Abstract
Angiotensin-converting enzyme 2 (ACE2) is the receptor of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) causing Coronavirus disease 2019 (COVID-19). Transmembrane serine protease 2 (TMPRSS2) is a coreceptor. Abnormal hepatic function in COVID-19 suggests specific or bystander liver disease. Because liver cancer cells express the ACE2 viral receptor, they are widely used as models of SARS-CoV-2 infection in vitro. Therefore, the purpose of this study was to analyze ACE2 and TMPRSS2 expression and localization in human liver cancers and in non-tumor livers. We studied ACE2 and TMPRSS2 in transcriptomic datasets totaling 1503 liver cancers, followed by high-resolution confocal multiplex immunohistochemistry and quantitative image analysis of a 41-HCC tissue microarray. In cancers, we detected ACE2 and TMPRSS2 at the biliary pole of tumor hepatocytes. In whole mount sections of five normal liver samples, we identified ACE2 in hepatocyte’s bile canaliculi, biliary epithelium, sinusoidal and capillary endothelial cells. Tumors carrying mutated β-catenin showed ACE2 DNA hypomethylation and higher mRNA and protein expression, consistently with predicted β-catenin response sites in the ACE2 promoter. Finally, ACE2 and TMPRSS2 co-expression networks highlighted hepatocyte-specific functions, oxidative stress and inflammation, suggesting a link between inflammation, ACE2 dysfunction and metabolic breakdown.
Subject terms: Bioinformatics, Epigenetics analysis, Gene expression analysis, Genomic analysis, Imaging, Immunological techniques, Microscopy, Sequencing
Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) causes the Coronavirus disease 2019 (COVID-19)1–3. SARS-CoV-2 is an enveloped RNA betacoronavirus phylogenetically similar to SARS-CoV and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV). Although ~ 80% of SARS-CoV-2-infected individuals develop asymptomatic self-limited upper airway forms of COVID-19, approximately 15% need hospitalization and 5% develop severe disease with acute respiratory distress syndrome, immune dysregulation and a “cytokine storm” with disseminated intravascular coagulation1,3,4. Approximately 50% of hospitalized patients present comorbidities: hypertension, type-2 diabetes and coronary heart disease1–3,5,6.
The baseline prevalence of chronic liver disease in COVID-19 was about three percent in two meta-analyses assembling 3301 patients7,8, but the mean prevalence of COVID-19 related liver dysfunction was about 20% (range 14–53%)1,6–9. Liver dysfunction correlates with the extent of pulmonary lesions10. Patients with hepatocyte-type dysfunction at admission are at higher risk of progressing to severe COVID-1911. After admission, antiviral drugs such as lopinavir and ritonavir are risk factors for liver damage11. Autopsies suggested that SARS-CoV-2 infects and replicates within hepatocytes in severe COVID-19 cases12–14 and SARS-CoV-2 RNA has been detected in the respiratory epithelium, kidney, liver, heart, brain, blood and stools in severe COVID-19 cases15,16. In addition, crown-like viral particles with complete envelope and typical spikes have been observed in lung, kidney16,17 and in hepatocytes14 suggesting multiorgan targeting of COVID-19.
Cell entry of SARS-CoV and SARS-CoV-2 depends on the binding of the viral spike (S) protein to the Angiotensin-converting enzyme 2 (ACE2) and on the serine protease TMPRSS2 for S protein fusion18,19. The rate of SARS-CoV-2 infection correlates with cell surface ACE2 expression18,19. Importantly, Huh-718,20 human hepatocellular carcinoma (HCC) and HepG2 hepatoblastoma20 cells are widely used models of SARS-CoV-2 infection because they express cell surface ACE2 and can be infected at high titers.
As liver cancer cells are in vitro models for the study of SARS-CoV-2 entry and cytopathic effects, we extensively studied mRNA, protein expression and localization of the viral receptors ACE2 and TMPRSS2 in human HCCs. This characterization will be useful to design in vitro studies on virus entry, replication and metabolic alterations in liver cells.
Results
Higher expression of ACE2 and TMPRSS2 mRNAs in HCCs with mutated β-catenin
We first searched for relationships of ACE2 and TMPRSS2 mRNA expression levels with HCC aggressiveness. To this end, overall and disease-free survival analyses were carried out in the TCGA dataset (n = 370). After applying the exclusion criteria described in Supplementary Fig. 1a, the survival dataset consisted of 256 patients. In consistency with previous reports21–23, higher ACE2 and TMPRSS2 mRNA expression were associated with better overall and disease-free survival (Supplementary Fig. 1b and 2a). ACE2 and TMPRSS2 mRNAs were also detected in 47 non-tumor livers in the TCGA dataset. Whereas ACE2 mRNA expression was 1.7-fold higher in non-tumors livers than in HCCs, TMPRSS2 expression did not differ between both groups (Supplementary Fig. 2b).
In HCCs, the expression of genes associated with relatively good prognosis is frequently preserved in well-differentiated tumors because they maintain the phenotype of adult hepatocytes24,25. To verify if this applied to ACE2 and TMPRSS2, we measured their expression levels in HCCs classified according to their likeness to normal hepatocytes. Thus classified, HCCs are divided into two major classes, namely “low-proliferation” and “high-proliferation”25. Four HCC subclasses result from the interaction of, on one hand, the metabolic phenotype and, on the other hand, the proliferation/differentiation ratios of tumor cells24,26. Hence, in the “low-proliferation” class of well-to-moderately differentiated HCCs, the periportal-type (PP) and perivenous-type (PV) subclasses refer to the preservation of the metabolic zonation functions of normal hepatocytes24,26. At the opposite end of the spectrum, in the “high-proliferation” class of moderately-to-poorly differentiated HCCs, the extracellular-matrix-type (ECM) and STEM-type subclasses, refer to tumors developing an important stromal and vascular support and expressing cancer stem cell markers24,26. In this context, we found higher ACE2 mRNA expression in perivenous-type HCCs than in the other subclasses (Fig. 1a; Supplementary Fig. 2c) in both the Désert’s cDNA microarray meta-dataset (1133 HCCs)26 (Fig. 1a) and in the TCGA dataset (370 HCCs) (Supplementary Fig. 2c). Of note, ACE2 mRNA expression was higher in HCCs expressing previously validated β-catenin pathway activation signatures26,27 (Fig. 1b) and in HCCs carrying sequenced β-catenin (CTNNB1) activating mutations (Supplementary Fig. 2c). In turn, TMPRSS2 was expressed at higher levels in periportal-type HCCs (Supplementary Fig. 2c, d), which we previously demonstrated to carry wild-type CTNNB1 and to belong to the “low-proliferation” class of well-to-moderately differentiated HCCs that preserve the periportal metabolic functions of normal hepatocytes26. The MERS-CoV receptor, DPP4, was also related to β-catenin pathway activation (Supplementary Fig. 2e–g). ACE2 DNA was hypomethylated in tumors carrying CTNNB1 mutations (Fig. 1c), which is consistent with transcriptional activation the ACE2 gene. By contrast, TMPRSS2 was hyper-methylated in HCCs carrying CTNNB1 mutations (Supplementary Fig. 3a; Supplementary Table 1), which is consistent with transcriptional repression. Neither ACE2 nor DPP4 mRNA expression were related TERT and TP53 gene mutations, which are the two most frequent mutations in HCCs (Supplementary Fig. 3c, d). In turn, TMPRSS2 mRNA levels were higher in HCCs carrying wild-type TP53 (Supplementary Fig. 3d), which is consistent with the above-described higher expression of TMPRSS2 in the well-differentiated, non-proliferative class of HCCs, where TP53 gene mutations are rare26,28. Indeed, TP53 mutations are most frequent in poorly-differentiated HCCs (S229; G330), showing an aggressive, cancer stem cell phenotype26,31.
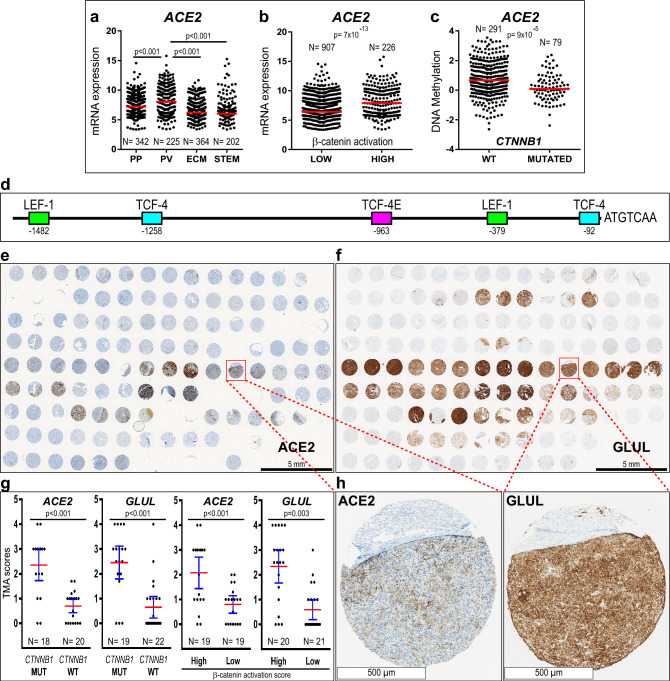
Figure 1.
High ACE2 levels in well-differentiated, non-proliferative HCCs, with mutated β-catenin (CTNNB1) and hypomethylated DNA. (a) and (b) ACE2 mRNA expression in the Désert’s meta-dataset of 1133 HCCs26. (a) HCC subclasses: PP, periportal-type; PV, perivenous-type; ECM, extracellular-matrix-type and STEM, stem-cell-type HCCs. (b) ACE2 mRNA expression according to β-catenin activation levels, assessed as described26. (c) ACE2 DNA is hypomethylated in HCCs carrying mutated CTNNB1 (TCGA dataset). (d) TCF/LEF-1 binding sites, responding to β-catenin transcriptional activation in the proximal ACE2 DNA sequence upstream the transcription start site, according to the PROMO program (TRANSFAC database). (e) and (f) Immunohistochemical detection (brown signal) of ACE2 (e) and GLUL (glutamine synthetase, (f) in an HCC tissue microarray (TMA). Slides were slightly counterstained with hematoxylin (blue). Three 1-mm in diameter spots were punched from each formalin-fixed paraffin-embedded routine liver tissue block (n = 41 HCCs; 2 normal liver controls). Digital slides were acquired with a 20X objective. (g) ACE2 and GLUL immunohistochemical signal scoring in HCCs, according to the tumor’s CTNNB1 mutational status, (MUT) versus wild-type (WT); and β-catenin activation scores, (High) versus (Low), using the median value as a cut-off (High, β-catenin activation score > 4). TMA scores are shown as mean (red bar) ± 95% confidence intervals (blue bars). Each dot corresponds to the average out of triplicate tissue cores from each HCC. Statistical differences between means calculated with Mann–Whitney U test. Tumor numbers for each group are indicated. Only were scored those HCCs for which at least two spots were exploitable. TMA33 and β-catenin activation26 scoring were performed as we previously described. Average GLUL and ACE2 scores, CTNNB1 mutational status and β-catenin activation scores for each HCC are provided in Supplementary Table 5. (h) Higher magnification from the indicated spots in (e) and (f).
In line with the higher ACE2 and DPP4 mRNA expression in HCCs showing increased β-catenin pathway activation and CTNNB1 activating mutations, in silico analysis of 5,000 base pairs upstream of the transcription start sites of ACE2 and DPP4 DNAs revealed putative T-cell factor 4/LEF-1 consensus transcription factor binding sites, respectively (Fig. 1d; Supplementary Fig. 4a, b).
ACE2 is predominantly immunodetected in perivenous-type HCCs carrying CTNNB1 mutations and preserving hepatocyte polarization
In consistency with the above findings, scoring of immunohistochemical signal for ACE2 and for the β-catenin target gene GLUL (glutamine synthetase) in a tissue microarray of 41 HCCs (Fig. 1e, f; Supplementary Table 5) revealed that the expression of ACE2 and GLUL were correlated (Spearman’s R = 0.62; p < 0.001; n = 38). Sanger sequencing of the mutational hotspot in the 3rd exon of CTNNB1 revealed that HCCs carrying CTNNB1 activating mutations expressed 5.1- and 3.3-fold higher levels of GLUL and ACE2 proteins than those carrying wild-type CTNNB1 (Fig. 1e–h; Supplementary Table 5). To confirm whether tumors expressing the highest levels of ACE2 protein corresponded to perivenous-type HCCs, we applied our previously described β-catenin pathway activation score26. It results from an equation based on mRNA expression data of five genes as follows: β-catenin activation score = [(GLUL × LGR5 ×ODAM) ÷ (VNN1 × HAL)]26. Thus, we measured mRNA expression for these five genes by real-time PCR on mirror frozen samples from the same HCCs as those included in the TMA, applying our previously described method26,32,33. GLUL and ACE2 protein expression were correlated with β-catenin activation scores (GLUL, R = 0.69, p < 0.001, n = 41; ACE2, R = 0.51, p = 0.001, n = 38). β-catenin activation scores ranged from − 2 to 20, median = 4 (Supplementary Table 5). As previously described26, high scores (above the median), define perivenous-type HCCs. On average, GLUL and ACE2 proteins were respectively 4.0- and 2.6-fold higher in HCCs with high β-catenin activation scores (Fig. 1g). These results indicate that HCCs preserving a perivenous-type metabolic phenotype express higher level of ACE2 protein26.
HCCs with mutated CTNNB1 belong to the class of well-to-moderately differentiated tumors that preserve adult hepatocyte features25. Thus, they are frequently cholestatic and contain pseudo-glandular structures with bile plugs, where hepatocytes are polarized and preserve the basal and apical poles31. Co-immunolabeling for ACE2 and ABCC2 (a.k.a. MRP2, a marker of the biliary pole in hepatocytes) in HCCs, detected ACE2 at the biliary pole and within the lumen of pseudo-glandular tumor structures and in slit-like bile canaliculi (Fig. 2a,b). By contrast, ACE2 was not detected at the basal hepatocyte pole, which was highlighted by the capillary endothelial cell marker CD34 (Fig. 2c). To quantify ACE2 protein expression at the biliary pole of HCC cells, we measured the fluorescence area for separate ACE2 and ABCC2 signals and for dual (ACE2 + ABCC2) staining in 96 TMA spots corresponding to 32 HCCs. To correct for differences in cell densities among HCCs, we plotted specific signal areas as the fraction of the DAPI area (i.e., nuclear staining) (Fig. 2d,e). Mean ± SD ABCC2/DAPI or ACE2/DAPI ratios were: ABCC2/DAPI, 33 ± 27%; ACE2/DAPI, 16 ± 34%; (ACE2 + ABCC2)/DAPI, 3 ± 6%. Of note, 19 ± 25% of all ACE2 expression (i.e., (ACE2 + ABCC2)/ACE2) was located at the biliary poles of HCC cells (Fig. 2d,e).
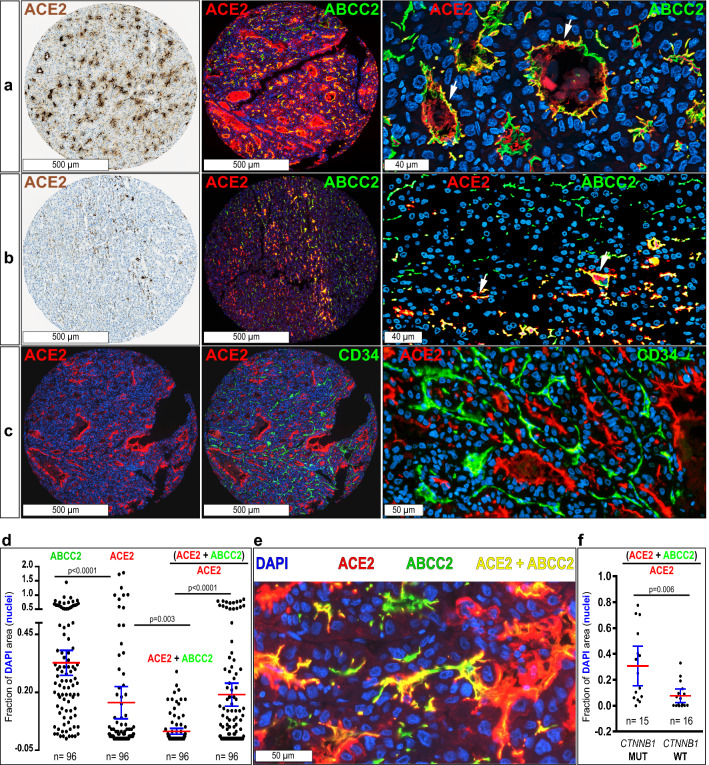
Figure 2.
ACE2 is detected in the biliary pole of moderately-to-well-differentiated HCCs with trabecular or pseudo-glandular patterns. Combined immunoperoxidase and immunofluorescence analysis of a tissue microarray containing 41 HCCs and two normal liver controls, spotted in triplicates. Representative images are shown. Immunoperoxidase signal (brown) is counterstained with hematoxylin (blue). ACE2 appears in red by immunofluorescence, other markers in green and nuclei in blue (DAPI staining). (a), (b) ACE2 colocalizes with the apical hepatocyte marker ABCC2 (a.k.a. MRP2) (arrows). (c) Tumor capillary vessels are marked with CD34. Confocal digital images were acquired with a 40X objective. Images are Z-stacks of four 500 nm focusing steps. (d) Quantification of ACE2, ABCC2, dual (ABCC2 + ACE2) staining and the ratio of (ACE2 + ABCC2) × ACE2–1, representing the fraction of ACE2 protein expression detected as dual (ACE2 + ABCC2) staining, i.e., the fraction of overlap of ACE2 with ABCC2. Values for individual TMA spots from 32 HCCs are expressed as the fraction of DAPI staining of nuclei, to correct for natural variations in cell density in each spot. Means ± 95% confidence intervals are shown for each condition. The statistical significance between groups was calculated with Kruskal–Wallis test (p < 0.0001), followed by the post-hoc Dunn’s test as indicated. (e) Sample scan from a TMA spot showing examples for the different signals quantified in (d). (f) The fraction of overlapping (ACE2 + ABCC2) over total ACE2 signal (i.e., HCC cells expressing ACE2 at the biliary pole), is higher in HCCs carrying CTNNB1 mutations. Each data point represents the mean of two to three cores for each tumor. Statistical significance calculated by Mann–Whitney U test.
In tumors without CTNNB1 mutations, only 8 ± 9% of all ACE2 was localized at the biliary poles. By contrast, in tumors with mutated CTNNB1, 31 ± 27% of all ACE2 was detected at the biliary poles (Fig. 2f) and the mean dual (ACE2 + ABCC2)/DAPI signal was 35 folds higher in HCCs carrying mutated than in tumors carrying wild-type CTNNB1 (mean ± SD: wild-type, 0.0018 ± 0.004; mutated, 0.063 ± 0.07; p = 0.002), thus confirming that HCCs carrying mutated CTNNB1 are enriched in ACE2 expression and indicating that ACE2 is particularly associated with the biliary pole in tumor cells preserving polarized hepatocyte-like features. Adding robustness to these data, dual (ACE2 + ABCC2) expression was correlated with β-catenin activation scores (Spearman’s R = 0.58, n = 31; p = 0.0005).
TMPRSS2 was also detected at the apical pole of some of the pseudo-glandular formations or within slit-like trabecular structures and in cell–cell borders (Supplementary Fig. 5). In contrast with ACE2, the signal for TMPRSS2 was mild in HCCs, suggesting low protein expression levels, which were unrelated to CTNNB1 mutations. Neither ACE2 nor TMPRSS2 (not shown) were detected in myofibroblasts as suggested by dual ACE2 and ACTA2 staining (Supplementary Fig. 5).
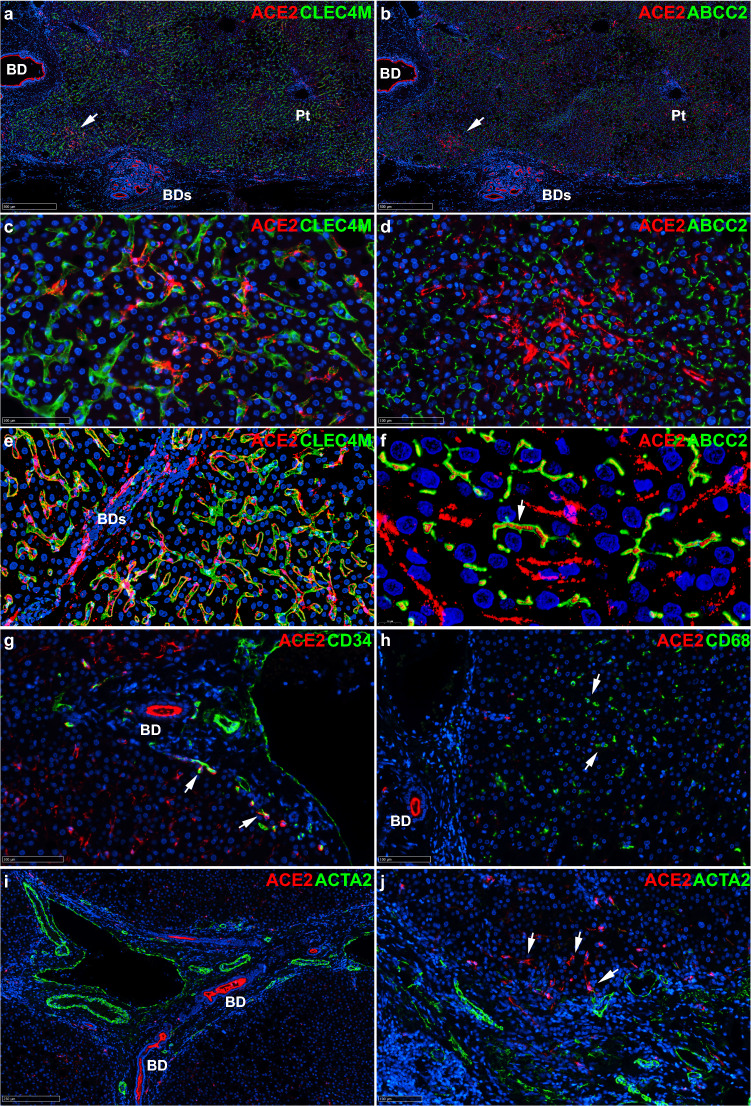
In five non-tumor samples from patients undergoing resection of colon cancer metastases and with minimal inflammatory changes in the liver, ACE2 was co-detected with the sinusoidal endothelial cell marker CLEC4M (a.k.a. DC-SIGNR) (Fig. 3a,c,e) and the bile canaliculi marker ABCC2 (a.k.a. MRP2) (Fig. 3b,d,f). The presence of ACE2 within bile canaliculi is consistent with the facts that the extracellular part of ACE2 can be cleaved off the cell surface by ADAM17, thus shedding functional ACE2 fragments34; and that ACE2 is an abundant component of the normal human bile proteome35. Also, ACE2 was co-detected with CD34 in capillary vessels of the periportal vascular plexus (Fig. 3g). By contrast, ACE2 was not detected in CD68-positive sinusoidal cells (Kupffer cells, Fig. 3h) or ACTA2-positive vascular smooth muscle cells or myofibroblasts (Fig. 3i,j). As expected, the antibodies used detected high levels of ACE2 in the apical intestinal epithelium, Bowman’s capsule and convoluted tubes in the kidney; in turn, TMPRSS2 was detected in the apical compartment of the epithelial lining of prostatic glands (Supplementary Fig. 5).
Figure 3.
ACE2 is detected in sinusoidal endothelial cells, bile ducts and capillaries of the periportal plexus in non-tumor livers. Co-immunostaining for ACE2 and the indicated markers in three non-tumor livers from patients undergoing resection of colon cancer metastasis. Representative images are shown. ACE2 appears in red by immunofluorescence, other markers in green and nuclei in blue (DAPI staining). (a), (b) Low power views (X5 magnification) show ACE2 in a bile duct (BD) and in a zone with mild inflammatory infiltration (arrow) and dystrophic bile ducts (BDs). The sinusoidal endothelial cell marker CLEC4M (a.k.a. DC-SIGNR) and the bile canaliculi marker ABCC2 (a.k.a. MRP2) show lobular staining at this magnification. Pt, portal tract. (c), (d) At X30 magnification, ACE is detected in sinusoidal endothelial cells, which can be identified by comparing CLEC4M (c) and ABCC2 (d) staining. Digital images were acquired with a microscope scanner and a X40 objective and exported at the indicated magnifications (Nanozoomer, Hamamatsu Photonics). (e), (f) Confocal scans of whole tissue sections as Z-stacks of 4 X 500 nm focusing steps at X40 (e) and X143 (f). ACE2 is detected in CLEC4M-positive sinusoidal endothelial cells (e) and within ABCC2-positive bile canaliculi (f). ACE2-positive bile ducts (BDs) are seen in (e). (g) At 28X magnification, ACE2 is sparsely detected in CD34-positive endothelial cells of the periportal vascular plexus (arrows). (h) At 24X magnification, no evidence of ACE2 detection in CD68-positive Kupffer cells. (i), (j) No evidence of ACE2 detection in vascular smooth muscle (i, 10X magnification) or in ACTA2 (a.k.a alpha smooth muscle actin)-positive cells within portal tract stroma (j, 20X magnification). ACE2-positive cells within the portal tract are ACTA2 negative (arrows).
In summary, dual labeling of HCC and of non-neoplastic liver with cell-type specific antibodies showed ACE2 in the epithelial lining of bile ducts and in bile canaliculi, i.e., the biliary pole of normal and differentiated tumor hepatocytes. In non-neoplastic liver, CLEC4M-positive sinusoidal endothelial cells and CD34-positive capillary endothelial cells of the periportal vascular plexus also contain ACE2.
ACE2 and TMPRSS2 co-expression networks highlight metabolic functions typical of normal hepatocytes
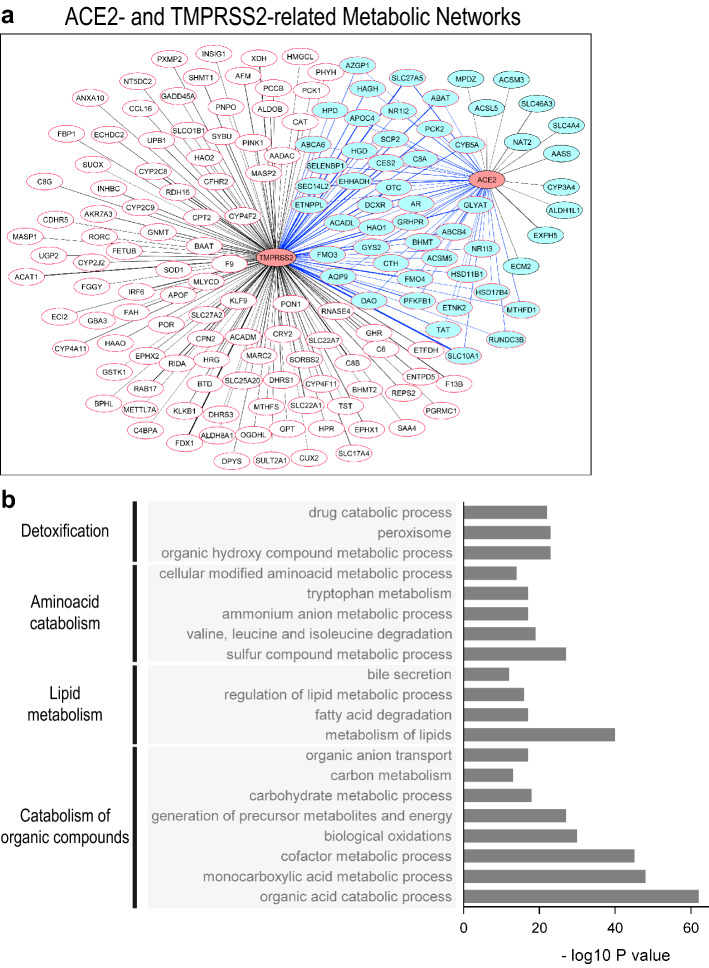
By Weighted Gene Correlation Network Analysis36 comparing periportal-type (n = 342) and perivenous-type (n = 225) HCCs within the Désert’s dataset (n = 1133), we found that ACE2 and TMPRSS2 were related through interacting metabolic functions (Fig. 4a), including detoxification, aminoacid catabolism, lipid metabolism, fatty acid oxidation and catabolism of organic compounds, which is probably related to the carboxypeptidase functions of ACE2, involved in the degradation of inflammatory polypeptides37,38 (Fig. 4b). These findings are in line with the above-described expression of ACE2 and TMPRSS2 in well-to-moderately differentiated HCCs preserving the metabolic programs of adult hepatocytes. Next, we specifically investigated the ACE2 co-expression network in HCCs and analyzed associated gene functions (Supplementary Tables 2, 3, 4). Not surprisingly, ACE2 was associated with hepatocyte-specific functions such as aminoacid metabolism, xenobiotic detoxification, fatty acid uptake and oxidation. High ACE2 expression was associated with low expression levels of oxidative stress markers and cytokine-mediated inflammatory signals (Supplementary Fig. 7a, b). These findings raise the hypothesis of a link between inflammation, ACE2 dysfunction and metabolic breakdown that may be relevant to the pathogenesis of COVID-19.
Figure 4.
(a) Connections between the ACE2 and the TMPRSS2 expression networks in the Désert’s meta-dataset26, in non-proliferative-moderately-to-well-differentiated, periportal-type and perivenous-type HCCs24. The co-expression network was identified by Weighted Gene Correlation Network Analysis and the WGCNA36 R package, followed by visualization with Cytoscape49. Genes co-expressed with ACE2, light-blue nodes with black borders and black links; genes co-expressed with TMPRSS2, white nodes with red borders and black links; genes co-expressed with both ACE2 and TMPRSS2, light-blue nodes with red borders and blue links. Link thickness is proportional to the correlation coefficient between a given node and ACE2 or TMPRSS2. (b) Functional analysis and gene ontology enrichment of the gene co-expression network shown in b are visualized with Express Analysis from Metascape50. Gene ontology (GO) terms, families and P values are indicated.
Discussion
In this work, we show that ACE2 and TMPRSS2 mRNAs and proteins are detected at higher levels in well-to-moderately differentiated HCCs than in poorly differentiated tumors. In particular, ACE2 is detected at the biliary pole in tumors preserving hepatocyte polarization. By contrast, in non-tumor livers, ACE2 is detected not only in the epithelial lining of bile ducts and within hepatocyte’s bile canaliculi, but also in the CLEC4M-positive sinusoidal endothelium and the CD34-positive capillary vessels of the periportal plexus. TMPRSS2, in turn, is not detected in normal liver, but is detected at the biliary pole in well-to-moderately differentiated HCCs, which is consistent with the functional association of ACE2 and TMPRSS2 through periportal and perivenous metabolic networks. Of note, high-resolution 500-nm optical sections revealed ACE2 within hepatocyte’s bile canaliculi in histologically normal liver. This finding agrees with the high content of N-glycosylated, lectin-binding ACE2 in the normal human bile proteome35 and with the role of ACE2 in aminoacid uptake in the gut and in the regulation of the splanchnic blood flow37.
In HCCs carrying activating β-catenin (CTNNB1) mutations, ACE2 DNA is hypomethylated, and ACE2 mRNA expression upregulated. Conversely, TMPRSS2 mRNA is lower in HCCs carrying mutated CTNNB1 and its DNA hypermethylated. This finding agrees with the fact that only ACE2 (and DPP4, but not TMPRSS2) contain TCF/LEF-1 binding sites that are predicted to respond to β-catenin transcriptional activation. Neither ACE2, nor DPP4 or TMPRSS2 are associated to TERT or TP53 gene mutations. On the contrary, HCCs containing mutated TP53 show relatively lower levels of TMPRSS2 (Supplementary Fig. 3D). This association does not necessarily imply a direct regulation of TMPRSS2 by TP53, but may be coincidental. In fact, well-differentiated, periportal-type HCCs express the highest levels of TMPRSS2 and, as we previously showed, have the lowest prevalence of TP53 mutations26. Nonetheless, the relationships of ACE2 and TMPRSS2 with histological, transcriptomic and genomic HCC subclasses converge to demonstrate that tumors preserving hepatocyte-like features, such as hepatocyte trabeculae with bile canaliculi and/or pseudo-glandular structures with an apical biliary pole, express higher levels of the SARS-CoV-2 receptor and co-receptor couple.
The above findings agree with further observations in this work: the ACE2 co-expression network included liver-enriched genes involved in hepatocyte-specific functions, such as aminoacid metabolism (GNMT), xenobiotic detoxification (CYP3A4), fatty acid uptake, bile acid reconjugation (SLC27A5), enterohepatic circulation of bile acids and cholesterol homeostasis (SLC10A1). ACE2 was also co-expressed with SIRT1 and CD36 and the fatty acid oxidation enzymes ACADL, EHHADH, ACADM and ECI2, and alternative glucose metabolism pathways (CRYL1), as well as enzymes involved in gluconeogenesis (PCK2). This network is in line with the role of ACE2 as a regulator of fatty acid uptake in hepatocytes through the angiotensin1-7-MAS1-SIRTUIN1-CD36 axis39. Indeed, ACE2 knock-out mice show low levels of peripheral lipid uptake, but high hepatocyte uptake of triglycerides, resulting in steatosis, lipid peroxidation, oxidative stress and inflammation39. In turn, the induction of oxidative stress by ACE2 deficiency may be related to its effects on glucose and lipid metabolism because the ACE2/Ang1-7/MAS axis increases glucose uptake, decreasing insulin resistance. Thus, ACE2 knock-out mice exhibit high oxidative stress, whereas exposure to Ang1-7 reduces oxidative stress in liver cells through insulin signaling40. Taken together, this body of evidence matches the metabolic landscape of HCCs carrying activating CTNNB1 mutations. These tumors oxidize fatty acids as a source of energy to feed the oxidative phosphorylation pathway41, but resist to oxidative stress42 and suppress inflammation43.
ACE2 expression was related to gene functions that smother inflammation. Indeed, ACE2 was related to low levels of the inflammation-related transcripts BIRC3, MSN, MMP9, LTB, TNFRSF4, IL4R, CCL20 and IL32, among others (Supplementary Table 2). ACE2 exerts anti-inflammatory effects through carboxyterminal cleavage of vasoactive polypeptides, such as Apelin-13, which induces vasodilation, and des-arg9-bradykinin, a neutrophil chemoattractant44. In turn, deletion of ACE2 leads to tissue injury and oxidative stress, through upregulation of the AT1 receptor and inflammatory cytokines like IL1β, IL6, IL17, RANTES, ICAM1, TNFα, TNFRSF1A and downregulation of the anti-inflammatory cytokine IL1045. This picture is surprisingly similar to that seen in the spectrum going from immune dysregulation to macrophage activation syndrome in moderate-to-severe COVID-194,46.
The anti-inflammatory role of ACE2 goes well with its up-regulation in response to liver injury, such as bile-duct ligation47 and with the evidence that ACE2 produced by hepatocytes limits fibrogenesis through degradation of AngII to Ang1-737. In human liver cirrhosis, ACE2 protein is increased by > 90 folds in hepatocytes. In vitro, hypoxia increases ACE2 activity in hepatocytes, enhancing acetylcholine-induced vasodilatation38. As hypoxia activates Wnt/β-catenin signaling, it would be interesting to investigate whether ACE2 expression in perivenous hepatocytes depends on β-catenin34,48.
This body of matching data suggests that ACE2 may regulate energy metabolism, protecting against oxidative stress and inflammation. As both TMPRSS2 and ACE2 are functionally connected through metabolic pathways in the liver, it may be reasonable to hypothesize that ACE2 depletion resulting from SARS-CoV-2 infection18 may contribute to metabolic dysfunction and inflammation. Our results point to the relevance of further studies on the signaling pathways and pathological contexts leading to cell surface ACE2 availability and to the possible impact of viral infection on the disruption of the ACE2 metabolic network.
Materials and methods
Patients, samples and datasets
Human hepatocellular carcinoma (HCC): immunohistochemistry was performed on a tissue microarray (TMA) built from formalin-fixed-paraffin-embedded (FFPE) routine tissue blocks of 41 HCC cases and two histologically normal liver controls, for which the exon 3 of β-catenin (CTNNB1) had been Sanger-sequenced, as we previously described26. In addition, three formalin-fixed, paraffin-embedded non-tumor liver tissue blocks from patients undergoing resection of liver metastases from extra-hepatic cancers were added to the immunohistochemistry analysis. To calculate β-catenin activation scores based on the mRNA expression of GLUL, LGR5, ODAM, VNN1 and HAL, we retrieved real-time PCR mRNA expression data from our previously published work26 corresponding to frozen fragments of the 41 tumors arrayed in the TMA. Quality control matching of frozen and FFPE fragments has been previously described in an 80 HCC dataset including the 41-tumor subset32,33. Tissue samples and patient data were collected and anonymized after obtaining written informed consent from the patients. De-identification was performed in accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. The study was approved by INSERM’s Institutional Advisory Board (approval number 19–630). Research involving human research participants has been performed in accordance with the Declaration of Helsinki. All experiments were performed in accordance with relevant guidelines and regulations. Two different immunohistochemical methods were used to detect primary antibodies: peroxidase-labeled secondary antibodies, revealed by a chromogenic substrate or multiplex immunofluorescence followed by analysis with a confocal slide scanner. The Désert’s microarray meta-dataset composed of 1133 human HCCs was previously described26. In this dataset, HCCs expressing a β-catenin-activated transcriptomic program were identified with a 5-gene signature composed of GLUL, LGR5, ODAM, VNN1 and HAL, which predicts activating CTNNB1 mutations with high sensitivity (0.86–0.91) and specificity (0.83–1.0)26. β-catenin pathway activation was confirmed with an independent 23-gene signature27. The publicly available TCGA dataset was composed of 370 HCCs and 47 matching non-tumor samples. RNA sequencing, mutation and methylation data were extracted as described26, using the TCGAbiolinks R package. Raw mRNA expression was median-normalized (DESeq R package). DNA methylation was quantile-normalized (preprocessCore R package); probes with B-value over 0.8 or under 0.2 were respectively considered as hypermethylated or hypomethylated. The R-package Limma was used to identify differential methylation.
See Supplementary Information for a detailed description of immunohistochemical methods and statistical analyses and Supplementary Table 6 for primary, secondary antibodies, fluorochromes used, epitope unmasking and incubation conditions.
Supplementary Information
Acknowledgements
We thank Caroline Aninat, Latifa Bousarghin, Pierre-Jean Ferron and Olivier Loréal for insightful discussions; Michèle Le Guennec, Patricia Jouas, Adina Pascu and Thomas Poussou for logistics and administrative support.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- Ang
Angiotensin
- CLEC4M
C-Type Lectin Domain Family 4 Member M
- DPP4
Dipeptidyl Peptidase 4
- COVID-19
Coronavirus disease 2019
- CTNNB1
β-Catenin
- HCC
Hepatocellular carcinoma
- LSECs, MERS-CoV
Middle East Respiratory Syndrome Coronavirus
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- TCGA
The Cancer Genome Atlas
- TMPRSS2
Transmembrane serine protease 2
Author contributions
Study design, O.M., L.D., L.C., A.C., B.C.; meta-datasets merge and pretreatment, L.D., R.D.; database search, L.D., L.C., C.L., O.M.; anatomic pathology analysis, B.T., L.C., O.M.; immunohistochemistry, G.G.; statistics, L.D., L.C., O.M.; image analysis, L.C., N.M., O.M.; data analysis, L.D., L.C., O.M.; manuscript preparation, L.D., L.C., O.M.; manuscript editing, all authors.
Funding
Inserm; Univ Rennes 1; Ministère de l’Enseignement Supérieur; Institut National du Cancer, grant # INCA_12688; Ligue Nationale Contre le Cancer 2018, Comités d’Ille-et-Vilaine et Vendée.
Data availability
Datasets analyzed in this study are available at The Cancer Genome Atlas or at the Gene Expression Omnibus databases. Accession numbers and standard, publicly available R packages used are provided in the main text and supplementary files. The procedures applied for merging nine publicly available transcriptomic datasets into a meta-dataset totaling 1133 hepatocellular carcinomas have been previously published26. A filtered, normalized and batch-effect-corrected user-friendly version of this meta-dataset is available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-03710-0.
References
- 1.Guan W-J, et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhatraju PK, et al. Covid-19 in critically Ill patients in the seattle region—case series. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2004500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grasselli G, et al. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy Region, Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giamarellos-Bourboulis EJ, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu C, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Int. Med. 2020 doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantovani A, Beatrice G, Dalbeni A. Coronavirus disease 2019 and prevalence of chronic liver disease: a meta-analysis. Liver Int. 2020 doi: 10.1111/liv.14465. [DOI] [PubMed] [Google Scholar]
- 8.Mao R, et al. Manifestations and prognosis of gastrointestinal and liver involvement in patients with COVID-19: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020 doi: 10.1016/S2468-1253(20)30126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Shi L, Wang FS. Liver injury in COVID-19: management and challenges. Lancet Gastroenterol. Hepatol. 2020;5:428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie H, et al. Clinical characteristics of non-ICU hospitalized patients with coronavirus disease 2019 and liver injury: A retrospective study. Liver Int. 2020 doi: 10.1111/liv.14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Q, et al. COVID-19: abnormal liver function tests. J. Hepatol. 2020 doi: 10.1016/j.jhep.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian S, et al. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Modern Pathol. 2020 doi: 10.1038/s41379-020-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J. Hepatol. 2020 doi: 10.1016/j.jhep.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg. Microbes Infections. 2020;9:469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puelles VG, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020 doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su H, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020 doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie J, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infections. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye G, et al. The association of renin-angiotensin system genes with the progression of hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2015;459:18–23. doi: 10.1016/j.bbrc.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 22.Dai YJ, et al. A profiling analysis on the receptor ACE2 expression reveals the potential risk of different type of cancers vulnerable to SARS-CoV-2 infection. Ann Transl Med. 2020;8:61. doi: 10.21037/atm.2019.12.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu J, et al. Expressions and significances of the angiotensin-converting enzyme 2 gene, the receptor of SARS-CoV-2 for COVID-19. Mol. Biol. Rep. 2020 doi: 10.1007/s11033-020-05478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desert R, Nieto N, Musso O. Dimensions of hepatocellular carcinoma phenotypic diversity. World J. Gastroenterol. 2018;24:4536–4547. doi: 10.3748/wjg.v24.i40.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 26.Desert R, et al. Human hepatocellular carcinomas with a periportal phenotype have the lowest potential for early recurrence after curative resection. Hepatology. 2017;66:1502–1518. doi: 10.1002/hep.29254. [DOI] [PubMed] [Google Scholar]
- 27.Abitbol S, et al. AXIN deficiency in human and mouse hepatocytes induces hepatocellular carcinoma in the absence of beta-catenin activation. J. Hepatol. 2018;68:1203–1213. doi: 10.1016/j.jhep.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 28.Guichard C, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat. Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshida Y, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Can. Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyault S, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 31.Calderaro J, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J. Hepatol. 2017;67:727–738. doi: 10.1016/j.jhep.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 32.Mebarki S, et al. De novo HAPLN1 expression hallmarks Wnt-induced stem cell and fibrogenic networks leading to aggressive human hepatocellular carcinomas. Oncotarget. 2016;7:39026–39043. doi: 10.18632/oncotarget.9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desert R, et al. "Fibrous nests" in human hepatocellular carcinoma express a Wnt-induced gene signature associated with poor clinical outcome. Int. J. Biochem. Cell Biol. 2016;81(Pt A):195–207. doi: 10.1016/j.biocel.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 34.Joshi S, Wollenzien H, Leclerc E, Jarajapu YP. Hypoxic regulation of angiotensin-converting enzyme 2 and Mas receptor in human CD34+ cells. J. Cell. Physiol. 2019;234:20420–20431. doi: 10.1002/jcp.28643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kristiansen TZ, et al. A proteomic analysis of human bile. Mol. Cell. Proteomics. 2004;3:715–728. doi: 10.1074/mcp.M400015-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osterreicher CH, et al. Angiotensin-converting-enzyme 2 inhibits liver fibrosis in mice. Hepatology. 2009;50:929–938. doi: 10.1002/hep.23104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paizis G, et al. Chronic liver injury in rats and humans upregulates the novel enzyme angiotensin converting enzyme 2. Gut. 2005;54:1790–1796. doi: 10.1136/gut.2004.062398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunes-Souza V, et al. CD36/Sirtuin 1 axis impairment contributes to hepatic steatosis in ACE2-deficient mice. Oxid. Med. Cell Longev. 2016;2016:6487509. doi: 10.1155/2016/6487509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao X, Yang FY, Xin Z, Xie RR, Yang JK. The ACE2/Ang-(1–7)/Mas axis can inhibit hepatic insulin resistance. Mol. Cell Endocrinol. 2014;393:30–38. doi: 10.1016/j.mce.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 41.Senni N, et al. beta-catenin-activated hepatocellular carcinomas are addicted to fatty acids. Gut. 2018 doi: 10.1136/gutjnl-2017-315448. [DOI] [PubMed] [Google Scholar]
- 42.Charawi S, et al. LKB1 signaling is activated in CTNNB1-mutated HCC and positively regulates β-catenin-dependent CTNNB1-mutated HCC. J. Pathol. 2019;247:435–443. doi: 10.1002/path.5202. [DOI] [PubMed] [Google Scholar]
- 43.Sia D, et al. Identification of an immune-specific class of hepatocellular carcinoma based on molecular features. Gastroenterology. 2017;153:812–826. doi: 10.1053/j.gastro.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arendse LB, et al. Novel therapeutic approaches targeting the renin-angiotensin system and associated peptides in hypertension and heart failure. Pharmacol. Rev. 2019;71:539–570. doi: 10.1124/pr.118.017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin H-Y, et al. Deletion of angiotensin-converting enzyme 2 exacerbates renal inflammation and injury in apolipoprotein E-deficient mice through modulation of the nephrin and TNF-alpha-TNFRSF1A signaling. J. Transl. Med. 2015;13:255. doi: 10.1186/s12967-015-0616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen SF, Ho Y-C. SARS-CoV-2: a storm is raging. J. Clin. Investig. 2020;130:2202–2205. doi: 10.1172/jci137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paizis G, et al. Up-regulation of components of the renin-angiotensin system in the bile duct-ligated rat liver. Gastroenterology. 2002;123:1667–1676. doi: 10.1053/gast.2002.36561. [DOI] [PubMed] [Google Scholar]
- 48.Zhan L, et al. Hypoxic postconditioning activates the Wnt/β-catenin pathway and protects against transient global cerebral ischemia through Dkk1 Inhibition and GSK-3β inactivation. FASEB J. 2019;33:9291–9307. doi: 10.1096/fj.201802633R. [DOI] [PubMed] [Google Scholar]
- 49.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets analyzed in this study are available at The Cancer Genome Atlas or at the Gene Expression Omnibus databases. Accession numbers and standard, publicly available R packages used are provided in the main text and supplementary files. The procedures applied for merging nine publicly available transcriptomic datasets into a meta-dataset totaling 1133 hepatocellular carcinomas have been previously published26. A filtered, normalized and batch-effect-corrected user-friendly version of this meta-dataset is available from the corresponding author upon reasonable request.