Abstract
We propose a model for high dimensional mediation analysis that includes latent variables. We describe our model in the context of an epidemiologic study for incident breast cancer with one exposure and a large number of biomarkers (i.e., potential mediators). We assume that the exposure directly influences a group of latent, or unmeasured, factors which are associated with both the outcome and a subset of the biomarkers. The biomarkers associated with the latent factors linking the exposure to the outcome are considered “mediators.” We derive the likelihood for this model and develop an expectation-maximization algorithm to maximize an L1-penalized version of this likelihood to limit the number of factors and associated biomarkers. We show that the resulting estimates are consistent and that the estimates of the nonzero parameters have an asymptotically normal distribution. In simulations, procedures based on this new model can have significantly higher power for detecting the mediating biomarkers compared with the simpler approaches. We apply our method to a study that evaluates the relationship between body mass index, 481 metabolic measurements, and estrogen-receptor positive breast cancer.
Keywords: direct effect, factor analysis, mediation analysis, oracle property, penalized likelihood
1 |. INTRODUCTION
In epidemiology, a mediation model aims to explain how an exposure (E) is associated with an outcome (Y). Traditionally, the model proposes that the exposure influences a single mediating variable (M), which, influences the outcome. More advanced models propose that the exposure influences a small set of mediating variables (M = (M1,...,Mp)), which, in turn influence the outcome (VanderWeele and Vansteelandt, 2014; Assi et al., 2015; Steen et al., 2017). Here, we consider the scenario where the set of (putative) mediators is large and a study aims to identify the true set of mediators and to describe the underlying mediation model. Our motivation is an estrogen-receptor positive (ER+) breast cancer case-control study that measured body mass index (BMI) and 481 serum metabolites. The objective is to identify those metabolites that mediate the well-established relationship between high BMI and an increased risk of ER+ breast cancer (Moore et al., 2018).
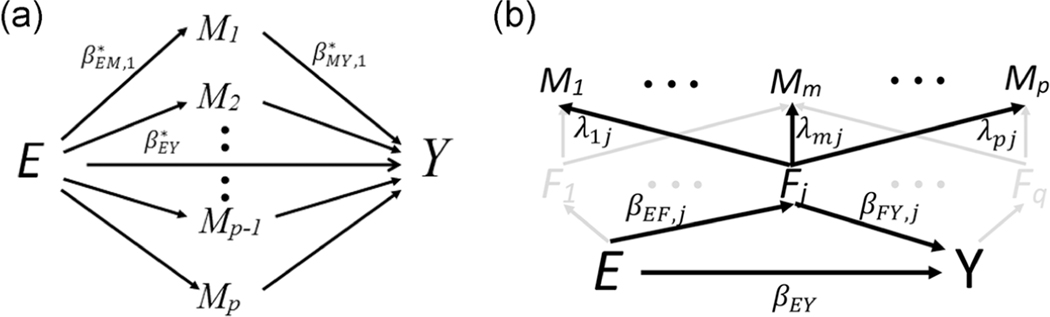
In most discussions of mediation, the presumption is that the exposure directly influences a subset of conditionally independent mediators (Figure 1A), with more recent discussions (Daniel et al., 2015) allowing the mediators to be causally ordered (Figure 2A). Here, following our beliefs about the underlying biology, we presume that the exposure directly influences a group of conditionally independent latent, or unmeasured, factors (F = (F1,...,Fq)) which, in turn, influence both a subset of “mediating” biomarkers and the outcome (Figure 1B). In our motivating breast cancer study, for example, we might expect BMI to reduce the level of the sex hormone-binding globulin (SHBG) protein (Calle and Kaaks, 2004), which increases the availability of many of the measured hormones listed in Table 1 and the unmeasured, carcinogenic, hormones (i.e., estrogens) that cause breast cancer. In this example, the factor is a well-defined but unmeasured protein level. A more heuristic example might be evaluating the relationship between poverty, metabolites, and cancer, where poverty influences a number of distinct factors (e.g., consumption of specific foods, hours of sleep, proximity to sources of pollution, etc) that are each known to affect the levels of multiple metabolites and the risk of cancer.
FIGURE 1.
Causal graphs of the mediation models. A, Traditional mediation analysis where the exposure influences the biomarkers. B, Latent-variable mediation analysis where the exposure influences a set of q latent, or unmeasured, factors and those factors influence both the biomarkers and the outcome. To simplify the figure, we highlight the arrows and notation for only a single factor. In the sparse scenario, we expect most effects to be 0 (i.e., λ, βEY, and βFY, are usually 0). We define the p × q matrix, Λ, so that the m, jth entry is λmj
FIGURE 2.
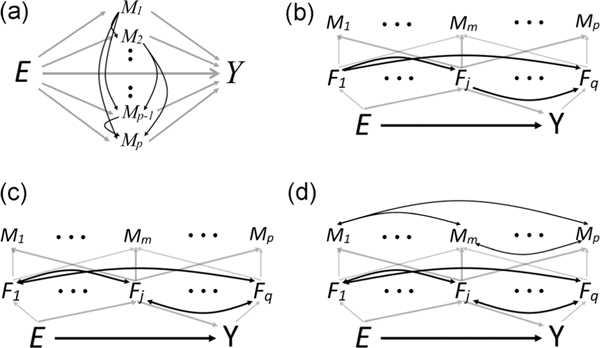
Alternative causal graphs of the mediation models. A, Causally ordered mediation analysis where the exposure influences the biomarkers. B, Latent-variable mediation analysis where the exposure influences a set of q causally ordered latent factors. C, Latent-variable mediation analysis where the exposure influences a set of q bidirectionally connected latent factors. D, Latent-variable mediation analysis where the exposure influences a set of q bidirectionally connected latent factors and those factors influence bidirectionally connected biomarkers
TABLE 1.
Metabolites linking BMI and breast cancer
| Metabolite | Loading (λmj) |
|---|---|
| 5α-Pregnan-3β,20-α-diol monosulfate (2) | 0.46 |
| 5α-Pregnan-3β,20-α-diol disulfate | 0.49 |
| Etiocholanolone glucuronide | 0.49 |
| 16α-Hydroxydehydroepiandrosterone 3-sulfate | 0.51 |
| Epiandrosterone sulfate | 0.60 |
| Androsterone sulfate | 0.61 |
| 4-Androsten-3β,17-β-diol monosulfate (2) | 0.61 |
| Pregnen-diol disulfate | 0.63 |
| 4-Androsten-3β,17-α-diol monosulfate (3) | 0.64 |
| 5α-Androstan-3β,17-β-diol disulfate | 0.65 |
| 21-Hydroxypregnenolone disulfate | 0.65 |
| Pregnen steroid monosulfate | 0.65 |
| 4-Androsten-3β,17-β-diol monosulfate (1) | 0.67 |
| 4-Androsten-3β,17-β-diol disulfate (2) | 0.70 |
| 4-Androsten-3β,17-β-diol disulfate (1) | 0.70 |
| Dehydroisoandrosterone sulfate (DHEA-S) | 0.75 |
This list includes those metabolites that were strongly affected (γ > 0.4) by the factor mediating increased BMI and ER+ breast cancer in PLCO.
Our goal is to formalize the latent variable model for mediation depicted in Figure 1B. We specify an L1-penalized version of the corresponding likelihood, propose an extension of the expectation-maximization (EM) algorithm used for sparse factor analysis (Hirose and Yamamoto, 2014; Srivastava et al., 2017) to obtain the maximum-likelihood estimates, and show that these estimates have the “oracle” property (Zou, 2006). We develop our estimation procedure for data from cohorts and retrospectively collected case-control studies. Furthermore, we show that accounting for latent variables, when the proposed model holds, can significantly increase a study’s power to detect “mediating” biomarkers (i.e., those biomarkers influenced by the mediating factors). Importantly, we note that our model is a simplification. The graph describing the true relationship between the exposure, biomarkers, and outcome is likely more complicated, where in addition to unmeasured confounders, the mediating factors can be causally ordered (Figure 2B), the graph can include bidirectional edges and cycles (Figure 2C), and the graph can include edges directly connecting the biomarkers (Figure 2D).
We note that the models in Figures 1B, 2B, and 2C are not distinguishable without imposing additional restrictions (Bai and Li, 2012) on the latent variables (e.g., conditional independence). Therefore, our independent factors are constructs of the model and we caution against the interpretation of the indirect effects through any specific factors. Nevertheless, with more modest assumptions, we can estimate and interpret the total indirect effect through all factors.
These methods extend procedures that handle latent factors affecting a small set of biomarkers (Muthén and Asparouhov, 2015; Albert et al., 2016) and add to the literature exploring high dimensional mediators. Zhang et al. (2016) model how epigenetic changes mediate the relationship between smoking and reduced lung function, assuming smoking directly affects methylation levels (i.e., Figure 1A). Huang and Pan (2016) test whether the expression levels of specific sets of genes mediate the relationship between miR-223 and glioblastoma by rotating the biomarkers and testing the resulting conditionally independent components. Chen et al. (2018) identify brain regions, from functional magnetic resonance imaging (fMRI) images, that link thermal response and self-reported pain by identifying orthogonal linear combinations of the biomarkers (i.e., fMRI voxels), known as “directions of mediation.” We contrast these methods with our own approach further in Section 6.
The remainder of the paper is organized as follows. In Section 2, we describe the statistical model and the proposed EM algorithm. In Section 3, we state the theoretical properties of the resulting estimates. In Section 4, we describe two alternative approaches for identifying mediating biomarkers and estimating relevant parameters. In Section 5, we study the properties of the estimates obtained using the different approaches and apply our method to the motivating study of breast cancer. Section 6 concludes with a brief discussion.
2 |. LATENT-VARIABLE MEDIATION ANALYSIS
2.1 |. Overview
Our first goal is to propose a mediation model, where mediators are latent variables (Figure 1B). Our second goal is to provide a procedure to estimate the parameters in this model.
2.2 |. Mediation model
We index subjects in the study by i, i = 1,...,N, and assume that the relationship between the exposure (Ei), factors (Fi), biomarkers (Mi), and outcome (Yi) can be described by the directed acyclic graph in Figure 1B. Moreover, although not pictured, we allow for a set of baseline covariates (Xi) that can influence Ei, Fi, and Yi. We then define , where Fij (e) is the value of the jth factor in subject i if Ei is set to e and is the value of Yi if Ei is set to e and the vector of factors Fi is set to . We further assume that sequential ignorability (Imai et al., 2010) holds, or more specifically, that
| (1) |
| (2) |
We note that, in contrast to standard models, our mediators are latent variables (Albert et al., 2016). These latent variables are unlikely to individually match up with the underlying, conditionally independent biologic variables (e.g., F1 is the unmeasured level of SHBG in the motivating example and is conditionally independent of all other mediating factors). In reality, it is more likely that there is a set (B = (Bi1,...,Biq)) of interrelated biologic mediators (e.g., levels of SBHG, insulin, and cytokines) and the independent factors represent weighted combinations of these biological quantities (e.g., ). This truth suggests that we should focus and interpret the combined (i.e., through all factors) indirect effect defined in Equation (5), as opposed to factor or path specific indirect effects.
We can now partition the total effect (TE) of changing the exposure from e to e′ into the natural direct effect (NDE) and natural indirect effect (NIE): TE = NDE + NIE, where the indirect effect passes through those pathways captured by the latent factors:
| (3) |
| (4) |
| (5) |
For binary Yi, we focus on the mediation effects defined on the odds ratio scale (VanderWeele and Vansteelandt, 2014), where ORTE = ORNDE × ORNIE,
and
We note that it has already been demonstrated (Albert et al., 2016) that these effects are not generally (e.g., nonparametrically) identifiable when the mediators are factors. However, these effects are identifiable under the parametric model represented in Figure 1B and discussed in the next section. Moreover, these effects are identifiable even without assumptions (e.g., independence) about the factors as discussed in Web Appendix A. Finally, path-specific or factor-specific indirect effects are not well-defined because the models of Figures 1b, 2a, 1b and 2c are not distinguishable.
2.3 |. Parametric assumptions
Recall our notation. For subject i, i = 1, ..., N, let Ei be the exposure, Yi be the outcome, and be a vector of biomarkers with p > > N, and be a vector of q latent mediators with q < < p. We assume that the distribution of Yi belongs to an exponential family,
| (6) |
with
| (7) |
We also assume that Fi and Mi are normally distributed:
| (8) |
where Iq is the q by q identity matrix and
| (9) |
Under retrospective sampling (i.e., for case-control data), we rely on the additional assumption that . We note that and are vectors of length q, Λ is a p × q matrix and the (m, j)th element, denoted by λmj, represents the effect of the jth factor on the mth biomarker. Moreover, we define “mediating” biomarkers to be the set {m: } and therefore, when trying to identify the mediators, select the set .
The proposed setup accommodates outcomes from a variety of distributions, but for ease of exposition we focus on outcomes from either a binomial or a normal distribution. For a continuous Y we assume , and Equation (6) simplifies to
| (10) |
In this scenario, the pathway specific effects can be related to model parameters by
and we note that these effects are identifiable. Similar conclusions were drawn by Albert et al. (2016) in the case of a single latent mediator. For a binary Y we assume and and rewrite Equation (6) in logistic form, . When the outcome is rare, we can approximate the pathway-specific effects on the OR scale by and , with modifications available to accommodate matched case-control studies (VanderWeele and Tchetgen Tchetgen, 2016) or interactions between the factors and exposure (VanderWeele and Vansteelandt, 2014).
In Web Appendix E, we consider extensions of Models (6–9) to accommodate additional covariates and extend the estimation procedure and theory presented to this setting. To estimate the vector of parameters using the observed data, for i = 1, ...,N, we first assume q, the number of latent factors, is known. In Section 2.5, we discuss how to choose q in practice.
2.4 |. Likelihood
Under prospective sampling, we derive the joint likelihood of (Yi, Mi, Fi) and (Yi, Mi) while conditioning on Ei to avoid modeling the distribution of the exposure. Under retrospective sampling (i.e., case-control data), we derive the joint likelihood of (Yi, Ei, Mi, Fi) and (Yi, Ei, Mi).
2.4.1 |. Prospective likelihood
Here the full data likelihood for (Y, M, F) is
| (11) |
where f is the product of the densities defined by Equations (6–9),
| (12) |
We use fM, fY, and fF to denote implied distribution of M, Y, and F, respectively. However, the factors Fi are not observed. The likelihood for the observed data, (Yi, Mi), is therefore
| (13) |
Although does not have a closed form in general, we show in the Web Appendix B that is the product of normal distributions when Yi is normally distributed.
2.4.2 |. Retrospective likelihood
Under retrospective sampling, N1 cases and N0 controls are drawn from the population of cases and controls, respectively, and biomarkers and exposures are observed (N1 + N0 = N). The corresponding likelihood is
| (14) |
Here, we assume in the overall population. Although closed forms of the conditional distributions of (Ei, Mi, Fi) are generally not available, they can be approximated well when the outcome is rare in the general population, that is,
| (15) |
Under the rare disease assumption, the distribution of (Ei, M i, Fi) in controls is approximately equal to the distribution in the general population. Thus, under Models (6–9)
| (16) |
where is a multivariate normal distribution with mean μ and covariance matrix ΣE,M,F. The covariance matrix ΣE,M,F is defined in the Web Appendix B.2 and . Note that, ΣE,M,F is a function of the parameters used in Models (6–9). The distribution of (Ei, Mi, Fi) in cases under Model (15) is
| (17) |
where
Note that the distributions of (Ei, Mi, Fi) in cases and controls differ only in their means.
Based on the above approximations, the likelihood for the full data (Ei, Mi, Fi) is
| (18) |
and the likelihood for the observed data is therefore easily shown to be
| (19) |
where , , and is the appropriate submatrix of ΣE,M,F. The effects of the exposure and the latent factors on the outcome are thus completely captured by the difference between the means of (Ei, Mi) in cases and controls.
2.5 |. Penalty to induce sparsity in the factors, F
To introduce sparseness in the factors F and the number of biomarkers associated with those factors, we maximize the penalized log-likelihood
| (20) |
where is the likelihood defined in (13) or (19) and P(·) is the chosen penalty function. We use the adaptive lasso penalty, where and is a square root consistent estimate of φ, but other options, such as SCAD or MC+ (Fan and Li, 2001; Zhang, 2010), are possible. In practice, we let , and be initial estimates from the observed likelihood LO (θ). We allow the penalties (ρ1N, ρ2N, ρ3N) to differ for each type of association. The penalized log-likelihood estimator is defined as
| (21) |
Because cannot be expressed in a closed from, we develop an (EM) algorithm building upon the methods for sparse factor analysis (Hirose and Yamamoto, 2014; Srivastava et al., 2017). Although the EM algorithm is a significant contribution of this paper, we provide details in Web Appendix C to keep the main text focused.
In practice, we specify the total number of factors, qmax, to be 40 and choose values of ρ1N, ρ2N, and ρ3N that minimize the extended Bayes information criterion (EBIC; Chen and Chen, 2008) defined as , where is the observed log-likelihood (Equations (13) or (19)), df is the number of parameters with nonzero estimates, and τ is the number of possible models with df nonzero parameters. We generally suggest that users start with a value of qmax that is likely to exceed the true number of factors influencing the biomarkers. In practice, we set γ = 0.5 in Equation (21). EBIC tends to outperform the more traditional selection criteria AIC and BIC in previous high dimensional settings (Chen and Chen, 2008; Srivastava et al., 2017) and in our simulations.
3 |. THEORETICAL PROPERTIES OF THE ESTIMATES
We highlight key properties of the model, the EM algorithm, and the estimates, , with proofs in the Supporting Information (see Web Appendix D). First, we show that the parameters θ are identifiable under the following condition for factor analysis presented in Anderson and Rubin (1956),
Condition 1.
If any row of the loading matrix Λ is deleted, there remain two disjoint submatrices of rank q.
Proposition 1 (Identifiability).
If Condition 1 holds, then θ is identifiable, and Λ, βFY, βEF are identifiable up to an orthogonal rotation.
We note that the products of parameters and mixed products and are uniquely identified. The identifiability of these terms is crucial when estimating direct and indirect effects (Section 2.2).
Our second property, building upon previous work of Hirose and Yamamoto (2014), Srivastava et al. (2017), states the properties the EM algorithm.
Proposition 2 (Convergence of the EM algorithm).
With each iteration of the proposed EM algorithm, the penalized log-likelihood (20) does not decrease,
and the sequence of EM estimates converges to a local maximum .
Our third property, building upon work of Zou (2006), is that the resulting estimates, , where W is the total number of parameters, have the oracle property. Let θ be the vector of all parameters (see Section 2.3), A = {j|θj ≠ 0} index the set of parameters not equal to 0, index the set of parameters with nonzero estimates; based on our dataset of N subjects and let be the vector of estimates for the nonzero parameters .
Proposition 3 (Oracle Property).
Suppose that and for k ∈ {1, 2, 3}. Then we obtain
- consistency of the selection of nonzero effects:
(22) - asymptotic normality for the nonzero effects:
where is Fisher’s information matrix for the true model (i.e., excluding zero coefficients).(23)
4 |. ALTERNATIVE METHODS TO IDENTIFY SUBSETS OF MEDIATORS
Here, we propose two alternative, two-step approaches to identify the subset of “mediating” biomarkers. Recall for latent variable mediation analysis (LVMA), we identify the mediating biomarkers to be the set .
4.1 |. Individual marker mediation analysis (IMA)
We first consider testing each biomarker individually using Sobel’s test (Sobel, 1982). For a continuous outcome, we can test biomarker m by fitting two linear regression models
and then calculating the P value, pm, for the test statistic , where is the estimated variance of the product and is calculated using the Taylor-Series expansion (Sobel, 1982). Logistic regression would be used when Y is binary. We can then identify the set of “mediating” biomarkers as those with a P value below a specified threshold.
4.2 |. Two-step mediation analysis (TMA)
We next consider a two-step approach where, in the first step, we identify the latent factors underlying the biomarkers and, in the second step, we test whether each of those latent factors are mediators. Specifically, in the original-version (TMAO), we first perform sparse factor analysis on M, ignoring E and Y :
| (24) |
where is a multivariate normal distribution with mean 0 and variance and obtain our estimated factors . Again, we choose the penalty, ρ, which minimizes the EBIC. In the second step, we use Sobel’s test (Sobel, 1982), to individually test each of the estimated factors. The “mediating” biomarkers are those with a nonzero loading on the selected factors.
In a modified version (TMAR), we perform sparse factor analysis (24) on MR, where the jth row of MR are residuals after regressing the jth row of M on E. Specifically, we substitute MR for M in Equation (24) to obtain and , and then let . Under prospective sampling, M, Y, and E need to be centered by their corresponding sample means because intercepts of are not identifiable directly from M or MR by these two approaches. Under retrospective sampling, M and the exposure E are centered by their means in the controls. TMAR is conceptually similar to the mediation approach proposed by Huang and Pan (2016) and to surrogate variable analysis (Leek and Storey, 2007); that it is MR, not M, that is accurately described by factor analysis.
4.2.1 |. Remarks
TMAO and TMAR are computationally faster than the full joint model LVMA and, for TMAO, it is straight-forward to calculate P values for the tested factors. However, by ignoring E and Y, less information is available for identifying factors and, ultimately, detecting mediators. Second, TMAO incorrectly assumes independence of factors. TMAR, or allowing for correlated factors (Hirose and Yamamoto, 2014), attempts to handle this issue, but these methods perform poorly in small samples. Third, in case-control studies, the assumption that the biomarkers are normally distributed is violated and Section 2.4.2 shows that much of the information is contained in the difference between the group means. Fourth, by not explicitly modeling the latent variables, the terms, βEY and used to estimate the direct and indirect effects are biased because the imputed mediators contain measurement error (Carroll et al., 2006; le Cessie et al., 2012; Valeri et al., 2014).
5 |. SIMULATIONS
5.1 |. Data generation
We compared the properties of LVMA, IMA, and TMA (O and R) in simulated data with N = 300 or N = 500 observations. We assumed that there were fifteen factors (q = 15) each affecting twenty unique biomarkers, M. The first factor was a mediator (i.e., βEF,1 ≠ 0 and βFY,1 ≠ 0), the next four factors were associated only with E (i.e., βEF,j ≠ 0 and βFY,j = 0 for j = 2,...,5) and the last ten factors were associated with neither E nor Y (i.e., βEF,j = 0 and βFY,j = 0 for j = 6,...,15). We then added an additional ninety independent normally distributed biomarkers so that p = 390. Data were simulated based on the model in Section 2.3, with binary outcomes assuming a logistic link and continuous outcomes assuming normality. For case-control studies, we sampled an equal number of cases and controls (i.e., N1 = N0 = 150 or N1 = N0 = 250) from a larger population prospectively simulated. We varied the effect of the exposure on the mediating factor (βEF,1 ∈ {0.4, 0.5}), the effect of exposure-related factors on their constituent biomarkers (λ1 ∈ {0.25, 0.3}), and the effect of exposure-unrelated factors on their constituent biomarkers (λ0 ∈ {0.4, 0.5}). Here, λmj = λ1 if j ≤ 5 (when λmj ≠ 0) and λmj = λ0 if 6 ≤ j ≤ 15 (when λmj ≠ 0). Other parameters/distributions were set as follows: βEY = βFY,1 = 0.3, βEF,2 = ··· = βEF,5 = 0.7, γM,1 = ··· = γM,p = 0.5, , γY = 4.6 (i.e., P(Y = 1|E = 0, F1 = 0) = 0.01 for binary outcomes), and E∼N (0.5, 1). In Web Appendix F, we describe the simulation setup and corresponding results for scenarios when there are no latent variables and the exposure directly affects individual biomarkers.
For each parameter and sample size combination, we simulated 1000 datasets. Then we applied each of the four methods and calculated the average number of true positives (TPs) selected, the average number of false positives (FPs) selected, and the average estimate of the conditional exposure effect, βEY. Here, the biomarker m is a “true positive” (TP) if it is identified as a mediator and . It is a “false positive” (FP) if it is identified as a mediator but . For valid comparison between LVMA and the other methods, we selected P value thresholds for these methods so that the FP remained constant. Note, the requirement for a biomarker to be “selected” as a mediator is defined in Section 2. For IMA, TMAO, and TMAR, we estimated the conditional exposure effect as the coefficient for the exposure in a model for the outcome that also included all selected biomarkers or factors.
5.2 |. Results
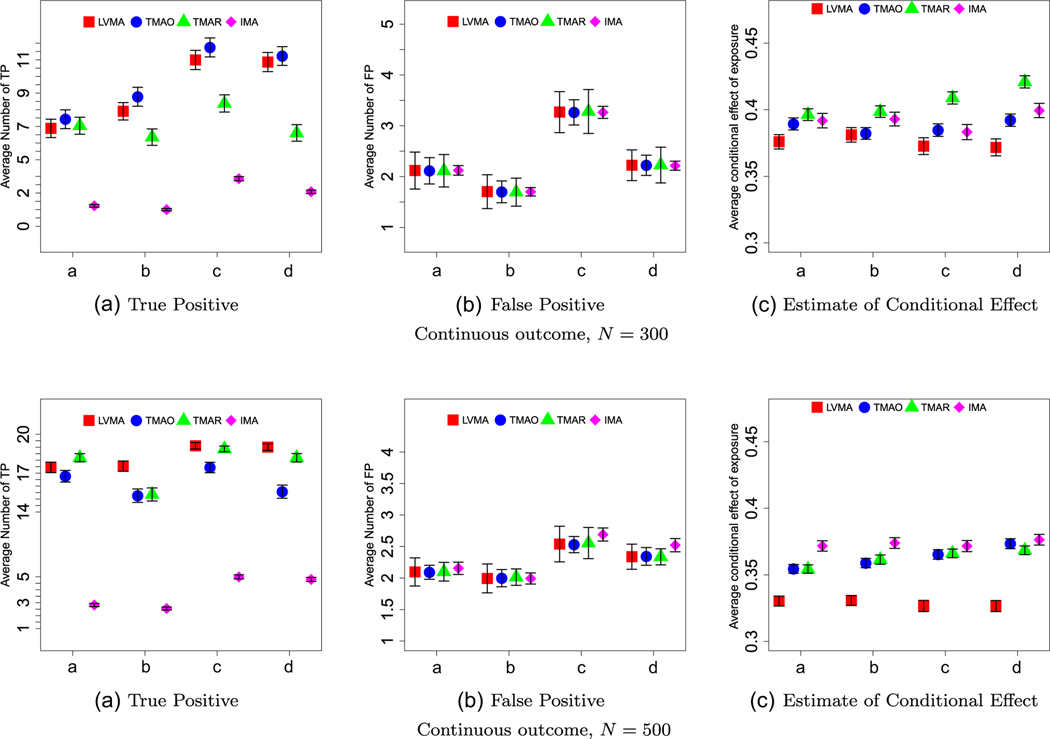
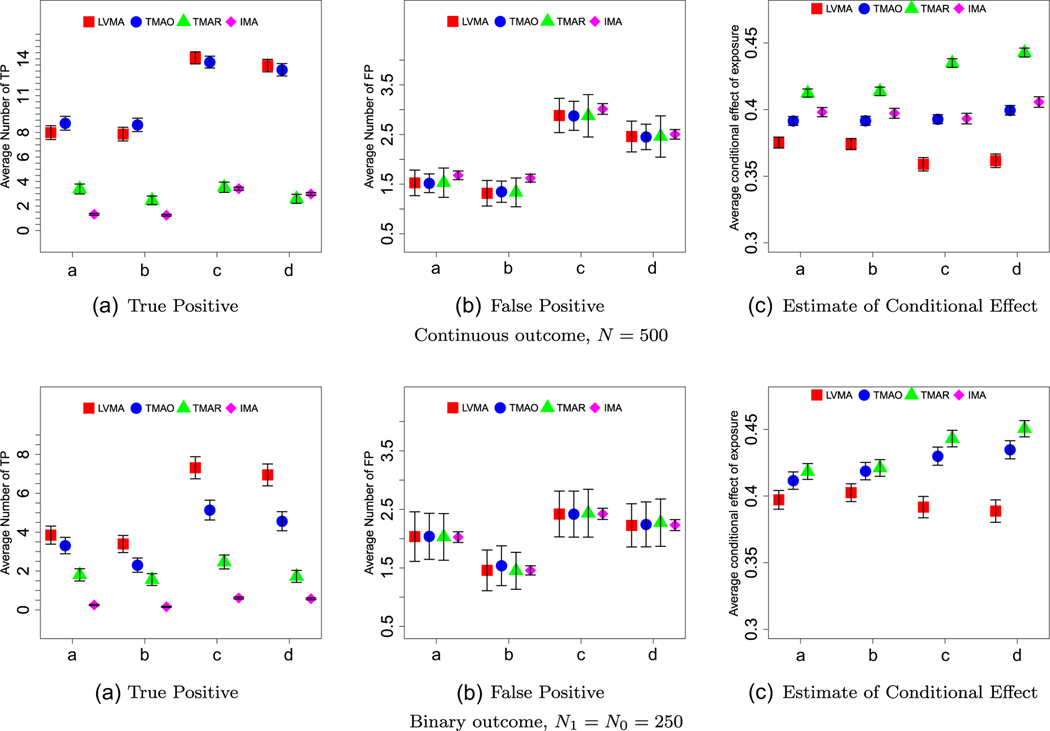
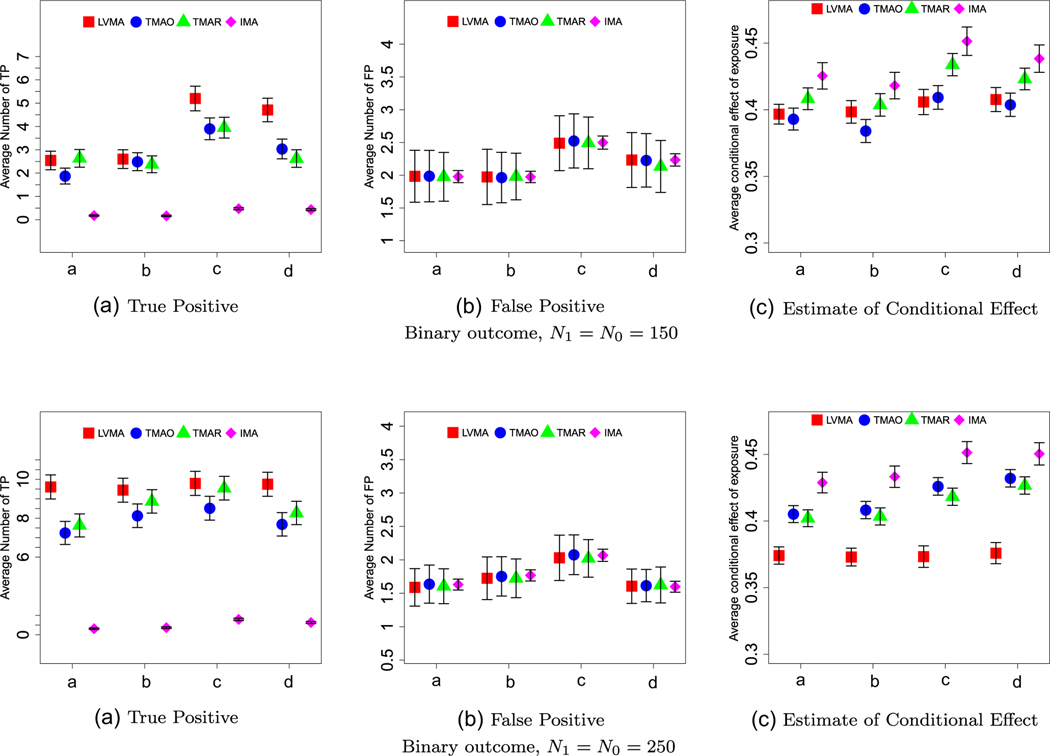
We summarize our results in Figures 3. First, for most settings, LMVA tended to have, the largest TP rate (panels (A) of Figures 3-5). Recall, we chose significance thresholds for the other three methods so that the average FP rate (see panels (B) of Figures 3-5) was similar across methods. TMAR and TMOR tended to perform similarly. The one exception is that TMAR performed poorly when we reduced the effect of exposure-related factors to λ1 = 0.25 (see Figure 5). IMA consistently performed poorly (e.g., on average it had four-five times lower TP rate). The latter reinforces the need to use some form of latent variable analysis when the exposure does not directly affect biomarkers individually.
FIGURE 3.
Continuous outcome and large factor effects (λ1 = 0.3). The panels, labeled A-C, show the average number of true positives (TP), the average number of false positives (FP), and the average estimate of the direct effect for the four methods (red = LVMA; blue = TMAO; green = TMAR; purple = IMA) and for four scenarios (a: βEF,1 = 0.4, λ0 = 0.4; b: βEF,1 = 0.4, λ0 = 0.5; c: βEF,1 = 0.5, λ0 = 0.4; d: βEF,1 = 0.5, λ0 = 0.5) based on 1000 simulations. Top and bottom panel are for studies with N = 300 and N = 500 subjects, respectively. The whiskers show two standard errors around the average estimates.
FIGURE 5.
Small factor effects (λ1 = 0.25). The panels, labeled A-C, show the average number of true positives (TP), the average number of false positives (FP), and the average estimate of the direct effect for the four methods (red = LVMA; blue = TMAO; green = TMAR; purple = IMA) and for four scenarios (a: βEF,1 = 0.4, λ0 = 0.4; b: βEF,1 = 0.4, λ0 = 0.5; c: βEF,1 = 0.5, λ0 = 0.4; d: βEF,1 = 0.5, λ0 = 0.5) based on 1000 simulations. Top and bottom panel are for studies with continuous and binary outcomes, respectively (N = 500). The whiskers show two standard errors around the average estimates.
The pronounced when the effect of factors on associated biomarkers is small (Figure 5) and when the outcome was a binary variable (Figures 4 and 5). When comparing TMAO to TMAR, neither clearly performed better. Their relative performance strongly depends on sample size, with TMAR performing better as the sample sizes increased and the effect of the exposure could be better estimated. This advantage was further magnified when the effects of the factor on the biomarkers increased.
FIGURE 4.
Binary outcome and large factor effects (λ1 = 0.3). The panels, labeled A-C, show the average number of true positives (TP), the average number of false positives (FP), and the average estimate of the direct effect for the four methods (red = LVMA; blue = TMAO; green = TMAR; purple = IMA) and for four scenarios (a: βEF,1 = 0.4, λ0 = 0.4; b: βEF,1 = 0.4, λ0 = 0.5; c: βEF,1 = 0.5, λ0 = 0.4; d: βEF,1 = 0.5, λ0 = 0.5) based on 1000 simulations. Top and bottom panel are for studies with N = 300 and N = 500 subjects, respectively. The whiskers show two standard errors around the average estimates.
As expected, TMAR, TMAO, and IMA produced biased estimates of the conditional effect of exposure; compare the results in panel (C) of Figures 3-5 to the true direct effect, βEF = 0.3. Although LVMA had smaller bias, the estimates of the conditional effect of exposure using LVMA still exceeded 0.35 in most scenarios. When we increased the sample size or the effect size so that TP = 20 (see Web Figures 5, 8, and 21), the bias in LVMA, but not for the other methods, disappeared.
LVMA was robust to the number of specified factors (see Web Figures 11 and 12). Therefore, in practice, we suggest specifying a relatively large number of factors. Furthermore, we found that using EBIC was slightly preferable to using AIC or BIC, but the two two-step methods, TMAO and TMAR, were far more sensitive to the choice of selection criteria (see Web Figures 5-10). LVMA was also robust to violations of the assumption that the error terms for the biomarkers were normally distributed (see Web Figures 13-18). Finally, when we decreased exposure effects on nonmediating factors to βEF,2 = ··· =βEF,5 = 0.4, the TP of LVMA, TMAO, and TMAR become similar. Web Figures 19-21 shows that, if we also decrease the effect of the mediating factors on the biomarkers to λ1 = 0.25, LVMA regains its advantages. Note, in this latter setting the exposure becomes important for identifying factors.
5.3 |. Data example
Our motivating study aims to identify metabolites that mediate the known relationship between high BMI and the increased risk of ER+ breast cancer. This study, nested inside the prostate, lung, colorectal, and ovarian cancer screening study (PLCO), includes 410 (ER+) breast cancers and 410 controls matched on study age (±2 years), date of blood collection (±3 months), and hormone therapy use at baseline. The study collected serum samples at the first follow-up visit, 1-year after baseline, and using these specimen, measured 481 known serum metabolites (<kDA). Metabolite levels were log transformed. Details on the study are in Moore et al. (2018).
We modeled the data using LVMA with qmax = 40 factors adjusting for the three matching variables (see Web Appendix E). The model identified only a single factor associated with both BMI ( = 0.035) and risk of breast cancer ( = 0.14). This factor had 111 nonzero loadings but only 16 of these loadings had an absolute value larger than 0.4, with the majority of remaining metabolites having loadings below 0.01. In Table 1, we list these 16 metabolites. In Web Figure 22, we display loadings for all metabolites from LVMA and standard factor analysis. Of interest, many of these metabolites are products of estrogen metabolism, suggesting estrogen metabolism does partially explain why increased BMI is associated with increased risk of ER+ breast cancer. However, most of the effect of BMI was not mediated by this factor. When estimating the TE, NDE, and NIE on the OR scale, we find ORTE = exp(0.039), ORNDE = exp(0.034), and ORNIE = exp(0.005) suggesting that the estrogen pathways explains only a small fraction of the TE of BMI.
We also applied TMAO, TMAR, and IMA to the data, adjusting for the matching variables. TMAO and TMAR did not detect statistically significant factors mediating the relationship between BMI and the risk of ER+ breast cancers (Web Figures 23 and 24). Similarly, IMA did not identify statistically significant metabolites mediating the relationship (Web Figure 25). Further details are in Web Appendix G.
6 |. DISCUSSION
We proposed a latent variable model for high dimensional mediation analysis (LVMA). Our theoretical results show that the model parameters are identifiable, and LVMA estimates those parameters that have the so-called oracle properties of consistency and efficiency. Our simulation results further show that using LVMA, when appropriate, can significantly increase the number of discovered mediators. LVMA, by considering all variables simultaneously, efficiently estimates all parameters in the model. Specifically, using LVMA, we better estimate the mediating factors by using additional information about the exposure and outcome, as opposed to only using the information about the biomarkers. However, under model misspecification, such as when the exposure directly affects individual biomarkers, the assumption of latent variables can reduce the power to detect such biomarkers.
We highlight a couple of features of our method. First, we extend current literature on mediation analysis with a single latent mediator (Muthén and Asparouhov, 2015; Albert et al., 2016) to handle multiple latent mediators. Second, although some recent studies have started exploring penalized structural equation modeling (Jacobucci et al., 2016), these methods were not designed to handle the p > > n setting. Third, we extend our mediation model and, more generally, sparse factor analysis to accommodate case-control sampling.
None of the previously published methods (Boca et al., 2013; Huang and Pan, 2016; Zhang et al., 2016; Zhao and Luo, 2016; Chen et al., 2018 explicitly assumed the latent structure illustrated in Figure 1, but two methods did so implicitly. Huang and Pan (2016) take an approach similar to TMAR but do not impose any sparsity on the factors. Their objective is also fundamentally different as they are testing whether the set of all biomarkers are mediators as opposed to trying to identify the subset that is mediators. Chen et al. (2018) aims to identify linear combinations of biomarkers that are associated with both the exposure and outcome. However, their approach does not allow for factors that are only associated with the exposure or the outcome. Therefore, the biomarkers associated with only one of those variables get mistakenly included in the “direction of mediation.”
Several problems remain to be addressed in future work. Our latent model in (6–9) does not detect the existence of biomarkers that directly mediate the effect of E on Y. As evidenced by a large number of nonzero loadings in our application, the shrinkage of loadings for unrelated biomarkers may not be satisfactorily using EBIC. Some of our assumptions could also be violated in real-world examples. The factors need not be independent conditional on the exposure; the biomarkers need not be normally distributed; for retrospective sampling, the exposure needs to be normally distributed. We note the latter may be accommodated by the semiparametric approach based on Qin (1998) but that the discussion is beyond the scope of this paper. Despite these limitations, we believe the newly proposed LVMA offers a novel important tool for detecting biological mediators.
Supplementary Material
ACKNOWLEDGMENTS
This study utilized the computational resources of the NIH HPC Biowulf cluster.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Albert JM, Geng C. and Nelson S. (2016) Causal mediation analysis with a latent mediator. Biometrical Journal, 58, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TW and Rubin H. (1956) Statistical Inference in Factor Analysis. Proceedings of the Third Berkeley Symposium on Mathematical Statistics and Probability, Volume 5: Contributions to Econometrics, Industrial Research, and Psychometry, 111–150, University of California Press, Berkeley, CA. https://projecteuclid.org/euclid.bsmsp/1200511860 [Google Scholar]
- Assi N, Fages A, Vineis P, Chadeau-Hyam M, Stepien M. and Duarte-Salles T et al. (2015) A statistical framework to model the meeting-in-the-middle principle using metabolomic data: Application to hepatocellular carcinoma in the epic study. Mutagenesis, 30, 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J. and Li K. (2012) Statistical analysis of factor models of high dimension. The Annals of Statistics, 40, 436–465. [Google Scholar]
- Boca SM, Sinha R, Cross AJ, Moore SC and Sampson JN (2013) Testing multiple biological mediators simultaneously. Bioinformatics, 30, 214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle EE and Kaaks R. (2004) Overweight, obesity and cancer: Epidemiological evidence and proposed mechanisms. Nature Reviews Cancer, 4, 579. [DOI] [PubMed] [Google Scholar]
- Carroll RJ, Ruppert D, Stefanski LA and Crainiceanu CM (2006) Measurement error in nonlinear models: A modern perspective. New York: CRC Press. [Google Scholar]
- Chen J. and Chen Z. (2008) Extended bayesian information criteria for model selection with large model spaces. Biometrika, 95, 759–771. [Google Scholar]
- Chen OY, Crainiceanu C, Ogburn EL, Caffo BS, Wager TD and Lindquist MA (2018) High-dimensional multivariate mediation with application to neuroimaging data. Biostatistics, 19, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R, De Stavola B, Cousens S. and Vansteelandt S. (2015) Causal mediation analysis with multiple mediators. Biometrics, 71, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. and Li R. (2001) Variable selection via nonconcave penalized likelihood and its oracle properties. Journal of the American Statistical Association, 96, 1348–1360. [Google Scholar]
- Hirose K. and Yamamoto M. (2014) Estimation of an oblique structure via penalized likelihood factor analysis. Computational Statistics & Data Analysis, 79, 120–132. [Google Scholar]
- Huang Y-T and Pan W-C (2016) Hypothesis test of mediation effect in causal mediation model with high-dimensional continuous mediators. Biometrics, 72, 402–413. [DOI] [PubMed] [Google Scholar]
- Imai K, Keele L. and Tingley D. (2010) A general approach to causal mediation analysis. Psychological Methods, 15, 309–334. [DOI] [PubMed] [Google Scholar]
- Jacobucci R, Grimm KJ and McArdle JJ (2016) Regularized structural equation modeling. Structural Equation Modeling: A Multidisciplinary Journal, 23, 555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Cessie S, Debeij J, Rosendaal FR, Cannegieter SC and Vandenbrouckea JP (2012) Quantification of bias in direct effects estimates due to different types of measurement error in the mediator. Epidemiology, 23, 551–560. [DOI] [PubMed] [Google Scholar]
- Leek JT and Storey JD (2007) Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genetics, 3, e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SC, Playdon MC, Sampson JN, Hoover RN, Trabert B. and Matthews CE et al. (2018) A metabolomics analysis of body mass index and postmenopausal breast cancer risk. Journal of the National Cancer Institute, 110, 588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén B. and Asparouhov T. (2015) Causal effects in mediation modeling: An introduction with applications to latent variables. Structural Equation Modeling: A Multidisciplinary Journal, 22, 12–23. [Google Scholar]
- Qin J. (1998) Inferences for case-control and semiparametric twosample density ratio models. Biometrika, 85, 619–630. [Google Scholar]
- Sobel ME (1982) Asymptotic confidence intervals for indirect effects in structural equation models. Sociological Methodology, 13, 290–312. [Google Scholar]
- Srivastava S, Engelhardt BE and Dunson DB (2017) Expandable factor analysis. Biometrika, 104, 649–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen J, Loeys T, Moerkerke B. and Vansteelandt S. (2017) Flexible mediation analysis with multiple mediators. American Journal of Epidemiology, 186, 184–193. [DOI] [PubMed] [Google Scholar]
- Valeri L, Lin X. and VanderWeele TJ (2014) Mediation analysis when a continuous mediator is measured with error and the outcome follows a generalized linear model. Statistics in Medicine, 33, 4875–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWeele TJ and Tchetgen Tchetgen EJ (2016) Mediation analysis with matched case-control study designs. American Journal of Epidemiology, 183, 869–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWeele T. and Vansteelandt S. (2014) Mediation analysis with multiple mediators. Epidemiologic Methods, 2, 95–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-H (2010) Nearly unbiased variable selection under minimax concave penalty. The Annals of Statistics, 38, 894–942. [Google Scholar]
- Zhang H, Zheng Y, Zhang Z, Gao T, Joyce B. and Yoon G et al. (2016) Estimating and testing high-dimensional mediation effects in epigenetic studies. Bioinformatics, 32, 3150–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, and Luo X. (2016). Pathway lasso: Estimate and select sparse mediation pathways with high dimensional mediators. arXiv preprint arXiv:1603.07749.
- Zou H. (2006) The adaptive lasso and its oracle properties. Journal of the American Statistical Association, 101, 1418–1429. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.