ABSTRACT
Long non-coding RNA (lncRNA) FGD5 antisense RNA 1 (FGD5-AS1) was reported to exert critical roles in multiple cancers. The current work aimed to determine the role of FGD5-AS1 in cisplatin (DDP) resistance of hepatocellular carcinoma (HCC). The levels of FGD5-AS1, miR-153-3p, and twinfilin actin binding protein 1 (TWF1) were analyzed using RT-qPCR. CCK-8, colony formation, Transwell, and TUNEL assays were used to examine the IC50 value of DDP, cell viability, invasion, and apoptosis. The interaction between miR-153-3p and TWF1 or FGD5-AS1 was determined by luciferase reporter and RIP assays. In our study, we found that FGD5-AS1 level was elevated in DDP-resistant HCC tissues and cell lines. FGD5-AS1 silencing improved the sensitivity of HCC cells to DDP. Moreover, FGD5-AS1 directly bound to miR-153-3p and FGD5-AS1 addition neutralized the inhibitory impacts of miR-153-3p supplementation on DDP resistance in the HCC cells. In addition, knockdown of TWF1 inhibited DDP resistance of HCC cells, which was reversed by miR-153-3p deletion. Lastly, FGD5-AS1 interference decreased TWF1 expression level, which was rescued by miR-153-3p inhibition. Our study exhibited that FGD5-AS1 promoted DDP resistance through modulating the miR-153-3p/TWF1 axis in HCC cells. This could be an effective treatment strategy for HCC patients.
KEYWORDS: Hepatocellular carcinoma, fgd5-as1, miR-153-3p, twf1, cisplatin resistance
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignant tumor and the third leading cause of cancer-associated mortality worldwide [1,2]. Although the clinical treatment of HCC vastly improved, the 5-year survival rate of HCC patients is still low because of recurrence and metastasis following surgery [3,4]. Thus, it is crucial to identify new treatment targets for HCC patients.
Extensive studies have shown that lncRNAs served as vital functions in the drug resistance of HCC [5,6]. For instance, LINC01234 accelerated HCC cell line progression and chemoresistance by targeting the miR-31-5p/MAGEA3 axis [7]. Enhanced CASC2 promoted DDP sensitivity in HCC cell lines by sponging miR-222 [8]. Several studies manifested that FGD5 antisense RNA 1 (FGD5-AS1) taken part in cancer malignant progression, including oral cancer and esophageal squamous cell carcinoma [9,10]. However, the effect of FGD5-AS1 on HCC chemoresistance is unclear.
MicroRNAs (miRNAs) are small non-coding RNAs (19–22 nts) that may cause cleavage or repress the translation of mRNAs by targeting their 3ʹ-UTR [11]. Accumulating evidence suggested that miRNAs participated in the development of multiple cancers, including HCC [12,13]. Moreover, miR-153-3p was uncovered to be abnormally expressed in some cancers. For example, circular RNA 0084043 increased melanoma cell metastasis via absorbing miR-153-3p and enhancing Snail [14]. miR-153-3p repressed breast cancer cell line progression by downregulating ROCK1 [15]. Nevertheless, the mechanism of miR-153-3p in HCC is still undetermined.
Our study aims to investigate the role and molecular mechanism of FGD5-AS1 in HCC, and we hypothesized that FGD5-AS1 enhanced DDP resistance in HCC cells. Our data demonstrated for the first time that FGD5-AS1 facilitated DDP resistance via the miR-153-3p/twinfilin actin binding protein 1 (TWF1) axis, which might provide a new treatment strategy for DDP-resistant HCC.
Materials and methods
Patients and specimens
DDP-resistant (n = 32) and DDP-sensitive HCC tissues (n = 28) were collected between September 2015 and January 2018. All patients provided written informed consent prior to enrollment, and the research was granted ethics approval by the Ethics Committee of the Third Affiliated Hospital of Soochow University. The patients’ clinical features have been displayed in Table 1.
Table 1.
Clinicopathological characteristics of patients with hepatocellular carcinoma
| Characteristic | Patients, n |
|---|---|
| Age, years | |
| ≤55 | 12 |
| >55 | 20 |
| Sex | |
| Male | 22 |
| Female | 10 |
| Tumor size, cm | |
| ≤5 | 11 |
| >5 | 21 |
| Histological grade | |
| Good | 6 |
| Moderate | 18 |
| Poor | 8 |
| Metastasis | |
| With | 13 |
| Without | 19 |
Cell culture
The human HCC cells (Huh7 and Hep3B) were gained from the American Type Culture Collection (ATCC, USA). The DDP-resistant cells (Huh7/DDP and Hep3B/DDP) were established by exposing their parental cells to increasing concentrations of DDP for 12 months. Then, cells were cultured in RPMI-1640 medium at 37°C with 5% CO2.
Cell transfection
shRNA targeting FGD5-AS1 (shFGD5-AS1), shTWF1, and negative control (shNC), pcDNA3.1/FGD5-AS1 and pcDNA3.1, miR-153-3p mimics/inhibitor, NC mimic/inhibitor were purchased from GenePharma. DDP-resistant HCC cell lines were transfected with the above vectors using Lipofectamine® 2000 (Invitrogen).
Reverse transcription-quantitative PCR (RT-qPCR). Total RNA was isolated using TRIzol (Invitrogen). The reverse transcription was done by a PrimeScript reagent kit (Invitrogen). qPCR was performed with SYBRGreen PCR Master Mix kit (Takara) with GAPDH/U6 as the endogenous control. Genes levels were verified by 2−ΔΔCq method [16]. The sequences of the primers were as follows: FGD5-AS1: forward, 5ʹ-AACAGTGCCTATGTGGACGG-3ʹ and reverse, 5ʹ-CCCATCACAGAGGTCCACAC-3ʹ; miR-153-3p: forward, 5ʹ-TGCGGGTGCTCGCTTCGCAGC-3ʹ and reverse, 5ʹ-CCAGTGCAGGGTCCGAGGT-3ʹ; TWF1: forward, 5ʹ-TGGAGTCGCTGGTTTTCGCG-3ʹ and reverse, 5ʹ-CTGTGCTTTACTGCGCGCTC-3ʹ; U6: forward, 5ʹ‑CTCGCTTCGGCAGCACA‑3ʹ and reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ; GAPDH: forward, 5ʹ-ACACCCACTCCTCCACCTTT‑3ʹ and reverse, 5ʹ‑TTACTCCTTGGAGGCCATGT‑3ʹ.
Cell counting kit 8 (CCK-8) assay
Cells were seeded into 96-well plates and treated with various concentrations of DDP (0, 0.625, 1.25, 2.5, 5, 10, 20, 40 or 60 μg/ml). The CCK-8 solution was added to the wells and incubated for 4 h. Then, OD value was examined by a microplate reader at 450 nm. The IC50 of DDP was determined to be the concentration of DDP to reduce viability by 50%.
Colony formation assay
HCC cells were plated in 6-well plates and incubated in RPMI-1640 with 10% FBS. After 2 weeks, the cells were fixed and stained with 0.1% crystal violet [17].
Transwell assay
Cell invasion was assessed via Transwell chambers (8.0-μm pore size) and Matrigel. A total of 2 × 105 cells was added to the upper chambers in serum-free medium. RPMI-1640 medium (600 µl) was added to the lower chambers. After incubation, the invasive cells were stained with 0.1% crystal violet and counted using a microscope [18,19].
TUNEL assay
A TUNEL Apoptosis kit (Roche Diagnostics GmbH) was used to analyze cell apoptosis [20]. After dehydrating with ethanol, the DDP-resistant HCC cells were stained and cultured with the TUNEL reaction mixture (Roche Diagnostics). A fluorescent microscope was used to observe the TUNEL-positive cells.
Bioinformatic analysis
Prediction of downstream target sites of FGD5-AS1 or miR-153-3p was performed using StarBase. Medium stringency (≥2) and strict stringency (≥5) of CLIP data were used to screen the downstream target of FGD5-AS1 and miR-153-3p, respectively.
Dual-luciferase reporter assay
The wild-type (WT) or mutant (Mut) binding sites of miR-153-3p and FGD5-AS1 or TWF1 were inserted into pmirGLO vector (Promega). Then, these were co-transfected into the HCC cells with miR-153-3p mimics or NC mimics using Lipofectamine® 2000 [21].
Western blotting
Total protein was extracted via RIPA buffer (Beyotime). Proteins were divided with 10% SDS-PAGE and transferred to PVDF membranes. Next, membranes were probed with primary antibodies targeted against: TWF1 and GAPDH (Abcam). Subsequently, membranes were incubated with secondary antibody (Abcam). Protein bands were visualized by an enhanced chemiluminescent kit (Applygen) [22].
RNA-binding protein immunoprecipitation (RIP) assay
RIP was performed with a Magna RIP kit (EMD Millipore). Briefly, the cells were lysed in RIP lysis buffer and the cell extracts were incubated with Ago2 or IgG antibodies. FGD5-AS1 enrichment was examined using RT-qPCR [23].
Statistical analysis
All quantitative data are presented as mean ± SD. The differences between groups were assessed using Student’s t-test or one-way ANOVA. P < 0.05 was regarded as statistically significant.
Results
The study was aimed to elucidate the role and mechanism of FGD5-AS1 in DDP-resistant HCC, and the functional experiments revealed that FGD5-AS1 facilitated DDP resistance in HCC via the miR-153-3p/TWF1 axis, which might provide a new theoretical basis for HCC treatment.
FGD5-AS1 level is elevated in DDP-resistant HCC tissues and cells. Firstly, FGD5-AS1 level was tested in HCC patients following DDP treatment. RT-qPCR analysis disclosed that FGD5-AS1 level was elevated in DDP-resistant HCC tissues (Figure 1a). Moreover, FGD5-AS1 was notably increased in DDP-resistant Huh7 and Hep3B cells (Figure 1b). All these discoveries concluded that FGD5-AS1 might be implicated in DDP resistance of HCC.
Figure 1.
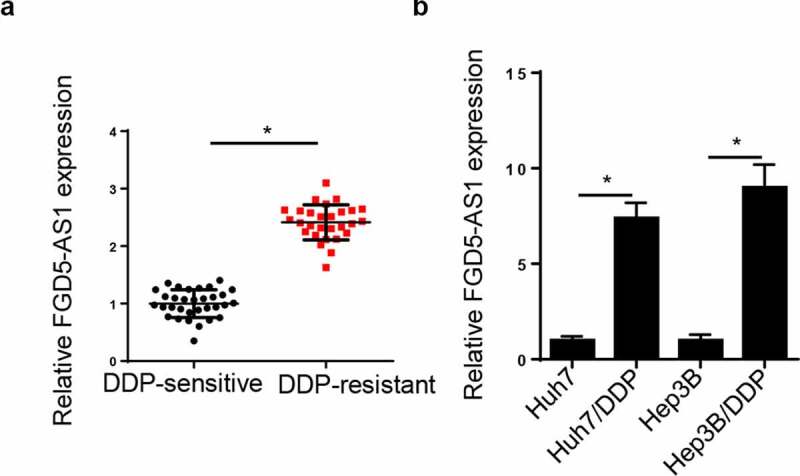
FGD5-AS1 expression levels are upregulated in DDP-resistant HCC tissues and cells. (a) RT-qPCR was used to determine relative FGD5-AS1 expression in DDP-resistant HCC tissues (n = 32) and DDP-sensitive HCC tissues (n = 28). (b) RT-qPCR was used to analyze the expression of FGD5-AS1 in Huh7, Hep3B, Huh7/DDP and Hep3B/DDP cell lines. *P < 0.05
FGD5-AS1 silence suppresses DDP resistance in the Huh7/DDP and Hep3B/DDP cells
To probe whether FGD5-AS1 mediated DDP resistance in HCC cells, we transfected shFGD5-AS1 into Huh7/DDP and Hep3B/DDP cells. The knockdown efficiency was shown in Figure 2a. Moreover, the IC50 of DDP was markedly reduced by FGD5-AS1 deletion in the Huh7/DDP and Hep3B/DDP cell lines (Figure 2b). Furthermore, CCK-8, colony formation, and Transwell assays implied that FGD5-AS1 deficiency notably suppressed the viability and invasion of Huh7/DDP and Hep3B/DDP cell lines (Figure 2c-f). Lastly, the results from the TUNEL assay indicated that FGD5-AS1 silence induced apoptosis in DDP-resistant HCC cells (Figure 2g). Taken together, FGD5-AS1 interference restrained DDP resistance in HCC.
Figure 2.
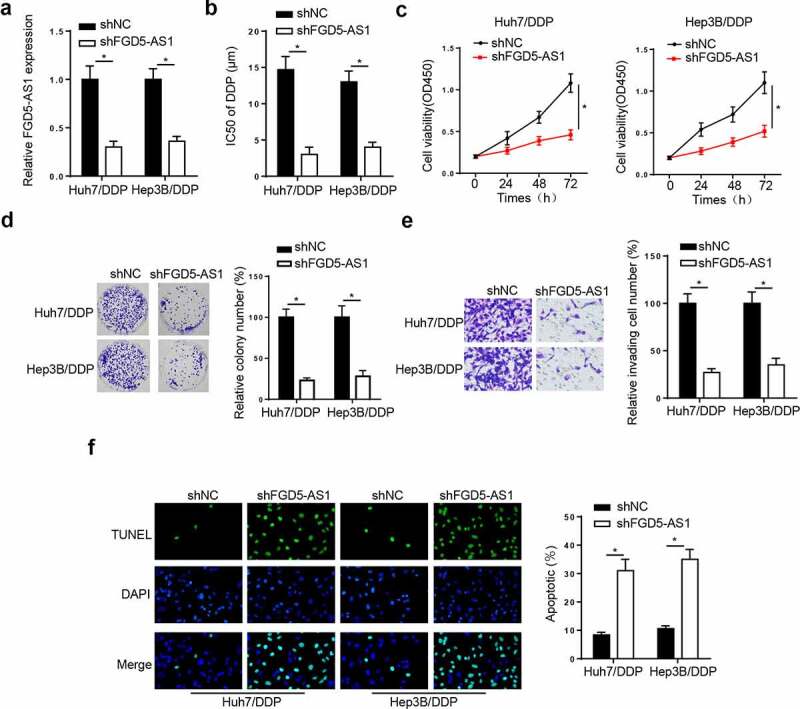
FGD5-AS1 knockdown suppresses DDP resistance in Huh7/DDP and Hep3B/DDP cells. (a) Reverse transcription-quantitative PCR analysis was used to determine relative FGD5-AS1 expression in Huh7/DDP and Hep3B/DDP cells transfected with shFGD5-AS1 or shNC. (b) CCK-8 assay was used to determine the IC50 value of Huh7/DDP and Hep3B/DDP cells transfected with shNC or shFGD5-AS1. (c-d) CCK-8, (e) colony formation and (f) Transwell assays were used to analyze Huh7/DDP and Hep3B/DDP cell proliferation and invasion, respectively, following the transfection with shFGD5-AS1 or shNC. (g) TUNEL assay was used to determine the apoptosis of Huh7/DDP and Hep3B/DDP cells transfected with shFGD5-AS1 or shNC. *P < 0.05
miR-153-3p is a target of FGD5-AS1 in DDP-resistant HCC cells
The StarBase website presented that miR-153-3p had binding sites with FGD5-AS1 (Figure 3a). Subsequently, miR-153-3p supplementation impeded the luciferase activity of FGD5-AS1-WT, however, no effect was observed in FGD5-AS1-Mut (Figure 3b and c). Moreover, the results from a RIP assay found that miR-153-3p supplementation increased FGD5-AS1 enrichment in the RIP-Ago2 group (Figure 3d). In addition, RT-qPCR results manifested that miR-153-3p was downregulated in the DDP-resistant HCC cells (Figure 3e). FGD5-AS1 overexpression blocked the amount of miR-153-3p, which was promoted by FGD5-AS1 knockdown (figure 3f). In sum, the above data manifested that FGD5-AS1 sponged miR-153-3p in the Huh7/DDP and Hep3B/DDP cell lines.
Figure 3.
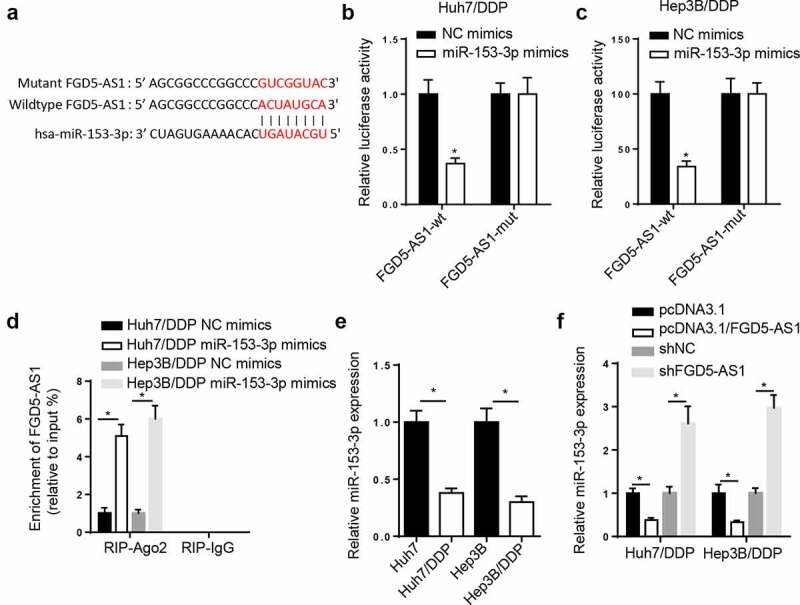
miR-153-3p is a target of FGD5-AS1 in DDP-resistant HCC cells. (a) StarBase database was used to predict the binding site between FGD5-AS1 and miR-153-3p. Dual luciferase reporter assay was used to determine the relative luciferase activity of FGD5-AS1-WT or FGD5-AS1-Mut vectors in (b) Huh7/DDP and (c) Hep3B/DDP cells co-transfected with NC mimic or miR-153-3p mimic. (d) RIP assay was used to measure the enrichment level of FGD5-AS1 in Huh7/DDP and Hep3B/DDP cells transfected with miR-153-3p mimic or NC mimic. (e) RT-qPCR was used to analyze the relative expression levels of miR-153-3p in DDP-resistant and -sensitive HCC cells. (f) RT-qPCR was used to determine relative miR-153-3p expression levels in Huh7/DDP and Hep3B/DDP cells transfected with shNC, shFGD5-AS1, pcDNA3.1 and pcDNA3.1/FGD5-AS1. *P < 0.05
FGD5-AS1 regulates DDP resistance in DDP-resistant HCC cell lines by targeting miR-153-3p. To validate whether miR-153-3p was involved in FGD5-AS1-mediated DDP resistance, a series of rescue assays were performed. First, the results revealed that FGD5-AS1 was elevated by FGD5-AS1 addition (Figure 4a). Next, miR-153-3p was elevated by miR-153-3p supplementation, which was weakened by FGD5-AS1 addition (Figure 4b). Then, the IC50 of DDP was markedly decreased following transfection of miR-153-3p mimics, which was restored by the transfection with FGD5-AS1 overexpression vector (Figure 4c). Lastly, miR-153-3p addition suppressed cell viability and invasion, and induced apoptosis, while these effects were counteracted by the overexpression of FGD5-AS1 (Figure 4d-g). In summary, FGD5-AS1 was associated with DDP resistance via modulating miR-153-3p in HCC cells.
Figure 4.
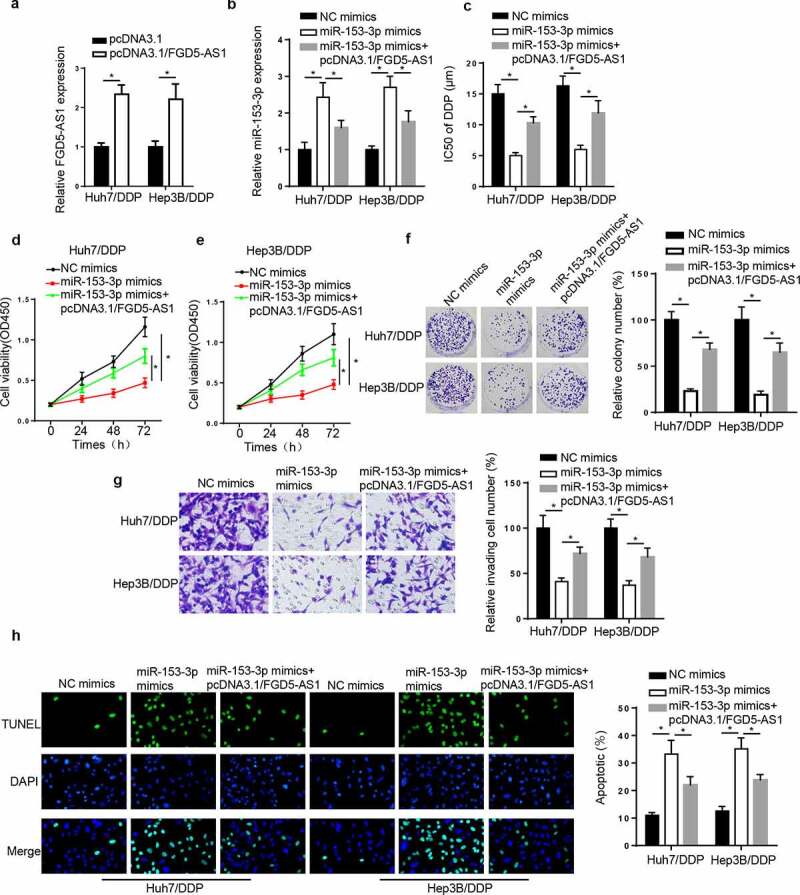
FGD5-AS1 regulates DDP resistance in DDP-resistant HCC cells by targeting miR-153-3p expression. (a) RT-qPCR was used to determine relative FGD5-AS1 expression in Huh7/DDP and Hep3B/DDP cells transfected with pcDNA3.1 or pcDNA3.1/FGD5-AS1. (b) RT-qPCR was used to analyze relative miR-153-3p expression in DDP-resistant HCC cells transfected with NC mimic, miR-153-3p mimic and miR-153-3p mimic + pcDNA3.1/FGD5-AS1. (c) CCK-8 assay was used to determine the IC50 value of DDP in Huh7/DDP and Hep3B/DDP cells transfected with NC mimic, miR-153-3p mimic and miR-153-3p mimic + pcDNA3.1/FGD5-AS1. (d and e) CCK-8, (f) colony formation, (g) Transwell and (h) TUNEL assays were used to analyze the proliferation, invasion and apoptosis, respectively, of Huh7/DDP and Hep3B/DDP cells transfected with NC mimic, miR-153-3p mimic and miR-153-3p mimic + pcDNA3.1/FGD5-AS1. *P < 0.05
TWF1 is a target of miR-153-3p. Through analyzing StarBase website, we uncovered that TWF1 had binding sites with miR-153-3p (Figure 5a). Moreover, the luciferase activity of TWF1-WT was inhibited in the Huh7/DDP and Hep3B/DDP cells following miR-153-3p addition, but there was no change in TWF1-Mut group (Figure 5b and c). Subsequently, RT-qPCR assay elaborated that TWF1 level was markedly enhanced in DDP-resistant HCC tissues (Figure 5d), and the mRNA and protein levels of TWF1 was uncovered to be elevated in the DDP-resistant HCC cells (Figure 5e and f). The results from the RT-qPCR and western blotting implied that TWF1 level was reduced following miR-153-3p overexpression and was elevated by miR-153-3p inhibition (Figure 5g and h). The results elaborated that miR-153-3p could interact with TWF1 in DDP-resistant HCC cell lines.
Figure 5.
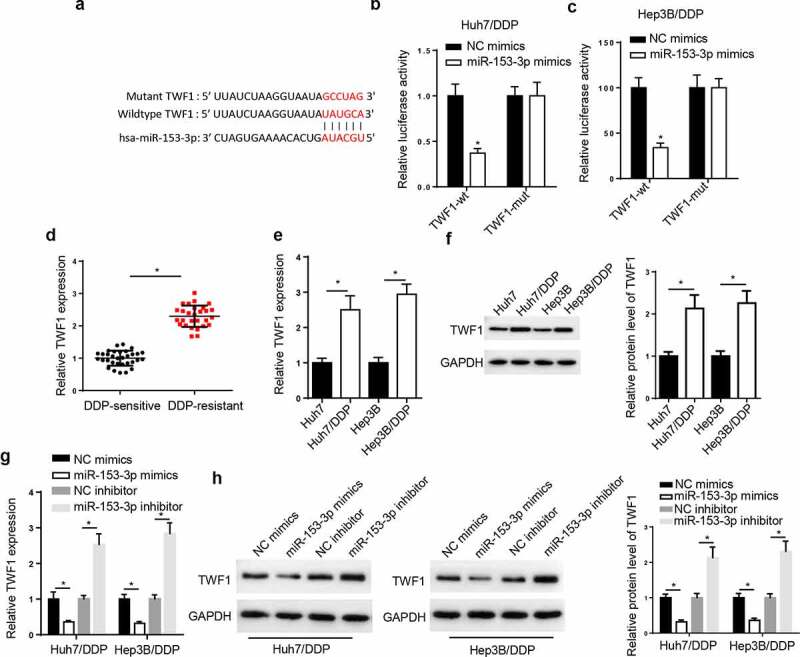
TWF1 is a target of miR-153-3p. (a) StarBase database was used to predict the binding site between TWF1 and miR-153-3p. Dual luciferase reporter assay was used to determine the relative luciferase activity of TWF1-WT or TWF1-Mut in (b) Huh7/DDP and (c) Hep3B/DDP cells co-transfected with NC mimic or miR-153-3p mimic. (d) RT-qPCR was used to analyze relative TWF1 expression in DDP-resistant HCC tissues (n = 32) and DDP-sensitive HCC tissues (n = 28). (e) RT-qPCR and (f) western blotting were used to determine the relative mRNA and protein expression of TWF1, respectively, in Huh7/DDP and Hep3B/DDP cells. (g) RT-qPCR and (h) western blotting were used to determine the relative mRNA and protein expression of TWF1 in Huh7/DDP and Hep3B/DDP cells transfected with NC mimic, miR-153-3p mimic, NC inhibitor and miR-153-3p inhibitor. *P < 0.05
TWF1 knockdown decreases DDP resistance in DDP-resistant HCC cell lines by modulating miR-153-3p. To probe whether TWF1 was involved in miR-153-3p-mediated DDP resistance, the Huh7/DDP and Hep3B/DDP cell lines were transfected with shNC, shTWF1, and shTWF1+ miR-153-3p inhibitor. RT-qPCR exhibited that miR-153-3p silence reduced miR-153-3p level in the DDP-resistant HCC cells (Figure 6a). Moreover, TWF1 knockdown reduced TWF1 mRNA expression, while inhibition of miR-153-3p rescued this effect (Figure 6b). In addition, miR-153-3p deficiency reversed the suppressive impacts of TWF1 silence on the IC50 value of DDP, cell viability, and invasion in the Huh7/DDP and Hep3B/DDP cell lines (Figure 6c-g). On the contrary, the TUNEL assay revealed that the knockdown of TWF1 promoted cell apoptosis, while miR-153-3p knockdown abrogated this effect (Figure 6h). Besides, the results elucidated that the interference with FGD5-AS1 reduced TWF1 mRNA and protein expression, which was counteracted by miR-153-3p inhibition (Figure 6i and j). In summary, TWF1 knockdown impeded DDP resistance in HCC cells by mediating miR-153-3p.
Figure 6.
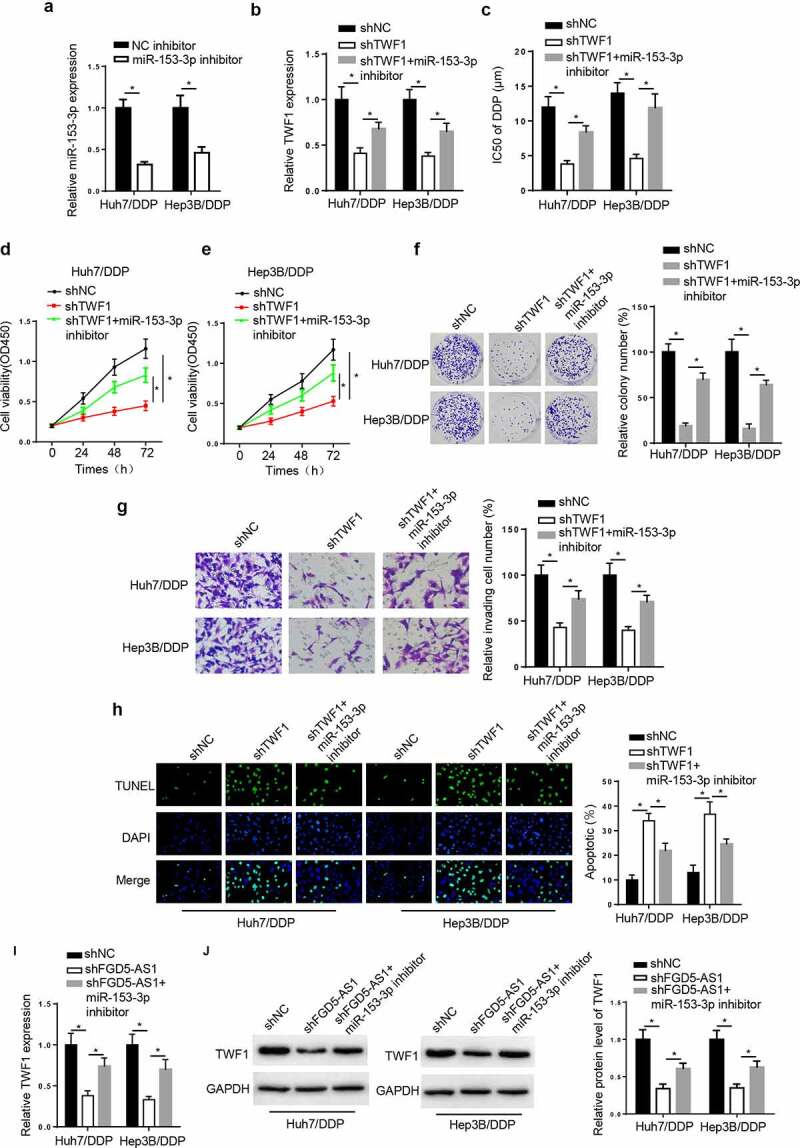
TWF1 knockdown decreases DDP resistance in DDP-resistant hepatocellular carcinoma cells by modulating miR-153-3p expression. (a) RT-qPCR was used to determine relative miR-153-3p expression in DDP-resistant cells transfected with NC inhibitor and miR-153-3p inhibitor. (b) RT-qPCR was used to determine relative TWF1 expression in Huh7/DDP and Hep3b/DDP cells transfected with shNC, shTWF1 or shTWF1 + miR-153-3p inhibitor. (c) CCK-8 assay was used to determine the IC50 value of DDP in DDP-resistant cells transfected with shNC, shTWF1 and shTWF1 + miR-153-3p inhibitor. (d and e) CCK-8, (f) colony formation and (g) Transwell assays were used to examine the proliferation and invasion, respectively, of Huh7/DDP and Hep3B/DDP cells transfected with shNC, shTWF1 and shTWF1 + miR-153-3p inhibitor. (h) TUNEL assays were used to assess the apoptosis of Huh7/DDP and Hep3B/DDP cells transfected with shNC, shTWF1 and shTWF1 + miR-153-3p inhibitor. (i) RT-qPCR and (j) western blotting were used to determine relative TWF1 expression in Huh7/DDP and Hep3B/DDP cells transfected with shNC, shFGD5-AS1 and shFGD5-AS1 + miR-153-3p inhibitor. *P < 0.05
Discussion
Mounting evidence indicated that lncRNAs were associated with drug resistance in various types of cancer [24]. This work is the first to elucidate FGD5-AS1 function in DDP resistance in HCC cells and reveal that FGD5-AS1 modulated TWF1 expression to aggravate DDP resistance in the HCC cell lines by targeting miR-153-3p.
Previous reports have probed that FGD5-AS1 served as an essential role in tumorigenesis and chemoresistance. For example, lncRNA FGD5-AS1 increased the metastasis by increasing the level of cell division cycle associated 7 by sponging miR-302e in colorectal cancer [25]. Overexpression of FGD5-AS1 was associated with DDP resistance by modulating the miR-140-5p/WEE1 axis in non-small cell lung cancer [26]. Herein, it was uncovered that FGD5-AS1 was distinctly increased in DDP-resistant HCC tissues and cells, and interference of FGD5-AS1 restrained the IC50 value of DDP, cell viability and invasion, and facilitated cell apoptosis. Therefore, these results revealed that FGD5-AS1 enhanced DDP resistance in HCC cells, suggesting that FGD5-AS1 might be a therapeutic target for HCC.
MiR-153-3p was previously reported as a tumor suppressor and involved in drug resistance in several types of malignant tumors [27]. For example, miR-153-3p targeted MCL1 apoptosis regulator, BCL2 family member to inhibit ovarian carcinoma progression [28]. miR-153-3p restrained DDP resistance by sponging Nrf-2 in esophageal squamous cell carcinoma [29]. The lncRNA ROR, a regulator of reprogramming modulated ABCB1 to promote DDP resistance in an osteosarcoma cell line via miR-153-3p [30]. The current study observed that miR-153-3p directly bound to FGD5-AS1. miR-153-3p addition repressed DDP resistance, cell viability and invasion, and induced apoptosis, however, FGD5-AS1 overexpression reversed these effects. The aforementioned results revealed that FGD5-AS1 modulated DDP resistance in HCC cell lines by sponging miR-153-3p.
TWF1, an actin-binding protein, is reported to modulate a diverse range of actin dynamics [31]. TWF1 was reported to be aberrantly expressed in multiple cancers. For example, miRNA-30 c reduced pancreatic cancer cell viability by regulating TWF1 [32]. lncRNA SBF2-AS1 acted as a ceRNA to reduce gemcitabine resistance in pancreatic cancer cells via the miRNA-142-3p/TWF1 axis [33]. miRNA-486-5p improved DDP sensitivity via modulating TWF1 in non-small cell lung cancer [34]. Here, we uncovered that miR-153-3p directly targeted TWF1, and TWF1 level was raised in DDP-resistant HCC cell lines. Then, functional assays determined that miR-153-3p deficiency neutralized the suppressive impacts of TWF1 depletion on the IC50 value of DDP, cell viability and invasion. These results manifested that FGD5-AS1 facilitated DDP resistance in HCC cell lines via the miR-153-3p/TWF1 axis. A previous research uncovered IL-11 was a downstream target of TWF1 and TWF1 facilitated breast cancer progression through increasing the level of IL-11 [35]. The failure to identify the downstream mechanism of TWF1 is a limitation to the present study. Therefore, further investigations to reveal the pathway involved in the FGD5-AS1/miR-153-3p/TWF1 axis are required.
Conclusion
The results from the current research revealed that FGD5-AS1 knockdown suppressed DDP resistance in the HCC cells via decreasing TWF1 by sponging miR-153-3p. These findings might provide a promising treatment for overcoming DDP resistance in HCC. In the future study, in vivo experiments should be performed to further investigate the role and mechanisms of FGD5-AS1/miR-153-3p/TWF1 axis in DDP-resistant HCC.
Funding Statement
This study was supported by 2019 high-level health talents ‘Six first Project’ scientific research project (LGY2019022).
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Consent for publication was obtained from the participants.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].El-Serag HB, Rudolph KL.. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. [DOI] [PubMed] [Google Scholar]
- [2].Hernandez-Gea V, Toffanin S, Friedman SL, et al. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Siegel RL, Miller KD, Jemal A.. Cancer statistics. CA Cancer J Clin. 2016;66(2016):7–30. [DOI] [PubMed] [Google Scholar]
- [4].Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. [DOI] [PubMed] [Google Scholar]
- [5].Chen J, Yuan D, Hao Q, et al. LncRNA PCGEM1 mediates oxaliplatin resistance in hepatocellular carcinoma via miR-129-5p/ETV1 axis in vitro. Adv Clin Exp Med. 2021;30(8) [DOI] [PubMed] [Google Scholar]
- [6].Zhao G, Zhang A, Sun S, et al. Long non-coding RNA LINC00173 enhances cisplatin resistance in hepatocellular carcinoma via the microRNA-641/RAB14 axis. Oncol Lett. 2021;21:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen Y, Zhao H, Li H, et al. LINC01234/MicroRNA-31-5p/MAGEA3 Axis Mediates the Proliferation and Chemoresistance of Hepatocellular Carcinoma Cells. Mol Ther Nucleic Acids. 2020;19:168–178. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [8].Liu Z, Dang C, Xing E, et al. Overexpression of CASC2 Improves Cisplatin Sensitivity in Hepatocellular Carcinoma Through Sponging miR-222. DNA Cell Biol. 2019;38:1366–1373. [DOI] [PubMed] [Google Scholar]
- [9].Liu L, Zhan Y, Huang Y, et al. LncRNA FGD5-AS1 can be predicted as therapeutic target in oral cancer. J Oral Pathol Med. 2020;49:243–252. [DOI] [PubMed] [Google Scholar]
- [10].Gao J, Zhang Z, Su H, et al. Long Noncoding RNA FGD5-AS1 Acts as a Competing Endogenous RNA on microRNA-383 to Enhance the Malignant Characteristics of Esophageal Squamous Cell Carcinoma by Increasing SP1 Expression. Cancer Manag Res. 2020;12:2265–2278. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [11].Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- [12].Wang Y, Li CF, Sun LB, et al. microRNA-4270-5p inhibits cancer cell proliferation and metastasis in hepatocellular carcinoma by targeting SATB2. Hum Cell. 2020;33(4):1155-1164. [DOI] [PubMed] [Google Scholar]
- [13].Zhang YM, Wu QM, Chang LY, et al. miR-34a and miR-125a-5p inhibit proliferation and metastasis but induce apoptosis in hepatocellular carcinoma cells via repressing the MACC1-mediated PI3K/AKT/mTOR pathway. Neoplasma. 2020;67(5):1042–1053. [DOI] [PubMed] [Google Scholar]
- [14].Luan W, Shi Y, Zhou Z, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun. 2018;502:22–29. [DOI] [PubMed] [Google Scholar]
- [15].Sun L, Wang H, Jiang J, et al. [miR-153-3p inhibits proliferation and migration of breast cancer cells via down-regulating ROCK1]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2020;36:138–144. [PubMed] [Google Scholar]
- [16].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- [17].Qin S, Ning M, Liu Q, et al. Knockdown of long non-coding RNA CDKN2B-AS1 suppresses the progression of breast cancer by miR-122-5p/STK39 axis. Bioengineered. 2021;12:5125–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Xu Y, Fu Z, Gao X, et al. Long non-coding RNA XIST promotes retinoblastoma cell proliferation, migration, and invasion by modulating microRNA-191-5p/brain derived neurotrophic factor. Bioengineered. 2021;12:1587–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen L, Zhu Q, Lu L, et al. MiR-132 inhibits migration and invasion and increases chemosensitivity of cisplatin-resistant oral squamous cell carcinoma cells via targeting TGF-beta1. Bioengineered. 2020;11:91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang CC, Li Y, Feng XZ, et al. Circular RNA circ_0001287 inhibits the proliferation, metastasis, and radiosensitivity of non-small cell lung cancer cells by sponging microRNA miR-21 and up-regulating phosphatase and tensin homolog expression. Bioengineered. 2021;12:414–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang H, Liu S, Tang L, et al. Long non-coding RNA (LncRNA) MRPL23-AS1 promotes tumor progression and carcinogenesis in osteosarcoma by activating Wnt/β-catenin signaling via inhibiting microRNA miR-30b and upregulating myosin heavy chain 9 (MYH9). Bioengineered. 2021;12:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cui N, Sun Q, Liu H, et al. Long non-coding RNA LINC00511 regulates the expression of microRNA-625-5p and activates signal transducers and activators of transcription 3 (STAT3) to accelerate the progression of gastric cancer. Bioengineered. 2021;12:2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jiang L, Ge W, Cui Y, et al. The regulation of long non-coding RNA 00958 (LINC00958) for oral squamous cell carcinoma (OSCC) cells death through absent in melanoma 2 (AIM2) depending on microRNA-4306 and Sirtuin1 (SIRT1) in vitro. Bioengineered. 2021;12:5085–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Malek E, Jagannathan S, Driscoll JJ. Correlation of long non-coding RNA expression with metastasis, drug resistance and clinical outcome in cancer. Oncotarget. 2014;5:8027–8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li D, Jiang X, Zhang X, et al. Long noncoding RNA FGD5-AS1 promotes colorectal cancer cell proliferation, migration, and invasion through upregulating CDCA7 via sponging miR-302e. In Vitro Cell Dev Biol Anim. 2019;55:577–585. [DOI] [PubMed] [Google Scholar]
- [26].Fu J, Cai H, Wu Y, et al. Elevation of FGD5-AS1 contributes to cell progression by improving cisplatin resistance against non-small cell lung cancer cells through regulating miR-140-5p/WEE1 axis. Gene. 2020;755:144886. [DOI] [PubMed] [Google Scholar]
- [27].Wu X, Ren Y, Yao R, et al. Circular RNA circ-MMP11 Contributes to Lapatinib Resistance of Breast Cancer Cells by Regulating the miR-153-3p/ANLN Axis. Front Oncol. 2021;11:639961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Li C, Zhang Y, Zhao W, et al. miR-153-3p regulates progression of ovarian carcinoma in vitro and in vivo by targeting MCL1 gene. J Cell Biochem. 2019;120:19147–19158. [DOI] [PubMed] [Google Scholar]
- [29].Zuo J, Zhao M, Fan Z, et al. MicroRNA-153-3p regulates cell proliferation and cisplatin resistance via Nrf-2 in esophageal squamous cell carcinoma. Thorac Cancer. 2020;11:738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cheng FH, Zhao ZS, Liu WD. Long non-coding RNA ROR regulated ABCB1 to induce cisplatin resistance in osteosarcoma by sponging miR-153-3p. Eur Rev Med Pharmacol Sci. 2019;23:7256–7265. [DOI] [PubMed] [Google Scholar]
- [31].Ydenberg CA, Johnston A, Weinstein J, et al. Combinatorial genetic analysis of a network of actin disassembly-promoting factors. Cytoskeleton (Hoboken). 2015;72:349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sun LL, Cheng M, Xu XD. MicroRNA-30c inhibits pancreatic cancer cell proliferation by targeting twinfilin 1 and indicates a poor prognosis. World J Gastroenterol. 2019;25:6311–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hua YQ, Zhu YD, Xie GQ, et al. Long non-coding SBF2-AS1 acting as a competing endogenous RNA to sponge microRNA-142-3p to participate in gemcitabine resistance in pancreatic cancer via upregulating TWF1. Aging (Albany NY). 2019;11:8860–8878. [DOI] [PubMed] [Google Scholar]
- [34].Jin X, Pang W, Zhang Q, et al. MicroRNA-486-5p improves nonsmall-cell lung cancer chemotherapy sensitivity and inhibits epithelial-mesenchymal transition by targeting twinfilin actin binding protein 1. J Int Med Res. 2019;47:3745–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bockhorn J, Dalton R, Nwachukwu C, et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.


