Significance
Successful microbial infections typically involve the subversion of host immune pathways by pathogen-specific virulence factors. Here, we uncovered that viruses and bacteria independently evolved effectors targeting the same conserved loop of the mitogen-activated protein kinase (MAPK) p90-ribosomal S6-kinases (RSKs). Kinase usurpation relies on a previously unidentified short linear motif (SLiM) shared by these virulence factors. Mechanistically, RSK kinase binding prevents its dephosphorylation, thus promoting its phosphorylating activity. Remarkably, while viruses and bacteria evolved an identical mechanism of kinase activation, downstream effect diverged, effectively disarming different arms of the immune system according to their own need. This is a prominent illustration of trans-kingdom convergent evolution across microbial pathogens to usurp key signaling kinases.
Keywords: innate immunity, type-3 secretion, short linear motif, picornavirus, host–pathogen interaction
Abstract
Microbes have been coevolving with their host for millions of years, exploiting host resources to their own benefit. We show that viral and bacterial pathogens convergently evolved to hijack cellular mitogen-activated protein kinase (MAPK) p90-ribosomal S6-kinases (RSKs). Theiler’s virus leader (L) protein binds RSKs and prevents their dephosphorylation, thus maintaining the kinases active. Recruitment of RSKs enables L-protein-mediated inhibition of eukaryotic translation initiation factor 2 alpha kinase 2 (EIF2AK2 or PKR) and stress granule formation. Strikingly, ORF45 protein of Kaposi’s sarcoma-associated herpesvirus (KSHV) and YopM protein of Yersinia use the same peptide motif as L to recruit and activate RSKs. All three proteins interact with a conserved surface-located loop of RSKs, likely acting as an allosteric regulation site. Some unrelated viruses and bacteria thus evolved to harness RSKs in a common fashion, yet to target distinct aspects of innate immunity. As documented for Varicella zoster virus ORF11, additional pathogens likely evolved to hijack RSKs, using a similar short linear motif.
In the course of infection, pathogens, such as viruses and bacteria, are confronted with multiple host defense mechanisms that need to be defeated to produce progeny and spread. To fulfill this, pathogens have evolved virulence factors that modulate key cellular processes, such as apoptosis, translation, and innate immune signaling. This cellular reprogramming heavily relies on the subversion of protein networks regulated by short linear motifs (SLiMs). SLiMs are short (3 to 12 amino acids long), contiguous regions of proteins mediating protein–protein interactions (1). These motifs, generally located in intrinsically disordered regions of proteins, are responsible for key biological processes, such as subcellular targeting, posttranslational modification, protein-complex scaffolding, and degradation (2). The low complexity of many linear motifs combined with the high plasticity of viral genomes promote SLiMs as a prevalent strategy for viral subversion (3). However, while prominent instances of SLiMs have been experimentally documented across a wide variety of virus families, their low complexity and binding affinity confound the identification of functional linear motifs and suggest that numerous instances are still to be discovered (4, 5).
Theiler’s murine encephalomyelitis virus (TMEV) is a positive-stranded RNA virus belonging to the Cardiovirus genus of the Picornaviridae family. The TMEV leader (L) protein is a 76-amino-acid-long peptide endowed with multiple activities related to innate immunity escape. Notably, L inhibits interferon genes transcription (6), disrupts the nucleo-cytoplasmic trafficking of host proteins (7), and prevents stress granule (SG) formation through inhibition of eukaryotic translation initiation factor 2 alpha kinase 2 (EIF2AK2 or PKR) (8, 9). Point mutations in the zinc-finger domain of L (LZn mutation), thought to destabilize the overall structure of L, as well as the M60V mutation (LM60V), which maps to the likely disordered C terminus of L, were shown to abrogate L activities (10). The related L protein encoded by encephalomyocarditis virus (EMCV) displays similar activities and was further reported to interfere with mitogen-activated protein kinase pathways (11).
In this study, we sought to investigate the mechanisms underlying the immune suppression elicited by the cardiovirus leader proteins. We found that L physically interacts with a conserved region of p90-ribosomal S6-kinase (RSK) kinases through an aspartate-aspartate-valine-phenylalanine linear amino acid motif. Epistatic analysis indicated that PKR inhibition was dependent on both RSK and leader proteins, identifying an additional mechanism of PKR evasion. Interestingly, our data unravel a common virulence mechanism shared by a set of unrelated viruses and bacteria.
Results
To identify cellular targets that may underlie L activities, BHK-21 cells were infected with a recombinant TMEV expressing an N-terminally FLAG-tagged L protein, and L binding partners were identified by coimmunoprecipitation and mass spectrometry (MS) analysis (SI Appendix, Fig. S1 A and B and Dataset S1). RSK was identified as an L target. All four RSK isoforms were coimmunoprecipitated with a virally expressed FLAG-L protein (Fig. 1A). L of representative mouse and human Theilovirus strains (12) readily interacted with RSK2. Moreover, endogenous RSK2 coimmunoprecipitated with virally expressed FLAG-L from TMEV, Saffold virus, and Mengo virus (an EMCV strain), showing that RSK binding is a conserved feature among Cardiovirus L proteins (SI Appendix, Fig. S1C).
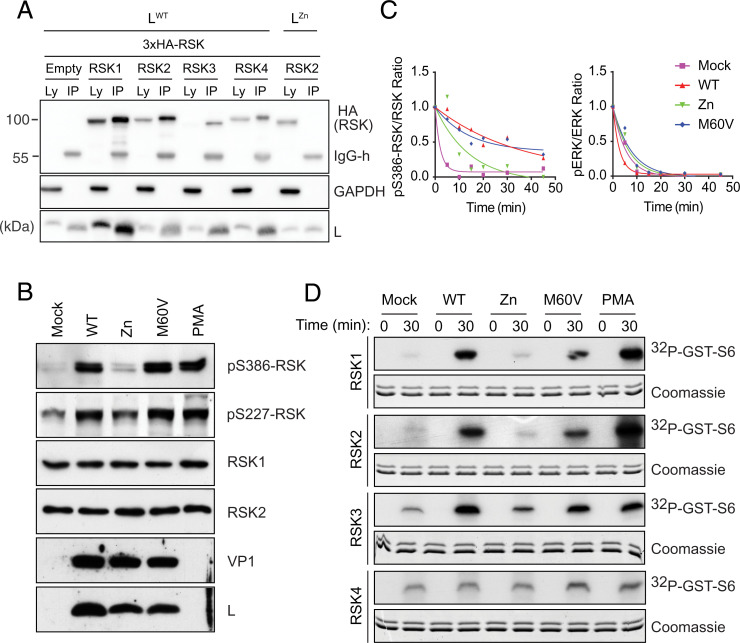
Fig. 1.
TMEV L protein binds to and activates RSK kinases through inhibition of RSK dephosphorylation. (A) L binds the four RSK isoforms. Immunoblots showing detection of L, GAPDH, and HA (RSK) in cell lysates (Ly) or after FLAG-L immunoprecipitation (IP) from HeLa-M cells transduced to express indicated HA-tagged human RSK isoforms and infected with TMEV carrying a FLAG-tagged L. Negative controls included cells transduced with an empty vector or infected with a FLAG-LZn TMEV mutant. (B) Immunoblots for RSK1/2 phosphorylation after L929 cell infection for 8 h with indicated viruses. (C) L inhibits RSK dephosphorylation by phosphatases. Plots show time-dependent dephosphorylation of PMA-activated RSK and ERK after ATP depletion, as measured by Western blotting from L929 mock- or virus-infected cells. (D) L promotes RSK catalytic activity. Kinase assays performed with immunoprecipitated HA-RSK from L929 cells infected with indicated LWT and mutant TMEV. Shown is the detection of 32P-labeled–GST-S6 by autoradiography at 0 and 30 min of reaction time and Coomassie-blue stained SDS-PAGE of reaction samples showing recombinant GST-S6 detection. Note that the lower band visible on Coomassie blue-stained gels likely results from partial GST-S6 proteolysis. Zn, zinc finger mutation thought to affect the overall conformation of L. M60V, mutation in the C-terminal region of L, which affects L activities, but not binding to RSK.
RSKs are directly phosphorylated by ERK1/2, ultimately leading to phosphorylation of RSK S386 and S227 residues and activation (13). As shown in Fig. 1B, infection with wild-type (WT) TMEV or with the LM60V mutant that retains RSK binding ability led to RSK S386 and S227 phosphorylation, whereas infection with a virus carrying a mutation in the Zn finger motif of L (LZn) expected to disrupt the overall L structure did not. After adenosine triphosphate (ATP) depletion, RSK phosphorylation decreased slower in cells infected with LWT or LM60V viruses than in mock-infected cells or in cells infected with the LZn mutant (Fig. 1C), suggesting that L binding protects RSK from dephosphorylation. ERK phosphorylation decay, analyzed as a control, was not affected by L. That L binding protects RSK from dephosphorylation was confirmed in an experiment where infected cell lysates were treated with recombinant λ-phosphatase (SI Appendix, Fig. S1D).
Sustained phosphorylation of RSK is expected to increase catalytic activity of RSK N-terminal kinase domain (NTKD), unless L binding affects ATP or substrate recruitment to the kinase active site. In vitro kinase assays confirmed that RSK activity was higher when RSK was immunoprecipitated from cells infected with LWT and LM60V viruses than from uninfected cells or from cells infected with the LZn virus (Fig. 1D and SI Appendix, Fig. S1E). Collectively, these results demonstrate that Cardiovirus L proteins interact with all four RSK isoforms and increase kinase activity by slowing down S227 and S386 dephosphorylation.
We next identified L residues involved in RSK binding. RSK2 coimmunoprecipitated with a truncated FLAG-L1–59 (SI Appendix, Fig. S2A). Coimmunoprecipitation of HA-RSK2 and FLAG-L mutants from either transfected 293T cells (SI Appendix, Fig. S2B) or infected L929 cells (SI Appendix, Fig. S2C) confirmed that mutations in the Zn finger affected RSK binding, whereas a M60V mutation did not. Importantly, mutations in a DDVF motif had the most impact on binding. Moreover, a biotinylated peptide encompassing this DDVF motif, but not an F48A mutant peptide, readily pulled down endogenous RSK from 293T cell lysates (Fig. 2A).
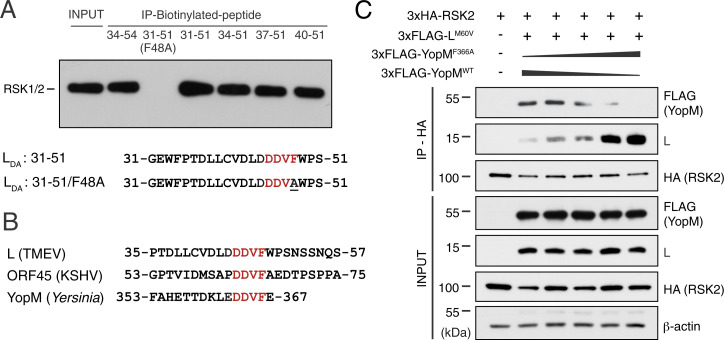
Fig. 2.
Proteins from different pathogens bind RSK through a DDVF motif. (A) L-derived biotinylated peptides encompassing the DDVF motif pull down endogenous RSK1/2 from 293T cell lysates. (B) Protein sequence alignments of L, ORF45, and YopM encompassing the conserved DDVF motif required for RSK binding. (C) Representative immunoblot (n = 2) showing competition of LM60V and YopM for coimmunoprecipitation with 3×HA-RSK2 from lysates of 293T cells cotransfected with indicated constructs and with varying ratios of plasmids encoding 3×FLAG-YopMWT and 3×FLAG-YopMF366A. LM60V was used instead of LWT because it is less toxic and does not inhibit expression of proteins in transfected cells.
Two other proteins encoded by pathogens, namely, ORF45 of Kaposi’s sarcoma-associated herpes virus (KSHV or HHV-8) and YopM of bacteria from the genus Yersinia, including Yersinia pestis, were reported to bind and activate RSK (14–17). Strikingly, the DDVF motif responsible for L binding to RSK is conserved in ORF45 and YopM sequences (Fig. 2B), and maps to regions of these proteins that were reported to be involved in RSK binding (18–23). Thus, proteins from at least three unrelated pathogens likely interact with RSK via the same DDVF motif. They also likely bind the same RSK region. That YopM and L bind the same cluster of residues in RSK is supported by the competition experiment presented in Fig. 2C, showing that YopMWT, but not YopMF366A (F > A mutation in the DDVF motif), competes with L for coimmunoprecipitation of RSK2.
To identify the RSK domain that is targeted by the DDVF motif, RSK2 deletion mutants were tested for interaction with L1–559 by coimmunoprecipitation. Results indicate that amino acids 166 to 367 of RSK2 (part of the NTKD and linker region) are sufficient for interaction with L (SI Appendix, Fig. S2D). Accordingly, complexes between RSK2 amino acids 44 to 367 (RSK244–367) and L were formed after expression in Escherichia coli (SI Appendix, Fig. S3A). Thus, we used an ultraviolet (UV)–cross-linking strategy based on parabenzoyl-l-phenylalanine (pBpA) incorporation in E. coli to identify RSK2 residues that are targeted by the DDVF motif of pathogens’ proteins (principle explained in SI Appendix, Fig. S3B). YopM was used for this experiment because its higher solubility facilitates cross-linking and subsequent purification steps. Thus, pBpA was substituted for F366 in the DDVF sequence of YopM, and cross-linking was performed in bacteria that coexpressed YopMF366#pBpA and RSK244–367. MS identified RSK2 residues L285, G286, and/or M287 as those contacted by YopM F366 (SI Appendix, Fig. S3 C and D). These residues belong to a well-conserved KAKLGM motif found in all four mammalian RSK isoforms and across evolution, from Caenorhabditis elegans to human (Fig. 3 A and B).
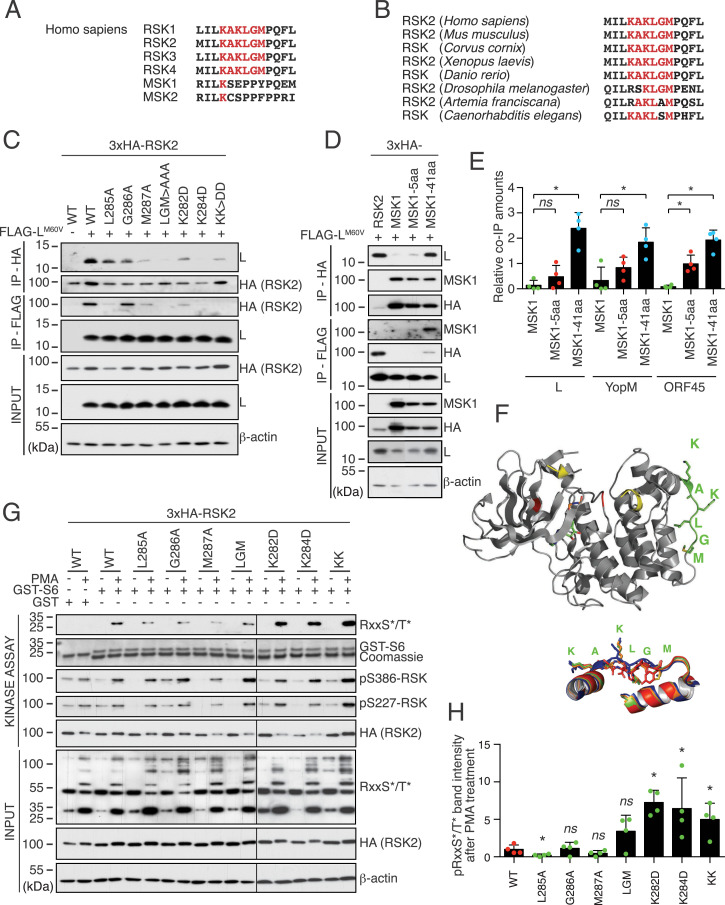
Fig. 3.
L, ORF45, and YopM bind a common motif that regulates RSK activity. (A) The KAKLGM motif is conserved in the NTKD of all RSK isoforms, but not in the NTKD of MSK. (B) The KAKLGM motif is conserved across evolution. (C) Mutations in the RSK2 KAKLGM motif affect L binding. Representative immunoblots (n = 2) for HA-RSK2 and L in cell lysates and immunoprecipitates (IP) of 293T cells cotransfected with plasmids expressing FLAG-LM60V and HA-RSK2 mutants. (D) RSK2/MSK1 chimeric constructs confirm KAKLGM motif involvement in L binding. Immunoblots showing L, HA (RSK2/MSK1), and MSK1 detection in cell lysates and HA- or FLAG-immunoprecipitates from 293T cells cotransfected with indicated plasmid. (E) Plots (mean ± SD) showing the relative abundance of MSK1 and MSK1/RSK2 chimeras that were coimmunoprecipitated with L, YopM, and ORF45 in blots, as presented in D and in SI Appendix, Fig. S3 G and H (n = 4). ns, not significant ; *P < 0.05. (F) Structure of RSK2 NTKD. Green, KAKLGM motif; yellow, activation loop (partial in the crystal structure); red, catalytic site (F, Upper). Superimposition of crystallographic structures derived from the KAKLGM RSK2 (blue) and KSEPPY MSK1 (red) motifs with corresponding regions of Alphafold-predicted structures of MSK1–RSK2-5aa (green) and MSK1–RSK2-41aa (orange) chimeras (F, Lower). PDB ID codes: RSK2, 3G51; MSK1, 1VZO. (G and H) Mutations in the KAKLGM motif modulate RSK activity. In vitro kinase assay performed using WT or mutant 3×HA-RSK2 immunoprecipitated with an anti-HA antibody from transfected 293T cells treated with dimethyl sulfoxide (−) or with PMA. Phosphorylation of the recombinant GST-S6 substrate was measured by immunoblot, using an anti–phospho-RxxS*/T* antibody (n = 4).
Mutations introduced in the KAKLGM sequence of RSK2 affected binding of L, YopM, and ORF45 (SI Appendix, Fig. S3E), the most critical residues for binding being K284, L285, and M287 (Fig. 3C and SI Appendix, Fig. S3F).
The closely related mitogen- and stress-activated protein kinase 1 (MSK1) lacks the KAKLGM sequence (Fig. 3A) and failed to bind L, YopM, or ORF45 (Fig. 3D and SI Appendix, Fig. S3 G and H). Substituting 41 residues of MSK1 by the homologous region of RSK2, including the KAKLGM motif, significantly induced binding of L, YopM, and ORF45 to chimeric MSK1 (Fig. 3 D and E and SI Appendix, Fig. S3 G and H). Accordingly, Alphafold2 prediction (24, 25) showed a good structural agreement of side-chain orientations in residues critical for L, YopM, and ORF45 binding between RSK2 and chimeric MSK1 proteins (Fig. 3F). Moreover, substitution of five residues (KAKLGM for KSEPPY) in MSK1 was sufficient to increase binding, though not significantly. Taken together, these data show that TMEV L, Yersinia YopM, and KSHV ORF45 use a common DDVF motif to bind the KAKLGM sequence of RSK, thereby promoting sustained kinase activation.
The KAKLGM motif of RSK is part of a surface-exposed loop at the basis of an α-helix (Fig. 3F). Binding of L, YopM, or ORF45 to this loop may indirectly stabilize the neighboring kinase activation loop in an active conformation. Interestingly, the KAKLGM motif is extremely well conserved across evolution (Fig. 3B), despite being part of a surface-exposed loop, a structure typically prone to variation. This suggests a role for this loop as an allosteric site for RSK kinase activity regulation by cellular factors. Kinase assays were performed by using lysates of 293T cells transfected to express RSK2 bearing mutations in the KAKLGM motif (Fig. 3 G and H). All tested mutants conserved their ability to be phosphorylated on S227 and S386 after stimulation with phorbol 12-myristate 13-acetate (PMA). Mutations K282D and K284D slightly, but significantly, increased, while mutations L285A and M287A decreased the ability of RSK2 to phosphorylate a glutathione S-transferase (GST)-S6 RSK substrate. These data support the idea that pathogen-encoded proteins evolved to harness such an accessible allosteric RSK regulation site. These observations suggest that physiological regulation of RSK may depend on cellular factors that interact with RSK in the same fashion as L, YopM, and ORF45.
Yersinia are extracellular bacteria that use a type III secretion system (26, 27) to inject YopM into the cytosol of eukaryotic cells. YopM contains a leucine-rich repeat (LRR) domain reported to recruit protein kinase N2 (PKN2 or PRK2) (19, 23). Formation of a trimeric YopM–RSK–PKN2 complex triggers PKN2 phosphorylation by RSK, which would lead to interleukin-10 expression up-regulation and pyrin inflammasome inhibition (17, 28–30). L does not contain a similar LRR domain and likely makes a different use of RSK recruitment. We reported that TMEV L inhibits SG formation in infected cells (8) through inhibition of PKR (9).
Interestingly, in HeLa-M cells, where RSK1 and RSK2 were knocked down using short hairpin RNAs or knocked out by CRISPR–Cas9, L no longer inhibited PKR activation and SG formation (Fig. 4 A and B and SI Appendix, Fig. S4 A–C). Re-expression of any of the four RSK isoforms in RSK1/2 knockout (KO) cells, by lentiviral transduction, fully restored inhibition of PKR and SG formation (SI Appendix, Fig. S4D). In contrast, re-expression of a KAKLGM–KAKAAA RSK2 mutant in the same cells did not restore SG formation inhibition in response to infection (SI Appendix, Fig. S4E), confirming the involvement of RSK in L-mediated PKR inhibition. PKR inhibition by L depends on both the DDVF motif and the C-terminal theilodomain that is not required for RSK binding and activation (Fig. 4 B and C and SI Appendix, Fig. S4 D and E), suggesting that L recruits RSK via the DDVF motif and recruits another target by the theilodomain, which could inhibit PKR after being phosphorylated by RSK. Accordingly, ectopic expression of L, but not of YopM or ORF45, inhibited polyinosinic:polycytidylic acid [poly(I:C)]-mediated PKR activation, although all three proteins readily activated RSK (Fig. 4 C and D).
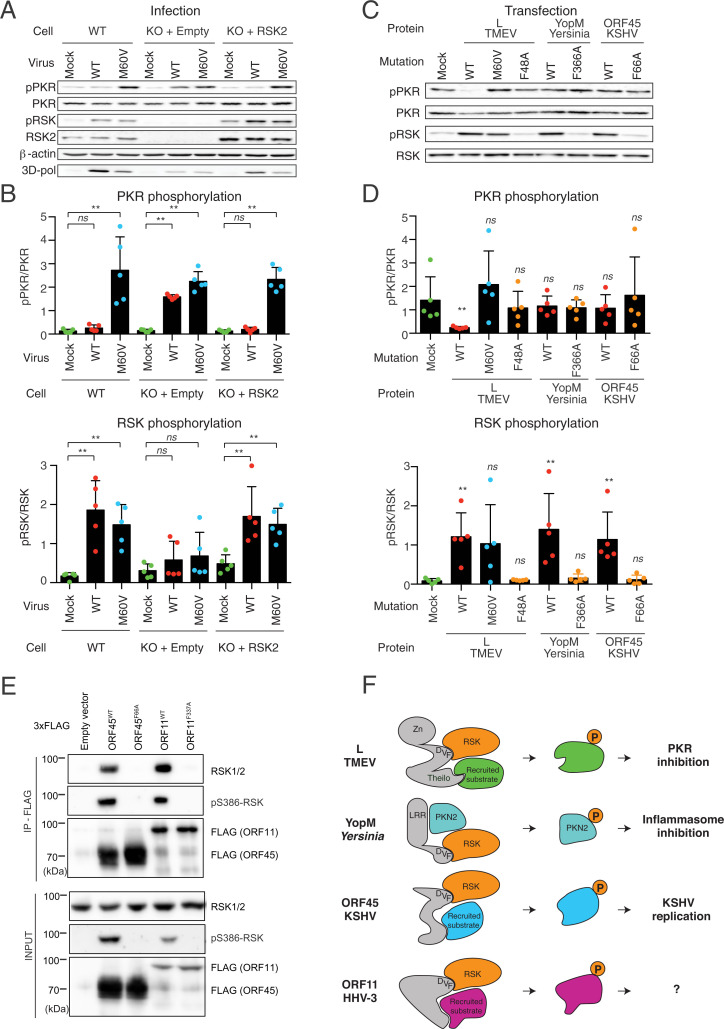
Fig. 4.
Pathogen-encoded proteins use a convergent RSK activation mechanism for distinct purposes. (A and B) TMEV L protein inhibits PKR activation in an RSK-dependent manner. (A) Immunoblots for RSK2, PKR and their phosphorylated forms (pS386-RSK and pT446-PKR) in mock-infected and infected HeLa-M cells. Cells were either WT HeLa-M or RSK1/2 double-KO derivatives that were transduced with the empty pTM952 lentiviral vector (KO+Empty) or with this vector expressing RSK2 to restore RSK2 expression (KO+RSK2). Transduced cell populations, selected with 1 mg/mL G418, were either mock-infected or infected for 12 h with 2 PFU per cell of TMEV KJ6 derivatives expressing either LWT or the LM60Vmutant protein, which conserves RSK binding, but lost described L activities. (B) Plots showing the quantification (mean ± SD) of phospho-PKR relative to PKR (Upper) and phospho-RSK relative to RSK2 (Lower; n = 5). (C and D) Ectopic expression of L, but not of YopM or ORF45, inhibits poly(I:C)-mediated activation of PKR. HeLa-M cells were transfected with plasmids expressing WT or DDV(F) mutants of L, YopM, and ORF45 or a control LM60V mutant that activates RSK, but fails to inhibit PKR activation. At 24 h posttransfection, cells were transfected with poly(I:C) to activate PKR. (C) Shown are immunoblots performed on cell lysates obtained 5 h after poly(I:C) transfection. (D) Plots showing the immunoblot quantification for phospho-PKR relative to total PKR (Upper) and phospho-RSK relative to total RSK (Lower) (n = 5). (E) Immunoblot showing the interaction of VZV ORF11WT, but not ORF11F337A, with endogenous RSK. ORF45 WT and F66A mutant were used as positive and negative controls for RSK coimmunoprecipitation and phosphorylation. (F) Model proposing how different pathogen-encoded proteins use a common mechanism to hijack RSKs for distinct purposes linked to replication and innate immunity escape. ns, not significant; **P < 0.01.
The low complexity of the DDVF motif strongly suggests that other viruses have evolved proteins targeting RSK. As a proof of principle, we screened human and veterinary viruses for the presence of conserved [DE]-[DE]-V-F linear motifs. Out of 33 positive proteins encoded by 21 different viruses, 4 proteins were selected on the basis of the availability of their genomic material. While UL21 (human alphaherpesvirus 1 [HHV-1]), NS3 (Dengue virus), and the large T antigen of the human polyomavirus 1 (BKV) did not immunoprecipitate endogenous RSKs, a clear interaction was detected between the ORF11 protein of Varicella zoster virus (VZV or HHV-3) and RSKs, leading to sustained RSK phosphorylation (Fig. 4E and SI Appendix, Fig. S4F).
Taken together, our results suggest a model (Fig. 4F) where YopM, L, ORF45, and ORF11 would recruit distinct RSK targets through a specific domain (LRR for YopM and theilodomain for L), while concomitantly activating RSK through their common [DE]-[DE]-V-F motif, thereby promoting phosphorylation of specific targets by the kinase. L, YopM, ORF45, and ORF11 proteins are encoded by highly divergent organisms, and their sequences are not homologous, making it very unlikely that they arose through horizontal genetic transfer, thus supporting the emergence of the RSK-binding mechanism by convergent evolution.
Discussion
In this study, we provide direct biochemical evidence that a set of unrelated proteins encoded by both bacteria and viruses bind the same interface of the RSK protein kinases. Remarkably, this interaction was found to be dependent on a DDVF linear motif shared by these proteins. RSK binding shielded the activation loop from dephosphorylation, resulting in an enhanced catalytic activity of the RSK kinases. Characterization of the RSK:DDVF interaction interface, using a genetically encoded photoreactive cross-linker coupled with MS analysis, pointed to a group of KAKLGM residues located in a solvent-accessible loop of the kinase. Protein sequence and phylogenetic analysis revealed a strong evolutionary conservation of this sequence across highly divergent eukaryote phyla, suggesting a critical function of this region in the regulation of its kinase activity. In line with this, mutational scanning of the DDVF-binding loop on RSK was found to regulate its enzymatic activity, suggesting a potential role of this conserved loop as an allosteric regulator of RSK activity.
Our data also support a two-step mechanism, whereby these pathogens activate RSKs via their DDVF motif, then phosphorylate pathogen-specific targets recruited by the remainder of the protein, allowing the subversion of cellular responses tailored to each pathogen requirement. This model is consistent with previous reports dissecting the ability of the adenovirus E1A and HPV16-E7 oncoproteins to bind and inactivate the retinoblastoma protein (Rb). While both proteins use a shared linear LxCxE motif for Rb binding, nonhomologous regions of these proteins were shown to be responsible for different posttranslational modifications, with E7 promoting ubiquitin-mediated degradation and E1A stimulating the acetylation of Rb, respectively (31, 32). Given the low sequence complexity of the DDVF motif, other pathogens are expected to have evolved a functional DDVF motif in order to subvert RSK and rewire cellular networks to their advantage. Accordingly, we observed a significantly greater number of DDVF occurrences than expected by chance in the proteome of human viruses compared to the human proteome (P = 0.0004, Fisher’s exact test).
It is becoming increasingly evident that microbial SLiMs often evolve to mimic and subvert functional host protein interfaces (4, 5, 33). Whether the present DDVF motif resembles an existing eukaryotic motif is not currently known. Of interest, a computational screen of linear motifs previously identified a highly similar DEVF sequence as significantly enriched in metazoan as well as in viral proteomes (1, 3). The low specificity of the RSK substrate recognition consensus (34) combined with the known use of bridging protein for its activation (35) raise the possibility that such cellular DDVF-containing proteins might act as scaffold proteins to direct RSK to specific substrates.
Taken together, our data identify a linear DDVF motif convergently evolved by unrelated pathogens to hijack cellular protein kinases, likely by mimicking the activity of a regulatory cell factor. They suggest a model where proteins encoded by these pathogens recruit specific targets that, once phosphorylated, counteract distinct arms of the host defenses.
Materials and Methods
RSK Dephosphorylation Assay.
L929 cells were mock-treated or infected with 2 plaque-forming units (PFU) per cell of indicated viruses. At 7 h postinfection, cells were washed and incubated with PMA at a concentration of 200 nM for 13 min in culture medium. Cells were then washed once with 200 nM PMA in phosphate-buffered saline (PBS) for 2 min and treated either with a mixture of 10 µM rotenone and 10 mM 2-deoxy-d-glucose in PBS at 37 °C or with 400 units of lambda phosphatase (New England Biolabs). At indicated time points, cellular proteins were extracted. RSK and ERK phosphorylation status was analyzed by Western blotting using an Odyssey infrared imaging system (Li-Cor).
Kinase Assay.
Anti-HA immunoprecipitates were incubated in 25 mM Hepes (pH 7.5), 50 mM NaCl, 20 mM β-glycerophosphate, 1 mM dithiothreitol, 20 mM MgCl2, 1 mM Na3VO4 and 2.5 µg of recombinant GST or GST-S6. The ATP concentration was 500 µM for cold labeling and 100 µM with 100 µCi/mL 32P-ATP for radiolabeling. After 0 or 30 min at 30 °C under 1,000 rpm mixing, reaction was stopped with the addition of Laemmli buffer. Detection of phosphorylated GST-S6 was done by autoradiography (for 32P labeling) or by immunoblot using an anti-phospho RxxS/T antibody.
Cross-Linking.
E. coli ER2566 carrying plasmids coding for 6×His-3×FLAG-YopM amber, 6×His-RSK244–367, and the transfer RNA (tRNA) synthetase/tRNA suppressor couple were grown overnight at 37 °C on agar plates containing 250 µg/mL ampicillin, 50 µg/mL kanamycin, and 30 µg/mL chloramphenicol. One colony was picked and cultured overnight at 37 °C in Luria-Bertani medium containing the same antibiotics. This culture was then used as an inoculum for 100-mL culture batches. When the optical density reached 0.6 to 0.8, 0.2 mM isopropyl β-d-1-thiogalactopyranoside was added to induce expression of 6×His-3×FLAG-YopM amber and 6×His-RSK244–367; 0.2% arabinose was added to induce expression of the tRNA synthetase/tRNA suppressor couple; 0.8 mM pBpA was added for incorporation by suppression of the amber codon (F366, V365, or E367) in YopM. Bacteria were cultured overnight at 25 °C. Culture samples were then transferred to 24-well plates and exposed to UV light (≥350 nm) for 10 min. Bacteria were then lysed with a French press (1,100 psi of pressure), and His-tagged proteins were enriched on His-Trap columns (GE Healthcare) according to the manufacturer’s recommendations.
MS.
After coimmunoprecipitation, samples were resolved by using a 10% Tris–glycine sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were visualized by using PageBlue (Thermo Scientific, catalog no. 24620). Bands of interest were cut out from the gel and digested with trypsin (50 ng/μL in 50 mM NH4HCO3 buffer, pH 8.0). The peptides were analyzed as described (36) by capillary liquid chromatography–tandem MS in an LTQ XL ion-trap mass spectrometer (ThermoScientific) fitted with a microelectrospray probe. The data were analyzed with the Proteome Discoverer software (ThermoScientific, version 1.4.1), and the proteins were identified with SequestHT against a target-decoy nonredundant mouse-protein database obtained from Uniprot. The following parameters were used: Trypsin was selected with proteolytic cleavage only after arginine or lysine; the number of internal cleavage sites was set to one; mass tolerance for precursors and fragment ions was 1.0 Da; and considered dynamic modifications were +15.99 Da for oxidized methionine. Peptide matches were filtered by using the q value and Posterior Error Probability calculated by the Percolator algorithm, ensuring an estimated false positive rate below 5%. The filtered Sequest HT output files for each peptide were grouped according to the protein from which they were derived. Cross-linked peptides were analyzed on a Q Exactive HF (Thermo) mass spectrometer as described (37) and mapped with the program “Crossfinder” as described (38, 39) using the following filter settings: number of fragment ions per spectrum > 5, number of fragment ions per peptide > 2, fractional intensity of assigned MS2 peaks > 0.05, and score > 2 00.
Supplementary Material
Acknowledgments
We are grateful to Graciela Andrei and Robert Snoeck (Katholieke Universiteit Leuven, Rega Institute, Leuven, Belgium) for the gift of VZV and HHV-1 DNA, Daniel Santos Mansur (Centro de Ciências Biológicas, Universidade Federal de Santa Catarina, Santa Catarina, Brazil) for the gift of Dengue virus cDNA, and Samira Fafi-Kremer (Virology Laboratory, Strasbourg University Hospital, Strasbourg, France) for the gift of JC and BK virus DNA. We thank Eric Freundt (University [Univ.] of Tampa, FL) and Guido Bommer (de Duve Institute, Univ. of Louvain, Belgium) for critical reading of the manuscript and Matvei Appraxine for the construction of L mutant plasmids. Funding: Excellence of Science (EOS) Joint Programme of Fonds de la Recherche Scientifique (FNRS) and Fonds Wetenschapellijk Onderzoek-Vlaanderen (EOS ID 30981113); Interuniversitary Attraction Poles programme (IAP-P7/45 Belgian Society for Virology); Belgian Fund for Medical Research (Projet de Recherche [PDR] T.0185.14 and Crédit de recherche [CDR] J.0143.18F); Actions de Recherches Concertées; Era-Net (Neuron); Deutsche Forschungsgemeinschaft (MU3613/3-1 and MU3613/1-2); Fund for Research Training in Industry and Agriculture fellowship (to F.B., M.D., and M.P.); FNRS aspirant fellowship (to T.C.); and Fondation Roi Baudoin (Fonds Maurange) (to N.C.). Y.H. received a salary from Otsuka Pharmaceutical Co., Ltd., Tokushima, Japan.
Footnotes
Competing interest statement: Y.H. received a salary from Otsuka Pharmaceutical Co., Ltd., Tokushima, Japan, while working as guest scientist at the Université Catholique de Louvain. The company had no influence on the research project.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114647119/-/DCSupplemental.
Data Availability
All sequence-read data have been deposited in the National Center for Biotechnology Information Sequence Read Archive Database (https://www.ncbi.nlm.nih.gov/sra; BioProject accession no. PRJNA768960) (40).
References
- 1.Davey N. E., et al. , Attributes of short linear motifs. Mol. Biosyst. 8, 268–281 (2012). [DOI] [PubMed] [Google Scholar]
- 2.van der Lee R., et al. , Classification of intrinsically disordered regions and proteins. Chem. Rev. 114, 6589–6631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagai T., Azia A., Babu M. M., Andino R., Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions. Cell Rep. 7, 1729–1739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chemes L. B., de Prat-Gay G., Sánchez I. E., Convergent evolution and mimicry of protein linear motifs in host-pathogen interactions. Curr. Opin. Struct. Biol. 32, 91–101 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Elde N. C., Malik H. S., The evolutionary conundrum of pathogen mimicry. Nat. Rev. Microbiol. 7, 787–797 (2009). [DOI] [PubMed] [Google Scholar]
- 6.van Pesch V., van Eyll O., Michiels T., The leader protein of Theiler’s virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75, 7811–7817 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delhaye S., van Pesch V., Michiels T., The leader protein of Theiler’s virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 78, 4357–4362 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borghese F., Michiels T., The leader protein of cardioviruses inhibits stress granule assembly. J. Virol. 85, 9614–9622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borghese F., Sorgeloos F., Cesaro T., Michiels T., The leader protein of Theiler’s virus prevents the activation of PKR. J. Virol. 93, e01010-19•• (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ricour C., et al. , Random mutagenesis defines a domain of Theiler’s virus leader protein that is essential for antagonism of nucleocytoplasmic trafficking and cytokine gene expression. J. Virol. 83, 11223–11232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter F. W., Brown B., Palmenberg A. C., Nucleoporin phosphorylation triggered by the encephalomyocarditis virus leader protein is mediated by mitogen-activated protein kinases. J. Virol. 84, 12538–12548 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freundt E. C., Drappier M., Michiels T., Innate immune detection of cardioviruses and viral disruption of interferon signaling. Front. Microbiol. 9, 2448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romeo Y., Zhang X., Roux P. P., Regulation and function of the RSK family of protein kinases. Biochem. J. 441, 553–569 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Hentschke M., et al. , Yersinia virulence factor YopM induces sustained RSK activation by interfering with dephosphorylation. PLoS One 5, e13165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuang E., Tang Q., Maul G. G., Zhu F., Activation of p90 ribosomal S6 kinase by ORF45 of Kaposi’s sarcoma-associated herpesvirus and its role in viral lytic replication. J. Virol. 82, 1838–1850 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuang E., Wu F., Zhu F., Mechanism of sustained activation of ribosomal S6 kinase (RSK) and ERK by Kaposi sarcoma-associated herpesvirus ORF45: Multiprotein complexes retain active phosphorylated ERK AND RSK and protect them from dephosphorylation. J. Biol. Chem. 284, 13958–13968 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald C., Vacratsis P. O., Bliska J. B., Dixon J. E., The Yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J. Biol. Chem. 278, 18514–18523 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Fu B., et al. , Activation of p90 ribosomal S6 kinases by ORF45 of Kaposi’s sarcoma-associated herpesvirus is critical for optimal production of infectious viruses. J. Virol. 89, 195–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Höfling S., et al. , Manipulation of pro-inflammatory cytokine production by the bacterial cell-penetrating effector protein YopM is independent of its interaction with host cell kinases RSK1 and PRK2. Virulence 5, 761–771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karijolich J., Zhao Y., Peterson B., Zhou Q., Glaunsinger B., Kaposi’s sarcoma-associated herpesvirus ORF45 mediates transcriptional activation of the HIV-1 long terminal repeat via RSK2. J. Virol. 88, 7024–7035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuang E., Fu B., Liang Q., Myoung J., Zhu F., Phosphorylation of eukaryotic translation initiation factor 4B (EIF4B) by open reading frame 45/p90 ribosomal S6 kinase (ORF45/RSK) signaling axis facilitates protein translation during Kaposi sarcoma-associated herpesvirus (KSHV) lytic replication. J. Biol. Chem. 286, 41171–41182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy M. W., Marré M. L., Lesser C. F., Mecsas J., The C-terminal tail of Yersinia pseudotuberculosis YopM is critical for interacting with RSK1 and for virulence. Infect. Immun. 78, 2584–2598 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McPhee J. B., Mena P., Bliska J. B., Delineation of regions of the Yersinia YopM protein required for interaction with the RSK1 and PRK2 host kinases and their requirement for interleukin-10 production and virulence. Infect. Immun. 78, 3529–3539 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirdita M., et al. , ColabFold—Making protein folding accessible to all. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2021.08.15.456425v2 (Accessed 29 September 2021).
- 26.Boland A., et al. , Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y.enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 15, 5191–5201 (1996). [PMC free article] [PubMed] [Google Scholar]
- 27.Evdokimov A. G., Anderson D. E., Routzahn K. M., Waugh D. S., Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: A leucine-rich repeat protein with the shortest repeating unit. J. Mol. Biol. 312, 807–821 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Berneking L., et al. , Immunosuppressive Yersinia effector YopM binds DEAD box helicase DDX3 to control ribosomal S6 kinase in the nucleus of host cells. PLoS Pathog. 12, e1005660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung L. K., et al. , The Yersinia virulence factor YopM hijacks host kinases to inhibit Type III effector-triggered activation of the pyrin inflammasome. Cell Host Microbe 20, 296–306 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratner D., et al. , The Yersinia pestis effector YopM inhibits pyrin inflammasome activation. PLoS Pathog. 12, e1006035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyer S. N., Wazer D. E., Band V., E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56, 4620–4624 (1996). [PubMed] [Google Scholar]
- 32.Chan H. M., Krstic-Demonacos M., Smith L., Demonacos C., La Thangue N. B., Acetylation control of the retinoblastoma tumour-suppressor protein. Nat. Cell Biol. 3, 667–674 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Davey N. E., Travé G., Gibson T. J., How viruses hijack cell regulation. Trends Biochem. Sci. 36, 159–169 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Bradley D., Beltrao P., Evolution of protein kinase substrate recognition at the active site. PLoS Biol. 17, e3000341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaidyanathan H., et al. , ERK MAP kinase is targeted to RSK2 by the phosphoprotein PEA-15. Proc. Natl. Acad. Sci. U.S.A. 104, 19837–19842 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreit M., Vertommen D., Gillet L., Michiels T., The interferon-inducible mouse Apolipoprotein L9 and Prohibitins cooperate to restrict Theiler’s virus replication. PLoS One 10, e0133190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrer N., et al. , Structural architecture of the nucleosome remodeler ISWI determined from cross-linking, mass spectrometry, SAXS, and modeling. Structure 26, 282–294 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Forné I., Ludwigsen J., Imhof A., Becker P. B., Mueller-Planitz F., Probing the conformation of the ISWI ATPase domain with genetically encoded photoreactive crosslinkers and mass spectrometry. Mol. Cell. Proteomics 11, 012088 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller-Planitz F., Crossfinder-assisted mapping of protein crosslinks formed by site-specifically incorporated crosslinkers. Bioinformatics 31, 2043–2045 (2015). [DOI] [PubMed] [Google Scholar]
- 40.F. Sorgeloos, Transcriptional profiling of HeLa M cells genetically modified using CRISPR sgRNA targeting RSK1, RSK2, RSK3 or PKR. NCBI SRA. https://www.ncbi.nlm.nih.gov/bioproject/768960. Deposited 6 October 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence-read data have been deposited in the National Center for Biotechnology Information Sequence Read Archive Database (https://www.ncbi.nlm.nih.gov/sra; BioProject accession no. PRJNA768960) (40).