Abstract
Liver fibrosis is a common pathologic stage of the development of liver failure. It has showed that exosomes loaded with therapeutic circRNAs can be manufactured in bulk by exosome secreted cells in vitro, thus enabling personalized treatment. This study aimed to investigate the role of exosome-based delivery of circDIDO1 in liver fibrosis. Levels of genes and proteins were examined by qRT-PCR and Western blot. Cell proliferation, apoptosis, and cell cycle were analyzed by using cell counting kit-8 (CCK-8) assay, EdU assay, and flow cytometry, respectively. The binding between circDIDO1 and miR-141-3p was confirmed by dual-luciferase reporter, RNA pull-down and RIP assays. Exosomes were isolated by ultracentrifugation, and qualified by transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA) and Western blot. CircDIDO1 overexpression or miR-141-3p inhibition suppressed the proliferation, reduced pro-fibrotic markers, and induced apoptosis as well as cell cycle arrest in hepatic stellate cells (HSCs) by blocking PTEN/AKT pathway. Mechanistically, circDIDO1 acted as an endogenous sponge for miR-141-3p, further rescue experiments showed that circDIDO1 suppressed HSC activation by targeting miR-141-3p. Extracellular circDIDO1 could be incorporated into exosomes isolated from mesenchymal stem cells (MSCs), and transmitted to HSCs to restrain HSC activation. Clinically, low levels of serum circDIDO1 in exosome were correlated with liver failure, and serum exosomal circDIDO1 had a well diagnostic value for liver fibrosis in liver failure patients. Transfer of circDIDO1 mediated by MSC-isolated exosomes suppressed HSC activation through the miR-141-3p/PTEN/AKT pathway, gaining a new insight into the prevention of liver fibrosis in liver failure patients.
Keywords: circDIDO1, miR-141-3p, mesenchymal stem cells, exosomes, liver fibrosis
Background
Liver fibrosis represents a common pathologic process of end-stage liver diseases, which may progress to cirrhosis, liver failure, hepatocellular carcinoma, and ultimately death (Zhang et al., 2016; Ma et al., 2017). It is featured by the excessive accumulation of extracellular matrix (ECM) proteins and decreased matrix remodeling (Lodder et al., 2015). Hepatic stellate cells (HSCs) are considered as the main source of fibrogenic cell type in the liver (Higashi et al., 2017). During liver fibrosis process, HSCs are activated by fibrogenic mediators and cytokines; moreover, activated HSCs lead to the production of α-smooth muscle actin (α-SMA) and the section of collagens, which is followed by matrix deposition and fibrosis (Zhang et al., 2016; Shi et al., 2017; Tsuchida & Friedman, 2017). Therefore, prevention of HSC activation is of great significance for attenuating liver fibrosis, and even liver failure.
Circular RNAs (circRNAs) are covalently closed single stranded circular transcripts that lack the 3′ and 5′ ends (Yu et al., 2018). CircRNAs are widespread expressed across various species, besides that, they are more stable, specific, and highly organized relative to other types of RNA (Vo et al., 2019). Moreover, it has been reported that circRNAs have crucial effects on the regulation of cellular biological processes (Hua et al., 2019; Kristensen et al., 2019; Wu et al., 2019). Therefore, circRNAs have obvious advantages as biomarkers for clinical diseases (Liu et al., 2020). Recently, mounting evidence shows the effect of circRNAs on HSC activation, which is essential for the pathogenesis of liver fibrosis. For example, hsa_circ_0070963 overexpression suppressed HSC activation with the reductions of collagen I and α-SMA by sponging miR-223-3p (Ji et al., 2020). Wang et al. showed that serum circMTO1 had a well diagnostic value for liver fibrosis in chronic hepatitis B (CHB) patients, and circMTO1 restoration restrained HSC activation by miR-17-5p/Smad7 axis (Wang et al., 2019). CircDIDO1 (ID: hsa_circ_0061137) is a circRNA derived from 2 to 6 exons of DIDO1 gene, a recent study showed that circDIDO1 expression was decreased in irradiated HSCs (Chen et al., 2017). However, the potential functions and underlying mechanisms of circDIDO1 in liver fibrosis remain elusive.
Exosomes are one of the vesicle types described as 40–150 nm diameter, which are actively released by different mammalian cell types to the extracellular environment (Zhou et al., 2018). Exosomes are natural information carriers that deliver exosomal surface proteins and bio-functional cargoes, including lipids, proteins, or nucleic acids, to the target-cells by docking and fusing to the membrane (Gonzalez-Begne et al., 2009). Recently, the use of exosomes as in vivo delivery vehicles loading with therapeutic circRNA is growing interest due to their universal and stable presence in body fluids and capability of crossing the blood–brain barrier (BBB) (Batrakova & Kim, 2015; Chen et al., 2020; Kalluri & LeBleu, 2020). Thus, exosome-based transfer of circRNA may be an ideal therapeutic strategy for disease treatment.
Here, this work aimed to investigate the functions and mechanisms of circDIDO1 in liver fibrosis. Moreover, we explored whether exosome-based delivery of circDIDO1 had the therapeutic potential for liver fibrosis.
Materials and methods
Patients and blood collection
The blood samples (7 mL) were collected from 30 patients with clinically diagnosed liver failure or 30 sex- and age-matched health individuals without hepatopathy at Henan Provincial People’s Hospital, Zhengzhou University People’s Hospital, Zhengzhou, China. After centrifugation at 3000×g for 10 min, serum samples were obtained and immediately preserved at −80 °C for exosome (Exo) isolation. This work was approved by the Ethics Committee of Henan Provincial People’s Hospital, Zhengzhou University People’s Hospital, Zhengzhou, China (approval number 2021215), and manipulated in line with the Declaration of Helsinki. Written informed consent was collected from all subjects before sample collection.
Cell culture
The human HSC cell line LX2 was purchased from the Cell Center of Shanghai Institutes for Biological Sciences, and then cultured in the Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies, Wuhan, China). Human bone marrow-derived mesenchymal stem cells (MSCs) were obtained from Jining Cell Culture Center (Shanghai, China) and grown in the α-modified Eagle’s medium (MEM) (Life Technologies, Wuhan, China). All mediums were supplemented with 10% fetal bovine serum (FBS; Life Technologies, Wuhan, China), and 1% streptomycin–penicillin antibiotics, and cells were then maintained in a humidified incubator with 5% CO2 at 37 °C. Four to seven passages of LX2 and MSCs were used for further experiments.
Total RNA isolation, RNase R digestion, and actinomycin D treatment
Total RNAs were isolated by Trizol reagent (Invitrogen, Carlsbad, CA) and qualified by NanoDrop-1000 apparatus (NanoDrop Technologies, Wilmington, DE). For actinomycin D treatment, LX2 cells were incubated with 5 μg/mL actinomycin D or dimethylsulfoxide (control) to block the de novo RNA synthesis, and half-life of mRNA was examined by transcript levels at indicated time points. For the RNase R digestion, approximately 3 µg of isolated RNAs were mixed with 4 U/μg of RNase R (Epicentre Biotechnologies, Madison, WI) for 3 h at 37 °C, then qRT-PCR was performed to determine the expression stability of circDIDO1 as compared to GAPDH mRNA.
Reverse transcription reaction and quantitative real-time PCR
For circRNA and mRNA analyses, random or Oligo(dT)18 primers were used for the synthesis of first strand cDNA with the Prime Script RT Reagent Kit (Takara, Dalian, China). For miRNA analysis, cDNA was synthesized by using the High Capacity cDNA Reverse Transcription Kit (Takara, Dalian, China). Then, qRT-PCR was performed by using the SYBR Premix Ex Taq II (Takara, Dalian, China) with GAPDH or U6 as an internal standard control. The specific primers used for qRT-PCR are shown in Table 1.
Table 1.
Primers sequences used for qRT-PCR.
| Name | Primers for qRT-PCR (5′–3′) | |
|---|---|---|
| hsa_circ_0061137 (circDIDO1) | Forward | CTTGTGAGAGCAGCACGC |
| Reverse | CGTGGAGTAAGCTCATTTATACAAC | |
| miR-141-3p | Forward | GCCGAGTAACACTGTCTGGT |
| Reverse | CTCAACTGGTGTCGTGGAGT | |
| miR-128-3p | Forward | TCCGAGTCACAGTGAACCG |
| Reverse | CTCAACTGGTGTCGTGGAGT | |
| miR-188-5p | Forward | TTCCGAGCATCCCTTGCATG |
| Reverse | CTCAACTGGTGTCGTGGAGT | |
| miR-942-5p | Forward | TCGGCAGGTCTTCTCTGTTTTG |
| Reverse | CTCAACTGGTGTCGTGGAGT | |
| miR-217 | Forward | TTCCGAGTACTGCATCAGGAAC |
| Reverse | CTCAACTGGTGTCGTGGAGT | |
| GAPDH | Forward | GACAGTCAGCCGCATCTTCT |
| Reverse | GCGCCCAATACGACCAAATC | |
| U6 | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT |
Subcellular fractionation
Cytoplasmic and nuclear circDIDO1 were determined following the instruction of the PARIS kit (Life Technologies, Wuhan, China). Then levels of circDIDO1 in nuclear and cytoplasmic fractions were detected by qRT-PCR with GAPDH or U6 as the cytoplasmic or nuclear control transcript.
Cell transfection
The full-length circDIDO1 was cloned into pLC5-ciR vector (Invitrogen, Carlsbad, CA) to overexpress circDIDO1 with empty pLC5-ciR as negative control (NC) (vector). The mimic or inhibitor of miR-141-3p and the corresponding NC (NC mimic or NC inhibitor) were provided by Genema (Shanghai, China). Then, transient transfection was performed to introduce plasmids or oligonucleotides into corresponding cells using lipofectamine 2000 provided by Invitrogen (Carlsbad, CA).
Isolation and identification of exosome (Exo)
The exosomes were isolated from the supernatants of MSC cultures by differential centrifugation. After 48 h of transfection, MSCs were cultured in DMEM-containing exosome-depleted serum. Seventy-two hours later, the conditioned culture medium was collected and centrifuged at 4000×g for 10 min at 4 °C to remove cell fragments, and then again at 17,000×g for 1 h at 4 °C. After filtering with a 0.22-μm pore filter, the supernatants were ultracentrifuged at 200,000×g for 1 h at 4 °C. The pellets were collected, washed using phosphate-buffered saline (PBS) followed by a second ultracentrifugation at 200,000×g for 1 h at 4 °C. Then, exosomes were collected and resuspended in ice-cold PBS. For exosome isolation from serum, serum samples were subjected to centrifugation at 3000×g for 15 min to eliminate cells and cellular debris, and then mixed with a quarter volume of Exoquick exosome precipitation solution (System Biosciences, Palo Alto, CA). After 24 h of refrigeration, the mixture was centrifuged at 1500×g for 30 min, the supernatant was removed, and exosome pellets were suspended in 1× PBS. The pelleted exosomes resuspended in 100 μL PBS were subjected to RNA extraction with mirVana miRNA isolation kit (Life Technologies, Wuhan, China), and transmission electron microscopy (TEM) for morphology analysis as described previously (FEI, Hillsboro, OR) (×200) (Cooks et al., 2018). The size distribution and concentration of exosomes were analyzed by nanoparticle-tracking analysis with a NanoSight NS300 instrument (Malvern Panalytical, Malvern, UK) following the manufacturer’s instructions. The exosome markers (TSG101, CD63, and CD81) were detected by Western blot analysis with RIPA lysis buffer.
For cell treatment, LX2 cells were placed at a six-well plate with exosome-depleted culture medium plus 10% FBS at a density of 5 × 105 cells/well, and then incubated with 50 μg/mL exosomes isolated from MSCs cells transfected with circDIDO1 or vector.
Cell counting kit-8 (CCK-8) assay
After indicated transfection or treatment, LX2 cells were seeded in 96-well plates at a density of 5 × 104 cells/well, and mixed with 10 μL CCK8 solution. 2 h later, cell viability was assessed by reading the absorbance value at 450 nm.
5-Ethynyl-2′-deoxyuridine (EDU) assay
LX2 cells were incubated with 50 mM EDU medium diluent (RiboBio, Guangzhou, China) in 96-well plates for 3 h. Following being fixed and permeabilized, 1 × Apollo® reaction cocktail (100 μL) was employed to react with the EdU for 30 min. Cell nuclei were stained with DAPI. Finally, EdU positive cells were imaged by the fluorescence microscopy (Bio-Rad, Hercules, CA) and analyzed by ImageJ software (Bethesda, MD).
Flow cytometry
For cell cycle analysis, LX2 cells were fixed by 70% ice-cold ethanol for 48 h at 4 °C and then stained with propidium iodide (PI). Thereafter, cell cycle was evaluated by FACSCanto II flow cytometry (BD Biosciences, Heidelberg, Germany).
For cell apoptosis analysis, LX2 cells were trypsinized, washed, and then stained with FITC-conjugated anti-Annexin V antibody and PI for 15 min under darkness. Finally, apoptotic cells were analyzed by using FACSCanto II flow cytometry.
Western blot
The isolation of proteins was conducted by using RIPA lysis buffer (Beyotime, Nantong, China), and protein concentration was determined by a BCA Protein Assay kit (Beyotime, Nantong, China). Then, 20 μg of total protein were separated by 10% SurePAGE (Genscript, Nanjing, China), and protein electrophoresis membranes were transferred. Next, the membranes were incubated with the primary antibodies at 4 °C overnight, followed by incubation with the secondary antibody at 37 °C for 1 h. The protein bands were analyzed using the ECL kit (Beyotime, Nantong, China). The primary antibodies included: TSG101 (1:5000, ab125011), CD81 (1: 2000, ab109201), CD63 (1:2000, ab68418), α-SMA (1:2000, ab5694), collagen I (1:2000, ab34710), PTEN (1:10,000, ab32199), and β-actin (1:1000, ab6276), were obtained from Abcam (Cambridge, UK), and protein kinase B (AKT) (1:1000, Cat# 9272) and p-AKT (1:1000, Cat# 9271) were purchased from Cell Signaling Technology (Boston, MA).
RNA immunoprecipitation (RIP) assay
We performed RIP assay by using the Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA). Human anti-Ago2 antibody or NC IgG was bounded on magnetic beads, and then incubated with the lysates of LX2 cells lysed by RIP lysis buffer. Then precipitated RNAs were purified, and qRT-PCR was used to detect the levels of molecules.
RNA pull-down assay
Biotin-labeled miR-141-3p probe (wt-bio-miR-141-3p and mut-miR-141-3p) and control probe (bio-NC) were synthesized by CloudSeq Biotech Inc. (Shanghai, China). LX2 cells were transfected with biotin-coupled probes, then the lysates were incubated with Dynabeads M-280 Streptavidin (Invitrogen, Carlsbad, CA) for 3 h at 4 °C. The bound RNAs were purified using TRIzol and subjected to qRT-PCR analysis.
Dual-luciferase reporter assay
The sequences of circDIDO1 possessing the wild-type miR-141-3p binding sites were predicted by using bioinformatics analysis. The sequences in mutated (MUT) miR-141-3p binding sites were provided by Genema (Shanghai, China). Then, the binding sites were amplified and cloned into the pmirGLO report luciferase vector (Promega, Fitchburg, WI). Next, the reporter plasmids were transiently transfected into LX2 cells together with miR-141-3p mimic or NC mimic. The relative luciferase activity was determined after 48 h of transfection.
Statistical analyses
The results in the bar graphs were expressed as mean ± standard deviation. Group comparison was conducted by Student’s t-test (two-sided) or analysis of variance. The GraphPad Prism 6.0 software (GraphPad, San Diego, CA) was used for statistical analyses. p < .05 indicated statistical differences.
Results
Characterization of circDIDO1 in LX2 cells
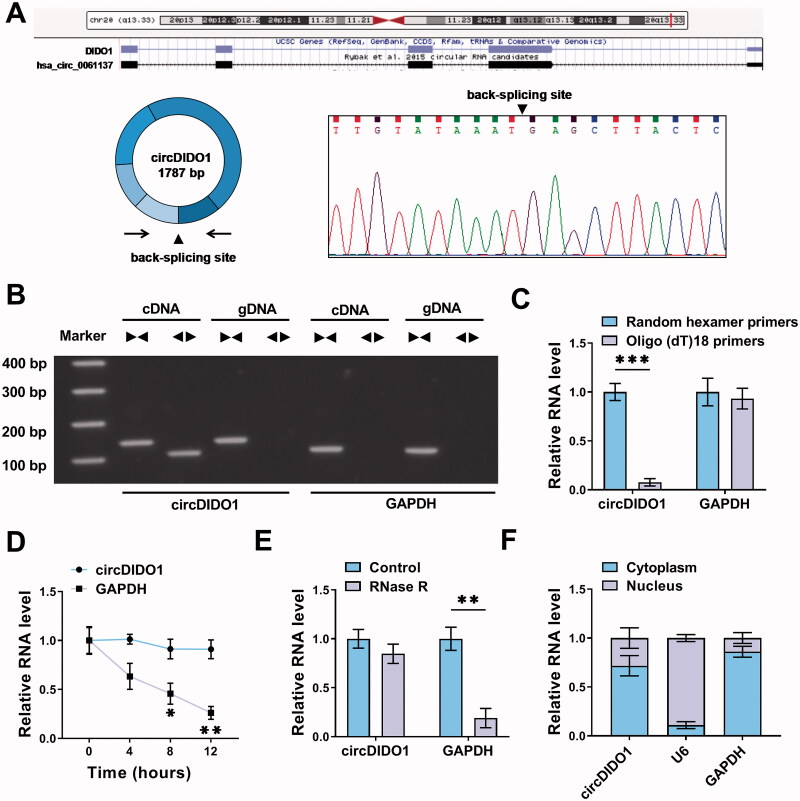
The characterization of circDIDO1 was first investigated in LX2 cells. CircDIDO1 is derived from exons (2–6) of DIDO1 gene, it is located at chr20: 61537238-61545758 strand, and the genomic length was 1781 bp, and the head-to-tail splicing was also confirmed by Sanger sequencing (Figure 1(A)). Besides that, we designed convergent primers and divergent primers for the amplification of linear mRNA (GAPDH) and circDIDO1 by using cDNA and gDNA as templates to rule out other potential factors that could induce such head-to-tail splicing. The results showed that circDIDO1 amplification products were only detected from cDNA instead of gDNA by divergent primers (Figure 1(B)). Subsequently, the circular characteristics of circDIDO1 were investigated. The Oligo(dT)18 primers in reverse transcription experiments were used, then it was found that the relative expression of circDIDO1 was significantly decreased relative to random hexamer primers, while linear mRNA (GAPDH) was not changed (Figure 1(C)). Additionally, the half-life of circDIDO1 exceeded 12 h after actinomycin D treatment, while that of GAPDH mRNA was about 8 h (Figure 1(D)). Moreover, RNase R treatment showed that circDIDO1 could resistant to the degradation by RNase R compared with the linear GAPDH, further indicating the stability of circDIDO1 (Figure 1(E)). In addition, subcellular fractionation assay suggested the predominant cytoplasmic distribution of circDIDO1 (Figure 1(F)). In all, circDIDO1 is a stable circular transcript in LX2 cells.
Figure 1.
Characterization of circDIDO1 in LX2 cells. (A) The Schematic illustration showing the production of circDIDO1 from its host gene, and the validation by Sanger sequencing. (B) The existence of circDIDO1 was validated using divergent and convergent primers from cDNA or genomic DNA (gDNA) of LX2 cells. (C) Random hexamer or Oligo(dT)18 primers were used in the reverse transcription experiments, and the products were examined in by qRT-PCR. (D) The relative RNA levels of circDIDO1 and GAPDH were determined by qRT-PCR after actinomycin D treatment at the indicated times. (E) The expression of circDIDO1 and GAPDH mRNA was examined in LX2 cells treated with or without RNase R. (F) Subcellular fractionation assay indicating the distribution of circDIDO1 in the cytoplasmic and nuclear fractions of LX2 cells. *p < .05, **p < .01, and ***p < .001.
CircDIDO1 suppresses the activation of HSCs by PTEN/AKT pathway
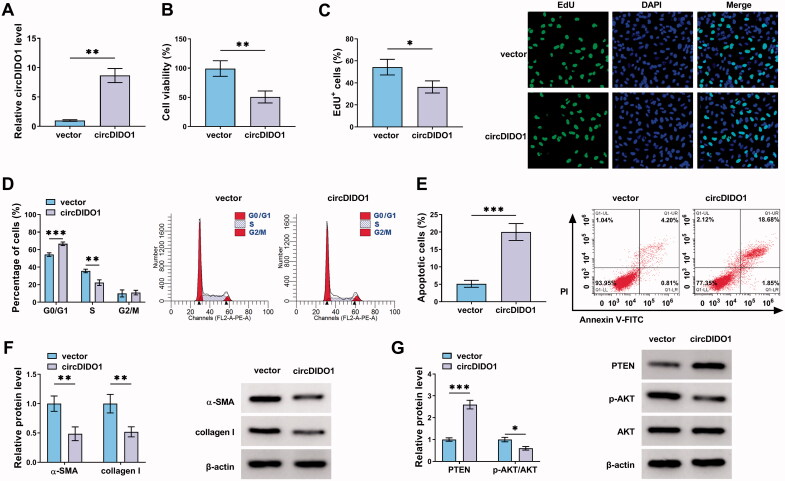
To explore the biological function of circDIDO1 in liver fibrosis, the expression of circDIDO1 was elevated by using circDIDO1 overexpression vector. The results of qRT-PCR validated the elevation efficiency of circDIDO1 vector in LX2 cells with the increased circDIDO1 expression after transfection (Figure 2(A)). Functionally, circDIDO1 overexpression suppressed LX2 cell viability ability and DNA synthesis activity, manifested by the CCK-8 and EdU assays (Figure 2(B,C)). Flow cytometry suggested that circDIDO1 overexpression delayed the progression of cell cycle and inhibited cell proliferation by arresting LX2 cells at G0/G1 phase (Figure 2(D)). Moreover, flow cytometry analysis also exhibited that overexpression of circDIDO1 significantly induced apoptosis in LX2 cells (Figure 2(E)), accompanied with the increase of cleaved-caspase3 and Bax, as well as decrease of Bcl-2 in LX2 cells (Fig. S1). Additionally, Western blot analysis revealed that circDIDO1 overexpression led to the decreases of the protein levels of α-SMA and collagen I in LX2 cells (Figure 2(F)). These results demonstrated that circDIDO1 repressed the growth and fibrosis of HSCs. It has been reported that PTEN negatively regulates the process of liver fibrosis (Hao et al., 2009), besides that, PTEN loss in the liver leads to hyperactivation of the PI3K/AKT pathway, which contributes to HSC activation, proliferation and hepatic fibrosis (Reif et al., 2003; He et al., 2016). Therefore, whether there was a change in the expression of PTEN and AKT after circDIDO1 overexpression was determined. Western blot analysis showed that circDIDO1 up-regulation elevated PTEN protein level and decreased the ratio of p-AKT/AKT in LX2 cells (Figure 2(G)). Taken together, circDIDO1 suppressed the activation of HSCs by PTEN/AKT pathway.
Figure 2.
CircDIDO1 suppresses the activation of HSCs by PTEN/AKT pathway. (A–G) LX2 cells were transfected with circDIDO1 or vector. (A) qRT-PCR analysis for the expression level of circDIDO1 in LX2 cells after transfection. (B, C) The proliferation ability analysis of LX2 cells using CCK-8 and EdU assays. (D, E) Flow cytometry for cell cycle and cell apoptosis in LX2 cells. (F, G) Western blot analysis for the protein levels of α-SMA, collagen I, PTEN, AKT, and p-AKT in LX2 cells. *p < .05, **p < .01, and ***p < .001.
MiR-141-3p is a target of circDIDO1
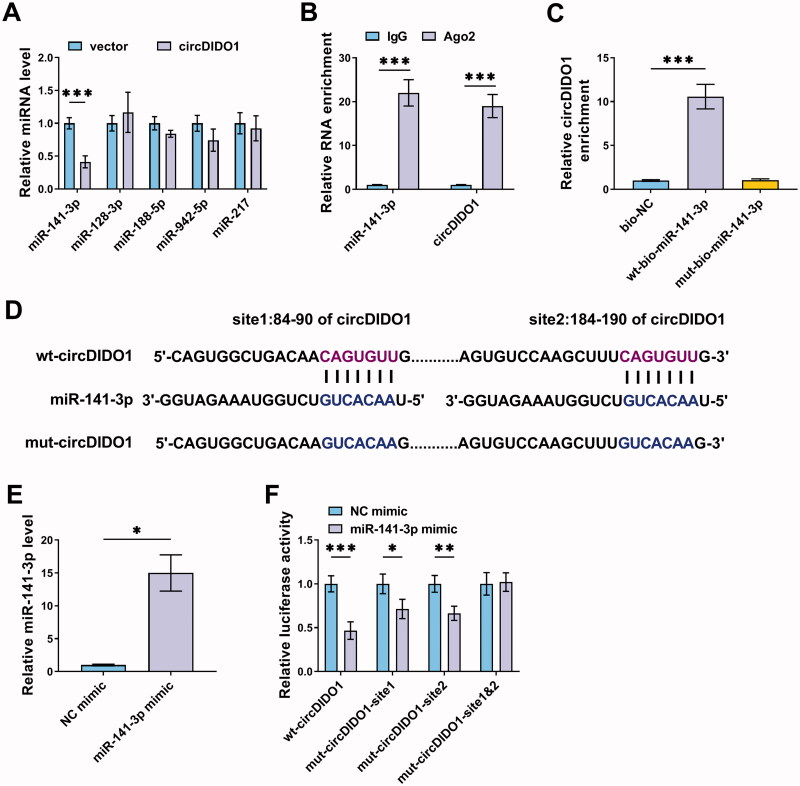
Given that circDIDO1 was predominantly distributed in the cytoplasm of LX2 cells, we then investigated whether circDIDO1 acted as a miRNA sponge to exert its effects on HSC activation. According to the prediction of starBase database, five miRNAs possessed the potential a binding site on circDIDO1 were selected, among them, miR-141-3p was found to be significantly decreased by circDIDO1 up-regulation in LX2 cells (Figure 3(A)). Thus, miR-141-3p was selected for binding analysis. RIP assay suggested that circDIDO1 and miR-141-3p were significantly enriched in the presence of Ago2 antibody relative to the NC IgG (Figure 3(B)), suggesting that circDIDO1 interacts and binds with miR-141-3p through Ago2 protein. Moreover, RNA pull-down with biotin-labeled wild- or mutated-type miR-141-3p in LX2 cells was performed. The results revealed that circDIDO1 was significantly pulled down by wild-type miR-141-3p probe (Figure 3(C)). Next, we mutated the response elements of circDIDO1 on miR-141-3p to conduct dual-luciferase reporter assay (Figure 3(D)). MiR-141-3p mimic significantly elevated miR-141-3p expression in LX2 cells (Figure 3(E)). Then, it was found that the luciferase activities of wt, mut1, and mut2 reporters were reduced by miR-141-3p mimic, while miR-141-3p overexpression has no effect on the luciferase activity of the reporter containing the co-mutant with the two mutated binding sites (Figure 3(F)). In all, circDIDO1 directly targeted miR-141-3p in LX2 cells.
Figure 3.
MiR-141-3p is a target of circDIDO1. (A) qRT-PCR analysis for the expression of miR-141-3p, miR-128-3p, miR-188-5p, miR-942-5p, and miR-217 in LX2 cells transfected with circDIDO1 or vector. (B) RIP experiment was performed with Ago2 antibody and negative control IgG, followed by qRT-PCR to detect the expression of miR-141-3p and circDIDO1. (C) RNA pull-down assay was performed using the miR-141-3p-specific probe and control probe, and the enrichment of circDIDO1 was detected by qRT-PCR. (D) The starBase database showing the binding site of miR-141-3p on circDIDO1, and the mutated sequence of circDIDO1 in the binding site was established. (E) qRT-PCR analysis for miR-141-3p expression level in LX2 cells after miR-141-3p mimic or NC mimic transfection. (F) Dual-luciferase reporter assay for the luciferase activity of wild and mutated circDIDO1 reporter after miR-141-3p overexpression in LX2 cells. *p < .05, **p < .01, ***p < .001.
MiR-141-3p knockdown suppresses the activation of HSCs by PTEN/AKT pathway
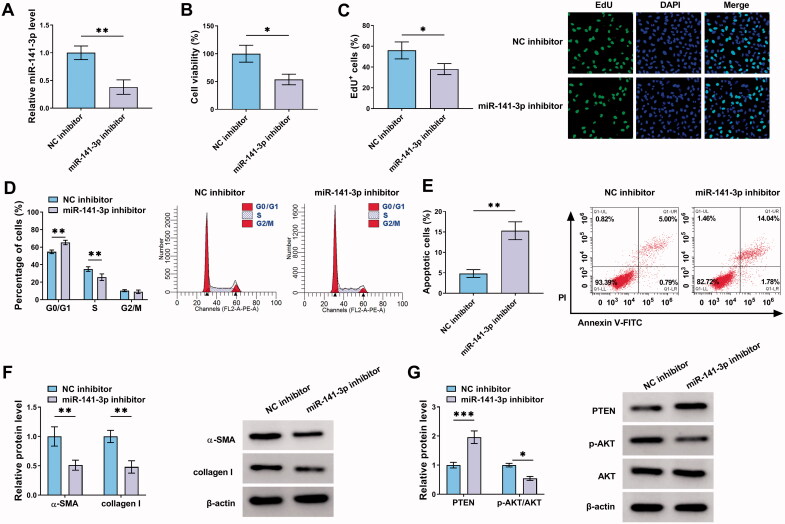
Subsequently, the function of miR-141-3p in liver fibrosis was investigated. After confirming the knockdown efficiency of miR-141-3p inhibitor (Figure 4(A)), it was proved that miR-141-3p inhibition suppressed LX2 cell proliferation (Figure 4(B,C)), resulted in cell cycle arrest (Figure 4(D)), and induced cell apoptosis (Figure 4(E)). Moreover, miR-141-3p inhibition reduced the protein levels of α-SMA and collagen I in LX2 cells (Figure 4(F)). Additionally, miR-141-3p inhibition mediated the increase of PTEN and decrease of the ratio of p-AKT/AKT in LX2 cells (Figure 4(G)). Thus, we demonstrated that miR-141-3p knockdown restrained the activation of HSCs by PTEN/AKT pathway.
Figure 4.
MiR-141-3p knockdown suppresses the activation of HSCs by PTEN/AKT pathway. (A–G) LX2 cells were transfected with miR-141-3p inhibitor or NC inhibitor. (A) qRT-PCR analysis for the expression level of miR-141-3p in LX2 cells after transfection. (B, C) The proliferation ability analysis of LX2 cells using CCK-8 and EdU assays. (D, E) Flow cytometry for cell cycle and cell apoptosis in LX2 cells. (F, G) Western blot analysis for the protein levels of α-SMA, collagen I, PTEN, AKT, and p-AKT in LX2 cells. *p < .05, **p < .01, and ***p < .001.
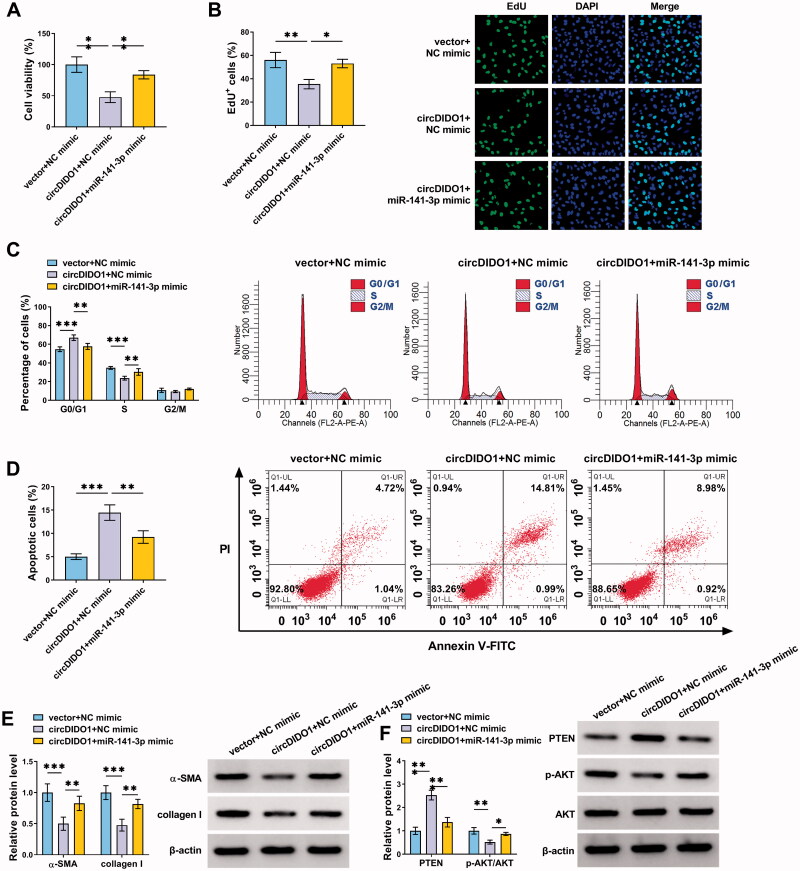
CircDIDO1 suppresses the activation of HSCs by miR-141-3p/PTEN/AKT pathway
To determine whether circDIDO1 mediated the activation of HSCs by targeting miR-141-3p, rescue experiments were carried out. LX2 cells were co-transfected with circDIDO1 and miR-141-3p mimic. After that, we demonstrated that miR-141-3p up-regulation attenuated circDIDO1 overexpression-induced inhibition of cell proliferation (Figure 5(A,B)), arrest of cell cycle (Figure 5(C)), enhancement of cell apoptosis (Figure 5(D)) in LX2 cells. Besides that, co-transfection of circDIDO1 and miR-141-3p mimic overturned the decreased effects of circDIDO1 alone on α-SMA and collagen I protein expression in LX2 cells (Figure 5(E)). In the meanwhile, we also observed that both the increase of PTEN and the decrease of p-AKT/AKT ratio in LX2 cells mediated by circDIDO1 were abolished by miR-141-3p mimic (Figure 5(F)). Altogether, circDIDO1/miR-141-3p axis was engaged in HSC activation through regulating PTEN/AKT pathway.
Figure 5.
CircDIDO1 suppresses the activation of HSCs by miR-141-3p/PTEN/AKT pathway. (A–F) LX2 cells were co-transfected with circDIDO1 and miR-141-3p mimic. (A, B) The proliferation ability analysis of LX2 cells using CCK-8 and EdU assays. (C, D) Flow cytometry for cell cycle and cell apoptosis in LX2 cells. (E, F) Western blot analysis for the protein levels of α-SMA, collagen I, PTEN, AKT, and p-AKT in LX2 cells. *p < .05, **p < .01, and ***p < .001.
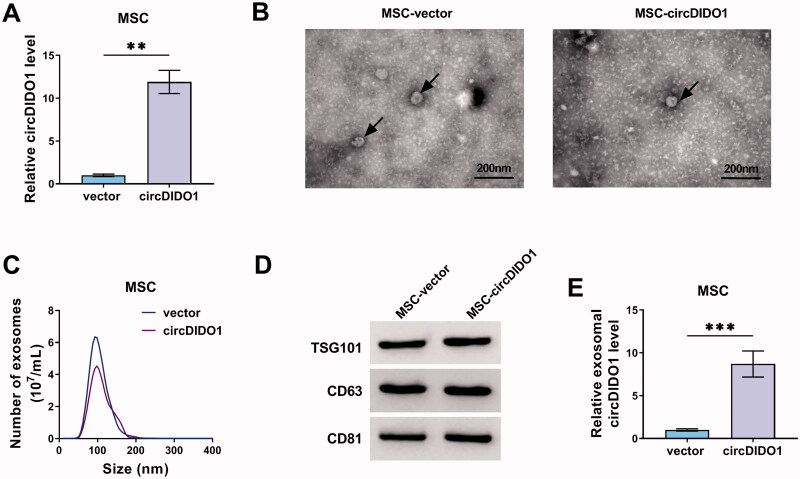
Identification of exosomes isolated from circDIDO1 transfected MSCs
Previous studies have uncovered that circRNAs can be packaged into exosomes and can be mass-produced by exosome secreted cells, thereby delivering circRNAs to target cells by exosomes to directly exert the therapeutic effects of circRNAs (Cao et al., 2020; Wang et al., 2020). MSCs can produce abundant amounts of exosomes (Pedrosa et al., 2020). Herein, we transfected circDIDO1 or vector into MSCs, qRT-PCT showed a significant elevation of circDIDO1 expression in MSCs after transfection (Figure 6(A)). Then, extracellular exosomes were isolated from the supernatants of MSC-vector or MSC-circDIDO1 group. TEM data showed the typical cup-shaped morphology of exosomes (Figure 6(B)). Nanoparticle tracking analysis (NTA) indicated a typical average exosome size distribution ranging from 90 to 120 nm (Figure 6(C)). Besides that, Western blot confirmed the presence of exosomal marker proteins, including TSG-101, CD63, and CD81 (Figure 6(D)). Interestingly, circDIDO1 expression level was higher in exosomes of MSC-circDIDO1 group than those in MSC-vector group (Figure 6(E)), indicating that circDIDO1 could be packaged into MSC isolated exosomes.
Figure 6.
Identification of exosomes isolated from circDIDO1 transfected MSCs. (A) qRT-PCR analysis of circDIDO1 expression level in MSCs transfected circDIDO1 or vector. (B) TEM images of exosomes isolated from circDIDO1 or vector transfected-MSCs. Scale bar, 200 nm. (C) Exosomes isolated from circDIDO1 or vector transfected-MSCs underwent an NTA to determine exosomal size distribution. (D) Western blot analysis for exosomes markers CD63, TSG101, and CD81. (E) qRT-PCR analysis of circDIDO1 expression level in exosomes isolated from circDIDO1 or vector transfected-MSCs. **p < .01, ***p < .001.
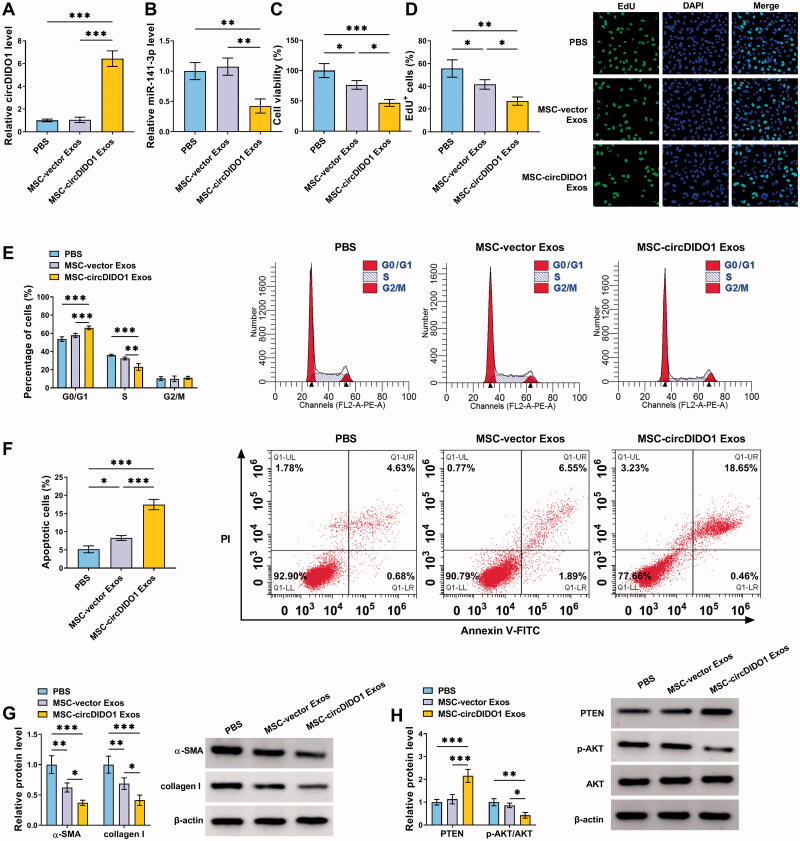
Transfer of circDIDO1 mediated by MSC isolated exosomes suppresses the activation of HSCs by PTEN/AKT pathway
To evaluate the role of exosome-transferred circDIDO1 in liver fibrosis, we incubated LX2 cells with exosomes isolated from circDIDO1 or vector transfected MSCs (MSC-circDIDO1 Exo or MSC-vector Exo). As shown in Figure 7(A), circDIDO1 expression was elevated in LX2 cells after MSC-circDIDO1 Exo incubation, while MSC-vector Exo did not affect circDIDO1 expression levels, suggesting the successful uptake of circDIDO1 in LX2 cells via MSC-exosomes. Besides that, exosomal circDIDO1 significantly reduced the expression levels of miR-141-3p in LX2 cells (Figure 7(B)). Functionally, we observed that the secreted exosomes of MSCs themselves without circDIDO1 overexpression could inhibit the proliferation (Figure 7(C,D)), mediate cell cycle arrest (Figure 7(E)), induce apoptosis, and decrease pro-fibrotic markers α-SMA and collagen I (Figure 7(G)) in LX2 cells compared with PBS treatment, but they did not affect the activation of PTEN/AKT pathway (Figure 7(H)). Importantly, compared with these circDIDO1-free exosomes of MSCs, exosomes isolated from MSCs with circDIDO1 overexpression plasmids had much greater effects on proliferation inhibition, cell cycle arrest, apoptosis enhancement, pro-fibrotic markers decrease (Figure 7(C–G)). Additionally, results of Western blot also showed that exosomal circDIDO1 caused the increase of PTEN and the decrease of p-AKT/AKT ratio in LX2 cells (Figure 7(H)). Collectively, these results confirmed that circDIDO1 delivered by MSC-exosomes suppressed the activation of HSCs by PTEN/AKT pathway.
Figure 7.
MSC-exosome-mediated transfer of circDIDO1 suppresses the activation of HSCs by PTEN/AKT pathway. (A–H) LX2 cells were co-cultured with PBS, MSC-circDIDO1 Exo or MSC-vector Exo. (A) qRT-PCR analysis of circDIDO1 and miR-141-3p expression level in LX2 cells after incubation. (C, D) The proliferation ability analysis of LX2 cells using CCK-8 and EdU assays. (E, F) Flow cytometry for cell cycle and cell apoptosis in LX2 cells. (G, H) Western blot analysis for the protein levels of α-SMA, collagen I, PTEN, AKT, and p-AKT in LX2 cells. *p < .05, **p < .01, and ***p < .001.
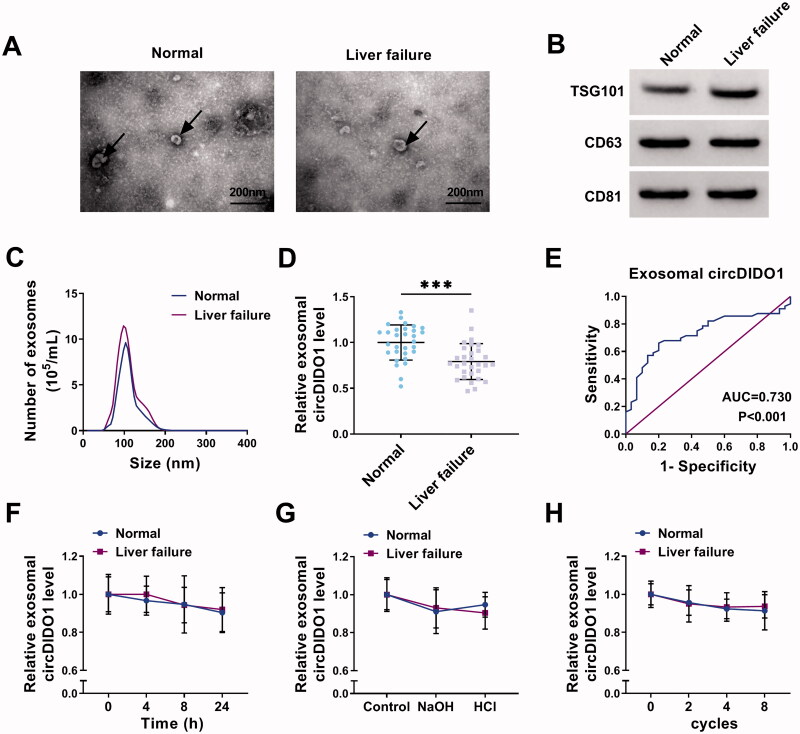
Serum exosomal circDIDO1 level is down-regulated in liver failure patients
Following this, we then explored circDIDO1 expression in exosomes of liver failure patients. The exosomes in the serum of healthy individuals and liver failure patients were isolated. TEM data verified exosome morphology (Figure 8(A)). Exosome markers were detected by Western blot (Figure 8(B)). NTA suggested the typical average exosome size distribution (Figure 8(C)). Afterwards, we proved that circDIDO1 expression was detectable in extracted serum exosomes, and was much lower in exosomes isolated from liver failure patients than those in exosomes of healthy individuals (Figure 8(D)). Additionally, ROC curve was performed to evaluate the diagnostic value of exosomal circDIDO1 in serum, the results showed an area under curve (AUC) of 0.730, with a diagnostic sensitivity and specificity reaching 80.0 and 66.1%, respectively (95% CI = 0.622–0.838) (Figure 8(E)). Furthermore, the stability of exosomal circDIDO1 in serum was investigated. Exosomes isolated from the serum of healthy individuals and liver failure patients were exposed to different conditions, including incubation at room temperature for 0, 4, 8, or 12 h, high (pH = 13) or low (pH = 1.0) pH solution treatment for 3 h at room temperature or repeat freezing and thawing for eight cycles, and our results confirmed that exosomal circDIDO1 expression did not affect by any of the experimental conditions (Figure 8(F–H)). In all, exosomal circDIDO1 in serum is stable and might be a promising diagnostic biomarker for live failure.
Figure 8.
Serum exosomal circDIDO1 level is down-regulated in liver failure patients. (A) TEM images of exosomes isolated from the serum of healthy individuals and liver failure patients. Scale bar, 200 nm. (B) Western blot analysis for exosomes markers CD63, TSG101, and CD81. (C) Exosomes isolated from the serum of healthy individuals and liver failure patients underwent an NTA to determine exosomal size distribution. (D) qRT-PCR analysis of exosomal circDIDO1 expression level in the serum of healthy individuals and liver failure patients. (E) ROC curve analysis of the diagnostic value of exosomal circDIDO1 in liver failure patients. (F–H) Exosomal circDIDO1 expression level was not significantly affected by long exposure (F), PH values (G), or repeat freezing and thawing (H). ***p < .001.
Discussion
Currently, more and more circRNAs are identified to act as crucial regulators in multiple pathological and physiological processes of many types of disease. Previous studies have showed that circRNAs can regulate the activation and proliferation of HSCs by usually acting as miRNA sponges, thus to involve in the process of liver fibrosis in liver failure (Ji et al., 2020; Li et al., 2020). In this study, we first investigated that circDIDO1 overexpression could suppress the proliferation, reduced pro-fibrotic markers α-SMA and collagen I, and induced apoptosis as well as cell cycle arrest in HSCs, thus repressing HSC activation. Given that circDIDO1 was predominantly distributed in the cytoplasm, based on the ceRNA hypothesis, the downstream miRNAs of circDIDO1 were investigated. Mechanically, circDIDO1 could sponge miR-141-3p. Yang et al. showed that miR-141 attenuated autophagy-mediated hepatitis B virus inhibition (Yang et al., 2017). Moreover, Liang’s team demonstrated that miR-141 was elevated in activated HSCs, and promoted HSC activation with the increased expression levels of pro-fibrotic markers by activating AKT/mTOR pathway through repressing PTEN (Liang et al., 2020). In the present study, we also confirmed that inhibition of miR-141-3p suppressed HSC activation by regulating the actions mentioned above. Importantly, our data further showed that circDIDO1 suppressed HSC activation by sponging miR-143-3p. The PTEN/AKT signal pathway constitutes a significant pathway modulating the signaling of multiple biological processes (Carnero et al., 2008). This study also confirmed that circDIDO1/miR-143-3p axis was responsible for the activation of PTEN/AKT pathway, which has been consistently showed in previous study (Liang et al., 2020).
Recent years, stem cell therapy has been proposed as a promising option for the treatment of liver diseases (Duan et al., 2017). Moreover, relative to the stem cells themselves, the secretomes of the stem cells appear to be of greater benefit on tissue regeneration and repair (Teixeira et al., 2013). MSCs are multipotent stem cells with the ability for differentiation into tissue-specific cells and self-renewal (Horwitz et al., 2005; Samsonraj et al., 2017). Exosomes are important components of MSCs, and MSCs-derived exosomes have been widely applied in clinical and scientific research in liver disease (Lou et al., 2017; Rong et al., 2019). Exosomes are involved in intercellular communication through paracrine actions, besides that, exosomes possess low-risk for cancer formation, low immunogenicity, long half-life in circulation, and the ability to cross the BBB, which make them as potential clinical diagnostic and therapeutic biomarkers (Kalani et al., 2014; Yang et al., 2017). Thus, we then investigated whether exosomes secreted by MSCs can be used to deliver circDIDO1 for liver fibrosis prevention. It was confirmed that transfer of circDIDO1 mediated by MSC-secreted exosomes also could restrain the activation of HSCs. In the meanwhile, a low expression of exosomal circDIDO1 was observed in the serum exosomes of liver failure patients, exosomal circDIDO1 in serum was stable and possessed well diagnostic value for live failure.
Nevertheless, we have found some interesting results in this work, the data presented are based on a limited number of cells in vitro. The in vivo assay using the animal model with circDIDO1 overexpression plasmids is essential to verify these conclusions in future research.
Conclusions
In conclusion, our findings suggest that the transfer of circDIDO1 mediated by MSC-secreted exosomes suppressed HSCs activation by elevating PTEN to suppress AKT pathway through sponging miR-143-3p, which provided a novel insight into the development of exosomal therapeutic delivery of RNAs in liver fibrosis prevention.
Authors contributions
Li Ma and Junfeng Wei conducted the experiments and drafted the manuscript. Yanli Zeng and Junping Liu collected and analyzed the data. Erhui Xiao contributed the methodology. Yuehua Kang operated the software and edited the manuscript. Yi Kang designed and supervised the study. All authors read and approved the final manuscript.
Supplementary Material
Disclosure statement
The authors declare that they have no conflicts of interest.
Data availability statement
The data used to support the findings of this study are included within the article.
References
- Batrakova EV, Kim MS. (2015). Using exosomes, naturally-equipped nanocarriers, for drug delivery. J Control Release 219:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Meng X, Han X, Li J. (2020). Exosomes derived from circRNA Rtn4-modified BMSCs attenuate TNF-α-induced cytotoxicity and apoptosis in murine MC3T3-E1 cells by sponging miR-146a. Biosci Rep 40:BSR20193436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnero A, Blanco-Aparicio C, Renner O, et al. (2008). The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets 8:187–98. [DOI] [PubMed] [Google Scholar]
- Chen W, Quan Y, Fan S, et al. (2020). Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett 475:119–28. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yuan B, Wu Z, et al. (2017). Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene 629:35–42. [DOI] [PubMed] [Google Scholar]
- Cooks T, Pateras IS, Jenkins LM, et al. (2018). Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun 9:771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Wang Y, He J, et al. (2017). [Progress of the application of stem cell therapy for end-stage liver disease]. Zhong Nan Da Xue Xue Bao Yi Xue Ban 42:457–62. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Begne M, Lu B, Han X, et al. (2009). Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J Proteome Res 8:1304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao LS, Zhang XL, An JY, et al. (2009). PTEN expression is down-regulated in liver tissues of rats with hepatic fibrosis induced by biliary stenosis. APMIS 117:681–91. [DOI] [PubMed] [Google Scholar]
- He L, Gubbins J, Peng Z, et al. (2016). Activation of hepatic stellate cell in Pten null liver injury model. Fibrogenesis Tissue Repair 9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Friedman SL, Hoshida Y. (2017). Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev 121:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz EM, Le Blanc K, Dominici M, et al. (2005). Clarification of the nomenclature for MSC: The International Society for Cellular Therapy Position Statement. Cytotherapy 7:393–5. [DOI] [PubMed] [Google Scholar]
- Hua X, Sun Y, Chen J, et al. (2019). Circular RNAs in drug resistant tumors. Biomed Pharmacother 118:109233. [DOI] [PubMed] [Google Scholar]
- Ji D, Chen GF, Wang JC, et al. (2020). Hsa_circ_0070963 inhibits liver fibrosis via regulation of miR-223-3p and LEMD3. Aging 12:1643–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalani A, Tyagi A, Tyagi N. (2014). Exosomes: mediators of neurodegeneration, neuroprotection and therapeutics. Mol Neurobiol 49:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, LeBleu VS. (2020). The biology, function, and biomedical applications of exosomes. Science 367:eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen LS, Andersen MS, Stagsted LVW, et al. (2019). The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:675–91. [DOI] [PubMed] [Google Scholar]
- Li S, Song F, Lei X, et al. (2020). hsa_circ_0004018 suppresses the progression of liver fibrosis through regulating the hsa-miR-660-3p/TEP1 axis. Aging 12:11517–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Wang X, Si C, et al. (2020). Downregulation of miR‑141 deactivates hepatic stellate cells by targeting the PTEN/AKT/mTOR pathway. Int J Mol Med 46:406–14. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yang Y, Wang Z, et al. (2020). Insights into the regulatory role of circRNA in angiogenesis and clinical implications. Atherosclerosis 298:14–26. [DOI] [PubMed] [Google Scholar]
- Lodder J, Denaës T, Chobert MN, et al. (2015). Macrophage autophagy protects against liver fibrosis in mice. Autophagy 11:1280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou G, Chen Z, Zheng M, Liu Y. (2017). Mesenchymal stem cell-derived exosomes as a new therapeutic strategy for liver diseases. Exp Mol Med 49:e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PF, Gao CC, Yi J, et al. (2017). Cytotherapy with M1-polarized macrophages ameliorates liver fibrosis by modulating immune microenvironment in mice. J Hepatol 67:770–9. [DOI] [PubMed] [Google Scholar]
- Pedrosa M, Gomes J, Laranjeira P, et al. (2020). Immunomodulatory effect of human bone marrow-derived mesenchymal stromal/stem cells on peripheral blood T cells from rheumatoid arthritis patients. J Tissue Eng Regen Med 14:16–28. [DOI] [PubMed] [Google Scholar]
- Reif S, Lang A, Lindquist JN, et al. (2003). The role of focal adhesion kinase-phosphatidylinositol 3-kinase-akt signaling in hepatic stellate cell proliferation and type I collagen expression. J Biol Chem 278:8083–90. [DOI] [PubMed] [Google Scholar]
- Rong X, Liu J, Yao X, et al. (2019). Human bone marrow mesenchymal stem cells-derived exosomes alleviate liver fibrosis through the Wnt/β-catenin pathway. Stem Cell Res Ther 10:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonraj RM, Raghunath M, Nurcombe V, et al. (2017). Concise review: multifaceted characterization of human mesenchymal stem cells for use in regenerative medicine. Stem Cells Transl Med 6:2173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao J, Zhang X, et al. (2017). Activated hepatic stellate cells impair NK cell anti-fibrosis capacity through a TGF-β-dependent emperipolesis in HBV cirrhotic patients. Sci Rep 7:44544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira FG, Carvalho MM, Sousa N, Salgado AJ. (2013). Mesenchymal stem cells secretome: a new paradigm for central nervous system regeneration? Cell Mol Life Sci 70:3871–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Friedman SL. (2017). Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol 14:397–411. [DOI] [PubMed] [Google Scholar]
- Vo JN, Cieslik M, Zhang Y, et al. (2019). The landscape of circular RNA in cancer. Cell 176:869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhan J, Lin X, et al. (2020). CircRNA-0077930 from hyperglycaemia-stimulated vascular endothelial cell exosomes regulates senescence in vascular smooth muscle cells. Cell Biochem Funct 38:1056–68. [DOI] [PubMed] [Google Scholar]
- Wang W, Dong R, Guo Y, et al. (2019). CircMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J Cell Mol Med 23:5486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Qi X, Liu L, et al. (2019). Emerging epigenetic regulation of circular RNAs in human cancer. Mol Ther Nucleic Acids 16:589–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu Y, Xue J, et al. (2017). MicroRNA-141 targets Sirt1 and inhibits autophagy to reduce HBV replication. Cell Physiol Biochem 41:310–22. [DOI] [PubMed] [Google Scholar]
- Yang Y, Ye Y, Su X, et al. (2017). MSCs-derived exosomes and neuroinflammation, neurogenesis and therapy of traumatic brain injury. Front Cell Neurosci 11:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Xu QG, Wang ZG, et al. (2018). Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol 68:1214–27. [DOI] [PubMed] [Google Scholar]
- Zhang CY, Yuan WG, He P, et al. (2016). Liver fibrosis and hepatic stellate cells: etiology, pathological hallmarks and therapeutic targets. World J Gastroenterol 22:10512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Chen KK, Zhang J, et al. (2018). The decade of exosomal long RNA species: an emerging cancer antagonist. Mol Cancer 17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are included within the article.