ABSTRACT
Numerous studies have examined the composition of and factors shaping the oral bacterial microbiota in healthy adults; however, similar studies on the less dominant yet ecologically and clinically important fungal microbiota are scarce. In this study, we characterized simultaneously the oral bacterial and fungal microbiomes in a large cohort of systemically healthy Chinese adults by sequencing the bacterial 16S rRNA gene and fungal internal transcribed spacer. We showed that different factors shaped the oral bacterial and fungal microbiomes in healthy adults. Sex and age were associated with the alpha diversity of the healthy oral bacterial microbiome but not that of the fungal microbiome. Age was also a major factor affecting the beta diversity of the oral bacterial microbiome; however, it only exerted a small effect on the oral fungal microbiome when compared with other variables. After controlling for age and sex, the bacterial microbiota structure was most affected by marital status, recent oral conditions and oral hygiene-related factors, whereas the fungal microbiota structure was most affected by education level, fruits and vegetables, and bleeding gums. Bacterial–fungal interactions were limited in the healthy oral microbiota, with the strongest association existing between Pseudomonas sp. and Rhodotorula dairenensis. Several bacterial amplicon sequence variants (ASVs) belonging to Veillonella atypica and the genera Leptotrichia, Streptococcus and Prevotella_7 and fungal ASVs belonging to Candida albicans and the genus Blumeria were revealed as putative pivotal members of the healthy oral microbiota. Overall, our study has facilitated understanding of the determining factors and cross-kingdom interactions of the healthy human oral microbiome.
IMPORTANCE Numerous studies have examined the bacterial community residing in our oral cavity; however, information on the less dominant yet ecologically and clinically important fungal members is limited. In this study, we characterized simultaneously the oral bacterial and fungal microbial communities in a large cohort of healthy Chinese adults, examined their associations with an array of host factors, and explored potential interactions between the two microbial groups. We showed that different factors shape the diversity and structure of the oral bacterial and fungal microbial communities in healthy adults, with, for instance, sex and age only associated with the diversity of the bacterial community but not that of the fungal community. Besides, we found that bacterial–fungal interactions are limited in the healthy oral cavity. Overall, our study has facilitated understanding of the determining factors and bacterial–fungal interactions of the healthy human oral microbial community.
KEYWORDS: oral microbiome, bacterial microbiota, mycobiome, bacterial–fungal interaction, keystone species
INTRODUCTION
The human oral cavity is among the most diverse microbial ecosystems in the human body, together with the gut (1), harboring over 600 bacterial species and 100 fungal species (2, 3). Microbes residing in the human oral cavity, collectively known as the oral microbiota, play crucial roles in health and disease (4). Numerous efforts have been made to characterize the composition and determinants of the normal oral bacterial microbiome in healthy adults (5–7). However, the oral fungal microbiome, also known as mycobiome, in healthy adults has received much less attention (3, 8), albeit their ecological and clinical significance (9). As a result, factors shaping the healthy oral mycobiome remain largely elusive to date. Besides, potential interactions between the two kingdoms in the oral microbiome of healthy individuals remain largely unexplored. A comprehensive understanding of the influence of the human oral microbiota on health and disease requires a holistic view of both intra- and cross-kingdom interactions among members of the oral microbiota (10). In fact, based on the analysis of ecological association networks, it has been illustrated that characterization of the relationships among members of a microbial ecosystem is biased unless members from other kingdom(s) are also included (11).
In this study, we characterized simultaneously for the first time the oral bacterial and fungal microbiomes in a large cohort of systemically healthy Chinese adults by using high-throughput next-generation sequencing. We examined the associations between 24 metadata variables related to sociodemographic characteristics, lifestyle, diet, oral hygiene, oral health, and oral intimate behavior and the oral bacterial and fungal microbiomes. We further elucidated bacterial–fungal interactions and identified potential keystone taxa of the healthy oral microbiome by constructing cross-kingdom association networks.
RESULTS
After sample selection and quality filtering, 16S rRNA gene sequence data from 664 healthy individuals were included in bacterial microbiome analysis and ITS sequence data from 304 healthy individuals were included in mycobiome analysis (Fig. 1). Both populations had a sex ratio of 1:1 and a mean age of around 45 yrs (range: 18–65 yrs) (Table S1 in the supplemental material). Detailed characteristics of the study cohort are given in Table S1. Rarefaction curve analysis showed that at a depth of 3,000 and 500 sequence reads, the majority of the bacterial and fungal diversity was captured, respectively (Fig. S1).
FIG 1.
Flow chart of sample selection for the current study. Quality-filtered 16S rRNA and internal transcribed spacer (ITS) sequence data sets were used in analysis of cohort characteristics, rarefaction curves, and taxonomic barplots, whereas subsets with complete metadata were used in diversity analysis, effect size analysis, and differential abundance analysis. Samples with both 16S and ITS sequence data were used in network analysis.
Composition of the healthy oral bacterial and fungal microbiota.
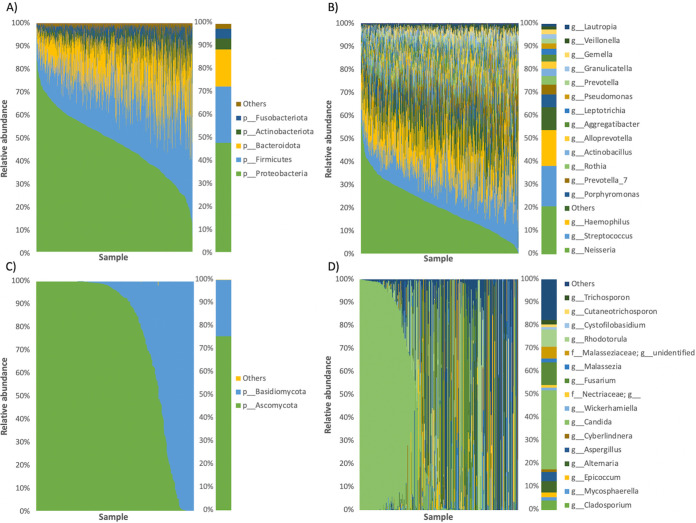
The oral bacterial microbiota in our healthy Chinese adult population was dominated by the phyla Proteobacteria (47.9%), Firmicutes (24.6%), Bacteroidota (16.2%), Actinobacteriota (4.8%), and Fusobacteriota (4.2%) (Fig. 2A). Top bacterial genera included Neisseria (20.8%), Streptococcus (17.4%), Haemophilus (15.6%), Porphyromonas (5.5%), and Prevotella_7 (4.2%) (Fig. 2B). Using a prevalence threshold of 90% and 70%, 19 core genera and 11 core ASVs were identified, respectively (Data set S1 in the supplemental material). In particular, the genera Neisseria, Streptococcus, Haemophilus, Leptotrichia, and Granulicatella were detected in all 664 samples and made up an average of 58.4% of the total microbial community. Core ASVs included those identified as Haemophilus parainfluenzae T3T1, Rothia mucilaginosa, Rothia aeria, and Rothia dentocariosa ATCC 17931.
FIG 2.
Taxonomic barplots of the bacterial (A-B) and fungal (C-D) oral microbiome at the phylum (A, C) and genus (B, D) levels. Taxa with a mean relative abundance < 1% were grouped into “Others”. The left panels show the results of each individual, whereas the right panels are the averaged values.
The oral fungal microbiota in our healthy population was composed of the phyla Ascomycota (75.5%) and Basidiomycota (24.5%) (Fig. 2C). It was most represented by the genus Candida (34.3%), followed by Fusarium (9.7%), Rhodotorula (7.5%), Alternaria (5.0%), and Cladosporium (4.1%) (Fig. 2D). Among all genera, Candida appeared in 65% of all 304 samples, representing a core genus of the healthy oral fungal microbiota.
Determinants of alpha diversity of the healthy oral microbiota.
Among the 24 metadata variables examined, sex was significantly associated with the number of observed ASVs, Shannon diversity and Pielou’s evenness of the healthy oral bacterial microbiome (P < 0.05), whereas age was significantly associated with Faith’s PD (P < 0.001, q < 0.001) (Fig. 3; Data set S2 in the supplemental material). After controlling for sex and age, significant associations were observed between alpha diversity metrics and nail-biting and recent oral conditions, namely, false teeth, mouth ulcer, and pharyngolaryngitis within 3 months (P < 0.05); however, none of them remained significant after controlling for multiple comparisons (q > 0.1) (Fig. 3; Data set S2).
FIG 3.
Boxplots showing the associations between metadata variables and alpha diversity in the bacterial (A–K) and fungal (L–M) oral microbiome. Apart from sex and age group, all other variables were controlled for sex and age. Only statistically significant variables are shown here. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Unlike in the case of bacterial microbiome, no significant associations were observed between sex or age and any of the alpha diversity metrics calculated for the oral mycobiome (P > 0.05) (Data set S3). Among all 24 metadata variables examined, only marital status was observed to be associated with Shannon diversity and Pielou's evenness of the healthy oral mycobiome (P < 0.05). The associations remained significant after controlling for sex and age (P < 0.05) but not multiple comparisons (q > 0.1) (Fig. 3; Data set S3).
Determinants of beta diversity of the healthy oral microbiota.
Similar to alpha diversity, sex and age were significantly associated with all three beta diversity metrics calculated for the oral bacterial microbiome (P < 0.01, q < 0.1) (Fig. S2, Data set S4 in the supplemental material). Age had a relatively large effect on the overall structure of the bacterial microbiome based on all three beta diversity metrics (Bray–Curtis, R2 = 0.0066; unweighted UniFrac, R2 = 0.0102; weighted UniFrac, R2 = 0.0096); however, a large effect of sex was only observed in the weighted UniFrac distance metric (Bray–Curtis, R2 = 0.0029; unweighted UniFrac, R2 = 0.0032; weighted UniFrac, R2 = 0.0077) (Fig. S3). After controlling for sex and age, largest impacts on the healthy oral bacterial microbiome were attributed to marital status (R2 = 0.0079, P < 0.05), oral hygiene-related variables, namely, brush before bed (R2 = 0.0045, P < 0.01) and mouthwash (R2 = 0.0042, P < 0.05), and false teeth (R2 = 0.0044, P < 0.01) based on Bray–Curtis dissimilarity; recent oral conditions, namely, bleeding gums (R2 = 0.0052, P < 0.01) and false teeth (R2 = 0.0051, P < 0.01) within 3 months based on unweighted UniFrac distance; and mouth ulcer (R2 = 0.0072, P < 0.05) and spicy food (R2 = 0.0062, P < 0.05) based on weighted UniFrac distance (Fig. 4A to C, Data set S4). Significant associations remained after controlling for multiple comparisons included those between Bray–Curtis dissimilarity and brush before bed and false teeth, and those between unweighted UniFrac distance and false teeth and bleeding gums (q < 0.1).
FIG 4.
Effect size of metadata variables on the oral bacterial (A–C) and fungal (D–E) microbiota composition based on Bray–Curtis dissimilarity (A, D), unweighted Unifrac (B), weighted Unifrac (C), and Jaccard (E) distances after controlling for sex and age. Bars are colored based on the category of the variables. *, P < 0.05; **, P < 0.01.
For fungi, beta diversity analysis revealed that both sex and age were significantly associated with Jaccard distance (P < 0.05, q < 0.1); however, for Bray–Curtis dissimilarity, significant association was only observed in age without controlling for multiple comparisons (P < 0.05, q > 0.1) (Fig. S4, Data set S5 in the supplemental material). Unlike in the case of bacterial microbiome, age only showed a relatively small effect (R2 < 0.0080) based on both beta diversity metrics (Fig. S5). After controlling for sex and age, largest impacts on the healthy oral mycobiome were attributed to education level (R2 = 0.0216, P < 0.05), followed by fruits and vegetables (R2 = 0.0127, P < 0.05) and bleeding gums (R2 = 0.0110, P < 0.05) based on Bray–Curtis dissimilarity; and fruits and vegetables (R2 = 0.0096, P < 0.05) based on Jaccard distance (Fig. 4D and E); however, none of them remained significant after controlling for multiple comparisons (q > 0.1) (Data set S5 in the supplemental material).
Differentially abundant bacterial and fungal ASVs across metadata groups.
Using the compositionality-aware tool Songbird, differentially abundant bacterial ASVs were identified between groups in nine metadata variables (Data set S6 in the supplemental material). Some ASVs were associated with multiple metadata variables. For instance, an ASV belonging to Aggregatibacter segnis was enriched in individuals with false teeth or bleeding gums or in the elderly and was depleted in those who brush before bed or with mouth ulcer (Fig. 5). Another ASV identified as Aggregatibacter aphrophilus was more abundant in those with mouth ulcer, bleeding gums or false teeth. Besides, an ASV belonging to Prevotella pallens was enriched in individuals with bleeding gums or mouth ulcer or in male and was depleted in those in middle age. Finally, an ASV identified as Neisseria subflava was more abundant in those who commonly use mouthwash or brush before bed and was less abundant in the elderly.
FIG 5.
Examples of differentially abundant ASVs as detected by Songbird. Only ASVs that can be identified to the species level and were differentially abundant across multiple metadata variables are shown here. ASV IDs are provided after each species name. For each ASV, a positive log(numerator/denominator) ratio for a particular metadata variable (blue bar) indicates an enrichment of the ASV in the numerator group compared to the denominator group, whereas a negative ratio (red bar) indicates an enrichment in the denominator group.
Similarly, differentially abundant fungal ASVs were identified between groups in four metadata variables (Data set S7 in the supplemental material). For bleeding gums, which also showed significant association in the beta diversity analysis, a Songbird Q2 value < 0 was obtained, indicating poor predictive accuracy and suggesting possible overfitting of model, and was thus removed from the analysis. Similar to the case of bacterial microbiome, some fungal ASVs were associated with multiple metadata variables. Example includes an ASV belonging to Candida parapsilosis, which was depleted in individuals who commonly consume fruits and vegetables or in male and enriched in the elderly or middle age or with a high education level (Fig. 5).
Bacterial–fungal interactions in the healthy oral microbiota.
High-quality 16S and ITS sequence data were available from 292 healthy individuals (Fig. 1). These data sets were used to study associations between the oral bacterial and fungal microbiota in healthy individuals. Bacterial–fungal associations were first examined at the community level based on alpha and beta diversity. No association was observed between the alpha diversity of the bacterial and fungal microbiomes (number of observed ASVs, Spearman’s ρ = -0.01, P = 0.91; Shannon diversity, Spearman’s ρ = -0.01, P = 0.89; Pielou’s evenness, Spearman’s ρ = -0.02, P = 0.70) (Fig. S6 in the supplemental material). Similarly, no association was observed between the beta diversity of the bacterial and fungal microbiomes (Bray–Curtis, Spearman’s ρ = 0.04, P = 0.10; Jaccard, Spearman’s ρ = 0.04, P = 0.04) (Fig. S7).
Bacterial–fungal interactions were then studied at the taxon level by constructing cross-kingdom association networks. The ASV-based network constructed was composed of 177 connected nodes, including 128 bacterial ASVs and 49 fungal ASVs (Fig. 6A). Forty-seven ASVs, 45 from bacteria and two from fungi, were unconnected and not shown in the final network. There were 383 interactions (as edges) in the network, 286 (74.7%) of which were positive and 97 (25.3%) of which were negative. The strongest positive interactions were mainly between fungal ASVs, whereas the strongest negative interactions were mainly between bacterial ASVs (Data set S8 in the supplemental material). Six interactions (1.57%) between bacterial and fungal ASVs were determined. All of them were positive but the strength of five of them were weak (edge weight < 0.05) (Data set S8). The strongest bacterial–fungal interaction (edge weight: 0.59) was observed between a bacterial ASV belonging to the genus Pseudomonas from the phylum Proteobacteria (prevalence: 16.72%, relative abundance: 2.21%) and a fungal ASV belonging to Rhodotorula dairenensis from the phylum Basidiomycota (prevalence: 16.61%, relative abundance: 6.90%) (Data set S8).
FIG 6.
Cross-kingdom association networks at the amplicon sequence variant (ASV) (A) and genus (B) levels constructed using SPIEC-EASI. Taxa, as nodes, from different kingdoms are in different shapes and those from different phyla are in different colors. Edges in blue represent positive interactions, whereas those in red are negative ones. The thickness of edges are proportional to the interaction strength. The strongest bacterial–fungal interaction in each network is marked with an asterisk. Only connected nodes are shown here.
The association network constructed at the genus level comprised of 148 connected nodes, including 96 bacterial genera and 52 fungal genera (Fig. 6B). There were 263 interactions in the network, majority (83.7%) of which were positive. Ten interactions (3.80%) between bacterial and fungal genera were predicted, nine of which were positive (Data set S8 in the supplemental material). Strongest bacterial–fungal interactions were observed between Pseudomonas and Rhodotorula (edge weight = 0.48), Roseburia and Rhizomucor (edge weight = 0.29), Citrobacter and Lasiodiplodia (edge weight = 0.20), and Enterobacter and Botrytis (edge weight = 0.16).
Potential keystone ASVs in the healthy oral microbiota.
Potential keystone ASVs were identified based on their degree and betweenness centrality in the network. Seven ASVs had a degree > 8 and betweenness centrality > 1,000 and were regarded as potential keystone ASVs here (Fig. 7, Table S2 in the supplemental material). These included four bacterial ASVs, belonging to Veillonella atypica and the genera Leptotrichia, Prevotella_7 and Streptococcus, and three fungal ASVs, belonging to Candida albicans, genus Blumeria, and phylum Ascomycota.
FIG 7.
Keystone species analysis. Potential keystone ASVs were identified as those with a high node degree (>8) and betweenness centrality (>1,000) in the association network. Four potential bacterial keystone ASVs and three potential fungal keystone ASVs were identified and labeled here. The dashed lines correspond to the cutoff values used here.
DISCUSSION
Although there are numerous studies on the healthy oral bacterial microbiome, our current understanding of the determinants of the healthy oral mycobiome remains limited, let alone knowledge on the bacterial–fungal interactions. In this study, we characterized the variations of the oral bacterial microbiome in 664 individuals and the oral mycobiome in 304 individuals of a systemically healthy Chinese population, examined their associations with a range of, mostly oral-related, metadata variables, and explored potential bacterial–fungal interactions and identified potential keystone species in the oral microbiome. To the best of our knowledge, this study represents one of the first population-based studies that characterized simultaneously the healthy oral bacterial and fungal microbiomes in the same cohort. It is also one of the largest studies on a healthy Chinese population (28, 29).
The oral bacterial microbiome in our healthy Chinese population was dominated by the genus Neisseria. This differs from findings from other populations. For instance, Veillonella dominates the salivary bacterial microbiome of a Canadian population (7), whereas Prevotella dominates that of a Qatari population (6). This points to a population-specific healthy oral bacterial microbiome, which may be attributed to factors such as host genetics and diet. However, genera such as Neisseria, Streptococcus, and Haemophilus were detected as core genera in all these studies as well as in ours, suggesting the presence of a global core bacterial microbiome in heathy adults. Additional studies on other underexplored populations are needed to verify this hypothesis. Compared with the bacterial counterpart, studies of the healthy oral mycobiome at the population level are scarce. Zakaria et al. (30) studied the salivary mycobiome in a healthy Japanese population of community-dwelling elderly and revealed the dominance of Candida albicans in most samples. Candida is also reported to be the predominant genus in the oral mycobiome of an American population comprising HIV-positive and HIV-negative individuals (31). The dominance of Candida in the oral mycobiome was also observed in our population, indicating its importance as a major member of the healthy oral mycobiome. However, as this most prevalent fungal genus revealed in our study was only detected in less than 70% of all individuals examined, it is unlikely for it, and other fungi alike, to represent a global core genus in the healthy oral mycobiome.
We examined the associations between 24 metadata variables related to sociodemographic characteristics, lifestyle, diet, oral hygiene, oral health, and oral intimate behavior and the oral bacterial and fungal microbiomes. We showed that sex and age were significantly associated with the structure of the oral bacterial microbiome, in agreement with findings from a healthy Canadian population (7). After controlling for sex and age, significant variables affecting the oral bacterial microbiome included marital status, oral hygiene-related variables, namely, brush before bed and mouthwash, recent oral conditions, namely, bleeding gums, false teeth, and mouth ulcer within 3 months, as well as spicy food. Marital status has been shown to associate with the taxonomic abundance of the oral bacterial microbiota in adult residents of New York City (32). Tooth brushing does not affect the bacterial community structure of subgingival plaque (33); however, tooth brushing frequency affects the relative abundance of periodontal bacteria in patients with intracranial aneurysms (34). Use of chlorhexidine mouthwash is associated with major shifts in the salivary bacterial microbiome (35). Bleeding gums, mouth ulceration and denture use have been reported to affect the salivary bacterial microbiome of a Qatari population (6). Although there is no specific report on the association of a spicy food diet on the oral microbiome, multiple studies have revealed a role of diet on the oral bacterial microbiome (36, 37). Overall, results from our current study are in concordance with previous reports, showing that the healthy oral bacterial microbiome is associated with marital status, oral hygiene, oral health, and diet.
Sex and age also significantly affected the structure of the oral mycobiome in our study population. However, unlike in the case of bacterial microbiome, age only showed a small effect compared to the other variables, indicating a smaller influence of it on the oral mycobiome than on the bacterial microbiome. After controlling for sex and age, significant variables affecting the oral mycobiome included education level, fruits and vegetables, and bleeding gums, in decreasing order of importance. Although information on the relationship between diet and oral mycobiome is lacking, several studies have revealed a role of diet on the gut mycobiome (reviewed in Ref. 38). Besides, research has shown that postpartum females with gingivitis, a symptom of which is bleeding gums, have altered abundance of numerous fungal genera compared to those with good oral health (39). The association between one's education level and their oral mycobiome is less clear; it is likely that a university degree may represent a composite variable of multiple factors that affect the oral mycobiome, including the choice of diet (40). While bleeding gums was also an explanatory variable for the oral bacterial microbiome, the other two were not. Besides, unlike in the case of bacterial microbiome, none of the oral hygiene-related variables were associated with the oral mycobiome. The presence of a different set of explanatory variables for the oral bacterial and fungal microbiomes suggests that the two microbial communities are shaped by different factors. However, it is noteworthy that while we focused mainly on oral-related variables in this study, the list of included variables is not exhaustive and has likely missed variables that might explain the variation of the microbiomes, such as fat-free mass (7) and flossing habit (5). In agreement with our findings, a study on the oral mycobiome of a population comprising HIV-positive and HIV-negative individuals based on ITS2 sequencing has identified sex as a significant variable affecting the oral mycobiome, with brushing frequency, mouthwash usage, frequency of alcoholic beverage intake, and frequency of oral sex being some of the insignificant variables (31). However, in contrast to our findings, the oral mycobiome in that study was also affected by caries status and missing teeth, and not by gingivitis and diet. Discrepancies in these findings could be due to differences in the study populations, sampling methods, design of questionnaire, choice of ITS regions, and/or bioinformatics approaches used.
We identified several ASVs that were differentially abundant in the oral microbiome across multiple metadata variables. A bacterial ASV belonging to Aggregatibacter segnis was found enriched in individuals with bleeding gums or false teeth, in the elderly, or in those who do not brush before bed. This is in concordance with previous suggestion of a potential etiological role of A. segnis, formerly Haemophilus segnis, in adult periodontitis (41). Another ASV belonging to A. aphrophilus was found more abundant in individuals with mouth ulcer, bleeding gums or false teeth. This supports previous suggestion that A. aphrophilus is a causative agent of periodontal disease (42). An ASV belonging to Prevotella pallens was enriched in individuals with bleeding gums or mouth ulcer, in male, or in young individuals. Although there are no previous reports on the association between P. pallens and bleeding gums or mouth ulcer, this bacterium together with Streptococcus mutans form a saliva-based diagnosis model for childhood caries (43). In contrast to the aforementioned potentially pathogenic oral bacteria, an ASV identified as generally nonpathogenic Neisseria subflava was found more abundant in those who commonly use mouthwash or brush before bed, or in young individuals (44). Regarding the fungal members, an ASV belonging to Candida parapsilosis was found enriched in the elderly and depleted in individuals who commonly consume fruits and vegetables. This agrees with previous reports on the increased Candida colonization in the oral cavity of the elderly (45). Our finding further suggests that regular consumption of fruits and vegetables may reduce C. parapsilosis colonization.
Bacterial–fungal interactions in the oral cavity of healthy individuals have been examined previously (46). However, the majority, if not all, of these studies predict potential interactions based on traditional statistical metrics such as Spearman's correlation. Since microbiome data are compositional in nature, these approaches can yield spurious results (26). Here, we studied the associations between the oral bacterial and fungal microbiota in healthy Chinese adults using SPIEC-EASI, a compositionally robust inference approach of cross-domain associations (11), based on 16S and ITS sequence data from 292 individuals. We showed no significant associations between the bacterial and fungal microbiota at the broad scale in terms of alpha and beta diversity. We then built cross-kingdom association networks to study bacterial–fungal interactions at the ASV and genus levels. Results showed that the majority (96%) of the fungal ASVs were connected to at least one bacterial/fungal member and that strongest positive interactions were mainly observed between fungal ASVs. These findings support previous postulations that fungi play a stabilizing role in the whole microbial community organization, as revealed by increased overall connectivity and stability in networks inferred for the human lung and skin microbiomes based on bacteria and fungi combined compared to those based on either domain (11). Cross-kingdom interactions made up only 1.6% and 3.8% of the total interactions at the ASV and genus level, respectively, in the healthy oral microbiome. These values are comparable to that (4.5%) obtained for the human lung microbiome (11) and indicate limited bacterial–fungal interactions in the oral microbiome of healthy adults.
Among the cross-kingdom interactions predicted, the strongest association was observed between a bacterial ASV belonging to the genus Pseudomonas and a fungal ASV belonging to Rhodotorula dairenensis. A strong association between Pseudomonas and Rhodotorula was also detected in the genus-level association network. Pseudomonas is a diverse and metabolically versatile genus of bacteria. While Pseudomonas members are not normally considered inhabitants of the healthy oral cavity, they can be enriched in individuals with oral conditions such as chronic periodontitis (47) or receiving orthodontic treatment (48). Rhodotorula is a dominant fungal genus in the oral cavity of healthy individuals. Although R. dairenensis has been detected in clinical blood culture samples (49), there is no report of its presence in the oral cavity, in particular of healthy individuals, in the literature. The high edge weight and relative abundance of both ASVs, and the low prevalence of both ASVs indicate that the Pseudomonas–R. dairenensis interaction is a strong cross-kingdom interaction between dominant ASVs of the oral microbiome that appears in a small subset of healthy individuals, possibly with undesirable oral health conditions. However, the inability to identify the Pseudomonas ASV down to the species level has hindered interpretation of the importance of the interaction from a biological perspective. Nonetheless, coculture experiments should be performed in the future to validate the predicted interactions (11).
Keystone species are species which have a disproportionately large effect on an ecosystem relative to their abundance (50). Here, four bacterial ASVs and three fungal ASVs were identified as potential keystone taxa of the healthy oral microbiota based on network analysis. An ASV belonging to Veillonella atypica had the highest degree and betweenness centrality, a low relative abundance and high prevalence, representing a strong candidate of keystone taxa. V. atypica and other Veillonella species are early colonisers in oral biofilm formation (51). They are suggested to play a key role in maintaining oral health due to their ability to produce nitrite from nitrate and convert lactic acid to weaker acids and thereby preventing oral diseases (52). Another potential keystone taxon detected here is an ASV belonging to the genus Streptococcus. Commensal Streptococcus species are initial colonisers of the oral cavity. They are considered key players in oral homeostasis due to their metabolic plasticity and abilities to colonize multiple oral surfaces, moderate biofilm acidification, generate hydrogen peroxide, and secrete antimicrobial compounds (53). An ASV belonging to the genus Prevotella_7 represents another potential keystone taxon. Prevotella_7 is a genus-level subdivision of the diverse genus Prevotella designated based on the degree of sequence divergence and includes species such as P. multiformis, P. albensis, and P. dentalis (54). Previous physical proximity analysis has revealed that members of the genus Prevotella, together with those of Actinomyces, display extensive inter-generic associations, suggesting a central role of them in establishing and maintaining oral biofilm complexity (55). However, despite their abundance and role in oral biofilm formation, Prevotella species in the oral cavity are understudied compared with other oral bacteria (56). The last potential bacterial keystone taxon detected here is an ASV belonging to the genus Leptotrichia. Leptotrichia species are primarily oral commensals but are also opportunistic pathogens (57). It has been reported that Leptotrichia species can trigger the transcription levels of both pro- and anti-inflammatory interleukins in oral epithelial cells, suggesting their crucial role in the “fine-tune” regulation of epithelial immune response (58).
Candida albicans is among one of the fungal keystone taxa identified in the healthy oral microbiota here. Based on reported physical, metabolic, and chemical interactions between C. albicans and a range of bacteria in the oral cavity, it has been argued that C. albicans should be regarded as a keystone commensal in the oral cavity (59). Young et al. (60) tested this hypothesis using in vitro polymicrobial oral biofilm models, showing that the presence of C. albicans increased the level of metabolic activity and biomass of bacteria in the biofilms. An ASV belonging to the genus Blumeria represents another potential fungal keystone taxon in the oral microbiome of healthy adults. An unidentified species of Blumeria has also been detected in high abundance in dental plaque of caries-free children (61). However, additional information of Blumeria species in the oral cavity are scarce to date. Overall, results of our current network analysis are in concordance with previous reports on the importance of well-known oral microbes, such as Streptococcus species and C. albicans, providing further support to their potential pivotal roles in the oral microbiome of healthy adults. Our results have also unveiled potential important roles of understudied taxa, such as Prevotella_7 and Blumeria, in the healthy oral microbiome, providing clues for future research directions on this subject.
There are several limitations to our study. First, data on sampling time of day (62), medication use (63), and anthropometric measurements such as fat-free mass and waist-hip ratio (7), all of which have been reported to affect the oral microbiome, are not available in our study. Second, most metadata collected in this study were self-reported. Lastly, the use of the 16S rRNA gene V1-V2 region, just like other regions, hinders accurate taxonomic identification down to the species or strain level (64).
CONCLUSIONS
To conclude, in this study, we characterized simultaneously for the first time the oral bacterial and fungal microbiomes in a large cohort of healthy Chinese adults. We examined the associations between 24 metadata variables and the oral microbiomes and showed that the oral bacterial and fungal microbiomes in healthy Chinese adults are shaped by a different set of factors. By performing network analysis, we suggest that bacterial–fungal interactions are limited in the healthy oral microbiome. Besides, we have identified Veillonella atypica and Candida albicans, among a few other taxa, as potential keystone taxa of the healthy oral microbiome. Overall, our study has facilitated understanding of the determining factors and cross-kingdom interactions of the healthy human oral microbiome. Future studies should validate interactions predicted here using techniques such as coculture.
MATERIALS AND METHODS
Study population.
The current study includes the analysis of a subset of oral rinse samples collected from our previous population-wide oral human papillomavirus (HPV) screening study in Hong Kong (12). In the original study, a cohort of 1,469 local residents were recruited through media advertisements and health talks in the community from 2015 to 2016. Each participant completed a questionnaire including information on sociodemographic characteristics, lifestyle, diet, oral hygiene, oral health, and oral intimate behavior (Table S1 in the supplemental material). The inclusion criteria for the current study were ethnically Chinese aged ≥ 18 years at the time of recruitment. Current smokers and subjects with diagnosis of cancer, diabetes, hypertension or sexually transmitted diseases, drug eruption, tonsillectomy, lichen planus, pemphigus, oral leukoplakia, Behcet's disease or erythema multiforme in the last 3 months, long-term use of steroid, and infection with high-risk HPV were excluded to compose a cohort of healthy subjects.
Sample collection, DNA extraction, and amplicon sequencing.
Oral rinse samples were collected in 20 ml 0.9% normal saline gargled twice, for 20 and 10 s. Approximately 1 ml of the oral rinse solution was centrifuged at 5,000 g for 5 min and DNA was extracted from the pellet using the QIAamp DNA Mini-Kit (Qiagen, Germany) following the manufacturer’s instructions. The bacterial 16S rRNA gene V1-V2 region was amplified with PCR using universal primers 27F-YM (5′-AGA GTT TGA TYM TGG CTC AG-3′) and 338R (5′-TGC TGC CTC CCG TAG GAG T-3′), whereas the fungal internal transcribed spacer 1 (ITS1) region was amplified with ITS1F (5′-CTT GGT CAT TTA GAG GAA GTA A-3′) and ITS2 (5′-GCT GCG TTC TTC ATC GAT GC-3′). A pair of dual 12 bp barcodes was indexed to each amplicon set through the forward and reverse primers modified from the Earth Microbiome Project protocol (13). PCR products were pooled and sequenced on an Illumina MiSeq instrument (Illumina, San Diego, CA) at The Genomics Core Facility of the Weill Cornell Medicine Core Laboratories Centre following the 2 × 300 bp paired-end sequencing protocol. Negative controls, positive controls (mock communities), and technical replicates were also sequenced for quality control.
Microbiome analysis.
Microbiome analysis was performed with QIIME2 2020.11 (14), unless specified otherwise. Primers were first trimmed from demultiplexed raw sequence data using the q2-cutadapt plugin (15). No insertions or deletions of bases were allowed when matching primers, and reads with no primer found were discarded. Paired-end reads were then joined using q2-vsearch (16), quality-filtered using q2-quality-filter, and denoised using q2-deblur (17). Taxonomy was assigned to amplicon sequence variants (ASVs) using Naïve Bayes classifiers trained on the V1-V2 region of the SILVA 138 SSU Ref NR 99 data set for 16S (18) and full-length UNITE ver. 8.3 dynamic data set for ITS (19) with q2-feature-classifier (20). ASVs with a total read count <10 or present in only one sample were removed. Archaeal, mitochondrial, chloroplast, and phylum-unclassified reads were also discarded. Samples with <3,000 or <500 quality-filtered sequence reads were removed from the 16S and ITS data sets, respectively. Feature tables were collapsed at different taxonomic levels using q2-taxa. Core bacterial ASVs and genera, defined here at a prevalence >70% and >90%, respectively, were identified using the core-features function of q2-feature-table.
Representative sequences of bacterial ASVs were inserted into the SILVA 128 SEPP reference phylogenetic tree with SATé-enabled phylogenetic placement (SEPP) using q2-fragment-insertion (21). After rarefying the samples to the smallest number of sequences, alpha and beta diversity metrics as well as principal coordinate analysis (PCoA) plots were generated using the q2-diversity plugin with the core-metrics-phylogenetic pipeline and core-metrics pipeline for 16S and ITS, respectively. Alpha diversity metrics computed for both 16S and ITS included the number of observed ASVs, Shannon diversity, and Pielou's evenness, whereas Faith’s phylogenetic diversity (PD) was also computed for 16S. Beta diversity metrics computed for 16S included Bray–Curtis dissimilarity and weighted and unweighted UniFrac distances, whereas Bray–Curtis dissimilarity and Jaccard distance were computed for ITS. Jaccard distance of 16S was additionally computed in the analysis of bacterial–fungal interactions. Alpha rarefaction curves were generated using the alpha-rarefaction function of q2-diversity. Associations between categorical metadata variables and alpha diversity metrics were tested using q2-diversity with the alpha-group-significance function. Individuals were binned into three age groups, namely, young (18–39 yr), middle (40–59 yr) and elderly (60 yr or above) (22). Samples with missing data in any metadata variable were removed and the effect size of each metadata variable was calculated using the adonis function of q2-diversity with 9,999 permutations. Sex- and age-controlled associations between metadata variables and sample composition were tested using the same function by adding sex and age into the model formula. Similarly, sex- and age-controlled associations between metadata variables and alpha diversity metrics were tested using the anova function of q2-longitudinal (23). For metadata variables showing significant difference in beta diversity, differentially abundant ASVs were identified using the compositionality-aware q2-Songbird plugin (24) in QIIME2 2019.7 while controlling for sex and age. Feature rankings and log-ratios produced were then visualized using q2-qurro (25).
Network analysis.
Cross-kingdom association networks were built from samples with both high-quality 16S and ITS sequence data. Feature tables generated from QIIME2 were imported into R as phyloseq objects using the R package qiime2R ver. 0.99.6 (https://github.com/jbisanz/qiime2R). Rare bacterial ASVs appearing in less than 20% of samples and fungal ASVs appearing in less than 5% of samples were filtered. Cross-kingdom networks were then constructed using the R package SpiecEasi ver. 1.1.1 with the Meinshausen–Bühlmann neighborhood selection method (11, 26). The strength of associations between members in the network was estimated using edge weights, which are the average covariations from the estimated covariance matrix between all members in concern; a higher edge weight indicates a stronger association (11). Degree (the number of edges a node has) and betweenness centrality (proportion of the shortest paths in the network that pass through a node) of nodes in the networks were computed using the R package igraph ver. 1.2.6 (27). Potential keystone ASVs were identified as nodes with a high degree and betweenness centrality (11). Cross-kingdom association networks were also built using feature tables collapsed at the genus level.
Statistical analysis.
Differences in alpha diversity among groups were tested using Kruskal–Wallis test, whereas differences in beta diversity were tested using permutational multivariate analysis of variance with 999 permutations. P values were adjusted using the Benjamini–Hochberg procedure to control for multiple comparisons. Beta diversity of the 16S and ITS data sets were compared using Mantel test in q2-diversity with 9,999 permutations. Differences were considered statistically significant when P < 0.05 or q < 0.1.
Data availability.
All sequence data generated from this study were deposited in the NCBI Sequence Read Archive under BioProject accession PRJNA778006.
ACKNOWLEDGMENTS
We thank Alexander C. Vlantis, Miaoyin Liang, Ryan Kin Ho Sze and Colette Leung for the assistance to collect questionnaires and sociodemographic information. The authors also thank the anonymous participants who provided oral rinse samples for this study.
This work was partially supported by the Research Grants Council of Hong Kong SAR, China (project number CUHK 14161017) to Z.C. and a seed fund for gut microbiota research from the Faculty of Medicine, The Chinese University of Hong Kong, to P.K.S.C. The funders had no role in the study design, data collection, analysis, interpretation, or writing of the report.
M.K.C.: formal analysis, visualization, writing - original draft; J.Y.K.C.: conceptualization; M.C.S.W.: resources; P.Y.W.: investigation; P.L.: investigation; L.C.: data curation; L.L.: investigation; W.C.S.H.: investigation; A.C.M.Y.: project administration; P.K.S.C.: conceptualization, funding acquisition, resources, writing - review & editing; Z.C.: conceptualization, funding acquisition, supervision, writing - review & editing.
Footnotes
Supplemental material is available online only.
Contributor Information
Paul K. S. Chan, Email: paulkschan@cuhk.edu.hk.
Zigui Chen, Email: zigui.chen@cuhk.edu.hk.
Kileen L. Shier, Quest Diagnostics Nichols Institute
REFERENCES
- 1.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J Bacteriol 192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wade WG. 2013. The oral microbiome in health and disease. Pharmacol Res 69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Burcham ZM, Garneau NL, Comstock SS, Tucker RM, Knight R, Metcalf JL, Genetics of Taste Lab Citizen Scientists. 2020. Patterns of oral microbiota diversity in adults and children: a crowdsourced population study. Sci Rep 10:2133. doi: 10.1038/s41598-020-59016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murugesan S, Al Ahmad SF, Singh P, Saadaoui M, Kumar M, Al Khodor S. 2020. Profiling the salivary microbiome of the Qatari population. J Transl Med 18:127. doi: 10.1186/s12967-020-02291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nearing JT, DeClercq V, Van Limbergen J, Langille MGI. 2020. Assessing the variation within the oral microbiome of healthy adults. mSphere 5:e00451-20. doi: 10.1128/mSphere.00451-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD. 2014. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One 9:e90899. doi: 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz PI, Dongari-Bagtzoglou A. 2021. Critically appraising the significance of the oral mycobiome. J Dent Res 100:133–140. doi: 10.1177/0022034520956975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker JL, Bor B, Agnello M, Shi W, He X. 2017. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol 25:362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tipton L, Müller CL, Kurtz ZD, Huang L, Kleerup E, Morris A, Bonneau R, Ghedin E. 2018. Fungi stabilize connectivity in the lung and skin microbial ecosystems. Microbiome 6:12. doi: 10.1186/s40168-017-0393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong MCS, Vlantis AC, Liang M, Wong PY, Ho WCS, Boon SS, Sze RKH, Leung C, Chan PKS, Chen Z. 2018. Prevalence and epidemiologic profile of oral infection with alpha, beta, and gamma papillomaviruses in an asian Chinese population. J Infect Dis 218:388–397. doi: 10.1093/infdis/jiy160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, Prill RJ, Tripathi A, Gibbons SM, Ackermann G, Navas-Molina JA, Janssen S, Kopylova E, Vázquez-Baeza Y, González A, Morton JT, Mirarab S, Zech Xu Z, Jiang L, Haroon MF, Kanbar J, Zhu Q, Jin Song S, Kosciolek T, Bokulich NA, Lefler J, Brislawn CJ, Humphrey G, Owens SM, Hampton-Marcell J, Berg-Lyons D, McKenzie V, Fierer N, Fuhrman JA, Clauset A, Stevens RL, Shade A, Pollard KS, Goodwin KD, Jansson JK, Gilbert JA, Knight R, Earth Microbiome Project Consortium. 2017. A communal catalogue reveals Earth's multiscale microbial diversity. Nature 551:457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 16.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amir A, McDonald D, Navas-Molina JA, Kopylova E, Morton JT, Zech Xu Z, Kightley EP, Thompson LR, Hyde ER, Gonzalez A, Knight R. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:e00191-16. doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K. 2019. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res 47:D259–D264. doi: 10.1093/nar/gky1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janssen S, McDonald D, Gonzalez A, Navas-Molina JA, Jiang L, Xu ZZ, Winker K, Kado DM, Orwoll E, Manary M, Mirarab S, Knight R. 2018. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. mSystems 3:e00021-18. doi: 10.1128/mSystems.00021-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh TS, Das M, Jeffery IB, O'Toole PW. 2020. Adjusting for age improves identification of gut microbiome alterations in multiple diseases. Elife 9:e50240. doi: 10.7554/eLife.50240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bokulich NA, Dillon MR, Zhang Y, Rideout JR, Bolyen E, Li H, Albert PS, Caporaso JG. 2018. q2-longitudinal: longitudinal and paired-sample analyses of microbiome data. mSystems 3:e00219-18. doi: 10.1128/mSystems.00219-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, Zengler K, Knight R. 2019. Establishing microbial composition measurement standards with reference frames. Nat Commun 10:2719. doi: 10.1038/s41467-019-10656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fedarko MW, Martino C, Morton JT, González A, Rahman G, Marotz CA, Minich JJ, Allen EE, Knight R. 2020. Visualizing 'omic feature rankings and log-ratios using Qurro. NAR Genom Bioinform 2:lqaa023. doi: 10.1093/nargab/lqaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. 2015. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol 11:e1004226. doi: 10.1371/journal.pcbi.1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Csardi G, Nepusz T. 2006. The igraph software package for complex network research. InterJournal Complex Systems :1695. https://igraph.org. [Google Scholar]
- 28.Debelius JW, Huang T, Cai Y, Ploner A, Barrett D, Zhou X, Xiao X, Li Y, Liao J, Zheng Y, Huang G, Adami HO, Zeng Y, Zhang Z, Ye W. 2020. Subspecies niche specialization in the oral microbiome is associated with nasopharyngeal carcinoma risk. mSystems 5:e00065-20. doi: 10.1128/mSystems.00065-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia YJ, Liao Y, He YQ, Zheng MQ, Tong XT, Xue WQ, Zhang JB, Yuan LL, Zhang WL, Jia WH. 2021. Association between oral microbiota and cigarette smoking in the Chinese population. Front Cell Infect Microbiol 11:658203. doi: 10.3389/fcimb.2021.658203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zakaria MN, Furuta M, Takeshita T, Shibata Y, Sundari R, Eshima N, Ninomiya T, Yamashita Y. 2017. Oral mycobiome in community-dwelling elderly and its relation to oral and general health conditions. Oral Dis 23:973–982. doi: 10.1111/odi.12682. [DOI] [PubMed] [Google Scholar]
- 31.Fidel PL, Jr, Thompson ZA, Lilly EA, Granada C, Treas K, Dubois KR, 3rd, Cook L, Hashmi SB, Lisko DJ, Mukherjee C, Vazquez JA, Hagensee ME, Griffen AL, Leys EJ, Beall CJ. 2021. Effect of HIV/HAART and other clinical variables on the oral mycobiome using multivariate analyses. mBio 12:e00294-21. doi: 10.1128/mBio.00294-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renson A, Jones HE, Beghini F, Segata N, Zolnik CP, Usyk M, Moody TU, Thorpe L, Burk R, Waldron L, Dowd JB. 2019. Sociodemographic variation in the oral microbiome. Ann Epidemiol 35:73–80.e2. doi: 10.1016/j.annepidem.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowicki EM, Shroff R, Singleton JA, Renaud DE, Wallace D, Drury J, Zirnheld J, Colleti B, Ellington AD, Lamont RJ, Scott DA, Whiteley M. 2018. Microbiota and metatranscriptome changes accompanying the onset of gingivitis. mBio 9:e00575-18. doi: 10.1128/mBio.00575-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyysalo MJ, Mishra PP, Sundström K, Lehtimäki T, Karhunen PJ, Pessi T. 2019. Increased tooth brushing frequency is associated with reduced gingival pocket bacterial diversity in patients with intracranial aneurysms. PeerJ 7:e6316. doi: 10.7717/peerj.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bescos R, Ashworth A, Cutler C, Brookes ZL, Belfield L, Rodiles A, Casas-Agustench P, Farnham G, Liddle L, Burleigh M, White D, Easton C, Hickson M. 2020. Effects of Chlorhexidine mouthwash on the oral microbiome. Sci Rep 10:5254. doi: 10.1038/s41598-020-61912-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen TH, Kern T, Bak EG, Kashani A, Allin KH, Nielsen T, Hansen T, Pedersen O. 2018. Impact of a vegan diet on the human salivary microbiota. Sci Rep 8:5847. doi: 10.1038/s41598-018-24207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson AC, Rothballer M, Altenburger MJ, Woelber JP, Karygianni L, Vach K, Hellwig E, Al-Ahmad A. 2020. Long-term fluctuation of oral biofilm microbiota following different dietary phases. Appl Environ Microbiol 86:e01421-20. doi: 10.1128/AEM.01421-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai GC, Tan TG, Pavelka N. 2019. The mammalian mycobiome: a complex system in a dynamic relationship with the host. Wiley Interdiscip Rev Syst Biol Med 11:e1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khadija B, Imran M, Faryal R. 2021. Keystone salivary mycobiome in postpartum period in health and disease conditions. J Mycol Med 31:101101. doi: 10.1016/j.mycmed.2020.101101. [DOI] [PubMed] [Google Scholar]
- 40.Ding T, Schloss PD. 2014. Dynamics and associations of microbial community types across the human body. Nature 509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petsios A, Nakou M, Manti F. 1995. Microflora in adult periodontitis. J Periodontal Res 30:325–331. doi: 10.1111/j.1600-0765.1995.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 42.Boyanova L, Setchanova L, Gergova G, Kostyanev T, Yordanov D, Popova C, Kotsilkov K, Mitov I. 2010. Microbiological diagnosis of the severe chronic periodontitis. JofIMAB 15, book 2:89–94. doi: 10.5272/jimab.1522009_89. [DOI] [Google Scholar]
- 43.Zhang L, Sun T, Zhu P, Sun Z, Li S, Li F, Zhang Y, Tan K, Lu J, Yuan R, Chen Z, Guo D, Guo Q, Teng F, Yang F. 2020. Quantitative analysis of salivary oral bacteria associated with severe early childhood caries and construction of caries assessment model. Sci Rep 10:6365. doi: 10.1038/s41598-020-63222-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Entesari-Tatafi D, Bagherirad M, Quan D, Athan E. 2014. Iatrogenic meningitis caused by Neisseria sicca/subflava after intrathecal contrast injection, Australia. Emerg Infect Dis 20:1023–1025. doi: 10.3201/eid2006.131117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lockhart SR, Joly S, Vargas K, Swails-Wenger J, Enger L, Soll DR. 1999. Natural defenses against Candida colonization breakdown in the oral cavities of the elderly. J Dent Res 78:857–868. doi: 10.1177/00220345990780040601. [DOI] [PubMed] [Google Scholar]
- 46.Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, Salata RA, Lederman MM, Gillevet PM, Ghannoum MA. 2014. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog 10:e1003996. doi: 10.1371/journal.ppat.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Souto R, Silva-Boghossian CM, Colombo AP. 2014. Prevalence of Pseudomonas aeruginosa and Acinetobacter spp. in subgingival biofilm and saliva of subjects with chronic periodontal infection. Braz J Microbiol 45:495–501. doi: 10.1590/S1517-83822014000200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun F, Ahmed A, Wang L, Dong M, Niu W. 2018. Comparison of oral microbiota in orthodontic patients and healthy individuals. Microb Pathog 123:473–477. doi: 10.1016/j.micpath.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 49.Nunes JM, Bizerra FC, Ferreira RC, Colombo AL. 2013. Molecular identification, antifungal susceptibility profile, and biofilm formation of clinical and environmental Rhodotorula species isolates. Antimicrob Agents Chemother 57:382–389. doi: 10.1128/AAC.01647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paine RT. 1969. A note on trophic complexity and community stability. Am Nat 103:91–93. doi: 10.1086/282586. [DOI] [Google Scholar]
- 51.Periasamy S, Kolenbrander PE. 2010. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol 192:2965–2972. doi: 10.1128/JB.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wicaksono DP, Washio J, Abiko Y, Domon H, Takahashi N. 2020. Nitrite production from nitrate and its link with lactate metabolism in oral Veillonella spp. Appl Environ Microbiol 86:e01255-20. doi: 10.1128/AEM.01255-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abranches J, Zeng L, Kajfasz JK, Palmer SR, Chakraborty B, Wen ZT, Richards VP, Brady LJ, Lemos JA. 2018. Biology of oral streptococci. Microbiol Spectr 6. doi: 10.1128/microbiolspec.GPP3-0042-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henderson G, Yilmaz P, Kumar S, Forster RJ, Kelly WJ, Leahy SC, Guan LL, Janssen PH. 2019. Improved taxonomic assignment of rumen bacterial 16S rRNA sequences using a revised SILVA taxonomic framework. PeerJ 7:e6496. doi: 10.7717/peerj.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valm AM, Mark Welch JL, Rieken CW, Hasegawa Y, Sogin ML, Oldenbourg R, Dewhirst FE, Borisy GG. 2011. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci U S A 108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tett A, Pasolli E, Masetti G, Ercolini D, Segata N. 2021. Prevotella diversity, niches and interactions with the human host. Nat Rev Microbiol 19:585–599. doi: 10.1038/s41579-021-00559-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eribe ERK, Olsen I. 2017. Leptotrichia species in human infections II. J Oral Microbiol 9:1368848. doi: 10.1080/20002297.2017.1368848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langfeldt D, Neulinger SC, Stiesch M, Stumpp N, Bang C, Schmitz RA, Eberhard J. 2014. Health- and disease-associated species clusters in complex natural biofilms determine the innate immune response in oral epithelial cells during biofilm maturation. FEMS Microbiol Lett 360:137–143. doi: 10.1111/1574-6968.12596. [DOI] [PubMed] [Google Scholar]
- 59.Janus MM, Willems HM, Krom BP. 2016. Candida albicans in multispecies oral communities; a keystone commensal? Adv Exp Med Biol 931:13–20. doi: 10.1007/5584_2016_5. [DOI] [PubMed] [Google Scholar]
- 60.Young T, Alshanta OA, Kean R, Bradshaw D, Pratten J, Williams C, Woodall C, Ramage G, Brown JL. 2020. Candida albicans as an essential “keystone” component within polymicrobial oral biofilm models? Microorganisms 9:59. doi: 10.3390/microorganisms9010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Jesus VC, Khan MW, Mittermuller BA, Duan K, Hu P, Schroth RJ, Chelikani P. 2021. Characterization of supragingival plaque and oral swab microbiomes in children with severe early childhood caries. Front Microbiol 12:683685. doi: 10.3389/fmicb.2021.683685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Skarke C, Lahens NF, Rhoades SD, Campbell A, Bittinger K, Bailey A, Hoffmann C, Olson RS, Chen L, Yang G, Price TS, Moore JH, Bushman FD, Greene CS, Grant GR, Weljie AM, FitzGerald GA. 2017. A pilot characterization of the human chronobiome. Sci Rep 7:17141. doi: 10.1038/s41598-017-17362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mishiro T, Oka K, Kuroki Y, Takahashi M, Tatsumi K, Saitoh T, Tobita H, Ishimura N, Sato S, Ishihara S, Sekine J, Wada K, Kinoshita Y. 2018. Oral microbiome alterations of healthy volunteers with proton pump inhibitor. J Gastroenterol Hepatol 33:1059–1066. doi: 10.1111/jgh.14040. [DOI] [PubMed] [Google Scholar]
- 64.Johnson JS, Spakowicz DJ, Hong BY, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM. 2019. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun 10:5029. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM02410-21_Supp_1_seq8.pdf, PDF file, 4.2 MB (4.1MB, pdf)
Supplemental material. Download SPECTRUM02410-21_Supp_2_seq9.xlsx, XLSX file, 0.01 MB (13.2KB, xlsx)
Supplemental material. Download SPECTRUM02410-21_Supp_3_seq10.xlsx, XLSX file, 0.01 MB (16.1KB, xlsx)
Supplemental material. Download SPECTRUM02410-21_Supp_4_seq11.xlsx, XLSX file, 0.01 MB (14.7KB, xlsx)
Supplemental material. Download SPECTRUM02410-21_Supp_5_seq12.xlsx, XLSX file, 0.01 MB (15.3KB, xlsx)
Supplemental material. Download SPECTRUM02410-21_Supp_6_seq13.xlsx, XLSX file, 0.01 MB (14KB, xlsx)
Supplemental material. Download SPECTRUM02410-21_Supp_7_seq14.xlsx, XLSX file, 0.03 MB (32.6KB, xlsx)
Supplemental material. Download SPECTRUM02410-21_Supp_8_seq15.xlsx, XLSX file, 0.01 MB (18.3KB, xlsx)
Supplemental material. Download SPECTRUM02410-21_Supp_9_seq16.xlsx, XLSX file, 0.01 MB (18.1KB, xlsx)
Supplemental material. Download SPECTRUM02410-21_Supp_10_seq17.xlsx, XLSX file, 0.01 MB (15.1KB, xlsx)
Data Availability Statement
All sequence data generated from this study were deposited in the NCBI Sequence Read Archive under BioProject accession PRJNA778006.