Abstract
Plant carotenoid cleavage oxygenase (CCO) is an enzyme that catalyzes the synthesis of carotenoids and participates in many important physiological functions. The plant CCOs exist in two forms, namely carotenoid cleavage dioxygenase (CCD) and nine-cis epoxide carotenoid dioxygenase (NCED). Although studies have shown that this gene family has been identified in many species, such as Arabidopsis, grape, and tomato, the evolutionary origin of the CCO family and the expression pattern of pepper genes in response to H2O2 and other abiotic stresses are still unclear. In this study, we used the bioinformatics method to identify and analyze the members of the CCO gene family from pepper and other 13 plants from lower to higher plant species based on the whole genome sequence. A total of 158 CCO genes were identified in different plant species and further divided into two groups (e.g., groups I and II). The former was subdivided into CCD7 and CCD8 and have independent evolutionary origins, respectively, while the latter was subdivided into CCD1, CCD4, CCD-like, and NCED, which may have come from a common ancestor. In addition, the results of RNA-seq showed that the expression patterns of pepper CaCCO genes were different in the tissues tested, and only few genes were expressed at high levels such as CaCCD1a, CaCCD4a, CaNCED3, and CaCCD1b. For hydrogen peroxide (H2O2) and other abiotic stresses, such as plant hormones, heat, cold, drought, and NaCl treatments, induction of about half of the CaCCO genes was observed. Moreover, the expression patterns of CaCCOs were further investigated under heat, cold, drought, and NaCl treatments using quantitative real-time PCR (qRT-PCR), and most members were responsive to these stresses, especially some CaCCOs with significant expression changes were identified, such as CaCCD4c, CaCCD-like1, CaCCD8, and CaCCD1b, suggesting the important roles of CaCCOs in abiotic stress responses. All these results will provide a valuable analytical basis for understanding the evolution and functions of the CCO family in plants.
Keywords: pepper, phylogenetic analysis, gene expression, abiotic stress, NCED
Introduction
As is known to all, due to the global climate change, plant resources and genetic diversity as well as the world food security has had a certain impact, so environmental stress has become the focus of attention of people (Raza et al., 2020). Plants are often subjected to various environmental stresses during their life, such as drought, high salt, high temperature, or invasion of pathogenic bacteria. These abiotic stresses or biotic stresses will have a certain impact on the growth and development of plants (Roychoudhury et al., 2013). Carotenoids are important lipid-soluble compounds, contain a large family over 700 types of structures (Britton et al., 2004), and perform a series of important biotic and abiotic stress functions. Apocarotenoids, or carotenoid cleavage products, were produced by carotenoid cleavage oxygenases (CCOs) of specific cleavage of carotenoids. Plant CCOs are a class of dioxygenases that catalyze the cleavage of carotenoids and their conjugate double bonds in plants. The CCOs can be further divided into nine-cis-epoxide carotenoid dioxygenase (NCED) and carotenoid cleavage dioxygenase (CCDs), based on their substrate informing an epoxy structure (Tan et al., 2003; Auldridge et al., 2006).
The first NCED gene vp14 was identified in maize (Schwartz et al., 1997; Tan et al., 1997). Since then, the function of NCED gene has been widely studied. The NCED genes in these species exist in plants in the form of gene family (Yang and Guo, 2007), and their expression sites and functions are also different (Lefebvre et al., 2010). It is reported that the NCED genes play a certain role in the growth and development of plants. For example, in Arabidopsis, the NCED1 gene can respond to water stress (Blankenship and Sisler, 1993). Similarly, according to the phenotypic analysis of NCED5, NCED6, and NCED9 mutants at specific time and tissue, the results show that the three genes work together to induce seed dormancy (Frey et al., 2012). Seo et al. (2009) showed that spatiotemporal expression of the NCED gene is particularly important for the regulation of abscisic acid (ABA) level, which affects the seed dormancy and germination (Nambara et al., 2010). Furthermore, the inhibition of far-red light on Arabidopsis seed germination was reported in part due to the photoreversible regulation of NCED6 to maintain the ABA levels (Seo et al., 2006), and Toh et al. (2008) suggested that three different NCED genes, namely, NCED2, NCED5, and NCED9, inhibited germination by increasing the ABA levels at high temperatures. Finally, two (PaNCED1 and PaNCED3) of three avocado NCED genes cloned were highly expressed at ripening (Chernys and Zeevaart, 2000).
In addition, studies have shown that the NCED gene not only involves in the growth and development of plants but also plays an important role in their stress tolerance (Gómez et al., 2002). It has been reported that the NCED gene can upregulate ABA biosynthesis (Chernys and Zeevaart, 2000; Rodrigo, 2006). Overexpression of the NCED gene can enhance the resistance of plants to abiotic stresses (Xian et al., 2014). For example, overexpression of the AtNCED3 gene can increase the content of ABA and increase the water loss tolerance of Arabidopsis thaliana. Previous reports have shown that overexpression of the LeNCED1 gene can improve the drought tolerance of tomato (Thompson et al., 2000). The heterologous overexpression of MhNCED3 in Malus crabapple alleviated oxidative damage and enhanced the tolerance of A. thaliana to chlorine stress (Zhang et al., 2014, 2015). The real-time PCR (RT-PCR) analysis of OsNCED3, OsNCED4, and OsNCED5 on the root of rice seedlings showed that the three NCED genes were induced by salt and ABA treatments (Welsch et al., 2008). More recently, Hwang et al. (2018) studied the sugar sensitivity and drought tolerance of the allograft expression of rice OsNCED4 in A. thaliana and found that the allograft expression of OsNCED4 increased ABA levels, changed plant size and leaf shape, and delayed seed germination, leading to oversensitivity to sugar after germination and enhanced tolerance to drought. There are few reports on the involvement of the NCED gene in disease resistance regulation, but some studies have found that AtNCED5 was overexpressed in A. thalli mutants, the endogenous ABA level was increased by about 2 times, and the susceptibility of this mutant was significantly improved. After inoculation with Pseudomonas syringa, AtNCED2, AtNCED3, and AtNCED5 were significantly induced, and ABA was also accumulated in large quantities (Fan et al., 2009).
Hydrogen peroxide (H2O2) and NO are important signals involved in plant growth and abiotic stress tolerance (Neill et al., 2002; Uchida et al., 2002). The exogenous ABA treatment can induce the production of H2O2 and NO, resulting in stomatal closure, and improve the expression and activity of antioxidant enzymes (Jiang and Zhang, 2003; Desikan et al., 2004; Zhou et al., 2005; Zhang et al., 2007). Especially, research reported that rice OsNECD3 regulates plant growth and enhances abiotic stress tolerance. Huang et al. (2018) showed that under H2O2 stress, OsNCED3 was expressed in different tissues, and the OsNCED3 gene avoided oxidative damage by increasing ABA biosynthesis when the plant was subjected to oxidative stress. Other studies have pointed out that the expression of the NCED gene increased the ABA and the H2O2 contents in roots in the non-saline side increased. Exogenous H2O2 can reduce ABA content by downregulating the NCED gene, indicating that there is a feedback mechanism between ABA and H2O2 (Kong et al., 2016).
Compared with the NCED gene family, only few studies were reported on the CCD gene family. Researchers reported that each member of this gene family had formed a functional carotenoid, such as β-ionone and geranylacetone, which were vital for the flavor of vegetable and fruit plants (Simkin et al., 2004a,b; Sun et al., 2008; Ilg et al., 2009). The CCD1 enzymes have cleavage activities on 5,6 (5’, 6’) double bonds position, while CCD4 were also found to have a 9,10 (9’, 10’) cleavage activity (Huang et al., 2009; Campbell et al., 2010; Brandi et al., 2011). In addition, the researchers reported that the cooperation of CCD7 and CCD8 can regulate plant architectures and reproductive development (Snowden et al., 2005; Liu et al., 2013).
Presently, nine members of the CCO gene family have been identified in Arabidopsis: four of which were designed as CCDs (CCD1, CCD4, CCD7, and CCD8) and the remaining genes were proven to be NCEDs (NCED2, NCED3, NCED5, NCED6, and NCED9) (Schwartz et al., 1997). In recent years, with the completion of plant genome sequencing, this gene family has been identified in different plant species, such as tomato (Burbidge et al., 1999), avocado (Chernys and Zeevaart, 2000), and bean (Qin and Zeevaart, 2002). Although identification and the roles of many CCOs in plants have been completed, members of the CCO gene family in pepper “zunla-1” have not been reported. Recently, the whole genome of “zunla-1” in pepper was sequenced (Qin et al., 2014). In this study, we systematically surveyed the CCO gene family from 14 plant species, ranging from lower to higher plant species, and analyzed their evolutionary origins. In addition, the expression profiles of the members of the pepper CCO gene family in response to H2O2 and other biotic stresses were also further investigated. These results will provide the value information of understanding of evolutionary modes and theoretical basis for exploring the functional roles of CCO in pepper.
Materials and Methods
Data Retrieval of Carotenoid Cleavage Oxygenase Gene Families in Different Plant Species
To identify the CaCCO gene family members, the predicted pepper gene sequences were downloaded from the Pepper Genome Database1 ,2. The genome information of Arabidopsis was derived from the database3. In addition, twelve plant species were also selected for analyses, including Volvox carteri, Chlamydomonas reinhardtii, Marchantia polymorpha, Physcomitrella patens, Selaginella moellendorffii, Oryza sativa, Zea mays, Solanum lycopersicum, Brachypodium distachyon, Setaria italic, Aquilegia coerulea, and Solanum tuberosum. Genome sequences of these plant species were downloaded from JGI4.
Identification and Chromosome Localization of the Carotenoid Cleavage Oxygenase Gene Family in Pepper
The CCO gene sequences were downloaded from A. thaliana genome database and used as a seed sequence to retrieve the pepper genome database to obtain the candidate genes. To confirm the members of the pepper CCO family, BlastP methods were performed using the conserved domain of Arabidopsis CCO protein as a query sequence in the local database. The pepper CCO genes with e-value less than 1e − 5 were used for further analysis. Then, the Hidden Markov Model (HMM) profile of the PF03055 (RPE65) conserved domain was downloaded from the Pfam protein family database5 with e-value < 1e − 5. Finally, all of the putative CaCCO sequences with incomplete domains were excluded by the HMM analysis. Genome sequences of other 12 plant species were used to construct the local database using Bioedit 7.0 software6.
The physicochemical properties of CaCCO genes were analyzed using the online ExPASy-ProtParam tool7, which included several amino acids, molecular weight (MW), and theoretical isoelectric point (pI) of deduced CaCCO proteins. According to the information in the pepper gene database, chromosome localization of the CCO genes was further performed using MapDrawV2.1 software.
Phylogenetic Analysis of Carotenoid Cleavage Oxygenase Gene Family in Pepper and Other Plant Species
To explore the phylogenetic relationships of the pepper CCO genes, multiple sequence comparison was conducted using CCO amino acid sequences of pepper and other plant species using ClustalW program, and MEGA5.0 software8 was used to construct neighbor-joining (NJ) tree (Ilg et al., 2009).
Spatiotemporal Expression Analysis of CaCCO Genes in Various Tissues of Pepper
To study the spatiotemporal expression patterns of the pepper CCO genes, data of reported RNA-seq were selected for analyses (see text footnote 2). Previously, a pepper variety (Line 6421) with good heat resistance, drought resistance, and disease resistance was selected as the preparation materials of various samples for RNA sequencing. The seeds were surface disinfected with 5% sodium hypochlorite solution for 15 min. After rinsing with water, the seeds were sown in 200-well vermiculite-filled seedling trays and placed in an environment of 25/18oC day/night temperature, 16/8 h light/dark cycle, 60--70% relative humidity, and 6,000 lux light intensity. The RNA-seq sequencing data of various tissue development stages of pepper, including seeds and placenta (ST1--ST2), seed (S3--S11), placenta (T3--T11), flower (F1--F9), petal (P10), ovary (O10), stamen (STA10), leaf (L1--L9), fruits (FST0--FST1), and peel (G1--G11), were used to make heat maps, and the heat map of tissue-specific expression patterns of the CCO genes was drawn using the MeV software9. All samples were collected in quadruplicate from each of the sampling points, and five seedlings were randomly picked and mixed as one biological replicate.
Expression Analysis of CaCCO Genes Under Abiotic Stresses
Similarly, to determine the response of the CaCCO gene to abiotic stresses (heat, cold, drought, and NaCl treatments), 40-day-old seedlings were treated and RNA samples were extracted from leaves and roots. Salt stress was carried out by adding NaCl with a final concentration of 200 mM to the nutrient solution, and osmotic stress was carried out by adding mannitol with a final concentration of 400 mM. For high/low-temperature treatment, the seedlings at the fourth true leaf stage will be transferred to the 42oC (high temperature) or 10oC (low temperature) of the growth chamber, photoperiod, and relative humidity consistent with the untreated plants. The control was treated with nutrient solution only. The leaves and roots were selected for RNA-seq analysis at 0, 1, 6, and 24 h after treatment.
Expression Analysis of CaCCO Genes in Response to Hormone Stresses
To study the expression of these genes under different hormone treatments, 30 μM ABA, 2 mM salicylic acid (SA), and 10 μM methyl jasmonic acid (MeJA) were used to treat the seedling for 40 days. These three different hormones (ABA, MeJA, and SA) were selected and added to the nutrient solution. The leaves and roots were selected for RNA-seq analysis (see text footnote 2) at 0, 1, 6, and 24 h after treatment. An isometric histogram was drawn using OriginPro7.510.
Expression Analysis of CaCCO Genes in Response to H2O2 Stress
The oxidative stress was also applied by adding H2O2 into the nutrient solution to a final concentration of 30 mM. Then, the leaves and roots were selected for RNA-seq analysis (see text footnote 2) at 0, 1, 6, and 24 h after treatment. The heat map of expression patterns of CCO genes in response to H2O2 treatment was drawn using the MeV software.
RNA Isolation and Quantitative Real-Time PCR Analysis
Total RNA was extracted from the pepper leaves using E.Z.N.A.® Plant RNA Kit (OMEGA, United States). The first strand cDNA was synthesized using FastKing RT Kit (with gDNase) (TIANGEN, China) according to the instructions of the manufacturer. Gene-specific primers were designed using the Genscript online tool11, and the detailed information is listed in Supplementary Table 2. The pepper GAPDH gene was utilized as an internal control for normalizing the expression levels. The use of primers was diluted according to the synthetic instructions. The reaction system followed the instructions of SYBR Green Master Mix reagent of Vazyme using CFX96 Real Time System (Bio-Rad, United States) with AceQ® qPCR SYBR® Green Master Mix with 20 μl reaction mixture of volume. The reaction volume consists of 10 μl SYBR® Green Master Mix, 0.4 μl of each primer (10 μM), 1 μl of the cDNA template, and 7.8 μl of RNase free H2O. Thermal cycling parameters for the amplification were as follows: 95°C, 10 min, followed by 40 cycles at 95°C, 15 s, and 55°C, 1 min. Three technical repeats were performed for each gene, and the data were analyzed according to the 2–ΔΔCt method.
Results
Identification of the CaCCO Gene Family in Pepper and Other Plant Species
To identify the members of the CCO gene family at the genome-wide levels, the whole genome sequence of pepper was searched using HMM and BLAST. The results showed that a total of 158 candidate CCO genes were identified in various plant species, each with a distinct number of variants. In addition, fourteen CCO DNA binding domains containing proteins were encoded in the pepper genome. The detailed information of these proteins is shown in Table 1, including gene name, genome location, coding sequence length, amino acid sequence length, MW, and isoelectric point. The predicted size of the CaCCO proteins was from 321 to 744 AA (for CaCCD4b and CaCCD7, respectively; Table 1). The MW of these genes ranged from 36.33 (CaCCD4b) to 83.79 kDa (CaCCD7), while their estimated pI values ranged from 5.16 (CaCCD4b) to 8.25 (CaNCED1). Most CaCCO proteins were acidic (pI-values 7) according to the pI values.
TABLE 1.
Information for the CaCCO gene family in pepper.
| Gene name | Gene ID | Protein length (AA) | Molecular weight (kD) | Isoelectric point | |
| CaCCD | CaCCD1a | Capana01g002623 | 581 | 65.81 | 6.49 |
| CaCCD1b | Capana01g002624 | 565 | 63.14 | 5.86 | |
| CaCCD4a | Capana01g000948 | 603 | 66.05 | 6.34 | |
| CaCCD4 | Capana01g000946/Capana01g000947 | 321 | 36.33 | 5.16 | |
| CaCCD4c | Capana01g004038 | 568 | 63.13 | 7.85 | |
| CaCCD7 | Capana01g003456 | 744 | 83.79 | 6.67 | |
| CaCCD8 | Capana08g000570 | 501 | 56.15 | 6.04 | |
| CaCCD-like1 | Capana08g000589 | 567 | 64.06 | 6.18 | |
| CaCCD-like2 | Capana11g000021 | 565 | 64.72 | 5.56 | |
| CaCCD-like3 | Capana11g000005 | 442 | 49.35 | 5.49 | |
| CaNCED | CaNCED1 | Capana05g002276 | 579 | 64.67 | 8.25 |
| CaNCED2 | Capana01g003704 | 596 | 66.49 | 6.64 | |
| CaNCED3 | Capana00g003114 | 506 | 56.47 | 5.6 | |
| CaNCED4 | Capana03g002722/Capana03g002723 | 353 | 40.04 | 6.16 |
Evolutionary Origin of Carotenoid Cleavage Oxygenase Gene Family in Plant Species
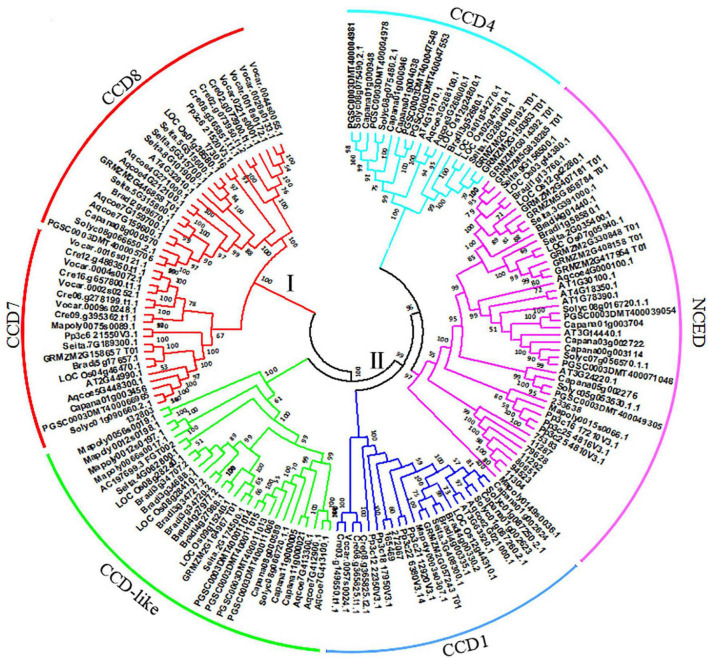
To investigate the evolutionary origin, a total of 158 CCOs (CCDs and NCEDs) from 14 plant species were identified (Supplementary Table 1). A phylogenetic tree was constructed based on the amino acid sequences of these genes using the NJ method in MEGA 5.0 software (Tamura et al., 2011). As shown in Figure 1, all of the CCOs in different plants ranging from green algae to higher plant species were divided into two groups (groups I and II), and the former was further separated into two subgroups (CCD7 and CCD8). Both of these two subgroups contained the members from the lower to higher plant species, suggesting that they have independent evolutionary origins, respectively. Inversely, the latter was subdivided into CCD1, CCD4, CCD-like, and NCED. The CCD1 subgroup was composed of members from green algae to higher plant species, both NCED and CCD-like subgroups contained CCO genes from moss to higher plant species, and CCD4 was composed of CCO genes from angiosperm. This showed that group II may have come from a common ancestor, and NCED, CCD-like, and CCD4 were produced from multiple duplication events in land plant species.
FIGURE 1.
Evolutionary origin of carotenoid cleavage oxygenase (CCO) proteins in plant species. The unrooted tree was constructed using the neighbor-joining (NJ) method in MEGA5.0 software. The parameters were 1,000 bootstraps. The CCO proteins were classified into two groups: I and II. The former was subdivided into CCD7 and CCD8, while the latter was subdivided into CCD1, CCD-like, CCD4, and NCED.
Chromosome Distribution of the CaCCO Gene Family Members
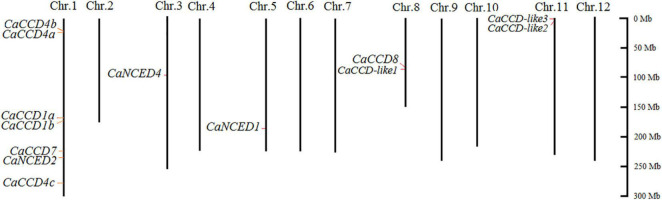
In this study, the chromosome distribution of the pepper CaCCO gene was analyzed based on the genome sequence of the pepper. Except for CaNCED3, the remaining thirteen genes in the fourteen CaCCO members were irregularly distributed on five of the twelve pepper chromosomes, according to the findings (Figure 2). Among them, most of the genes are located on the first chromosome (Chr1), and there are seven genes, namely, CaCCD1a, CaNCED2, CaCCD4a, CaCCD1b, CaCCD4c, CaCCD7, and CaCCD4b. The third and fifth chromosomes had only one member (CaNCED4 and CaNCED1, respectively). Furthermore, two genes are found to be present on each of the two chromosomes (Chr8 and 11). None of the CaCCO genes was mapped on Chr2, 4, 6, 7, 9, 10, and 12. Subsequently, we further analyzed the tandem and segmental duplications of the CaCCO genes in pepper. We found that both of the tandem duplication and segmental duplication events were not found in the CaCCO gene family.
FIGURE 2.
Chromosomal mapping of the CCO genes in Capsicum annuum. The chromosome number is shown at the top of each bar chart, while the size of the chromosome is shown in its relative length. The scale on the right is in MB, and the short line represents the approximate positions of the CaCCO genes on the respective chromosome.
Expression Analysis of CaCCO Genes in Different Tissues
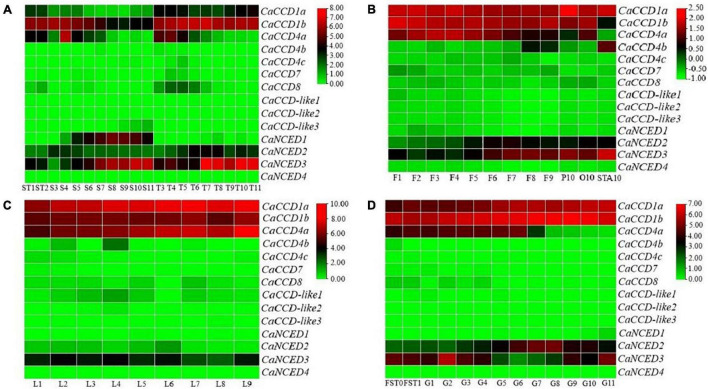
It is well known that RNA-seq data have a wide range of applications and can be used to conveniently analyze the differential expression of genes in higher plant species (del Río et al., 2006). In this study, the expression patterns of fourteen CaCCO genes were analyzed using RNA-seq data from seeds, leaves, flowers, and fruits. The results revealed that most of the CaCCO genes in these organs were completely unexpressed in all of the examined tissues (Figure 3). In different developmental stages of leaves from L1 to L9, expression levels of 10 of 14 CaCCO genes were not detected, including CaNCED1, CaNCED2, CaCCD4c, CaCCD7, CaCCD-like1, CaCCD8, CaCCD-like2, CaCCD-like3, CaNCED4, and CaCCD4b. Three genes (CaCCD1a, CaCCD4a, and CaCCD1b) were expressed constitutively in all of the stages that were analyzed. The expression of CaNCED3 is weak and gradually decreases along with the development.
FIGURE 3.
Expression patterns of CaCCOs in different tissues of pepper. Key: red, strong expression; black, weak expression; green, no expression. The expression data were hierarchically clustered based on MeV software. (A) Seeds and placenta, (B) flowers, (C) leaf, and (D) fruit and peel.
The expression patterns of all CaCCO genes in the flower organs were explored, including twelve different tested stages (F1–F9), petals (P10), ovary (O10), and stamens (STA10). As shown in Figure 3, among 14 CaCCO genes, three members (CaCCD1a, CaNCED3, and CaCCD1b) were expressed in all twelve tested stages. The expression levels of CaNCED2 and CaNCED3 increased gradually in different developmental stages, but those of CaCCD4a and CaCCD1b decreased gradually in different stages, and CaCCD4a was not expressed in stamens. CaCCD1a has the highest expression in all periods, while CaCCD4b only has the expression in F8, F9, and STA10, and the expression in STA10 is the highest.
In pepper fruit, three tissues (placenta, seeds, and pericarp) were selected for analyzing the expression patterns of the CaCCO genes. The developmental stages in each of the tissues (placenta: T3–T11, seed: S3–S11, pericarp: G1–G11, fruits at days 3 and 7: FST0, FST1, and seeds and placenta at days 10 and 15: ST1, ST2) were included. As shown in Figure 3, half members of the CaCCO gene family have relatively high expression levels, and the mean of the log-signal values of each gene is within the range of 10 to 20. On the contrary, nine genes show relatively low expression levels with the mean of the log-signal values of each gene ranging from 0 to 1.
In the placenta and seed, the results showed that most of the CaCCO genes were expressed in different developmental stages. Some genes such as CaCCD1a, CaNCED2, CaCCD4a, CaNCED3, and CaCCD1b were highly expressed in the seeds and placenta of capsicum, indicating that these genes may play an important role in the development of capsicum plants. The gene CaCCD1a was highly expressed in T3–T11, and CaNCED1 was in S5–S11. The CaNCED3 and CaCCD1b were highly expressed in ST1–T11. The transcript levels of CaCCD1b were highly expressed in S3. In pericarp, there were four members with high expression, namely, CaCCD1a, CaCCD4a, CaNCED3, and CaCCD1b. Among them, CaCCD1a and CaCCD1b have the highest expression in each stage of fruit development; CaCCD4a has a higher expression in the early stage of fruit development and a lower expression in the later stage, but not in the G11 stage. CaCCD4c, CaCCD7, CaCCD-like1, CaCCD8, CaCCD-like2, CaCCD-like3, CaCCD4b, and CaNCED1 gene has a similar expression in different period of time, such as FST0–G11. The CaCCO gene has a lower expression. These results indicate that half of CaCCO genes are involved in plant growth and development.
Expression of CaCCOs in Response to Hormone Treatments
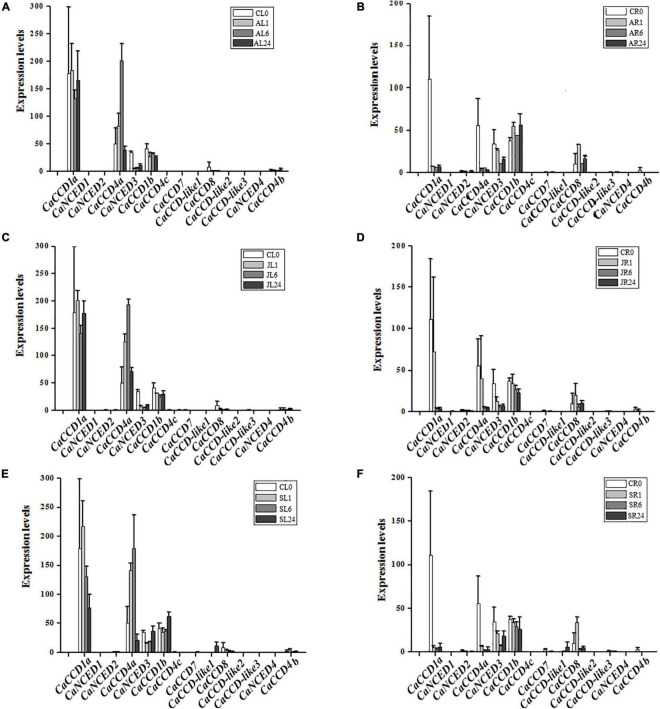
Previous studies have shown that hormones, including cytokinins (CKs), gibberellins (GAs), salicylic acid (SA), jasmonic acid (MeJA), ethylene (ET), ABA, and brassinosteroids (BRs), play a key role in regulating plant growth and development, adapting to environmental stress and complex signal networks (Vallabhaneni et al., 2010). To further analyze the expression patterns of CaCCO genes under stress-related stimulation, the expression profiles of leaves and roots of CaCCO genes under ABA, MeJA, and SA-hormone treatments were analyzed.
As illustrated in Figure 4, we found that fourteen CaCCO genes have different expression patterns in roots and leaves under three different hormone treatments. Among them, five CaCCO genes had high expression levels, while the remaining nine genes had low or even no expression levels. The five genes were induced significantly by ABA, MeJA, and SA treatments. Among the five genes, the root expressions of CaCCD1a and CaCCD4a significantly decreased after treatment with ABA, MeJA, and SA for 1, 6, and 24 h, while the expression levels in leaves showed a trend of first increasing and then decreasing. Among them, the expression levels of CaCCD4a under treatment with different hormones were the highest at 6 h for ABA, MeJA, and SA, while the expressions of CaCCD1a and CaNCED3 were in reverse manner. Expression levels were decreased at 6 h for ABA, MeJA, and SA.
FIGURE 4.
Expression of CaCCO genes under different hormone treatments. (A) Leaves under abscisic acid (ABA) treatment, (B) roots under ABA treatment, (C) leaves under jasmonic acid (MeJA) treatment, (D) roots under MeJA treatment, (E) leaves under salicylic acid (SA) treatment, and (F) roots under SA treatment.
Expression of CaCCOs in Response to Cold, Heat, Drought, and NaCl Treatments
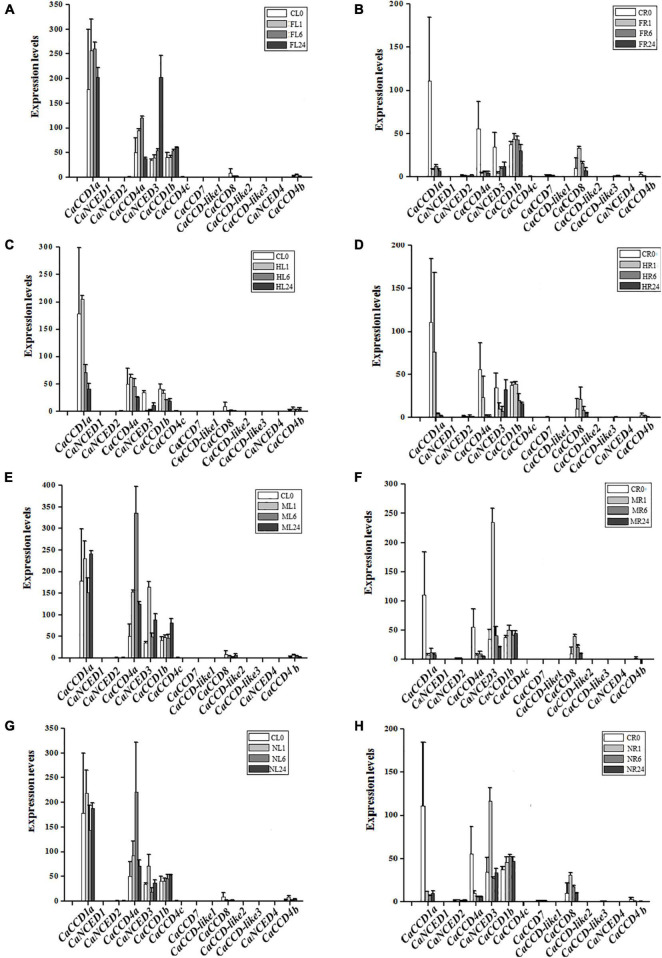
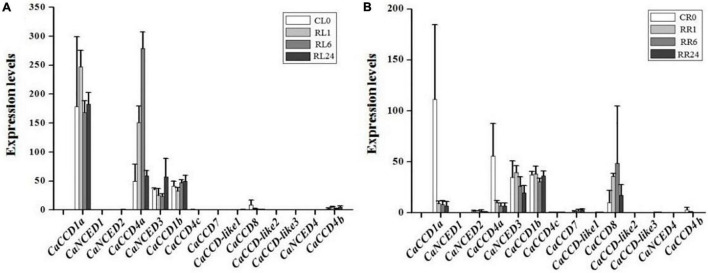
Abiotic stresses (heat, cold, drought, and salt) affected the plant growth and development and were the major factors limiting crop production (Simkin et al., 2004a; Ohmiya et al., 2006; Sun et al., 2008; Adami et al., 2013; Liu et al., 2013; Bai et al., 2017). To illustrate the roles of CaCCO gene family members in abiotic stresses, the RNA-seq data of pepper under four treatments (heat, cold, drought, and NaCl) have been selected. Interestingly, similar expression patterns of most of the CaCCO genes were observed in pepper, whereas some differences in expression patterns can be carefully shown. The results showed that at least four genes were significantly induced under different stresses, and these genes have similar expression patterns, such as CaCCD1a, CaCCD4a, CaNCED3, CaCCD1b, and CaCCD8. The remaining CaCCO genes are not regulated under low temperature, high temperature, drought, and salt stresses.
For cold stress (Figure 5), CaCCD1a and CaCCD4a share a similar expression pattern, and the upregulated expression was followed by a downregulated expression in FL1–FL24. However, in roots, the expression of these two genes was downregulated after cold treatment. The expressions of CaNCED3 and CaCCD1b were upregulated in leaves under low-temperature treatment, especially the expression level of CaNCED3 was increased about 200 times after 24 h of low-temperature treatment. Under heat treatment conditions, the CaCCD1a and CaCCD4a slightly upregulated the stage of CL0 and HL1, then the CaCCD1a and CaCCD4a gene was downregulated in all the stages analyzed. However, in roots, the expression of these two genes was significantly decreased after high-temperature treatment.
FIGURE 5.
Expression patterns of CaCCO genes under low/high temperature, drought, and salt stresses. (A) Leaves under cold stress, (B) roots under cold stress, (C) leaves under heat stress, (D) roots under heat stress, (E) leaves under drought stress, (F) roots under drought stress, (G) leaves under NaCl stress, and (H) roots under NaCl stress.
Under drought stress (Figure 5), the expression of CaCCD4a in leaves was upregulated first and then downregulated, and the highest expression level was reached after 4 h of drought treatment. However, the expression of this gene was significantly downregulated in roots. The expression of CaNCED3 was the highest in leaves and roots at 1 h after drought treatment and then decreased. Of the fourteen genes, about half responded to salt stress, suggesting that these genes were involved in coping with environmental changes. In leaves, a total of 6 genes were upregulated, among which the most significant ones were CaCCD1a and CaCCD4a, followed by CaNCED3 and CaNCCD1b, and the expressions of CaCCD8 and CaCCD4b were the lowest. The expression pattern of the CaCCO gene family in roots under salt stress was the same as that in roots under drought treatment, the expressions of CaCCD1a and CaCCD4a were significantly downregulated, and the expressions of CaNCED3, CaCCD1b, and CaCCD8 were upregulated first and then downregulated.
Expression of CaCCOs in Response to H2O2 Treatments
Under the H2O2 treatment condition (Figure 6), expressions of 6 CaCCO genes were induced in pepper leaf, including CaCCD1a, CaCCD4a, CaNCED3, CaCCD1b, CaCCD8, and CaCCD4b. Among them, the expression levels of CaCCD1a and CaCCD4a were first increased and then decreased. The expression level of CaCCD1a was the highest at 1 h of H2O2 treatment, while CaCCD4a reached the maximum at 6 h of H2O2 treatment. In addition, the trend of CaNCED3 and CaCCD1b treated with H2O2 was opposite to the former. The expression levels reached a peak at 24 h after treatment. In root, expressions of 8 CaCCO genes were induced, namely CaCCD1a, CaNCED2, CaCCD4a, CaNCED3, CaCCD1b, CaCCD7, CaCCD8, and CaCCD4b. Among them, the expression levels of CaCCD1a, CaCCD4a, and CaNCED3 decreased gradually, while CaCCD1b and CaCCD8 increased first and then decreased. The expression levels of CaCCD1b reached the highest at 1 h and CaCCD8 reached the peak at 6 h.
FIGURE 6.
Expression of CaCCO genes under hydrogen peroxide (H2O2) treatment. (A) Leaves under H2O2 stress and (B) roots under H2O2 stress.
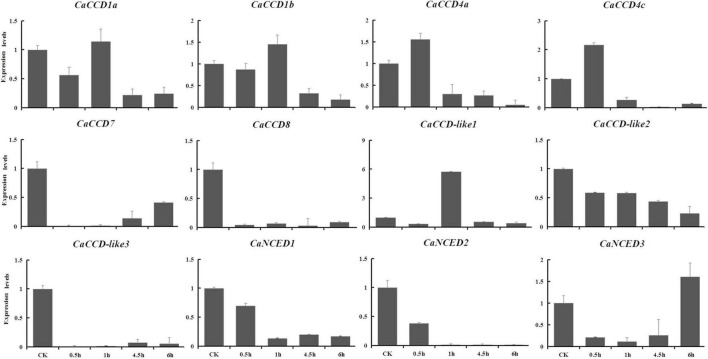
Quantitative RT-PCR Analysis of Members of CaCCO Gene Family in Response to Abiotic Stresses
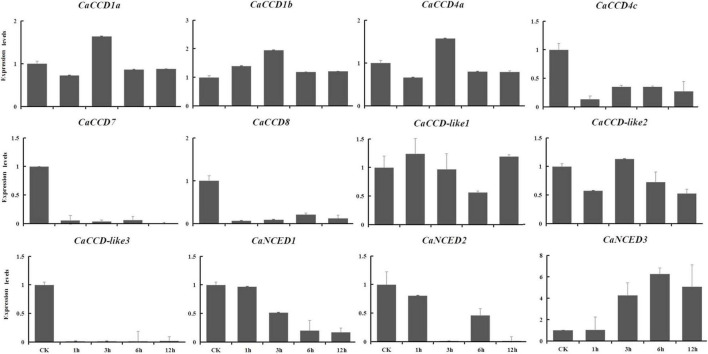
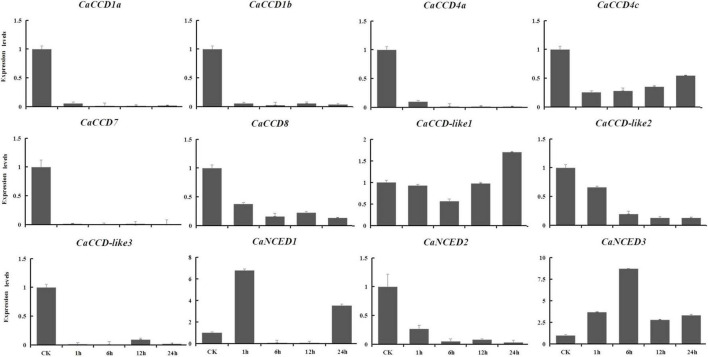
The expression levels of the CaCCOs were investigated under heat, cold, drought, and NaCl treatments using qRT-PCR to understand their possible roles in responses to abiotic stresses (Table 2). Under heat stress, the expression levels of half of the genes were significantly downregulated with large-fold changes at the 0.5, 1, 4.5, and 6 h, respectively, while CaCCD4a, CaCCD4c, and CaCCD-like1 were first upregulated and then downregulated at three of the four time points (Figure 7). Notably, some CaCCDs exhibited significant differences across the four time points. CaCCD4a and CaCCD4c reached the maximum value at 0.5 h. Three members (CaCCD1a, CaCCD1b, and CaCCD-like1) significantly elevated at 1 h. CaNCED3 expression was significantly downregulated at 0.5 h, but significantly upregulated at 6 h after heat treatment. Under cold stress, we found that expression levels of CaCCD4c, CaCCD7, CaCCD8, CaCCD-like3, CaNCED1, and CaNCED2 were significantly downregulated at all of the four time points (Figure 8). In addition, the expression levels of four genes, namely, CaCCD1a, CaCCD1b, CaCCD4a, and CaNCED3, showed a trend of upregulation and then downregulation, respectively. Notably, CaCCD1a, CaCCD1b, CaCCD4a, and CaCCD-like2 all peaked at 3 h after cold treatment.
TABLE 2.
Sequences of the primers used in quantitative real-time PCR (qRT-PCR) analysis in this study.
| Gene name | Forward primers | Reverse primers |
| GAPDH | ATGATGATGTGAAAGCAGCG | TTTCAACTGGTGGCTGCTAC |
| CaCCD1a | GCGTCATGCAAGATCCAGTT | AAAGCGAGCCTTCTTTGTGG |
| CaCCD1b | TCGTCCGAAGGAAATGGTGA | CCAAGCGTTGGCATTATGGA |
| CaCCD4a | TCTTCCACGTGGTCCTTACC | AGGCAGAGAGACCGTTGAAA |
| CaCCD4c | TGGACCTAATTCGCCCAAGA | AGGGTTCCCAATAGCTGCAT |
| CaCCD7 | GGATATGTTGCTCCCTCGGA | TGCTGTCATTGACCCAGGAA |
| CaCCD8 | GTGTAAAGCCAGTGGCAACA | CGGAGATTGTCAAGGCGAAG |
| CaCCD-like1 | GTGTTGAGGTTTGGCGTGAT | GCTTTCTTTGGGTGGCTTGT |
| CaCCD-like2 | AAGATGAGGTGGTGGTGAGG | GAAGTTTGGCTAGCCCTCCA |
| CaCCD-like3 | TGCGAGAATTGGAATAATGCCT | CTACTGCGATAGCCGGTCTT |
| CaNCED1 | CTGCTTCTGCATGCATCTGT | CCGTTTGGTTGACTCACCTG |
| CaNCED2 | GAAGAGCCAGAGACGGATGA | TCGTTTCCTCGACTCTCCAG |
| CaNCED3 | TGGCATGGTTCATGCTGTTC | CCGAAGAGTCCACGAGCATA |
FIGURE 7.
Expression patterns of CaCCO genes in response to heat stress at different time points.
FIGURE 8.
Expression patterns of CaCCO genes in response to cold stress at different time points.
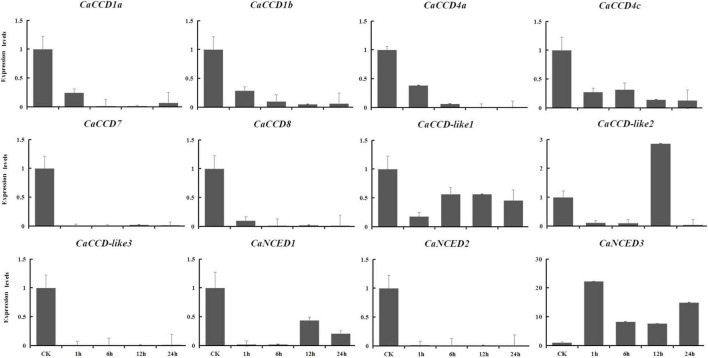
To explore the expression patterns of CCO gene family in pepper in response to drought and NaCl stresses, based on the RNA-seq data, we showed the upregulated in leaves, selected to verify using qRT-PCR analysis at 1, 6, 12, and 24 h expression patterns after treatments. For drought stress, nine genes were downregulated (Figure 9), including CaCCD1a, CaCCD1b, CaCCD4a, CaCCD4c, CaCCD7, CaCCD8, CaCCD-like2, CaCCD-like3, and CaNCED2. In addition, only CaNCED1 and CaNCED3 showed upregulation before downregulation among the twelve genes. However, the difference between the two groups was that the expression level of the former reached its peak at 1 h under drought treatment, increased by 5.7 times compared with the control, while the latter reached its peak at 6 h and increased by 7.7 times compared with the control. In addition, under 400 mM NaCl treatment (Figure 10), our results found that the expression levels of most genes were downregulated, including CaCCD1a, CaCCD1b, CaCCD4a, CaCCD4c, CaCCD7, CaCCD8, CaCCD-like1, CaCCD-like3, CaNCED1, and CaNCED2. However, only CaCCD-like2 and CaNCED3 were upregulated and then downregulated. Notably, the expression of CaCCD-like2 was the highest after 12 h of stress treatment, while the expression of CaNCED3 was the highest after 1 h of stress treatment. These results showed that the CaCCD gene family has a certain function in plant response to abiotic stress, but its molecular mechanism may be different among different genes.
FIGURE 9.
Expression patterns of CaCCO genes in response to drought stress at different time points.
FIGURE 10.
Expression patterns of CaCCO genes in response to salt stress at different time points.
Discussion
Plant CCO gene family, a class of specific enzymes, can catalyze conjugated double-bond system of carotenoids and their apocarotenoids to form smaller compounds (Ilg et al., 2009; Walter et al., 2010). These CCO proteins catalyze the cleavage of carotenoids and help to adjust the plant responses to stress (Espasandin et al., 2014). Therefore, it is very important to understand the evolutionary relationship and function of CCO genes in plant species. In this study, our results showed that plant CCO genes could be divided into two subfamilies (groups I and II). Group I was subdivided into CCD7 and CCD, and group II was composed of CCD1, CCD4, CCD-like, and NCED. We found that orthology groups (CCD7 and CCD8) appeared to be conserved only as single- or low-copy genes in all plants, whereas group II genes underwent several duplication events, resulting in multiple gene copies. These duplication events were due to whole-genome duplications in plants. Our results indicate that group II genes had strikingly different patterns of gene duplication from genes from CCD7 and CCD8. The CCD1 genes expanded during the histories of land plants (Figure 1). Unlike CCO genes from group II, other types of plant CCO genes (CCD7 and CCD8) are single- or low-copy genes. The stably maintained low-copy numbers for these CCO genes suggest the functional conservation during the process of evolution in plants. These results also suggested that differential evolution of members of the CCO gene family with conservative and divergent patterns were observed.
The gene expression profiles often have some connection with gene function. To date, although the expression patterns of CCO genes have been determined in other plants, there are few detailed studies on the expression of CCO genes in pepper. To determine whether there were differences in the expression of CaCCO genes, the transcriptional levels of each member in pepper were analyzed spatially and temporally. In four different organs, about half of the CaCCO genes were not fully expressed at all stages of detection. In leaf, the CaCCD1a, CaCCD4a, CaNCED3, and CaCCD1b were expressed in all of the detected stages. Compared with the other three genes, the expression of CaNCED3 was lower at all stages. We concluded that they are likely to act as a housekeeping gene of pepper cells under normal growth conditions. In flower, high expression levels of four genes (CaCCD1a, CaCCD4a, CaNCED3, and CaCCD1b) were observed in all the stages analyzed (F1–F9), indicating that these genes were involved in the growth and development of the flower. The CaNCED2 and CaNCED3 are transcribed at the late stage (F5–F9), while CaCCD1a, CaCCD4a, and CaCCD1b are transcribed in the whole development stage (F1–F9). These results suggest that these genes may play different functional roles at different stages of development.
We further analyzed the expression profiles of these CaCCO genes in the placenta, seed, and pericarp. In the placenta, all genes were expressed at different stages. Among them, the expressions of CaCCD1a, CaNCED2, CaNCED3, and CaCCD1b were the highest (T3–T11), and the expression of CaNCED1 was the lowest in the whole placenta development period. The expression levels of CaCCD4a and CaCCD8 were highest in the early placental stage (T1–T6) and decreased in the late placental stage (T7–T11). In seeds, CaCCD4a was expressed preferentially in the early stage (S1–S7), while CaNCED1 was transcribed in the late stage (S5–S11). The expression level of CaCCD1b was the highest in each period, and the expression level of CaNCED3 was the highest in all periods except S3–S4. The results indicated that these genes are related to seed development and maturation in pepper plants. The expression levels of two genes (CaCCD1a and CaCCD1b) in the pericarp were the highest (FST0–G11) during the whole development period. CaCCD4a and CaNCED3 were expressed in the early stage (G1–G4), and CaCCD4c and CaCCD1b were expressed in the late stage (G5–G11). We concluded that these genes play important roles in the formation and maturity of pepper pericarp. In conclusion, these CaCCO genes may have specific functions in different organs and stages.
Under abiotic stress, the effects of heat and cold stress on plant growth induced the expression of multiple CaCCO genes in leaves. Under heat stress, CaCCD1a, CaCCD4a, and CaCCD1b genes were downregulated at different stages. Under cold stress, four CaCCO genes (CaCCD1a, CaCCD4a, CaNCED3, and CaCCD1b) were obviously induced, while CaCCD8 and CaCCD4b were slightly induced. Drought stress and salt stress also induced this trend.
The NCED, a key rate-limiting enzyme in ABA biosynthesis, plays an important role in regulating plant growth and development and stress responses. Some studies have reported that ABA synthesis in plants can respond to oxidative stress, and oxidative reaction has the function of reactive oxygen species (ROS) clearance. ROS are biological molecules that are necessary for seed dormancy and germination. Among them, H2O2 can maintain cell homeostasis and participate in the regulation of photosynthesis, seed germination, and stomatal characteristics, and other physiological activities. O2– and other ROS such as hydroxyl radical (OH), perhydroxy radical (HO2), alkoxy radicals (RO), H2O2, and singlet oxygen (1O2) are produced continuously as byproducts of various metabolic pathways that are localized in different cellular compartments including mitochondria, chloroplast, and peroxisomes (del Río et al., 2006; Navrot et al., 2007). Under stable conditions, ROS can be removed and maintained in equilibrium utilizing antioxidants or antioxidant enzymes (Gill and Tuteja, 2010). In the study, the results indicate that about half of the CCO genes can be induced by H2O2 stress in both roots and leaves, and their expression levels in leaves are upregulated first and downregulated later, which may be related to ABA synthesis and stress regulation. The specific mechanism of CCO genes needs to be confirmed by further experiments.
Conclusion
The RNA-seq analysis of the CaCCO gene family provides a solid foundation for us to fully understand the functions of the CCO gene in pepper growth and development and abiotic stress. However, the functions of these genes in each genome are still unclear and need to be verified by further experiments.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YXY, LJ, and HW conceived and designed the research. YC, MR, QY, RW, ZY, and GZ performed the experiments. JL, JY, PZ, and YHY analyzed the data and wrote the manuscript. YXY, WD, and HW revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This research was partially supported by the National Key Research and Development Program of China (2018YFD1000800 and 2017YFE0114500), China Agriculture Research System of MOF and MARA (CARS-23-G-44), National Natural Science Foundation of China (31772294, 31960601, and 32060446), and Natural Science Foundation of Inner Mongolia Autonomous Region of China (2021MS03106 and 2021MS03071).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.792832/full#supplementary-material
References
- Adami M., Franceschi P., Brandi F., Liverani A., Giovannini D., Rosati C., et al. (2013). Identifying a carotenoid cleavage dioxygenase (CCD4) gene controlling yellow/white fruit flesh color of peach. Plant Mol. Biol. Rep. 31 1166–1175. 10.1007/s11105-013-0628-6 [DOI] [Google Scholar]
- Auldridge M. E., Block A., Vogel J. T., Dabney-Smith C., Klee H. J. (2006). Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 45 982–993. 10.1111/j.1365-313X.2006.02666.x [DOI] [PubMed] [Google Scholar]
- Bai G., Yang D. H., Yao H., Xie H. (2017). Identification and analysis of NtNCED gene in tobacco. Mol. Plant Breed. 10 73–78. 10.13271/j.mpb.015.003907 [DOI] [Google Scholar]
- Blankenship S. M., Sisler E. C. (1993). Response of apples to diazocyclopentadiene inhibition of ethylene binding. Postharvest Biol. Technol. 3 95–101. 10.1016/0925-5214(93)90001-J [DOI] [Google Scholar]
- Brandi F., Bar E., Mourgues F., Horváth G., Turcsi E., Giuliano G., et al. (2011). Study of’ ‘Redhave’ peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism. BMC Plant Biol. 11:24. 10.1186/1471-2229-11-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton G., Liaaen-Jensen S., Pfander H. P. (2004). Handbook of Carotenoids. Basel: Birkhäuser [Google Scholar]
- Burbidge A., Grieve T. M., Jackson A., Thompson A., McCarty D. R., Taylor I. B. (1999). Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. Plant J. 17 427–431. 10.1046/j.1365-313x.1999.00386.x [DOI] [PubMed] [Google Scholar]
- Campbell R., Ducreux L. J., Morris W. L., Morris J. A., Suttle J. C., Ramsay G., et al. (2010). The metabolic and developmental roles of carotenoid cleavage dioxygenase 4 from potato. Plant Physiol. 154 656–664. 10.1104/pp.110.158733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernys J. T., Zeevaart J. A. (2000). Characterization of the 9-cis-epoxycarotenoid dioxygenase gene family and the regulation of abscisic acid biosynthesis in avocado. Plant Physiol. 124 343–353. 10.1104/pp.124.1.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río L. A., Sandalio L. M., Corpas F. J., Palma J. M., Barroso J. B. (2006). Reactive oxygen species and reactive nitrogen species in peroxisomes. production, scavenging, and role in cell signaling. Plant Physiol. 141 330–335. 10.1104/pp.106.078204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R., Cheung M. K., Bright J., Henson D., Hancock J. T., Neill S. J. (2004). ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 55 205–212. 10.1093/jxb/erh033 [DOI] [PubMed] [Google Scholar]
- Espasandin F. D., Maiale S. J., Calzadilla P., Ruiz O. A., Sansberro P. A. (2014). Transcriptional regulation of 9-cis-epoxycarotenoid dioxygenase (NCED) gene by putrescine accumulation positively modulates ABA synthesis and drought tolerance in Lotus tenuis plants. Plant Physiol. Biochem. 76 29–35. 10.1016/j.plaphy.2013.12.018 [DOI] [PubMed] [Google Scholar]
- Fan J., Hill L., Crooks C., Doerner P., Lamb C. (2009). Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 150 1750–1761. 10.1104/pp.109.137943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A., Effroy D., Lefebvre V., Seo M., Perreau F., Berger A., et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70 501–512. 10.1111/j.1365-313X.2011.04887.x [DOI] [PubMed] [Google Scholar]
- Gill S., Tuteja N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 909–930. 10.1016/j.plaphy.2010.08.016 [DOI] [PubMed] [Google Scholar]
- Gómez C. A., Arbona V., Jacas J., Primo-Millo E., Talon M. (2002). Abscisic acid reduces leaf abscission and increases salt tolerance in citrus plants. J. Plant Growth Regul. 21 234–240. 10.1007/s00344-002-0013-4 [DOI] [Google Scholar]
- Huang F. C., Molnár P., Schwab W. (2009). Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 60 3011–3022. 10.1093/jxb/erp137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Guo Y., Liu Y., Zhang F., Wang Z., Wang H., et al. (2018). 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front. Plant Sci. 9:162. 10.3389/fpls.2018.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. G., Lee C. Y., Tseng C. S. (2018). Heterologous expression of rice 9–cis–epoxycarotenoid dioxygenase 4 (OsNCED4) in Arabidopsis confers sugar oversensitivity and drought tolerance. Bot. Stud. 59:2. 10.1186/s40529-018-0219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilg A., Beyer P., Babili S. (2009). Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J. 276 736–747. 10.1111/j.1742-4658.2008.06820.x [DOI] [PubMed] [Google Scholar]
- Jiang M., Zhang J. (2003). Cross-talk between calcium and reactive oxygen species originated from NADPH oxidase in abscisic acid-induced antioxidant defense in leaves of maize seedlings. Plant Cell Environ. 26 929–939. 10.1046/j.1365-3040.2003.01025.x [DOI] [PubMed] [Google Scholar]
- Kong X., Luo Z., Dong H., Eneji A. E., Li W. (2016). H2O2 and ABA signaling are responsible for the increased Na+ efflux and water uptake in Gossypium hirsutum L. roots in the non-saline side under non-uniform root zone salinity. J. Exp. Bot. 8:2247. 10.1093/jxb/erw026 [DOI] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., et al. (2010). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45 309–319. 10.1111/j.1365-313X.2005.02622.x [DOI] [PubMed] [Google Scholar]
- Liu J., Novero M., Charnikhova T., Ferrandino A., Schubert A., Ruyter Spira C., et al. (2013). Carotenoid cleavage dioxygenase 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J. Exp. Bot. 64 1967–1981. 10.1093/jxb/ert056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Okamoto M., Tatematsu K., Yano R., Seo M., Kamiya Y. (2010). Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 20 55–67. 10.1017/S0960258510000012 [DOI] [Google Scholar]
- Navrot N., Rouhier N., Gelhaye E., Jaquot J. P. (2007). Reactive oxygen species generation and antioxidant systems in plant mitochondria. Physiol. Plant. 129 185–195. 10.1111/j.1399-3054.2006.00777.x [DOI] [Google Scholar]
- Neill S. J., Desikan R., Clarke A., Hurst R. D., Hancock J. T. (2002). Hydrogen peroxide and nitric oxide as signaling molecules in plants. J. Exp. Bot. 53 1237–1247. 10.1093/jexbot/53.372.1237 [DOI] [PubMed] [Google Scholar]
- Ohmiya A., Kishimoto S., Aida R., Yoshioka S., Sumitomo K. (2006). Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 142 1193–1201. 10.1104/pp.106.087130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Yu C. S., Shen Y. O., Fang X. D., Chen L., Min J. M., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 14 5135–5140. 10.1073/pnas.1400975111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X., Zeevaart J. A. D. (2002). Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol. 128 544–551. 10.1104/pp.010663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A., Ashraf F., Zou X. L., Zhang X. K., Tosif H. (2020). “Plant adaptation and tolerance to environmental stresses: mechanisms and perspectives,” in Plant Ecophysiology and Adaptation Under Climate Change: Mechanisms and Perspectives I, ed. Hasanuzzaman M. (Berlin: Springer; ), 117–145. [Google Scholar]
- Rodrigo M. J. (2006). Cloning and characterization of two 9-cis-epoxycarotenoid dioxygenase genes, differentially regulated during fruit maturation and under stress conditions, from orange (Citrus sinensis L. Osbeck). J. Exp. Bot. 57 633–643. 10.1093/jxb/erj048 [DOI] [PubMed] [Google Scholar]
- Roychoudhury A., Paul S., Basu S. (2013). Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 32 985–1006. 10.1007/s00299-013-1414-5 [DOI] [PubMed] [Google Scholar]
- Schwartz S. H., Tan B. C., Gage D. A., Zeevaart J. A., Mccarty D. R. (1997). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276 1872–1874. 10.1126/science.276.5320.1872 [DOI] [PubMed] [Google Scholar]
- Seo M., Hanada A., Kuwahara A., Endo A., Okamoto M., Yamauchi Y., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48 354–366. 10.1111/j.1365-313X.2006.02881.x [DOI] [PubMed] [Google Scholar]
- Seo M., Nambara E., Choi G., Yamaguchi S. (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69 463–472. 10.1007/s11103-008-9429-y [DOI] [PubMed] [Google Scholar]
- Simkin A. J., Schwartz S. H., Auldridge M., Taylor M. G., Klee H. J. (2004a). The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J. 40 882–892. 10.1111/j.1365-313X.2004.02263.x [DOI] [PubMed] [Google Scholar]
- Simkin A. J., Underwood B. A., Auldridge M., Loucas H. M., Shibuya K., Schmelz E., et al. (2004b). Circadian regulation of the PhCCD1 carotenoid cleavage dioxygenase controls emission of β-ionone, a fragrance volatile of petunia flowers. Plant Physiol. 136 3504–3514. 10.1104/pp.104.049718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden K. C., Simkin A. J., Janssen B. J., Templeton K. R., Loucas H. M., Simons J. L., et al. (2005). The decreased apical dominance 1/petunia hybrida carotenoid cleavage dioxygenase 8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17 746–759. 10.1105/tpc.104.027714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Hans J., Walter M. H., Matusova R., Beekwilder J., Verstappen F. W., et al. (2008). Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta 228 789–801. 10.1007/s00425-008-0781-6 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B. C., Joseph L. M., Deng W. T., Liu L., Li Q., Cline K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis-epoxycarotenoid dioxygenase gene family. Plant J. 35 44–56. 10.1046/j.1365-313x.2003.01786.x [DOI] [PubMed] [Google Scholar]
- Tan B. C., Schwartz S. H., Zeevaart J. A., McCarty D. R. (1997). Genetic control of abscisic acid biosynthesis in maize. Proc. Natl. Acad. Sci. U.S.A. 94 12235–12240. 10.1073/pnas.94.22.12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. J., Jackson A. C., Symonds R. C., Mulholland B. J., Dadswell A. R., Blake P. S., et al. (2000). Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 23 363–374. 10.1046/j.1365-313x.2000.00789.x [DOI] [PubMed] [Google Scholar]
- Toh S., Imamura A., Watanabe A., Nakabayashi K., Okamoto M., Jikumaru Y., et al. (2008). High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146 1368–1385. 10.1104/pp.107.113738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida A., Jagendorf A. T., Hibino T., Takabe T., Takabe T. (2002). Effect of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 163 515–523. 10.1016/S0168-9452(02)00159-0 [DOI] [Google Scholar]
- Vallabhaneni R., Bradbury L. M., Wurtzel E. T. (2010). The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch. Biochem. Biophys. 504 104–111. 10.1016/j.abb.2010.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M. H., Floss D. S., Strack D. (2010). Apocarotenoids: hormones, mycorrhizal metabolites and aroma volatiles. Planta 232 1–17. 10.1007/s00425-010-1156-3 [DOI] [PubMed] [Google Scholar]
- Welsch R., Wüst F., Bär C., Al-Babili S., Beyer P. (2008). A third phytoene synthase is devoted to abiotic stress-induced abscisic acid formation in rice and defines functional diversification of phytoene synthase genes. Plant Physiol. 147 367–380. 10.1104/pp.108.117028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian L. H., Sun P. P., Hu S. S., Wu J., Liu J. H. (2014). Molecular cloning and characterization of CrNCED1, a gene encoding 9-cis-epoxycarotenoid dioxygenase in Citrus reshni, with functions in tolerance to multiple abiotic stresses. Planta 239 61–77. 10.1007/s00425-013-1963-4 [DOI] [PubMed] [Google Scholar]
- Yang J., Guo Z. (2007). Cloning of a 9-cis-epoxycarotenoid dioxygenase gene (SgNCED1) from Stylosanthes guianensis and its expression in response to abiotic stresses. Plant Cell Rep. 26 1383–1390. 10.1007/s00299-007-0325-8 [DOI] [PubMed] [Google Scholar]
- Zhang A., Jiang M., Zhang J., Ding H., Xu S., Hu X., et al. (2007). Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol. 175 36–50. 10.1111/j.1469-8137.2007.02071.x [DOI] [PubMed] [Google Scholar]
- Zhang W. W., Yang H. Q., You S. Z., Fan S. L., Ran K. (2015). MhNCED3, a gene encoding 9-cis-epoxycarotenoid dioxygenase in Malus hupehensis Rehd., enhances plant tolerance to Cl- stress by reducing Cl- accumulation. Plant Physiol. Biochem. 89 85–91. 10.1016/j.plaphy.2015.02.012 [DOI] [PubMed] [Google Scholar]
- Zhang W. W., Yang H. Q., You S. Z., Xu Y. T., Ran K., Fan S. (2014). Cloning, characterization and functional analysis of the role MhNCED3, a gene encoding 9-cis-epoxycarotenoid dioxygenase in Malus hupehensis Rehd. plays in plant tolerance to osmotic and Cd2+ stresses. Plant Soil 381 143–160. 10.1007/s11104-014-2120-y [DOI] [Google Scholar]
- Zhou B., Guo Z., Xing J., Huang B. (2005). Nitric oxide is involved in abscisic acid-induced antioxidant activities in Stylosanthes guianensis. J. Exp. Bot. 56 3223–3228. 10.1093/jxb/eri319 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.