Abstract
We sought to determine the relationship between age-related clonal hematopoiesis (CH) and chronic kidney disease (CKD). CH, defined as mosaic chromosome abnormalities (mCA) and/or driver mutations was identified in 5449 (2.9%) eligible UK Biobank participants (n = 190,487 median age = 58 years). CH was negatively associated with glomerular filtration rate estimated from cystatin-C (eGFR.cys; β = −0.75, P = 2.37 × 10–4), but not with eGFR estimated from creatinine, and was specifically associated with CKD defined by eGFR.cys < 60 (OR = 1.02, P = 8.44 × 10–8). In participants without prevalent myeloid neoplasms, eGFR.cys was associated with myeloid mCA (n = 148, β = −3.36, P = 0.01) and somatic driver mutations (n = 3241, β = −1.08, P = 6.25 × 10–5) associated with myeloid neoplasia (myeloid CH), specifically mutations in CBL, TET2, JAK2, PPM1D and GNB1 but not DNMT3A or ASXL1. In participants with no history of cardiovascular disease or myeloid neoplasms, myeloid CH increased the risk of adverse outcomes in CKD (HR = 1.6, P = 0.002) compared to those without myeloid CH. Mendelian randomisation analysis provided suggestive evidence for a causal relationship between CH and CKD (P = 0.03). We conclude that CH, and specifically myeloid CH, is associated with CKD defined by eGFR.cys. Myeloid CH promotes adverse outcomes in CKD, highlighting the importance of the interaction between intrinsic and extrinsic factors to define the health risk associated with CH.
Subject terms: Haematological diseases, Cancer genomics
Introduction
Clonal hematopoiesis (CH) is an age-related phenomenon characterised by a gradual replacement of polyclonal leucocytes by one or more clones marked by somatic mutations [1, 2] or mosaic chromosomal alterations (mCA) [3, 4]. CH is associated with an elevated relative risk of developing haematological malignancies compared to age and sex-matched controls without CH [5], and also an elevated risk of developing non-malignant, immune and inflammatory disorders [6, 7] such as atherosclerotic cardiovascular disease (CVD) [2, 8], chronic obstructive pulmonary disease [9] and premature menopause [10].
Chronic kidney disease (CKD) is a common worldwide health problem defined by low estimated glomerular filtration rate (eGFR) and/or elevated urine albumin to creatinine ratio (uACR) [11]. Patients with CKD experience a gradual and progressive loss of kidney function, but only small minority progress to end-stage kidney disease (ESKD) and require kidney replacement therapy. The majority of cases are at an early stage of the disease process [12], which remains incompletely defined due to variation in eGFR and albuminuria measurements [13–17].
Like CH, CKD is associated with an elevated risk of CVD and mortality [18]. Atherosclerotic risk factors for CVD, such as diabetes, smoking, hypertension and dyslipidemia, are prevalent in individuals with CKD, but there is an excess risk of CVD associated with CKD that is over and above that captured by atherosclerotic risk factors alone. In addition to sharing some risk factors, CH, CKD and CVD are characterised by persistent low-grade inflammation [19–22]. however, a specific relationship between CH and CKD has not been defined. In this study, we sought to assess the relationship between CH and CKD in UK Biobank (UKB), an ongoing, prospective UK cohort study of approximately 500,000 community-dwelling participants aged 40–69 years when recruited between 2006 and 2010.
Methods
Study cohort
UKB participants provided comprehensive demographic, psychosocial and medical information during an initial assessment along with baseline blood and urine samples for genomic, biochemical, and other laboratory tests. Long-term follow-up was provided via linked medical records [23]. We focused on participants with both genome wide single nucleotide polymorphism (SNP) array and whole exome sequence (WES) data (n = 200,361; median age = 58 years, median follow up = 11 years) [24]. All participants provided informed consent according to the Declaration of Helsinki; UKB received ethical approval from the North West multi-centre Research Ethics Committee (REC reference 11/NW/0382).
Identification of CH
We previously described the identification of myeloid, lymphoid or other mCA in UKB from SNP array data [25]. The process for identifying likely somatic driver variants is described in the Supplementary Methods. Mutated genes were defined as myeloid-neoplasia related (‘myeloid’) according to previously published criteria [8], other genes were defined as ‘lymphoid’. The complete list of unique putative somatic driver variants (n = 1611) is shown in Supplementary Table 1. CH was defined as participants with any mCA and/or any somatic driver mutation; myeloid CH was defined as the presence of myeloid mCA and/or a myeloid somatic driver mutation(s); lymphoid was defined by lymphoid mCA and/or lymphoid mutations, without myeloid mutations or myeloid mCA [8].
Kidney function
The eGFR in units of mL/min/1.73 m2 was calculated in R using the Nephro package [26] and three different formulae as defined by the Chronic Kidney Disease Epidemiology Collaboration: creatinine (UKB field: 30700, eGFR.creat), cystatin-C (UKB field: 30720, eGFR.cys) or creatinine and cystatin-C (eGFR.creat.cys) [27]. The creatinine-based scores included ethnicity as recorded in UKB field: 21000. With respect to CKD, patients were considered as healthy (≥90), mild (≥60 and <90), moderate (≥15 and <60) or end stage (<15) for each eGFR score [27]. In addition, uACR in mg/mmol was calculated as a further measure of kidney disease using albumin in urine (UKB field: 30,500) and creatinine in urine (UKB field: 30,510). Shrunken pore syndrome (SPS) is typically defined by an eGFR.cys/eGFR.creat ratio of ≤0.6 in the absence of factors that interfere with cystatin C or creatinine measurement, such as high muscle mass [28]. We defined potential SPS as an eGFR.cys/eGFR.creat ratio of ≤0.6.
Discovery and validation cohorts
To investigate the relationship between CKD and either CH or myeloid neoplasia, the data were split randomly into equally sized discovery and validation cohorts. Results from the discovery and validation cohorts were combined using a fixed effect inverse variance weighted meta-analysis using STATA version 16 (StataCorp LLC, College Station, TX) and Cochran’s Q test to measure heterogeneity.
The relationship between CH and CKD
To study the association between CH and CKD we excluded 10,144 participants due to (i) missing creatinine or cystatin-C data (n = 9913), (ii) any form of ESKD (n = 231) that was either diagnosed before study entry according to relevant ICD10 codes or interventions and procedures as detailed in the Supplementary Methods, or if any of the three eGFR scores was <15. Participants with ESKD were excluded due to the possibility of dialysis and/or erythropoietin treatment that would influence their eGFR scores and blood counts, and because the relationship between ESKD and CVD is well characterised. Individuals with diabetes or hypertension were not excluded. The relationship between CH and CKD was tested using multivariable logistic regression in R where CKD was used as the dependant and CH as a binary predictor. CKD was coded into cases (1) and controls (0) using the eGFR thresholds of <60 or ≥60, respectively [29] and the analysis was repeated for each eGFR score (eGFR.creat, eGFR.cys, and eGFR.creat.cys). Logistic regressions were adjusted for potential confounding variables: age, sex, smoking status, systolic blood pressure, diastolic blood pressure, cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL), body mass index (BMI), glycated haemoglobin (HbA1c) as an indicator of diabetes, high-sensitivity C-reactive protein (hs-CRP) as a marker of inflammation and the first 10 genetic principal components. Effect sizes were reported as odds ratios (OR) with 95% confidence intervals (CI). The relationship between eGFR scores and CH was tested using multivariable linear regression in R where eGFR status was treated as the dependant and CH as a binary predictor and correcting for same confounding variables. UKB did not include follow-up biochemical assessments for the great majority of participants and so incident ESKD was inferred from recorded hospital episodes as indicated above. Prevalent and incident myeloid neoplasia are defined in the Supplementary Methods.
Mendelian randomisation (MR)
MR was used to assess the possibility of a causal relationship between CH and CKD by using germline SNPs associated with the development of CH as instrumental variables. Following the STROBE guidelines [30], we investigated the use of two significance thresholds for selecting instrumental variables based on their association with CH defined by driver somatic mutations in a subset of the TOPMed cohort (n = 65,405 total participants; n = 3831 CHIP cases) [31]. The first used a modest threshold (P < 0.001) to select 380 SNPs with MAF ≥ 0.01 and SNPs clumped (r2 > 0.001, within 10 Mb) for a liberal analysis which aimed to investigate the evidence for a true null relationship. In the second, conservative, analysis we used a stricter threshold (P < 1 × 10–5) to select a subset of 28 SNPs that were strongly associated with CH and would provide more robust evidence of causality. The effect sizes on CKD were obtained from a meta-analysis of 60 GWAS from the CKDgene consortium (n = 625,219, including 64,164 CKD cases) [32]. We estimated that ~2.4% of individuals from the TOPMed cohort are also included in the CKDgene consortium which could inflate false-positive findings [33]. To mitigate against this, we performed a sensitivity analysis using the estimated effect sizes in a subset of patients from the CKDgene cohort with European ancestry (n = 480,698, including 41,395 cases). Detailed information for the SNPs used in both analyses is shown in Supplementary Table 2. MR was performed using the TwoSamplesMR package in R [34] to apply the Robust Adjusted Profile Score (MR-RAPS) methodology which enables the use of weak instrumental variables, is robust to pleiotropy and considers measurement error in the exposure estimate [35]. Additional sensitivity analyses were performed using methods that test the different assumptions of MR, specifically the inverse-variance weighted (IVW) method which performs a meta-analysis for the estimates of the instrumental variants [36], the MR-Egger method which uses the average pleiotropic effect as the intercept that allow the use of instrumental variables with pleiotropic effects [37], and the weighted median method which allows for a subset of instrumental variables to be invalid [38].
Prediction of adverse outcomes
A Cox proportional hazard model (survival package in R) [39] was used to determine if the risk of adverse outcome was associated with CH, CKD defined by each eGFR score or the urine albumin-to-creatinine ratio (uACR). Adverse outcomes were defined by a composite endpoint of either death (UKB data release April 2020), myocardial infarction (MI, field 40002, February 2018) or stroke (field 40006, February 2018). Participants who suffered MI or stroke before entering UKB were excluded. Follow-up times were calculated using the lubridate package [40] to determine the duration between study entry and the earliest of date of death (UKB field 40000), date of MI (UKB field 40002) or date of stroke (UKB field 40006). Patients without an adverse outcome were censored at the date of last follow-up for MI and stroke or the date of they were lost to follow-up (UKB field 191). Univariate survival analyses were performed for all traditional risk factors (age, sex, smoking status, LDL, HDL, cholesterol, HbA1c, BMI, hs-CRP, systolic and diastolic blood pressure). Variables with P < 0.2 were entered into a multivariate survival analysis in a backward stepwise manner and retained if they reached nominal significance (P < 0.05).
To assess the potential for a non-linear relationship between eGFR scores and adverse outcomes, we used a restricted cubic spline function [41] to transform and segment the eGFR scores. Separate curves were fitted to each segment to generate a smooth fitted curve. The method was used to transform each eGFR score using the rms package in R [42] and default values for the number of knots (n = 5) and degrees of freedom (n = 4). The regression included the covariates described above. The adjusted spline values were plotted with 95% CI.
Receiver operating characteristic curves (ROC) and area under the curve (AUC) metrics [43] were used to evaluate the prediction accuracy of the multivariable survival models. AUCs were reported for three pairs of prediction models with and without CH: (i) traditional risk factors, (ii) traditional risk factors and eGFR.cys and (iii) traditional risk factors and uACR. Where relevant, P values for all tests were corrected for multiple testing using the false discovery rate (FDR).
Results
Definition and breakdown on CH in UKB
In a previous analysis of SNP array data from the entire UKB cohort, we identified 8203 mCA larger than 2 Mb in 5040 participants [44]. In the subset of participants with available WES data (n = 200,631), we identified 3085 mCA in 2016 participants, of which 197 (185 participants) were associated with myeloid neoplasms and 278 (237 participants) were associated with lymphoid neoplasms. Analysis of the WES data identified 4137 putative somatic driver mutations (1611 unique variants) in 3863 participants (Supplementary Table 3). In total, 5718 (2.9%) participants had CH defined by one or more mCA and/or driver mutations and 194,913 participants were considered as CH-free controls. For further analysis, these data were split randomly into discovery and validation cohorts (Table 1 and Supplementary Table 4).
Table 1.
CH defined by both acquired mCA and/or driver somatic mutations.
| Participants | Discovery cohort | Validation cohort | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Males | Female | Total | Males | Females | Total | ||||||
| N | % | N | % | N | % | N | % | ||||
| Total number | 45,198 | 45 | 55,118 | 55 | 100,316 | 44,956 | 45 | 55,359 | 55 | 100,315 | 200,631 |
| All CH | 1286 | 45 | 1582 | 55 | 2868 | 1335 | 47 | 1515 | 53 | 2850 | 5718 |
| Myeloid CHa | 831 | 46 | 960 | 54 | 1791 | 885 | 49 | 912 | 51 | 1797 | 3588 |
| Lymphoid CHb | 135 | 48 | 146 | 52 | 281 | 140 | 46 | 167 | 54 | 307 | 588 |
| All mCA | 431 | 42 | 585 | 58 | 1016 | 439 | 44 | 561 | 56 | 1000 | 2016 |
| Myeloid mCA | 48 | 53 | 42 | 47 | 90 | 54 | 57 | 41 | 43 | 95 | 185 |
| Lymphoid mCA | 57 | 50 | 56 | 50 | 113 | 60 | 48 | 64 | 52 | 124 | 237 |
| Other mCA | 326 | 40 | 487 | 60 | 813 | 325 | 42 | 456 | 58 | 781 | 1594 |
| All driver mutations | 894 | 47 | 1027 | 53 | 1921 | 941 | 48 | 1001 | 52 | 1942 | 3863 |
| Myeloid genesc,d | 805 | 46 | 933 | 54 | 1738 | 854 | 49 | 890 | 51 | 1744 | 3482 |
| DNMT3A | 295 | 39 | 470 | 61 | 765 | 348 | 45 | 419 | 55 | 767 | 1532 |
| TET2 | 193 | 46 | 225 | 54 | 418 | 188 | 46 | 217 | 54 | 405 | 823 |
| ASXL1 | 103 | 64 | 59 | 36 | 162 | 92 | 64 | 51 | 36 | 143 | 305 |
| JAK2 | 37 | 58 | 27 | 42 | 64 | 46 | 57 | 35 | 43 | 81 | 145 |
| Other myeloid genes | 220 | 54 | 190 | 46 | 410 | 225 | 52 | 207 | 48 | 432 | 842 |
| Lymphoid genes | 89 | 49 | 94 | 51 | 183 | 87 | 44 | 111 | 56 | 198 | 381 |
| Control (CH-free) | 43,912 | 45 | 53,536 | 55 | 97,448 | 43,621 | 45 | 53,844 | 55 | 97,465 | 194,913 |
a79 participants had both myeloid mutations and myeloid mCA.
bLymphoid CH was defined by lymphoid mCA and/or lymphoid mutations, without myeloid mutations or myeloid mCA. 30 participants had both lymphoid mutations and lymphoid mCA.
c14 participants had both myeloid and lymphoid mutations and were classed as myeloid.
d218 participants had more than one myeloid gene mutation.
Assessment of the relationship between CH and CKD
We compared eGFR.cys, eGFR.creat and eGFR.creat.cys in participants with or without CH after excluding 10,144 ineligible participants with pre-existing ESKD or missing biochemistry measures. After excluding ineligible cases, the discovery cohort consisted of 2735 participants with CH and 92,457 CH-free controls, and the validation cohort compromised of 2714 participants with CH and 92,581 CH-free controls. As expected, the cystatin-C-derived eGFR score was lower than the scores that included creatine [45] and consequently fewer participants were determined to have moderate CKD, defined by an eGFR score between 15 and 60, according to eGFR.creat (n = 4194) and eGFR.creat.cys (n = 4433) compared with eGFR.cys (n = 8304). The median for all three eGFR scores was lower in participants with CH compared to those without CH (Fig. 1) and the median uACR was higher (1.2 with CH versus 1.05 without CH; P < 0.001) indicating impairment of kidney function in association with CH. Participants with lower eGFR scores tended to be older, male, smokers, with low HDL, high LDL, high BMI, high systolic and diastolic blood pressure, and high albuminuria (Supplementary Table 4).
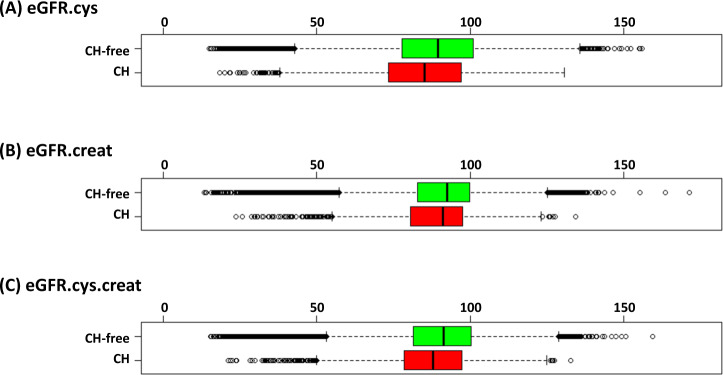
Fig. 1. CH is associated with lower eGFR scores.
Meta-analysis of discovery and validation cohorts (cases with CH, n = 5449; controls without CH, n = 185,038). A eGFR.cys: CH, median = 84.4; CH-free, median = 88.6 (P < 0.001; Mann–Whitney test), B eGFR.creat: CH median = 88.7; CH-free, median = 90.7 (P < 0.001), C eGFR.creat.cys: CH, median = 87.2; CH-free, median = 90.4 (P < 0.001).
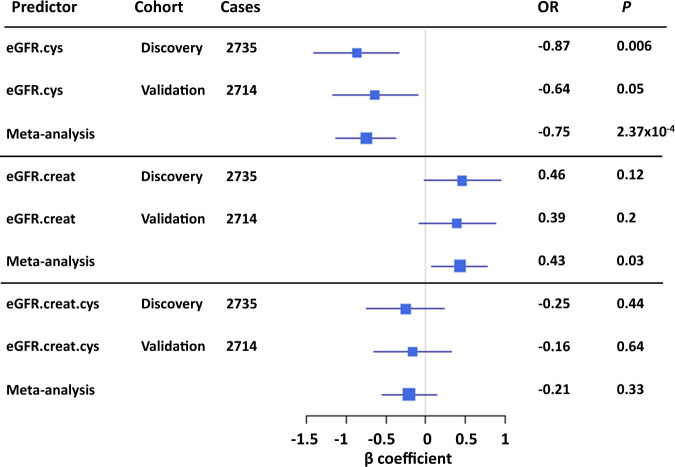
To determine the association between CH and CKD, we performed logistic and linear regression analyses where CKD was coded as either a binary (1 = moderate CKD eGFR > 15 and <60, 0 = eGFR ≥60) or as a continuous trait based on each eGFR score and adjusted for potential confounding variables (Supplementary Table 5). In the logistic models, CH was associated with an increased risk of moderate CKD estimated from cystatin-C scores (eGFR.cys, OR = 1.02 [95% CI: 1.01–1.02], P = 8.44 × 10−8). A weaker association was observed for eGFR.creat.cys (OR = 1.01 [95% CI: 1.00–1.01], P = 0.04) and there was no association with eGFR.creat (OR = 1.00 [95% CI: 0.995–1.004], P = 0.93) (Supplementary Table 6). Similar results were obtained from linear regression analysis where eGFR scores estimated from cystatin-C were negatively associated with CH (eGFR.cys, β = −0.75, P = 2.37 × 10−4) but not eGFR.creat.cys (β = −0.21, P = 0.33), or eGFR.creat (β = 0.43, P = 0.03, not significant in the discovery and validation cohorts) (Fig. 2). For all tests there was no evidence for heterogeneity between the discovery and validation cohorts (P > 0.05, Cochran’s Q test).
Fig. 2. CH is specifically and negatively associated with eGFR estimated from cystatin-C.
eGFR.cys: eGFR estimated from cystatin-C, eGFR.creat: eGFR estimated from creatinine, eGFR.creat.cys: estimated from both creatinine and cystatin-C. Square sizes represent the precision of each eGFR score.
To investigate the relationship between CH and CKD in more detail, we tested the constituent components of CH for association with eGFR.cys as a continuous trait using linear regression. Low eGFR.cys scores were associated with myeloid mCA (β = −4.44, P = 8.90 × 10–5) but not lymphoid mCA (β = −1.7, P = 0.12) or other mCA (β = 0.61, P = 0.15). Alterations involving chr9p were the most strongly associated subtype of myeloid mCA (β = −8.06, P = 8.80 × 10−5). For CH defined by somatic mutations, myeloid neoplasia-associated genes were strongly associated with lower levels of eGFR.cys (β = −1.33, P = 5.52 × 10−7), whereas lymphoid genes were not significant (β = −1.31, P = 0.125). At the gene level, the relationship was significant for CH defined by JAK2 (n = 139, β = −1.03, P < 1 × 10–300) and TET2 (n = 788, β = −1.94, P = 4.50 × 10–4) variants but not DNMT3A or ASXL1. Again, for all tests there was no evidence for heterogeneity between the discovery and validation cohorts (P > 0.05, Cochran’s Q test). Full results for the discovery and validation cohorts are presented in Supplementary Table 7. The median VAF of CH defined by myeloid neoplasia associated genes was higher in participants with CKD (eGFR < 60) defined by eGFR.cys (median VAF = 0.24) compared to other participants (eGFR ≥ 60) (median VAF = 0.21, P = 1.71 × 10–7) but no difference was seen for CKD defined by eGFR.creat (median VAF = 0.23 vs. 0.21, P = 0.12) (Supplementary Fig. 1). At the level of individual genes, a significant difference was only seen for JAK2 with a median VAF of 0.56 in cases with CKD defined by eGFR.cys compared to other participants (VAF = 0.20, P = 4.70 × 10–6).
The link between myeloid neoplasms and reduced kidney function is well established and was replicated in our subset of UKB participants which included 320 participants with a prevalent myeloid neoplasms (diagnosed before or within a year of study entry) that was associated with lower eGFR.cys score (β = −5.22, P = 7.77 × 10–10). Excluding these cases, eGFR.cys was still associated with myeloid CH (n = 3,330, β = −1.05, P = 8.80 × 10–5), including both myeloid mCA (n = 148, β = −3.36, P = 0.01) and myeloid related-genes (n = 3241, β = −1.08, P = 6.25 × 10–5). Stratification at the gene level identified associations between eGFR.cys and mutations in CBL, TET2, JAK2, PPM1D and, to a lesser degree, GNB1 (Table 2; Supplementary Table 8).
Table 2.
Association between myeloid CH subtypes and eGFR.cys score after removing participants with prevalent myeloid neoplasms.
| Predictor | Cases | β | CI 2.5% | CI 97.5% | P |
|---|---|---|---|---|---|
| myeloid CH | 3330 | −1.05 | −1.54 | −0.57 | 8.80 × 10–5 |
| myeloid mCA | 148 | −3.36 | −5.65 | −1.07 | 9.11 × 10–3 |
| myeloid genes | 3241 | −1.08 | −1.57 | −0.60 | 6.25 × 10–5 |
| DNMT3A | 1446 | 0.14 | −0.59 | 0.87 | 0.73 |
| TET2 | 778 | −1.74 | −2.74 | −0.73 | 1.84 × 10–3 |
| ASXL1 | 283 | −1.59 | −3.21 | 0.03 | 0.09 |
| JAK2 | 92 | −4.69 | −7.56 | −1.82 | 3.21 × 10–3 |
| GNB1 | 86 | −3.51 | −6.40 | −0.62 | 0.04 |
| SRSF2 | 67 | −2.42 | −5.94 | 1.11 | 0.27 |
| TP53 | 62 | −0.01 | −3.30 | 3.29 | 1.00 |
| PPM1D | 61 | −5.87 | −9.43 | −2.31 | 3.08 × 10–3 |
| SF3B1 | 52 | −2.37 | −6.27 | 1.52 | 0.32 |
| FLT3 | 36 | −0.65 | −5.49 | 4.19 | 0.81 |
| GNAS | 33 | 1.89 | −2.61 | 6.40 | 0.49 |
| NF1 | 28 | −1.03 | −6.07 | 4.01 | 0.73 |
| CBL | 28 | −12.14 | −17.40 | −6.88 | 3.44 × 10–5 |
| STAG2 | 27 | 3.90 | −1.37 | 9.16 | 0.23 |
| PRPF40B | 26 | 2.96 | −2.29 | 8.21 | 0.35 |
| CREBBP | 24 | −3.10 | −8.49 | 2.30 | 0.34 |
| KDM6A | 22 | −2.68 | −8.49 | 3.14 | 0.45 |
| BRCC3 | 21 | −1.72 | −7.70 | 4.27 | 0.66 |
| IDH2 | 14 | −5.56 | −13.78 | 2.67 | 0.28 |
| KMT2D | 13 | −3.43 | −10.54 | 3.69 | 0.44 |
59 participants had both myeloid mutated genes and mCA.
We assessed the relationship between myeloid CH and the risk of developing ESKD in participants without prevalent myeloid neoplasms or prior ESKD. Myeloid CH (n = 3330) was weakly but significantly associated with ESKD incidence (n = 307, β = 0.002, P = 0.006). Specifically, 0.33% (11 out of 3330) of participants with myeloid CH developed ESKD after study entry compared with 0.16% of controls (296 of 184,811).
MR analysis to test causal effect of CH on kidney function
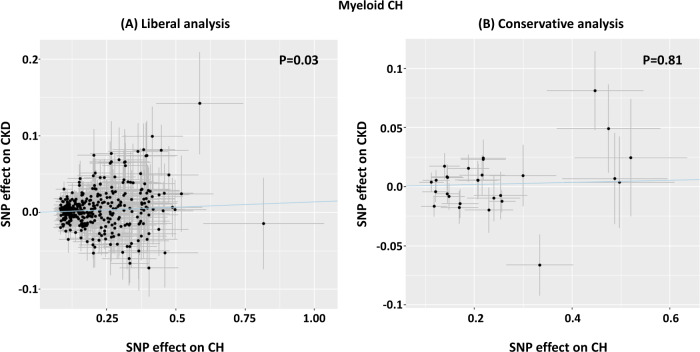
The possibility of a causal relationship between CH and kidney function was assessed using MR. In a liberal analysis, 380 independent SNPs associated with CH at (P < 0.001) [31] were used to estimate the effect of CH on CKD (Supplementary Table 2). To test the different assumptions and scenarios, several MR methods were used as recommended and the results corrected for multiple testing [33]. Only the MR-RAPS method, which is adapted to test weak instrumental variables as applicable to our study, identified a positive causal relationship [OR = 1.01; P = 0.029]. However, this relationship failed to reach significance (P = 0.81) in a more conservative analysis that applied stricter threshold (P < 1 × 10–5) to select 28 SNPs associated with CH (Fig. 3). Due to the potential limited overlap between cohorts used to select instrumental variables, we performed a sensitivity analysis using a subset of samples with European American ancestry which yielded similar results for the causal association between CH and CKD [OR = 1.02; P = 0.029]. Detailed results are presented in Supplementary Table 9.
Fig. 3. Mendelian randomisation using robust adjusted profile score (MR-RAPS) to estimate the effect of SNPs associated with CH against their effect in relation to CKD.
A Liberal analysis using 380 independent SNPs associated with CH at P < 0.001. The MR-RAPS test estimated a significant positive effect of CH on CKD (OR = 1.014, CI 95%:1.003–1.024; P = 0.03). B Conservative analysis using 28 SNPs associated with CH at P < 1 × 10−5. The line of regression is indicated in blue and the axes show β coefficients for SNP effects on CH and CKD.
Prediction of adverse outcomes by myeloid CH in CKD
As expected, established risk factors (myeloid CH, age, sex, ethnicity, smoking status, cholesterol, HbA1C, HDL, LDL, blood pressure, BMI, uACR, hs-CRP and eGFR scores) were associated on univariate analysis with an adverse outcome as defined by a composite endpoint of death, MI, or stroke (Supplementary Table 10).
To understand the influence of myeloid CH and CKD on adverse outcomes, we focused on participants without prevalent myeloid neoplasms (n = 320) or any prior history of CVD (n = 8459). Initially, Cox proportional-hazard analysis was used to identify risk factors unrelated to CH and CKD (Supplementary Table 11), and then these factors were added into the model. To determine which of the three eGFR scores was most appropriate to use in the model, we tested the linearity of each score in relation to outcome using a restricted cubic spline test, as described previously [45]. Although all three scores were associated with adverse outcomes, eGFR.cys was more linear and negative compared to the scores that used creatinine in both the discovery and validation cohorts (Supplementary Fig. 2). Focusing on eGFR.cys, the risk of adverse outcomes was higher in subjects who had CKD (HR = 1.9, n = 1180/6970) compared to CKD free participants (n = 8295/172,857; P = 8.4 × 10–65) (Supplementary Table 12). The risk of adverse outcomes was estimated to be 1.56-fold higher (P = 1.4 × 10–11) in cases with myeloid CH (n = 338/3078) compared to myeloid CH-free participants (n = 9137/176,749). Testing each component of adverse outcomes confirmed the previously reported features of UKB cohort [46] that CH was associated with all-cause mortality (HR = 1.91, P = 2.5 × 10–10) but did not reach significance for MI (HR = 1.13, P = 0.38) or stroke (HR = 1.28, P = 0.15) considered independently, in accordance with previous findings [44, 47] (Supplementary Table 12).
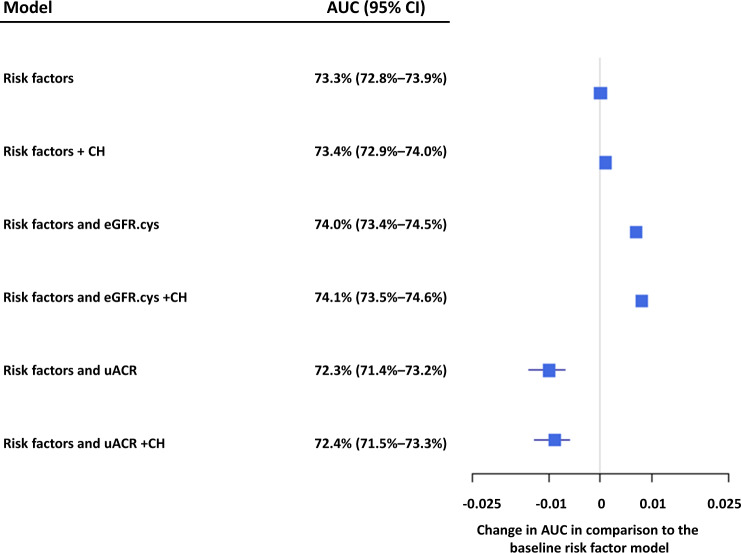
ROC analysis was used to assess the predictiveness of multivariable models that incorporated myeloid CH, eGFR.cys and uACR. The baseline model consisting of age, sex, smoking status, HDL, HbA1c, systolic blood pressure, hs-CRP, BMI (Supplementary Table 12) and corrected for 10 genetic principal components had an AUC of 73.3% (72.8–73.9%). The addition of myeloid CH as a binary factor or eGFR.cys as a continuous trait improved the predictiveness of the model to an AUC of 73.4% and 74%, respectively, and including both further improved the AUC to 74.1% (73.5–74.6%), with very similar results achieved in both the discovery and validation cohorts (Fig. 4, Supplementary Table 13).
Fig. 4. Risk factors for adverse outcome.
The baseline risk factors included age, sex, smoking status, HDL, HbA1c, systolic blood pressure, hs-CRP, BMI and was corrected for 10 genetic principal components. The effect on AUC of adding in CH, eGFR.cys, and uACR relative to the baseline model is shown (meta-analysis of discovery and validation cohorts).
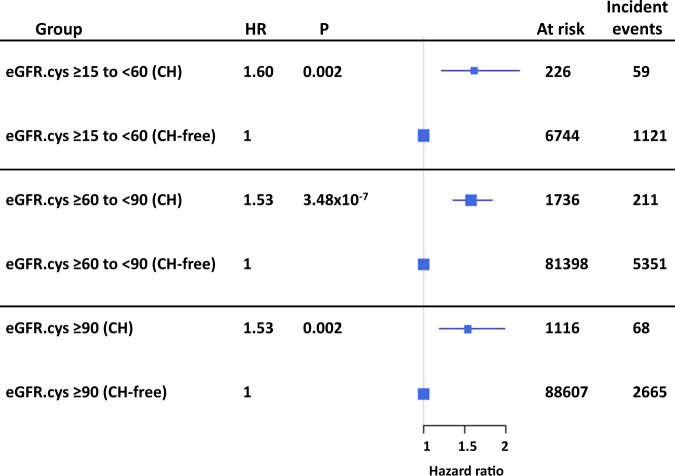
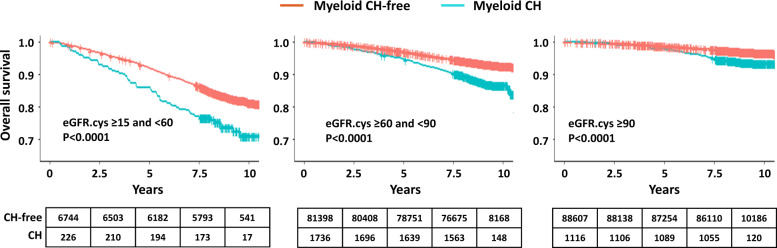
To further investigate the relationship between CH and adverse outcome in participants with CKD, we stratified the cohort (excluding prior CVD and prevalent myeloid malignancies), into participants with moderate renal impairment (eGFR.cys ≥15 to <60), mild impairment (eGFR.cys ≥60 to <90) and normal kidney function (eGFR.cys ≥90). We then tested the effect of CH in each subset using Kaplan–Meier survival analysis. CH increased the risk of adverse outcome in all groups but was particularly marked (HR = 1.6, 95% CI 1.2–2.14, P = 0.002) for participants with moderate CKD (n = 59/226 with myeloid CH compared to n = 1121/6744 without myeloid CH) (Figs. 5 and 6; Supplementary Table 14). Much of the risk of adverse outcomes was related to incident myeloid neoplasms which were diagnosed in 19 participants at a median of 3.6 years after study entry. Of these, 11 (58%) had adverse outcomes in comparison to 48/207 (23%) who did not develop a myeloid neoplasm during the study period. Excluding the incident cases reduced but did not eliminate the risk of adverse outcomes (HR = 1.4, P = 0.05).
Fig. 5. Myeloid CH predicts adverse outcomes in CKD.
The forest plots show data stratified according to eGFR.cys as healthy (≥90), mild CKD (≥60 to <90) and moderate CKD (≥15 to <60). The risk of adverse outcomes was predicted by myeloid CH in all groups but was particularly marked (HR = 1.6, P = 0.002) for participants with moderate CKD.
Fig. 6. Kaplan–Meier survival estimates for the three CKD groups according to absence or presence of myeloid CH.
Log-rank test P values are reported for each group, and numbers at risk at 0, 2.5, 5, 7.5, and 10 years after study entry.
Relationship between myeloid CH and SPS
We identified 966 (0.5%) UKB participants with potential SPS (eGFR.cys/eGFR.creat ratio ≤0.6). Of these, 58 (6.0%) had myeloid CH compared to 2.9% (n = 5391) of participants with eGFR.cys/eGFR.creat ratio > 0.6 (OR = 2.2, 95% CI = 1.6–2.9; P = 2.9 × 10–7 Fisher’s exact test), but after eliminating these cases myeloid CH was still associated with an adverse prognosis in CKD (HR = 1.61, 95% CI 1.17–2.21, P = 0.003) and remained most pronounced for participants with moderate renal impairment (Supplementary Fig. 3).
Discussion
In this study we identified that CH, and specifically myeloid CH, is associated with CKD. The association was not seen with all markers of CH and, strikingly, not with mutations in DNMT3A or ASXL1, two of the most common drivers of clonality, although there was an overall association with clone size. These findings confirm previous observations that that not all CH is equal [9, 25, 31], as well as the importance of having sufficiently large studies to understand the granularity of CH with respect to clinical outcomes.
We found that myeloid CH is specifically associated with eGFR.cys but not eGFR.creat and only marginally with eGFR.cys.creat. Similarly, recent studies have reported the superior utility of eGFR.cys in predicting the incidence of CVD and mortality in patients with CKD [45, 48, 49]. In the UK, the cost to measure cystatin C is 10-fold higher than that to measure serum creatinine, and consequently eGFR.creat is widely used for initial assessment of possible CKD. Although eGFR.cys is recommended to confirm CKD, this is not believed to be common practice, at least in the UK [45]. Our findings provide further weight to the argument that eGFR.cys is more informative than eGFR.creat to define CKD.
The finding that myeloid CH is associated with eGFR.cys also provides further evidence for the importance of chronic inflammation in CH-related disorders. Levels of cystatin C correlate generally with oxidative stress and inflammation [45, 50], a well-recognized feature of CKD [19] that is also associated with an elevated risk of development of CVD [51, 52]. Other biomarkers of chronic inflammation have been associated with CH, e.g. C-reactive protein and IL-6. [20, 31] CH predisposes to haematological malignancies, particularly myeloid neoplasms [5], and both CKD and chronic inflammation have been described as features of myeloproliferative neoplasms [53, 54]. Our data show that myeloid CH increases the risk of adverse outcomes in the context of CKD and that this increase is only partly explained by incident myeloid neoplasms or SPS, a recently described phenomenon that may be observed in both children or adults with normal or reduced eGFR and is associated with increased mortality and morbidity in a variety of settings [28]. Although our analysis was corrected for hs-CRP, it is possible that part of the increase in adverse outcomes is due to chronic inflammation induced by CH.
MR uses genetic variation as a natural experiment to estimate causality in observational data [33] and has, for example, been used to detect a causal effect of cystatin C on risk of stroke [55]. Our initial analysis of 380 SNPs that predispose to CH provided suggestive evidence for a causal relationship between CH and CKD (P = 0.03), but this link was not supported by a more conservative analysis of 28 SNPs that are more strongly associated with CH. Given that two of the most common CH genes (DNMT3A and ASXL1) were not associated with CKD, and that the 380 SNPs only explain 3.6% of the heritability of CH [31], the use of MR in this context is clearly challenging, and may be compounded the possibility of other factors such as horizontal pleiotropy but these concerns are partly mitigated by the large sample size of the GWAS used for CH and CKD.
In summary, the role of CH in the pathogenesis of benign diseases varies widely and depends on intrinsic factors that define the clone as well as extrinsic factors that impact the inflammatory environment [25, 56]. In this study, we have shown that CH is associated with CKD and confers an adverse prognosis over and above conventional risk factors for this common disorder. Our findings suggest that screening for CH in CKD may be of clinical value to help predict outcomes.
Supplementary information
Acknowledgements
AAZD was supported by a Lady Tata International Award; NCPC and WJT were supported by Blood Cancer UK. The authors acknowledge the use of the IRIDIS High Performance Computing Facility and associated support services at the University of Southampton.
Author contributions
The study was designed by AAZD, WJT and NCPC. AAZD performed the data analysis in conjunction with WJT. All four authors refined the analysis and contributed to writing the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: William J. Tapper, Nicholas C.P. Cross.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-021-01382-3.
References
- 1.Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–87. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–98. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zekavat SM, Lin S-H, Bick AG, Liu A, Paruchuri K, Wang C, et al. Hematopoietic mosaic chromosomal alterations increase the risk for diverse types of infection. Nat. Med. 2021;27:1012–24. [DOI] [PMC free article] [PubMed]
- 7.Bick AG, Popadin K, Thorball CW, Uddin MM, Zanni M, Yu B, et al. Increased CHIP prevalence amongst people living with HIV. MedRxiv: 11.06.20225607 [Preprint]. 2020.
- 8.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–21. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buscarlet M, Provost S, Zada YF, Barhdadi A, Bourgoin V, Lépine G, et al. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130:753–62. doi: 10.1182/blood-2017-04-777029. [DOI] [PubMed] [Google Scholar]
- 10.Honigberg MC, Zekavat SM, Niroula A, Griffin GK, Bick AG, Pirruccello JP, et al. Premature menopause, clonal hematopoiesis, and coronary artery disease in postmenopausal women. Circulation. 2021;143:410–23. doi: 10.1161/CIRCULATIONAHA.120.051775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jha V, Garcia-Garcia G, Iseki K, Li Z, Naicker S, Plattner B, et al. Chronic kidney disease: global dimension and perspectives. Lancet. 2013;382:260–72. doi: 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- 12.Hamm LL, McCullough PA, Kasiske BL, Kelepouris E, Klag MJ, Parfrey P, et al. Kidney disease as a risk factor for development of cardiovascular disease. Circulation. 2003;108:2154–69. doi: 10.1161/01.CIR.0000095676.90936.80. [DOI] [PubMed] [Google Scholar]
- 13.Levey AS, Coresh J, Bolton K, Culleton B, Harvey KS, Ikizler TA, et al. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(2 Suppl 1):S1–266. [PubMed]
- 14.Group KDIGOCW. KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int Suppl. 2013;3:1–150. doi: 10.1038/ki.2013.243. [DOI] [PubMed] [Google Scholar]
- 15.Israni A, Snyder J, Skeans M, Peng Y, Maclean J, Weinhandl E, et al. Predicting coronary heart disease after kidney transplantation: Patient Outcomes in Renal Transplantation (PORT) Study. Am J Transplant. 2010;10:338–53. doi: 10.1111/j.1600-6143.2009.02949.x. [DOI] [PubMed] [Google Scholar]
- 16.Gansevoort RT, Correa-Rotter R, Hemmelgarn BR, Jafar TH, Heerspink HJL, Mann JF, et al. Chronic kidney disease and cardiovascular risk: epidemiology, mechanisms, and prevention. Lancet. 2013;382:339–52. doi: 10.1016/S0140-6736(13)60595-4. [DOI] [PubMed] [Google Scholar]
- 17.Kasiske BL, Guijarro C, Massy ZA, Wiederkehr MR, Ma JZ. Cardiovascular disease after renal transplantation. J Am Soc Nephrol. 1996;7:158–65. doi: 10.1681/ASN.V71158. [DOI] [PubMed] [Google Scholar]
- 18.Levey AS, Coresh J. Chronic kidney disease. Lancet. 2012;379:165–80. doi: 10.1016/S0140-6736(11)60178-5. [DOI] [PubMed] [Google Scholar]
- 19.Oberg BP, McMenamin E, Lucas F, McMonagle E, Morrow J, Ikizler T, et al. Increased prevalence of oxidant stress and inflammation in patients with moderate to severe chronic kidney disease. Kidney Int. 2004;65:1009–16. doi: 10.1111/j.1523-1755.2004.00465.x. [DOI] [PubMed] [Google Scholar]
- 20.Busque L, Sun M, Buscarlet M, Ayachi S, Zada YF, Provost S, et al. High-sensitivity C-reactive protein is associated with clonal hematopoiesis of indeterminate potential. Blood Adv. 2020;4:2430. doi: 10.1182/bloodadvances.2019000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hojs R, Ekart R, Bevc S, Hojs N. Markers of inflammation and oxidative stress in the development and progression of renal disease in diabetic patients. Nephron. 2016;133:159–62. doi: 10.1159/000447434. [DOI] [PubMed] [Google Scholar]
- 22.Mihai S, Codrici E, Popescu ID, Enciu AM, Albulescu L, Necula LG, et al. Inflammation-related mechanisms in chronic kidney disease prediction, progression, and outcome. J Immunol Res. 2018;2018:2180373. doi: 10.1155/2018/2180373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szustakowski JD, Balasubramanian S, Kvikstad E, Khalid S, Bronson PG, Sasson A, et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat. Genet. 2021;53:942–8. [DOI] [PubMed]
- 25.Dawoud AAZ, Tapper WJ, Cross NCP. Clonal myelopoiesis in the UK Biobank cohort: ASXL1 mutations are strongly associated with smoking. Leukemia. 2020;34:2660–72. doi: 10.1038/s41375-020-0896-8. [DOI] [PubMed] [Google Scholar]
- 26.Pattaro C, Riegler P, Stifter G, Modenese M, Minelli C, Pramstaller PP. Estimating the glomerular filtration rate in the general population using different equations: effects on classification and association. Nephron Clin Pract. 2013;123:102–11. doi: 10.1159/000351043. [DOI] [PubMed] [Google Scholar]
- 27.Levey AS, Stevens LA, Schmid CH, Zhang Y, Castro AF, III, Feldman HI, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–12. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grubb A. Shrunken pore syndrome—a common kidney disorder with high mortality. Diagnosis, prevalence, pathophysiology and treatment options. Clin Biochem. 2020;83:12–20. doi: 10.1016/j.clinbiochem.2020.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Kdigo A. Work Group. KDIGO clinical practice guideline for acute kidney injury. Kidney Int Suppl. 2012;2:1–138. [Google Scholar]
- 30.Smith GD, Davies NM, Dimou N, Egger M, Gallo V, Golub R, et al. STROBE-MR: guidelines for strengthening the reporting of Mendelian randomization studies: PeerJ Preprints; 2019. Report no. 2167-9843.
- 31.Bick AG, Weinstock JS, Nandakumar SK, Fulco CP, Bao EL, Zekavat SM, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature. 2020;586:763–8. doi: 10.1038/s41586-020-2819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet. 2019;51:957. doi: 10.1038/s41588-019-0407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess S, Smith GD, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2019;4. [DOI] [PMC free article] [PubMed]
- 34.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann Stat. 2020;48:1742–69. [Google Scholar]
- 36.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658–65. doi: 10.1002/gepi.21758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowden J, Davey, Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Therneau TM, Grambsch PM. The Cox Model. In: Therneau TM, Grambsch PM (eds). Modeling Survival Data: Extending the Cox Model. Springer New York: New York, NY, 2000, pp 39–77.
- 40.Grolemund G, Wickham H. Dates and times made easy with lubridate. J Stat Softw. 2011;40:1–25. [Google Scholar]
- 41.Croxford R Restricted cubic spline regression: a brief introduction. Toronto: Institute for Clin Eval Sci 2016:1–5.
- 42.Harrell Jr FE. Regression modeling strategies: with applications to linear models, logistic and ordinal regression, and survival analysis. Springer, Cham;; 2015.
- 43.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997;30:1145–59. [Google Scholar]
- 44.Dawoud AAZ, Tapper WJ, Cross NCP. Clonal myelopoiesis in the UK Biobank cohort: ASXL1 mutations are strongly associated with smoking. Leukemia. 2020;34:2660–72. [DOI] [PubMed]
- 45.Lees JS, Welsh CE, Celis-Morales CA, Mackay D, Lewsey J, Gray SR, et al. Glomerular filtration rate by differing measures, albuminuria and prediction of cardiovascular disease, mortality and end-stage kidney disease. Nat Med. 2019;25:1753–60. doi: 10.1038/s41591-019-0627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bick AG, Pirruccello JP, Griffin GK, Gupta N, Gabriel S, Saleheen D, et al. Genetic Interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141:124–31. doi: 10.1161/CIRCULATIONAHA.119.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pedersen KM, Çolak Y, Ellervik C, Hasselbalch HC, Bojesen SE, Nordestgaard BG. Loss-of-function polymorphism in IL6R reduces risk of JAK2V617F somatic mutation and myeloproliferative neoplasm: A Mendelian randomization study. EClinicalMedicine 2020;21:100280. [DOI] [PMC free article] [PubMed]
- 48.Grubb A. Cystatin C is indispensable for evaluation of kidney disease. eJIFCC. 2017;28:268–76. [PMC free article] [PubMed] [Google Scholar]
- 49.Nowak C, Ärnlöv J. Kidney disease biomarkers improve heart failure risk prediction in the general population. Circulation. 2020;13:e006904. doi: 10.1161/CIRCHEARTFAILURE.120.006904. [DOI] [PubMed] [Google Scholar]
- 50.Zi M, Xu Y. Involvement of cystatin C in immunity and apoptosis. Immunol Lett. 2018;196:80–90. doi: 10.1016/j.imlet.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiner DE, Tighiouart H, Elsayed EF, Griffith JL, Salem DN, Levey AS, et al. The relationship between nontraditional risk factors and outcomes in individuals with stage 3 to 4 CKD. Am J Kidney Dis. 2008;51:212–23. doi: 10.1053/j.ajkd.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zimmermann J, Herrlinger S, Pruy A, Metzger T, Wanner C. Inflammation enhances cardiovascular risk and mortality in hemodialysis patients. Kidney Int. 1999;55:648–58. doi: 10.1046/j.1523-1755.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- 53.Christensen AS, Møller JB, Hasselbalch HC. Chronic kidney disease in patients with the Philadelphia-negative chronic myeloproliferative neoplasms. Leuk Res. 2014;38:490–5. doi: 10.1016/j.leukres.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Koschmieder S, Chatain N. Role of inflammation in the biology of myeloproliferative neoplasms. Blood Rev. 2020;42:100711. doi: 10.1016/j.blre.2020.100711. [DOI] [PubMed] [Google Scholar]
- 55.Sinnott-Armstrong N, Tanigawa Y, Amar D, Mars N, Benner C, Aguirre M, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021;53:185–94. [DOI] [PMC free article] [PubMed]
- 56.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science 2019;366. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.