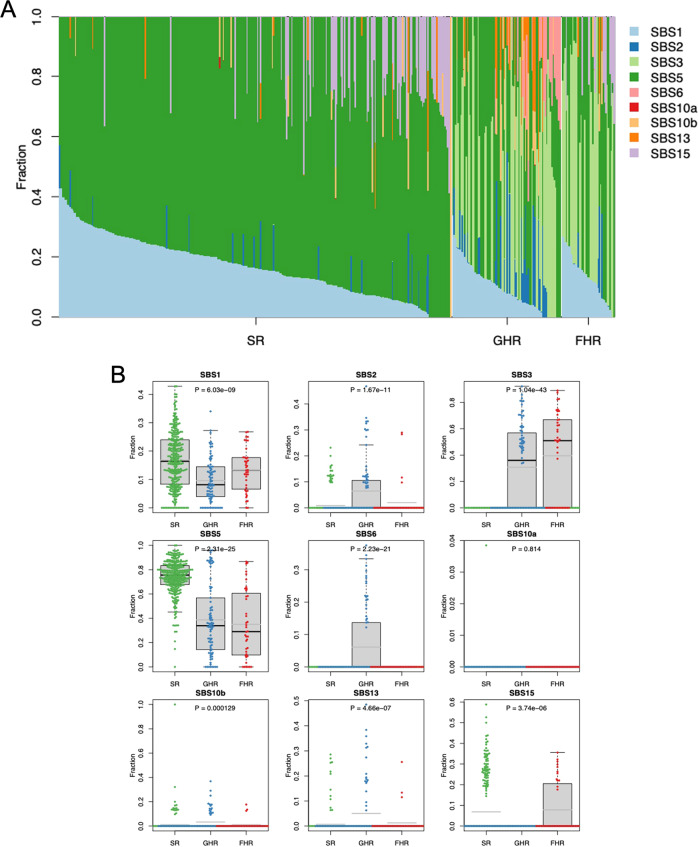
We also analyzed the MS using the SigProfiler with Catalogue Of Somatic Mutations in Cancer (COSMIC) reference catalogue [38]. SBS1 and SBS5 were highly specific to SR. SBS3 was highly specific to the GHR and FHR groups. SBS6 was very specific to GHR (Fig. 4). There was therefore no MS specific only to FHR.
Fig. 4. Mutational Signatures for SR, GHR, and FHR groups.
A For each patient, respective contributions of component MSs are indicated with different color codes. B For individual MSs, respective level of contributions among all MSs per patient are compared among the three risk groups using Kruskal-Wallis test. Here, whether the distribution of respective level of contributions in one risk group is particularly different from those of the other risk groups is tested. SBS5 is more prominent in the SR group than in the GHR and FHR groups (p = 2.31 × 10−25), while SBS3 is more prominent in the GHR and FHR groups (p = 1.04 × 10−43) . SBS2 (p = 1.67 × 10−11) and SBS6 (p = 2.23 × 10−21) are more prominent in the GHR group.
Machine learning based predictor for FHR patients
It is clear that none of the current signatures or high-risk features can identify majority of these FHR patients very well. We therefore used machine learning methodology to develop a classifier.
Our machine learning based predictor showed that performance of the individual model was not optimal (Supplementary Table 2). In order to improve the model and to obtain results that are more robust and useful, we decided to stack multiple models (Supplementary Table 3).
For the highest accuracy, a model with the combination of mutation matrix, gep_normed and clinical parameters, with an accuracy of 0.75, specificity of 0.76, sensitivity of 0.67, false negative rate of 0.33, false positive rate of 0.23, Area Under the Receiver Operating Curve (AUC-ROC) of 0.71, F1 score of 0.34, and MCC score of 0.28, can be used (Supplementary Fig. 5). To ensure that we do not give unnecessary treatment to non-FHR patients and avoid harm, we may want use the model with highest specificity with lowest false positive rate with a combination of cna_by_gene_reduced, mutation_matrix, gep_normed, and clinical parameters, which has an accuracy of 0.78, specificity of 0.80, sensitivity of 0.60, false negative rate of 0.4, false positive rate of 0.2, AUC-ROC of 0.70, F1 score of 0.35, and MCC score of 0.28. However, if the treatment proposed for the FHR patients is not likely to increase harm and can also benefit non-FHR patients, we may want to use a model that gives maximum sensitivity with lowest false negative rate, using a combination of gep_normed and mutation matrix. This model has an accuracy of 0.65, specificity of 0.63, sensitivity of 0.87, false negative rate of 0.13, false positive rate of 0.37, AUC-ROC of 0.75, F1 score of 0.33, and MCC score of 0.30.
Discussion
In this study, we showed that the FHR MM patients that do not have any of the known clinically applied high-risk genetic factors have very poor outcomes. Most of these patients also do not harbor other high-risk characteristics that have been published. As these FHR MM patients are defined based on poor response to induction treatment and early disease progression, there is currently no easy way to identify them at diagnosis. There is much interest to design clinical trials specifically targeting high-risk patients as they need a different therapeutic strategy. However, our current definition of high-risk would have failed to identify these FHR patients. In this study, we developed a machine learning classifier that allows us to identify FHR patients a priori.
Understanding the genomics and biology of these FHR patients may also provide insights into potential therapeutic strategy. New therapeutic approaches are needed for these patients as current approaches have not improved their outcomes significantly. FHR patients seem to have increased mutations affecting the IL-6/JAK/STAT3 signaling pathway. The IL-6/JAK/STAT3 signaling has been shown to drive the proliferation, survival, invasiveness, and metastasis of cancer cells, while suppressing the antitumor immune response [39]. The IL-6/JAK/STAT pathway is also important in myeloma and may be a good therapeutic target in myeloma [40]. We have previously shown that IL6-STAT activation may drive high-risk phenotypes via promotion of aberrant RNA editing through upregulation of ADAR1 [41] and also upregulation of a high-risk phosphatase, PRL-3 [42]. Some studies showed that MM cells with an IL-6-activated JAK/STAT3 pathway are particularly sensitive to heat shock protein 90 (Hsp90) inibitors [43], making this a potential therapeutic target for the FHR MM patients.
The DEG and MS point to the importance of genomic instability, aberrant centromere, mitosis, and abnormal DNA damage repair. Chromosome instability has been known to be a hallmark in MM [44]. Centromeres and their associated kinetochores play an important role in affecting cell mitosis and therefore chromosomal integrity [45]. Along with the context of mitosis, centrosome amplification has also been shown to have prognostic implication in MM. A gene expression-based centrosome index (CI) of more than 4, which was calculated by adding the normalized expression value of the expression levels of genes encoding for the proteins in the centrosomes, has been previously shown to be associated with short survival in MM [46]. The Intergroupe Francophone du Myelome (IFM) from France showed that overexpression of genes involved in mitosis was associated with high-risk disease resulting in poor survival [24]. Similarly, a study from the United Kingdom showed that mutations in the DNA damage pathways are associated with poor outcomes [47]. Recently, we have showed that NEIL1, a gene involved in DNA damage repair, is hyperedited in MM patients with poor outcome and leads to aberrant DNA damage response in these cells [48]. Synthetic lethal approach to exploit DNA damage repair abnormalities in MM has been studied, showing an addiction in these cells to ATR inhibition [49, 50]. In our recent studies, we showed that targeting CHEK1 is also a synthetic lethal approach in high-risk disease with abnormal DNA damage repair phenotype [51].
However, what is most striking is how little distinguished FHR patients are from GHR and SR patients genomically in terms of transcriptomics, MS, copy number, and genes affected by somatic mutations. The lack of distinguishing genomic profile in FHR patients might also suggest that factors outside of the tumor cells such as the immune dysregulation or tumor microenvironment may play an important role in FHR patients. This will need to be addressed in future studies. There are two important implications. First, understanding the differences outside of the tumor cells may allow the development of more effective strategies against the FHR patients as current therapies targeting the traditional myeloma vulnerabilities are ineffective. Second, while our current artificial intelligence (AI) model is better than existing tools in identifying FHR patients, the additional knowledge about the immune dysregulation and tumor microenvironment may add to the model to make the model even better or in fact may simplify the model if the differentiating power is greater.
In summary, we have shown that FHR MM patients, even without any high-risk genetic factors, have very poor outcomes, and we developed a machine learning based classifier that can identify most of these patients at diagnosis. These patients are characterized by increased mutations affecting the IL-6/JAK/STAT3 pathway, and a gene expression profile associated with aberrant mitosis and DNA damage response. This is also corroborated by the association with the MS associated with abnormal DNA damage response. Targeting the STAT pathway and taking advantage of synthetic lethal addictions to the abnormal DNA damage response may lead to novel therapeutic strategies to explore for these patients.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary figures, tables and details of supplementary files
Acknowledgements
The CoMMpass study is sponsored by the Multiple Myeloma Research Foundation (MMRF), which had no role in the interpretation, writing, or publication of this manuscript. WJC is supported by NMRC Singapore Translational Research (STaR) Investigatorship. This research is partly supported by the National Research Foundation Singapore and the Singapore Ministry of Education under the Research Centers of Excellence initiative as well as the RNA Biology Center at the Cancer Science Institute of Singapore, NUS, as part of funding under the Singapore Ministry of Education’s Tier 3 grants, grant number MOE2014- T3-1-006.
Author contributions
CYS and WJC designed the study. CYS, THC, MSF, and WJC analyzed the data. CYS and WJC wrote the manuscript. All authors approved the final version of the manuscript before submission.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cinnie Yentia Soekojo, Tae-Hoon Chung.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-021-00576-3.
References
- 1.Chng WJ, Dispenzieri A, Chim CS, Fonseca R, Goldschmidt H, Lentzsch S, et al. IMWG consensus on risk stratification in multiple myeloma. Leukemia. 2014;28:269–77. doi: 10.1038/leu.2013.247. [DOI] [PubMed] [Google Scholar]
- 2.Kumar SK, Dispenzieri A, Lacy MQ, Gertz MA, Buadi FK, Pandey S, et al. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia. 2014;28:1122–8. doi: 10.1038/leu.2013.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.John RJ, Niels W, Cody A, Brian AW, Chris W, Charlotte P, et al. Clonal evolution in myeloma: the impact of maintenance lenalidomide and depth of response on the genetics and sub-clonal structure of relapsed disease in uniformly treated newly diagnosed patients. Haematologica. 2019;104:1440–50.. doi: 10.3324/haematol.2018.202200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol. 2020;95:548–67. doi: 10.1002/ajh.25791. [DOI] [PubMed] [Google Scholar]
- 5.Palumbo A, Avet-Loiseau H, Oliva S, Lokhorst HM, Goldschmidt H, Rosinol L, et al. Revised international staging system for multiple myeloma: a report from international myeloma working group. J Clin Oncol. 2015;33:2863–9. doi: 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker BA, Mavrommatis K, Wardell CP, Ashby TC, Bauer M, Davies F, et al. A high-risk, Double-Hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia. 2019;33:159–70.. doi: 10.1038/s41375-018-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuiper R, Broyl A, de Knegt Y, van Vliet MH, van Beers EH, van der Holt B, et al. A gene expression signature for high-risk multiple myeloma. Leukemia. 2012;26:2406–13. doi: 10.1038/leu.2012.127. [DOI] [PubMed] [Google Scholar]
- 8.Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–84. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 9.Usmani SZ, Heuck C, Mitchell A, Szymonifka J, Nair B, Hoering A, et al. Extramedullary disease portends poor prognosis in multiple myeloma and is over-represented in high-risk disease even in the era of novel agents. Haematologica. 2012;97:1761–7. doi: 10.3324/haematol.2012.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nowakowski GS, Witzig TE, Dingli D, Tracz MJ, Gertz MA, Lacy MQ, et al. Circulating plasma cells detected by flow cytometry as a predictor of survival in 302 patients with newly diagnosed multiple myeloma. Blood. 2005;106:2276–9. doi: 10.1182/blood-2005-05-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan R, Apewokin S, Grazziutti M, Yaccoby S, Epstein J, van Rhee F, et al. Renal insufficiency retains adverse prognostic implications despite renal function improvement following Total Therapy for newly diagnosed multiple myeloma. Leukemia. 2015;29:1195–201. doi: 10.1038/leu.2015.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimopoulos MA, Sonneveld P, Leung N, Merlini G, Ludwig H, Kastritis E, et al. International myeloma working group recommendations for the diagnosis and management of myeloma-related renal impairment. J Clin Oncol. 2016;34:1544–57. doi: 10.1200/JCO.2015.65.0044. [DOI] [PubMed] [Google Scholar]
- 13.Spencer A, Mollee P, Blacklock HA, Harrison S, Quach H, Ho PJ, et al. Real-world outcome for newly diagnosed patients with functional high-risk myeloma - a myeloma and related diseases registry analysis. Blood. 2019;134:269. [Google Scholar]
- 14.Kumar SK, Dispenzieri A, Fraser R, Mingwei F, Akpek G, Cornell R, et al. Early relapse after autologous hematopoietic cell transplantation remains a poor prognostic factor in multiple myeloma but outcomes have improved over time. Leukemia. 2018;32:986–95. doi: 10.1038/leu.2017.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chng WJ, Braggio E, Mulligan G, Bryant B, Remstein E, Valdez R, et al. The centrosome index is a powerful prognostic marker in myeloma and identifies a cohort of patients that might benefit from aurora kinase inhibition. Blood. 2008;111:1603–9. doi: 10.1182/blood-2007-06-097774. [DOI] [PubMed] [Google Scholar]
- 19.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 20.Chibon F, Lagarde P, Salas S, Pérot G, Brouste V, Tirode F, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010;16:781–7. doi: 10.1038/nm.2174. [DOI] [PubMed] [Google Scholar]
- 21.Chung TH, Mulligan G, Fonseca R, Chng WJ. A novel measure of chromosome instability can account for prognostic difference in multiple myeloma. PLoS ONE. 2013;8:e66361. doi: 10.1371/journal.pone.0066361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreaux J, Klein B, Bataille R, Descamps G, Maïga S, Hose D, et al. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica. 2011;96:574–82. doi: 10.3324/haematol.2010.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickens NJ, Walker BA, Leone PE, Johnson DC, Brito JL, Zeisig A, et al. Homozygous deletion mapping in myeloma samples identifies genes and an expression signature relevant to pathogenesis and outcome. Clin Cancer Res. 2010;16:1856–64. doi: 10.1158/1078-0432.CCR-09-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Decaux O, Lodé L, Magrangeas F, Charbonnel C, Gouraud W, Jézéquel P, et al. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myélome. J Clin Oncol. 2008;26:4798–805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- 25.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 26.Shaughnessy JD, Jr., Qu P, Usmani S, Heuck CJ, Zhang Q, Zhou Y, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood. 2011;118:3512–24. doi: 10.1182/blood-2010-12-328252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–57.. [Google Scholar]
- 29.Blagus R, Lusa L. SMOTE for high-dimensional class-imbalanced data. BMC Bioinform. 2013;14:106. doi: 10.1186/1471-2105-14-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fallahi A, Jafari S. An expert system for detection of breast cancer using data preprocessing and bayesian network. Int J Adv Sci Technol. 2011;34:65–70. [Google Scholar]
- 31.Batuwita R, Palade V. microPred: effective classification of pre-miRNAs for human miRNA gene prediction. Bioinformatics. 2009;25:989–95. doi: 10.1093/bioinformatics/btp107. [DOI] [PubMed] [Google Scholar]
- 32.Xiao J, Tang X, Li Y, Fang Z, Ma D, He Y, et al. Identification of microRNA precursors based on random forest with network-level representation method of stem-loop structure. BMC Bioinform. 2011;12:165. doi: 10.1186/1471-2105-12-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacIsaac KD, Gordon DB, Nekludova L, Odom DT, Schreiber J, Gifford DK, et al. A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics. 2005;22:423–9. doi: 10.1093/bioinformatics/bti815. [DOI] [PubMed] [Google Scholar]
- 34.Furqan MS, Siyal MY. Random forest Granger causality for detection of effective brain connectivity using high-dimensional data. J Integr Neurosci. 2016;15:55–66. doi: 10.1142/S0219635216500035. [DOI] [PubMed] [Google Scholar]
- 35.Zhou L-T, Cao Y-H, Lv L-L, Ma K-L, Chen P-S, Ni H-F, et al. Feature selection and classification of urinary mRNA microarray data by iterative random forest to diagnose renal fibrosis: a two-stage study. Sci Rep. 2017;7:39832. doi: 10.1038/srep39832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toth R, Schiffmann H, Hube-Magg C, Büscheck F, Höflmayer D, Weidemann S, et al. Random forest-based modelling to detect biomarkers for prostate cancer progression. Clin Epigenet. 2019;11:148. doi: 10.1186/s13148-019-0736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sievers QL, Petzold G, Bunker RD, Renneville A, Słabicki M, Liddicoat BJ, et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science. 2018;362:eaat0572. [DOI] [PMC free article] [PubMed]
- 38.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578:94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–48.. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong PSY, Chng W-J, de Mel S. STAT3: a promising therapeutic target in multiple myeloma. Cancers. 2019;11:731. doi: 10.3390/cancers11050731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phaik JuT, Tae-Hoon C, Pamela YZC, Sabrina HMT, Wee Joo C. IL6R-STAT3-ADAR1 (P150) interplay promotes oncogenicity in multiple myeloma with 1q21 amplification. Haematologica. 2020;105:1391–404. doi: 10.3324/haematol.2019.221176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chong PSY, Zhou J, Lim JSL, Hee YT, Chooi JY, Chung TH, et al. IL6 Promotes a STAT3-PRL3 Feedforward Loop via SHP2 Repression in Multiple Myeloma. Cancer Res. 2019;79:4679–88. doi: 10.1158/0008-5472.CAN-19-0343. [DOI] [PubMed] [Google Scholar]
- 43.Kolosenko I, Grander D, Tamm KP. IL-6 activated JAK/STAT3 pathway and sensitivity to Hsp90 inhibitors in multiple myeloma. Curr Med Chem. 2014;21:3042–7. doi: 10.2174/0929867321666140414100831. [DOI] [PubMed] [Google Scholar]
- 44.Soekojo CY, de Mel S, Ooi M, Yan B, Chng WJ. Potential clinical application of genomics in multiple myeloma. Int J Mol Sci. 2018;19:1721. [DOI] [PMC free article] [PubMed]
- 45.Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–21. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 46.Chng WJ, Ahmann GJ, Henderson K, Santana-Davila R, Greipp PR, Gertz MA, et al. Clinical implication of centrosome amplification in plasma cell neoplasm. Blood. 2006;107:3669–75. doi: 10.1182/blood-2005-09-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33:3911–20. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teoh PJ, An O, Chung T-H, Chooi JY, Toh SHM, Fan S, et al. Aberrant hyperediting of the myeloma transcriptome by ADAR1 confers oncogenicity and is a marker of poor prognosis. Blood. 2018;132:1304–17. doi: 10.1182/blood-2018-02-832576. [DOI] [PubMed] [Google Scholar]
- 49.Cottini F, Hideshima T, Suzuki R, Tai YT, Bianchini G, Richardson PG, et al. Synthetic lethal approaches exploiting dna damage in aggressive myeloma. Cancer Discov. 2015;5:972–87. doi: 10.1158/2159-8290.CD-14-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Botrugno OA, Bianchessi S, Zambroni D, Frenquelli M, Belloni D, Bongiovanni L, et al. ATR addiction in multiple myeloma: synthetic lethal approaches exploiting established therapies. Haematologica. 2020;105:2440–7. doi: 10.3324/haematol.2018.215210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teoh PJ, Chung T-H, An O, Chng P, Jeyasekharan A, Yang H, et al. Synthetic lethality in multiple myeloma harboring double oncogenic hits of 17p13(del) and 1q21(amp) Clin Lymphoma Myeloma Leuk. 2019;19:e50–e1. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures, tables and details of supplementary files