Abstract
We conducted a prospective evaluation of the diagnostic performance of high-resolution microendoscopy (HRME) to detect cervical intraepithelial neoplasia (CIN) in women with abnormal screening tests. Study participants underwent colposcopy, HRME, and cervical biopsy. The prospective diagnostic performance of HRME using an automated morphologic image analysis algorithm was compared to colposcopy using histopathologic detection of CIN as the gold standard. To assess the potential to further improve performance of HRME image analysis, we also conducted a retrospective analysis assessing performance of a multi-task convolutional neural network to segment and classify HRME images. 1,486 subjects completed the study; 435 (29%) subjects had CIN grade 2 or more severe (CIN2+) diagnosis. HRME with morphologic image analysis for detection of CIN grade 3 or more severe diagnoses (CIN3+) was similarly sensitive (95.6% vs. 96.2%, p=0.81) and specific (56.6% vs. 58.7%, p=0.18) as colposcopy. HRME with morphologic image analysis for detection of CIN2+ was slightly less sensitive (91.7% vs. 95.6%, p<0.01) and specific (59.7% vs. 63.4%, p=0.02) than colposcopy. Images from 870 subjects were used to train a multi-task convolutional neural network-based algorithm and images from the remaining 616 were used to validate its performance. There were no significant differences in the sensitivity and specificity of HRME with neural network analysis vs. colposcopy for detection of CIN2+ or CIN3+. Using a neural network-based algorithm, HRME has comparable sensitivity and specificity to colposcopy for detection of CIN2+. HRME could provide a low-cost, point-of-care alternative to colposcopy and biopsy in the prevention of cervical cancer.
Keywords: cervical cancer prevention, diagnostic imaging, high-resolution microendoscopy, point-of-care, deep learning
Introduction
Cervical cancer remains a major global health problem; women living in low-income countries and rural areas are disproportionately affected.1 In Brazil, there are an estimated 16,590 new cases of cervical cancer per year, with disproportionately higher incidence rates in underserved populations.2 Barretos Cancer Hospital (BCH) in São Paulo state has implemented innovative mobile screening programs to reach women living in rural areas3,4, but diagnostic follow-up of women who screen positive remains challenging. The standard of care for screen-positive women in Brazil and many other countries is to undergo colposcopy. The provider performs a tissue biopsy of any visually abnormal area and submits the sample for examination by a pathologist. Yet, the availability of colposcopy and pathology services are often limited in low-resource settings, resulting in long delays between visits, high cost, and loss to follow-up.5 For example, over one-third of women with abnormal screening tests did not return for their follow-up visit when referred for colposcopy in a previous study at BCH.6
In 2018, the World Health Organization (WHO) called for the elimination of cervical cancer as a public health problem. The WHO proposed screening 70% of women at ages 35 and 45 using a high-precision test, such as HPV DNA testing.5 HPV screening tests identify women at risk of cervical cancer with high sensitivity but low specificity.7 Given the limitations of performing colposcopy and histopathology in low-resource settings, effective strategies are urgently needed to triage women with abnormal cervical screening tests and ensure they receive appropriate management.8,9
In vivo microscopy has the potential to aid clinical decision-making by providing high resolution images of biological tissues in real time, obviating the need for colposcopy and cervical biopsy.10 We developed a low-cost, high-resolution microendoscope (HRME) capable of imaging cervical epithelium with sub-cellular resolution for real-time, in vivo diagnosis.11 Previous smaller clinical studies have shown HRME to be feasible for real-time detection of high-grade cervical abnormalities.6,12-14 HRME images can be interpreted rapidly and objectively using a morphologic image analysis algorithm.6,15 Here we report results of the CLARA (Cervical Lesion Assessment with Real-time microendoscopy image Analysis) study. The primary objective of this study was to prospectively evaluate diagnostic performance of HRME with morphologic image analysis in women with abnormal cervical cancer screening tests and compare accuracy to that of colposcopy. The secondary objective was to use the acquired dataset to explore whether the diagnostic accuracy of HRME could be further improved using a multi-task convolutional neural network trained to segment and classify HRME images.
Materials and Methods
Study design and participants
We conducted an unblinded, single arm trial to prospectively assess the diagnostic performance of HRME with morphologic image analysis to detect high-grade cervical abnormalities in women with abnormal screening tests. The primary endpoint was a comparison of the diagnostic performance of HRME with morphologic image analysis and colposcopy for detection of high-grade cervical abnormalities. Sample size calculations were performed to determine the number of subjects needed to estimate 95% confidence intervals of both sensitivity and specificity within a 4% margin of error. Based on a 25% prevalence of high-grade cervical abnormalities, a 12.5% drop-out rate, and the diagnostic accuracy of HRME reported in a prior study6, the necessary sample size was calculated to be 1,600 subjects.
Potential subjects were identified through a regional screening program operated by BCH. Per BCH standard-of-care practices, women who had abnormal cervical cytology (atypical squamous cells of undetermined significance [ASC-US] or more severe abnormality [ASC-US+]) or tested positive for hrHPV by the cobas 4800 HPV test were referred for colposcopy. Women scheduled for colposcopy at BCH were assessed for eligibility using the following inclusion criteria: 1) abnormal cervical screening test, 2) at least 18 years old, 3) intact uterine cervix, 4) not pregnant (negative urine pregnancy test required for subjects with child-bearing potential), 5) no known allergy to the fluorescent dye used for HRME imaging (proflavine or acriflavine), 6) does not belong to an indigenous Brazilian population, and 7) willing and able to provide written informed consent.
This study was approved by the BCH Ethics Research Committee, the Brazilian National Ethics Research Commission / CONEP (CAAE: 61743416.1.0000.5437), and the Institutional Review Boards of Rice University (ID#2017–293) and The University of Texas MD Anderson Cancer Center (ID#2017–0096). Written informed consent was obtained from all participants. The protocol was registered at ClinicalTrials.gov (NCT03195218).
High-resolution microendoscope
The HRME instrument is a portable, fiber-optic fluorescence microscope designed to obtain high-resolution images of cervical tissue in vivo with sub-cellular resolution.11,15,16 The system has been described in detail previously.15 Images are obtained by placing the fiber optic probe (Fujikura FIGH–30–850N , Myriad Fiber Imaging Tech. Inc., NJ, USA) in gentle contact with the uterine cervix; the field-of-view is 790 microns and the lateral spatial resolution is approximately 4 microns. The current cost of goods for the HRME is US$1500/unit with an additional US$745 per optical probe.
HRME is performed following topical application of proflavine (0.01% concentration), a fluorescent antiseptic that labels cell nuclei in the superficial epithelium. Proflavine has a long history of safe clinical use.17,18 A retrospective case-control analysis of screen positive women previously exposed to proflavine during HRME found no significant differences in disease progression between the proflavine exposed and control groups.19
The HRME morphologic image analysis software allows clinicians to acquire and automatically analyze images to assess the likelihood that a cervical site has high-grade abnormalities. The diagnostic algorithm evaluated prospectively in this study was trained using data collected in a prior study at BCH.6 The algorithm uses morphological analysis to segment and categorize individual cell nuclei as normal or abnormal based on predefined shape and size criteria. The software reports a morphologic abnormality score for each image analyzed, which is calculated as the number of abnormal nuclei per square millimeter in the image. Samples with a morphologic abnormality score below 120 were classified as non-neoplastic while those equal to or greater than 120 were classified as neoplastic.
Diagnostic exam procedure
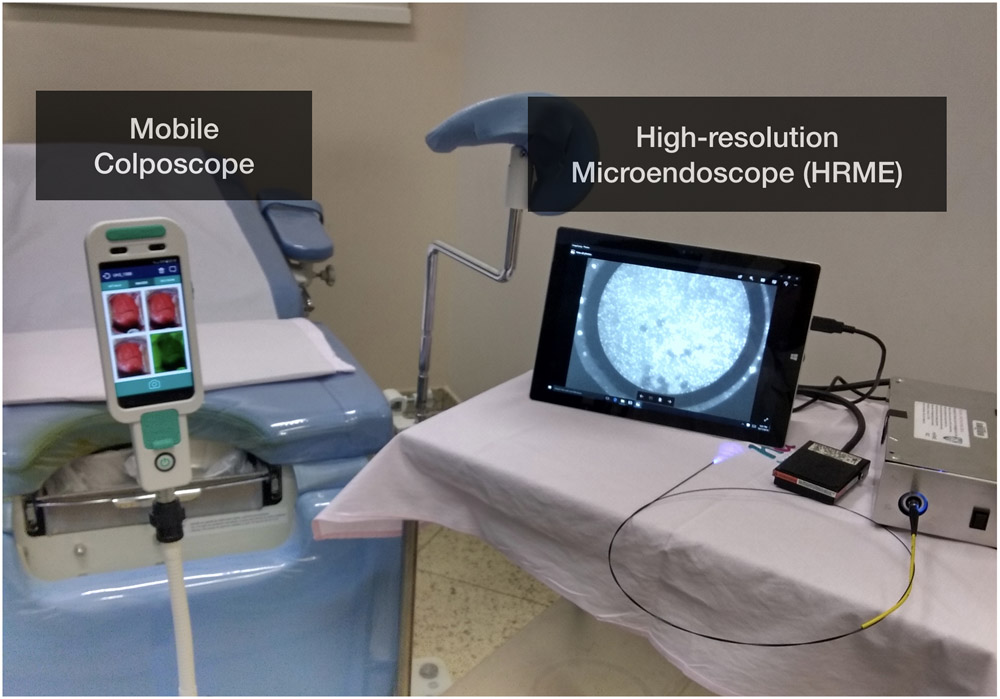
All diagnostic exams and study procedures were performed by one of three study colposcopists in the Cancer Prevention department at BCH (Fig. 1). All three had prior experience using HRME for diagnostic assessment of the uterine cervix. Clinicians first performed general visual inspection of the cervix and collected cervical swabs for cytology and HPV DNA testing. Then, colposcopy was performed after application of 5% acetic acid followed by Lugol’s iodine. Colposcopy was performed using either a traditional stationary colposcope (CP-M1255 colposcope, D.F. Vasconcellos, Brazil) or a mobile colposcope (EVA 3 Plus, MobileODT, Israel). The clock position and colposcopic impression of any abnormal areas (low-grade, high-grade, or suspected cancer) were indicated on the case report form.
Figure 1:
Photograph of high-resolution microendoscope (HRME) and colposcope in the clinic.
Following colposcopy, proflavine was applied to the uterine cervix and HRME imaging was performed. HRME images were acquired from areas noted as abnormal by colposcopy and from each quadrant with no lesions. In each quadrant without a lesion, HRME images were acquired from a randomly selected colposcopically normal site at the squamocolumnar junction. For each site where an HRME image was acquired, the following information was recorded: site number, clock position, HRME result (morphologic abnormality score and classification), colposcopist impression of tissue type (squamous, columnar, or metaplasia), and colposcopy impression (normal, low-grade, high-grade, or suspected cancer).
Sites identified as abnormal by colposcopy and/or by HRME were biopsied. In cases where a large lesion spanning multiple quadrants was present, the colposcopist biopsied the most severe area within the lesion based on colposcopic impression. If no abnormal sites were identified by either method, then a single biopsy was taken from a clinically normal site imaged by the clinician. Endocervical curettage (ECC) was performed if indicated per local standard of care.
Histopathology & follow-up
Histopathology was used as the gold standard to assess the diagnostic performance of colposcopy and HRME for detection of high-grade cervical abnormalities. Two experienced pathologists, blinded to the results of HRME, independently reviewed the histologic slides from each biopsy categorizing the diagnosis for the specimen into one of twelve categories using criteria defined by the WHO20 . These include the following: atrophy, inflammation, metaplasia, cervical intraepithelial neoplasia (CIN) 1, CIN 2, CIN 3, adenocarcinoma in situ (AIS), squamous cell carcinoma, adenocarcinoma, adenosquamous carcinoma, indeterminate (insufficient for diagnosis), or ‘other’ (in which case the pathologist specified a classification category not included in this list). Discrepant results were resolved by consensus review between the two pathologists. Based on final histopathology results, participants were treated or scheduled for follow-up per BCH standard of care practices. This included excision with loop electrosurgical excision or cold knife conization for women with CIN2/3 and referral to gynecologic oncology for those diagnosed with AIS or invasive cancer.
Prospective analysis of diagnostic accuracy of HRME with morphologic image analysis
Study data were collected on a paper case report form and subsequently entered into an electronic study database using REDCap (Research Electronic Data Capture).21 De-identified colposcopy and HRME images and de-identified lab report documents for all other diagnostic test results including cytology, HPV testing, and histopathology were uploaded to the database. Both CIN 2 or more severe diagnoses (CIN 2+) and CIN 3 or more severe diagnoses (CIN 3+) were used as endpoints to evaluate the diagnostic performance of colposcopy and HRME. HRME was recorded as a binary result (positive or negative) and colposcopy positivity was defined as low-grade or more severe impression.
Study data including age, colposcopy, HRME morphologic image analysis, and pathology results were exported from REDCap into Mathematica 12.0 for cross-tabulation of contingency tables and calculation of descriptive statistics. Contingency tables were entered into GraphPad Prism 8.3 for statistical analyses. Sensitivity and specificity with 95% confidence intervals (95% CI) were calculated on a per-site basis and a per-patient basis. In the per-patient analysis, the most abnormal colposcopy result, the most abnormal HRME result (maximum morphologic abnormality score for all images), and the most abnormal histopathology result were used. 95% CI of sensitivity and specificity were generated by a modified Wald method.22 Significance testing comparing the sensitivity and specificity of colposcopy vs. HRME with morphologic image analysis was performed using McNemar’s test.23 A non-parametric test for trend was used to assess differences in sensitivity and specificity by age group.24 HRME results were also stratified by tissue type (squamous vs. columnar/metaplasia) using colposcopic impression of tissue type. One-way analysis of variance (ANOVA) on ranks was performed with Dunn’s multiple comparison tests to evaluate differences in morphologic abnormality score distributions by histopathology diagnosis and by tissue type impression. In addition to evaluating diagnostic performance using the prospective binary HRME threshold, receiver operator characteristic (ROC) analysis was performed using the morphologic abnormality score to assess the trade-off in sensitivity and specificity of this metric at all possible thresholds for positivity. Area under the ROC curve was calculated using the trapezoidal rule. Significance testing for differences in area under the curve for squamous vs columnar/metaplasia was performed using DeLong’s test.25
Retrospective performance analysis using a multi-task convolutional neural network
To explore whether deep learning methods could improve the diagnostic accuracy of HRME, a subset of HRME images collected in this study were used to train a multi-task convolutional neural network (multi-task CNN) to classify HRME images. Using study data withheld from training, the diagnostic accuracy of HRME using a multi-task CNN was compared to that of colposcopy and to HRME using morphologic image analysis.
Study data were randomly divided by patient into training (~60%), validation (~20%), and test sets (~20%), ensuring that each partition had an equal distribution of pathology outcomes. The dataset partitions were as follows: 870 patients in the training set (516 [59%] negative, 98 [11%] CIN 1, 55 [6%] CIN 2, 189 [22%] CIN 3, and 12 [1%] invasive carcinoma), 302 patients in the validation set (181 [60%] negative, 33 [11%] CIN 1, 20 [7%] CIN 2, 64 [21%] CIN 3, and 4 [1%] invasive carcinoma), and 314 patients in the test set (185 [59%] negative, 38 [12%] CIN 1, 19 [6%] CIN 2, 66 [21%] CIN 3, and 6 [2%] invasive carcinoma).
The multi-task CNN model utilized the architecture proposed by Mehta et al. to perform joint segmentation and classification of histopathology images.26 The multi-task CNN was trained in two-stages: (i) optimization of nuclear segmentation on a pixel-by-pixel basis and (ii) optimization of image classification accuracy (<CIN 2 vs CIN 2+). Prior to input into the CNN model, the circular region of HRME images corresponding to the fiber bundle was cropped to yield four non-overlapping, square sub-images. Nuclear segmentation masks generated using morphologic image analysis were used as a weakly supervised ground truth for pixel-level segmentation. Once segmentation performance was optimized, the diagnostic classification branch was appended to the architecture and the network was trained to perform simultaneous image segmentation and classification. Diagnostic performance for images in the validation set was monitored during training to avoid overfitting; the model with the best performance for images in the validation set was selected and used to prospectively analyze images in the test set.
The final model was used to assess per-site and per-patient diagnostic performance in the validation and test sets by comparing results to histologic diagnosis. A multi-task CNN score was calculated for each site image by averaging the network output (probability of CIN 2+) across the four sub-images classified by the network. The per-patient CNN score was defined as the maximum multi-task CNN score for all images. One-way ANOVA on ranks was again utilized to evaluate differences in multi-task CNN score distributions by histopathology diagnosis. A threshold for positivity by the multi-task CNN was determined as the point on the ROC curve which minimized the Euclidean distance with respect to the operating point of colposcopy (low-grade or more severe). The performance of the multi-task CNN at this cutoff was then compared to colposcopy for both CIN 2+ and CIN 3+ endpoints with significance testing again performed using McNemar’s test.23 Significance testing for differences in area under the curve for the multi-task CNN vs morphologic image analysis was performed using DeLong’s test.25
Results
Figure 2 shows the number of participants at each stage of the study. 2,028 screen positive women were interviewed to assess eligibility prior to their scheduled colposcopy. 1,821 eligible women were invited to participate in the study. Two hundred twenty-one subjects declined to participate, and the remaining 1,600 provided written informed consent. 1,523 subjects completed the diagnostic examination. An additional 37 participants were excluded from analysis for reasons outlined in Figure 2, leaving 1,486 participants with complete diagnostic information.
Figure 2:
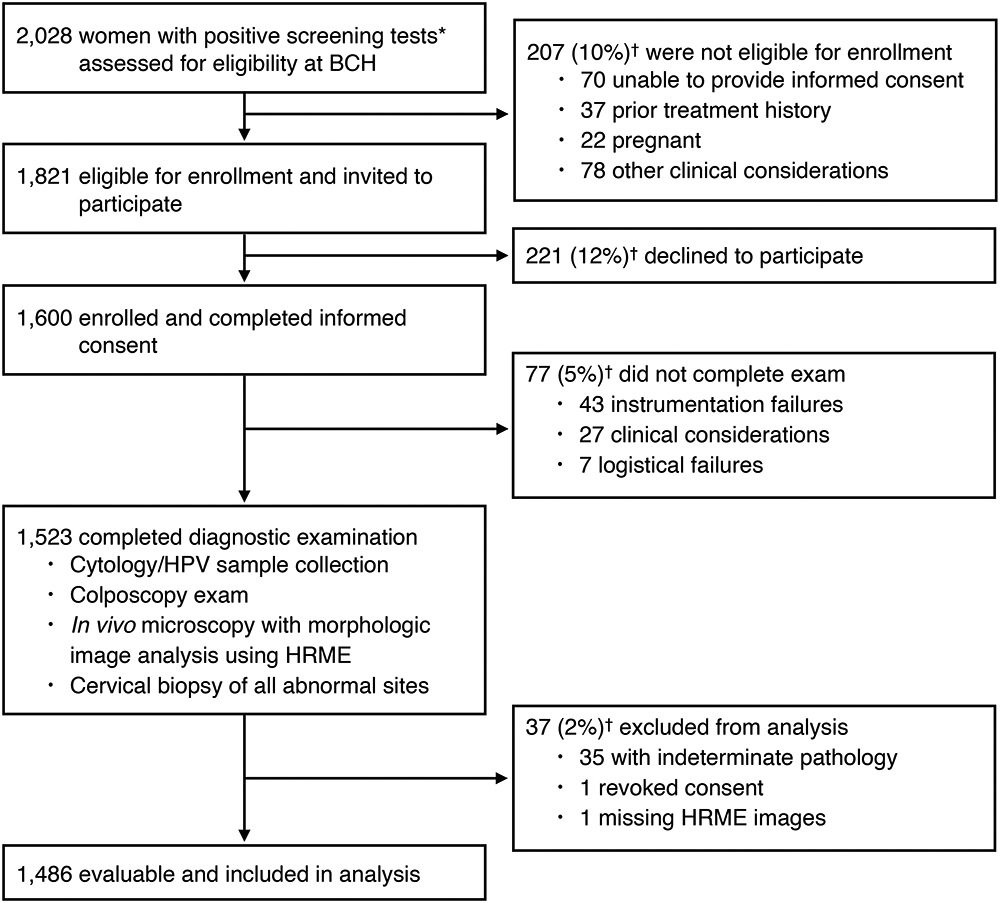
Flow diagram of subjects assessed for study eligibility and participants enrolled in and completing the study.
* Positive cytology test results included the following: ASC-US, ASC-H, LSIL, HSIL, and AGC, positive HPV test results included: HPV16 positive, HPV18 positive, pooled hrHPV positive (31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68)
† Exclusions are reported as n (%), with percentages based on the total from the previous box.
Table 1 summarizes the age, cytology and HPV results, and final histopathology diagnosis for the 1,486 subjects included in the analysis. The mean age of participants was 40.0 years old (SD=12.1), with 796 (54%) between the ages of 30 to 49. The distribution of patient pathology diagnoses was as follows: 882 (59%) negative, 169 (11%) CIN 1, 94 (6%) CIN 2, 319 (21%) CIN 3, and 22 (1%) cervical cancer. The prevalence of high-grade cervical abnormalities (CIN 2+) was 29%.
Table 1:
Age, cytology/HPV testing results, and final pathology diagnosis for 1,486 subjects included in analysis.
| No. patients | 1,486 |
| Age (years) | |
| 18-29 | 323 (22%) |
| 30-39 | 434 (29%) |
| 40-49 | 362 (24%) |
| 50-59 | 289 (19%) |
| 60 and older | 78 (5%) |
| Mean | 40.0 |
| Cytology | |
| Normal | 609 (41%) |
| ASC-US | 188 (13%) |
| ASC-H | 264 (18%) |
| LSIL | 79 (5%) |
| HSIL | 244 (16%) |
| AGC | 59 (4%) |
| AIS | 4 (<1%) |
| Carcinoma | 12 (1%) |
| Unsatisfactory/Not collected | 27 (2%) |
| HPV | |
| hrHPV Negative | 561 (38%) |
| hrHPV Positive | 910 (61%) |
| HPV16/18 Positive | 389 (26%) |
| Other hrHPV* Positive | 521 (35%) |
| Invalid/Not collected | 15 (1%) |
| Final pathology diagnosis | |
| Negative | 882 (59%) |
| CIN 1 | 169 (11%) |
| CIN 2 | 94 (6%) |
| CIN 3 | 319 (21%) |
| Invasive carcinoma | 22 (1%) |
Data are n (%) with percentages based on total number of patients.
Abbreviations: ASC-US (atypical squamous cells of undetermined significance), ASC-H (atypical squamous cells, cannot exclude HSIL), LSIL (low grade squamous intraepithelial lesion), HSIL (high grade squamous intraepithelial lesion), AGC (atypical glandular cells), AIS (adenocarcinoma in-situ), hrHPV (high-risk Human Papillomavirus), CIN (cervical intraepithelial neoplasia).
Pooled Other hrHPV positive for COBAS HPV test includes HPV31/33/35/39/45/51/52/56/58/59/66/68. If patients were positive for HPV16 and/or HPV18 in addition to Other hrHPV, they were categorized as HPV16/18 positive.
A total of 45 imaging probes were used throughout the study, with an average of 33 uses per probe (min-max: 2-95 uses).
Prospective analysis of diagnostic accuracy of HRME with morphologic image analysis
Figure 3 summarizes the per-patient diagnostic performance of HRME with morphologic image analysis and that of colposcopy for 1,486 patients included in the analysis. Figure 3A shows the maximum morphologic abnormality score for each patient stratified by histopathology result. Figures 3B and 3C show ROC curves and resulting AUC of HRME with morphologic image analysis for detection of CIN 2+ (AUC=0.83; 95% CI: 0.81 to 0.85) and CIN 3+ (AUC=0.84; 95% CI: 0.82 to 0.86) using all possible thresholds for positivity. Figure 3D shows a contingency table comparing diagnostic results for colposcopy (low-grade or more severe) and HRME with morphologic image analysis by histopathology result. Percent agreement of colposcopy and HRME with morphologic image analysis was 79%. Figures 3E and 3F compare the sensitivity and specificity of colposcopy and HRME with morphologic image analysis for CIN 2+ and CIN 3+ cut-points. Using a cut-point of CIN 3+, there were no statistically significant differences in the sensitivity and specificity of colposcopy and HRME with morphologic image analysis. However, using a cut-point of CIN 2+, the sensitivity and specificity of colposcopy were higher than that of HRME with morphologic image analysis (sensitivity: 95.6% vs 91.7%, p<0.01; specificity: 63.4% vs 59.7%, p=0.02). The difference between colposcopy and HRME on CIN 2 biopsies was further investigated by examining additional indicators of high-grade disease for concordant (Colpo+/HRME+) and discordant (Colpo+/HRME−) cases (Supplementary Table 1). Discordant cases were less likely to have high-grade cytology (Fisher’s exact test: OR=2.7, p=0.13), high-grade colposcopy impression (OR=11, p<0.001), as well as CIN 3+ upon histopathology review of tissue excised during treatment (OR=4.8, p=0.05).
Figure 3:
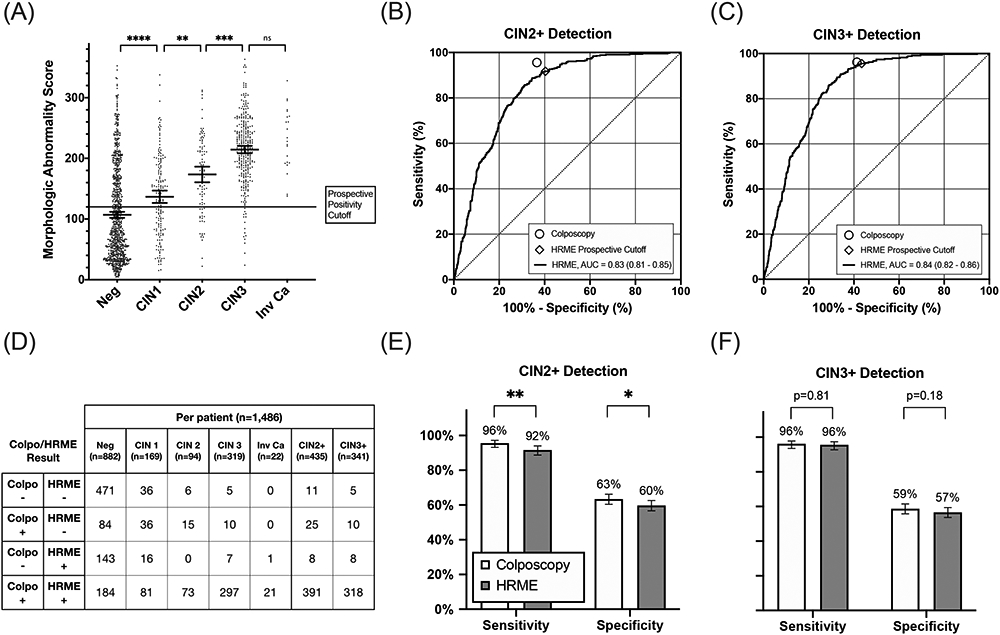
Diagnostic performance of colposcopy and HRME with morphologic image analysis using histopathology as the gold standard. (A) Per-patient HRME morphologic abnormality scores stratified by histopathology result. The solid line represents the prospective cutoff for positivity by morphologic image analysis. Bars overlaid on scatter plots indicate mean and 95% confidence intervals within each category. (B-C) Receiver operator characteristic curves of HRME with morphologic image analysis for detection of CIN2+ and CIN3+. Prospective performance of colposcopy (low-grade or more severe) and HRME with morphologic image analysis using the prospective positivity threshold are plotted on ROC curves for reference. (D) Cross tabulation of outcomes for colposcopy (low-grade or more severe) and HRME with morphologic image analysis by pathology result for each patient. (E-F) Comparisons of the sensitivity and specificity of colposcopy and HRME with morphologic image analysis for detection of CIN2+ and CIN3+. Error bars represent 95% confidence intervals. The following symbols indicate p-value results as follows: ns (P>0.05), *(P≤0.05), **(P≤0.01), ***(P≤0.001), ****(P≤0.0001).
Given the WHO recommendation of prioritizing screening for women ages 30 to 49 year, per-patient diagnostic performance of HRME with morphologic image analysis was examined for the subset of participants in this recommended age bracket. Similar results were obtained (Supplementary Figure 1). Analysis of diagnostic performance for both older and younger age groups was also performed, including a non-parametric test for trend (Supplementary Figure 2). Sensitivity of colposcopy decreased with increasing age (p=0.02) whereas the performance of HRME was less age dependent (p=0.76). Additionally, the specificity of each diagnostic increased with older ages, but there was a greater improvement for colposcopy than HRME (colposcopy: p<0.001, HRME: p=0.42).
As some participants underwent multiple biopsies, a per-site analysis in which diagnostic performance was evaluated for each individual biopsy result was also conducted (Supplementary Figure 3). The distribution of colposcopy, HRME, and histopathology results for 1,901 biopsied sites are provided (Supplementary Table 2). On a per-site basis, HRME with morphologic image analysis had comparable sensitivity and specificity to colposcopy for detection of CIN3+, but had lower sensitivity and specificity for detection of CIN2+ (sensitivity: 93.2% vs 89.0%, p<0.01; specificity: 58.3% vs 54.2%, p=0.01) (Supplementary Figure 4).
Diagnostic performance of HRME with morphologic image analysis was stratified by colposcopic impression of tissue type. The distribution of colposcopic tissue type for the 1,901 biopsied sites are provided (Supplementary Table 2). As shown in Supplementary Figure 5A, for sites with negative or CIN 1 biopsy results, the mean morphologic abnormality score is significantly lower for sites with a colposcopic impression of squamous tissue than for sites with a colposcopic impression of columnar tissue or metaplasia. As a result, the accuracy of HRME with morphologic image analysis is significantly higher for sites with a colposcopic impression of squamous tissue than for sites with a colposcopic impression of columnar tissue or metaplasia (CIN 2+: AUC=0.80 vs. 0.62, p<0.001; CIN 3+: AUC=0.81 vs 0.67, p<0.001).
Retrospective performance analysis using a multi-task convolutional neural network
Figure 4 summarizes the per-patient diagnostic performance of HRME with multi-task CNN analysis and that of colposcopy for the 616 patients in the validation and test sets. Figure 4A shows the maximum multi-task CNN score for each patient stratified by histopathology result. Figures 4B and 4C show ROC curves and resulting AUC of HRME with multi-task CNN analysis for detection of CIN 2+ (AUC=0.86; 95%CI: 0.83 to 0.89) and CIN3+ (AUC=0.85; 95%CI: 0.82 to 0.88). Figure 4D shows a contingency table comparing diagnostic results for colposcopy (low-grade or more severe) and HRME with multi-task CNN analysis by histopathology result. Percent agreement of colposcopy and HRME with multi-task CNN analysis was 79%. Figures 4E and 4F compare the sensitivity and specificity of colposcopy and HRME with multi-task CNN analysis for CIN 2+ and CIN 3+ cut-points. At both CIN 2+ and CIN 3+ cut-points, there were no statistically significant differences in the sensitivity and specificity of colposcopy and HRME with multi-task CNN analysis (CIN 2+: 96.1% vs. 92.7% sensitivity, p=0.11; 61.3% vs. 59.3% specificity, p=0.46; CIN 3+: 96.4% vs. 95.7% sensitivity, p=1.00; 56.7% vs. 55.9% specificity, p=0.79).
Figure 4: Diagnostic performance of HRME using a multi-task CNN.
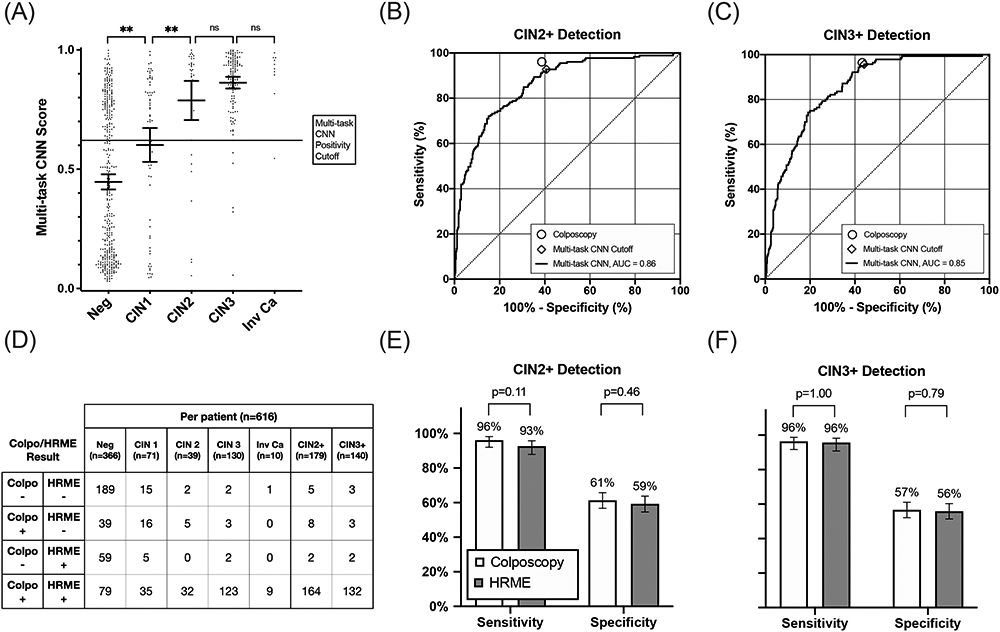
(A) Per-patient HRME multi-task CNN scores stratified by histopathology result. The solid line represents the cutoff for positivity by multi-task CNN for comparison with colposcopy (low-grade or more severe). Bars overlaid on scatter plots indicate mean and 95% confidence intervals within each column. (B-C) Receiver operator characteristic curves of HRME with multi-task CNN analysis for detection of CIN2+ and CIN3+. Performance of colposcopy (low-grade or more severe) are HRME with multi-task CNN analysis using the post-hoc positivity threshold are plotted on ROC curves for reference. (D) Cross tabulation of outcomes for colposcopy (low-grade or more severe) and multi-task CNN HRME classification by pathology result for each patient. (E-F) Comparisons of the sensitivity and specificity of colposcopy and HRME with multi-task CNN analysis for detection of both CIN2+ and CIN3+. Error bars represent 95% confidence intervals. The following symbols indicate p-value results as follows: ns (P>0.05), *(P≤0.05), **(P≤0.01).
Figure 5 compares diagnostic performance of HRME with morphologic image analysis to that of HRME with multi-task CNN analysis for data in the validation and test sets. Overall, the AUC for detecting CIN 2+ was significantly higher when images were analyzed using a multi-task CNN than with morphologic analysis (0.83 vs. 0.76, p<0.001). Diagnostic performance using the multi-task CNN was improved for all tissue types, but especially for sites with a colposcopic impression of columnar tissue or metaplasia. For images with a colposcopic impression of squamous tissue, the multi-task CNN improved AUC from 0.78 to 0.83 (p=0.003); whereas for images with a colposcopic impression of columnar tissue or metaplasia, AUC was improved from 0.64 to 0.78 (p<0.001).
Figure 5: Diagnostic performance of HRME using morphologic image analysis and the multi-task CNN.
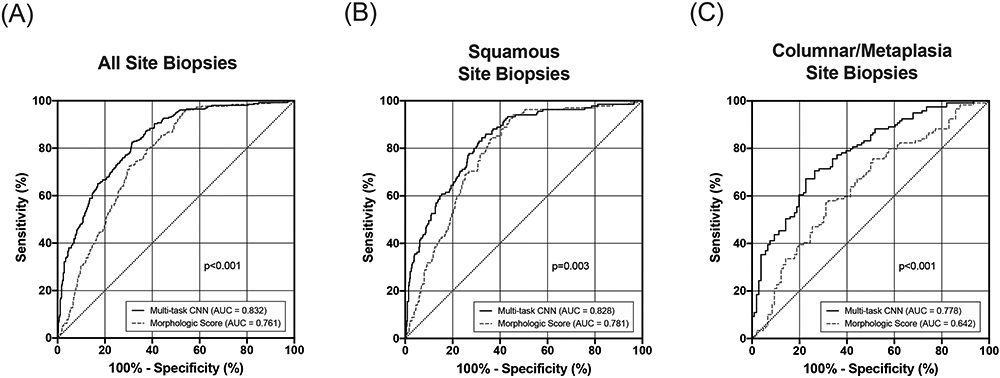
ROC analysis comparing CIN2+ detection using morphologic image analysis with the multi-task CNN. (A) ROC curves for all 789 images in the validation/test sets. (B) ROC curves for 559 images with a colposcopic impression of squamous tissue. (C) ROC curves for 225 images with a colposcopic impression of columnar tissue or metaplasia.
Figure 6 shows three example HRME images from different tissue types. Figure 6A shows an image of colposcopically normal squamous epithelium; the image shows small, round, uniform nuclei throughout the field of view as is characteristic for normal, squamous cervical tissue.20 Using both real-time morphologic image analysis and multi-task CNN this site was classified as negative, in agreement with the histologic diagnosis of benign. Figure 6B shows an HRME image of colposcopically abnormal squamous epithelium; the image shows enlarged, pleomorphic nuclei throughout the field-of-view. Using both real time morphologic image analysis and the multi-task CNN, this site was classified as positive, consistent with the histopathologic diagnosis of CIN3. Figure 6C shows an HRME image of colposcopically abnormal metaplastic epithelium; the image shows moderate nuclear enlargement, with nuclei arranged in glandular patterns. Histologic assessment revealed low-grade dysplasia (CIN1) with columnar tissue present. This site was incorrectly classified as positive using morphologic image analysis but was correctly classified as negative by multi-task CNN analysis.
Figure 6: Example HRME images.
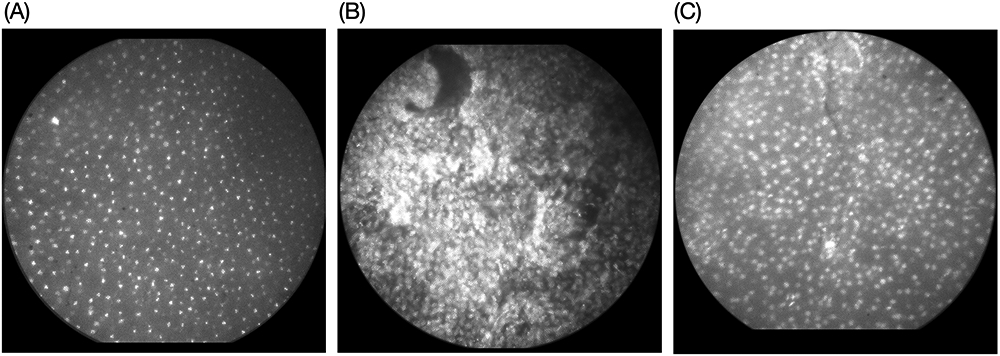
(A) Colposcopically normal, squamous epithelium with small, round, uniform nuclei. The image was classified as negative by real-time morphologic image analysis as well as histopathology. (B) Colposcopically abnormal squamous epithelium exhibiting enlarged, crowded, pleomorphic nuclei. The image was classified as positive by morphologic image analysis and the multi-task CNN; the histopathology result was high-grade dysplasia (CIN3). (C) Colposcopically abnormal, metaplastic epithelium with moderate nuclei crowding. The histopathology result was low-grade dysplasia (CIN1) with columnar tissue present. This image was incorrectly classified as positive by the prospective morphologic image analysis but correctly classified as negative by multi-task CNN analysis.
Discussion
In this large, prospective analysis of HRME with morphologic image analysis, we observed that colposcopy outperformed HRME by a small but statistically significant difference for detection of CIN 2+, whereas no statistical differences were observed for detection of CIN 3+. However, additional analysis the CIN 2 cases concordant and discordant colposcopy and HRME outcomes suggests that most of the few additional pick-ups by colposcopy alone represented low-grade cervical abnormalities which were less likely to cause cervical cancer (Supplementary Table 1). HRME performance also appeared to be less affected by age when compared with colposcopy (Supplementary Figure 2). Notably, HRME was more specific than colposcopy in the youngest age group (52% vs 38%, McNemar’s test: p<0.001), whereas colposcopy was more specific in the oldest age group (81% vs 59%, McNemar’s test: p<0.001). Inferences from these data are that: (1) benign HPV infections are more likely to be detected by colposcopy than HRME, and (2) atrophy of the cervix, which is increasingly likely with older age, decreases the sensitivity of colposcopy and the specificity of HRME for CIN2+.
The retrospective analysis portion of this study is the first large scale evaluation of deep learning for HRME image analysis, and represents a promising area for further improving HRME performance. The multi-task CNN analysis increased the AUC of HRME, particularly for sites with a colposcopic impression of columnar/metaplasia. Although both morphologic analysis and multi-task CNN analysis approaches perform nuclear segmentation, the multi-task CNN model parameters are simultaneously optimized to perform segmentation and classification. We hypothesize that this joint optimization results in a more robust feature representation which can better account for columnar/metaplasia tissue morphologies. As machine learning is a very dynamic field of research, future developments could supersede this multi-task CNN and further improve automated image analysis with HRME. In addition to improved image analysis, a low-cost confocal HRME has recently been demonstrated to improve the image contrast of nuclear morphometry in columnar cervical tissue.27 Coupling multi-task CNN analysis with confocal imaging has potential to further diagnostic accuracy of HRME with automated image analysis.
A limitation of this study was that the field of view of the HRME probe is smaller than the punch biopsy specimens analyzed by histopathology. Therefore, even when the biopsy is acquired at the precise location where the optical probe was placed, the tissue examined by the pathologist may include areas outside the field of view of the associated HRME image. This imposes limitations on our ability to perfectly correlate HRME findings to pathology diagnosis. Additionally, not all sites imaged by HRME were biopsied. Sites lacking a gold-standard diagnosis were excluded from analysis (Supplementary Figure 2).
Given its cost and required operator skill, current HRME instrumentation is suited for use in several low- and middle-income countries, but is likely not yet appropriate for very low-resource areas. In this study, colposcopic guidance was used to direct probe placement and to facilitate a direct comparison between colposcopy and HRME. Additionally, the colposcopy device utilized was not standardized for all patients (approximately 25% of patients underwent colposcopy with the MobileODT system). These are both limitations of this study. The usability of the HRME system by non-specialists and with mobile colposcopy has been demonstrated in other low-resource settings including El Salvador (general practitioner doctor) and the Rio Grande Valley along the Texas-Mexico border (nurse practitioner and physician assistant).14,28 However, in order for HRME to be effectively utilized in very low-resource settings, improved methods to guide probe placement may be needed. Recent advances in visual evaluation using deep learning could potentially provide automated wide-field assessment of the uterine cervix and highlight suspected lesions for subsequent high-resolution imaging.29,30 Ongoing studies to evaluate HRME with automated visual assessment are under way. Additional studies to further assess the safety of proflavine use for HRME imaging as well as explore alternative fluorescent dyes will be useful moving forward.
HRME optical instrumentation functioned well throughout the course of the study with periodic maintenance. Optical probes were replaced if they became scratched or chipped, which likely reduced the average number of uses per probe. Recycling worn probes on site by polishing the tip is one strategy which could be utilized to further extend probe lifetimes. Image quality was regularly monitored using an image quality control (QC) check built into the HRME software based on the signal to background of morphologic image analysis. When low-quality images were detected by the automated QC, users were prompted to take another image. Eighty six percent of all study images acquired passed the automated QC check, and 98% of all sites imaged had at least one image that passed QC. When reduced image quality was observed, it was either due to degradation of the optical probes (related to repeated exposure to disinfection detergents) and/or degradation of an optical filtering component (related to sustained humidity exposure).
In order to eliminate cervical cancer as a public health problem, effective strategies for management of screen-positive women are urgently needed. The clinical and infrastructural resource requirements of colposcopy and biopsy remain prohibitive in low- and middle-income countries.31,32 In this study, automated, in vivo assessment of cervical tissues using HRME was demonstrated to have equivalent sensitivity and specificity as expert colposcopy for detection of high-grade cervical abnormalities. The potential to further optimize real-time image analysis approaches for HRME using deep learning was also demonstrated. HRME may be a viable alternative to colposcopy and biopsy for low-resource healthcare settings, providing a point-of-care diagnosis and allowing for immediate treatment of pre-cancerous cervical lesions.
Supplementary Material
Novelty and Impact:
High-resolution microendoscopy (HRME) is a promising, non-invasive diagnostic imaging method with potential for more rapid and objective triage of women with abnormal cervical cancer screening tests. However, large-scale prospective evaluations of HRME with automated image classifiers have yet to be reported. Our study prospectively evaluated the diagnostic performance of HRME at a Brazilian cancer hospital. Additionally, we explore the potential for deep learning image analysis to further improve HRME diagnostic performance using study data.
Acknowledgements
The authors thank all the women who volunteered to participate in the study as well as the following individuals for their contributions to the study: Karen Cristina Borba Souza, M.S., Viviane Andrade, M.S., Elisa Alves Messias Silva, M.S., Livia Loami Ruyz Jorge de Paula, Ph.D., and Naitielle de Paula Pantano, M.S., and Gisele da Rocha Sant’ana (clinical team/data management); Dr. Ligia Kerr (study pathologist); Fernanda de Paula Cury, M.S. (HPV testing); Katelin Cherry, M.S. and Jennifer Carns, Ph.D. (instrumentation); Mark F. Munsell, M.S. (database development and management); Jessica R. Gallegos, M.S. (protocol/ data management).
Funding
Research reported in this publication was supported by the NCI of the NIH under Award Numbers UH2/3 CA189910 and R01 CA251911. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funder of the study had no role in study design, data collection, data analysis, or writing of this manuscript. The corresponding author had full access to all of the data and the final responsibility to submit for publication. The opinions expressed by the authors are their own and this material should not be interpreted as representing the official viewpoint of the U.S. Department of Health and Human Services, the National Institutes of Health or the National Cancer Institute.
Abbreviations:
- AIS
adenocarcinoma in situ
- ANOVA
analysis of variance
- AUC
area under the curve
- BCH
Barretos Cancer Hospital
- CI
confidence interval
- CIN
cervical intraepithelial neoplasia
- CNN
convolutional neural network
- ECC
endocervical curettage
- HPV
human papillomavirus
- HRME
high-resolution microendoscopy
- QC
quality control
- ROC
receiver operator characteristic
- WHO
world health organization
Footnotes
Conflict of interest
R. Richards-Kortum is an inventor on patents owned by the University of Texas licensed to Remicalm LLC. P. E. Castle has received HPV tests and assays for research at a reduced or no cost from Roche, Becton Dickinson, Cepheid, and Arbor Vita Corporation. No potential conflicts of interest were disclosed by the other authors.
Ethics statement
This study was approved by the BCH Ethics Research Committee, the Brazilian National Ethics Research Commission / CONEP (CAAE: 61743416.1.0000.5437), and the Institutional Review Boards of Rice University (ID#2017–293) and The University of Texas MD Anderson Cancer Center (ID#2017–0096). Written informed consent was obtained from all participants. The protocol was registered at ClinicalTrials.gov (NCT03195218).
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request through a data-sharing agreement that provides for: 1) a commitment to securing the data only for research purposes and not to identify any individual participant; 2) a commitment to securing the data using appropriate computer technology; and 3) a commitment to destroying or returning the data after analyses are completed.
References
- 1.Arbyn M, Weiderpass E, Bruni L, et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Glob Health 2020; 8(2): e191–e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brasil. Estimativa 2020: Incidência de câncer no Brasil.
- 3.Fregnani JH, Scapulatempo C, Haikel RL Jr., et al. Could alarmingly high rates of negative diagnoses in remote rural areas be minimized with liquid-based cytology? Preliminary results from the RODEO Study Team. Acta Cytol 2013; 57(1): 69–74. [DOI] [PubMed] [Google Scholar]
- 4.Lorenzi AT, Fregnani JH, Possati-Resende JC, et al. Can the careHPV test performed in mobile units replace cytology for screening in rural and remote areas? Cancer Cytopathol 2016; 124(8): 581–8. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. Accelerating the elimination of cervical cancer as a global public health problem: World Health Organization. Regional Office for South-East Asia, 2019. [Google Scholar]
- 6.Hunt B, Fregnani JHT, Schwarz RA, et al. Diagnosing cervical neoplasia in rural Brazil using a mobile van equipped with in vivo microscopy: A cluster-randomized community trial. Cancer Prev Res (Phila) 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arbyn M, Ronco G, Anttila A, et al. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine 2012; 30 Suppl 5: F88–99. [DOI] [PubMed] [Google Scholar]
- 8.Denny L, De Sousa M, Kuhn L, Pollack A, Wright TC. Cervical cancer prevention—A paradigm shift? Gynecologic oncology 2005; 99(3): S12. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn L, Saidu R, Boa R, et al. Clinical evaluation of modifications to a human papillomavirus assay to optimise its utility for cervical cancer screening in low-resource settings: a diagnostic accuracy study. The Lancet Global Health 2020; 8(2): e296–e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Loewke NO, Mandella MJ, et al. Real-time pathology through in vivo microscopy. Stud Health Technol Inform 2013; 185: 235–64. [PubMed] [Google Scholar]
- 11.Muldoon TJ, Pierce MC, Nida DL, Williams MD, Gillenwater A, Richards-Kortum R. Subcellular-resolution molecular imaging within living tissue by fiber microendoscopy. Opt Express 2007; 15(25): 16413–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce MC, Guan Y, Quinn MK, et al. A pilot study of low-cost, high-resolution microendoscopy as a tool for identifying women with cervical precancer. Cancer Prev Res (Phila) 2012; 5(11): 1273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinn MK, Bubi TC, Pierce MC, Kayembe MK, Ramogola-Masire D, Richards-Kortum R. High-resolution microendoscopy for the detection of cervical neoplasia in low-resource settings. PLoS One 2012; 7(9): e44924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parra SG, Rodriguez AM, Cherry KD, et al. Low-cost, high-resolution imaging for detecting cervical precancer in medically-underserved areas of Texas. Gynecologic oncology 2019; 154(3): 558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quang T, Schwarz RA, Dawsey SM, et al. A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointest Endosc 2016; 84(5): 834–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierce M, Yu D, Richards-Kortum R. High-resolution fiber-optic microendoscopy for in situ cellular imaging. JoVE (Journal of Visualized Experiments) 2011; (47): e2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell G, Buttle G. Proflavine in closed wounds. The Lancet 1943; 242(6276): 749. [Google Scholar]
- 18.Rahman G, Adigun I, Yusuf I, Ofoegbu C. Wound dressing where there is limitation of choice. Nigerian Journal of Surgical Research 2006; 8(3-4): 151–4. [Google Scholar]
- 19.Pantano N, Hunt B, Schwarz RA, et al. Is Proflavine Exposure Associated with Disease Progression in Women with Cervical Dysplasia? A Brief Report. Photochemistry and Photobiology 2018; 94(6): 1308–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurman RJ, Carcangiu ML, Harrington CS, Young RH. WHO Classification of Tumours of Female Reproductive Organs.
- 21.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42(2): 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agresti A, Coull BA. Approximate is better than “exact” for interval estimation of binomial proportions. The American Statistician 1998; 52(2): 119–26. [Google Scholar]
- 23.Fay MP. Two-sided exact tests and matching confidence intervals for discrete data. R journal 2010; 2(1): 53–8. [Google Scholar]
- 24.Cuzick J A Wilcoxon-type test for trend. Stat Med 1985; 4(1): 87–90. [DOI] [PubMed] [Google Scholar]
- 25.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics 2011; 12(1): 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta S, Mercan E, Bartlett J, Weaver D, Elmore JG, Shapiro L. Y-Net: joint segmentation and classification for diagnosis of breast biopsy images. International Conference on Medical Image Computing and Computer-Assisted Intervention; 2018: Springer; 2018. p. 893–901. [Google Scholar]
- 27.Tang Y, Kortum A, Parra SG, et al. In vivo imaging of cervical precancer using a low-cost and easy-to-use confocal microendoscope. Biomed Opt Express 2020; 11(1): 269–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parra SG, López-Orellana LM, Molina Duque AR, et al. Cervical cancer prevention in El Salvador: A prospective evaluation of screening and triage strategies incorporating high-resolution microendoscopy to detect cervical precancer. International Journal of Cancer In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu L, Bell D, Antani S, et al. An observational study of deep learning and automated evaluation of cervical images for cancer screening. JNCI: Journal of the National Cancer Institute 2019; 111(9): 923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan C, Yao Y, Cheng B, et al. The application of deep learning based diagnostic system to cervical squamous intraepithelial lesions recognition in colposcopy images. Sci Rep 2020; 10(1): 11639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue P, Ng MTA, Qiao Y. The challenges of colposcopy for cervical cancer screening in LMICs and solutions by artificial intelligence. BMC Med 2020; 18(1): 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu ES, Jeronimo J, Feldman S. Barriers and challenges to treatment alternatives for early-stage cervical cancer in lower-resource settings. Journal of global oncology 2017; 3(5): 572–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request through a data-sharing agreement that provides for: 1) a commitment to securing the data only for research purposes and not to identify any individual participant; 2) a commitment to securing the data using appropriate computer technology; and 3) a commitment to destroying or returning the data after analyses are completed.