Abstract
Trichosporon asahii, which is distributed in the environment, is the major causative agent of the opportunistic infection trichosporonosis, and it also causes summer-type hypersensitivity pneumonitis (SHP). Random amplification of polymorphic DNA analysis was used to determine the intraspecies diversity of 39 T. asahii isolates from clinical specimens, SHP patients' houses, and environmental materials. The three primers used revealed 46 polymorphic bands. A phenogram was generated by the unweighted pair-group method with arithmetic mean. Clinical isolates formed a cluster, characterized by a 90% matching coefficient, but they did not cluster with strains isolated from SHP patients' houses or environmental sources. In addition, the biochemical characteristics of 86 strains from three sources were examined with 31 compounds using an ID32C kit, and a phenogram was constructed. The phenogram consisted of three major clusters. Cluster I included most of the clinical SHP isolates, and cluster II included most of the environmental isolates. Cluster III contained only one strain. A remarkable difference was found in the abilities of the strains belonging to clusters I and II to utilize six compounds. These results suggest that the genetic diversity and biochemical characteristics of T. asahii seem to be related to the source of the isolate. We also found a specific DNA fragment for the clinical isolates and strains isolated from SHP patients' houses.
The basidiomycetous anamorphic yeast Trichosporon asahii Akagi ex Sugita et al. is the major causative agent of fungemia due to Trichosporon species in immunocompromised patients (7, 8). This infection is associated with a high mortality rate and a poor prognosis (20). Neutropenia due to cytotoxic chemotherapy is the most common risk factor for deep-seated trichosporonosis, and neutropenic patients are more likely to have fungemia or disseminated infection than nonneutropenic patients. T. asahii also causes summer-type hypersensitivity pneumonitis (SHP) (1, 2). SHP follows the development of type III or type IV allergies by repeated inhalation of Trichosporon arthroconidia, which often contaminate home environments during the summer months. In western and southern Japan, the summer is hot, humid, and rainy. Such conditions favor the growth of Trichosporon species, and most patients initially experience symptoms during the summer. Although SHP is considered peculiar to Japan, a case was recently reported in a neighboring country (22). Trichosporon is widely distributed throughout the environment, especially in soil. We have previously reported that T. asahii is a common environmental pathogen (19).
Many investigations of the intraspecies diversity and epidemiology of Candida albicans and Cryptococcus neoformans using random amplified polymorphic DNA (RAPD) analysis, hybridization with specific probes such as Ca3, and multilocus enzyme electrophoresis (MLEE) have been reported (3–5, 12–15, 21). They include studies examining the origins of nosocomial infection (12), a generic comparison between bloodstream and nonbloodstream isolates (5), a study of the hospital specificity or regional specificity of the isolates (14), and comparison of the genotypes and fluconazole susceptibilities of the isolates (13, 21). In contrast, there are only a few reports for Trichosporon species. Isoenzyme profiles, restriction fragment length polymorphisms (RFLP) of ribosomal DNA (rDNA), and analysis of glucuronoxylomannan polysaccharide antigen revealed that blood, superficial, and environmental isolates of Trichosporon beigelii (synonymous with Trichosporon cutaneum) were distinct from each other (9, 10). However, T. beigelii is a taxonomically highly heterogeneous species; at present, this species has been reclassified into more than 10 species (6).
In this study, we examined the genetic diversity and biochemical characteristics of T. asahii strains isolated from various sources, including clinical specimens, SHP patients' houses, and environmental materials.
MATERIALS AND METHODS
T. asahii isolates.
Of the 86 isolates identified as T. asahii, 42 were derived from clinical specimens (ascites, blood, feces, lung, pleural fluid, skin, and urine). They were obtained from eight centers in Japan and the United States. Seventeen were from the homes of 17 SHP patients, and 27 were from soil (Table 1). They were identified by PCR with T. asahii-specific primers or by direct sequence analysis of internal transcribed spacer (ITS) regions of the rRNA gene (17, 18)
TABLE 1.
T. asahii isolates and their sources, API profiles, and fingerprinting genotypes
| Strain | Site | Sourcea | API profile | Fingerprinting genotypeb with primer:
|
||
|---|---|---|---|---|---|---|
| M13 | OPE1 | RC8 | ||||
| Clinical isolates | ||||||
| C.I.1 | Urine | JU | 7 7 5 7 6 7 5 3 3 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 1 1 0 0 1 0 | 1 0 0 0 0 0 0 0 1 0 1 0 0 0 |
| C.I.2 | Urine | JU | 7 7 5 7 6 6 5 2 2 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 0 0 0 0 1 0 | 1 0 0 0 1 0 0 0 1 0 1 0 1 0 |
| C.I.3 | Blood | TMBH | 7 7 5 7 6 7 1 3 2 7 | 0 1 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 1 1 1 0 0 1 0 | 1 1 0 0 1 0 0 0 1 0 1 0 1 0 |
| C.I.4 | Sputum | NU | 7 7 5 7 6 4 5 2 2 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 1 1 1 0 0 1 0 | 1 0 0 0 0 0 0 0 1 0 1 0 1 0 |
| C.I.5 | Pleural fluid | NU | 5 7 5 7 6 7 5 3 2 5 | 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 | 0 0 0 0 0 0 0 0 1 0 | 0 0 0 0 0 0 0 0 1 0 1 0 1 0 |
| C.I.6 | Urine | NU | 7 7 5 7 6 6 5 2 2 5 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 1 1 0 0 1 0 | 0 0 0 0 0 0 0 0 1 0 1 0 1 0 |
| C.I.7 | Sputum | OMU | 7 7 5 7 6 4 5 2 2 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 1 1 0 0 1 0 | 1 0 0 0 0 0 0 0 1 0 1 0 1 0 |
| C.I.8 | Urine | OMU | 7 7 5 7 6 4 5 2 2 7 | 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 0 1 0 0 1 0 | 0 0 0 0 0 0 0 0 1 0 1 0 1 0 |
| C.I.9 | Ascites | OMU | 7 7 5 7 6 4 5 3 2 5 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 1 1 0 1 1 0 | 0 0 0 0 1 0 0 0 1 0 1 0 1 0 |
| C.I.10 | Feces | OMU | 7 7 5 7 6 4 5 2 2 7 | 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 0 0 0 0 1 0 | 0 0 0 0 0 0 0 0 1 0 1 0 1 0 |
| C.I.11 | Lung | KU | 7 7 5 7 6 4 5 2 2 5 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 1 1 1 0 0 1 0 | 1 0 0 0 1 0 0 0 1 0 1 0 1 0 |
| C.I.12 | Urine | KULM | 7 7 5 7 6 4 5 2 2 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 1 1 1 0 0 1 0 | 1 0 0 0 1 0 0 0 1 0 1 0 1 0 |
| C.I.13 | Ascites | JU | 7 7 5 7 6 4 5 2 2 7 | 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 1 0 0 1 1 1 0 0 1 0 | 1 0 0 0 1 0 0 0 1 0 1 0 1 0 |
| C.I.14 | Feces | NYSDH 297-87c | 7 7 5 7 6 4 5 3 2 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 1 1 1 0 1 1 0 | 1 0 0 1 1 0 0 0 1 0 1 0 1 0 |
| C.I.15 | Blood | UMSMT2c | 7 7 5 7 7 6 5 2 3 5 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 1 1 1 0 0 1 0 | 1 0 0 0 0 0 0 0 1 0 1 0 1 0 |
| C.I.16 | Blood | JU | 7 7 5 7 7 6 5 3 3 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 0 1 0 0 1 0 | 0 0 0 0 0 0 0 0 1 0 1 0 1 0 |
| C.I.17 | Blood | JU | 7 7 5 7 6 4 5 2 2 5 | 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 0 0 0 0 1 0 | 0 0 0 0 0 0 0 0 1 0 1 0 0 0 |
| C.I.18 | Blood | JU | 7 7 5 7 6 6 5 2 2 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 0 1 0 0 1 0 | 0 0 0 0 0 0 0 0 1 0 1 0 0 0 |
| C.I.19 | Urine | OMU | 7 7 5 7 6 4 5 2 2 5 | |||
| C.I.20 | Feces | OMU | 7 7 5 6 6 4 5 3 2 5 | |||
| C.I.21 | Blood | OMU | 5 7 5 7 6 5 5 3 2 5 | |||
| C.I.22 | Urine | OMU | 7 7 5 7 6 4 5 2 2 5 | |||
| C.I.23 | Urine | OMU | 7 7 5 7 6 4 5 2 2 5 | |||
| C.I.24 | Urine | NU | 7 7 5 7 6 6 5 2 2 5 | |||
| C.I.25 | Skin | NU | 7 7 5 7 6 7 5 3 2 5 | |||
| C.I.26 | Feces | NU | 7 7 5 7 7 7 5 2 3 5 | |||
| C.I.27 | Skin | NU | 7 7 5 7 6 7 5 2 2 5 | |||
| C.I.28 | Blood | NU | 3 7 1 3 6 0 5 2 2 7 | |||
| C.I.29 | Blood | NU | 7 7 5 7 2 4 5 2 2 5 | |||
| C.I.30 | Blood | NU | 7 7 5 7 6 6 5 2 2 5 | |||
| C.I.31 | Pleural fluid | NU | 7 7 5 7 6 6 5 3 2 5 | |||
| C.I.32 | Urine | NU | 7 7 5 7 6 4 1 3 2 5 | |||
| C.I.33 | Urine | NU | 7 7 1 7 6 4 5 2 2 5 | |||
| C.I.34 | Urine | NU | 5 7 5 7 6 4 5 2 2 5 | |||
| C.I.35 | Urine | NU | 5 7 5 7 6 5 5 3 2 5 | |||
| C.I.36 | Urine | NU | 7 7 5 7 6 7 5 3 2 5 | |||
| C.I.37 | Urine | NU | 7 7 5 7 6 6 5 2 2 5 | |||
| C.I.38 | Blood | JU | 7 7 5 7 6 4 5 2 2 7 | |||
| C.I.39 | Blood | JU | 7 7 5 7 6 6 5 3 2 5 | |||
| C.I.40 | Sputum | NYSDH958-85c | 7 7 5 7 7 6 5 3 2 5 | |||
| C.I.41 | Blood | UMSMT3c | 7 7 5 7 7 6 5 2 3 5 | |||
| C.I.42 | Urine | KULM | 7 7 5 7 6 4 5 2 2 7 | |||
| Strains isolated from SHP patients' houses | ||||||
| SHP 1 | SHP patient's house | Tokyo, Japan | 7 7 5 7 7 6 5 3 2 5 | 0 0 0 0 0 0 0 0 1 0 0 0 0 1 0 0 1 0 0 0 0 0 | 0 0 0 0 0 0 0 0 1 1 | 0 0 0 0 0 0 1 0 0 1 0 0 0 0 |
| SHP 2 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 4 5 3 2 5 | 1 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 1 1 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 0 1 0 1 |
| SHP 3 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 6 5 3 2 5 | 0 1 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 1 0 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 0 1 0 1 |
| SHP 4 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 6 5 3 2 5 | 0 0 0 1 0 0 1 1 0 0 1 0 0 0 1 0 0 1 1 1 0 0 | 0 0 0 1 1 0 1 0 0 0 | 0 0 0 1 0 1 0 0 1 0 0 1 0 1 |
| SHP 5 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 4 5 3 2 5 | 0 0 0 0 0 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 0 0 | 0 0 0 0 0 0 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 0 1 0 1 |
| SHP 6 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 4 5 3 3 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 1 1 1 0 0 1 0 | 0 0 1 0 0 1 0 0 1 0 1 0 0 0 |
| SHP 7 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 7 4 5 3 3 5 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 1 1 0 0 1 0 | 0 0 0 0 0 1 0 0 1 0 1 0 0 0 |
| SHP 8 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 7 4 5 3 3 5 | 0 1 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 1 1 1 0 0 1 0 | 0 0 0 0 1 0 0 0 1 0 1 0 0 0 |
| SHP 9 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 6 5 2 2 7 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 0 0 1 1 1 1 0 0 | 0 0 0 0 1 0 0 0 1 0 1 0 0 0 |
| SHP 10 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 7 6 5 3 2 5 | |||
| SHP 11 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 4 5 3 2 5 | |||
| SHP 12 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 7 4 5 3 3 5 | |||
| SHP 13 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 6 5 3 2 5 | |||
| SHP 14 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 6 5 3 2 5 | |||
| SHP 15 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 4 5 3 3 7 | |||
| SHP 16 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 4 5 3 2 5 | |||
| SHP 17 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 7 4 5 3 3 5 | |||
| Environmental | ||||||
| SHP 10 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 7 6 5 3 2 5 | |||
| SHP 11 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 4 5 3 2 5 | |||
| SHP 12 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 7 4 5 3 3 5 | |||
| SHP 13 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 6 5 3 2 5 | |||
| SHP 14 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 6 5 3 2 5 | |||
| SHP 15 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 4 5 3 3 7 | |||
| SHP 16 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 6 4 5 3 2 5 | |||
| SHP 17 | SHP patient's house | Kumamoto, Japan | 7 7 5 7 7 4 5 3 3 5 | |||
| Environmental isolatesd | ||||||
| E.I.1 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 2 5 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 0 0 1 0 | 0 0 1 1 1 1 1 1 0 0 | 0 0 0 0 1 0 0 0 1 0 1 0 0 0 |
| E.I.2 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | 0 1 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 0 0 1 0 | 0 0 0 0 1 0 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 1 1 0 1 |
| E.I.3 | Soil | Tokyo, Japan | 7 7 5 7 7 6 5 2 3 5 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 1 0 1 0 | 0 0 1 1 1 1 1 0 0 0 | 0 0 0 0 1 0 0 0 1 0 1 0 0 0 |
| E.I.4 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | 0 0 0 0 1 0 0 0 0 0 0 0 1 0 1 0 0 0 0 0 1 0 | 0 0 0 1 1 1 0 0 1 0 | 0 0 0 0 0 0 0 0 1 0 1 0 0 0 |
| E.I.5 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | 0 0 0 1 0 0 1 1 0 0 1 0 0 1 1 0 0 1 0 1 0 0 | 0 0 0 1 1 0 0 1 1 0 | 0 0 1 0 1 0 0 0 0 0 1 0 0 0 |
| E.I.6 | Soil | Tokyo, Japan | 7 7 5 7 6 4 5 3 2 5 | 0 0 0 1 0 0 0 1 0 0 1 0 0 0 1 0 0 0 0 1 0 0 | 0 0 0 1 1 0 0 1 1 0 | 0 0 0 1 0 0 0 1 0 0 0 1 0 0 |
| E.I.7 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | 0 0 1 0 0 1 0 0 0 1 0 0 1 0 0 1 0 1 0 0 1 0 | 0 0 0 0 1 0 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 0 1 0 0 |
| E.I.8 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | 0 0 1 1 0 1 0 0 0 1 0 1 1 0 0 1 0 1 0 0 1 1 | 0 1 0 1 1 0 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 0 1 0 0 |
| E.I.9 | Soil | Tokyo, Japan | 7 7 5 7 7 6 5 2 3 5 | 0 0 1 1 0 1 0 0 0 1 0 1 1 0 0 1 0 1 0 0 1 1 | 0 1 0 1 1 0 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 0 1 0 1 |
| E.I.10 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | 0 0 1 1 0 1 0 0 0 1 0 1 1 0 0 1 0 1 0 0 1 1 | 0 1 0 1 1 0 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 0 1 0 1 |
| E.I.11 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | 0 0 1 1 0 1 0 0 0 1 0 0 1 0 0 1 0 1 0 0 1 0 | 0 0 0 1 1 0 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 0 1 0 1 |
| E.I.12 | Soil | Tokyo, Japan | 7 7 5 7 7 6 5 2 3 5 | 0 0 1 1 0 1 0 0 0 1 0 0 1 0 0 1 0 1 0 0 1 0 | 0 1 0 1 1 0 0 1 0 0 | 0 0 0 0 0 0 0 0 0 0 0 1 0 1 |
| E.I.13 | Soil | Tokyo, Japan | 7 7 5 7 7 6 5 2 3 5 | |||
| E.I.14 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | |||
| E.I.15 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | |||
| E.I.16 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | |||
| E.I.17 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | |||
| E.I.18 | Soil | Tokyo, Japan | 7 7 5 7 6 6 5 2 2 5 | |||
| E.I.19 | Soil | Tokyo, Japan | 7 7 5 7 6 4 5 2 2 5 | |||
| E.I.20 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | |||
| E.I.21 | Soil | Tokyo, Japan | 7 7 5 7 6 4 5 2 2 5 | |||
| E.I.22 | Soil | Tokyo, Japan | 7 7 5 7 7 7 5 3 3 5 | |||
| E.I.23 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 2 5 | |||
| E.I.24 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 3 5 | |||
| E.I.25 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 2 5 | |||
| E.I.26 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 2 5 | |||
| E.I.27 | Soil | Tokyo, Japan | 7 7 5 3 7 6 5 2 2 5 | |||
JU, Juntendo University School of Medicine, Tokyo, Japan; KULM, Kitazato University Laboratory of Mycology, Kanagawa, Japan; KU, Kumamoto University School of Medicine, Kumamoto, Japan; NU, Nagasaki University School of Medicine, Nagasaki, Japan; OMU, Oita Medical University, Oita, Japan; TMBH, Tokyo Metropolitan Bokuto Hospital, Tokyo, Japan; NYSDH, New York State Department of Health, Albany; UMSMT, University of Maryland School of Medical Technology, Baltimore.
1, PCR product present; 0, fragment absent.
See Kemker et al. (9).
RAPD analysis.
Nuclear DNA was extracted by the method of Makimura et al. (11). Eleven oligonucleotides (15, 21) were preliminarily investigated for reactivity and reproducibility using T. asahii DNA: M13 (GAGGGTGGCGGTTCT), T3B (AGGTCGCGGGTTCGAATCC), (GACA)4 (GACAGACAGACAGACA), TELO1 (TGGGTGTGTGGGTGTGTGGGTGTG), (CAG)4 (CAGCAGCAGCAG), OPE1 (CCCAAGGTCC), OPE2 (GGTGCGGGAA), OPE3 (CCAGATGCAC), OPE4 (GTGACATGCC), R28 (ATGGATCCGC), and RC8 (GGATGTCGAA). Three oligonucleotides, M13, OPE1, and RC8, were selected as single primers for PCR fingerprinting of 39 representative isolates. The three PCRs were individually optimized, and the reaction parameters for each primer were critical. Amplifications were performed in a total buffer volume of 50 μl containing 5 μl of 10× PCR buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2; Nippon Gene, Toyama, Japan), 4 μl of 200 μM deoxynucleoside triphosphates (equimolar dNTPs; Nippon Gene), 30 pmol of each primer, and 2.5 U of Gene Taq DNA polymerase (Nippon Gene). Gene Taq DNA polymerase was developed for RAPD analysis. For primer M13, PCR was performed in a thermocycler (model 9700; Perkin-Elmer Applied Biosystems, Foster City, Calif.) with an initial denaturation at 94°C for 3 min, followed by 40 cycles of 20 s at 94°C, 60 s at 50°C, and 20 s at 72°C, and a final extension at 72°C for 10 min. For primers OPE1 and RC8, PCR was performed with an initial denaturation at 94°C for 3 min, followed by 40 cycles consisting of 20 s at 94°C, 60 s at 36°C, and 20 s at 72°C, and a final cycle of 10 min at 72°C. Amplification products were separated by 1.5% agarose gel electrophoresis in 1× TAE (Tris-acetate EDTA) buffer. Electrophoretic bands were sized automatically using Digital Science Image Analysis Software (Eastman Kodak, Rochester, N.Y.). Each DNA fragment was scored as present or absent. The intensities of the PCR fragments were not measured. A phenogram showing the similarities of isolates was generated by the unweighted pair group method with arithmetic mean (UPGMA phenogram), based on the pairwise similarity coefficient matrix. The PAUP program (version 4.0b2; Phylogenetic Analysis Using Parsimony; David L. Swofford, Laboratory of Molecular Systematics, National Museum of Natural History, Smithsonian Institution) was used to calculate similarity values and to generate the UPGMA phenogram. A similarity value (SAB) was calculated for each pair of patterns, based on matching fragment positions. Eighteen clinical isolates from eight centers, 9 isolates from SHP patients' houses, and 12 environmental isolates were analyzed by RAPD analysis.
Origin-specific DNA sequences from RAPD fingerprinting.
Forty-six polymorphic bands were used to determine whether there was an origin-specific DNA band. Although three origin-specific DNA bands were not found, we observed a specific 330-bp band (see Fig. 1) for the clinical isolates and the strains obtained from the homes of SHP patients in RAPD fingerprinting using primer M13. The 330-bp DNA fragment was extracted from an agarose gel using a NucleoSpin kit (Clontech Laboratories Inc., Palo Alto, Calif.) according to the manufacturer's instructions. The fragment was cloned into pCR-2.1 using a TA cloning kit (Invitrogen Corp., Carlsbad, Calif.) and was sequenced with an ABI PRISM Cycle-Sequencing kit (Applied Biosystems). From these DNA sequences, two oligonucleotide primers were designed: M13-7F (TGCGCTCATGCGCTCATGAC) and M13-7R (TCCGCTGAGGAAGGAAGAGC). The PCR conditions were as follows: initial denaturation at 94°C for 3 min, followed by 30 cycles of 20 s at 94°C, 60 s at 50°C, and 20 s at 72°C, and a final cycle of 10 min at 72°C. For this PCR, Takara (Shiga, Japan) Ex Taq polymerase was used. Table 2 shows the strains used for specificity of PCR.
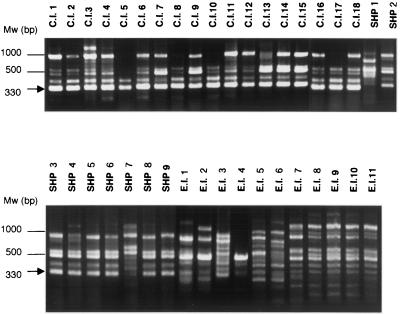
FIG. 1.
Representative electrophoresis gel of PCR fingerprints obtained from T. asahii isolates using primer M13. Mw, molecular weight marker. A 330-bp DNA fragment (arrow) shows specificity for the clinical isolates and strains isolated from SHP patients' houses.
TABLE 2.
Specificity of primers M13-7F and M13-7R for Trichosporon species and related species
| Species | Straina | PCR productb |
|---|---|---|
| Trichosporon coremiiforme | CBS 2482 | — |
| Trichosporon faecale | CBS 4828 | — |
| Trichosporon asteroides | CBS 2481 | — |
| Trichosporon aquatile | CBS 5973 | — |
| Trichosporon brassicae | CBS 6382 | — |
| Trichosporon cutaneum | CBS 2466 | — |
| Trichosporon dulcitum | CBS 8257 | — |
| Trichosporon inkin | CBS 5585 | — |
| Trichosporon gracile | CBS 8189 | — |
| Trichosporon montevideense | CBS 6721 | — |
| Trichosporon mucoides | CBS 7625 | — |
| Trichosporon ovoides | CBS 5585 | — |
| Trichosporon pullulans | CBS 2532 | — |
| Candida albicans | CBS 562 | — |
| Candida glabrata | IFO 0622 | — |
| Candida guilliermondii | CBS 566 | — |
| Candida kefyr | JCM 9559 | — |
| Candida lusitaniae | CBS 4413 | — |
| Candida parapsilosis | ATCC 22019 | — |
| Candida tropicalis | ATCC 7349 | — |
| Cryptococcus neoformans var. neoformans | CBS 132 | — |
| Cryptococcus neoformans var. gattii | NIH 191 | — |
| Cryptococcus albidus | CBS 142 | — |
| Malassezia furfur | CBS 1878 | — |
| Rhodotorula mucilaginosa | CBS 17 | — |
Abbreviations: CBS, Centraal Bureau voor Schimmelcultures, Baarn, The Netherlands; IFO, Institute for Fermentation, Osaka, Japan; JCM, Japan Collection of Microorganisms, Saitama, Japan; NIH, National Institutes for Health, Bethesda, Md.
—, no product obtained.
Biochemical characteristics.
The isolates were examined with an ID32C kit (bioMérieux SA, Marcy I'Etoile, France) in accordance with the manufacturer's instructions. A total of 42 clinical isolates, 17 SHP isolates, and 27 environmental isolates were examined. The PAUP program was used to calculate similarity values and to construct a UPGMA phenogram.
Nucleotide sequence accession number.
The sequences of specific DNA fragments obtained by PCR of T. asahii clinical isolates and strains isolated from SHP patients' houses have been deposited in the DNA Data Bank of Japan under accession number AB049759.
RESULTS
RAPD analysis.
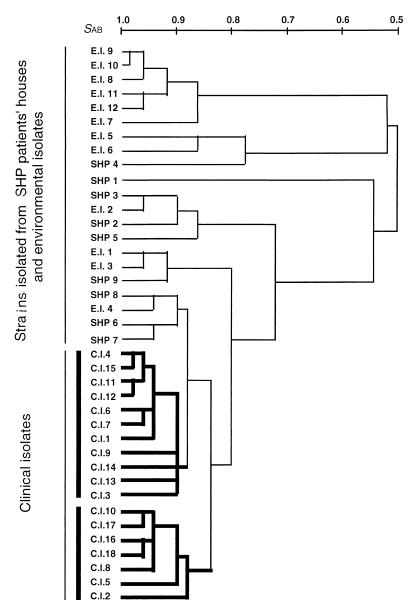
The three primers produced 46 polymorphic bands, as shown in Table 1. A representative photograph of the PCR products for primer M13 is presented in Fig. 1. The similarities of the polymorphic bands within the clinical isolates, strains isolated from SHP patients' houses, and environmental isolates were 91.3, 74.5, and 72.0%, respectively (Table 3). Clinical isolates were more similar to each other than to the strains of the two other origins (P < 0.01). and they formed two clusters (Fig. 2). Although the clinical isolates were obtained from eight different Japanese and U.S. centers, there was no relationship between the RAPD fingerprinting pattern and the source hospital. Neither environmental isolates nor strains isolated from SHP patients' houses formed a cluster (P > 0.1).
TABLE 3.
Mean similarity between pairs of T. asahii isolates from three different origins
| Comparison | Mean similarity (%) ± SD |
|---|---|
| Within clinical isolates | 91.3 ± 4.5 |
| Within strains isolated from SHP patients' houses | 74.5 ± 12.5 |
| Within environmental isolates | 72.0 ± 14.9 |
| Between clinical isolates and strains isolated from SHP patients' housesa | 78.0 ± 12.0 |
| Between clinical isolates and environmental isolatesa | 66.9 ± 13.7 |
| Between strains isolated from SHP patients' houses and environmental isolatesb | 68.9 ± 12.4 |
Significant difference (P < 0.01).
No significant difference (P > 0.1).
FIG. 2.
UPGMA phenogram of T. asahii isolates calculated from DNA fingerprinting patterns obtained with primers M13, OPE1, and RC8.
Origin-specific DNA fragment.
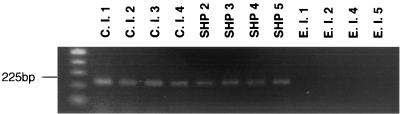
An origin-specific 330-bp DNA fragment was obtained. Newly designed oligonucleotide primers amplified all T. asahii clinical isolates. PCRs were positive for the strains isolated from SHP patients' houses, with the exception of strain SHP 1. DNA from environmental isolates, excluding strain E.I.3, was amplified by PCR and was negative for all other Trichosporon species and other medically relevant yeasts (Table 2). A representative electrophoretic gel of the PCR product is shown in Fig. 3. The sequences obtained from specific DNA fragments have been deposited in the DNA Data Bank of Japan (accession number AB049759).
FIG. 3.
Representative electrophoretic gel of PCR products using primers M13-7F and M13-7R. A 225-bp PCR product was amplified.
Biochemical characteristics determined by using the ID32 kit.
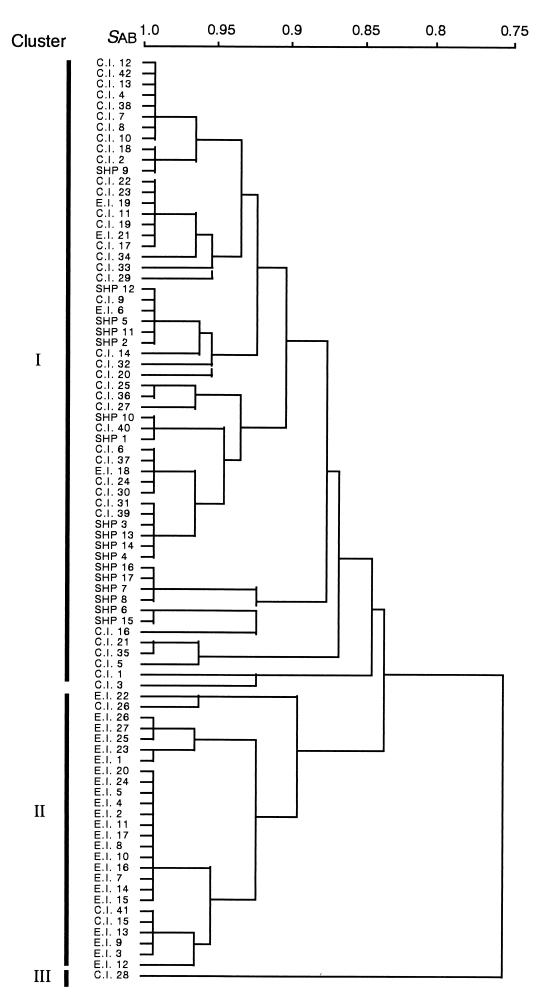
The biochemical characteristics of the isolates are listed as an API profile in Table 1. The phenogram shown in Fig. 4 was generated using 31 characteristics and consists of three major clusters: I, II, and III. Cluster I includes most of the clinical and SHP strains (93%; 55 of 59), and cluster II comprises most of the environmental isolates (85%; 23 of 27). Cluster III contains only one strain, C.I.28. The ability to utilize 25 of 31 compounds was almost the same for strains belonging to clusters I and II; however, remarkable differences were found in the ability to utilize the other 6 compounds, as shown in Table 4.
FIG. 4.
UPGMA phenogram of T. asahii isolates calculated from the ability to utilize 31 compounds.
TABLE 4.
Characteristics differentiating strains belonging to clusters I and II
| Cluster | % of strains able to utilize the indicated compounda
|
|||||
|---|---|---|---|---|---|---|
| MDG | SOR | RHA | MLZ | SBE | MAN | |
| I (n = 59) | 100 | 14 | 41 | 56 | 29 | 14 |
| II (n = 26) | 31 | 100 | 100 | 4 | 0 | 81 |
MDG, α-methyl-d-glucoside; SOR, sorbitol; RHA, rhamnose; MLZ, melezitose; SBE, sorbose; MAN, mannitol.
DISCUSSION
Because T. asahii is responsible for both opportunistic fungal infections and allergies and is also distributed in the environment, it is interesting to examine the genetic diversity of strains obtained from different sources. RAPD analysis suggested that the clinical isolates were distinct from isolates obtained from SHP patients' houses and the environmental isolates. While the assimilation patterns of clinical isolates and strains obtained from SHP patients' houses were similar, they were notably different from those of environmental isolates. Comparison of Fig. 2 and 4 shows that the figures are not entirely correlated with each other in the phenogram. However, it is obvious that the clinical and environmental isolates have distinct RAPD profiles and biochemical characteristics. A similar finding was reported by Bertout et al. (3), who found that C. neoformans isolates formed three clusters in an MLEE data analysis. The first cluster contained clinical isolates, the second included environmental isolates, and the third contained strains isolated from either patients or the environment. In 1991, blood, superficial, and environmental isolates of T. beigelii were reported to be distinct, based on RFLP of rDNA and isoenzyme profiles (9). Subsequently, T. beigelii has been divided into more than 10 species, and some of the superficial and environmental isolates have been reidentified as Trichosporon aquatile, T. cutaneum, T. domesticum, and T. ovoides according to current taxonomical criteria (16; see also the American Type Culture Collection [ATCC, Rockville, Md.] catalog at http://www.atcc.org/). In addition to the correlation between the genotype and the origin of a strain, RAPD can be used to differentiate genotype and drug susceptibility. Xu et al. (21) showed that fluconazole-resistant C. albicans strains isolated from patients infected with human immunodeficiency virus formed a cluster distinct from that of fluconazole-sensitive strains by RAPD. Strains C.I.16, C.I.17, and C.I.18 were resistant to fluconazole (data not shown). We have not yet determined the drug susceptibilities of all T. asahii isolates, but they were in the same subcluster. No correlation between the RAPD fingerprinting pattern and the hospital was found for T. asahii isolates, whereas for C. albicans, there is hospital, regional, and country specificity among isolates. In this study, 11 oligonucleotides that had been widely used for C. albicans and C. neoformans were tested against T. asahii DNA. Of these 11 oligonucleotides, PCR parameters could not be optimized for 8. The development of a highly or specifically reactive oligonucleotide primer would permit an intensive epidemiological study of disease due to T. asahii.
In this study, we obtained DNA fragments specific for clinical isolates and strains isolated from SHP patients' houses. We previously reported species-specific DNA sequences derived from T. asahii by ITS sequence analysis (17, 18). Since Trichosporon species are phylogenetically closely related, it is difficult to design species-specific primers. Our T. asahii-specific primers designed from ITS-derived sequences amplify DNA of T. faecale (Trichosporon asahii var. faecalis), and T. coremiiforme (Trichosporon asahii var. coremiiformis). The latter two species are nonpathogenic and are not known to cause clinical problems. If new oligonucleotide primers are designed, it should be possible to detect T. asahii with high specificity. No similar sequence was found in GenBank by a BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/).
ACKNOWLEDGMENTS
This study was supported in part by a Grant for the Promotion of the Advancement of Education and Research in Graduate Schools from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
We thank the physicians who provided us with T. asahii isolates.
REFERENCES
- 1.Ando M, Sakata T, Yoshida K, Yamasaki H, Araki S, Onoue K, Shinoda T. Serotype-related antigen of Trichosporon cutaneum in the induction of summer-type hypersensitivity pneumonitis: correlation between serotype of inhalation challenge-positive antigen and that of the isolates from patients' homes. J Allergy Clin Immunol. 1990;85:36–44. doi: 10.1016/0091-6749(90)90218-s. [DOI] [PubMed] [Google Scholar]
- 2.Ando M, Arima K, Yoneda R, Tamura M. Japanese summer-type hypersensitivity pneumonitis: geographic distribution, home environment, and clinical characteristics of 621 cases. Am Rev Respir Dis. 1991;144:765–769. doi: 10.1164/ajrccm/144.4.765. [DOI] [PubMed] [Google Scholar]
- 3.Bertout S, Renaud F, Swinne D, Mallie M, Bastide J M. Genetic multilocus studies of different strains of Cryptococcus neoformans: taxonomy and genetic structure. J Clin Microbiol. 1999;37:715–720. doi: 10.1128/jcm.37.3.715-720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemons K V, Feroze F, Holmberg K, Stevens D A. Comparative analysis of genetic variability among Candida albicans isolates from different geographic locales by three genotypic methods. J Clin Microbiol. 1997;35:1332–1336. doi: 10.1128/jcm.35.6.1332-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalle F, Franco N, Lopez J, Vagner O, Caillot D, Chavanet P, Cuisenier B, Aho S, Lizard S, Bonnin A. Comparative genotyping of Candida albicans bloodstream and nonbloodstream isolates at a polymorphic microsatellite locus. J Clin Microbiol. 2000;38:4554–4559. doi: 10.1128/jcm.38.12.4554-4559.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guého E, Smith M T, de Hoog G S, Billon-Grand G, Christen R, Batenburg-van der Vegte W H. Contributions to a revision of the genus Trichosporon. Antonie Leeuwenhoek. 1992;61:289–316. doi: 10.1007/BF00713938. [DOI] [PubMed] [Google Scholar]
- 7.Guého E, Improvisi L, de Hoog G S, Dupont B. Trichosporon on humans: a practical account. Mycoses. 1994;37:3–10. doi: 10.1111/j.1439-0507.1994.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 8.Herbrecht R, Koening H, Waller K, Liu L, Guého E. Trichosporon infections: clinical manifestations and treatment. J Mycol Med. 1993;3:129–136. [Google Scholar]
- 9.Kemker B J, Lehmann P F, Lee J W, Walsh T J. Distinction of deep versus superficial clinical and nonclinical isolates of Trichosporon beigelii by isoenzymes and restriction fragment length polymorphisms of rDNA generated by polymerase chain reaction. J Clin Microbiol. 1991;29:1677–1683. doi: 10.1128/jcm.29.8.1677-1683.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyman C A, Devi S J, Nathanson J, Frasch C E, Pizzo P A, Walsh T J. Detection and quantitation of the glucuronoxylomannan-like polysaccharide antigen from clinical and nonclinical isolates of Trichosporon beigelii and implications for pathogenicity. J Clin Microbiol. 1995;33:126–130. doi: 10.1128/jcm.33.1.126-130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makimura K, Murayama Y S, Yamaguchi H. Detection of a wide range of medically important fungal species by polymerase chain reaction (PCR) J Med Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 12.Marco F, Lockhart S R, Pfaller M A, Pujol C, Rangel-Frausto M S, Wiblin T, Blumberg H M, Edwards J E, Jarvis W, Saiman L, Patterson J E, Rinaldi M G, Wenzel R P, Soll D R. Elucidating the origins of nosocomial infections with Candida albicans by DNA fingerprinting with the complex probe Ca3. J Clin Microbiol. 1999;37:2817–2828. doi: 10.1128/jcm.37.9.2817-2828.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaller M A, Jones R N, Doern G V, Sader H S, Hollis R J, Messer S A. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and antifungal susceptibilities of isolates collected in 1997 in the United States, Canada, and South America for the SENTRY Program. The SENTRY Participant Group. J Clin Microbiol. 1998;36:1886–1889. doi: 10.1128/jcm.36.7.1886-1889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller M A, Lockhart S R, Pujol C, Swails-Wenger J A, Messer S A, Edmond M B, Jones R N, Wenzel R P, Soll D R. Hospital specificity, region specificity, and fluconazole resistance of Candida albicans bloodstream isolates. J Clin Microbiol. 1998;36:1518–1529. doi: 10.1128/jcm.36.6.1518-1529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pujol C, Joly S, Lockhart S R, Noel S, Tibayrenc M, Soll D R. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J Clin Microbiol. 1997;35:2348–2358. doi: 10.1128/jcm.35.9.2348-2358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugita T, Nishikawa A, Shinoda T, Kume H. Taxonomic position of deep-seated, mucosa-associated, and superficial isolates of Trichosporon cutaneum from trichosporonosis patients. J Clin Microbiol. 1995;33:1368–1370. doi: 10.1128/jcm.33.5.1368-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugita T, Nishikawa A, Shinoda T. Identification of Trichosporon asahii by PCR based on sequences of the internal transcribed spacer regions. J Clin Microbiol. 1998;36:2742–2744. doi: 10.1128/jcm.36.9.2742-2744.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugita T, Nishikawa A, Ikeda R, Shinoda T. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J Clin Microbiol. 1999;37:1985–1993. doi: 10.1128/jcm.37.6.1985-1993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugita T, Nishikawa A, Ichikawa T, Ikeda R, Shinoda T. Isolation of Trichosporon asahii from environmental materials. Med Mycol. 2000;38:27–30. doi: 10.1080/mmy.38.1.27.30. [DOI] [PubMed] [Google Scholar]
- 20.Tashiro T, Nagai H, Kamberi P, Goto Y, Kikuchi H, Nasu M, Akizuki S. Disseminated Trichosporon beigelii infection in patients with malignant diseases: immunohistochemical study and review. Eur J Clin Microbiol Infect Dis. 1994;13:218–224. doi: 10.1007/BF01974540. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, Ramos A R, Vilgalys R, Mitchell T G. Clonal and spontaneous origins of fluconazole resistance in Candida albicans. J Clin Microbiol. 2000;38:1214–1220. doi: 10.1128/jcm.38.3.1214-1220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo C G, Kim Y W, Han S K, Nakagawa K, Suga M, Nishiura Y, Ando M, Shim Y S. Summer-type hypersensitivity pneumonitis outside Japan: a case report and the state of the art. Respirology. 1997;2:75–77. doi: 10.1111/j.1440-1843.1997.tb00057.x. [DOI] [PubMed] [Google Scholar]