Abstract
Appearance of plaques on a bacterial lawn is a sign of successive rounds of bacteriophage infection. Yet, mechanisms evolved by bacteria to limit plaque spread have been hardly explored. Here, we investigated the dynamics of plaque development by lytic phages infecting the bacterium Bacillus subtilis. We report that plaque expansion is followed by a constriction phase owing to bacterial growth into the plaque zone. This phenomenon exposed an adaptive process, herein termed "phage tolerance response", elicited by non‐infected bacteria upon sensing infection of their neighbors. The temporary phage tolerance is executed by the stress‐response RNA polymerase sigma factor σX (SigX). Artificial expression of SigX prior to phage attack largely eliminates infection. SigX tolerance is primarily conferred by activation of the dlt operon, encoding enzymes that catalyze D‐alanylation of cell wall teichoic acid polymers, the major attachment sites for phages infecting Gram‐positive bacteria. D‐alanylation impedes phage binding and hence infection, thus enabling the uninfected bacteria to form a protective shield opposing phage spread.
Keywords: Bacillus subtilis, bacteriophage, cell wall teichoic acid, dlt operon, plaque formation
Subject Categories: Microbiology, Virology & Host Pathogen Interaction
Phage‐infected bacteria send signals to neighbouring cells inducing cell wall modifications that lead to an adaptive phage tolerance response.

Introduction
Plaques, reporting bacterial clearance, are the hallmark of bacteriophage (phage) infection since the pioneering discoveries made by Twort and d'Herelle at the beginning of the 20th century (Twort, 1914; d'Hérelles, 1917; Chanishvili, 2012). Plaques are basically visible holes, formed on a lawn of bacteria grown on a solid surface, that report bacterial clearance following successive cycles of infection, including phage adsorption, replication, and spread to nearby hosts. Intriguingly, plaques exhibit a considerable variation in shape according to the host and the infecting phage and are predominantly restricted in size (Abedon & Yin, 2009). It has been proposed that such size limitation is achieved, at least in part, by the entry of bacteria into stationary phase, which frequently restrains phage replication (Abedon & Yin, 2009). However, although plaque employment as a method for monitoring phage infection began decades ago, relatively little is known about the kinetics of plaque development. Furthermore, factors limiting plaque size and expansion are mostly unrevealed.
In this study, we investigated the dynamics of plaque formation, utilizing the Gram‐positive soil bacterium Bacillus subtilis (B. subtilis) and its lytic phages SPP1 and Phi29. Binding of phages to B. subtilis is commonly mediated by wall teichoic acid (WTA) polymers, a diverse family of cell surface glycopolymers containing phosphodiester‐linked glycerol repeat units poly(Gro‐P), decorated by glucose and D‐alanine moieties, and anchored to peptidoglycan (PG) through an N‐acetylmannosaminyl (Brown et al, 2013). WTA polymers were found to be crucial surface components, required for invasion by manifold phages into Gram‐positive bacteria such as Bacilli, Listeria, and Staphylococci (Lindberg, 1973; Habusha et al, 2019; Ingmer et al, 2019; Sumrall et al, 2020). SPP1, a double‐stranded DNA (dsDNA) phage (44 kb) and a member of the Siphoviridae family, characterized by a long noncontractile tail (Alonso et al, 1997), initiates infection by reversible binding of the tail tip to poly‐glycosylated WTA (gWTA). Subsequently, SPP1 binds irreversibly to its membrane receptor protein YueB, resulting in DNA injection into the bacterium cytoplasm (Sao‐Jose et al, 2004; Baptista et al, 2008). Phi29 phage, which is significantly smaller (19.3 kb) and belongs to the Podoviridae family, harboring a short noncontractile tail (Salas, 2012), was shown to entirely rely on intact gWTA for infection (Young, 1967). The fact that phages assigned to distinct families utilize gWTA to invade the host strengthens the vital role of these polymers in host recognition by phages and implicates them as major elements for host cell vulnerability. Consistent with this notion, we have shown that a mutant bacteriophage capable of bypassing the need for binding the glucosyl residues, decorating the WTA polymers, gained a broader host range, as it could infect non‐host bacterial species presenting dissimilar glycosylation patterns (Habusha et al, 2019).
Previously, we demonstrated that phages could occasionally invade resistant cells that acquire phage receptors from their sensitive neighbors, highlighting the importance of understanding infection dynamics in a temporal and spatial fashion (Tzipilevich et al, 2017). Here, we visualized plaques formed on a lawn of B. subtilis bacteria. We revealed that plaque spreading is followed by a phase of constriction mediated by bacterial regrowth into the plaque zone. Characterization of the plaque constriction phase uncovered a temporary immunity mechanism, propelled by a programed transcriptional response, enabling bacteria to tolerate infection by remodeling WTA polymers. This modification reduces phage binding and restricts phage spread. Unlike other mechanisms affording long‐term bacterial immunity to phages, namely, restriction enzymes and CRISPR (Labrie et al, 2010), this tolerance mechanism confers a transient adaptive response, providing protection to the uninfected bacterial population subsequent to infection of their neighbors.
Results
Plaque expansion is followed by a phase of plaque constriction
To explore the bacterial population dynamic during phage attack on solid surfaces, we followed the process of plaque formation by SPP1 on a lawn of mCherry‐labeled B. subtilis cells at high resolution, using time‐lapse confocal microscopy. Maximal SPP1 plaque size was detected approximately eight hours postinfection, but remarkably the spread was counteracted by bacteria growing into the plaque area, limiting plaque expansion (Fig 1A; Movie EV1). Consequently, the final plaque diameter measured after overnight incubation was significantly smaller than the maximal size reached during plaque development (Fig 1A). To further explore this phenomenon, we followed the kinetics of plaque formation on agar plates, the typical methodology used over the years for estimation of plaque‐forming units (PFU) [e.g., Abedon and Yin (2009), Ellis and Delbruck (1939)]. Consistent with the confocal microscopy results, plate monitoring revealed a steep expansion phase that was proceeded by a gradual decrease in plaque size, with the final zone being approximately 50% of the maximal plaque area measured during the process (Fig 1B and C; Movie EV2). This plaque constriction occurrence was also evident when bacteria were infected with the distinct lytic phage Phi29 (Fig 1D), suggesting that such a kinetic pattern is widespread.
Figure 1. Plaque formation dynamics reveals phases of expansion and constriction.
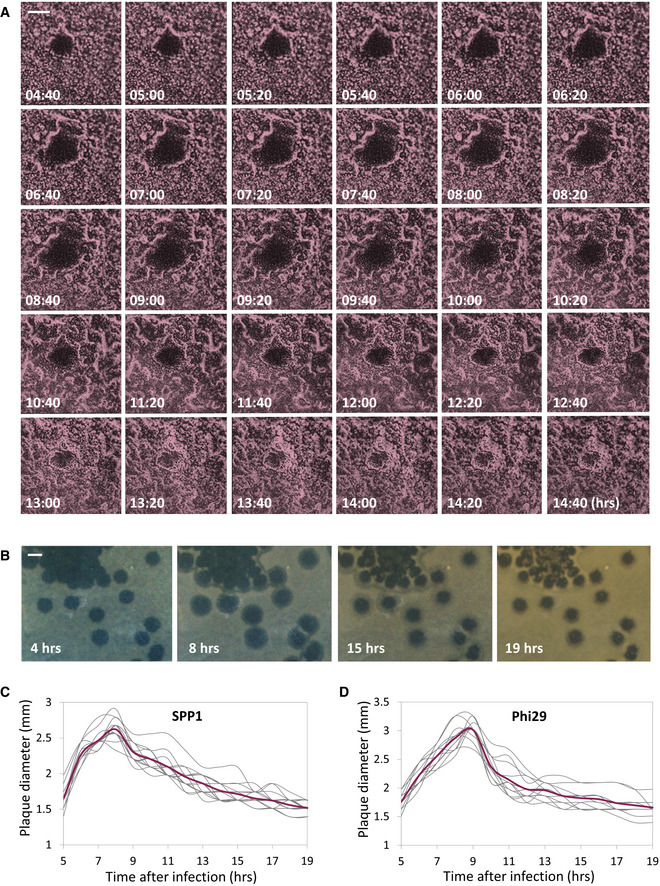
-
ABDR2637 (P veg‐mCherry) cells were infected with low concentrations (10‐8 PFU/ml) of SPP1, placed on an agarose pad, and plaque formation was followed by time‐lapse confocal microscopy. Shown are overlay images from mCherry signal (purple) and phase contrast (gray) of the bacterial lawn captured at the indicated time points (h). The plaque is seen as a hole formed on the bacterial lawn. Scale bar, 150 µm. Corresponds to Movie EV1.
- B
-
C, DThe dynamic of SPP1 (C) and Phi29 (D) plaque formation following PY79 (WT) infection was monitored as described in (B). Shown is the diameter of individual plaques for each phage (n ≥ 12), with the average highlighted in red.
To concomitantly track plaque development and phage localization, we followed plaque formation by SPP1 harboring its lysin gene (gp51) fused to a yellow fluorescent protein (YFP) as a sole copy (SPP1‐lysin‐yfp), marking active infection (Tzipilevich et al, 2017). Fluorescence from YFP demarcated the plaque periphery even during the constriction phase, signifying the presence of actively infected cells that release phage particles (Fig EV1A–C; Movie EV3). This observation implies that bacteria at the rim could withstand the presence of phages. Isolating bacteria from the edge of 30 different plaques subsequent to the constriction phase and re‐plating them over plate‐containing phages revealed all tested bacteria to remain phage sensitive (Fig EV1D). We refer to the phenomenon of phage‐sensitive bacteria that can confront phages at the plaque circumference as “phage tolerance”.
Figure EV1. Evidence for the presence of phages at the plaque periphery during constriction.
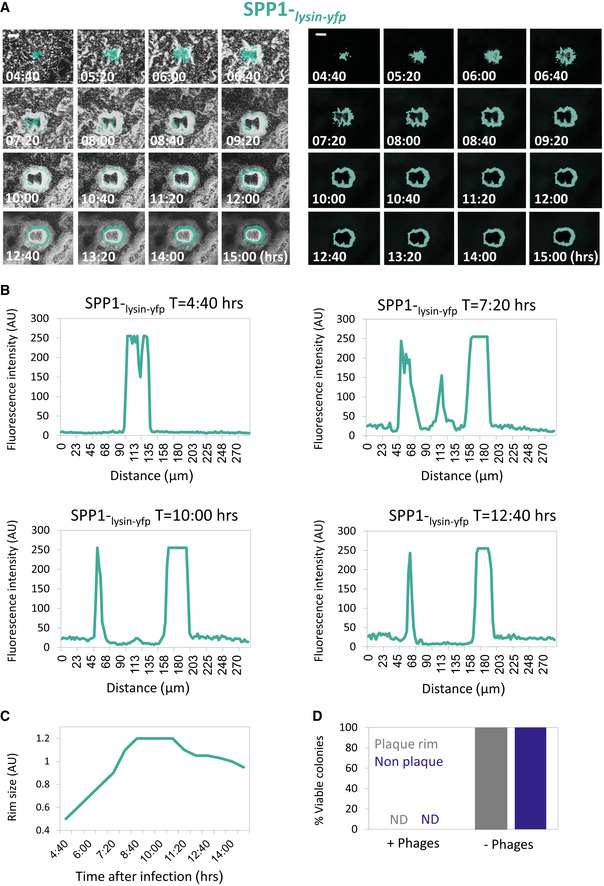
- PY79 (WT) cells were infected with low concentrations (10‐8 PFU/ml) of SPP1‐lysin‐yfp, placed on an agarose pad, and plaque formation was followed by time‐lapse confocal microscopy. Shown are overlay images of phase contrast (gray) and signal from Lysin‐SPP1‐YFP (cyan) captured at the indicated time points (h) postinfection (left panels). Corresponding signal from Lysin‐SPP1‐YFP (cyan) is shown separately (right panels). Scale bars, 50 µm. Corresponds to Movie EV3.
- Quantification of the SPP1‐lysin‐yfp fluorescence intensity (AU) at the indicated time points. Fluorescence from Z sections that include the plaque region and flanking area was measured. Corresponds to EV1A.
- Quantification of the diameter of the YFP fluorescence (AU) ring, derived from SPP1‐lysin‐yfp. Corresponds to EV1A.
- Screening for phage‐resistant bacteria at the plaque rim. PY79 (WT) cells were infected with SPP1 (10‐6 PFU/ml) and plated for plaque formation. At t = 18 h, similar numbers of bacteria were collected from 30 "non‐plaque" and 30 "plaque rim" regions, and bacterial smears were plated over plates with (10‐4 PFU/ml) or without phages. No phage‐resistant colonies were detected in both populations.
Source data are available online for this figure.
SigX is necessary for plaque constriction
The phenomenon of plaque constriction directed by phage‐sensitive bacteria prompted us to postulate that bacteria residing at the plaque periphery could mount a transient phage tolerance response. We further reasoned that such a response could be orchestrated by one of the B. subtilis extracytoplasmic function (ECF) RNA polymerase sigma (σ) factors. These factors are activated in response to stress imposed on the cell envelope (Helmann, 2016) that could stem from remains of surrounding lysed cells. To examine this premise, we deleted each of the seven known ECF sigma factors of B. subtilis and assayed their impact on the final plaque size. Intriguingly, ∆sigX strain exhibited significantly larger plaques in comparison with wild‐type (WT) when challenged with SPP1 or Phi29 phages (Fig 2A; Appendix Fig S1A). Furthermore, monitoring plaque dynamics revealed that this size difference is due to the substantial attenuation of the plaque constriction phase (Fig 2B and C). Importantly, when grown in liquid cultures, ∆sigX cells propagated with kinetics similar to that of WT cells (Appendix Fig S1B), indicating that the observed phenotype was not due to growth perturbation. To further elucidate the role of SigX in counteracting phage spread, we challenged ∆sigX cells with SPP1 or Phi29 phages in liquid cultures. No difference in lysis kinetics was observed when bacteria were infected at 1:1 (phage:bacteria) multiplicity of infection (MOI). However, while infected with low MOI (phage:bacteria, 1:20), ∆sigX cells lysed significantly faster than WT cells (Fig 2D; Appendix Fig S1C), a phenotype that could be reversed by ectopic expression of sigX (Appendix Fig S1D). The low MOI might be equivalent to the bacteria:phage ratio reached during the phase of plaque constriction. These results are consistent with the view that uninfected bacteria induce a SigX‐regulated defense response, capable of tempering future phage infections.
Figure 2. SigX is required for plaque constriction.
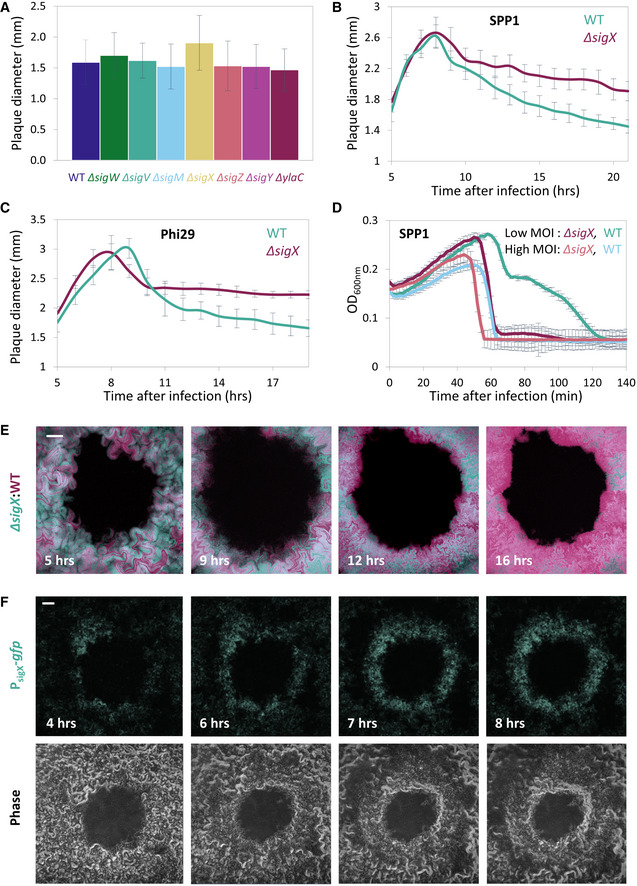
-
AThe indicated strains were infected with SPP1 (10‐6 PFU/ml), spread over MB plates, and plaque diameter was monitored after 20 h of incubation. Shown is average plaque diameter and SD for each strain (n ≥ 54).
-
B, CPlaque formation dynamic of SPP1 (B) and Phi29 (C) was monitored by automated scanning (Levin‐Reisman et al, 2010) on a lawn of infected (10‐6 PFU/ml) PY79 (WT) or ET19 (∆sigX) cells spread over an MB agar plate. Shown are average values and SD from kinetic of random plaques for each strain (n ≥ 7).
-
DPY79 (WT) and ET19 (∆sigX) cells were infected with SPP1 at either high (phages:bacteria 1:1) or low (1:20) MOI, and OD600nm was followed at 2‐min intervals. Shown is a representative experiment out of 6 biological repeats, and the average values and SD of 8 technical repeats.
-
EBDR2637 (Pveg ‐mCherry) (WT, purple) and ET191 (PrrnE ‐gfp, ∆sigX) (∆sigX, cyan) cells were mixed, infected with low concentrations (10‐8 PFU/ml) of SPP1, placed on an agarose pad, and plaque formation was followed by time‐lapse confocal microscopy. Shown are overlay images from mCherry (purple) and GFP (cyan) signals of the bacterial lawn captured at the indicated time points (h). The plaque is seen as a hole formed on the bacterial lawn. Scale bar, 100 µm. Corresponds to Movie EV4.
-
FET27 (PsigX ‐gfp) cells were infected with low concentrations (10‐8 PFU/ml) of SPP1, placed on an agarose pad, and plaque formation was followed by time‐lapse confocal microscopy. Shown are fluorescence from GFP (upper panels) and corresponding phase contrast images (lower panels), captured at the indicated time points (h). The plaque is seen as a hole formed on the bacterial lawn. Scale bar, 100 µm.
To compare the response to phage attack of WT and ∆sigX cells in real time, we mixed mCherry‐labeled WT cells with GFP‐labeled ∆sigX cells and monitored plaque dynamics following infection with SPP1, utilizing time‐lapse confocal microscopy. At the initial stages of phage spreading, both strains appeared to be infected and to be lysed equally (Fig 2E, t = 5, 9 h; Appendix Fig S2A; Movie EV4). However, during the constriction phase, when the bacteria re‐grew into the plaque zone, WT cells outcompeted the ∆sigX cells, as signified by the dominant colonization of the mCherry‐labeled WT cells at the plaque rim (Fig 2E, t = 12, 16 h; Fig EV2; Appendix Fig S2A, Movie EV4). When GFP‐ and mCherry‐labeled WT cells were mixed as a control, both strains were evenly distributed at the plaque edge even 16 hours postinfection (Fig EV2). Moreover, WT and ∆sigX cells equally occupied regions located remotely from visible plaque sites (Fig EV2), and, in accord, the growth rate of ∆sigX cells was not significantly affected by the presence of WT cells in a co‐culture (Appendix Fig S2C). Consistent with these results, fusion of the sigX promoter to gfp (PsigX‐gfp) specified that cells located at the plaque rim produced GFP chiefly during the constriction phase (Fig 2F and Appendix Fig S2B). Interestingly, infection at 48°C, a temperature shown to activate sigX expression (Huang et al, 1997), led to a significant reduction in plaque size in a sigX‐dependent manner (Fig EV3A). Of note, although no measurable difference between ∆sigX and WT growth kinetics was seen at 48°C (Fig EV3B), phage manufacture could be alleviated at high temperatures (Schachtele et al, 1970). In sum, we conclude that ∆sigX cells are deficient in inducing a defense mechanism that enables bacteria to tolerate the presence of phages and invade into the plaque zone.
Figure EV2. ∆sigX cells are excluded from the plaque rim during constriction.
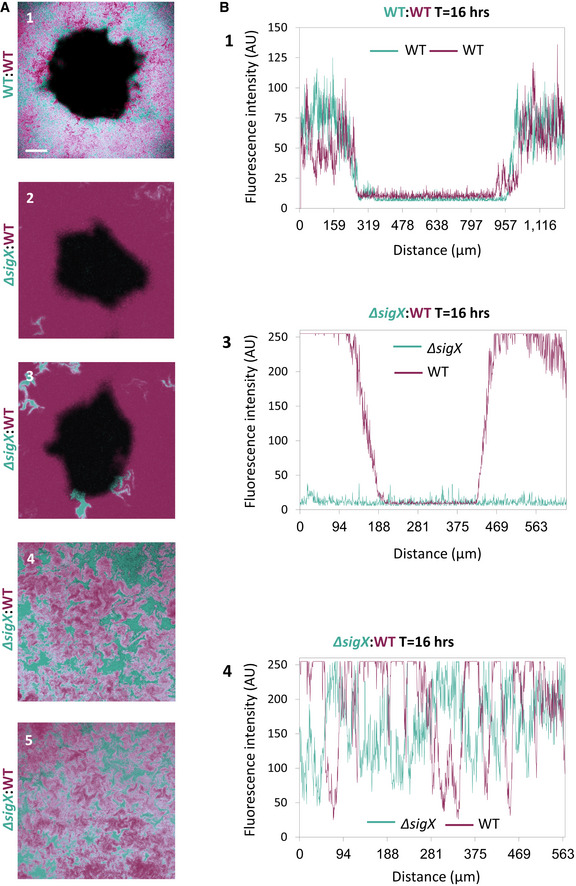
- BDR2637 (Pveg ‐mCherry) (WT) (purple) cells were mixed with AR16 (PrrnE ‐gfp) (WT) (cyan) (1) or with ET191 (∆sigX, PrrnE ‐gfp) (cyan) (2‐5) cells. The mixtures were infected with low concentrations (10‐8 PFU/ml) of SPP1, placed on an agarose pad, and plaque formation was followed by time‐lapse confocal microscopy. Shown are overlay images of mCherry (purple) and GFP (cyan) signals captured 16 h postinfection. (1‐3) show plaque regions, whereas (4‐5) show regions remote from any visible plaque site. Scale bar, 100 µm.
- Quantification of images 1, 3, and 4 presented in EV2A. Fluorescence intensity (AU) of the plaques formed by phages infecting the corresponding cells is shown. Fluorescence from Z sections that include the plaque region and flanking area or control areas was measured.
Source data are available online for this figure.
Figure EV3. Monitoring SigX activation during phage infection.
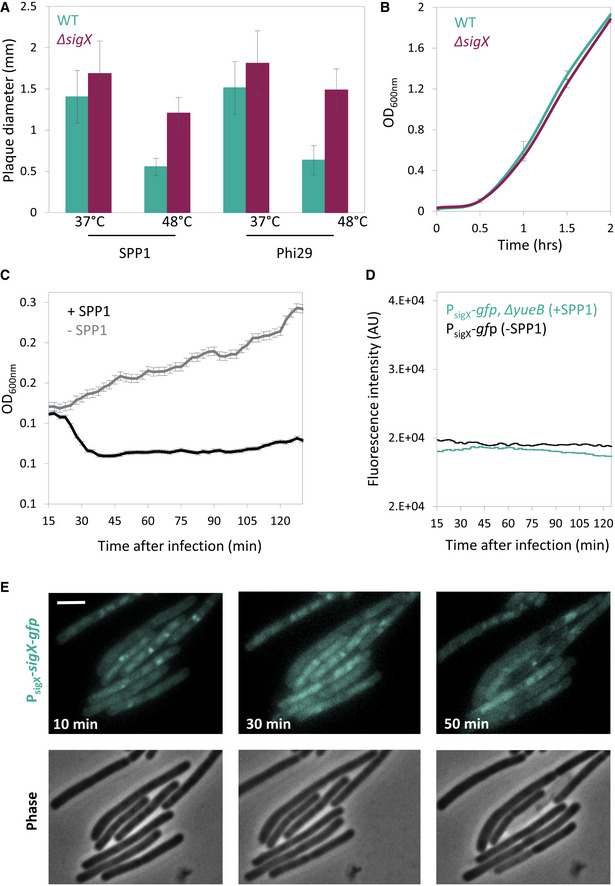
- PY79 (WT) and ET19 (∆sigX) cells were infected with SPP1 or Phi29 (10‐6 PFU/ml), spread over MB agar plates, and incubated at either 37°C or 48°C. Plaque diameter was monitored after 20 h of incubation (n ≥ 50). Shown are average values and SD of 3 independent repeats.
- PY79 (WT) and ET19 (∆sigX) were grown in liquid LB medium at 48°C and OD600nm monitored. Shown are average values and SD of 3 biological repeats.
- Corresponds to the experiment presented in Fig 3A. Phage‐sensitive PY79 (WT) cells were mixed with phage‐resistant BS12 (ΔyueB, PsigX‐gfp) cells and the mixture was infected with SPP1 at 2:1 (phages:bacteria) MOI, and OD600nm was followed at 2.5‐min intervals. Uninfected mixed population served as a control (‐SPP1). Shown is a representative experiment out of 3 biological repeats, and the average values and SD of n ≥ 3 technical repeats.
- BS12 (PsigX ‐gfp, ∆yueB) cells were infected with SPP1 at 2:1 (phages:bacteria) MOI, and fluorescence intensity from PsigX ‐gfp (AU) was followed at 2.5‐min intervals. Uninfected BS4 (PsigX ‐gfp) cells served as a control. Shown is a representative experiment out of 3 biological repeats, and the average values and SD of 3 technical repeats.
- ET26 (PsigX ‐sigX‐gfp) cells were infected with SPP1 at 5:1 (phages:bacteria) MOI, placed on an agarose pad, and followed by time‐lapse fluorescence microscopy. Shown are signal from SigX‐GFP (upper panels), and corresponding phase contrast images (lower panels), captured at the indicated time points postinfection. Scale bar, 1 µm.
Source data are available online for this figure.
SigX is activated in non‐infected bacteria following infection of their neighbors
The results so far raised the prospect that bacteria could sense a danger signal, emanating from nearby infected bacteria, and in turn activate SigX‐dependent phage tolerance response. To assay sigX induction in uninfected bacteria, SPP1 was added to WT cells that were co‐cultured with SPP1‐resistant bacteria, lacking the phage receptor (ΔyueB), and harboring the PsigX‐gfp reporter. Indeed, monitoring GFP fluorescence showed a continuous increase in sigX expression that reached the maximal level approximately 35 min postinfection and started to decline at t = 60 min (Figs 3A and EV3C and D). A similar fluorescence profile was obtained when the two strains were separated by a membrane that allows the passage of small molecules while compartmentalizing the cells (Fig 3B), suggesting that the sigX‐inducing factor is secreted into the shared medium. To further substantiate the ability of bacteria to activate SigX in response to nearby infected cells, we followed the production and localization of SigX protein during infection at the cellular level. In the absence of phages, a functional SigX‐GFP fusion (P sigX‐sigX‐gfp) mainly localized onto the membrane, frequently forming focal assemblies in proximity to the cell circumference and at septal positions (Fig 3C and Appendix Fig S1E). This localization pattern is consistent with previous reports, showing that SigX is sequestered to the plasma membrane by its anti‐sigma factor as a way to halt its action (Ho & Ellermeier, 2012). To assay SigX activity in uninfected bacteria, we added SPP1 phage to mCherry‐labeled WT bacteria mixed with ΔyueB phage‐resistant bacteria, harboring sigX‐gfp. Time‐lapse microscopy revealed repositioning of SigX‐GFP from membrane and foci locations to massive nucleoid deployment in the resistant bacteria (Fig 3D; t = 35 min), indicating a switch from an inactive to an active mode. Noticeably, this shift in localization occurred prior to lysis of nearby infected sensitive bacteria and corresponded to the increase in PsigX‐GFP signal observed (Fig 3A and B). SigX‐GFP level was dropped, and its localization into foci was largely restored in the resistant bacteria 95 min postinfection (Fig 3D), in line with the decline in sigX expression (Fig 3A and B), presumably corresponding to conclusion of the phage‐sensing response. Taken together, SigX appears to be activated in phage‐resistant bacteria upon sensing a danger signal from nearby infected sensitive cells. Notably, infecting sigX‐gfp‐sensitive cells with SPP1 showed that SigX‐GFP largely displaces its position form the membrane to the nucleoid in the course of infection (Fig EV3E), denoting that also infected cells activate the SigX response.
Figure 3. SigX is activated in non‐infected cells upon infection of their neighbors.
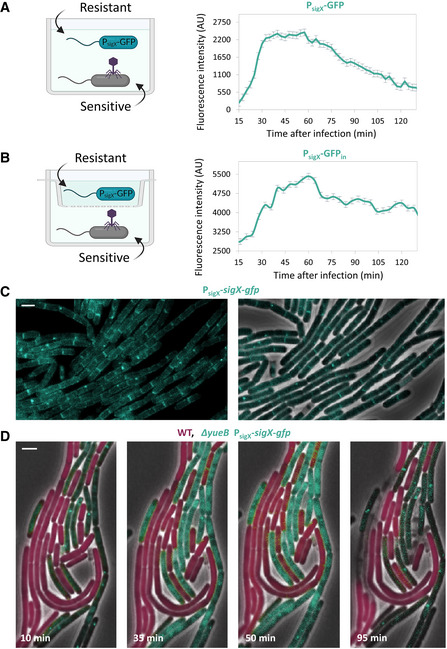
- Phage‐sensitive PY79 (WT) cells were mixed with phage‐resistant BS12 (ΔyueB, PsigX‐gfp) cells and the mixture was infected with SPP1, as illustrated. Infection was conducted at 2:1 (phages:bacteria) MOI, and fluorescence intensity from PsigX‐gfp (AU) was followed at 2.5‐min intervals. Uninfected mixed population served as a control, and its fluorescence was subtracted from the overall GFP signal of the infected culture. Shown is a representative experiment out of 3 biological repeats, and the average values and SD of 8 technical repeats.
- Phage‐sensitive PY79 (WT) cells were infected with SPP1 at 2:1 (phages:bacteria) MOI, and placed in the outer ring of a transwell, as illustrated. Phage‐resistant BS12 (ΔyueB, PsigX‐gfp) were placed in the inner ring. Fluorescence intensity from PsigX‐gfp (AU) of the inner compartment was followed at 2.5‐min intervals. Uninfected population served as a control, and its fluorescence was subtracted from the overall GFP signal of the infected culture. Shown is a representative experiment out of 3 biological repeats, and the average values and SD of 8 technical repeats.
- ET26 (PsigX ‐sigX‐gfp) cells were visualized by fluorescence microscopy. Shown are signal from SigX‐GFP (cyan) (left panel), and an overlay image of phase contrast (gray) and signal from SigX‐GFP (cyan) (right panel). Scale bar, 1 μm.
- BDR2637 (Pveg ‐mCherry) (WT, purple) and ET261 (∆yueB, PsigX ‐sigX‐gfp) (cyan) cells were mixed, infected with SPP1 at 5:1 (phages:bacteria) MOI, placed on an agarose pad, and followed by time‐lapse fluorescence microscopy. Shown are overlay images from mCherry (purple), SigX‐GFP (cyan), and phase contrast (gray), captured at the indicated time points postinfection. Scale bar, 1 μm.
Expression of SigX protects from phage infection
The impact of SigX on phage infection was further explored by constructing bacteria artificially expressing SigX under an IPTG‐inducible promoter. Remarkably, expressing sigX prior to phage addition markedly attenuated both SPP1 and Phi29 infections, with the cells being capable of extending the infection process (Fig 4A). Next, mCherry‐labeled cells, over‐expressing SigX (PIPTG ‐sigX), were incubated with non‐labeled WT cells, and the mixture was infected with SPP1‐lysin‐yfp. Consistent with the above observations, WT cells were rapidly infected and lysed, while cells over‐expressing SigX appeared to be infected at slower kinetics and to a lesser extent (Fig 4B and C), a phenomenon that was also observed during infection with Phi29 (Fig EV4A).
Figure 4. SigX expression confers phage tolerance.
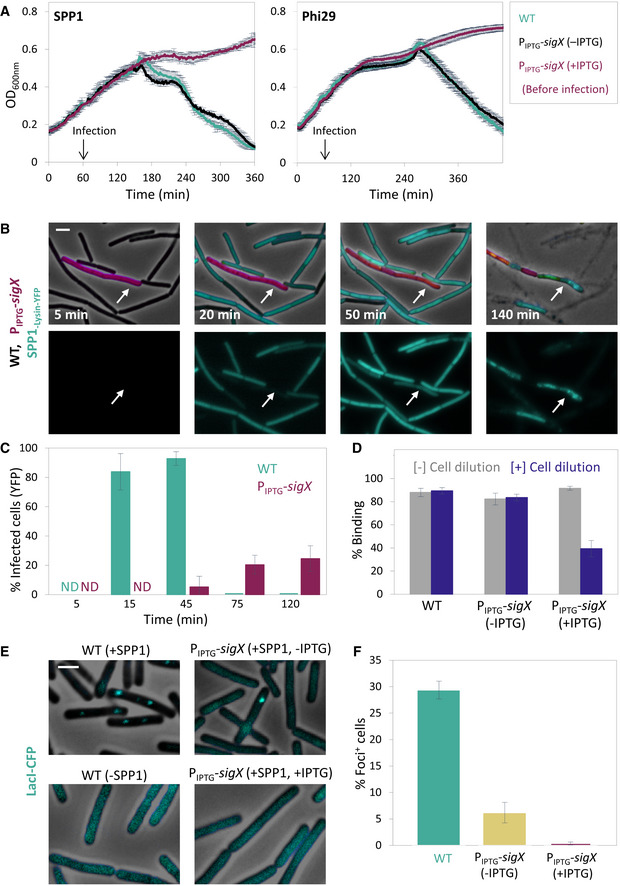
- PY79 (WT) and ET28 (PIPTG ‐sigX) cells were infected with SPP1 or Phi29 (t = 60 min) at 1:20 (phages:bacteria) MOI, and OD600nm was followed at 2‐min intervals. IPTG was added 30 min before infection (t = 30 min). Shown is a representative experiment out of 3 biological repeats, and the average values and SD of 4 technical repeats.
- ET29 (Pveg ‐mCherry, PIPTG ‐sigX) (purple) cells were grown in the presence of IPTG and mixed with PY79 (WT) cells. The mixture was infected with SPP1‐lysin‐yfp 5:1 (phages:bacteria) MOI, placed on an IPTG‐containing agarose pad, and followed by time‐lapse fluorescence microscopy. Shown are overlay images of phase contrast (gray), signal from mCherry‐labeled cells (purple), and signal from Lysin‐SPP1‐YFP (cyan), captured at the indicated time points postinfection (upper panels). Corresponding signal from Lysin‐SPP1‐YFP (cyan) is shown separately (lower panels). Arrows highlight the delayed infection of ET29 cells. Scale bar, 1 μm.
- Quantification of the experiment described in (B). Shown is the percentage of phage infected PY79 (WT) and ET29 (Pveg ‐mCherry, PIPTG ‐sigX) cells at the indicated time points, scored by the Lysin‐SPP1‐YFP signal, with average values and SD (n ≥ 200 cells for each time point). Of note, the majority of WT cells were lysed at t = 75 min postinfection.
- PY79 (WT) and ET28 (PIPTG ‐sigX) cells, grown in the presence or absence of IPTG, were infected with SPP1 (1:1 MOI) for 10 min. Next, phage adsorption was monitored before and after cell dilution (×100 fold). Percentage of phage adsorption was calculated as follows: (P0−P1) × 100/P0, where P0 is the initial phage input in the lysate (PFU/ml), and P1 is the titer of free phages (PFU/ml) 10 min after infection. Shown are average values and SD of a representative experiment out of 3 independent experiments.
- OF83 (Ppen ‐lacIΔ11‐cfp) (WT) and ET40 (Ppen ‐lacIΔ11‐cfp, PIPTG ‐sigX) cells, grown in the presence or absence of IPTG, were infected with SPP1‐ delX110lacO64 at 5:1 (phages:bacteria) MOI. The formation of LacI‐CFP foci on phage DNA was monitored 10 min postinfection. Non‐infected OF83 cells were used for comparison. Shown are overlay images of phase contrast (gray) and signal from LacI‐CFP (cyan). Scale bar, 1 µm.
- Quantification of the experiment described in (E). Shown is the percentage of LacI‐CFP foci 10 min postinfection of OF83 and ET40 cells by SPP1, with average values and SD (n ≥ 850 cells for each population). This is a representative experiment out of 3 biological repeats.
Source data are available online for this figure.
Figure EV4. SigX over‐expression interferes with phage infection.
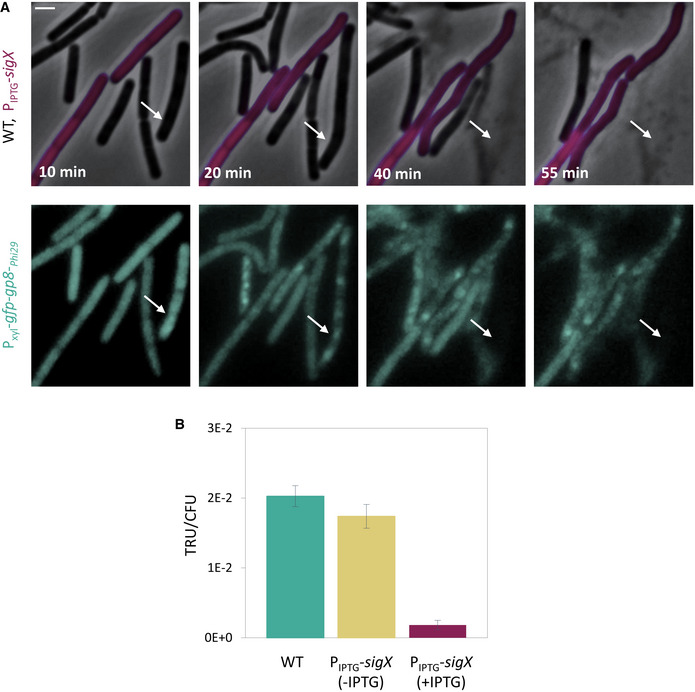
- ET9 (Pxyl ‐gfp‐gp8‐ Phi29 ) (WT) and ET44 (Pveg ‐mCherry, PIPTG ‐sigX, Pxyl ‐gfp‐gp8‐Phi29 ) (PIPTG ‐sigX, purple) cells were grown in the presence of IPTG and xylose, mixed, and infected with Phi29 at 5:1 (phages:bacteria) MOI. The mixture was placed on an IPTG and xylose‐containing agarose pad and followed by time‐lapse fluorescence microscopy. Gp8‐Phi29 is the major Phi29 capsid protein that localizes into discrete foci during Phi29 infection (Tzipilevich et al, 2017). Shown are overlay images of phase contrast (gray) and signal from mCherry (purple) (upper panels), and signal from GFP‐Gp8‐Phi29 (cyan) (lower panels), captured at the indicated time points. Arrows highlight GFP‐Gp8‐Phi29 foci appearance in ET9 cells. Scale bar, 1 µm.
- PY79 (WT) and ET28 (PIPTG ‐sigX), grown in the presence or absence of IPTG, were transduced with SPP1‐pBT163 lysate, and the number of transductants was monitored by plating the cells on corresponding selective plates. Transduction unit (TRU) was calculated as the number of transductant colonies /total CFU. Shown are average values and SD of 3 biological replicates.
Source data are available online for this figure.
To define the specific stage at which phage infection was interrupted by SigX activity, we followed the adsorption of phages to cells over‐expressing sigX. A standard adsorption assay yielded no significant difference in SPP1 adsorption rate between WT and SigX expressing cells (Fig 4D). Nonetheless, since SPP1 phage exhibits two modes of cell surface binding, reversible in which it associates with gWTA polymers, and irreversible, through interaction with the YueB receptor (Baptista et al, 2008), it was still conceivable that the irreversible mode of binding was impaired. To inspect this possibility, cells were diluted after an initial phage adsorption period to enable reversibly adsorbed phages to detach from the host (Baptista et al, 2008). Indeed, a large fraction of phages, adsorbed to sigX over‐expressing cells, were liberated after dilution, whereas no significant release of WT‐attached phages was detected (Fig 4D), indicating that SigX expression delays SPP1 irreversible binding. To corroborate this finding, we investigated whether phage DNA injection is consequently delayed by induced expression of SigX. To monitor phage DNA injection, we utilized SPP1‐ delX110lacO64 phage, which contains 64 repeats of lacO (Jakutyte et al, 2012), and infected bacteria chromosomally expressing lacI‐cfp. The presence of phage DNA within the host cytoplasm was visualized subsequent to injection by the formation of LacI‐CFP foci (Fernandes et al, 2016; Tzipilevich et al, 2017). Infection of WT cells resulted in foci appearance within 10 min after infection, while no foci were observed in sigX over‐expressing cells at the same time point (Fig 4E and F), indicating a delay in phage DNA penetration into the latter cell population. Consistent with this possibility, a significant decrease in SPP1‐mediated plasmid transduction rate into SigX‐producing cells was monitored (Fig EV4B). Thus, we surmise that sigX expression interferes with SPP1 irreversible binding and consequently delays phage DNA injection.
The dlt operon mediates the SigX tolerance response to phage infection
To identify the gene(s) required for SigX‐mediated phage tolerance, we mutated known genes in the SigX regulon (Huang & Helmann, 1998) and tested their impact on the phage protection phenotypes. Out of the mutants tested, only the disruption of ywbO, encoding a predicted disulfide oxidoreductase, and that of the dlt operon (∆dltA), largely countered the tolerance to phage infection conferred by SigX over‐expression (Figs 5A and EV5A). The dlt operon encodes an enzymatic pathway that is known to ligate D‐alanine moieties to TA polymers (Perego et al, 1995), the major phage surface attachment components, and was therefore selected for further investigation. Examination of infected ΔdltA mutant cells harboring inducible sigX by fluorescence microscopy substantiated that they were lysed with kinetics similar to that of WT cells in the presence of the inducer (Fig EV5B). Furthermore, deletion of dltA restored the capacity of SPP1 to bind irreversibly to the surface of sigX over‐expressing cells, and consistently increased the level of SPP1 DNA injection into these cells, as detected by transduction assay (Fig 5B and C). Lastly, deletion of dltA, in an otherwise WT background, resulted in enhanced sensitivity to infection at low MOI, whereas over‐expression of the dlt operon could increase resistance to phages compared to WT (Fig EV5C and D).
Figure 5. The dlt operon mediates the SigX‐induced phage tolerance response.
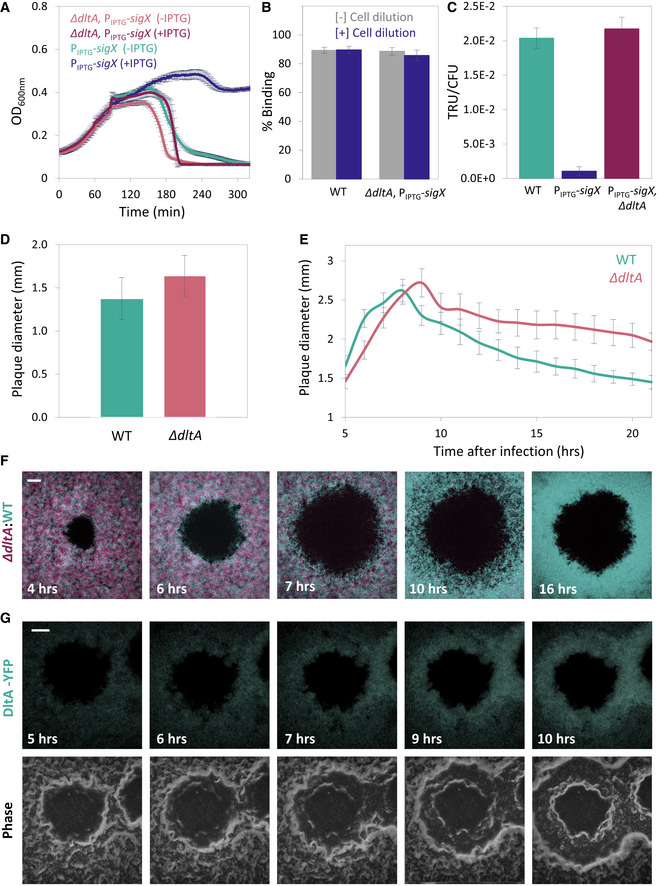
- ET28 (PIPTG ‐sigX) and ET42 (∆dltA, PIPTG ‐sigX) cells, grown in the presence or absence of IPTG, were infected with SPP1 at 1:20 (phages:bacteria) MOI, and OD600nm was followed at 2‐min intervals. Shown is a representative experiment out of 6 biological repeats, with the average values and SD of 8 technical repeats.
- PY79 (WT) and ET42 (ΔdltA, PIPTG ‐sigX) cells, grown in the presence of IPTG, were infected with SPP1 (1:1 MOI) for 10 min. Next, phage adsorption was monitored before and after cell dilution (×100‐fold). Percentage of phage adsorption was calculated as follows: (P0−P1) × 100/P0, where P0 is the initial phage input in the lysate (PFU/ml), and P1 is the titer of free phages (PFU/ml) 10 min after infection. Shown are average values and SD of 5 biological repeats.
- PY79 (WT), ET28 (PIPTG ‐sigX), and ET42 (ΔdltA, PIPTG ‐sigX), grown in the presence of IPTG, were transduced with SPP1‐pBT163 lysate, and the number of transductants was monitored by plating the cells on selective plates. Transduction unit (TRU) was calculated as the number of transductant colonies /total colony‐forming unit (CFU). Shown are average values and SD of 3 biological repeats.
- PY79 (WT) and ET41 (ΔdltA) cells were infected with SPP1 (10‐6 PFU/ml), spread over an MB agar plate, and plaque diameter was monitored after 20 h of incubation. Shown is plaque diameter distributions for each strain (n ≥ 40).
- Plaque formation dynamic of SPP1 was monitored by automated scanning (Levin‐Reisman et al, 2010) on a lawn of infected (10‐6 PFU/ml) PY79 (WT) or ET41 (ΔdltA) cells grown on an MB agar plate. Shown are average values and SD from kinetics random plaques for each strain (n ≥ 10).
- AR16 (PrrnE ‐gfp) (WT, cyan) and ET411 (Pveg ‐mCherry, ∆dltA) (∆dltA, purple) cells were mixed, infected with low concentrations (10‐8 PFU/ml) of SPP1, placed on an agarose pad, and plaque formation was followed by time‐lapse confocal microscopy. Shown are overlay images from GFP (cyan) and mCherry (purple) signals of the bacterial lawn captured at the indicated time points (h). The plaque is seen as a hole formed on the bacterial lawn. Scale bar, 150 µm.
- ET43 (dltA‐yfp) cells were infected with low concentrations (10‐8 PFU/ml) of SPP1, placed on an agarose pad, and plaque formation was followed by time‐lapse confocal microscopy. Shown are fluoresce from DltA‐YFP signal (upper panels) and corresponding phase contrast images (lower panels), captured at the indicated time points (h). The plaque is seen as a hole formed on the bacterial lawn. Scale bar, 100 µm.
Figure EV5. SigX impact on phage tolerance is Dlt‐mediated.
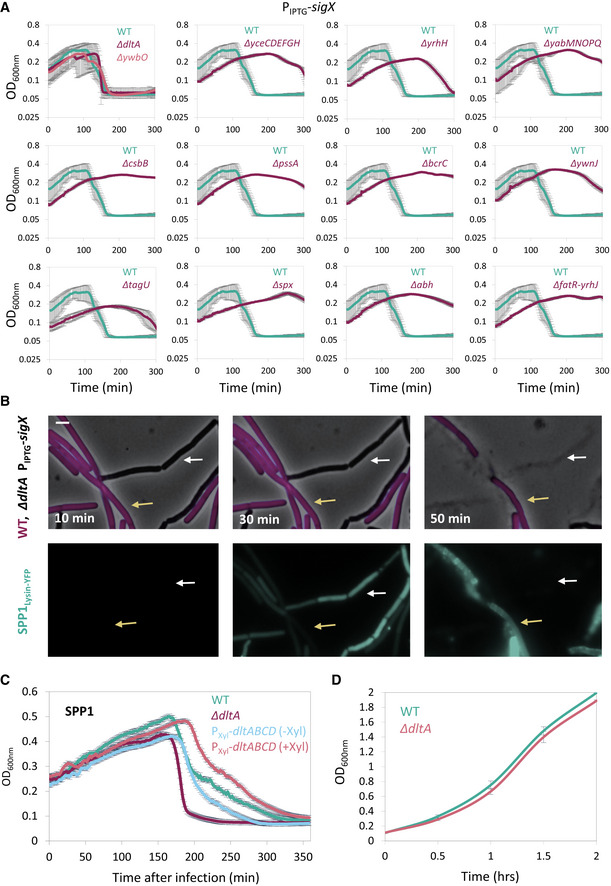
- Bacterial strains harboring PIPTG ‐sigX as well as the indicated gene deletions were grown in the presence of IPTG. At t = 60 min, cells were infected with SPP1 at low (phages:bacteria 1:20) MOI, and OD600nm was followed at 2‐min intervals. PY79 (WT) was infected in parallel for comparison. Knockout of ywbO and the dlt operon (ΔdltA) largely abolished the tolerance to phage infection conferred by SigX over‐expression. Shown is a representative experiment out of 3 independent biological repeats, with average values and SD of 3 technical repeats.
- ET42 (ΔdltA, PIPTG ‐sigX) cells were grown in the presence of IPTG and mixed with BDR2637 (Pveg ‐mCherry) (WT, purple) cells. The mixture was infected with SPP1‐lysin‐yfp at 5:1 (phages:bacteria) MOI, placed on an IPTG‐containing agarose pad, and followed by time‐lapse fluorescence microscopy. Shown are overlay images of phase contrast (gray) and signal from mCherry‐labeled cells (purple) (upper panels), and the corresponding signal from Lysin‐SPP1‐YFP (green) (lower panels), captured at the indicated time points. Yellow arrows denote infected WT cells, whereas white arrows highlight infected ET42 cells that lysed rapidly. Scale bar, 1 μm.
- PY79 (WT), ET41 (∆dltA), and ET72 (PXyl‐dltABCD) cells, grown with or without xylose as indicated, were infected with SPP1 at low (1:20) MOI, and OD600nm was followed at 2‐min intervals. Shown is a representative experiment out of 2 biological repeats, and the average values and SD of 4 technical repeats.
- PY79 (WT) and ET41 (ΔdltA) cells were grown in LB liquid medium, and OD600nm was followed. Shown are average values and SD of 3 biological repeats.
Source data are available online for this figure.
To substantiate the role of the dlt operon in the phage tolerance response, we examined the phenotype of ∆dltA during plaque generation. ∆dltA cells exhibited plaques larger than that of the WT, along with prominent deficiency in plaque constriction phase (Fig 5D and E), similar phenotypes to those observed for the ∆sigX cells (Fig 2A and B). Next, we mixed differentially labeled ∆dltA and WT cells, and followed plaque formation dynamics by time‐lapse confocal microscopy. While the two populations were evenly distributed at early stages of plaque generation, WT cells were manifestly dominating the plaque rim during constriction (Fig 5F and Appendix Fig S3A), indicating a clear deficiency of the mutant cells in opposing infection. Monitoring DltA expression by following DltA‐YFP showed an enrichment of the fusion protein preferentially at the edge of the constricting plaque (Fig 5G and Appendix Fig S3B), supporting a role in mediating this process. In sum, the majority of the phage protection phenotypes conferenced by SigX can be assigned to the dlt operon, encoding enzymes that modify the TA surface polymers.
Discussion
By following the dynamics of plaque formation on lawns of the soil bacterium B. subtilis, we discovered that plaque development includes a phase of constriction, typified by bacterial growth into the plaque zone, counteracting plaque expansion. Examination of bacteria located at the plaque rim subsequent to constriction revealed that they are not genetically resistant to phages, but instead elicit a temporary phage tolerance response, activated by the stress‐induced RNA polymerase sigma factor σX. We further uncovered that the impact of SigX on tolerance is not restricted to plaques, as uninfected bacteria activated sigX expression following infection of their neighbors in a co‐culture. Furthermore, pre‐expression of sigX prior to phage addition was found to protect from phage attack. SigX tolerance is mostly attributed to the action of the dlt operon, encoding for WTA modifying enzymes, thereby altering the polymer properties. Based on our results, we propose that uninfected bacteria can sense infection of their neighbors, and in turn trigger a tolerance response, modifying their phage attachment surface components to antagonize phage penetration (Fig 6). As such, the cells at the plaque rim form a protective barrier that locally constrains plaque spread, shielding the non‐infected population.
Figure 6. A model for eliciting phage tolerance response by uninfected bacteria.
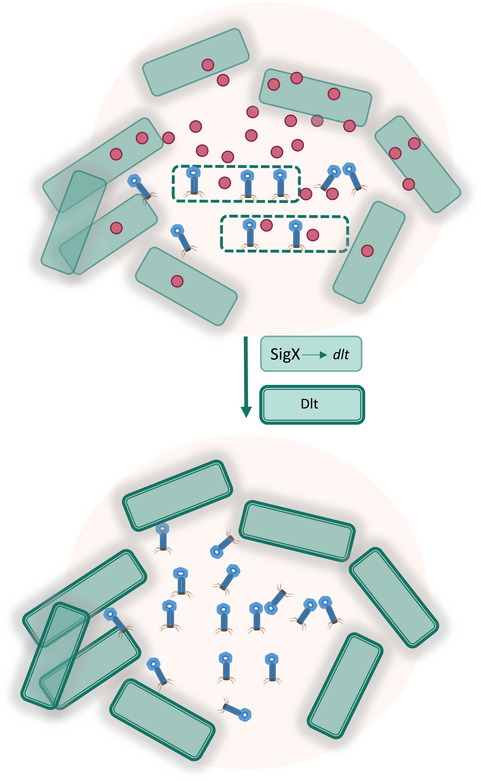
Upon phage infection, bacteria (dashed‐line cells) generate a yet unidentified "danger signal" (red circles) that is received by uninfected neighboring cells. In turn, the latter cells activate SigX that induces the transcription of the dlt operon. The produced Dlt enzymes modulate the phage receptor WTA polymers to reduce phage binding, providing temporary protection against phage attack and limiting phage spread.
This scenario resembles the eukaryotic innate immune response, with the hallmark being interferon released by infected cells and received by neighbors to activate an anti‐viral response (Isaacs & Lindenmann, 1957; McNab et al, 2015). In addition, endogenous danger signals, such as extracellular ATP and DNA, released by infected eukaryotic cells, stimulate innate immunity (Gallucci & Matzinger, 2001), a program that could be applicable for activating the phage tolerance response. Importantly, damaged‐self recognition stimulated by similar factors exists in plants and even in algae and fungi (Heil & Land, 2014). Interestingly, it has been shown that bacteria possess the cyclic GMP–AMP synthase (cGAS)–STING pathway, a central component of the mammalian innate immune system, as part of an anti‐phage defense mechanism (Cohen et al, 2019; Morehouse et al, 2020). The observation that sigX expression in non‐infected bacteria can be stimulated by compartmentally separated infected cells suggests that the signaling factor is a small molecule, capable of passing thought a small sized pore. Such molecule might determine a threshold level required for robust and efficient activation of the tolerance response seen during advanced stages of plaque development. The effectiveness of tolerance was also MOI‐dependent, suggesting that phage to bacteria ratio at the plaque rim decreases eventually to allow a powerful response. It is thus possible that phages exhibiting large burst size are more adapted to antagonize tolerance.
Our data indicate that bacteria acquire phage tolerance chiefly by remodeling their cell surface, decorating WTA polymers with D‐alanine residues (Brown et al, 2013). D‐alanylation of TA polymers is executed by a series of chemical reactions performed by the Dlt enzymes (Perego et al, 1995; Percy & Grundling, 2014; Ma et al, 2018). It has been shown that TA D‐alanylation contributes to resistance against cationic antimicrobial peptides and lysozyme (Kovacs et al, 2006; Kingston et al, 2013), presumably due to changes in the cell surface electric charge or increase in peptidoglycan density (Saar‐Dover et al, 2012; Percy & Grundling, 2014). D‐alanylation of TA was also shown to enhance host cell adhesion and virulence of Gram‐positive pathogens such as Staphylococcus aureus and B. anthracis (Simanski et al, 2013; Percy & Grundling, 2014). Since TA glycosylation is known to serve as a pervasive phage binding molecule (Young, 1967; Habusha et al, 2019), it is plausible that WTA‐D‐alanylation masks WTA glycosylated sites, and/or interferes with bacteriophage access to the membrane. Notably, apart from the dlt operon, ywbO appears to contribute to phage tolerance. Recently, mutations in ywbO promoter region were linked to impediment in TA glycosylation, pointing at an additional path through which phage tolerance by surface modulation might be achieved (Tzipilevich & Benfey, 2021).
The induction of the phage tolerance response was shown to be mediated by SigX, belonging to the ECF family that monitors cell wall integrity. SigX is recruited to the membrane by its cognate anti‐sigma factor and is liberated to the nucleoid in response to envelope stress to transcribe downstream target genes, with one of the most prominent being the dlt operon (Cao & Helmann, 2004; Helmann, 2016). A sigX mutant strain was shown to be sensitive to heat and oxidative stress, and to be susceptible to cationic antimicrobial peptide (Huang et al, 1997; Cao & Helmann, 2004). Indeed, we found that elevated growth temperature seems to protect bacteria from phage attack in a SigX‐dependent manner, hinting that cross‐activation of ECF enables bacteria to simultaneously resist multiple stress conditions, similarly to SOS response (Storz, 2016; Gottesman, 2019). Still, the nature of the activating signals and how they are transduced to release the membrane‐attached SigX have yet to be elucidated. Since ECFs are widespread among bacteria (Helmann, 2002), it is tempting to assume that, similarly to B. subtilis, many species have the capacity to execute such a defense strategy, following infection of nearby bacteria. We postulate that such phage tolerance might expedite the acquisition of permanent phage resistance mutations in nature, similarly to tolerance to antibiotic stress that facilitates emergence of antibiotic resistance mutants (Liu et al, 2020).
Bacteria have evolved numerous remarkable phage resistance strategies to counteract infection, including restriction‐modification systems, abortive infection, CRISPR‐Cas immunity (Labrie et al, 2010; Salmond & Fineran, 2015), and a plethora of recently identified additional exciting systems [e.g., Cohen et al (2019), Doron et al (2018), Gao et al (2020), Makarova et al (2011)]. Still, relatively little is known about the dynamics of the activation of these systems within populations. For instance, it is not entirely understood how activation of the various CRISPR‐Cas systems is prompted. There is evidence for constitutive activity of Cas genes in some bacteria, and for quorum sensing‐mediated transcription of Cas genes in others (Patterson et al, 2017; Hampton et al, 2020). Consistently, proteomic analysis revealed that phage infection elevates Cas production in Streptococcus thermophilus (Young et al, 2012). Knowledge concerning population dynamics of other phage defense systems is even more fragmentary. Here, we uncovered a general phage tolerance mechanism that provides only temporary protection by phenotypic modulation of the bacterial cell surface. To the best of our knowledge, this is the first detailed characterization of a phage defense system in space and time.
Materials and Methods
Strains and plasmids
Bacillus subtilis strains were derivatives of the wild‐type PY79 (Youngman et al, 1984). All bacterial strains and phages are listed in Appendix Table S1. Plasmid constructions were performed in E. coli DH5α using standard methods and are listed in Appendix Table S1. All primers used in this study are listed in Appendix Table S2.
General growth conditions
Bacterial cultures were inoculated at OD600nm 0.05 from an overnight culture and growth was carried out at 37°C, unless indicated otherwise, in LB medium supplemented with 5 mM MgCl2 and 0.5 mM MnCl2 (MB). Fluorescence measurement experiments in plate reader were conducted in CH (10% casein hydrolysate) medium supplemented with 5 mM MgCl2 and 0.5 mM MnCl2 (Harwood & Cutting, 1990). For induction, all P hyper‐spank controlled genes were induced with isopropyl‐β‐D‐thiogalactopyranoside (IPTG) at concentration of 0.5 mM.
General phage infection and transduction methodologies
Phage lysate was prepared by adding approximately 109 phages to 1 ml mid‐log cells grown in MB until the culture was completely cleared. Next, the lysate was filtered through 0.45‐µm or 0.22‐µm Millipore filter. For lysis dynamic experiments, phages were added to mid‐log growing cells at the indicated MOI, and OD600nm was monitored. For SigX‐induction experiments, cells were grown for 30 min in the absence of inducer. Subsequently, inducer was added and cells were grown for additional 30 min. Phages were added after overall growth of 60 min, and OD600nm was followed by Spark 10M (Tecan) multiwell fluorometer set at 37°C with a constant shaking.
For transduction experiments, lysates were prepared from a given donor strain as describe above. All lysates for transduction were treated with DNase I (Sigma‐Aldrich) 200 ng/ml for 20 min at RT. Recipient strains were grown to 1.0 OD600nm, and cells (1 ml) were mixed with 100 µl of lysate and 9 ml of MB, and incubated at 37°C without shaking for 20 min. Subsequently, cells were centrifuged and spread on selective plates supplemented with 10 mM sodium citrate. For burst size measurement, 1:104 phages:bacteria were added to a mid‐log culture, and 30 min later, the culture was lysed with chloroform and the number of phages in the culture was measured.
Phage attachment assay
Indicated bacterial strains were grown in a liquid culture to 0.6 OD600 nm, and phage adsorption to the cells was measured at 10 min postinfection, by titrating the free phages present in the supernatant as previously described (Ellis & Delbruck, 1939). In brief, logarithmic cells were grown in MB at 37°C till 0.8 OD600nm; then, 15 mM CaCl2 and 50 µg chloramphenicol/ml were added to the medium and cells were incubated for 10 min. Next, cells were infected with phages (107 PFU/ml), and samples (0.5 ml) were collected at 10 min postinfection and centrifuged for 1 min, and 50 µl of the supernatant was diluted and plated, and PFU/ml was determined. To measure reversible attachment, 10 min postinfection, cells were diluted 100 fold in fresh MB and incubated for 2 min in 37°C, and free phages present in the supernatant were plated and PFU/ml was determined.
Plaque size determination
For final plaque size determination, bacteria from mid‐log culture were infected with low concentration of phages (10‐6 PFU/ml) and spread over 1.5% MB agar plates at 37°C or 48°C for 20 h. Next, plates were photographed and plaque size was determined by measuring the size of random plaques. Image processing was performed using MetaMorph 7.4 software (Molecular Devices). For analyzing plaque growth dynamics on agarose plates, bacteria were infected as describe above, and 1.5% MB agar plates were incubated at 37°C on top of a scanner and covered with a dark paper cover. An automated scanning program (Levin‐Reisman et al, 2010) was utilized for time‐lapse imaging of the plates. Plaque size was determined by measuring the size of random plaques at intervals of 1 h. Image processing was performed using MetaMorph 7.4 software (Molecular Devices).
Fluorescence microscopy
For fluorescence microscopy, bacterial cells (0.5 ml, OD600nm 0.5) were centrifuged and suspended in 50 μl of MB. For time‐lapse microscopy, bacteria were placed over 1.5% MB agarose pad and incubated in a temperature controlled chamber at 37°C. Infection experiments were carried out at 5:1 (phages:bacteria) MOI. Samples were photographed using Axio Observer Z1 (Zeiss) or Eclipse Ti microscope (Nikon, Japan), equipped with CoolSnap HQII camera (Photometrics, Roper Scientific, USA). System control and image processing were performed using MetaMorph 7.4 software (Molecular Devices) or NIS Elements AR 4.3 (Nikon, Japan).
Live imaging of developing plaques by confocal microscopy
A custom designed construct (Mamou et al, 2016) was used to monitor bacterial plaques by confocal microscopy. Accordingly, a 40‐mm metal ring was filled with 1.5% MB agarose and assembled, and the bacterial cells were infected at low MOI (10‐8 PFU/ml) and spotted over the agarose pad. Plaque growth construct was covered with a 35‐mm cultFoil membrane (Pecon) to reduce agar dehydration and incubated in Lab‐Tek S1 heating insert (Pecon) placed inside an incubator XL‐LSM 710 S1 (Pecon). Initial plaques could be observed under the microscope at t = 4 h. Developing plaques were visualized and photographed by CLSM LSM700 (Zeiss). Cells expressing GFP or YFP were irradiated using 488 nm laser beam, while mCherry‐expressing cells were irradiated using 555 nm laser beam. For each experiment, both transmitted and reflected light were collected from 6 consecutive Z positions by 10‐µm steps. System control, image processing, and fluorescence quantifications were carried using Zen software version 5.5 (Zeiss).
Fluorescence intensity measurements by a plate reader
For continuous measurements of fluorescence intensity and OD600nm, cells were grown in CH (due to a low background fluorescence of the medium), at 37°C until 0.2‐0.5 OD600nm, and infected with SPP1 or Phi29. Fluorescence intensity (AU) and OD600nm were measured by Spark 10M (Tecan) multiwell fluorometer plate reader every 2/2.5 min at 37°C with constant shaking. A 12‐well Transwell Permeable Supports plates with 12‐mm‐diameter inserts was used for the transwell assay. In transwell experiments, sensitive and resistant cells were separated by a 0.4‐µm polycarbonate membrane. The plate was read either at a setting to read the full well or only the center (insert). OD600nm cannot be faithfully read by Spark 10M in transwells due to light distortion. Thus, for transwell experiments, samples were tested for OD600nm in a spectrophotometer.
Statistical analysis
Unless stated otherwise, bar charts and graphs display a mean ± SD from at least 3 repeats. Quantifications of CPU, PFU, and infected cells were done manually. MS Excel was used for all statistical analysis, data processing, and presentation.
Author contributions
ET and OPF involved in conception and design, acquisition, analysis, interpretation of data, writing, and revising the article; BS performed acquisition of data, interpretation and analysis of data, and revising the article; and SBY performed conception and design, analysis and interpretation of data, writing and revising the article, supervision, and funding.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Source Data for Expanded View and Appendix
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgments
We thank R. Losick (Harvard University, USA), I. Rosenshine and A. Rouvinski (Hebrew University, Israel), and members of the Ben‐Yehuda laboratory for valuable discussions. We thank N. Balaban and her laboratory members (Hebrew University, Israel) for assistance with the automated plaque scanning procedure. We are grateful to P. Tavers (Gif‐sur‐Yvette, France), J. Helmann (Cornell University, USA), and D. Rudner (Harvard University, USA) for providing strains. Artwork in Fig 3 and synopsis was prepared using BioRender (Agreement Number II2338U308, CN2368MHCR). This work was supported by the NSF/BSF‐United States‐Israel Binational Science Foundation (2017672), and the ERC Synergy grant (810186) awarded to S. B.‐Y.
Conflict of interest
The authors declare that they have no conflict of interest.
The EMBO Journal (2022) 41: e109247
See also: EE Maffei & A Harms (February 2022)
Data availability
There are no public datasets associated with the results presented in this manuscript.
References
- Abedon ST, Yin J (2009) Bacteriophage plaques: theory and analysis. Methods Mol Biol 501: 161–174 [DOI] [PubMed] [Google Scholar]
- Alonso JC, Luder G, Stiege AC, Chai S, Weise F, Trautner TA (1997) The complete nucleotide sequence and functional organization of Bacillus subtilis bacteriophage SPP1. Gene 204: 201–212 [DOI] [PubMed] [Google Scholar]
- Baptista C, Santos MA, Sao‐Jose C (2008) Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J Bacteriol 190: 4989–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S, Santa Maria JP Jr, Walker S (2013) Wall teichoic acids of gram‐positive bacteria. Annu Rev Microbiol 67: 313–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Helmann JD (2004) The Bacillus subtilis extracytoplasmic‐function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J Bacteriol 186: 1136–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanishvili N (2012) Phage therapy‐history from Twort and d'Herelle through Soviet experience to current approaches. Adv Virus Res 83: 3–40 [DOI] [PubMed] [Google Scholar]
- Cohen D, Melamed S, Millman A, Shulman G, Oppenheimer‐Shaanan Y, Kacen A, Doron S, Amitai G, Sorek R (2019) Cyclic GMP‐AMP signalling protects bacteria against viral infection. Nature 574: 691–695 [DOI] [PubMed] [Google Scholar]
- d'Hérelles F (1917) Sur un microbe invisible antagoniste des bacilles dysentériques. C R Acad Sci Paris 165: 373–375 [Google Scholar]
- Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R (2018) Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359: eaar4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis EL, Delbruck M (1939) The growth of bacteriophage. J Gen Physiol 22: 365–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes S, Labarde A, Baptista C, Jakutyte L, Tavares P, Sao‐Jose C (2016) A non‐invasive method for studying viral DNA delivery to bacteria reveals key requirements for phage SPP1 DNA entry in Bacillus subtilis cells. Virology 495: 79–91 [DOI] [PubMed] [Google Scholar]
- Gallucci S, Matzinger P (2001) Danger signals: SOS to the immune system. Curr Opin Immunol 13: 114–119 [DOI] [PubMed] [Google Scholar]
- Gao L, Altae‐Tran H, Bohning F, Makarova KS, Segel M, Schmid‐Burgk JL, Koob J, Wolf YI, Koonin EV, Zhang F (2020) Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369: 1077–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S (2019) Trouble is coming: signaling pathways that regulate general stress responses in bacteria. J Biol Chem 294: 11685–11700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habusha M, Tzipilevich E, Fiyaksel O, Ben‐Yehuda S (2019) A mutant bacteriophage evolved to infect resistant bacteria gained a broader host range. Mol Microbiol 111: 1463–1475 [DOI] [PubMed] [Google Scholar]
- Hampton HG, Watson BNJ, Fineran PC (2020) The arms race between bacteria and their phage foes. Nature 577: 327–336 [DOI] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM (1990) Molecular biological methods for Bacillus . Chichester, New York: Wiley; [Google Scholar]
- Heil M, Land WG (2014) Danger signals – damaged‐self recognition across the tree of life. Front Plant Sci 5: 578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD (2002) The extracytoplasmic function (ECF) sigma factors. Adv Microb Physiol 46: 47–110 [DOI] [PubMed] [Google Scholar]
- Helmann JD (2016) Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr Opin Microbiol 30: 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TD, Ellermeier CD (2012) Extra cytoplasmic function sigma factor activation. Curr Opin Microbiol 15: 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Decatur A, Sorokin A, Helmann JD (1997) The Bacillus subtilis sigma(X) protein is an extracytoplasmic function sigma factor contributing to survival at high temperature. J Bacteriol 179: 2915–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Helmann JD (1998) Identification of target promoters for the Bacillus subtilis sigma X factor using a consensus‐directed search. J Mol Biol 279: 165–173 [DOI] [PubMed] [Google Scholar]
- Ingmer H, Gerlach D, Wolz C (2019) Temperate phages of Staphylococcus aureus . Microbiol Spectr 7: GPP3‐0058‐2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs A, Lindenmann J (1957) Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci 147: 258–267 [PubMed] [Google Scholar]
- Jakutyte L, Lurz R, Baptista C, Carballido‐Lopez R, Sao‐Jose C, Tavares P, Daugelavicius R (2012) First steps of bacteriophage SPP1 entry into Bacillus subtilis . Virology 422: 425–434 [DOI] [PubMed] [Google Scholar]
- Kingston AW, Liao X, Helmann JD (2013) Contributions of the sigma(W), sigma(M) and sigma(X) regulons to the lantibiotic resistome of Bacillus subtilis . Mol Microbiol 90: 502–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Bruckner R (2006) A functional dlt operon, encoding proteins required for incorporation of D‐alanine in teichoic acids in gram‐positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae . J Bacteriol 188: 5797–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8: 317–327 [DOI] [PubMed] [Google Scholar]
- Levin‐Reisman I, Gefen O, Fridman O, Ronin I, Shwa D, Sheftel H, Balaban NQ (2010) Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nat Methods 7: 737–739 [DOI] [PubMed] [Google Scholar]
- Lindberg AA (1973) Bacteriophage receptors. Annu Rev Microbiol 27: 205–241 [DOI] [PubMed] [Google Scholar]
- Liu J, Gefen O, Ronin I, Bar‐Meir M, Balaban NQ (2020) Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367: 200–204 [DOI] [PubMed] [Google Scholar]
- Ma D, Wang Z, Merrikh CN, Lang KS, Lu P, Li X, Merrikh H, Rao Z, Xu W (2018) Crystal structure of a membrane‐bound O‐acyltransferase. Nature 562: 286–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Snir S, Koonin EV (2011) Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol 193: 6039–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamou G, Malli Mohan GB, Rouvinski A, Rosenberg A, Ben‐Yehuda S (2016) Early developmental program shapes colony morphology in bacteria. Cell Rep 14: 1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F, Mayer‐Barber K, Sher A, Wack A, O'Garra A (2015) Type I interferons in infectious disease. Nat Rev Immunol 15: 87–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morehouse BR, Govande AA, Millman A, Keszei AFA, Lowey B, Ofir G, Shao S, Sorek R, Kranzusch PJ (2020) STING cyclic dinucleotide sensing originated in bacteria. Nature 586: 429–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AG, Yevstigneyeva MS, Fineran PC (2017) Regulation of CRISPR‐Cas adaptive immune systems. Curr Opin Microbiol 37: 1–7 [DOI] [PubMed] [Google Scholar]
- Percy MG, Grundling A (2014) Lipoteichoic acid synthesis and function in gram‐positive bacteria. Annu Rev Microbiol 68: 81–100 [DOI] [PubMed] [Google Scholar]
- Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W (1995) Incorporation of D‐alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J Biol Chem 270: 15598–15606 [DOI] [PubMed] [Google Scholar]
- Saar‐Dover R, Bitler A, Nezer R, Shmuel‐Galia L, Firon A, Shimoni E, Trieu‐Cuot P, Shai Y (2012) D‐alanylation of lipoteichoic acids confers resistance to cationic peptides in group B streptococcus by increasing the cell wall density. PLoS Pathog 8: e1002891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas M (2012) My life with bacteriophage phi29. J Biol Chem 287: 44568–44579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmond GP, Fineran PC (2015) A century of the phage: past, present and future. Nat Rev Microbiol 13: 777–786 [DOI] [PubMed] [Google Scholar]
- Sao‐Jose C, Baptista C, Santos MA (2004) Bacillus subtilis operon encoding a membrane receptor for bacteriophage SPP1. J Bacteriol 186: 8337–8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtele CF, Oman RW, Anderson DL (1970) Effect of elevated temperature on deoxyribonucleic acid synthesis in bacteriophage phi‐29‐infected Bacillus amyloliquefaciens . J Virol 6: 430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanski M, Glaser R, Koten B, Meyer‐Hoffert U, Wanner S, Weidenmaier C, Peschel A, Harder J (2013) Staphylococcus aureus subverts cutaneous defense by D‐alanylation of teichoic acids. Exp Dermatol 22: 294–296 [DOI] [PubMed] [Google Scholar]
- Storz G (2016) New perspectives: Insights into oxidative stress from bacterial studies. Arch Biochem Biophys 595: 25–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrall ET, Keller AP, Shen Y, Loessner MJ (2020) Structure and function of Listeria teichoic acids and their implications. Mol Microbiol 113: 627–637 [DOI] [PubMed] [Google Scholar]
- Twort FW (1914) An investigation on the nature of ultra‐microscopic viruses. Lancet 186: 1241–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzipilevich E, Benfey PN (2021) Phage‐Resistant Bacteria Reveal a Role for Potassium in Root Colonization. MBio 12: e0140321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzipilevich E, Habusha M, Ben‐Yehuda S (2017) Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell 168: 186–199.e12 [DOI] [PubMed] [Google Scholar]
- Young FE (1967) Requirement of glucosylated teichoic acid for adsorption of phage in Bacillus subtilis 168 . Proc Natl Acad Sci U S A 58: 2377–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Dill BD, Pan C, Hettich RL, Banfield JF, Shah M, Fremaux C, Horvath P, Barrangou R, Verberkmoes NC (2012) Phage‐induced expression of CRISPR‐associated proteins is revealed by shotgun proteomics in Streptococcus thermophilus . PLoS One 7: e38077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P, Perkins JB, Losick R (1984) A novel method for the rapid cloning in Escherichia coli of Bacillus subtilis chromosomal DNA adjacent to Tn917 insertions. Mol Gen Genet 195: 424–433 [DOI] [PubMed] [Google Scholar]


