Visual Abstract
Keywords: clinical nephrology, APOL1 protein, human, apolipoprotein L1, area under curve, biologic specimen banks, diabetes mellitus, type 2, electronic health records, follow-up studies, glomerular filtration rate, HAVCR1 protein, human, hepatitis A virus cellular receptor 1, prognosis, receptors, tumor necrosis factor, type I, TNFRSF1A protein, human, tumor necrosis factors
Abstract
Background
Individuals with type 2 diabetes (T2D) or the apolipoprotein L1 high-risk (APOL1-HR) genotypes are at increased risk of rapid kidney function decline (RKFD) and kidney failure. We hypothesized that a prognostic test using machine learning integrating blood biomarkers and longitudinal electronic health record (EHR) data would improve risk stratification.
Methods
We selected two cohorts from the Mount Sinai BioMe Biobank: T2D (n=871) and African ancestry with APOL1-HR (n=498). We measured plasma tumor necrosis factor receptors (TNFR) 1 and 2 and kidney injury molecule-1 (KIM-1) and used random forest algorithms to integrate biomarker and EHR data to generate a risk score for a composite outcome: RKFD (eGFR decline of ≥5 ml/min per year), or 40% sustained eGFR decline, or kidney failure. We compared performance to a validated clinical model and applied thresholds to assess the utility of the prognostic test (KidneyIntelX) to accurately stratify patients into risk categories.
Results
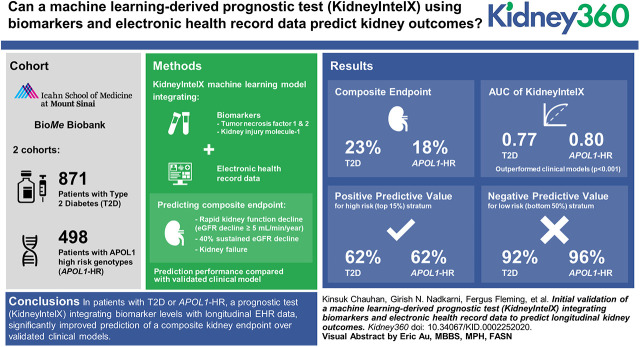
Overall, 23% of those with T2D and 18% of those with APOL1-HR experienced the composite kidney end point over a median follow-up of 4.6 and 5.9 years, respectively. The area under the receiver operator characteristic curve (AUC) of KidneyIntelX was 0.77 (95% CI, 0.75 to 0.79) in T2D, and 0.80 (95% CI, 0.77 to 0.83) in APOL1-HR, outperforming the clinical models (AUC, 0.66 [95% CI, 0.65 to 0.67] and 0.72 [95% CI, 0.71 to 0.73], respectively; P<0.001). The positive predictive values for KidneyIntelX were 62% and 62% versus 46% and 39% for the clinical models (P<0.01) in high-risk (top 15%) stratum for T2D and APOL1-HR, respectively. The negative predictive values for KidneyIntelX were 92% in T2D and 96% for APOL1-HR versus 85% and 93% for the clinical model, respectively (P=0.76 and 0.93, respectively), in low-risk stratum (bottom 50%).
Conclusions
In patients with T2D or APOL1-HR, a prognostic test (KidneyIntelX) integrating biomarker levels with longitudinal EHR data significantly improved prediction of a composite kidney end point of RKFD, 40% decline in eGFR, or kidney failure over validated clinical models.
Introduction
CKD affects >35 million individuals in the United States. Diabetic kidney disease due to type 2 diabetes (T2D) accounts for 44% of patients with ESKD and is a major independent risk factor for other complications. Those with African ancestry have higher rates of ESKD compared with European Americans across all baseline eGFR levels (1,2). Of relevance, genetic studies demonstrated that two distinct alleles in the apolipoprotein L1 (APOL1) gene confer increased risk for many kidney diseases in those with African ancestry. The APOL1 high-risk (APOL1-HR) genotypes (i.e., two copies of risk allele) are associated with increased risk of ESKD, CKD incidence/progression (3), and eGFR decline (4,5).
Although these populations are on average at higher risk than the general population, accurate prediction of who will have rapid kidney function decline (RKFD), defined as eGFR decline >5 ml/min per 1.73 m2 per year, and worse kidney outcomes is lacking (6,7). A current standard for ESKD prediction in CKD stages 3–5 is the Kidney Failure Risk Equation, where clinical variables are assigned standard weights for a recursive score calculation. However, the Kidney Failure Risk Equation has not been validated in individuals with relatively preserved kidney function at baseline (8). Recent work has shown other recursive scores may aide in risk prediction of kidney outcomes in patients with preserved kidney function (9).
Several biomarkers have been investigated to aide in the prediction of kidney outcomes (10). Three of the most extensively studied and strongly associated biomarkers with kidney disease progression in several settings are soluble tumor necrosis factor receptors (TNFR) 1 and 2 and plasma kidney injury molecule-1 (KIM-1) (11–19). Although these markers have uniformly shown an independent association with kidney outcome risk along with specific clinical variables such as eGFR and urinary albumin-creatinine ratio (UACR), the implementation of accurate models that combine clinical data with these plasma biomarkers to predict progression of kidney disease is unavailable.
Widespread electronic health record (EHR) usage provides the potential to leverage thousands of clinical features. Standard statistical approaches are inadequate to leverage this data due to feature volume, unaligned nature of data, and correlation structure (3). However, contemporary machine learning approaches have improved the capacity of analytical model development to combine both biomarkers and longitudinal EHR data for improved prediction.
In this study, we used retrospectively collected plasma samples linked to longitudinal clinical data from the Icahn School of Medicine at Mount Sinai (ISMMS) BioMe Biobank to examine the ability of a prognostic test (KidneyIntelX) that uses machine learning algorithms to predict RKFD and kidney outcomes in two discrete, high-risk patient populations, T2D and APOL1-HR.
Materials and Methods
The BioMe Biobank at ISMMS
The BioMe Biobank at ISMMS is an institutional review board–approved biorepository that includes consented access to the patients’ EHR from a diverse community in New York City, New York (10,11). Operations were initiated in 2007 and include direct recruitment from >30 broadly selected clinical sites. For the purpose of this study, we selected two subpopulations: (1) T2D, enrollment eGFR 45–90 ml/min, and ≥3 years of follow-up data; and (2) APOL1-HR, enrollment eGFR >30 ml/min and ≥3 years of follow-up data. We included all patients from these two biobanks meeting these criteria (n=1369).
Ascertainment and Definition of the Kidney End Point
We determined eGFR using the CKD–Epidemiology Collaboration equation and eGFR slope using a minimum of three values from baseline. The primary composite outcome was comprised of three components: RKFD, defined as an eGFR slope decline of ≥5 ml/min per 1.73 m2 per year (20–23), or a sustained (confirmed ≥3 months later) decline in eGFR of ≥40% (20) from baseline, or “kidney failure” defined by sustained eGFR <15 ml/min per 1.73 m2 confirmed at least 30 days later or long-term maintenance dialysis or kidney transplant (i.e., ESKD) (23–28).
Ascertainment of Clinical Variables in BioMe Biobank
Sex and race were obtained from an enrollment questionnaire. Clinical data were extracted for all continuous variables at the time of and before baseline from the EHR with concurrent time stamps. Hypertension and T2D were determined using phenotyping algorithms (29–31). Cardiovascular disease and heart failure were determined by a validated algorithm and International Classification of Diseases, Ninth/Tenth Revision codes, respectively. We considered a participant to be on an angiotensin-converting enzyme inhibitor or angiotensin receptor blocker if they had a concurrent prescription at enrollment. We calculated follow-up time from enrollment to the latest visit. Only variables present in >70% of subjects (except UACR/BP due to their established clinical importance) were included and used for training of the KidneyIntelX algorithm.
Biospecimen Storage and Analyte Measurement
Plasma specimens collected on the day of BioMe enrollment were stored continuously at −80°C. The biomarkers were measured in a multiplex format using the Meso Scale platform (Meso Scale Diagnostics, Gaithersburg, MD), employing proprietary electrochemiluminescence detection methods combined with patterned arrays allowing for analyte multiplexing. The intra- and interassay coefficient of variation for quality control samples with known low, moderate, and high concentrations of each biomarker run on each plate were 3.5%, 3.9%, and 4.5%; and 12.4%, 10.8%, and 7.7% for TNFR1, TNFR2, and KIM-1, respectively. The laboratory personnel were blinded to clinical information.
Statistical Analyses
We expressed descriptive results for the participants’ baseline characteristics and biomarkers via means and SD or, for skewed variables, medians and interquartile ranges (IQRs).
For the random forest model, we considered two inputs: (1) biomarker concentrations/ratios; and (2) EHR features including laboratory values, diagnosis/procedure codes, demographics (age, sex, and listed race), medications, and healthcare encounter history. Missing UACR values were imputed to 10 mg/g (9), missing BP values were imputed using multiple predictors (age, sex, race, and antihypertensive medications) (32), and median value imputation was used for other missing values. We then created meta-features from these variables including maximum, minimum, median, variability, and change over time to account for their longitudinal aspect and repeated nature. For model development, the clinical data was randomly and demographically split to create an 80%:20% training and test set, respectively, with 10-fold cross-validation on all candidate models.
We then performed further iterations of the random forest model by tuning three hyperparameters. Hyperparameter 1 is the number of decision trees, hyperparameter 2 is the number of variables randomly selected for splitting at each node, and hyperparameter 3 is the minimum size of terminal nodes. The final model that had the best area under the receiver operator characteristic curve (AUC) was chosen.
We generated risk probabilities for the composite kidney end point using the final model on all subjects from both cohorts (T2D and APOL1-HR) and then scaled to generate a continuous score. We compared KidneyIntelX to a published validated clinical model consisting of a regression equation for 40% eGFR decline prediction (9) including age, sex, race, eGFR, cardiovascular disease, smoking, hypertension, body mass index, and UACR in individuals who are nondiabetics and the aforementioned variables plus insulin, diabetes medications, and hemoglobin A1c for patients with T2D (eTable11 in (9)). We compared all differences between AUCs using the DeLong test for comparisons.
We examined the thresholds of the risk score to define low-, intermediate-, and high-risk strata in each cohort. The low-risk stratum was set to encompass 50% of the study population, and the thresholds for the high-risk stratum were assessed to classify the top 10%, 15%, and 20% highest risk in each cohort. The remaining population was defined as the intermediate-risk stratum. We calculated sensitivity, specificity, and positive predicted values/negative predicted values (PPV/NPV) for the high-risk and low-risk cutoffs and compared these to the clinical model. The goodness-of-fit statistics (Hosmer–Lemeshow) was used to assess calibration.
We then conducted subgroup/sensitivity analyses: (1) in individuals with existing CKD (eGFR <60 ml/min per 1.73 m2 and/or UACR >30 mg/g at baseline), and (2) using only data from ≤1 year before biomarker measurement (i.e., “contemporary data,” to ensure that KidneyIntelX was robust in advanced stages of the disease and performed equally well with clinical data limited to a year before biomarker measurement). (3) We generated a trained and tested random forest model that did not include any of the biomarkers (TNFR1, TNFR2, or KIM-1) in both cohorts. (4) We evaluated the performance of the full KidneyIntelX model for the individual components of the composite kidney end point. (5) We conducted Kaplan–Meier survival analyses for time-dependent outcomes of 40% decline and kidney failure with hazard ratios using the Cox proportional hazards method for the high-risk (top 15%) versus the intermediate- and low-risk strata (bottom 50%). All analyses were performed with R software (www.rproject.org).
Results
Baseline Characteristics of Cohorts
Patients with T2D (n=871)
The median age was 60 years, 507 (58%) were female, and the median eGFR was 68 ml/min per 1.73 m2 (Table 1). The most common comorbidities were hypertension (93%), coronary heart disease (50%), and heart failure (22%). The majority (77%) were on angiotensin-converting enzyme inhibitors or angiotensin receptor blockers. Patient characteristics including events between the training and test cohorts were balanced (Supplemental Table 1).
Table 1.
Clinical characteristics at baseline
| Characteristics | Type 2 DM (n=871) | APOL1-HR (n=498) |
| Clinical characteristics | ||
| Age in yr, median (IQR) | 60 (53–66) | 56 (46–66) |
| Female, n (%) | 507 (58.2) | 337 (67.6) |
| Race | ||
| White | 62 (7.1) | 0 |
| African ancestry | 346 (39.7) | 498 (100) |
| Hispanic/Latino | 412 (47.3) | 0 |
| Other | 51 (5.9) | 0 |
| Body Mass index in kg/m2, median (IQR) | 30.9 (26.6–36.1) | 30.5 (26.2–35.8) |
| Hypertension, n (%) | 813 (93.3) | 220 (44.2) |
| Coronary artery disease, n (%) | 432 (49.6) | 39 (7.8) |
| Heart failure, n (%) | 192 (22.0) | 15 (3) |
| Systolic BP in mm Hg, median (IQR) | 131.9 (123.7–143.4) | 129 (117–140.5) |
| Diastolic BP in mm Hg, median (IQR) | 73.4 (68.7–79.3) | 77 (69.5–84.5) |
| Follow-up time in years, median (IQR) | 4.5 (3.3–6.1) | 5.9 (3.9–7.1) |
| Laboratory characteristics | ||
| Baseline eGFR in ml/min per 1.73 m2, median (IQR) | 68.4 (55.3–80.0) | 83.3 (68.9–99.4) |
| Baseline UACR in mg/g, median (IQR) | 13.0 (4.0–66.3) | 11 (4.5–55) |
| UACR available, n (%) | 486 (56%) | 112 (23%) |
| Baseline hemoglobin A1C, median (IQR) | 7.0 (6.2–8.7) | 5.9 (5.5–6.4) |
| Medications | ||
| ACE/ARB at baseline, n (%) | 675 (77.5) | 122 (24.5) |
| Plasma biomarker concentrations | ||
| TNFR1, in pg/ml, median (IQR) | 6057.0 (4764.9–8224.4) | 2465 (1988–3266) |
| TNFR2, in pg/ml, median (IQR) | 6914.2 (5332.9–9832.9) | 4215 (3234–5654) |
| KIM-1, in pg/ml, median (IQR) | 323.3 (196.8–592.1) | 154 (96–269) |
DM, diabetes mellitus; IQR, interquartile range; UACR, urinary albumin-creatinine ratio; ACE, angiotensin-converting enzyme; ARB, angiotensin receptor blocker; TNFR1 and TNFR2, tumor necrosis factor receptors 1 and 2; KIM-1, kidney injury molecule-1.
Patients with APOL1-HR (n=498)
The median age was 56 years, 337 (67.6%) were female, and the median eGFR was 83.3 ml/min per 1.73 m2 (Table 1). The prevalence of comorbidities were lower than the T2D cohort: hypertension (44%), coronary heart disease (8%), and heart failure (3%). Patient characteristics including events between the training and test cohorts were comparable (Supplemental Table 2).
Composite Kidney End Point
For participants with T2D, 201 of the 871 (23%) experienced the composite kidney end point over a median follow-up of 4.6 (IQR, 3.4–5.6) years. In participants with the APOL1-HR genotypes, 90 of the 498 (18%) experienced the composite kidney end point over a median follow-up of 5.9 (IQR, 3.9–7.1) years.
Machine Learning (Random Forest) Model for Prediction of the Composite Kidney End Point
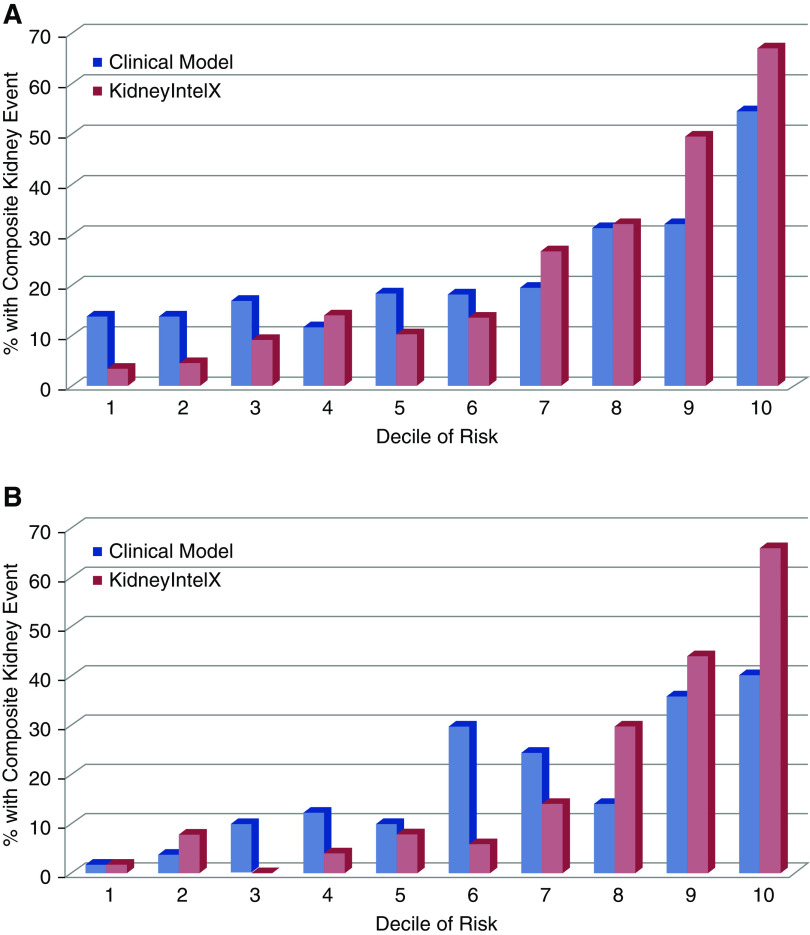
The observed composite kidney event by deciles of risk with KidneyIntelX versus the standard clinical model (9) are shown in Figure 1. For patients with T2D, applying 10-fold cross-validation, the KidneyIntelX AUC in the training set (80%, n=697) for the composite kidney end point was 0.81 (95% CI, 0.80 to 0.82) and 0.77 (95% CI, 0.75 to 0.79) in the test set (20%, n=174). By comparison, the clinical model (9) had an AUC of 0.66 (95% CI, 0.65 to 0.67) in the entire T2D cohort (n=871).
Figure 1.
Stratification by KidneyIntelX predicted risk classified more patients correctly for the composite kidney end point than stratification with predicted risk derived from the clinical model in both T2D and APOL1-HR population. (A) T2D cohort and (B) APOL1 –HR cohort. In those with T2D, a higher proportion of patients in the top 2 deciles of KidneyIntelX predicted risk experienced the composite kidney end point than those in the top 2 deciles of the clinical model and a lower proportion of patients with T2D classified in the bottom 3 deciles of risk experienced the composite kidney end point than those in the bottom 3 deciles of the clinical model. In those with APOL1-HR genotypes, a higher proportion in the top 3 deciles of KidneyIntelX predicted risk experienced the composite kidney end point than those in the top 3 deciles of the clinical model and a lower proportion of those in the lower 7 deciles of KidneyIntelX experienced the composite kidney end point compared to the clinical model.
For the patients with APOL1-HR genotypes, applying 10-fold cross-validation, the AUC for KidneyIntelX in the training set (80%, n=398) was 0.86 (95% CI, 0.84 to 0.87) and 0.80 (95% CI, 0.77 to 0.83) in the test set (20%, n=99). The clinical model (9) had an AUC of 0.72 (95% CI, 0.71 to 0.73) in the APOL1-HR cohort (n=498).
In both the T2D and APOL1-HR cohorts, the features noted to contribute most to performance were the three plasma biomarkers (TNFR1, TNFR2, and KIM-1) or their ratios of individual biomarker values to each other (i.e., three ratios) and laboratory values or vital signs (either baseline or changes over time) that are linked to kidney disease (Supplemental Figure 1). The P values of the Hosmer–Lemeshow goodness-of-fit test for the prognostic models were 0.15 and 0.11, indicating there was no significant difference between the predicted and observed outcomes (Supplemental Figure 2).
KidneyIntelX Cutoffs for the Composite Kidney End Point (Entire T2D [n=871] and APOL1-HR [n=498] Cohorts)
The PPVs of KidneyIntelX were 58%, 62%, and 68% in the top 20%, 15%, and 10% highest risk of the T2D population versus 43%, 46%, and 54% in the top 20%, 15%, and 10% of highest risk as classified by the clinical model (P<0.01 for all comparisons; Table 2) (9). The PPVs of KidneyIntelX were 56%, 62%, and 66% in the top 20%, 15%, and 10% highest risk of APOL1-HR population versus PPV of 38%, 39%, and 40% of the highest risk as classified by the clinical model (P<0.01 for all comparisons) (9). When applying cutoffs for the lowest 50% of risk in the T2D cohort, the NPV for KidneyIntelX compared with the clinical model was 92% versus 85% (P=0.76). Similarly, for the APOL1-HR cohort, the NPV for KidneyIntelX compared with the clinical model was 96% versus 93% (P=0.93).
Table 2.
KidneyIntelX thresholds for the composite kidney end point with sensitivity, specificity, PPV and NPV for T2D and APOL1-HR populations in high- and low-risk strata
| Threshold | Risk Cutoff | Sensitivity | Specificity | PPV | NPV |
| T2D KidneyIntelX | |||||
| Bottom 50% | 0.192 | 0.82 | 0.59 | 0.38 | 0.92 |
| Top 20% | 0.444 | 0.50 | 0.89 | 0.58 | 0.86 |
| Top 15% | 0.555 | 0.40 | 0.93 | 0.62 | 0.84 |
| Top 10% | 0.707 | 0.29 | 0.96 | 0.68 | 0.82 |
| T2D clinical model | |||||
| Bottom 50% | 0.148 | 0.68 | 0.55 | 0.31 | 0.85 |
| Top 20% | 0.240 | 0.38 | 0.85 | 0.43 | 0.82 |
| Top 15% | 0.278 | 0.30 | 0.89 | 0.46 | 0.81 |
| Top 10% | 0.338 | 0.23 | 0.94 | 0.54 | 0.80 |
| APOL1-HR KidneyIntelX | |||||
| Bottom 50% | 0.209 | 0.88 | 0.58 | 0.32 | 0.96 |
| Top 20% | 0.438 | 0.60 | 0.89 | 0.56 | 0.91 |
| Top 15% | 0.489 | 0.52 | 0.93 | 0.62 | 0.90 |
| Top 10% | 0.546 | 0.36 | 0.96 | 0.66 | 0.87 |
| APOL1-HR clinical model | |||||
| Bottom 50% | 0.151 | 0.79 | 0.57 | 0.29 | 0.93 |
| Top 20% | 0.322 | 0.42 | 0.85 | 0.38 | 0.87 |
| Top 15% | 0.387 | 0.32 | 0.87 | 0.39 | 0.85 |
| Top 10% | 0.448 | 0.22 | 0.93 | 0.40 | 0.84 |
Risk Cutoff, predicted probability of the composite kidney end point; PPV, positive predictive value; NPV, negative predictive value; T2D, type 2 diabetes; APOL1-HR, high-risk APOL1.
Supplementary and Sensitivity Analyses
Prevalent CKD
When we stratified the performance of KidneyIntelX by baseline CKD (i.e., eGFR ≤60 ml/min per 1.73 m2 and/or UACR ≥30 mg/g at baseline, n=366), 27.6% experienced the primary composite kidney end point during follow-up, compared with 12.5% in those without baseline CKD (n=505). The AUC was 0.84 (95% CI, 0.81 to 0.87) in individuals with prevalent CKD versus 0.79 (95% CI, 0.75 to 0.83) in those without CKD (Table 3). For APOL1-HR individuals, 112 had baseline prevalent CKD, of which 31.2% experienced the composite kidney end point. In this subgroup, the KidneyIntelX model produced an AUC of 0.88 (95% CI, 0.84 to 0.92) versus the 386 without baseline CKD, the AUC was 0.79 (95% CI, 0.77 to 0.82; Table 3).
Table 3.
AUCs (95% CI) for KidneyIntelX versus clinical model for subgroups
| Subgroup | KidneyIntelX | Clinical Model |
| T2D | ||
| Prevalent CKD (n=366) | 0.84 (0.81 to 0.87) | 0.70 (0.69 to 0.71) |
| No CKD (n=505) | 0.79 (0.75 to 0.83) | 0.63 (0.61 to 0.64) |
| Contemporary data (n=871) | 0.78 (0.77 to 0.80) | 0.66 (0.65 to 0.67) |
| APOL1-HR | ||
| Prevalent CKD (n=112) | 0.88 (0.84 to 0.92) | 0.59 (0.56 to 0.61) |
| No CKD (n=386) | 0.79 (0.76 to 0.83) | 0.74 (0.73 to 0.75) |
| Contemporary data (n=498) | 0.79 (0.77 to 0.82) | 0.72 (0.71 to 0.73) |
AUC, area under the receiver operator characteristic; T2D, type 2 diabetes; APOL1-HR, high-risk APOL1.
Contemporary Data
Using contemporary data only (data within 1 year before enrollment and biomarker measurement), the discriminatory performance of the KidneyIntelX model in both the T2D (AUC, 0.78; 95% CI, 0.77 to 0.80) and APOL1-HR (AUC, 0.79; 95% CI, 0.77 to 0.82) cohorts were similar when all clinical data were available, demonstrating KidneyIntelX is not dependent on multiyear history to provide accurate prognostic information (Table 3).
Random Forest Model with and without Biomarkers
A newly created random forest model with different clinical features that did not include any of the biomarkers (TNFR1, TNFR2, or KIM-1) in both cohorts had lower training and test AUCs than the full KidneyIntelX model with plasma biomarkers and their ratios (Supplemental Table 3).
Discrimination for Individual Components of the Composite Kidney End Point
The discriminatory performance of KidneyIntelX (trained for the entire composite end point) for the individual components of the composite end point (RKFD alone, sustained 40% decline alone, or kidney failure alone) in the test cohorts for T2D and APOL1-HR did not vary substantially (Supplemental Table 4).
Time-to-Event Analyses for 40% Sustained Decline in eGFR or Kidney Failure
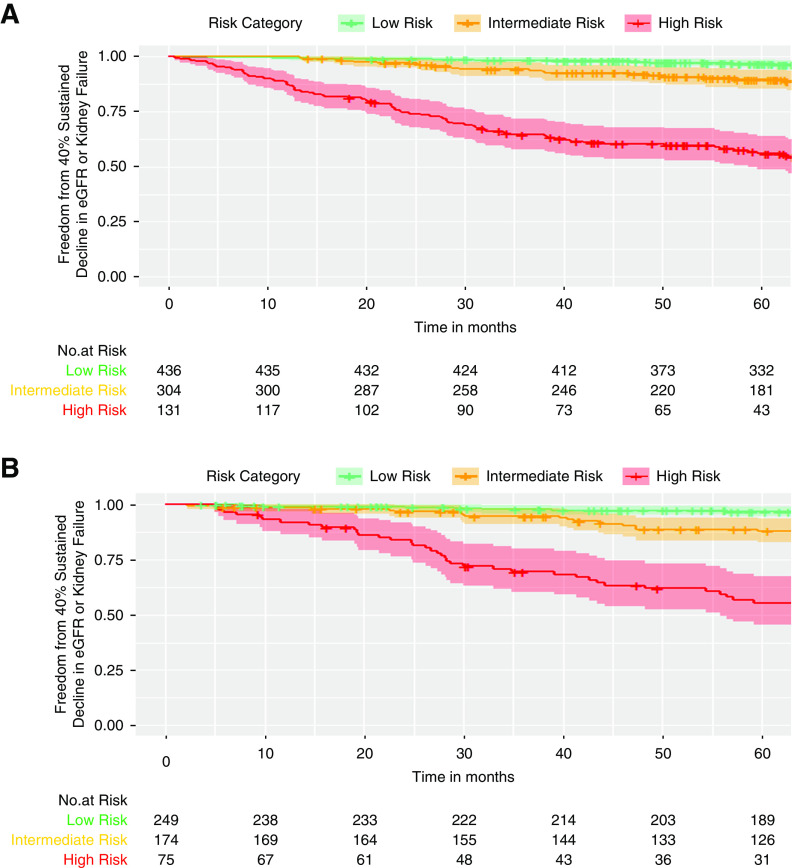
Patients with high-risk KidneyIntelX scores (top 15% in T2D and in APOL1-HR) had a greater risk of progression to time-to-event categoric outcomes of 40% sustained decline or kidney failure than patients in the low- or medium-risk strata combined (hazard ratios 9.9 [95% CI, 6.7 to 14.6] and 9.1 [95% CI, 5.8 to 14.3]), respectively. Separation of the high-risk stratum Kaplan–Meier curve occurred within the first year and progressively declined over time (Figure 2).
Figure 2.
Patients with T2D or APOL1-HRclassified as high-risk by KidneyIntelX, experienced faster progression to end point of sustained 40% decline in eGFR or kidney failure. (A) T2D cohort and (B) APOL1 –HR cohort. Separation of the high-risk stratum Kaplan–Meier curve occurred within the first year and progressively declined over time. The proportion of population in each risk stratum were 50% in low-risk, 35% in intermediate-risk, and 15% in high-risk in both T2D and APO1-HR. In the T2D cohort, the hazard ratio for high- versus low-risk strata was 16.8 (95% CI, 9.7 to 29.3) and was 9.9 (95% CI 6.7-14.6) for high- versus low- and intermediate-risk strata combined. In the APOL1-HR cohort, the hazard ratio for high- versus low-risk strata was 20.2 (95% CI, 9.8 to 41.2) and was 9.1 (95% CI, 5.8 to 14.3) for high-risk versus the low- and intermediate-risk strata combined.
Discussion
Using two large cohorts (T2D and APOL1-HR) of patients at high risk for progressive kidney function decline, with banked plasma samples linked to the corresponding EHR data, we developed a prognostic model combining EHR data and three previously validated plasma biomarkers to predict a composite kidney end point, which included RKFD, 40% sustained decline, or kidney failure. The KidneyIntelX prognostic model was more accurate for predicting the risk of kidney function decline than a validated clinical model in this study population (9). The ability to identify a distinct patient group with the composite kidney end point with a PPV of >55% allows for more appropriate future patient management including nephrologist referral, improved awareness of kidney health, and guidance toward more targeted, intensive therapies to slow progression. The demonstrated PPV in the high-risk stratum represents a 3-fold improvement over the observed baseline event rate in the two populations.
CKD is a complex, common problem challenging modern healthcare. In the absence of specific therapies, early identification of patients more likely to experience RKFD and adverse kidney outcomes is paramount. Early identification would help in the allocation of limited resources as well as implementation or intensification of proven interventions to slow kidney function decline. In real-world practice, the prediction of kidney disease progression in patients with T2D and/or APOL1-HR is challenging, particularly in patients with largely preserved kidney function. There are two major problems contributing to the difficulties in early identification and prediction: (1) serum creatinine/eGFR and UACR are relatively insensitive and nonspecific biomarkers, with significant fluctuations and variability in early stages of CKD; and (2) the prevalent standard includes recursive scores incorporating only a single (baseline) value of a selected predictive feature and does not include longitudinal data.
Recently, several biomarkers representing injury and inflammation have been the subject of an intense research focus. Among these, three biomarkers (soluble TNFR1, soluble TNFR2, and plasma KIM-1) have been extensively validated in multiple studies to support their translation to clinical use in a range of CKD settings, including patients with and without T2D, as well as those with the APOL1-HR genotypes (10,12–17,19,33,34). The individual biomarkers have added a significant improvement to clinical metrics, and the combination of all three, perhaps because of different pathophysiologic pathways, appears to be synergistic (12,33). We have demonstrated that combining these biomarkers with clinical information using machine learning techniques can significantly improve the discrimination/prediction of composite kidney end points.
Biomarkers that can be measured during a routine clinical encounter can be combined with longitudinal EHR data present in most healthcare systems for optimal prediction. We have previously shown that the addition of longitudinal data using supervised machine learning significantly outperforms “baseline” clinical models and also has utility for subtyping disease trajectories (35,36). Thus, we hypothesized that combining biomarker information and extant longitudinal EHR data would improve prediction of future kidney progression.
This integrated approach has near-term clinical implications, especially when linked to clinical decision support and embedded care pathways within the EHR. For example, patients with a high KidneyIntelX risk score, with a probability of >50% for the kidney end point, should be referred to a nephrologist, which has been associated with improved outcomes (37). In addition, referral of high-risk patients to a dietician and the delivery of educational materials regarding the importance and consequences of CKD should increase awareness and facilitate motivation for changes in lifestyles and behavior. Finally, the optimization of medical therapy including renin-angiotensin-aldosterone system inhibitors, statins for cardiovascular risk management, and intensification of antihypertensive medication to meet guideline-recommended BP targets can be pursued. The application of sodium glucose transporter-2 inhibitors might also be advantageous in the high KidneyIntelX score group with T2D given recent data on robust renoprotection (38–40).
Alternatively, patients with a low-risk score could be clinically managed by their primary care provider and have a standard-of-care treatment with scheduled monitoring of their KidneyIntelX results. Finally, patients with an intermediate-risk score would be recommended for the standard of care and retesting longitudinally. Such patients may demonstrate changes in KidneyIntelX based on behavioral changes, clinical parameters, and treatment adjustments over time, with appropriate clinical actions as necessary. This overall approach would not only benefit individual patient outcomes but also positively affect health systems where there is uncertainty about which patients to refer to a limited number of subspecialists.
Our study is not without limitations. Although we used a multiethnic data set, further validation in geographically diverse populations is necessary. Secondly, data structures/relationships change over time due to adjusted practice patterns and thus the algorithm may not perform similarly if the full complement of longitudinal data is not available or if the clinical practice is altered. We conducted a sensitivity analysis with only 1 year of data available before baseline, and the loss of performance was minimal; however, this should be evaluated further in additional validation studies. We have imputed the missing values for some features using different imputation methods which can have an effect on the correlations between features in the random forest model. Because the training and testing were performed within a single cohort, the potential for overfitting exists. However, we plan to expand our analysis to independent cohorts for external validation before prospective testing. Finally, this analysis does not address implementation or utility. Therefore, a prospective clinical utility trial to assess the decision effect of the KidneyIntelX risk score when provided to primary care physicians and patients is underway.
In conclusion, we have demonstrated that machine learning techniques (random forest models) combining longitudinal EHR information with three plasma biomarkers improved prediction of RKFD or kidney failure over validated clinical models in two distinct clinical settings and populations. With the advent of advanced high-performance computing, validated biomarkers, and an integrated EHR, the ability to improve outcomes with the integration of the KidneyIntelX into routine patient care should be assessed.
Disclosures
J. Bonventre is a coinventor on KIM-1 patents assigned to Partners Healthcare. He is a consultant to Aldeyra, Angion, Cadent, Praxis, and Seattle Genetics, and owns equity in DxNow, Goldfinch, Innoviva, MediBeacon, Sensor Kinesis, Sentien, and Verinano. J. Bonventre is also a nonpaid advisory board member for Renalytix. S. Coca has received consulting fees from Bayer, Boehringer-Ingelheim, CHF Solutions, Relypsa, and Takeda Pharmaceuticals in the past 3 years. S. Coca, C. He, B. Murphy, G. Nadkarni, and J. Quackenbush receive financial compensation as consultants and advisory board members for RenalytixAI, Inc., and own equity in Renalytix. S. Coca and G. Nadkarni are scientific cofounders of RenalytixAI. M. Donovan, F. Fleming, J. McCullough, and B. Murphy are officers of RenalytixAI. G. Nadkarni has received consulting fees from BioVie Inc and GLG consulting and has received operational funding from Goldfinch Bio in the past 3 years. All remaining authors have nothing to disclose.
Funding
This research was supported, in part, by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grant R01DK096549 (to S. Coca). G. Nadkarni is supported by a National Institutes of Health (NIH) career development award K23DK107908 and is also supported by NIH grants R01DK108803, U01HG007278, U01HG009610, and 1U01DK116100. S. Coca and J. Bonventre are members of and are supported in part by the Chronic Kidney Disease Biomarker Consortium, grant U01DK106962. S. Coca is also supported by NIH grants R01DK115562, R01HL85757, R01DK112258, U01OH011326, and R01DK126477. J. Bonventre is also supported by NIH grants R37 DK39773 and RO1 DK072381.
Supplementary Material
Author Contributions
J. Bonventre, K. Chauhan, S. Coca, F. Fleming, C. He, B. Murphy, G. Nadkarni, and J. Quackenbush reviewed and edited the manuscript; K. Chauhan, S. Coca, M. Donovan, F. Fleming, and G. Nadkarni were responsible for formal analysis and wrote the original draft; K. Chauhan and G. Nadkarni were responsible for methodology; S. Coca, M. Donovan, and F. Fleming were responsible for funding acquisition; S. Coca, M. Donovan, F. Fleming, J. McCullough, and G. Nadkarni provided supervision; S. Coca, F. Fleming, J. McCullough, and G. Nadkarni conceptualized the study; K. Chauhan, S. Coca, and G. Nadkarni had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis; and G. Nadkarni was responsible for investigation.
Footnotes
This article contains supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0002252020/-/DCSupplemental.
References
- 1.Klag MJ, Whelton PK, Randall BL, Neaton JD, Brancati FL, Stamler J: End-stage renal disease in African-American and white men. 16-year MRFIT findings. JAMA 277: 1293–1298, 1997 [PubMed] [Google Scholar]
- 2.Choi AI, Rodriguez RA, Bacchetti P, Bertenthal D, Hernandez GT, O’Hare AM: White/black racial differences in risk of end-stage renal disease and death. Am J Med 122: 672–678, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, Feldman HI, Parekh RS, Kusek JW, Greene TH, Fink JC, Anderson AH, Choi MJ, Wright JT Jr., Lash JP, Freedman BI, Ojo A, Winkler CA, Raj DS, Kopp JB, He J, Jensvold NG, Tao K, Lipkowitz MS, Appel LJ; AASK Study Investigators; CRIC Study Investigators : APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 369: 2183–2196, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayek SS, Koh KH, Grams ME, Wei C, Ko YA, Li J, Samelko B, Lee H, Dande RR, Lee HW, Hahm E, Peev V, Tracy M, Tardi NJ, Gupta V, Altintas MM, Garborcauskas G, Stojanovic N, Winkler CA, Lipkowitz MS, Tin A, Inker LA, Levey AS, Zeier M, Freedman BI, Kopp JB, Skorecki K, Coresh J, Quyyumi AA, Sever S, Reiser J: A tripartite complex of suPAR, APOL1 risk variants and αvβ3 integrin on podocytes mediates chronic kidney disease. Nat Med 23: 945–953, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grams ME, Rebholz CM, Chen Y, Rawlings AM, Estrella MM, Selvin E, Appel LJ, Tin A, Coresh J: Race, APOL1 risk, and eGFR decline in the general population. J Am Soc Nephrol 27: 2842–2850, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunkler D, Gao P, Lee SF, Heinze G, Clase CM, Tobe S, Teo KK, Gerstein H, Mann JF, Oberbauer R; ONTARGET and ORIGIN Investigators : Risk prediction for early CKD in type 2 diabetes. Clin J Am Soc Nephrol 10: 1371–1379, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jardine MJ, Hata J, Woodward M, Perkovic V, Ninomiya T, Arima H, Zoungas S, Cass A, Patel A, Marre M, Mancia G, Mogensen CE, Poulter N, Chalmers J; ADVANCE Collaborative Group : Prediction of kidney-related outcomes in patients with type 2 diabetes. Am J Kidney Dis 60: 770–778, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Tangri N, Stevens LA, Griffith J, Tighiouart H, Djurdjev O, Naimark D, Levin A, Levey AS: A predictive model for progression of chronic kidney disease to kidney failure. JAMA 305: 1553–1559, 2011 [DOI] [PubMed] [Google Scholar]
- 9.Nelson RG, Grams ME, Ballew SH, Sang Y, Azizi F, Chadban SJ, Chaker L, Dunning SC, Fox C, Hirakawa Y, Iseki K, Ix J, Jafar TH, Köttgen A, Naimark DMJ, Ohkubo T, Prescott GJ, Rebholz CM, Sabanayagam C, Sairenchi T, Schöttker B, Shibagaki Y, Tonelli M, Zhang L, Gansevoort RT, Matsushita K, Woodward M, Coresh J, Shalev V; CKD Prognosis Consortium : Development of risk prediction equations for incident chronic kidney disease. JAMA 322: 2104–2114, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tummalapalli L, Nadkarni GN, Coca SG: Biomarkers for predicting outcomes in chronic kidney disease. Curr Opin Nephrol Hypertens 25: 480–486, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlsson AC, Nordquist L, Larsson TE, Carrero JJ, Larsson A, Lind L, Ärnlöv J: Soluble tumor necrosis factor receptor 1 is associated with glomerular filtration rate progression and incidence of chronic kidney disease in two community-based cohorts of elderly individuals. Cardiorenal Med 5: 278–288, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coca SG, Nadkarni GN, Huang Y, Moledina DG, Rao V, Zhang J, Ferket B, Crowley ST, Fried LF, Parikh CR: Plasma biomarkers and kidney function decline in early and established diabetic kidney disease. J Am Soc Nephrol 28: 2786–2793, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gohda T, Niewczas MA, Ficociello LH, Walker WH, Skupien J, Rosetti F, Cullere X, Johnson AC, Crabtree G, Smiles AM, Mayadas TN, Warram JH, Krolewski AS: Circulating TNF receptors 1 and 2 predict stage 3 CKD in type 1 diabetes. J Am Soc Nephrol 23: 516–524, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niewczas MA, Gohda T, Skupien J, Smiles AM, Walker WH, Rosetti F, Cullere X, Eckfeldt JH, Doria A, Mayadas TN, Warram JH, Krolewski AS: Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J Am Soc Nephrol 23: 507–515, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krolewski AS, Niewczas MA, Skupien J, Gohda T, Smiles A, Eckfeldt JH, Doria A, Warram JH: Early progressive renal decline precedes the onset of microalbuminuria and its progression to macroalbuminuria. Diabetes Care 37: 226–234, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nowak N, Skupien J, Niewczas MA, Yamanouchi M, Major M, Croall S, Smiles A, Warram JH, Bonventre JV, Krolewski AS: Increased plasma kidney injury molecule-1 suggests early progressive renal decline in non-proteinuric patients with type 1 diabetes. Kidney Int 89: 459–467, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavkov ME, Nelson RG, Knowler WC, Cheng Y, Krolewski AS, Niewczas MA: Elevation of circulating TNF receptors 1 and 2 increases the risk of end-stage renal disease in American Indians with type 2 diabetes. Kidney Int 87: 812–819, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nowak N, Skupien J, Smiles AM, Yamanouchi M, Niewczas MA, Galecki AT, Duffin KL, Breyer MD, Pullen N, Bonventre JV, Krolewski AS: Markers of early progressive renal decline in type 2 diabetes suggest different implications for etiological studies and prognostic tests development. Kidney Int 93: 1198–1206, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabbisetti VS, Waikar SS, Antoine DJ, Smiles A, Wang C, Ravisankar A, Ito K, Sharma S, Ramadesikan S, Lee M, Briskin R, De Jager PL, Ngo TT, Radlinski M, Dear JW, Park KB, Betensky R, Krolewski AS, Bonventre JV: Blood kidney injury molecule-1 is a biomarker of acute and chronic kidney injury and predicts progression to ESRD in type I diabetes. J Am Soc Nephrol 25: 2177–2186, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grams ME, Sang Y, Ballew SH, Matsushita K, Astor BC, Carrero JJ, Chang AR, Inker LA, Kenealy T, Kovesdy CP, Lee BJ, Levin A, Naimark D, Pena MJ, Schold JD, Shalev V, Wetzels JFM, Woodward M, Gansevoort RT, Levey AS, Coresh J: Evaluating glomerular filtration rate slope as a surrogate end point for ESKD in clinical trials: An individual participant meta-analysis of observational data. J Am Soc Nephrol 30: 1746–1755, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inker LA, Heerspink HJL, Tighiouart H, Levey AS, Coresh J, Gansevoort RT, Simon AL, Ying J, Beck GJ, Wanner C, Floege J, Li PK, Perkovic V, Vonesh EF, Greene T: GFR slope as a surrogate end point for kidney disease progression in clinical trials: A meta-analysis of treatment effects of randomized controlled trials. J Am Soc Nephrol 30: 1735–1745, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kidney Disease: Improving Global Outcomes (KDIGO) CKD Work Group : Clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int Suppl 3: 1–163, 2012 [Google Scholar]
- 23.Krolewski AS, Skupien J, Rossing P, Warram JH: Fast renal decline to end-stage renal disease: An unrecognized feature of nephropathy in diabetes. Kidney Int 91: 1300–1311, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shlipak MG, Katz R, Kestenbaum B, Siscovick D, Fried L, Newman A, Rifkin D, Sarnak MJ: Rapid decline of kidney function increases cardiovascular risk in the elderly. J Am Soc Nephrol 20: 2625–2630, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young BA, Katz R, Boulware LE, Kestenbaum B, de Boer IH, Wang W, Fülöp T, Bansal N, Robinson-Cohen C, Griswold M, Powe NR, Himmelfarb J, Correa A: Risk factors for rapid kidney function decline among African Americans: The Jackson heart study (JHS). Am J Kidney Dis 68: 229–239, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirahatake KM, Jacobs DR, Gross MD, Bibbins-Domingo KB, Shlipak MG, Mattix-Kramer H, Odegaard AO: The association of serum carotenoids, tocopherols, and ascorbic acid with rapid kidney function decline: The coronary artery risk development in young adults (CARDIA) study. J Ren Nutr 29: 65–73, 2019 [DOI] [PubMed] [Google Scholar]
- 27.Peters KE, Davis WA, Ito J, Winfield K, Stoll T, Bringans SD, Lipscombe RJ, Davis TME: Identification of novel circulating biomarkers predicting rapid decline in renal function in type 2 diabetes: The Fremantle Diabetes Study Phase II. Diabetes Care 40: 1548–1555, 2017 [DOI] [PubMed] [Google Scholar]
- 28.Inker L, Levey A, Coresh J, Greene T, Heerspink HL: How can we better define outcomes in progression of CKD? Presented at the Kidney Disease Improving Global Outcomes Controversies Conference on Challenges in the Conduct of Clinical Trials in Nephrology, Paris, France, September 8–11, 2016. Available at: https://kdigo.org/wp-content/uploads/2017/02/Inker-_CKD-outcomes_final.pdf. Accessed March 10, 2019
- 29.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Li R, Masys DR, Ritchie MD, Roden DM, Struewing JP, Wolf WA; eMERGE Team : The eMERGE network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 4: 13, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, Denny JC, Peissig PL, Miller AW, Wei WQ, Bielinski SJ, Chute CG, Leibson CL, Jarvik GP, Crosslin DR, Carlson CS, Newton KM, Wolf WA, Chisholm RL, Lowe WL: Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc 19: 212–218, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nadkarni GN, Gottesman O, Linneman JG, Chase H, Berg RL, Farouk S, Nadukuru R, Lotay V, Ellis S, Hripcsak G, Peissig P, Weng C, Bottinger EP: Development and validation of an electronic phenotyping algorithm for chronic kidney disease. AMIA Annu Symp Proc 2014: 907–916, 2014 [PMC free article] [PubMed] [Google Scholar]
- 32.De Silva AP, Moreno-Betancur M, De Livera AM, Lee KJ, Simpson JA: A comparison of multiple imputation methods for handling missing values in longitudinal data in the presence of a time-varying covariate with a non-linear association with time: A simulation study. BMC Med Res Methodol 17: 114, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadkarni GN, Chauhan K, Verghese DA, Parikh CR, Do R, Horowitz CR, Bottinger EP, Coca SG: Plasma biomarkers are associated with renal outcomes in individuals with APOL1 risk variants. Kidney Int 93: 1409–1416, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhatraju PK, Zelnick LR, Shlipak M, Katz R, Kestenbaum B: Association of soluble TNFR-1 concentrations with long-term decline in kidney function: The multi-ethnic study of atherosclerosis. J Am Soc Nephrol 29: 2713–2721, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huopaniemi I, Nadkarni G, Nadukuru R, Lotay V, Ellis S, Gottesman O, Bottinger EP: Disease progression subtype discovery from longitudinal EMR data with a majority of missing values and unknown initial time points. AMIA Annu Symp Proc 2014: 709–718, 2014 [PMC free article] [PubMed] [Google Scholar]
- 36.Singh A, Nadkarni G, Gottesman O, Ellis SB, Bottinger EP, Guttag JV: Incorporating temporal EHR data in predictive models for risk stratification of renal function deterioration. J Biomed Inform 53: 220–228, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P, Quinn RR, Karim ME, Bello A, Tam-Tham H, Weaver R, Ronksley PE, Quan H, Strippoli GFM, Manns B, Hemmelgarn BR, Tonelli M, Ravani P: Nephrology consultation and mortality in people with stage 4 chronic kidney disease: A population-based study. CMAJ 191: E274–E282, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neal B, Perkovic V, Mahaffey KW, de Zeeuw D, Fulcher G, Erondu N, Shaw W, Law G, Desai M, Matthews DR; CANVAS Program Collaborative Group : Canagliflozin and cardiovascular and renal events in type 2 diabetes. N Engl J Med 377: 644–657, 2017 [DOI] [PubMed] [Google Scholar]
- 39.Wanner C, Inzucchi SE, Lachin JM, Fitchett D, von Eynatten M, Mattheus M, Johansen OE, Woerle HJ, Broedl UC, Zinman B; EMPA-REG OUTCOME Investigators : Empagliflozin and progression of kidney disease in type 2 diabetes. N Engl J Med 375: 323–334, 2016 [DOI] [PubMed] [Google Scholar]
- 40.Toyama T, Neuen BL, Jun M, Ohkuma T, Neal B, Jardine MJ, Heerspink HL, Wong MG, Ninomiya T, Wada T, Perkovic V: Effect of SGLT2 inhibitors on cardiovascular, renal and safety outcomes in patients with type 2 diabetes mellitus and chronic kidney disease: A systematic review and meta-analysis. Diabetes Obes Metab 21: 1237–1250, 2019 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.