Abstract
Infant electroencephalography (EEG) presents several challenges compared with adult data: recordings are typically short and heavily contaminated by motion artifacts, and the signal changes throughout development. Traditional data preprocessing pipelines, developed mainly for event-related potential analyses, require manual steps. However, larger datasets make this strategy infeasible. Moreover, new analytical approaches may have different preprocessing requirements. We propose an Automated Pipeline for Infants Continuous EEG (APICE). APICE is fully automated, flexible, and modular. The use of multiple algorithms and adaptive thresholds for artifact detection makes it suitable across age groups and testing procedures. Furthermore, the preprocessing is performed on continuous data, enabling better data recovery and flexibility (i.e., the same preprocessing is usable for different analyzes). Here we describe APICE and validate its performance in terms of data quality and data recovery using two very different infant datasets. Specifically, (1) we show how APICE performs when varying its artifacts rejection sensitivity; (2) we test the effect of different data cleaning methods such as the correction of transient artifacts, Independent Component Analysis, and Denoising Source Separation; and (3) we compare APICE with other available pipelines. APICE uses EEGLAB and compatible custom functions. It is freely available at https://github.com/neurokidslab/eeg_preprocessing, together with example scripts.
Keywords: Preprocessing, EEG, MEG, ERP, Infant, Development
1. Introduction
Electroencephalography (EEG) is a valuable tool for developmental cognitive studies as it provides a non-invasive, direct, and low-cost measure of neural activity with high temporal resolution. However, the employment of this technique embeds two major challenges. First, the EEG signal is unavoidably contaminated by many artifacts from different sources, such as environmental factors (e.g., line noise), physiological phenomena (e.g., ocular movements, heartbeats, muscle activity), and movements, whose amplitudes are often much larger than the neural signal. Second, the neural signal relative to the cognitive processes under investigation is lost among many other computations overlapping in time and space due to the wide diffusion of the electrical fields. One successful solution to isolate a cognitive process is to average across many trials to recover a reproducible neural activity time-locked to the stimulus presentation, i.e., the event-related potential (ERP). For the averaging method to be successful, many trials are needed, and those trials should not be contaminated by high amplitude events, whose impact on the average could not be eliminated without thousands of trials. Other EEG analysis techniques (e.g., multivariate pattern decoding, time-frequency analyses) have similar constraints, requiring a high number of trials without too large variability. Critically, these demands stand at odds with the testing circumstances encountered with infants (short recordings often heavily contaminated by motion), calling for a specific approach to obtain a sufficiently good signal-to-noise ratio despite these challenging recording conditions.
Simple steps such as filtering can remove some artifacts, e.g., line noise, but correcting physiological artifacts requires more sophisticated methods (Islam et al., 2016). Fortunately, when EEG data is acquired with high-density systems, high redundancy in the signal (caused by the diffusion of the electric field) allows the implementation of different signal reconstruction techniques (Jiang et al., 2019). In this regard, several pipelines and physiological artifact removal algorithms have been developed for adult EEG (e.g., PREP (Bigdely-Shamlo et al., 2015), Automagic (Pedroni et al., 2019), FASTER (Nolan et al., 2010), ADJUST (Mognon et al., 2011), MARA (Winkler et al., 2011)). However, these tools are not well suited to the challenging infant data for several reasons. First, the correction methods currently available for physiological artifacts require long recordings without high amplitude artifacts (Onton and Makeig, 2006), a condition difficult to achieve in infants. Second, the power spectrum (Eisermann et al., 2013, Marshall et al., 2002) and the properties of the evoked responses (Kushnerenko et al., 2002, Nelson and Monk, 2001) evolve throughout development due to maturational changes. More specifically, infant background activity is rich and ample in low frequencies, and the signal variability between trials is much higher in infants than in adults (Naik et al., 2021). Third, exogenous artifacts vary according to infants’ age (e.g., fewer blinks and less motion in younger infants). Lastly, most developmental datasets do not include electrocardiogram (ECG) and electromyogram (EMG) recordings usually used to identify non-cortical activity contaminating the EEG. Due to all these factors, the methods developed for adult EEG are ill-suited for infant studies, and no agreement has been reached on the most appropriate preprocessing procedures for infant EEG.
Traditional infant preprocessing relies on manually identifying non-functional channels and data segments contaminated by motion artifacts, which are subsequently discarded. However, high-density recording systems (64, 128, 256 electrodes) and longer recording sessions make this approach time-consuming and inefficient, revealing the need for automated pipelines. As a solution to automate the process and easily rule out high-amplitude events, a standard procedure is to determine thresholds below which the voltage should remain. Then, non-functional channels are either rejected or interpolated, but no additional correction of specific artifacts, such as physiological artifacts, is applied (e.g., Adibpour et al., 2018; Friedrich et al., 2015; Kabdebon and Dehaene-Lambertz, 2019; Winkler et al., 2009). Although straightforward, this method presupposes setting arbitrarily fixed thresholds, weighing the risk of a high rejection rate that might leave too few trials to obtain an ERP free of background activity and a low rejection rate that might retain an artifactual signal in the ERP. Setting an adequate fixed threshold is usually not possible because the signal’s amplitude depends on the distance between the channel and the reference; thus, fixed thresholds are more or less able to detect artifacts, likely being too sensitive for distant electrodes and not enough for close electrodes. Furthermore, given the changes in EEG amplitude as a function of age and sleep-wakefulness stages, fixed thresholds need to be re-adjusted for each particular dataset. Another drawback of the traditional approach is that artifacts’ detection is done on segmented data, limiting its applicability on analysis requiring longer data segments.
Nowadays, more complex paradigms are implemented to explore infants’ rich cognition (Friedrich et al., 2015, Kabdebon and Dehaene-Lambertz, 2019). The impatience of young subjects, the lack of verbal instructions, and the common need for familiarization periods mean that the amount of data in relevant conditions is scarce, such that maximum data retention without data quality loss becomes crucial. Furthermore, new analysis techniques with different requirements are now combined in the same study. For example, frequency tagging may require segmenting the data in longer epochs (e.g., de Heering and Rossion, 2015; Kabdebon et al., 2015) or multivariate decoding to retain as many trials as possible (Gennari et al., 2021), two demands hard to reconcile when there is a high degree of artifact contamination.
A few papers proposing automatic and more complex pipelines for developmental data have been recently published, HAPPE (Gabard-Durnam et al., 2018), MADE (Debnath et al., 2020), EEG-IP-IL (Desjardins et al., 2021), and EPOS (Rodrigues et al., 2021). All these pipelines include Independent Component Analysis (ICA) as a fundamental step for removing physiological artifacts providing different strategies for its proper application on infant data. However, in HAPPE, MADE, and EPOS, artifacts rejection is based on fixed thresholds, meaning they do not provide a specific way to deal with the high amount of motion artifacts present in infants’ recordings, which drastically affects ICA or any other blind separation technique. EEG-IP-IL (Desjardins et al., 2021) is, to our knowledge, the only pipeline offering more sophisticated methods to identify data contaminated by motion artifacts: after robust average reference, artifacts are individuated over short data segments based on too high voltage variance using relative thresholds.
Here we propose an Automated Pipeline for Infants Continuous EEG (APICE), in which automatized artifact detection is performed on the continuous data before any further preprocessing step. This pipeline stems from the needs we have encountered in our long practice of cognitive studies in infants. EEG consists of the superposition of multiple electrical sources, some relevant, some not, which are hard to disentangle. Consequently, our philosophy in preprocessing the data is to exclude outlier values to eliminate strong artifacts (mainly due to motion). To do so, we based our approach on the distribution of the voltage values of a specific recording (i.e., a specific channel in a specific individual). Furthermore, because in infants, data are scarce, but EEG is redundant, it is also possible to reconstruct transient artifacts instead of rejecting the recording. Finally, as experiments become more complex with several steps within a trial, we often have to work with epochs of different lengths depending on the type of analysis, which required re-doing the preprocessing. The solution is thus to preprocess the whole recording before epoching.
We developed APICE in a modular manner to provide high versatility and remarkable flexibility, such that it is suitable for a broad range of analyzes. Crucially, we aimed APICE to offer good data recovery while ensuring data quality across different developmental populations and inform the experimenter on the quality of the recording. Relatively to previous work, the key innovations we propose with APICE are (1) an iterative artifact detection procedure based on multiple algorithms applied on continuous data, (2) the use of automatically adapted thresholds applicable on non-average referenced data, and (3) the correction of transient artifacts on continuous data.
The use of multiple algorithms and adaptive thresholds makes APICE applicable without modifications across different ages and protocols. It also makes it easily adaptable to adult EEG datasets. The early detection of artifacts in the continuous data enables the experimenter to decide how to deal with artifacts before further preprocessing steps (e.g., re-referencing the data to the average, blind source separation methods). Additionally, the detection and correction of artifacts on continuous data increase data recovery by avoiding rejecting data segments containing transient voltage jumps. Finally, APICE is highly flexible, allowing the experimenter to use the same preprocessed data for many kinds of analysis.
Note that we built this pipeline to suit the needs of cognitive studies using high-density nets. Processing clinical data with a few electrodes has neither the same goal of high robustness at the individual level nor the same spatial redundancy to enable data interpolation. Nevertheless, we believe that most of the solutions proposed in APICE can be employed in many experimental and clinical situations if we consider the signal features and the purpose of the EEG recording. Therefore, we made all parameters adjustable, but we provided default values based on our experience.
In this paper, we first describe the general logic behind the pipeline and its different steps. Then, we validate APICE’s performance in terms of data quality and data recovery. Specifically, we evaluate the effect of the relative thresholds used for rejection by comparing three different values. We also compare APICE with a reduced version of it, in which we kept the automatic detection of artifacts in continuous data but removed the correction of transient artifacts. Then, we evaluate whether incorporating additional data cleaning methods described in the literature provides any improvement. Specifically, we tested ICA coupled with automatic rejection of components using iMARA (Haresign et al., 2021) and Denoising Source Separation (DSS) (De Cheveigné and Parra, 2014, de Cheveigné and Simon, 2008). Finally, we validated APICE by comparing it with a standard widely used preprocessing pipeline and with MADE (Debnath et al., 2020). We chose MADE between the available pipelines for developmental EEG because it allows ERPs analysis and has already been implemented in published infant EEG studies (e.g., Hwang et al., 2021; Troller-Renfree et al., 2020). We performed the validation on two datasets with very different properties, an auditory experiment in asleep neonates and a visual experiment in awake 5-month-olds.
APICE is implemented in MATLAB, and it uses the EEGLAB toolbox and custom functions compatible with the EEGLAB structure (Delorme and Makeig, 2004). It is freely available at https://github.com/neurokidslab/eeg_preprocessing, together with example scripts. APICE is modular, allowing functions to be easily recombined to meet different requirements. APICE also provides toolkit functions to modify events, correct event timings using Digital Input events (DINs), add information about trial features, and obtain average ERPs.
2. Pipeline general description
APICE uses EEGLAB (Delorme and Makeig, 2004) functions for standard processing steps (e.g., importing the data, filtering, epoching) and includes new functions for more specific steps. APICE includes the following crucial additions. (1) The identification of motion artifacts on continuous data using relative thresholds applicable on single electrodes. (2) The correction of artifacts in the continuous data when they involve a few channels or affect a brief period. (3) The possibility to define contaminated samples and non-functional channels based on the rejected data. Independently from our work, a similar procedure has been recently developed for EEG-IP-L (Desjardins et al., 2021). (4) Once recordings are segmented into epochs, the definition of bad epochs is based on the amount of rejected data within the epoch, specifically, in terms of time samples and electrodes rejected.
Additionally, APICE includes functions that allow applying other standard data cleaning methods. For example, we provide a function to perform ICA, which omits the samples and channels previously identified as containing artifacts, uses Wavelet-thresholding before performing the ICA, and automatically identifies components associated with artifacts using iMARA (Haresign et al., 2021). We also provide a function to apply DSS, a method to clean ERP proposed by De Cheveigné and Parra, 2014, de Cheveigné and Simon, 2008.
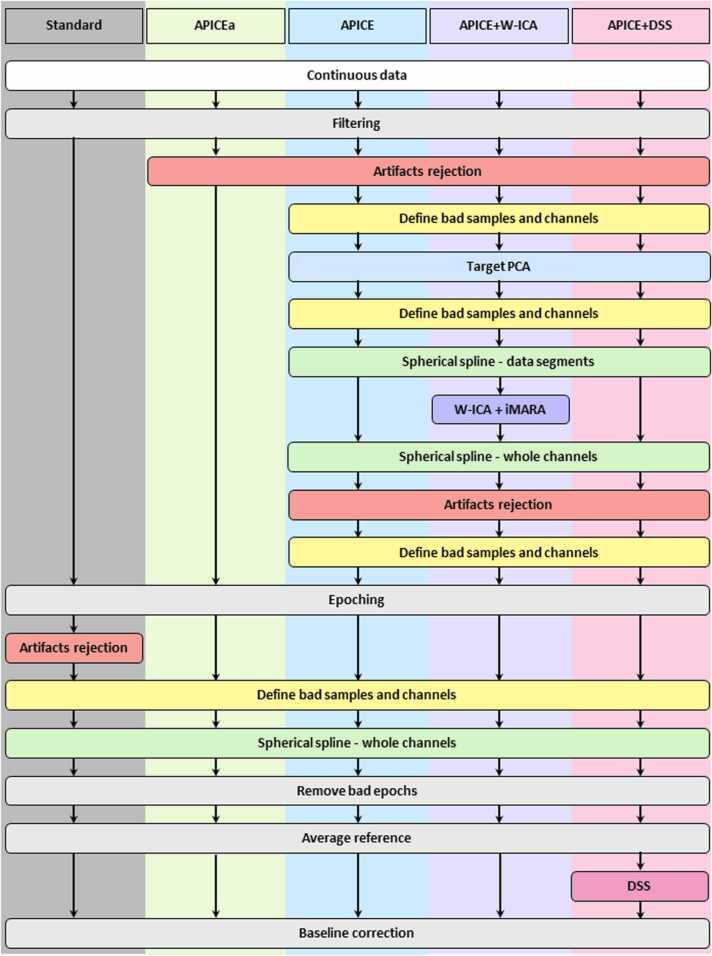
It is important to note that all the functions and steps are modular, offering flexibility to the user. Although we propose a recipe to perform the different steps in a specific order to obtain optimal results, the functions can be re-combined according to particular needs. Based on our research and clinical experience in infant EEG data, we propose the following steps for preprocessing infant EEG data (Fig. 1).
-
1.
Importing the data to EEGLAB.
-
2.
Minimal filtering of the data.
-
3.
Detecting artifacts.
-
4.
Correcting artifacts when possible.
-
5.
Re-detecting artifacts.
-
6.
Applying ICA (optional).
-
7.
Epoching.
-
8.
Rejecting bad epochs.
-
9.
Applying DSS (optional).
-
10.
Re-referencing (averaging reference), data normalization, baseline correction (optional).
Fig. 1.
Schematic description of different preprocessing pipelines. Standard corresponds to a common basic preprocessing pipeline usually used on infant experiments. APICEa is a reduced version of our pipeline in which artifacts detection is performed on continuous data using multiple algorithms and relative thresholds (see Section 2.2). APICE is our full preprocessing pipeline, including correcting transient artifacts on continuous data (see Section 2.4). APICE + W-ICA is APICE with the addition of ICA. APICE+DSS is APICE with the addition of DSS.
Note that the steps after epoching (step 7) are specific to ERP analyses or other analyzes involving evoked activity (i.e., an averaging process across trials to recover reproducible time-locked activity).
2.1. Importing the data
APICE is based on EEGLAB (a free MATLAB toolbox) (Delorme and Makeig, 2004). We provide the codes to import the data exported from the EGI system (Electrical Geodesics, Inc) in the EEGLAB format. However, APICE can be implemented on any data imported to the EEGLAB format. It is worth noticing that in many systems, such as EGI, voltages of all electrodes are recorded relative to a single electrode, the reference (e.g., the mastoids, the vertex). With systems using an active and a passive electrode to generate a common-mode voltage (e.g., Biosemi), the data needs to be referenced to one of the electrodes (e.g., the mastoids, the vertex) when imported to remove the common-mode signal.
2.2. Filtering
As the first preprocessing step, data are filtered to remove common environmental artifacts. We use a low-pass filter below the line noise frequency (e.g., with a line noise at 50 Hz, we low-pass filter the data at 40 Hz). Unless high-frequency activity needs to be investigated, low-pass filtering is, in our experience, the most effective way to remove line noise.
High pass filtering enables the removal of drifts and slow activity in the data. Low-frequency noise strongly contaminates high impedance EEG recordings, mainly due to skin potential (Kappenman and Luck, 2010). Furthermore, slow waves are common in very young infants' EEG recordings (Eisermann et al., 2013, Marshall et al., 2002, Selton et al., 2000) due to immaturity and partially to the brain's movement in the skull following respiration. For most EEG analyses, it is imperative to reduce this contamination. However, extensive filtering (above 0.1 Hz) can introduce critical distortions in the data (de Cheveigné and Nelken, 2019). While alternatives to high-pass filtering exist, such as local detrending methods (de Cheveigné and Arzounian, 2018), we have not observed better performance of these methods. Therefore, we apply a high pass filter at 0.1 Hz at this early stage of preprocessing, which should not introduce critical distortions in the data but remove the main drifts. Using a very low high-pass filter at the initial stage brings flexibility since the same continuous preprocessed data can be used for analysis requiring different filtering levels.
We use the pop_eegfiltnew EEGLAB function, which uses the FIRfilt plugin, to perform a non-causal Finite Impulse Response (FIR) filter. We first apply a low-pass filter at 40 Hz with a transition band of 10 Hz. We then apply a high-pass filter at 0.1 Hz with a transition band of 0.1 Hz.
It is worth noticing that while low-pass filters can be applied at any preprocessing stage, high-pass filters should always be performed on continuous data to avoid edge effects.
2.3. Artifacts detection
One of APICE’s key innovations is detecting artifacts, which is done automatically. Artifacts are identified on the continuous recording before re-referencing the data to the average and through adaptive rather than absolute thresholds set per subject and electrode. As we discuss below, these features are decisive for an adequate and versatile preprocessing pipeline for infant recordings.
Non-working electrodes have a response that deviates radically from the rest of the channels, with amplitudes much higher or lower than expected. However, in unipolar recordings (i.e., the signal is recorded as the difference of potential between each electrode and a single reference), the amplitude of the signal varies in function of the distance of each electrode to the reference electrode — with electrodes closer to the reference having smaller amplitudes than more distant ones. Thus, absolute thresholds (classically used in standard pipelines) differently penalize each channel: when the signal’s amplitude is already large, a slight supplementary increase might be interpreted as an artifact, whereas the same deviation in electrodes close to the reference might remain undetected. Therefore, the same noise level is not similarly detected across electrodes, and the percentage of data rejected in each channel might be very different at the end of the process.
A solution to homogenize the voltage in high-density recordings is to measure the potential relative to an ideal reference, such as the average reference (Bertrand et al., 1985). The integrated scalp potential must be null and can be approximated by the average over numerous homogeneously distributed channels. Re-referencing the data to the average results in a null integrated potential by subtracting the average voltage at each time point to each electrode. Crucially, the data must be clean to avoid affecting functional channels with contaminated data through the average process.
We thus face a circular problem: to estimate non-functional channels based on their amplitude, we need to re-reference the data to the average, but to obtain a proper estimation of the average reference, we need clean data. Pipelines, such as PREP (Bigdely-Shamlo et al., 2015), overcome this problem by computing a robust average reference. This procedure consists of computing a first average across channels, then detecting and interpolating bad channels, and computing a new average reference, steps that can be iterated to obtain more accurate results. However, this procedure does not properly deal with artifacts contaminating only subgroups of electrodes within a limited portion of time (e.g., electrodes that dry or lose contact because the infant touches them), a common situation in developmental studies employing wet high-density nets. In APICE, we use multiple algorithms sensitive to different signal features that allow detecting artifacts without an initial estimation of the average reference. This is possible because some algorithms are independent of the signal amplitude (e.g., the correlation among electrodes, or the proportion of power in certain frequency bands), while for others, thresholds are adapted to the signal properties of each electrode, allowing the identification of channels not working during restricted periods. Once artifactual data has been identified, it can be excluded to obtain a first yet robust average reference before applying algorithms requiring average referenced data. Later, aberrant data can be interpolated before computing the final robust estimation of the average reference based on clean data.
Adaptive thresholds present another crucial advantage; they do not have to be customized to each population or testing procedure (i.e., age group, resting-state, or active task), providing and standard procedure to all datasets. Since the amplitude and properties of the signal radically change during development, and between-subjects variability is considerable, using adaptive thresholds is an essential feature for infant EEG.
The detection of artifacts in continuous data rather than in segmented epochs also has its advantages. First, the properties of the EEG recording and the threshold for the different artifacts can be better estimated. Second, some transient artifacts that may lead to the rejection of the epoch can be corrected, facilitating the recovery of more data. Third, it provides more flexibility, enabling performing multiple types of analysis using a common preprocessing pipeline.
All the algorithms used in APICE compute a measure for each sample or in a sliding time window. Then, the algorithms reject the data when the measure is above or/and below a threshold. While all our functions allow using absolute thresholds, we advise using relative thresholds. Relative thresholds are determined based on the distribution of the measure throughout the entire recording. More precisely, a threshold is computed as, , and/or , where and are the first and third quartiles of the distribution. should be provided as an input (by default 3). Because the measures used by the algorithms usually have a normal distribution, this threshold definition successfully identifies extreme values (i.e., outliers). The functions allow defining a single threshold for all electrodes or individual thresholds per electrode. We also provide the option of computing the algorithms in data z-scored per channel or on average reference data. While the algorithms might implement some data transformations for detection, the output data is never modified.
Different algorithms are sensitive to different artifacts in the data. Using a collection of algorithms and methods enables a large and complete detection of artifacts of all kinds. We can distinguish three groups of algorithms based on their sensitivity. (1) Algorithms that are particularly apt to individuate non-functional channels. Specifically, one of them looks at the power spectrum of the different channels across frequency bands, and another detects channels with very low activity correlation with other channels (similar to PREP (Bigdely-Shamlo et al., 2015)). (2) Algorithms sensitive to motion artifacts, resulting in high amplitudes and signal variance. These algorithms detect when the signal’s amplitude, temporal variance, or running average are too high. (3) An algorithm that identifies when the signal changes too rapidly and serves to detect jumps or discontinuities in the signal.
All these artifact detection algorithms define for each sample and channel if the data is clean or contaminated, and the information is stored in a logical matrix, BCT, of the size of the recording (i.e., channels × samples × epochs (epochs is equal to 1 in case of continuous recording)), where a true value indicates the presence of artifacts. Being the outcome of heterogeneous detection strategies, the rejection matrix obtained at this point is likely to present a “salt and pepper” structure. According to the neighborhood context, a final group of algorithms (4) refines this rejection pattern through further (minor) exclusions or data reintegration. For example, they rule out short data segments (shorter than 2 s) sandwiched between rejected segments or re-include very short rejected data segments (shorter than 20 ms). A full description of all the algorithms and functions is provided in Appendix A.
APICE detects artifacts through multiple cycles of rejection, and the data rejected in one cycle is no longer entered into the signal estimation used to construct adaptive rejection thresholds for subsequent cycles. Considering that the distribution for the different measures in the absence of artifacts is normal, once extreme outliers are rejected in the first cycle of rejection, subsequent cycles reject very little or no data. In brief, these multiple cycles allow a progressive skimming of the signal. We propose the following rejection cycles:
-
•
Rejection cycle 1 includes the algorithms that primarily identify channels not working based on their power spectrum and their absence of correlation with other channels.
-
•
Rejection cycle 2 rejects all data with an amplitude higher than 500 μV (non-average referenced data). This absolute threshold is very high and is only used to avoid taking very large amplitude data into account. This step accelerates the reiteration procedure but can be skipped without substantial changes.
-
•
Rejection cycles 3a and 3b apply twice the algorithms sensitive to motion artifacts using relative thresholds per electrode and non-average referenced data. Specifically, the algorithms reject data with a too high amplitude, too high variance, or too high running average relative to the distribution of each electrode.
-
•
Rejection cycles 4a and 4b apply the same algorithms as in the third cycle twice, but this time on average referenced data and using a single relative threshold across all electrodes. Note that outlier values identified in the previous cycles are not considered for the estimation of the average reference (robust average referencing).
-
•
Rejection cycles 5a and 5b detect fast transient changes in the signal, once using one threshold per electrode on non-average reference data (5a) and one on average reference data and defining a single threshold across all electrodes (5b).
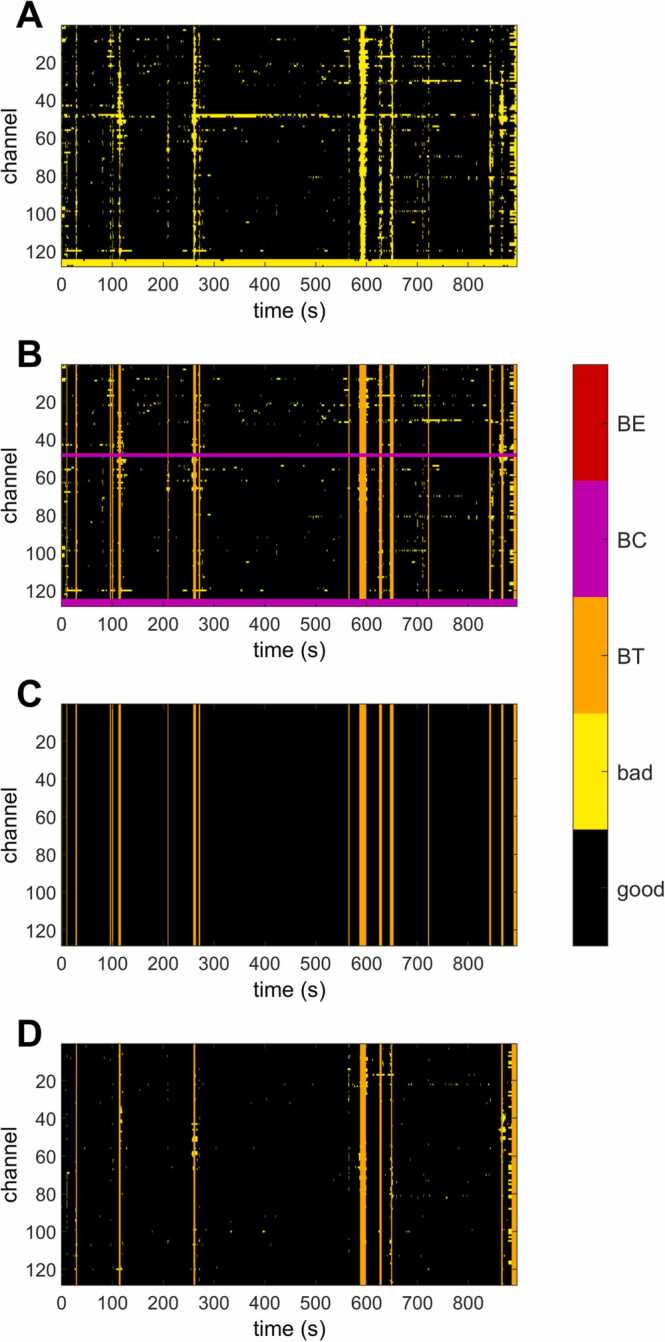
After artifact detection (Fig. 2A), segments heavily contaminated by artifacts (bad times or BT) and non-functional channels (bad channels or BC) are defined (Fig. 2B) (see Section 2.4). Then, artifacts are corrected (Fig. 2C) (see Section 2.5). Finally, the rejection matrix is reset, and the artifact detection algorithms are rerun to detect artifacts in the “clean” data (Fig. 2D). Only the algorithms to detect motion artifacts (cycles 2–4) are rerun in the final artifacts rejection step, as the non-functional channels and jumps in the signal, when possible, were already corrected.
Fig. 2.
Example of the rejection matrixes (channels × time-samples) for one subject obtained during successive preprocessing stages. BE = Bad Epochs, BC = Bad Channels, BT = Bad Times. Bad refers to epochs/channels/times identified as containing artifacts. Notice that the example refers to continuous data; thus, there are no bad epochs. (A) Rejection matrix after the artifacts’ detection. (B) Identification of bad channels (pink) and bad time samples (orange). (C) Rejection matrix with bad times and channels after interpolating localized artifacts (bad channels and transient artifacts). Notice that all rejected samples, except bad times, have been corrected. (D) Rejection matrix after applying the artifacts detection algorithm again. Notice that most of the corrected transient artifacts (difference between B and C) are no longer identified as artifacts when the artifact detection algorithms are applied again (D), meaning that the correction of transient artifacts was successful.
2.4. Definition of bad samples and channels
The rejection matrix is a logical matrix of the size of the data indicating good and artifacted data (i.e., outliers). However, thresholds, even relative, are only a workaround method to reject contaminated data. Thus, some data points not identified as outliers can still contain artifacts, or the data quality of the neighboring points can be too strongly contaminated to be trustable. For example, it is probably the case of short “good” periods sandwiched between “bad” periods and of samples where most of the channels, but not all, are considered “bad”. Therefore, we defined bad-times (BT) to refer to time samples strongly contaminated by artifacts (i.e., those for which most of the channels are identified as “bad”) and bad-channels (BC) non-functional channels (i.e., channels for which most of the time was identified as “bad”). These definitions allow to easily exclude from future analysis samples containing large artifacts that cannot be corrected and reconstruct non-functional channels. A similar logic has been recently also implemented in EEG-IP-L (Desjardins et al., 2021). The functions used to define BT and BC are described in Appendix B. In Fig. 2, we present an example of a rejection matrix and the definition of BT and BC.
From our practice, we advise defining periods in which artifacts affect more than 30% of functional electrodes as bad times. Afterward, we remove the bad time tag of segments shorter than 100 ms. Next, we tag as bad the 500 ms before and after any bad time segment and short time segments of less than 1 s. Two reasons motivate these last choices: first, because data samples surrounding motion artifacts tend to be partially contaminated, and second, to avoid interpolating non-functional channels close to motion artifacts. As a result, we obtain BT, a logical matrix of size 1 × samples × epochs signaling strongly contaminated data segments.
The channels with a proportion of bad tags in BCT relative to the number of good times (i.e., the total number of samples excluding the ones previously identified as bad in BT) higher than a certain threshold are marked as bad. The process is applied either at the epoch level or across all epochs. At the epoch level, it means that the total number of good times is computed for each epoch, and bad channels are marked in BC, a logical matrix of size channels × 1 × epochs. During the whole recording implies that the number of good samples is computed across all epochs, and bad channels are therefore identified across the whole data and marked in Bcall, a logical matrix of size channels × 1 × 1. Notice that the distinction between BC and Bcall is only necessary when working with epoched data. In continuous data, we usually define BCall as those channels with bad tags during more than 30% of the good times. In a single epoch, we defined as BC those channels with bad tags during good times lasting more than 100 ms. We chose to use 100 ms for consistency (i.e., it is the same limit value we use for the definition of bad times because periods shorter than this are probably false positives). However, a change in this parameter within certain limits (from 100 ms to ~ 1 s) does not affect the results because electrodes usually stop working for periods much longer than 100 ms.
2.5. Correction of localized artifacts
Artifact correction is a critical point because it implies data reconstruction, in other words, the estimation and removal of the artifacts or the interpolation of the contaminated segments. Data segments containing large artifacts (i.e., motion artifacts affecting most electrodes during a period) are very common in infant recordings but unfortunately cannot be reconstructed and need to be discarded. Thus, the best we can do is to identify them accurately. These periods correspond to the bad times defined in the previous section.
By contrast, other types of artifacts can be corrected. For example, we have already described the removal of electrical noise by filtering, and many methods exist to correct physiological noise (Islam et al., 2016, Jiang et al., 2019). We will specifically discuss the use of ICA to clean this type of artifact in the next section. This section discusses data reconstruction when artifacts are localized in either time or space (channels). A detailed description of the function for data correction is presented in Appendix C.
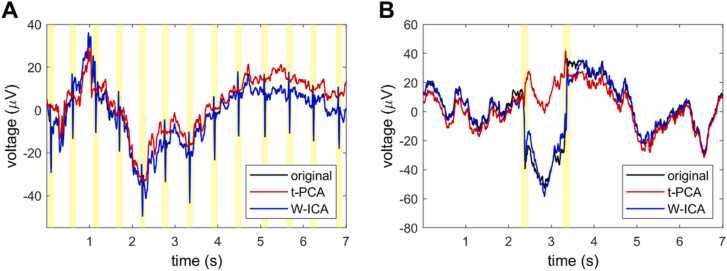
Transient artifacts, like jumps and discontinuities, frequently contaminate the EEG signal. To remove this type of artifact, we apply a target PCA and remove the first components, a procedure already implemented on Near-Infrared Spectroscopy data (Yucel et al., 2014). The underlying assumption is that, during these periods, the majority of the variance can be attributed to the artifact. Crucially, this approach is restricted to very brief data segments showing big amplitude jumps in the signal. Therefore, the first components carrying the higher variance mainly contain the artifact. Indeed, even if all the variance is removed (i.e., the data is replaced by a flat segment), this approach remains better than losing a longer data segment or keeping the artifact because the artifact is brief but of high amplitude. Contrary to blind source separations methods, as ICA or PCA applied to the whole recording, in this procedure, the PCA is restricted to specific events, limiting the undesired removal of neural activity. We used it for segments shorter than 100 ms and removed the first components carrying 90% of the variance (see Appendix C for more details). The time limit for the length of the artifacts circumscribes this correction to jumps in the signal and other fast events such as heartbeats (see Fig. 3).
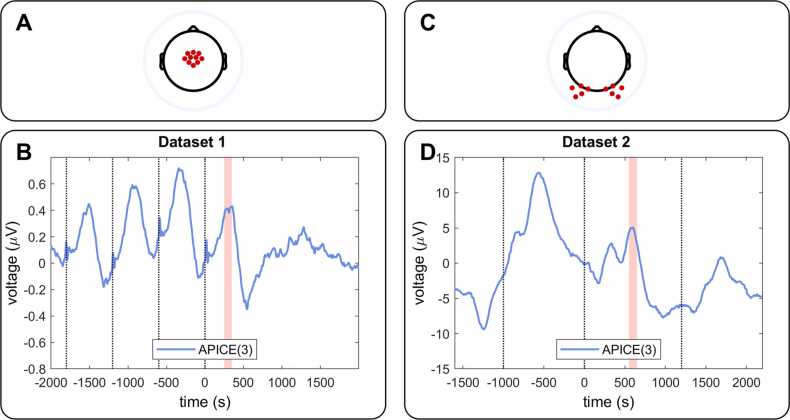
Fig. 3.
Example of heartbeat (A) and a jump in the signal (B) artifact correction by target-PCA and ICA. The shaded area represents the samples identified as having artifacts and on which target-PCA is applied. The figure shows the original data (original: black), the data preprocessed using APICE(3) + W-ICA (W-ICA: blue), and the data preprocessed APICE(3) (t-PCA: red). The original data and the W-ICA data overlap, depicting how W-ICA fails to remove the heartbeat and the jump in the signal. The target-PCA removes the artifacts but introduces drifts in the signal affecting low-frequency activity (below the high-pass filter used to remove the drifts).
Another possible scenario is that artifacts affect a small number of electrodes. For these cases, a widely used reconstruction method is the spatial interpolation of the channels using a spherical spline (Perrin et al., 1989). In adult experiments, electrodes are identified as non-working during the whole recording and eventually spatially interpolated. However, with infant high-density systems, it is common that some channels stop working during limited periods (e.g., the infant touches some electrodes, making them lose contact, or after a child’s movement, the electrode moves before returning to its original position). To account for these scenarios and recover as much data as possible, we reconstruct the channels identified as bad during the whole recording and restricted periods using spatial interpolation. It is important to highlight that while the reconstruction of non-functional channels is a common practice that can simplify the between-subjects analysis, the signal is estimated as a weighted sum of the good electrodes. Thus, it does not add new information, and the dataset becomes rank deficient.
In our practice, we apply the artifacts correction as follows. First, we correct transient artifacts using target PCA. Second, we spatially interpolate channels not working during a certain period. We restrict this interpolation to good times, i.e., samples with less than 30% of the channels tagged as bad. The underlying assumption is that when too many channels are affected by an artifact at a given time, the spatial interpolation is ineffective. Finally, we interpolate channels rejected during the whole recording (i.e., BCall). By applying the functions in this order, transient artifacts are corrected before non-functional channels, minimizing the use of contaminated data to reconstruct non-functional channels. Fig. 2C shows the rejection matrix after interpolation. Observe how bad data remains only during bad times.
2.6. Independent Component Analysis
ICA is commonly applied on multichannel recordings of EEG data to remove physiological noise (e.g., eye blinks, eye-motions, muscle activity, heartbeat). The recording is decomposed into temporally independent components (IC). Then, the components related to artifacts are identified either automatically or manually by their topography, temporal profile, and power spectrum and removed from the data. Ideally, each IC is related to a neural or non-neural source. Therefore, ICA removes specific artifacts (sources) from the data without discarding the EEG segments affected by that artifact. Removing a specific IC alters the entire recording, potentially eliminating genuine neural activity. Thus, the successful application of ICA for EEG data cleaning depends on (1) an appropriate separation of non-neural sources in distinct components and (2) their proper identification.
ICA is a common practice in the preprocessing of adult data. However, its implementation in infant data is not widespread, and its benefits are unclear. The poor performances of the method are partially due to the nature of infant EEG data. Young infants’ recordings contain more slow waves than adult data (Eisermann et al., 2013, Marshall et al., 2002), ERPs are less precise in time (Kushnerenko et al., 2002), and more variable (Naik et al., 2021). The nature of the artifacts is also different in infants and adults. Adults are often quiet and attentive, with a low voltage EEG. Thus, artifacts are rare and have a markedly different signal, contrary to infants. All these factors hamper a successful separation of neural and non-neural sources. A second reason for the lower performances is that algorithms for automatic identification of artifact-related IC have been developed primarily for adult data. Recently, some algorithms have been adapted for developmental data. For example, the ADJUST algorithm (Mognon et al., 2011) has been optimized into adjusted-ADJUST (Leach et al., 2020) using 6-year-old children recordings and the MARA algorithm (Winkler et al., 2011) into iMARA (Haresign et al., 2021) using training recordings from 10-month-old infants. However, the continuous developmental change of the signal (e.g., changes in the power spectrum profile and ERP due to maturation of the neural circuits, changes in the diffusive properties of the skull due to its maturation) may hinder the classification and degrade the algorithm’s performance.
A good ICA decomposition requires several considerations. In order to obtain a reliable separation in ICs, the data must be high-pass filtered at least at 1 Hz and should not contain high amplitude noise (e.g., motion artifacts) (Winkler et al., 2015). However, high-pass data filtering may not be suitable for many EEG analyses (e.g., ERPs are distorted, and slow waves may be lost). We implemented a standard solution consisting of high-pass filtering and applying ICA to a copy of the data. In this way, the non-neural components are estimated and subtracted from the original data (Debnath et al., 2020).
To avoid high amplitude noise on the data, we restricted the ICA to the time samples tagged good and set the remaining bad data points to zero. Next, we performed a wavelet-thresholding on a first ICA (Geetha and Geethalakshmi, 2011, Johnstone and Silverman, 1997) to remove potentially remaining transient high amplitude artifacts. Then, on the clean data, we applied ICA again. The use of wavelet-thresholding on a first ICA decomposition improves the final ICA decomposition (Gabard-Durnam et al., 2018, Rong-Yi and Zhong, 2005).
Another important consideration is that the recording should fulfill , where m is the number of samples and n is the number of channels (Onton and Makeig, 2006). Unfortunately, infants’ recordings are generally not long enough to guarantee this condition. There are two alternatives to overcome this issue. One is to reduce the analysis to a subset of channels. The second possibility is applying PCA and retaining only the first components to reduce the problem’s dimensionality.
We do not regularly apply ICA in APICE (see the pipeline validation section for further discussion). When we apply it, we recommend that data are high-pass filtered at 2 Hz (Winkler et al., 2015), and we use PCA first to reduce the dimensionality of the problem. We have noticed that the variance lost by keeping only the first ~ 50 components is minimal with high-density nets, and results are more accurate than by reducing the number of channels because fewer channels entail a loss of spatial resolution and a decrease in the performance of the classification algorithms. Moreover, reducing the number of channels implies either not analyzing some of them or analyzing the data in multiple loops. Therefore, we opted to use PCA instead for short recordings.
Notice that for ICA, the signals need to be independent. Thus, in principle, ICA should be applied before the spatial interpolation of the non-functional channels. However, if PCA is applied first to reduce the dimensionality, ICA can be performed before or after interpolating non-functional channels. We use the iMARA algorithm (Haresign et al., 2021) to identify components associated with non-neural artifacts automatically. In Appendix D, we describe the function that performs ICA in APICE.
2.7. Definition of bad epochs
Once continuous recordings are segmented into epochs, we can define bad epochs that should not be considered in subsequent analyses. An epoch is defined as bad if any of the following three criteria is present: 1) it contains any bad time; 2) it contains more than 30% of bad channels; 3) if more than 50% of the data was interpolated. Note that 30% is the limit in the proportion of channels to define bad times; thus, the first two criteria overlap. The function to define bad epochs is described in Appendix E.
2.8. Denoising Source Separation (DSS)
Spatial filters are linear combinations of the sensors designed to partition the signal between components carrying the signal of interest from non-interest. In the particular case of the DSS, the spatial filter is designed to select components carrying evoked activity, meaning activity that is reproducible across trials from non-evoked activity. Thus, the method has been proposed as an alternative to clean ERPs (De Cheveigné and Parra, 2014, de Cheveigné and Simon, 2008). This data cleaning method is specific to the study of evoked activity because the activity that is not phase-locked to the stimuli is partially removed.
In Appendix D, we describe the function provided in APICE to perform DSS.
2.9. Preprocessing report
Within the EEGLAB structure, we register a report containing the size of the data, the amount of rejected data, and the amount of interpolated data after each data processing step. More specifically, we retain the number of channels, samples, epochs, the number of rejected and interpolated points, and the number of bad times, bad channels, and bad epochs. We also provide a function that prints a table in the command window and a text file to summarize these measures. The information is collected for all subjects at critical points during the pipeline. Specifically, it summarizes the rejection percentages before data epoching, before the rejection of bad epochs, and at the final stage. This summary information should enable evaluating the pipeline’s performance and detect possible problems in some participants.
A description of these functions can be found in Appendix E together with the report for the analysis performed using the APICE pipeline on datasets 1 and 2 as examples (Tables S1 and S2).
3. Pipeline validation
Since the “real” ERP is unknown, it is not banal to evaluate different pipelines. We can distinguish two ways in which different preprocessing steps can influence data quality. On the one hand, some processes might introduce a systematic bias in the measurement; thus, affecting accuracy. For example, this is the case of filtering that can modify the timing and shape of the response (de Cheveigné and Nelken, 2019) or blind source separation methods that might result in the removal of neural signal attenuating the ERPs (Haresign et al., 2021). On the other hand, other processes might affect the variability across different measurements of the same response (i.e., trials); thus, the precision, for example, when motion or physiological artifacts are not adequately removed. While a bias in the measurement results in a change of the grand average ERP, a loss of precision results in an increase in the error of the ERPs but not necessarily in a change in the grand average response (there is no systematic error across trials and participants). In brief, an increased error due to a loss of precision means a loss of statistical power but not a distortion of the evoked response.
Ideally, the goal is not to introduce biases in the signal and retain as much data as possible without losing precision. Therefore, we verified that no systematic biases were introduced by any method, and we compared the pipelines performances based on: (1) the proportion of retained epochs and (1) the standardized measurement error (SME) proposed by Luck et al. (2021). The SME measures the variability across epochs, with a lower SME implying that the responses across epochs are closer to each other. A smaller SME and more trials retained translate into a lower error for the ERP obtained for each subject and, therefore, an increase in statistical power in eventual statistical analysis.
To validate APICE, we first identified the relative thresholds for artifact detection that provides minimal data loss (high trial retention) with good data quality (low SME). To do so, we ran APICE with three different values: 2, 3, and 4 interquartile ranges (see Section 2.3), and we compared the performance. Then, we investigated whether different modifications of APICE bring any improvement. Specifically, we validated the interpolation of localized artifacts in the continuous data by comparing APICE with a reduced version of it, APICEa, which includes the artifacts’ detection in the continuous data, but not the interpolation of localized artifacts (Fig. 1). Additionally, we compared APICE with two other versions, including W-ICA and DSS filters, to evaluate if these cleaning algorithms improve the pipeline performance (Fig. 1). Finally, we validated APICE by comparing its performance with a Standard preprocessing pipeline (STD) and MADE (Debnath et al., 2020). The STD pipeline is a preprocessing consisting of minimal steps widely used in infants studies (e.g., Adibpour et al., 2018; Friedrich and Friederici, 2017; Kabdebon and Dehaene-Lambertz, 2019; Winkler et al., 2009). The data is filtered, epoched, and bad channels are identified on the segmented data using absolute thresholds and, when possible, reconstructed using spline-interpolation; thus, the procedure implies a minimal number of steps. MADE is a pipeline optimized for developmental data using ICA combined with adjusting-ADJUST (Leach et al., 2020) for automatic IC removal.
While APICE can be used to preprocess data that might be used in its continuous form, we decided to validate it using ERPs for several reasons. First, ERPs remain the most frequent method to study infant cognition. Second, data quality measures have been described for ERPs (Luck et al., 2021). Third, data quality measures for ERPs should be valid for any other type of analysis based on stimuli evoked responses (e.g., decoding, fast periodic stimulation).
To have a more robust validation, we analyzed two very distinct datasets. Significant changes occur in the EEG features during infant development (Eisermann et al., 2013, Marshall et al., 2002, Nelson and Monk, 2001), and the level of contamination by motion artifacts can vary considerably according to infants' age, vigilance, and the type of task. Therefore, we decided to use two datasets differing in infants’ age and vigilance, stimulation modality, and type of task. The first dataset corresponds to an auditory experiment in sleeping neonates, and the task consisted of passive listening to syllables. Motion artifacts were thus minimally contaminating the data. The second experiment corresponded to a visual task where 5-month-old infants looked at a sequence of images on the screen. The infants were awake and actively engaged in the task, resulting in strong motion artifacts in the data.
3.1. Datasets
3.1.1. Dataset 1: neonates dataset
The neonate’ dataset corresponds to an auditory experiment. During each trial, infants heard 4 or 5 syllables lasting 250 ms presented every 600 ms. 216 trials were presented to each infant. Scalp electrophysiological activity was recorded using a 128-electrode net (Electrical Geodesics, Inc.) referred to the vertex with a sampling frequency of 250 Hz. Neonates were tested in a soundproof booth while sleeping or during quiet rest. Participants were 24 (11 males), healthy-full-term neonates, with normal pregnancy and birth (gestational age > 38 weeks of gestation, Apgar score ≥ 7/8 at 1 and 5 min, and cranial perimeter ≥ 33.0 cm). All participants were tested at the Port Royal Maternity (AP-HP) in Paris, France. Parents provided informed consent.
3.1.2. Dataset 2: 5-month-old dataset
The 5-month-old infants’ dataset was a study investigating their capacity to associate two sets of images. During each trial, an attention grabber appeared on the center of the screen for 0.6 s, followed by a first image lasting 1 s, a second image lasting 1.2 s, and the attention grabber again during other 1.0–1.2 s. The experiment lasted until the infants were fussy (80–140 trials per participant). Scalp electrophysiological activity was recorded using a 128-electrode net (Electrical Geodesics, Inc.) referred to the vertex with a sampling frequency of 500 Hz. Infants were tested in a soundproof shielded booth while sitting in their parents’ lap. Participants were 26 (12 males), 22.98-weeks-old infants (SD 1.41, min 20.86, max 27). All participants were tested at NeuroSpin, in Gif/Yvette, France. Parents provided informed consent.
3.2. Preprocessing
We included different preprocessing approaches. The final steps were the same for all methods. After bad epochs were removed, data were average referenced, and baseline corrected over [− 100, 100] ms.
3.2.1. APICE pipeline
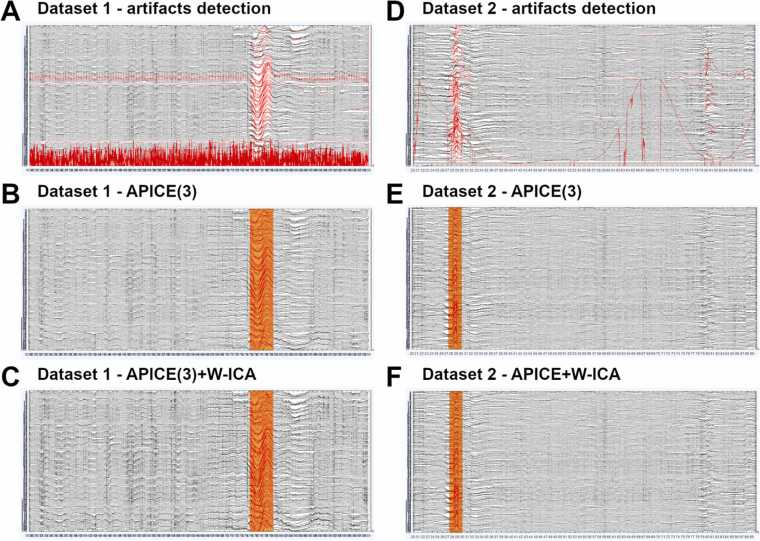
The APICE pipeline consisted of the steps described in each of the corresponding sections above. Data were filtered (low-pass filter at 40 Hz and high-pass filter at 0.1 Hz), and artifacts were detected on the continuous data (see Section 2.3). Afterward, bad times and channels were defined (see Section 2.4). Bad times were identified as those with more than 30% of the good channels rejected and lasting at least 100 ms. Bad channels were those presenting artifacts during more than 30% of the good times. Artifacts were corrected using target PCA on segments shorter than 100 ms and spatial spherical spline to interpolate bad channels (see Section 2.5). Finally, artifacts were detected again, and bad times and channels were re-defined. Fig. 4 shows an example of preprocessed data.
Fig. 4.
Examples of 70 s of preprocessed continuous data. (A) Data from one subject of dataset 1 after the artifacts’ detection. Data tagged as “bad” is shown in red. Y-axis scale 30 μV. (B) Data from the same subject of dataset 1 after the artifacts’ detection and correction (APICE(3) pipeline). Bad times are shown in orange. Y-axis scale 30 μV. (C) Data from one subject of dataset 1 after the artifacts detection, correction, and W-ICA (APICE(3) + W-ICA pipeline). Bad times are shown in orange. Y-axis scale 30 μV. (D) Analog than (A) for data from one subject of dataset 2. Y-axis scale 120 μV. (E) Analog than (B) for data from the same subject of dataset 2. Y-axis scale 120 μV. (F) Analog than (C) for data from the same subject of dataset 2. Y-axis scale 120 μV.
We used three different relative thresholds for artifact detection. As described in Section 2.3, the relative thresholds are fixed based on a certain number of interquartile ranges from the first and third quartiles. We fixed this value to 2 (APICE (2)), 3 (APICE (3)), and 4 (APICE (4)) in different runs of the preprocessing to evaluate how it affects the rejection percentage. A lower value (2) detects more artifacts but discards more data. A higher value (4) misses some artifacts but keeps more data.
To obtain ERPs, the continuous preprocessed data was further high-pass filtered at 0.2 and epoched. Then, bad times and channels were re-defined on the epoched data based on the data already rejected. A sample was defined as bad as explained on continuous data. A channel in a given epoch was defined as bad if it presented any artifact lasting more than 100 ms. Notice that some channels may present artifact events during periods not defined as bad times because we re-detected artifacts after the correction of transient artifacts (see Fig. 2D). If less than 30% of the channels were bad, they were interpolated using spherical splines. Epochs were rejected based on the amount of bad data: either when more than 30% of the channels were bad channels or when the epoch contained any bad time. Finally, the rejected epochs were removed, data was average referenced, and the average over the period [− 100, 100] ms was used as the baseline. All epochs were averaged in each infant and then across infants to create a grand average ERP.
3.2.2. APICEa pipeline (reduced version)
APICEa is a reduced version of APICE(3) in which we removed the reconstruction of transient artifacts in the continuous data using PCA and spatial spherical spline interpolation. All the other steps were the same. Notice that in this case, interpolation was done only after segmenting the data into epochs as it is usually do all the other available pipelines.
3.2.3. APICE + W-ICA pipeline
The APICE + W-ICA pipeline was primarily the same as the APICE(3) pipeline using a relative threshold equal to 3 for the artifacts rejection steps. After artifacts identification and correction in the continuous data, ICA was applied as described in Section 2.5. The steps to obtain the ERP are the same as in the APICE pipeline.
3.2.4. APICE + DSS pipeline
The APICE +DSS pipeline is the same as the APICE(3) pipeline, with a relative threshold equal to 3 for the artifacts rejection steps. The only difference is that the DSS filter was applied to the remaining trials after bad epochs were removed and the data were average-referenced. In the first PCA, we retained 50 components, and in the second PCA, 15. Finally, data were baseline corrected as in the other pipelines.
3.2.5. Standard pipeline
We based the STD pipeline on a preprocessing procedure widely used in infant studies (e.g., Adibpour et al., 2018; Friedrich and Friederici, 2017; Kabdebon and Dehaene-Lambertz, 2019; Winkler et al., 2009), consisting of minimal steps. The data were filtered (low-pass filter at 40 Hz and high-pass filter at 0.2 Hz) and epoched. Note that the same 0.2 Hz high pass-filtered is applied in all the pipelines before epoching. Afterward, we defined bad channels per epoch based on three criteria. First, we discarded channels for which the 5% stronger correlations with the other channels were lower than 0.4. Then, we rejected channels with an amplitude bigger than 500 μV on non-average reference data. Finally, we rejected channels with a fast running average bigger than 250 μV or a difference between the fast and slow running average bigger than 150 μV on average reference data. If less than 30% of the channels were rejected, they were interpolated using spherical splines. If more than 30% of the channels contained artifacts, the epoch was rejected. The retained epochs were average referenced, and baseline corrected using the average over [− 100, 100]. All epochs were averaged in each infant and then across infants to create a grand average ERP.
3.2.6. MADE pipeline
We implemented the MADE pipeline (Debnath et al., 2020) using the codes available at https://github.com/ChildDevLab/MADE-EEG-preprocessing-pipeline. In the MADE pipeline, the data is first filtered. Afterward, not working channels are identified using FASTER (Nolan et al., 2010). Then ICA is performed on a copy of the data high-passed filtered at 1 Hz. Before applying ICA, the data is epoched in one-second non-overlapping epochs, and electrodes and epochs containing artifacts are removed. After ICA decomposition, components associated with artifacts are automatically removed using adjusting-ADJUST (Leach et al., 2020). Then, data is segmented and baseline corrected. Epochs containing residual ocular artifacts (high amplitude on predefined frontal channels) are removed. If any remaining channels show high amplitude activity, they are reconstructed using spherical spline interpolation. Epochs with too many interpolated channels are rejected.
Here we filter the data between 0.2 Hz and 40 Hz. To detect residual ocular artifacts and non-functional channels, the authors recommend an amplitude threshold of 150 μV for infant data. However, this value rejected almost all the data for most of the subjects of Dataset 2 (5-month-old infants). We, therefore, increased it to 500 μV to Dataset 2. In the MADE pipeline, a limit of 10% of interpolated channels is used to reject epochs. We increased this value to 30% because otherwise, the rejection was very high (even when higher amplitude thresholds were used). Moreover, 30% is the limit used by APICE to define bad times; thus, the two pipelines become easy to compare. We want to point out that applying the MADE pipeline with the same values as proposed by the authors (Debnath et al., 2020) gave much worse results (see Appendix F); thus, we tried to optimize the absolute threshold used for rejection for both datasets. See Fig. 4 for an example of preprocessed data.
3.3. Pipelines evaluation
To control for systematic biases introduced by the different preprocessing approaches, we report the grand average responses and statistically compare them. To compare the pipeline’s performance in terms of data quality, we report two metrics: (1) the proportion of retained epochs and (2) the SME (Luck et al., 2021).
The proportion of retained epochs was computed as the number of epochs after rejection divided by the number of epochs before rejection. The SME was computed for the average response over a region of interest and time window corresponding to the auditory (Dataset 1) or visual ERPs (Dataset 2) using bootstrap. First, we randomly sampled with replacement N responses for each subject, where N is the number of retained epochs. Then, we computed the mean in time and space. We repeated the process 1000 times, and the standard deviation of the measure across all iterations corresponded to the SME for each subject ERP (Luck et al., 2021). Higher SMEs denote nosier data and smaller SMEs cleaner data. The SME for Dataset 1 was computed over central electrodes in the time window 250–350 ms (Fig. 5), which corresponds to the auditory response (Dehaene-Lambertz and Pena, 2001). The SME for Dataset 2 was computed over occipital electrodes in the time window 550–650 ms (Fig. 5), which corresponds to the P400 visual ERP (de Haan and Nelson, 1999). We used a time window centered at the peak of the ERP of the same length for both datasets; however, a change in the time window length does not affect the pattern of results.
Fig. 5.
Grand average ERPs obtained using APICE with a threshold for artifact rejection of 3. (A) Central electrodes (in red) considered for dataset 1. (B) Grand average ERP for the electrodes of interest for dataset 1. The peaks after the dotted lines (syllables’ onset) correspond to the auditory ERP following each syllable. The shaded area shows the time windows where the SME was computed (250–350 ms, peak of the auditory response to the last syllable of the epoch). (C) Occipital electrodes (in red) considered for dataset 2. (D) Grand average ERP for the electrodes of interest for dataset 2. The first two dotted lines indicate the onset of two images, and the third dotted line the appearance of the attention grabber. P1 and P400 are visible after the onset of the images, followed by the visual response to the attention grabber. The shaded area shows the time windows where the SME was computed (550–650 ms, P400 to the second image).
We statistically compared the two metrics across the different preprocessing approaches. When an ANOVA is used for comparison, the partial-eta-squared (η2p) and the generalized-eta-squared (η2G) are reported. Post-hoc pairwise comparisons were Bonferroni corrected. For pair comparisons, Cohen’s d effect sizes were computed based on the means and standard deviations.
3.4. Results
The grand average ERPs obtained with APICE and the regions and time windows of interest used on the two datasets are illustrated in Fig. 5. The grand average responses for each of the pipelines are reported in Appendix F (Figs. S1–S3). No main differences were observed between the grand-average responses across pipelines, suggesting no substantial biases were introduced by any of the methods (or the bias was comparable across all the pipelines).
3.4.1. APICE rejection level
Before comparing the SME and the percentage of retained epochs with varying thresholds, it is worth noticing that, as expected, the level of artifact contamination in the two datasets was considerably different. By using APICE(3) the average percentage of data tagged as bad in Dataset 1 was 4.19% (SD 3.84%, min 0.19%, max 12.92%), and in Dataset 2 12.19% (SD 8.79%, min 1.88%, max 30.40%). Regarding the amount of bad times, it was for Dataset 1 6.44% (SD 5.46%, min 0.29%, max 18.30%), and for Dataset 2 17.06% (SD 11.93%, min 1.93%, max 43.93%). These results illustrate the differences in terms of artifacts contamination between datasets. The tables summarizing the rejection obtained from the continuous preprocessed data are presented in Appendix E.
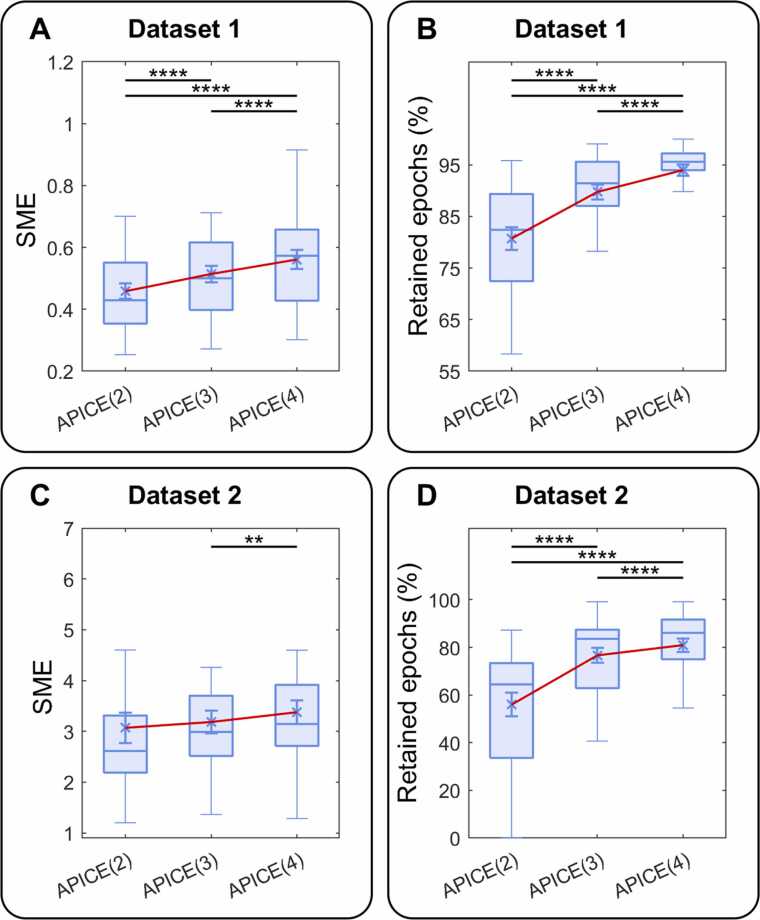
We ran two 1-way-ANOVAs to test the effect of three artifact-rejection levels in APICE (2, 3, and 4) on the SME and the percentage of retained epochs. For Dataset 1, on the SME, we observed a main effect of threshold (F(2,46) = 34.83; p = 6.16 × 10−10; η2p = 0.60; η2G = 0.11). Pairwise comparisons, Bonferroni corrected, showed that the SME was lower in APICE(2) than in APICE(4) (p = 7.8 × 10−6, d = 0.82) and APICE(3) (p = 3.0 × 10−5, d = 0.46), and in APICE(3) than in APICE(4) (p = 7.2 ×10−5, d = 0.40) (Fig. 6A). The effect of threshold on the amount of retained data was also significant (F(2,46) = 69.31; p < 1.3 × 10−14; η2p = 0.75; η2G = 0.34). Pairwise comparisons, Bonferroni corrected, showed that, as expected, more epochs were retained in APICE(4) than APICE(2) (p = 4.7 × 10−8, d = 1.59) and APICE(3) (p = 3.8 × 10−7, d = 0.68), and in APICE(3) than APICE(2) (p = 7.9 × 10−8, d = 1.01) (Fig. 6B).
Fig. 6.
Effect of the threshold level used for artifact detection in APICE. The boxplot shows the median, 25 and 75 percentiles, and the whiskers 1.5 interquartile ranges. The cross shows the mean and the error bar the standard error. (A) SME for Dataset 1. (B) Percentage of retained epochs for Dataset 1. (C) SME for Dataset 2. (D) Percentage of retained epochs for Dataset 2. A good pipeline performance should result in a small SME (little noise) and a high percentage of epochs retained. Asterisks indicate significant differences (Bonferroni corrected).
For Dataset 2, the threshold level 2 rejected all epochs for two subjects; thus, the SME could not be estimated, resulting in a smaller n. The effect of threshold on the SME was marginally significant (F(2,48) = 3.19, p = 0.05; η2p = 0.12; η2G = 0.01). Pairwise comparisons, Bonferroni corrected, showed no significant difference between APICE(2), and APICE(3) (p > 0.1, d = 0.12) and APICE(4) (p > 0.1, d = 0.25), and a significant difference between APICE(4) and APICE (3) (p = 0.01, d = 0.17) (Fig. 6C). The effect of threshold on the amount of retained epochs was significant (F(2,50) = 48.68; p = 1.84 × 10−12; η2p = 0.66; η2G = 0.25). Pairwise comparisons, Bonferroni corrected, showed that the proportion of epochs retained was larger in APICE(4) than APICE(2) (p = 6.2 × 10−7, d = 1.21) and APICE(3) (p = 3.1 × 10−5, d = 0.28), and in APICE(3) than APICE(2) (p = 8.6 × 10−7, d = 0.97) (Fig. 6D).
3.4.2. APICE modifications
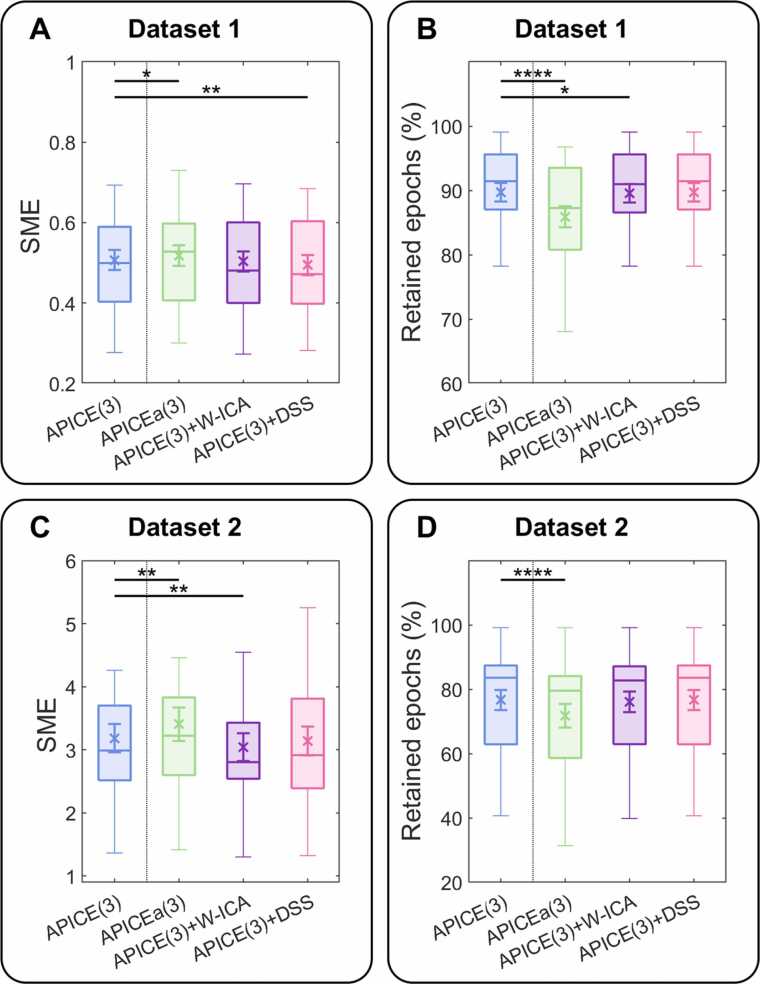
We evaluated the effect of interpolating localized artifacts by comparing our two metrics on the full and reduced pipelines, APICE vs. APICEa (Fig. 7). For both Datasets, APICE was better than APICEa. In Dataset 1, the SME was marginally lower in APICE (t(23) = − 2.11; p = 0.046; mean of the difference − 0.011; d = 0.09); and more data was retained after APICE (t(23) = 6.038; p = 3.7 × 10−6; mean of the difference 3.84%; d = 0.50). In dataset 2, the SME was lower with APICE (t(25) = − 3.1488; p = 0.0042; mean of the difference − 0.22; d = 0.18), and more data retained after APICE (t(25) = 6.12; p = 2.12 × 10−6; mean of the difference 4.84%; d = 0.28).
Fig. 7.
Comparison between APICE (blue) and some variations of it. In particular, a reduced version, APICEa, in which artifacts were detected in the continuous data but without interpolation of artifacts in the continuous data (green); APICE + W-ICA, in which wavelet-thresholding ICA and iMARA for automatic components classification was applied to removed physiological artifacts; and APICE + DSS, in which denoising source separation was used to remove the non-evoked activity. The boxplot shows the median, 25 and 75 percentiles, and the whiskers 1.5 interquartile ranges. The cross shows the mean and the error bar the standard error. (A) SME for dataset 1. (B) Retained epochs for dataset 1. (C) SME for dataset 2. (D) Retained epochs for dataset 2. Asterisks indicate significant differences between APICE and its variations.
To evaluate the effect of the two supplementary cleaning methods, ICA and DSS, we compared them with APICE. The ICA step applied on Dataset 1 removed on average 11.9% of the components (SD 8.37%, min 0%, max 30%) and 0.57% of the total variance (SD 0.46%, min 0%, max 2.09%). On Dataset 2, it removed on average 48.3% of the components (SD 17.18%, min 12%, max 75%) and 2.89% of the total variance (SD 1.71%, min 0.07%, max 7.30%). The DSS filter on Dataset 1 removed 13.18% of the total variance (SD 4.13%, min 6.51%, max 23.11%). On Dataset 2, it removed 33.46% of the total variance (SD 5.31%, min 19.48%, max 44.43%).
ICA and DSS slightly improve the validation metrics on specific datasets. In Dataset 1, the SME did not differ between APICE and APICE + W-ICA (p > 0.1; d = 0.03), and the amount of retained epochs was slightly higher for APICE than APICE + W-ICA (t(23) = 2.58; p = 0.017, mean of the difference 0.17%; d = 0.02), denoting that ICA did not improved data quality. In Dataset 2, APICE resulted in a slightly higher SME than APICE + W-ICA (t(25) = 3.55; p = 0.0016; mean of the difference 0.14; d = 0.12), and there was not significant difference in the amount of retained epochs (p > 0.1; d = 0.03), which means a modest increase in data quality.
On dataset 1, a slightly higher SME was observed for APICE than APICE+DSS (t(23) = 2.86; p = 0.0090; mean of the difference 0.013; d = 0.10). On dataset 2, the SME did not significantly differ when APICE and APICE + DSS were applied (p > 0.1; d = 0.04). Note that the percentage of retained epochs is not affected since the DSS is applied after bad epochs are removed.
3.4.3. APICE compared to other pipelines
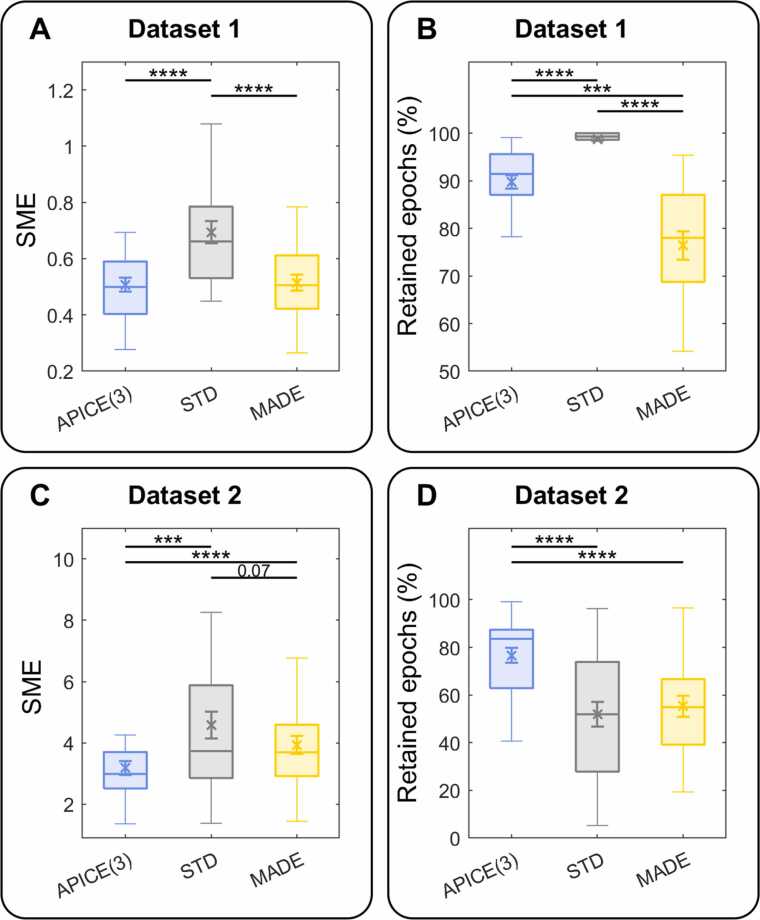
For Dataset 1, we compared the results obtained with MADE using the threshold for artifact rejection suggested by the authors for infant data (150 μV) (Debnath et al., 2020). For Dataset 2, since too much data was rejected using this threshold, we tested different thresholds and used the one providing the best performance. Figs. S4 and S5 show how the SME and percentage of retained epochs for MADE vary for different thresholds.
To compare the performances of APICE, STD, and MADE pipelines, we run two 1-way-ANOVAs on the SME and the percentage of retained epochs. On dataset 1, there was a main effect of pipeline on the SME (F(2,46) = 28.66; p = 8.28 × 10−9; η2p = 0.55; η2G = 0.24). Pairwise comparisons, Bonferroni corrected, showed that the SME was lower in APICE(3) than in STD (p = 1.4 × 10−5; d = 1.15) and in MADE than in STD (p = 9.9 × 10−5; d = 1.06), while there was no significant difference between APICE(3) and MADE (p > 0.1; d = 0.06). The main effect of the pipeline was also significant for the percentage of retained epochs (F(2,46) = 46.22; p = 9.89 × 10−12; η2p = 0.67; η2G = 0.50). Pairwise comparisons, Bonferroni corrected, showed more retained epochs for STD than APICE (p = 5.1 × 10−7; d = 1.76), and MADE (p = 1.7 × 10−7; d = 2.17), and more retained epochs for APICE than MADE (p = 0.00011; d = 1.17).
On dataset 2, on the SME, we observed a main effect of pipeline (F(2,50) = 16.38; p = 3.38 × 10−6; η2p = 0.440; η2G = 0.11). Pairwise comparisons, Bonferroni corrected, revealed that it was due to lower SME for APICE than STD (p = 0.00024; d = 0.79) and MADE (p = 2.7 × 10−5; d = 0.57), and a marginally significant lower SME for MADE than for STD (p = 0.073; d = 0.34). The effect on percentage of retained epochs was also significant (F(2,50) = 18.23; p = 1.13 × 10−6; η2p = 0.42; η2G = 0.20). Pairwise comparisons, Bonferroni corrected, showed a significantly higher retention for APICE than STD (p = 1.5 × 10−6; d = 1.13) and MADE (p = 1.7 × 10−5; d = 1.09), and no difference between STD and MADE (p > 0.1; d = 0.14).
3.5. Discussion
We tested the effect of the rejection level used for artifacts detection, the validity of the different steps composing APICE, and its performance relative to other available pipelines on two different infant datasets. We evaluated the pipelines by the stability and reproducibility of the obtained ERPs as measured by the SME, a procedure proposed by Luck et al. (2021) to quantify ERP quality. This procedure computes the subject ERP through multiple random draws of the epochs and examines the distribution of the ERP values across draws. When the averaging process is successful at neutralizing unwanted “noise,” that is, if no unexpected large-amplitude non-evoked activity remains in the preprocessed data, the values of the SME are low. Further, we examined the number of kept epochs, crucial for complex paradigms where pertinent conditions comprise only a few trials.
3.5.1. APICE rejection level
We validated APICE on three threshold levels for artifact detection to find the best compromise between quality and retained data. Results show that decreasing the threshold from 4 to 3 minimally changes the percentage of rejected data while reducing the SME. On the other hand, a further decrease to 2 reduces the SME at the cost of a considerable increase in the rejection rate. In other words, while the change in SME shows a linear decrease with threshold decrease, the change in rejection shows a logarithmic trend. Therefore, using a too low threshold implies losing too much data relative to the gain in data quality, while too high thresholds result in a too high loss in data quality compared to the gain in data retention. Accordingly, we recommend using a threshold of 3 as default to keep enough epochs per subject. Nevertheless, the threshold can be adjusted depending on the analysis requirements –either more data but noisier or less data but cleaner.
3.5.2. APICE modifications
Besides detecting artifacts on the continuous data, APICE also corrects localized artifacts. We tested if this step brings any improvement by comparing APICE with APICEa, a reduced version in which we removed the interpolation of transient artifacts. Results show that while in APICE, the SME is only marginally lowered relatively to APICEa, the amount of retained epochs is considerably higher. Thus, the interpolation of transient artifacts enables the recovery of otherwise lost epochs without loss of data quality.
Finally, we tested whether the addition of ICA or DSS, two data cleaning methods extensively used in adult studies, can improve data quality. For ICA, we implemented the latest proposals offered in the literature. To achieve a better ICA decomposition, we high-pass filtered a copy of the data and applied a combination of ICA with wavelet-thresholding (Rong-Yi and Zhong, 2005), as it was also applied in HAPPE (Gabard-Durnam et al., 2018). For the IC automatic classification, we used iMARA (Haresign et al., 2021), a recent modification of MARA (Winkler et al., 2011), adapted to infant data. Finally, the activity separated as non-neural was removed from the original data. For dataset 1 (neonates), ICA did not produce any improvement: the SME did not change, and it slightly reduced the number of retained epochs. For dataset 2 (5-month-old), ICA decreased the SME, but the size of the improvement was small, indicating that the obtained noise reduction was consistent across infants but meager in magnitude. The average number of components and the total variance removed was much bigger for dataset 2 than 1 (percentage of removed components: 48.3% vs. 11.9%; total variance: 2.89% vs. 0.57%). The difference in the number of removed components, together with the no change between APICE and APICE+W-ICA in terms of SME for dataset 1, suggest that the ICA less successfully identifies physiological artifacts in neonates’ data.
Overall, despite our effort to optimize ICA, its effectiveness on young infants’ ERP remains weak. iMARA performs better than the original MARA algorithm on classifying non-neural IC in 10–13-month-old infants (Haresign et al., 2021), and authors prove substantial improvements in data quality in an ERP study at this age. Nevertheless, our results suggest that this improvement might be less substantial in younger populations. Given the high computational cost of applying ICA and the risk of removing neural activity, we recommend evaluating the cost/benefit of implementing this routine on a case-by-case basis. For example, for sleeping neonates for whom physiological artifacts as ocular movements are less prominent, and the ICA algorithms seem to be less efficient at separating them, its use does not seem to be justified. Instead, for older infants involved in an active task, ICA might improve data quality by removing physiological artifacts that otherwise remain in the data.
The application of DSS on the retained epochs led to a similar outcome: a statistically significant but minimal reduction of the SME, specifically in Dataset 1 (neonates). Thus, we do not recommend it as default for infant ERPs studies, even though it might provide more substantial benefits if numerous trials are available.
Possible explanations for the modest improvements observed following the application of ICA and DSS might reside in the intrinsic properties of the infant EEG. For example, high inter-trial variability (Naik et al., 2021) is likely to compromise the efficiency of DSS filters, as this method is based on the presence of highly reproducible activity across trials. In addition, developmental changes and variability of both neural responses and physiological artifacts might result in poor decomposition during ICA for infant data. Indeed, the authors of the iMARA algorithm report more variability in the manual coding of infant IC than adult IC (Haresign et al., 2021). While substantial improvements have been recently made in the implementation of blind source separation techniques in pediatric recordings (Haresign et al., 2021, Leach et al., 2020), more research is needed to adapt these techniques to younger populations and better characterize infant EEG recordings.
3.5.3. APICE compared to other pipelines
Results show that the APICE(3) pipeline outperformed the STD and MADE pipelines (Fig. 8). While the STD pipeline retained more epochs than APICE(3) for Dataset 1 (an auditory experiment in sleeping neonates), this was at the cost of a much higher SME. If a greater number of epochs is a priori an advantage to recover the ERP through the average process, this is no longer the case when it implies a decrease in data quality. The neonates were tested asleep, which means that the recording was only mildly contaminated by motion and that the EEG amplitude was low; thus, high-amplitude artifacts exceeding the absolute thresholds used in the standard pipeline were rare. The implementation of algorithms based on various signal features makes APICE more sensitive to outlier signals. In Dataset 2 (a visual experiment in awake 5-month-old infants), where the contamination by motion artifacts was substantial, APICE(3) retained more data and yielded a smaller SME than STD.
Fig. 8.
Comparison of APICE’s performance with the Standard pipeline (STD) and the MADE pipeline. The boxplot shows the median, 25 and 75 percentiles, and the whiskers 1.5 interquartile ranges. The cross shows the mean and the error bar the standard error. (A) SME for dataset 1. (B) Retained epochs for dataset 1. (C) SME for dataset 2. (D) Retained epochs for dataset 2. Asterisks indicate significant differences (Bonferroni corrected).
APICE(3) also outperformed MADE for both datasets. For Dataset 1, the SME was comparable between pipelines, but APICE retained a much higher number of epochs than MADE. For Dataset 2, APICE(3)’s performance was better in terms of both SME and epochs retention. The retention of more trials and smaller SMEs achieved by APICE than other pipelines entails smaller ERP errors, implying more statistical power (Luck et al., 2021).
It is worth noticing that we needed to adjust the MADE rejection threshold in Dataset 2 to achieve a reasonable performance (Fig. S5). In APICE, thresholds are based on the distribution of the voltage values in each subject (and electrode), thereby ensuring optimal sensitivity across subjects and datasets and making APICE robust across different populations without main adjustments. Pipelines for which the artifacts’ detection relies on fixed thresholds need to be adjusted for each population, a costly and time-consuming process if it has to be done each time a dataset has to be analyzed. Moreover, even if the thresholds are adjusted, their performance remains poorer because differences in signal amplitude also exist between testing systems and subjects (e.g., due to differences in resistance). One could argue that also APICE requires the experimenter to set an overall rejection level at the beginning of the analysis. However, the results across the two tested datasets suggest that the effects of the rejection level are considerably stable across different populations. Moreover, APICE outperformed STD and MADE irrespective of the particular choice at hand (see Fig. 6, Fig. 7). In brief, the automatic detection of contaminated data through adaptive thresholds and the correction of localized artifacts on the continuous recording, implemented in APICE, results in better recovery of good quality data with a wide range of relative rejection levels.
4. Conclusion
EEG is a widely used technique in developmental studies. Nevertheless, no standard procedure exists for the preprocessing of infant data, partly because of the small size of the research community and partly because of the many challenges preprocessing infant EEG entails. For example, infants’ recordings are shorter and more heavily contaminated by motion artifacts. Moreover, the types of artifacts and the features of the EEG signal change during development (Eisermann et al., 2013, Kushnerenko et al., 2002, Marshall et al., 2002, Nelson and Monk, 2001), making the approaches optimized for adult data ineffective and the design of methods applicable across age groups challenging.
APICE can successfully identify artifacts across different ages and experimental conditions by employing multiple algorithms and adaptive thresholds for artifacts detection and the interpolation of transient artifacts on the continuous data. The approach we propose improves data recovery and data quality relative to other pipelines. Moreover, it brings flexibility because the same preprocessed data can serve to perform analyzes requiring different segmentation strategies. Furthermore, accurately detecting artifacts allows one to decide how to handle them in subsequent processes. For example, the amount of data with artifacts in an epoch can be used as a criterion to reject it, and detected motion artifacts could be excluded before performing ICA (or any other blind source separation method), a fundamental step for good sources separation.
Nevertheless, crucial challenges remain. Many physiological artifacts (e.g., eye movements, muscle artifacts, skin potential) present amplitude and spectral properties similar to those characterizing the neural signal. Therefore, algorithms based on local properties might fail in disentangling the two. For example, with APICE, we can identify the heartbeat using an algorithm that detects fast changes in the signal, and we can correct them using target PCA. However, APICE does not explicitly search for artifacts like blinks or eye movements. Moreover, even if all physiological artifacts could be identified, efficient removal of physiological artifacts cannot reside on the rejection of any segment contaminated by them. Instead, it would require blind source separation methods (Islam et al., 2016, Jiang et al., 2019) as ICA and proper individuation of the irrelevant sources. APICE can be combined with some of the latest ICA and automatic components classification methods available for infant data to deal with physiological artifacts. Nevertheless, our results suggest that the benefits for young infant EEG in the current state of the arts are still limited. Considering the computational cost of ICA and the risk of removing neural activity, the use of ICA should be evaluated case-by-case, taking into account the level of physiological artifacts as eye movements expected in the data.
We created APICE to be fully automated and flexible. Automation guarantees replicability and scalability for growing data sets (without increasing the human workload). APICE performs all the artifact detection steps in the continuous data to ensure flexibility and better data recovery. Consequently, the same preprocessed data is ready for different types of analysis. Furthermore, APICE is modular, allowing it to be easily modified to meet specific needs and incorporate new steps. Additionally, APICE includes functions for renaming events, correcting their timing, organizing epochs by condition, and computing the average ERP. APICE is freely available at https://github.com/neurokidslab/eeg_preprocessing, with example scripts illustrating its application. APICE is currently limited to MATLAB. However, we are currently working on (Savalle, 2022) transferring it to Python and integrating it into MNE (Gramfort et al., 2013).
CRediT authorship contribution statement
Ana Fló: conceived the pipeline, wrote the software, and wrote the manuscript. Giulia Gennari: contributed to testing the software, revised the manuscript and the pipeline validation. Lucas Benjamin: contributed to testing the software, revised the manuscript and the pipeline validation. Ghislaine Dehaene-Lambertz: conceived the pipeline, revised the manuscript and the pipeline validation.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Ghislaine Dehaene-Lambertz reports financial support was provided by European Research Council.
Acknowledgments
This research has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant agreement no. 695710).
We thank the families that participated in the studies. We would also like to thank Marie Palu and Chanel Valera for their help in recruiting and testing the infants and the entire Neurokids group at Neurospin for their feedbacks.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dcn.2022.101077.
Appendix A. Supplementary material
Supplementary material
Data statement
The datasets analyzed during the current study are available from the corresponding author.
References
- Adibpour P., Dubois J., Dehaene-Lambertz G. Right but not left hemispheric discrimination of faces in infancy. Nat. Hum. Behav. 2018;2(1):67–79. doi: 10.1038/s41562-017-0249-4. [DOI] [PubMed] [Google Scholar]
- Bertrand O., Perrin F., Pernier J. A theoretical justification of the average reference in topographic evoked potential studies. Electroencephalogr. Clin. Neurophysiol./Evoked Potentials Sect. 1985;62(6):462–464. doi: 10.1016/0168-5597(85)90058-9. [DOI] [PubMed] [Google Scholar]
- Bigdely-Shamlo N., Mullen T., Kothe C., Su K.-M., Robbins K.A. The PREP pipeline: standardized preprocessing for large-scale EEG analysis. Front. Neuroinform. 2015;9(June):1–20. doi: 10.3389/fninf.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cheveigné A., Arzounian D. Robust detrending, rereferencing, outlier detection, and inpainting for multichannel data. NeuroImage. 2018;172:903–912. doi: 10.1016/j.neuroimage.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cheveigné A., Nelken I. Filters: when, why, and how (not) to use them. Neuron. 2019;102(2):280–293. doi: 10.1016/j.neuron.2019.02.039. [DOI] [PubMed] [Google Scholar]
- De Cheveigné A., Parra L.C. Joint decorrelation, a versatile tool for multichannel data analysis. NeuroImage. 2014;98:487–505. doi: 10.1016/j.neuroimage.2014.05.068. [DOI] [PubMed] [Google Scholar]
- de Cheveigné A., Simon J.Z. Denoising based on spatial filtering. J. Neurosci. Methods. 2008;171(2):331–339. doi: 10.1016/j.jneumeth.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan M., Nelson C.A. Brain activity differentiates face and object processing in 6-month-old infants. Dev. Psychol. 1999;35(4):1113–1121. doi: 10.1037/0012-1649.35.4.1113. [DOI] [PubMed] [Google Scholar]
- de Heering A., Rossion B. Rapid categorization of natural face images in the infant right hemisphere. ELife. 2015;4(JUNE):1–14. doi: 10.7554/eLife.06564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath R., Buzzell G.A., Morales S., Bowers M.E., Leach S.C., Fox N.A. The Maryland analysis of developmental EEG (MADE) pipeline. Psychophysiology. 2020;57(6) doi: 10.1111/psyp.13580. [DOI] [PubMed] [Google Scholar]
- Dehaene-Lambertz G., Pena M. Electrophysiological evidence for automatic phonetic processing in neonates. Neuroreport. 2001;12(14):3155–3158. doi: 10.1097/00001756-200110080-00034. [DOI] [PubMed] [Google Scholar]
- Delorme A., Makeig S. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods. 2004;134(1):9–21. doi: 10.1016/j.jneumeth.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Desjardins J.A., van Noordt S., Huberty S., Segalowitz S.J., Elsabbagh M. EEG Integrated Platform Lossless (EEG-IP-L) pre-processing pipeline for objective signal quality assessment incorporating data annotation and blind source separation. J. Neurosci. Methods. 2021;347 doi: 10.1016/j.jneumeth.2020.108961. [DOI] [PubMed] [Google Scholar]
- Eisermann M., Kaminska A., Moutard M.-L., Soufflet C., Plouin P. Normal EEG in childhood: from neonates to adolescents. Neurophysiol. Clin./Clin. Neurophysiol. 2013;43(1):35–65. doi: 10.1016/j.neucli.2012.09.091. [DOI] [PubMed] [Google Scholar]
- Friedrich M., Friederici A.D. The origins of word learning: brain responses of 3-month-olds indicate their rapid association of objects and words. Dev. Sci. 2017;20(2) doi: 10.1111/desc.12357. [DOI] [PubMed] [Google Scholar]
- Friedrich M., Wilhelm I., Born J., Friederici A.D. Generalization of word meanings during infant sleep. Nat. Commun. 2015;6:6004. doi: 10.1038/ncomms7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabard-Durnam L.J., Mendez Leal A.S., Wilkinson C.L., Levin A.R. The Harvard Automated Processing Pipeline for Electroencephalography (HAPPE): standardized processing software for developmental and high-artifact data. Front. Neurosci. 2018;12:12. doi: 10.3389/fnins.2018.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha G., Geethalakshmi S.N. EEG denoising using SURE thresholding based on Wavelet Transforms. Int. J. Comput. Appl. 2011;24(6):29–33. doi: 10.5120/2948-3935. [DOI] [Google Scholar]
- Gennari G., Marti S., Palu M., Fló A., Dehaene-Lambertz G. Orthogonal neural codes for speech in the infant brain. Proc. Natl. Acad. Sci. USA. 2021;118(31) doi: 10.1073/pnas.2020410118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramfort A., Luessi M., Larson E., Engemann D., Strohmeier D., Brodbeck C., Goj R., Jas M., Brooks T., Parkkonen L., Hämäläinen M. MEG and EEG data analysis with MNE-Python. Front. Neurosci. 2013;7:267. doi: 10.3389/fnins.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haresign I.M., Phillips E., Whitehorn M., Noreika V., Jones E.J.H., Leong V., Wass S.V. Automatic classification of ICA components from infant EEG using MARA [Preprint] Neuroscience. 2021 doi: 10.1101/2021.01.22.427809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H.G., Debnath R., Meyer M., Salo V.C., Fox N.A., Woodward A. Neighborhood racial demographics predict infants’ neural responses to people of different races. Dev. Sci. 2021;24(4) doi: 10.1111/desc.13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.K., Rastegarnia A., Yang Z. Methods for artifact detection and removal from scalp EEG: a review. Neurophysiol. Clin./Clin. Neurophysiol. 2016;46(4):287–305. doi: 10.1016/j.neucli.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Jiang X., Bian G.-B., Tian Z. Removal of artifacts from EEG signals: a review. Sensors. 2019;19 doi: 10.3390/s19050987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone I.M., Silverman B.W. Wavelet threshold estimators for data with correlated noise. J. R. Stat. Soc.: Ser. B (Stat. Methodol.) 1997;59(2):319–351. doi: 10.1111/1467-9868.00071. [DOI] [Google Scholar]
- Kabdebon C., Dehaene-Lambertz G. Symbolic labeling in 5-month-old human infants. Proc. Natl. Acad. Sci. USA. 2019;116(12):5805–5810. doi: 10.1073/pnas.1809144116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabdebon C., Pena M., Buiatti M., Dehaene-Lambertz G. Electrophysiological evidence of statistical learning of long-distance dependencies in 8-month-old preterm and full-term infants. Brain Lang. 2015;148:25–36. doi: 10.1016/j.bandl.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Kappenman E.S., Luck S.J. The effects of electrode impedance on data quality and statistical significance in ERP recordings. Psychophysiology. 2010;47(5):888–904. doi: 10.1111/j.1469-8986.2010.01009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnerenko E., Ceponienė R., Balan P., Fellman V., Näätänen R. Maturation of the auditory change detection response in infants: a longitudinal ERP study. Neuroreport. 2002;13(15):3–8. doi: 10.1097/00001756-200210280-00002. [DOI] [PubMed] [Google Scholar]
- Leach S.C., Morales S., Bowers M.E., Buzzell G.A., Debnath R., Beall D., Fox N.A. Adjusting ADJUST: optimizing the ADJUST algorithm for pediatric data using geodesic nets. Psychophysiology. 2020;57(8) doi: 10.1111/psyp.13566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck S.J., Stewart A.X., Simmons A.M., Rhemtulla M. Standardized measurement error: a universal metric of data quality for averaged event-related potentials. Psychophysiology. 2021;58(6) doi: 10.1111/psyp.13793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall P.J., Bar-Haim Y., Fox N.A. Development of the EEG from 5 months to 4 years of age. Clin. Neurophysiol. 2002;113(8):1199–1208. doi: 10.1016/S1388-2457(02)00163-3. [DOI] [PubMed] [Google Scholar]
- Mognon A., Jovicich J., Bruzzone L., Buiatti M. ADJUST: an automatic EEG artifact detector based on the joint use of spatial and temporal features. Psychophysiology. 2011;48(2):229–240. doi: 10.1111/j.1469-8986.2010.01061.x. [DOI] [PubMed] [Google Scholar]
- Naik, S., Adibpour, P., Dubois, J., Dehaene-Lambertz, G., Battaglia, D., 2021. Structured modulations of ongoing variability by task and development. BioRxiv, 2021.03.07.434162. 〈 10.1101/2021.03.07.434162〉. [DOI]
- Nelson C.A., Monk C.S. Handbook of Developmental Cognitive Neuroscience. MIT Press; 2001. The use of event-related potentials in the study of cognitive development. [Google Scholar]
- Nolan H., Whelan R., Reilly R.B. FASTER: fully automated statistical thresholding for EEG artifact rejection. J. Neurosci. Methods. 2010;192(1):152–162. doi: 10.1016/j.jneumeth.2010.07.015. [DOI] [PubMed] [Google Scholar]
- Onton J., Makeig S. In: Neuper C., Klimesch W., editors. Vol. 159. Elsevier; 2006. Information-based modeling of event-related brain dynamics; pp. 99–120. (Progress in Brain Research). [DOI] [PubMed] [Google Scholar]
- Pedroni A., Bahreini A., Langer N. Automagic: standardized preprocessing of big EEG data. NeuroImage. 2019;200:460–473. doi: 10.1016/j.neuroimage.2019.06.046. [DOI] [PubMed] [Google Scholar]
- Perrin F., Pernier J., Bertrand O., Echallier J.F. Spherical splines for scalp potential and current density mapping. Electroencephalogr. Clin. Neurophysiol. 1989;72(2):184–187. doi: 10.1016/0013-4694(89)90180-6. [DOI] [PubMed] [Google Scholar]
- Rodrigues J., Weiß M., Hewig J., Allen J.J.B. EPOS: EEG processing open-source scripts. Front. Neurosci. 2021:0. doi: 10.3389/fnins.2021.660449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong-Yi Y., Zhong C. Blind source separation of multichannel electroencephalogram based on wavelet transform and ICA. Chin. Phys. 2005;14(11):2176–2180. doi: 10.1088/1009-1963/14/11/006. [DOI] [Google Scholar]
- Savalle E. Paris-Saclay University; 2022. Master 2 CNN Supervised Project Report Implementation of APICE Algorithms in Python for Integration into MNE. [Google Scholar]
- Selton D., Andre M., Hascoët J.M. Normal EEG in very premature infants: reference criteria. Clin. Neurophysiol. 2000;111(12):2116–2124. doi: 10.1016/S1388-2457(00)00440-5. [DOI] [PubMed] [Google Scholar]
- Troller-Renfree S.V., Brito N.H., Desai P.M., Leon-Santos A.G., Wiltshire C.A., Motton S.N., Meyer J.S., Isler J., Fifer W.P., Noble K.G. Infants of mothers with higher physiological stress show alterations in brain function. Dev. Sci. 2020;23(6) doi: 10.1111/desc.12976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, I., Debener, S., Müller, K., Tangermann, M., 2015. On the influence of high-pass filtering on ICA-based artifact reduction in EEG-ERP. In: Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 4101–5. 〈 10.1109/EMBC.2015.7319296〉. [DOI] [PubMed]
- Winkler I., Haden G.P., Ladinig O., Sziller I., Honing H. Newborn infants detect the beat in music. Proc. Natl. Acad. Sci. USA. 2009;106(7):2468–2471. doi: 10.1073/pnas.0809035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler I., Haufe S., Tangermann M. Automatic classification of artifactual ICA-components for artifact removal in EEG signals. Behav. Brain Funct. 2011;7(1):30. doi: 10.1186/1744-9081-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel M.A., Selb J., Cooper R.J., Boas D.A. Targeted principle component analysis: a new motion artifact correction approach for near-infrared spectroscopy. J. Innov. Opt. Health Sci. 2014;7(2):1–8. doi: 10.1142/S1793545813500661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The datasets analyzed during the current study are available from the corresponding author.