Abstract
Purpose:
To develop a cardiac T1 mapping method for free-breathing 3D T1 mapping of the whole heart at 3T with transmit B1 (B1+) correction
Methods:
A free-breathing, ECG-gated inversion recovery sequence with spoiled gradient-echo readout was developed and optimized for cardiac T1 mapping at 3T. High-frame rate dynamic images were reconstructed from sparse (k,t)-space data acquired along a stack-of-stars trajectory using a subspace-based method for accelerated imaging. Joint T1 and flip-angle (FA) estimation was performed in T1 mapping to improve its robustness to B1+ inhomogeneity. Subject-specific timing of data acquisition was utilized in the estimation to account for natural heart-rate variations during the imaging experiment.
Results:
Simulations showed that accuracy and precision of T1 mapping can be improved with joint T1 and FA estimation and optimized ECG-gated SPGR-based IR acquisition scheme. The phantom study showed good agreement between the T1 maps from the proposed method and the reference method. 3D cardiac T1 maps (40 slices) were obtained at a 1.9 mm in-plane and 4.5 mm through-plane spatial resolution from healthy subjects (n=6) with an average imaging time of 14.2 ± 1.6 min (heartbeat rate: 64.2±7.1 bpm), showing myocardial T1 values comparable to those obtained from MOLLI. The proposed method generated B1+ maps with spatially smooth variation showing 21–32% and 11–15% variations across the septal-lateral and inferior-anterior regions of the myocardium in the left ventricle.
Conclusion:
The proposed method allows free-breathing 3D T1 mapping of the whole-heart with transmit B1 correction in a practical imaging time.
Keywords: Cardiac T1 Mapping, Myocardial T1 Mapping, Free-breathing, Inversion Recovery, Transmit B1 Inhomogeneity, Low-Rank, Spoiled Gradient-echo, 3T
Introduction
Cardiac T1 mapping is a powerful cardiovascular magnetic resonance imaging (MRI) technique that allows quantitative assessment of tissue characteristics and underlying pathology of myocardium. Native (i.e.,without the usage of exogenous contrast agent) myocardial T1 characterizes alterations in the structure and intra-/extra-cellular components of myocardium. Native myocardial T1 is a well-recognized biomarker for quantitative assessment of diseases that alter tissue composition, such as iron deposition, amyloid disease, Anderson-Fabry disease, and myocarditis(1–3). Extracellular volume fraction (ECV), measured from pre- and post-contrast T1 values, provides quantitative measurement of interstitial expansion and associated diseases, such as amyloidosis, fibrosis, or myocardial edema(2,3). ECV is an emerging biomarker for diffuse fibrosis (e.g.,in heart failure, dilated cardiomyopathy, and amyloidosis)(3), which is challenging to detect using qualitative late gadolinium enhancement (LGE) methods.
Modified Look-Locker inversion-recovery (MOLLI)(4) is a widely used method for 2D cardiac T1 mapping which utilizes adiabatic inversion pulses for magnetization preparation and performs electrocardiogram (ECG)-gated balanced steady-state free precession (bSSFP) acquisitions through multiple cardiac cycles with breath-holding. Although MOLLI produces myocardial T1 maps with high precision(5), the method is limited to a single-slice imaging per breath-hold. Methods have been developed to extend conventional 2D MOLLI method to multi-slice 2D or 3D acquisitions with breath-holding by leveraging the state-of-the-art parallel imaging, simultaneous multi-slice acquisition, compressed sensing, and non-Cartesian sampling techniques(6,7). However, these methods suffer from limited through-plane resolution and coverage, often involving long or repetitive breath-holds to obtain volumetric T1 maps of the heart which imposes significant burden on patients.
Various methods have been developed to overcome the limitations of breath-holding and allow 3D cardiac T1 mapping with free-breathing acquisitions. Respiratory and cardiac gating-based T1 mapping methods have been developed to acquire interleaved multi-slice 2D(8–10) or segmented 3D k-space data(11–13) at end-diastole with free-breathing, where effects from respiratory motion were mitigated by prospectively tracking respiratory motion using navigators or self-navigation techniques. MR fingerprinting approaches have been combined with free-breathing ECG gated acquisitions for multi-parametric cardiac MRI(14). However, most of these methods are limited by spatial coverage, resolution in slice direction, or imaging time due to the low data acquisition efficiency of gating. Recently, free-running (i.e., no cardiac or respiratory gating) continuous acquisition methods have been proposed for 2D or 3D cardiac T1 mapping(15–20). Of note, Qi et al has reported a free-running 3D whole-heart T1 mapping method(15), which uses translational respiratory motion correction and a patch-based low-rank tensor model to reconstruct 3D T1 maps with isotropic resolution. The T1 maps obtained by this method, however, were from 1.5T and the method may not translate well to 3T for reasons discussed below.
While 3D cardiac T1 mapping methods developed up to date have been applied mostly at 1.5T, unique technical challenges arising from more severe B0 and transmit B1(B1+) inhomogeneities need to be addressed at 3T. For instance, spoiled gradient-echo (SPGR) readout with small flip angle (FA) is often used in 3D cardiac imaging at 3T to avoid B0 inhomogeneity-caused banding-artifacts associated with bSSFP readout(7,16,17,19). However, T1 mapping with SPGR readout is known to be sensitive to errors in FA caused by imperfect RF pulses and B1+ inhomogeneities(21). The latter is particularly problematic at 3T, where B1+ variation over the left ventricle with body-coil transmission is reported on the order of 30–60%(22), leading to bias in T1 estimation. Robustness of cardiac T1 mapping methods with SPGR acquisitions needs to be thoroughly investigated in the presence of B1+ inhomogeneity(23,24).
In this work, we present a new cardiac T1 mapping method for rapid 3D T1 mapping of the heart at 3T. A free-breathing, ECG-gated inversion recovery (IR) sequence with SPRG readout was developed and optimized in terms of acquisition protocol and excitation FA for accurate and precise cardiac T1 mapping at 3T. The optimized scheme was combined with sparse (k,t)-space sampling along a stack-of-stars trajectory to accelerate imaging. A subspace-based image reconstruction method was used to recover high frame-rate dynamic images from highly under-sampled (k,t)-space data. The effects of FA errors on T1 mapping were mitigated by joint estimation of T1 and FA, where the reconstructed dynamic images were first binned to different respiratory motion phases and then fitted voxel-by-voxel to a signal dictionary generated using Bloch equation simulations. The effects of heart-rate variations on T1 mapping were reduced by generating signal dictionary with subject-specific timing of data acquisition recorded during imaging experiment. The performance of the proposed method was characterized and validated through numerical simulations, phantom studies, and in vivo experiments on healthy human subjects (n=6). Preliminary accounts of this work have been presented previously in the form of conference abstracts(25–27).
Methods
Data Acquisition
The proposed ECG-gated cardiac T1 mapping sequence is shown in Figure 1. A non-selective inversion pulse was applied every N+M heartbeats with two different inversion times (TI). This scheme is referred to as N-(M)-N-(M) protocol for simplicity, where N denotes number of cardiac cycles for acquisition and M denotes number of cardiac cycles for signal recovery. Data were acquired at end-diastole period using SPGR readout. A special data acquisition scheme was employed for sparse sampling (k,t)-space data along a stack-of-stars trajectory. A limited number of “training” data (e.g., along the kx, ky, and kz directions across the center of the k-space) were acquired with high sampling rate to determine the temporal changes of the underlying signal. Data at all other k-space locations were sparsely-sampled over the entire (k,t)-space to ensure sufficient number of measurements were acquired at each k-space location for subspace-based image reconstruction.
Figure 1.
Schematic diagram of the proposed data acquisition scheme. N-(M)-N-(M) protocol is shown with non-selective inversion pulse applied every N+M heartbeats (where N denotes the number of cardiac cycles for acquisition and M denotes the number of cardiac cycles for signal recovery) with two different inversion times (TI). Data acquisition consists of “training” dataset acquiring a limited number of k-space lines with high sampling rate and “imaging” dataset sparsely-sampling all other k-space locations for subspace-based image reconstruction. To track respiratory motion, 1D respiratory navigator signals were acquired in the sagittal plane at the dome of the right hemi-diaphragm after a spatially-selective inversion pulse was applied in the same sagittal plane to invert the magnetization signals back to the equilibrium state.
To track respiratory motion, 1D respiratory navigator signals were acquired in the sagittal plane at the dome of the right hemi-diaphragm at the beginning and end of data acquisition of each cardiac cycle. A spatially-selective inversion pulse was applied right after the non-selective inversion pulse to invert the magnetization signals in the same sagittal plane back to the equilibrium state and, therefore, to mitigate the contrast changes caused by the non-selective inversion pulses in the navigator signals.
Image Reconstruction
Image reconstruction of sparsely-sampled data was performed by solving the following constrained optimization problem:
| (1) |
where d(k,t) denotes the measured data, the Fourier transform in the spatial domain, Ω the sampling mask in the (k,t)-space, and λ the regularization parameter, which was chosen based on the discrepancy principle(28). The first term of the cost function in Eq.(1) penalizes data inconsistency while second term promotes sparsity of the reconstructed dynamic images ρ(x,t) in the spatio-spectral domain(29). The constraint in Eq.(1) represents ρ(x,t) as a partially-separable (PS) function(30,31), where vl(t) denotes temporal basis function, ul(x) denotes corresponding spatial coefficients, and L is model order. In this work, vl(t) was estimated separately from training data using singular value decomposition (SVD) for simplified computation(29). Image reconstruction problem was then reduced to determining the spatial coefficients ul(x) from measured data. The optimization problem in Eq.(1) was solved using an alternating direction method of multipliers (ADMM)(32) based algorithm(33). For fast computation of , 1D Fourier transform was applied along the kz direction first and non-uniform fast Fourier transform(34) was applied in the kx-ky plane for slice-by-slice reconstruction. The dynamic images were reconstructed in a coil-by-coil fashion and then combined using the sum-of-squares method to form the final reconstruction. We implemented the image reconstruction algorithm in Python and utilized the SigPy package(35) to accelerate the computation using Graphical Processing Units (GPUs). Reconstructions were performed on four NVIDIA Tesla V100 SXM2 GPUs (parallelized over slice and coil dimensions) with reconstruction time around one minute for each slice and coil.
Estimation of T1 and FA
Prior to parameter estimation, the reconstructed dynamic images were binned to different respiratory motion phases based on diaphragm position information extracted from navigator signals. Let denote the reconstructed dynamic signals at a voxel in a selected respiratory phase m and denote the nonlinear parameters (i.e., T1 and FA). We estimated ηn,m that best fit the dynamic signals using variable projection (VARPRO) method(36):
| (2) |
where a(·) denotes an atom of a dictionary of signals calculated using Bloch equation simulation with varying T1 and FA values defined on a discrete 2D grid and the actual timing of acquisition recorded during the imaging experiment. The evolution of the bulk magnetization vector was calculated excitation-by-excitation over the course of the entire scan by solving the Bloch equation numerically. The signal bases were formed using the simulated signals associated with the excitations where the spoke at the center of the k-space (e.g., kz =0) was acquired at each frame. The bases were then binned to a motion phase to form the dictionary for joint T1 and FA fitting. For simulation and phantom studies, dictionary was generated for a range of T1 from 1–3000ms in increments of 1ms and for a range of FA from 0 to 2 × FAo in increments of 0.01 × FAo, where FAo is the nominal FA, in case of joint estimation. For the in vivo study, the dictionary was generated for a range of T1 from 500–2500ms in increments of 10ms and for a range of FA from 0.2 × FAo to 1.5 × FAo in increments of 0.01 × FAo in case of joint estimation, in consideration of expected smaller range of T1 and FA values and for the sake of reducing computation time in dictionary generation and fitting. The time to generate the dictionary and to fit the data was 6.7±2.1min and 51.1±2.8s, respectively, using 8 Intel Xeon 2.4GHz CPUs (4-core per CPU) on a workstation.
Simulation Study
We performed simulation studies to optimize the proposed data acquisition scheme in Fig. 1. Bloch equation simulations were performed for various ECG-gated IR schemes with following parameters in common unless otherwise mentioned: heart rate=80 bpm, acquisition window=180 ms, inversion delay times=100/180 ms (i.e., delays from the inversion pulse to the beginning of the first acquisition), FA=6°, and SPGR readout. The effect of B1+ inhomogeneity on the accuracy of T1 estimation was investigated for 8–8, 5-(3)-5-(3), and 10-(3)-10-(3) protocols without noise at different B1+ scenarios (i.e., B1+=0.8/1/1.2). Relative difference (in relation to the ground truth T1 value) was used to assess any bias in T1 estimation. The precision of T1 estimation was investigated for N-(M)-N-(M) protocols with noise and perfect B1+ (i.e., B1+=1) using Monte Carlo simulations (i.e., 10,000 noise realizations). Normalized standard deviation (nSD) (i.e., standard deviation of estimated T1 values normalized by standard deviation of noise and inverse of square root of acquisition time) was used to assess the precision of the T1 estimation. The standard deviation of noise was assumed to be constant for all considered scenarios. Additionally, the effect of nominal FA on T1 mapping was investigated for 8–8, 5-(3)-5-(3), and 10-(3)-10-(3) protocols with FAs ranging from 1 to 15° in 1° increments. Lastly, the effect of heart rate variation on T1 mapping was investigated for 10-(3)-10-(3) protocol with heart rate ranging from 50 to 120bpm in 5bpm increments.
Note that the (k,t)-space is highly undersampled and the dynamic images are reconstructed with an explicit low-rank constraint in the proposed method. Therefore, different from MOLLI where the k-space of each frame can be fully sampled, the imaging time of the proposed method is given by , where Ps and Pz denotes the number of spokes in the kx−ky plane and phase encoding steps in the kz axis, respectively, which are determined by the required spatial resolution; L is the rank of the dynamic images; tRR denotes a fixed cardiac cycle duration; and Qc denotes the number of spokes acquired per cardiac cycle. Here, PsPzL is the total number of unknowns of the low-rank model when the temporal basis is predetermined. Note that the inherent rank of the dynamic images reflects the spatial-temporal correlations of the temporal signal variations of all the voxels. It depends on the respiratory motion pattern and the T1 value distributions rather than protocol parameters N and M. Since tRR, Ps, Pz, L and Qc were the same for different imaging protocols, the imaging time of the proposed method was calculated as in our simulation study for simplicity.
Phantom Study
A structured phantom consisting of 21-vials of deionized water doped with concentrations of gadolinium (Dotarem®) varying from 0 to 0.5mmol/L was built to validate the performance of the proposed method. Imaging experiment was performed using a 3T MR scanner (MAGNETOM Trio, Siemens Healthcare, Erlangen, Germany) with a body-coil for transmission and a 12-channel head coil for reception. Acquisitions were performed using 8–8, 5-(3)-5-(3), and 10-(3)-10-(3) protocols for a simulated heart rate of 80bpm. Common imaging parameters were: field-of-view (FOV)=360×304mm2, matrix-size=192×162, slice-thickness=6mm, FA=9°, TR/TE=3.0/1.5ms, inversion delay times=100/180ms, and SPGR readout. Fully-sampled (k,t)-space data were acquired on a Cartesian grid with a temporal resolution of 30 ms (i.e., 10 phase-encoding [PE] lines per frame) to evaluate the performance of different acquisition protocols. The 8–8, 5-(3)-5-(3), and 10-(3)-10-(3) protocols were repeated over 400, 400, 640 cardiac cycles, respectively, to ensure full-sampling of the (k,t)-space data. An IR sequence with fast spin echo (FSE) readout was performed to obtain reference T1 maps with the following imaging parameters: FOV=360×304mm2, matrix-size=192×162, slice-thickness=6mm, TR=10,000ms, echo-train-length=7, and TI=50/100/250/500/750/1000/1500/2000/2500/3000ms. An additional scan with MOLLI (37) was performed for comparison with the following parameters: FOV=360×304mm2, matrix-size=192×162, slice-thickness=6mm. Region-of-interests (ROIs) were drawn within each vial and the average T1 value within each vial was used for analysis. Scatter plots were generated to show the correlation between the T1 values from the different methods and those from the reference IR-FSE method. Bland-Altman analysis was performed to analyze the agreement between the T1 values from the different methods and those from the reference IR-FSE method.
In Vivo Study
Six healthy volunteers (four males and two females; 32 ± 3 years) were recruited under a study protocol approved by our local Institutional Review Board (IRB). Written informed consent was obtained from all subjects before study participation. Imaging experiments were performed using a 3T MR scanner (MAGNETOM Trio, Siemens Healthcare, Erlangen, Germany) with a body-coil for transmission and spine and surface coils for reception. Imaging was performed using 10-(3)-10-(3) protocol with data sampling following a stack-of-stars trajectory. Two frames were acquired per cardiac cycle, each consisting of k-space spokes along the same angle in the kx-ky plane over all kz encodings and 3 additional training lines at the center of the k-space along the kx, ky, and kz direction, respectively (Fig.1). The spoke angle varied randomly from frame to frame following uniform random distribution. The other imaging parameters were: FOV=308×308×180mm3, matrix-size=160×160×40, image orientation=short-axis view, FA=9°, TR/TE=3.4/1.7ms, and inversion delay times=100/180ms. A relatively large through-slice coverage was chosen to mitigate errors in FA due to imperfect slab excitation profile in the presence of both respiratory and cardiac motions. Data acquired over the first 800 cardiac cycles (corresponding to anticipated scan time of 10-min considering average adult heart rate of 80 bpm) were used for reconstruction and analysis. The dynamic images were reconstructed using temporal basis functions vl(t) estimated from training data with model order L=15. The model order was chosen based on the singular value decay of the Casorati matrix formed by the training data as in the previous work on using low-rank constraints for image reconstruction(29,33). The reconstructed dynamic images were then binned into 8 respiratory motion bins using the acquired 1D respiratory navigator signals. The number of respiratory bins was chosen to be 8 based on our previous experience on using MR for respiratory motion correction in PET(33). The diaphragm position was first estimated for each frame by fitting a logistic function to the 1D spatial profile near the interface between the liver and lung. Frames were then grouped into bins according to the estimated diaphragm position while ensuring a similar number of frames within each bin. Results obtained from respiratory motion phase at or near end-exhalation were used for analysis. The short-axis view slices were divided into ROIs of 16 segments according to the AHA recommendations(38) for analysis. For comparison, 2D T1 maps were acquired using MOLLI(37) for five slices in the short-axis view over the apical, mid-cavity, and basal regions of the heart and for one slice in the long-axis 4-chamber view with the following parameters: FOV=360×304mm2, matrix-size=192×162, slice-thickness=4.5mm. All the five short-axis slices were categorized into apical, mid-cavity, and the basal regions based on location and were used for analysis. The mean and standard deviation of the T1 and B1+ (defined as the ratio between the measured and nominal FAs) were calculated for each ROI and were visualized through bull’s-eye plot and bar plot. Statistical analysis was performed using Wilcoxon signed rank test to compare T1 values obtained by MOLLI and the proposed method.
Results
Results from the simulation study are shown in Figures 2 and 3. When estimating T1 only with the assumption of perfect B1+ (i.e., B1+=1), noticeable bias in T1 estimation was found in all the investigated imaging protocols in the presence of typical B1+ inhomogeneities at 3T (the blue dashed lines in Fig.2). The 8–8 protocol, which has the highest data acquisition efficiency (i.e., number of k-space lines acquired per unit time) among all the schemes, showed the highest sensitivity to B1+ inhomogeneity (Fig.2a). Insertion of cardiac cycles for signal recovery reduced this bias in T1 estimation at the cost of imaging time (Figs.2b and c). Joint T1 and FA estimation led to unbiased T1 estimation in the simulation study as expected (the red solid lines in Fig.2). Figure 3a shows the precision of T1 estimation for different imaging protocols. The nSD of the proposed method with joint T1 and FA estimation was minimized and plateaued around N=13, 14, 14, and 13 for N-N, N-(1)-N-(1), N-(2)-N-(2), and N-(3)-N-(3) protocols, respectively. The nSD of the proposed method with T1 estimation only was minimized and plateaued around N=12, 8, 10, and 12 for N-N, N-(1)-N-(1), N-(2)-N-(2), and N-(3)-N-(3) protocols, respectively. While resulting in unbiased estimation of T1, joint T1 and FA estimation led to larger nSD than the case of estimating T1 only. Increasing the number of cardiac cycles for signal recovery reduced the nSD observed in joint T1 and FA estimation, eventually to a level similar to those observed in the case of estimating T1 only with 10-(3)-10-(3) protocol. Note that joint T1 and FA estimation was unstable for 8–8 protocol despite the desired data acquisition efficiency. Figure 3b shows effects of FAs on the precision of T1 estimation for different imaging protocols. The nSD was minimized and plateaued around FA of 9° in the case of joint T1 and FA estimation for both 5-(3)-5-(3) and 10-(3)-10-(3) protocols. Based on the results, FA of 9° was used for the ECG-gated IR sequence with SPGR readout in the phantom and in vivo experiments. Figure 3c further shows that heartbeat rate has only marginal effects on precision of T1 estimation for 10-(3)-10-(3) protocol, which is the protocol selected for in vivo experiments.
Figure 2.
Simulation results showing the effect of B1+ inhomogeneity on the accuracy of T1 estimation. Relative difference plots from the 8–8 (A), 5-(3)-5-(3) (B), and 10-(3)-10-(3) (C) protocols are shown for typical B1+ inhomogeneities at 3T (i.e., B1+ = 0.8, 1, and 1.2). T1 values obtained by joint T1 and FA estimation (red solid line) and T1 estimation only with the assumption of perfect B1+ (blue dashed line) are shown. Small fluctuations are presumed to be due to numerical errors.
Figure 3.
Simulation results showing the precision of T1 estimation for different N-(M)-N-(M) protocols. A: Effect of N-(M)-N-(M) protocols on normalized standard deviation (nSD). B: Effect of flip angle (FA) on nSD for the 8–8, 5-(3)-5-(3), and 10-(3)-10-(3) protocols. C: Effect of heart rate variation on nSD for the 10-(3)-10-(3) protocol. T1 values obtained by joint T1 and FA estimation (red solid line) and T1 estimation only with the assumption of perfect B1+ (blue dashed line) are shown.
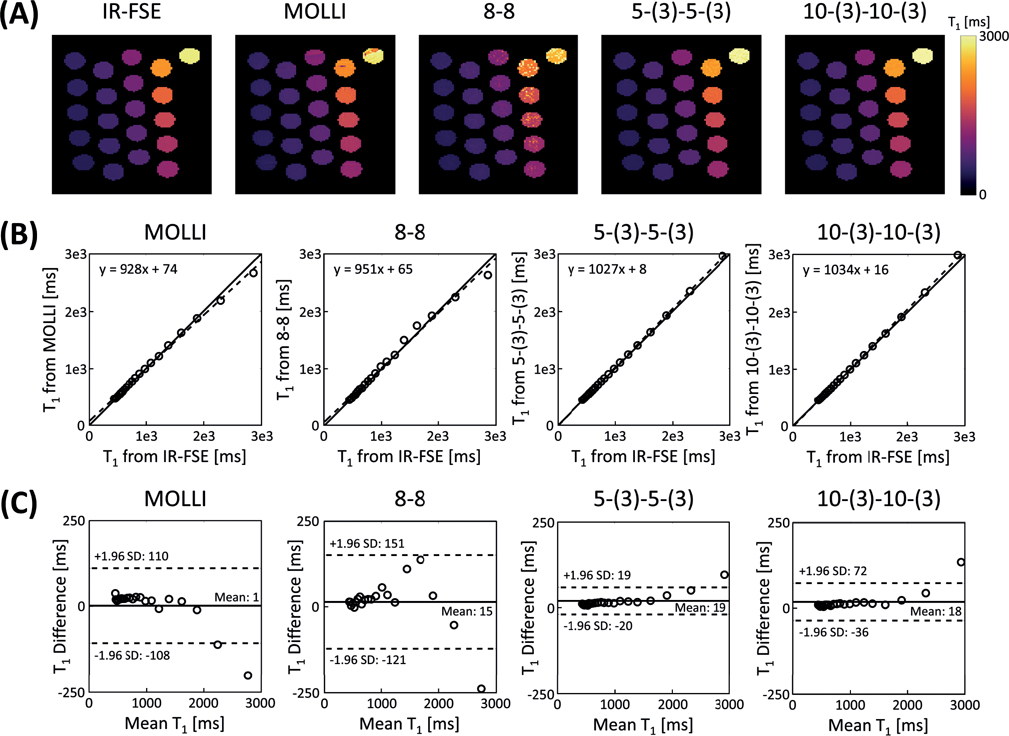
The results from phantom studies are shown in Figure 4 and Supporting Information Figures S1 and S2. Banding artifacts were observed in the estimated T1 maps from MOLLI, whereas no noticeable artifacts were shown in the estimated T1 maps from proposed method (Fig.4a). 5-(3)-5-(3) and 10-(3)-10-(3) protocols both showed accurate T1 mapping in relation to the reference IR-FSE method when T1 and FA were estimated jointly, as shown in the correlation plots in Fig.4b and Bland-Altman plots in Fig.4c. 8–8 protocol produced T1 maps with large variations when T1 and FA were estimated jointly (Fig.4a), which matched with the simulation results in Fig.3. Supporting Information Figure S1 shows the results obtained from the same experiment but with estimation of T1 only assuming prefect B1+ (i.e., B1+ = 1). B1+ inhomogeneity caused bias was found in the estimated T1 maps from IR sequence with SPGR readout as expected. Compared to the case of estimation of T1 only, joint T1 and FA estimation reduced the limits of agreement by 77.7, 44.7, and 49.4 ms for the T1 mapping experiment with 8–8, 5-(3)-5-(3), and 10-(3)-10-(3) protocol, respectively (Figs.4c and S1c). Supporting Information S2 shows results from another phantom study with variation in B1+ field strength via control of transmitter voltage. 5-(3)-5-(3) protocol achieved robust T1 mapping when T1 and FA were estimated jointly, despite the variation in B1+ field strength. Estimated T1 from each vial were in good agreement with those estimated from reference IR-FSE method. Ratios between different nominal FAs and estimated average FAs from each vial were also in good agreement.
Figure 4.
Phantom results of ECG-gated IR schemes with SPGR readout and joint T1 and FA estimation. A: Estimated T1 maps from different methods. B: Scatter plots showing comparison of estimated T1 from different methods with those from IR-FSE. Solid line represents line of identity and dashed line represents line of regression. C: Bland-Altman plots showing comparison of estimated T1 from different methods with those from IR-FSE. Solid line represents the mean difference and dashed line represents the 95% confidence interval for limits of agreement.
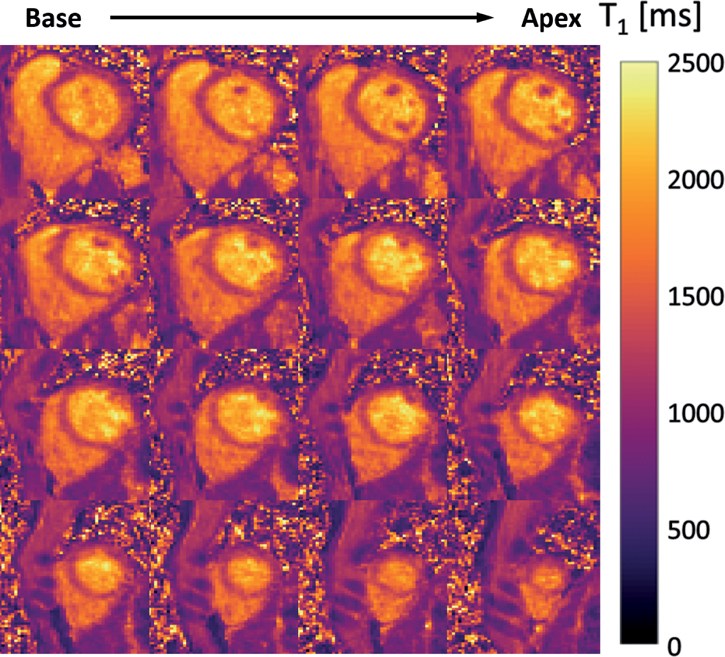
Results from the in vivo study are shown in Figures 5 to 10 and Supporting Information Figures S3 and S4. The average heartrate of the 6 volunteers was 64.2±7.1bpm (min:53.7bpm, max:73.5bpm). The acquisition time for the 6 volunteers was 14.2±1.6min (min:12.2min, max:16.4min). Figure 5 shows representative reconstructed images at various slice positions and inversion times from Subject 1 using the proposed method. No significant artifacts were seen in the reconstructed images across different slices and TI times. After respiratory motion binning, an average of 153.8±23.4 inversion times were observed per respiratory motion bin over the course of 800 heartbeats across all subjects. Figure 6 shows representative short-axis-view T1 maps from Subject 1 and 2 obtained by the proposed method. Figure 6 also shows 4-chamber-view T1 maps which were generated by re-slicing the 3D T1 map from the proposed method. For comparison, T1 maps from MOLLI at the same slice position and orientation are shown at the bottom of each subfigure. Overall, the T1 maps from the proposed method were comparable to those from MOLLI. Note that the nominal in- and through-plane resolution from the proposed method was 1.9mm and 4.5mm, respectively. Figure 7 further shows 3D T1 maps from Subject 2, covering the whole left ventricle from base to apex. As can be seen, high-quality 3D T1 maps of the heart were produced using the proposed method. Figure 8 shows a quantitative comparison of T1 maps from all subjects using the proposed method (with and without joint T1 and FA estimation) and MOLLI, respectively. The bull’s-eye plot and bar plot of the mean and standard deviation of T1 values from each ROI show a very good agreement between the two methods. This observation is also supported by the Bland-Altman plots shown in Supporting Information Figure S3. Compared to the case of estimation of T1 only, joint T1 and FA estimation reduced the standard deviation of T1 values across 16 myocardial segments by 30.5ms. Statistical test showed that the T1 values of the 16 myocardial segments from the proposed method with joint T1 and FA estimation were not statistically different from MOLLI at 5% significance level (P=0.08). The mean and standard deviation of septal T1 values between MOLLI and the proposed method across subjects is compared in Supporting Information Table S1.
Figure 5.
In vivo study results of reconstructed images from the proposed method. Representative reconstructed images from Subject 1 are shown at various slice positions for a fixed TI of 1343 ms (top row) and various TI times for slice position index of 18 (bottom row). Note that the images were selected from reconstructed images in clock time and may be at different respiratory motion phase.
Figure 10.
In vivo study results of estimated B1+ (defined as the ratio between the measured and nominal FAs) from all the subjects. A: 16-segment AHA bull’s eye plot of normalized mean B1+ (i.e., B1+ normalized by the mean B1+ estimated from the mid-cavity anterior region of the myocardium in the left ventricle for each subject) estimated from the proposed method. Notice the difference in estimated B1+ between the septal-lateral and inferior-anterior regions of the myocardium. B: Bar plot showing mean and standard deviation of normalized B1+ estimated from the proposed method.
Figure 6.
T1 maps from Subject 1 and 2 obtained by the proposed method. Representative T1 maps are shown for various slice positions in the short-axis view and 4-chamber view. T1 maps from MOLLI at the same slice position and orientation are shown at the bottom of each subfigure for comparison. Note that for the 4-chamber view, the T1 map from MOLLI were acquired with in-plane resolution of 1.5 mm, whereas the T1 map from the proposed method was generated by re-slicing the 3D T1 map acquired with through-plane resolution of 4.5 mm.
Figure 7.
Representative 3D T1 map in the short-axis view covering the whole left ventricle from base to apex.
Figure 8.
In vivo study results showing quantitative comparison of estimated T1 from MOLLI and those from the proposed method. A: 16-segment AHA bull’s eye plots of mean T1 from MOLLI, proposed method with T1 estimation only assuming perfect B1+ (i.e., B1+ = 1), and proposed method with joint T1 and FA estimation. B: Bar plots showing mean and standard deviation of estimated T1 from MOLLI and proposed method. Cases of proposed method with T1 estimation only assuming perfect B1+ (i.e., B1+ = 1) and joint T1 and FA estimation are shown. BA, BAS, BIS, BI, BIL, BAL, MA, MAS, MIS, MA, MIL, MAL, AA, AS, AI, AL each denotes basal anterior, basal anteroseptal, basal inferoseptal, basal inferior, basal inferolateral, basal anterolateral, mid-cavity anterior, mid-cavity anteroseptal, mid-cavity inferoseptal, mid-cavity inferior, mid-cavity inferolateral, mid-cavity anterolateral, apical anterior, apical septal, apical inferior, and apical lateral regions of the myocardium in the left ventricle.
Figure 9 shows representative T1 and B1+ maps from Subject 2 obtained by proposed method. Notice the similarity in estimated T1 values for each tissue type (e.g., myocardium, liver, and muscle) even with significant variations in estimated B1+ across different regions (Fig. 9a). The 3D B1+ maps of the heart (Fig. 9b) show larger B1+ values in lateral/anterior regions than septal/inferior regions, which is consistent with literature(39). Group analysis of B1+ maps acquired from all subjects are shown in Fig. 10. B1+ variation ranged from 21–32% and 11–15% across the septal-lateral and inferior-anterior regions of the myocardium in the left ventricle, respectively. When such B1+ inhomogeneities were ignored in T1 estimation, T1 values in the septal and inferior regions were overestimated (Supporting Information Figure S4). This was consistent with simulation results in Figure 2.
Figure 9.
Representative 3D B1+ map (defined as the ratio between the measured and nominal FAs) obtained by the proposed method. A: Short-axis view T1 and B1+ maps at two slice positions. Notice the smooth variation of B1+ across regions. Also notice the similarity in estimated T1 for each tissue type even with variation in the estimated B1+ across different regions. B: Short-axis view B1+ maps of the heart at various slice positions from base to apex.
Discussion
In this work, a new free-breathing cardiac T1 mapping method is proposed for robust T1 mapping of the heart at 3T. The T1 maps obtained using proposed method have strong correlation and good agreement compared to reference and comparison methods in both phantom experiments with various conditions (Fig.4 and Supporting Information Fig.S2) and in vivo experiments across all subjects (Figs.6, 8 and Supporting Information Fig.S3). Robust T1 mapping was achieved despite significant B1+ variations at 3T. This is most noticeable by the fact that uniform T1 distributions for each tissue type (e.g., myocardium, liver, and muscle) were achieved across the entire FOV (Fig.9a) and is further supported by the close agreement of T1 maps between proposed method and MOLLI (Fig.8), despite the observed B1+ variation across septal-lateral and inferior-anterior regions of the myocardium in the left ventricle (Fig.10). Noticeable bias in T1 estimation was otherwise observed when B1+ inhomogeneities were ignored in T1 estimation (Figs.2,4,8,10, Supporting Information Figs.S1 and S4). These observations were consistent throughout simulation, phantom, and in vivo results. The estimated B1+ values of the myocardium show spatial variations which are consistent with those reported in literature(39), i.e., the B1+ values in the lateral/anterior regions were 10–30% larger than the septal/inferior regions. However, the B1+ distributions within the blood pool should be carefully interpreted since flow effects were not considered in joint T1 and FA estimation. As a result, the estimated T1 and FA values of blood may be biased.
This method has several novel features. First, the proposed method mitigates bias in T1 estimation caused by errors in FA via joint estimation of T1 and FA. We carried out systematic numerical simulation studies to optimize the ECG-gated IR sequence with SPGR readout in terms of acquisition protocols and nominal FAs with the goal of minimizing the standard deviation of the estimated T1. Second, the proposed method utilizes special (k,t)-space sampling scheme and subspace-based image reconstruction to recover dynamic images from under-sampled data, i.e., two 3D volumes for every cardiac cycle with data acquisition. This allows mitigating the effects of natural heart-rate variations on T1 mapping by fitting the reconstructed dynamic signals to a signal dictionary generated with subject-specific timing of data acquisition recorded during imaging experiment. Third, the proposed method is robust to B0 inhomogeneities since it utilizes adiabatic non-selective pulse for inversion and SPGR acquisitions. Altogether, the proposed method achieves free-breathing T1 mapping in the presence of B1+ and B0 inhomogeneity at 3T in a practical imaging time.
The proposed method may be potentially useful for quantification of post-contrast T1 and ECV mapping where accurate and precise estimation of T1 is important. Although results from simulation and phantom studies show that proposed method can estimate short T1 values with accuracy and precision, further investigation is necessary to evaluate the performance of proposed method for post-contrast T1 estimation. Since T1 relaxation in the tissue changes over time in vivo after contrast agent is injected, the performance of proposed method needs to be carefully examined for these applications, including investigations in the context of subspace-based reconstruction. An interesting next step would be to investigate the feasibility as well as the performance of the proposed method for these applications in vivo.
In this work, a subspace-based image reconstruction method was used to recover dynamic images in clock time for ECG-gated acquisitions and respiratory motion was resolved by subsequently binning reconstructed images to different respiratory motion phases. A potentially interesting future work would be to investigate possibility of treating respiratory motion as an additional temporal dimension using low-rank tensor model(16) for ECG-gated acquisitions. In the ideal case with fixed heartrate, this would be feasible since T1-weighted contrast changes can then be modeled by inversion recovery at a small number of inversion delay times. In reality, however, natural variations of heartbeat rate will require modeling the T1-weighted contrast changes in clock time for ECG-gated acquisitions over the entire imaging experiment. This can impose technical challenges when attempting to represent the underlying dynamic images using a low-rank tensor model.
Results from in vivo study showed that T1 values from proposed method were not statistically different from MOLLI at 5% significance level (P=0.08). This indicates that the proposed method has similar bias as MOLLI in in vivo study. Several factors contribute to the apparent underestimation of T1 when proposed method is used for in vivo cardiac T1 mapping. First, magnetization transfer effects can lead to underestimated T1 values in the proposed method as in MOLLI since both methods use inversion-recovery based acquisition schemes for T1 mapping. Second, the proposed method currently assumes perfect inversion pulse. Adding inversion efficiency to parametric fitting may potentially improve accuracy of T1 estimation at the cost of computation time (i.e., a larger dictionary of bases needed for parametric fitting). Further investigation is necessary to study the potential source of bias in proposed method in in vivo settings.
The current work has several limitations that warrant further investigation. First, the proposed method involves ECG-gated acquisition, which is susceptible to ECG mis-triggering and may suffer from image blurring due to cardiac motion. The former could be addressed by adaptive heartbeat rate prediction as in double-gating technique(40). The latter can be mitigated by retrospectively discarding k-space data acquired outside the end-diastole window based on recorded ECG signals. Free-running (i.e., no cardiac or respiratory gating)-based continuous acquisition scheme may be preferable over ECG-gated acquisition schemes for maximizing data acquisition efficiency. However, in free-running continuous acquisition scheme, apparent inversion recovery rate is strongly coupled with FA. Therefore, accurate estimation of FA is expected to be critical for accurate T1 estimation with free-running acquisition in the presence of B1+ inhomogeneity at 3T. This research direction is currently under investigation. Second, FA estimation by proposed method was validated using a phantom experiment with varying B1+ field strength via changing transmitter voltage (Supporting Information Fig.S2). Validation with a reference cardiac FA or B1+ mapping method, e.g., actual flip-angle imaging method(41), in human subject studies is necessary to further verify the performance of FA mapping results from proposed method. Third, performance of proposed method was validated in vivo with a small number of healthy subjects (n=6). Studies with a larger number of healthy subjects and patients are necessary to assess the accuracy and reproducibility of proposed method and to evaluate its value in clinical applications. Future work can also include investigation with different sparsity constraints (e.g., finite difference or wavelet transform), investigation with different sampling schemes, and multi-site/multi-vendor validations of proposed method and findings from this work.
Conclusions
A new free-breathing cardiac T1 mapping method was proposed and optimized for fast 3D T1 mapping of the whole-heart at 3T with transmit B1 correction in practical imaging time.
Supplementary Material
Figure S1. Phantom results of ECG-gated IR schemes with SPGR readout and T1 estimation only assuming perfect B1+ (i.e., B1+ = 1). A: Estimated T1 maps from different methods. B: Scatter plots showing comparison of estimated T1 from different methods with those from IR-FSE. Solid line represents line of identity and dashed line represents line of regression. C: Bland-Altman plots showing comparison of estimated T1 from different methods with those from IR-FSE. Solid line represents the mean difference and dashed line represents the 95% confidence interval for limits of agreement.
Figure S2. Phantom study results with variation in B1+ field strength via control of transmitter voltage. A: Estimated T1 maps from IR-FSE and 5-(3)-5-(3) protocol with 80, 100, and 120% of reference transmitter voltage. B: Vial number positions. C: Estimated B1+ maps (defined as the ratio between the measured and nominal FAs) from 5-(3)-5-(3) protocol with 80, 100, and 120% of reference transmitter voltage. D: Estimated average FA within each vial from 5-(3)-5-(3) protocol with 80 (green line and circle), 100 (blue line and circle) and 120% (red line and circle) of reference transmitter voltage. Note that the ratio between the different B1+ field strength and the estimated average FAs from each vial are in good agreement.
Figure S3. In vivo study results of Bland-Altman plots comparing estimated T1 from the proposed method with those from MOLLI. Results from the anterior, septal, inferior, and lateral regions of the myocardium in the left ventricle are shown. Solid line represents the mean difference and dashed line represents the 95% confidence interval for limits of agreement.
Table S1. Comparison of mean and standard deviation of septal T1 values between MOLLI and proposed method across subjects
Acknowledgements
This work was supported in part by the National Institutes of Health (P41EB022544, R01CA165221, R01HL137230, R01HL118261, T32EB013180, and K01EB030045).
References
- 1.Schelbert EB, Messroghli DR. State of the art: Clinical applications of cardiac T1 mapping. Radiology 2016;278:658–676 doi: 10.1148/radiol.2016141802. [DOI] [PubMed] [Google Scholar]
- 2.Haaf P, Garg P, Messroghli DR, Broadbent DA, Greenwood JP, Plein S. Cardiac T1 Mapping and Extracellular Volume (ECV) in clinical practice: A comprehensive review. J. Cardiovasc. Magn. Reson. 2016;18:1–12 doi: 10.1186/s12968-016-0308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messroghli DR, Moon JC, Ferreira VM, et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2 and extracellular volume: A consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imagin. J. Cardiovasc. Magn. Reson. 2017;19:1–24 doi: 10.1186/s12968-017-0389-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified look-locker inversion recovery (MOLLI) for high-resolution T 1 mapping of the heart. Magn. Reson. Med. 2004;52:141–146 doi: 10.1002/mrm.20110. [DOI] [PubMed] [Google Scholar]
- 5.Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J. Cardiovasc. Magn. Reson. 2014;16:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weingärtner S, Moeller S, Schmitter S, et al. Simultaneous multislice imaging for native myocardial T1 mapping: Improved spatial coverage in a single breath-hold. Magn. Reson. Med. 2017;78:462–471 doi: 10.1002/mrm.26770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Lo WC, Hamilton JI, et al. Single breath-hold 3D cardiac T1 mapping using through-time spiral GRAPPA. NMR Biomed. 2018;31:1–13 doi: 10.1002/nbm.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weingärtner S, Roujol S, Akçakaya M, Basha TA, Nezafat R. Free-breathing multislice native myocardial T1 mapping using the slice-interleaved T1 (STONE) sequence. Magn. Reson. Med. 2015;74:115–124 doi: 10.1002/mrm.25387. [DOI] [PubMed] [Google Scholar]
- 9.Guo R, Cai X, Kucukseymen S, et al. Free-breathing whole-heart multi-slice myocardial T1 mapping in 2 minutes. Magn. Reson. Med. 2021;85:89–102 doi: 10.1002/mrm.28402. [DOI] [PubMed] [Google Scholar]
- 10.Jang J, Bellm S, Roujol S, et al. Comparison of spoiled gradient echo and steady-state free-precession imaging for native myocardial T1 mapping using the slice-interleaved T1 mapping (STONE) sequence. NMR Biomed. 2016;29:1486–1496 doi: 10.1002/nbm.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weingärtner S, Akçakaya M, Roujol S, et al. Free-breathing combined three-dimensional phase sensitive late gadolinium enhancement and T1 mapping for myocardial tissue characterization. Magn. Reson. Med. 2015;74:1032–1041 doi: 10.1002/mrm.25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nordio G, Henningsson M, Chiribiri A, Villa ADM, Schneider T, Botnar RM. 3D myocardial T1 mapping using saturation recovery. J. Magn. Reson. Imaging 2017;46:218–227 doi: 10.1002/jmri.25575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo R, Chen Z, Herzka DA, Luo J, Ding H. A three-dimensional free-breathing sequence for simultaneous myocardial T1 and T2 mapping. Magn. Reson. Med. 2019;81:1031–1043 doi: 10.1002/mrm.27466. [DOI] [PubMed] [Google Scholar]
- 14.Cruz G, Jaubert O, Qi H, et al. 3D free-breathing cardiac magnetic resonance fingerprinting. NMR Biomed. 2020;33:1–16 doi: 10.1002/nbm.4370. [DOI] [PubMed] [Google Scholar]
- 15.Qi H, Jaubert O, Bustin A, et al. Free-running 3D whole heart myocardial T1 mapping with isotropic spatial resolution. Magn. Reson. Med. 2019;82:1331–1342 doi: 10.1002/mrm.27811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christodoulou AG, Shaw JL, Nguyen C, et al. Magnetic resonance multitasking for motion-resolved quantitative cardiovascular imaging. Nat. Biomed. Eng. 2018;2:215–226 doi: 10.1038/s41551-018-0217-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw JL, Yang Q, Zhou Z, et al. Free-breathing, non-ECG, continuous myocardial T1 mapping with cardiovascular magnetic resonance multitasking. Magn. Reson. Med. 2019;81:2450–2463 doi: 10.1002/mrm.27574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milotta G, Bustin A, Jaubert O, Neji R, Prieto C, Botnar RM. 3D whole-heart isotropic-resolution motion-compensated joint T1/T2 mapping and water/fat imaging. Magn. Reson. Med. 2020;84:3009–3026 doi: 10.1002/mrm.28330. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton JI, Jiang Y, Eck B, Griswold M, Seiberlich N. Cardiac cine magnetic resonance fingerprinting for combined ejection fraction, T1 and T2 quantification. NMR Biomed. 2020;33:1–17 doi: 10.1002/nbm.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaubert O, Cruz G, Bustin A, et al. Free-running cardiac magnetic resonance fingerprinting: Joint T1/T2 map and Cine imaging. Magn. Reson. Imaging 2020;68:173–182 doi: 10.1016/j.mri.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman AJ, Gowland PA, Mansfield P. Optimization of the ultrafast look-locker echo-planar imaging T1 mapping sequence. Magn. Reson. Imaging 1998;16:765–772 doi: 10.1016/S0730-725X(98)00011-3. [DOI] [PubMed] [Google Scholar]
- 22.Sung K, Nayak KS. Measurement and characterization of RF nonuniformity over the heart at 3T using body coil transmission. J. Magn. Reson. Imaging 2008;27:643–648 doi: 10.1002/jmri.21253. [DOI] [PubMed] [Google Scholar]
- 23.Zhou R, Weller DS, Yang Y, et al. Dual-excitation flip-angle simultaneous cine and T1 mapping using spiral acquisition with respiratory and cardiac self-gating. Magn. Reson. Med. 2021;86:82–96 doi: 10.1002/mrm.28675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serry FM, Ma S, Mao X, et al. Dual flip-angle IR-FLASH with spin history mapping for B1+ corrected T1 mapping: Application to T1 cardiovascular magnetic resonance multitasking. Magn. Reson. Med. 2021:1–10 doi: 10.1002/mrm.28935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han PK, Horng DE, Petibon Y, et al. Free-Breathing 3D T1 Mapping of the Whole-Heart Using Low-Rank Tensor Modeling. In: Proceedings of the ISMRM. Montreal, Canada; 2019. p. 4396. [Google Scholar]
- 26.Han PK, Horng DE, Marin T, et al. Free-Breathing Three-Dimensional T1 Mapping of the Heart Using Subspace-Based Data Acquisition and Image Reconstruction. In: The 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Berlin, Germany; 2019. p. 2496. [DOI] [PubMed] [Google Scholar]
- 27.Han PK, Marin T, Vanessa L, Djebra Y, El Fakhri G, Ma C. Optimization of Spoiled GRE-based IR Acquisition Scheme for 3D Cardiac T Mapping at 3T. In: Proceedings of the ISMRM. Virtual; 2021. p. 3288. [Google Scholar]
- 28.Vogel CR. Computational methods for inverse problems. Society for Industrial and Applied Mathematics; 2002. [Google Scholar]
- 29.Zhao B, Haldar JP, Christodoulou AG, Liang ZP. Image reconstruction from highly under sampled (k, t)-space data with joint partial separability and sparsity constraints. IEEE Trans. Med. Imaging 2012;31:1809–1820 doi: 10.1109/TMI.2012.2203921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang Z-P. SPATIOTEMPORAL IMAGING WITH PARTIALLY SEPARABLE FUNCTIONS. IEEE Int. Symp. Biomed. Imaging 2007;2:988–991. [Google Scholar]
- 31.Haldar JP, Liang Z-P. Spatiotemporal imaging with partially separable functions: A matrix recovery approach. In: 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE; 2010. pp. 716–719. doi: 10.1109/ISBI.2010.5490076. [Google Scholar]
- 32.Boyd S, Parikh N, Chu E, Peleato B, Eckstein J. Distributed optimization and statistical learning via the alternating direction method of multipliers. Found. Trends Mach. Learn. 2010;3:1–122 doi: 10.1561/2200000016. [DOI] [Google Scholar]
- 33.Marin T, Djebra Y, Han PK, et al. Motion correction for PET data using subspace-based real-time MR imaging in simultaneous PET/MR. Phys. Med. Biol. 2020;65 doi: 10.1088/1361-6560/abb31d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fessler JA, Sutton BP. Nonuniform fast fourier transforms using min-max interpolation. IEEE Trans. Signal Process. 2003;51:560–574 doi: 10.1109/TSP.2002.807005. [DOI] [Google Scholar]
- 35.Ong F, Lustig M. SigPy: a python package for high performance iterative reconstruction. In: Proceedings in: the 27th Annual Meeting of International Society of Magnetic Resonance in Medicine.; 2019. [Google Scholar]
- 36.Golub G, Pereyra V. Separable Nonlinear Least Squares: the Variable Projection Method and its Applications. Inverse Probl. 2003;19:R1. [Google Scholar]
- 37.Kellman P, Arai AE, Xue H. T1 and extracellular volume mapping in the heart: Estimation of error maps and the influence of noise on precision. J. Cardiovasc. Magn. Reson. 2013;15:1–12 doi: 10.1186/1532-429X-15-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerqueira MD, Weissman NJ, Dilsizian V, et al. Standardized myocardial sementation and nomenclature for tomographic imaging of the heart: A Statement for Healthcare Professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation 2002;105:539–542 doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 39.Rincón-Domínguez T, Menini A, Solana AB, et al. Accelerated multi-snapshot free-breathing B1+ mapping based on the dual refocusing echo acquisition mode technique (DREAM): An alternative to measure RF nonuniformity for cardiac MRI. J. Magn. Reson. Imaging 2019;49:499–507 doi: 10.1002/jmri.26234. [DOI] [PubMed] [Google Scholar]
- 40.Do HP, Yoon AJ, Fong MW, Saremi F, Barr ML, Nayak KS. Double-gated myocardial ASL perfusion imaging is robust to heart rate variation. Magn. Reson. Med. 2017;77:1975–1980 doi: 10.1002/mrm.26282. [DOI] [PubMed] [Google Scholar]
- 41.Yarnykh VL. Actual flip-angle imaging in the pulsed steady state: A method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn. Reson. Med. 2007;57:192–200 doi: 10.1002/mrm.21120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Phantom results of ECG-gated IR schemes with SPGR readout and T1 estimation only assuming perfect B1+ (i.e., B1+ = 1). A: Estimated T1 maps from different methods. B: Scatter plots showing comparison of estimated T1 from different methods with those from IR-FSE. Solid line represents line of identity and dashed line represents line of regression. C: Bland-Altman plots showing comparison of estimated T1 from different methods with those from IR-FSE. Solid line represents the mean difference and dashed line represents the 95% confidence interval for limits of agreement.
Figure S2. Phantom study results with variation in B1+ field strength via control of transmitter voltage. A: Estimated T1 maps from IR-FSE and 5-(3)-5-(3) protocol with 80, 100, and 120% of reference transmitter voltage. B: Vial number positions. C: Estimated B1+ maps (defined as the ratio between the measured and nominal FAs) from 5-(3)-5-(3) protocol with 80, 100, and 120% of reference transmitter voltage. D: Estimated average FA within each vial from 5-(3)-5-(3) protocol with 80 (green line and circle), 100 (blue line and circle) and 120% (red line and circle) of reference transmitter voltage. Note that the ratio between the different B1+ field strength and the estimated average FAs from each vial are in good agreement.
Figure S3. In vivo study results of Bland-Altman plots comparing estimated T1 from the proposed method with those from MOLLI. Results from the anterior, septal, inferior, and lateral regions of the myocardium in the left ventricle are shown. Solid line represents the mean difference and dashed line represents the 95% confidence interval for limits of agreement.
Table S1. Comparison of mean and standard deviation of septal T1 values between MOLLI and proposed method across subjects