Abstract
Machine learning uses historical data to make predictions about new data. It has frequently been applied in healthcare to optimize diagnostic classification through discovery of hidden patterns in data, that may not be obvious to the human brain. Congenital heart defect (CHD) machine learning research entails one of the most promising clinical applications, in which timely and accurate diagnosis is essential. The objective of this scoping review is to summarize the application and clinical utility of machine learning techniques used in pediatric cardiology research, specifically focusing on approaches aiming to optimize diagnosis and assessment of underlying CHD. Out of 50 full-text articles identified between 2015 and 2021, 40% focused on optimizing the diagnosis and assessment of CHD. Deep learning and support vector machine were the most commonly used algorithms, accounting for an overall diagnostic accuracy > 0.80. Clinical applications primarily focused on the classification of auscultatory heart sounds, transthoracic echocardiograms, and cardiac magnetic resonance images. The range of these applications and directions of future research are discussed in this scoping review.
Keywords: congenital heart defect, machine learning
INTRODUCTION
Clinical intuition is commonly characterized as a “feeling”. This feeling of subconscious pattern divergence, can be applied to the diagnosis of complex illnesses or impending clinical deterioration. Clinical intuition is derived from repeated exposures to similar events, that are stored in the human brain over time. This library of events can fine tune intuition so that when a future clinical event occurs, the clinician can anticipate or predict what will happen next. Humans are relatively efficient at taking into account multidimensional data (i.e., laboratory results, monitor data, diagnostic imaging). Importantly, machine learning can closely replicate human intuition and support the deep infrastructure that goes into diagnosing complex illnesses or prediction of clinical deterioration, however remains unbiased by the emotions and recent experiences that so often cloud our human judgement.
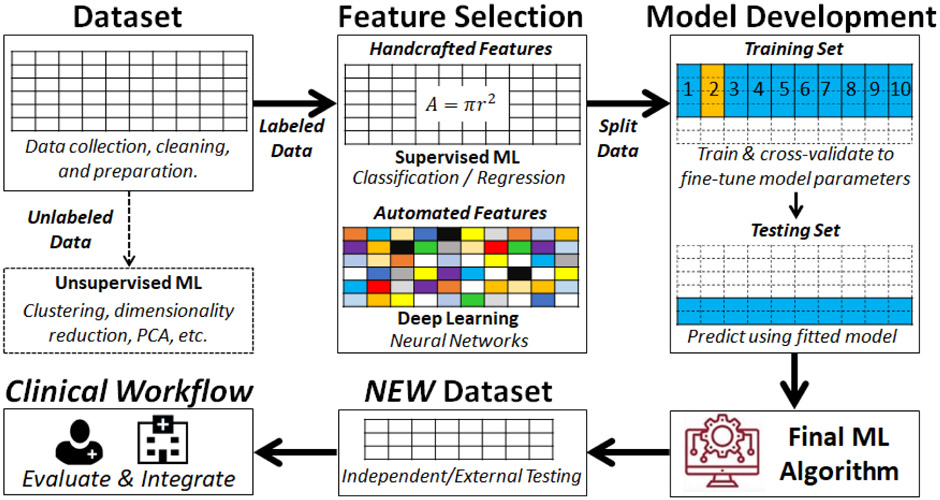
Machine learning is a discipline at the intersection of mathematics, statistics, and computer science that provides a powerful catalogue of techniques used to make predictions about future events. Machine learning implies training a computer algorithm on historical data stored in a large dataset in order to “learn” how to make predictions about future events. Advances in computational power to handle complex amounts of reference data at high speed has led to the observed exponential growth of machine learning applications in healthcare in recent years. Such machine learning applications are conceptually different from computerized algorithms based on classical statistical modeling. The latter follows a rule-based logic where a programmer decides a set of conditional statements derived from domain knowledge to automate “human-like” clinical decision making (i.e., if body mass index > 30 then class = “obese”). In contrast, a machine learning algorithm “learns” the modeling parameters from historical data to develop decision rules for future predictions. This “learning” process performs in a fashion completely unbiased by existing domain-knowledge given that there are hidden patterns in the data that might not be obvious to humans. Furthermore, since machine learning algorithms use a data-driven logic independent from that of clinicians, it has been shown that machine learning algorithms outperform clinicians in some scenarios.1 Figure 1 summarizes the machine learning pipeline, emphasizing its role in “data-driven” decision making.
Figure 1.
Summary of a Typical Machine Learning Pipeline Application
*Machine Learning (ML), Principal Component Analysis (PCA)
Machine learning has been successfully applied to medicine in many fields, such as advanced cardiac imaging.2 In this context, medical images are processed and compiled to extract features used by the algorithm to fine tune its classification of outcomes. The majority of advances in medical imaging machine learning applications (i.e., artifact removal, augmentation of disease classification accuracy, etc.) have been developed in various adult populations.3-8
Neonatal and pediatric populations, especially the most vulnerable like those with congenital heart defects (CHD), could greatly benefit from machine learning-based improved diagnostic accuracy and early disease detection. One in 100 live births in the United States is diagnosed with CHD every year, of which nearly 7,200 have critical CHD.9-12 These life-threatening structural malformations of the heart are present at birth and require intervention in the first year of life.13,14 Delays in timely diagnosis in neonates with CHD, and limited access to specialized cardiac programs, could result in preventable morbidity or mortality in some cases. Securing access to specialized neonatal or pediatric cardiac programs preemptively, although challenging, has been shown to play an instrumental role in decreasing risk of CHD infant mortality.15 The observed benefits of machine learning applications in healthcare are promising for optimizing timing and accuracy of CHD diagnoses, thereby providing early targeted access to highly specialized cardiac care. The objective of this scoping review is to describe the application and clinical utility of machine learning techniques used for diagnosing and assessing underlying critical and non-critical CHD. In this review, we will briefly define the emerging research applications where machine learning has been applied in pediatric cardiology, describe the various machine learning techniques used in these categories, and summarize the specific applications used for diagnosis and assessment of critical and non-critical CHD.
MATERIALS & METHODS
Literature Search Strategy
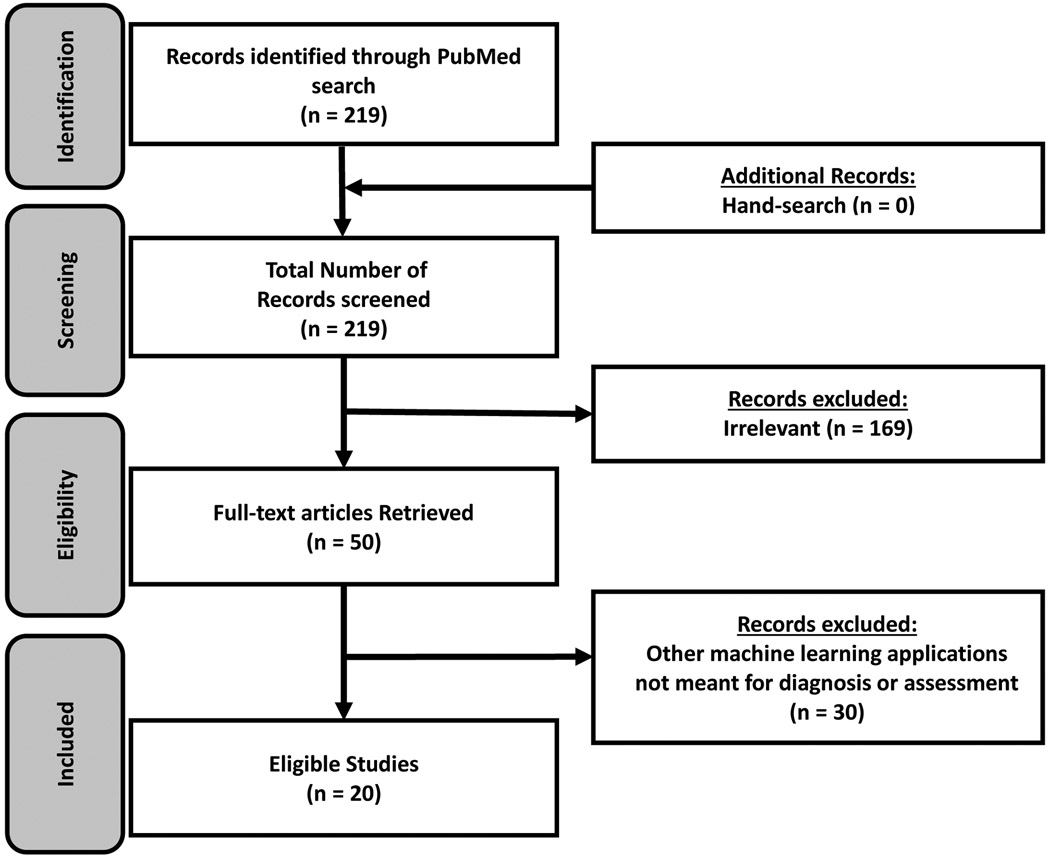
We followed the guidelines for Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews.16 All original, peer-reviewed studies published in PubMed database between January 2015 and February 2021 that described the use of machine learning or predictive analytics for predicting diagnostic outcomes in patients with critical and non-critical CHD were included. The most recent search was conducted on February 20, 2021. The search terms were “(machine learning) AND ((congenital heart disease) OR (cardiovascular disease in children))”. Studies focused on populations without a CHD diagnosis were excluded. The search was limited to English-language articles. This search yielded 219 journal articles. After screening the titles and abstracts, we excluded articles that were irrelevant (n= 169). Among the 50 full-text articles reviewed, 20 articles focusing on CHD diagnosis and assessment were retained for inclusion in this scoping review. Figure 2 briefly summarizes the work flow for article search and selection.
Figure 2.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses Flow Diagram of Included Articles
Data Coding Scheme
Full-text articles were analyzed using the matrix method as per recommendations by Whittemore and Knafl.17 A single reviewer first sorted each article into a table using ascending chronological order with the following eight domains: journal / author information, purpose, design, sample, variables, results, limitations, and implications for future research. These domains were selected after discussions between the coauthors, then the information related to each domain were abstracted by a single reviewer. In addition to these general domains, we defined a data dictionary for the abstracted machine learning elements necessary for this study. These elements included: class of machine learning approach (supervised vs. unsupervised, regression vs. classification, traditional learning vs. deep learning); specific algorithm used (logistic regression, support vector machine, etc.); techniques used for testing and cross-validation; and the use of independent external validation.
Synthesis of Findings
Results were reported using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Extension for Scoping Reviews guidelines.16 Each article was assessed for its standard of documentation using the Minimum Information About Clinical Artificial Intelligence Modeling checklist.18 Simple descriptive statistics and pie charts were used to report frequencies of different machine learning algorithms applied across eligible studies. Next, the content of the abstracted domains of eligible studies were qualitatively synthesized. Based on domain expertise and discussions among coauthors, three categories of machine learning applications for the diagnosis and assessment of CHD emerged and results were summarized for each of these categories.
RESULTS
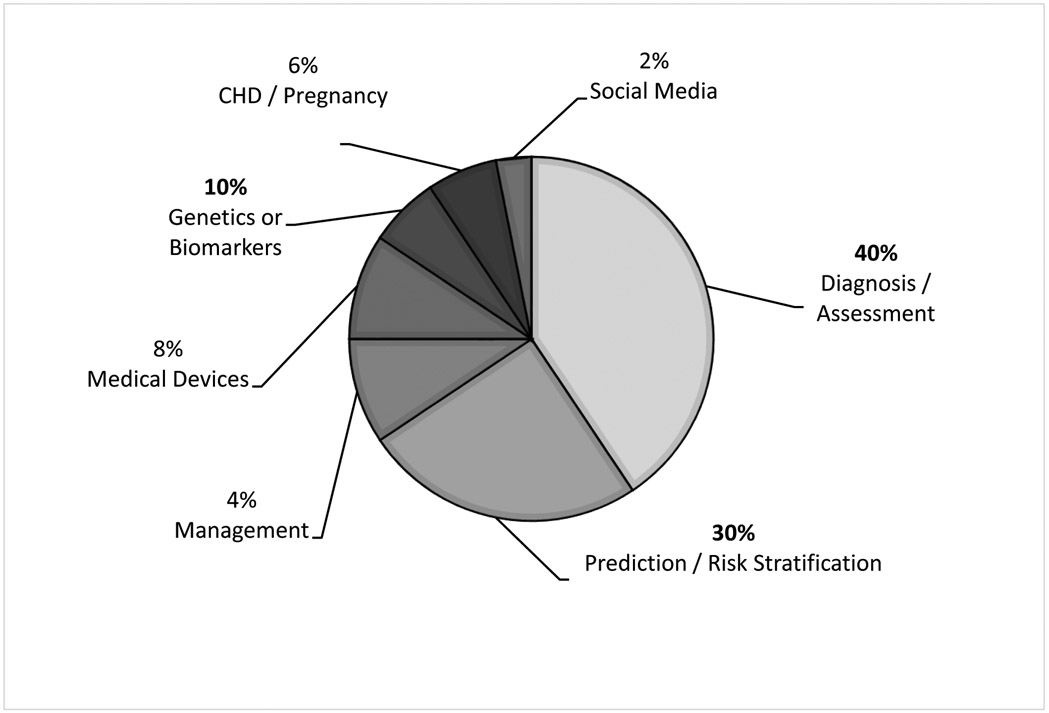
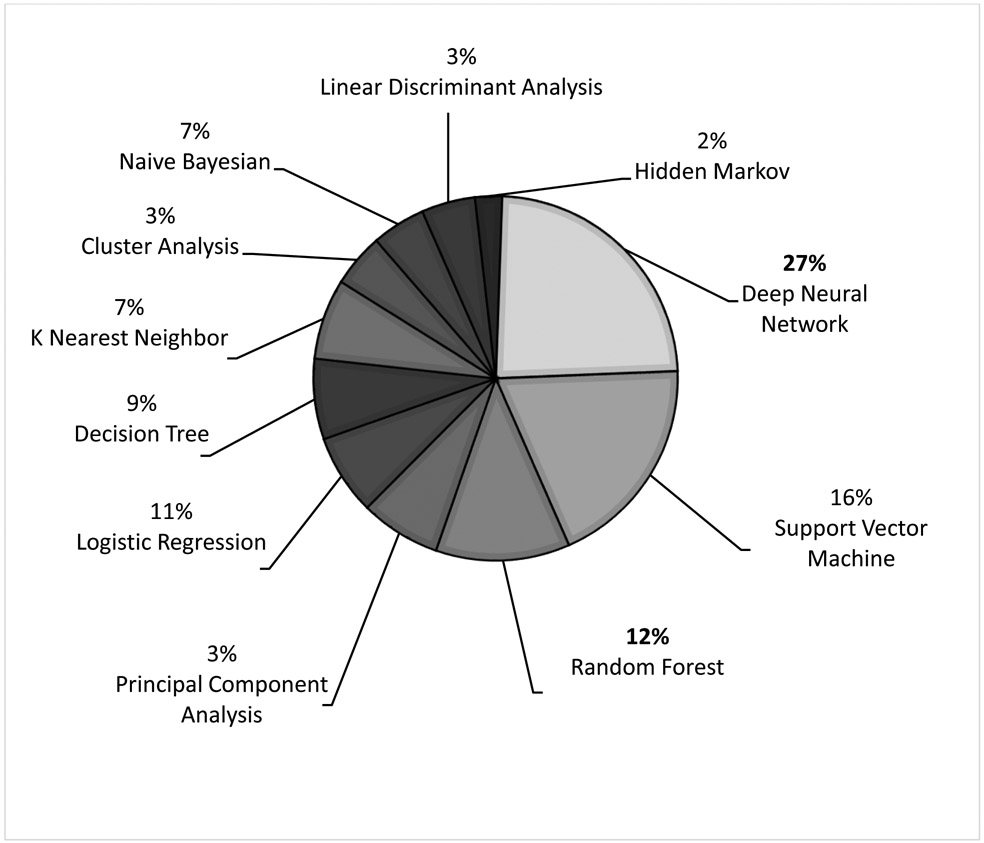
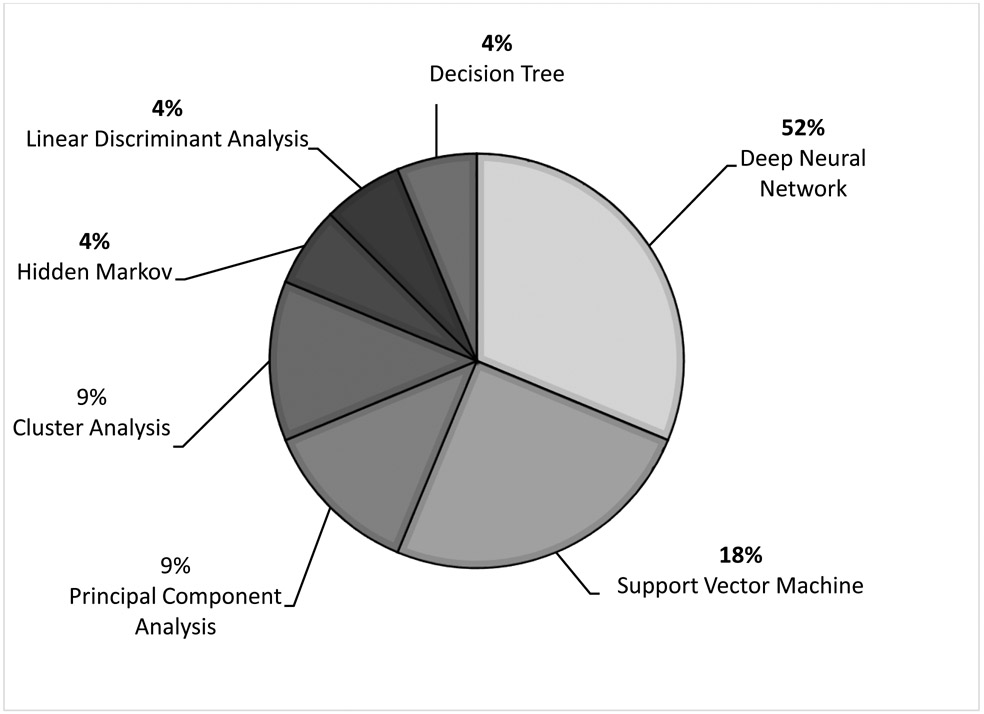
There were 50 studies that broadly focused on the application of machine learning in pediatric cardiology research. These studies were categorized and focused on various intentions for clinical use (Figure 3): diagnosis and assessment of underlying critical and non-critical CHD (n= 20),1,19-37 prediction and risk stratification of outcomes in CHD (n= 15),38-52 management of patients with CHD (n= 2),53,54 medical device research (n= 4),55-58 novel genetics and biomarkers in CHD (n= 5),59-63 CHD in pregnancy (n= 3),64-66 and social media research (n= 1).67 The distribution of various machine learning algorithms used in these studies is summarized in Figure 4. The two most common algorithms were deep neural networks (deep learning) and support vector machines. Hidden Markov models and linear discriminant analysis were the least common algorithms.
Figure 3.
Distribution of Machine Learning Applications and Uses in Pediatric Cardiology Research
*Congenital Heart Defect (CHD)
Figure 4.
Distribution of Machine Learning Algorithms Used in Pediatric Cardiology Research in General
There were 20 studies focusing on the diagnosis and assessment of critical and non-critical CHD. The Table summarizes the details of these studies, including purpose, design, sample, machine learning technique, and primary findings. All studies were observational, generally of small sample size. None of the studies provided 100% of the Minimum Information About Clinical Artificial Intelligence Modeling checklist items (standard documentation guidelines).18 Included studies applied machine learning to auscultation of heart sounds in patients with CHD, interpreting transthoracic echocardiogram data, or processing medical images (cardiovascular magnetic resonance imagining). As shown in Figure 5, deep neural networks and support vector machines were also the most commonly used classification algorithms in these studies. More importantly, the Table highlights that using cross validation on existing retrospective data, the overall accuracy of the various machine learning algorithms exceeded 80%, with some techniques reaching 95% to 100%. With the exception of two studies, none of these models were externally validated on independent datasets. The remainder of this scoping review will focus on the qualitative synthesis of 20 studies that focused on machine learning applications for the diagnosis and assessment of CHD.
Table:
Summary of Studies Focusing on Machine Learning Techniques Applied to Diagnosing and Assessing CHD in Neonates, Infants, and Children
| Study Citation | Study Aim and Design | Machine Learning Approach | Main Findings |
|---|---|---|---|
| Auscultation of Heart Sounds | |||
| Gomez-Quintana S, et.al. (2021).34 | Case-control study (n= 265 newborns [39 Other CHD, 89 PDA, 137 healthy], gestational ages 35 to 42 weeks). PCG recordings in the first 6 days of life used to create a decision support system that can detect sound signatures with and without PDA and CHD. Two clinical sites were used. | A boosted decision tree classifier with 10-fold cross validation using a training and testing 90/10 split dataset was used to estimate the probability of PDA or CHD. No external validation. | The classifier achieved an AUC of 78% for detecting CHD and 77% for detecting PDA. |
| Lv Jingjing, et. al. (2021).37 | Case-control study (n= 1362 [1194 abnormal heart sounds and 168 normal heart sounds], mean age 2.4 years [SD 3.1, median 0.9, and age range 1 day to 15.9 years]). To compare detection of abnormal heart sounds via remote and automated artificial intelligence auscultation with face-to-face auscultations by experienced cardiologists (gold standard). One clinical site was used. | CNN with 5-fold cross validation. No external validation. | Remote auscultations compared to face-to-face auscultations- sensitivity, specificity, and accuracy of 98% (95% CI 97-99%), 91% (95% CI 87-95%), and 97% (95% CI 96-98%), respectively. Automated artificial intelligence auscultations compared to face-to-face auscultations- sensitivity, specificity, and accuracy of 97% (95% CI 96-98%), 89% (95% CI 84-94%), and 96% (95% CI 95-97%), respectively. |
| Aziz S, et. al. (2020).29 | Case-control study (n= 56 [17 ASD, 11 VSD, and 28 healthy], to automate detection and classification of CHD through use of pattern recognition techniques. One clinical site was used. | SVM with 10-fold cross validation. Quadratic, cubic, and Gaussian kernels were applied to the SVM classifier. No external validation. | SVM classifier with a cubic kernel function using a subset of fused frequency and temporal based features, had the best performance for binary and multiclass experiments. Accuracy 95.24%, sensitivity 95.24%, specificity 95.24%, PPV 86.96%, NPV 98.36%, and error 4.76%. |
| Gharehbaghi A, et. al. (2020).30 | Case-control study (n= 115 [10 ASD, 25 healthy with innocent murmur, 25 healthy with no murmur, 15 MR, 15 TR, and 25 VSD], average age 3.9 – 2.4 to 12.6 ± 4.4) to diagnose children with a septal defect versus children with valvular leakage. Both of these conditions are known to have a systolic murmur. One clinical site was used. | TGNN with K-fold validation where there are different values of K, ranging from 2 to half of the minimum group size (A-test method), using a training and testing 70/30 spilt dataset. Repeated random sub-sampling was applied, as well. No external validation. | Average accuracy 88.4% ± 3.9, sensitivity 91.6% ± 5.7, classification error 9.89% using the A-test method (evaluates structural risk). |
| Elgendi M, et. al. (2018).27 | Cohort study (n=60, median age 7 years [range 3 months to 78 years]) to classify pulmonary artery hypertension using sound signatures from non-invasive pulmonary circulation vibrations. One clinical site was used. | LDA with leave one out cross validation. No external validation. | Sensitivity 84%, specificity 88.57% for entropy (disorder of heart sound pattern) of the first sinusoid formant (frequency resonance) of heart sounds. |
| Sun S, et. al. (2018).20 | Case-control study (n= 227 [60 VSD and 167 healthy], VSD group average age 2 years and healthy group average age 21 years) toclassify small, medium, and large VSD based on heart sound feature extraction. Two databases (3M and Michigan), 1 cohort of undergraduate students, and 1 clinical site were used. | PCA for feature generation and ellipse model, and SVM for classification using 2,276 heart sounds (22% positive cases). A Gaussian kernel was applied to the SVM classifier. No external validation. | The ellipse model used for classification, had the highest performance with accuracy 95.5%, 92.1%, and 96.2% for small, medium, and large VSD, sensitivity of 94.9%, 93.8%, and 95.3%, and specificity 95.6%, 91.9%, and 96.3%, respectively. |
| Thompson W, et. al. (2018).36 | Case-control study (n=603 cases [374 abnormal confirmed by echocardiogram and pathologic murmur, and 229 normal confirmed by echocardiogram and innocent murmur (90) or no murmur (139)], median age 8.8 ± 0.1 to 80.9 years) to compare classification of heart rate by murmur detection algorithm and gold standard 3-lead electrocardiogram. One clinical site was used. | Murmur detection algorithm that performs a signal quality check, heart sounds are segmented (S1 systole, S2, and diastole), feature vectors emerge, and then these feature vectors are used to build a non-linear artificial intelligence classifier. The patients included in the Johns Hopkins Cardiac Auscultatory Database were not used for training, only for testing. | Murmur detection algorithm sensitivity and specificity for detecting pathologic cases was 93% (CI 90-95%) and 81% (CI 75-85%), with accuracy of 88% (CI 85-91%). |
| Gharehbaghi A, et. al. (2017).22 | Case-control study (n=90 [55 healthy and 35 CHD], age 6.6± 1.2 to 11.8± 4.1 years) to classify BAV and mitral regurgitation using recorded heart sounds. One clinical site was used. | Hidden Markov Model and SVM using repeated random sub-sampling with 5-fold cross validation training / testing 50/50 split dataset. A quadratic kernel was applied to the SVM classifier. No external validation. | Accuracy 86.4%, sensitivity 85.6%, and specificity 87%. |
| Gharehbaghi A, et. al. (2017).26 | Case-control study (n=90 [30 VSD, age 3.6 ±1.2 years; 30 valvular regurgitation, age 11.8 ± 4.1 and 12.6 ± 4.4 years; 30 healthy, age 6.7 ± 3.7 years]) to classify phonocardiography recordings and distinguish between VSD and AV valve regurgitation. One clinical site was used. | TGNN with leave one out validation method. No external validation. | Accuracy 86.7% and sensitivity 83.3%. |
| Gharehbaghi A, et. al. (2015).1 | Case-control study (n=50 [22 BAV and 28 healthy], median age 7 years [range 2.5 to 12 years]) to classify BAV through use of recorded heart sounds. One clinical site was used. | Statistical TGNN, and SVM using 856 cardiac cycles (45% positive cases) with cross validation on training / testing 50/50 split dataset. A linear kernel was applied to the SVM classifier. No external validation. | The statistical TGNN on average had better performance than the other models with classification rate 87.4%, sensitivity 86.5%, and specificity 88.4%. |
| Transthoracic Echocardiogram | |||
| Wang J, et. al. (2021) 33 | Case-control study (n= 1308 children [823 healthy, 209 VSD, 276 ASD]) designed to automatically interpret five-view echocardiograms. One clinical site was used. | A multi-channel CNN was applied to the dataset with a training / testing 90/10 split. No external validation. | The video-based model diagnosed the binary classification problem (positive or negative) with 93.9% accuracy, and the 3-class classification problem (negative, ASD, VSD) with 92.1% accuracy. This model was also able to achieve an AUC for binary classification of 0.922. This model did not use a ground truth label or key-frame annotation. |
| Diller GP, et. al. (2019).19 | Case-control study (n= 267 [152 CHD and 155 healthy], mean age 39±16 years) to remove artifacts from transthoracic echocardiograms by estimating cross-entropy (a loss function that measures differences between two probability distributions-original image vs. reconstructed image)19,73 and sum of squared differences (measures quality between healthy and CHD images). One clinical site was used. | DNN with an autoencoder applied to 153,420 apical 4-chamber views from CHD subjects and 24,354 from healthy subjects with 70/30 training/testing split. No external validation. | Autoencoders trained significantly better on CHD samples than healthy samples (cross-entropy- healthy: 0.2649 ± 0.0369 vs. 0.2597 ± 0.0327 for CHD), and (mean squared difference- healthy: 133.89 ± 79.06 vs. 118.86 ± 61.52 for CHD). A lower cross-entropy indicates a closer representation of the underlying distribution. |
| Diller GP, et. al. (2019).25 | Case-control study (n= 199 [132 CHD and 67 healthy], mean age 38±12 to classify transposition of the great arteries after arterial switch procedure vs. congenitally corrected or healthy subjects. Two clinical sites were used. | CNN on 4-chamber apical view images with 80/20 training/testing split. No external validation. | Model accuracy 95% in the training set and 94.4% in the testing set. |
| Meza JM, et. al. (2018).21 | Cohort study of neonates with critical left heart obstruction (n=651, median gestational age 38 [38-39] weeks) to phenotype clinically meaningful clusters of baseline and pre-intervention disease. 21 clinical sites enrolled in the Congenital Heart Surgeons’ Society Data Center were used. | Unsupervised hierarchical, non-overlapping, agglomerative cluster analysis used 136 baseline quantitative and qualitative morphologic and functional variables from baseline echocardiograms. No external validation. | Three distinct groups emerged (C1=215, C2=338, and C3=98). Aortic valve atresia and LV end diastolic volume were significantly different between groups (11%, 87%, and 8% for aortic atresia and 1.35, 0.69, and 2.47 cm2 for median LV end diastolic area between the three clusters, respectively). |
| Pereira F, et. al. (2017).23 | Case-control study (n= 90 [26 CoA and 64 healthy], neonatal mean age 7 days) to classify CoA and healthy hearts from 2-D echocardiograms. One clinical site was used. | SVM using 5-fold cross-validation on training and testing datasets of ~80/20. A Gaussian kernel was applied to the SVM classifier. No external validation. | The parasternal long axis view had the lowest false negative error rate (end diastolic phase [7.7], end systolic phase [11.5]), and the lowest total error rates (end diastolic phase [18.9], end systolic phase [20.0]). |
| Advanced Medical Imaging | |||
| Tandon A, et. al. (2021) 35 | Cohort study (n= 87 patients with repaired tetralogy of Fallot and pulmonary stenosis or atresia, age in the training dataset was 13.5 years [IQR 10-17.5], and age in the testing dataset was 13.9 [IQR 11.7-18]) to automate ventricular contouring during CMR of repaired tetralogy of Fallot patients. One clinical site and one scanner type was used. | CNN using training / testing datasets ~70/30. These datasets were not randomly split, the groups were separated by time. The earlier enrolled cases were assigned to the training dataset. No external validation. | This study was a continuation of previously established research. The retrained contouring algorithm included mostly structural normal hearts with the addition of the repaired tetralogy of Fallot patients. Spatial metrics were used to evaluate algorithm performance (Dice Similarity Coefficient- shows spatial overlap in 3-dimensions. A Dice of 1 = perfect spatial overlap and 0 = no spatial overlap). The LV endocardial, LV epicardial, and RV endocardial at end diastole all had improved Dice metrics in the retrained algorithm 0.903 (0.875, 0.920) p = 0.0248; 0.905 (0.881, 0.937) p < 0.0001; and 0.894 (0.855, 0.907) p < 0.0001. |
| Lu Y, et. al. (2020).32 | Cohort study (n= 3 ASD patients [550 images, 275 before and 275 after atrial septal occlusion surgery) to segment right atrium CMR images to aid in determining surgical outcomes. One clinical site and one scanner type was used. | U-net deep CNN was compared to an active contour model with cross validation using training and testing datasets in a 3:1 ratio. No external validation. | The proposed technique outperformed the traditional active contour model when accurately segmenting the atria. The U-net mean and SD reported for the Dice Similarity Index, Jaccard Index, and Hausdorff Distance were 0.9488 (± 0.0209), 0.9033 (± 0.0374), and 7.5625 (± 4.4549). |
| Karimi-Bidhedi S, et. al. (2020).31 | Case-control study (n= 64 patients, age range of 2 to 18 years; [20 tetralogy of Fallot, 9 double outlet right ventricle, 9 transposition of the great arteries - repaired arterial switch operation, 8 cardiomyopathy, 9 coronary artery anomaly, 4 pulmonary stenosis or atresia, 3 truncus arteriosus, and 2 aortic arch anomaly]). Developed synthetically segmented CMR images to produce a large training dataset used for automated detection of complex heart disease. One clinical site and two scanner types were used. | A generative adversarial network was used to augment the training dataset. A fully convolutional network was used to segment the CMR images. The sample was split randomly, 26 patients were assigned to the training dataset and 38 patients to the testing dataset. The training dataset was split further 80/20 for training and validation. The framework was externally validated on second dataset. | The fully convolutional network (automated) produced average Dice Similarity Index metrics of 91% and 86.8% for LV at end-diastole and end-systole; and 87.4% and 80.6% for RV at end-diastole and end-systole, respectively. |
| Hauptmann A, et. al. (2019).28 | Cohort study (n=250 [retrospective data, mean age 22±13 years] and n=10 [prospective data, mean age 34±17 years]). For the prospective study, one clinical site and one scanner type was used. | CNN used to de-noise CMR images of free-breathing individuals with cross-validation on retrospective data and external validation on the prospective data. | RMSE and SSIM error rates of SNR, acceleration factor, and image cropping features were computed on the reconstructed image of the test dataset. The continuously rotating tiny golden angle CMR sampling pattern had the lowest RMSE and highest SSIM compared to all other sampling methods. SNR decreased from 20 dB to 10 dB, and the acceleration factor increased from 10x to 16x. |
| Bruse JL, et. al. (2017).24 | Case-control study (n= 60 [20 healthy aged 15±2 years, 20 with surgical aortic arch reconstruction aged 23±7 years, and 20 with Lecompte maneuver reconstruction aged 14±3 years) to identify meaningful clusters within anatomical shape data. One clinical site and one scanner type was used. | Agglomerative hierarchical clustering was performed to subdivide groups. Followed by PCA with leave one out strategy and 10-fold cross-validation. No external validation. | The best performing distance/linkage combination had correlation coefficient scores > 0.8 and an F score ~0.9. Classification accuracy for healthy arches, CoA shapes, and arterial switch shapes were 83%, 85%, and 100%, respectively. |
Area Under the Curve (AUC), Atrial Septal Defect (ASD), Atrioventricular (AV), Bicuspid Aortic Valve (BAV), Cardiovascular Magnetic Resonance Imagining (CMR), Coarctation of the aorta (CoA), Confidence Interval (CI), Congenital Heart Defect (CHD), Convolutional Neural Network (CNN), Deep Neural Network (DNN), Linear Discriminant Analysis (LDA), Interquartile Range (IQR), Left Ventricle (LV), Mitral Regurgitation (MR), Negative Predictive Value (NPV), Patent Ductus Arteriosus (PDA), Positive Predictive Value (PPV), Principal Component Analysis (PCA), Right Ventricle (RV), Root Mean-Square Error (RMSE), Signal-to-Noise Ratio (SNR), Standard Deviation (SD), Structural Similarity Index (SSIM), Support Vector Machine (SVM), Time Growing Neural Network (TGNN), Tricuspid Regurgitation (TR), Ventricular Septal Defect (VSD)
Figure 5.
Distribution of Machine Learning Algorithms Used in Pediatric Cardiology Research Focusing on the Diagnosis and Assessment of Critical and Non-Critical CHD
The most common neural network types were the convolutional neural network, time growing neural network, autoencoder, and generative adversarial network.
The most common kernel among the studies who used support vector machine was a Gaussian kernel.
*Congenital Heart Defect (CHD)
SYNTHESIS OF LITERATURE
It is apparent that machine learning in pediatric cardiology research is an evolving field. Diagnosing and assessing patients with CHD can typically be done non-invasively, using examination findings and diagnostic tools based on auditory or visual pattern recognition. However, such medical images and signal data are considered unstructured; they are not stored as tabular data or in formatted fields.68 This type of data historically requires the time-consuming process of clinician review and interpretation. Thus, in individuals with CHD, machine learning techniques described in the Table have focused on accurate diagnosis and assessment based on the classification of auscultatory heart sounds, transthoracic echocardiograms, or cardiovascular magnetic resonance images.
Auscultation
Aortic valves are normally tricuspid in nature and provide an outlet for blood flow from the heart to the body. Bicuspid aortic valves occur in 0.5% to 2% of children.1,69-72 A subset of these patients develop progressive valve disease and/or aortic dilatation, with risk of life-threatening aortic aneurysm and dissection. Children with known bicuspid aortic valves must be monitored throughout their lives to measure risk for aortic aneurysm. Diagnosis of bicuspid aortic valves can be done via phonocardiogram, which records cyclical sounds produced by the heart.1 Patients with a bicuspid aortic valve typically have a systolic ejection click. Identification of this click through traditional auscultation methods can be limited due to provider expertise and skill, as well as rapid heart rates in young children. Gharehbaghi and colleagues performed a study that collected phonocardiogram data prospectively.1 This study created a statistical time growing neural network that automatically classified bicuspid aortic valves through use of recorded heart sounds produced by the phonocardiogram.1 The phonocardiogram recordings were preprocessed, so that the cardiac cycle could be segmented to recognize the additional heart sound (systolic ejection click). This segmentation was used to build a classifier to identify healthy subjects and those with a bicuspid aortic valve. The model was able to classify the 865 cardiac cycles with 98.5% accuracy. This improved diagnostic sensitivity for a subtle exam finding stands to bring many previously unrecognized children and adults to appropriate cardiac care.
Several other investigators are also using non-invasively recorded heart sounds to diagnose cardiac disease.29,30,34 For example, Elgendi and colleagues27 used linear discriminant analysis to detect pulmonary arterial hypertension using digital auscultation to record the unique vibrations of the hypertensive pulmonary circulation. Their model performed well, with a sensitivity of 84% and specificity of 88.6% for entropy (disorder of heart sound pattern) of the first sinusoid formant (frequency resonance of heart sounds). Sun and colleagues,20 aimed to diagnose small, medium, and large ventricular septal defects based on heart sound feature extraction, using classification boundary curves and ellipse models. The ellipse model outperformed five other models used in the study for normal, small, medium, and large ventricular septal defect classification (accuracy 99%, 95.5%, 92.1%, 96.2%). Cardiac auscultation is nuanced, but clearly machine learning holds promise for improving diagnostic accuracy for providers at all experience levels.
Transthoracic Echocardiogram
Echocardiography is the mainstay of non-invasive assessment for CHD, but requires experienced reviewers that remain susceptible to biases. Coarctation of the aorta, a common form of critical CHD characterized by narrowing of the thoracic aorta, is particularly well-suited for echocardiographic diagnosis, though poses challenges when it comes to image interpretation.23 Narrowing of the aorta causes obstruction of normal blood flow to the body, and excessive pressure to be generated by the left ventricle. If the obstruction is not diagnosed in a timely manner it can lead to heart failure and poor systemic perfusion. Pereira and colleagues23 retrospectively collected 2-dimensional echocardiographic images of the aortic arch. The aim of this study was to develop a fully automated algorithm to detect coarctation of the aorta from 2-dimensional echocardiographic images using standard view planes (suprasternal, apical, and parasternal windows). Static images representing a single cardiac cycle (end diastolic and end systolic phase) were pre-selected for model development. Neonates born with coarctation of the aorta alone, and healthy neonates born without coarctation of the aorta were included in the sample. A stacked denoising autoencoder neural network was used for feature extraction over predefined image regions (sectors). A support vector machine classifier was trained on a random subset of training data features. The parasternal long axis view had the lowest coarctation error rate (end diastolic phase [7.7], end systolic phase [11.5]), and the apical view had the lowest healthy error rate (end diastolic phase [20.0], end systolic phase [20.0]).23 When the views were combined, more undecided cases resulted. The increase in undecided cases with inclusion of multiple views is not surprising given the increased model complexity, and therefore difficulty for the model to classify outcomes.
Further research has focused on image denoising, automatic detection, and clustering subjects based on quantitative image data to further support clinical decision making and diagnosis of critical and non-critical CHD.33 Diller et al,19 aimed to remove acoustic shadowing artifacts that occur during transthoracic echocardiograms through use of a deep neural network and autoencoder. Cross-entropy (a loss function that measures differences between two probability distributions-original image vs. reconstructed image)19,73 and sum of squared differences (measures image quality) were performance evaluation metrics used on the test dataset. Autoencoders extracted features and trained significantly better on CHD samples compared to healthy samples, represented by a lower cross-entropy and lower mean squared difference (0.2597 ± 0.0327 and 118.86 ± 61.52).19 Finally, Meza et al,21 used an unsupervised hierarchical cluster analysis in 651 neonates with critical left heart obstruction to determine if subjects could be defined using more clinically meaningful clusters. Images were derived from transthoracic echocardiograms, and three distinct groups emerged (n= 215, 338, 98). Aortic valve atresia and left ventricular end diastolic volume variables significantly distinguished the groups. Median left ventricular end diastolic area for groups 1, 2, 3 was 1.35, 0.69, and 2.47 cm2 (p < 0.0001). Aortic atresia in groups 1, 2, 3 was present in 11%, 87%, and 8%, (p < 0.0001).21 The authors suggest that clustering analyses yield more reliable delineation of subject heart structure characteristics. These data support the use of clustering approaches to more accurately diagnose CHD. The added complexity and volume of data in diagnostic imaging poses a challenge to machine learning, but these investigators demonstrate the potential value machine learning brings to clinical pattern-fitting and decision making.
Cardiovascular Magnetic Resonance Imagining
Cardiovascular magnetic resonance imaging is considered the clinical gold standard for accurate assessment of ventricular volumes and function. Obtaining quality images requires repetitive breath holding, which can be a challenge for patients with CHD who may suffer from shortness of breath at baseline or who are simply too young to comply with breath-holding instructions. The investigators of the next study explored alternative ways to denoise cardiovascular magnetic resonance images that were captured during free-breathing. Specifically, they aimed to use real-time imaging, while applying reconstruction techniques to denoise the images (artifact versus artifact free images).28 Metrics used to evaluate the performance of their convolutional neural network were the signal-to-noise ratio, acceleration factor, and image cropping. The root-mean square error and the structural similarity index were used to evaluate the test dataset reconstructed image accuracy. The continuously rotating tiny golden angle sampling pattern had the lowest root-mean square error, and the highest structural similarity index (p < 0.0001) compared to all other sampling methods. The signal-to-noise ratio decreased from 20 dB to 10 dB, and the acceleration factor increased from 10x to 16x. Finally, the reconstruction time for all slices originating from raw data was 5.6x faster for the convolutional neural network (22 seconds).28 This study demonstrates that machine learning models have the potential to successfully remove artifact, while decreasing reconstruction times to produce better quality images and more accurate measurements in real time. More importantly, if these models become widely used, they have the potential to improve patient comfort or decrease the need for sedation in infants and younger children during cardiovascular magnetic resonance imaging, as the need for frequent breath holding is no longer required.
Further, clustering subjects based on their cardiovascular magnetic resonance imaging data is a very popular approach presented in the literature. While cardiac imaging has already been established as a standard and definitive way to diagnose complex heart disease, investigators are now taking advantage of the machine learning applications that can unveil hidden patterns in these data to boost diagnostic accuracy.31,32,35 Bruse and colleagues24 used agglomerative hierarchical clustering and principal component analysis to detect clinically meaningful shape clusters using anatomical cardiovascular magnetic resonance image data. Subjects with and without surgically corrected coarctation of the aorta, and subjects with healthy aortic arches were included. For each cross-validated run, 83% of healthy aortic arches were assigned to the healthy group, 85% of the coarctation shapes were correctly assigned, and 100% of the surgically corrected shapes were accurately assigned. Human clinicians would ideally use the results provided by the machine learning model to either compare or validate their own interpretation of the image data. This suggests that clustering techniques have the potential to inform clinical decision making at the time of diagnosis, thus improving accuracy and efficiency.
Finally, cardiac segmentation applied to cardiovascular magnetic resonance images is a process that takes a complex multidimensional image of the heart and separates its major sections, for example, the ventricles and coronary arteries. Segmentation is necessary because each ventricle or vessel can then be assessed quantitatively. Particularly, ventricular mass, volume, or ejection fraction can be quantitively measured. Segmentation driven by deep learning; specifically convolutional neural networks require large amounts of training data. Research studies aiming to optimize cardiac segmentation driven by deep learning in patients with CHD are faced with data accessibility challenges related to the extreme heterogeneity of cardiac anatomy and rarity of disease within the CHD population. Generative adversarial networks learn from real images in order to generate synthetic image data. The generative adversarial network has two networks that compete with one another. A generator network creates false images that the discriminator network will use to decipher between real and false images.74 Through these networks, new synthetic images are produced and can increase the size of training datasets in populations with limited image data.75 Investigators used these methods to create synthetic image data in patients with Tetralogy of Fallot, an extremely rare heart condition and found the images to be anatomically accurate.76 Generative adversarial networks may have major implications in CHD diagnostic image research, as they can potentially expand training datasets and allow models to predict rare and life-threatening diseases.
DISCUSSION
This scoping review demonstrates that machine learning is a rapidly evolving field of pediatric cardiology with a myriad of potential functions. The majority of these applications focused on the diagnosis and assessment of underlying CHD, including classification of auscultatory heart sounds, transthoracic echocardiograms, or cardiovascular magnetic resonance images.
Deep neural networks and support vector machines were commonly used algorithms for such tasks. Deep neural networks are popular to use when analyzing human data because they are robust and can handle inconsistent data. They are designed to model human cognitive abilities, which process, store, and retrieve information. Deep neural networks are extremely complex and often not interpretable to clinicians; therefore, their utility is somewhat controversial when applied to clinical decision making tasks. On the other hand, support vector machines are a popular technique used for binary outcome classification (i.e.; diseased versus healthy). Support vector machines use features and can separate classes by maximizing the distance between data points in each class. For healthcare related classification problems, support vector machines have high accuracy and do not suffer from multicollinearity (highly correlated features), which is an issue with human data as many features are often highly correlated (i.e.; systolic, diastolic, mean blood pressure values). Although support vector machines have their advantages, this technique is computationally intensive, with the non-linear support vector machine being more exhaustive than linear.
Implementing and translating machine learning results into clinically meaningful tools is a necessary path to improving diagnostic accuracy and efficiency.
Importantly, there are techniques to improve deep neural network interpretability, which is one of the most used algorithm types for diagnosing and assessing CHD. Heat maps are one example that can ensure models are capturing valid image signatures to further expand clinical usefulness. The integrated gradient method is another technique used to explain how a deep neural network predicted an outcome by visualizing input feature importance. The linear interpretable model-agnostic explanation is an application used with convolutional neural networks to discover what the convolutional neural network learns while deriving predictions, providing more interpretability to clinical end-users. Investigators have used this technique after comparing diagnostic performance of a convolutional neural network versus trained cardiologists and MUSE (GE Healthcare) automated analysis. The linear interpretable model-agnostic explanation technique presented physiologically relevant electrocardiogram segments chosen by the convolutional neural network when predicting diagnostic classes.77 Implementing any of these techniques is equivalent to feature importance ranking in random forest or coefficients in support vector machine for interpretability purposes. These methods can be applied to validate and corroborate that a model is predicting a legible signal, further supporting clinical end-user interpretability. For clinicians, the algorithm outputs must be relevant and trustworthy to garner clinician support. If implemented, clinicians will use machine learning models to classify healthy individuals versus those with diseases. Clinicians have a desire to understand how classification results are derived. Achieving this understanding will support machine learning prediction translation and clinical uptake.
To date, machine learning has been successfully explored and applied to support clinicians in many ways including, early recognition of cardiorespiratory instability. Bose and colleagues found that in 634 individuals with in-hospital cardiac arrest, 79% of these patients also had cardiorespiratory instability four to 24-hours prior to arrest.78,79 Predicting cardiorespiratory instability risk is significant because patients who experience this type of instability, if not attended to, can progress to in-hospital cardiac arrest. However, if cardiorespiratory instability is recognized early, cardiac arrest may be prevented.
Machine learning based technologies have the opportunity to not only advance expert care at tertiary centers, but to provide access to quality care in remote areas without local subspecialty expertise. Udine and colleagues found that CHD infant mortality was associated with higher poverty levels.15 The American Academy of Family Physicians’ position paper on poverty and health describes that poverty affects the built environment, which includes buildings, infrastructure, and services.80,81 In the case of limited access to expert care, machine learning can be used as a tool to connect specialized cardiac programs with distant primary care providers. Machine learning can systematically be applied to clinical data, process it, and provide improved accuracy and timely diagnosis of CHD for patients in various clinical settings. For patients in remote clinical settings, diagnostic data could be sent to a tertiary care center for consultation (machine learning output analysis and patient triage). Even for patients with access to specialized cardiac programs, they too may benefit from having their clinical data processed by machine learning algorithms. Specifically, advanced cardiac imaging data could result in automated detection of cardiac diseases. Machine learning models are likely to save time, while boosting diagnostic accuracy.
Although, machine learning appears to be an evolving and promising tool for CHD diagnostics, there are still several limitations to consider. General limitations of machine learning in healthcare include lack of diverse and large datasets, poor standardization across hospital systems, expertise and time challenges related to ground-truth labeling, lack of comparable testing sets, poor transparency of algorithm design, and inadequate prospective integration into clinical workflow. Specifically for the models addressed in this scoping review, their limitations are as follows: 1.) deep neural network requires large amounts of data, and carries the black box concept (you don’t know how or why the network came up with the output); 2.) support vector machine is computationally exhaustive and not ideal for problems with many training examples; 3.) principal component analysis reduces variables and dimensionality to improve interpretability, but will sacrifice prediction accuracy in doing so; 4.) cluster analyses are used when the outcome is unknown, so accuracy cannot be determined, and results tend to not be representative of real-world problems; 5.) Hidden Markov models require a priori knowledge about the problem, otherwise severe overfitting will result; 6.) linear discriminant analyses require a normal distribution, but do not impose assumptions which will increase bias. Linear discriminant analysis also suffers from issues with multicollinearity; 7.) decision trees are considered an unstable classifier, meaning small changes in data can cause large changes in decision tree structure. Decision trees are expensive and time consuming as it takes a lot of time to train the model. Overfitting can also be a problem with this type of classifier.
Further limitations related to this scoping review include using a broad literature search strategy. Our goal was to describe the comprehensive applications of machine learning used in pediatric cardiology research, and then focus on machine learning techniques used for diagnosis and assessment. This approach required screening an extensive amount of journal article titles and abstracts. In the future, it will be interesting to explore the predictive capabilities of other non-invasive diagnostic technologies, such as the 12-lead electrocardiogram. Few pediatric focused studies have considered the disease detection capabilities of this technology.82 In adult focused machine learning cardiac research, 12-lead electrocardiograms are already being leveraged to detect acute coronary syndrome.83,84
In conclusion, these findings indicate that machine learning is a very promising tool for diagnosing and assessing critical and non-critical CHD, yet extensive research is still needed to build robust and generalizable models for clinical use, especially considering the extreme heterogeneity of complex CHD.
FINANCIAL SUPPORT
This work was supported by grants from the National Institute of Health (S.H., grant number T32NR008857, 1F31NR019725-01A1).
Footnotes
CONFLICTS OF INTEREST
The Role of Machine Learning Applications in Critical and Non-Critical Congenital Heart Disease Diagnosis and Assessment: A Scoping Review. None.
ETHICAL STANDARDS
This study was exempt from Institutional Review Board review.
REFERENCES
- 1.Gharehbaghi A, Dutoit T, Sepehri AA, Kocharian A, Lindén M. A Novel Method for Screening Children with Isolated Bicuspid Aortic Valve. Cardiovascular engineering and technology. 2015;6(4):546–556. [DOI] [PubMed] [Google Scholar]
- 2.Yasaka K, Abe O. Deep learning and artificial intelligence in radiology: Current applications and future directions. PLoS medicine. 2018;15(11):e1002707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bien N, Rajpurkar P, Ball RL, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS medicine. 2018;15(11):e1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosny A, Parmar C, Coroller TP, et al. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS medicine. 2018;15(11):e1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kather JN, Krisam J, Charoentong P, et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS medicine. 2019;16(1):e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajpurkar P, Irvin J, Ball RL, et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS medicine. 2018;15(11):e1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor AG, Mielke C, Mongan J. Automated detection of moderate and large pneumothorax on frontal chest X-rays using deep convolutional neural networks: A retrospective study. PLoS medicine. 2018;15(11):e1002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS medicine. 2018;15(11):e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oster ME, Lee KA, Honein MA, Riehle-Colarusso T, Shin M, Correa A. Temporal trends in survival among infants with critical congenital heart defects. Pediatrics. 2013;131(5):e1502–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, Correa A. Prevalence of congenital heart defects in metropolitan Atlanta, 1998-2005. The Journal of pediatrics. 2008;153(6):807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman JI, Kaplan S. The incidence of congenital heart disease. Journal of the American College of Cardiology. 2002;39(12):1890–1900. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention Congential Heart Defects 2019; https://www.cdc.gov/ncbddd/heartdefects/data.html. Accessed February 28, 2020.
- 13.CDC. National Birth Defects Prevention Study. https://www.cdc.gov/ncbddd/birthdefects/nbdps.html. Accessed July 24, 2020.
- 14.Ailes EC, Gilboa SM, Riehle-Colarusso T, et al. Prenatal diagnosis of nonsyndromic congenital heart defects. Prenatal diagnosis. 2014;34(3):214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Udine MEF, Burns K, Pearson G, Kaltman J. 269 - Geographic Variation in Infant Mortality Due to Congenital Heart Disease. Paper presented at: American Heart Association- Scientific Sessions; November 13, 2020; Virtual. [Google Scholar]
- 16.Tricco AC, Lillie E, Zarin W, et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Annals of internal medicine. 2018;169(7):467–473. [DOI] [PubMed] [Google Scholar]
- 17.Garrard J. Health sciences literature review made easy: The matrix method. 5th ed. Burlington, MA: Jones & Bartlett; 2017. [Google Scholar]
- 18.Norgeot B, Quer G, Beaulieu-Jones BK, et al. Minimum information about clinical artificial intelligence modeling: the MI-CLAIM checklist. Nat Med. 2020;26(9):1320–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diller GP, Lammers AE, Babu-Narayan S, et al. Denoising and artefact removal for transthoracic echocardiographic imaging in congenital heart disease: utility of diagnosis specific deep learning algorithms. The international journal of cardiovascular imaging. 2019;35(12):2189–2196. [DOI] [PubMed] [Google Scholar]
- 20.Sun S, Wang H. Principal component analysis-based features generation combined with ellipse models-based classification criterion for a ventricular septal defect diagnosis system. Australasian physical & engineering sciences in medicine. 2018;41(4):821–836. [DOI] [PubMed] [Google Scholar]
- 21.Meza JM, Slieker M, Blackstone EH, et al. A novel, data-driven conceptualization for critical left heart obstruction. Computer methods and programs in biomedicine. 2018;165:107–116. [DOI] [PubMed] [Google Scholar]
- 22.Gharehbaghi A, Lindén M, Babic A. A Decision Support System for Cardiac Disease Diagnosis Based on Machine Learning Methods. Studies in health technology and informatics. 2017;235:43–47. [PubMed] [Google Scholar]
- 23.Pereira F, Bueno A, Rodriguez A, et al. Automated detection of coarctation of aorta in neonates from two-dimensional echocardiograms. Journal of medical imaging (Bellingham, Wash). 2017;4(1):014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruse JL, Zuluaga MA, Khushnood A, et al. Detecting Clinically Meaningful Shape Clusters in Medical Image Data: Metrics Analysis for Hierarchical Clustering Applied to Healthy and Pathological Aortic Arches. IEEE transactions on bio-medical engineering. 2017;64(10):2373–2383. [DOI] [PubMed] [Google Scholar]
- 25.Diller GP, Babu-Narayan S, Li W, et al. Utility of machine learning algorithms in assessing patients with a systemic right ventricle. European heart journal cardiovascular Imaging. 2019;20(8):925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gharehbaghi A, Sepehri AA, Lindén M, Babic A. Intelligent Phonocardiography for Screening Ventricular Septal Defect Using Time Growing Neural Network. Studies in health technology and informatics. 2017;238:108–111. [PubMed] [Google Scholar]
- 27.Elgendi M, Bobhate P, Jain S, et al. The Voice of the Heart: Vowel-Like Sound in Pulmonary Artery Hypertension. Diseases (Basel, Switzerland). 2018;6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauptmann A, Arridge S, Lucka F, Muthurangu V, Steeden JA. Real-time cardiovascular MR with spatio-temporal artifact suppression using deep learning-proof of concept in congenital heart disease. Magnetic resonance in medicine. 2019;81(2):1143–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aziz S, Khan MU, Alhaisoni M, Akram T, Altaf M. Phonocardiogram Signal Processing for Automatic Diagnosis of Congenital Heart Disorders through Fusion of Temporal and Cepstral Features. Sensors (Basel, Switzerland). 2020;20(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gharehbaghi A, Sepehri AA, Babic A. Distinguishing Septal Heart Defects from the Valvular Regurgitation Using Intelligent Phonocardiography. Studies in health technology and informatics. 2020;270:178–182. [DOI] [PubMed] [Google Scholar]
- 31.Karimi-Bidhendi S, Arafati A, Cheng AL, Wu Y, Kheradvar A, Jafarkhani H. Fully-automated deep-learning segmentation of pediatric cardiovascular magnetic resonance of patients with complex congenital heart diseases. Journal of cardiovascular magnetic resonance : official journal of the Society for Cardiovascular Magnetic Resonance. 2020;22(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y, Fu X, Li X, Qi Y. Cardiac Chamber Segmentation Using Deep Learning on Magnetic Resonance Images from Patients Before and After Atrial Septal Occlusion Surgery. Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual International Conference. 2020;2020:1211–1216. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Liu X, Wang F, et al. Automated interpretation of congenital heart disease from multi-view echocardiograms. Medical image analysis. 2020;69:101942. [DOI] [PubMed] [Google Scholar]
- 34.Gómez-Quintana S, Schwarz CE, Shelevytsky I, et al. A Framework for AI-Assisted Detection of Patent Ductus Arteriosus from Neonatal Phonocardiogram. Healthcare (Basel, Switzerland). 2021;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tandon A, Mohan N, Jensen C, et al. Retraining Convolutional Neural Networks for Specialized Cardiovascular Imaging Tasks: Lessons from Tetralogy of Fallot. Pediatric cardiology. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson WR, Reinisch AJ, Unterberger MJ, Schriefl AJ. Artificial Intelligence-Assisted Auscultation of Heart Murmurs: Validation by Virtual Clinical Trial. Pediatric cardiology. 2019;40(3):623–629. [DOI] [PubMed] [Google Scholar]
- 37.Lv J, Dong B, Lei H, et al. Artificial intelligence-assisted auscultation in detecting congenital heart disease. European Heart Journal - Digital Health. 2021;2(1):119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz VM, Saenz L, Lopez-Magallon A, et al. Early prediction of critical events for infants with single-ventricle physiology in critical care using routinely collected data. The Journal of thoracic and cardiovascular surgery. 2019;158(1):234–243.e233. [DOI] [PubMed] [Google Scholar]
- 39.Luo Y, Li Z, Guo H, et al. Predicting congenital heart defects: A comparison of three data mining methods. PloS one. 2017;12(5):e0177811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gharehbaghi A, Borga M, Sjöberg BJ, Ask P. A novel method for discrimination between innocent and pathological heart murmurs. Medical engineering & physics. 2015;37(7):674–682. [DOI] [PubMed] [Google Scholar]
- 41.Miller R, Tumin D, Cooper J, Hayes D Jr., Tobias JD. Prediction of mortality following pediatric heart transplant using machine learning algorithms. Pediatric transplantation. 2019;23(3):e13360. [DOI] [PubMed] [Google Scholar]
- 42.Jalali A, Simpao AF, Gálvez JA, Licht DJ, Nataraj C. Prediction of Periventricular Leukomalacia in Neonates after Cardiac Surgery Using Machine Learning Algorithms. Journal of medical systems. 2018;42(10):177. [DOI] [PubMed] [Google Scholar]
- 43.Samad MD, Wehner GJ, Arbabshirani MR, et al. Predicting deterioration of ventricular function in patients with repaired tetralogy of Fallot using machine learning. European heart journal cardiovascular Imaging. 2018;19(7):730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diller GP, Orwat S, Vahle J, et al. Prediction of prognosis in patients with tetralogy of Fallot based on deep learning imaging analysis. Heart (British Cardiac Society). 2020;106(13):1007–1014. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz-Fernández D, Monsalve Torra A, Soriano-Payá A, Marín-Alonso O, Triana Palencia E. Aid decision algorithms to estimate the risk in congenital heart surgery. Computer methods and programs in biomedicine. 2016;126:118–127. [DOI] [PubMed] [Google Scholar]
- 46.Dimopoulos AC, Nikolaidou M, Caballero FF, et al. Machine learning methodologies versus cardiovascular risk scores, in predicting disease risk. BMC medical research methodology. 2018;18(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liem DA, Murali S, Sigdel D, et al. Phrase mining of textual data to analyze extracellular matrix protein patterns across cardiovascular disease. American journal of physiology Heart and circulatory physiology. 2018;315(4):H910–h924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boskovski MT, Homsy J, Nathan M, et al. De Novo Damaging Variants, Clinical Phenotypes, and Post-Operative Outcomes in Congenital Heart Disease. Circulation Genomic and precision medicine. 2020;13(4):e002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang L, Li J, Huang M, et al. Prediction of pulmonary pressure after Glenn shunts by computed tomography-based machine learning models. European radiology. 2020;30(3):1369–1377. [DOI] [PubMed] [Google Scholar]
- 50.Jalali A, Lonsdale H, Do N, et al. Deep Learning for Improved Risk Prediction in Surgical Outcomes. Scientific reports. 2020;10(1):9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toba S, Mitani Y, Yodoya N, et al. Prediction of Pulmonary to Systemic Flow Ratio in Patients With Congenital Heart Disease Using Deep Learning-Based Analysis of Chest Radiographs. JAMA cardiology. 2020;5(4):449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cainelli E, Bisiacchi PS, Cogo P, et al. Detecting neurodevelopmental trajectories in congenital heart diseases with a machine-learning approach. Scientific reports. 2021;11(1):2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diller GP, Kempny A, Babu-Narayan SV, et al. Machine learning algorithms estimating prognosis and guiding therapy in adult congenital heart disease: data from a single tertiary centre including 10 019 patients. European heart journal. 2019;40(13):1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf MJ, Lee EK, Nicolson SC, et al. Rationale and methodology of a collaborative learning project in congenital cardiac care. American heart journal. 2016;174:129–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Javadekar N, Rajagopalan A, et al. Eligibility for subcutaneous implantable cardioverter-defibrillator in congenital heart disease. Heart rhythm. 2020;17(5 Pt B):860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma Y, Alhrishy M, Narayan SA, Mountney P, Rhode KS. A novel real-time computational framework for detecting catheters and rigid guidewires in cardiac catheterization procedures. Medical physics. 2018;45(11):5066–5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kan CD, Wang JN, Lin CH, et al. Handmade trileaflet valve design and validation for patch-valved conduit reconstruction using generalized regression machine learning model. Technology and health care : official journal of the European Society for Engineering and Medicine. 2018;26(4):605–620. [DOI] [PubMed] [Google Scholar]
- 58.Liu X, Aslan S, Hess R, et al. Automatic Shape Optimization of Patient-Specific Tissue Engineered Vascular Grafts for Aortic Coarctation. Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual International Conference. 2020;2020:2319–2323. [DOI] [PubMed] [Google Scholar]
- 59.Liu M, Zhao L, Yuan J. Establishment of Relational Model of Congenital Heart Disease Markers and GO Functional Analysis of the Association between Its Serum Markers and Susceptibility Genes. Computational and mathematical methods in medicine. 2016;2016:9506829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gopalakrishnan V, Menon PG, Madan S. cMRI-BED: A novel informatics framework for cardiac MRI biomarker extraction and discovery applied to pediatric cardiomyopathy classification. Biomedical engineering online. 2015;14 Suppl 2(Suppl 2):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H, Zhang CH, Ammanamanchi N, et al. Control of cytokinesis by β-adrenergic receptors indicates an approach for regulating cardiomyocyte endowment. Science translational medicine. 2019;11(513). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Troisi J, Cavallo P, Richards S, et al. Noninvasive screening for congenital heart defects using a serum metabolomics approach. Prenatal diagnosis. 2021. [DOI] [PubMed] [Google Scholar]
- 63.Qi H, Zhang H, Zhao Y, et al. MVP predicts the pathogenicity of missense variants by deep learning. Nature communications. 2021;12(1):510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ren Z, Zhu J, Gao Y, et al. Maternal exposure to ambient PM(10) during pregnancy increases the risk of congenital heart defects: Evidence from machine learning models. The Science of the total environment. 2018;630:1–10. [DOI] [PubMed] [Google Scholar]
- 65.Chu R, Chen W, Song G, et al. Predicting the Risk of Adverse Events in Pregnant Women With Congenital Heart Disease. Journal of the American Heart Association. 2020;9(14):e016371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dozen A, Komatsu M, Sakai A, et al. Image Segmentation of the Ventricular Septum in Fetal Cardiac Ultrasound Videos Based on Deep Learning Using Time-Series Information. Biomolecules. 2020;10(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klein AZ, Sarker A, Cai H, Weissenbacher D, Gonzalez-Hernandez G. Social media mining for birth defects research: A rule-based, bootstrapping approach to collecting data for rare health-related events on Twitter. Journal of biomedical informatics. 2018;87:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kagiyama N, Shrestha S, Farjo PD, Sengupta PP. Artificial Intelligence: Practical Primer for Clinical Research in Cardiovascular Disease. Journal of the American Heart Association. 2019;8(17):e012788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahle WT, Sutherland JL, Frias PA. Outcome of isolated bicuspid aortic valve in childhood. The Journal of pediatrics. 2010;157(3):445–449. [DOI] [PubMed] [Google Scholar]
- 70.Marinho J, Pires A, Sousa G, Castela E. Right subclavian artery aneurysm in an adolescent with a bicuspid aortic valve. Pediatric cardiology. 2013;34(8):1952–1954. [DOI] [PubMed] [Google Scholar]
- 71.Siu SC, Silversides CK. Bicuspid aortic valve disease. Journal of the American College of Cardiology. 2010;55(25):2789–2800. [DOI] [PubMed] [Google Scholar]
- 72.Spaziani G, Ballo P, Favilli S, et al. Clinical outcome, valve dysfunction, and progressive aortic dilation in a pediatric population with isolated bicuspid aortic valve. Pediatric cardiology. 2014;35(5):803–809. [DOI] [PubMed] [Google Scholar]
- 73.Rubinstein R. The Cross-Entropy Method for Combinatorial and Continuous Optimization. Methodology And Computing In Applied Probability. 1999;1(2):127–190. [Google Scholar]
- 74.Chen C, Qin C, Qiu H, et al. Deep Learning for Cardiac Image Segmentation: A Review. Frontiers in Cardiovascular Medicine. 2020;7(25). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeleznik R, Weiss J, Taron J, et al. Deep-learning system to improve the quality and efficiency of volumetric heart segmentation for breast cancer. NPJ digital medicine. 2021;4(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Diller GP, Vahle J, Radke R, et al. Utility of deep learning networks for the generation of artificial cardiac magnetic resonance images in congenital heart disease. BMC Med Imaging. 2020;20(1):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hughes JW, Olgin JE, Avram R, et al. Performance of a Convolutional Neural Network and Explainability Technique for 12-Lead Electrocardiogram Interpretation. JAMA cardiology. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bose E, Hoffman L, Hravnak M. Monitoring cardiorespiratory instability: Current approaches and implications for nursing practice. Intensive & critical care nursing. 2016;34:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kause J, Smith G, Prytherch D, et al. A comparison of antecedents to cardiac arrests, deaths and emergency intensive care admissions in Australia and New Zealand, and the United Kingdom--the ACADEMIA study. Resuscitation. 2004;62(3):275–282. [DOI] [PubMed] [Google Scholar]
- 80.Czapp P, Kovach K. Poverty and Health- The Family Medicine Perspective (Position Paper). 2015; https://www.aafp.org/about/policies/all/poverty-health.html. Accessed January 2, 2021. [Google Scholar]
- 81.Macintyre S, Ellaway A, Cummins S. Place effects on health: how can we conceptualise, operationalise and measure them? Social science & medicine (1982). 2002;55(1):125–139. [DOI] [PubMed] [Google Scholar]
- 82.Du Y, Huang S, Huang C, Maalla A, Liang H. Recognition of Child Congenital Heart Disease using Electrocardiogram based on Residual of Residual Network. Paper presented at: 2020 IEEE International Conference on Progress in Informatics and Computing (PIC); 18-20 Dec. 2020, 2020. [Google Scholar]
- 83.Bouzid Z, Faramand Z, Gregg RE, et al. In Search of an Optimal Subset of ECG Features to Augment the Diagnosis of Acute Coronary Syndrome at the Emergency Department. Journal of the American Heart Association. 2021;10(3):e017871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Al-Zaiti S, Besomi L, Bouzid Z, et al. Machine learning-based prediction of acute coronary syndrome using only the pre-hospital 12-lead electrocardiogram. Nature communications. 2020;11(1):3966. [DOI] [PMC free article] [PubMed] [Google Scholar]