Summary
Background
Multiple sclerosis (MS) has a complex genetic, immune and metabolic pathophysiology. Recent studies implicated the gut microbiome in MS pathogenesis. However, interactions between the microbiome and host immune system, metabolism and diet have not been studied over time in this disorder.
Methods
We performed a six-month longitudinal multi-omics study of 49 participants (24 untreated relapse remitting MS patients and 25 age, sex, race matched healthy control individuals. Gut microbiome composition and function were characterized using 16S and metagenomic shotgun sequencing. Flow cytometry was used to characterize blood immune cell populations and cytokine profiles. Circulating metabolites were profiled by untargeted UPLC-MS. A four-day food diary was recorded to capture the habitual dietary pattern of study participants.
Findings
Together with changes in blood immune cells, metagenomic analysis identified a number of gut microbiota decreased in MS patients compared to healthy controls, and microbiota positively or negatively correlated with degree of disability in MS patients. MS patients demonstrated perturbations of their blood metabolome, such as linoleate metabolic pathway, fatty acid biosynthesis, chalcone, dihydrochalcone, 4-nitrocatechol and methionine. Global correlations between multi-omics demonstrated a disrupted immune-microbiome relationship and a positive blood metabolome-microbiome correlation in MS. Specific feature association analysis identified a potential correlation network linking meat servings with decreased gut microbe B. thetaiotaomicron, increased Th17 cell and greater abundance of meat-associated blood metabolites. The microbiome and metabolome profiles remained stable over six months in MS and control individuals.
Interpretation
Our study identified multi-system alterations in gut microbiota, immune and blood metabolome of MS patients at global and individual feature level. Multi-OMICS data integration deciphered a potential important biological network that links meat intakes with increased meat-associated blood metabolite, decreased polysaccharides digesting bacteria, and increased circulating proinflammatory marker.
Funding
This work was supported by the Washington University in St. Louis Institute of Clinical and Translational Sciences, funded, in part, by Grant Number # UL1 TR000448 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award (Zhou Y, Piccio, L, Lovett-Racke A and Tarr PI); R01 NS10263304 (Zhou Y, Piccio L); the Leon and Harriet Felman Fund for Human MS Research (Piccio L and Cross AH). Cantoni C. was supported by the National MS Society Career Transition Fellowship (TA-180531003) and by donations from Whitelaw Terry, Jr. / Valerie Terry Fund. Ghezzi L. was supported by the Italian Multiple Sclerosis Society research fellowship (FISM 2018/B/1) and the National Multiple Sclerosis Society Post-Doctoral Fellowship (FG-190734474). Anne Cross was supported by The Manny & Rosalyn Rosenthal-Dr. John L. Trotter MS Center Chair in Neuroimmunology of the Barnes-Jewish Hospital Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Keywords: Microbiome, Multi-omics, Diet, Multiple sclerosis
Abbreviations: MS, Multiple Sclerosis; RRMS, Relapsing-remitting MS; DMT, Disease modifying therapy; PCA, Principal component analysis; CNS, central nervous system; mWGS, metagenomic shotgun sequencing; FDR, false discovery rate; EDSS, expanded disability status scale; KOs, KEGG ortholog; GC–MS, gas chromatography/mass spectrometry; SAM, S-adenosyl-L-methionine
Research in context.
Evidence before this study
Previous studies have focused on single aspects of the complex MS pathophysiology. Interactions among the gut microbiome, immune system, metabolism and diet have never been investigated in patients with MS, not to mention the longitudinal stability of these profiles.
Added value of this study
Leveraging advanced multi-omics technologies, our study revealed distinct interaction patterns between immune, metabolic, and gut microbiome domains in people with MS compared to healthy controls. We specifically identified a correlation network linking meat servings with decreased gut microbe B. thetaiotaomicron, increased Th17 cell and greater abundance of meat-associated blood metabolites in MS patients. We demonstrated that these OMICS profiles were stable over a six-month period.
Implications of all the available evidence
Detection of multi-system alterations and interaction patterns in MS patients suggests that an integrated approach is needed for better MS diagnosis and treatment. Our study also underscores the power of using multi-OMICS to dissect the pathophysiology of this and other complex human diseases.
Alt-text: Unlabelled box
Introduction
Multiple sclerosis (MS) is a chronic, autoimmune disease characterized by central nervous system (CNS) inflammation, demyelination, and axonal loss. MS affects 2·5 million people worldwide, and imposes major burdens on individuals and society.1 The aetiology of MS remains elusive, but has been postulated to result from host genetics and environmental factors.2 Dysregulation of immune responses and abnormal metabolism in MS patients suggest that multiple systems are involved in its pathophysiology.3, 4, 5, 6
Gut bacterial communities modulate extra-intestinal immune and metabolic responses in experimental autoimmune encephalomyelitis (EAE),7 a commonly used mouse model of MS. Recent human studies have shown slight to moderate differences at the whole gut microbiome community level between MS patients and healthy controls.7, 8, 9, 10, 11 Intriguingly, specific microbes from MS patients or controls can either adversely or beneficially influence EAE development.12, 13, 14, 15 However, confounding factors such as demographics and diet that might influence gut microbial community have not been well addressed in previous microbiome studies in MS patients; their cross-sectional design is another common limitation. Most importantly, interactions between the gut microbiome, host immune responses and metabolism, and diet have not been evaluated in an integrated manner over time in MS patients. Thus, a holistic and dynamic view of host-microbiome interactions is critically needed, given the multi-factorial nature of MS pathophysiology.
We recently demonstrated the significance of applying multi-omics in studying complex diseases.16 Here, we present a longitudinal deep multi-omics profiling of host immune status, metabolome, gut microbiome, and dietary habits in MS patients and healthy controls. This will generate novel, comprehensive, and dynamic insights of host-microbiome interactions in this complex disease.
Methods
Ethics
The study was approved by the Human Research Protection Office at Washington University in St. Louis School of Medicine (WUSM) (approval number: 201,502,105). All patients gave informed consent to participation.
Study participants
This is a matched case and control longitudinal study. MS patients were recruited at the John L. Trotter MS Center of WUSM from 2015–2018. Inclusion criteria for MS patients were: (1) diagnosis of MS using the 2010 revision of the McDonald criteria;17 (2) no DMT or steroid treatments in the past 3 months; (3) ages 18 to 50 years; and (4) not in clinical relapse at study enrolment. Exclusion criteria were: (1) coexistence of other chronic inflammatory (e.g., asthma, chronic hepatitis, inflammatory bowel disease, celiac disease, etc.) and autoimmune (e.g., rheumatoid arthritis, SLE, type I diabetes, etc.), or metabolic (e.g., type II diabetes, familial hypercholesterolemia, etc.) diseases; (2) antibiotics or steroid therapy in the past 3 months; (3) history of immunosuppressive or chemotherapeutic treatment; (4) history of chronic infectious disease (e.g., TBC, HIV, HBV, HCV, etc.); (5) neoplastic disease not in complete remission, and (6) pregnancy. Age, sex, BMI and ethnicity-matched healthy controls were enrolled using the same exclusion criteria. Table 1 details the demographic and clinical characteristics for all participants at enrolment.
Table 1.
Demographics and clinical characteristics of MS patients and healthy controls.
| Healthy control | RRMS | P value | |
|---|---|---|---|
| Number | 25 | 24 | |
| Sex M:F | 3:22 | 3:21 | P=0·5 |
| Age, Y, mean±SD | 38·9±7·3 | 40·2±8·76 | P=0·5 |
| BMI, kg/m2 | 26·9 (6·3) | 27·3 (6·7) | P=0·8 |
| Caucasian n (%n) | 20 (80%) | 23 (95·83%) | P=0·1 |
| Age at first symptoms Y, mean±SD | NA | 32·9±5·9 | NA |
| Age at MS diagnosis Y, mean±SD | NA | 33·9±5·9 | NA |
| OCB+ n (%n) | NA | 10 (45·5%) | NA |
| EDSS median (min-max) | NA | 2·5 (0·0–4·5) | NA |
| Tobacco use# | 0 | 9 (37·5%) | P<0·0005 |
| Started DMTs during the study n (%n) | NA | 8 (33·3%) | NA |
| Probiotic n (%n) | 3 (12%) | 4 (16·6%) | P=0·7 |
| Supplements n (%n) | 9 (36%) | 9 (37·5%) | P=0·9 |
Data are provided as n (%), mean (SD) or range; # including cigars, pipes or electronic cigarettes. BMI: body mass index; EDSS: expanded disability status scale; OCB: oligoclonal bands. DMTs: disease modifying therapies; SD=standard deviation.
Differences between groups were compared using Pearson chi-square test for categorical data and the t-test or non-parametric Mann-Whitney test as appropriate for continuous data.
MS and healthy control participants were studied again six months after baseline. No relapse was reported in any of the MS patients during the study. Although DMTs commencement was strongly recommended to the 24 MS patients by their clinicians, only 8 started DMT treatment during the six-month study period. The DMTs started were glatiramer acetate and fingolimod (n=1 each), interferon-β1a (n=3), and dimethyl fumarate (n=3).
Sample collection
Stool and blood of all participants were collected at the time of enrolment and six months later. Collection of stool and blood samples were done concomitantly (less than 24 h difference). Stools were self-collected and placed on frozen gel packs and shipped overnight to the research laboratory. Upon receipt, stools were immediately stored at -80 °C until further processing. Stools from baseline and six months were processed at the same time for DNA extraction and microbiome sequencing to minimize batch effects among the specimens.
Blood was collected in heparinized tubes, insulated, and shipped at room temperature overnight to Ohio State University for immunophenotyping. Peripheral blood mononuclear cells (PBMCs) were isolated immediately on arrival and analyzed by flow cytometry. Serum samples were stored frozen at -80 °C and sent to University of Massachusetts for metabolomics.
Stool DNA extraction and microbiome sequencing
16S rRNA gene sequencing permits deep microbiota profiling, especially of low abundance taxa. Metagenomic whole genome shotgun sequencing (mWGS) classifies to species levels but may not enumerate low abundance bacteria. We applied these complementary platforms to sequencing the gut microbiome.
Stool DNA extraction and sequencing were performed as we have done previously.16 Briefly, stool DNAs were extracted using the MOBIO PowerSoil DNA Extraction kit. For 16S rRNA gene sequencing, hyper-variable regions V1–V3 of 16S gene were amplified using primers 27F and 534R (27F: 5′-AGAGTTTGATCCTGGCTCAG-3′ and 534R: 5′- ATTACCGCGGCTGCTGG-3′). 16S libraries were prepared and sequenced on the Illumina MiSeq sequencing platform using a V3 2 × 300 bp paired end sequencing protocol with a target read depth of 10,000 reads/sample. Illumina's software handles the initial processing of all the raw sequencing data. One mismatch in primer and zero mismatch in barcodes were applied to sample deconvolution. Reads were further processed by removing sequences with low quality (average qual <35) and ambiguous bases (Ns). Chimeric amplicons were removed using UChime (v4·2·40), and amplicon sequences were clustered and operational taxonomic units (OTU) picked by Usearch against SILVA_132_SSURef_Nr99 database at 97% threshold. Final taxonomic assignment was conducted using RDP-classifier (v2·11) with 0·8 confidence value as cut-off. Potential contaminant taxa present in DNA extraction control and PCR controls were removed for downstream analysis from the sequenced samples. mWGS libraries were prepared using the Nextera XT DNA Library Preparation kit (Illumina), according to the manufacturer's standard protocol. Pooled libraries were sequenced on the Illumina HiSeq2500 instrument using a 2 × 150 bp paired end sequencing protocol, targeting 3 G bp/sample. We demultiplexed the raw reads, and further processed them by (a) removing human reads using NCBI's BMTagger (v3·101) (ftp://ftp.ncbi.nlm.nih.gov/pub/agarwala/bmtagger); (b) removing duplicated reads using GATK-Picard 4·1·0 (MarkDuplicates); (c) trimming low-quality bases and low-complexity screening using PRINSEQ (v0·20·4). We used MetaPhlAn2 to classify species and HUMAnN2 for microbiome functional potential inference (gene ortholog or KEGG ortholog and metabolic pathways),18 and Lefse (v1·0) to determine the statistical difference of KOs or metabolic potentials between MS patients and controls.19 We performed sequencing and processed data at The Jackson Laboratory for Genomic Medicine.
Blood sample preparation and processing for non-targeted metabolome
Serum samples were vortexed with 80% chilled methanol aqueous in the ratio of 1:60 (v/v) to precipitate protein, following by centrifugation (14,000 g, 4 °C, 10 min). The supernatants were collected and dried under vacuum. The dried residues were reconstituted with 50% methanol aqueous and centrifuged (14,000 g, 4 °C, 15 min). We performed ultraperformance liquid chromatography-mass spectrometry (UPLC-MS) at the University of Massachusetts Amherst mass spectrometry facility to analyse serum for untargeted metabolomics. Equal volume of each serum sample was pooled to prepare the quality control (QC) sample. Five identical QC sample runs were conducted prior to running test samples, and one QC sample run was performed for every eight sample runs throughout the experiment. Acquity UPLC HSS T3 column (2·1 mm × 100 mm, 1·8 µm, Waters Co., MA, USA) was used to obtain the chromatographic separation by injecting 5 µL aliquots of each sample. The column was maintained at 40 °C, and the flow rate was 0·5 mL/min. Solvent A was 95% water with 5% ACN and 0·1% formic acid and solvent B was 100% ACN with 0·1% formic acid. The gradient started at 2% of solvent B and linearly increased to 95% of solvent B at 8 min; held at 95% of solvent B for 2 min. The column was equilibrated at 2% of solvent B for 5 min before the next run. MS was conducted using the Thermo Fisher Orbitrap-Fusion in negative electrospray ionization mode at the detection range of 120–1000 m/z with 60,000 full width at half maximum resolution. The following conditions were used for MS: spray voltage 3500 V, sheath gas flow rate 15 (arbitrary units), auxiliary gas flow rate 6 (arbitrary units), sweep gas flow rate 3 (arbitrary units), vaporizer temperature 275 °C, ion transfer tube temperature 325 °C.20
Short-chain fatty acids determination by GC chromatography
SCFAs in stool samples were extracted by rigorous vortex with 0·05% phosphoric acid at the ratio of 1:15 (w/v), followed by centrifugation. Supernatants were mixed with ethyl acetate at the ratio of 1:1 (v/v), the SCFAs were transferred to ethyl acetate, and the supernatants were obtained after centrifugation. SCFAs were measured by gas chromatography with a flame ionization detector (Schimadzu GC-QP2010 SE, Tokyo, Japan). Acetate, propionate, butyrate, isobutyrate, valerate, isovalerate were used as standards for the identification, and 2-ethylbutyrate was used as the internal standard.
Blood sample preparation for immune profiling
PBMCs were isolated on a Ficoll gradient, washed in PBS, and resuspended in RPMI1640 medium (Corning) containing 5% heat-inactivated-human serum (Sigma-Aldrich), 1% HEPES containing 1% L- glutamine, and 1% penicillin/streptomycin. We used seven flow cytometry antibody panels (from A to G) to analyze lymphocyte and monocyte populations. Panel A included: V450-CD3 (UCHT1), PECy7-CD19 (SJ25C1), FITC-CD56 (NCAM16·2), and APC-H7-CD27 (M-T271). Panel B included: V450-CD3 (UCHT1), V500-CD4 (L200), PECy7-CD8 (RPA-T8), APC-CD45RA (HI100), and APC-H7-CD27 (M-T271). Panel C included: V450-CD3 (UCHT1), V500-CD4 (L200), FITC-CD25 (M-A251), and PE-FOXP3 (PCH101). Panel D included: V450-CD3 (UCHT1), V500-CD4 (L200), PECy7-CD8 (RPA-T8), APC-CD45RA (HI100), FITC-Tbet (4B10), PE- IFNγ (4S.B3), PerCP-Cy5·5-GMCSF (BVD2–21C11), and APC-Cy7-IL17 (BL168). Panel E included: V450-CD3 (UCHT1), PECy7-CD19 (SJ25C1), PerCP-Cy5·5-CD5 (L17F12), PE-CD1d (42·1), APC-H7-CD27 (M-T271), FITC-IL10 (BT-10), and APC-IL35/27(EB13). Panel F included: V450-CD3 (UCHT1), V450-CD19 (SJ25C1), PE-HLA-DR (G46–6), PE-Cy7-CD11c (B-ly6), PerCP-CD14 (MφP9), APC-CD123 (7G3), and FITC-BDCA2 (201A). Panel G included: V450-CD3 (UCHT1), V450-CD19 (SJ25C1), PE-HLA-DR (G46–6), PE-Cy7-CD11c (B-ly-6), PerCP-CD14 (MφP9), APC-CD16 (B73·1), and APC-H7-CD80 (L307·4). For panel D, the PBMCs were stimulated with 50 ng/ml of PMA (Sigma cat#16,561–29–8) and 1 μg/ml of ionomycin (Sigma cat#I0634), and treated with 0·2 ul of Golgi plug (BD cat#555,029) for 4 h before staining. For Panel E, we stimulated PBMCs with 50 ng/ml of PMA (Sigma cat#16,561–29–8), 1 μg/ml of ionomycin (Sigma cat#I0634), and CpG for 4 h before staining. We performed staining as previously described.21 Briefly, 1 × 106 cells were washed and resuspended in cold PBS/1% BSA, incubated with FcR Blocking Reagent (Miltenyi) for 10 min, and incubated with antibody cocktails for 30 min. Samples were washed and fixed with either PFA (0·5%), Cytofix/Cytoperm Solution Kit (BD Biosciences), or Foxp3 Transcription Factor Staining Set (eBioscience). Intracellular molecules were stained in their appropriate permeabilization wash buffers for 30 min (cytokines) or 45 min (Foxp3). After a final wash with PBS/1% BSA, we acquired and analyzed cytometric data using BD FACSCanto II, FACSDiva, and FlowJo Software v.10·7 (Becton, Dickinson and Company, 2019). Table S2 and Figure S5 describe the gating strategy for each panel, and the percentage of each cell subset population within the parent gate.
Food diary and conversion to food serving
A four consecutive days food diary was self-recorded to provide qualitative dietary information before the stool sample collection at baseline and at six months. The four days included two weekend days and two weekdays.22, 23, 24 The participants were asked to continue their usual eating habits before the initial stool collection and for the next six months.
We analyzed the food diaries using the Nutrition Coordinating Center (NCC) Food Group Serving Count System which estimates intake of food groups (e.g., daily servings of sugar sweetened beverages).25 This system assigns foods in the Nutrition Data System for Research (NDSR) database (2017 version) to food groups that fit within nine major categories. Eighteen food groups including average daily servings of fruit, vegetable, whole grain, refined grain, meat, poultry, fish and shellfish, cold cuts and sausage, eggs, nuts and seeds, butter and animal fats, plain and flavored cow's milk, dairy cheese, yogurt live active cultures, vegetable oils, salad dressings, beer and liquor and wine, sugar sweetened soft drinks (soda, punch, tea), and their daily serving sizes were used for our study. Serving sizes are assigned to each NDSR food based on the 2000 Dietary Guidelines for Americans recommendations when available.26 For foods not included in recommendations (e.g., cookies, fruit drinks), United States Food and Drug Administration (FDA) serving sizes are used.
Statistical analysis of the microbiome data
We performed both formal and exploratory statistical testing of the microbiome data. To visualize differences in overall microbial community structure, principal component analysis (PCA) was conducted after log-ratio transformation of the microbiome data and scaling using ‘Compositions’ package from R. To determine if the overall microbiome differs significantly between MS patients and controls, we employed permutational multivariate analysis of variance (PERMANOVA) using the ‘vegan’ package, after which we tested the homogeneity of dispersion among groups after controlling for tobacco use, given tobacco use is statistically different between MS and controls. To identify specific microbes (>0·1% relative abundance) that statistically differ between MS patients and controls, we performed differential analysis based on the negative binomial distribution using DESeq2 software packages.27 All P values were two sided. Adjusted P values with a False Discovery Rate (FDR) of <0·05 was considered as statistically significant. The FDR statistical adjustment of P values and their cut-offs were applied to other omics data analysis involving multiple comparisons unless otherwise noted. Because of potential high false positive rate of DESeq,28 we further inspected the results by plotting raw and relative abundance data. We removed results that are low in relative abundance (<0·1%) from final reporting. As a sensitivity analysis, DESeq was also run after excluding these low abundance taxa.
We conducted PERMANOVA with Bray-Curtis distance to quantify the microbiome variance explained by individual factors, as described previously.29 We tested age, BMI, smoking, family history of autoimmune diseases, probiotics intake and diagnosis (MS vs controls) individually in the PERMANOVA model. Note the total variance explained by each variable was calculated independent of other variables and should therefore be considered the total variance explainable by that variable.
Statistical analysis of immune profile
We present immune cell populations from the flow cytometry as proportions and scaled for PCA analysis to estimate the overall similarity of the immune profile among all participants. Wilcoxon Rank Sum Test was used to identify significantly different immune cell populations between MS and controls. P values were further adjusted by FDR approach. All p-values were two-tailed.
Statistical analysis of blood metabolome
The ion data set was subjected to peak detection, nonlinear alignment, and integration by XCMS (https://xcmsonline.scripps.edu). The data set in the mzXML data matrix converted by XCMS was normalized by sum of total of the observed peaks. The peaks with RSD > 20% in QC samples was excluded to guarantee the quality of data set, following by the univariate and multivariate analysis to differentiate the unbiased metabolites. We subjected resulting m/z values to the “MS peaks to pathways” analysis in Metaboanalyst (https://www.metaboanalyst.ca/) to analyze pathways and identify metabolites with a maximum error of 5 ppm using KEGG and Metlin databases. Welch's t-test was used to determine significant changes between the control and MS groups. Previous studies supported that parametric and non-parametric univariate tests result in very similar results for metabolome data.30 P values were further adjusted by FDR approach. All p-values were two-tailed.
Mantel correlation and multi-OMIC feature-feature correlation
We quantified covariation between multi-omics using Mantel tests (Pearson correlation between distances of two matrices). A pair-wised inter-participant variation/distance matrix was first computed for each OMIC dataset, with Bray-Curtis dissimilarity for the microbiome data and Euclidean distance for the immune profile, blood metabolome and food intakes. Inter-participant dissimilarity matrices were then compared using the mantel function in the vegan package. Mantel correlation analysis was also conducted in similar manner to quantify longitudinal covariation for two given omics data. The significance of the statistic is produced by permuting rows and columns of the first dissimilarity matrix for 1000 times.
We performed feature-feature correlations within and between omics datasets using Pearson correlation with cor.test function in the stats package in R. Because of potential for different interactions in MS patients and controls, all correlations were performed separately for the two groups, accounting for BMI and age. P values were corrected based on FDR approach. FDR <0·2 was considered significant for correlations among features from the microbiome, immune profile and food intakes and FDR <0·05 was considered significant for correlations with features from the blood metabolome. To further remove the false positive or negative correlations that were driven by single values, we generated x-y scatter plots all statistically significant correlations, and confirmed them by manual inspection. The resulting correlations with absolute correlation coefficient >0·7 were considered strong correlations and the network of correlation was illustrated using Cytoscape. A hub in the correlation network was defined as nodes with at least 20 connections. All correlation results including before and after FDR corrections and after manual inspections are summarized in Table S4.
MS classification using machine learning models
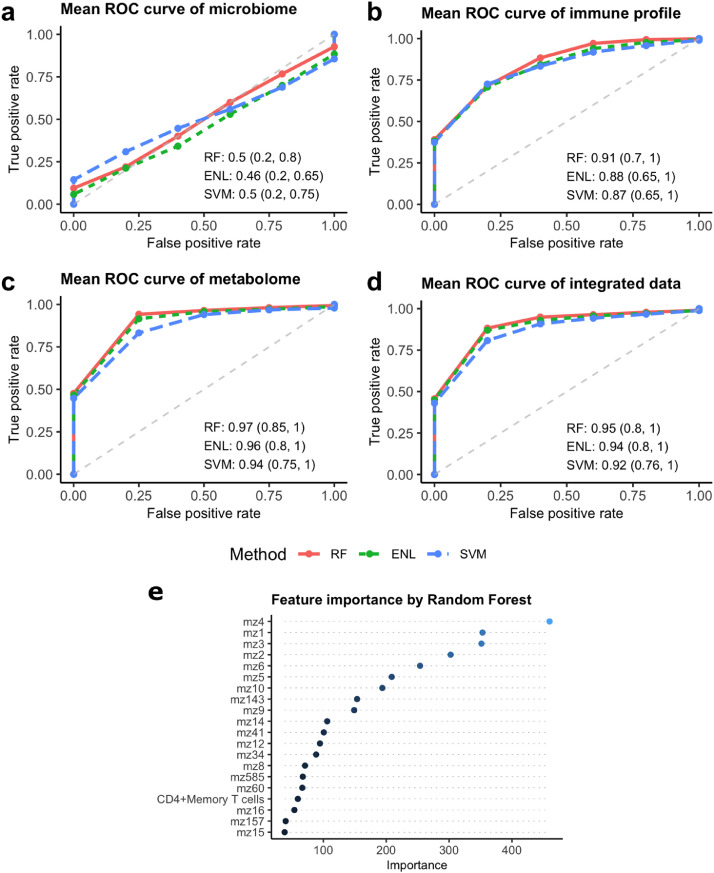
We tested three machine learning models (random forest (RF), elastic net regularized linear regression (ENL), and elastic net regularized support vector machine (SVM)) to classify MS patients and controls. All three models can be used to analyze high-dimensional data (when the number of features is larger than the sample size) and to generate measures of feature importance.31, 32, 33 The models were trained by each individual omics to determine the importance of a given omics data in classification performance (Figure 3a, b, c), or by the combination of all the omics to determine whether it achieves a better classification performance (Figure 3d). For the microbiome data, OTUs with 0 abundances were firstly replaced by a small value 1e-5. Then the centred log-ratio transformation was applied,34 so that the transformed data obey the Euclidean geometry. The number of raw features could greatly exceed the sample size and many of them are either redundant or irrelevant in distinguishing MS patients and controls. Hence, we reduced the size of the feature set before fitting any machine learning model, by applying a statistical marginal screening procedure through multiple hypothesis testing with false discovery rate control.35 Such a hybrid “marginal screening + machine learning” approach facilitated the model training and consistently improved the performance of the classifiers in our study.
Figure 3.
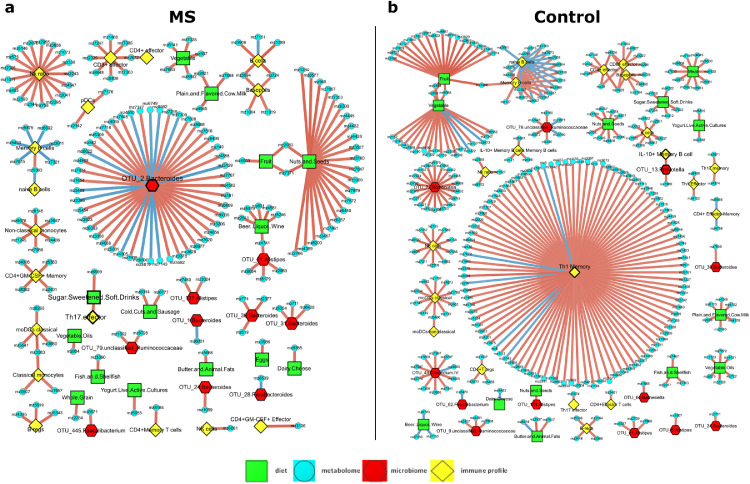
Feature-feature correlations from multiple-omics in MS patients and controls. Three-hundred thirty-two (controls) and 185 (MS patients) statistically significant correlations are illustrated using Cytoscape. Nodes–Red pentagons-Microbiome; yellow squares-immune profile; blue circles-blood metabolome; green squares- food serving; Edges–Blue- negative correlations between compared pairs; Red-positive correlations between compared pairs.
We randomly divided the data into a training set with ∼80% samples to build a classifier and a testing set with the remaining ∼20% of the samples to evaluate the performance of the resulting classifier. Under each setting, this random-splitting procedure were repeated 200 times in order to stably assess the out-of-sample predictability of a classifier and its associated feature importance. Specifically, in each random split, when training the ENL and the SVM, we used Leave-one-out Cross Validation (LOOCV) to tune their regularization parameters.36 For RF, we set the maximum number of features allowed to try in an individual tree as the square root of the number of features, and the number of trees as 30,000, a sufficiently large number. The feature importance in the RF is measured by the Mean Decrease Gini. After building a classifier using the training samples, we used the testing samples to compute its pairs of out-of-sample true positive rate (TPR) and false positive rate (FPR), based on which we constructed the sample receiver operating characteristic (ROC) curve and calculated the corresponding area under the ROC curve (AUC) value. By aggregating these results from 200 random splits, we draw the average ROC curve and computed the average AUC and its 90% confidence interval for each classifier. Missing data were imputed based on group mean.
Justification for statistical approaches used in single and multi-OMICS analysis
All the analysis were conducted using a complete case analysis except for the multi-omics predictive modelling analysis, in which multi-OMICS data were combined to evaluate classification accuracy for MS and controls. There was no missing in microbiome and metabolome data. In the immune data, 36.0% of the subjects have missing values. This is largely attributed by one immune feature that has very low level of immune cells. In the metadata, 14.0% of subjects have missing values. In the nutrition data, 27 % of subjects have missing values. In the multi-omics predictive modeling analysis, as much as 82.7% of subjects have at least one missing value. Therefore, due to the already limited sample size, it is not practical to run a complete case study. On the other hand, the overall missing rate (total missing values among all value) of the multi-omics data is very low: it is 4.6%. We thus chose the simple imputation method of using group means (by MS/Control).
For our immune data analysis, some immune cell populations are not normally distributed, thus we chose a robust non-parametric Wilcoxon Rank Sum test for two group comparisons. For metabolome data, because of large variation of the data, log transformation is conventionally encouraged and performed, followed by Welch's t-test. Mantel correlations between two matrices were based on continuous data derived from Euclidean distance or Bray-Curtis dissimilarity and showed good linear relationships as demonstrated by scatter plots. Generally, if the scatter plot shows linear trend, instead of other curvature trend, ex quadratic trend, the data satisfies the linearity assumption. Due to the limited sample size, our general strategy is to use parametric methods to gain more power when their underlying assumptions are not violated, otherwise non-parametric tests are adopted to ensure robustness.
Role of funding source
The funders had no role in the conceptualization, study design, data collection, analysis, interpretation of data, in writing the paper, or in the decision to submit the paper for publication.
Results
Baseline characteristics of the study population
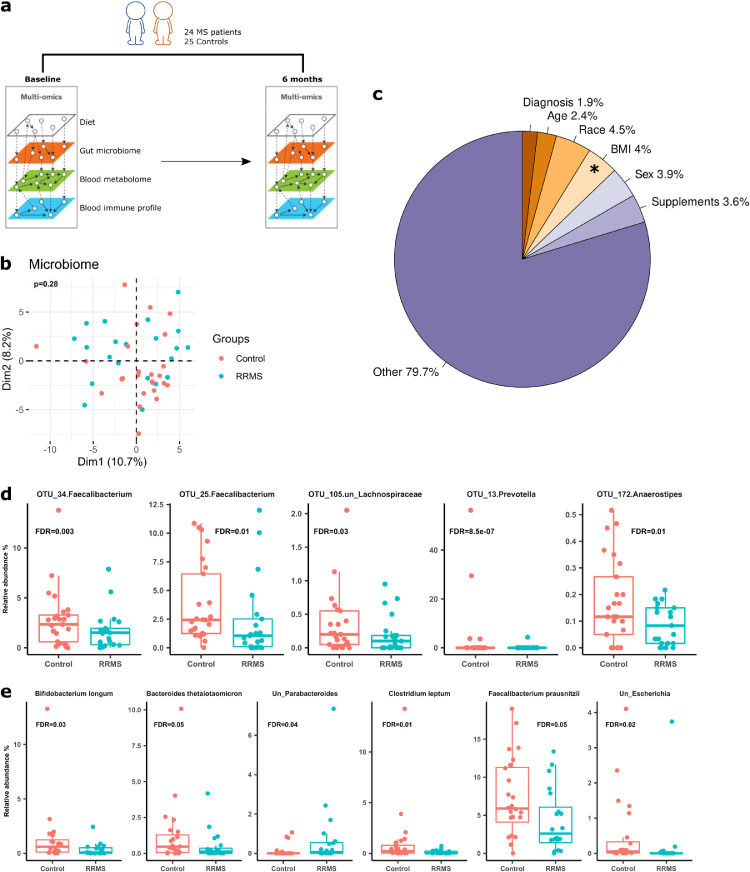
Twenty-four relapsing-remitting MS (RRMS) patients and 25 unrelated healthy controls matched for age, gender and other important clinical variables were enrolled in this study. The majority of the participants are females. At baseline, no MS patient was in active relapse or had received any disease modifying therapies (DMTs) in the prior three months. Mean disease duration at study entry was 6·4 (SD 1·5) years. Table 1 summarizes the characteristics of the participants at baseline. The MS group used more tobacco (P<0·0005, chi-square test), but other characteristics of the control and MS groups did not differ significantly. Stool and blood samples were collected at entry (baseline) and six months later for gut microbiome, blood metabolome and blood immune cell analyses (Figure 1a). Participants collected four-day food diaries to provide qualitative dietary information.
Figure 1.
The gut microbiome in MS and control individuals at baseline. (a) Study design–Stool and blood samples were collected from RRMS patients (n=24) and healthy controls (n=25) at baseline and six months later. Stools were used for gut microbiome characterization, and blood samples were used for immunophenotyping and global metabolome characterization. 4-day food diary was obtained to inform on habitual dietary patterns of study participants. (b) Principal component analysis (PCA) of the gut microbiome in MS patients and controls using baseline 16S rRNA data. The microbiome proportional data was subjected to log-ratio transformation. The resulting data were used for PCA analysis to view inter-participant variation in MS patients and controls. The first two principal components and their corresponding proportion of variance explained are shown. (c) Variance of baseline microbiome explained by clinical and demographic factors. BMI significantly contributed to microbiome variance, accounting for 4·0% of total variance. Status (MS vs controls) did not have significant impact on the microbiome variation. (d). Taxa that are significantly different between MS patients and controls at baseline in 16S rRNA gene sequencing (FDR <0·05). Differential taxa were identified by DESeq2. (e) Taxa that are significantly different between MS patients and controls at baseline in metagenomic whole genome shotgun sequencing (FDR <0·05, DESeq2).
Overall gut microbiota profile and factors underlying microbial variation in MS patients and controls
We first compared gut microbiome profiles at baseline in MS and control groups using 16S rRNA gene sequencing. Principal component analysis (PCA) plot (Figure 1b) demonstrated no clustering that distinguished the MS microbiome from that of controls at the operational taxonomic unit (OTU) level. PERMANOVA analysis after adjusting for confounding factor (tobacco use) further confirmed no statistically significant difference between groups (P=0·28), suggesting similar overall gut bacterial community structures in the two groups. Alpha and beta diversity of the gut microbiome were also similar between groups (Figure S1a). We additionally conducted metagenomic whole genome shotgun (mWGS) sequencing to map bacterial community structure at species level (Figure S1b-c). Similar to what observed for 16S sequencing, PCA analysis and PERMANOVA test using mWGS data failed to define a distinctive MS microbiome community structure at the species level (P=0·20) (Figure S1d). These findings are consistent with prior studies that showed only modest gut microbiome difference between MS patients and healthy controls.7,37
Next, we quantified the variance explained by individual host characteristics and other factors that might influence the microbiome community structure (Figure 1c). By PERMANOVA, the tested variables explained only a small proportion of microbiome variance. Body mass index (BMI) accounted for 4·0% of total variance in the gut microbiome (P=0·03), whereas participant status (MS or control), age, race, sex and use of oral supplements (i.e., vitamins) accounted for small proportions of total microbiome variation (Figure 1c), none of which achieved statistical significance. Hence, BMI is modestly associated with the configuration of the gut microbial community in MS and controls, but other yet-to-be determined factors may play larger roles.
Specific gut microbiota associated with MS
Next, we sought to identify differences in specific microbes that might be associated with MS. To do so, we compared the gut microbiota compositions between MS and controls at baseline by differential microbiome abundance analysis using DESeq2. 16S rRNA gene sequencing demonstrated that the relative abundances of two Faecalibacterium OTUs, one Prevotella, one unclassified Lachnospiraceae, and one Anaerostipes OTU were significantly decreased in MS patients after multiple comparison correction by false discovery rate (FDR) (Figure 1d, FDR < 0·05). The relative abundance of Prevotella were bimodally distributed, with high abundances in only several healthy controls, in accordance with Human Microbiome Project data, in which only a small fraction of healthy American adults harbours this genus in high abundance.29 mWGS data identified six species that are significantly lower in abundance in MS patients than in controls, three of which have known immunomodulatory properties (Bifidobacterium longum, Clostridium leptum, Faecalibacterium prausnitzii) (FDR <0·05).38, 39, 40 Bacteroides thetaiotaomicron and two unclassified Parabacteroides and Escherichia species were also significantly different between two groups (Figure 1e). Thus, lower abundance of Faecalibacterium species was consistently detected by both sequencing technologies in MS patients compared to healthy controls. The average relative abundance of B. fragilis, which is protective in the EAE model,41 was 0·34% at baseline, with no statistically significant differences between MS and controls by univariate analysis (Figure S2). In summary, decreased relative abundances of bacteria with immunomodulatory properties seem to characterize gut microbiome changes in MS patients vs. controls.
Among MS patients, the gut microbiome differed significantly by the degree of disability at baseline (P=0·035, PERMANOVA), as measured using the expanded disability status scale (EDSS) (mean 2·9, range 0–6·5). The difference maintained statistically significance after controlling for BMI (P= 0·045). EDSS was positively correlated with BMI (Spearman correlation r=0·80, P=4·4e-05) (Figure S3) and was greater in MS patients who smoked, but the difference did not achieve statistical significance (p=0·06). Correlation analysis using mWGS data showed that Collinsella aerofaciens (r=0·49, FDR=0·046), Coprococcus comes (r=0·62, FDR=0·016), Phascolarctobacterium succinatutens (r=0·63, FDR=0·016), Sutterella wadsworthensis (r=0·56, FDR=0·025) were positively correlated with the EDSS. Notably, Collinsella is increased in patients with rheumatoid arthritis and is associated with increased gut permeability.42 Sutterella, a Gram-negative genus from Proteobacteria, has been associated with various diseases including autism and inflammatory bowel disease (IBD).43 Eubacterium siraeum (r=-0·47, FDR=0·049) was negatively correlated with EDSS. These findings suggest that specific gut microbes may be associated with the degree of disability in MS patients.
Next, we inferred the metabolic potentials of the gut microbiome for all study participants using mWGS data by HUMAnN2 and LEfSe. Sixty-six metabolic pathways and 276 gene Ortholog or KEGG ortholog (KOs) significantly differed between MS patients and controls before adjusting for multiple comparisons (Table S1). Interestingly, most differentiating pathways (64/66=96·9%), which included glycolysis, glutamate degradation, fermentation pathways or phospholipid biosynthesis and KOs (244/276=88·4%), were under-represented in MS patients compared to controls. However, after adjusting for multiple comparisons, no KO or pathway differed significantly between the two groups (all FDR > 0·3).
Additionally, we measured concentrations of the short chain fatty acids (SCFAs) including acetic acid and butyric acid in stool by gas chromatography/mass spectrometry (GC-MS) and found a trend of lower level of SCFAs in stools of MS patients than in those of controls (Figure S4), but the trend did not attain statistical significance (P>0·05, Wilcoxon Rank Sum Test) and the inter-individual variations of SCFAs in both groups were quite high.
Loss of the microbiome-immune homeostatic interaction and establishment of an immune-metabolome association in MS
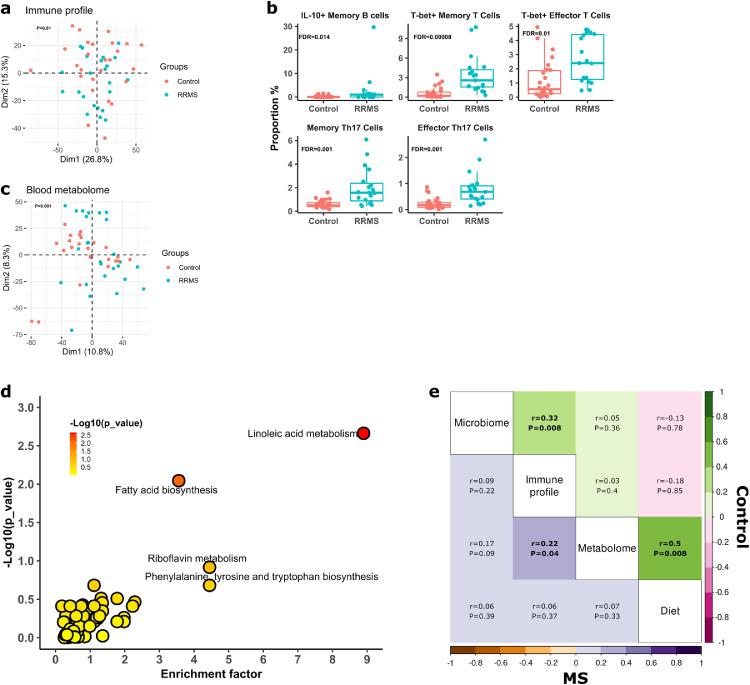
We next asked if and to what extent the gut microbiome is associated with peripheral blood immune and metabolome profiles. PCA analysis of 42 blood immune cell populations identified using flow cytometry (Figure S5) and intracellular cytokines at baseline indicated an overall significant difference between MS and controls (P=0·01, PERMANOVA, Figure 2a). Specifically, immune cell subset analysis by Wilcoxon Rank Sum tests after multiple comparison correction showed that the percentages of peripheral blood IL-10+ memory B cells, T-bet+ memory and effector T cells, memory and effector Th17 cells were significantly greater in MS than in controls (Figure 2b; Table S2), suggesting an overactive peripheral pro-inflammatory response in untreated MS patients. We found no significant differences in T regulatory immune cells between MS and controls.
Figure 2.
Host-microbiome interaction in MS patients at baseline. (a) PCA analysis of blood immune profiles in MS patients and controls. The two groups show a distinct immune profile, as indicated by separation in first dimension of PCA. The overall blood immune profile statistically differs between MS patients and controls (P=0·01, by PERMANOVA). (b) Specific immune cell populations that significantly differ between MS patients and controls (FDR <0·05, Wilcoxon Rank Sum Test). (c) PCA analysis of blood metabolome in MS patients and controls. The global metabolome profiles were statistically different between MS patients and control (P=0·001, by PERMANOVA). (d) Pathway enrichment analysis. x-axis is the enrichment (impact) factor, which is determined by the pathway topology analysis (the importance of a metabolite within a pathway). –log (P) in y-axis refers to negative logarithmic value of the original P value from statistical analysis of pathway difference between MS patients and controls. The colour of each dot is positively correlated with the P values. The black dashed line indicates the cut off of -log (P) value of 2 as statistical significance. (e) Multi-omics correlation by Mantel test. Mantel correlations were performed based on distance matrix of any two of omics datasets including the gut microbiome (Bray-Curtis distance), blood immune profile (Euclidian distance), metabolome (Euclidian distance) and food servings (Euclidian distance). Correlation matrix generated from paired omics are presented for MS patients (lower triangle) and controls (upper triangle) separately. Rho (r) and P represented correlation co-efficient and statistical significance, respectively, of mantel tests, and their exact values are listed in the correlation matrix. r values were also indicated by colour gradient for MS or controls.
Untargeted metabolomics analysis of serum from MS and controls at baseline identified 8857 potential metabolites, with overall metabolome distinguished between MS patients from controls (P=0·001, PERMANOVA, Figure 2c). One hundred and eighteen metabolites were differentially represented in MS and controls by Welch's t-test after correcting for multiple comparison by FDR (FDR<0·05, Table S3). Interestingly, the preponderance of these metabolites was significantly greater in MS patients than in controls. Not surprisingly, most metabolites were not annotated in reference databases, but among those that were, we found that methionine and S-adenosylmethionine (SAM), which are involved in meat metabolism, mercaptopyruvate, leukotriene B4,44 dihydrochalcone, 3-mercaptolactate guanine 1-naphthaldehyde, riboflavin, 4-hydroxy lauric acid and chalcone were significantly enriched in the MS patients. Blood bile acid metabolism has been reported to be decreased in MS.45 Our study did not find statistically significant differences in primary and secondary bile acid metabolism between MS and controls, though we did identify a trend towards decreased glycocholate, taurodeoxycholate, glycochenodeoxycholate, and taurohyocholate, and increased deoxycholic acid and other bile acid metabolites in MS. Pathway analysis showed that pathways involved in linoleate metabolism, fatty acid metabolism were altered in MS patients (Figure 2d).
Overall dietary patterns did not differ significantly by PERMANOVA between MS patients and controls. The MS group had a higher median meat intake compared to controls before (P=0·006, Wilcoxon Rank Sum Test) (Figure S6), but not after FDR adjustment (FDR=0·10).
We next sought overall correlations between the gut microbiome, peripheral blood immune and metabolome profiles, and diet, in MS and controls at baseline. The gut microbiome and host blood immune profiles were positively correlated in controls (r=0·32, P=0·0008, Mantel test) (Figure 2e, Figure S7), suggesting a close interaction between the gut microbiome and peripheral immune profiles in healthy controls. However, this association was absent in MS patients (r=0·09, P=0·22, Mantel Test), suggesting that MS might dissociate immune-microbiome homeostatic interactions. Strikingly, we found a positive correlation between peripheral immune and metabolome profiles in MS patients (r=0·22, P=0·04, Mantel test), but not in controls (r=0·03, P=0·40). The association between immune and metabolome profiles signifies potentially concomitant changes of blood immune cell populations and metabolism in MS patients. In addition, diet was positively correlated with the blood metabolome in controls (r=0·50, P=0·008, Mantel Test), while this association was lacking in MS patients (r=0·07, P=0·33, Mantel Test), suggesting that diet significantly affects the blood metabolome in health status. We did not find significant associations between diet and the gut microbiome or the peripheral immune profile by Mantel correlations in either MS patients, controls or the two groups combined. Taken together, the multi-omics analysis demonstrates a disruption of the gut microbiome-immune homeostatic relationship in MS participants and a positive interaction between the peripheral immune profile and blood metabolome in MS.
We next performed large-scale association analyses to identify specific correlated features within and between omics datasets by Pearson correlation, using rigorous multiple comparison adjustment (Table S4). Within and between group comparisons contained 332 and 185 significant correlations for controls and MS patients, respectively (Table S4), a nearly two-fold decrease of molecular correlations in MS patients. Strong and significant correlations (FDR <0·05 for the metabolome and FDR <0·2 for other omics, r >0·7 or r<-0·7) are presented as complex networks in Figure 3. Specifically, in controls, OTU_13_ Prevotella (P. copri, top right in Figure 3b) was strongly and positively correlated with circulating proportions of IL-10+ memory B cells and one metabolite. Importantly, we identified a correlation hub with memory Th1 cells being the node positively connecting to a variety of blood metabolites in controls. This Th1-metabolite correlation pattern was not evident in MS patients (Figure 3a). Routine dietary components have strong and positive correlations with a large number of blood metabolites, particularly in controls. Fruits and vegetable servings and specific microbes or immune cell populations also formed a number of satellite-style connections with various metabolites. One particularly notable gut microbiota and blood metabolite hub identified in MS participants involved OTU_2_Bacteroides (B. uniformis, in the centre in Figure 3a), a highly abundant and prevalent human gut bacterium. Interestingly, we also identified a tandem and positive association between sugar-sweetened soft drinks with effector Th17 cells (middle left) and the latter was further correlated with a blood metabolite. This finding is consistent with the exacerbation of EAE associated with increased Th17 cells following long-term consumption of high sucrose beverages.46 Together, our data infer distinct, diverse, and cross-system interrelationships of key pathways in MS patients and controls, providing a compendium of potential targets for future studies of pathogenic mechanisms underlying MS.
A potential pathway linking meat serving, gut microbiome, Th17 cells and blood metabolites
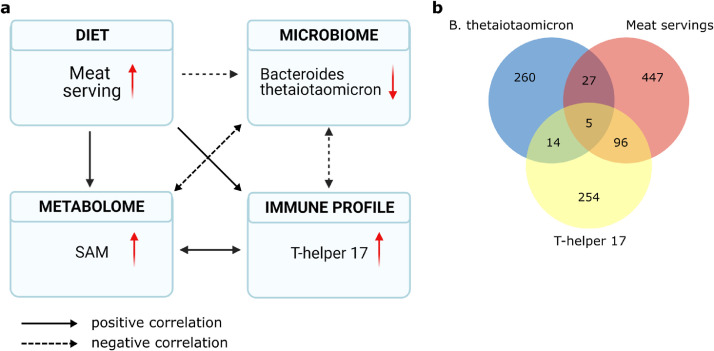
To gain in-depth understanding of biological processes deciphered by multi-omics datasets, we next tested the hypothesis that an interactive pathway could link diet, gut microbiome, immune response and metabolome. The observation that MS patients had significant more meat servings and correspondingly greater concentrations of circulating Th17 cells prompted us to explore a correlation network focusing on meat serving, Th17 cells, gut microbiome and blood metabolome. Combining the data from the MS and control cohorts, we found that meat servings were negatively correlated with the relative abundance of B. thetaiotaomicron (r=-0·42, p=0·01) (Figure 4a), a common gut bacterium with high genetic capacity (18% of its genes) to digest polysaccharides.47 B. thetaiotaomicron was strongly negatively correlated with proportions of circulating Th17 cells (r=-0·40, p=0·01), while Th17 cells were positively correlated with meat servings (r=0·50, p=0·003). As diet, the gut microbiome and immune response all potentially affect blood metabolites, we next correlate blood metabolites with B. thetaiotaomicron, Th17 cells and meat servings and found five blood metabolites significantly correlated with all three measurements (Figure 4b, Figure S8). Interestingly, metabolite mz34, annotated as SAM, was positively correlated to meat servings (r=0·42, p=0·01), Th17 cell proportion (r=0·75, p=1·7e-07), and negatively correlated with relative abundance of B. thetaiotaomicron (r=-0·40, p=0·006) (Figure 4a). Taken together, our multi-omics analysis suggests a correlation network involving dietary meat serving, gut microbiome, Th17 cells and blood metabolites. However, our analyses do not indicate the directionality of regulation between each of the aforementioned correlative pair. These results highlight a discovery process driven by omics analysis and provide an interesting hypothesis that now warrants further validation.
Figure 4.
Pathway linking meat servings in the diet with gut microbiome, immune response and blood metabolome. (a) Higher meat serving is negatively correlated with relative abundances of B. thetaiotaomicron, a fibre digesting bacterium. The latter is negatively correlated with proportions of Th17 cells. Increased Th17 cells are positively correlated with methyl donor S-adenosyl-L-methionine (SAM) that is a metabolic product from methionine, an amino acid enriched in meat. SAM was negatively correlated with B. thetaiotaomicron. (b) Correlation between blood metabolites and B. thetaiotaomicron, meat serving and Th17 cells. Pearson correlations were performed between B. thetaiotaomicron, meat servings and Th17 cells and all blood metabolites from MS and controls, with 306 significant correlations between metabolites and B. thetaiotaomicron, 575 between metabolites and meat servings, and 369 between metabolites and Th17 cells. There are 5 metabolites correlated with all three type of measures.
Host-microbiome multi-omics capacity to classify MS patients and controls
To investigate the power of individual and multi-omics to classify MS patients and controls, we applied random forest (RF), elastic net regularized linear regression (ENL) and elastic net regularized support vector machine (SVM), which are suited for high dimension data. We applied different regularization approaches to control model complexity to avoid overfitting. The classification of MS patients and controls based on either all or a subset of microbiome features after marginal screening generated an Area Under the Curve (AUC) with a wider range (0·2–0·8 for RF) (Figure 5a), indicating unstable classification performance using the gut microbiome alone. By contrast, the three classifiers constructed based on blood metabolome and immune profile had the greatest out-of-sample classification performance, with mean AUC close to, or exceeding, 0·90 (Figs. 5b, c). To determine if integration of data types improve classification performance, we trained three classification models with all the features of diet, microbiome, blood metabolome and immune profile. The mean AUC obtained with integrated data was comparable to that obtained from the blood metabolome data (Figure 5d). The three classifiers trained with blood metabolome, immune profile or integrated data performed similarly, though RF performed best in classifying MS patients and controls. To identify the top predictive features, we examined the feature importance measures from the RF model trained with the integrated data (averaged over 200 random splits). As expected, the top 20 variable-of-importance consisted of blood metabolites such as mz4-dihydrochalcone and immune cell populations, such as CD4+ memory T cells (Figure 5e). Notably, these highest-ranking features achieved similar classification accuracy as using all the omics features (Figure S9).
Figure 5.
Out-of-sample ROC curves of three classifiers to discriminate MS patients and Controls. The mean ROC curves generated from 200 iterations of model validation for the microbiome (a), immune profile (b), blood metabolome (c), and a combination of all the datasets (d). Means and 90% confidence intervals of AUC scores of ROC curves were listed at the bottom right for classification accuracy. (e) Top 20 important features from random forest (RF) model. Nineteen blood metabolites (indicated by mz) and memory Th17 cells were ranked as the top 20 features of important from RF model in classification of MS patients and controls.
Longitudinal changes of the gut microbiome and host peripheral immune and metabolome profiles in MS patients and controls
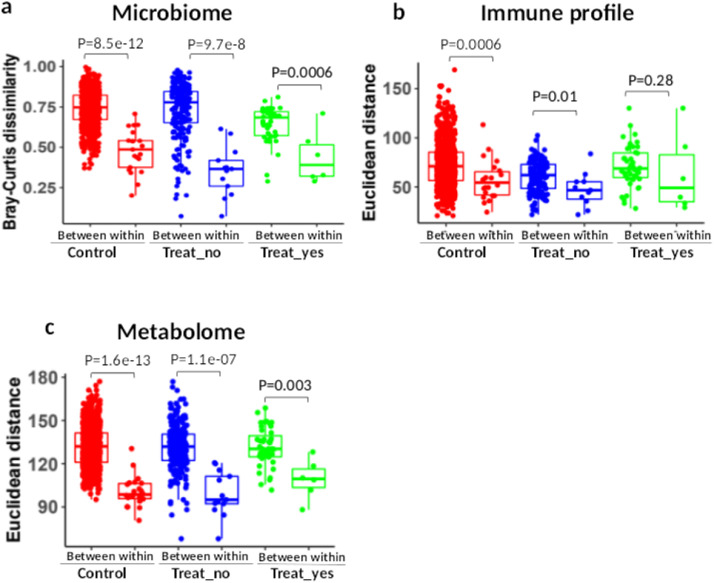
To measure the temporal stability of each omics over time (6 months), we computed pair-wised dissimilarity between and within the controls and MS patients who did and did not receive DMTs. Within- and between-participant dissimilarity refers to the dissimilarity of baseline and six month timepoints for the same individuals, and dissimilarity between different individuals at each time point, respectively (Figure S10). Without challenge or intervention, within-participant variations are significantly smaller than between participant variations,48,49 indicating temporal stability. Compared to between-participant variation, within-participant variations of the microbiome and metabolome were significantly lower for all MS patients and controls (Figs. 6a, c), and the within-participant variations of the immune profile were also significantly lower than between-participant variations in controls (Figure 6b). This suggests a relatively stable overall microbiome and metabolome for MS patients and controls as well as the immune profile in controls during the study period. In contrast, between- and within-participant variations of the immune profile in MS patients who received treatment with DMTs were not statistically different (P=0·28, Wilcoxon-rank test, Figure 6b). To determine if DMTs altered the immune profile and subsequently affect the within-participant dissimilarity, we performed Wilcoxon Rank Sum test for MS patients before and after these interventions. We found that proportions of memory Th17 cells and GM-CSF+ memory T Cells were significantly reduced at six months compared to baseline in MS patients who initiated DMTs (FDR=0·05, Figure S11).
Figure 6.
Temporal stability of the gut microbiome, blood immune profile and blood metabolome within six months in MS patients and controls. The stability of multi-omics was evaluated by measuring the between-participant dissimilarity and within-participant dissimilarity (samples collected at both baseline and after six months) of the gut microbiome (Bray-Curtis dissimilarity) (a), immune profile (Euclidian distance) (b), blood metabolome (Euclidian distance) (c) in controls (n=22), MS patients who initiated DMTs (Treat_Yes, n=6), and MS patients who did not initiate DMTs (Treat_No, n=14). Difference of between- and within-participant dissimilarity was tested by Wilcox Rank Sum test.
We further performed a Mantel correlation to test if between-participant similarities were maintained over the study interval based on distance measures of gut microbiome, blood immune cell and metabolome profiles. We found significant correlations of the gut microbiome (r=0·69, P=0·001 for MS; r=0·4, P=0·005 for controls) and blood metabolome (r=0·39, p=0·001 for MS; r=0·27, p=0·005 for controls) between baseline and six-month samples. In contrast, the blood immune profile showed no correlation between baseline and six-month for MS patients (r=0·05, P=0·32), while controls demonstrated significant correlation (r=0·33, p=0·01). These findings suggest that between-participant relationships were maintained over the course of the study in gut microbiome and metabolome profiles for both MS and controls, and in the immune profiles for controls, but not for MS participants.
We did not identify specific gut microbiome, metabolome or food servings that significantly changed between the beginning of the study and six-month follow-up in the MS patients. We also did not identify microbiota that differed between treated and untreated MS patients at six months. Strikingly, 41·9% of the blood metabolites that were significantly different between MS patients and controls at baseline still maintained similar differences at six months follow-up. Therefore, the analysis of the data obtained at six months follow-up validated our baseline findings. Accordingly, machine learning models constructed using six-months follow-up data consistently showed the best classification accuracy in differentiating MS patients from controls based on metabolome features, compared to those based on immune and microbiome profiles. In summary, host peripheral immune profile, blood metabolites and the gut microbiome of MS participants remain relatively stable over six months in those patients who remained untreated, while initiation of treatment with DMTs affected specific immune cell populations.
Discussion
We present the first integrated characterization of gut microbiome alterations and host-microbiome interactions in MS patients compared to healthy control individuals using advanced multi-omics technologies and a longitudinal study design that offers insight into the stability of these factors over time. We have confirmed prior findings that the gut microbiome community structure in MS patients largely resembles that of controls.9,10,37 We also extended the microbiome analyses by seeking factors that might drive microbiome variations between MS patients and controls, and validated the small impact from the disease itself. Interestingly, BMI contributed significantly to microbiome variation among all tested variables, and positively correlated with EDSS in MS patients. Hence, BMI should be considered in future inter-group microbiome comparisons in MS studies.
The under-representation of Faecalibacteria, Prevotella, Lachnospiraceae and Anaerostipes species in MS patients compared to controls aligns with previous findings,10 and is biologically plausible. Faecalibacteria, Lachnospiraceae, and Anaerostipes produce butyrate, which acts via G-protein coupled receptors activation and histone deacetylase inhibition to suppress CNS demyelination,50 the main pathological feature in MS. Indeed, concentrations of fecal SCFAs (i.e., acetate, butyrate and propionate) were decreased in RRMS patients, compared to healthy controls.51, 52, 53 Blood SCFAs were significantly decreased in long-term active progressive MS patients.54 Propionic acid, but not butyrate and acetate, was significantly reduced in blood and stool in MS patients with all disease subtypes, particular after relapse. Supplementation of propionic acid promoted Treg cell function, and in long-term administration reduced relapse rate, disability and brain atrophy.55 In our study, we found a trend toward decreased concentrations of butyrate in the stools of MS patients, consistent with decreased SCFA-producing bacteria in MS. Notably, SCFA levels can also be affected by diet, and higher meat servings in MS patients may also contribute to the observed reduction of SCFAs.
Prevotella histicola reduces EAE severity by inducing FoxP3+ regulatory T cells and decreasing pro-inflammatory Th1 and Th17 cells in the CNS.15,56 We found reduced abundance of a different Prevotella species, P. copri, in stools of MS patients. P. copri is a dominant Prevotella species in healthy American adults,57 and is more prevalent in non-western populations.58 Moreover, P. copri was highly correlated with IL10+ memory B cells in our control group, providing a potential novel microbiome-driven immune pathway to test in future.
In this study, the MS group was compared to unrelated healthy controls, matched for age, gender and other important variables. An alternative approach would have been to obtain same household controls, to account for environmental influences.59 However, it is possible that the same household controls may decrease the sensitivity to detect MS associated microbes, because individuals from the same household tend to share gut microbes, and a shared microbe may still influence MS development in genetically predisposed individuals. Changes in gut microbiota observed in MS participants in our study resembled previous studies conducted in different geographical locations, offering credence to our findings. MS-associated microbes are also over-represented in other autoimmune and metabolic diseases as well as cancer that are associated with inflammation.8,60,61 This finding argues against a unique microbiome signature for MS patients but supports a common microbiome dysbiosis indicator for extra-intestinal pathophysiology associated with inflammation.
Much work has shown that the immune system confines the gut microbiome within its physical niche and shapes microbial compositions in animal models,62 and our findings now link gut microbiome and systemic cellular immune profiles in healthy human adults. Our data recapitulate in a human cohort the inter-relatedness of the gut microbiome, diet, immune system and host metabolome, a relation that has been reported mostly in germ-free mice.62 Specifically, we now demonstrate an association between the gut microbiome and peripheral immune phenotype in healthy participants, implying that healthy people with similar gut microbiome tend to have similar immune phenotype. In contrast, gut microbiome-immune haemostatic interactions were disrupted in the MS cohort we studied. Immune cell phenotypes of MS patients significantly differed from healthy controls in our dataset. However, we wish to note that the degree of dysbiosis of gut microbes in MS was modest. The discordance of the changes in immune phenotypes and microbiome may explain the lack of correlation between the microbiome and peripheral blood immune profiles in MS. It will be of great interest to elucidate the directionality and time-to-response of microbiome-immune regulation in MS. Future work might also be directed towards microbiota and immune response at the gut mucosa,63 as the site of the systemic changes that ultimately affect the CNS.
Interestingly, blood metabolites best differentiate MS patients from controls in our models. Indeed, MS patients experience metabolic alteration in different tissues,64,65 and dysfunctional lipid metabolism.66 The observed enrichment of circulating novel metabolites and multiple pathways in MS patients requires future validation to address their role in MS. We found that methionine was significantly enriched in MS patients, which is consistent with higher meat consumption in the patients we studied. Methionine drives T cell proliferation and differentiation.67 Our data are also in accord with recent findings that methionine activates Th17 cells through epigenetic modification.68 Methionine is an essential metabolite for methyl donor SAM synthesis, and SAM promotes Th17 cell activation through methylation.68 Reduction of dietary methionine ameliorated EAE through reprogramming pathogenic Th17 cells. Our data, together with Roy et al., prompt the hypotheses that meat or methionine restriction might beneficially decrease the number of circulating inflammatory Th17 cells in MS patients. Future studies of metabolites in MS should consider specific dietary nutrients and gut microbiome derived metabolites, as these factors play large roles in ordaining human metabolism.
Global correlation among multi-omics provides a powerful tool to understand systemic interactions across organs. However, among hundreds or thousands of correlations, in the context of under-annotated features such as blood metabolites, mining biology from global correlations is challenging. Consequently, multi-omics has often been criticized for generating massive amount of data but providing little biology. We highlighted the significant discovery power of multi-omics by deeply delving into a pathway linking meat serving, B. thetaiotaomicron, Th17 cell, and SAM. We not only confirmed the relationship between meat consumption, SAM, and Th17 cells,68 we identified their connections with a common gut commensal bacterium, B. thetaiotaomicron. This opens new research directions to elucidate regulatory pathways among diet, metabolites, microbiome, and immune response, and may identify therapeutic targets for MS.
Interestingly, routine dietary intake had small overall impact on gut microbiome variation, as is also reported in inflammatory bowel disease.69 However, differential microbiome responses to dietary intake in different individuals might explain the lack of strong correlation between diet and gut microbiome in human studies.70 We also found that food composition did not correlate with circulating immune phenotypes. Nonetheless, lack of systemic associations does not exclude the possibility of specific feature-feature correlations, as we have identified associations between food compositions and the microbiome, as well as food compositions and immune cell populations. Feature-feature correlations were reported conservatively, and only significant associations after multiple comparison corrections were included in this final report, thereby strengthening confidence in the correlations.
Our longitudinal study design provided a unique opportunity to evaluate the stability of multi-omics data in MS patients over a period of six months, during which most DMTs take effect. Baseline and six-month follow-up measures for microbiome, metabolome components, immune-phenotypes and diet were similar, except for MS patients who began DMTs, which showed a decrease in memory Th17 cells and GM-CSF+ T cells at the six-month time point. These findings suggest an overall stability of the different systems in this defined interval in humans without strong exogenous influences. In addition, the consistent results between baseline and six-months strengthen our findings related to differential features between MS and controls, suggesting that these differences are not likely spurious.
While our longitudinal study offers a highly textured view of microbial-host interactions in MS, we acknowledge several limitations. First, the relatively small sample size increases the risk of type II error. To avert this, we carefully chose analytical tools and statistical tests suitable for high dimension data analysis. Second, food diary in our study was self-recorded, which potentially pose selection bias since it is less likely that all food consumptions are completed by all participants. Third, the majority of study participants are female because MS is more prevalent in women. It would be interesting to research next whether these findings still hold true in male MS patients. Lastly, we could not determine the causal connection or directionality of feature-feature interactions. Nonetheless, we provide data on multiple novel molecules that could be plausibly implicated in MS pathogenesis that obligate a more in-depth analysis in the future. A larger study with multiple sampling points that capture both relapse and remitting stages of MS will demonstrate a more complete dynamic picture of multiple-omics interactions in MS.
Conclusions
Multi-omics integration provides a holistic view of the pathophysiology of MS, laying a foundation for precision medicine that combines advanced technologies with clinical practice to improve our understanding of MS aetiology and patient care.
Declaration of interests
Dr. Evans has been a paid consultant and/or speaker for the following: Biogen, EMD Serono, National MS Society, Genentech/Roche, Novartis, Sanofi/Genzyme & Teva.
Dr. Cross has done paid consulting for: Biogen, Celgene, EMD Serono, Genentech/Roche, Greenwich Biosciences, Janssen and Novartis, and has contracted research funded by EMD Serono and Genentech.
Dr. Tarr is a consultant to, a member of the Scientific Advisory Board of, and a holder equity in, MediBeacon Inc, which is developing a method to test intestinal permeability in humans. He might receive royalty payments if the product generates revenues.
Acknowledgments
Contributors
YZ, LP, ALR designed the study. YZ, CC, QL, LG, ZL, YP, KC, YH, HX, AL performed the omics analysis. ALR, MG, YL, CC, LG, performed flow cytometry analysis. YD, SB, HP, CS, EE, LD, RB, KO, PN contributed to the collection and assembly of data. YZ, LP, ALR, PIT, YW, AS, AHC, contributed to the writing of the grant proposal and/or data interpretation. YZ wrote the manuscript, PIT, CC, LG, LP and YD edit the manuscript. YZ and ZL verified underlying data. All authors read and approved the final manuscript.
Acknowledgments
We thank the Microbial Genomic Services Core at Jackson Laboratory and Metabolome core at University of Massachusetts for data generation.
This work was supported by the Washington University in St. Louis Institute of Clinical and Translational Sciences, funded, in part, by Grant Number # UL1 TR000448 from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award (Zhou Y, Piccio, L, Lovett-Racke A and Tarr PI); R01 NS102633–04 (Zhou Y, Piccio L); the Leon and Harriet Felman Fund for Human MS Research (Piccio L and Cross AH). Cantoni C. was supported by the National MS Society Career Transition Fellowship (TA-1805–31003) and by donations from Whitelaw Terry, Jr. / Valerie Terry Fund. Ghezzi L. was supported by the Italian Multiple Sclerosis Society research fellowship (FISM 2018/B/1) and the National Multiple Sclerosis Society Post-Doctoral Fellowship (FG-1907–34474). Anne Cross was supported by The Manny & Rosalyn Rosenthal-Dr. John L. Trotter MS Center Chair in Neuroimmunology of the Barnes-Jewish Hospital Foundation.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data sharing statement
The raw reads of the microbiome data were deposited in the short reads archive database (SRA) (accession no. PRJNA634779).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103798.
Contributor Information
Laura Piccio, Email: picciol@wustl.edu.
Yanjiao Zhou, Email: yazhou@uchc.edu.
Appendix. Supplementary materials
References
- 1.Wallin MT, Culpepper WJ, Campbell JD, et al. The prevalence of MS in the United States–A population-based estimate using health claims data. Neurology. 2019;92:e1029–e1040. doi: 10.1212/WNL.0000000000007035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ascherio A. Environmental factors in multiple sclerosis. Expert Rev Neurother. 2013;13:3–9. doi: 10.1586/14737175.2013.865866. [DOI] [PubMed] [Google Scholar]
- 3.Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2 Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costantino CM, Baecher-Allan C, Hafler DA. Multiple sclerosis and regulatory T cells. J Clin Immunol. 2008;28:697–706. doi: 10.1007/s10875-008-9236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funaro M, Messina M, Shabbir M, et al. The role of B cells in multiple sclerosis–More than antibodies. Discov Med. 2016;22:251–255. [PubMed] [Google Scholar]
- 6.Pazhouhandeh M, Sahraian M-A, Siadat SD, et al. A systems medicine approach reveals disordered immune system and lipid metabolism in multiple sclerosis patients. Clin Exp Immunol. 2018;192:18–32. doi: 10.1111/cei.13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantarel BL, Waubant E, Chehoud C, et al. Gut microbiota in multiple sclerosis–Possible influence of immunomodulators. J Investig Med. 2015;63:729–734. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vieira SM, Pagovich OE, Kriegel MA. Diet, microbiota and autoimmune diseases. Lupus. 2014;23:518–526. doi: 10.1177/0961203313501401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyake S, Kim S, Suda W, et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to clostridia XIVa and IV clusters. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J, Chia N, Kalari KR, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takewaki D, Suda W, Sato W, et al. Alterations of the gut ecological and functional microenvironment in different stages of multiple sclerosis. Proc Natl Acad Sci U S A. 2020;117:22402–22412. doi: 10.1073/pnas.2011703117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berer K, Gerdes LA, Cekanaviciute E, et al. Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A. 2017;114:10719–10724. doi: 10.1073/pnas.1711233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cekanaviciute E, Yoo BB, Runia TF, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci U S A. 2017;114:10713–10718. doi: 10.1073/pnas.1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cignarella F, Cantoni C, Ghezzi L, et al. Intermittent fasting confers protection in CNS autoimmunity by altering the gut microbiota. Cell Metab. 2018;27:1222–1235. doi: 10.1016/j.cmet.2018.05.006. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangalam A, Shahi SK, Luckey D, et al. Human gut-derived commensal bacteria suppress CNS inflammatory and demyelinating disease. Cell Rep. 2017;20:1269–1277. doi: 10.1016/j.celrep.2017.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W, Sailani MR, Contrepois K, et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature. 2019;569:663–671. doi: 10.1038/s41586-019-1236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polman CH, Reingold SC, Banwell B, et al. Diagnostic criteria for multiple sclerosis–2010 revisions to the McDonald criteria. Ann Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franzosa EA, McIver LJ, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15:962–968. doi: 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcobal A, Kashyap PC, Nelson TA, et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovett-Racke AE, Gormley M, Liu Y, et al. B cell depletion with ublituximab reshapes the T cell profile in multiple sclerosis patients. J Neuroimmunol. 2019;332:187–197. doi: 10.1016/j.jneuroim.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 22.Basiotis PP, Welsh SO, Cronin FJ, Kelsay JL, Mertz W. Number of days of food intake records required to estimate individual and group nutrient intakes with defined confidence. J Nutr. 1987;117:1638–1641. doi: 10.1093/jn/117.9.1638. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y, Olendzki BC, Pagoto SL, et al. Number of 24 h diet recalls needed to estimate energy intake. Ann Epidemiol. 2009;19:553–559. doi: 10.1016/j.annepidem.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Racette SB, Weiss EP, Schechtman KB, et al. Influence of weekend lifestyle patterns on body weight. Obesity (Silver Spring) 2008;16:1826–1830. doi: 10.1038/oby.2008.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bingham SA, Gill C, Welch A, et al. Comparison of dietary assessment methods in nutritional epidemiology: weighed records v. 24 h recalls, food-frequency questionnaires and estimated-diet records. Br J Nutr. 1994;72:619–643. doi: 10.1079/bjn19940064. [DOI] [PubMed] [Google Scholar]
- 26.Johnson RK, Kennedy E. The 2000 dietary guidelines for Americans–What are the changes and why were they made? The dietary guidelines advisory committee. J Am Diet Assoc. 2000;100:769–774. doi: 10.1016/s0002-8223(00)00225-x. [DOI] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss S, Xu ZZ, Peddada S, et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Mihindukulasuriya KA, Gao H, et al. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014;15:R66. doi: 10.1186/gb-2014-15-5-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vinaixa M, Samino S, Saez I, Duran J, Guinovart JJ, Yanes O. A guideline to univariate statistical analysis for LC/MS-based untargeted metabolomics-derived data. Metabolites. 2012;2:775–795. doi: 10.3390/metabo2040775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 32.Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc. 2005;67:301–320. [Google Scholar]
- 33.Wang L, Zhu J, Zou H. The doubly regularized support vector machine. Statistica Sinica. 2006;16:589–615. [Google Scholar]
- 34.Aitchison J. The statistical analysis of compositional data. J R Stat Soc B. 1982;44:139–177. [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the false discovery rate–A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 36.Stone M. Cross-validatory choice and assessment of statistical predictions. J R Stat Soc. 1974;36:111–133. [Google Scholar]
- 37.Jangi S, Gandhi R, Cox LM, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plaza-Díaz J, Ruiz-Ojeda FJ, Vilchez-Padial LM, Gil A. Evidence of the anti-inflammatory effects of probiotics and synbiotics in intestinal chronic diseases. Nutrients. 2017;9:E555. doi: 10.3390/nu9060555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabeerdoss J, Sankaran V, Pugazhendhi S, Ramakrishna BS. Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease–A case-control study in India. BMC Gastroenterol. 2013;13:20. doi: 10.1186/1471-230X-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Telesford KM, Ochoa-Repáraz J, et al. An intestinal commensal symbiosis factor controls neuroinflammation via TLR2-mediated CD39 signaling. Nat Commun. 2014;5:4432. doi: 10.1038/ncomms5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Wright K, Davis JM, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiippala K, Kainulainen V, Kalliomäki M, Arkkila P, Satokari R. Mucosal prevalence and interactions with the epithelium indicate commensalism of Sutterella spp. Front Microbiol. 2016;7:1706. doi: 10.3389/fmicb.2016.01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gladue RP, Carroll LA, Milici AJ, et al. Inhibition of leukotriene B4-receptor interaction suppresses eosinophil infiltration and disease pathology in a murine model of experimental allergic encephalomyelitis. J Exp Med. 1996;183:1893–1898. doi: 10.1084/jem.183.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhargava P, Smith MD, Mische L, et al. Bile acid metabolism is altered in multiple sclerosis and supplementation ameliorates neuroinflammation. J Clin Invest. 2020;130:3467–3482. doi: 10.1172/JCI129401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao G, Wang Q, Huang W, et al. Long-term consumption of caffeine-free high sucrose cola beverages aggravates the pathogenesis of EAE in mice. Cell Discov. 2017;3:17020. doi: 10.1038/celldisc.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu J, Bjursell MK, Himrod J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 48.Brodin P, Davis MM. Human immune system variation. Nat Rev Immunol. 2017;17:21–29. doi: 10.1038/nri.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen T, Noto D, Hoshino Y, Mizuno M, Miyake S. Butyrate suppresses demyelination and enhances remyelination. J Neuroinflammation. 2019;16:165. doi: 10.1186/s12974-019-1552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng Q, Gong null Junli, Liu X, et al. Gut dysbiosis and lack of short chain fatty acids in a Chinese cohort of patients with multiple sclerosis. Neurochem Int. 2019;129 doi: 10.1016/j.neuint.2019.104468. [DOI] [PubMed] [Google Scholar]
- 52.Zheng P, Zeng B, Zhou C, et al. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol Psychiatry. 2016;21:786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 53.Trend S, Leffler J, Jones AP, et al. Associations of serum short-chain fatty acids with circulating immune cells and serum biomarkers in patients with multiple sclerosis. Sci Rep. 2021;11:5244. doi: 10.1038/s41598-021-84881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park J, Wang Q, Wu Q, Mao-Draayer Y, Kim CH. Bidirectional regulatory potentials of short-chain fatty acids and their G-protein-coupled receptors in autoimmune neuroinflammation. Sci Rep. 2019;9:8837. doi: 10.1038/s41598-019-45311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duscha A, Gisevius B, Hirschberg S, et al. Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell. 2020;180:1067–1080. doi: 10.1016/j.cell.2020.02.035. e16. [DOI] [PubMed] [Google Scholar]
- 56.Shahi SK, Freedman SN, Murra AC, et al. Prevotella histicola, a human gut commensal, is as potent as COPAXONE® in an animal model of multiple sclerosis. Front Immunol. 2019;10:462. doi: 10.3389/fimmu.2019.00462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Integrative HMP (iHMP) research network consortium. the integrative human microbiome project. Nature. 2019;569:641–648. doi: 10.1038/s41586-019-1238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tett A, Huang KD, Asnicar F, et al. The prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe. 2019;26:666–679. doi: 10.1016/j.chom.2019.08.018. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.The iMSMS Consortium. Household paired design reduces variance and increases power in multi-city gut microbiome study in multiple sclerosis. Mult Scler. 2020 doi: 10.1177/1352458520924594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Luca F, Shoenfeld Y. The microbiome in autoimmune diseases. Clin Exp Immunol. 2019;195:74–85. doi: 10.1111/cei.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ou J, Carbonero F, Zoetendal EG, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98:111–120. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cosorich I, Dalla-Costa G, Sorini C, et al. High frequency of intestinal TH17 cells correlates with microbiota alterations and disease activity in multiple sclerosis. Sci Adv. 2017;3 doi: 10.1126/sciadv.1700492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reinke SN, Broadhurst DL, Sykes BD, et al. Metabolomic profiling in multiple sclerosis–Insights into biomarkers and pathogenesis. Mult Scler. 2014;20:1396–1400. doi: 10.1177/1352458513516528. [DOI] [PubMed] [Google Scholar]
- 65.Kim H-H, Jeong IH, Hyun J-S, Kong BS, Kim HJ, Park SJ. Metabolomic profiling of CSF in multiple sclerosis and neuromyelitis optica spectrum disorder by nuclear magnetic resonance. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kooij G, Troletti CD, Leuti A, et al. Specialized pro-resolving lipid mediators are differentially altered in peripheral blood of patients with multiple sclerosis and attenuate monocyte and blood-brain barrier dysfunction. Haematologica. 2020;105:2056–2070. doi: 10.3324/haematol.2019.219519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Klein Geltink RI, Pearce EL. The importance of methionine metabolism. Elife. 2019;8:e47221. doi: 10.7554/eLife.47221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roy DG, Chen J, Mamane V, et al. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab. 2020;31:250–266. doi: 10.1016/j.cmet.2020.01.006. e9. [DOI] [PubMed] [Google Scholar]
- 69.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johnson AJ, Vangay P, Al-Ghalith GA, et al. Daily sampling reveals personalized diet-microbiome associations in humans. Cell Host Microbe. 2019;25:789–802. doi: 10.1016/j.chom.2019.05.005. e5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.