Abstract
The ongoing severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) evolution has resulted in many variants, contributing to the striking drop in vaccine efficacy and necessitating the development of next-generation vaccines to tackle antigenic diversity. Herein we developed a multivalent Semliki Forest virus replicon-based mRNA vaccine targeting the receptor binding domain (RBD), heptad repeat domain (HR), membrane protein (M), and epitopes of non-structural protein 13 (nsp13) of SARS-CoV-2. The bacteria-mediated gene delivery offers the rapid production of large quantities of vaccine at a highly economical scale and notably allows needle-free mass vaccination. Favorable T-helper (Th) 1-dominated potent antibody and cellular immune responses were detected in the immunized mice. Further, immunization induced strong cross-protective neutralizing antibodies (NAbs) against the B.1.617.2 delta variant (clade G). We recorded a difference in induction of immunoglobulin (Ig) A response by the immunization route, with the oral route eliciting a strong mucosal secretory IgA (sIgA) response, which possibly has contributed to the enhanced protection conferred by oral immunization. Hamsters immunized orally were completely protected against viral replication in the lungs and the nasal cavity. Importantly, the vaccine protected the hamsters against SARS-CoV-2-induced pneumonia. The study provides proof-of-principle findings for the development of a feasible and efficacious oral mRNA vaccine against SARS-CoV-2 and its variants.
Keywords: SARS-CoV-2, variants, COVID-19, oral, mRNA, vaccine, Salmonella-mediated, alphaviruses, replicon-based, mucosal vaccine
Graphical abstract
Exploiting Semliki Forest virus replicon allows the development of bacteria-mediated oral mRNA vaccine. The vaccine induces potent Th1 polarized humoral and cellular immune responses. Oral administration elicits robust mucosal IgA response in lung and intestine. Oral replicon-based mRNA vaccine protects hamsters against parental and delta variant SARS-CoV-2 challenge.
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has witnessed the development of vaccines at an unprecedented rate, with several vaccines receiving emergency use authorization in various countries. Two of the most widely used vaccines from Pfizer-BioNTech and Moderna demonstrated >94% efficacy in preventing COVID-19.1,2 However, the emergence of novel severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants that exhibit increased ability to escape the vaccine-induced immunity has raised a great concern on future vaccine efficacy. Indeed, recently concluded clinical trials have observed a staggering drop in efficacy from 96% to 48% for Novavax vaccine (NVX-CoV2373), 85% to 57% for Johnson & Johnson vaccine (JNJ-78436,735), and 62% to 10% for Oxford-AstraZeneca vaccine (ChAdOx1d) against the B.1.351 variant.3,4
The World Health Organization (WHO) classified the SARS-CoV-2 variants as variants of interest (VOIs) and variants of concern (VOCs).5 As of September 2021, four variants fall into the category of VOC, and they rapidly become dominant in the reported countries. The VOCs are B.1.1.7 (Alpha), B.1.351 (501Y.V2, Beta), P1 (Gamma), and B.1.617.2 (Delta). The VOIs include B.1.525 (Eta), B.1.526 (Iota), B.1.617.1 (Kappa), and C.37 (Lambda). As many as 48% of sera/plasma samples from COVID-19 patients failed to neutralize the B.1.351 variant,6 highlighting the ability of the variants to escape natural immunity. Sera from individuals vaccinated with BBIBP-CorV vaccine, the BNT162b2 vaccine, and the mRNA-1273 exhibited reduced neutralizing activity against the B.1.351 variant, while an 86-fold to complete immune escape was observed for AZD1222 vaccine.7 Further, BNT162b2 and mRNA-1273 vaccinee sera had a reduced neutralizing activity by a factor of 6.7 and 4.5, respectively, against the P1 variant7. The VOCs showed a reduction of 1.2- to 8.4-fold neutralization titers against mRNA-1273-elicited sera.8 Along with evading the pre-existing immunity, these variants demonstrate enhanced infectiousness and transmissibility, highlighting the urgent need for next-generation vaccines to fight the evolving variants.
The genome of SARS-CoV-2 encodes four structural proteins; namely, spike (S), envelope (E), membrane (M), and nucleocapsid (N). The vaccine landscape against COVID-19 comprises over 300 vaccines with over 100 candidates undergoing clinical trial.9 Currently, 19 vaccines are being used globally to curtail the disease. Virtually all vaccines, barring inactivated viral vaccines, rely on the spike protein to generate an anti-SARS-CoV-2 immune response. However, variants with spike protein mutations acquire the ability to evade the vaccine-elicited immunity, leading to a striking drop in their efficacy.3,4 Thus, it necessitated the development of next-generation vaccines that induce broad protective immune responses. Hence, we hypothesized that targeting multiple SARS-CoV-2 proteins in a single vaccine would induce the protective response against ancestral virus and the variants. To this end, we designed a multicistronic vaccine candidate targeting the receptor binding domain (RBD), heptad repeat domain (HR), membrane protein (M), and epitopes of non-structural protein 13 (nsp13) of SARS-CoV-2 (V-P2A)10. The current vaccine exploits Semliki Forest virus (SFV) replicon, and Salmonella bactofection, respectively, for mRNA amplification and gene delivery.
Although an unparalleled vaccine development against COVID-19 resulted in the global rollout of vaccines in less than a year, inequitable distribution, particularly to middle- and low-income countries, is glaringly visible.11 An analysis of the recent data as of September 2021 showed that ≥50% of people living in upper-middle- and high-income countries are fully vaccinated, while only 10.3% and 0.6% of people living in low-middle- and low-income countries, respectively, have been vaccinated.12 Therefore, the bacteria-mediated vaccine development offers a unique platform for rapid and inexpensive production of large quantities of vaccine and, thus, may play an instrumental role in increasing the global reach of the COVID-19 vaccine. Moreover, the vaccine has the advantage of being administered orally and, in case of emergencies, can be easily controlled through antibiotics. Most importantly, oral delivery can be exploited to develop mucosal vaccines that elicit potent responses at respiratory mucosa along with systemic immunity.13 Further, mucosal vaccines are better suited to achieve protection against respiratory infectious diseases.14 Indeed, an intranasal vaccine induced robust mucosal immune responses and durably protected the mice against SARS-CoV-2 challenge.15,16 Herein we evaluated the immune response and protection conferred by the Salmonella-enabled oral mRNA vaccine and compared results with the intramuscular delivery.
The vaccine elicited neutralizing antibodies upon both systemic and oral delivery against the parental and delta variant SARS-CoV-2. Most importantly, the oral delivery induced complete protection against both ancestral (clade L) and delta variant (clade G) SARS-CoV-2 strains, with infectious virus completely cleared from the lungs and nasal wash of vaccinated hamsters. Moreover, the vaccine protected the hamsters against SARS-CoV-2-induced weight loss and lung pathology. The data suggest the prospect of exploiting bacteria-mediated gene delivery to develop effective oral replicon-based mRNA vaccines.
Results
We designed a multicistronic vaccine candidate targeting SARS-CoV-2 RBD, HR, M, and epitopes of nsp13 (RNA helicase) (Figure S1). To improve the transgene expression, SFV replicon-based vector was exploited, which drives the gene expression by a self-replicating mechanism of mRNA.17 Further, a Salmonella delivery vehicle was employed for efficient DNA delivery. Figure 1 describes the mechanism of transgene expression and the generation of an immune response. We previously demonstrated that the vaccine was safe and elicited potent virus-neutralizing antibodies coupled with robust cellular response in mice.10 In the present study, we extend our findings to determine the immune response and protective efficacy in mouse and hamster models of SARS-CoV-2. In particular, we evaluated the elicitation of the cross-protective immune response against the parental virus (clade L) and the delta variant (B.1.617.2, clade G) SARS-CoV-2 both in vitro and in vivo.
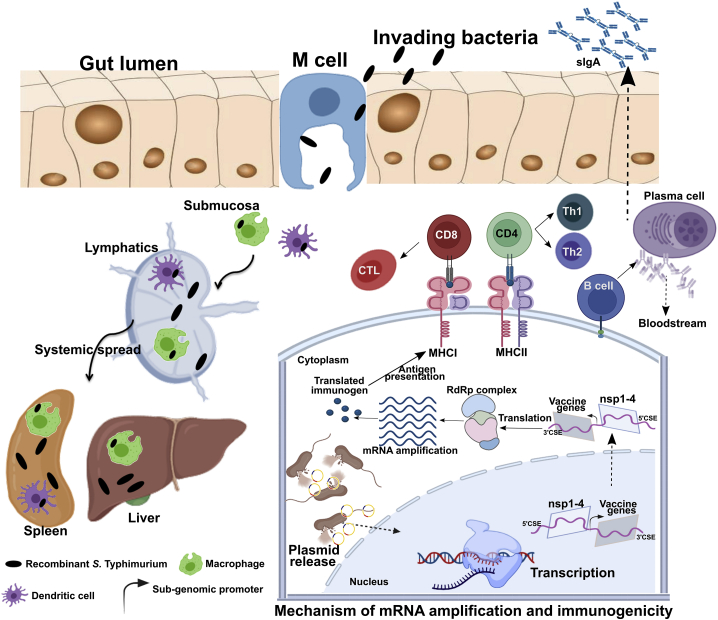
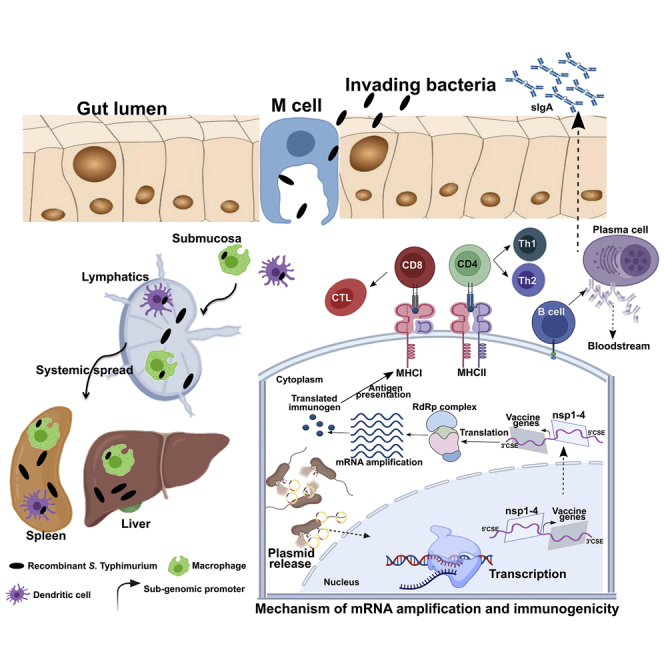
Figure 1.
Mechanism of gene delivery, mRNA amplification, and initiation of immune response
Upon oral administration, Salmonella is translocated from the luminal surface to submucosa by specialized M cells in the gut epithelium. Bacteria then invade APCs such as macrophages and DCs and are spread to different organs, like the liver and spleen, through lymphatics and the bloodstream. The vector encoding SFV replicon (nsp1-4) and SARS-CoV-2 immunogens is released within the host cell cytoplasm through bacterial lysis. Transcription of the delivered plasmid takes place in the cell nucleus and, following in situ translation, the nsp1-4 proteins form an RNA-dependent RNA polymerase (RdRp) complex. The RdRp complex then recognizes the sub-genomic promoter and flanking conserved sequence elements (CSEs) leading to enhanced mRNA amplification of vaccine genes. The resulting mRNAs are translated to produce immunogenic proteins. The APCs process and present antigen to CD8 and CD4 T cells via the MHC I and MHC II molecules, respectively, leading to the elicitation of T cell response. DCs can present antigen directly to B cells or follicular DCs (FDCs). FDCs store antigen for a longer time, periodically displaying the antigen to cognate B cells. B cells then differentiate to specific antibody-secreting plasma cells and memory B cells. nsp, non-structural protein; CD, cluster of differentiation; CTL, cytotoxic T cell. The figure was created with the help of the BioRender online tool (https://app.biorender.com/).
The expressed protein was antigenically intact
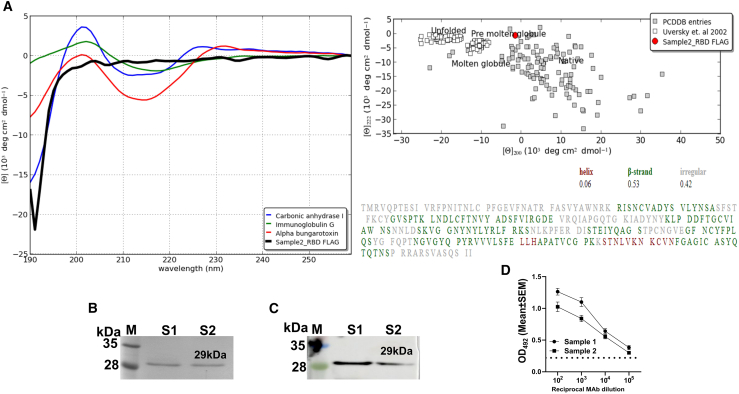
The protein conformation plays a vital role in the efficient presentation of antigenic peptides on major histocompatibility complex (MHC) molecules required for eliciting potent immune responses.18 Therefore, we analyzed the secondary structure of a protein by circular dichroism (CD) spectroscopy to know whether the expressed protein is in its native conformation using RBD, one of the target proteins of our vaccine candidate. The RBD was cloned into the pJHL204 vector, and a FLAG peptide was placed at the N terminus for purification of the protein. The resulting clone was electroporated into Salmonella and was used to infect the macrophage-like (RAW) cells for protein expression. The anti-FLAG resin enabled the purification of RBD to over 90% purity (Figure 2B). CD spectral data in mean residue ellipticity (deg cm2 dmol−1) was analyzed by an online-based server CAPITO, and no conformational changes were observed in the RBD protein compared with the other published proteins (Figure 2A). Analysis of CD spectra from two independent protein purifications revealed similar results. To further verify that the expressed protein was antigenically intact, reactivity of purified RBD against a known neutralizing monoclonal antibody (mAb) from mouse was evaluated by western blot (Figure 2C) and ELISA. The results indicated that RBD expressed in our system was highly reactive and exhibited a titer of >105 in an ELISA (Figure 2D). The findings collectively suggest the antigenically intact nature of the expressed protein.
Figure 2.
Analysis of the secondary structure of RBD by CD spectroscopy and verification of antigenic intactness
(A) Representative CD spectral data as analyzed by online server CAPITO. (B) SDS-PAGE analysis of the purified RBD. RBD was purified using anti-FLAG resin from RAW cells infected with Salmonella carrying pJHL204-FLAG-RBD. (C) Western blot and (D) ELISA results demonstrate reactivity of purified RBD against a known spike neutralizing mAb from the mouse (clone 57, RBD-mFc immunogen). Lane M, protein molecular weight marker; lane S1 and S2, respectively, denote purified protein samples 1 and 2. For ELISA, serial dilutions of RBD MAb ranging from 102 to 105 were reacted with purified RBD for 1 h at 37°C. Data in (D) represent mean ± SEM of OD492 values derived from two protein sample preparations. pJHL204, SFV replicon-based vector.
pJHL204-V-P2A elicits potent Th1-dominated humoral and cellular immune responses in mice
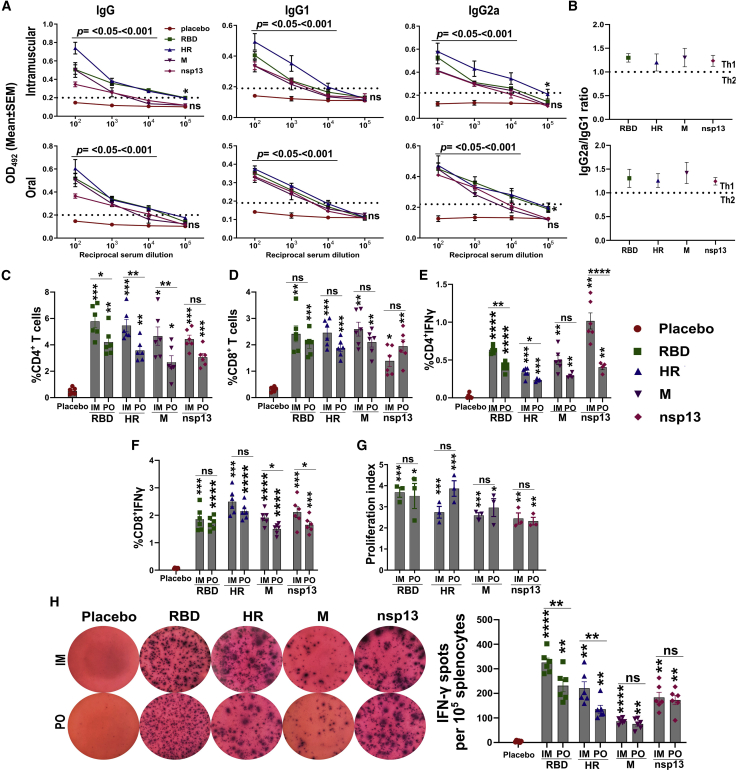
As tools do not exist for hamsters, we evaluated the humoral and cellular immune responses in BalB/c mouse immunized via the intramuscular and oral routes. The intramuscular injection consisted of a single dose of 1 × 107 colony-forming units (CFU), whereas the oral route of administration consisted of two doses of 1 × 108 CFU at a 2-week interval. The immune response was evaluated 3 weeks after the final immunization, and, at this point, all mice receiving the vaccine had seroconverted with robust antigen-specific immunoglobulin (Ig) G, IgG1, and IgG2a (Figure 3A). An IgG and IgG2a titer of >104 was recorded for RBD and HR, whereas it was >103 for IgG1. Similarly, IgG, IgG1, and IgG2a titers of 103 to 104 were observed for M and nsp13 proteins. Further, the ratio of IgG2a to IgG1 revealed a T-helper (Th) 1-dominated immune response (Figure 3B).
Figure 3.
Evaluation of humoral and cellular immune responses in mice
BalB/c mice were immunized via intramuscular and oral routes. The intramuscular injection consisted of a single dose of 1 × 107 CFU. The oral route of administration consisted of two doses of 1 × 108 CFU at a 2-week interval. The immune responses were evaluated 3 weeks after the final inoculation. (A) IgG, IgG1, and IgG2a response in mice sera determined by ELISA. Data denote the OD492 values against the corresponding sera dilution, collected from five biologically independent mice per group. Dotted lines indicate the cutoff value determined by multiplying the mean OD492 values of healthy sera by 2.1. (B) The ratio of IgG2a to IgG1, compared with a theoretical Th1 or Th2 immune response. The ratio was calculated from the data obtained at 1:100 sera dilution. Percentages of (C) CD4+, (D) CD8+, (E) CD4+IFNγ, and (F) CD8+IFNγ T cells. Data represent the total change in cell population in response to stimulation of splenocytes with respective immunogens. (G) Splenocyte proliferation index in immunized mice compared with the controls. (H) Representative photographs of IFN-γ ELISpot in splenocytes stimulated with respective proteins. Images were taken at a magnification of 50×. The total number of spots counted per 1 × 105 splenocytes is presented in the right panel. IM, intramuscular; PO, per os; M, membrane glycoprotein, Data in (A) were analyzed by unpaired Student's t test. Data in (C)–(H) were analyzed by two-way ANOVA using Šídák's or Tukey's multiple comparisons test. Data in (C)–(H) represent six biologically independent mice per group. The data points represent the individual value from each animal and error bars denote the SEM at 95% confidence interval (CI). ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
To assess the cellular immune response, splenocytes were stimulated with individual recombinant proteins as a measure of recall immunity. We determined the changes in CD3+CD4+ and CD3+CD8+ T cell subpopulation by fluorescence-activated cell sorting (FACS). Flow cytometric data were analyzed by gating lymphocytes (P1) to identify CD3+ T cells (P2), and, from P2, the percentage of T cells expressing CD4 and CD8 was measured (Figure S2). Immunization resulted in a significant increase in the proportions of CD4+ and CD8+ T cells in response to protein stimulation compared with the placebo controls (Figures 3C and 3D). The highest expansion in the CD4+ T cells was recorded for RBD, whereas M protein resulted in the maximum increase in CD8+T cells. Intracellular cytokine staining revealed a higher number of interferon (IFN)-γ-positive CD4+ and CD8+ T cells in splenocytes from immunized mice stimulated with respective proteins (Figures 3E and 3F). Contrarily, very few cells (<0.1%) expressing interleukin (IL)-4 were detected in the stimulated splenocytes (data not shown). Further, the proliferation index in stimulated splenocytes was determined by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. A proliferation index of 2.44–3.68 and 2.32–3.86 was recorded, respectively, in mice immunized by intramuscular and oral routes (Figure 3G). The Th1-dominated cellular response was further substantiated by an IFN-γ ELISpot assay. We detected a significantly higher number of IFN-γ-secreting cells in response to stimulation of splenocytes with respective vaccine immunogens (Figure 3H). The results support the elicitation of strong humoral and cellular immune responses with polarization toward antiviral Th1 immunity by the replicon-based mRNA vaccine.
pJHL204-V-P2A induces potent cross-protective neutralizing antibodies in hamsters
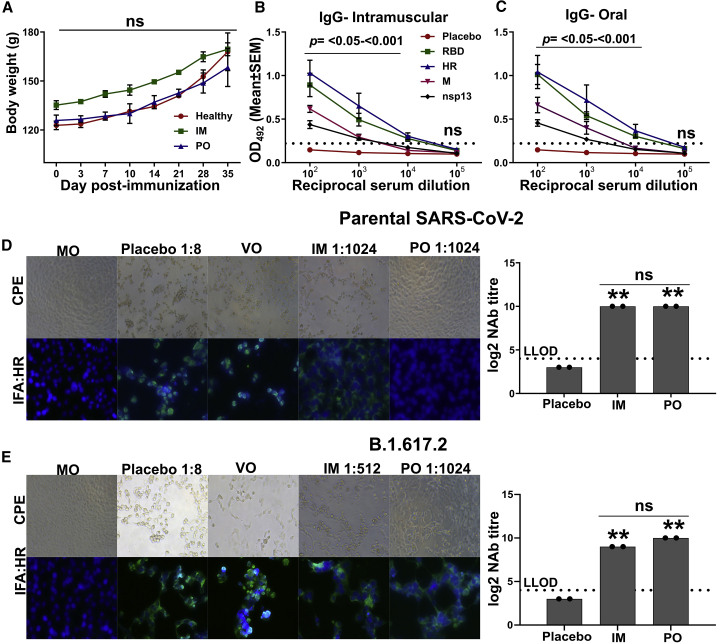
First, we evaluated the safety and immune response in hamsters immunized with 2 × 107 and 2 × 108 CFU, respectively, via the intramuscular and oral routes. The intramuscular injection consisted of a single dose, whereas the oral route of administration consisted of two doses at a 2-week interval. We assessed the safety by examining the animals physically and measuring the body weights at regular intervals. Either systemic or oral delivery did not cause any untoward symptoms in the immunized hamsters, and immunization did not affect the weight gain (Figure 4A), thus confirming the highly safe nature of the vaccine. Owing to the limited or unavailability of reagents to study the immunological parameters, we only determined the serum IgG levels in hamsters. The vaccine elicited a potent systemic IgG response in hamsters against all four target antigens. At week 3 post-immunization, all hamsters had seroconverted with high titers of antigen-specific IgG (Figures 4B and 4C). We recorded a titer of >104 for RBD and HR, and two animals in the oral group showed a titer of ≥105. A titer of >103 was observed for M and nsp13 proteins.
Figure 4.
Analysis of humoral and cross-protective neutralizing antibodies in hamster
Syrian hamsters were immunized via intramuscular and oral routes. The intramuscular injection consisted of a single dose of 2 × 107 CFU, whereas the oral route of administration consisted of two doses of 2 × 108 CFU at a 2-week interval. The IgG and NAb titers were evaluated 3 weeks after the final immunization. (A) Body weights of hamster post-immunization. Data denote two biologically independent hamsters per group. (B and C) IgG response in sera of hamsters immunized through the intramuscular and oral routes, respectively. Data were derived from six biologically independent hamsters per group. Dotted lines indicate the cutoff value determined by multiplying the mean OD492 values of healthy sera by 2.1. (D and E) Representative CPE and IFA images showing the viral replication by detecting the expression of HR protein of SARS-CoV-2. One of the wells of a serum dilution recorded as NAb titer is shown. NAb titer quantified by CPE and IFA analysis is presented in the right panel. (D) SARS-CoV-2 parental strain and (E) SARS-CoV-2 (B)1.617.2 variant. The dashed line in (D) and (E) represents the lower limit of detection (LLOD). MO, medium-only control; VO, virus-only control. Data in (A) were analyzed by two-way ANOVA using Bonferroni post test. Data in (B) and (C) were analyzed by unpaired Student's t test. Data in (D and E) were analyzed by one-way ANOVA using Bonferroni's multiple comparison test. The data points represent the individual value from each animal and error bars denote the SEM at 95% CI. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
The multivalent vaccine was designed to elicit broad neutralizing antibodies (NAbs) against SARS-CoV-2. To test the hypothesis, we next analyzed the ability of hamster sera to neutralize the live SARS-CoV-2 parental strain (clade L) and the B.1.617.2 delta variant (clade G). The delta variant was chosen as it is the most widely circulating SARS-CoV-2 VOC across the globe. The levels of NAbs were measured by microneutralization, and the titers were expressed as MN50. The sera exhibited a potent cross-neutralizing activity, with the sera from animals immunized intramuscularly and orally inhibiting the viral replication with great potency (Figures 4D and 4E). A log2-transformed microneutralization-50 (MN50) titer of 10 was recorded against both the strains; however, an MN50 titer of 9 was observed in the hamsters immunized via the intramuscular route against the delta variant. None of the hamsters in the placebo control had neutralizing antibodies. The results were corroborated by analyzing the viral replication by immunofluorescence assay (IFA). The results of the neutralization assay validated the elicitation of potent cross-protective NAbs by the vaccine.
pJHL204-V-P2A confers cross-protection against SARS-CoV-2 in hamsters
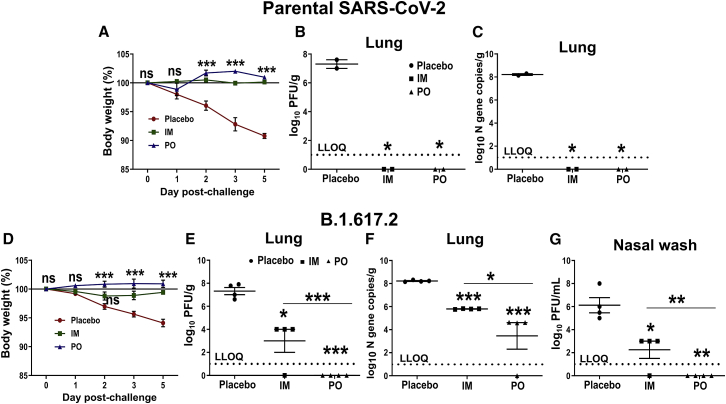
Hamster is the more relevant model to study vaccine efficacy because it mimics the COVID-19 pathology caused by SARS-CoV-2 in humans and, therefore, we assessed the efficacy of the vaccine in hamsters. A separate group of hamsters were immunized through the intramuscular and oral routes. The intramuscular injection consisted of a single dose of 2 × 107 CFU, whereas the oral route of administration consisted of two doses of 2 × 108 CFU at a 2-week interval. The hamsters were challenged 3 weeks after the final immunization with 1 × 104 plaque-forming units (PFU) of SARS-CoV-2 and sacrificed at 5 days post-challenge (DPC). The immunization protected hamsters against weight loss (Figures 5A, 5D, and S3). The lung samples collected at 5 DPC were subjected to virological and histopathological analysis. We detected a high mean log10-transformed viral load of 7.3 PFU/g in the lungs of placebo controls infected with the parental strain, while the mean log10-transformed viral N gene copies were recorded as 8.21/g (Figures 5B and 5C). On the contrary, the immunized hamsters were protected against the challenge infection, and neither live virus nor the viral RNA was detected in the lungs.
Figure 5.
Evaluation of protection conferred by the vaccine in a hamster model
Syrian hamsters were immunized via intramuscular and oral routes. The intramuscular injection consisted of a single dose of 2 × 107 CFU, whereas the oral route of administration consisted of two doses of 2 × 108 CFU at a 2-week interval. Hamsters were challenged 3 weeks after the final immunization with 1 × 104 PFU of either parental strain (clade L) or delta variant (clade G). Animals were sacrificed on day 5 post-challenge, and lung samples were collected for virological and histopathological analysis. Viral load was determined by plaque assay and N gene-specific qPCR. (A and D) Hamster body weights following viral challenge. Lung viral measurements in hamsters challenged with (B and C) parental SARS-CoV-2 and (E and F) B.1.617.2 SARS-CoV-2. (G) Viral load in the nasal wash of hamsters challenged with B.1.617.2 SARS-CoV-2. Data are presented as log10-transformed PFU/g or PFU/mL and log10-transformed viral N gene copies/g. The dashed line represents the lower limit of quantification (LLOQ). (Data denote two and four biologically independent hamsters per group for challenge studies with parental and (B)1.617.2 SARS-CoV-2, respectively. Body weight data in (A) and (D) were analyzed by two-way ANOVA using Bonferroni post test. Viral load data were analyzed by repeated-measures ANOVA using Tukey's multiple comparisons test. The data represent the individual value from each animal and error bars denote the SEM at 95% CI. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
For further protection analysis, we evaluated the cross-protective efficacy of the vaccine against the B.1.617.2 variant (delta, clade G). The hamsters were immunized and challenged as earlier. The mean log10-transformed viral load in placebo controls at 5 DPC reached 7.58 PFU/g and 7.3 PFU/mL in lung and nasal wash, respectively (Figures 5E, 5G, and S3). No infectious virus was detected in the hamsters immunized orally, while a mean log10-transformed viral load of 4.0 PFU/g and 3.0 PFU/mL was recorded, respectively, in the lung and nasal wash of the intramuscular group. Mean log10-transformed viral N gene copies/g were recorded as 8.22, 5.79 and 3.4 in the lungs of the placebo, intramuscular, and oral groups, respectively (Figure 5F). No RNA was detected in one of the four orally immunized hamsters. The ability of the oral vaccine to prevent the replication of the virus in the nasal cavity suggested induction of robust protection in the upper respiratory tract as well.
pJHL204-V-P2A protects against SARS-CoV-2-induced lung pathology in hamsters
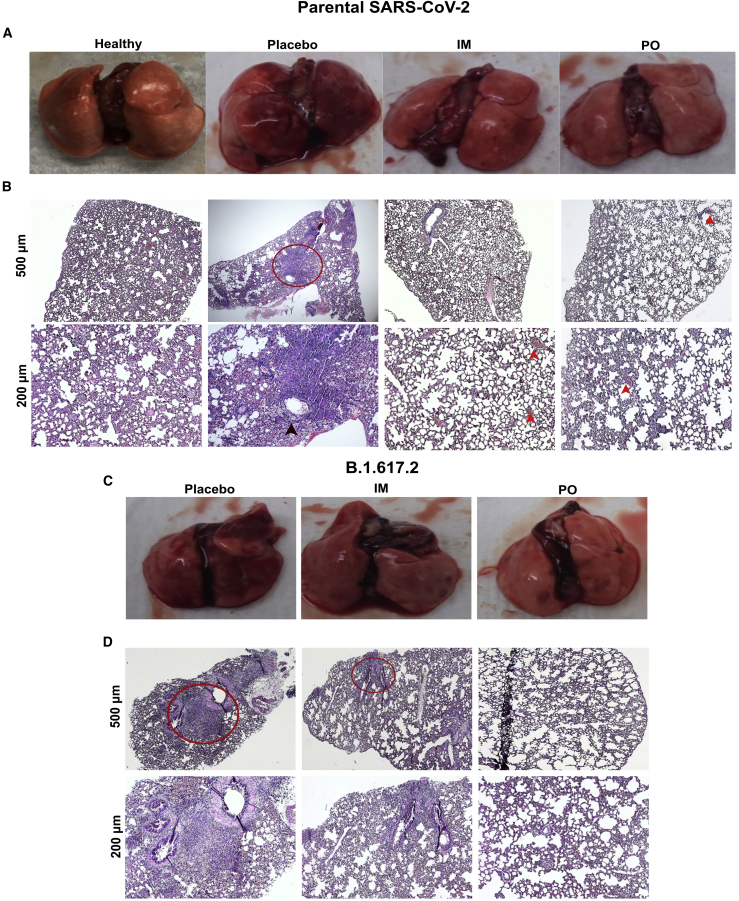
SARS-CoV-2 in humans causes a severe form of pneumonia, and related lung pathology has been documented as one of the common causes of COVID-19-associated mortality19. Hence, we subjected the lung samples from hamsters to gross and histopathological analysis to assess the protection conferred by the vaccine against SARS-CoV-2-induced lung pathology. On gross examination, the lungs of the placebo group exhibited diffuse areas of consolidation and congestion, whereas no such abnormalities were evident in the immunized animals (Figures 6A and 6C). Further, tissues were analyzed for histopathological changes after hematoxylin-eosin (H&E) staining. The lesions in unimmunized placebo controls were characteristic of an acute viral interstitial pneumonia (Figures 6B and 6D). We observed congestion, inflammatory cell infiltrate chiefly composed of lymphocytes, consolidation of air spaces with eosinophilic exudate, and perivascular cuffing. The intramuscularly immunized group exhibited mild inflammatory infiltrate and focal congestion, with one of the hamsters infected with delta variant developing mild interstitial pneumonia. However, the animals immunized orally were completely protected against viral pneumonia, except for a few focal lesions of congestion.
Figure 6.
Evaluation of protection against SARS-CoV-2-induced lung pathology
Syrian hamsters were immunized via intramuscular and oral routes. The intramuscular injection consisted of a single dose of 2 × 107 CFU, whereas the oral route of administration consisted of two doses of 2 × 108 CFU at a 2-week interval. Hamsters were challenged 3 weeks after the final immunization with 1 × 104 PFU of either parental strain (clade L) or delta variant (clade G). (A) Gross and (B) H&E-stained lung images from hamsters challenged with parental SARS-CoV-2 at day 5 post-challenge. Each image is representative of a group of two hamsters. (C) Gross and (D) H&E-stained lung images from hamsters challenged with B.1.617.2 SARS-CoV-2 at day 5 post-challenge. Each image is representative of a group of four hamsters. Infected lung tissue sections showing the characteristic lesion of interstitial pneumonia (red circle), perivascular cuffing (black arrowhead), and congestion (red arrowhead).
Oral route elicits potent mucosal and systemic IgA
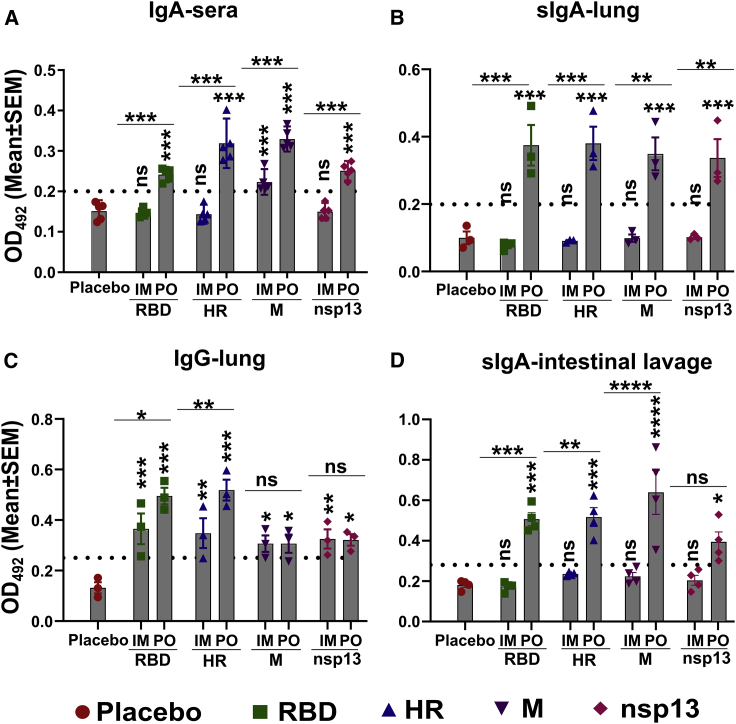
Even though there was no significant difference in serum IgG and neutralizing antibody titer in animals immunized orally and intramuscularly, the detection of infectious delta virus in the lung and nasal wash of the intramuscular group was surprising. Previously, the enhanced ability of the dimeric IgA to neutralize the SARS-CoV-2 has been documented,20 and superior protection was achieved by the intranasal immunization against variants.15,16 Therefore, we asked whether the route of immunization affected IgA response. Mice were immunized by oral and intramuscular routes, and sera, lung, and intestinal lavage samples were collected at 3 weeks. The IgA response in sera was determined by ELISA using 1:50 and 1:100 diluted sera. We detected robust IgA response in the sera and lung homogenates of orally immunized mice, while little or no IgA was detected in the intramuscular group (Figures 7A and 7B). Mean serum IgA titers of >100 were recorded in the orally immunized animals against all four target antigens, with the maximum response being observed for the M protein. Mice in the intramuscular group did not show serum IgA response even at dilutions of 1:50. However, three mice in the intramuscular group exhibited serum IgA response against M protein, albeit of a low level. A pronounced effect of the route of immunization was observed on the secretory IgA (sIgA) response in the lung homogenates. We detected the sIgA with a titer of >200 in the oral group, whereas no sIgA was detected in the lung homogenates of intramuscular group (Figure 7B). On the contrary, we detected a strong IgG response in the lung homogenates of both intramuscular and orally immunized mice (Figure 7C). Additionally, evaluation of sIgA response in the intestinal lavage revealed similar findings (Figure 7D). The findings highlight the role of sIgA in protection against rapidly replicating variants.
Figure 7.
Evaluation of mucosal immune response in lung and intestine
BalB/c mice were immunized via intramuscular and oral routes. The intramuscular injection consisted of a single dose of 1 × 107 CFU, whereas the oral route of administration consisted of two doses of 1 × 108 CFU at a 2-week interval. The immunoglobulin response in sera, lung homogenates, and intestinal lavage samples was evaluated 3 weeks after the final inoculation by ELISA. (A) IgA response in sera of immunized mice at a sera dilution of 1:100. (B) sIgA and (C) IgG response in the lung homogenates at a sample dilution of 1:200. (D) sIgA response in the intestinal lavage at a sample dilution of 1:1,000. Dotted lines indicate the cutoff value determined by multiplying the mean OD492 values of healthy samples by 2.1. Data derived from three to five biologically independent mice per group. Data were analyzed by two-way ANOVA using Tukey's multiple comparisons test. The data points represent the individual value from each animal and error bars denote the SEM at 95% CI. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Discussion
In the present communication, we expand upon our previously reported preliminary immunogenicity studies in mice, which showed the elicitation of high neutralizing antibody titers against the parental SARS-CoV-2.10 Herein, we tested the vaccine efficacy in a hamster model as SARS-CoV-2 replication and lung pathology in hamsters are akin to the observations made in natural human infections.21,22 Further, due to the lack of tools for hamsters, we evaluated the humoral and cellular responses in parallel in mice. The bacteria-mediated gene delivery offers a novel and highly versatile platform for the development of mucosal vaccines. The vaccines developed by Pfizer-BioNTech and Moderna have been instrumental in limiting COVID-19 and constitute the first successful mRNA vaccines. However, to date, the development of the oral mRNA vaccine has not been explored.23 Herein, we provide findings exploiting the Salmonella-mediated oral replicon-based mRNA vaccine delivery as a proof of principle for the development of a feasible mRNA vaccine. The self-amplifying mRNA vaccine design was enabled by the SFV replicon-based vector with elements for plasmid maintenance in Salmonella and expression within eukaryotic cells.10 Through its innate ability to replicate within the professional antigen-presenting cells (APCs), Salmonella delivers the gene directly to macrophages and dendritic cells (DCs),24,25 thereby eliciting potent cellular and humoral immune responses. Of note, Salmonella Typhimurium serves as a preclinical organism to study Salmonella Typhi in small laboratory rodents.26 Because a plethora of studies have supported the use of S. Typhi as a vaccine vector in human volunteers,27, 28, 29 and live-attenuated S. Typhi is available as a licensed vaccine,30 the use of S. Typhi for further vaccine development provides the possibility of direct translation to humans. The findings have implications for the development of bacteria-enabled replicon-based oral mRNA vaccines against infectious diseases not limited to SARS-CoV-2.
The appearance of SARS-CoV-2 variants with mutations in the spike protein, particularly the RBD and N-terminal domain (NTD), has been a great concern due to their ability to evade the neutralizing antibodies.6, 7, 8,31, 32, 33 Indeed, a staggering drop in efficacy has been observed for NVX-CoV2373, JNJ-78436735, and ChAdOx1d vaccines against the B.1.351 variant.3,4 Therefore, we designed a multicistronic vaccine incorporating multiple SARS-CoV-2 protein targets to achieve broader protection. To test the hypothesis, we analyzed the induction of cross-protective NAb against the B.1.617.2 delta variant, the most widely circulating variant globally and accounting for the most reported severe COVID-19 cases.34, 35, 36 The sera from orally immunized hamsters potently neutralized both the parental strain (clade L) and delta variant (clade G) SARS-CoV-2, and no reduction in NAbs was observed. The finding is important considering the reduction of NAbs observed with LNP-mRNA and other vaccines against SARS-CoV-2 spike variants.37,38 Of note, BNT162b2 and mRNA-1273 vaccinee sera had a reduced neutralizing activity by a factor of 6.7 and 4.5, respectively, against the P1 variant.7 The mRNA vaccines from Pfizer-BioNTech/Moderna encode the full-length spike gene alone, and the development of variants with mutations in the spike region might have contributed to the drop in NAb and efficacy. Although our vaccine encodes only RBD and the heptad repeat region from the spike, it also includes membrane protein and epitopes from nsp13. Thus, incorporation of other antigenic targets, in particular membrane protein, might have contributed to the detection of potent NAb titer against the delta variant. The nsp13 that encodes RNA helicase was included in the vaccine design to complement the cellular response considering the role of non-structural proteins in protection against other viruses, such as HIV, dengue, and Japanese encephalitis.39, 40, 41 However, whether nsp13 alone has any immuno-protective role against SARS-CoV-2 needs to be assessed. On the contrary, sera from the intramuscular group exhibited a 2-fold reduction in NAb titer against the delta variant. This discrepancy of the effect of route of immunization on NAb may be attributed to the two-dose scheme employed for the oral route, while only a single dose was administered via the intramuscular route. Further, the NAb detected in our study exceeded the NAb titers recorded in convalescent sera.42,43 Although correlates of protection are not completely established, recent studies have highlighted the levels of neutralizing antibodies as a likely protective correlate against SARS-CoV-2.44,45
Although only a single dose was administered intramuscularly, the magnitude of systemic antibody response induced was not significantly different compared with the two-dose oral dosing. Further, intramuscular immunization induced a higher number of IFN-γ-secreting splenocytes. However, two-dose oral immunization protected hamsters against viral replication and lung disease better than the intramuscular route. Therefore, we studied the effect of route of administration on induction of mucosal sIgA response as the superior ability of dimeric IgA to neutralize SARS-CoV-2 was documented recently.20 Oral immunization resulted in a significant increase in the antigen-specific sIgA in both lungs and intestines, whereas no sIgA response was elicited by the intramuscular route. The finding was not surprising as Salmonella invasion and multiplication within the Peyer patches is required for the induction of sIgA response,46 and the DCs in Peyer patch have the unique ability to drive IgA secretion from B cells.47 It should be noted that the gut bacteria not only affect the sIgA production in the gut lumen but also influence sIgA production in the lungs via the CD103+ DCs.48 In agreement, a recombinant ST-expressing hepatitis B virus core antigen (HBc) induced sIgA in lungs upon oral delivery.13 Consistent with the induction of sterilizing protection by the mucosal vaccine delivered intranasally,15,16 our findings reinforce the contribution of sIgA in inducing sterilizing immunity against rapidly replicating SARS-CoV-2 variants.
In summary, our studies demonstrated the generation of potent antigen-specific humoral and cellular immune responses coupled with cross-protective NAbs. The oral route of immunization conferred robust protection in both lung and the nasal cavity of hamsters against SARS-CoV-2 challenge, including the B.1.617.2 delta variant. The selective induction of sIgA likely contributed to the superior protection conferred by oral immunization. Collectively, our proof-of-concept study establishes the feasibility of bacteria-mediated gene delivery for the development of a safe and efficacious oral replicon-based mRNA vaccine to prevent SARS-CoV-2 transmission and curtail the pandemic. Additionally, findings have implications for the development of replicon-based oral mRNA vaccines against infectious agents.
Materials and methods
Experimental animals and ethics statement
Female BALB/c mice, aged 5 weeks and specific pathogen free (SPF), were obtained from Koatech in Pyeongtaek, Korea. SPF hamsters, female, aged 5 weeks, were used. Animals were maintained on a standard feeding regimen with a 12-h light-dark cycle at the Animal Housing Facility of the College of Veterinary Medicine, Jeonbuk National University. Animal experiments were approved by the Jeonbuk National University Animal Ethics Committee (JBNU 2021-027) under the Korean Council on Animal Care and the Korean Animal Protection Law, 2001, Article 13. The experiments involving live SARS-CoV-2 were carried out at BSL3 and ABSL3 facilities of the Korea Zoonosis Research Institute, South Korea.
Cell lines and viruses
HEK293T and Vero E6 cell lines procured from ATCC (American Type Culture Collection) were maintained in DMEM (Lonza, Switzerland) supplemented with 10% FBS (Gibco, USA) and 1× penicillin-streptomycin at 37°C in 5% CO2. Vero E6 cells were used to propagate the SARS-CoV-2 clinical isolate BetaCoV/Korea/KCDC/03/2020 (parental strain, clade L) and hCoV/Korea/KDCA119861/2021 (B.1.617.2, delta variant, clade G). The viral titers were calculated by a standard plaque assay. SARS-CoV-2 stocks were stored at −80°C until further use and passage 3 virus was used in all the experiments.
Bacterial strains, plasmids, and primers
Bacteria, plasmids, and primers used in this study are summarized in Table 1. The bacteria were grown in Luria-Bertani (LB) broth (BD, USA) with agitation at 37°C using appropriate selection markers whenever applicable.
Table 1.
List of bacterial strains, plasmids, and primers used in the present investigation
| Bacteria/Plasmid | Genotypic characteristics | Reference |
|---|---|---|
| S. Typhimurium | ||
| JOL3000 | Δlon ΔcpxR ΔrfaL ΔpagL::lpxE Δasd | laboratory stock |
| JOL3014 | JOL3000 carrying pJHL204-V-P2A | 10 |
| JOL3015 | JOL3000 carrying pJHL204 | 10 |
| JOL2829 | JOL3000 carrying pJHL204-FLAG-RBD | this study |
| Escherichia coli | ||
| E. coli 232 | F− λ− φ80 Δ(lacZYA-argF) endA1 recA1 hadR17 deoR thi-1 glnV44 gyrA96 relA1 ΔasdA4 | laboratory stock |
| JOL3013 | E. coli 232 carrying pJHL204-V-P2A | 10 |
| JOL2765 | E. coli 232 carrying pJHL204-FLAG-RBD | this study |
| Plasmids | ||
| pSFV3-lacZ | ampR,SP6 promoter, pBR322 ori | Addgene, USA |
| pJHL204 | SFV replicon, asd+, CMV promoter, SV40 promoter, pBR322 ori | laboratory stock |
| FLAG cloning primer | ||
| FLAG-RBD | forward, GGGCCCATGGACTACAA AGACGATGACGACAAGACCATG AGAGTCCAAC reverse, GGCGCGC CTTAAATGATGGATTGACTAG CTACACTACGTG |
this study |
| Construct primer | ||
| V-P2A | forward, GGGCCCGCCACCATGA GAGTC reverse, GGCGCGCCTTA TATTTGTGGCCTG |
10 |
| qRT-PCR primers | ||
| SARS-CoV-2 N gene | forward, CACATTGGCACCCGCAATC reverse, GAGGAACGAGAAGAGGCTTG |
51 |
pJHL204-V-P2A, the vaccine candidate
The construction and characterization of the vaccine candidate have been described previously.10 The vaccine encompasses RBD, HR, M, and epitopes of nsp13 of SARS-CoV-2 parental strain, each separated by a self-cleaving peptide, p2A, for multicistronic expression. The vaccine construct is based on the SFV replicon vector, which functions as a self-replicating mRNA molecule leading to enhanced transgene expression (Figure S1). The immunogens do not encode any secretory tags. JOL3000, an S. Typhimurium mutant with the genotype Δlon ΔcpxR ΔrfaL, ΔpagL::lpxE, and Δasd, served as a biological vaccine delivery vehicle.
Vaccine production
JOL3014, S. Typhimurium carrying the pJHL204-V-P2A. The vaccine strain was grown overnight in LB broth. Next morning, the strain was sub-cultured and allowed to grow for 4–5 h. The bacteria were pelleted and washed twice in sterile phosphate-buffered saline (PBS), pH 7.4. The pellet was resuspended in a final volume of 2 mL, and the bacterial number was enumerated by measuring the optical density 600 (OD600) (OD600 of 1.0 = 8×108 bacteria/mL) (https://www.agilent.com/store/biocalculators/calcODBacterial.jsp). The prepared vaccine was used fresh.
Immunization and sampling
Mice were immunized with 1 × 107 and 1 × 108 CFU, respectively, via the intramuscular and oral routes, while hamsters were immunized with 2 × 107 and 2 × 108 CFU, respectively, through intramuscular and oral routes. A single dose was administered via the intramuscular route, whereas two doses at a 2-week interval were administered via the oral route. Animals were monitored for Salmonella-induced clinical signs such as ruffled fur, weight loss, and mortality.49 Samples such as sera, splenocytes, lung homogenate, and intestinal lavage were collected from mice, whereas only sera samples were obtained from hamsters. The humoral and cellular immune responses were evaluated 3 weeks after the final inoculation.
Intestinal wash/lavage fluid and IgA determination
Intestinal wash samples were collected by adopting the method from Lycke et al. (1999) with minor modifications50. Briefly, mice were sacrificed under chloroform anesthesia (Sigma, USA), and small intestines were collected and rinsed with PBS to remove fecal material. After ligating one end of the intestine, 2 mL of 50 mM EDTA (Samchun chemicals, South Korea) in PBS containing 1 mM PMSF (catalog no. 10837091001; Sigma, USA) as protease inhibitor was carefully injected, and the other end of the intestine was also clamped. The contents were collected following 10 min of incubation at room temperature. The contents were vortexed, subjected to six cycles of 10 s of sonication with a 10 s gap at 50% amplitude, and clarified by centrifugation for 10 min at 13,000 ×g at 4°C. The supernatant was collected into a 1.5-mL microcentrifuge tube, 1 mM PMSF was added, it was mixed, and centrifuged for 15 min at 13,000 ×g at 4°C. The resulting supernatant was added with 1 mM PMSF and 0.001% sodium azide and incubated at 4°C for 30 min. Next, 50 μL of FCS was added to every milliliter of solution, mixed, and centrifuged as before. The intestinal wash fluids so collected were used freshly in an ELISA assay or stored at −80°C until use. The samples were diluted at 1:100 and 1:1,000 to determine the sIgA by ELISA.
Intracellular cytokine staining, flow cytometry, and splenocyte proliferation
BalB/c mice were immunized via intramuscular and oral routes. The intramuscular injection consisted of a single dose of 1 × 107 CFU, whereas the oral route of administration consisted of two doses of 1 × 108 CFU at a 2-week interval. Splenocytes were collected 3 weeks after the final immunization. The splenocytes were cultured in 10% RPMI (catalog no. AL028A; HiMedia, India) containing 2-mercaptoethanol (catalog no. 21985–023; 1,000×; Gibco, USA) in a 96-well plate at 1 × 105 cells/well and stimulated with 400 ng of individual recombinant proteins for 48 h.10 Brefeldin A (catalog no. 00-4506-51; Thermo Scientific, USA) was added to inhibit the intracellular protein transport 5 h before processing the cells. After blocking the Fc receptors, splenocytes were stained for surface markers with FITC-anti-CD8a (1:125 dilution; clone 53-6.7), PerCPVio700-anti-CD4 (1:125 dilution; clone GK1.5), and/or PE-anti-CD3e antibodies (1:125 dilution; clone 17A2) from Miltenyi Biotec for 30 min at 4°C. Cells were subsequently fixed, permeabilized (Intracellular Fixation & Permeabilization Buffer, Thermo Scientific, USA), and stained for PE-IFN-γ (1:100 dilution; Clone XMG1.2) and APC-IL-4 (1:100 dilution; Clone 11B11) from BioLegend. The cells were acquired on a Miltenyi MACSQuant Flow Cytometer (Miltenyi Biotec, Germany). The gating strategy is depicted in Figure S2. The final population of cells was calculated by subtracting from the unstimulated population. Splenocyte proliferation was determined by a standard MTT assay, and proliferation index was calculated by dividing A570 values from immunized mice with that of control mice.
IFN-γ ELISpot
ELISpot assay was carried out using the mouse IFN-γ ELISpot Kit (catalog no. EL485; R&D Systems, USA) and by following the manufacturer's instructions. The splenocytes collected 3 weeks after the final immunization were cultured (1 × 105 cells/well) in 10% RPMI containing 2-mercaptoethanol in a 96-well polyvinylidene fluoride (PVDF)-backed microplate pre-coated with mouse IFN-γ capture antibody and stimulated with 400 ng of individual recombinant proteins for 48 h. Spots were counted under a microscope. The total number of spots in wells stimulated with proteins were calculated by subtracting from the unstimulated cells.
Lung homogenate preparation and IgA and IgG determination
Lung samples weighing 100 mg were isolated into tubes containing 0.5 mL of RIPA buffer supplemented with protease inhibitor cocktail (catalog no 87786; Thermo Fisher Scientific, USA). Lung samples were processed freshly or kept at −80°C until use. Lung samples in RIPA buffer were lysed by two cycles of sonication for 10 s with a 60-s gap at 50% amplitude. The lysates were kept on a rocking platform for 1 h for complete lysis followed by two more cycles of sonication. The lysates were clarified by centrifugation at 13,000 ×g for 10 min at 4°C and the supernatants were preserved at −80°C until use. The samples were handled on the ice at every step of the experiment. The samples were diluted at 1:50 and 1:200 to determine the sIgA and IgG by ELISA.
ELISA
ELISA was performed as described previously.10 Briefly, 96-well high-binding polystyrene plates (Greiner Bio-One, Austria) were coated with recombinant proteins at 2.5 μg mL−1 in carbonate-bicarbonate buffer, pH 9.6, at 4°C overnight. The wells were then blocked with 5% skim milk. Different sera dilutions were added to the wells and allowed to interact at 37°C for 1 h. Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (catalog no. 1030-05), IgG1 (catalog no. 1070-05), or IgG2a (catalog no. 1080-05) antibody at 1:5,000 or 1:3,000 was added, and the plates were incubated at 37°C for 1 h. The goat anti-hamster IgG HRP (catalog no. 6060-05) was used at a dilution of 1:10,000. At each step, plates were stringently washed with 0.1% PBST. The assay was developed using OPD substrate for 5–10 min in the dark. The ODs were read at 492 nm in a microplate reader (Tecan, Switzerland) after stopping the reaction with 3 M H2SO4. Additionally, IgA response in sera of immunized was mice was also determined by using goat anti-mouse IgA HRP (catalog no. 1040-05) at 1:3,000. All HRP-conjugated secondary antibodies used were from Southern Biotech, USA.
Live virus neutralization assays
NAb titer in hamster sera was determined by microneutralization (MN) assay.10 Two-fold serial sera dilutions were made and incubated with 50 PFU of either parental SARS-CoV-2 or delta variant at 37°C for 2 h. Antibody-virus complexes were added onto Vero E6 cell monolayers in 96-well plates and incubated for 72 h. Plates were observed under a microscope for cytopathic effect (CPE) daily for 3 days. Plates were also analyzed by IFA. The highest sera dilution that resulted in complete inhibition of CPE in two of the four wells was recorded as neutralizing antibody (MN50) titer.
IFA
The cells were fixed for 10 min with chilled 80% acetone at −20°C. Cells were permeabilized using 0.1% PBS-Triton X-100 and then blocked with 3% BSA. A 1:500 dilution of SARS-CoV-2 HR primary antibody10 was added and incubated overnight at 4°C. After washing, cells were stained with Alexa Fluor 488-conjugated donkey anti-rabbit IgG (catalog no. A21206; Invitrogen, USA) at 1:5,000 as a secondary antibody, and DAPI (catalog no. D9542-1MG; Sigma, USA) was used to stain the nucleus. Cells were observed under a Leica fluorescence microscope (Leica Biosystems, Germany).
Challenge study
The hamsters were challenged with 1 × 104 PFU of either SARS-CoV-2 parental strain or the delta variant via the intranasal route under general anesthesia. General anesthesia was achieved by injecting a mixture of 5 mg/kg xylazine and 80 mg/kg ketamine through the intraperitoneal route. The volume of the challenge inoculum was 30 μL. The body weights were monitored daily, and all animals were sacrificed at day 5 post-challenge. The blood, sera, and lung samples were collected.
Viral burden
The viral burden in the lung samples was determined by plaque assay and qRT-PCR.51,52 The lung samples were weighed and homogenized in DMEM or Trizol for plaque and qRT-PCR assays, respectively. Lung homogenates were clarified by centrifugation at 13,000 ×g for 10 min and stored at −80°C. RNA was extracted by a commercial kit (catalog no. 305-101; GeneAll, South Korea) and cDNA was prepared (catalog no. EBT-1515; Elpis Biotech, South Korea).
Lung histopathology
Formalin-fixed paraffin-embedded (FFPE) lung tissue sectioning followed by H&E staining was carried out to study the histopathological changes.
RBD expression and CD
FLAG tag was inserted into the 5′ end of RBD by PCR using the primer pair listed in Table 1. FLAG-RBD was cloned into pJHL204 plasmid using ApaI (catalog no. R0114S, NEB) and AscI (catalog no. R0558S, NEB) restriction enzymes. The RAW cell was used for the protein expression and purification following infection with Salmonella carrying pJHL204-RBD-FLAG. RAW cells growing in T75 flasks were infected at an MOI of 50. The external bacteria were killed by adding 150 μg/mL gentamicin (1 h) after allowing the bacteria to invade the cells (3 h). The cells were then maintained on 2% DMEM without antibiotics. The cells were harvested 48 h post-bactofection and lysed in 3 mL of RIPA buffer per flask. The FLAG-RBD was purified using anti-FLAG resin following the manufacturer's instructions (catalog no. A36801, Thermo Fisher Scientific, USA). The purified protein was analyzed by SDS-PAGE. The antigenic intactness of the purified protein was evaluated by determining its reactivity against known spike-neutralizing mAb from mouse (catalog no. 40592-MM57; clone 57; Sino Biological, China).53,54 The immunogen used in mAb production was recombinant SARS-CoV-2 spike RBD-mFc (https://www.sinobiological.com/antibodies/cov-spike-40592-mm57). The reactivity was assayed by western blot and ELISA.
CD spectra were collected on a Chirascan CD spectrometer (Applied Photophysics, UK) at The National Instrumentation Center for Environmental Management (NICEM), Seoul National University College of Agriculture and Life Sciences, Republic of Korea. Data was collected between 190 and 260 nm with the appropriate buffer and solvent background subtraction. Spectral data in mean residue ellipticity (deg cm2 dmol−1) was analyzed by the online-based server CAPITO (https://data.nmr.uni-jena.de/capito/index.php).55 Additionally, an amino acid sequence was submitted for the prediction of secondary structures.
Statistical analysis
Data analysis was performed by Student's t test and ANOVA using GraphPad Prism 9.0 software (GraphPad, USA) and IBM SPSS. A p value <0.05 was considered significant. Data in graphs are presented as the mean ± SEM with ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001. The statistical test employed and the number of animals is indicated in the figure legends.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2019R1A6A1A03033084).
Authors contributions
V.J. and P.K. conceived and designed the study, performed the experiments, wrote the manuscript, analyzed data, and prepared the figures. V.J., C.H., and J.-Y.P performed ABSL3 experiments for viral challenge, sacrifice, and sampling. M.S., B.K., V.J., and P.K. performed histopathology. J.H.L. supervised the project, acquired funding, and commented on the manuscript. All authors approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.01.042.
Supplemental information
References
- 1.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B. 1.351 variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreano E., Rappuoli R. SARS-CoV-2 escaped natural immunity, raising questions about vaccines and therapies. Nat. Med. 2021;27:759–761. doi: 10.1038/s41591-021-01347-0. [DOI] [PubMed] [Google Scholar]
- 5.Konings F., Perkins M.D., Kuhn J.H., Pallen M.J., Alm E.J., Archer B.N., Barakat A., Bedford T., Bhiman J.N., Caly L. SARS-CoV-2 variants of interest and concern naming scheme conducive for global discourse. Nat. Microbiol. 2021;6:821–823. doi: 10.1038/s41564-021-00932-w. [DOI] [PubMed] [Google Scholar]
- 6.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., De Oliveira T., Vermeulen M., Van der Berg K. SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 7.Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants-clinical, public health, and vaccine implications. N. Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi A., Koch M., Wu K., Dixon G., Oestreicher J., Legault H., Stewart-Jones G.B.E., Colpitts T., Pajon R., Bennett H. Serum neutralizing activity of mRNA-1273 against SARS-CoV-2 variants. Preprint at bioRxiv. 2021 doi: 10.1128/JVI.01313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrotri M., Swinnen T., Kampmann B., Parker E.P.K. An interactive website tracking COVID-19 vaccine development. Lancet Glob. Heal. 2021;9:E590–E592. doi: 10.1016/S2214-109X(21)00043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jawalagatti V., Kirthika P., Park J., Hewawaduge C., Lee J. Highly feasible immunoprotective multicistronic SARS-CoV-2 vaccine candidate blending novel eukaryotic expression and Salmonella bactofection. J. Adv. Res. 2021;36:211–222. doi: 10.1016/j.jare.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asundi A., O’Leary C., Bhadelia N. Global COVID-19 vaccine inequity: the scope, the impact, and the challenges. Cell Host Microbe. 2021;29:1036–1039. doi: 10.1016/j.chom.2021.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maxmen A. The fight to manufacture COVID vaccines in lower-income countries. Nature. 2021;597:455–457. doi: 10.1038/d41586-021-02383-z. [DOI] [PubMed] [Google Scholar]
- 13.Hopkins S., Kraehenbuhl J.-P., Schodel F., Potts A., Peterson D., De Grandi P., et al. A recombinant Salmonella Typhimurium vaccine induces local immunity by four different routes of immunization. Infect. Immun. 1995;63:3279–3286. doi: 10.1128/iai.63.9.3279-3286.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Ginkel F.W., Nguyen H.H., McGhee J.R. Vaccines for mucosal immunity to combat emerging infectious diseases. Emerg. Infect. Dis. 2000;6:123. doi: 10.3201/eid0602.000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan A.O., Shrihari S., Gorman M.J., Ying B., Yaun D., Raju S., Chen R.E., Dmitriev I.P., Kashentseva E., Adams L.J., et al. An intranasal vaccine durably protects against SARS-CoV-2 variants in mice. Cell Rep. 2021;36:109452. doi: 10.1016/j.celrep.2021.109452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., Chen R.E., Winkler E.S., Wessel A.W., Case J.B. A single-dose intranasal Chad vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–184. doi: 10.1016/j.cell.2020.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundstrom K. Alphaviruses in gene therapy. Viruses. 2015;7:2321–2333. doi: 10.3390/v7052321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheiblhofer S., Laimer J., Machado Y., Weiss R., Thalhamer J. Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev. Vaccin. 2017;16:479–489. doi: 10.1080/14760584.2017.1306441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elezkurtaj S., Greuel S., Ihlow J., Michaelis E.G., Bischoff P., Kunze C.A., Sinn B.V., Gerhold M., Hauptmann K., Ingold-Heppner B. Causes of death and comorbidities in hospitalized patients with COVID-19. Sci. Rep. 2021;11:1–9. doi: 10.1038/s41598-021-82862-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.-H., Oliveira T.Y., Oren D.A. Enhanced SARS-CoV-2 neutralization by dimeric IgA. Sci. Transl. Med. 2021;13:eabf1555. doi: 10.1126/scitranslmed.abf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sia S.F., Yan L.-M., Chin A.W.H., Fung K., Choy K.-T., Wong A.Y.L., Kaewpreedee P., Perera R.A.P.M., Poon L.L.M., Nicholls J.M. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583:834–838. doi: 10.1038/s41586-020-2342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coffey J.W., Gaiha G.D., Traverso G. Oral biologic delivery: advances toward oral subunit, DNA, and mRNA vaccines and the potential for mass vaccination during pandemics. Annu. Rev. Pharmacol. Toxicol. 2021;61:517–540. doi: 10.1146/annurev-pharmtox-030320-092348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wick M.J. The role of dendritic cells during Salmonella infection. Curr. Opin. Immunol. 2002;14:437–443. doi: 10.1016/s0952-7915(02)00364-3. [DOI] [PubMed] [Google Scholar]
- 25.Gog J.R., Murcia A., Osterman N., Restif O., McKinley T.J., Sheppard M., Achouri S., Wei B., Mastroeni P., Wood J.L.N. Dynamics of Salmonella infection of macrophages at the single cell level. J. R. Soc. Interf. 2012;9:2696–2707. doi: 10.1098/rsif.2012.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santos R.L., Zhang S., Tsolis R.M., Kingsley R.A., Adams L.G., Baumler A.J. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 2001;3:1335–1344. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- 27.Kantele A., Makela P.H. Different profiles of the human immune response to primary and secondary immunization with an oral Salmonella Typhi Ty21a vaccine. Vaccine. 1991;9:423–427. doi: 10.1016/0264-410x(91)90129-t. [DOI] [PubMed] [Google Scholar]
- 28.Hone D.M., Tacket C.O., Harris A.M., Kay B., Losonsky G., Levine M.M. Evaluation in volunteers of a candidate live oral attenuated Salmonella Typhi vector vaccine. J. Clin. Invest. 1992;90:412–420. doi: 10.1172/JCI115876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forrest B.D., LaBrooy J.T., Beyer L., Dearlove C.E., Shearman D.J.C. The human humoral immune response to Salmonella Typhi Ty2la. J. Infect. Dis. 1991;163:336–345. doi: 10.1093/infdis/163.2.336. [DOI] [PubMed] [Google Scholar]
- 30.Syed K.A., Saluja T., Cho H., Hsiao A., Shaikh H., Wartel T.A., Mogasale V., Lynch J., Kim J.H., Excler J.-L. Review on the recent advances on typhoid vaccine development and challenges ahead. Clin. Infect. Dis. 2020;71:S141–S150. doi: 10.1093/cid/ciaa504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuzmina A., Khalaila Y., Voloshin O., Keren-Naus A., Boehm-Cohen L., Raviv Y., Shemer-Avni Y., Rosenberg E., Taube R. SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe. 2021;29:522–528. doi: 10.1016/j.chom.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L. Increased resistance of SARS-CoV-2 variant P. 1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Twohig K.A., Nyberg T., Zaidi A., Thelwall S., Sinnathamby M.A., Aliabadi S., Seaman S.R., Harris R.J., Hope R., Lopez-Bernal J. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B. 1.617. 2) compared with alpha (B. 1.1. 7) variants of concern: a cohort study. Lancet Infect. Dis. 2021;22:35–42. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campbell F., Archer B., Laurenson-Schafer H., Jinnai Y., Konings F., Batra N., Pavlin B., Vandemaele K., Van Kerkhove M.D., Jombart T. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Eurosurveillance. 2021;26:2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheikh A., McMenamin J., Taylor B., Robertson C. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397:2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tada T., Zhou H., Samanovic M.I., Dcosta B.M., Cornelius A., Mulligan M.J., Landau N.R. Comparison of neutralizing antibody titers elicited by mRNA and adenoviral vector vaccine against SARS-CoV-2 variants. Preprint at bioRxiv. 2021 [Google Scholar]
- 38.Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Y.-L., Chen L.-K., Liao C.-L., Yeh C.-T., Ma S.-H., Chen J.-L., Huang Y.-L., Chen S.-S., Chiang H.-Y. DNA immunization with Japanese encephalitis virus nonstructural protein NS1 elicits protective immunity in mice. J. Virol. 1998;72:191–200. doi: 10.1128/jvi.72.1.191-200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koup R.A., Douek D.C. Vaccine design for CD8 T lymphocyte responses. Cold Spring Harb. Perspect. Med. 2011;1:a007252. doi: 10.1101/cshperspect.a007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henriques H.R., Rampazo E.V., Goncalves A.J.S., Vicentin E.C.M., Amorim J.H., Panatieri R.H., Amorim K.N.S., Yamamoto M.M., Ferreira L.C.S., Alves A.M.B. Targeting the non-structural protein 1 from dengue virus to a dendritic cell population confers protective immunity to lethal virus challenge. PLoS Negl. Trop. Dis. 2013;7:e2330. doi: 10.1371/journal.pntd.0002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau E.H.Y., Tsang O.T.Y., Hui D.S.C., Kwan M.Y.W., Chan W., Chiu S.S., Ko R.L.W., Chan K.H., Cheng S.M.S., Perera R.A.P.M. Neutralizing antibody titres in SARS-CoV-2 infections. Nat. Commun. 2021;12:1–7. doi: 10.1038/s41467-020-20247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang Q., Ji K., Tian S., Wang F., Huang B., Tong Z., Tan S., Hao J., Wang Q., Tan W. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat. Commun. 2021;12:1–10. doi: 10.1038/s41467-021-21037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Earle K.A., Ambrosino D.M., Fiore-Gartland A., Goldblatt D., Gilbert P.B., Siber G.R., Dull P., Plotkin S.A. 2021. Evidence for Antibody as a Protective Correlate for COVID-19 Vaccines. Vaccine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 46.Martinoli C., Chiavelli A., Rescigno M. Entry route of Salmonella Typhimurium directs the type of induced immune response. Immunity. 2007;27:975–984. doi: 10.1016/j.immuni.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 47.Sato A., Hashiguchi M., Toda E., Iwasaki A., Hachimura S., Kaminogawa S. CD11b+ Peyer’s patch dendritic cells secrete IL-6 and induce IgA secretion from naive B cells. J. Immunol. 2003;171:3684–3690. doi: 10.4049/jimmunol.171.7.3684. [DOI] [PubMed] [Google Scholar]
- 48.Lycke N.Y., Bemark M. The regulation of gut mucosal IgA B-cell responses: recent developments. Mucosal Immunol. 2017;10:1361–1374. doi: 10.1038/mi.2017.62. [DOI] [PubMed] [Google Scholar]
- 49.Kirthika P., Senevirathne A., Jawalagatti V., Park S., Lee J.H. Deletion of the lon gene augments expression of Salmonella Pathogenicity Island (SPI)-1 and metal ion uptake genes leading to the accumulation of bactericidal hydroxyl radicals and host pro-inflammatory cytokine-mediated rapid intracellular clearance. Gut Microbes. 2020;11:1695–1712. doi: 10.1080/19490976.2020.1777923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lycke N., Erlandsson L., Ekman L., Schon K., Leanderson T. Lack of J chain inhibits the transport of gut IgA and abrogates the development of intestinal antitoxic protection. J. Immunol. 1999;163:913–919. [PubMed] [Google Scholar]
- 51.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W., Bleicker T., Brunink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hewawaduge C., Senevirathne A., Jawalagatti V., Kim J.W., Lee J.H. Copper-impregnated three-layer mask efficiently inactivates SARS-CoV2. Environ. Res. 2021;196:110947. doi: 10.1016/j.envres.2021.110947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorshkov K., Susumu K., Chen J., Xu M., Pradhan M., Zhu W., Hu X., Breger J.C., Wolak M., Oh E. Quantum dot-conjugated SARS-CoV-2 spike pseudo-virions enable tracking of angiotensin converting enzyme 2 binding and endocytosis. ACS Nano. 2020;14:12234–12247. doi: 10.1021/acsnano.0c05975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Asaka M.N., Utsumi D., Kamada H., Nagata S., Nakachi Y., Yamaguchi T., Kawaoka Y., Kuba K., Yasutomi Y. Highly susceptible SARS-CoV-2 model in CAG promoter-driven hACE2-transgenic mice. JCI insight. 2021;6:e152529. doi: 10.1172/jci.insight.152529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiedemann C., Bellstedt P., Gorlach M. CAPITO-a web server-based analysis and plotting tool for circular dichroism data. Bioinformatics. 2013;29:1750–1757. doi: 10.1093/bioinformatics/btt278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.