Abstract
SARS-CoV-2 infects less than 1% of cells in the human body, yet it can cause severe damage in a variety of organs. Thus, deciphering the non-cell-autonomous effects of SARS-CoV-2 infection is imperative for understanding the cellular and molecular disruption it elicits. Neurological and cognitive defects are among the least understood symptoms of COVID-19 patients, with olfactory dysfunction being their most common sensory deficit. Here, we show that both in humans and hamsters, SARS-CoV-2 infection causes widespread downregulation of olfactory receptors (ORs) and of their signaling components. This non-cell-autonomous effect is preceded by a dramatic reorganization of the neuronal nuclear architecture, which results in dissipation of genomic compartments harboring OR genes. Our data provide a potential mechanism by which SARS-CoV-2 infection alters the cellular morphology and the transcriptome of cells it cannot infect, offering insight to its systemic effects in olfaction and beyond.
Keywords: COVID-19, anosmia, nuclear architecture
Graphical abstract
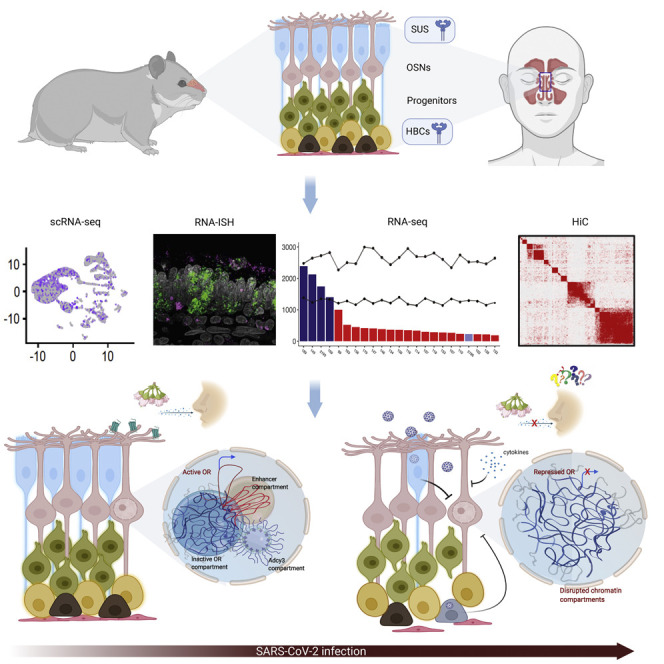
SARS-CoV-2 induces non-cell-autonomous effects in olfactory epithelium that disrupts nuclear architecture and downregulates olfactory receptor expression in olfactory sensory neurons.
Introduction
Neurological symptoms in COVID-19 patients have immense importance due to their role in exacerbating initial disease presentation and their persistence (Chippa et al., 2021; Ellul et al., 2020; Proal and VanElzakker, 2021). Anosmia emerged as one of the most common and yet heterogeneous neurological symptoms (Nalbandian et al., 2021). Early studies correlated higher propensity for acute olfactory loss with a more indolent course, but subsequent work suggested elevated prevalence of smell loss across most COVID-19 cases (Garrigues et al., 2020; Graham et al. 2021). Usually, smell loss is transient, with patients recovering over 6 weeks. However, for ∼10% of patients, this resolution is elusive, resulting in persistent olfactory dysfunction (Boscolo-Rizzo et al., 2022, 2021b; Butowt and von Bartheld, 2020; Gerkin et al., 2020; Hornuss et al., 2020; Luers et al., 2020; Tong et al., 2020). Although olfactory deficits are common in upper respiratory infections, these symptoms are accompanied by rhinorrhea and nasal congestion that insulate olfactory sensory neurons (OSNs) from odorants. In contrast, anosmia in COVID-19 is independent from conductive interference. Thus, the association between COVID-19 and anosmia raises mechanistic questions, as OSNs do not express host cell entry proteins (Bilinska et al., 2020; Brann et al., 2020; Chen et al., 2020), and they are not infected by SARS-CoV-2 (Khan et al., 2021).
To gain insight into COVID-19-induced anosmia, we explored the consequences of SARS-CoV-2 infection in hamster and human autopsies of the olfactory epithelium (OE). Experiments in hamsters revealed transient recruitment of various immune cells to the OE and rapid upregulation of antiviral genes in OSNs. Further, scRNA-seq revealed preferential SARS-CoV-2 infection and transient depletion of sustentacular (SUS) cells, followed by their restoration by day 10 post-infection (dpi). Although we do not detect OSN depletion, we report significant downregulation of olfactory receptor (OR) genes and of key genes of the OR signaling pathway. OR gene downregulation is preceded by rapid and persistent reorganization of nuclear architecture and disruption of genomic OR compartments. Analysis of human OE autopsies confirmed that SARS-CoV-2 infection correlates with significant decrease of OR and OR signaling gene transcription and reduction of interchromosomal OR contacts. Effects of SARS-CoV-2 infection in nuclear architecture are non-cell autonomous and can be induced by neutralized serum from SARS-CoV-2 infected hamsters. Our data provide a potential explanation for the neurological symptoms caused by a virus with no tropism for neurons.
Results
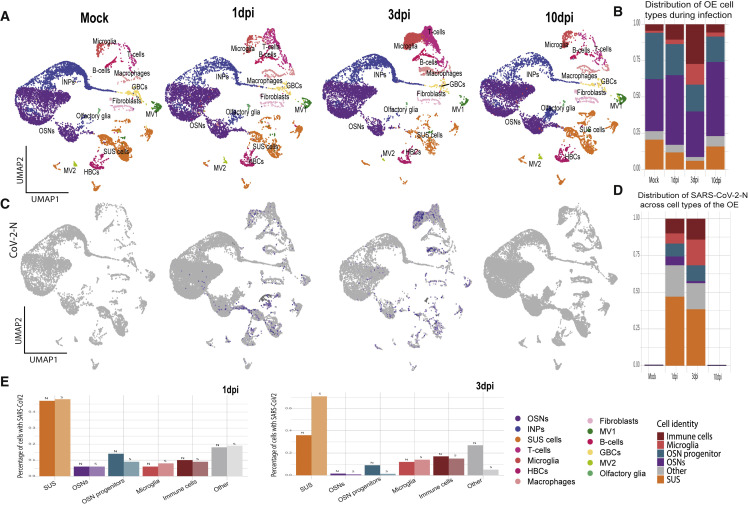
To explore mechanisms of COVID-19-induced anosmia, we infected golden hamsters (M. auratus) with SARS-CoV2 and monitored changes over a period of 10 days post-infection (dpi) by scRNA-seq. This rodent species is a good animal model for SARS-CoV-2 infection owing to sequence homology between hamster and human ACE2 and similarity in pathogenesis and immunological responses (Cleary et al., 2020; Hoagland et al., 2021; Imai et al., 2020; Sia et al., 2020). We performed scRNA-seq at mock and SARS-CoV-2-infected OEs at 1, 3, and 10 dpi. We analyzed a total of 68,951 cells and identified 13 cell types (Figure S1 A) using previously described markers (Durante et al., 2020; Fletcher et al., 2017). We detect a decrease of SUS cells at 1 dpi that is exaggerated at 3 dpi (Figures 1A and 1B), when SUS representation decreases from 20.6% in mock-infected hamsters to 6% at 3 dpi. SUS diminution coincides with increase of microglia and other immune cells (Figures 1A and 1B). Both SUS and microglia return to pre-infection representation in the hamster OE by 10 dpi (Figures 1A and 1B). In contrast, OSN representation is stable throughout the infection (Figures 1A and 1B).
Figure S1.
SARS-CoV-2 infects hamster OSNs very infrequently, related to Figure 1
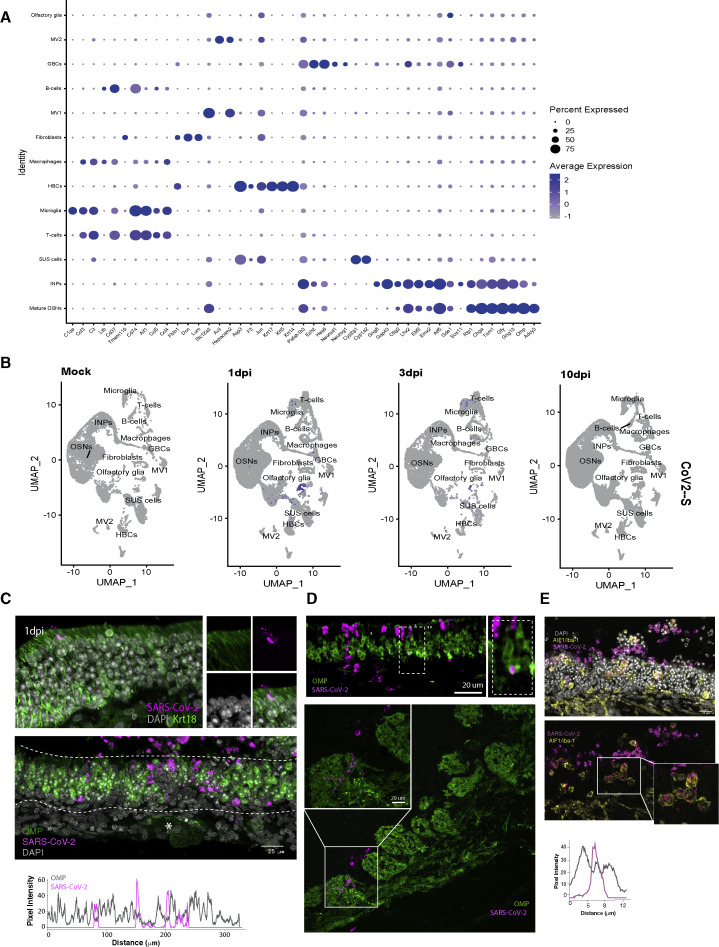
(A) Dot plot showing expression of cell markers across clusters. Cell types are listed on y axis showing expression of 45 selected genes identified by log fold change; genes are listed along x axis. Dot size reflects percentage of cells in a cluster expressing each gene, and dot color represents expression level. The plot shows clusters from 68,951 combined cells extracted from eight OEs with two biological replicates per condition.
(B) Feature plot depicting expression of S SARS-CooV-2 transcript in hamster olfactory epithelium. Cell types are same as in Figure 1A (n = 2 biological replicates for each sample).
(C) Representative confocal micrograph of IF-FISH experiment labeling RNA-FISH SARS-CoV-2 (magenta) and OMP protein (green) in hamster OE at 4 dpi. Rarely OMP-positive cells colocalize with SARS-CoV-2. No viral particles are detected in the axon bundles (asterisk). The line intensity scan drawn at the center of the OE section shows a discrete distribution of the pixel intensity of the two channels.
(D) Top, IF-FISH confocal micrograph of OE tissue section showing colocalization of RNA FISH signal of SARS-CoV-2 (magenta) and antibody staining for OMP (green), suggesting that a small percentage of OSNs is infected. Bottom, IF-FISH in hamster OB section at 4 dpi shows rare RNA-FISH SARS-CoV-2 (magenta) detected in proximity to glomeruli labeled by OMP protein (green).
(E) RNA-FISH SARS-CoV-2 signal (magenta) in the OE and lamina propria strongly correlates with the microglia marker AIF1/Iba1 (yellow), as highlighted by the enlarged picture in the white box. The line intensity scan of one single cell shows that SARS-CoV-2 signal is correlated with AIF1/Iba1 suggesting engulfment of viral particles by microglia.
Figure 1.
SARS-CoV-2 infects and transiently depletes hamster sustentacular cells
(A) UMAP plots of hamster OEs for mock- and SARS-CoV-2-infected hamsters at 1, 3, and 10 dpi. See also Figure S1A for the distribution of cell-specific markers.
(B) Representation of cell types in mock- and SARS-CoV-2-infected hamster OEs at 1, 3, and 10 dpi. HBCs, GBCs, and INPs combined as OSN progenitor cells, macrophages, T cells, B cells combined as immune cells, MV1, MV2, olfactory glia, and fibroblasts combined as other.
(C) Feature plot showing expression of N SARS-CoV-2 transcript in hamster OEs. See also Figure S1B for expression of S SARS-CoV-2 transcript.
(D) Bar charts depicts proportion of SARS-CoV-2 N transcript across the cell types of the OE. Color code of cell types same as B. See also Figures S1C and S1E for histological confirmation of SARS-CoV-2 tropism.
(E) Percentage of cells with N and S transcripts in the annotated clusters (HBCs, GBCs, and INPs combined as OSN progenitor cells; macrophages, T cells, B cells combined as immune cells; MV1, MV2, olfactory glia, and fibroblasts combined as other) at 1 and 3 dpi. Each panel (A–E) represents two combined biological replicates, per condition.
At 1 and 3 dpi, we detect the viral RNA in ∼5% of the cells, followed by complete elimination of the virus at 10 dpi. At 1 dpi, 47% of the total infected cells are SUS, and ∼40% of which are infected (Figures 1C–1E and S1B). In contrast, only 6% of the infected cells are OSNs (Figures 1C, 1E, and S1B). At 3 dpi, microglia and other immune cells also frequently contain SARS-CoV-2 transcripts (Figures 1C–1E and S1B). Consistent with this, we detect colocalization of spike protein with Krt18 and with Aif1/lba-1, SUS and microglia markers, respectively (Figures S1C and S1E). Spike colocalization with OSN markers is rare and occurs at OE regions of viral shedding and structural damage (Figures S1C and S1E). Finally, SARS-CoV-2 presence in OSN axons innervating the olfactory bulb (OB) is rare, consistent with the infrequent OSN infection in the OE (Figure S1D).
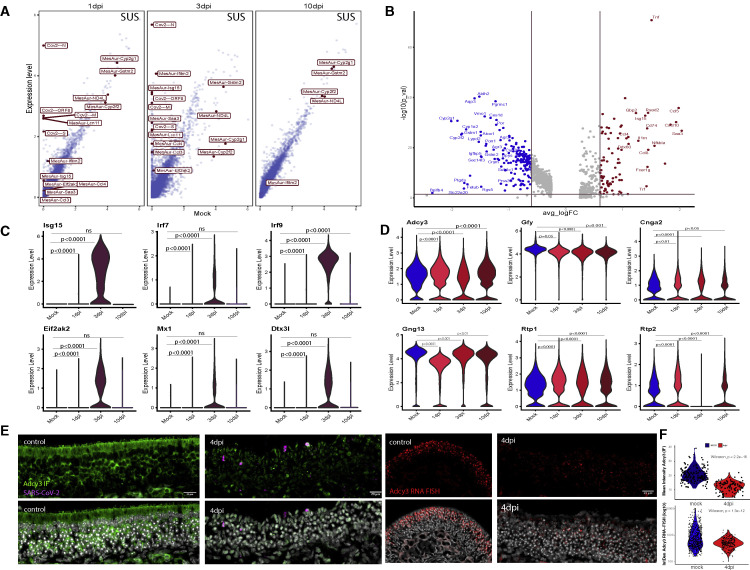
To identify transcriptional changes caused by SARS-CoV-2 infection, we first analyzed SUS cells, which are directly infected by this virus. There are significant differences between mock and infected SUS cells at 1 and 3 dpi, with the viral RNAs representing the most enriched genes in the infected samples (Figure 2 A). If we split the SUS cells of the infected OEs into SARS-CoV-2+ and SARS-CoV-2− populations, we detect upregulation of cytokines and chemokines and downregulation of SUS-specific markers in the SARS-CoV-2+ cells (Figure 2B), consistent with cell-autonomous transcriptional consequences induced by infection.
Figure 2.
SARS-CoV-2 infection induces non-cell-autonomous changes in hamster OSNs
(A) Scatter plots showing average expression of mock- and SARS-CoV-2-infected SUS cells at 1, 3, and 10 dpi. Top differentially expressed genes are shown in red boxes, hamster genes highlighted with “MesAur-” prefix, SARS-CoV-2 transcripts with “Cov2-.”
(B) Volcano plot showing upregulated and downregulated genes in SARS-CoV-2+ vs SARS-CoV-2− SUS cells from infected OEs at 1 and 3 dpi. Significantly upregulated genes are shown in red, downregulated in blue; top 1% of differentially expressed genes highlighted on the plot.
(C) Violin plots representing the log-normalized expression of antiviral genes in OSNs from mock- and SARS-CoV-2-infected OEs at 1, 3, and 10 dpi. See also Figure S2 for feature plots.
(D) Violin plots representing the log-normalized expression of key OSN genes from mock- and SARS-CoV-2-infected OEs at 1, 3, and 10 dpi.
(E) Confocal micrographs of IF-FISH in hamster OE, mock and 4 dpi, shows decreased ADCY3 protein and mRNA levels in tissues infected with SARS-CoV-2. Left, ADCY3 Ab (green) and SARS-CoV-2 gRNA (magenta). The SARS-CoV-2 probe targets the antisense strand of the S gene, detecting replicating virus. Right, Adcy3 mRNA FISH (red) is reduced in 4 dpi samples compared with mock.
(F) ACDY3 protein and mRNA quantifications show significantly lower levels in hamster OE 4 dpi compared with mock controls. Top, distribution of mean intensity ACDY3 antibody staining for individual cells in in control and infected hamster OE sections. Bottom, distribution of integrated density (mean intensity × DAPI area) of ADCY3 mRNA FISH signal in control and infected hamster OE sections.
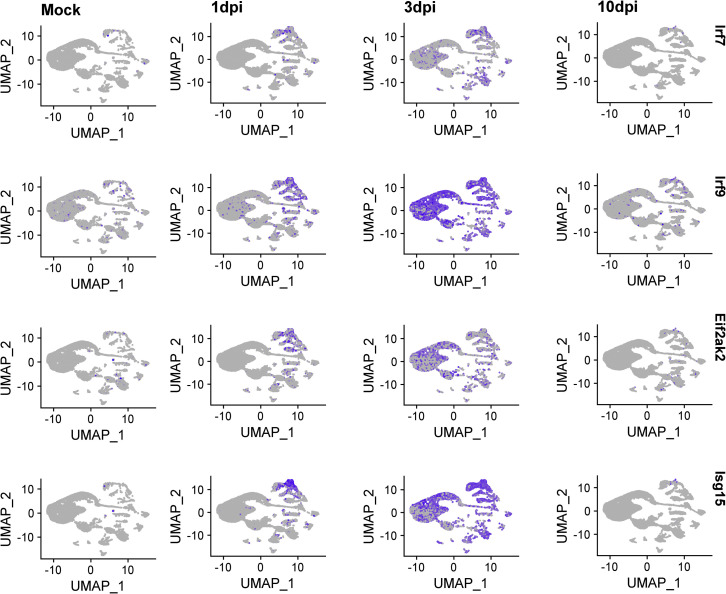
Since SARS-CoV-2 infects OSNs infrequently, we asked if the infection elicits non-cell-autonomous transcriptional changes in these neurons. Indeed, OSNs activate antiviral responses at 3 dpi (Figures 2C and S2 ). By 10 dpi, transcription of antiviral genes is reduced, concomitantly with the clearance of the virus from the OE. Importantly, genes essential for the sense of smell, such as Adcy3, are significantly downregulated in OSNs at 3 dpi (Figure 2D). Consistent with this, Adcy3 RNA ISH and IF show significant reduction of Adcy3 mRNA and protein at infected hamster OEs, even in regions with little detectable virus (Figures 2E and 2F).
Figure S2.
Evidence for induction of antiviral programs in OSNs upon SARS-CoV-2 infection of hamster OEs, related to Figure 2
Feature plot of Irf7, Irf9, Isg15, and Eif2ak2 (PKR) expression across clusters. Expression of these genes starts at microglia and immune cells at 1 dpi but expands on OSNs and other OE-resident cells by 3 dpi (two biological replicates per condition combined).
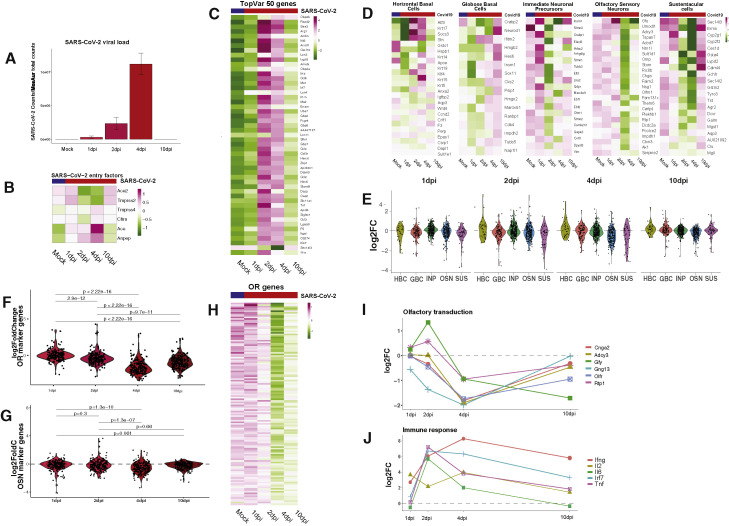
COVID-19 causes downregulation of OR and OR signaling genes in hamster OEs
Our scRNA-seq analysis could not provide insight to the effects of the virus into OR expression because we used a 5′-based cDNA synthesis approach. This approach is most sensitive for the detection of the SARS-CoV-2 but inadequate for detection OR mRNAs due to poor annotation in the hamster genome. To overcome this and to quantify rigorously transcriptional changes, we complemented our analysis with bulk RNA-seq. We collected infected OEs at 1, 2, 4, and 10 dpi. This approach confirmed high viral loads in the hamster OE that increase till 4 dpi before complete elimination by day 10 (Figure 3 A). Downregulation of SARS-CoV-2 host entry factors suggests depletion of cells that can be infected by this virus (Figure 3B). Further, we detect strong upregulation of antiviral genes that last until 10 dpi (Figure 3C), consistent with observations in other tissues (Blanco-Melo et al., 2020; Hoagland et al., 2021).
Figure 3.
SARS-CoV-2 infection causes downregulation of hamster OR and OR signaling genes
(A) SARS-CoV-2 genomic counts in hamster OE following intranasal inoculation of SARS-CoV-2 and harvest at 1, 2, 4, and 10 dpi. SARS-CoV-2 raw counts were normalized to the MesAur1.0 genome reads and plotted as DESeq2’s median ratio normalization (MRN). No mapped counts were found in the mock-infected OE.
(B) Z-scored expression of SARS-CoV-2 entry genes across infection time course.
(C) Z-scored expression of the 50 genes with highest variance.
(D) Z-scored expression of HBC, GBC, INP, OSN, and SUS markers (left to right) across SARS-CoV-2 infection time course.
(E) Distribution of log2FC for cell-type-specific markers in the OE during SARS-CoV-2 infection time course.
(F) Distribution of log2FC for OR genes during SARS-CoV-2 infection time course.
(G) Distribution of log2FC for OSN markers during SARS-CoV-2 infection time course.
(H) Z-scored expression of OR genes during SARS-CoV-2 infection time course.
(I) Aggregate expression of ORs and OR signaling transduction genes during SARS-CoV-2 infection time course. See also Figure S3.
(J) Antiviral gene expression during SARS-CoV-2 infection time course. Data for each panel (A–J) represent averages from three biological replicates per condition, except for 1 dpi, which is the average of two biological replicates.
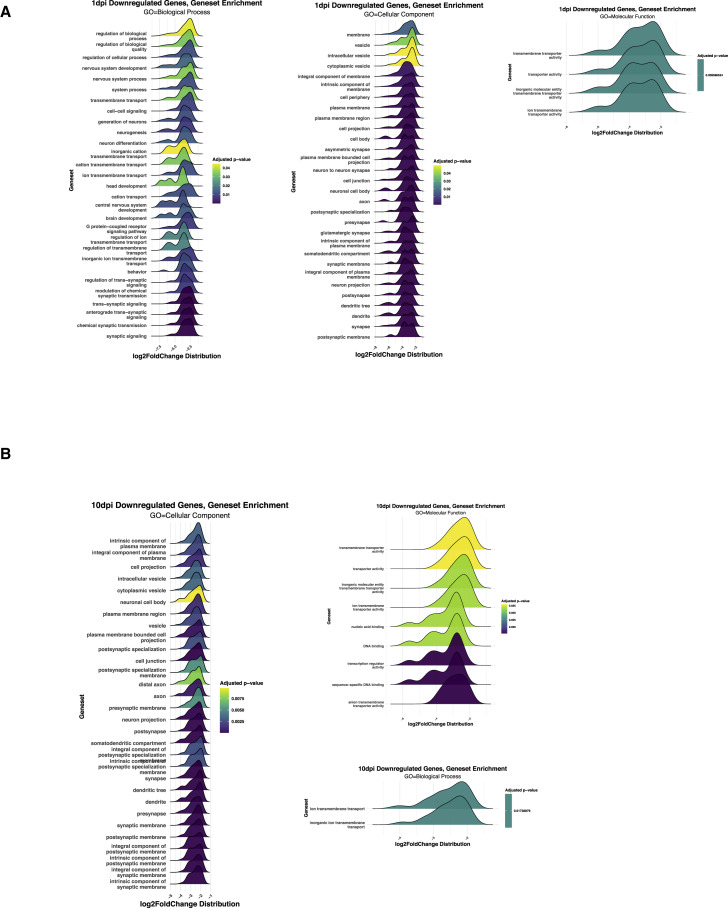
In addition to the aforementioned alterations, we detect an early wave of transient transcriptional changes in SUS cells and immediate neuronal precursors (INPs), followed by delayed, transcriptional changes in OSNs and globose basal cells (GBCs). Reduction in INP markers is mostly restricted to 2 dpi, including the downregulation of Lhx2, Ebf1, and Ebf2, transcription factors with key roles in expression of OR and OR signaling genes (Hirota and Mombaerts, 2004; Monahan et al., 2019; Monahan et al., 2017; Wang et al., 2004; Wang et al., 1997). Downregulation of SUS markers starts at 2 dpi and peaks at 4 dpi before being restored to pre-infection levels by day 10 (Figures 3D and 3E). In contrast, OSN responses are delayed and persistent, with key molecules for olfaction, such as Adcy3 (Wong et al., 2000), remaining downregulated through day 10 (Figures 3D and 3E). Finally, markers of OSN progenitor cells increase, with GBC markers peaking at 10 dpi (Figures 3D and 3E). This may reflect progenitor cell activation toward the replenishment of infected cells of the OE (Fletcher et al., 2017; Gadye et al., 2017). A summary of other processes that may be affected at the early and late stages of the infection is shown in GSEA plots for 1 and 10 dpi (Figure S3 ), and all the significant transcriptional changes are listed in Table S1.
Figure S3.
GSE analysis of enriched genes 1 and 10 days post SARS-CoV-2 infection in hamsters, related to Figure 3
(A) GSE analysis for enriched genes at 1 dpi in hamster reveals consequences for neurogenesis and OSNS activity at 1 dpi for three GO domains (biological process, cellular component, and molecular function).
(B) GSE analysis for enriched genes at 10 dpi in hamster for three GO domains.
The most striking transcriptional change observed in infected OEs is the widespread downregulation of OR genes. Significant OR downregulation is first observed at 2 dpi, peaks at 4 dpi, and continues through 10 dpi (Figures 3F and 3H), when other OSN markers have recovered (Figure 3G). This pattern is distinct from the changes in the most variable genes in the OE, whose expression is fully restored by 10 dpi, or the changes observed in INPs and SUS cells (Figures 3C and 3D). Genes with critical role in olfaction follow the pattern of OR gene expression, as we also detect strong and significant downregulation of Adcy3, Gng13, Cnga2, Rtp1, and Gfy, at 4 dpi that is partially preserved till 10 dpi (Figure 3I). Some antiviral responses are also sustained till day 10 (Figure 3J).
SARS-CoV-2 infection induces reorganization of OSN nuclear architecture
To decipher mechanisms responsible for widespread and sustainable OR downregulation, we directed our studies to a known regulator of OR expression, the OSN nuclear architecture (Bashkirova and Lomvardas, 2019; Clowney et al., 2012; Markenscoff-Papadimitriou et al., 2014; Monahan et al., 2019). OR gene clusters from multiple chromosomes converge to OSN-specific genomic compartments, which facilitate stable and singular OR transcription (Clowney et al., 2012). We therefore asked if disruption of OR compartments is the cause of OR downregulation upon SARS-CoV-2 infection.
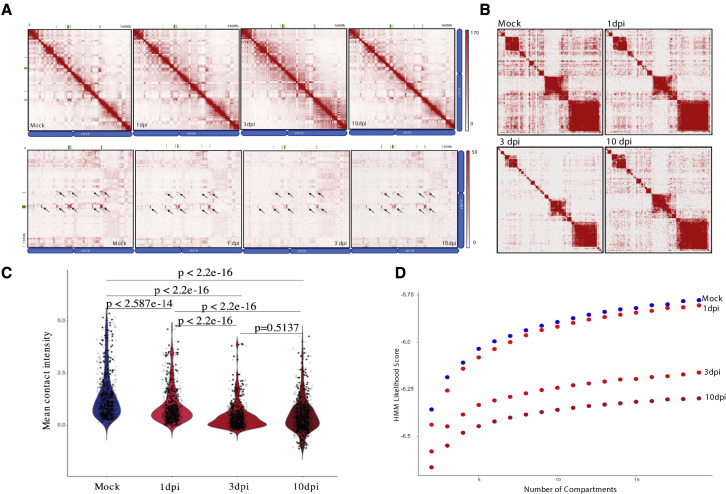
We performed in situ HiC in hamster OEs from control (mock-infected) and SARS-CoV-2-infected samples at 1, 3, and 10 dpi. OR gene clusters form robust long-range cis and trans genomic contacts in hamster OSNs, as shown for OR genes from chromosomes 16 and 17 (Figure 4 A). SARS-CoV-2 infection reduces these contacts, starting at 1 dpi and peaking at 3 dpi (Figure 4A). A contact matrix for all the hamster OR clusters arranged by chromosome shows strong long-range cis interactions and widespread trans contacts between them in control samples (Figure 4B). However, these interactions become reduced as early as 1 dpi and remain low by 10 dpi (Figures 4B and 4C). Hidden Markov model (HMM) calculation of genomic compartment scores shows widespread reduction of most compartments by 3 dpi, revealing a delayed disruption of genome-wide nuclear architecture compared with the disruption of OR compartments (Figure 4D). However, genomic compartmentalization remains disrupted 10 dpi, when the virus is already cleared from the OE.
Figure 4.
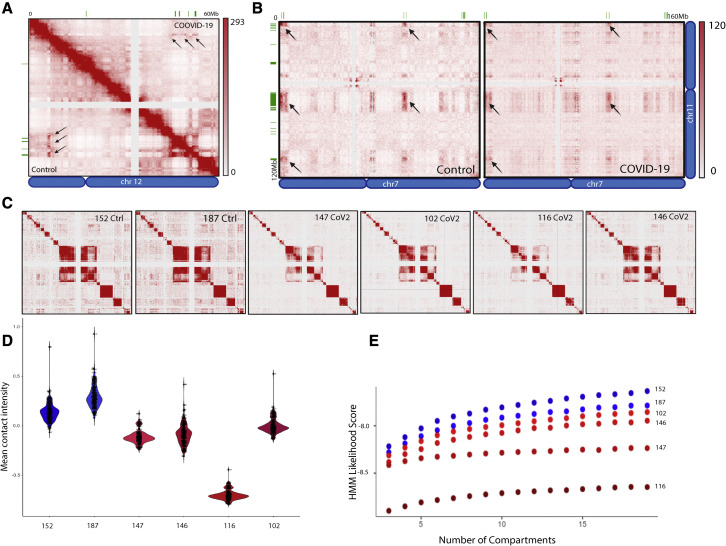
SARS-CoV-2 infection disrupts interchromosomal OR compartments
(A) In situ HiC maps of contacts between OR clusters in cis (top) or trans (bottom) for mock, 1, 3, and 10 dpi hamster from pooled in situ HiC data. Pixel intensity represents normalized number of contacts between pair of loci. Maximum intensity indicated at the top of each scale bar. Genomic position of OR clusters indicated as green bars; arrows indicate the same OR compartments for both conditions.
(B) Pairwise heatmap shows reduction of in situ HiC contacts between OR clusters (n = 46 clusters) that increases as SARS-CoV-2 infection progresses.
(C) Violin plot depicting the mean number of normalized trans in situ HiC contacts between OR clusters genome wide at 100-kb resolution for mock, 1, 3, and 10 dpi. Every dot indicates aggregated contacts for each OR-to-OR cluster pair in trans; p value was computed using Wilcoxon rank test.
(D) HMM score for a given number of compartments indicating differences in genomic compartmentalization for mock (blue) and SARS-CoV-2-infected hamsters at 1, 3, and 10 dpi (shades of red). For each panel (A–D), data represent averages from two biological replicates per condition.
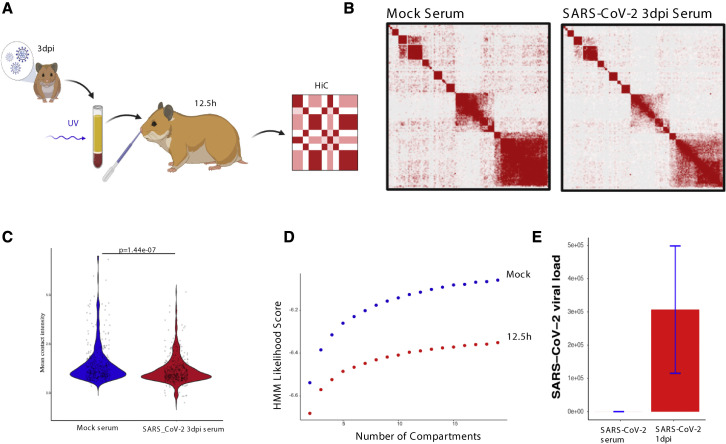
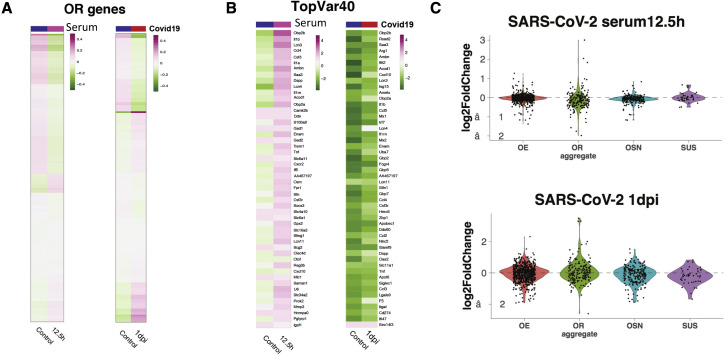
Observations of widespread and persistent disruption of OSN genomic compartments is consistent with a non-cell-autonomous mechanism of nuclear reorganization. Since previous reports implicate cytokines and antiviral responses in olfactory deficits and OR downregulation (Lane et al., 2005, 2010; Rodriguez et al., 2020), we hypothesized that we could disrupt trans OR contacts by imitating the systemic effects of SARS-CoV-2 infection without the virus. We collected serum from mock and SARS-CoV-2-infected hamsters at 3 dpi and inactivated the circulating virus by UV irradiation. We applied these sera to the OEs of naive hamsters by intranasal inoculation (Figure 5 A). Strikingly, in situ HiC revealed significant reduction of trans OR contacts upon OE exposure to infected sera for 12.5 h (Figures 5B and 5C). HMM confirmed genome-wide changes in compartment scores between the two sample groups (Figure 5D). RNA-seq did not detect the viral genome on serum exposed OEs, confirming that we did not transfer active SARS-CoV-2 from infected to naive hamsters (Figure 5E). As expected from the infection time course, there are no significant changes in OR transcription at this time point (Figure S4 A). However, there is a trend of OR downregulation across most OR genes, which is more pronounced than the effects observed at 1 dpi upon SARS-CoV-2 infection (Figure S4A). Thus, by providing sera from the peak of the inflammatory response, we likely accelerated the molecular changes observed during viral infection, consistent with the upregulation of genes induced 2 days post-SARS-CoV-2 infection (Figure S4B). SUS markers, however, are nonresponsive to the signals from the infected sera (Figure S4C), supporting the notion that SUS cells only exhibit mostly cell-autonomous transcriptional changes.
Figure 5.
Serum from SARS-CoV2-infected hamsters disrupts genomic OR compartments
(A) The experimental pipeline used to expose naive hamsters OEs serum from SARS-CoV-2 or mock-infected hamsters prior to in situ HiC analysis. Serum was collected 3 dpi from mock or SARS-CoV-2-infected hamsters, centrifuged, and UV-irradiated before intranasal inoculation to naive hamster OEs for 12.5 h. See also Figure S4.
(B) Pairwise heatmap of in situ HiC contacts between OR clusters (n = 46 clusters) from hamster OEs. The heatmap on the left is from OEs exposed to serum from mock-infected hamsters, whereas on the right is from OEs exposed to serum from SARS-CoV-2-infected hamsters.
(C) The mean number of normalized trans in situ HiC contacts between OR clusters genome wide at 100-kb resolution for mock and 12.5 h SARS-CoV-2 serum-treated hamster. Every dot indicates aggregated contacts for each OR-to-OR cluster pair in trans; p value was computed using Wilcoxon rank test.
(D) HMM score for a given number of compartments indicating differences in genomic compartmentalization upon OE exposure for 12.5 h to serum from mock (blue) and SARS-CoV-2-infected hamsters at 3 dpi (red).
(E) SARS-CoV-2 genomic counts in inactivated serum of 3 dpi hamster applied to naive hamster compared with the viral load at 1 dpi hamster. SARS-CoV-2 raw counts were normalized to the MesAur1.0 genome reads and plotted as DESeq2’s median ratio normalization (MRN). No mapped counts were found in the mock-infected OE. For each panel (A–D), data represent averages of three biological replicates per condition.
Figure S4.
Subtle transcriptional changes in the hamster OE upon exposure to UV-neutralized serum from infected hamsters, related to Figure 5
(A) Z-scored expression of OR genes from OEs exposed to mock- versus SARS-CoV-2-infected serum for 12.5 h (left) or from OEs that were mock infected versus SARS-CoV-2 infected for 1 day (right).
(B) Z-scored expression of the top 40 most variable genes upon serum exposure (left). For comparison, Z-scored expression of top 40 genes at 1 dpi identified in SARS-CoV-2-infected hamster and harvested at different time points (mock, 1 dpi, 2 dpi, 4 dpi, and 10 dpi) (right).
(C) Violin plots showing the effect of exposure to serum from SARS-CoV2 versus mock-infected hamsters for 12.5 h at the transcription of OR genes, OE, OSN, and SUS markers. For comparison, we plot the same groups at SARS-CoV-2-infected (1 dpi) versus mock-infected hamsters. For each panel (A–C), data represent averages of three biological replicates per condition.
SARS-CoV-2 infection induces downregulation of OR and OR signaling genes in humans
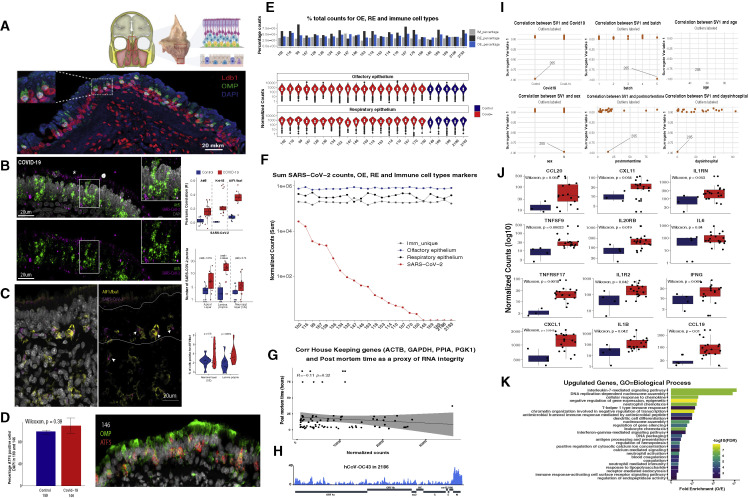
To explore whether our observations from hamsters apply to humans, we analyzed the consequences of SARS-CoV-2 infection of human OE autopsies. We identified a region at the roof of the nasal cavity bridging the superior septum and middle turbinate bones that is highly enriched for OSNs as demonstrated by detection of the mature OSN-specific olfactory marker protein (OMP) and the OSN-enriched LDB1 (Figure S5 A). This is further supported by scRNA-seq analysis on an autopsy from a control (noninfected) sample (Zazhytska et al., 2021). RNA ISH in section of OE autopsies from infected patients shows enrichment of the SARS-CoV-2 RNA at the non-neuronal layers of the OE (Figure S5B), consistent with recent observations (Khan et al., 2021). We also detect SARS-CoV-2 RNA in microglia cells recruited to the infected human OEs (Figure S5C), replicating observations from hamster OEs. Finally, as in hamsters, we did not observe OSN depletion in infected OEs (Figure S5D).
Figure S5.
Quality control analyses of human OE autopsies, related to Figure 6
(A) (Top) En bloc resection of the cribriform plate along with underlying mucosa from the olfactory cleft, which contains OE more superiorly and respiratory epithelium below. (Bottom) Section of this human olfactory epithelium stained for OMP (green) and LDB1 (red), OSN-specific and OSN-enriched markers, respectively. Nuclei are labeled with DAPI (blue).
(B) Confocal micrograph of RNA FISH for SARS-CoV-2 gRNA (magenta) and OSN/OSN progenitor marker ATF-5 (green) in COVID-19+ human OE. The SARS-CoV-2 probe targets the antisense strand of the S gene, detecting replicating virus. Nuclei are stained with DAPI. SARS-CoV-2 signal is detected in the apical layers of the epithelium (asterisk), proximal to SUS cells, and in the basal layer where HBCs reside. Correlation of the RNA-FISH SARS-CoV-2 signal and markers for OSNs (ATF5), sustentacular cells (Krt-18), and microglia (AIF1/Iba1) is measured by Pearson’s correlation coefficient (R) in COVID-19 (red) and control (blue) human OE autopsies (top right panel). Quantification of RNA-FISH signal was measured as local maxima at the apical, neuronal, and basal layers for a total of 2,140 cells in infected and 1,819 cells in control OEs. Only apical and basal layers have significantly enriched signal in infected OEs.
(C) RNA-FISH SARS-CoV-2 signal (magenta), detected in the neuronal layer (OE), marked in between the two white lines, and lamina propria. S probe signal (magenta) strongly correlates with microglia marker AIF1/Iba1 (yellow) immunofluorescence (IF) signal, as indicated by arrows. The number of AIF1/Iba1-positive cells is measured over the number of total cells counted (right panel). In COVID-19 patients, a significantly enrichment of microglia is observed in the lamina propria, while in the neuronal layer (OE), more variability between images is observed.
(D) Bar plot of the percentage of Atf5 RNA+ cells over total number of DAPI-positive cells on sections of COVID-19 (146) and control (189) human OEs. No significant difference between samples is detected (n = 508 for 146, n = 458 for 189). Cells with more than two RNA puncta were counted as positive. On the right, a representative image of sample 146 for Atf5 RNA-FISH (red) and OMP protein (green).
(E) Percentage of total counts of OE, RE, and immune-cell-type genes. Pairwise Wilcoxon test shows no significant difference between samples. Distribution of normalized counts for OE and RE genes for each sample (bottom panels).
(F) SARS-CoV-2 counts plotted in decreasing order (red) together with normalized counts for OE (blue), RE (black), and immune cells (gray).
(G) Correlation of house-keeping genes (ACTB, GAPDH, PPIA, and PGK1) and post-mortem times were used as a proxy of RNA integrity. Pearson’s test shows no significant correlation.
(H) Coverage map for hCoV-OC43 in sample 2,186.
(I) Surrogate variable analysis reveals sample 205 as an extreme outlier. SV1 does not correlate with known variables (COVID-19, processing batch, age, sex, post-mortem time, or days after symptoms onset), but rather in all cases distinguishes sample 205 from other samples.
(J) Box plot depicting normalized counts between pooled control and infected samples for antiviral genes.
(K) GO analysis for upregulated genes depicting significant enrichment for genes involved in immune/antiviral responses.
Upon establishing histological similarities between hamster and human SARS-CoV-2 infection, we performed bulk RNA-seq in 6 control and 18 infected human OE autopsies. Autopsies were donated from both male and female patients, representing a variety of ages, duration of infection, hospitalization, treatment, and post-mortem intervals (Table S2). These variations did not influence cellular constitution, as quantification of OE, respiratory epithelium (RE), and immune cells show consistency between samples (Figures S5E and S5F). Post-mortem time (PMT) had no obvious effect on the quality of these libraries (Figure S5G). Surrogate variable analysis (Leek, 2014) identified one control sample as extreme outlier, resulting in its removal from further analyses (Figure S5I). The remaining samples were subjected to batch adjustment corrections using ComBat-seq (Love et al., 2014; Zhang et al., 2020) and then were further analyzed.
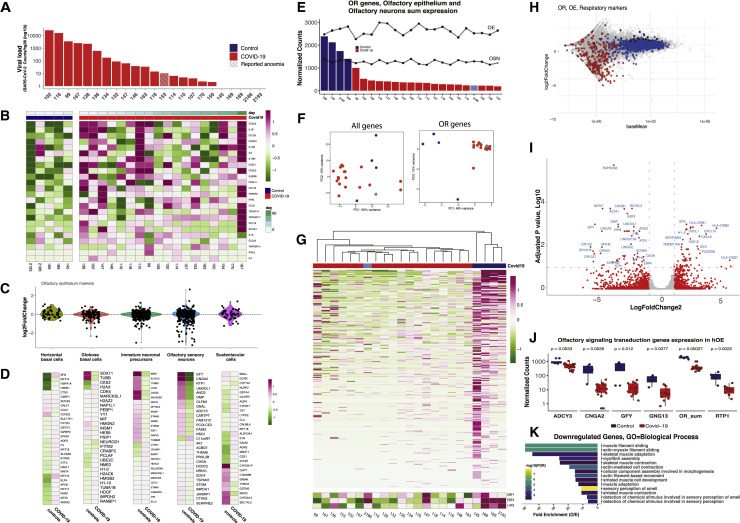
SARS-CoV-2 RNA is detected in every infected OE, with variable amounts between samples (Figure 6 A). However, representation of OE and RE markers did not change with viral load (Figures S5E and S5F). Unexpectedly, in one of the remaining five control samples, we detected the RNA genome of a non-SARS coronavirus, hCoV-OC43 (Figure S5H), which was previously shown to infect the RE and OE (Dubé et al., 2018). This sample, highlighted with light blue, was not pooled with control samples. We have information only on one patient about olfactory deficits (highlighted with striped bar) due to limited solicitation of these symptoms at the early phase of the pandemic. However, based on current reports, >60% of these subjects may have experienced olfactory deficits (Butowt and von Bartheld, 2020; Wang et al., 2020).
Figure 6.
SARS-CoV-2 infection of human OEs coincides with downregulation of OR/OR signaling genes
(A) SARS-CoV-2 genomic counts from the OE of 18 COVID-19 patients (red) and 4 controls (blue). SARS-CoV-2 raw counts were normalized to the hg38 genome reads using DESeq2’s median ratio normalization (MRN). The striped bar highlights the only sample with known anosmia. See also Figure S5 for histological confirmation of SARS-CoV-2 detection.
(B) Z score for inflammatory makers for each sample shows variability of inflammatory response among patients. See also Figure S4J for aggregate analysis of log2FC of antiviral/inflammatory markers.
(C) Distribution of log2FC for cell-type-specific markers in the OE. A subset of OSN markers is downregulated.
(D) Z-scored expression of inflammatory makers calculated across samples shows variability of inflammatory response among COVID-19 patients. Samples are ordered according to the number of days after symptoms onset (top). See also Figure S4J.
(E) Distribution of aggregated normalized OR genes counts across all human OEs. Transformed aggregate expression of OE and OSN markers are plotted for each sample as dots. Infected samples depicted in red and control samples in blue. The self-reported anosmic patient marked with stripes (153) and hCoV-OC43+ is marked in light blue (2186).
(F) COVID-19 samples (red) and controls (blue) do no cluster in PCA analysis with all genes (left) but do cluster when only OR genes (right) are considered. Sample with coronavirus hCoV-OC43 is highlighted in light blue (2186).
(G) Z-scored expression of OR genes (top). Unsupervised clustering using only OR genes distinguish COVID-19 and control samples. Sample 2186 (light blue, hCov-OC43+), clusters with COVID-19 samples. (Bottom) Z score for Ebf1, Ebf2, and Lhx2, transcription factors with known role in the expression of OR/OR signaling genes.
(H) MA-plot with OE genes (blue), OR genes (red), and RE genes (black).
(I) Volcano plot of COVID-19 versus control RNA-seq data. Log2FC genes with abs(log2FC) ≥ 1 highlighted in red. Gene with padj < 0.05 are identified with blue fonts.
(J) Boxplot representation of the normalized counts (MRN) grouped in COVID-19 positive and control specimens for Adcy3, Cnga2, Gfy, Gng13, aggregate OR, and Rtp1. Significance was calculated using Wilcoxon test. Significance value for OR downregulation does not change if we omit the most lowly expressed ORs (shown in the chevron-shaped distribution of the MA plot).
(K) GO analysis for downregulated genes reveals enrichment for genes involved in sensory perception of smell. See also Figure S5K for GO analysis of upregulated genes.
In most infected samples, we detected elevated levels of cytokines and antiviral genes such as IFN-γ (Figures 6B and S5J). GO analysis of significantly upregulated genes between infected autopsies is overrepresented with terms related to immune response (Figure S5K). We did not detect an overall downregulation of SUS markers (Figure 6C), although some SUS-enriched genes are downregulated in the infected samples (Figure 6D). Similarly, we did not detect depletion of OSN markers, but selective reduction of OSN-enriched genes with established critical roles in olfaction, such as Adcy3 (Figures 6C and 6D). Control OEs have higher OR mRNA levels than the infected OEs (Figure 6E), except for the hCoV-OC43-infected control. PCA analysis using only OR genes, segregates control from infected samples, while PCA with the whole transcriptome does not (Figures 6F and 6G), suggesting that reduced OR transcription constitutes one of the few distinctive features between infected OEs. OR downregulation generally tracks with downregulation of Lhx2 and Ebf1/2, but two samples with low OR expression have high Lhx2 and Ebf1/2 expression (Figure 6G).
Comparisons between individual autopsies are replicated in comparisons of pooled control and infected samples. MA plots depicting the levels of OR genes (red), OE genes (blue), and RE genes (black) in control and infected samples, supports a bona fide transcriptional downregulation of OR genes, as OE and respiratory markers are not changing upon infection (Figure 6H). Volcano and boxplots showing the transcriptional effects of SARS-CoV-2 infection in aggregate further support significant downregulation of OR and OR signaling genes (Figures 6I and 6J). Finally, GO analysis of downregulated genes in infected autopsies shows that “sensory perception of smell” constitutes the most significantly enriched GO term (Figure 6K).
SARS-CoV-2 infection disrupts interchromosomal OR compartments in humans
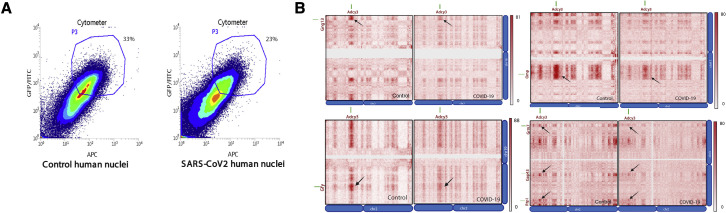
To ask whether OR downregulation in SARS-CoV-2-infected OEs is caused by changes in nuclear architecture, we established a protocol for the isolation of OSN nuclei from OE autopsies by FACS (Figure S6 A). In situ HiC on FAC-sorted nuclei confirmed that the OR-specific long-range cis and trans genomic contacts are conserved in humans (Figures 7A and 7B). Contact matrixes depicting human OR genes from every chromosome confirms that OR genes form interchromosomal compartments (Figures 7C and 7D) that are disrupted in SARS-CoV-2-infected OEs (Figures 7A–7D), independently of genome-wide changes in nuclear architecture (Figure 7E). Finally, we identified interchromosomal compartments containing Adcy3 and other genes with key functions in olfaction that also dissipate in infected samples (Figure S6B).
Figure S6.
FACS and HiC of OSN nuclei from human OE autopsies, related to Figure 7
(A) FACS data for control and COVID-19 human OE. Fixed DAPI positive, Lhx2/Atf5 double positive nuclei were collected for in situ HiC.
(B) Reduction in HiC contacts in compartments formed by Adcy3, Gng3, Gfy, OMP, Grm7, Gap43, and Rtp1 genes.
Figure 7.
SARS-CoV-2 infection of human OEs disrupts genomic OR compartments
(A) In situ HiC maps from human OSNs depicting contacts between OR clusters in cis. Control is the lower triangle below the diagonal, and COVID-19 the upper triangle. Pixel intensity represents normalized number of contacts between pair of loci. Maximum intensity indicated at the top of each scale bar. Genomic position of OR clusters indicated as green bars; arrows indicate the same OR compartments for both conditions.
(B) Contact maps revealing decrease in trans in situ HiC contacts in COVID-19+ OE versus control. Pixel intensity represents normalized number of contacts between pair of loci. Maximum intensity indicated at the top of each scale bar. Genomic position of OR clusters indicated as green bars; arrows indicate the same OR compartments for both conditions.
(C) Heatmap depicting contacts between every human OR gene cluster (n = 82 OR clusters) arranged by chromosome. In situ HiC was performed on FAC-sorted OSNs from two control and four infected human OE autopsies. Reduction in OR contacts is observed both in trans and in cis.
(D) Violin plot depicting the mean number of normalized trans HiC contacts between OR clusters genome wide at 100-kb resolution for each sample. Every dot indicates aggregated contacts for each OR-to-OR cluster pair in trans; p value < 0.05 was computed using Wilcoxon rank test.
(E) HMM score for a given number of compartments indicating differences in genomic compartmentalization between two control (blue) and four infected samples (red).
Discussion
We provide a molecular explanation for SARS-CoV-2-induced anosmia and a mechanism by which this virus can alter the identity and function of cells that lack entry receptors. Consistent with absence of ACE2 and TMPRSS2 from OSNs (Bilinska et al., 2020; Brann et al., 2020; Chen et al., 2020), and recent histological analyses (Khan et al., 2021), our data suggest that OSN infection by SARS-CoV-2 is too infrequent to account for the reported smell loss. Moreover, the cell-autonomous effects of SUS infection may be too transient to account for long-lasting olfactory deficits reported by COVID-19 patients, which have a mean duration of ∼20 days (Chapurin et al., 2022). Thus, the most likely explanation for COVID-19-induced anosmia is the non-cell-autonomous, widespread, and persistent downregulation of OR and OR signaling genes. The ability of the virus to alter the OSN transcriptome solves a puzzle that emerged from numerous studies in various organs: the virus is only infecting a small fraction of cells, yet it elicits devastating and often life-threatening physiological disruption (Thakur et al., 2021). The demonstration that UV-neutralized serum from infected hamsters induces significant and rapid changes in OSN nuclear architecture suggests that systemic changes caused by SARS-CoV-2 infection alter the physiology and function of the cells that this virus cannot infect.
Comparing the effects of SARS-CoV-2 infection in hamster and human OEs
Hamster scRNA-seq shows that SARS-CoV-2 predominantly infects SUS cells, resulting in cell-autonomous transcriptional changes and transient depletion of this cell population. In human OEs, where the viral load is lower, cell-autonomous transcriptional changes could not be detected by RNA-seq, probably due to infrequent SUS infection. However, these changes were detected by spatial transcriptomics that compared human OE regions with different viral loads (Khan et al., 2021). Moreover, in both hamster and human Oes, we detect strong, persistent, and widespread downregulation of OR genes as well as downregulation of Adcy3 and other key genes for odor perception, providing a plausible explanation for COVID-19-induced anosmia.
The insight afforded by hamster studies explains why spatial transcriptomics detected downregulation of SUS markers in human OE regions with high SARS-CoV-2 load but no reduction of OR and OR signaling molecules (Khan et al., 2021). SUS marker downregulation is cell autonomous, thus expected to be stronger in regions with high viral load. In contrast, transcriptional changes in OSNs occur independently of direct infection and do not corelate with the viral load in the OE, hindering their elucidation by transcriptomic comparison within a sample. Furthermore, there is a delay in OSN transcriptional changes compared to SUS marker downregulation. Thus, autopsies corresponding to longer infection periods and comparison with noninfected samples may be required for the detection of OSN transcriptional changes by this elegant approach.
Disruption of genome architecture as a “nuclear memory” for persistent anosmia
COVID-19-induced downregulation of Lhx2 and Ebf, key transcription factors for OSN physiology, explains the downregulation of a plethora of genes involved in odor perception. In hamsters, however, disruption of OR compartments precedes Lhx2/Ebf downregulation and persists after their restoration. Further, in two infected human OEs (146, 147), both OR transcription and OR compartmentalization are disrupted, while Lhx2 and Ebf levels are near control levels. Thus, although COVID-19-induced Lhx2/Ebf downregulation is likely to have major role in the downregulation of OR and OR signaling genes, widespread disruption of OR compartments may be the first insult in OSN physiology and, importantly, a form of “nuclear memory” that delays restoration of OR transcription. This is because OR compartments may form only during differentiation, and, thus, their disruption in mature OSNs may be irreversible. If OSNs cannot reactivate OR transcription, then the sense of smell in COVID-19 patients will recover only after these OSNs are replaced, a process that takes from weeks to months.
If OR contacts could be restored after the elimination of the virus, their pre and post-infection patterns may be different, due to the inherent stochasticity of trans OR interactions (Bashkirova et al., 2020; Tan et al., 2019). Thus, OSNs that were already innervating a glomerulus may activate a different OR from the one originally chosen, resulting in odor misrepresentation in the OB and altered odor perception. This sensory confusion may also be exacerbated by Adcy3 downregulation as this molecule plays important roles in OSN axon guidance and the stabilization of OR expression (Imai et al., 2006; Lyons et al., 2013; Zou et al., 2007). Long-term deficits in nuclear architecture could be applicable to other neuronal populations since adult CNS neurons also assemble long-range cis and trans genomic compartments between OR genes and other neuronal gene families (Jiang et al., 2017; Tan et al., 2021). Additional mechanisms, such as sustained expression of antiviral programs (Frere et al., 2022), damage in tissue vasculature, and hypoxia (Thakur et al., 2021), could also contribute to long-lasting neurological deficits, including the loss of smell (Lane et al., 2010). In either case, the realization that the sense of smell relies on extremely “fragile” genomic interactions between chromosomes has important implications: if OR expression ceases every time maladaptive physiological responses disrupt interchromosomal OR contacts, then olfaction may act as the “canary in the coal mine” for a variety of human conditions, from viral infections to neurodegeneration (Albers et al., 2006).
Limitations of the study
We did not identify the circulating molecule(s) that induce reorganization of OSN nuclear architecture and the OSN signaling pathway responsible for it. Thus, currently, we can only speculate that similar mechanisms apply to other neuronal populations, a concept that we have not explored. Furthermore, we did not establish that the reported downregulation in OR and OR signaling genes is responsible for COVID-19-induced anosmia, but we infer this from the phenotypes of knockout mice. Reduced expression of genes involved in every step of odor detection, such as receptor proteins (ORs) (Buck and Axel, 1991), olfactory receptor chaperones (Rtp1 and Rtp2) (Saito et al., 2004), olfactory receptor signaling molecules (Adcy3 and Gng13) (Liu et al., 2018; Wong et al., 2000), and ion channels generating odor-evoked axon potential (Cnga2) (Brunet et al., 1996), provides the most likely explanation for COVID-19-induced anosmia. Finally, we can only deduce that COVID-19 infection caused OR and OR signaling gene downregulation in humans, as we cannot measure the expression of these genes before the infection. Although experiments in hamsters support this hypothesis, we cannot exclude rodent-specific mechanisms that preclude direct comparisons between species.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-OMP | Chen et al. (2005) | N/A |
| Anti-Adcy3 | Abcam | Cat no. ab123803; RRID:AB_10973698Anti-Iba1; RRID:AB_2636859 |
| Anti-Iba1 | Abcam | Cat no. ab178846 |
| Anti-Cytokeratin 18 | EMD Millipore | Cat no. MAB3234; RRID:AB_94763Anti-Atf5; RRID:AB_205876 |
| Anti-Atf5 | Santa Cruz Biotechnology | Cat no. sc-46934 |
| anti-mouse IgG conjugated to Alexa-488 | Thermo Fisher Scientific | Cat no. A-21202; RRID:AB_141607 |
| anti-rabbit IgG conjugated to Alexa-555 | Thermo Fisher Scientific | Cat no. A-31572, RRID:AB_162543 |
| anti-chicken IgG conjugated to Alexa-488 | Jackson ImmunoResearch | Cat no. 703-545-155; RRID:AB_2340375 |
| Molecular Probes | ||
| RNAscope Probe for Hs-ADCY3 | Acdbio | Cat no. 441671 |
| RNAscope Probe for Hs-ATF5 | Acdbio | cat no. 507471 |
| Opal dye 520 | Akoya Biosciences | FP1487001KT |
| Opal dye 570 | Akoya Biosciences | FP1488001KT |
| Opal dye 690 | Akoya Biosciences | FP1497001KT |
| Chemicals, peptides, and recombinant proteins | ||
| OptiPrep density Gradient Medium | Sigma-Aldrich | cat no. D1556-250ML |
| Mse I | NEB | cat no. R0525M |
| Triton X-100 | Sigma | cat no. 93443 |
| BSA | NEB | cat no. B9000S |
| Biotin-11-dUTP | Thermo Fisher Scientific | cat no. R0081 |
| Quick ligase kit | NEB | cat no. M2200L |
| Proteinase K | NEB | cat no. P8107S |
| AMPure XP beads | Beckman Coulter | A63881 |
| Dynabeads MyOne Streptavidin T1 beads | Thermo Fisher Scientific | cat no. 65602 |
| T4 DNA ligase reaction buffer | NEB | cat no. B0202S |
| 16% Formaldehyde Solution | Thermo Sciientific | cat no. 28906 |
| Critical commercial assays | ||
| Truseq RNA Library Prep Kit v2 | Illumina | 20020597 |
| Direct-zol RNA kits from Zymo Research | Zymo Research | cat no. R2052 |
| RNAscope Multiplex Fluorescenct v2 Assay | Acdbio | cat no. 323135 |
| Ovation Ultralow System V2 32 | Tecan Genomics | cat no. 0344-32 |
| Next GEM Single Cell 5′ GEM Kit v2 | 10x Genomics | cat no.1000265 |
| Papain Dissociation system | Worthington Biochemical | cat no. LK003178 |
| Deposited data | ||
| All sequencing data for hamster and human samples | This manuscript, 4DN portal | [https://doi.org/10.1101/2021.02.09.430314] |
| Experimental models: Organisms/strains | ||
| Golden Syrian hamsters (Mesocricetus auratus) | Charles River Laboratories | LVG hamsters, strain code: 049 |
| Software and algorithms | ||
| ImageJ | Schneider et al. (2012) | https://imagej.nih.gov/ij/ |
| R version 4.0.5 base packages | The R Foundatiion | https://www.rstudio.com/products/rstudio/download/ |
| Subread | Liao et al. (2014) | http://subread.sourceforge.net |
| Deseq2 | Love et al. (2014) | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| STAR | Dobin et al. (2013) | https://github.com/alexdobin/STAR |
| Samtools | Li et al. (2009) | http://samtools.sourceforge.net/ |
| sva package in R | Zhang et al. (2020) | https://bioconductor.org/packages/release/bioc/html/sva.html |
| Blast | Camacho et al. (2009) | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| Megahit | Li et al. (2015) | https://github.com/voutcn/megahit |
| IGV | Robinson et al. (2011) | https://software.broadinstitute.org/software/igv/ |
| R version 4.0.5 ggplot2 package | The R Foundation | https://cran.r-project.org/web/packages/ggplot2/index.html |
| R version 4.0.5 Seurat package | The R Foundation | https://cran.r-project.org/web/packages/Seurat/index.html |
| Cellranger 5.0.1 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/release-notes |
| CellBender | https://doi.org/10.1101/791699 | https://cellbender.readthedocs.io/en/latest/ |
| Juicer | Durand et al. (2016) | https://github.com/aidenlab/juicer/ |
| Juicebox | Robinson et al. (2018) | https://github.com/aidenlab/Juicebox |
| Python version 3.7.10 | Python Software Foundation | https://www.python.org |
Resource availability
Lead contact
Further information and requests for resources and raw data should be directed to and addressed to Stavros Lomvardas (sl682@cumc.columbia.edu)
Materials availability
This study did not generate any materials and unique reagents.
Experimental model and subject details
Hamsters
LVG Golden Syrian hamsters (Mesocricetus auratus) were treated and euthanized in compliance with the rules and regulations of IACUC under protocol number PROTO202000113-20-0743. Only adult male hamsters were used for experiments. All experiments were performed on dissected olfactory epithelium tissue or on dissociated cells prepared from whole olfactory epithelium tissue. Dissociated cells were prepared using papain (Worthington Biochemical) and FAC-sorted as previously described.
Virus stock and propagation
Infectious work was performed at a CDC/USDA-approved BSL-3 facility at the Icahn School of Medicine at Mount Sinai. SARS-CoV-2 (clinical isolate UAS/WA1/2020) virus was propagated in Vero E6 cells in DMEM supplemented with 0.35% BSA. Infectious titer of virus was determined by plaque assay in Vero E6 cells using an overlay of Modified Eagle Medium (Gibco), 0.2%BSA (MP Biomedicals), 4mM L-glutamine (Gibco), 10mM HEPES (Fisher Scientific), 0.12% NaHCO3, 1% heat-inactivated FBS, and 0.7% Oxoid agar (Thermo Scientific). SARS-CoV-2 virus stocks used for hamster experiments were passage 3.
Human samples
25 patients previously diagnosed with COVID-19 at symptoms presentation and postmortem by SARS-CoV-2 RT-PCR analysis underwent full body autopsy at Columbia University Irving Medical Center (New York, NY, USA). The study was approved by the ethics and Institutional Review Board of Columbia University Medical Center (IRB AAAT0689, AAAS7370). Specimens noted to have metastatic cancer and non-SARS coronavirus were removed from further analysis. Brain tissue and nasal epithelium, including the olfactory region, were retrieved under a collaborative effort by the Department of Neuropathology and the Department of Otolaryngology. Tissues were obtained and preserved for histological, molecular, and microscopic evaluation using separate surgical instruments to prevent cross-contamination. 7 control specimens were collected in similar fashion from deceased individuals who had no clinical history of COVID-19 and had negative SARS-CoV-2 PCR at the time of their presentation and again prior to post-mortem dissection. Nasal tissues, including olfactory and respiratory epithelium were harvested from the skull base using an en-bloc resection of the anterior skull base including the cribriform plate. Olfactory epithelium was isolated from the olfactory cleft, spanning turbinate and adjacent septal mucosa prior to being preserved in 1% paraformaldehyde (for HiC), 4% paraformaldehyde (for RNA ISH/IF), or Trizol (for RNA-seq).
Method details
SARS-CoV-2 inoculation
All hamster infections were performed in a BSL-3 animal facility at the Center for Comparative Medicine and Surgery at the Icahn School of Medicine at Mount Sinai (New York, NY) using 4-6-week-old male golden hamsters purchased from Charles River Laboratories. Hamsters were intraperitonially administered anesthesia of ketamine/xylazine (3:1), [100mg/kg] before inoculation. Inoculations were performed by intranasally administering 100 plaque-forming units (pfu) in a total volume of 100ul per hamster, diluted in PBS. For infected serum experiments whole blood of mock and infected animal at 3dpi was centrifuged to extract serum following UV neutralization of any viral particles remained. Total volume of 100ul per animal was intranasally inoculated in the same fashion as virus administration. Golden hamsters were provided thermal support after infection until recovery from anesthesia. Before sacrifice, the animals were anesthetized and then perfused with 60mL of PBS through the heart.
Serum collection, inactivation and intranasal inoculation
The blood from a mock and 3dpi infected hamster with 100pfu of SARS-CoV-2 virus was collected from aorta upon euthanasia. Serum was separated via centrifugation following subsequent UV-C inactivation at dose of 100J/m2. Inoculations were performed into naïve hamsters by intranasally administering 100ul of mock or 3dpi inactivated serum per hamster upon anesthesia. 12.5h after serum inoculation hamsters were sacrificed, OE was dissociated and subjected for HiC and bulk RNA-seq.
RNA-seq
RNA was extracted using Direct-zol RNA kits from Zymo Research. 50ng-1ug of total RNA was used to prepare DNA libraries with Truseq RNA Library Prep Kit v2 followed by 75 HO paired-end and multiplexed sequencing. Reads were aligned to human genome (hg38), Mesocricetus auratus (MesAur1.0) and SARS-CoV-2 (wuhCor1) using Subread (Liao et al., 2013) and the raw read counts were assembled using featureCounts pipeline(Liao et al., 2014). Deseq2 was used to detect differences between conditions from the human samples and from the hamster biological replicates. Because of the inherent sources of biological and technical variability, we performed surrogate variable analysis to identify outliers between our samples for human samples. The Surrogate Variable Analysis (SVA) was performed using the “sva” package in R(Leek, 2014). The number of surrogate variables were estimated with the num.sv() function using the DESeq2-generated normalized counts and “model = ∼Covid19" (plus or minus)(Love et al., 2014). One variable was estimated using the default method. The svseq() was thus run with n.sv=1, and all samples exhibited a tight distribution, except sample “205”, which was an extreme outlier, and thus was excluded from subsequent analysis. The remaining 23 samples were subject to batch correction using Combat-seq(Zhang et al., 2020). Subsequently, Deseq2 was used to determine the transcriptional consequences of COVID-19 infection in these autopsies and hamster samples. Two biological replicates were used for 1dpi and three biological replicates were used for control, 2dpi, 4dpi and 10 dpi. Z-score expression was calculated for each gene on DeSeq VS-transformed data (Variance Stabilizing Transformation) across samples in humans and across all time points in hamster. Heatmaps were generated using R function pheatmap(). Aggregated OR expression refers to the sum of all counts for annotated OR genes in each species.
hCoV-OC43 Identification
Sequencing reads from sample 2186 were mapped to the human genome using STAR, and unmapped reads were output using the “--outReadsUnmapped Fastx” parameter(Dobin et al., 2013). These reads were then provided as input into “megahit”, a short read assembly algorithm using default settings. The resultant contig assemblies were blasted against the NCBI nucleotide collection using “blast+” command line tools. Blast results were filtered for “virus” in the “sskingdom” output format variable. hCoV-OC43 was the only human virus identified in the sample. Unmapped reads from STAR were then aligned to the hCoV-OC43 genome (ACC: NC_006213) using bwa, converted to bam, sorted, and indexed using SAMtools, and visualized using IGV.
Single cell RNA-seq and analysis
Cells were dissociated according to the Worthington Papain Dissociation System by incubating fresh olfactory tissue with papain and Calcein VIolet for 40 min at 37 °C. Following dissociation, the live Celcein Violet-positive cells were sorted (MACSQuant Tyto cell sorter - Miltenyi Biotech) and assayed for scRNA-seq. Library preparation was performed accordingly to Chromium Single Cell 3ʹ v.3 Protocol for human samples and Chromium Next GEM Single Cell 5’ v.2 Protocol for hamster, respectively, and sequenced on NextSeq. Cell Ranger pipelines were used to generate fastq files which subsequently were aligned against hybrid hg38/ wuhCor1 and MesAur1.0/ wuhCor1 genomes. After alignment resulting in 8 datasets Cellbender (Fleming et al., 2019) was used to model and remove systematic biases and background noise, and to remove empty droplets. Post Cellbender h5 matrixes of 8 samples were aggregated into Seurat (Stuart et al., 2019) object using Read10X() function. Cells with more than 400 UMIs, expressed 500 and 6000 genes and less than 5% of mitochondrial genes were kept for further analysis. Data were normalized using LogNormalize() function with scale factor of 10,000. FindVariableFeatures() function with 2000 genes and the selection method set to “vst” was used to find variable features. To identify integration anchor genes among the 8 samples the FindIntegrationAnchors() function was used with 30 principal components and 2000 genes, then with IntegrateData() all data was combined into one Seurat object. The data was scaled using the ScaleData() function. Then PCA analysis was performed to reduce dimensionality and the first 30 principal components were used UMAP plots. The number pf PC was chosen based on JackStraw and elbow plots. Clustering was performed using FindClusters() function. Identified 13 clusters were visualized with UMAP (tSNE in case of human samples) and annotated using known marker genes for each cell type. Differential expression analysis was performed using the default two-sided non-parametric Wilcoxon rank sum test with Bonferroni correction using all genes in the dataset.
RNAscope in hOE
Dissected tissue was fixed in freshly prepared 4% PFA for 24 hrs at 4C and soaked sequentially at 10%, 20% and 30% sucrose 1X PBS for cryopreservation. The tissue was embedded in OCT and 10 um thick sections were mounted on SUPERFROS Plus Gold slides. To detect the S gene transcripts of SarsCov2, RNAscope® Probe - V-nCoV2019-S-sense, cat no. 845708, was incubated for 2 hr at 40C, in pre-treated sections as indicated by the RNAscope Multiplex Fluorescenct v2 Assay kit. Zeiss Zen2012 SP1 (v8.1.9.484) was used for capturing confocal images. Same conditions were applied for detection of RNAscope® Probe for Hs-ADCY3(cat no. 441671) and Hs-ATF5 (cat no. 507471). Autofluorescence of the human OE sections was removed post-acquisition using ImageJ add-on function Autofluorescence Identifier (AFid) (Baharlou et al., 2019). If followed by immunofluorescence, tissue was permeabilized with 1XPBS 0.1% Triton X 100 and blocked in a solution of 4% Donkey serum and 1x PBS 0.1% Triton X 100 for 30 minutes at RT, before incubation with primary antibodies for 2 hrs at RT at the concentrations described below.
Immunofluorescence
Dissected tissue was fixed in freshly prepared 4% PFA for 24 hrs at 4C. OE was embedded in OCT and coronal cryosections were collected at a thickness of 12μm in human specimens. In hamster, OE and OB were similarly embedded in OCT and sagittal cryosections were collected at a thickness of 8-12μm. Antigen retrieval was performed with 0.01M citric acid buffer (pH 6.0) for 10 minutes at 99C. Sections were rinsed in PBS and after permeabilization with 1x PBS 0.1% Triton X 100, slides were incubated in blocking solution (4% donkey serum +5% nonfat dry milk + 4% BSA + 0.1% Triton X-100) for 30 minutes at RT. Tissue sections were stained with primary antibodies against OMP(Chen et al., 2005) (1:50 dilution) and NP (1:200 dilution, MyBiosource cat no. MBS8574840), Anti-Iba1 (1:250 dilution, Abcam cat no. ab178846). Nuclei were labeled with DAPI (2.5 μg/ml, Thermo Fisher Scientific cat no. D3571), Anti-Cytokeratin 18 (1:250, EMD Millipore cat no. MAB3234), Anti-Adcy3 (1:250, Abcam cat no. ab123803). Primary antibodies were labeled with the following secondary antibodies: for OMP, anti-chicken IgG conjugated to Alexa-488 (2 μg/ml, Jackson ImmunoResearch cat no. A-11055, RRID:AB_2534102), for Adcy3, anti-rabbit IgG conjugated to Alexa-555 (2 μg/ml, Thermo Fisher Scientific cat no. 703-545-155, RRID:AB_2340375), for cyto-Krt18, anti-mouse IgG conjugated to Alexa-488 (2 μg/ml, Thermo Fisher Scientific cat no. A-212-2, RRID:AB_2340375). Confocal images were collected with a Zeiss LSM 700 and image processing was carried out with Fiji (NIH).
Fluorescence-activated nuclei sorting
Frozen 1% PFA-fixed tissue was mechanically crushed using Covaris Impactor and then nuclei were extracted with OptiPrep Density Gradient Medium according to the Sigma Millipore protocol. Following extraction and filtering two times through a 35-μm cell strainer, nuclei were stained with Lhx2/Atf5 antibodies for human samples. Next DAPI/Lhx2/Atf5 triple positive nuclei were sorted on a BD Aria II or BD Influx cell sorter for HiC experiments.
In situ Hi-C
Depending on the sample, between 30 and 100 thousand nuclei were used for in situ Hi-C. Sorted nuclei were lysed and processed through an in situ Hi-C protocol as previously described with a few modifications. In brief, cells were lysed with 10 mM Tris pH 8 0.2% Igepal, 10 mM NaCl. Pelleted intact nuclei were then resuspended in 0.5% SDS and incubated for 20 min at 62 °C for nuclear permeabilization. After being quenched with 1.1% Triton-X for 10 min at 37 °C, nuclei were digested with 25 U/μl MseI in 1× CutSmart buffer for 1.5 hours at 37 °C. Following digestion, the restriction enzyme was inactivated at 62 °C for 20 min. For the 45-min fill-in at 37 °C, biotinylated dUTP was used instead of dATP to increase ligation efficiency. Ligation was performed at 25 °C for 30 min with rotation after which nuclei were centrifuges. To degrade proteins and revers crosslinks pellets were incubated overnight at 75 °C with proteinase K. Each sample was transferred to Pre-Slit Snap-Cap glass mictoTUBE and sonicated on a Covaris S220 for 90 sec.
Hi-C library preparation and sequencing
Sonicated DNA was purified with 2× Ampure beads following the standard protocol and eluted in 300 μl water. Biotinylated fragments were enriched as previously described using Dynabeads MyOne Strepavidin T1 beads. The biotinylated DNA fragments were prepared for next-generation sequencing directly on the beads by using the Nugen Ovation Ultralow kit protocol as described (Monahan et al., 2019). DNA was amplified by 7 cycles of PCR. Beads were reclaimed and amplified unbiotinylated DNA fragments were purified with 1× Ampure beads. The quality and concentration of libraries were assessed using Agilent Bioanalyzer and Qubit Quantification Kit. Hi-C libraries were sequenced paired-end on NextSeq 500 (2 × 75 bp), or NovaSeq 6000 (2 × 150 bp).
Hi-C data processing and analysis
Raw fastq files were processed using the Juicer single CPU BETA version on AWS. Human data were aligned against hg19 and hamster reads were aligned to MesAur1.0_HiC.fasta.gz using BWA 0.7.17 mem algorithm. Hamster genome assembly was obtained from the DNA Zoo Consortium(Dudchenko et al., 2017) and polished with generated HiC data for mock hamster. After reads are aligned, merged, and sorted, chimaeras and duplicates are removed, and finally Hi-C contact matrices are generated by binning at various resolutions and matrix balancing. In this paper we present data with stringent cutoff of MAPQ >30. Hi-C matrices used in this paper were matrix-balanced using Juicer’s built-in Knight-Ruiz (KR) algorithm. Matrices were visualized using Juicebox(Robinson et al., 2018).
Cumulative interchromosomal contacts at the 100kb resolution were constructed by calling Juicer Tools dump function to extract genome wide normalized data from a.hic file and subsequently analyzed as previously described(Monahan et al., 2019). Briefly, we counted all OR-OR cluster combinations and measured the interchromosomal contacts that map within OR clusters. These counts were then aggregated per genomic bin. The same was done for any genomic contact outside the region of interest to estimate average 'background' contact intensity which was subsequently subtracted from intensity observed within the OR cluster. Obtained value for each cluster was used for visualization on violin plots. Of note, the intensity of random contacts was higher in COVID-19 samples that resulted in negative values on violin plots.
For human OR cluster annotation, we used genes annotated in HORDE database. For the hamster, firstly, we annotated OR genes via alignment of transcripts present in Ensemble against MesAur1.0_HiC.fasta.gz; next, we defined OR cluster as a stretch of OR genes in vicinity of 50 kb not interrupted by another non OR gene. 82 OR clusters were annotated for human, while for the hamster we were able to annotate only 46 clusters due to poor OR gene annotation in hamster genome in general.
A hidden Markov model (HMM) was used to assess the presence of genomic compartments(Rao et al., 2014). As we previously described(Monahan et al., 2019), we extracted subset of normalized interchromosomal contacts to construct 500 kb contact matrix in a manner that 500 kb loci on odd chromosomes emerged as rows while the same size loci on even chromosomes appeared on the columns. We tested 2-21 components to construct HMMs for odd vs even chromosomes; we found that 9 components reveal the existence of trans OR cluster specific compartments. A score was calculated using hmmlearn to deduce the likelihood of the given number of compartments. The same was done for the transposed even vs odd chromosome matrix. The mean value for given compartment was used for graphical visualization.
Quantification and statistical analysis
Statistical tests were performed in R using base packages for statistical analysis and ggplot2 for visualization. For all data, a p-value < 0.05 was considered to be statistically significant. Statistical details for each experiment including statistical tests applied and number of replicates can be found in the figure legends and methods details, p-values are indicated on figures.
Acknowledgments
We thank members of the Lomvardas lab, Konstantin Popadin, and Muhammad Saad Shamim for helpful analysis notes; David Weisz for assistance with software; and Gary Struhl for the help with imaging. We thank Drs. Axel, Zuker, Maniatis, and Rizvi for helpful comments and suggestions. We are grateful to the patients with COVID-19 and their families for giving their consent to autopsies and neuropathologists and staff of the CU biobank for assistance in acquiring OE autopsies. The study was approved by the ethics and Institutional Review Board of CIUMC (IRB AAAT0689 and AAAS7370). LVG hamsters (Mesocricetus auratus) were treated in compliance with the rules and regulations of IACUC under protocol number PROTO202000113-20-0743. Funding: NIDCD 3R01DC018744-01S1 (S.L. and J.B.O.), U01DA052783 (S.L.), NIA 3R01AG065582-01S1 (P.R.), 3R01AG067025-02S3 (P.R.), HHMI Faculty Scholar Award (S.L.), Zegar Family Foundation (S.L.), and Marc Haas Foundation (BRT). Graphical abstract and schemes were created with BioRender.com.
Author contributions
Conceptualization: M.Z., A.K., M.E.G., J.B.O., and S.L. Methodology: M.Z., A.K., D.A.H., B.R.T., J.B.O., and S.L. Software: A.D.O. Formal analysis: M.Z., A.K., and S.H. Investigation: M.Z., A.K., D.A.H., J.F., J.F.F., R.M., S.U., and N.G.M. Resources: J.F.F., H.S., S.F., Q.G., P.D.C., J.E.G., P.R., B.R.T., and J.B.O. Data curation: M.Z. and A.K. Writing: M.Z., A.K., J.B.O., and S.L. Supervision: B.R.T., J.B.O., and S.L. Funding acquisition: P.R., B.R.T., J.B.O., and S.L.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We worked to ensure sex and racial balance in the collection of human OE autopsies. We worked to ensure gender balance in our reference list while citing work relevant to this study.
Published: February 2, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2022.01.024.
Supplemental information
Data and code availability
Human and hamster RNAseq, scRNAseq, in situ HiC, supplemental spreadsheets and detailed experimental protocols have been deposited to 4DN portal at https://doi.org/10.1101/2021.02.09.430314.
This study did not generate custom computer code.
Any additional information is available from the Lead Contact upon request.
References
- Albers M.W., Tabert M.H., Devanand D.P. Olfactory dysfunction as a predictor of neurodegenerative disease. Curr. Neurol. Neurosci. Rep. 2006;6:379–386. doi: 10.1007/s11910-996-0018-7. [DOI] [PubMed] [Google Scholar]
- Baharlou H., Canete N.P., Bertram K.M., Sandgren K.J., Cunningham A.L., Harman A.N., Patrick E. AFid: A tool for automated identification and exclusion of autofluorescent objects from microscopy images. bioRxiv. 2019 doi: 10.1093/bioinformatics/btaa780. [DOI] [PubMed] [Google Scholar]
- Bashkirova E., Lomvardas S. Olfactory receptor genes make the case for inter-chromosomal interactions. Curr. Opin. Genet. Dev. 2019;55:106–113. doi: 10.1016/j.gde.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirova E., Monahan K., Campbell C.E., Osinski J.M., Tan L., Schieren I., Barnea G., Xie X.S., Gronostajski R.M., Lomvardas S., et al. Homeotic regulation of olfactory receptor choice via NFI-dependent heterochromatic silencing and genomic compartmentalization. bioRxiv. 2020 [Google Scholar]
- Bilinska K., Jakubowska P., Von Bartheld C.S., Butowt R. Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: Identification of cell types and trends with age. ACS Chem. Neurosci. 2020;11:1555–1562. doi: 10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscolo-Rizzo P., Guida F., Polesel J., Marcuzzo A.V., Antonucci P., Capriotti V., Sacchet E., Cragnolini F., D'Alessandro A., Zanelli E., et al. Self-reported smell and taste recovery in coronavirus disease 2019 patients: A one-year prospective study. Eur. Arch. Otorhinolaryngol. 2022;279:515–520. doi: 10.1007/s00405-021-06839-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscolo-Rizzo P., Menegaldo A., Fabbris C., Spinato G., Borsetto D., Vaira L.A., Calvanese L., Pettorelli A., Sonego M., Frezza D., et al. Six-month psychophysical evaluation of olfactory dysfunction in patients with COVID-19. Chem. Senses. 2021;46 doi: 10.1093/chemse/bjab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brann D.H., Tsukahara T., Weinreb C., Lipovsek M., Van den Berge K., Gong B., Chance R., Macaulay I.C., Chou H.J., Fletcher R.B., et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abc5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet L.J., Gold G.H., Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17:681–693. doi: 10.1016/s0896-6273(00)80200-7. [DOI] [PubMed] [Google Scholar]
- Buck L., Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Butowt R., von Bartheld C.S. Anosmia in COVID-19: Underlying mechanisms and assessment of an olfactory route to brain infection. Neuroscientist. 2020 doi: 10.1177/1073858420956905. 1073858420956905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapurin N., Totten D.J., Chaballout B., Brennan J., Dennis S., Lubner R., Chowdhury N.I., Turner J.H., Trone T., Chandra R.K. Differential olfactory outcomes in COVID-19: A large healthcare system population study. Int. Forum Allergy Rhinol. 2022;12:108–111. doi: 10.1002/alr.22870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Kohno K., Gong Q. Conditional ablation of mature olfactory sensory neurons mediated by diphtheria toxin receptor. J. Neurocytol. 2005;34:37–47. doi: 10.1007/s11068-005-5046-8. [DOI] [PubMed] [Google Scholar]
- Chen M., Shen W., Rowan N.R., Kulaga H., Hillel A., Ramanathan M., Jr., Lane A.P. Elevated ACE-2 expression in the olfactory neuroepithelium: implications for anosmia and upper respiratory SARS-CoV-2 entry and replication. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.01948-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippa V., Aleem A., Anjum F. StatPearls (Treasure Island (FL) 2021. Postacute coronavirus (COVID-19) syndrome. [Google Scholar]
- Cleary S.J., Pitchford S.C., Amison R.T., Carrington R., Robaina Cabrera C.L., Magnen M., Looney M.R., Gray E., Page C.P. Animal models of mechanisms of SARS-CoV-2 infection and COVID-19 pathology. Br. J. Pharmacol. 2020;177:4851–4865. doi: 10.1111/bph.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney E.J., LeGros M.A., Mosley C.P., Clowney F.G., Markenskoff-Papadimitriou E.C., Myllys M., Barnea G., Larabell C.A., Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubé M., Le Coupanec A., Wong A.H.M., Rini J.M., Desforges M., Talbot P.J. Axonal transport enables neuron-to-neuron propagation of human coronavirus OC43. J. Virol. 2018;92 doi: 10.1128/JVI.00404-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O., Batra S.S., Omer A.D., Nyquist S.K., Hoeger M., Durand N.C., Shamim M.S., Machol I., Lander E.S., Aiden A.P., et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand N.C., Shamim M.S., Machol I., Rao S.S., Huntley M.H., Lander E.S., Aiden E.L. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante M.A., Kurtenbach S., Sargi Z.B., Harbour J.W., Choi R., Kurtenbach S., Goss G.M., Matsunami H., Goldstein B.J. Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat. Neurosci. 2020;23:323–326. doi: 10.1038/s41593-020-0587-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellul M.A., Benjamin L., Singh B., Lant S., Michael B.D., Easton A., Kneen R., Defres S., Sejvar J., Solomon T. Neurological associations of COVID-19. Lancet Neurol. 2020;19:767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming S.J., J. C.M., Babadi M. CellBender remove-background: A deep generative model for unsupervised removal of background noise from scRNA-seq datasets. bioRxiv. 2019 [Google Scholar]
- Fletcher R.B., Das D., Gadye L., Street K.N., Baudhuin A., Wagner A., Cole M.B., Flores Q., Choi Y.G., Yosef N., et al. Deconstructing olfactory stem cell trajectories at single-cell resolution. Cell Stem Cell. 2017;20:817–830.e8. doi: 10.1016/j.stem.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frere J.J., Serafini R.A., Pryce K.D., Zazhytska M., Oishi K., Golynker I., Panis M., Zimering J., Horiuchi S., Hoagland D.A., et al. SARS-CoV-2 infection results in lasting and systemic perturbations post recovery. bioRxiv. 2022 doi: 10.1101/2022.01.18.476786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadye L., Das D., Sanchez M.A., Street K., Baudhuin A., Wagner A., Cole M.B., Choi Y.G., Yosef N., Purdom E., et al. Injury activates transient olfactory stem cell states with diverse lineage capacities. Cell Stem Cell. 2017;21:775–790.e9. doi: 10.1016/j.stem.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigues E., Janvier P., Kherabi Y., Le Bot A., Hamon A., Gouze H., Doucet L., Berkani S., Oliosi E., Mallart E., et al. Post-discharge persistent symptoms and health-related quality of life after hospitalization for COVID-19. J. Infect. 2020;81:e4–e6. doi: 10.1016/j.jinf.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerkin R.C., Ohla K., Veldhuizen M.G., Joseph P.V., Kelly C.E., Bakke A.J., Steele K.E., Farruggia M.C., Pellegrino R., Pepino M.Y., et al. Recent smell loss is the best predictor of COVID-19: A preregistered, cross-sectional study. medRxiv. 2020 [Google Scholar]
- Graham E.L., Clark J.R., Orban Z.S., Lim P.H., Szymanski A.L., Taylor C., DiBiase R.M., Jia D.T., Balabanov R., Ho S.U., et al. Persistent neurologic symptoms and cognitive dysfunction in non-hospitalized COVID-19 “long haulers. Ann. Clin. Transl. Neurol. 2021;8:1073–1085. doi: 10.1002/acn3.51350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota J., Mombaerts P. The LIM-homeodomain protein Lhx2 is required for complete development of mouse olfactory sensory neurons. Proc. Natl. Acad. Sci. USA. 2004;101:8751–8755. doi: 10.1073/pnas.0400940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D.A., Møller R., Uhl S.A., Oishi K., Frere J., Golynker I., Horiuchi S., Panis M., Blanco-Melo D., Sachs D., et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity. 2021;54:557–570.e5. doi: 10.1016/j.immuni.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornuss D., Lange B., Schröter N., Rieg S., Kern W.V., Wagner D. Anosmia in COVID-19 patients. Clin. Microbiol. Infect. 2020;26:1426–1427. doi: 10.1016/j.cmi.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Suzuki M., Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314:657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Loh Y.E., Rajarajan P., Hirayama T., Liao W., Kassim B.S., Javidfar B., Hartley B.J., Kleofas L., Park R.B., et al. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat. Genet. 2017;49:1239–1250. doi: 10.1038/ng.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Yoo S.J., Clijsters M., Backaert W., Vanstapel A., Speleman K., Lietaer C., Choi S., Hether T.D., Marcelis L., et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell. 2021;184:5932–5949.e15. doi: 10.1016/j.cell.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane A.P., Turner J., May L., Reed R. A genetic model of chronic rhinosinusitis-associated olfactory inflammation reveals reversible functional impairment and dramatic neuroepithelial reorganization. J. Neurosci. 2010;30:2324–2329. doi: 10.1523/JNEUROSCI.4507-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane A.P., Zhao H., Reed R.R. Development of transgenic mouse models for the study of human olfactory dysfunction. Am. J. Rhinol. 2005;19:229–235. [PubMed] [Google Scholar]
- Leek J.T. svaseq: Removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079. [DOI] [PMC free article] [PubMed]
- Liao Y., Smyth G.K., Shi W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu Q., Li S., Lu C., Yu C.R., Huang L. G protein gamma subunit Ggamma13 is essential for olfactory function and aggressive behavior in mice. NeuroReport. 2018;29:1333–1339. doi: 10.1097/WNR.0000000000001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luers J.C., Rokohl A.C., Loreck N., Wawer Matos P.A., Augustin M., Dewald F., Klein F., Lehmann C., Heindl L.M. Olfactory and gustatory dysfunction in coronavirus Disease 19 (COVID-19) Clin. Infect. Dis. 2020;71:2262–2264. doi: 10.1093/cid/ciaa525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons D.B., Allen W.E., Goh T., Tsai L., Barnea G., Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markenscoff-Papadimitriou E., Allen W.E., Colquitt B.M., Goh T., Murphy K.K., Monahan K., Mosley C.P., Ahituv N., Lomvardas S. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 2014;159:543–557. doi: 10.1016/j.cell.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K., Horta A., Lomvardas S. LHX2− and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature. 2019;565:448–453. doi: 10.1038/s41586-018-0845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan K., Schieren I., Cheung J., Mumbey-Wafula A., Monuki E.S., Lomvardas S. Cooperative interactions enable singular olfactory receptor expression in mouse olfactory neurons. eLife. 2017;6 doi: 10.7554/eLife.28620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbandian A., Sehgal K., Gupta A., Madhavan M.V., McGroder C., Stevens J.S., Cook J.R., Nordvig A.S., Shalev D., Sehrawat T.S., et al. Post-acute COVID-19 syndrome. Nat. Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proal A.D., VanElzakker M.B. Long COVID or post-acute sequelae of COVID-19 (PASC): An overview of biological factors that may contribute to persistent symptoms. Front. Microbiol. 2021;12:698169. doi: 10.3389/fmicb.2021.698169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative Genomics Viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Turner D., Durand N.C., Thorvaldsdóttir H., Mesirov J.P., Aiden E.L. Juicebox.js provides a cloud-based visualization system for Hi-C data. Cell Syst. 2018;6:256–258.e1. doi: 10.1016/j.cels.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez S., Cao L., Rickenbacher G.T., Benz E.G., Magdamo C., Ramirez Gomez L.A., Holbrook E., Dhilla Albers A., Gallagher R., Westover M.B., et al. Innate immune signaling in the olfactory epithelium reduces odorant receptor levels: modeling transient smell loss in COVID-19 patients. medRxiv. 2020 [Google Scholar]
- Saito H., Kubota M., Roberts R.W., Chi Q., Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119:679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Schneider C., Rasband W., Eliceiri K. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia S.F., Yan L.M., Chin A.W.H., Fung K., Choy K.T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Ma W., Wu H., Zheng Y., Xing D., Chen R., Li X., Daley N., Deisseroth K., Xie X.S. Changes in genome architecture and transcriptional dynamics progress independently of sensory experience during post-natal brain development. Cell. 2021;184:741–758.e17. doi: 10.1016/j.cell.2020.12.032. [DOI] [PubMed] [Google Scholar]
- Tan L., Xing D., Daley N., Xie X.S. Three-dimensional genome structures of single sensory neurons in mouse visual and olfactory systems. Nat. Struct. Mol. Biol. 2019;26:297–307. doi: 10.1038/s41594-019-0205-2. [DOI] [PubMed] [Google Scholar]
- Thakur K.T., Miller E.H., Glendinning M.D., Al-Dalahmah O., Banu M.A., Boehme A.K., Boubour A.L., Bruce S.S., Chong A.M., Claassen J., et al. COVID-19 neuropathology at Columbia University Irving Medical Center/New York Presbyterian Hospital. Brain. 2021;144:2696–2708. doi: 10.1093/brain/awab148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J.Y., Wong A., Zhu D., Fastenberg J.H., Tham T. The prevalence of olfactory and gustatory dysfunction in COVID-19 patients: A systematic review and meta-analysis. Otolaryngol. Head Neck Surg. 2020;163:3–11. doi: 10.1177/0194599820926473. [DOI] [PubMed] [Google Scholar]
- Wang S.S., Lewcock J.W., Feinstein P., Mombaerts P., Reed R.R. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- Wang S.S., Tsai R.Y., Reed R.R. The characterization of the Olf-1/EBF-like HLH transcription factor family: Implications in olfactory gene regulation and neuronal development. J. Neurosci. 1997;17:4149–4158. doi: 10.1523/JNEUROSCI.17-11-04149.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.Z., Sell, J., Weiss, D., and Lall, R. (2020). COVID-19 presenting as anosmia and dysgeusia in New York City emergency departments, March - April 2020. medRxiv, 2020.2007.2006.20147751.
- Wong S.T., Trinh K., Hacker B., Chan G.C., Lowe G., Gaggar A., Xia Z., Gold G.H., Storm D.R. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27:487–497. doi: 10.1016/s0896-6273(00)00060-x. [DOI] [PubMed] [Google Scholar]
- Zazhytska M., Kodra A., Hoagland D.A., Fullard J.F., Shayya H., Omer A., Firestein S., Gong Q., Canoll P.D., Goldman J.E., et al. Disruption of nuclear architecture as a cause of COVID-19 induced anosmia. bioRxiv. 2021 [Google Scholar]
- Zhang Y., Parmigiani G., Johnson W.E. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020;2:lqaa078. doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D.J., Chesler A.T., Le Pichon C.E., Kuznetsov A., Pei X., Hwang E.L., Firestein S. Absence of adenylyl cyclase 3 perturbs peripheral olfactory projections in mice. J. Neurosci. 2007;27:6675–6683. doi: 10.1523/JNEUROSCI.0699-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Human and hamster RNAseq, scRNAseq, in situ HiC, supplemental spreadsheets and detailed experimental protocols have been deposited to 4DN portal at https://doi.org/10.1101/2021.02.09.430314.
This study did not generate custom computer code.
Any additional information is available from the Lead Contact upon request.