Abstract
Chromatin features are thought to have a role in the epigenetic transmission of transcription states from one cell generation to the next. It is unclear how chromatin structure survives disruptions caused by genomic replication or whether chromatin features are instructive of the transcription state of the underlying gene. We developed a method to monitor budding yeast replication, transcription, and chromatin maturation dynamics on each daughter genome in parallel, with which we identified clusters of secondary origins surrounding known origins. We found a difference in the timing of lagging and leading strand replication on the order of minutes at most yeast genes. We propose a model in which the majority of old histones and RNA polymerase II (RNAPII) bind to the gene copy that replicated first, while newly synthesized nucleosomes are assembled on the copy that replicated second. RNAPII enrichment then shifts to the sister copy that replicated second. The order of replication is largely determined by genic orientation: If transcription and replication are codirectional, the leading strand replicates first; if they are counterdirectional, the lagging strand replicates first. A mutation in the Mcm2 subunit of the replicative helicase Mcm2-7 that impairs Mcm2 interactions with histone H3 slows down replication forks but does not qualitatively change the asymmetry in nucleosome distribution observed in the WT. We propose that active transcription states are inherited simultaneously and independently of their underlying chromatin states through the recycling of the transcription machinery and old histones, respectively. Transcription thus actively contributes to the reestablishment of the active chromatin state.
All eukaryotic genomic processes happen in the context of chromatin. The smallest repeating subunit of chromatin is the nucleosome: A 147-bp DNA segment wrapped 1.65 turns around a histone octamer core, consisting of one H3/H4 tetramer and two H2A/H2B dimers (Luger et al. 1997). Since architectural features of chromatin limit the accessibility of the DNA substrate to DNA processing enzymes involved in replication, transcription, or repair, chromatin—in addition to being a genome packaging system—is the foremost regulatory system for all DNA-based processes.
Chromatin configuration is transiently disrupted with every round of genome replication. Nucleosomes bound to DNA, which are referred to here as old nucleosomes, are disassembled ahead of the replication fork and recycled behind it on the two newly formed daughter chromatids (Foltman et al. 2013). Concomitantly with old nucleosome recycling, new histones are assembled on daughter chromatids to restore optimal nucleosome density after genome duplication (for review, see Alabert and Groth 2012). The process of nucleosome reassembly on daughter chromatids and the (re)establishment of a specific chromatin architecture on replicated gene copies is called chromatin maturation. In theory, chromatin maturation should result in the accurate “inheritance” of a specific chromatin feature, which should then help preserve the expression state of the underlying gene for the next cell generation. Following the same logic, if the chromatin state of a gene is modified during genome replication, the transcription state of that gene should also be changed. This epigenetic transformation is thought to be at the heart of cellular differentiation. Whether specific chromatin configurations are indeed accurately inherited and whether they are instructive of the underlying gene expression state are, however, still open questions.
The cell is faced with two problems after every replication event: (1) It has to either restore its chromatin configuration to its prereplication state on both daughter genomes in order to maintain the same transcription program or use the disruption caused by replication as an “opportunity” to modulate chromatin configuration (symmetrically on both genomes or asymmetrically only on one replicated copy) and change the transcription program in response to developmental or environmental signals. (2) It has to globally regulate transcription levels in response to gene copy number doubling after genome replication, until cellular division restores the original gene copy number.
While it is largely accepted that old and new histones bind to both daughter chromatids after replication (Cusick et al. 1984; Gruss et al. 1990), the precise distribution pattern is still not clear. Old and new histones could bind completely symmetrically and randomly to either chromatid or in locally asymmetrical segments, thus forming contiguous alternating “patches” of old and new nucleosomes on the same chromatid. The mode of old and new histone distribution has implications for the mechanism of restoration of chromatin states. A map of old nucleosome distribution on replicated chromatids would thus provide clues on how and whether potential epigenetic information “embedded” in old nucleosomes might be “copied” to new nucleosomes. It would also shed light on how replicated gene copies might end up in different gene expression states, which presumably occurs during differentiation. Most importantly, it is still not known whether and how the distribution pattern of old and new histones affects transcription after replication. While recent studies have observed asymmetrical binding of old and newly synthesized histones to replicated daughter chromatids, (Gan et al. 2018; Petryk et al. 2018; Yu et al. 2018), it is still not clear how RNA polymerase II (RNAPII) binding and consequently transcription of replicated gene copies might be affected by such asymmetrical chromatin assembly.
The goal of this study is to better understand the molecular mechanisms responsible for chromatin maturation and to find out whether and how chromatin maturation affects the transmission of transcription states to replicated gene copies. We have therefore combined ChIP with our method for chromatin mapping on newly replicated DNA—NChAP (nascent chromatin avidin pull-down) (Vasseur et al. 2016)—and measured the distribution of RNAPII, histone H3, H3K56ac (new histones), and H3K4me3 and H3K36me3 (these marks of active transcription were used as proxies for old histones on replicated daughter genomes as in Yu et al. 2018). In addition to WT yeast, our assay was carried out in mutants that compromise new nucleosome assembly (rtt109Δ) (Kaplan et al. 2008) or are thought to impair old histone recycling (mcm2-3A) (Foltman et al. 2013; Gan et al. 2018; Petryk et al. 2018).
Results
RNAPII ChIP-NChAP
How does the cellular transcription machinery transition from transcribing only one gene copy to activating two copies after replication has doubled the gene copy number? Does the transcriptional output increase twofold immediately after gene copy number doubling and are both gene copies transcribed equally? Previous studies have shown that there is a significant delay between gene copy number doubling and the expected twofold increase in gene expression, which typically happens at the end of S phase when the whole genome has finished replicating (Voichek et al. 2016; Topal et al. 2019). This effectively means that, for the entire duration of S phase, two copies of a gene that replicated early produce as much mRNA as one copy had been producing before replication. Our findings on the kinetics of nucleosome positioning maturation, which suggest that one replicated gene copy is more expressed than its sister copy, are also consistent with this observation (Vasseur et al. 2016).
We expect that postreplicative gene expression levels are most likely directly linked to the amount of RNAPII that was bound to the gene locus at the time of replication. Our conjecture is based on three previous findings: (1) The rate of transcription by individual RNAPII complexes at any given gene does not decrease after replication (Topal et al. 2019). (2) mRNA degradation rates of genes that are not regulated by the cell cycle do not increase after replication (Eser et al. 2014). (3) Bulk RNAPII density on actively transcribed genes correlates with mRNA levels and mRNA synthesis rates (Miller et al. 2011). We therefore hypothesize that there is a limited amount of RNAPII that is available to rebind to replicated gene copies immediately after replication and that the majority of RNAPII complexes that bind to replicated gene copies immediately after the passage of the replication fork are the ones that were bound to that same gene before replication. Unless the transcription rates of individual RNAPII complexes bound to replicated genes increase—a possibility for which there is currently no experimental evidence—gene expression cannot go up even when DNA content has doubled if the total RNAPII level bound to two replicated gene copies is the same as the amount of RNAPII that was bound to only one gene copy before replication. If our hypothesis is correct, there should be a detectable lag between gene copy number doubling and the increase in RNAPII density on replicated genes. During that period there would be just “enough” RNAPII on replicated gene copies to produce the same amount of mRNA as before replication.
We therefore tested whether RNAPII is limiting relative to DNA content after replication. We measured RNAPII/DNA ratios for all Saccharomyces cerevisiae genes during S phase using ChIP-on-chip with two channel DNA microarrays. RNAPII enrichment on gene bodies was monitored by immunoprecipitation of the HA-tagged RNAPII core subunit 3 (Rpb3) from a synchronized cell population 25 min (early S phase) and 32 min (early-mid S phase) after release from α factor-induced G1 arrest. The isolated DNA fragments from ChIP (first channel) and input (second channel) fractions were then mixed and hybridized to whole genome yeast DNA microarrays.
Note that this type of experiment allows us to directly monitor how RNAPII enrichment in gene bodies (promoters and transcription start sites were excluded from the analysis) changes relative to the postreplicative increase in gene copy number because the final readout of the experiment is the ratio of the amount of gene-bound RNAPII versus DNA content of every gene at a specific point in S phase. We see that, as more genes in the population are replicated, the ratios of RNAPII (ChIP DNA) to DNA content (input DNA) progressively decrease in replicated genes compared with unreplicated genes (Supplemental Fig. S1A). Genes that replicated earlier (genes with replication timing < 55 min) have less RNAPII per gene copy than late genes that have not yet been replicated (replication timing ≥ 55 min). This is consistent with our initial hypothesis: mRNA production does not increase immediately after gene copy number doubling because the quantity of available RNAPII complexes in proximity to replicated genes is not yet sufficient to sustain transcription of both gene copies at prereplication levels. Conversely, RNAPII occupancy in asynchronous cells (Kim et al. 2010) shows the expected pattern of higher occupancy in early genes compared with late genes, as early genes are known to have, on average, a higher transcriptional activity than late replicating genes.
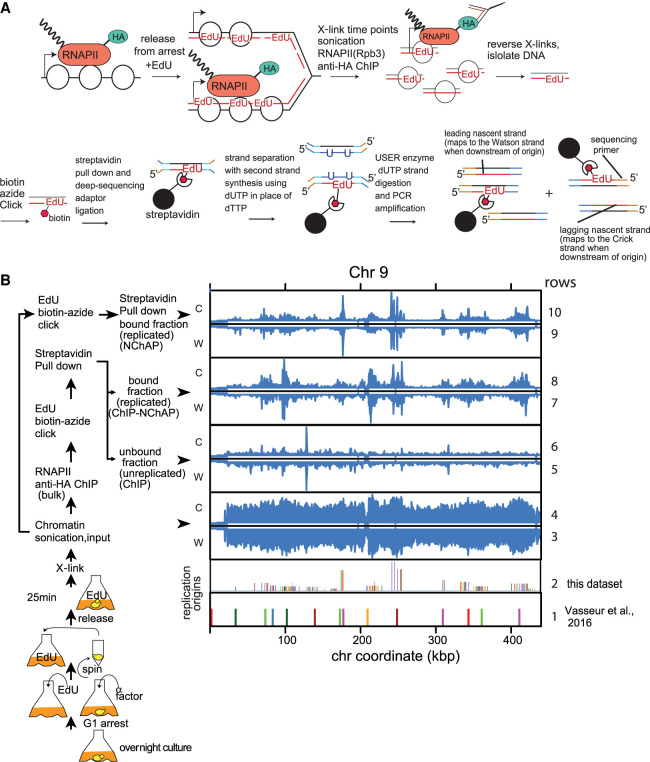
Next, we measured how RNAPII complexes partition between the two replicated gene copies. We combined RNAPII ChIP with our NChAP assay developed in Vasseur et al. (2016) to measure the differences in RNAPII enrichment between two newly replicated daughter chromatids. The strategy is to grow cells in the presence of the thymidine analog EdU, which labels newly replicated DNA strands, fix the culture with formaldehyde at different time points in S phase, and then perform ChIP of RNAPII with an HA-tagged Rpb3 subunit. This is followed by the isolation of nascent DNA fragments from ChIP-ed DNA using streptavidin pull-down after biotinylation of incorporated EdU. The purified replicated DNA is then used to make strand-specific deep-sequencing NChAP libraries as described previously (Vasseur et al. 2016). Thus, sequencing reads that map to the Watson or the Crick strand will have originated from one or the other replicated daughter chromatid, respectively (Fig. 1A). RNAPII enrichment peaks from the ChIP fraction on replicated chromatin (ChIP-NChAP, rows 7 and 8) are localized in regions that replicated early in S phase (NChAP, rows 9 and 10) and are centered at origins of replication (rows 1 and 2). Conversely, RNAPII peaks from unreplicated chromatin (unreplicated ChIP; i.e., the fraction that did not bind to streptavidin beads, rows 5 and 6) are detected throughout the chromosome and are not concentrated around origins of replication. This confirms that we are indeed able to isolate replicated DNA specifically bound by RNAPII (Fig. 1B).
Figure 1.
RNAPII ChIP-NChAP in early S phase. (A) Diagram of the RNAPII ChIP-NChAP experiment. (B) RNAPII distribution on Chromosome 9 from chromatin fractions diagrammed on the left, in early S phase 25 min after release from G1 arrest. The positions of replication origins are shown in rows 1 and 2—(1) identified in Vasseur et al. (2016); and (2) from this data set. Read counts from all fractions were grouped in 50-bp bins and first normalized to the genome average read count and then to the highest peak value in each chromosome. W and C are reads from Watson and Crick strands, respectively.
Asymmetric distribution of RNAPII on daughter chromatids
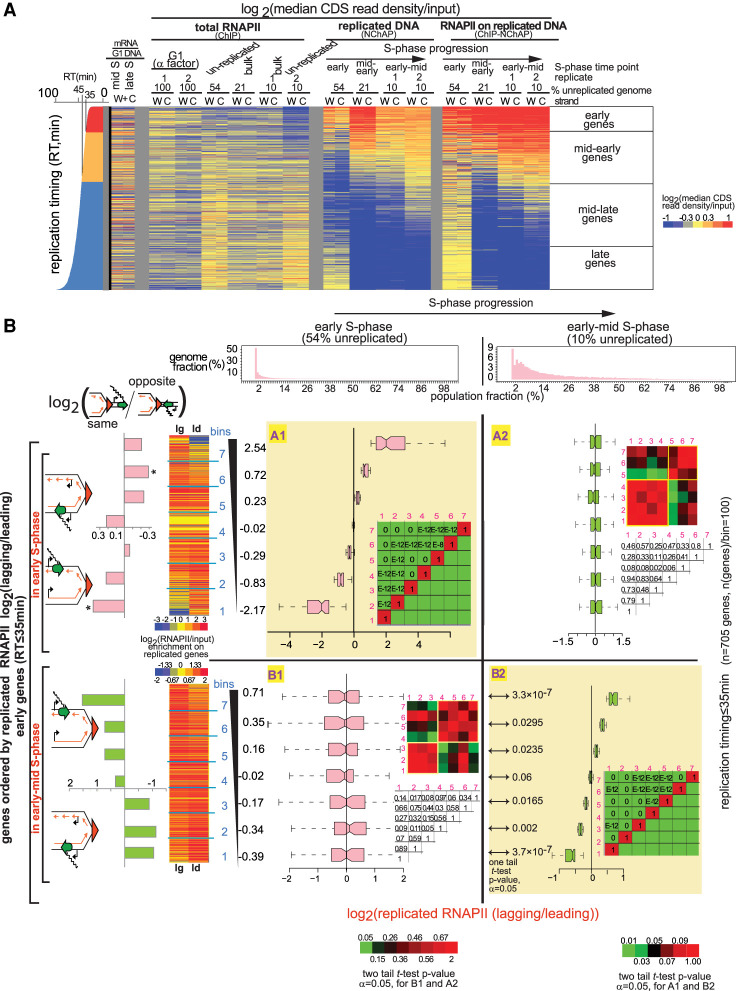
What is the distribution pattern of RNAPII complexes on replicated gene copies? The median read density in the coding region of each gene from Watson (W) or Crick (C) reads was used as a measure of relative RNAPII occupancy on each gene copy (Fig. 2A). The heat map in Figure 2A shows median read densities for the coding regions (promoters and transcription start sites were excluded from our analysis) of all yeast genes that are not regulated by the cell cycle, from fractions representing bulk RNAPII (ChIP), replicated DNA (NChAP), and RNAPII on replicated DNA (ChIP-NChAP) at different time points before and during S phase: (1) late G1, (2) early S phase, (3) mid-early S phase, and (4) early-mid S phase. The heat map confirms the specificity and reproducibility of our assay, as RNAPII enrichment in the ChIP-NChAP fraction is only detected on replicated genes (cf. ChIP-NChAP and NChAP fractions).
Figure 2.
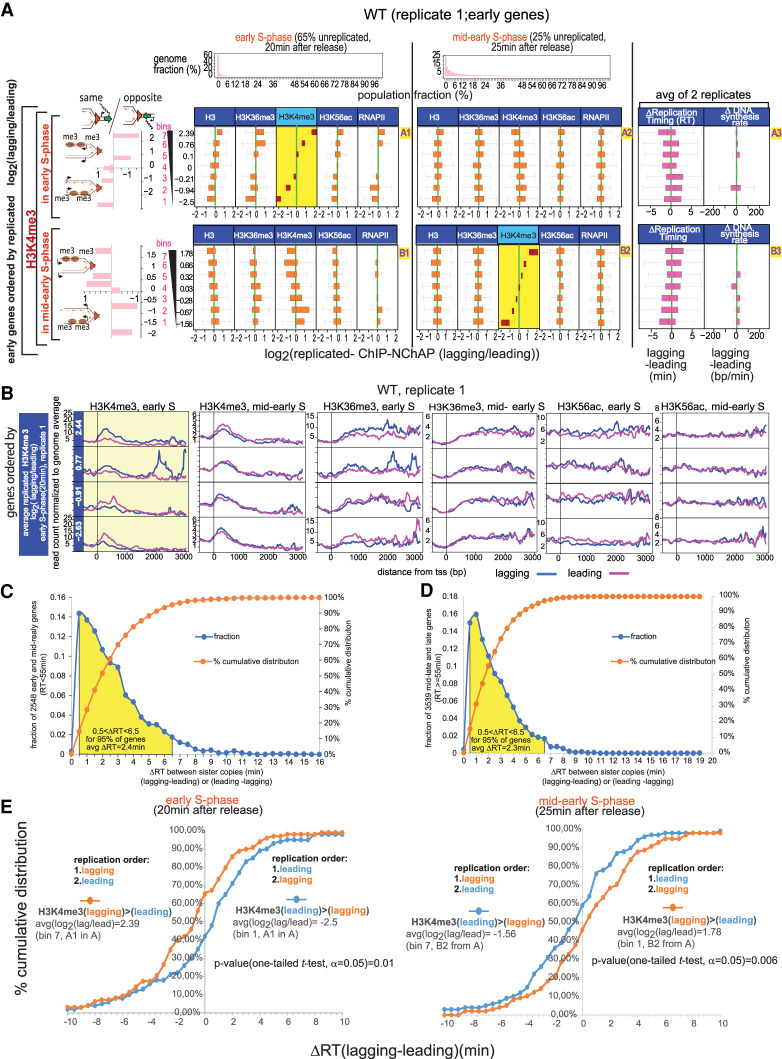
RNAPII is distributed asymmetrically on replicated gene copies. (A) Heat map of median RNAPII occupancies in coding regions (CDS) of all yeast genes that are not cell cycle-regulated. Reads from promoter regions have been excluded from median read density calculations. Each line is an individual gene and columns represent occupancy values for (W)atson and (C)rick gene copies for different G1- and S-phase time points. S-phase time points are defined by the fraction of the genome that has not yet been replicated: (Early S phase) 54% of the genome is unreplicated over the whole cell population; (mid-early S phase) 21% unreplicated; (early-mid S phase; replicates 1 and 2) 10% unreplicated. The represented fractions are bulk RNAPII (ChIP), replicated DNA (NChAP), and RNAPII on replicated DNA (ChIP-NChAP). The first two columns on the left represent mRNA enrichment over G1 genomic DNA in mid and late S (in the absence of EdU) (Vasseur et al. 2016). Genes are ordered by replication timing of the Watson strand, shown in the bar graph on the left (RT) (Vasseur et al. 2016). (B) Box plot distributions of lagging/leading RNAPII ratios. The figure is organized into a grid. Rows A and B represent the data sets whose lagging/leading ratios were used to sort early genes: (row A) early genes in early S phase; (row B) early genes in early-mid S phase. Columns 1 and 2 represent S-phase time points: (1) early; (2) early-mid S phase. The header row shows the distribution of replicated genome fractions as described in the “Data Analysis” section of Supplemental Material. The 54% unreplicated and the 10% unreplicated (replicate 1) RNAPII ChIP-NChAP from A are, respectively, used as reference points for early and early-mid S phase throughout the article. Early replicating genes in fields A1 and B2 have been sorted by decreasing lagging/leading RNAPII occupancy in early S phase and early-mid S phase, respectively, and divided into seven bins (y-axis) (yellow background), and box plot distribution of RNAPII lagging/leading ratios (x-axis) have been determined for each bin in each time point. The average lagging/leading ratios for each gene bin are indicated in the y-axis on the left. For example, the bottom group of genes (bin 1) in A1 has, on average, 5.6 times more RNAPII on the leading copy than on the lagging in early S phase. The heat maps on the left show RNAPII enrichment on replicated DNA (ChIP-NChAP fractions) for the lagging (lg) and leading (ld) copies of the genes in the seven bins. The bar graphs on the left show the enrichment of “same” genes for each bin indicated in the y-axis calculated as the ratio of “same” orientation genes versus “opposite” genes for each bin normalized to the same/opposite ratio of all 705 early genes (rows A and B; [*] P-value of hypergeometric test < 0.05). The heat map insets in each field show the P-values of the pairwise two tailed t-test for two independent samples (α = 0.05) for each bin pair. The color bars on the bottom left or bottom right correspond to heat maps for fields B1 and A2 or A1 and B2, respectively. The corresponding P-values are also shown in the tables on the right of the box plot graphs within each field (E–12 = 10−12 and E–8 = 10−8). The P-values of the pairwise one-tailed t-test for two independent samples (α = 0.05) between early and early-mid S phase for each bin in row B are shown in B2 on the left of the box plots.
The median occupancy of total RNAPII in the gene body correlates well with mRNA abundance in S phase (cf. the bulk RNAPII ChIP fractions with mRNA enrichment in Fig. 2A; see Supplemental Fig. S1B). We can therefore reasonably conclude that the median occupancy of total RNAPII in the gene body is a good measure of transcription activity and that a significant portion of all RNAPII complexes located exclusively in the gene body is actively engaged in transcription. Supplemental Figure S1C shows that the unreplicated ChIP fractions are indeed enriched for genes that have not yet been replicated compared with the bulk ChIP fractions. Late genes with replication timing >55 min that are not yet replicated in early-mid and mid-early S phase (late genes in Fig. 2A) have, on average, a 50%–74% higher RNAPII signal, respectively, compared with genes that replicated early, with replication timing under 35 min (early genes in Fig. 2A). Conversely, the RNAPII signal from late genes is just ∼15% higher than the signal from early genes in the bulk ChIP fractions. This indicates that we are successfully separating RNAPII bound to replicated chromatin from RNAPII bound to unreplicated chromatin.
Next, we asked whether there is a difference in RNAPII occupancy between replicated gene copies. The linear fit of the scatterplot comparing median read densities of W and C gene copies of 705 early replicating genes shows that only 34% of the differences in RNAPII occupancy between W and C copies on replicated chromatin (the ChIP-NChAP fraction) can be explained by the linear fit model (R2 =0.34), as opposed to 52% for unreplicated chromatin (R2 = 0.52), which suggests that most of the observed differences in RNAPII occupancy between replicated W and C gene copies are caused by replication (Supplemental Fig. S1D). The observed asymmetry in RNAPII occupancy is also not due to sequencing or a strand bias in EdU incorporation because differences in W and C read densities are smaller (R2= 0.66) in the replicated chromatin fraction (NChAP fraction) for which we used the same library construction protocol as for the ChIP-NChAP fraction (Supplemental Fig. S1D).
We then determined the pattern of RNAPII distribution between leading and lagging gene copies. Lagging and leading annotations were assigned as described previously (Vasseur et al. 2016): Watson reads upstream of the closest replication origin (see Supplemental Tables S1, S2; “Replication Origins Mapping” in the “Data Analysis” section of Supplemental Material) originate from the lagging strand copy, while the complementary Crick reads are from the leading copy. The opposite is true for reads located downstream from origins.
In order to assess how RNAPII is distributed between the leading and the lagging copy, we first calculated the lagging to leading ratio of RNAPII occupancies for all 705 genes that replicate in early S phase, sorted the ratios in ascending order, and then divided the set into seven bins of ∼100 genes each (row A, Fig. 2B). We then determined box plot distributions of lagging/leading RNAPII occupancy ratios for each bin in early (field A1, Fig. 2B) and early-mid S phase (field A2, Fig. 2B). Next, we ordered the same 705 early genes by lagging/leading RNAPII ratios in early-mid S phase and performed the same analysis as above (row B, Fig. 2B). The same analysis was also done for genes replicating in mid-early S phase with replication timings between 35 min and 45 min (rows A and B, Supplemental Fig. S3A).
As shown in Figure 2B, RNAPII distribution has an apparent bias for the lagging or the leading strand in at least half of early replicating genes. In early S phase, when 54% of the genome is still unreplicated, ∼28% of early and mid-early genes have, on average, approximately three times more RNAPII on the leading strand (bins 1 and 2, field A1, Fig. 2B; Supplemental Fig. S3A) and another 28% have approximately three times more RNAPII on the lagging strand (bins 6 and 7, field A1, Fig. 2B; Supplemental Fig. S3A).
Moreover, all early (field A1, Fig. 2B) and mid-early (field A1, Supplemental Fig. S3A) gene bins are significantly different from each other, with two-tailed t-test P-values ranging from 0 to 10−8. The biased segregation of RNAPII enrichments between the lagging and the leading gene copies mostly “disappears” later in S phase (cf. distributions in A1 with A2 in Fig. 2B; Supplemental Fig. S3A), as differences in RNAPII bias in different gene bins become insignificant.
We also found a large number of genes with significantly asymmetrical RNAPII binding when we sort gene bins by lagging/leading RNAPII ratios in early-mid S phase (P-values from 0 to 10−12) (field B2, Fig. 2B; Supplemental Fig. S3A). The differences between average lagging and leading RNAPII occupancy in early-mid S phase (B2, Fig. 2B; Supplemental Fig. S3A) are, however, smaller than in early S phase (A1, Fig. 2B; Supplemental Fig. S3A). Approximately 28% of early and mid-early genes have, on average, 35% more RNAPII on the leading strand (bins 1 and 2, B2, Fig. 2B; Supplemental Fig. S3A) and another 28% has 45% more RNAPII on the lagging strand (bins 6 and 7, B2, Fig. 2B; Supplemental Fig. S3A). Genes with the biggest bias in RNAPII binding in early-mid S phase are different from the genes that had the biggest bias in early S phase. Although most gene bins that were ordered by their RNAPII lagging/leading ratio in early-mid S phase (B2) do not have significant differences (i.e., a P-value < 0.05 in Fig. 2B) in their lagging/leading RNAPII ratios in early S phase (see B1 in Fig. 2B; Supplemental Fig. S3A), there is a slight anticorrelation between lagging/leading ratios of early and mid-early genes in early S phase (B1 in Fig. 2B; Supplemental Fig. S3A) compared with early-mid S phase (B2 in Fig. 2B; Supplemental Fig. S3A). Additionally, the average change in RNAPII lagging/leading ratios from early to early-mid S phase for almost all bins in row B is statistically significant (see P-values of one-tail pairwise t-tests in B2 in Fig. 2B; Supplemental Fig. S3A). The only exceptions are bin 4 for early genes (Fig. 2B) and bins 3 and 4 for mid-early genes (Supplemental Fig. S3A), in which RNAPII distribution on either strand stays symmetrical. So, it appears that RNAPII distribution on the lagging and leading strands changes significantly as chromatin maturation progresses and that RNAPII enrichment switches from one strand (lagging or leading) in early S phase (step 1) to the other (leading or lagging, respectively) in early-mid S phase (step 2).
We recently showed that nucleosome positioning maturation after replication depends on transcription, and we measured faster nucleosome repositioning on the copy replicated by the leading strand when gene transcription travels in the same direction as the replication fork (“same” orientation genes). Conversely, nucleosome repositioning was faster on the lagging strand copy for “opposite” orientation genes (Vasseur et al. 2016). We consequently predicted that the leading copy will be more transcribed than the lagging copy when transcription and replication are codirectional, while the lagging copy will be more expressed when they are counterdirectional.
In order to test this prediction, we calculated the ratio of “same” versus “opposite” orientation genes for each bin in which genes were organized by increasing lagging/leading ratios of RNAPII occupancy in early S phase (A1, Fig. 2B; Supplemental Fig. S3A) or early-mid S phase (B2, Fig. 2B; Supplemental Fig. S3A) and normalized it to the “same”/“opposite” ratio of all 705 early (Fig. 2B) or 1868 mid-early genes (Supplemental Fig. S3A). As predicted in Vasseur et al. (2016), “same” genes in early S phase tend to indeed have more RNAPII on the leading copy and “opposite” genes tend to have more RNAPII on the lagging copy (row A, Fig. 2B; Supplemental Fig. S3A). Later, in early-mid S phase, RNAPII “polarity” switches and RNAPII becomes more abundant either on lagging copies when transcription and replication travel in the same direction or on leading copies when they are opposite (row B, Fig. 2B; Supplemental Fig. S3A). The switch in genic orientation “preference” and the slight anticorrelation of the bias in RNAPII occupancy between early and early-mid S phase suggest that RNAPII is enriched on one replicated gene copy first (step 1: RNAPII binds first to the leading copy on “same” genes or the lagging copy on “opposite genes), then switches to the other later (step 2: RNAPII switches to the lagging copy on “same” genes or the leading copy on “opposite” genes). Indeed, EdU pulse and thymidine chase experiments in asynchronous cultures confirmed that RNAPII enrichment does shift from the leading to the lagging copy at “same” genes and from the lagging to the leading copy at “opposite” genes (Supplemental Fig. S3B)
Note that this switch in RNAPII enrichment from one replicated copy to the other explains why the differences in RNAPII enrichment between the lagging and the leading copy are, on average, smaller later in S phase compared with early S phase (∼30% to ∼45% in early-mid S phase vs. threefold in early S phase). Since the replication program is at a slightly different stage in each cell, the recorded average lagging/leading ratios represent a mixture of the configuration when RNAPII is first preferentially bound to one gene copy in a subset of the population (step 1) and the configuration when it has switched to the other copy in another subset of the population (step 2). The prevalence of one configuration over the other in the population varies depending on how long after the passage of the replication fork we managed to capture a specific group of genes in most cells in the population.
This bias in RNAPII enrichment is specific to replicated chromatin, as the differences in RNAPII occupancies on lagging and leading strands in bulk or unreplicated chromatin are significantly smaller than in replicated chromatin (Supplemental Fig. S2). More importantly, the asymmetrical pattern of RNAPII distribution observed in early-mid S phase is highly reproducible (Supplemental Figs. S3C, S4). We detect the same RNAPII distribution pattern in the same gene bins in four biological replicates of the mid-early S-phase time point from Figure 2B (two are shown in Fig. 2A; Supplemental Fig. S3C; the other two are shown in fields D3 and D4 in Supplemental Fig. S4B). This suggests that the configuration when RNAPII is mostly enriched on the lagging strand of “same” genes or on the leading strand of “opposite” genes that is detected later in S phase represents the final step in the process of RNAPII (re)binding to replicated genes.
The patterns of RNAPII distribution and genic orientation in the early S-phase replicate from Supplemental Figure S4B are also similar to the early S-phase replicate from Figure 2. These patterns are, however, found in different gene groups in the two replicates. We propose that this is a consequence of the transient and short-lived nature of the first step in the RNAPII (re)binding process. Due to the stochastic nature of replication origin activation and nonsynchronous fork progression in different cells in the population, the same genes from different early S-phase replicates shown in Supplemental Figure S4 were captured at different distances from their respective replication forks when cells were fixed in early S phase. Consequently, these genes were at different stages of their chromatin maturation process in different cells. Finally, it all comes down to the length of time RNAPII spends in the first configuration after any particular gene has been replicated. According to our results, each gene in each cell can be either in the first intermediate short-lived configuration with more RNAPII on the leading copy for “same” genes or the lagging copy for “opposite” genes, or in the second final configuration when RNAPII enrichment has already shifted to the other replicated gene copy (leading for “opposite” genes or lagging for “same” genes). The RNAPII lagging/leading ratio for any gene at a given time point in S phase will therefore represent the average of all ratios from each cell in the population. If the time point was taken early in S phase (15–25 min after release from G1 arrest), the average lagging/leading ratio will skew toward the first configuration for a good number of genes that have just finished replicating or are still being replicated in a majority of cells in the population at that particular time. This first configuration is very short-lived, and since replication origin firing is stochastic at the single cell level, it is technically impossible to fix the cells at the exact time in S phase when this same group of genes has just finished replicating or is still being replicated and would consequently predominantly be in that configuration in a majority of cells when we repeat the experiment. Later in S phase (26–32 min after release from arrest), when most early genes in most cells have been replicated for a while, the average lagging/leading ratio will skew toward the second final configuration, and because this final configuration is long-lived, we can record it reproducibly on the same gene groups every time we fix cells in early-mid S phase.
Thus, short of monitoring chromatin maturation dynamics in real time and in single cells, the stochasticity of chromatin maturation dynamics in the cell population and the very short-lived nature of the first configuration (step 1) makes it technically impossible to pinpoint the exact moment in the replication program when a particular group of genes is mostly at the same intermediate short-lived early step of the RNAPII rebinding process even if we fix cells at the same time in early S phase (i.e., 25 min after release from G1 arrest) every time we repeat the experiment. Consequently, even though the RNAPII lagging/leading ratios do not correlate in the two early S-phase replicates (cf. A1 with A2 and B2 with B1 in Supplemental Fig. S4B), we did observe the same pattern of RNAPII distribution in each replicate: prevalence of RNAPII on the leading copy of “same” genes or on the lagging copy of “opposite” genes in early S phase (see same/opposite bar graphs in rows A and B in Supplemental Fig. S4B).
Pulse chase experiments such as the ones shown in Supplemental Figure S3B could have solved this problem in theory but only on the condition that the half-life of the first intermediate step in a particular group of genes (i.e., more RNAPII on leading copies of “same” genes and more RNAPII on lagging copies of opposite genes) lasted longer than the time required for the cell to process the excess thymidine added in the chase in order to stop EdU incorporation. In practice, however, the thymidine chase stops EdU incorporation 5–10 min after its addition, as demonstrated in Vasseur et al. (2016). Since the first step of the RNAPII (re)binding process to replicated genes is very short-lived (probably <2 min), there is a 5- to 10-min window after the EdU pulse and during the thymidine chase in which RNAPII will have switched to the second configuration in cells in which a particular gene has replicated during the EdU pulse but will still be in the first intermediate step on that same gene in cells that are replicating that locus 5–10 min after the pulse (and still incorporating EdU). Since the rates of thymidine uptake and processing are also stochastic at the single-cell levels (different cells process thymidine at slightly different rates), the combination of the short half-life of the first RNAPII configuration and the cell-to-cell variability in the length of time that it takes to stop EdU incorporation after thymidine addition makes it technically impossible to get the same groups of genes with the same configurations during a pulse chase time course (Supplemental Fig. S3B). Thus, both biological replicates in Supplemental Figure S3B show the same pattern: a switch of RNAPII from one strand to the other during chromatin maturation, with “same” genes mostly switching from the leading to the lagging copy and “opposite” genes mostly switching from the lagging to the leading copy. This switching pattern is, however, seen on different genes in each replicate, just as we have seen in the experiments with synchronized cell cultures described above.
Since we repeatedly found that same pattern on different groups of genes in different early S-phase replicates and in different replicates of the EdU pulse chase experiment, we conclude that all genes undergo the same steps in the process of RNAPII rebinding to replicated gene copies: a first short-lived intermediate step with more RNAPII bound to the leading or the lagging gene copy of same or opposite genes, respectively, followed by the second final step with a “switch” of RNAPII enrichment to the other gene copy—lagging or leading for same or opposite genes, respectively.
The asymmetric distribution of RNAPII on daughter chromatids is independent of the asymmetric distribution of new histones
Does RNAPII binding to replicated gene copies correlate with and is it dependent on nucleosome binding? This section describes the dynamics of new nucleosome binding, and the distribution pattern of old nucleosomes is discussed in the next section. Acetylation of Lysine 56 on Histone H3 with the Rtt109 histone acetyl-transferase marks newly synthesized histones in yeast (Masumoto et al. 2005; Kaplan et al. 2008). It is consequently enriched at promoters with high H3 turnover rates and on newly replicated DNA (Kaplan et al. 2008). In order to assess whether and how new nucleosome assembly influences RNAPII binding to replicated genes, we checked whether H3K56ac distribution on daughter chromatids correlates with the asymmetric distribution of RNAPII described above.
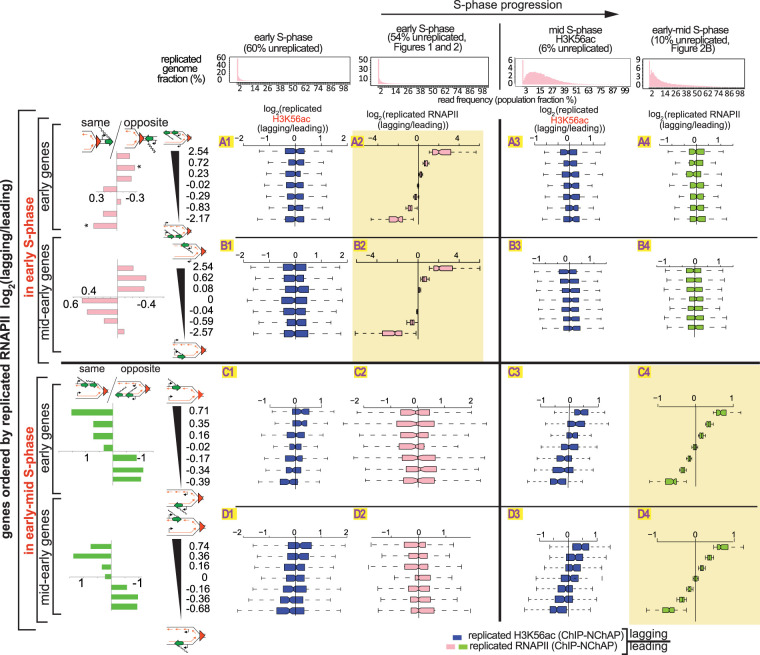
H3K56ac ChIP-NChAP for a synchronized cell population in mid S phase shows that the distribution of “new” histones indeed correlates with the RNAPII distribution from early-mid S phase (Fig. 3, cf. fields C3 to C4 and D3 to D4): RNAPII is enriched on the gene copy that also contains more new acetylated histones. Conversely, H3K56ac and RNAPII lagging/leading ratios do not correlate in early S phase (Fig. 3, cf. A1 to A2, B1 to B2, C1 to C2, and D1 to D2) and even appear to be anticorrelated at some genes (cf. D1 to D2 in Fig. 3). The lack of correlation between the distribution patterns of new histones and RNAPII in early S phase was also confirmed in a replicate experiment where H3K56ac and RNAPII ChIP-NChAP were performed in parallel from the same cell culture in each replicate (cf. RNAPII [left] and H3K56ac [right] lagging/leading ratios in B2, C2, D3, and D4 in Supplemental Fig. S4B). H3K56ac is nevertheless distributed asymmetrically in early S phase when the genome is 45% unreplicated, with an approximately threefold bias for the leading strand in 28% of genes (bottom bins 1–2, C2, Supplemental Fig. S4B) or for the lagging strand in another 28% (top bins 6–7, C2, Supplemental Fig. S4B). Moreover, these gene bins, sorted by lagging/leading H3K56ac ratios, have a similar genic orientation pattern as when genes are sorted by RNAPII lagging/leading ratios in early S phase (cf. rows B and C in Supplemental Fig. S4B).
Figure 3.
The asymmetric distribution of RNAPII on daughter chromatids is independent of the asymmetric distribution of new histones. Box plot distributions of lagging/leading ratios for replicated H3K56ac (dark blue) and replicated RNAPII (pink and green for early and early-mid S phase, respectively) in early (columns 1 and 2, respectively) and mid and early-mid (columns 3 and 4, respectively) S phase for early (rows A and C) and mid-early genes (rows B and D). (Header) Distribution of genome read densities, columns: (1) early S phase (MNase-NChAP, 60% unreplicated); (2) early S phase from Figure 2 (54% unreplicated); (3) bulk H3K56ac ChIP, mid S phase (6% unreplicated); (4) early-mid S phase (NChAP) from Figure 2B (10% unreplicated). (Rows A and B) Early and mid-early genes have been sorted by increasing lagging/leading RNAPII ratios from early S phase (Fig. 2B), respectively, and then divided into seven bins as in Figure 2B (y-axis), and box plot distributions of H3K56ac lagging/leading ratios (x-axis) from early (left) and mid (right) S phase have been determined (dark blue boxes) and compared with RNAPII lagging/leading ratios from early and early-mid S phase (pink and green boxes, respectively). The bar graphs on the left show the “same” gene enrichment calculated as in Figure 2B for gene bins indicated in the y-axis of each row on the right. (Rows C and D) As rows A and B but sorted by increasing lagging/leading RNAPII occupancy from early-mid S phase (Fig. 2B).
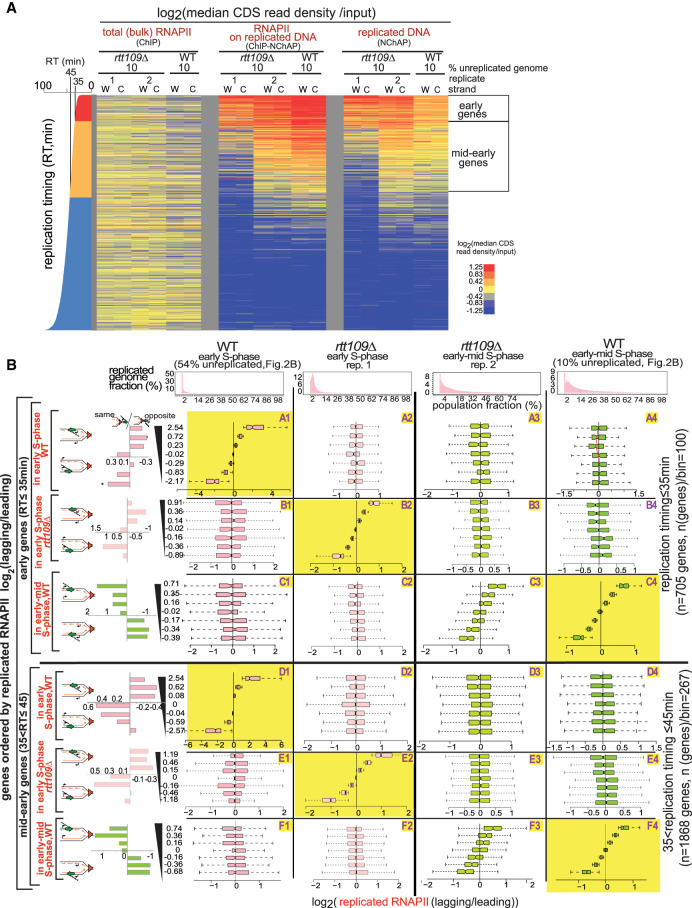
Taken together, the lack of correlation between RNAPII and H3K56ac binding patterns in early S phase and a similar dependence on genic orientation for either feature suggest that new nucleosomes and RNAPII follow the same order of binding to daughter chromatids but that the two processes are independent of each other. The coincidence of H3K56ac and RNAPII enrichments later in S phase is consequently due to the convergence of these two independent pathways. RNAPII follows the same asymmetric distribution pattern that correlates with genic orientation and that appears to switch from one gene copy to the other even in the absence of H3K56ac in rtt109Δ cells (Fig. 4B). Genic orientation trends for genes with asymmetric RNAPII distribution in early S phase are similar in WT (54% unreplicated) (Fig. 2) and mutant cells (replicate 1, Fig. 4A; cf. the same/opposite graphs in rows A, B, D, and E in Fig. 4B). Later in S phase, RNAPII distribution patterns in the mutant (replicate 2, Fig. 4A) correlate well with WT (10% unreplicated replicate 1) (Figs. 2A, 4A) distribution patterns and are indistinguishable from WT replicates at a comparable stage of genome replication (cf. C3 to C4 and F3 to F4 in Fig. 4B; cf. A1 and A3 to A2 or B1 and B3 to B2 in Supplemental Fig. S3C). Additionally, a duplicate RNA-seq experiment using spike-in normalization with total mRNA from Schizosaccharomyces pombe, shows an ∼30% genome-wide reduction of mRNA levels in rtt109Δ mutants compared with WT cells. Consequently, since H3K56ac stimulates transcription globally and the pattern of RNAPII binding to replicated gene copies is independent of H3K56ac, we conclude that H3K56ac is unlikely to directly suppress transcription of newly replicated genes as recently proposed (Supplemental Fig. S5; Voichek et al. 2016).
Figure 4.
RNAPII is distributed asymmetrically in the absence of H3K56ac in rtt109Δ cells. (A) Heat map of median RNAPII occupancies in coding regions of yeast genes not regulated by the cell cycle, as in Figure 2A. (Columns, from left) Early-mid S phase after release from G1 arrest in rtt109Δ (early S phase [rep.1] and early-mid S phase [rep. 2]) and WT (10% unreplicated, from Fig. 2B), from total RNAPII (ChIP), replicated RNAPII (ChIP-NChAP), and replicated DNA (NChAP) fractions. Even though both rtt109Δ time points have 10% of their genome still unreplicated, replicate1 is at an earlier stage of genome replication than replicate 2 because a greater number of genes have been replicated in more cells in replicate 2 than in replicate 1 (mid-early genes are more “red” and “yellow” in the NChAP fraction of replicate 2 compared with replicate 1; also compare the distributions of percentage replicated genome fractions between replicate 1 and 2 in B). (B) Box plot distributions of replicated RNAPII lagging/leading ratios from early S phase (WT) to early-mid S phase (WT and rtt109Δ, columns 1–4) for early (rows A–C) and mid-early genes (rows D–F). (Header) Distribution of genome read densities as in Figure 2B. (Rows A and D) Genes have been sorted by increasing lagging/leading RNAPII occupancy in early S phase (WT, Fig. 2B) and divided into seven bins, as in Figure 2B (y-axis), and box plot distributions of replicated RNAPII lagging/leading ratios (x-axis) have been determined for each bin at indicated time points. (Rows B, E and C, F) As rows A and C except that genes have been ordered by increasing lagging/leading RNAPII ratios from rtt109Δ (early S phase, replicate 1) or early-mid S phase (WT) (Fig. 2B), respectively. The bar graphs on the left show “same” gene enrichments calculated as in Figure 2B for gene bins indicated in the y-axis of each row on the right. The RNAPII distribution pattern between leading and lagging strand gene copies in rtt109Δ cells from replicate 2 (fields C3 and F3) correlates with WT (fields C4 and F4), indicating that the asymmetric distribution of RNAPII on replicated DNA is independent of the distribution of H3K56ac.
Differences in the timing of leading and lagging strand replication direct the distribution of “old” and “new” histones, and RNAPII on replicated gene copies
Our next question was whether the distribution pattern of old histones influences RNAPII binding to replicated genes. In order to establish a comprehensive timeline of nucleosome assembly on replicated DNA and explore how the reassembly of old nucleosomes influences RNAPII distribution on replicated DNA, we performed parallel ChIP-NChAP experiments for H3, H3K4me3 and H3K36me3 (these methylated histones were used as “proxies” for old histones, as in Gan et al. 2018; Yu et al. 2018), H3K56ac, and RNAPII in two biological replicates at 20 and 25 min after release from G1 arrest (Fig. 5; Supplemental Fig. S6 for replicate 1; Supplemental Fig. S7 for replicate 2). H3K4me3 and H3K36me3 are marks of “active” transcription that are deposited cotranscriptionally at the beginning and the middle and end of genes, respectively. A direct comparison of their inheritance pattern with the binding pattern of RNAPII will therefore help us answer whether these “active” marks influence RNAPII binding on replicated gene copies and consequently whether they are instructive of transcription, as epigenetic marks, in the strict sense, are expected to be.
Figure 5.
Old nucleosomes are recycled to the daughter chromatid that replicated first. (A) Box plot distributions of lagging/leading ChIP-NChAP ratios for H3, H3K36me3, H3K4me3, H3K56ac, and RNAPII from early (column 1) and mid-early S phase (column 2) for early genes from the same WT culture (biological replicate 1). The histograms in the header show the distribution of genome read densities for each NChAP (replicated DNA) fraction at indicated time points in S phase, as in Figure 2B. (Rows A and B) Genes have been sorted by decreasing lagging/leading ratios of H3K4me3 in early S phase (row A) and mid-early S phase (row B) (yellow background), and divided into seven bins (y-axis on the left), as in Figure 2B. Box plot distributions of lagging/leading ratios (x-axis) for the chromatin features indicated in the headers have been determined for each bin. Average lagging/leading H3K4me3 ratios in early S phase for each bin are shown on the y-axis on the left. The bar graphs on the left show the “same” gene enrichment for gene bins indicated in the y-axis of each row on the right. Column 3 shows box plot distributions for the difference in replication timing (ΔRT) between the lagging and the leading strand for each gene (RT for each gene is the median RT of all 50-bp segments in the CDS of any given gene, averaged from two replicate time courses in Supplemental Fig. S10) (left) and ΔDNA synthesis rate (average difference between lagging and leading DNA synthesis rates for each gene in the bin) (right). Synthesis rates were calculated as in Supplemental Figure S11 using replication timing from Supplemental Figure S10 and ROADs (replication origin-associated domains) determined from NChAP fractions of the 20- and 25-min time points of two biological replicates (replicate 1 from Fig. 5A and replicate 2 from Supplemental Fig. S7A). (B) Average TSS (transcription start site)-centered metagene profiles of ChIP-NChAP fractions indicated in the header, from WT cells (replicate 1) from gene bins from A sorted according to the average log2 (lagging/leading) ratios for H3K4me3 in early S phase. Only the two bottom (bins 1–2) and top (bins 6–7) bins are shown. The value of the average ratio for each bin is indicated in the blue strip on the left. (C) Distribution of the differences in replication timing between sister gene copies (ΔRT, lagging-leading and leading-lagging) in WT cells for all early and mid-early genes. The yellow surface shows 95% of early and mid-early genes whose ΔRT is between 0.5 and 6.5 min. “avgΔRT = 2.4 min” is the average of the 95% of the gene population represented under the yellow surface (n = 0.95 × 2548 = 2421). (D) As in C, for mid-late and late genes with RT ≥ 55 min. (E) Cumulative distribution of differences in replication timing between the lagging and the leading sister gene copy (ΔRT) for the top bin 7 (orange) and bottom bin 1 (blue) from field A3 (ordered by H3K4me3 log2[lagging/leading] in early S phase, left panel) and field B3 (ordered by H3K4me3 log2[lagging/leading] in mid-early S phase, right panel).
Our experiments show that H3K4me3 and H3K36me3 distributions follow a similar pattern to the one seen for H3K56ac in early S phase (Fig. 5; Supplemental Figs. S6, S7). Twenty minutes after release from arrest in early S phase of replicate 1, when the genome is 65% unreplicated, H3K4me3 has an approximately threefold bias for either the leading or the lagging copy in 56% of early genes: 28% has a lagging strand bias (bins 6–7, A1, Fig. 5A) and 28% has a leading bias (bins 1–2, A1, Fig. 5A). This bias reproducibly correlates with the binding patterns of H3, H3K36me3, H3K56ac, and RNAPII (see A1 in Fig. 5A; Supplemental Figs. S6A, S7A).
Also, as observed earlier for H3K56ac and RNAPII, “same” genes tend to have more H3/H3K36me3/H3K4me3/H3K56ac/RNAPII on the leading copy and “opposite” genes tend to have more of these chromatin features on the lagging copy in early S phase in both biological replicates (row A in Fig. 5A; rows A, C, and E in Supplemental Figs. S6A, S7A). We also observed a switch in the genic orientation bias for H3K4me3 in mid-early S phase of the first biological replicate (25-min time point, row B, Fig. 5A; Supplemental Fig. S6A), as we did previously for H3K56ac and RNAPII. Thus, H3K4me3 enrichment shifts from the leading to the lagging gene copy at “same” genes or from the lagging to the leading copy at “opposite” genes. This switch can be directly observed in Figure 5A, where the H3K4me3 lagging/leading ratios from mid-early and from early S phase are anticorrelated (cf. H3K4me3 distributions in A1 with A2, and B1 with B2, Fig. 5A).
The 5′ enrichment of H3K4me3 and 3′ enrichment of H3K36me3 that is characteristic of coding regions of actively transcribed genes is already apparent in the earliest time point (Fig. 5B; Supplemental Fig. S6B). This is consistent with an earlier finding that old histones are recycled close to the site to which they were bound before replication (Radman-Livaja et al. 2011).
The polarity shift for H3K4me3 was, however, not observed in the second biological replicate (Supplemental Fig. S7A,B) because the 25-min time point in the second replicate represents an earlier stage of S phase, when 39% of the genome is still not replicated compared with the replicate in Figure 5 and Supplemental Figure S6A, where only 25% of the genome has not yet replicated. Consequently, the 20- and 25-min points of the second replicate have the same H3/ H3K36me3/ H3K4me3/ H3K56ac/ RNAPII distribution bias for early and mid-early genes (cf. columns 1 and 2 in Supplemental Fig. S7, A and B, respectively).
At 25 min after release from arrest, chromatin maturation has progressed further in the first replicate than in the second replicate. At this more advanced stage of chromatin maturation, the correlation in occupancy bias between all measured chromatin features that was observed in early S phase “disappears” in the later time point. All chromatin features in mid-early S phase appear to be symmetrical at the cell population level except for the feature used to sort genes into bins of increasing lagging/leading ratios (fields with the yellow background in Fig. 5A; Supplemental Fig. S6A). We thus detect equal numbers of genes with significantly asymmetrical partitioning of each measured chromatin feature, but the asymmetrical distribution of any one of these features is not found on the same genes as the asymmetrical distribution of any other feature. As replication and chromatin maturation gradually become less synchronized at the cell population level, different chromatin features will have matured at different times in different cells by mid-early S phase. In some cells, histones and/or RNAPII will be on the first step of chromatin maturation and, depending on genic orientation, preferentially occupy the leading or the lagging strand. Meanwhile, in other cells, the shift in new histone and/or RNAPII enrichment to the sister strand will have already taken place. RNAPII and histones thus bind to chromatin with different kinetics and independently of each other even as they follow the same steps in the chromatin maturation process. Consequently, when we look at the cell population level, we detect apparently symmetrical histone distributions on genes that have asymmetrical RNAPII distributions, and vice versa, we see symmetrical RNAPII distributions on genes that have asymmetrical histone distributions.
What determines how old histones will be distributed on replicated gene copies? The “choice” of the leading or the lagging gene copy in gene bins with a significant bias for one or the other strand, respectively, correlates strongly with genic orientation; that is, with the direction of transcription relative to the direction of the replication fork. We therefore assumed that the recycling of old histones will be influenced by local replication dynamics of the leading and lagging strand, which are in turn probably influenced by the interactions of the replication fork with the transcription machinery. We therefore measured replication timing (RT) of leading and lagging copies of all genes and calculated the difference in RT (ΔRT) between lagging and leading copies for all gene copy pairs, as described in Supplemental Figure S10 (see also Supplemental Table S4 that lists ΔRT values for all yeast genes; “Replication Timing” in the “Data Analysis” section of Supplemental Material).
Our replication timing measurements reveal unexpected differences between sister gene copies that explain the observed pattern of nucleosome binding to newly replicated DNA (column 3 in Fig. 5A; Supplemental Fig. S6A for replicate 1; Supplemental Fig. S7A,B for replicate 2). As shown in Figure 5, C and D, one sister copy (lagging or leading) replicates 0.5–6.5 min before the other at ∼95% of yeast genes, with an average difference in replication timing of 2.35 min. Moreover, the lagging copy or the leading copy replicate first in an equal number of genes (Supplemental Fig. S7C).
The order of replication of sister copies correlates with the observed nucleosome and RNAPII binding bias. As shown in Figure 5E, the leading strand has replicated 0.5–10 min before the lagging strand (ΔRT [lagging-leading] > 0) at 58% of genes that had, on average, 5.5 times more H3K4me3 on the leading strand in early S phase. At the same time, the lagging strand has replicated 0.5–10 min before the leading strand (ΔRT [lagging-leading] < 0) at 65% of genes that had, on average, five times more H3K4me3 on the lagging strand in early S phase. The trend is reversed later in mid-early S phase: The lagging strand has replicated before the leading strand at 59% of genes that had, on average, three times more H3K4me3 on the leading strand in mid-early S phase, while the leading strand has replicated before the lagging strand at 54% of genes that had, on average, 3.4 times more H3K4me3 on the lagging strand in mid-early S phase. This is consistent with a shift in H3K4me3 enrichment from one sister copy to the other between early and mid-early S phase (see Fig. 5A,B).
It follows from the above analysis that old and new nucleosomes and RNAPII simply bind first to the gene copy that replicated before its sister. If the leading gene copy has an earlier (lower) replication time than the lagging copy of the same gene, old and new nucleosomes and RNAPII will bind to that copy first. Conversely, if the lagging copy replicates before the leading copy, the lagging copy will be “chromatinized” first. The bias in total H3 distribution for the strand that replicated first supports our hypothesis that the strand that replicated later is transiently “underchromatinized” shortly after the passage of the replication fork. If the difference in replication timing is insignificant, nucleosomes and RNAPII will randomly go to one or the other or to both copies, resulting in an average log2(lagging/leading) ratio of 0. Incidentally, total RNAPII density in the gene body does not have an effect on the difference in replication timing between the lagging and the leading gene copy as there is no significant difference in RNAPII density between gene bins with high ΔRT and the ones with low ΔRT (Supplemental Fig. S7C).
Are differences in replication timing between lagging and leading gene copies due to differences in DNA synthesis rates on either copy? The replication timing of genes that are replicated by the same replication fork (i.e., on the same ROAD [replication origin-associated domain], determined as described in the “Data Analysis” section of Supplemental Material; Supplemental Fig. S11A) gradually increases as genes get further away from the origin. We can consequently use the differences in replication timing between genes on the same ROAD to calculate average lagging and leading DNA synthesis rates on each replicated gene. Since DNA synthesis rates on any given gene are directly dependent on the average fork velocity at that locus, we can also use DNA synthesis rates as a measure of the average fork speed through that gene. If we plot leading and lagging strand replication timing versus the chromosomal coordinates of genes within the same ROAD upstream of and downstream from each origin, we can estimate replication fork velocity from the slope of the linear fit for every replicated gene copy (Supplemental Fig. S11B). It is then straightforward to calculate the differences in DNA synthesis rates between lagging and leading gene copies of all replicated genes (Fig. 6C,D; Supplemental Table S5).
Figure 6.
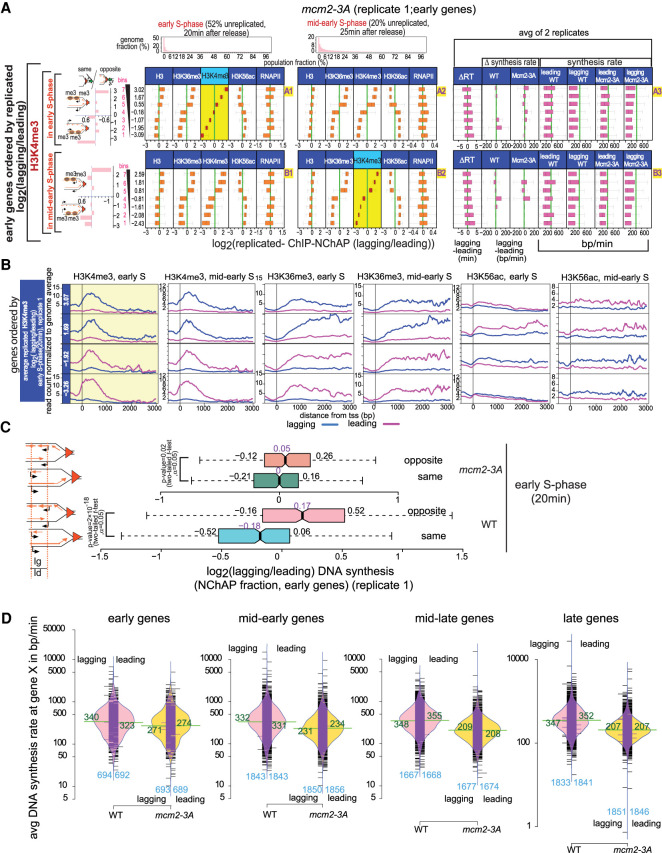
Chromatin maturation in mcm2-3A cells. (A) Box plot distributions of lagging/leading ChIP-NChAP ratios for H3, H3K36me3, H3K4me3, H3K56ac, and RNAPII from early (column 1) and mid-early S phase (column 2) for early genes measured in the same culture of mcm2-3A mutant cells (biological replicate 1). The histograms on top show the distribution of genome read densities for each NChAP (replicated DNA) fraction at indicated time points in S phase. (Rows A and B) Genes have been sorted by decreasing lagging/leading occupancy of H3K4me3 in early S phase (row A) and mid-early S phase (row B), respectively (yellow background), and divided into seven bins (y-axis on the left). Box plot distribution of lagging/leading ratios (x-axis) for the chromatin features indicated in the header have been determined for each bin, as in Figure 5A. The bar graphs on the left show the “same” gene enrichment for gene bins indicated in the y-axis of each row on the right. Column 3 shows box plot distributions for, from left to right, the difference in replication timing (ΔRT) between the lagging and the leading strand for each gene in mcm2-3A cells; ΔDNA synthesis rates (lagging-leading) in WT and mcm2-3A cells for each gene in the bin; and average leading and lagging DNA synthesis rates in WT and mcm2-3A cells used to obtain the ΔDNA synthesis rates in Figure 5A for WT and in this figure for mutant cells. Synthesis rates for mcm2-3A were calculated as in Supplemental Figure S11 using replication timing from Supplemental Figure S10 and ROADs determined from NChAP fractions of the 20- and 25-min time points of two biological replicates (replicate 1 from Fig. 6A; replicate 2 from Supplemental Fig. S9A). (B) Average TSS-centered metagene profiles of ChIP-NChAP fractions indicated in the header from mcm2-3A cells (replicate 1) from gene bins from A sorted according to the average log2(lagging/leading) ratios for H3K4me3 in early S phase. Only the two bottom (bins 1,2) and top (bins 6,7) bins are shown. The value of the average ratio for each bin is indicated in the blue strip on the left. (C) Box plot distribution of DNA synthesis bias (log2[lagging/leading] of the NChAP fraction) for early replicating “opposite” and “same” genes in early S phase (20 min after release from G1 arrest) for WT replicate 1 (Fig. 5A) and mcm2-3A replicate 1 (Fig. 6A). (D) Bean plot of lagging and leading strand synthesis rate distribution (from Supplemental Figs. S10, S11) at early, mid-early, mid-late, and late genes for WT and mcm2-3A strains. The mean for each distribution is shown in green. The number of genes in each distribution is shown in blue on the bottom of the plot. The black bars represent individual data points. The bulk of synthesis rates in mcm2-3A are lower than in WT. Rates are constant in WT for all genes, but they gradually decrease as a function of replication timing in mcm2-3A mutants.
Even though lagging DNA synthesis rates of early genes are, on average, somewhat slower than the leading DNA synthesis rates in each gene bin (cf. box plot distribution for leading and lagging synthesis rates in column 3 in Supplemental Fig. S7A,B), the differences between leading and lagging synthesis rates on any individual gene are small and do not appear to influence where chromatin will be assembled first (see the “ΔDNA synthesis rate” panel in column 3, Fig. 5A; Supplemental Figs. S6A, S7A). This means that sister copies start replicating at different times, but once replication has started, it proceeds at similar rates for either sister chromatid.
Genic orientation is a better predictor of which copy is more likely to replicate first and thus direct the order of chromatin assembly and RNAPII binding after replication. When transcription and replication go in “opposite” directions, lagging strand replication tends to be favored. Conversely, the leading gene copy replicates before the lagging copy for genes where transcription travels in the “same” direction as the replication fork (bar graphs on the left in Fig. 5A; Supplemental Figs. S6A, S7A).
Globally slower replication forks in mcm2-3A mutants bring into focus the genome-wide asymmetry of old histone recycling
Recent studies have hypothesized that the Mcm2 subunit of the replication fork helicase Mcm2-7 acts as a histone chaperone that is responsible for the transfer of old histones specifically to the lagging strand. This hypothesis was based on observations that alanine substitutions of tyrosines 79, 82, and 91 in the mcm2-3A mutant impair the interaction of Mcm2 with histones ahead of the replication fork (Foltman et al. 2013) and cause an increase in asymmetric recycling of old histones with an apparent bias for the leading daughter chromatid (Gan et al. 2018; Petryk et al. 2018).
Since we found that the binding preference of RNAPII and nucleosomes for the leading or lagging strands depends on their respective order of replication, we wanted to further explore the mechanism that regulates the replication timing of each strand. Considering Mcm2's possible involvement in histone recycling and the likelihood that a mutant of Mcm2 would affect replication fork velocity, we decided that the mcm2-3A mutant would be a good candidate to probe the relationship between replication timing of each strand and the binding patterns of RNAPII and histones to replicated genes.
Our results reveal that the general trends for the distribution of new and old histones and RNAPII are qualitatively similar in WT and mcm2-3A cells. The difference lies mostly in the apparent magnitude of asymmetrical nucleosome recycling. More than 80% of genes in the cell population appear to have, on average, more than a threefold difference in histone distributions between the lagging and leading sister copies in the mcm2-3A mutant, whereas in WT cells, ∼50% of genes exhibit that same level of asymmetry (cf. A1 in Fig. 5A with A1 in Fig. 6A). The number of genes with a leading strand bias is nevertheless comparable with the number of genes with a lagging strand bias in the mutant and WT cells alike (Fig. 6A,B; Supplemental Figs. S8A,B, S9A). This is not consistent with the hypothesis that Mcm2 recycles old histones specifically to the lagging strand as suggested recently (Gan et al. 2018; Petryk et al. 2018). We observed instead that the dynamics of histone and RNAPII distribution on replicated daughter chromatids follow a similar pattern but with different kinetics in mcm2-3A and WT cells. The positive correlation in the distribution of H3K4me3, H3K36me3, and H3 persists for a longer period in the mutants compared with WT and is still apparent even when 80% of the genome has been replicated in the whole population (A2–B2, Fig. 6A; Supplemental Fig. S8A). Unlike in WT cells, H3K56ac is already anticorrelated with the H3 methyl marks in the earliest time point in the mcm2-3A mutant (A1–B1, Fig. 6A; Supplemental Fig. S8A for replicate 1; A1–D1, Supplemental Fig. S9A for replicate 2). We also did not observe the shift in H3K4me3 enrichment from one strand to the other that we detected in WT cells (cf. A1 with A2 and B1 with B2 in Fig. 5B [WT] and 6B [mcm2-3A]).
The genic orientation pattern of RNAPII distribution in the later time point is the same as in WT: There is more RNAPII on the lagging strand at “same” orientation genes and more RNAPII on the leading strand at “opposite” genes (row D, Supplemental Fig. S8A). The genic orientation pattern in the earlier time point is not as clear-cut as for WT cells (row C, Supplemental Fig. S8A). It appears that, according to our chromatin maturation model, the bottom bin in row C is populated with a mixture of “same” genes where RNAPII binds to the leading strand first (step 1) and “opposite” genes where RNAPII binds after switching from the lagging strand (step 2). The idea that we are “catching” genes with asymmetrical RNAPII distribution that are either in step 1 or step 2 of the RNAPII binding process is supported by the distribution pattern of ΔRT (C3, Supplemental Fig. S8A), which shows no clear bias for either strand.
Unlike in WT cells, RNAPII binding in the mutant appears to be out of sync with nucleosome binding even in the earlier time point of replicate 1 (cf. the RNAPII distributions with H3K4me3/H3K36me3/H3K56ac in C1,2 and D1,2, Supplemental Fig. S8A), because genes with the most significant asymmetry in RNAPII occupancy show no asymmetry for new or old nucleosomes. Indeed, the correlation with histone binding patterns is weak and heterogeneous: On some genes, the RNAPII binding bias correlates better with H3K4me3/H3K36me3/H3, and on others, it is better correlated with H3K56ac (cf. A1 and B1 in Fig. 6A with A1 and B1 in Supplemental Fig. S8A). RNAPII binding on genes with the largest bias for one or the other daughter chromatid does not correlate at all with the binding pattern of old or new histones in replicate 1 (rows C and D, Supplemental Fig. S8A) and has a weak correlation with H3K56ac and a weak anticorrelation with H3K4me3 and H3K36me3 in replicate 2 (rows E and F in Supplemental Fig. S9A). This is further evidence that RNAPII binding to newly replicated DNA is independent of H3K4me3, H3K36me3, or H3K56ac enrichment and that any correlation or anticorrelation between the RNAPII and histone binding patterns is coincidental.
As with WT cells, the determining factor in the “choice” of daughter chromatid to which old nucleosomes are preferentially recycled in mcm2-3A mutants is replication timing. Like in the WT, sister gene copies do not replicate simultaneously in mcm2-3A mutants (Supplemental Fig. S9B). One sister copy (lagging or leading in equal proportion) replicates 0.5–8.5 min before the other at ∼95% of early and mid-early genes, with an average difference in replication timing of 3.5 min. Consistent with the chromatin maturation timeline that we proposed for WT cells, old nucleosomes bind first to the strand that replicated earlier and new nucleosomes then bind to the sister strand that replicated later. Genes in the mutant whose transcription goes in the same direction as the replication fork tend to replicate the leading strand first and “opposite” genes tend to replicate the lagging strand first, as in WT cells (Supplemental Fig. S12D). This is supported by the lagging/leading strand ratios of replicated DNA (NChAP fraction) for “same” and “opposite” genes in early S phase (Fig. 6C): There are more replicated lagging gene copies than their leading sisters on opposite genes and leading copies are, on average, more replicated than their lagging counterparts on same genes. The trend is similar in WT and mutant cells.
Why did we observe more genes with an asymmetrical nucleosome distribution in the mutant than in the WT? Our replication timing measurements reveal that mid-late and late genes replicate later in the mutant than in the WT (Supplemental Fig. S10D,E). This is probably linked to the gradual slowing down of DNA synthesis rates in the mutant (Fig. 6D; Supplemental Table S5), since replication origin usage in the mutant is indistinguishable from WT (Supplemental Fig. S8C,D). Indeed, DNA synthesis rates in WT cells are similar for early genes and late genes (∼340 bp/min, on average), while they are markedly slower for late genes relative to early genes in the mutant (∼270 bp/min for early genes compared with ∼200 bp/min for late genes). DNA synthesis rates are between 200 and 800 bp/min for most genes in WT and between 100 and 500 bp/min for most genes in mcm2-3A cells. Our fork velocity estimates in WT cells are lower than the previously estimated velocity of 1–2 kb/min (Sekedat et al. 2010; Yang et al. 2010), but they do match recent measurements from in vitro replication experiments done with single chromatinized substrates (Gruszka et al. 2020). Our calculations are based on synthesis rates specifically in gene bodies, while those older estimates are averages based on genome-wide replication rates. This may explain the discrepancy, as replication forks are likely to be slowed down by the transcription machinery when they advance through genes.
Origins are stochastically activated at different times in each cell, which means that replication forks will be in different positions from cell to cell at any given point in S phase. NChAP will consequently produce an ensemble average of all fork positions in the population at that point in S phase. We suspect that histone distributions still appear asymmetrical until late in S phase on most genes in mcm2-3A mutants because replication forks are globally slower in the mutant. Fork positions in different cells that started from the same origin, which fired at different times in different cells, will be more spread out if forks move faster, as we see in WT cells. At lower fork speeds, on the other hand, fork positions from different cells will be closer together and therefore appear better synchronized, as is the case in mcm2-3A mutants. Consequently, genes that are at a comparable distance from the fork in different mcm2-3A cells will be approximately at the same stage of their chromatin maturation process. On the other hand, genes in WT cells will be located at more variable distances from the fork in different cells and will therefore be captured at different stages of chromatin maturation. The observed histone distribution pattern captured in WT cells will come from a mixture of step 1 (when nucleosomes and RNAPII are enriched on the strand that replicated first) and step 2 (when new nucleosomes and RNAPII are enriched on the strand that replicated second), as is observed for approximately half of the genes in WT cell populations.
In the mutant, however, most genes are already at the second step in early S phase, with H3K4me3 and H3K36me3 enriched on the strand that replicated first and H3K56ac enriched on the strand that replicated second (cf. rows A and B in Fig. 6A with rows A and B in Supplemental Fig. S8A and rows A and B with rows C and D in Supplemental Fig. S9A). We suspect that the slowing down of replication forks in the mcm2-3A mutant effectively shortens the half-life of the transient first step of the chromatin maturation process and favors the accumulation of chromatin configurations from step 2. This is supported by the fact that ΔRT distributions for genes ordered by lagging/leading ratios of H3K56ac enrichment are inversely proportional to ΔRT distributions of genes that are ordered by lagging/leading ratios of H3K4me3 enrichment in early S phase (cf. ΔRT in A3 with ΔRT in C3 in Supplemental Fig. S9A). The leading strand has replicated 0.5–10 min before the lagging strand (ΔRT [lagging-leading] > 0) at 59% of genes that had, on average, 2.3 times more H3K4me3 on the leading strand in early S phase. At the same time, the lagging strand has replicated 0.5–10 min before the leading strand (ΔRT[lagging-leading] < 0) at 56% of genes that had, on average, 2.5 times more H3K4me3 on the lagging strand in early S phase. The trend is reversed if we consider genes ordered by H3K56ac lagging/leading ratios: The lagging strand has replicated before the leading strand at 51% of genes that had, on average, three times more H3K56ac on the leading strand in early S phase, whereas the leading strand has replicated before the lagging strand at 66% of genes that had, on average, 3.7 times more H3K56ac on the lagging strand in early S phase (Supplemental Fig. S9C).
A model of nucleosome assembly and RNAPII binding to daughter genomes based on the order of replication of sister chromatids
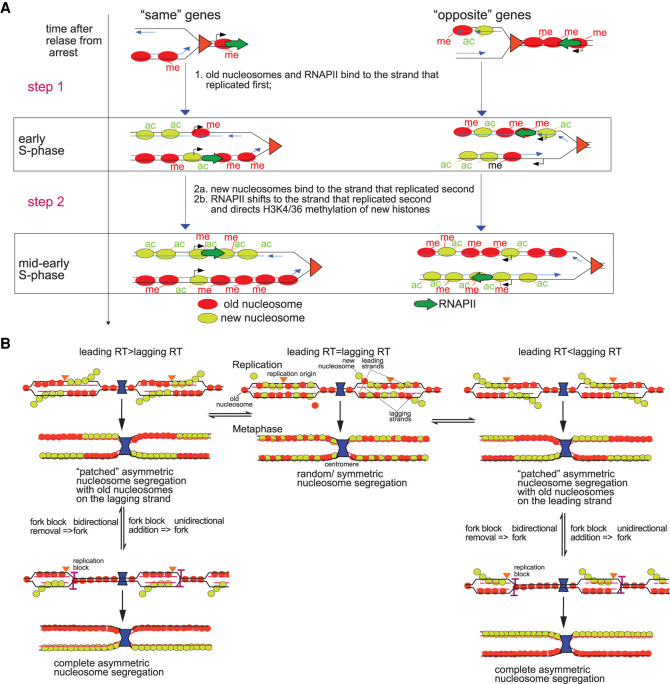
We propose the following model for chromatin assembly on sister chromatids (Fig. 7A). Chromatin maturation consists of three parallel independent processes: (1) old histone recycling; (2) new histone assembly; and (3) RNAPII binding to replicated gene copies. These processes follow similar steps, but each step proceeds at different rates in each process. Old nucleosomes with H3K4me3 and H3K36me3 are first recycled very close behind the fork, which explains why the typical pattern of H3K4me3 and H3K36me3 distribution on coding regions is largely preserved on newly replicated genes (Figs. 5B, 6B; Supplemental Figs. S6B, S8B). Old nucleosomes preferentially bind to the daughter chromatid that is replicated first: leading for “same” genes or lagging for “opposite” genes. If both chromatids are replicated almost simultaneously, old nucleosomes and new nucleosomes will bind to one or the other chromatid randomly and in equal proportions. On the other hand, when the difference in replication timing between the two chromatids is significant, new histones and old histones will compete for binding to the only chromatid that has already completed its synthesis. Since the concentration of old nucleosomes at the fork is initially higher than the concentration of new nucleosomes, the daughter chromatid that replicated first will mostly be populated with recycled old nucleosomes (step 1). We propose that RNAPII will at this point also be recycled from ahead of the fork and will also end up on the chromatid that replicated first or randomly on one or the other if they replicated nearly simultaneously. New nucleosomes will bind to the sister chromatid that replicated later because the majority of old nucleosomes have already been recycled to the chromatid that replicated earlier (step 2). RNAPII will then shift to the second daughter chromatid and direct Set1p and Set2p to, respectively, methylate new H3 histones on K4 and K36. Note that H3K4/K36 methylation of new histones on the strand that replicated second can result in a shift in H3K4me3 or H3K36me3 enrichment from the strand that replicated first to the strand that replicated second if the second strand has a higher nucleosome density than the first strand, respectively, at the 5′ end or in the middle and 3′ end of its coding sequence.
Figure 7.
Differences in replication timing between lagging and leading gene copies shape the distribution of old and new nucleosomes along sister chromatids. (A) Model for chromatin assembly on daughter chromatids. Nucleosome deposition follows a two-step process. (Step 1) “Old” nucleosomes (red) and RNAPII (green arrow) bind first to the leading strand behind the fork while the lagging strand is still replicating when transcription and replication travel in the same direction. When transcription and replication travel in opposite directions, old nucleosomes and RNAPII are deposited on the lagging strand that replicated first. New nucleosomes (light green) will be incorporated into the strand that replicated first mostly at promoters and ends of genes through replication independent turnover, although some will outcompete old nucleosomes for binding to other sites in the CDS. (Step 2) When replication of the other strand catches up, it will be mostly populated by new nucleosomes (2a) and RNAPII will then “switch” from the early replicating strand to the late one and direct H3K4 and H3K36 methylation of new histones by Set1p and Set2p, respectively, on the second gene copy (2b). (B) Modulation of the replication timing of replicated gene copies determines the pattern of old and new nucleosome segregation.
If we consider that replication forks advance through genes at an average speed of 340 bp/min in WT cells (Fig. 6D) and that the difference in replication timing between sister copies is, on average, 2.5 min (Fig. 5C), approximately five mostly old nucleosomes (850 bp/160 bp = 5.3) will have already assembled on the sister copy that replicated first before the same sequence on the other sister copy has even finished replicating. The number of nucleosomes that can bind to the sister copy that replicated first before the other copy has finished replicating is also approximately five in mcm2-3A cells: 250 bp/min (average fork velocity) (Fig. 6D) × 3.5 min (average ΔRT) (Supplemental Fig. S9B) = 875 bp; 875 bp/160 bp = 5.5. Incidentally, a previous study that measured where old nucleosomes end up after replication determined that they mostly rebind to replicated DNA within 400 bp upstream of or downstream from their original binding site (i.e., within an 800-bp region) (Radman-Livaja et al. 2011), which is in perfect agreement with our calculations above.
By mid-early S, the asymmetry in RNAPII, H3K4me3, H3K36me3, and H3K56ac distributions is not detectable or appears diminished on genes that replicated the earliest because these genes now form a mixed population of step 1 and step 2 configurations due to the imperfect synchronization of replication forks in different cells.
Discussion
Using ChIP-NChAP, we were able to show that all examined histone marks and RNAPII are initially enriched on either the leading or the lagging strand copy depending on which strand replicated first. We found that RNAPII, H3K4me3, and H3K56ac enrichments shift to the other daughter chromatid as chromatin matures following replication. We also show that H3K56ac has no direct role in RNAPII binding to replicated gene copies. Likewise, the asymmetrical distributions of H3K4me3 and H3K36me3 do not correlate with the asymmetrical distribution of RNAPII later in S phase, suggesting that these histone marks do not carry or transmit epigenetic information on the transcriptional activity of the underlying gene and do not influence RNAPII (re)binding after replication.
In addition to temporal maps of nucleosome and RNAPII binding dynamics after replication, our analysis of NChAP data sets has also uncovered new information on replication dynamics in S. cerevisiae. NChAP fractions revealed dozens of discrete smaller peaks that surround one or two dominant peaks that are, as a rule, the ones closest to previously identified origins (cf. rows 2 and 3 with row 1 starting from the bottom in Supplemental Fig. S8C). These smaller peaks are spaced every ∼500 bp and are localized within ∼10 kbp upstream of and downstream from the dominant peak(s) (Supplemental Tables S1, S2). Historically, the chromosomal coordinates of replication origins were determined from the center of the region that had the earliest replication timing within a given locus (see “valleys” in the replication timing rows in Supplemental Figs. S10A, S8C). The precision with which the earliest replicating regions were mapped in the early DNA microarray-based studies was limited by the distance between the chromosomal coordinates of 50-bp probes printed on the microarray (500 bp–1 kbp) and that typically narrowed the location of a replication origin to ∼2 kb (Raghuraman et al. 2001; Yabuki et al. 2002).
Massively parallel sequencing improved mapping precision in cell populations, which is now limited by the size of genome fragments in sequencing libraries. By keeping all DNA fragments after chromatin sonication—including the ones smaller than 100 bp—for NChAP library construction, we were able to detect dozens of new replication initiation sites that are located upstream of and downstream from previously mapped replication origins (Nieduszynski et al. 2007; Vasseur et al. 2016). Most cells probably initiate replication at the dominant origin, but secondary initiation sites can be activated in a subset of cells in addition to or instead of the dominant site. Consequently, the NChAP signals from cells in which these additional sites were activated will overlap with the signals from cells in which replication initiated at the dominant site, which means that the signal from these less frequent initiation sites would have been “masked” by the replication signal that originated from the dominant origin if we had not kept small fragments and increased the resolution of our origin maps. Incidentally, single molecule detection of replication origins by nanopore sequencing revealed that replication initiates at different positions on each molecule and that these discrete initiation events are clustered ±2 kbp around mapped replication origins (Hennion et al. 2020). Single molecule sequencing thus suggests that the replication initiation clusters we detect with our population-based sequencing probably represent single initiation events on chromosomes from different cells.
Our results also suggest that H3K56ac has no direct role in RNAPII binding to replicated genes and is therefore unlikely to act as a “buffer” for the expected up-regulation of transcription caused by gene copy number doubling, as recently proposed (Voichek et al. 2016, 2018). Even though H3K56ac globally stimulates transcription (Supplemental Fig. S5A), the asymmetric binding and switching of RNAPII enrichment from the copy that replicated first to the other copy, which is mostly occupied by new nucleosomes, is independent of rtt109-mediated H3K56 acetylation (Fig. 4B). RNAPII may still switch to the gene copy enriched for new nucleosomes due to their generally hyperacetylated state (Jackson et al. 1976; Sobel et al. 1995; Ge et al. 2013), although further experiments are needed to test this assumption. The transient increase in transcription shortly after replication that was observed in rtt109Δ cells (Voichek et al. 2016; Topal et al. 2019) was the basis for the hypothesis that H3K56ac directly attenuates transcription of replicated genes during S phase (Voichek et al. 2016). Topal et al. (2019) show, however, that gene expression goes back to a “buffered” state after an initial burst in transcription immediately after replication even when H3K56ac is depleted. This led them to suggest that the observed transcription burst on replicated genes in rtt109Δ cells is probably caused by low nucleosome density on replicated DNA immediately after replication because of impaired assembly of new nucleosomes caused by the absence of H3K56 acetylation (Li et al. 2008).
We now propose that replicated genes produce the same amount of mRNA as before replication despite gene copy number doubling, because transcription factors (TFs) and RNAPII that are present at the locus at the time of replication are not available in sufficient quantities to double mRNA output (Blank et al. 2020). Our model predicts that transcription resumes at “half capacity” shortly after replication, using mostly recycled TFs and RNAPII that were bound to genes before replication. Consequently, the delay in the twofold increase in gene expression should last for a period of time that is needed to accumulate sufficient quantities of transcription machinery components required to double mRNA production.
Only genes with a low to moderate total RNAPII enrichment (log2[enrichment] ≤ 1) show differences in RNAPII occupancy between lagging and leading gene copies that are >16-fold (Supplemental Fig. S1E). This is consistent with the idea that newly synthesized RNAPII complexes are limiting in the early period after replication (Supplemental Fig. S1A) and that most RNAPII complexes that bind to replicated genes shortly after replication are more likely to be immediately recycled from ahead of the replication fork rather than originate from a common nuclear pool where they would have presumably gone after they were displaced from chromatin by the replisome. A small number of RNAPII complexes that were bound to genes before replication are more likely to partition asymmetrically between the two replicated gene copies than large numbers of RNAPII complexes, which are more likely to be evenly distributed. Also, if displaced RNAPII complexes are not immediately recycled onto DNA and join a common pool of “free” RNAPII complexes in the nucleoplasm first, one would expect that there would be a delay between the time we first detect RNAPII bound to replicated genes and the time when these genes have finished replicating. Since RNAPII can be detected practically as soon as genes are replicated (cf. ChIP-NChAP and NChAP fractions from the same time point in Fig. 2A), we propose that RNAPII complexes are recycled behind the fork together with or shortly after old nucleosomes.
The distribution of RNAPII complexes on daughter chromatids is not random and follows the same pattern as the distribution of old nucleosomes. Our time course experiments spanning early to mid S phase with parallel monitoring of the dynamics of five different chromatin features on thousands of gene copies from replicated sister chromatids in WT and mutant yeast cells reveal that all nucleosomes (old and new) and RNAPII are initially enriched on the strand that replicates first. Genic orientation seems to be the main factor that determines which strand will replicate first: the leading strand when replication and transcription go in the same direction or the lagging strand when they go in opposite directions. Immediately after the passage of the fork, old nucleosomes and some new nucleosomes are preferentially assembled on the early replicating strand (step 1). Most new nucleosomes then bind to the other strand after it has finished replicating because the strand that replicated first is by then mostly occupied by old nucleosomes (step 2). RNAPII binding to replicated genes follows the same steps as nucleosome binding although with somewhat of a lag behind nucleosome assembly.
Old nucleosomes are thus inherited asymmetrically at most yeast genes, with a bias for the leading or the lagging strand copy depending on which copy replicated first. This inheritance is accurate since the canonical distributions of H3K4me3 and H3K36me3 on gene bodies of recently replicated genes is preserved even in the earliest time points in WT and mcm2-3A cells (Figs. 5B, 6B; Supplemental Figs. S6B, S8B). In order for chromatin features to be truly epigenetic, however, in addition to accurate transmission after cell division, they also have to be instructive of the transcription state at their genomic location. Our results suggest that the initial correlation between RNAPII and H3K4me3/H3K36me3/H3K56ac/H3 binding patterns in early S phase is coincidental. RNAPII and histones are initially enriched on the gene copy that replicated first simply because they have nowhere else to go. The correlation between RNAPII binding and H3K4me3/K36me3 enrichment is lost later in S phase because replication becomes progressively less synchronized in the population and also because the second step in the chromatin maturation process proceeds at a different rate for histones and RNAPII. Despite having been accurately inherited, the H3K4me3 and H3K36me3 marks that are recycled with old histones consequently do not dictate where RNAPII will bind. This is direct evidence that H3K4me3 and H3K36me3—long thought of as “epigenetic” marks of active transcription, merely because they are enriched on actively transcribed genes (Bannister and Kouzarides 2011)—are actually not epigenetic marks in the strict sense. Their “inheritance” to replicated gene copies does not have an effect on RNAPII occupancy and is thus unlikely to have a role in the postreplicative re-establishment of transcriptional activity of their underlying genes. Consequently, H3K4me3 and H3K36me3 do not transmit information on prereplicative transcriptional activity of their underlying gene to replicated gene copies.
“Active” chromatin and transcription states are therefore inherited independently from each other. According to our model, after RNAPII density shifts from the copy populated with old histones carrying “active” marks to its sister copy with new nucleosomes, RNAPII recruits the Set1p and Set2p methyltransferases, which then put the “active” H3K4me3 and H3K36me3 marks on new histones (Buratowski and Moazed 2005; Bae et al. 2020). Active transcription states are therefore not restored by a copying mechanism that uses recycled old nucleosomes as a template. We propose instead that the recycling of the transcription machinery itself, which sequentially activates both sister gene copies, ensures the faithful transmission of active chromatin states and transcription states to both daughter chromatids after disruptions caused by the passage of the replication fork. Nucleosome configurations on the two sister copies eventually become indistinguishable after global H3K56 deacetylation in late S (Celic et al. 2006) because both copies carry the H3K4me3 and H3K36me3 marks characteristic of transcribed genes by the time the cycle arrives at the G2 phase. Consequently, mother and daughter cells inherit gene copies with identical chromatin configurations. The shift in RNAPII occupancy from one gene copy to the other ensures that both gene copies are transcribed in S phase and that the same chromatin architecture is established on either copy even when RNAPII is limiting.
How are old nucleosomes recycled to replicated DNA? In order to explain the observed bias in old nucleosome deposition, three recent studies proposed a system of competing chaperone complexes—Mcm2 with DNA polymerase alpha and Dpb3/4—that specialize in the preferential deposition of old histones on lagging or leading strands, respectively (Gan et al. 2018; Petryk et al. 2018; Yu et al. 2018). Two of these studies—one in yeast and one in mouse ES cells—have postulated that Mcm2-mediated recycling of old nucleosomes to the lagging strand counteracts the “natural” tendency of old nucleosomes to rebind to the leading strand (Gan et al. 2018; Petryk et al. 2018). The third study—in yeast—has hypothesized that this “natural” predilection of old histones for the leading strand is actually orchestrated by the Dpb3/4 subunits of the leading strand DNA polymerase epsilon (Yu et al. 2018).
Models that are based on such highly specialized nucleosome deposition systems are, however, not entirely satisfactory because they do not explain why one chaperone would consistently, ubiquitously, and ever so slightly outcompete the other. According to the conclusions of the above studies, Mcm2 in WT yeast seems to always “win” over Dpb3/4, resulting in a slight bias for old nucleosome deposition on the lagging strand, whereas in mouse ES cells, Mcm2 consistently “loses” and old nucleosome bias leans more toward the leading strand. It is, however, not clear why a molecular mechanism based on specialized nucleosome chaperones would create such opposing “natural” tendencies in two organisms that essentially have the same replication machinery.
Our experiments with the Mcm2 mutant now show that old nucleosomes can be preferentially deposited on the leading or the lagging strand in equal proportions both in WT and mcm2-3A cells alike. Mcm2 is therefore not likely to be a nucleosome chaperone that exclusively recycles old nucleosomes to the lagging strand since old nucleosomes are still recycled to the lagging strand in mutant mcm2-3A cells. We consequently propose a model in which the strand to which old nucleosomes are recycled is determined by local replication dynamics and not by competing chaperone complexes that are specialized for deposition on the leading or the lagging strand.
We suspect that there are two reasons why previous studies might have “missed” that old nucleosomes can be recycled to either strand in WT and mcm2-3A cells and that the enrichment in new nucleosomes shifts from one strand to the other: (1) they analyzed only one time point after EdU/BrdU labelling; and (2) they broadly averaged signals from regions around replication origins that fired at different times and were consequently at different points of the chromatin maturation process, instead of first sorting genes or chromosome regions by replication timing, analyzing them separately, and then grouping them according to their “preference” for the leading or the lagging strand as we have done here.
What is then the role of Mcm2 in old nucleosome recycling? The major effect of the mcm2-3A mutation appears to be a global and progressive slowing down of replication forks. We hypothesize that this slowing down creates better synchrony between replication forks in different cells in the population, which produces a “more focused” snapshot of step 2 of the chromatin maturation process for three different chromatin features (H3K4me3, H3K36me3, and H3K56ac) on almost all replicated genes.
Why does a mutation in Mcm2 that impairs the interaction between the helicase and histone H3 slow down replication forks? We speculate that the interaction between Mcm2 and H3 facilitates the removal of old nucleosomes ahead of the fork. If nucleosome removal is necessary for optimal DNA unwinding by the Mcm2-7 helicase, a defect in nucleosome removal would disrupt the optimal progression of the replication fork. So, when nucleosome removal is impaired, fork progression also slows down.
Along the same line of reasoning, we also suspect that the preferential deposition of old histones to the lagging strand observed in dpb3/4 deletion mutants (Yu et al. 2018) is a consequence of a decrease in the rate of leading strand replication. We hypothesize that Dpb3/4, as subunits of the DNA polymerase epsilon, probably stimulate the rate of leading strand synthesis. Leading strand synthesis should therefore slow down in the absence of Dpb3/4 and old nucleosomes should be recycled to the lagging strand, which would have replicated first in these conditions.
Our model of chromatin structure re-establishment after DNA replication also explains how the inherent asymmetry of old nucleosome and RNAPII distribution on replicated gene copies could either maintain or modulate gene expression states and chromatin configuration from one cell generation to the next (Fig. 7B). The model predicts that daughter chromatids will be “decorated” with contiguous alternating “patches” of old and new nucleosomes (left and right panels in Fig. 7B), because replication origins are evenly distributed along yeast chromosomes and replication forks are bidirectional. The other prediction of our model is that the recycling of old nucleosomes should be less biased toward either daughter chromatid if replication of both gene copies is nearly simultaneous. In that case, old nucleosomes should partition more randomly and symmetrically, as shown in the middle panel of Figure 7B. Complete asymmetrical segregation of old and new histones could theoretically be achieved if replication fork barriers were introduced on the same side of all or most replication origins from the same chromosome and if the replication of one strand happened consistently earlier than the replication of its sister strand throughout the chromosome (left and right panels, Fig. 7B). Our model thus provides a mechanistic blueprint for asymmetric nucleosome deposition that could explain even the most extreme case of nucleosome segregation bias, like the one recorded in Drosophila male germline stem cells (Tran et al. 2012; Xie et al. 2015). There, the full complement of old nucleosomes is retained in the stem cell. We speculate that such completely asymmetric segregation could be achieved with unidirectional replication forks, as recent imaging data suggests (Wooten et al. 2019), and transcription that mostly goes in the same direction throughout the chromosome.
We observed that the replication machinery favors leading strand replication on “same” genes and “lagging” strand replication on “opposite” genes. Faster and earlier lagging replication on “opposite” genes is unexpected. If the progress of the two DNA polymerases is always coupled during “regular” replication, as currently believed, lagging strand synthesis should “naturally” lag behind leading strand synthesis. The same genome location should be replicated on the leading strand slightly before it is replicated on the lagging strand because the DNA strand that serves as a template for lagging strand synthesis has to fold into a loop to allow the leading and lagging strand DNA polymerases to travel in the same direction (Lewis et al. 2020). Recent examination of the role of the Rad53 checkpoint kinase in replication stress uncovered that lagging strand synthesis outpaces leading strand synthesis in Rad53 checkpoint kinase mutants when forks are stalled in the presence of hydroxyurea (Gan et al. 2017). Leading and lagging strand replication can therefore be uncoupled under certain conditions. We propose that direct encounters of the replication machinery with the transcription machinery, which are more likely to occur on “opposite” genes when the two complexes go towards each other, cause a progressive slowing down of the fork and fork pausing as observed previously (Prado and Aguilera 2005). We surmise that temporary fork pausing then transiently uncouples leading and lagging strand synthesis and favors lagging synthesis. The obstacles to replisome progression created by counterdirectional transcription could be physical or topological. For example, there is an intriguing possibility that counterdirectional transcription and replication create conditions that favor the appearance of R-loops ahead of the fork (Niehrs and Luke 2020).
We made several observations that support the hypothesis that forks become progressively slower when they go through a succession of “opposite” genes. First, the difference in replication timing between “opposite” and “same” genes increases with S-phase progression (Supplemental Fig. S12A). Second, genes of the same genic orientation tend to be replicated in succession by the same replication fork (Supplemental Fig. S12B). Consequently, a replication fork passing through an array of “opposite” genes is more likely to slow down compared with a fork replicating an array of “same” genes, presumably because of repeated disruptive “head on” encounters with the transcription machinery in “opposite” gene clusters. Finally, leading strand synthesis is, on average, faster on same genes than on opposite genes, while lagging strand synthesis tends to be faster on opposite genes compared with same genes (Supplemental Fig. S12C).
To summarize, we show that the recycling of chromatin features characteristic of actively transcribed chromatin is independent of the (re)binding of the transcription machinery to daughter chromatids. We suggest that their distribution pattern on replicated gene copies is constrained by the mechanics and interactions of replication and transcription ahead of the replication fork that determine which strand replicates first and then direct old nucleosomes and the transcription machinery to that strand. We also propose that RNAPII is recycled behind the fork. As RNAPII amounts at the replicated gene locus are initially limiting, only the gene copy on which RNAPII landed first is initially transcribed. The transcription machinery switches to the sister strand later on after that strand has finished replicating and directs the establishment of histone marks characteristic of active transcription onto new nucleosomes bound to that strand. Consequently, both replicated gene copies end up with the same chromatin features: one through inheritance of old histones and the other after the “inherited” RNAPII that binds there has recruited the relevant histone methyltransferases. So, the principal “epigenetic” factors that restore “active” chromatin features on most new histones are not old histones that serve as templates for a hypothetical copying mechanism that would put those marks on neighboring new histones. Our results are consistent with the idea that the epigenetic factor that perpetuates active transcription states and their underlying “active” chromatin states on newly replicated genes is simply the recycled/inherited RNAPII that recruits histone methyltransferases that then deposit “active” marks on new histones.
Methods
Yeast strains
All yeast strains are listed in Supplemental Table S3.
Cell culture
Cells were grown at 30°C in synthetic complete − URA + dextrose (SCD−URA) media until they reached exponential growth and were then arrested with α factor (0.15 μg/mL). Cells were released into fresh media supplemented with Pronase (20 μg/mL, Sigma-Aldrich) and 10 μM EdU (Carbosynth), and aliquots were taken at indicated times and fixed with 1% formaldehyde for 15–30 min (depending on the experiment, as detailed in Supplemental Material) and quenched with 125 mM glycine.
Chromatin immunoprecipitation (ChIP)
After mechanical cell wall disruption, chromatin was either digested with MNase (Fig. 3; see “MNase Digestion” in Supplemental Material for details) or sonicated (for all the other experiments, see “Chromatin Sonication” in Supplemental Material) and immunoprecipitated according to standard protocols (see “Chromatin Immunoprecipitation” in Supplemental Material). The specificity of H3K56ac antibodies was tested by Western blot using an H3K56A mutant strain (Supplemental Fig. S13).
Biotin conjugation to EdU with the Click reaction
Biotin-azide was conjugated to EdU from ChIP-ed and Input DNA using Click chemistry as described previously in Vasseur et al. (2016).
Illumina sequencing library construction
Libraries were constructed as described in Vasseur et al. (2016) or according to the manufacturer's protocols for the TrueSeq V2 LT Sample prep kit (Illumina) or the NEBNext Ultra II DNA library prep kit for Illumina (NEB) (see “Illumina Sequencing Library Construction” in Supplemental Material).
Illumina sequencing
Library pools were sequenced on a HiSeq 2000 or NovaSeq 6000 (2 × 75-bp) (Illumina) at the CNAG, Barcelona, Spain, or the NextSeq550 (2 × 75-bp) (Plateforme Transcriptome, IRMB, Montpellier, France).
RNA-seq with spike-in control (Supplemental Fig. S5)
Exponentially growing S. cerevisiae (YPD) and S. pombe (YES) cells were flash-frozen in liquid N2, and total RNA was isolated from frozen cell pellets with TRIzol. Each DNase I-treated total RNA extract from S. cerevisiae was mixed with the S. pombe total RNA extract at a mass ratio of 10:1. The mixed RNA samples were then used for second-generation sequencing library preparation using the Illumina TruSeq Stranded mRNA kit according to the manufacturer's protocol (see “RNA-Seq with Spike-In Control” in Supplemental Material).
ChIP DNA microarray hybridization (Supplemental Fig. S1A)
ChIP-ed DNA and their corresponding input samples were amplified using the DNA linear amplification method described previously (Liu et al. 2003, 2005).
Labeled probes (a mixture of Cy5-labeled input and Cy3-labeled ChIP-ed material or their corresponding dye flips) were hybridized onto an Agilent yeast 4 × 44 K whole genome array (ref. G4810A-14810) (see “ChIP DNA Microarray Hybridization” in Supplemental Material).
Data Access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo) under accession number GSE160509. The Perl scripts used for origin mapping and to calculate replication timing and DNA synthesis rates are provided as Supplemental Code.
Supplementary Material
Acknowledgments
We thank Susan Forsburg, Paul Kaufman, Traci Lee, Etienne Schwob, Karim Labib, and Zhiguo Zhang for yeast strains. We also thank Marta Gut and Julie Blanc from CNAG (Barcelona, Spain) and Veronique Pantesco (Plateforme Transcriptome-IRMB Montpellier) for massive parallel sequencing services, and Robert Feil for critical reading of the manuscript. This work was supported with the ERC-Consolidator grant- NChIP 647618 (M.R.L.).
Author contributions: R.Z. optimized ChIP-NChAP and performed all the experiments except the ones in Figure 3 (done by M.R.L.) and Supplemental Figures S5 and S13 (done by A.C.). A.C. assisted R.Z. in the cell culture and ChIP steps of the experiments. R.Z. and A.C. constructed the yeast strains. M.R.L., R.Z., and A.C. designed the experiments. M.R.L. conceived and developed ChIP-NChAP, wrote the Perl and R scripts for analysis, analyzed the data, and wrote the manuscript with input from R.Z. and A.C.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275387.121.
Competing interests
The authors declare no competing interests.
References
- Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13: 153–167. 10.1038/nrm3288 [DOI] [PubMed] [Google Scholar]
- Bae HJ, Dubarry M, Jeon J, Soares LM, Dargemont C, Kim J, Geli V, Buratowski S. 2020. The Set1 N-terminal domain and Swd2 interact with RNA polymerase II CTD to recruit COMPASS. Nat Commun 11: 2181. 10.1038/s41467-020-16082-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank HM, Papoulas O, Maitra N, Garge R, Kennedy BK, Schilling B, Marcotte EM, Polymenis M. 2020. Abundances of transcripts, proteins, and metabolites in the cell cycle of budding yeast reveal coordinate control of lipid metabolism. Mol Biol Cell 31: 1069–1084. 10.1091/mbc.E19-12-0708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S, Moazed D. 2005. Gene regulation: expression and silencing coupled. Nature 435: 1174–1175. 10.1038/4351174b [DOI] [PubMed] [Google Scholar]
- Celic I, Masumoto H, Griffith WP, Meluh P, Cotter RJ, Boeke JD, Verreault A. 2006. The sirtuins hst3 and Hst4p preserve genome integrity by controlling histone h3 lysine 56 deacetylation. Curr Biol 16: 1280–1289. 10.1016/j.cub.2006.06.023 [DOI] [PubMed] [Google Scholar]
- Cusick ME, DePamphilis ML, Wassarman PM. 1984. Dispersive segregation of nucleosomes during replication of simian virus 40 chromosomes. J Mol Biol 178: 249–271. 10.1016/0022-2836(84)90143-8 [DOI] [PubMed] [Google Scholar]
- Eser P, Demel C, Maier KC, Schwalb B, Pirkl N, Martin DE, Cramer P, Tresch A. 2014. Periodic mRNA synthesis and degradation co-operate during cell cycle gene expression. Mol Syst Biol 10: 717. 10.1002/msb.134886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltman M, Evrin C, De Piccoli G, Jones RC, Edmondson RD, Katou Y, Nakato R, Shirahige K, Labib K. 2013. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep 3: 892–904. 10.1016/j.celrep.2013.02.028 [DOI] [PubMed] [Google Scholar]
- Gan H, Yu C, Devbhandari S, Sharma S, Han J, Chabes A, Remus D, Zhang Z. 2017. Checkpoint kinase Rad53 couples leading- and lagging-strand DNA synthesis under replication stress. Mol Cell 68: 446–455.e3. 10.1016/j.molcel.2017.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan H, Serra-Cardona A, Hua X, Zhou H, Labib K, Yu C, Zhang Z. 2018. The Mcm2-Ctf4-Polα axis facilitates parental histone H3-H4 transfer to lagging strands. Mol Cell 72: 140–151.e3. 10.1016/j.molcel.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Nair D, Guan X, Rastogi N, Freitas MA, Parthun MR. 2013. Sites of acetylation on newly synthesized histone H4 are required for chromatin assembly and DNA damage response signaling. Mol Cell Biol 33: 3286–3298. 10.1128/MCB.00460-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss C, Gutierrez C, Burhans WC, DePamphilis ML, Koller T, Sogo JM. 1990. Nucleosome assembly in mammalian cell extracts before and after DNA replication. EMBO J 9: 2911–2922. 10.1002/j.1460-2075.1990.tb07482.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruszka DT, Xie S, Kimura H, Yardimci H. 2020. Single-molecule imaging reveals control of parental histone recycling by free histones during DNA replication. Sci Adv 6: eabc0330. 10.1126/sciadv.abc0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennion M, Arbona JM, Lacroix L, Cruaud C, Theulot B, Tallec BL, Proux F, Wu X, Novikova E, Engelen S, et al. 2020. FORK-seq: replication landscape of the Saccharomyces cerevisiae genome by nanopore sequencing. Genome Biol 21: 125. 10.1186/s13059-020-02013-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V, Shires A, Tanphaichitr N, Chalkley R. 1976. Modifications to histones immediately after synthesis. J Mol Biol 104: 471–483. 10.1016/0022-2836(76)90282-5 [DOI] [PubMed] [Google Scholar]
- Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, Kaufman PD, Friedman N, Rando OJ. 2008. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet 4: e1000270. 10.1371/journal.pgen.1000270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Liu CL, Yassour M, Holik J, Friedman N, Buratowski S, Rando OJ. 2010. RNA polymerase mapping during stress responses reveals widespread nonproductive transcription in yeast. Genome Biol 11: R75. 10.1186/gb-2010-11-7-r75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JS, Spenkelink LM, Schauer GD, Yurieva O, Mueller SH, Natarajan V, Kaur G, Maher C, Kay C, O'Donnell ME, et al. 2020. Tunability of DNA polymerase stability during eukaryotic DNA replication. Mol Cell 77: 17–25.e15. 10.1016/j.molcel.2019.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134: 244–255. 10.1016/j.cell.2008.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Schreiber SL, Bernstein BE. 2003. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics 4: 19. 10.1186/1471-2164-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol 3: e328. 10.1371/journal.pbio.0030328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Hawke D, Kobayashi R, Verreault A. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436: 294–298. 10.1038/nature03714 [DOI] [PubMed] [Google Scholar]
- Miller C, Schwalb B, Maier K, Schulz D, Dümcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dölken L, et al. 2011. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol 7: 458. 10.1038/msb.2010.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Hiraga S, Ak P, Benham CJ, Donaldson AD. 2007. OriDB: a DNA replication origin database. Nucleic Acids Res 35: D40–D46. 10.1093/nar/gkl758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs C, Luke B. 2020. Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol 21: 167–178. 10.1038/s41580-019-0206-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryk N, Dalby M, Wenger A, Stromme CB, Strandsby A, Andersson R, Groth A. 2018. MCM2 promotes symmetric inheritance of modified histones during DNA replication. Science 361: 1389–1392. 10.1126/science.aau0294 [DOI] [PubMed] [Google Scholar]
- Prado F, Aguilera A. 2005. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 24: 1267–1276. 10.1038/sj.emboj.7600602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. 2011. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol 9: e1001075. 10.1371/journal.pbio.1001075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. 2001. Replication dynamics of the yeast genome. Science 294: 115–121. 10.1126/science.294.5540.115 [DOI] [PubMed] [Google Scholar]
- Sekedat MD, Fenyö D, Rogers RS, Tackett AJ, Aitchison JD, Chait BT. 2010. GINS motion reveals replication fork progression is remarkably uniform throughout the yeast genome. Mol Syst Biol 6: 353. 10.1038/msb.2010.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci 92: 1237–1241. 10.1073/pnas.92.4.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topal S, Vasseur P, Radman-Livaja M, Peterson CL. 2019. Distinct transcriptional roles for Histone H3-K56 acetylation during the cell cycle in yeast. Nat Commun 10: 4372. 10.1038/s41467-019-12400-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran V, Lim C, Xie J, Chen X. 2012. Asymmetric division of Drosophila male germline stem cell shows asymmetric histone distribution. Science 338: 679–682. 10.1126/science.1226028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur P, Tonazzini S, Ziane R, Camasses A, Rando OJ, Radman-Livaja M. 2016. Dynamics of nucleosome positioning maturation following genomic replication. Cell Rep 16: 2651–2665. 10.1016/j.celrep.2016.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voichek Y, Bar-Ziv R, Barkai N. 2016. Expression homeostasis during DNA replication. Science 351: 1087–1090. 10.1126/science.aad1162 [DOI] [PubMed] [Google Scholar]
- Voichek Y, Mittelman K, Gordon Y, Bar-Ziv R, Lifshitz Smit D, Shenhav R, Barkai N. 2018. Epigenetic control of expression homeostasis during replication is stabilized by the replication checkpoint. Mol Cell 70: 1121–1133.e9. 10.1016/j.molcel.2018.05.015 [DOI] [PubMed] [Google Scholar]
- Wooten M, Snedeker J, Nizami ZF, Yang X, Ranjan R, Urban E, Kim JM, Gall J, Xiao J, Chen X. 2019. Asymmetric histone inheritance via strand-specific incorporation and biased replication fork movement. Nat Struct Mol Biol 26: 732–743. 10.1038/s41594-019-0269-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Wooten M, Tran V, Chen BC, Pozmanter C, Simbolon C, Betzig E, Chen X. 2015. Histone H3 threonine phosphorylation regulates asymmetric histone inheritance in the Drosophila male germline. Cell 163: 920–933. 10.1016/j.cell.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K. 2002. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7: 781–789. 10.1046/j.1365-2443.2002.00559.x [DOI] [PubMed] [Google Scholar]
- Yang SC, Rhind N, Bechhoefer J. 2010. Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol Syst Biol 6: 404. 10.1038/msb.2010.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Serra-Cardona A, Zhang L, Gan S, Sharma S, Johansson E, Chabes A, Xu RM, Zhang Z. 2018. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361: 1386–1389. 10.1126/science.aat8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.