Abstract
Background:
Chromosomal abnormalities are important causes of ventriculomegaly (VM). In mild and isolated cases of fetal VM, obstetricians rarely give clear indications for pregnancy termination. We aimed to calculate the incidence of chromosomal abnormalities and incremental yield of chromosomal microarray analysis (CMA) in VM, providing more information on genetic counseling and prognostic evaluation for fetuses with VM.
Methods:
The Chinese language databases Wanfang Data, China National Knowledge Infrastructure, and China Biomedical Literature Database (from January 1, 1991 to April 29, 2020) and English language databases PubMed, Embase, and Cochrane Library (from January 1, 1945 to April 29, 2020) were systematically searched for articles on fetal VM. Diagnostic criteria were based on ultrasonographic or magnetic resonance imaging (MRI) assessment of lateral ventricular atrium width: ≥10 to <15 mm for mild VM, and ≥15 mm for severe VM. Isolated VM was defined by the absence of structural abnormalities other than VM detected by ultrasonography or MRI. R software was used for the meta-analysis to determine the incidence of chromosomal abnormalities and incremental yield of CMA in VM, and the combined rate and 95% confidence interval (CI) were calculated.
Results:
Twenty-three articles involving 1635 patients were included. The incidence of chromosomal abnormalities in VM was 9% (95% CI: 5%–12%) and incremental yield of CMA in VM was 11% (95% CI: 7%–16%). The incidences of chromosomal abnormalities in mild, severe, isolated, and non-isolated VM were 9% (95% CI: 4%–16%), 5% (95% CI: 1%–11%), 3% (95% CI: 1%–6%), and 13% (95% CI: 4%–25%), respectively.
Conclusions:
Applying CMA in VM improved the detection rate of abnormalities. When VM is confirmed by ultrasound or MRI, obstetricians should recommend fetal karyotype analysis to exclude chromosomal abnormalities. Moreover, CMA should be recommended preferentially in pregnant women with fetal VM who are undergoing invasive prenatal diagnosis. CMA cannot completely replace chromosome karyotype analysis.
Keywords: Ventriculomegaly, Chromosome, Karyotype, Chromosomal microarray analysis, Meta-analysis
Introduction
Fetal ventriculomegaly (VM) is the most common abnormality that is detected during prenatal ultrasonographic assessment, with an incidence rate of approximately 1%.[1] VM may be caused by infections, malformations, chromosomal abnormalities, disorders of cerebrospinal fluid circulation, and injuries around the ventricles, and the prognosis varies from normal to very severe mental and behavioral disorders. The incidence of chromosomal abnormalities in VM remains controversial, and opinions on whether invasive prenatal diagnostic testing is required, particularly for mild and isolated VM, are divided. Conventional chromosome karyotype analysis such as G-banding has been the standard method for detecting a wide range of chromosomal abnormalities for several decades. However, this technique is limited to the detection of aneuploidies and chromosomal alterations >5 to 10 Mb; further, submicroscopic duplications and deletions, which are often associated with mental retardation and malformations, are not detectable by conventional karyotyping but can be successfully identified by chromosomal microarray analysis (CMA).[2]
This meta-analysis was conducted to investigate the incidence of chromosomal abnormalities in VM cases in China and internationally by traditional karyotype analysis. We also examined the incremental yield of CMA in VM.
Methods
Literature search
In April 2020, the Chinese language databases Wanfang Data, China National Knowledge Infrastructure, and China Biomedical Literature Database were searched by three researchers using the Chinese terms for “ventriculomegaly,” “fetal,” “chromosomes,” and “chromosomal microarray analysis.” Additionally, the English language databases PubMed, Embase, and Cochrane Library were searched by three researchers using combinations of the following keywords: “fetal,” ”fetus,” “Ventriculomegaly,” “Cerebral Ventriculomegaly,” “hydrocephaly,” and “chromosomal microarray analysis.” For all databases, the last search was run on April 29, 2020.
Study selection
The articles were screened by two researchers independently according to the inclusion and exclusion criteria. Any disagreements were resolved by discussion with the third researcher. Inclusion criteria are as follows: (1) Patients with a confirmed diagnosis of fetal VM included those with a singleton pregnancy, who underwent fetal ultrasonography or magnetic resonance imaging (MRI) assessment at mid- or late-gestation, and for whom the width of the fetal lateral ventricular atrium was ≥10 mm. (2) VM was classified as mild VM (width of one or both lateral ventricles ≥10 and <15 mm) and severe VM (≥15 mm). Isolated VM was defined by the absence of other abnormalities assessed by ultrasonography or MRI; otherwise, the case was defined as a non-isolated VM.[3] (3) Patients who underwent prenatal fetal karyotyping or CMA via amniocentesis or cordocentesis. (4) Articles in English or Chinese. Exclusion criteria are as follows: (1) Reviews and case reports; (2) studies performed on animals; (3) unconfirmed diagnosis and classification of VM; and (4) karyotyping or CMA was not performed.
Data extraction
Data were extracted and verified. In cases of disagreement, the decision was made after a consensus was reached between the three researchers. The characteristics of the included studies were recorded as follows: diagnostic criteria and classification of VM, name of first author, published year, country, number of VM, number of chromosome abnormalities (CAs), number of mild VM, severe VM, isolated VM, non-isolated VM, isolated mild VM, non-isolated mild VM, isolated severe VM, non-isolated severe VM and incremental yield of CMA. Type and number of chromosomal abnormalities.
Quality assessment of the selected articles
The Newcastle-Ottawa scale was used to evaluate the quality of the included literatures which mainly includes three categories of selection, comparability and outcomes, and eight items. The total score was nine stars, and a study with at least six stars was graded as high quality [Supplementary Table 1].
Statistical analysis
Meta-analysis of the incidence of chromosomal abnormalities and incremental yield of CMA in VM were performed using R software (R version 4.0.2) and meta package, and the combined rate and 95% confidence interval (CI) were calculated. The detection rate of CMA in VM in this study refers to the total detection rate consisting of pathogenic submicroscopic chromosomal abnormalities (copy number variations [CNVs]) and variants of unknown clinical significance. There are five methods for estimating sample rate in R software, including untransformed proportions, log transformation, logit transformation, arcsine transformation, Freeman-Tukey double arcsine transformation. Before meta-analysis, normality test is performed on the original rate after conversion according to the above estimation method, and the method close to the normal distribution is selected according to the result. Q and I2 tests were performed to detect heterogeneity. The meta-analysis was performed using the fixed-effects model and the random-effects model, when P > 0.10 and I2 ≤ 50%, we choose the result of the fixed-effects model, otherwise we choose the result of the random-effects model. Forest plots were used to describe the statistical results of the meta-analyses. Egger test was used to detect publication bias, P < 0.05 indicated the potential of publication bias. To further investigate the potential influencing factors of heterogeneity, we conducted a subgroup analysis of the whole study based on publication year and different countries. Furthermore, sensitivity analysis was performed by leaving out one study at a time when the heterogeneity was significant (I2 > 50%) to identify outlying studies.
Results
Characteristics of the included studies
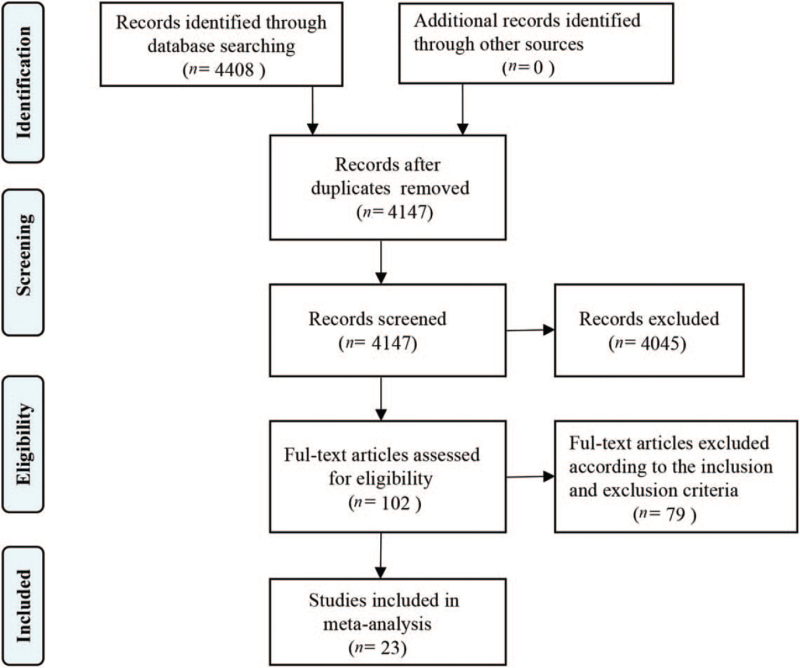
Articles were retrieved based on predefined search terms; among the 4408 Chinese and English language articles identified, we excluded 261 duplicated articles, 1360 reviews and case reports, and 2685 articles whose abstracts and titles were not relevant, finally selecting the remaining 102 articles. Upon further review, 23 articles were included in the final analysis [Figure 1].[4–26]Table 1 lists the number of cases and chromosomal abnormalities of the 17 included studies of conventional chromosome karyotype analysis. Table 2 describes the number of cases and the incremental yield of the eight included studies of CMA in VM. Among the 1635 patients evaluated in studies of conventional chromosome karyotype analysis, mild VM was observed in 784, severe VM in 138, isolated VM in 201, non-isolated VM in 316, isolated mild VM in 365, non-isolated mild VM in 200, isolated severe VM in 13, and non-isolated severe VM in 49.
Figure 1.
Flowchart of the study selection on chromosomal microarray analysis vs. karyotyping for fetal ventriculomegaly.
Table 1.
The number of cases and chromosomal abnormalities included in the studies of karyotyping.
| Mild VM | Severe VM | |||||
| First author year | N of cases | N of CAs | N of IVM (CA) | N of NIVM (CA) | N of IVM (CA) | N of NIVM (CA) |
| Tomlinson 1997[4] | 25 | 3 | 25 (3) | – | – | – |
| Vergani 1998[5] | 39 | 9 | 5 (2) | 34 (7) | – | – |
| Greco 2001[6] | 14 | 1 | 14 (1) | – | – | – |
| Gaglioti 2005[7] | 152 | 11 | – | – | – | – |
| Breeze 2005[8] | 18 | 2 | 18 (2) | – | – | – |
| Ouahba 2006[9] | 167 | 4 | 167 (4) | – | – | – |
| Weichert 2010[10] | 60 | 5 | – | – | – | – |
| Madazli 2011[11] | 67 | 2 | – | – | – | – |
| Sethna 2011[12] | 154 | 39 | – | – | – | – |
| Gezer 2014[13] | 140 | 7 | 24 (1) | 72 (3) | 11 (2) | 33 (1) |
| Chu 2016[14] | 57 | 5 | – | – | – | – |
| Gezer 2016[15] | 27 | 0 | – | – | – | – |
| Chang 2013[16] | 92 | 5 | 54 (3) | 34 (2) | 1 (0) | 3 (0) |
| Song 2010[17] | 132 | 25 | 58 (3) | 60 (20) | 1 (0) | 13 (2) |
| Hannon 2012[18] | 57 | 5 | – | – | – | – |
| Li 2017[19] | 341 | 21 | – | – | – | – |
| Donnelly 2014[20] | 93 | 12 | – | – | – | – |
Data are shown as number of cases. CAs: Chromosome abnormalities; IVM: Isolated VM; NIVM: Non-isolated VM; VM: Ventriculomegaly; –: Not mentioned.
Table 2.
The number of cases and ventriculomegaly status reported in the studies of CMA.
| First author year | N of cases | N of CMA | Mild VM | Severe VM | Total | |||
| N of IVM (CMA) | N of NIVM (CMA) | N of IVM (CMA) | N of NIVM (CMA) | N of IVM (CMA) | N of NIVM (CMA) | |||
| Li 2017[19] | 179 | 12 | – | – | – | – | 98 (6) | 81 (6) |
| Donnelly 2014[20] | 81 | 12 | – | – | – | – | – | – |
| Peng 2018[21] | 95 | 15 | – | – | – | – | – | – |
| Zhang 2015[22] | 50 | 13 | – | – | – | – | 21 (2) | 29 (11) |
| Shaffer 2012[23] | 272 | 14 | 84 (3) | 88 (7) | 66 (3) | 34 (1) | 150 (6) | 122 (8) |
| Hu 2017[24] | 154 | 13 | – | – | – | – | – | – |
| Duan 2019[25] | 101 | 8 | 101 (8) | – | – | – | 100 (8) | – |
| Bardin 2018[26] | 14 | 2 | – | – | – | – | – | – |
Data are shown as number of cases. CMA: chromosomal microarray analysis; IVM: Isolated VM; NIVM: Non-isolated VM; VM: Ventriculomegaly; –: Not mentioned.
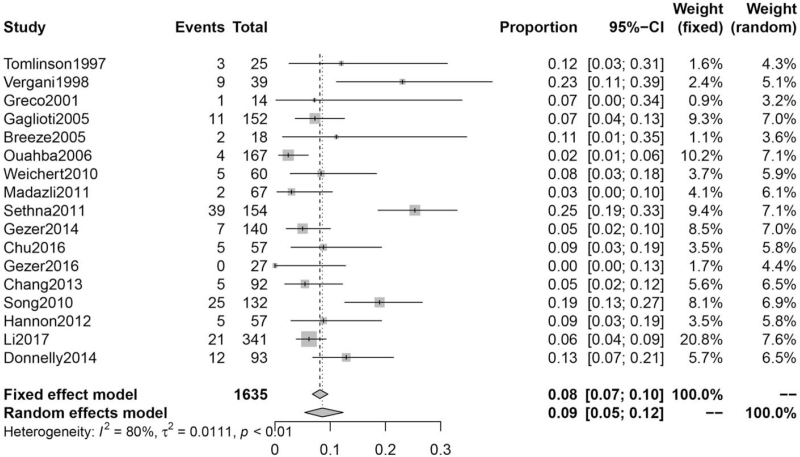
Incidence of chromosomal abnormalities analyzed by karyotype in VM
Seventeen articles were included. Heterogeneity testing across all studies yielded I2 = 80%. We choose the result of the random-effects model. The incidence of chromosomal abnormalities in VM was 9% (95% CI: 5%–12%) [Figure 2].
Figure 2.
Incidence of chromosomal abnormalities in VM. CI: Confidence interval; VM: Ventriculomegaly.
Incidence of chromosomal abnormalities in mild VM
A total of eleven articles with karyotype analyses for mild VM was included. Heterogeneity testing across all studies yielded I2 = 86%. We choose the result of the random-effects model. The incidence of chromosomal abnormalities in mild VM was 9% (95% CI: 4%–16%) following meta-analysis [Supplementary Figure 1].
Incidence of chromosomal abnormalities in severe VM
Six articles with karyotype analyses for severe VM were included. Heterogeneity testing across all studies yielded I2 = 0%. We choose the result of the fixed-effects model. The incidence of chromosomal abnormalities in severe VM was 5% (95% CI: 1%–11%) following meta-analysis [Supplementary Figure 2].
Incidence of chromosomal abnormalities in isolated VM
Five articles with karyotype analyses for isolated VM were included. Heterogeneity testing across all studies revealed I2 = 0%. We choose the result of the fixed-effects model. The incidence of chromosomal abnormalities in isolated VM was 3% (95% CI: 1%–6%) following meta-analysis [Supplementary Figure 3].
Incidence of chromosomal abnormalities in non-isolated VM
Five articles with karyotype analyses for non-isolated VM were included. Heterogeneity testing across all studies revealed I2 = 85%. We choose the result of the random-effects model. The incidence of chromosomal abnormalities in non-isolated VM was 13% (95% CI: 4%–25%) [Supplementary Figure 4].
Incidence of chromosomal abnormalities in isolated mild VM
A total of eight articles with karyotype analyses for isolated mild VM were included. Heterogeneity testing revealed I2 = 58%. We choose the result of the random-effects model. The incidence of chromosomal abnormalities in isolated mild VM was 8% (95% CI: 4%–15%) [Supplementary Figure 5].
Incidence of chromosomal abnormalities in non-isolated mild VM
Four articles with karyotype analyses for non-isolated mild VM were included. Heterogeneity testing revealed I2 = 88%. We choose the result of the random-effects model. The incidence of chromosomal abnormalities in non-isolated mild VM was 14% (95% CI: 3%–31%).
Incidence of chromosomal abnormalities in isolated severe VM
Three articles with karyotype analyses for isolated severe VM were included. Heterogeneity testing revealed I2 = 0%. We choose the result of the fixed-effects model. The incidence of chromosomal abnormalities in isolated severe VM was 14% (95% CI: 0%–34%).
Incidence of chromosomal abnormalities in non-isolated severe VM
Three articles with karyotype analyses for non-isolated severe VM were included. Heterogeneity testing revealed I2 = 23%. We choose the result of the fixed-effects model. The incidence of chromosomal abnormalities in non-isolated severe VM was 5% (95% CI: 1%–13%).
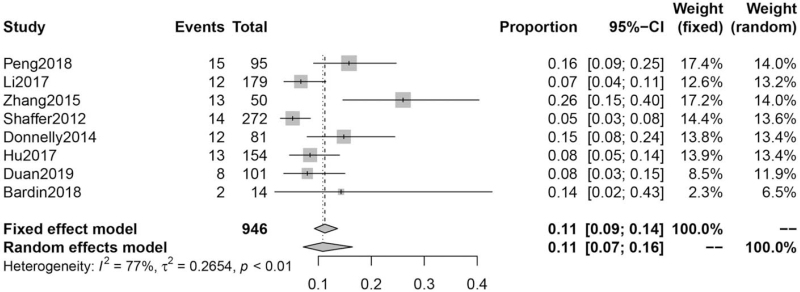
Incremental yield of CMA in VM
A total of eight articles were included. Heterogeneity testing revealed I2 = 77%. We choose the result of the random-effects model. The incremental yield of CMA in VM was 11% (95% CI: 7%–16%) [Figure 3].
Figure 3.
Incremental yield of CMA in VM. CI: Confidence interval; CMA: Chromosomal microarray analysis; VM: Ventriculomegaly.
Analysis of publication bias
The publication bias was detected by Egger test and the P value of Egger test in each analysis was all >0.05, thereby indicating an absence of publication bias in the included studies.
Subgroup analysis
Subgroup analysis based on publication year and different countries showed that the heterogeneity was not related to publication year and different countries [Supplementary Figure 6–8].
Sensitivity analysis
Sensitivity analyses were conducted by excluding each of the included studies individually to determine whether the combined results of other studies were stable. The original rate conforms to normal distribution after conversion according to arcsine transformation and Freeman-Tukey double arcsine transformation. The results revealed no significant change in the combined results with two different data conversions, indicating that the meta-analysis results were stable and reliable [Supplementary Figure 9–10].
Discussion
China has a high incidence of congenital anomalies and the incidence of birth defects in my country was 5.6%.[27] Particularly, anomalies of the central nervous system constitute the leading cause of fetal malformations. Their prognosis varies from normal to very severe mental and behavioral disorders. The most common cause of ventricular enlargement is dilatation of the lateral ventricles, which is closely related to the development of mental and psychomotor skills in children.[28] As a result, the lateral ventricles represent a routine parameter in fetal ultrasound examination. Chromosomal abnormalities, particularly trisomy 21, are important factors for ventricular enlargement.[29] Karyotype analysis is the preferred cytogenetic test in prenatal screening, as it can detect aneuploidy and structural abnormalities involving large chromosomal fragments. The incidence of chromosomal abnormalities in VM remains controversial both in China and internationally. Previous reports involving instances of karyotyping performance in fetal VM cases showed that the incidence of aneuploidy in VM was between 0% and 14%.[30]
Huang et al[29] retrospectively analyzed 150 cases of VM identified via ultrasonography and for which karyotyping data were available, revealing a chromosomal abnormality rate of 8%. This is close to the results of the present study, in which the rate of VM chromosomal abnormalities was determined to be 9%. Peng et al[21] conducted karyotyping in 109 women with a singleton pregnancy complicated by VM and showed that the rate of chromosomal abnormalities in the fetuses was 13%. Gezer et al[13] showed that the detection rate of abnormalities using karyotyping was 7% in fetuses with severe VM, and 4% in those with mild VM. Additionally, they reported a higher incidence of chromosomal abnormalities in fetuses with isolated VM (9%) than in those with non-isolated VM (4%). These results differ from those of the present study, in which the incidence of chromosomal abnormalities in mild VM was 9% and hence higher than the 5% observed in severe VM. The reason for this may be that the sample size of severe VM was small. In addition, the incidence of chromosomal abnormalities in non-isolated VM was 13%, which was higher than that in isolated VM (3%). Specifically, the incidence of chromosomal abnormalities was higher in non-isolated mild VM (14%) than in isolated mild VM (8%), these results indicate that patients with other structural abnormalities detected by ultrasonography or MRI are at an increased risk of chromosomal abnormalities. Furthermore, we found that the most common abnormal karyotype was an abnormal chromosome number, with an incidence of 63.1%. Most cases were due to trisomy 21 (32.5%) and, to a lesser extent, trisomy 18 and trisomy 13 (5.7% each). These results are consistent with the findings reported by Song et al.[17]
CMA is a molecular genetic testing method that has rapidly advanced in recent years. This method can detect not only subtle chromosomal structural abnormalities such as chromosome microdeletion and microduplication but also partial or entire chromosome uniparental diploidy, overcoming the limitations of traditional karyotyping.[31] CMA includes comparative genomic hybridization and single-nucleotide polymorphism arrays, and both techniques enable detection of CNVs at high resolution.
CMA can significantly improve the detection rate of chromosomal diseases in the prenatal diagnosis of B-ultrasound abnormality.[32,33] In the Shaffer et al's[23] model, as comparative genomic hybridization was performed to detect 272 fetuses with normal karyotype of lateral ventricle dilation, the rate of chromosomal abnormalities increased by 5%, whereas in that evaluated by Zhang et al,[22] there was an increase by 26%. The reason for the high detection rate may be the small sample size. Eight articles in the present study focused on the association between CNVs and VM, and R software was used for meta-analysis of the incremental yield of CMA in VM; the combined rate was 11% (95% CI: 7%–16%).
CMA has been used as a prenatal diagnostic tool in many genetic centers in recent years to confirm the diagnosis of derivative chromosomal abnormalities that cannot be diagnosed by karyotype analysis, determine the size, nature, and source of abnormal fragments, and provide a more accurate genetic basis for prenatal counseling and fetal prognosis evaluation.
In 2013, the American College of Obstetricians and Gynecologists recommended that CMA tests should be used as an alternative to traditional karyotype analysis when fetal ultrasound abnormalities are present and prenatal diagnosis is required.[34] However, CMA cannot detect fetal chromosomal structural abnormalities, and all methods have some limitations. The combined application of multiple technologies can provide doctors with more objective and comprehensive clinical information. Therefore, CMA cannot completely replace chromosome karyotype analysis.
In an article jointly published by the American Academy of Medical Genetics and Clinical Genome Resources in 2020, CNV results were classified into five types: pathogenic CNV, likely pathogenic CNV, variants of uncertain significance (VOUS), likely benign CNV, and benign CNV.[35] Since prenatal diagnosis is limited by limited clinical information, it is difficult to explain the clinical relevance of CNV. Many CNVs with unclear correlation with clinical phenotypes were detected. Fragments of CNVs involve genes with unclear clinical phenotype function or unclear dose effect, or both exist in the normal phenotype population and have reports of pathological phenotypes.[36] The emergence of VOUS has brought tremendous pressure to pregnant women and their families, as well as significant challenges to clinicians. Therefore, genetic counseling is related to the survival of the fetus. This crucial link must be handled carefully. Understanding the clinical relevance of CNVs is a complex and continually evolving process. A single formula or algorithm for CNV interpretation cannot be a substitute for adequate training in genetics and sound clinical judgment. Clinical reporting of constitutional CNVs should be performed by individuals with appropriate professional training and certification. In addition, given the complexity of CNV interpretation, the different laboratory methodologies utilized for CNV characterization, as well as the evaluation of additional family members, in an ideal laboratory setting for CNV analysis, should include both cytogenetic and molecular genetic expertise.[35] At present, the relevant clinical data in China are insufficient, and interpretation of the results is limited by medical progress. It remains difficult to judge the clinical significance of micro-deletion or micro-duplication of certain chromosomal segments. Therefore, more data must be accumulated to provide guidelines that enable pregnant women to make the most suitable choices and contribute to the reduction of birth defects.
The strengths of the present study are as follows. First, sufficient literature retrieval and comprehensive research were performed. Compared with a single original study, the sample size was greatly increased, thus the estimated incidence of chromosomal abnormalities is more accurate. Second, the included 23 studies are all high-quality studies, which increase the evidence-based value of the study.
A limitation of this study is represented by the heterogeneity of the included articles, as the original study involved a single group, and stability differed in the research with two groups. In addition to changing the effect model, we conducted a sensitivity analysis, which showed that when the included studies were individually excluded, there was no significant change in the combined results or total combined values, indicating that our results were stable and reliable. In addition, subgroup analysis was performed according to the year of publication of the included studies and different countries, and the I2 suggested that the heterogeneity was unrelated to the publication year and countries. All subjects included in the study were identified as singleton pregnancy patients with fetal VM. Since various data for each individual were unavailable, some influencing factors (such as maternal age and gestational age at diagnosis) were not thoroughly analyzed. Large-scale multicenter studies based on individual case data are needed to confirm our findings.
Chromosomal abnormalities are one of the most important causes of VM. The effects of VM can last until the postpartum period and affect brain development in newborns, leading to varying degrees of developmental abnormalities in the nervous system. For mild and isolated cases of VM, obstetricians rarely give clear indications for the termination of pregnancy. In this study, the incidence of CAs in traditional karyotype analysis in VM and incremental yield of CMA in VM were analyzed by meta-analysis. The results showed that chromosomal abnormalities exist in mild to severe VM, and that CMA increases their detection rate. In addition, CMA can further clarify the diagnosis of derivative chromosomal abnormalities that cannot be diagnosed by karyotyping, as well as clarify the size, nature, and source of abnormal fragments. At present, karyotype analysis technology cannot meet the clinical needs of prenatal diagnosis of chromosomal diseases. Thus, it is necessary to combine CMA technology with prenatal diagnosis to carry out a timely and accurate diagnosis of chromosomal diseases such as chromosome segment duplication and loss, chromosome number abnormality, pathogenic genome CNVs, etc. Therefore, when VM is confirmed by ultrasound or MRI, obstetricians should recommend fetal karyotype analysis to exclude chromosomal abnormalities. Moreover, follow-up ultrasound examinations should be performed to observe the progression or resolution of VM. Our results suggest that the incremental yield of CMA in VM was 11%. For fetuses with VM, further CMA analysis is recommended preferentially in pregnant women with fetal VM who are undergoing invasive prenatal diagnosis. This is expected to increase the survival rate of fetuses and newborns and improve overall population health. However, CMA increases the opportunity to detect VOUS. Therefore, professional geneticists should provide genetic counseling for the advantages and limitations of CMA and the test results. Finally, because of the inherent limitations of meta-analysis, such as the heterogeneity of individual studies, large-scale multi-center studies are required to confirm the current findings and provide a more accurate genetic basis for prenatal consultation and fetal prognosis assessment.
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Sun Y, Zhang W, Wang Z, Guo L, Shi S. Chromosomal microarray analysis vs. karyotyping for fetal ventriculomegaly: a meta-analysis. Chin Med J 2022;135:268–275. doi: 10.1097/CM9.0000000000001683
Supplemental digital content is available for this article.
References
- 1.Benkarim OM, Hahner N, Piella G, Gratacos E, González Ballester MA, Eixarch E, et al. Cortical folding alterations in fetuses with isolated non-severe ventriculomegaly. NeuroImage Clin 2018; 18:103–114. doi: 10.1016/j.nicl.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evangelidou P, Sismani C, Ioannides M, Christodoulou C, Koumbaris G, Kallikas I, et al. Clinical application of whole-genome array CGH during prenatal diagnosis: study of 25 selected pregnancies with abnormal ultrasound findings or apparently balanced structural aberrations. Mol Cytogenet 2010; 3:24.doi: 10.1186/1755-8166-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pisapia JM, Sinha S, Zarnow DM, Johnson MP, Heuer GG. Fetal ventriculomegaly: diagnosis, treatment, and future directions. Childs Nerv Syst 2017; 33:1113–1123. doi: 10.1007/s00381-017-3441-y. [DOI] [PubMed] [Google Scholar]
- 4.Tomlinson MW, Treadwell MC, Bottoms SF. Isolated mild ventriculomegaly: associated karyotypic abnormalities and in utero observations. J Matern Fetal Med 1997; 6:241–244. doi: 10.1002/(SICI)1520-6661(199707/08)6:4<241::AID-MFM11>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 5.Vergani P, Locatelli A, Strobelt N, Cavallone M, Ceruti P, Paterlini G, et al. Clinical outcome of mild fetal ventriculomegaly. Am J Obstet Gynecol 1998; 178:218–222. doi: 10.1016/S0002-9378(98)80003-3. [DOI] [PubMed] [Google Scholar]
- 6.Greco P, Vimercati A, De Cosmo L, Laforgia N, Mautone A, Selvaggi L. Mild ventriculomegaly as a counselling challenge. Fetal Diagn Ther 2001; 16:398–401. doi: 10.1159/000053947. [DOI] [PubMed] [Google Scholar]
- 7.Gaglioti P, Danelon D, Bontempo S, Mombrò M, Cardaropoli S, Todros T. Fetal cerebral ventriculomegaly: outcome in 176 cases. Ultrasound Obstet Gynecol 2005; 25:372–377. doi: 10.1002/uog.1857. [DOI] [PubMed] [Google Scholar]
- 8.Breeze ACG, Dey PK, Lees CC, Hackett GA, Smith GCS, Murdoch EM. Obstetric and neonatal outcomes in apparently isolated mild fetal ventriculomegaly. J Perinat Med 2005; 33:236–240. doi: 10.1515/JPM.2005.043. [DOI] [PubMed] [Google Scholar]
- 9.Ouahba J, Luton D, Vuillard E, Garel C, Gressens P, Blanc N, et al. Prenatal isolated mild ventriculomegaly: outcome in 167 cases. Br J Obstet Gynaecol 2006; 113:1072–1079. doi: 10.1111/j.1471-0528.2006.01050.x. [DOI] [PubMed] [Google Scholar]
- 10.Weichert J, Hartge D, Krapp M, Germer U, Gembruch U, Axt-Fliedner R. Prevalence, characteristics and perinatal outcome of fetal ventriculomegaly in 29,000 pregnancies followed at a single institution. Fetal Diagn Ther 2010; 27:142–148. doi: 10.1159/000304735. [DOI] [PubMed] [Google Scholar]
- 11.Madazli R, Sal V, Erenel H, Gezer A, Ocak V. Characteristics and outcome of 102 fetuses with fetal cerebral ventriculomegaly: experience of a university hospital in Turkey. J Obstet Gynaecol 2011; 31:142–145. doi: 10.3109/01443615.2010.541304. [DOI] [PubMed] [Google Scholar]
- 12.Sethna F, Tennant PWG, Rankin J, Robson SC. Prevalence, natural history, and clinical outcome of mild to moderate ventriculomegaly. Obstet Gynecol 2011; 117:867–876. doi: 10.1097/AOG.0b013e3182117471. [DOI] [PubMed] [Google Scholar]
- 13.Gezer C, Ekin A, Ozeren M, Taner CE, Ozer O, Koc A, et al. Chromosome abnormality incidence in fetuses with cerebral ventriculomegaly. J Obstet Gynaecol 2014; 34:387–391. doi: 10.3109/01443615.2014.896885. [DOI] [PubMed] [Google Scholar]
- 14.Chu N, Zhang Y, Yan Y, Ren Y, Wang L, Zhang B. Fetal ventriculomegaly: pregnancy outcomes and follow-ups in ten years. Biosci Trends 2016; 10:125–132. doi: 10.5582/bst.2016.01046. [DOI] [PubMed] [Google Scholar]
- 15.Gezer NS, Gezer C, Ekin A, Yesilirmak DC, Solmaz U, Dogan A, et al. Obstetric and neurodevelopmental outcome in fetal cerebral ventriculomegaly. Clin Exp Obstet Gynecol 2016; 43:490–494. doi: 10.12891/ceog.2138.2016. [PubMed] [Google Scholar]
- 16.Chang QX, Xiong L, Qiu Y, Chen C, Yu Y. Clinical significance and outcomes of fetal ventriculomegaly (in Chinese). Chin J Perinat Med 2013; 16:142–147. doi: 10.3760/cma.j.issn.1007-9408.2013.03.003. [Google Scholar]
- 17.Song HL, Chen BJ, Fang Q, Luo YM, Chen YZ, Chen YH. Relationship between fetal ventriculomegaly and chromosomal abnormalities (in Chinese). Chin J Pract Gynecol Obstet 2010; 26:925–928. doi: 10.3969/j.issn.1674-1870.2012.01.022. [Google Scholar]
- 18.Hannon T, Tennant PWG, Rankin J, Robson SC. Epidemiology, natural history, progression, and postnatal outcome of severe fetal ventriculomegaly (in Chinese). Obstet Gynecol 2012; 120:1345–1353. doi: 10.1097/aog.0b013e3182732b53. [DOI] [PubMed] [Google Scholar]
- 19.Li ZZ, Fu F, Lei TY, Li R, Jing XY, Yang X, et al. Application of chromosome microarray analysis for the delineation of pathogenesis for fetal, ventriculomegaly (in Chinese). Chin J Med Genet 2017; 34:576–582. doi: 10.3760/cma.j.issn.1003-9406.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 20.Donnelly JC, Platt LD, Rebarber A, Zachary J, Grobman WA, Wapner RJ. Association of copy number variants with specific ultrasonographically detected fetal anomalies. Obstet Gynecol 2014; 124:83–90. doi: 10.1097/AOG.0000000000000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng YX, Qiu YW, Chang QX, Yu YH, Zhong M, Li KR. Clinical value of genome-wide chromosome microarray technique in diagnosis of fetal cerebral ventriculomegaly (in Chinese). J South Med Univ 2018; 38:353–357. doi: 10.3969/j.issn.1673-4254.2018.03.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang ZQ, Xie JJ, Wu JZ, Chen XD, Lin SB, Ji YJ, et al. Chromosomal microarray analysis for lateral ventriculomegaly in fetus (in Chinese). Chin J Med Genet 2015; 32:789–792. doi: 10.3760/cma.j.issn.1003-9406.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Shaffer LG, Rosenfeld JA, Dabell MP, Coppinger J, Bandholz AM, Ellison JW, et al. Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenat Diagn 2012; 32:986–995. doi: 10.1002/pd.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu P, Wang Y, Sun R, Cao L, Chen X, Liu C, et al. Copy number variations with isolated fetal ventriculomegaly (in Chinese). Curr Mol Med 2017; 17:133–139. doi: 10.2174/1566524017666170303125529. [DOI] [PubMed] [Google Scholar]
- 25.Duan HL, Zhu XY, Zhu YJ, Wu X, Zhao GF, Wang WJ, et al. The application of chromosomal microarray analysis to the prenatal diagnosis of isolated mild ventriculomegaly. Taiwan J Obstet Gynecol 2019; 58:251–254. doi: 10.1016/j.tjog.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Bardin R, Hadar E, Haizler-Cohen L, Gabbay-Benziv R, Meizner I, Kahana S, et al. Cytogenetic analysis in fetuses with late onset abnormal sonographic findings. J Perinat Med 2018; 46:975–982. doi: 10.1515/jpm-2017-0071. [DOI] [PubMed] [Google Scholar]
- 27.Li FJ, Yao XY, Zhang YP. Research progress of copy number variation sequencing in prenatal diagnosis (in Chinese). J Int Obstet Gynecol 2021; 48:75–78. doi: 10.12280/gjfckx.20200527. [Google Scholar]
- 28.Li R, Chen YH, Gao XC, Zhao WH, Tian GY, Li XN. Effects of isolated fetal ventriculomegaly on mental and psychomotor development after birth (in Chinese). Chin J Child Health Care 2016; 24:655–658. doi: 10.11852/zgetbjzz2016-24-06-30. [Google Scholar]
- 29.Huang YM, Huang DP, Zhong W, Jiang W, Chen Y, Chen YR. Diagnostic ultrasonography of fetal ventricle dilatation combined with malformations and its relationship with chromosomal abnormalities (in Chinese). Chin J Med Imaging 2017; 25:617–622. doi: 10.3969/j.issn.1005-5185.2017.08.015. [Google Scholar]
- 30.Lam SJ, Kumar S. Evolution of fetal ventricular dilatation in relation to severity at first presentation. J Clin Ultrasound 2014; 42:193–198. doi: 10.1002/jcu.22124. [DOI] [PubMed] [Google Scholar]
- 31.Song TT, Wan SN, Li Y, Xu Y, Zheng YY, Dang YH, et al. Application value of chromosomal microarray analysis in prenatal diagnosis of lateral ventriculomegaly fetuses (in Chinese). Med J Chin PLA 2017; 42:902–908. doi: 10.11855/j.issn.0577-7402.2017.10.12. [Google Scholar]
- 32.Wapner RJ, Martin CL, Levy B, Ballif BC, Eng CM, Zachary JM, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med 2012; 367:2175–2184. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pons L, Till M, Alix E, Abel C, Boggio D, Bordes A, et al. Prenatal microarray comparative genomic hybridization: experience from the two first years of activity at the Lyon university-hospital. J Gynecol Obstet Hum Reprod 2017; 46:275–283. doi: 10.1016/j.jogoh.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 34.American College of Obstetricians and Gynecologists Committee on Genetics. Committee opinion no. 581: the use of chromosomal microarray analysis in prenatal diagnosis. Obstet Gynecol 2013; 122:1374–1377. doi: 10.1097/01.AOG.0000438962.16108.d1. [DOI] [PubMed] [Google Scholar]
- 35.Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med 2020; 22:245–257. doi: 10.1038/s41436-019-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang YL, Wei QQ, Meng H, Zhou XY, Hao N, Xu ZH, et al. Analysis of variations with clinically unambiguous significance in prenatal CMA for 308 patients with high-risk pregnancy (in Chinese). J Reprod Med 2017; 26:863–868. doi: 10.3969/j.issn.1004-3845.2017.09.003. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.