Abstract
Background
Human monoclonal antibody (mAb) treatments are promising for COVID-19 prevention or therapy. The pre-exposure prophylactic efficacy of neutralizing antibodies that are engineered with mutations to extend their persistence in human serum and the neutralizing antibody titer in serum required for protection against SARS-CoV-2 infection remain poorly characterized.
Methods
The Fc region of two neutralizing mAbs (COV2-2130 and COV2-2381) targeting non-overlapping epitopes on the receptor binding domain of SARS-CoV-2 spike protein was engineered to extend their persistence in humans and reduce interactions with Fc gamma receptors. We assessed protection by individual antibodies or a combination of the two antibodies (designated ADM03820) given prophylactically by an intravenous or intramuscular route in a non-human primate (NHP) model of SARS-CoV-2 infection.
Findings
Passive transfer of individual mAbs or ADM03820 conferred virological protection in the NHP respiratory tract in a dose-dependent manner, and ADM03820 potently neutralized SARS-CoV-2 variants of concern in vitro. We defined a protective serum-neutralizing antibody titer and concentration in NHPs for passively transferred human antibodies that acted by direct viral neutralization.
Conclusions
In summary, we demonstrate that neutralizing antibodies with extended half-life and lacking Fc-mediated effector functions are efficient for pre-exposure prophylaxis of SARS-CoV-2 infection in NHPs. These results support clinical development of ADM03820 for COVID-19 prevention.
Funding
This research was supported by a contract from the JPEO-CBRND (W911QY-20-9-003, 20-05); the Joint Sciences and Technology Office and Joint Program Executive Office (MCDC-16-01-002 JSTO, JPEO); a DARPA grant (HR0011-18-2-0001); an NIH grant (R01 AI157155); and the 2019 Future Insight Prize from Merck KGaA.
Keywords: SARS-CoV-2, aerosol, human monoclonal antibody, neutralizing antibodies, cynomolgus macaques, antibody therapeutics
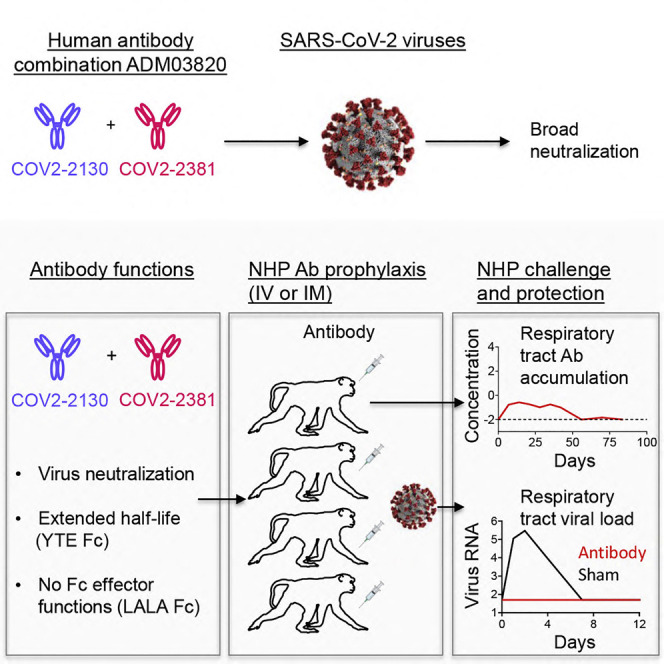
Graphical abstract
Context and significance
Antibodies are a principal mediator of protective immunity to SARS-CoV-2 infections, and human monoclonal antibodies are promising for COVID-19 prophylaxis or therapy. Defining antibody protective efficacy and establishing the titer of neutralizing antibodies required for protection are imperative for the development of efficient antibody-based therapeutics against COVID-19. Here, studies reveal that passive transfer of individual neutralizing human monoclonal antibodies or combinations of the two given prophylactically by an intravenous or intramuscular route protected non-human primates against SARS-CoV-2 infection. Efficient antibody-mediated prophylaxis protection was achieved principally via direct virus neutralization and defined a protective titer of neutralizing antibodies in serum. These results support clinical development of the tested antibody combination as a biologic for COVID-19 prevention.
In this work, Cobb et al. described a combination of two human neutralizing antibodies, engineered with extended half-life and lacking Fc effector functions, which protected non-human primates against SARS-CoV-2 infection when given prophylactically by an intravenous or intramuscular route. The serum antibody titer required for virological protection was defined.
Introduction
In the past decades, two pathogenic human coronaviruses, severe acute respiratory syndrome (SARS-CoV) and Middle East respiratory syndrome CoV (MERS-CoV), have been reported to cause severe respiratory tract disease associated with high morbidity and mortality. In December 2019, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, Hubei province, China.1 SARS-CoV-2 is the causative agent of the current worldwide coronavirus 2019 (COVID-19) outbreak. The pandemic caused by COVID-19 has made the development of countermeasures an urgent global priority.2, 3, 4, 5, 6 Safe and effective vaccines and therapeutics are essential to combat this global pandemic.
Initial work identified that SARS-CoV-2 uses the angiotensin-converting enzyme 2 (ACE2) protein from bats, civet cats, swine, non-human primate (NHPs), or humans as an attachment and entry receptor.6, 7, 8 As with related coronaviruses, interaction with ACE2 is mediated principally through the viral spike (S) protein. Hence, S on the surface of the virion is the main target for neutralizing antibodies on these coronaviruses. This homotrimeric glycoprotein is anchored in the viral membrane and consists of two subunits: S1, containing the N-terminal domain (NTD) and host cell receptor binding domain (RBD), and S2, which contains the fusion peptide.9 , 10 The S protein RBD directly interacts with the peptidase domain of ACE2.6, 7, 8 , 10 Recent studies of the S protein structure have shown that the protein exists in different conformations.9 , 11 Initially, the RBD switches from a closed conformation to an open conformation to allow human ACE2 (hACE2) interaction. Upon interaction with the hACE2 receptor and TMPRSS2 priming, S2 undergoes a dramatic conformational change to trigger host membrane fusion.12
The RBD is the primary target of most potently neutralizing anti-SARS-CoV-2 antibodies identified to date.13, 14, 15, 16, 17, 18, 19 The RBD is also the main antigenic site for neutralizing antibody responses in current and experimental COVID-19 vaccines.20, 21, 22 Previous studies established an NHP model for SARS-COV-2 infection,23 , 24 demonstrating protection from viral infection by transfer of a high dose of ACE2-blocking monoclonal antibodies (mAbs).19 All available antibody therapeutics that have received Emergency Use Authorization (EUA) from the Food and Drug Administration (FDA) except for tixagevimab/cilgavimab were approved for post-exposure treatment, not for pre-exposure prophylaxis.25, 26, 27 Prophylaxis with passive antibody therapy is important as an option for individuals at high risk of disease from SARS-CoV-2 infection who cannot be adequately vaccinated, including immunocompromised individuals or others who respond poorly to vaccination.28 , 29
In NHP studies, the prophylactic efficacy of a single mAb or a combination of two mAbs that were produced as a recombinant immunoglobulin (IgG) 1 with a conventional Fc region has been demonstrated previously.19 , 30 , 31 Here, we evaluated the prophylactic efficacy of low or moderate doses of two different human mAbs targeting non-overlapping neutralization epitopes in the RBD domain19 , 32 engineered with variant Fc regions associated with extended half-life, which we assessed individually or in combination. It has been previously shown that antibody combinations can limit the risk of viral mutations that escape antibody neutralization more efficiently than monotherapy.33, 34, 35 The antibody COV2-2381 binds directly to the receptor binding motif on the RBD on an S protomer in the open position. In contrast, the antibody COV2-2130 binds a non-overlapping site on the RBD that is accessible in either the open or closed S protomer conformation. We engineered the Fc portion of these antibodies to contain mutations that extend half-life (M252Y/S254T/T256E, designated YTE)36, 37, 38 and also to reduce Fcγ receptor binding (L234A/L235A, designated LALA).39, 40, 41 One conceptual advantage of this approach is that the use of these antibodies lacking Fc-mediated effects allowed us to assess the level of serum neutralizing activity needed in vivo to achieve efficacy in the absence of confounding variables. The resulting recombinant mAbs were designated COV2-2130-YTE-LALA and COV2-2381-YTE-LALA, and a two-mAb combination that is a 1:1 mixture of the two was designated ADM03820. The results demonstrate that ADM03820 protects against challenge with SARS-CoV-2 in the lungs and nasopharynx in a dose-dependent manner and define titers of passively transferred neutralizing antibodies that are necessary for protection in NHPs. In addition, our results support the use of an antibody combination that could be administered by either an intravenous or intramuscular route and that neutralizes SARS-CoV-2 variants of concern. This work provides evidence for developing a combination of antibodies as prophylaxis against SARS-CoV-2 in high-risk individuals.
Results
Human antibodies are detected at primary sites of SARS-CoV-2 infection in NHPs when ADM03820 is administered by IV or IM routes (study 1)
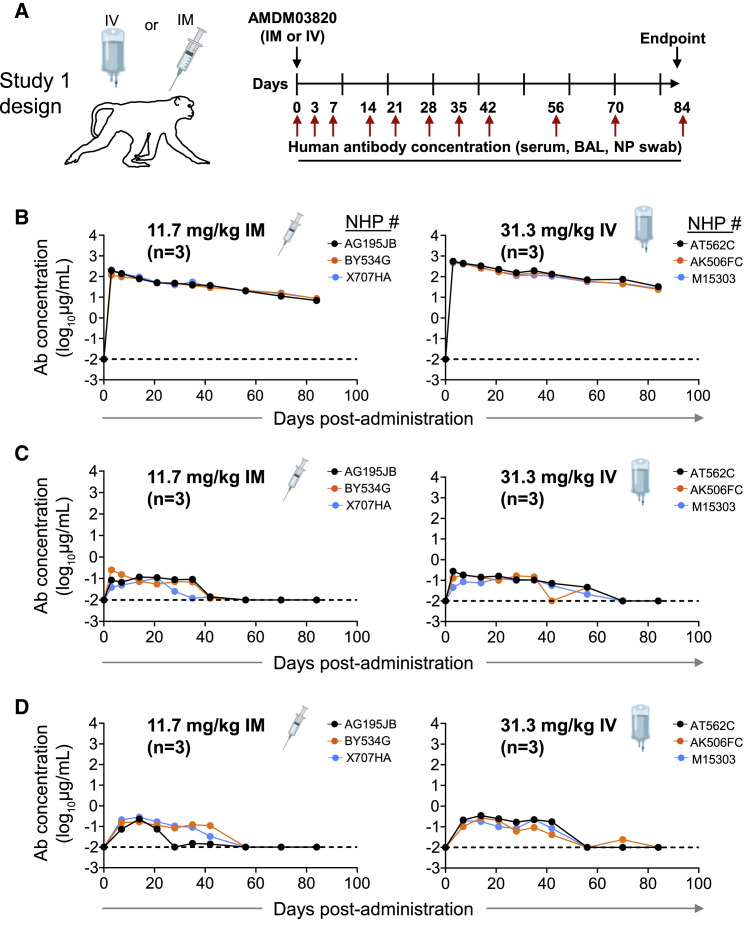
In this study, we used a cynomolgus macaque SARS-CoV-2 challenge model for pre-clinical development studies of a prophylactic combination ADM03820 comprising two engineered mAbs, COV2-2130-YTE-LALA and COV2-2381-YTE-LALA. We first assessed the human antibody concentration in serum and at primary sites of infection (e.g., upper and lower respiratory tract mucosa) after 11.7 mg/kg intramuscular (IM) or 31.3 mg/kg intravenous (IV) administration of ADM03820 in cynomolgus monkeys (Figure 1A). Circulating human mAbs were detected at high levels in serum on day 0 after administration (median 193 μg/mL after IM or 520 μg/mL after IV administration) and persisted in serum for >80 days, exhibiting a slow and gradual decline. The median human IgG serum concentration was 9 μg/mL on day 84 after IM or 26 μg/mL after IV administration (Figure 1B). Notably, ADM03820 antibodies also were detected in respiratory tract secretions, including bronchoalveolar lavage (BAL) fluids and nasopharyngeal (NP) swab samples up to 60 days after administration and at concentrations ranging from 10 (the assay limit of detection) to 270 ng/mL (Figures 1C–1D). The concentration of human antibodies in these secretions in vivo prior to collection is expected to be higher, given that specimen collection from the mucosa sites with saline washes resulted in antibody dilution.
Figure 1.
Pharmacokinetics and biodistribution of ADM03820
(A) Schema of study design. Different doses of antibody combination ADM03820 (containing COV2-2130-YTE-LALA and COV2-2381-YTE-LALA at a 1:1 ratio) were administered to cynomolgus macaques (n = 3 per group) by IV (11.7 or 31.3 mg/kg) or IM (11.7 or 31.3 mg/kg) route. Human antibody concentration was assessed by ELISA in (B) serum, (C) BAL, or (D) nasal swab eluate samples at indicated time points after ADM03820 administration. The dotted horizontal line depicts the assay limit of detection.
ADM03820 antibody combination potently neutralizes variants of concern
ADM03820 exhibited broad and potent neutralizing activity in vitro with half-maximal inhibitory concentration values <25 ng/mL, including potent neutralization of viruses representing wild-type SARS-CoV-2 WA1/2020 with or without D614G mutation, authentic B.1.1.7 virus, authentic B.1.617.2 virus, and chimeric Wash-B.1.351 and Wash-B.1.1.28 viruses, which contain an S gene from B.1.351 or B.1.1.28, respectively, in the backbone of WA1/202042 (Table 1 ). Collectively, these results showed prolonged persistence of administered human antibodies in serum and respiratory mucosa at concentrations sufficient for neutralization of currently circulating viral variants.
Table 1.
Neutralization breadth of ADM03820 against SARS-CoV-2 variants of concerna (IC50 [ng/mL] against indicated virus)b
| WA1/2020 | D614G | B.1.1.7 (alpha) | Wash-B.1.351 (beta) | B.1.617.2 (delta) | Wash-B.1.1.28 (gamma) |
|---|---|---|---|---|---|
| 28 | 21 | 20 | 19 | 25 | 8 |
Neutralizing activity of ADM03820 against authentic SARS-CoV-2 WA1/2020, authentic SARS-CoV-2 WA1/2020 bearing D614G mutation, or authentic B.1.1.7, authentic B.1.617.2, chimeric Wash-B.1.351, and chimeric Wash-B.1.1.28 viruses was assessed using a focus reduction neutralization test (FRNT).
Half-maximal inhibitory concentration (IC50) values are shown and represent the average of technical duplicates and two independent experiments.
Protective efficacy of ADM03820 in non-human primates (study 2)
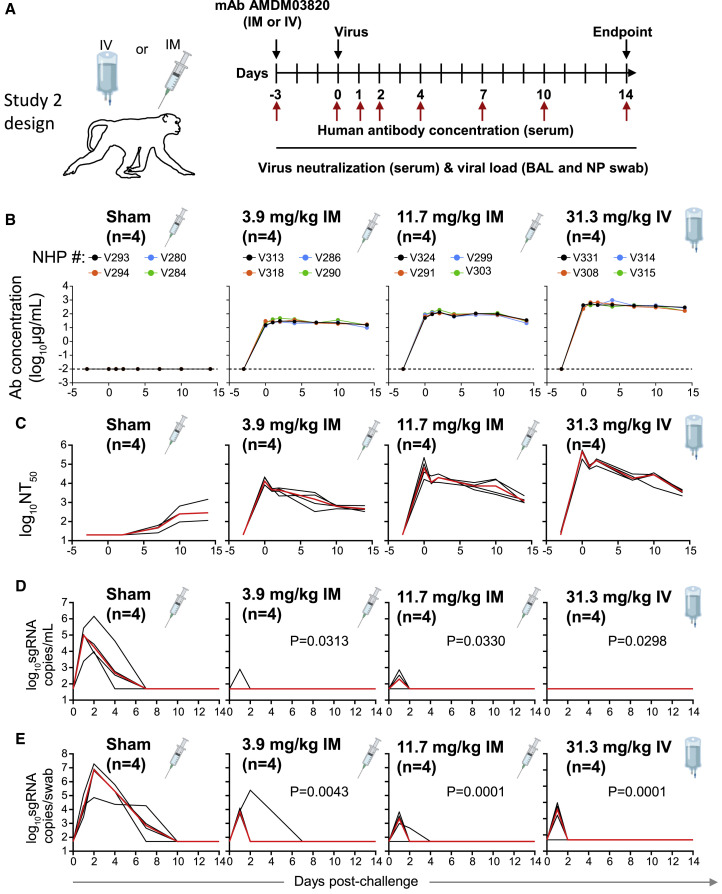
To evaluate the protective efficacy of ADM03820, animals received various doses (3.9–31.3 mg/kg) of the ADM03820 by either an IM or IV route followed by a viral challenge with 105 tissue culture infectious dose (TCID50) 3 days later (Figure 2A). We then measured the circulating human antibody concentration in serum and serum neutralizing titers up to day 14 following IM or IV administration. While antibody concentration was below the limit of detection in the sham-treated group, animals in the antibody treatment groups exhibited mAb levels proportional to the dose and route of administration of the combination product (Figure 2B). The antibody concentration in serum peaked approximately 3 days post-administration and remained constant throughout the remaining 14 days of the study.
Figure 2.
Pharmacokinetics, antibody neutralizing titers, and prophylactic efficacy of ADM03820 mAbs in SARS-CoV-2-challenged NHPs
(A) Schema of study design. Different doses of ADM03820 were administered to cynomolgus macaques (day −3) by IM (3.9 or 11.7 mg/kg) or IV (31.3 mg/kg) route (n = 4 per group). One group of NHPs was left untreated (sham; n = 4) and served as a control. Animals in all groups were challenged with 105 TCID50 SARS-CoV-2 by the intranasal and intratracheal routes on day 0.
(B) Human antibody concentration in serum was assessed by ELISA at indicated time points after ADM03820 administration and viral challenge.
(C) Total neutralizing antibody titers were assessed in serum at indicated time points using pseudovirus neutralization assay. The red line indicates the median titer of neutralizing antibodies in each group.
(D) Subgenomic RNA (sgRNA) levels were assessed at various time points after viral challenge in bronchoalveolar lavage (BAL) samples using qRT-PCR.
(E) sgRNA levels were assessed at various time points after viral challenge in nasopharyngeal (NP) swab samples. Each black curve shows the measurements from individual animals, with red lines indicating the median values of measurements for animals within each treatment group. Neutralization assay limit of detection = 50 copies/mL or 50 copies/swab for (D) and (E). For statistical analysis, refer to STAR Methods section.
We observed high circulating neutralizing antibody titers by pseudovirus neutralization assays in all ADM03820 treatment groups but not in the sham-treated control group. However, sham-treated control animals developed low-level neutralizing titers beginning around day 6, presumably due to the induction of natural host immunity (Figure 2C). In general, the overall neutralizing antibody titers were consistent with the pharmacokinetic data for the same treatment groups.
We assessed the kinetics of viral loads up to day 14 following viral challenge in BAL and NP swab samples by determining the levels of SARS-CoV-2 subgenomic RNA (sgRNA), which distinguishes replicating virus from input challenge virus, using RT-PCR23 , 24 , 43 (Chandrashekar et al., 2020; Wolfel et al., 2020; Yu et al., 2020). High levels of sgRNA were observed in the sham controls (Figures 2D–2E), with a median peak of 5.0 (range = 3.3 to 5.4) log10 sgRNA copies/mL in BAL fluid and 6.9 (range = 4.9 to 7.3) log10 copies per swab of sgRNA in NP swab samples. As expected, peak viral loads occurred between days 1 and 4 after challenge. All treatment groups showed nearly full protection from viral replication in the BAL fluid, although individual animals displayed low-level, transient viral replication on day 1, which was eliminated by day 2 (Figure 2D). Although somewhat higher sgRNA levels were observed in some animals in the NP swab samples on day 1, similar to BAL fluid, most treated animals quickly eliminated detectable virus by day 2 (Figure 2E), with the exception of 1 animal in the group receiving the lowest dose (3.9 mg/kg IM) and 1 animal in the group receiving 11.7 mg/kg dose.
Protective efficacy of individual mAbs of the combination in non-human primates (study 3)
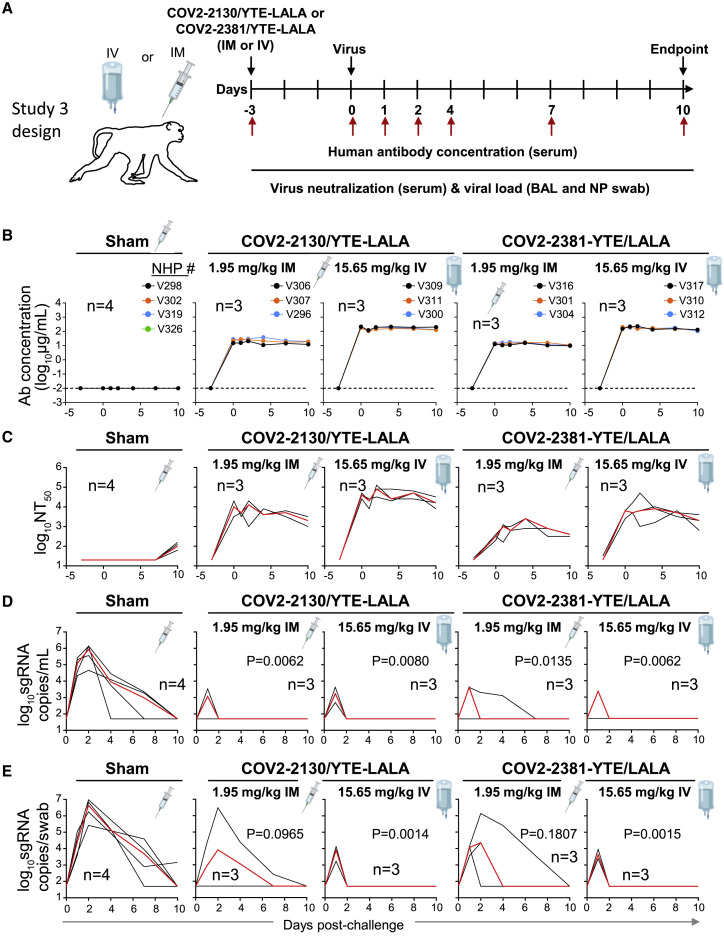
The next challenge study was conducted after prophylactic administration by either the IM or IV route of the individual 2130-YTE-LALA or 2381-YTE-LALA antibodies (1.95–15.65 mg/kg) and was followed by quantitative serum antibody levels and virological protection measurements as in the challenge study 2 (Figure 3A). As expected, the concentration of circulating human antibodies was below the level of detection in the sham-treated group. In contrast, animals that received either mAb demonstrated concentrations in serum proportional to the administered dose (Figure 3B). Peak antibody concentration was observed within 3 days of administration and remained constant throughout the study. Serum neutralizing titers of administered individual mAbs showed similar peak and kinetics to those seen with the ADM03820 combination (Figure 3C). Sham-treated animals showed low levels of neutralizing antibody activity by day 6 due to the host immune response to SARS-CoV-2 infection (Figure 3C).
Figure 3.
Pharmacokinetics, neutralizing titers, and prophylactic efficacy of individual mAbs of the combination in SARS-CoV-2-challenged NHPs
(A) Schema of study design. Individual mAbs COV2-2130/YTE-LALA or COV2-2381/YTE-LALA (n = 3 NHPs per group) were administered to cynomolgus macaques (day −3) at different doses (1.95 or 15.65 mg/kg) and routes (IM or IV), as indicated. One group of NHPs was left untreated (sham; n = 4) to serve as controls. Animals in all groups were challenged with SARS-CoV-2 by the intranasal and intratracheal routes on day 0.
(B) Human antibody concentration was assessed by ELISA in serum at indicated time points after indicated mAb administration and viral challenge.
(C) Total neutralizing antibody titers were assessed in serum at indicated time points using a pseudovirus neutralization assay. Each black curve shows the measurements from an individual animal, with red lines indicating the median values of measurements for animals within each treatment group.
(C) sgRNA levels were assessed after viral challenge at various time points in BAL samples using qRT-PCR.
(D) sgRNA levels were assessed after viral challenge at various time points in NP swab samples.
The red line depicts the median levels of sgRNA in each group. Each black curve shows an individual animal’s measurements, with red lines indicating the median values of measurements for animals within each treatment group. Neutralization assay limit of detection = 50 copies/mL or 50 copies/swab. For statistical analysis, refer to STAR Methods section.
As evidenced by sgRNA levels, viral infection again was observed in all the sham-treated control animals in both BAL fluid and NP swab samples (Figures 3D and 3E). For both treatment mAbs, most animals quickly cleared virus by day 2 post-challenge after transient viral replication regardless of dose or route of administration, except for one animal in the 1.95 mg/mL 2381-YTE-LALA IM group (Figure 3D).
Similar levels of viral protection were observed in NP swab samples with the 15.65 mg/mL dose of either individual antibody (Figure 3E), as was observed with similarly high tested doses of the ADM03820 combination (Figure 2E). However, higher median viral loads were observed in the NP swab samples for both antibody treatments at the low dose of 1.95 mg/mL. This dose is 2-fold lower than the lowest dose tested for the combination and likely represents viral breakthrough due to insufficient neutralizing antibody levels.
Protective efficacy of ADM03820 administered by IM route at low doses (study 4)
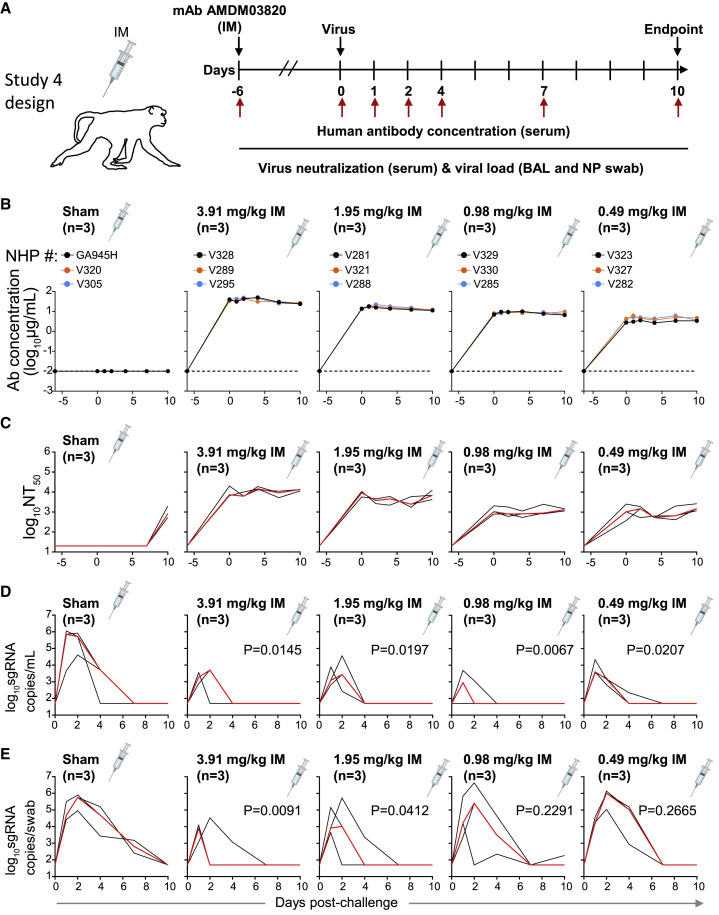
To determine the minimally protective dose of the ADM03820 combination, animals were treated with 2-fold decreasing doses of the antibody combination across 4 treatment groups, from 3.91 to 0.49 mg/kg by the IM route (Figure 4A). Circulating human antibody titers were not present in sham-treated animals and were consistent with the administered dose in the treatment groups (Figure 4B). The serum neutralizing antibody titer decrease was proportional to the administered ADM03820 dose and was observed across all 4 treatment groups but not observed in sham group animals (Figure 4C). BAL fluid viral load measurement suggested protection in the lower airways at all tested antibody doses, including at the 0.49 mg/kg dose (Figure 4D). However, increases in NP swab sample viral loads were seen across decreasing dose conditions, with no protection observed in the 0.98 or 0.49 mg/mL groups (Figure 4E).
Figure 4.
Pharmacokinetics, neutralizing titers, and prophylactic efficacy of ADM03820 in a dose de-escalation study and IM antibody administration in NHPs
(A) Schema of study design. Different doses of ADM03820 were administered to cynomolgus macaques (day −6) by IM route (3.91, 1.95, 0.98, and 0.49 mg/kg; n = 3 NHP per group). One group of NHPs was left untreated (sham; n = 3) and served as a control. Animals in all groups were challenged with SARS-CoV-2 by the intranasal and intratracheal routes at day 0.
(B) Human antibody concentration was assessed by ELISA in serum at indicated time points after ADM03820 administration and viral challenge.
(C) Total neutralizing antibody titers in serum were assessed at indicated time points using a pseudovirus neutralization assay. The red line shows median titer of neutralizing antibodies in each group.
(D) sgRNA levels were assessed at various time points after viral challenge in BAL samples using qRT-PCR.
(E) sgRNA levels were assessed at various time points after viral challenge in NP swab samples. The red line depicts the median levels of sgRNA in each group. Each black curve shows measurements from an individual animal, with red lines indicating the median values of measurements for animals within each treatment group. Assay limit of detection = 50 copies/mL or 50 copies/swab. For statistical analysis, refer to STAR Methods section.
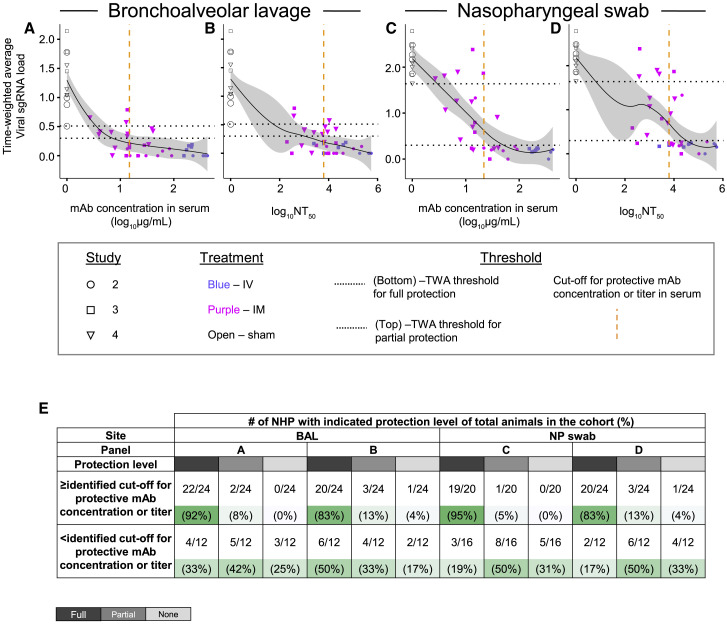
Defining protective serum antibody concentration and neutralizing antibody titer in NHP SARS-CoV-2 challenge model
We next estimated a protective threshold for prophylaxis with potent YTE-LALA-Fc-region-engineered human antibodies that acted principally via direct virus neutralization in vivo. We performed an overall analysis using data from challenge studies 2, 3, and 4, above, by comparing human mAb concentration in serum or half-maximal neutralizing titer values at the time of challenge with the time-weighted average values for the change of sgRNA viral load in BAL fluid or NP swab samples from day 1 to 10 after viral challenge (see STAR Methods and Tables S1 and S2). A threshold for virological protection in BAL fluid and NP swab samples was estimated to be equal to or higher than 20 μg/mL for circulating human antibody concentration and equal to or higher than 6,000 for serum neutralizing antibody titer (NT50) (Figures 5A–5D). Antibody levels above these thresholds conferred full protection in 83%–93% of challenged NHPs, which contrasted with 17%–50% fully protected animals with antibody levels below these estimated protective thresholds (Figure 5E). Spearman rank correlation analysis revealed a strong negative correlation between serum antibody concentration and time-weighted average viral load measurements in the upper airways when considering both antibody administration routes or the IM route only (Figure S1). These results suggested that a higher antibody dose would be necessary to control viral replication in the upper airways following IM or IV administration. Overall, our results suggested that high prophylaxis efficacy can be achieved with the combination of two YTE-LAL-Fc-region-engineered human antibodies formulated as the combination ADM03820 and demonstrated the potential for IM delivery of human antibody-based therapeutics for COVID-19.
Figure 5.
Human antibody concentration and antibody neutralizing titer in NHP serum associated with protection against viral challenge in BAL fluid or NP swab samples
(A–D) The time-weighted average (TWA) values for the change of sgRNA viral load in BAL fluid or NP swab samples from day 1 to 10 after viral challenge were compared with antibody concentration in serum or serum NT50 value for each animal from studies 2, 3, and 4, described in Figure 2, Figure 3, Figure 4. The fitting curves were estimated using the locally weighted scatterplot smoothing (LOWESS) method and are shown in black, and gray shading indicates the CI. Shapes indicate individual animals, colors indicate route of antibody treatment, and animals from separate studies are shown with different shapes, as detailed in the figure. Horizontal black dotted lines indicate designated TWA thresholds for full (bottom line) and partial (top line) protection. Vertical dotted orange dashed line in the graphs indicates designated estimated optimal cutoff for protective antibody concentration or titer in NHP serum. For calculation of TWA and cutoff values, refer to STAR Methods section.
(E) Percent animals that fully protected, partially protected, or non-protected determined using the estimated thresholds for protection as in (A–D). Gradient of green shading visualize percent of protected animals, in which dark green indicates higher percent of protected animals and light green indicates lower percent of protected animals for each described condition.
Discussion
Numerous groups have reported the isolation of potently neutralizing antibodies from survivors that target the RBD of SARS-CoV-2 S protein.13 , 16, 17, 18 , 47, 48, 49 These studies provide insights into both quantitative and qualitative aspects of the use of human mAbs as medical countermeasures for COVID-19. First, we demonstrate the principle that prophylaxis against infection in NHPs can be achieved using neutralizing antibodies engineered to lack Fc-mediated functions. These data extend previous findings that demonstrated prophylaxis efficacy for neutralizing mAbs with intact Fc-mediated functions in NHPs.19 , 30 , 31 Second, the data show excellent protection by antibodies acting only by direct neutralization of virus and define the protective level of serum neutralizing activity in the absence of confounding variables of Fc-mediated effects. A threshold for virological protection in BAL fluid and NP swab samples was estimated to be equal to or higher than 6,000 for serum NT50, because antibody levels above these thresholds conferred full protection in 83%–93% of challenged NHPs. Although it remains to be determined if the protective titer level observed in the current study is generalizable to other prophylactic candidate anti-SARS-CoV-2 human mAbs, the quantitative determination of a neutralizing titer as a direct mechanistic correlate of protection has implications for estimating the durability of protection conferred by passive immunization with antibodies29 or active immunization with vaccines. With the caveat that the results may be specific to these mAbs, the failure to achieve serum neutralizing titers above the threshold reported in this study may provide a plausible explanation for the limited efficacy observed in most clinical trials of COVID-19 convalescent plasma.44, 45, 46 Also, this quantitative threshold for correlate of protection sheds light on the somewhat limited magnitude and durability of the humoral immunity component of protection following natural infection or immunization. Third, the studies also support a public health strategy of prophylaxis of high-risk individuals who cannot be adequately vaccinated by using administration of neutralizing mAbs instead. Engineering of the Fc region to accomplish long half-life extends the prophylactic efficacy of the antibodies, predicted to last up to 12 months in humans.29 Fourth, we also assessed IM and IV administration and found that IM administration was effective, which could allow a much easier and more practical approach to administration of these antibodies at large scale in populations at risk.
The studies here support the further development of a two-mAb prophylactic anti-SARS-CoV-2 combination (ADM03820) incorporating mAbs that target non-overlapping regions of the RBD.19 , 32 The combination of engineered antibodies possesses desirable features consistent with the objectives above, including long half-life, an effective IM formulation, accumulation at respiratory mucosa following systemic administration, and a clear mechanism of action purely through direct virus neutralization. The combination was shown effective in a stringent rhesus macaque model for SARS-CoV-2 we previously developed with high viral loads in the upper and lower respiratory tract, cellular and humoral immune responses, and pathogenic evidence of viral pneumonia.23 , 24 In the present study, we demonstrated that prophylactic administration of the two-mAb combination ADM03820 for protection against SARS-CoV-2 infection in the cynomolgus macaque model reduced viral loads in the upper and lower airways and accelerated virus clearance.
These antibodies include the YTE mutations in Fc region, which increase the serum half-life of the mAbs,50, 51, 52 and the LALA Fc mutations that were designed to decrease the Fc effector function by reducing interaction with Fcγ receptors.39 , 40 , 52 , 53 Studies in murine SARS-CoV-2 challenge models have demonstrated equivalently high prophylactic efficacy by potently neutralizing RBD-specific mAb variants with intact or abrogated Fc region-mediated effector functions.54 Previous studies in a similar NHP model have shown that COV2-2381 IgG with a conventional Fc region cleared the virus infection, and no virus was observed when given at 50 mg/kg.19 Here, the addition of YTE and LALA mutations did not appear to reduce the ability of these mAbs to clear SARS-CoV-2 infection in either the BAL fluid or NP swab samples in cynomolgus macaques when administered 3 days prior to challenge. Thus, prophylaxis with ADM03820 containing antibodies with extended half-life and lacking Fc effector functions conferred a high level of protection in NHPs at the highest tested antibody dose, 31 mg/kg, with no virus detected in BAL fluid and only transient virus detection at day 1 in NP swab samples. A comparison with previous prophylactic studies in NHPs with casirivimab/imdevimab (no sgRNA detected in NP swab samples at a 50 mg/kg antibody dose after challenge with regular titer viral inoculum and high lasting virus shedding at a 150 mg/kg antibody dose with high titer viral inoculum),30 LY-CoV555 (no sgRNA detected in NP swab samples at a 50 mg/kg antibody dose and transient virus shedding at a 15 mg/kg mAb dose),31 and COV2-2381 or COV-2196 (no sgRNA detected in NP swab samples at a 50 mg/kg mAb dose)19 suggested at least a similar potency of ADM03820. More NHP studies are needed to compare the efficacy of extended-half-life IgG lacking Fc effector functions to IgG with a conventional Fc region to prevent respiratory tract infection in different prophylaxis settings.
A lower serum antibody neutralizing titer (>100) was associated with protection by vaccines in NHP SARS-CoV-2 challenge models24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56 and in human clinical trials57, 58, 59 relative to the protective titer associated with mAbs (∼6,000) that we defined here. However, a similar protective titer against SARS-CoV-2 was identified in NHPs for another combination of two neutralizing human mAbs in clinical use—tixagevimab/cilgavimab.29 Future studies are needed to determine if the lower serum neutralizing antibody protective titer for COVID-19 vaccines relative to that achieved by passive mAb transfer is due to targeting of multiple epitopes on the SARS-CoV-2 spike protein, different anatomical distribution of antibody responses, a contribution of Fc-mediated effector functions in the polyclonal response, or complementary mechanisms of protection that are mediated by vaccine-induced T cells.
The RBD sequence is highly variable in SARS-CoV-2, which may represent a selective adaptation.60, 61, 62, 63 Our approach, to use a combination of two antibodies that do not compete for the same epitope, could prevent the selection of escape mutant viruses that are likely inherent in monotherapy approaches. Recent work in the context of SARS-CoV-2 has demonstrated that combinations of two antibodies that do not compete for binding to the same region of the S protein offer higher resistance to escape mutations while protecting animals from SARS-CoV-2 challenge.19 , 30 , 33 , 42 , 64
In prior NHP studies, mAbs typically were infused via IV administration. The studies presented here demonstrate the efficacy of these antibodies either administered as a combination or alone when administered by the IM route This approach could provide a more broadly deployable route of administration for these antibodies to patients in clinical settings. In addition, the doses that were efficacious in these studies translate to very low doses in humans compared with conventional antibody therapies. The data generated in these studies provide strong evidence for the continued development of these antibodies for clinical use.
Limitations of the study
The estimated thresholds of protection for human mAbs cannot be generalized and should be confirmed through other studies using different mAbs. More studies are needed to compare the efficacy of extended-half-life IgG lacking Fc effector functions to IgG with a conventional Fc region to prevent respiratory tract infection in different prophylaxis settings.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SARS2-2 | VanBlargan et al.65 | N/A |
| SARS2-11 | VanBlargan et al.65 | N/A |
| SARS2-16 | VanBlargan et al.65 | N/A |
| SARS2-31 | VanBlargan et al.65 | N/A |
| SARS2-38 | VanBlargan et al.65 | N/A |
| SARS2-57 | VanBlargan et al.65 | N/A |
| SARS2-71 | VanBlargan et al.65 | N/A |
| ADM03826 drug product | Ology Bioservices | N/A |
| Goat Anti-Mouse IgG Antibody, HRP conjugate | Sigma | Cat # 12-349; RRID: AB_390192 |
| Bacterial and virus strains | ||
| SARS-CoV-2 WA1/2020 | CDC | N/A |
| SARS-CoV-2 D614G | Chen et al.42 | N/A |
| Wash-B.1.351 | Chen et al.42 | N/A |
| Wash-B.1.1.28 (or Wash-BR-B.1.1.248) | Chen et al.42 | N/A |
| B.1.1.7 | Chen et al.42 | N/A |
| B.1.617.2 | Human isolate | N/A |
| Biological samples | ||
| NHP serum | This study | N/A |
| NHP BAL | This study | N/A |
| NHP NP swabs | This study | N/A |
| Experimental models: Cell lines | ||
| Monkey: Vero E6 | ATCC | N/A |
| Monkey: Vero-TMPRSS2 | Zang et al., 2020 | N/A |
| Monkey: MA104 | American Type Culture Collection | CRL-2378.1 RRID: CVCL_3845 |
| Experimental models: Organisms/strains | ||
| Cynomolgus monkeys (Macaca fascicularis) | Bioqual, Inc. | N/A |
| Software and algorithms | ||
| GraphPad Prism 8.4.3 | GraphPad Software, Inc. | GraphPad Prism, RRID:SCR_002798 |
| R software | R Core Team, 2021 | R software, RRID: SCR_001905 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, James E. Crowe, Jr. (james.crowe@vumc.org).
Materials availability
Materials described in this paper are available for distribution for nonprofit use using templated documents from Association of University Technology Managers "Toolkit M.T.A.s", available at: https://autm.net/surveys-and-tools/agreements/material-transfer-agreements/mta-toolkit.
Experimental model and subject details
Animals
Cynomolgus monkeys (Macaca fascicularis) were maintained at Bioqual, Inc. (Rockville, MD) which is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) and approved by the Office of Laboratory Animal Welfare (NIH/PHS assurance number D16-00052). Studies were conducted in compliance with all relevant local, state, and federal regulations and were approved by the Bioqual Institutional Animal Care and Use Committee (IACUC).
Viruses
The SARS-CoV-2 USA-WA1/2020 strain was obtained from BEI Resource (NR-52281; Lot #7003175). The viral stocks were expanded using Vero E6 cells and harvested on day 5 following inoculation. To confirm the viral identity, complete genome sequencing was performed and was shown to be 100% identical to the parent virus sequence. The D614G virus was produced by introducing the mutation into an infectious clone of WA1/2020, and the B.1.351 and B.1.1.28 spike genes were cloned into the WA1/2020 infectious clone to produce Wash-B.1.351 and Wash-B.1.1.28 (P.1 lineage) chimeric viruses, as described previously.42 B.1.1.7 and B.1.617.2 were isolated from infected individuals. D614G, Wash-B.1.351, Wash-B.1.1.28, B.1.1.7, and B.1.617.2 viruses were propagated on Vero-TMPRSS2 cells and subjected to deep sequencing.
Monoclonal antibodies
The antibody COV2-2381 and COV2-2130 sequences have been previously described.19 , 32. The antibodies were produced and purified as previously described.66 Briefly, stably transfected CHO cells expressing either COV2-2130-YTE-LALA or COV2-2381-YTE-LALA were generated using Leap-In transposon vectors (ATUM) containing the respective antibody heavy and light chain genes and a glutamine synthetase gene as a selectable marker. Leap-In vectors were transfected into a CHO-K1 GS knockout cell line (HD-BIOP3; from Horizon Discovery) and stably transfected pools were selected using medium lacking L-glutamine. Manufacturing was performed under Good Manufacturing Practices using stably transfected pools in large-scale bioreactors, and antibody material was purified from harvested supernatants. The downstream processes consisted of 3 chromatography steps: 1) Protein A chromatography, 2) cation exchange chromatography, and 3) mixed mode anion exchange/hydrophobic interaction chromatography. Both individual antibodies and the combination were generated as cGMP-grade drug substance and drug product materials, were provided at a concentration of 52 mg/mL and were stored at −80°C until day of administration.
Method details
Antibody administration
On the day of administration, the stock vials were thawed at room temperature (RT) and gently inverted 6 to 10 times to mix the contents. After thawing, the vials were stored at RT until use. Based on individual animal weights and dose required, the purified antibody stock for each NHP was diluted to 1 mL in 0.9% normal saline diluent (Baxter) for IM injections and 10 mL in the same diluent for IV infusions. IM injections were delivered bilaterally in the upper quadriceps at 0.5 mL/quadriceps. IV infusions were performed at a rate of 1 to 2 mL/min over 5 to 10 min/animal for a total of 10 mL infused per animal.
Animal studies
Cynomolgus monkeys (2.2 – 5.8 kg body weight; 6 to 12 years old) were mixed male and female and randomly assigned to groups. In Study 1 (n=3/group), experimental animals received the ADM03820 cocktail of COV2-2130-YTE-LALA and COV2-2381-YTE-LALA at either 11.7 mg/kg IM or 31.3 mg/kg IV and were followed for 12 weeks for antibody pharmacokinetics only without any SARS-CoV-2 challenge. In Study 2 (n=4/group), sham control animals received no mAb while 12 experimental animals were administered the ADM03820 cocktail at varying doses and administration routes three days before challenge as described in Figure 2. Animals then were challenged with 105 TCID50 SARS-CoV-2 USA-WA1/2020. These doses were administered as 0.5 mL per nare intranasally and 1 mL intratracheally on day 0. In Study 3, four sham-treated controls received no mAb while 12 experimental animals (n=3/group) were administered three days prior to challenge with either COV2-2130-YTE-LALA or COV2-2381-YTE-LALA separately at varying doses and administration routes as described in Figure 3A. Animals were then challenged with 105 TCID50 SARS-CoV-2 similarly as in the first study. In Study 4 (n=3/group), sham control animals received no mAb while experimental animals were administered the ADM03820 cocktail IM at varying low doses three days before challenges performed similarly to studies 2 and 3 (Figures 2 and 3). In all studies, antibody doses were selected based on material availability and to approximate the various dosing regimens to achieve a wide concentration of circulating mAb in NHP plasma representative of low, intermediate or high levels. Macaques in all four studies were monitored daily with an internal scoring protocol approved by the IACUC. These studies were not blinded.
Quantification of circulating human mAbs and serum neutralization activity
The quantification of infused/injected human SARS-CoV-2 mAbs in NHP serum at multiple time points was performed as previously described.19 Additionally, the serum neutralization activities of infused or injected mAbs were also monitored at the same time points using a pseudovirus neutralization assay as previously described.23 , 24
BAL and NP swab collection
Collection of mucosal secretions was performed on sedated NHPs using cotton swabs (COPAN flocked swab) or nasosorption FX-I devices (Hunt Developments Ltd.). The swabs were inserted into the nasal cavity and rotated gently. Following collection, the swabs were placed into a collection vial containing 1 mL of phosphate buffered saline (PBS). All vials were stored at ≤ −70°C until viral load testing (or antibody quantification if required).
The bronchoalveolar lavage (BAL) collection procedure was performed on anesthetized animals by the “chair method”. In brief, each animal was placed in dorsal recumbency in a chair channel and a red rubber feeding tube inserted into the trachea via a laryngoscope during inspiration. A total of 10 mL PBS was flushed through the tube and the volume instilled and recovered from each animal recorded. The collected BAL samples were placed immediately onto wet ice and processed for isolation of fluid by centrifugation at 4°C followed by supernatant removal. BAL aliquots were stored at ≤ −70°C until viral load testing (or antibody quantification if required).
Focus reduction neutralization test
Serial dilutions of mAbs were incubated with 102 FFU of different strains or variants of SARS-CoV-2 for 1 h at 37 °C. Antibody–virus complexes were added to Vero-TMPRSS2 cell monolayers in 96-well plates and incubated at 37 °C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM. Plates were collected 30 h later by removing overlays and fixed with 4% PFA in PBS for 20 min at room temperature. Plates were washed and sequentially incubated with an oligoclonal pool of SARS2-2, SARS2-11, SARS2-16, SARS2-31, SARS2-38, SARS2-57 and SARS2-71 anti-S65 antibodies and HRP-conjugated goat anti-mouse IgG (Sigma, 12-349) in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies).
Subgenomic mRNA assay
The subgenomic mRNA of SARS-CoV-2 was assessed by RT-PCR as previously described.23 , 24 , 43 The standard curve is based on the SARS-CoV-2 E gene. Prior to PCR, cDNA was generated from each animal using Superscript III VILO (Invitrogen) according to the manufacturer’s instructions. Using the sequences targeting the E gene mRNA, a TaqMan custom gene expression assay (Thermo Fisher Scientific) was designed43 and reactions were carried out using a QuantStudio 6 and 7 Flex Real-Time PCR system (Applied Biosystems) according to the manufacturer’s instructions. Standard curves were generated to calculate sgRNA/mL or per swab. Viral load for each timepoint tested per NHP was reported as the average of two replicates. The sensitivity of this assay was 50 copies per mL of BAL or per swab.
Quantification and statistical analysis
The average change in viral load (log10 sgRNA copies/mL or swab) was assessed from day 1 to day 14 (Study 2), or from day 1 to day 10 (Study 3 and 4). The time-weighted average (TWA) values for the change of sgRNA viral load in BAL or NP from day 1 to day 10 after viral challenge were calculated as the area under the curve (AUC) of the change in viral load in Prism (version 9.1.2; GraphPad) and then divided by 10 as described previously30 (Table S1). The TWA values of each treatment group were compared to those of the sham group using Welch’s t-test. The significance level alpha of 10% was pre-specified, and estimated P-values are indicated in the figures. TWA threshold was set up to ≤ 0.3 for full protection, ≤ 0.51 (the lower sham point) for partial protection, and > 0.51 for no protection in BAL, and ≤ 0.3 for full protection, < 1.638 (the lower sham point), for partial protection, and > 1.638 for no protection in NP. To estimate protective antibody concentration or neutralizing titer in serum, the optimal thresholds that maximizes the sum of sensitivity and specificity for full protection were calculated and reported in Table S2. Sensitivity is the proportion above the threshold in the fully-protected subjects, and specificity is the proportion below the threshold in partially- or non-protected subjects. The fitting curves and confidence intervals to visualize the relationship between TWA and antibody levels were estimated using the Lowess curve smoothing method using ggplot2 in R software.67 Spearman's rank correlation analysis that assumes a monotonic (as X increases, Y decrease) rather than linear relationship was used to determine the relationship between serum antibody concentration and time-weighted average viral load measurements in the upper airways. The other data visualization was performed using Prism software.
Acknowledgments
We thank E. Bondzie, N. Mercado, V. Giffin, D. Hope, and F. Nampanya for their assistance. This research was supported by the Joint Program Executive Office for Chemical, Biological, Radiological and Nuclear Defense (JPEO-CBRND), contract number W911QY-20-9-003, 20-05; the Joint Sciences and Technology Office and Joint Program Executive Office, contract number MCDC-16-01-002 JSTO, JPEO; Defense Advanced Research Projects Agency (DARPA) grant HR0011-18-2-0001; and National Institutes of Health (NIH) grant R01 AI157155. J.E.C. is a recipient of the 2019 Future Insight Prize from Merck KGaA. The content is solely the responsibility of the authors and does not represent the official views of the US government or other sponsors.
Author contributions
R.R.C., M.S.D., R.H.C., D.H.B., and J.E.C. planned the studies. J.N., P.G., A.C., J.Y., R.V.H., C.G.E., N.M.D., S.A.H., D.M.S., R.E.C., L.A.V., M.H., B.H., L.C., M.T.T., G.H.N., K.H., J.C.S., M.L., H.A., A.C., and D.H.B. conducted experiments, analyzed data, and interpreted the results. J.C.S. and P.G. performed and oversaw the statistical analysis. D.H.B. and P.G. had unrestricted access to all the data in the paper. J.E.C, M.S.D., and D.H.B. obtained funding. R.R.C., J.N., P.G., and J.E.C. wrote the first draft of the paper, with the other authors providing editorial comments. All authors agreed to submit the manuscript, read and approved the final draft, and take full responsibility for its content, including the accuracy of the data.
Declaration of interests
R.R.C., R.V.H., D.M.S., M.H., B.H., L.C., G.H.N., M.T.T., and K.H. are employees of Ology Bioservices. C.G.E. and N.M.D. are employees of the Joint Program Executive Office for Chemical, Biological, Radiological and Nuclear Defense for the US Department of Defense (JPEO-CBRND). S.A.H. is an employee of Logistics Management Institute (LMI), performing technical contract support for JPEO-CBRND. J.E.C. has served as a consultant for Luna Innovations, is a member of the scientific advisory board of Meissa Vaccines, and is founder of IDBiologics. The Crowe laboratory at Vanderbilt University Medical Center has received sponsored research agreements from Takeda, IDBiologics, and AstraZeneca. Vanderbilt University has applied for patents related to antibodies studied in this paper. M.S.D. is a consultant for InBios, Vir Biotechnology, Senda Biosciences, and Carnival Corporation and is on the scientific advisory boards of Moderna and Immunome. The laboratory of M.S.D. has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, Kaleido, and Emergent BioSolutions.
Published: March 11, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.medj.2022.01.004.
Supplemental information
Data and code availability
-
•
All data needed to evaluate the conclusions in the paper are present in the paper or the supplemental information; source data for each of the display items is provided in key resources table.
-
•
No custom computer code or algorithms to report.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., et al. Early transmission dynamics in Wuhan, China, of Novel coronavirus-infected pneumonia. New Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the Novel Coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan X., Cao D., Kong L., Zhang X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat. Commun. 2020;11:3618. doi: 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 15.Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 16.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schafer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W.H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 2020:1–4. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 22.Zang J., Gu C., Zhou B., Zhang C., Yang Y., Xu S., Bai L., Zhang R., Deng Q., Yuan Z., et al. Immunization with the receptor-binding domain of SARS-CoV-2 elicits antibodies cross-neutralizing SARS-CoV-2 and SARS-CoV without antibody-dependent enhancement. Cell Discov. 2020;6:61. doi: 10.1038/s41421-020-00199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandrashekar A., Liu J., Martinot A.J., McMahan K., Mercado N.B., Peter L., Tostanoski L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., Nkolola J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.FDA Coronavirus (COVID-19) Update: FDA authorizes monoclonal antibodies for treatment of COVID-19. 2020. https://wwwfdagov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19
- 26.FDA Coronavirus (COVID-19) Update: FDA authorizes additional monoclonal antibody for treatment of COVID-19. 2021. https://wwwfdagov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-additional-monoclonal-antibody-treatment-covid-19
- 27.FDA FDA authorizes REGEN-COV monoclonal antibody therapy for post-exposure prophylaxis (prevention) for COVID-19. 2021. https://wwwfdagov/drugs/drug-safety-and-availability/fda-authorizes-regen-cov-monoclonal-antibody-therapy-post-exposure-prophylaxis-prevention-covid-19
- 28.AstraZeneca AZD7442 PROVENT Phase III prophylaxis trial met primary endpoint in preventing COVID-19. 2021. https://wwwastrazenecacom/media-centre/press-releases/2021/azd7442-prophylaxis-trial-met-primary-endpointhtml
- 29.Loo Y.M., McTamney P.M., Arends R.H., Gasser R.A.J., Abram M.E., Aksyuk A., Diallo S., Flores D.J., Kelly E.J., Ren K., et al. AZD7442 demonstrates prophylactic and therapeutic efficacy in non-human primates and extended half-life in humans. medRxiv. 2021 doi: 10.1101/2021.08.30.21262666. [DOI] [Google Scholar]
- 30.Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen R.E., Winkler E.S., Case J.B., Aziati I.D., Bricker T.L., Joshi A., Darling T.L., Ying B., Errico J.M., Shrihari S., et al. In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains. Nature. 2021;596:103–108. doi: 10.1038/s41586-021-03720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards J., Auger J., Peace D., Gale D., Michel J., Koons A., Haverty T., Zivin R., Jolliffe L., Bluestone J.A. Phase I evaluation of humanized OKT3: toxicity and immunomodulatory effects of hOKT3gamma4. Cancer Res. 1999;59:2096–2101. [PubMed] [Google Scholar]
- 37.Uppal H., Doudement E., Mahapatra K., Darbonne W.C., Bumbaca D., Shen B.Q., Du X., Saad O., Bowles K., Olsen S., et al. Potential mechanisms for thrombocytopenia development with trastuzumab emtansine (T-DM1) Clin. Cancer Res. 2015;21:123–133. doi: 10.1158/1078-0432.CCR-14-2093. [DOI] [PubMed] [Google Scholar]
- 38.Wang R., Xiao H., Guo R., Li Y., Shen B. The role of C5a in acute lung injury induced by highly pathogenic viral infections. Emerg. Microbes Infect. 2015;4:e28. doi: 10.1038/emi.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lund J., Winter G., Jones P.T., Pound J.D., Tanaka T., Walker M.R., Artymiuk P.J., Arata Y., Burton D.R., Jefferis R., et al. Human Fc gamma RI and Fc gamma RII interact with distinct but overlapping sites on human IgG. J. Immunol. 1991;147:2657–2662. [PubMed] [Google Scholar]
- 40.Wines B.D., Powell M.S., Parren P.W., Barnes N., Hogarth P.M. The IgG Fc contains distinct Fc receptor (FcR) binding sites: the leukocyte receptors Fc gamma RI and Fc gamma RIIa bind to a region in the Fc distinct from that recognized by neonatal FcR and protein A. J. Immunol. 2000;164:5313–5318. doi: 10.4049/jimmunol.164.10.5313. [DOI] [PubMed] [Google Scholar]
- 41.Xu D., Alegre M.L., Varga S.S., Rothermel A.L., Collins A.M., Pulito V.L., Hanna L.S., Dolan K.P., Parren P.W., Bluestone J.A., et al. In vitro characterization of five humanized OKT3 effector function variant antibodies. Cell Immunol. 2000;200:16–26. doi: 10.1006/cimm.2000.1617. [DOI] [PubMed] [Google Scholar]
- 42.Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;4:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 44.Begin P., Callum J., Jamula E., Cook R., Heddle N.M., Tinmouth A., Zeller M.P., Beaudoin-Bussieres G., Amorim L., Bazin R., et al. Convalescent plasma for hospitalized patients with COVID-19: an open-label, randomized controlled trial. Nat. Med. 2021;27:2012–2024. doi: 10.1038/s41591-021-01488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradfute S.B., Hurwitz I., Yingling A.V., Ye C., Cheng Q., Noonan T.P., Raval J.S., Sosa N.R., Mertz G.J., Perkins D.J., et al. Severe acute respiratory syndrome coronavirus 2 neutralizing antibody titers in convalescent plasma and recipients in New Mexico: an open treatment study in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:1620–1628. doi: 10.1093/infdis/jiaa505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janiaud P., Axfors C., Schmitt A.M., Gloy V., Ebrahimi F., Hepprich M., Smith E.R., Haber N.A., Khanna N., Moher D., et al. Association of convalescent plasma treatment with clinical outcomes in patients with COVID-19: a systematic review and meta-analysis. JAMA. 2021;325:1185–1195. doi: 10.1001/jama.2021.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dall'Acqua W.F., Kiener P.A., Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn) J. Biol. Chem. 2006;281:23514–23524. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- 51.Dall'Acqua W.F., Woods R.M., Ward E.S., Palaszynski S.R., Patel N.K., Brewah Y.A., Wu H., Kiener P.A., Langermann S. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: biological consequences. J. Immunol. 2002;169:5171–5180. doi: 10.4049/jimmunol.169.9.5171. [DOI] [PubMed] [Google Scholar]
- 52.Yu X.Q., Robbie G.J., Wu Y., Esser M.T., Jensen K., Schwartz H.I., Bellamy T., Hernandez-Illas M., Jafri H.S. Safety, tolerability, and pharmacokinetics of MEDI4893, an investigational, extended-half-life, anti-Staphylococcus aureus alpha-toxin human monoclonal antibody, in healthy adults. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01020-16. e01020–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodle E.S., Bluestone J.A., Zivin R.A., Jolliffe L.K., Auger J., Xu D., Thistlethwaite J.R. Humanized, nonmitogenic OKT3 antibody, huOKT3 gamma(Ala-Ala): initial clinical experience. Transpl. Proc. 1998;30:1369–1370. doi: 10.1016/s0041-1345(98)00278-4. [DOI] [PubMed] [Google Scholar]
- 54.Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820.e6. doi: 10.1016/j.cell.2021.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. New Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McMahan K., Yu J.U., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J.Y., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. New Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. New Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 60.Demogines A., Farzan M., Sawyer S.L. Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J. Virol. 2012;86:6350–6353. doi: 10.1128/JVI.00311-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frank H.K., Enard D., Boyd S.D. Exceptional diversity and selection pressure on SARS-CoV and SARS-CoV-2 host receptor in bats compared to other mammals. bioRxiv. 2020 doi: 10.1101/2020.04.20.051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacLean O.A., Orton R.J., Singer J.B., Robertson D.L. No evidence for distinct types in the evolution of SARS-CoV-2. Virus Evol. 2020;6 doi: 10.1093/ve/veaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310 e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. 2020;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.VanBlargan L.A., Adams L.J., Liu Z., Chen R.E., Gilchuk P., Raju S., Smith B.K., Zhao H., Case J.B., Winkler E.S., et al. A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity. 2021;54:2399–2416.e6. doi: 10.1016/j.immuni.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomic M.T., Espinoza Y., Martinez Z., Pham K., Cobb R.R., Snow D.M., Earnhart C.G., Pals T., Syar E.S., Niemuth N., et al. Monoclonal antibody combinations prevent serotype A and serotype B inhalational botulism in a Guinea pig model. Toxins. 2019;11:208. doi: 10.3390/toxins11040208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R Core Team . R foundation for statistical Computing; 2021. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data needed to evaluate the conclusions in the paper are present in the paper or the supplemental information; source data for each of the display items is provided in key resources table.
-
•
No custom computer code or algorithms to report.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.