Abstract
Granzyme A (GZMA) is a serine protease secreted by cytotoxic lymphocytes, with Gzma-/- mouse studies having informed our understanding of GZMA’s physiological function. We show herein that Gzma-/- mice have a mixed C57BL/6J and C57BL/6N genetic background and retain the full-length nicotinamide nucleotide transhydrogenase (Nnt) gene, whereas Nnt is truncated in C57BL/6J mice. Chikungunya viral arthritis was substantially ameliorated in Gzma-/- mice; however, the presence of Nnt and the C57BL/6N background, rather than loss of GZMA expression, was responsible for this phenotype. A new CRISPR active site mutant C57BL/6J GzmaS211A mouse provided the first insights into GZMA’s bioactivity free of background issues, with circulating proteolytically active GZMA promoting immune-stimulating and pro-inflammatory signatures. Remarkably, k-mer mining of the Sequence Read Archive illustrated that ≈27% of Run Accessions and ≈38% of BioProjects listing C57BL/6J as the mouse strain had Nnt sequencing reads inconsistent with a C57BL/6J genetic background. Nnt and C57BL/6N background issues have clearly complicated our understanding of GZMA and may similarly have influenced studies across a broad range of fields.
Research organism: Mouse, Viruses
Introduction
Granzyme A (GZMA) is a granule trypsin-like serine protease (trypase) secreted by cytotoxic lymphocytes such as NK cells (Fehniger et al., 2007; Wu et al., 2019), NKT cells (Gordy et al., 2011), and CD8+ cytotoxic T lymphocytes (Suhrbier et al., 1991). The traditional view has been that GZMA is a cytotoxic mediator that is secreted into the immunological synapse, entering the target cell via perforin pores, whereupon certain cytoplasmic proteins are cleaved, resulting in the initiation of cell death pathway(s) (Liesche et al., 2018; Martinvalet et al., 2008; Wu et al., 2019; Zhou et al., 2020). A key tool in the quest to understand the physiological role of GZMA has been the use of Gzma-/- mice (Ebnet et al., 1995). For instance, control of viral infections can be compromised in Gzma-/- mice (Loh et al., 2004; Müllbacher et al., 1996; Pereira et al., 2000; Riera et al., 2000), with cytotoxic lymphocytes from these mice reported to be less able to kill target cells (Pardo et al., 2002; Pardo et al., 2004; Shresta et al., 1999; Susanto et al., 2013). Like granzyme B (GzmB), GZMA has thus been classified as a cytotoxic granzyme (Golstein and Griffiths, 2018; Mpande et al., 2018; Muraro et al., 2017; Zhou et al., 2020), although in several studies a role for GZMA in mediating cellular cytotoxicity was not observed (Ebnet et al., 1995; Joeckel and Bird, 2014; Regner et al., 2009; Regner et al., 2011; Smyth et al., 2003). In a range of settings, GZMA has also been associated with the promotion of inflammation, providing an additional or alternative view of its physiological role, although consensus on mechanisms has remained elusive (Metkar et al., 2008; Park et al., 2020; Santiago et al., 2020; Santiago et al., 2017; Schanoski et al., 2019; Shimizu et al., 2019; van Daalen et al., 2020; Wensink et al., 2015; Wilson et al., 2017), with a number of potential intracellular and extracellular targets for GZMA reported. These include pro-IL-1β (Hildebrand et al., 2014), SET complex proteins (Mandrup-Poulsen, 2017; Mollah et al., 2017), gasdermin B (Zhou et al., 2020), mitochondrial complex I protein NDUFS3 (Martinvalet et al., 2008), protease activated receptors (Hansen et al., 2005; Sower et al., 1996; Suidan et al., 1994; Suidan et al., 1996), TLR2/4 (van Eck et al., 2017), and TLR9 (Shimizu et al., 2019). Gzma-/- mice have also been used to show a role for GZMA in inter alia diabetes (Mollah et al., 2017), cancer (Santiago et al., 2020), bacterial infections (García-Laorden et al., 2016; García-Laorden et al., 2017; van den Boogaard et al., 2016), and arthritis (Santiago et al., 2017). GZMA’s bioactivity has generally (Plasman et al., 2014; Schanoski et al., 2019; Zhou et al., 2020), but not always (Shimizu et al., 2019; van Eck et al., 2017), been associated with GZMA’s protease activity, with circulating GZMA in humans shown to be proteolytically active (Spaeny-Dekking et al., 1998).
After infection with the arthritogenic alphavirus, chikungunya virus (CHIKV) (Suhrbier, 2019), infected Gzma-/- mice showed a substantially reduced overt arthritic foot swelling when compared to infected C57BL/6J (6J) mice (Wilson et al., 2017). However, we show herein that active site mutant GzmaS211A mice generated by CRISPR on a 6J background showed no significant differences in CHIKV arthritic foot swelling when compared with 6J mice. The apparent contradiction was resolved when it emerged that Gzma-/- mice had a mixed 6J and C57BL/6N (6N) background. Gzma-/- mice retained expression of the full-length nicotinamide nucleotide transhydrogenase (Nnt) gene, whereas 6J mice have a truncated Nnt with a 5 exon deletion (Fontaine and Davis, 2016). The presence of full-length Nnt and the mixed 6N/6J genetic background (rather than absence of GZMA expression) emerged to be responsible for amelioration of CHIKV arthritic foot swelling in Gzma-/- mice. As much of our understanding of the physiological role of GZMA comes from studies in Gzma-/- mice, we used the GzmaS211A mice to gain new insights into GZMA function that were not compromised by genetic background.
The enzyme, nicotinamide nucleotide transhydrogenase (NNT), also known as proton-translocating NAD(P)+ transhydrogenase (EC 7.1.1.1), is located in the inner mitochondrial membrane and catalyzes the conversion of NADH plus NADP+ to NAD+ plus NADPH, while H+ is pumped from the inter-membrane space into the mitochondrial matrix (Rydström, 2006). NNT thereby sustains mitochondrial antioxidant capacity through generation of NADPH (Ward et al., 2020), with the loss of active NNT in 6J mice associated with reduced ability to detoxify reactive oxygen species (ROS) via the glutathione and thioredoxin pathways (McCambridge et al., 2019; Meimaridou et al., 2018; Ronchi et al., 2013; Rydström, 2006). As redox regulation is involved in many cellular processes (Gambhir et al., 2019; Lingappan, 2018; Sun et al., 2020) and genetic backgrounds are known to affect phenotypes (Leskov et al., 2017; Morales-Hernandez et al., 2018; Rao et al., 2020; Salerno et al., 2019; Vozenilek et al., 2018; Williams et al., 2021; Wolf et al., 2016), we sought to determine how many other studies in GMO mice might have been affected by 6J vs. 6N background differences using the Nnt gene as a genetic marker. k-mer mining of RNA-Seq datasets deposited in the NCBI Sequence Read Archive (SRA) revealed that ≈27% of Run Accessions and ≈38% of BioProjects listing the mouse strain as ‘C57BL/6J’ had Nnt reads inconsistent with a 6J background. Although reported as underappreciated in the metabolism literature (Fontaine and Davis, 2016), potential problems associated with differences in Nnt and/or other background genes clearly extends well beyond this field and are not restricted to Gzma-/- mice.
Results
CHIKV inflammatory arthritis in GzmaS211A mice
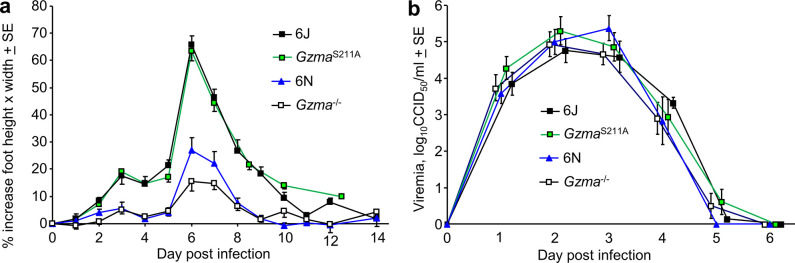
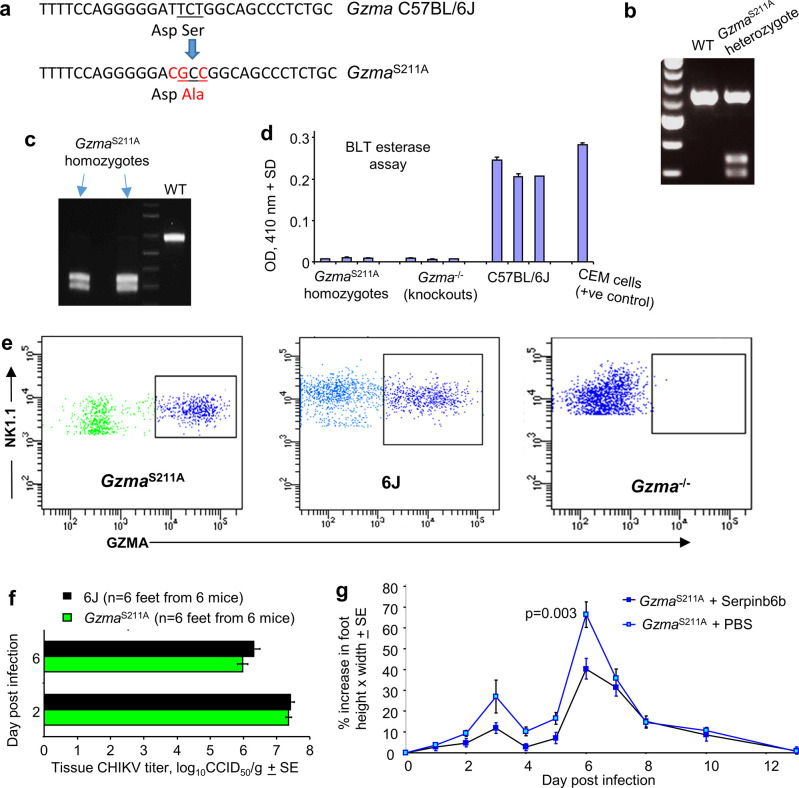
We reported previously that the inflammatory arthritis induced by CHIKV infection (manifesting as overt foot swelling) was significantly lower in Gzma-/- mice than in C57BL/6J (6J) mice (Wilson et al., 2017); an observation we confirm herein (Figure 1a). Injection of proteolytically active, but not proteolytically inactive, recombinant mouse GZMA induced inflammatory foot swelling, illustrating directly that GZMA’s protease activity is able to drive pro-inflammatory responses (Schanoski et al., 2019). To confirm and extend these findings, a new homozygous GZMA active site mutant mouse was generated using CRISPR technology in 6J mice, with the reactive site serine changed to alanine (GzmaS211A) (Susanto et al., 2013; Figure 1—figure supplement 1a–c). Loss of enzyme activity was confirmed by BLT assays (Suhrbier et al., 1991; Figure 1—figure supplement 1d). Intracellular staining (Schanoski et al., 2019) showed that expression of the GZMA proteins in resting splenic NK cells (Fehniger et al., 2007) was comparable for GzmaS211A and 6J mice (Figure 1—figure supplement 1e).
Figure 1. Chikungunya virus (CHIKV) infection in Gzma-/-, GzmaS211A, 6N, and 6J mice.
(a) Percent increase in foot swelling for the indicated mouse strains. Data is from 2 to 4 independent experiments with 5–6 mice (10–12 feet) per group per experiment. From day 3 to day 10, feet from GzmaS211A and 6J mice were significantly more swollen than feet from Gzma-/- and 6N mice (Kolmogorov–Smirnov tests, p<0.002). (b) Viremia for the mice in (a) (6N n = 12, GzmaS211A n = 18, Gzma-/- n = 15, 6J n = 17).
Figure 1—figure supplement 1. Construction and characterization of GzmaS211A mice.
When infected with CHIKV, viral titers in feet were not significantly different for GzmaS211A and 6J mice (Figure 1—figure supplement 1f), arguing that enzymically active GZMA has no significant antiviral activity against CHIKV. This is consistent with our previous study using Gzma-/- mice that also concluded that GZMA has no important antiviral function (Wilson et al., 2017). Surprisingly, however, foot swelling following CHIKV infection was not significantly different between the GzmaS211A and 6J mice (Figure 1a). If GZMA’s pro-inflammatory bioactivity (Wilson et al., 2017) depends on its protease activity (Schanoski et al., 2019), the results from Gzma-/- and GzmaS211A mice (Figure 1a) would appear to provide contradictory results.
The effective amelioration of CHIKV arthritic foot swelling in 6J mice following treatment with Serpinb6b (an inhibitor of GZMA) also supported the view that GZMA promotes inflammation in this setting (Wilson et al., 2017). However, Serpinb6b also inhibited CHIKV foot swelling in GzmaS211A mice (Figure 1—figure supplement 1g), arguing that Serpinb6b can inhibit other unknown proteases involved in promoting arthritic inflammation. The contention is supported by the broad inhibitory activity of the human orthologue, SerpinB6 (Strik et al., 2004).
Taken together, these new data argued that the role of GZMA in promoting CHIKV arthritis and the results obtained from Gzma-/- mice (Wilson et al., 2017) required re-evaluation.
Gzma-/- mice have a mixed C57BL/6N-C57BL/6J genetic background
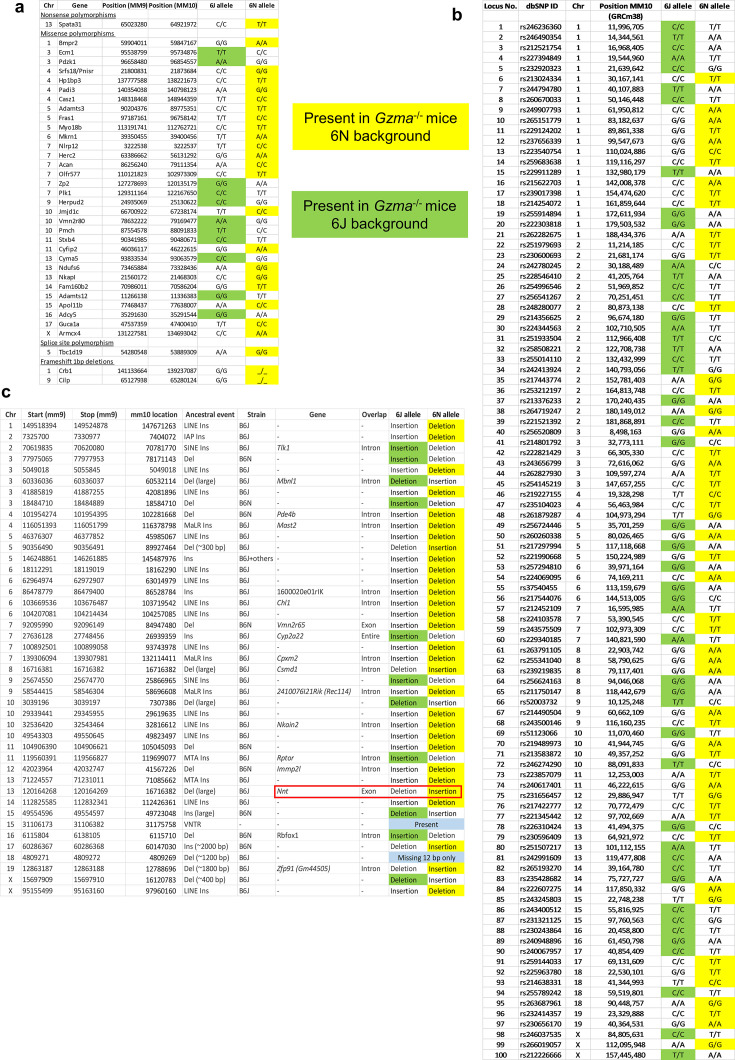
To reconcile the apparent contradictory data from GzmaS211A and Gzma-/- mice, and cognizant of previously described issues with knockout mice (Teoh et al., 2014), whole-genome sequencing (WGS) of Gzma-/- mice was undertaken (NCBI SRA; PRJNA664888). This analysis unequivocally demonstrated that Gzma-/- mice have a mixed genetic background, with ≈60% of the genome showing single-nucleotide polymorphisms (SNPs) and indels present in the C57BL/6N (6N) genome (Mekada et al., 2015; Simon, 2013), with the rest reflecting a 6J background (Figure 3—figure supplement 1). The strain origin of the BL/6-III ES cells used to generate Gzma-/- mice was only reported as C57BL/6 (Ebnet et al., 1995). As ES cells from 6J mice have a low rate of germline transmission, ES from 6N mice cells were frequently used to generate knockout mice (Fontaine and Davis, 2016), with our studies clearly arguing that BL/6-III ES cells also have a 6N background. The reported backcrossing of Gzma-/- mice onto C57BL/6 mice (Müllbacher et al., 1999) also clearly did not involve extensive backcrossing onto 6J mice.
Importantly, a large body of literature has resulted from the use of inbred Gzma-/- mice (and Gzma-/-/Gzmb-/- double knockout mice derived from them) with 6J mice used as controls, without being aware of the presence and potential confounding influence of the 6N background (Supplementary file 1).
Gzma-/- mice and 6N (Gzma+/+) mice both have reduced foot swelling after CHIKV infection
An explanation for the significantly lower CHIKV-induced foot swelling seen in Gzma-/- mice is that the partial 6N background is influencing the foot swelling phenotype. To test this contention, 6J and 6N mice (both Gzma+/+) were infected with CHIKV. 6N mice showed a significant reduction in foot swelling when compared with 6J mice (Figure 1a), arguing that the 6N background, rather than loss of GZMA expression, was primarily responsible for amelioration of foot swelling in Gzma-/- mice.
The reduced foot swelling in 6N and Gzma-/-mice was unlikely to be due to reduced viral loads as there were no significant differences in viremia for 6N, 6J, Gzma-/-, or GzmaS211A mice (Figure 1b).
RNA-Seq of CHIKV foot swelling for Gzma-/- vs. 6J mice
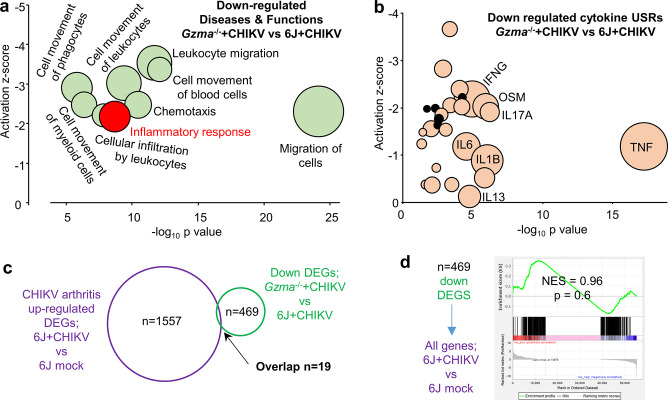
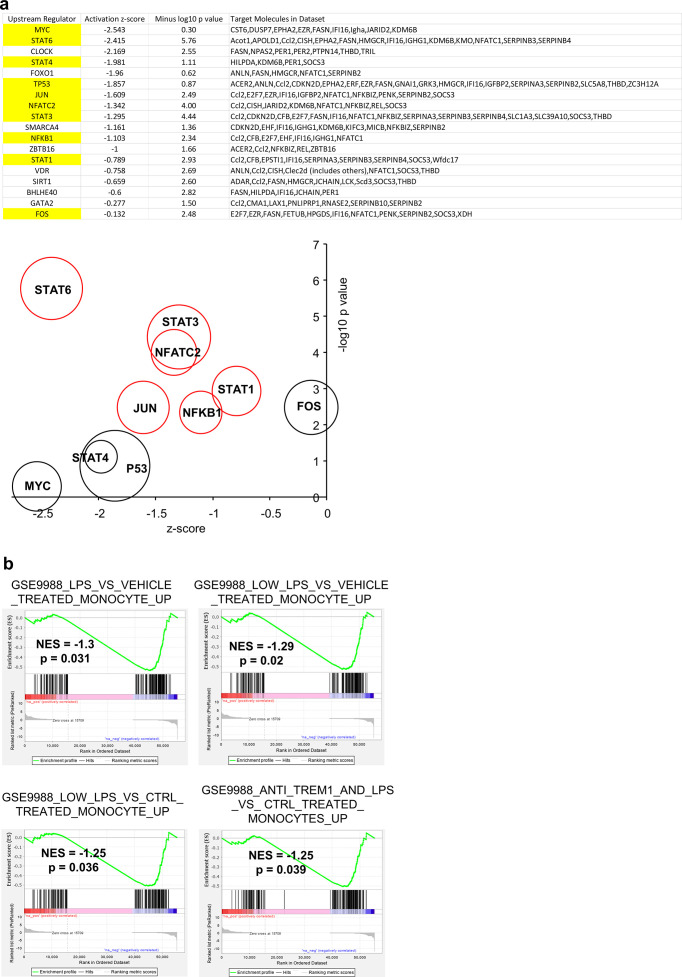
To gain insights into the reduced foot swelling in Gzma-/- mice (Figure 1a), RNA-Seq was undertaken on day 6 (peak arthritis) to compare gene expression in feet from CHIKV-infected Gzma-/- mice vs. 6J mice (NCBI BioProject PRJNA664644; full gene list in Supplementary file 2a). Differentially expressed genes (DEGs) (n = 1073) were generated after application of a q < 0.01 filter (Supplementary file 2b). When the 1073 DEGs (for Gzma-/- + CHIKV vs. 6J + CHIKV; Supplementary file 2b) were analyzed for Diseases and Functions using Ingenuity Pathway Analysis (IPA), the dominant annotations were associated with decreased cell movement (often leukocyte migration) (Figure 2a, Supplementary file 2c). These findings are consistent with immunohistochemistry data showing significantly reduced T cell and NK cells in the arthritic infiltrates in feet after CHIKV infection of Gzma-/- mice when compared with infected 6J mice (Wilson et al., 2017). IPA upstream regulator (USR) analysis of the 1073 DEGs (Supplementary file 2d) also illustrated that a series of pro-inflammatory cytokine USRs were downregulated in the infected feet of Gzma-/- + CHIKV vs. 6J + CHIKV mice (Figure 2b, Supplementary file 2e), consistent with the reduced foot swelling seen in infected Gzma-/- mice when compared with infected 6J mice (Wilson et al., 2017).
Figure 2. RNA-Seq of Gzma-/- + chikungunya virus (CHIKV) vs. 6J + CHIKV day 6 feet.
(a) Selected Ingenuity Pathway Analysis (IPA) Diseases and Function annotations for the 469 downregulated differentially expressed genes (DEGs) for Gzma-/- + CHIKV vs. 6J + CHIKV (full set of annotations shown in Supplementary file 2c). (b) IPA cytokine upstream regulators (USRs) downregulated in CHIKV-infected feet of Gzma-/- mice (Gzma-/-+CHIKV vs. 6J + CHIKV; Supplementary file 2e) plotted by p-value and z-score. Black circles – minor USRs for Gzma-/- + CHIKV vs. 6J + CHIKV not identified for 6J + CHIKV vs. 6J mock (see also Figure 4e). (c) RNA-Seq identified 1557 DEGs upregulated in feet for 6J + CHIKV vs. 6J mock infection (Supplementary file 2g). RNA-Seq of Gzma-/- + CHIKV vs. 6J + CHIKV day 6 feet identified 469 downregulated DEGs in Gzma-/- mice associated with the reduced foot swelling (Supplementary file 2b). Only 19 of these DEGs were shared by these datasets. (d) Gene Set Enrichment Analysis (GSEA) of downregulated DEGs from Gzma-/- + CHIKV vs. 6J + CHIKV day 6 feet (Supplementary file 2b) vs. all genes (preranked by fold change) from feet 6J + CHIKV vs. 6J mock infection (Supplementary file 2f).
We previously characterized the CHIKV arthritis signature by undertaking RNA-Seq of infected feet during peak foot swelling relative to control uninfected feet (6J + CHIKV vs. 6J mock infection) (Wilson et al., 2017). We reanalyzed the data (FastQ files NCBI BioProject PRJNA431476) using STAR aligner, RSEM EdgeR, and the more recent mouse genome build (GRCm38 Gencode vM23); all genes are shown in Supplementary file 2f, and a DEG list (with filters q < 0.01, fold change >2) is shown in Supplementary file 2g. The latter 2201 DEGs (for 6J + CHIKV vs. 6J mock infection) were analyzed by IPA, with the USRs shown in Supplementary file 2h. Of these USRs, 103 cytokine USR annotations showed significant upregulation (positive z-scores) after CHIKV infection (Supplementary file 2i). As might be expected, key pro-inflammatory cytokine USRs such as TNF, IFNG, IL6, and IL1B (Suhrbier, 2019) that were upregulated during CHIKV arthritis (Supplementary file 2i) were downregulated (negative z-scores) in the ameliorated foot swelling seen in Gzma-/- mice (Figure 2b, Supplementary file 2e). Curiously, however, five minor USRs (PPBP, CTF1, WNT7A, NAMPT, TIMP1) were downregulated in Gzma-/- mice, but were not upregulated during CHIKV infection (Figure 2b, black circles; Supplementary file 2e, yellow). Furthermore, of the 469 DEGs downregulated during CHIKV arthritis in Gzma-/- mice (Supplementary file 2b), only 19 were upregulated DEGs for CHIKV arthritis (Figure 2c, Supplementary file 2g). Gene Set Enrichment Analysis (GSEA) similarly revealed that DEGs downregulated in Gzma-/- mice were not significantly enriched in the upregulated genes for CHIKV arthritis in 6J mice (Figure 2d). Thus, although feet from CHIKV-infected Gzma-/- mice showed annotations associated with reduced cellular infiltrates and pro-inflammatory cytokines, this amelioration of arthritic signatures was associated with downregulation of genes largely not associated with CHIKV arthritis in 6J mice. In summary, arthritis amelioration in Gzma-/- mice was due to the downregulation of a largely distinct set of genes (and some distinct pathways), again arguing that the 6N background plays a key role in this phenotype.
Gzma-/- mice have an intact nicotinamide nucleotide transhydrogenase gene
There are multiple genes associated with inflammation and/or arthritis that differ between Gzma-/- (mixed 6N/6J background) and 6J mice (Supplementary file 3). One gene that has been highlighted as a key difference between 6J and 6N mice is Nnt (Freeman et al., 2006; Mekada et al., 2009; Ripoll et al., 2012; Ronchi et al., 2013; Vozenilek et al., 2018; Figure 3—figure supplement 1c, red box). The function of NNT is primarily to sustain mitochondrial antioxidant capacity through the generation of NADPH, which supports the antioxidant capacity of the glutathione and thioredoxin systems (McCambridge et al., 2019; Meimaridou et al., 2018; Ronchi et al., 2013; Rydström, 2006; Ward et al., 2020). These systems are generally viewed as having broad anti-inflammatory activities (Ghezzi, 2021; Yodoi et al., 2017).
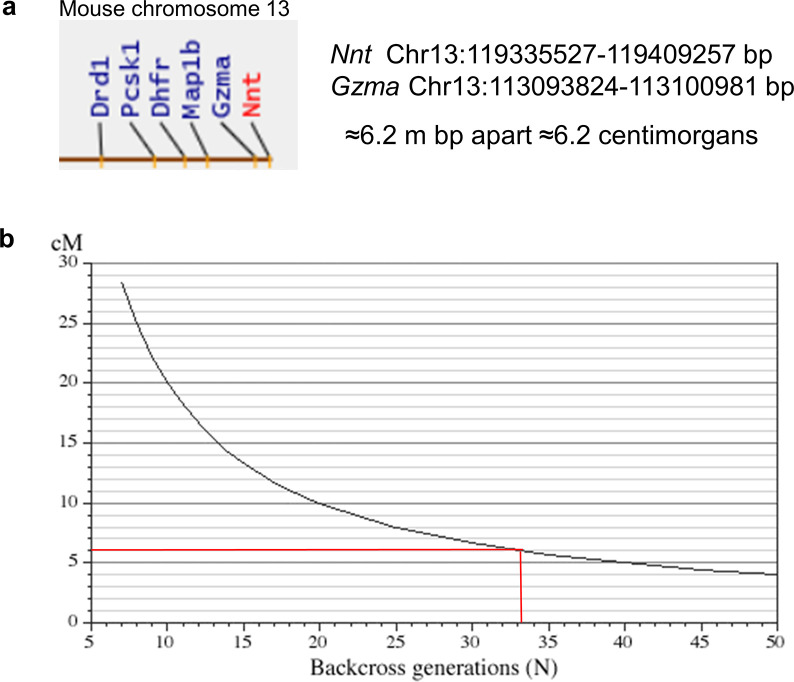
6N mice have a full-length Nnt gene with 21 protein-coding exons, whereas 6J mice have an in-frame 5-exon deletion removing exons 7–11 (Freeman et al., 2006). Confusingly, the MM10 mouse build numbers the Nnt exons differently and includes the noncoding exon 1 that is located before the ATG start site. According to this numbering (which is used herein), 6J mice have lost exons 8–12 of 22 exons. The Nnt gene is located only ≈6.2 megabases from the Gzma gene on mouse chromosome 13, so >30 backcrosses would be required to segregate these two loci (Silver, 2008; Figure 3—figure supplement 2). The close association of Nnt and Gzma genes also means Gzma-/-Gzmb-/- double-knockout mice (Supplementary file 1) would also very likely have full-length Nnt. Another Gzma-/- mouse generated using 129/SvJ ES cells (Shresta et al., 1997) would likely have the same issue as 129/SvJ mice also have a full-length Nnt gene.
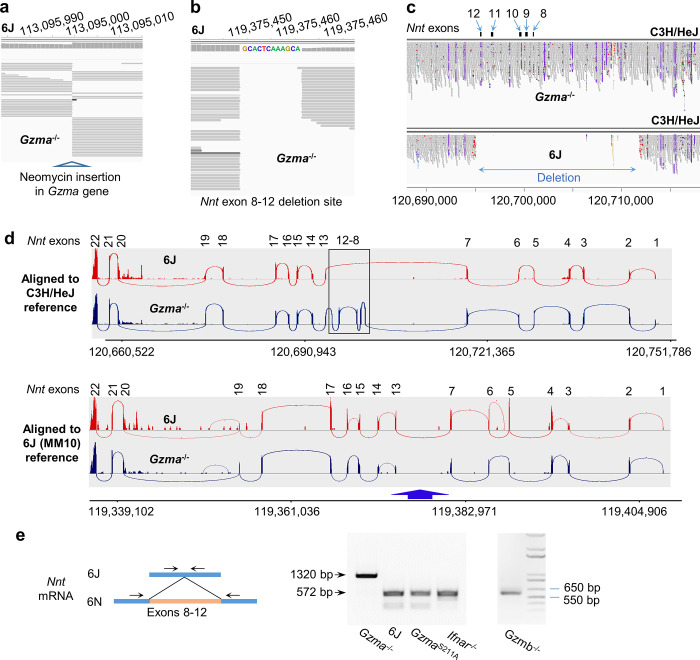
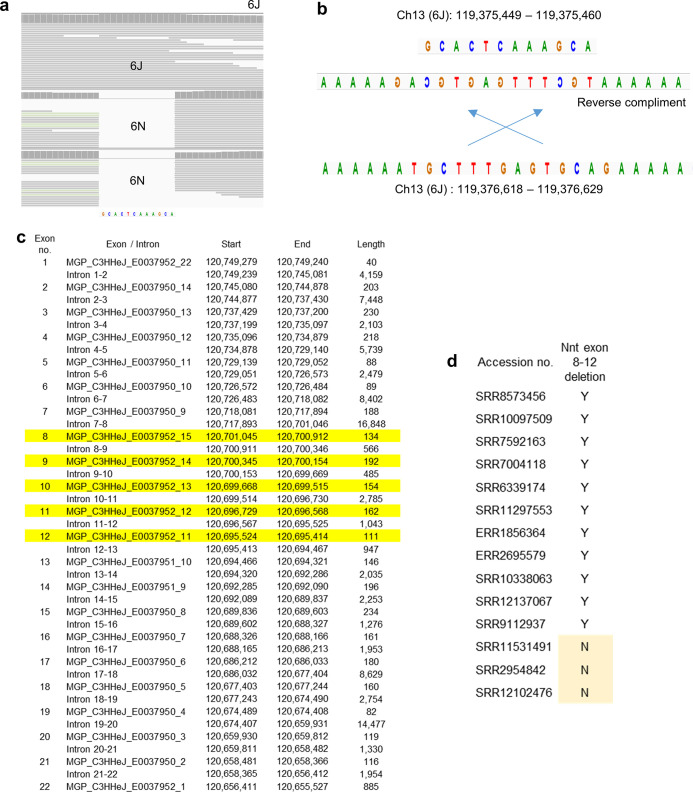
Alignment of the WGS of Gzma-/- mice (PRJNA664888) to the standard 6J MM10 mouse genome build allowed identification of the neomycin cassette insertion site into the Gzma gene that was used to generate the Gzma-/- mice (Ebnet et al., 1995; Figure 3a). Curiously, this alignment shows a 12-nucleotide insertion in the 6J genome at the Nnt exon 8–12 deletion junction (Figure 3b). The 12 nucleotides are also absent in other 6N WGS data (Figure 3—figure supplement 3a), indicating this is not a unique feature of Gzma-/- mice. This insertion in 6J may have accompanied deletion of Nnt exons 8–12 during the generation of 6J mice (Fontaine and Davis, 2016; Figure 3—figure supplement 3b).
Figure 3. Nnt deletion in 6J but not Gzma-/- mice.
(a) Whole-genome sequencing (WGS) of Gzma-/- mice aligned to the 6J (MM10) reference genome build, illustrating the insertion site of the neomycin cassette into the Gzma gene to create the knockout. (b) As for (a) but showing the site of the Nnt deletion, with the additional 12 nucleotides present in the 6J genome. (c) WGS of Gzma-/- and 6J mice aligned to the C3H/HeJ reference genome, illustrating that the Nnt deletion present in 6J mice is absent in Gzma-/- mice. The deletion is in chromosome 13; position 120,695,141–120,711,874 (C3H/HeJ numbering). (d) Reads from RNA-Seq of chikungunya virus (CHIKV)-infected 6J and Gzma-/- mice aligned to the C3H/HeJ and 6J (MM10) reference genomes showing the Sashimi plot (Integrative Genomics Viewer) for the Nnt gene. (e) RT-PCR of testes using primers either side of exons 8–12 in the Nnt mRNA.
Figure 3—figure supplement 1. Gzma-/- mice have a mixed 6N (yellow)/6J (green) background.
Figure 3—figure supplement 2. Nnt and Gzma gene locations and backcrossing requirements.
Figure 3—figure supplement 3. Nnt locus inserts and alignments.
Although a 6N genome sequence is available, it is poorly annotated, hence the C3H/HeJ genome build was used for alignments as it also has a full-length Nnt gene (Figure 3—figure supplement 3c).
Alignment of WGS reads from Gzma-/- mice to the C3H/HeJ genome clearly showed that Gzma-/- mice had a full-length Nnt gene, whereas 6J mice had the expected ≈16 kb deletion (Figure 3c). The approach (Figure 3c) was further validated using other 6J and non-6J WGS submissions (Figure 3—figure supplement 3d).
Sashimi plots of RNA-Seq reads aligned to the C3H/HeJ build clearly illustrated that Nnt mRNA from 6J mice was missing exons 8–12, whereas in Gzma-/- mice the full-length Nnt mRNA was expressed (Figure 3d, top). Alignment to the 6J genome and viewed by Sashimi plot showed that exons 7 and 13 are linked in the Nnt mRNA from 6J mice, consistent with expression of a truncated Gzma mRNA species. In contrast, exons 7 and 13 are not linked in the Nnt mRNA from Gzma-/- mice (Figure 3d, blue arrow) as the mRNA, but not the MM10 genome build, contains exons 8–12.
These results were confirmed by RT-PCR using primers located on either side of the exon 8–12 deletion (Huang et al., 2006; Figure 3e). Gzma-/- mice have the longer 6N Nnt PCR product as this Nnt mRNA includes exons 8–12, whereas 6J mice, GzmaS211A (generated by CRISPR on a 6J background), and type I IFN receptor knockout (Ifnar-/-) mice (also on a 6J background Swann et al., 2007), all showed a shorter PCR product, consistent with the deletion of exons 8–12 in the Nnt mRNA (Figure 3e). In a separate RT-PCR run, Gzmb-/- mice (Wilson et al., 2017) were shown to be missing Nnt exons 8–12, consistent with a 6J background (Figure 3e).
The Nnt deletion promotes CHIKV-induced foot swelling
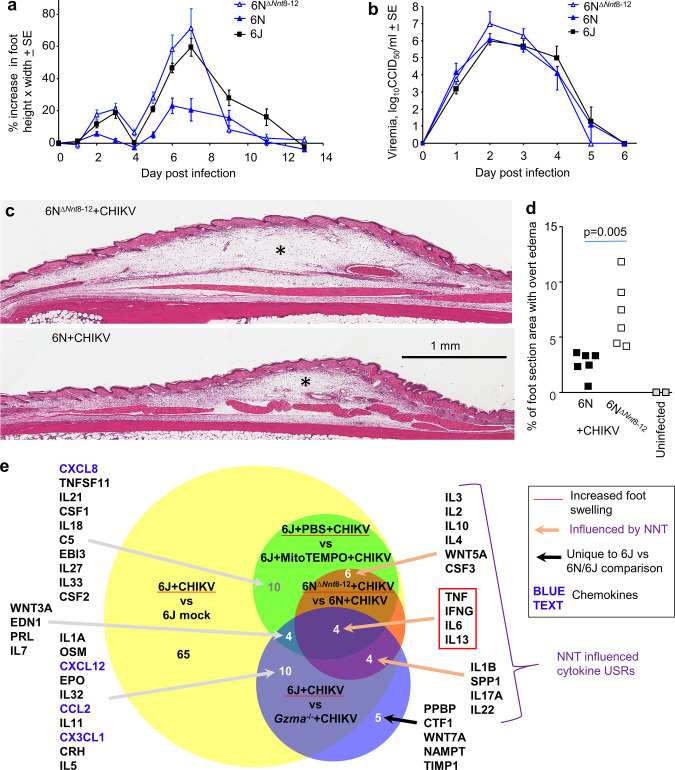
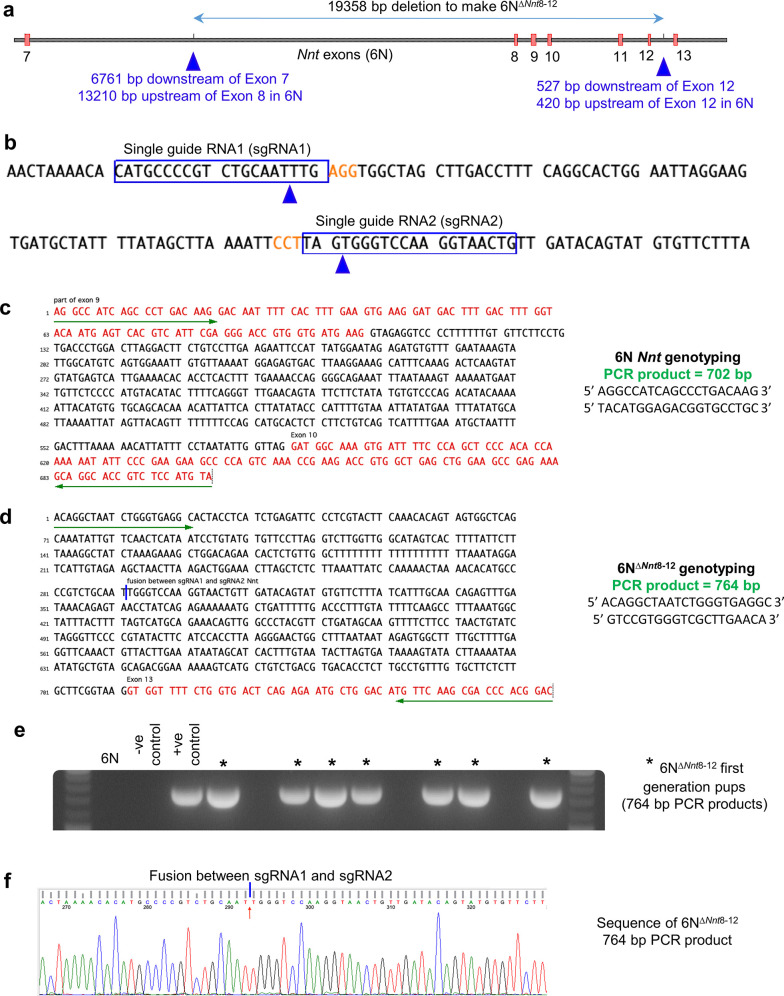
To determine whether the Nnt gene deletion seen in 6J mice might be responsible for promoting the arthritic foot swelling in CHIKV-infected mice, we generated 6N∆Nnt8-12 mice wherein exons 8–12 of Nnt were deleted from 6N mice using CRISPR (Figure 4—figure supplement 1). 6N∆Nnt8-12 mice thus have the same deletion of Nnt exons as 6J mice. As before, CHIKV-induced foot swelling was significantly higher in 6J mice when compared with 6N mice (Figure 4a). Importantly, foot swelling in 6N∆Nnt8-12 mice was significantly higher than in 6N mice, with the Nnt exon 8–12 deletion increasing foot swelling to levels comparable to those seen in 6J mice (Figure 4a). There were no significant differences in viremia between the mouse strains (Figure 4b). This data illustrates that the absence of a functional Nnt gene (6N∆Nnt8-12) can by itself promote overt foot swelling after CHIKV infection. The data also argues that the presence of functional Nnt gene in Gzma-/- mice likely contributes to the ameliorated foot swelling seen in Gzma-/- mice.
Figure 4. 6N∆Nnt8-12 mice.
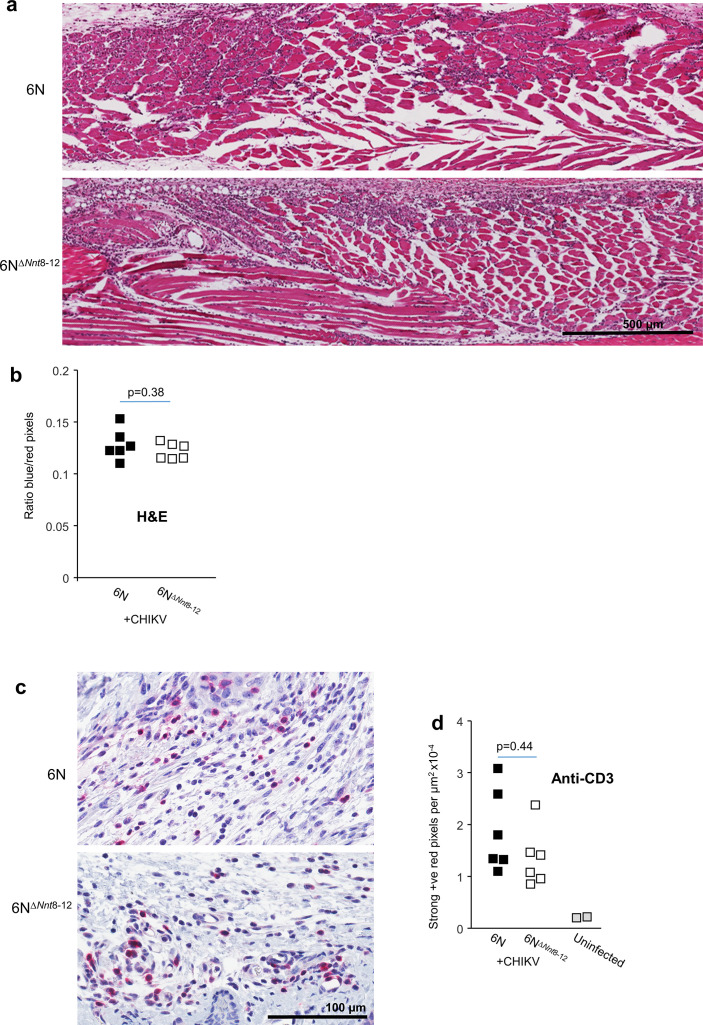
(a) 6N∆Nnt8-12 mice have the same Nnt exon deletion as 6J mice. Age-matched female 6N∆Nnt8-12, 6N, and 6J mice were infected with chikungunya virus (CHIKV) and foot swelling measured over time (n = 5 mice and 10 feet per group). Foot swelling was significantly higher in 6N∆Nnt8-12 mice when compared with 6N mice on days 2–7 (day 2 p=0.0026, day 7 p=0.0027, t-tests, parametric data distributions; days 3–6 p=0.003, Kolmogorov–Smirnov tests, nonparametric data distributions). Foot swelling was significantly lower in 6N mice when compared with 6J mice (day 2 p=0.042, day 6 p=0.001, day 7 p=0.0005, t-tests, parametric data distributions; days 3 and 5, p=0.002, Kolmogorov–Smirnov tests, nonparametric data distributions). (b) Viremia for the same mice as in (a). (c) H&E staining of feet from 6N∆Nnt8-12 and 6N mice day 6 post infection showing subcutaneous edema (*). (d) Percentage of foot section area showing overt subcutaneous edema (statistics by Kolmogorov–Smirnov test). (e) RNA-Seq data for four comparisons was analyzed by Ingenuity Pathway Analysis (IPA) and cytokine upstream regulator (USR) overlaps shown. Only cytokine USRs with positive z-scores associated with increased foot swelling are shown.
Figure 4—figure supplement 1. Generation of 6N∆Nnt8-12 mice.
Figure 4—figure supplement 2. Histological analyses for 6N∆Nnt8-12 mice feet.
Histology and H&E staining of arthritic feet illustrated that the increased foot swelling seen in 6N∆Nnt8-12 mice (compared with 6N mice) was due primarily to increased edema (Figure 4c and d), with no significant differences in cellular infiltrates (Figure 4—figure supplement 2a and b). Edema is a recognized feature of alphaviral arthritides and is well described in CHIKV mouse models (Gardner et al., 2010; Poo et al., 2014; Prow et al., 2019). Immunohistochemistry with anti-CD3 also showed no significant differences in T cell numbers in the inflammatory infiltrates (Figure 4—figure supplement 2c and d). To further characterize the role of Nnt in CHIKV arthritis, day 6 feet from 6N∆Nnt8-12 + CHIKV vs. 6N + CHIKV were compared using RNA-Seq (Supplementary file 4a and b). IPA USR analysis (Supplementary file 4c) provided 14 cytokine annotations with positive z-scores (Supplementary file 4d) that were associated with the increased foot swelling in 6N∆Nnt8-12 mice (Figure 4e, brown circle). These 14 cytokine USRs were also upregulated during CHIKV arthritis (Figure 4e, yellow brown overlap). The IPA cytokine classification also includes chemokines, with no chemokine USRs identified within these 14 annotations (Figure 4e, brown circle), consistent with the lack of a significant cell migration phenotype (Figure 4—figure supplement 2). In contrast, several chemokine annotations were identified for the Gzma-/- mice (Figure 4e, blue circle, blue text) consistent with the reduced inflammatory infiltrate (Wilson et al., 2017). Furthermore, only 8/27 cytokine USRs identified for Gzma-/- + CHIKV vs. 6J + CHIKV were also identified for the 6N∆Nnt8-12 + CHIKV vs. 6N + CHIKV comparison (Figure 4e, blue-brown overlap), arguing that other 6N background genes apart from Nnt ameliorated inflammatory cytokine activity in the feet of CHIKV-infected Gzma-/- mice.
Perhaps consistent with the arthritis literature in general (Kato, 2020; Ma and Xu, 2013; Ogata et al., 2019; Suhrbier, 2019), pronounced foot swelling in all comparisons (including MitoTEMPO, see below) was associated with upregulation of TNF, IFNG, and IL-6 USRs (Figure 4e, red box). IL-13 was also a consistently upregulated USR, with IL-13 associated with resolution of arthritic inflammation (Schett, 2019), with peak CHIKV arthritis in 6J mice associated with a significant resolution phase signature (Prow et al., 2019).
MitoTEMPO ameliorates CHIKV arthritis in 6J mice
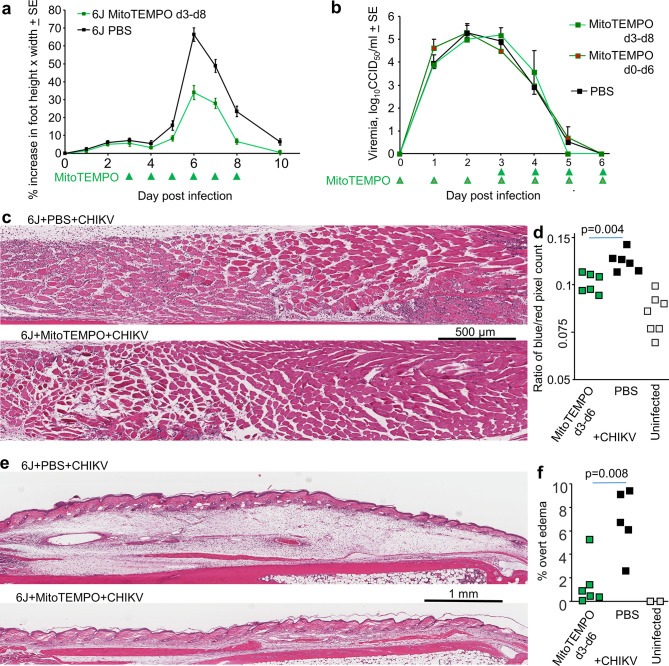
As NNT’s primary function is to sustain mitochondrial antioxidant capacity (McCambridge et al., 2019; Meimaridou et al., 2018; Ronchi et al., 2013; Rydström, 2006; Ward et al., 2020), the data argues that the reduced arthritic foot swelling in 6N mice (and to some extent in Gzma-/- mice) is due to increased mitochondrial antioxidant capacity. MitoTEMPO has been widely used as an experimental antioxidant treatment to scavenge mitochondrial ROS in a variety of disease settings (Aoyama et al., 2012; Li et al., 2018; To et al., 2020; Vincent et al., 2020; Wu et al., 2020). Treatment of CHIKV arthritis in 6J mice with MitoTEMPO from day 3 to 8 post infection significantly ameliorated peak foot swelling (Figure 5a). Viremia was not significantly affected by MitoTEMPO treatment, even when treatment was initiated on day 0 (Figure 5b).
Figure 5. MitoTEMPO treatment.
(a) 6J mice were infected with chikungunya virus (CHIKV) and then treated with MitoTEMPO or PBS i.v. daily on days 3–8 post infection and foot swelling measured (n = 5/6 mice and 10/12 feet per group). Statistics by t-tests, days 6, 7, and 8, p < 0.001. (b) Viremia for the mice in (a), with an additional group of six mice treated daily with MitoTEMPO from day 0 till day 6. (c) H&E staining of muscle tissues in feet of mice day 6 post CHIKV infection for mice treated with PBS or MitoTEMPO from day 3 to day 6. Clusters of small blue spots in and around the muscle bundles illustrate nuclei of infiltrating leukocytes. (d) Aperio pixel count of blue (nuclear) over red (cytoplasmic) pixels; a measure of leukocyte infiltration (statistics by t-test). (e) H&E staining of feet as in (c) but showing overt subcutaneous edema. (f) Percentage of foot section areas showing overt subcutaneous edema (statistics by t-test).
Figure 5—figure supplement 1. Additional H&E images of feet subcutaneous edema (as in Figure 5e).
Histology and H&E staining revealed that MitoTEMPO treatment significantly reduced the cellular infiltrate in feet day 6 post CHIKV infection of 6J mice when compared with PBS treatment (Figure 5c and d). Edema was also significantly reduced by MitoTEMPO treatment (Figure 5e and f); additional images are shown in Figure 5—figure supplement 1. RNA-Seq analysis (Supplementary file 5a and b) and IPA USR analysis (Supplementary file 5c) illustrated that MitoTEMPO treatment was associated with downregulation of 24 cytokine USRs (Figure 4e, green circle; Supplementary file 5d), with 10 of these also downregulated in 6N vs. 6N∆Nnt8-12 mice (Figure 4e, green and brown circle overlap). MitoTEMPO treatment in 6J mice and an intact Nnt gene in 6N mice thus provided overlapping anti-inflammatory activities. However, MitoTEMPO was also able to inhibit a series of additional pro-inflammatory arthritic responses including the chemokine CXCL8 and complement factor 5 (C5), with complement promoting arthritic infiltrates in a related alphavirus, Ross River virus (Morrison et al., 2007). Inhibition of CXCL8 and/or C5 is consistent with the reduced cellular infiltrate (Figure 5c and d).
Reinvestigation of the physiological role of GZMA using poly(I:C)
Our current understanding of the physiological function of GZMA comes, to a large extent, from multiple studies comparing Gzma-/- (and Gzma-/-/Gzmb-/-) mice with 6J mice (Supplementary file 1). Given the results herein, many of the reported phenotypes are likely to have arisen, at least in part, from an intact Nnt gene and/or the 6N background, complicating any conclusions regarding the physiological role of GZMA.
To gain new insights into GZMA’s activity in vivo, we sought to find an experimental setting where high levels of GZMA are secreted. We have shown previously that humans, nonhuman primates, and mice have elevated serum GZMA levels after infection with CHIKV (Schanoski et al., 2019; Wilson et al., 2017). Infection of mice with a series of other RNA viruses also resulted in high serum GZMA levels early in infection, with NK cells identified as the likely source (Schanoski et al., 2019). Resting NK cells constitutively contain abundant levels of GZMA protein (Fehniger et al., 2007), which is usually stored in granules as a mature protease, with the low pH of the granule preventing (premature) proteolytic activity (Stewart et al., 2012).
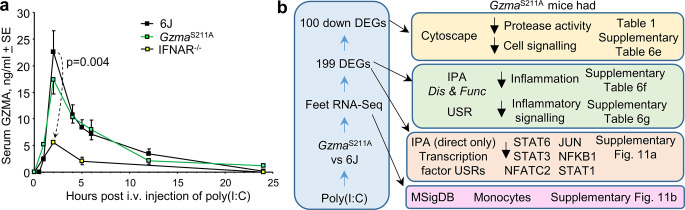
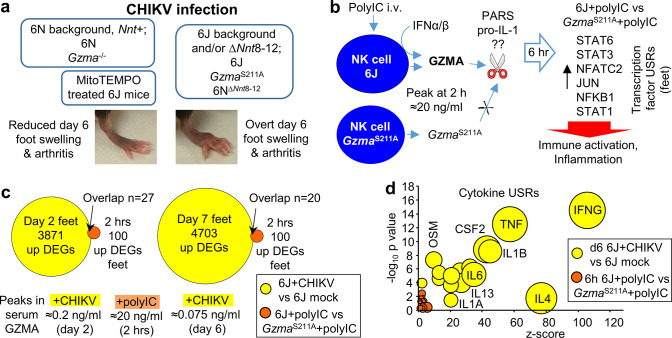
Polyinosinic:polycytidylic acid (poly(I:C)) can often mimic aspects of the innate responses to RNA viruses (Prow et al., 2017). We thus injected poly(I:C) i.v. into 6J mice and showed that serum GZMA levels reached mean peak serum levels of ≈20 ng/ml of serum after 2 hr, with levels dropping to baseline after 24 hr (Figure 6a). Although poly(I:C) treatment is known to activate NK cells (Djeu et al., 1979; Fehniger et al., 2007; Miyake et al., 2009; Ngoi et al., 2008), this rapid and prodigious poly(I:C)-induced release of GZMA into the circulation, to the best of our knowledge, has hitherto not been reported. Type I interferons (IFNs) are also rapidly induced by poly(I:C) (Dempoya et al., 2012; Santiago-Raber et al., 2003), and NK cells express the type I IFN receptor and can respond to type I IFNs (Madera et al., 2016; Mizutani et al., 2012). Injection of poly(I:C) i.v. into Ifnar-/- mice resulted in a significantly blunted elevation of GZMA (Figure 6a). Type I IFNs thus appear to augment this rapid GZMA release; however, the absence of type I IFN signaling does not prevent GZMA secretion, consistent with previous data (Schanoski et al., 2019). Poly(I:C) treatment of GzmaS211A mice resulted in serum GZMA levels not significantly different from those seen in 6J mice (Figure 6a), illustrating that the active site mutation does not significantly affect production, secretion, or stability. Finally, although GZMB and GZMA are often considered to be co-expressed (Supplementary file 1), in this setting no serum GZMB was detected (Figure 6—figure supplement 1). GZMB and perforin proteins are not expressed in resting NK cells and appear only after ≈24 hr of appropriate stimulation (Fehniger et al., 2007).
Figure 6. Polyinosinic:polycytidylic acid (poly(I:C)) injection into GzmaS211A, 6J mice, and Ifnar-/-.
(a) GzmaS211A, 6J, and Ifnar-/- mice were injected i.v. with 250 µg of poly(I:C) in 150 µl of PBS, and serum samples were taken at the indicated times and assayed for GZMA concentration using a capture ELISA kit (6J n = 5–8, Ifnar-/- n = 5–6 and GzmaS211A n = 3 mice per time point). (b) GzmaS211A and 6J mice were injected i.v. with 250 µg of poly(I:C) and feet removed 6 hr later and analyzed by RNA-Seq. The differentially expressed genes (DEGs) (Supplementary file 6b) were analyzed by Cytoscape and Ingenuity Pathway Analysis (IPA). The full gene list (Supplementary file 6a) was analyzed using the Molecular Signature Database (MSigDB).
Figure 6—figure supplement 1. Serum granzyme B (GZMB) levels after i.v. injection of polyinosinic:polycytidylic acid (poly(I:C)).
Figure 6—figure supplement 2. RNA-Seq for polyinosinic:polycytidylic acid (poly(I:C)) injection into GzmaS211A vs. 6J mice.
Figure 6—figure supplement 3. The gene expression signatures in feet seen 6 hr after injection of polyinosinic:polycytidylic acid (poly(I:C)) in GzmaS211A vs. 6J mice are not significantly recapitulated in spleen.
Figure 6—figure supplement 4. Ingenuity Pathway Analysis (IPA) and Gene Set Enrichment Analysis (GSEA) for polyinosinic:polycytidylic acid (poly(I:C)) injection into GzmaS211A vs. 6J mice.
RNA-Seq after poly(I:C) injection in GzmaS211A vs. 6J mice
GzmaS211A and 6J mice are both on a 6J genetic background, are both missing Nnt exons 8–12, and both show similar levels of serum GZMA secretion after poly(I:C) treatment (Figure 6a). Thus, the only difference between these strains is that GZMA in GzmaS211A mice is enzymically inactive (Figure 1—figure supplement 1d). GzmaS211A and 6J mice were injected with poly(I:C) (as in Figure 6a), with feet and spleens removed 6 hr later and analyzed by RNA-Seq (NCBI BioProject PRJNA666748). The rationale for this time point was to capture early transcriptional events after the peak of serum GZMA. The sample preparation strategy and RNA-Seq data overview is provided in Figure 6—figure supplement 2. The full gene list for feet is provided in Supplementary file 6a, with the 199 DEGs listed in Supplementary file 6b. For spleen, the full gene list is shown in Supplementary file 6c and the four DEGs in Supplementary file 6d. This represents the first study of GMO mice targeting Gzma that is free from the potentially confounding influence of the mixed 6J/6N background.
Immune/inflammation signatures stimulated by circulating proteolytically active GZMA
Circulating proteolytically active GZMA would appear to have limited influence in the spleen as only four DEGs were identified in the spleen of poly(I:C)-treated GzmaS211A vs. 6J mice (Supplementary file 6d). Interestingly, the only significantly upregulated DEG in GzmaS211A spleens was Mid1, a gene involved in controlling granule exocytosis by cytotoxic lymphocytes (Boding et al., 2014; Boding et al., 2015). GSEA also illustrated that neither up- nor downregulated DEGs identified in the feet were enriched in spleen (Figure 6—figure supplement 3), arguing that in this setting the activity of GZMA in the periphery is not significantly recapitulated in spleen.
Of the 199 DEGs identified in the feet of poly(I:C)-treated GzmaS211A vs. 6J mice, 100 were downregulated, with the top annotations associated with negative regulation of protease activity and negative regulation of cell signaling after Cytoscape analysis (Figure 6b, Table 1, Supplementary file 6). This result supports the view that GZMA is proteolytically active in vivo (Spaeny-Dekking et al., 2000) and that GZMA’s proteolytic activity mediates cell signaling events under physiological conditions. IPA Disease and Functions analysis of the 199 DEGs from feet (GzmaS211A + polyIC vs. 6J + polyIC; Supplementary file 6b) identified downregulation (negative z-scores) of a series of inflammation and leukocyte activation signatures (Figure 6b, Supplementary file 6f). IPA USR analysis (core analysis with direct and indirect interactions) indicated downregulation of a series of cytokine, immune receptor, and transcription factor USRs (Figure 6b, Supplementary file 6g). An IPA USR analysis using the direct-only interaction option, which largely limits the analysis to transcription factors, showed STAT6, STAT3, NFATC2, JUN, NFKB1, and STAT1 to be the dominant downregulated transcription factor signatures in GzmaS211A mice by z-score and p-values (Figure 6b, Figure 6—figure supplement 4a). These transcription factors are associated with various innate and adaptive immune responses, with NFATC2 playing a central role in the activation of T cells during the development of an immune response. STAT3 and NF-κB have previously been shown to be activated in macrophages by recombinant GZMA in vitro (Santiago et al., 2020), with monocytes/macrophages reported as targets for GZMA activity in a variety of settings (Garzón-Tituaña et al., 2021; Metkar et al., 2008; Santiago et al., 2017; Spencer et al., 2013; Uranga et al., 2016). Interrogation of the Molecular Signature Database (MSigDB) (Subramanian et al., 2005) using GSEAs also identified gene sets associated with activated monocytes (GSE19888) that were significantly enriched in the downregulated genes for GzmaS211A + polyIC vs. 6J + polyIC (Figure 6—figure supplement 4b). This observation supports the contention that monocytes/macrophages are activated by circulating GZMA (Figure 6b). IPA of the 150 core enriched genes from these GSEAs also identified STAT6, STAT3, NFATC2, JUN, NFKB1, and STAT1 as significant USRs, even though only 10 of these 150 genes were significant DEGs for GzmaS211A vs. 6J (Supplementary file 6h). These USR signatures would thus appear to be a consistent feature of the RNA-Seq data for GzmaS211A + polyIC vs. 6J + polyIC.
Table 1. Cytoscape analysis of downregulated differentially expressed genes (DEGs) in GzmaS211A mice.
RNA-Seq of feet taken from GzmaS211A vs. 6J mice 6 hr after polyinosinic:polycytidylic acid (poly(I:C)) injection provided 199 differentially expressed genes (DEGs), of which 100 were downregulated in GzmaS211A mice (Supplementary file 6b). When analyzed by Cytoscape, the top annotations were associated with negative regulation (underlined) of protease activities (bold) or negative regulation of protein metabolism (which includes anabolism and catabolism). Also significant were a series of annotations associated with negative regulation of cell signaling (italics). The complete list is shown in Supplementary file 6e; top annotations are shown here with duplicates removed.
| Category | Description | FDR value |
|---|---|---|
| GO Process | Negative regulation of cellular protein metabolic process | 1.91E-06 |
| UniProt Keywords | Protease inhibitor | 9.20E-06 |
| SMART Domains | SERine Proteinase INhibitors | 2.60E-05 |
| GO Process | Negative regulation of catalytic activity | 6.73E-05 |
| GO Process | Negative regulation of nitrogen compound metabolic process | 7.86E-05 |
| GO Process | Negative regulation of cellular metabolic process | 7.86E-05 |
| InterPro Domains | Serpin superfamily | 1.10E-04 |
| GO Process | Negative regulation of molecular function | 1.30E-04 |
| GO Function | Enzyme inhibitor activity | 1.50E-04 |
| UniProt Keywords | Serine protease inhibitor | 1.50E-04 |
| GO Process | Negative regulation of macromolecule metabolic process | 1.90E-04 |
| GO Process | Negative regulation of protein modification process | 2.60E-04 |
| GO Process | Negative regulation of hydrolase activity | 2.80E-04 |
| GO Function | Serine-type endopeptidase inhibitor activity | 6.60E-04 |
| GO Process | Negative regulation of phosphate metabolic process | 7.40E-04 |
| GO Function | Endopeptidase inhibitor activity | 8.50E-04 |
| GO Process | Negative regulation of endopeptidase activity | 0.001 |
| GO Process | Negative regulation of intracellular signal transduction | 0.0014 |
| GO Process | Negative regulation of protein phosphorylation | 0.0014 |
| GO Process | Regulation of protein metabolic process | 0.0015 |
| GO Process | Negative regulation of MAPK cascade | 0.0061 |
| GO Process | Negative regulation of cellular process | 0.0063 |
| GO Process | Negative regulation of biological process | 0.0071 |
Summary of CHIKV foot swelling results and GZMA’s protease bioactivity
The CHIKV foot swelling data so far is summarized in Figure 7a and argues that the 6N background, which includes a functional Nnt gene, rather than the absence of GZMA expression, causes the ameliorated foot swelling in Gzma-/- mice (Figure 1a). The presence of an intact Nnt gene can itself reduce foot swelling, although other 6N background genes in Gzma-/- mice also contribute (Figure 4e). MitoTEMPO treatment and NNT activity share anti-inflammatory activities (Figure 4e), presumably because both are involved in mitochondrial ROS mitigation. As CHIKV-infected GzmaS211A mice (on a 6J background) show no reduction in foot swelling, the data argues that GZMA is not a major player in CHIKV arthritis.
Figure 7. Summary of chikungunya virus (CHIKV) foot swelling and granzyme A (GZMA) bioactivity, and the poor concordance between the two.
(a) Summary of foot swelling results. 6N and Gzma-/- mice have a 6N or mixed 6J/6N background and have an intact Nnt gene and show reduced CHIKV-induced foot swelling. MitoTEMPO treatment also reduced foot swelling. 6J, GzmaS211A, and 6N∆Nnt8-12 mice are all missing exons 8–12 of Nnt and show increased foot swelling. (b) Summary of GZMA bioactivity. Using polyinosinic:polycytidylic acid (poly(I:C)) to induce high levels of GZMA secretion from NK cells, studies in GzmaS211A mice illustrated that proteolytically active circulating GZMA promotes certain immune-stimulating/pro-inflammatory responses (dominant transcription factor upstream regulators (USRs) are shown; Figure S11a). No clear consensus has emerged regarding the molecular target(s) of GZMA (??); two potential extracellular candidate targets are shown; protease activated receptors and pro-IL-1. (c) Low overlap between CHIKV and GZMA induced differentially expressed genes (DEGs). DEGs upregulated in feet by CHIKV infection of 6J mice on days 2 and 7 (a low stringency filter of q < 0.05 was applied to the all gene lists in Supplementary file 2f to provide these DEGs) were compared with the DEGs upregulated in feet by active GZMA in 6J mice 6 hr after poly(I:C) treatment (i.e., downregulated in GzmaS211A mice; Supplementary file 6b). Overlapping genes (n = 27 and 20) are listed in Supplementary file 6i. The mean peak levels of serum GZMA for each group are shown below the Venn diagrams. (d) Cytokine USRs for GZMA vs. CHIKV. A series of cytokine USRs were induced by proteolytically active GZMA (i.e., upregulated for 6J + polyIC vs. GzmaS211A + polyIC; Supplementary file 6g, column R). The same USRs were significantly more upregulated by CHIKV infection (Supplementary file 2i). Data for GZMA (orange) and CHIKV (yellow) are plotted with bubble size representing number of molecules in dataset.
The GzmaS211A poly(I:C) data is summarized in Figure 7b. Poly(I:C) induces high levels of circulating GZMA, which is potentiated by type I IFNs, with NK cells the likely source of GZMA (Schanoski et al., 2019). IPA of RNA-Seq data comparing GzmaS211A + poly(I:C) vs. 6J + poly(I:C) supports the view that GZMA’s proteolytic function is required for its bioactivity (Schanoski et al., 2019; Figure 6b, Table 1). Circulating proteolytically active GZMA promotes certain immune-stimulating and pro-inflammatory activities (Schanoski et al., 2019; Shimizu et al., 2019; van Daalen et al., 2020; Wensink et al., 2015), with STAT6, STAT3, NFATC2, JUN, NFKB1, and STAT1 identified as dominant transcription factor USRs (Figures 1 and 7b; Figure 6—figure supplement 4a). Consensus regarding the molecular target(s) of extracellular GZMA’s protease activity (Figure 7b, indicated as ??) remains elusive, but may include protease-activated receptors (Hansen et al., 2005; Sower et al., 1996; Suidan et al., 1994; Suidan et al., 1996) and/or pro-IL-1 Hildebrand et al., 2014; the latter can become extracellular when cells lyse (Afonina et al., 2015).
Minor role for GZMA in CHIKV infection and arthritis
If proteolytically active GZMA is present and has immune-stimulating and pro-inflammatory activities (Figure 7b), why does it have no significant role in driving the overt CHIKV arthritic foot swelling (with no ameliorated foot swelling in GzmaS211A mice, Figure 7a)? Firstly, the serum GZMA levels during CHIKV infection and arthritis (≈0.2 and ≈0.075 ng/ml) were substantially lower than those seen after poly(I:C) treatment (≈20 ng/ml) (Figure 7c), and very much lower than the ≈5 µg of recombinant GZMA-injected intraplantar to generate overt foot swelling in the absence of any other stimuli (Schanoski et al., 2019). Secondly, when the DEGs (q < 0.05) that were upregulated in feet during CHIKV peak viremia (day 2) and peak arthritis (day 7 in the Wilson et al. study) (6J + CHIKV vs. 6J mock infection) were compared with DEGs upregulated by proteolytically active GZMA (6J + poly(I:C) vs. GzmaS211A + poly(I:C); Supplementary file 6b), only small overlaps were evident, 27 genes for day 2 and 20 genes for day 7 (Figure 7c, Supplementary file 6i). Thirdly, although CHIKV infection showed upregulation of many transcription factor and cytokine USRs (Supplementary file 2h), with some of these also upregulated by poly(I:C) treatment (Supplementary file 6g), the magnitude of the effects (by p-value and z-scores) was very much smaller for poly(I:C) treatment (shown for cytokine USRs, Figure 7d). So CHIKV infection (day 2) and arthritis (day 6) stimulate immune and inflammation pathways that overlap with those stimulated by GZMA, but GZMA only plays a minor role in stimulating these pathways during CHIKV viremia (day 2) and arthritis (day 6).
6J SRA accessions with Nnt exon reads inconsistent with a 6J background
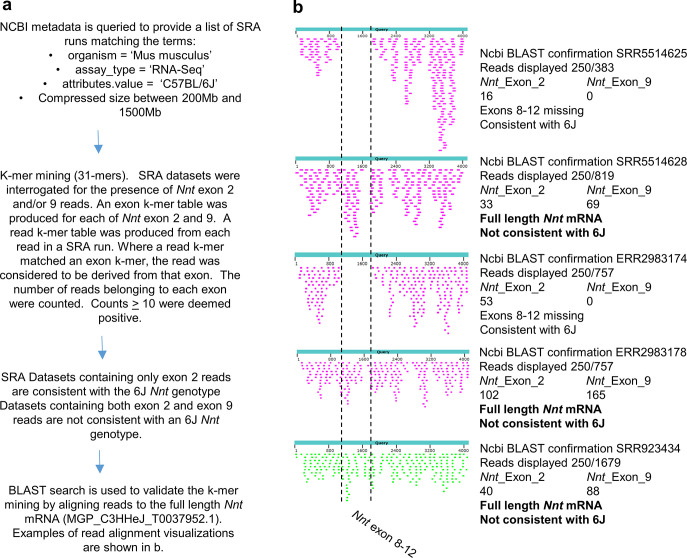
Given the data presented herein and elsewhere (Bourdi et al., 2011; Fontaine and Davis, 2016; Leskov et al., 2017; McCambridge et al., 2019; Mekada et al., 2009; Rao et al., 2020; Ripoll et al., 2012; Rydström, 2006; Toye et al., 2005; Vozenilek et al., 2018) and given that redox regulation affects many cellular processes (Gambhir et al., 2019; Lingappan, 2018; Sun et al., 2020), Nnt emerges as a legitimate focus of concern. In addition, the presence of Nnt exons 8–12 provides a useful genetic marker to illustrate that a mouse strain is not on a pure 6J background, with genetic backgrounds, as shown herein and elsewhere, able to have a profound influence on phenotype (Leskov et al., 2017; Morales-Hernandez et al., 2018; Salerno et al., 2019; Vozenilek et al., 2018; Williams et al., 2021; Wolf et al., 2016). We thus undertook a k-mer mining approach to interrogate the NCBI SRA (Figure 8—figure supplement 1a), which (at the time of analysis) contained 61,443 RNA-Seq Run Accessions listing ‘C57BL/6J’ in the strain field of the metadata.
For ‘C57BL/6J’ Run Accessions, k-mer mining was used to count the number of RNA-Seq reads with sequence homology to Nnt exon 2 or exon 9, with these two exons being of similar length (203 bp for exon 2 and 192 bp for exon 9). RNA-Seq analysis of 6J tissues would ordinarily provide reads for exon 2, whereas the presence of exon 9 reads would be inconsistent with a pure 6J background. A conservative k-mer mining approach was used; (i) only an exact match for ‘C57BL/6J’ in the strain field was allowed, (ii) Run Accessions with small or large compressed file sizes (<200 Mb and >1500 Mb) were excluded, (iii) nucleotide mismatches for the 31-nucleotide k-mers were disallowed, (iv) where there were technical replicates, only one was mined, and (v) a read count of >10 per exon was used as a cutoff. This k-mer mining analysis revealed that 1008 Run Accessions had reads aligning to both Nnt exons 2 and 9, indicating full-length Nnt (Nnt+), which is not consistent with a 6J background (Supplementary file 7a). In contrast, 2469 had exon 2 reads, but no exon 9 reads indicating truncated Nnt (Nnt-), which is consistent with a 6J background (Supplementary file 7b). Lastly, 267 Run Accessions had equivocal results (Supplementary file 7c). Therefore ≈27% (1008 of 3744) of Run Accessions listing ‘C57BL/6J’ in the strain field had sequencing reads not consistent with a 6J background. The k-mer mining approach was validated for a selected group of Run Accessions using NCBI BLAST alignments, which illustrated excellent concordance with the k-mer mining read count data (Supplementary file 7d).
The startlingly high percentage (≈27%) of SRA Accessions listing ‘C57BL/6J’ but having Nnt reads inconsistent with a 6J genetic background argues that the Nnt gene and associated mixed 6N/6J genetic backgrounds are widely underappreciated in a broad range of research areas. It should be noted that a large number of Run Accessions (n = 206,586) list ‘C57BL/6’ in the strain field and thus do not provide information on the substrain (Mekada et al., 2009) being used.
BioProjects comparing mice with truncated Nnt to mice with full-length Nnt
Based on the results of k-mer mining of ‘C57BL/6J’ Run Accessions, BioProjects (n = 373) were grouped into three categories: (i) BioProjects where all the Run Accessions with ‘C57BL/6J’ in the strain field had RNA-Seq reads that were consistent with 6J (Nnt-) (62%), (ii) BioProjects where all the Run Accessions with ‘C57BL/6J’ in the strain field were not consistent with 6J (Nnt+) (23%), and (iii) BioProjects where some Run Accessions with ‘C57BL/6J’ in the strain field were Nnt- and others were Nnt+ (n = 57; 15%). Thus, 38% (15% plus 23%) of BioProjects had Run Accessions with ‘C57BL/6J’ strain listings not compatible with a 6J background.
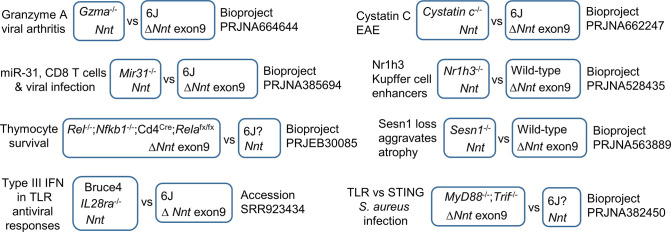
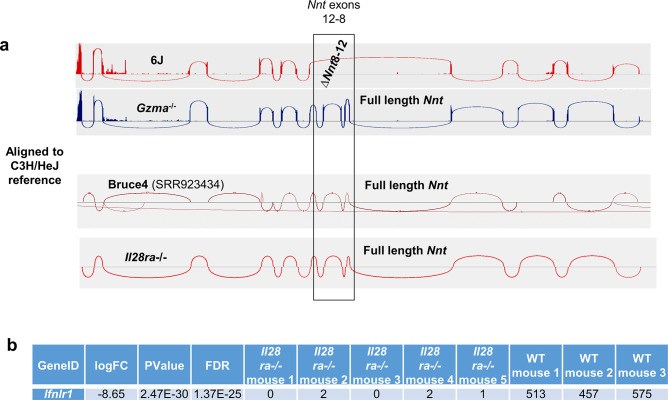
Of the 57 aforementioned BioProjects, 43 had at least one publication associated with the study. These BioProjects were then manually interrogated to identify studies where it was clear (from the paper and the metadata) that comparisons had been made between two groups, where all the Run Accessions in one group were Nnt+, and all the Run Accessions in the other group were Nnt- (Figure 8, Figure 8—figure supplement 1b, Supplementary file 7d and e). Aside from the CHIKV BioProject described herein, several others emerged (Figure 8). For example, Mir31-/- mice showed reduced CD8 T cell dysfunction during chronic viral infection when compared to 6J mice (Moffett et al., 2017); however, Mir31-/- mice were Nnt+ (Figure 8, BioProject PRJNA385694; Supplementary file 7d). Rel-/-;Nfkb1-/-;Cd4Cre;Relafx/fx mice were compared with 6J mice to implicate RIPK1 and IKK in thymocyte survival (Webb et al., 2019); however, the control 6J mice were Nnt+ (Figure 8, BioProject PRJEB30085; Supplementary file 7d). Bruce4 ES cells were reported to be on a 6J background (Ank et al., 2008) and were derived from a B6 mouse strain congenic for the Thy1.1 allele from an NZB mouse (Hughes et al., 2007; Köntgen et al., 1993), with these ES cells clearly Nnt+ (Figure 8, accession SRR923434; Supplementary file 7d). The reported differences between Bruce4 and 6J genomes were thus likely more to do with the background than with genetic instability (Hughes et al., 2007). Il28ra-/- mice were generated using Bruce4 cells and were compared with 6J mice (Ank et al., 2008); however, RNA-Seq analysis of Il28ra-/- mice showed that (like Bruce4 cells) these mice had full-length Nnt (Figure 8—figure supplement 2). Myd88-/-;Trif-/- double knockout mice were compared with 6J mice during infection with Staphylococcus aureus (Scumpia et al., 2017); however, the 6J (wild-type) mice were Nnt+ (Figure 8, BioProject PRJNA382450; Supplementary file 7e). Female Cystatin C-/- mice display significantly lower clinical signs of experimental autoimmune encephalomyelitis (EAE) when compared with 6J mice (Hoghooghi et al., 2020); however, Cystatin C-/- mice were all Nnt+ (Figure 8, BioProject PRJNA662247; Supplementary file 7d). Deletion of Nr1h3 resulted in reduced chromatin access at a large fraction of Kupffer-cell-specific enhancers (Sakai et al., 2019); however, Nr1h3-/-, but not the wild-type control, were all Nnt+ (Figure 8, BioProject PRJNA528435; Supplementary file 7d). Sesn1-/- mice were used to show that loss of Sestrin1 aggravates disuse-induced muscle atrophy when compared with 6J mice (Segalés et al., 2020); however, Sesn1-/- mice were all Nnt+ (Figure 8, BioProject PRJNA563889; Supplementary file 7d).
Figure 8. k-mer mining of BioProjects where Nnt+ mice were compared with Nnt- mice.
The NCBI Sequence Read Archive (SRA) database was interrogated by k-mer mining for BioProjects where (i) some Run Accessions (listing 6J as the mouse strain) had reads compatible with a 6J background (reads for Nnt exon 2, but not exon 9) and (ii) other Run Accessions in that BioProject (listing 6J as the mouse strain) had reads not compatible with a 6J background (reads for Nnt exons 2 and 9). The methodology is described in Figure 8—figure supplement 1a, validated by BLAST alignments (Figure 8—figure supplement 1b), with raw data in Supplementary file 7d and e.
Figure 8—figure supplement 1. k-mer mining methodology and validation.
Figure 8—figure supplement 2. Bruce4 ES cell line and IL28RA-/- mouse Nnt genotypes.
Of the 57 BioProjects containing Nnt+ and Nnt+ Run Accessions, several contained comparisons in which one group contained a combination of Nnt+ and Nnt- Run Accessions, while the other group(s) contained either all Nnt+ or all Nnt- Run Accessions (Supplementary file 7f).
Whether the Nnt differences (or other background gene differences) would have significantly affected the interpretation of phenotypes in the aforementioned studies remains to be established. It is worth noting that herein we have only compared phenotypes of homozygotes (Figure 7a), with intermediate phenotypes potentially seen for heterozygotes (Ronchi et al., 2013). Our k-mer mining analysis also did not distinguish between Nnt+/+ and Nnt+/-, doing so would require extraction of Nnt reads, alignment to the C3H/HeJ genome, and analysis using Sashimi plots (as in Figure 3d). Nevertheless, the data argues that differences in Nnt or other background gene differences are widely underappreciated in a range of research fields and have the potential to compromise a wide range of studies.
Discussion
Despite a large body of literature, no clear consensus has emerged regarding the physiological function of GZMA (see ‘Introduction’). This lack of consensus might now, at least in part, be explained by the extensive use of Gzma-/- mice (Supplementary file 1). We show herein that this mouse strain is on a mixed 6J/6N genetic background and contains a full-length Nnt gene, with both Nnt and other 6N background genes, rather than loss of GZMA expression, responsible for the ameliorated CHIKV arthritis phenotype. Whether all the phenotypes reported for Gzma-/- mice (Supplementary file 1) are compromised by Nnt and/or the mixed background remains unclear and would require new experiments to resolve, similar to those described herein for CHIKV arthritis. However, Nnt has been reported to affect redox regulation and activation in macrophages (Ripoll et al., 2012; Salerno et al., 2019), with an intact Nnt gene conceivably reducing cross-presentation (Dingjan et al., 2016; Nalle et al., 2020) and CD8 T cell responses (Oberkampf et al., 2018). In addition, 6N vs. 6J background differences (as herein) have shown clear phenotypes in a wide range of settings (Leskov et al., 2017; Morales-Hernandez et al., 2018; Rao et al., 2020; Salerno et al., 2019; Vozenilek et al., 2018; Williams et al., 2021; Wolf et al., 2016).
The data from GzmaS211A mice (that were generated using CRISPR on a 6J background) represents the first in vivo assessment of the physiological function of GZMA without the confounding influence of differences in Nnt or other genes associated with the mixed genetic background. The results from this analysis support the view that the physiological activity of GZMA is mediated by its protease activity (Figure 7b; Schanoski et al., 2019). This is an important point because protease-independent functions have been documented for several proteases (Calhan and Seyrantepe, 2017; McNutt et al., 2007; Pu et al., 2019; Szabo et al., 2016). One of the proposed activities for GZMA, the binding to TLR9 and potentiation of TLR9 signaling (Shimizu et al., 2019), was not reported to require GZMA’s protease activity. However, using mice defective in TLR9 signaling, we were unable to find evidence that TLR9 is required for GZMA’s pro-inflammatory activity (Supplementary file 8). Our studies also support the view that secreted extracellular GZMA has biological activity in the absence of perforin (or GZMB) (Figure 6—figure supplement 1). This contrasts with the traditional view of GZMA as a mediator of cell death, which requires perforin to deliver GZMA to the cytoplasm, where a series of target molecules are cleaved (Liesche et al., 2018; Wu et al., 2019; Zhou et al., 2020). Cleavage of SET complex proteins (Mandrup-Poulsen, 2017; Mollah et al., 2017) would similarly require delivery of GZMA to the cytoplasm. Although we cannot formally exclude translocation of circulating GZMA into the cytoplasm via some unknown mechanism, the GzmaS211A RNA-Seq experiment does ostensibly exclude perforin as NK cells only produce perforin (and GZMB) protein after ≈24 hr of appropriate stimulation (Fehniger et al., 2007). Extracellular target candidates for GZMA include protease-activated receptors (Hansen et al., 2005; Sower et al., 1996; Suidan et al., 1994; Suidan et al., 1996) and may also include pro-IL-1 (Hildebrand et al., 2014; Figure 7c) as pro-IL-1 can become extracellular when cells lyse (Afonina et al., 2015). Overall one might speculate that in such settings NK-derived GZMA synergizes with type I IFN responses to act as a systemic alarmin (Figure 7b). GzmaS211A mice will provide an invaluable tool for future studies into further refining our understanding of the role and molecular targets of GZMA.
The ability to reduce CHIKV arthritis in 6J mice with MitoTEMPO might suggest such antioxidant drugs have potential utility as anti-inflammatory treatments for alphaviral arthritides (Suhrbier et al., 2012; Zaid et al., 2021). However, MitoTEMPO treatment may simply be correcting (at least in part, Figure 4e) the Nnt defect in 6J mice by scavenging excess mitochondrial ROS arising from the loss of functional NNT (Ward et al., 2020). The argument that similar antioxidant treatments would be effective in human diseases may thus not be overly compelling, given that most humans have a functional Nnt gene. Perhaps noteworthy is that >300 papers listed in PubMed use MitoTEMPO in 6J mice, with many reporting effective disease amelioration with MitoTEMPO treatments, for example (Aoyama et al., 2012; Li et al., 2018; To et al., 2020; Vincent et al., 2020; Wu et al., 2020). Unfortunately, antioxidants have not shown clear benefits in human clinical trials (Casas et al., 2020; Jiang et al., 2020; Kovacic, 2020; Steinhubl, 2008).
The Jackson Laboratory generated the 6J inbred mouse strain in the 1920s–1930s, with this mouse strain the most frequently used mouse strain in biomedical research. Although differences in the Nnt gene (or other background genes) have previously been reported as underappreciated in metabolism research (Fontaine and Davis, 2016), the data herein and elsewhere argue that this issue extends to other areas of research (Bourdi et al., 2011; Leskov et al., 2017; McCambridge et al., 2019; Morales-Hernandez et al., 2018; Rao et al., 2020; Ripoll et al., 2012; Salerno et al., 2019; Vozenilek et al., 2018; Williams et al., 2021; Wolf et al., 2016), with age effects also reported (Ghosh et al., 2014; Ubaida-Mohien et al., 2019). Of concern was that ≈27% of SRA Run Accessions and ≈38% of BioProjects listing C57BL/6J as the mouse strain had Nnt sequence data not consistent with a pure 6J background. Mouse strain listing errors or inadequate backcrossing to 6J would thus appear to be common for SRA RNA-Seq submissions. The full extent to which Nnt and/or genetic backgrounds have complicated interpretation of knockout mouse studies remains to be addressed, but may require extensive new experiments such as those described herein for GZMA.
Materials and methods
Key resources table.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (chikungunya virus) | CHIKV | Dr. P. Roques (CEA, Fontenay-aux-Roses, France) | KT449801.1 | Isolate LR2006-OPY1 |
| Chemical compound, drug | TRIzol | Sigma-Aldrich | Cat# 15596026 | |
| Chemical compound, drug | MitoTEMPO | Sigma-Aldrich | Cat#1334850-99-5 | |
| Commercial assay, kit | TruSeq RNA Sample Prep Kit (v2) | Illumina | SCR_010233 | |
| Commercial assay, kit | TruSeq Stranded mRNA library preparation kit | Illumina | SCR_010233 | |
| Commercial assay, kit | QIAamp DNA Micro Kit | QIAGEN | Cat# 56304 | |
| Commercial assay, kit | iScript cDNA Synthesis Kit | Bio-Rad | Cat# 1708890 | |
| Commercial assay, kit | Q5 Hot Start High-Fidelity DNA Polymerase | NEB | Cat# M0493S | Enzyme |
| Other | Illumina HiSeq 2000 Sequencer | Illumina | RRID:SCR_010233 | Sequencing platform |
| Other | NextSeq 550 | Illumina | RRID:SCR_016381 | Sequencing platform |
| Other | NovaSeq 6000 | Illumina | RRID:SCR_016387 | Sequencing platform |
| Software, algorithm | k-mer_mining_SRA | GitHub | https://github.com/CameronBishop/k-mer_mining_SRA | |
| Cell line (Cercopithecus aethiops) | Vero cells | ATCC | RRID:CVCL_0059 | |
| Cell line (Aedes albopictus) | C6/36 cells | ATCC | RRID:CVCL_Z230 | |
| Strain, strain background (Mus musculus) | C57BL/6J | Animal Resources Centre (Canning Vale, WA, Australia) | IMSR_JAX:000664 | |
| Strain, strain background (M. musculus) | C57BL/6N | The Jackson Laboratory | Stock no. 005304 | |
| Strain, strain background (M. musculus) | C57BL/6-Gzma-/- | Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia | Knockout mouse | |
| Strain, strain background (M. musculus) | C57BL/6J-Gzmb-/- | Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia | Knockout mouse | |
| Strain, strain background (M. musculus) | C57BL/6J-GzmaS211A | The Australian Phenomics Network, Monash University, Melbourne, Australia (this paper) | Mutant mouse | |
| Strain, strain background (M. musculus) | C57BL/6N∆Nnt8-12 | The Australian Phenomics Network, Monash University, Melbourne, Australia (this paper) | Knockout mouse | |
| Strain, strain background (M. musculus) | C57BL/6J-Ifnar-/- | Dr P. Hertzog (Monash University, Melbourne, Australia) | Knockout mouse | |
| Strain, strain background (M. musculus) | C57BL/6-Il28ra-/- | Bristol-Myers Squibb (PMID:25901316) | Knockout mouse | |
| Sequence-based reagent | Nnt_RTPCR_F1 | This paper | PCR primers | AACAGTGCAAGGAGGTGGAC |
| Sequence-based reagent | Nnt_RTPCR_R1 | This paper | PCR primers | GTGCCAAGGTAAGCCACAAT |
| Software, algorithm | FastQC | Babraham Institute | RRID:SCR_014583 | |
| Software, algorithm | MultiQC | PMID:27312411 | RRID:SCR_014982 | |
| Software, algorithm | Cutadapt | DOI: https://doi.org/10.14806/ej.17.1.200 | RRID:SCR_011841 | |
| Software, algorithm | STAR | PMID:23104886 | RRID:SCR_004463 | |
| Software, algorithm | RSEM | PMID:21816040 | RRID:SCR_013027 | |
| Software, algorithm | EdgeR | PMID:27280887 | RRID:SCR_012802 | |
| Software, algorithm | ‘Ingenuity Pathway Analysis’ (IPA) | QIAGEN | RRID:SCR_008653 | |
| Software, algorithm | Cytoscape | PMID:14597658 | RRID:SCR_003032 | |
| Software, algorithm | STRING | PMID:30476243 | RRID:SCR_005223 | |
| Software, algorithm | ‘Gene Set Enrichment Analysis’ (GSEA) | PMID:16199517 | RRID:SCR_003199 | |
| Software, algorithm | ‘Integrative Genomics Viewer‘ (IGV) | PMID:21221095 | RRID:SCR_011793 | |
| Software, algorithm | minimap2 | PMID:29750242 | RRID:SCR_018550 | |
| Software, algorithm | BigQuery | Google Cloud Platform | RRID:SCR_001011 | |
| Software, algorithm | fasterq-dump | SRA tool kit | sra-tools v 2.9.1 |
Cell lines and CHIKV
Vero (ATCC#: CCL-81) and C6/36 cells (ATCC# CRL-1660) were cultured as described (Nguyen et al., 2020). Cells were checked for mycoplasma using MycoAlert Mycoplasma Detection Kit (Lonza, Basel, Switzerland). FBS was checked for endotoxin contamination using RAW264.7-HIV-LTR-luc cells (Johnson et al., 2005) before purchase. CHIKV (isolate LR2006-OPY1; GenBank KT449801.1; DQ443544.2) was a kind gift from Dr. P. Roques (CEA, Fontenay-aux-Roses, France), was propagated in C6/36 cells, and titers determined by CCID50 assays (Nguyen et al., 2020). Virus was also checked for mycoplasma (La Linn et al., 1995).
Mice and CHIKV infections
C57BL/6J mice were purchased from Animal Resources Centre (Canning Vale, WA, Australia). C57BL/6N mice were purchased from The Jackson Laboratory (stock no. 005304). Gzma-/- mice were generated as described (Ebnet et al., 1995) and were provided by the Peter MacCallum Cancer Centre, Melbourne, Victoria, Australia. Gzmb-/- mice were generated as described (Müllbacher et al., 1999) and were backcrossed onto C57BL/6J mice a total of 12 times and were provided by the Peter MacCallum Cancer Centre. The Australian Phenomics Network, Monash University, Melbourne, Australia, used CRISPR to generate (i) GzmaS211A mice on a 6J background and (ii) 6N∆Nnt8-12 mice on a 6N background. Ifnar-/- mice (Yan et al., 2020) were kindly provided by Dr P. Hertzog (Monash University). Il28ra-/- mice (Ank et al., 2008) were kindly provided by Bristol-Myers Squibb (Souza-Fonseca-Guimaraes et al., 2015). All GMO mice were bred in-house at QIMR Berghofer MRI.
Female mice 6–8 weeks old were infected with 104 CCID50 CHIKV (isolate LR2006 OPY1) s.c. into each hind foot, with foot measurements and viremia determined as described (Gardner et al., 2010; Nguyen et al., 2020; Prow et al., 2018).
RNA-Seq of feet of CHIKV-infected GMO mice
Mice were infected with CHIKV, feet collected on day 6, and RNA samples prepared as described previously (Hazlewood et al., 2021; Rawle et al., 2021; Wilson et al., 2017) with minor modifications. Briefly, library preparation and sequencing were conducted by the Australian Genome Research Facility (Melbourne, Australia) (Hazlewood et al., 2021) or were conducted in-house (Rawle et al., 2021). RNA concentration and quality was measured using TapeStation D1K TapeScreen assay (Agilent). cDNA libraries were prepared using a TruSeq RNA Sample Prep Kit (v2) (Illumina Inc, San Diego, USA), which included isolation of poly-adenylated RNA using oligo-dt beads or total RNA library preparation with rRNA depletion (NEBNext Ultra II Directional RNA Library Prep Kit for Illumina and NEBNext rRNA Depletion Kit v2). Paired end reads were generated using the Illumina HiSeq 2000 Sequencer (Illumina Inc) (100 bp) or Illumina NextSeq 550 platform (75 bp). Per-base sequence quality for >90% bases was above Q30 for all samples. Raw sequencing reads were assessed using FastQC (v0.11.8) (Simons, 2010) and MultiQC (v1.7) (Ewels et al., 2016) and trimmed using Cutadapt (v2.3) (Martin, 2011) to remove adapter sequences and low-quality bases. Trimmed reads were aligned to the GRCm38 primary assembly reference and the GENCODE M23 gene model using STAR aligner (v2.7.1a) (Dobin et al., 2013), with more than 95% of reads mapping to protein coding regions. Counts per gene were generated using RSEM (v1.3.1) (Li and Dewey, 2011), and differential expression analysis was undertaken using EdgeR in Galaxy (Varet et al., 2016) with default settings and a count sum >1 filter applied. Counts were normalized using the TMM method and modeled using the likelihood ratio test, glmLRT().
Bioinformatic analyses
Pathway analysis of DEGs in direct and indirect or direct-only interactions was investigated using IPA (QIAGEN; Shannon et al., 2003). Enrichment for biological processes, molecular functions, KEGG pathways, and other gene ontology categories in DEG lists was elucidated using the STRING database (Szklarczyk et al., 2019) in Cytoscape (v3.7.2) (Shannon et al., 2003). GSEA (Subramanian et al., 2005) was performed on a desktop application (GSEA v4.0.3) (http://www.broadinstitute.org/gsea/) to look for enrichment of DEGs in full gene sets preranked by fold change.
WGS of Gzma-/- mice
QIAamp DNA Micro Kit (QIAGEN) was used to purify genomic DNA from Gzma-/- mice spleen as per the manufacturer’s instructions. DNA was sent to the Australian Genome Research Facility (AGRF) for WGS using the Illumina NovaSeq platform with 150 bp paired end reads. The primary sequence data was generated using the Illumina bcl2fastq 2.20.0.422 pipeline. Reads were mapped to the mm10 genome assembly (GRCm38) using BWA-mem, and .bam files were provided. Mapped reads were viewed in Integrative Genomics Viewer (IGV) (Robinson et al., 2011) and 6N features identified manually based on previous publications (Mekada et al., 2015; Simon, 2013).
Alignment to mouse genomes
FastQ files were generated as described or were downloaded from the SRA using Aspera. Reads were trimmed using Cutadapt (Martin, 2011) and mapped using STAR aligner (v2.7.1a) for RNA-Seq or minimap 2 (Li, 2018) for WGS data. IGV was used to visualize reads mapping to the Nnt gene after mapping to the GRCm38 primary assembly reference for the truncated version of the gene and to the mouse C3H_HeJ_v1 reference (GCA_001632575.1) to observe full-length Nnt.
Nnt RT-PCR
RT-PCR was undertaken essentially as described using the primer set (F1 AACAGTGCAAGGAGGTGGAC, R1 GTGCCAAGGTAAGCCACAAT) (Integrated DNA Technologies; Huang et al., 2006). RNA was extracted from testes using TRIzol (Sigma-Aldrich) according to the manufacturer’s instructions. cDNA was generated using iScript cDNA Synthesis Kit (Bio-Rad) and Q5 Hot Start High-Fidelity DNA Polymerase (NEB) was used for PCR.
Histology
Histology and H&E staining were undertaken as described (Rawle et al., 2021). Sections were scanned using Aperio AT Turbo (Aperio, Vista, CA) and analyzed using Aperio Image-Scope software (Leica Biosystems, Mt Waverley, Australia) (v10). Quantitation using Positive Pixel Count v9 was used to generate blue/red pixel ratios as a measure of leukocyte infiltrates, as described (Poo et al., 2014).
MitoTEMPO treatment
Mice were injected i.v. daily, on the indicated day post CHIKV infection, with 62.5 µg of MitoTEMPO (Sigma-Aldrich) in 150 µl of PBS.
RNA-Seq of poly(I:C) injection for GzmaS211A vs. 6J mice
Age-matched female GzmaS211A and 6J mice were injected i.v. with 250 µg of poly(I:C) in 150 µl of PBS. After 6 hr, mice were euthanized, spleen and whole feet were harvested, and RNA isolated as described previously (Prow et al., 2019; Wilson et al., 2017). Three RNA pools were generated for each mouse strain, whereby each pool contained equal amounts of RNA from four feet from four mice, or two spleens from two mice. RNA-Seq of polyadenylated RNA was then undertaken in-house at QIMR Berghofer MRI. RNA integrity was assessed using the TapeStation system (Agilent Technologies), and libraries were prepared using the TruSeq Stranded mRNA library preparation kit (Illumina). Sequencing was performed on the Illumina NextSeq 550 platform with 75 bp paired end reads. Per-base sequence quality for >92% bases was above Q30 for all samples. Raw sequencing reads were then processed as above.
k-mer mining
An exact-match (31 mer) k-mer mining approach was used to identify RNA-Seq read files (Accessions) with C57BL/6J mice listed as the mouse strain, but where Nnt reads were incompatible with 6J background. Metadata associated with the National Center for Biotechnology Information’s SRA was screened using the Google Cloud Platform’s BigQuery service with the Structured Query Language (SQL) command: SELECT m.bioproject, m.acc, m.sample_name, m.platform, m.mbytes, m.mbases FROM nih-sra-datastore.sra.metadata as m, UNNEST (m.attributes) as a WHERE m.organism = ‘Mus musculus’ and m.assay_type = ‘RNA-Seq’ and a.v = ‘C57BL/6J.’ Technical replicates for the same sample were collapsed by taking only the first accession for each Biosample. Accessions were then filtered on the basis of their compressed size so that only those between 200 Mb and 1500 Mb were retained; we found that read files of this size provided adequate sequencing depth to detect Nnt exon reads. Accessions were sorted according to BioProject and used as input for a bioinformatics pipeline executed on the Google Cloud Platform, which allowed access to the ‘SRA in the cloud’ database. A copy of our Bash script to automate the pipeline is available at https://github.com/CameronBishop/k-mer_mining_SRA (Bishop, 2021). Accession read files were converted to FastQ format using fasterq-dump (SRA tool kit). BBduk version 38.87 (Bushnell, 2020) was used with default parameters to screen each read for sequence homology to exons 2 and 9 of the Nnt gene. Reads sharing at least one 31-mer with either exon were counted as a ‘match’ for that exon. FastQ files with at least 10 matches to exon 2 and 0 matches to exon 9 were classed as consistent with a 6J genotype (truncated Nnt), while FastQ files with at least 10 matches to each exon were classed as not consistent with a 6J genotype. Results were curated using BigQuery to confirm that for each Accession’s metadata entry the ‘strain_sam’ field (or equivalent) of the metadata ‘Attributes’ table was listed as C57BL/6J.
BioProjects were identified where some read files contained exon 2 reads and no exon 9 reads, whereas others contained both exon 2 and exon 9 reads. The literature and Gene Expression Omnibus submissions associated with these BioProjects were then consulted to identify BioProjects where mice with full-length Nnt had been compared with mice with truncated Nnt.
Determination of GZMA levels in mouse serum samples
Mouse serum was collected in Microvette 500 Z gel tubes (Sarstedt) with GZMA levels determined using a GZMA ELISA kit (MyBioSource, San Diego, CA, MBS704766) according to the manufacturer’s instructions.
Statistics
Statistical analysis of experimental data was performed using IBM SPSS Statistics for Windows, version 19.0. The t-test was performed when the difference in variances was <4, skewness was >-2, and kurtosis was <2, otherwise the Kolmogorov–Smirnov test was used.
Data, code, and GMO mouse availability
All data are provided in the article and accompanying supplementary files. Raw sequencing data generated for this publication has been deposited in the NCBI SRA. RNA-Seq NCBI BioProjects: (i) 6J + CHIKV vs. 6J mock infection, day 2 and day 7 feet (PRJNA431476); Gzma-/- + CHIKV vs. 6J + CHIKV, day 6 feet (PRJNA664644); (ii) 6N∆Nnt8-12 + CHIKV vs. 6N + CHIKV, day 6 feet (PRJNA779556); (iii) 6J + MitoTEMPO + CHIKV vs. 6J + PBS + CHIKV (PRJNA779556); and (iv) 6J GzmaS211A + poly(I:C) vs. 6J + poly(I:C) feet and spleen 6 hr (PRJNA666748). WGS of Gzma-/- mice NCBI BioProject PRJNA664888.
A copy of our code to automate the k-mer mining pipeline is available at https://github.com/CameronBishop/k-mer_mining_SRA (Bishop, 2021; copy archived at swh:1:rev:372d29b02972d96e8eff7b6c431883ea8dfb5c50). CRISPR GMO mouse lines 6N∆Nnt8-12 and GzmaS211A are available on request.
Acknowledgements
We thank the following staff at QIMR Berghofer MRI for their assistance; animal house staff, Dr I Anraku (BSL3 facility manager), Dr R Johnston (Bioinformatics), and Dr Viviana Lutzky (for proof reading). We thank Dr Dion Kaiserman (Monash University, Australia) for supplying recombinant GZMA. We also thank Dr Mark Heise (University of North Carolina) for valuable discussions.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Andreas Suhrbier, Email: Andreas.Suhrbier@qimrberghofer.edu.au.
Hiroshi Takayanagi, The University of Tokyo, Japan.
Carlos Isales, Medical College of Georgia at Augusta University, United States.
Funding Information
This paper was supported by the following grants:
National Health and Medical Research Council APP1141421 to Andreas Suhrbier.
National Health and Medical Research Council APP1173880 to Andreas Suhrbier.
Additional information
Competing interests
No competing interests declared.
No competing interests declared.
Author contributions
Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Visualization, Writing – review and editing.
Investigation.
Data curation, Formal analysis, Visualization, Writing – review and editing.
Data curation, Formal analysis, Visualization, Writing – review and editing.
Investigation.
Investigation, Conceptualization.
Conceptualization, Funding acquisition, Methodology, Resources, Writing – review and editing.
Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Visualization, Writing – original draft, Writing – review and editing.
Ethics
All mouse work was conducted in accordance with the "Australian code for the care and use of animals for scientific purposes" as defined by the National Health and Medical Research Council of Australia. Mouse work was approved by the QIMR Berghofer Medical Research Institute animal ethics committee (P2235 A1606-618M), with infectious CHIKV work conducted in a biosafety level-3 (PC3) facility at the QIMR Berghofer MRI (Australian Department of Agriculture, Water and the Environment certification Q2326 and Office of the Gene Technology Regulator certification 3445).
Additional files
Compilation of studies employing Gzma-/- or Gzma-/- Gzmb-/- double KO mice that reveal a phenotype or do not show a phenotype.
Datasets for RNA-Seq and Ingenuity Pathway Analysis (IPA) of mice feet day 6 post infection from CHIKV-infected Gzma-/- mice vs. CHIKV-infected 6J mice and of mice feet day 7 post infection from 6J + CHIKV vs. 6J mock infection.
Genetic differences potentially involved in inflammation or arthritis are indicated. Nnt not included. Differences in introns not included.
Datasets for RNA-Seq and Ingenuity Pathway Analysis (IPA) of mice feet day 6 post infection; 6NNnt∆8-12+ CHIKV vs. 6N + CHIKV.
Datasets for RNA-Seq and Ingenuity Pathway Analysis (IPA) of mice feet day 6 post infection; 6J + MitoTEMPO + CHIKV vs. 6J + PBS + CHIKV.
Datasets for RNA-Seq, Ingenuity Pathway Analysis (IPA), and Cytoscape analyses of mice feet and spleen taken 6 hr after injection with poly(I:C); GzmaS211A vs. 6J.
Datasets for exact-match (31 mer) k-mer mining approach to identify full-length Nnt reads (6N background) in accessions listing C57BL/6J mice as the mouse strain.
(a) Tlr9-/- and 6J mice were injected intraplantar into the feet with 5 µg recombinant mouse granzyme A (GZMA) in 20 µl and foot swelling measured over time as described (Schanoski et al., 2019) (n = 4 mice and four feet per group; statistics by Kolmogorov–Smirnov tests). Tlr9-/- mice were derived from 129/Ola × C57BL/6F1 progeny (http://www.myv.ne.jp/obs/index.files/tlr_eng.htm). (b) As for (a) using Tlr9M7Btlr/Mmjax and 6J mice. Tlr9M7Btlr/Mmjax mice have a Tlr9 missense point mutation and do not respond to oligonucleotides containing CpG motifs (https://www.jax.org/strain/014534). (c) Tlr9-/- mice (like 6J) do not encode the full Nnt gene. (d) Female 8–10-week-old Tlr9-/- and 6J mice (n = 6 mice and 12 feet per group) were infected with chikungunya virus (CHIKV) and feet measured over time. Statistics by Kolmogorov–Smirnov tests. (e) Female C57BL/6J-Tlr9M7Btlr/Mmjax mice (n = 6 mice and 12 feet per group) were infected as for (d). (f) Viremia for the mice in (d). After GZMA injection, Tlr9-/- mice showed increased foot swelling (a), whereas C57BL/6J-Tlr9M7Btlr/Mmjax mice showed no significant difference (b). Tlr9-/- mice also have the Nnt deletion (c); however, they are on a mixed 129/Ola and C57BL/6 background (Hemmi et al. 2000), with 129/SvJ mice showing increased inflammatory infiltrates in certain settings (Hoover-Plow et al., 2008). After CHIKV infection, foot swelling was again increased in Tlr9-/- (d), but not Tlr9M7Btlr/Mmjax mice (e). Tlr9-/- mice did not show an increased viremia (f). These data do not support a contention that TLR9 is required for GZMA’s bioactivity.
Source data for DNA gel images in Figure 3e, Figure 1—figure supplement 1b,c, and Figure 4—figure supplement 1e.
Data availability
Five supplementary files have been provided which constitute source data for all the results cited in the manuscript. Raw sequencing data was uploaded to SRA: BioProject accessions: PRJNA666748, PRJNA664888, PRJNA664644, PRJNA779556.
The following datasets were generated:
Suhrbier A. 2020. RNA-Seq of Granzyme A proteolytic site mutant mice injected with Poly (I:C) NCBI BioProject. PRJNA666748
Suhrbier A. 2020. Whole genome sequencing of a Granzyme A knock out mouse. NCBI BioProject. PRJNA664888
Suhrbier A. 2020. RNA-Seq of Granzyme A knockout mice infected with chikungunya Virus. NCBI BioProject. PRJNA664644
Suhrbier A. 2021. RNA_Seq of C57BL/6N Nnt knockout mice and MitoTEMPO treatment of C57BL/6J mice infected with CHIKV. NCBI BioProject. PRJNA779556
References
- Afonina IS, Müller C, Martin SJ, Beyaert R. Proteolytic Processing of Interleukin-1 Family Cytokines: Variations on a Common Theme. Immunity. 2015;42:991–1004. doi: 10.1016/j.immuni.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H, Klucher K, Paludan SR. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. Journal of Immunology. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- Aoyama T, Paik Y-H, Watanabe S, Laleu B, Gaggini F, Fioraso-Cartier L, Molango S, Heitz F, Merlot C, Szyndralewiez C, Page P, Brenner DA. Nicotinamide adenine dinucleotide phosphate oxidase in experimental liver fibrosis: GKT137831 as a novel potential therapeutic agent. Hepatology. 2012;56:2316–2327. doi: 10.1002/hep.25938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop C. k-mer_mining_SRA. swh:1:rev:372d29b02972d96e8eff7b6c431883ea8dfb5c50Software Heritage. 2021 https://archive.softwareheritage.org/swh:1:dir:439009609bd1ec2052fb1f2cbceab60dc48f5622;origin=https://github.com/CameronBishop/k-mer_mining_SRA;visit=swh:1:snp:0cb753db1380263f4ef875b7e076de0778a176d5;anchor=swh:1:rev:372d29b02972d96e8eff7b6c431883ea8dfb5c50
- Boding L, Hansen AK, Meroni G, Johansen BB, Braunstein TH, Bonefeld CM, Kongsbak M, Jensen BAH, Woetmann A, Thomsen AR, Odum N, von Essen MR, Geisler C. Midline 1 directs lytic granule exocytosis and cytotoxicity of mouse killer T cells. European Journal of Immunology. 2014;44:3109–3118. doi: 10.1002/eji.201344388. [DOI] [PubMed] [Google Scholar]
- Boding L, Hansen AK, Meroni G, Levring TB, Woetmann A, Ødum N, Bonefeld CM, Geisler C. MID2 can substitute for MID1 and control exocytosis of lytic granules in cytotoxic T cells. APMIS. 2015;123:682–687. doi: 10.1111/apm.12402. [DOI] [PubMed] [Google Scholar]
- Bourdi M, Davies JS, Pohl LR. Mispairing C57BL/6 substrains of genetically engineered mice and wild-type controls can lead to confounding results as it did in studies of JNK2 in acetaminophen and concanavalin A liver injury. Chemical Research in Toxicology. 2011;24:794–796. doi: 10.1021/tx200143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B. BBMap. Sourceforge. 2020 https://jgi.doe.gov
- Calhan OY, Seyrantepe V. Mice with Catalytically Inactive Cathepsin A Display Neurobehavioral Alterations. Behavioural Neurology. 2017;2017:4261873. doi: 10.1155/2017/4261873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas AI, Nogales C, Mucke HAM, Petraina A, Cuadrado A, Rojo AI, Ghezzi P, Jaquet V, Augsburger F, Dufrasne F, Soubhye J, Deshwal S, Di Sante M, Kaludercic N, Di Lisa F, Schmidt HHHW. On the Clinical Pharmacology of Reactive Oxygen Species. Pharmacological Reviews. 2020;72:801–828. doi: 10.1124/pr.120.019422. [DOI] [PubMed] [Google Scholar]
- Dempoya J, Matsumiya T, Imaizumi T, Hayakari R, Xing F, Yoshida H, Okumura K, Satoh K. Double-stranded RNA induces biphasic STAT1 phosphorylation by both type I interferon (IFN)-dependent and type I IFN-independent pathways. Journal of Virology. 2012;86:12760–12769. doi: 10.1128/JVI.01881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingjan I, Verboogen DR, Paardekooper LM, Revelo NH, Sittig SP, Visser LJ, Mollard GF, Henriet SS, Figdor CG, Ter Beest M, van den Bogaart G. Lipid peroxidation causes endosomal antigen release for cross-presentation. Scientific Reports. 2016;6:22064. doi: 10.1038/srep22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djeu JY, Heinbaugh JA, Holden HT, Herberman RB. Augmentation of mouse natural killer cell activity by interferon and interferon inducers. Journal of Immunology. 1979;122:175–181. [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebnet K, Hausmann M, Lehmann-Grube F, Müllbacher A, Kopf M, Lamers M, Simon MM. Granzyme A-deficient mice retain potent cell-mediated cytotoxicity. The EMBO Journal. 1995;14:4230–4239. doi: 10.1002/j.1460-2075.1995.tb00097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, Ley TJ. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Fontaine DA, Davis DB. Attention to Background Strain Is Essential for Metabolic Research: C57BL/6 and the International Knockout Mouse Consortium. Diabetes. 2016;65:25–33. doi: 10.2337/db15-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman H, Shimomura K, Cox RD, Ashcroft FM. Nicotinamide nucleotide transhydrogenase: a link between insulin secretion, glucose metabolism and oxidative stress. Biochemical Society Transactions. 2006;34:806–810. doi: 10.1042/BST0340806. [DOI] [PubMed] [Google Scholar]
- Gambhir L, Sharma V, Kandwal P, Saxena S. Perturbation in cellular redox homeostasis: Decisive regulator of T cell mediated immune responses. International Immunopharmacology. 2019;67:449–457. doi: 10.1016/j.intimp.2018.12.049. [DOI] [PubMed] [Google Scholar]
- García-Laorden MI, Stroo I, Blok DC, Florquin S, Medema JP, de Vos AF, van der Poll T. Granzymes A and B Regulate the Local Inflammatory Response during Klebsiella pneumoniae Pneumonia. Journal of Innate Immunity. 2016;8:258–268. doi: 10.1159/000443401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Laorden MI, Stroo I, Terpstra S, Florquin S, Medema JP, van T Veer C, de Vos AF, van der Poll T. Expression and Function of Granzymes A and B in Escherichia coli Peritonitis and Sepsis. Mediators of Inflammation. 2017;2017:4137563. doi: 10.1155/2017/4137563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, Schroder WA, Higgs S, Suhrbier A. Chikungunya virus arthritis in adult wild-type mice. Journal of Virology. 2010;84:8021–8032. doi: 10.1128/JVI.02603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzón-Tituaña M, Sierra-Monzón JL, Comas L, Santiago L, Khaliulina-Ushakova T, Uranga-Murillo I, Ramirez-Labrada A, Tapia E, Morte-Romea E, Algarate S, Couty L, Camerer E, Bird PI, Seral C, Luque P, Paño-Pardo JR, Galvez EM, Pardo J, Arias M. Granzyme A inhibition reduces inflammation and increases survival during abdominal sepsis. Theranostics. 2021;11:3781–3795. doi: 10.7150/thno.49288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezzi P. Redox regulation of immunity and the role of small molecular weight thiols. Redox Biology. 2021;44:102001. doi: 10.1016/j.redox.2021.102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Levault KR, Brewer GJ. Relative importance of redox buffers GSH and NAD(P)H in age-related neurodegeneration and Alzheimer disease-like mouse neurons. Aging Cell. 2014;13:631–640. doi: 10.1111/acel.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golstein P, Griffiths GM. An early history of T cell-mediated cytotoxicity. Nature Reviews. Immunology. 2018;18:527–535. doi: 10.1038/s41577-018-0009-3. [DOI] [PubMed] [Google Scholar]
- Gordy LE, Bezbradica JS, Flyak AI, Spencer CT, Dunkle A, Sun J, Stanic AK, Boothby MR, He Y-W, Zhao Z, Van Kaer L, Joyce S. IL-15 regulates homeostasis and terminal maturation of NKT cells. Journal of Immunology. 2011;187:6335–6345. doi: 10.4049/jimmunol.1003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KK, Sherman PM, Cellars L, Andrade-Gordon P, Pan Z, Baruch A, Wallace JL, Hollenberg MD, Vergnolle N. A major role for proteolytic activity and proteinase-activated receptor-2 in the pathogenesis of infectious colitis. PNAS. 2005;102:8363–8368. doi: 10.1073/pnas.0409535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlewood JE, Dumenil T, Le TT, Slonchak A, Kazakoff SH, Patch AM, Gray LA, Howley PM, Liu L, Hayball JD, Yan K, Rawle DJ, Prow NA, Suhrbier A, McFadden G. Injection site vaccinology of a recombinant vaccinia-based vector reveals diverse innate immune signatures. PLOS Pathogens. 2021;17:e1009215. doi: 10.1371/journal.ppat.1009215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand D, Bode KA, Rieß D, Cerny D, Waldhuber A, Römmler F, Strack J, Korten S, Orth JHC, Miethke T, Heeg K, Kubatzky KF. Granzyme A produces bioactive IL-1β through a nonapoptotic inflammasome-independent pathway. Cell Reports. 2014;9:910–917. doi: 10.1016/j.celrep.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Hoghooghi V, Palmer AL, Frederick A, Jiang Y, Merkens JE, Balakrishnan A, Finlay TM, Grubb A, Levy E, Gordon P, Jirik FR, Nguyen MD, Schuurmans C, Visser F, Dunn SE, Ousman SS. Cystatin C Plays a Sex-Dependent Detrimental Role in Experimental Autoimmune Encephalomyelitis. Cell Reports. 2020;33:108236. doi: 10.1016/j.celrep.2020.108236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-T, Naeemuddin M, Elchuri S, Yamaguchi M, Kozy HM, Carlson EJ, Epstein CJ. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Human Molecular Genetics. 2006;15:1187–1194. doi: 10.1093/hmg/ddl034. [DOI] [PubMed] [Google Scholar]
- Hughes ED, Qu YY, Genik SJ, Lyons RH, Pacheco CD, Lieberman AP, Samuelson LC, Nasonkin IO, Camper SA, Van Keuren ML, Saunders TL. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mammalian Genome. 2007;18:549–558. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Yin J, Chen J, Ma X, Wu M, Liu G, Yao K, Tan B, Yin Y. Mitochondria-Targeted Antioxidants: A Step towards Disease Treatment. Oxidative Medicine and Cellular Longevity. 2020;2020:8837893. doi: 10.1155/2020/8837893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joeckel LT, Bird PI. Are all granzymes cytotoxic in vivo? Biological Chemistry. 2014;395:181–202. doi: 10.1515/hsz-2013-0238. [DOI] [PubMed] [Google Scholar]
- Johnson BJ, Le TTT, Dobbin CA, Banovic T, Howard CB, Flores F, Vanags D, Naylor DJ, Hill GR, Suhrbier A. Heat shock protein 10 inhibits lipopolysaccharide-induced inflammatory mediator production. The Journal of Biological Chemistry. 2005;280:4037–4047. doi: 10.1074/jbc.M411569200. [DOI] [PubMed] [Google Scholar]
- Kato M. New insights into IFN-γ in rheumatoid arthritis: role in the era of JAK inhibitors. Immunological Medicine. 2020;43:72–78. doi: 10.1080/25785826.2020.1751908. [DOI] [PubMed] [Google Scholar]
- Köntgen F, Süss G, Stewart C, Steinmetz M, Bluethmann H. Targeted disruption of the MHC class II Aa gene in C57BL/6 mice. International Immunology. 1993;5:957–964. doi: 10.1093/intimm/5.8.957. [DOI] [PubMed] [Google Scholar]
- Korablev A, Lukyanchikova V, Serova I, Battulin N. On-Target CRISPR/Cas9 Activity Can Cause Undesigned Large Deletion in Mouse Zygotes. International Journal of Molecular Sciences. 2020;21:3604. doi: 10.3390/ijms21103604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacic H. 2020, A Decisive Decade for NADPH Oxidases Inhibitors. Antioxidants & Redox Signaling. 2020;33:329–331. doi: 10.1089/ars.2020.8069. [DOI] [PubMed] [Google Scholar]
- La Linn M, Bellett AJ, Parsons PG, Suhrbier A. Complete removal of mycoplasma from viral preparations using solvent extraction. Journal of Virological Methods. 1995;52:51–54. doi: 10.1016/0166-0934(94)00136-5. [DOI] [PubMed] [Google Scholar]
- Leskov I, Neville A, Shen X, Pardue S, Kevil CG, Granger DN, Krzywanski DM. Nicotinamide nucleotide transhydrogenase activity impacts mitochondrial redox balance and the development of hypertension in mice. Journal of the American Society of Hypertension. 2017;11:110–121. doi: 10.1016/j.jash.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xu M, Wang M, Wang L, Wang H, Zhang H, Chen Y, Gong J, Zhang JJ, Adcock IM, Chung KF, Zhou X. Roles of mitochondrial ROS and NLRP3 inflammasome in multiple ozone-induced lung inflammation and emphysema. Respiratory Research. 2018;19:230. doi: 10.1186/s12931-018-0931-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesche C, Sauer P, Prager I, Urlaub D, Claus M, Eils R, Beaudouin J, Watzl C. Single-Fluorescent Protein Reporters Allow Parallel Quantification of Natural Killer Cell-Mediated Granzyme and Caspase Activities in Single Target Cells. Frontiers in Immunology. 2018;9:1840. doi: 10.3389/fimmu.2018.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingappan K. NF-κB in Oxidative Stress. Current Opinion in Toxicology. 2018;7:81–86. doi: 10.1016/j.cotox.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J, Thomas DA, Revell PA, Ley TJ, Virgin HW. Granzymes and caspase 3 play important roles in control of gammaherpesvirus latency. Journal of Virology. 2004;78:12519–12528. doi: 10.1128/JVI.78.22.12519-12528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Xu S. TNF inhibitor therapy for rheumatoid arthritis. Biomedical Reports. 2013;1:177–184. doi: 10.3892/br.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madera S, Rapp M, Firth MA, Beilke JN. Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. The Journal of Experimental Medicine. 2016;213:225–233. doi: 10.1084/jem.20150712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrup-Poulsen T. Granzyme A in the Pathogenesis of Type 1 Diabetes: The Yes and the No. Diabetes. 2017;66:2937–2939. doi: 10.2337/dbi17-0037. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Martinvalet D, Dykxhoorn DM, Ferrini R, Lieberman J. Granzyme A cleaves a mitochondrial complex I protein to initiate caspase-independent cell death. Cell. 2008;133:681–692. doi: 10.1016/j.cell.2008.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCambridge G, Agrawal M, Keady A, Kern PA, Hasturk H, Nikolajczyk BS, Bharath LP. Saturated Fatty Acid Activates T Cell Inflammation Through a Nicotinamide Nucleotide Transhydrogenase (NNT)-Dependent Mechanism. Biomolecules. 2019;9:E79. doi: 10.3390/biom9020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt MC, Lagace TA, Horton JD. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. The Journal of Biological Chemistry. 2007;282:20799–20803. doi: 10.1074/jbc.C700095200. [DOI] [PubMed] [Google Scholar]
- Meimaridou E, Goldsworthy M, Chortis V, Fragouli E, Foster PA, Arlt W, Cox R, Metherell LA. NNT is a key regulator of adrenal redox homeostasis and steroidogenesis in male mice. The Journal of Endocrinology. 2018;236:13–28. doi: 10.1530/JOE-16-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekada K, Abe K, Murakami A, Nakamura S, Nakata H, Moriwaki K, Obata Y, Yoshiki A. Genetic Differences among C57BL/6 Substrains. Experimental Animals. 2009;58:141–149. doi: 10.1538/expanim.58.141. [DOI] [PubMed] [Google Scholar]
- Mekada K, Hirose M, Murakami A, Yoshiki A. Development of SNP markers for C57BL/6N-derived mouse inbred strains. Experimental Animals. 2015;64:91–100. doi: 10.1538/expanim.14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metkar SS, Menaa C, Pardo J, Wang B, Wallich R, Freudenberg M, Kim S, Raja SM, Shi L, Simon MM, Froelich CJ. Human and Mouse Granzyme A Induce a Proinflammatory Cytokine Response. Immunity. 2008;29:720–733. doi: 10.1016/j.immuni.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Miyake T, Kumagai Y, Kato H, Guo Z, Matsushita K, Satoh T, Kawagoe T, Kumar H, Jang MH, Kawai T, Tani T, Takeuchi O, Akira S. Poly I:C-Induced Activation of NK Cells by CD8α + Dendritic Cells via the IPS-1 and TRIF-Dependent Pathways. The Journal of Immunology. 2009;183:2522–2528. doi: 10.4049/jimmunol.0901500. [DOI] [PubMed] [Google Scholar]
- Mizutani T, Neugebauer N, Putz EM, Moritz N, Simma O, Zebedin-Brandl E, Gotthardt D, Warsch W, Eckelhart E, Kantner HP, Kalinke U, Lienenklaus S, Weiss S, Strobl B, Müller M, Sexl V, Stoiber D. Conditional IFNAR1 ablation reveals distinct requirements of Type I IFN signaling for NK cell maturation and tumor surveillance. Oncoimmunology. 2012;1:1027–1037. doi: 10.4161/onci.21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett HF, Cartwright ANR, Kim H-J, Godec J, Pyrdol J, Äijö T, Martinez GJ, Rao A, Lu J, Golub TR, Cantor H, Sharpe AH, Novina CD, Wucherpfennig KW. The microRNA miR-31 inhibits CD8+ T cell function in chronic viral infection. Nature Immunology. 2017;18:791–799. doi: 10.1038/ni.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollah ZUA, Quah HS, Graham KL, Jhala G, Krishnamurthy B, Dharma JFM, Chee J, Trivedi PM, Pappas EG, Mackin L, Chu EPF, Akazawa S, Fynch S, Hodson C, Deans AJ, Trapani JA, Chong MMW, Bird PI, Brodnicki TC, Thomas HE, Kay TWH. Granzyme A Deficiency Breaks Immune Tolerance and Promotes Autoimmune Diabetes Through a Type I Interferon–Dependent Pathway. Diabetes. 2017;66:3041–3050. doi: 10.2337/db17-0517. [DOI] [PubMed] [Google Scholar]
- Morales-Hernandez A, Martinat A, Chabot A, McKinney-Freeman G. Elevated Oxidative Stress Impairs Hematopoietic Progenitor Function in C57BL/6 Substrains. Stem Cell Reports. 2018;11:334–347. doi: 10.1016/j.stemcr.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison TE, Fraser RJ, Smith PN, Mahalingam S, Heise MT. Complement Contributes to Inflammatory Tissue Destruction in a Mouse Model of Ross River Virus-Induced Disease. Journal of Virology. 2007;81:5132–5143. doi: 10.1128/JVI.02799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpande CAM, Dintwe OB, Musvosvi M, Mabwe S, Bilek N, Hatherill M, Nemes E, Scriba TJ, The SATVI Clinical Immunology Team Functional, Antigen-Specific Stem Cell Memory (TSCM) CD4+ T Cells Are Induced by Human Mycobacterium tuberculosis Infection. Frontiers in Immunology. 2018;9:324. doi: 10.3389/fimmu.2018.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllbacher A, Ebnet K, Blanden RV, Hla RT, Stehle T, Museteanu C, Simon MM. Granzyme A is critical for recovery of mice from infection with the natural cytopathic viral pathogen, ectromelia. PNAS. 1996;93:5783–5787. doi: 10.1073/pnas.93.12.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllbacher A, Waring P, Tha Hla R, Tran T, Chin S, Stehle T, Museteanu C, Simon MM. Granzymes are the essential downstream effector molecules for the control of primary virus infections by cytolytic leukocytes. PNAS. 1999;96:13950–13955. doi: 10.1073/pnas.96.24.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro E, Merlo A, Martorelli D, Cangemi M, Dalla Santa S, Dolcetti R, Rosato A. Fighting Viral Infections and Virus-Driven Tumors with Cytotoxic CD4+ T Cells. Frontiers in Immunology. 2017;8:197. doi: 10.3389/fimmu.2017.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalle SC, Barreira da Silva R, Zhang H, Decker M, Chalouni C, Xu M, Posthuma G, de Mazière A, Klumperman J, Baz Morelli A, Fleire SJ, Verkman AS, Trombetta ES, Albert ML, Mellman I. Aquaporin-3 regulates endosome-to-cytosol transfer via lipid peroxidation for cross presentation. PLOS ONE. 2020;15:e0238484. doi: 10.1371/journal.pone.0238484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoi SM, Tovey MG, Vella AT. Targeting poly(I:C) to the TLR3-independent pathway boosts effector CD8 T cell differentiation through IFN-alpha/beta. Journal of Immunology. 2008;181:7670–7680. doi: 10.4049/jimmunol.181.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen W, Nakayama E, Yan K, Tang B, Le TT, Liu L, Cooper TH, Hayball JD, Faddy HM, Warrilow D, Allcock RJN, Hobson-Peters J, Hall RA, Rawle DJ, Lutzky VP, Young P, Oliveira NM, Hartel G, Howley PM, Prow NA, Suhrbier A. Arthritogenic Alphavirus Vaccines: Serogrouping Versus Cross-Protection in Mouse Models. Vaccines. 2020;8:E209. doi: 10.3390/vaccines8020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberkampf M, Guillerey C, Mouriès J, Rosenbaum P, Fayolle C, Bobard A, Savina A, Ogier-Denis E, Enninga J, Amigorena S, Leclerc C, Dadaglio G. Mitochondrial reactive oxygen species regulate the induction of CD8+ T cells by plasmacytoid dendritic cells. Nature Communications. 2018;9:2241. doi: 10.1038/s41467-018-04686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata A, Kato Y, Higa S, Yoshizaki K. IL-6 inhibitor for the treatment of rheumatoid arthritis: A comprehensive review. Modern Rheumatology. 2019;29:258–267. doi: 10.1080/14397595.2018.1546357. [DOI] [PubMed] [Google Scholar]
- Pardo J, Balkow S, Anel A, Simon MM. The differential contribution of granzyme A and granzyme B in cytotoxic T lymphocyte-mediated apoptosis is determined by the quality of target cells. European Journal of Immunology. 2002;32:1980–1985. doi: 10.1002/1521-4141(200207)32:7<1980::AID-IMMU1980>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Pardo J, Bosque A, Brehm R, Wallich R, Naval J, Müllbacher A, Anel A, Simon MM. Apoptotic pathways are selectively activated by granzyme A and/or granzyme B in CTL-mediated target cell lysis. The Journal of Cell Biology. 2004;167:457–468. doi: 10.1083/jcb.200406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Griesenauer B, Jiang H, Adom D, Mehrpouya-Bahrami P, Chakravorty S, Kazemian M, Imam T, Srivastava R, Hayes TA, Pardo J, Janga SC, Paczesny S, Kaplan MH, Olson MR. Granzyme A-producing T helper cells are critical for acute graft-versus-host disease. JCI Insight. 2020;5:124465. doi: 10.1172/jci.insight.124465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira RA, Simon MM, Simmons A. Granzyme A, a noncytolytic component of CD8(+) cell granules, restricts the spread of herpes simplex virus in the peripheral nervous systems of experimentally infected mice. Journal of Virology. 2000;74:1029–1032. doi: 10.1128/jvi.74.2.1029-1032.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasman K, Demol H, Bird PI, Gevaert K, Van Damme P. Substrate specificities of the granzyme tryptases A and K. Journal of Proteome Research. 2014;13:6067–6077. doi: 10.1021/pr500968d. [DOI] [PubMed] [Google Scholar]
- Poo YS, Rudd PA, Gardner J, Wilson JAC, Larcher T, Colle M-A, Le TT, Nakaya HI, Warrilow D, Allcock R, Bielefeldt-Ohmann H, Schroder WA, Khromykh AA, Lopez JA, Suhrbier A. Multiple immune factors are involved in controlling acute and chronic chikungunya virus infection. PLOS Neglected Tropical Diseases. 2014;8:e3354. doi: 10.1371/journal.pntd.0003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prow NA, Tang B, Gardner J, Le TT, Taylor A, Poo YS, Nakayama E, Hirata TDC, Nakaya HI, Slonchak A, Mukhopadhyay P, Mahalingam S, Schroder WA, Klimstra W, Suhrbier A. Lower temperatures reduce type I interferon activity and promote alphaviral arthritis. PLOS Pathogens. 2017;13:e1006788. doi: 10.1371/journal.ppat.1006788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prow NA, Liu L, Nakayama E, Cooper TH, Yan K, Eldi P, Hazlewood JE, Tang B, Le TT, Setoh YX, Khromykh AA, Hobson-Peters J, Diener KR, Howley PM, Hayball JD, Suhrbier A. A vaccinia-based single vector construct multi-pathogen vaccine protects against both Zika and chikungunya viruses. Nature Communications. 2018;9:1230. doi: 10.1038/s41467-018-03662-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prow NA, Hirata TDC, Tang B, Larcher T, Mukhopadhyay P, Alves TL, Le TT, Gardner J, Poo YS, Nakayama E, Lutzky VP, Nakaya HI, Suhrbier A. Exacerbation of Chikungunya Virus Rheumatic Immunopathology by a High Fiber Diet and Butyrate. Frontiers in Immunology. 2019;10:2736. doi: 10.3389/fimmu.2019.02736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu H, Shi Y, Zhang L, Lu Z, Ye Q, Leak RK, Xu F, Ma S, Mu H, Wei Z, Xu N, Xia Y, Hu X, Hitchens TK, Bennett MVL, Chen J. Protease-independent action of tissue plasminogen activator in brain plasticity and neurological recovery after ischemic stroke. PNAS. 2019;116:9115–9124. doi: 10.1073/pnas.1821979116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao KNS, Shen X, Pardue S, Krzywanski DM. Nicotinamide nucleotide transhydrogenase (NNT) regulates mitochondrial ROS and endothelial dysfunction in response to angiotensin II. Redox Biology. 2020;36:101650. doi: 10.1016/j.redox.2020.101650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawle DJ, Le TT, Dumenil T, Yan K, Tang B, Nguyen W, Watterson D, Modhiran N, Hobson-Peters J, Bishop C, Suhrbier A. ACE2-lentiviral transduction enables mouse SARS-CoV-2 infection and mapping of receptor interactions. PLOS Pathogens. 2021;17:e1009723. doi: 10.1371/journal.ppat.1009723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regner M, Pavlinovic L, Koskinen A, Young N, Trapani JA, Müllbacher A. Cutting edge: rapid and efficient in vivo cytotoxicity by cytotoxic T cells is independent of granzymes A and B. Journal of Immunology. 2009;183:37–40. doi: 10.4049/jimmunol.0900466. [DOI] [PubMed] [Google Scholar]
- Regner M, Pavlinovic L, Young N, Müllbacher A. In vivo elimination of MHC-I-deficient lymphocytes by activated natural killer cells is independent of granzymes A and B. PLOS ONE. 2011;6:e23252. doi: 10.1371/journal.pone.0023252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera L, Gariglio M, Valente G, Müllbacher A, Museteanu C, Landolfo S, Simon MM. Murine cytomegalovirus replication in salivary glands is controlled by both perforin and granzymes during acute infection. European Journal of Immunology. 2000;30:1350–1355. doi: 10.1002/(SICI)1521-4141(200005)30:5<1350::AID-IMMU1350>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Ripoll VM, Meadows NA, Bangert M, Lee AW, Kadioglu A, Cox RD. Nicotinamide nucleotide transhydrogenase (NNT) acts as a novel modulator of macrophage inflammatory responses. FASEB Journal. 2012;26:3550–3562. doi: 10.1096/fj.11-199935. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi JA, Figueira TR, Ravagnani FG, Oliveira HCF, Vercesi AE, Castilho RF. A spontaneous mutation in the nicotinamide nucleotide transhydrogenase gene of C57BL/6J mice results in mitochondrial redox abnormalities. Free Radical Biology and Medicine. 2013;63:446–456. doi: 10.1016/j.freeradbiomed.2013.05.049. [DOI] [PubMed] [Google Scholar]
- Rydström J. Mitochondrial NADPH, transhydrogenase and disease. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2006;1757:721–726. doi: 10.1016/j.bbabio.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Sakai M, Troutman TD, Seidman JS, Ouyang Z, Spann NJ, Abe Y, Ego KM, Bruni CM, Deng Z, Schlachetzki JCM, Nott A, Bennett H, Chang J, Vu BT, Pasillas MP, Link VM, Texari L, Heinz S, Thompson BM, McDonald JG, Geissmann F, Glass CK. Liver-Derived Signals Sequentially Reprogram Myeloid Enhancers to Initiate and Maintain Kupffer Cell Identity. Immunity. 2019;51:655–670. doi: 10.1016/j.immuni.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno AG, Rentz T, Dorighello GG, Marques AC, Lorza-Gil E, Wanschel ACBA, de Moraes A, Vercesi AE, Oliveira HCF. Lack of mitochondrial NADP(H)-transhydrogenase expression in macrophages exacerbates atherosclerosis in hypercholesterolemic mice. Biochemical Journal. 2019;476:3769–3789. doi: 10.1042/BCJ20190543. [DOI] [PubMed] [Google Scholar]
- Santiago L, Menaa C, Arias M, Martin P, Jaime-Sánchez P, Metkar S, Comas L, Erill N, Gonzalez-Rumayor V, Esser E, Galvez EM, Raja S, Simon MM, Sprague SM, Gabay C, Martinez-Lostao L, Pardo J, Froelich CJ. Granzyme A Contributes to Inflammatory Arthritis in Mice Through Stimulation of Osteoclastogenesis. Arthritis & Rheumatology. 2017;69:320–334. doi: 10.1002/art.39857. [DOI] [PubMed] [Google Scholar]
- Santiago L, Castro M, Sanz-Pamplona R, Garzón M, Ramirez-Labrada A, Tapia E, Moreno V, Layunta E, Gil-Gómez G, Garrido M, Peña R, Lanuza PM, Comas L, Jaime-Sanchez P, Uranga-Murillo I, Del Campo R, Pelegrín P, Camerer E, Martínez-Lostao L, Muñoz G, Uranga JA, Alcalde A, Galvez EM, Ferrandez A, Bird PI, Metkar S, Arias MA, Pardo J. Extracellular Granzyme A Promotes Colorectal Cancer Development by Enhancing Gut Inflammation. Cell Reports. 2020;32:107847. doi: 10.1016/j.celrep.2020.107847. [DOI] [PubMed] [Google Scholar]
- Santiago-Raber M-L, Baccala R, Haraldsson KM, Choubey D, Stewart TA, Kono DH, Theofilopoulos AN. Type-I Interferon Receptor Deficiency Reduces Lupus-like Disease in NZB Mice. Journal of Experimental Medicine. 2003;197:777–788. doi: 10.1084/jem.20021996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanoski AS, Le TT, Kaiserman D, Rowe C, Prow NA, Barboza DD, Santos CA, Zanotto PMA, Magalhães KG, Aurelio L, Muller D, Young P, Zhao P, Bird PI, Suhrbier A. Granzyme A in Chikungunya and Other Arboviral Infections. Frontiers in Immunology. 2019;10:3083. doi: 10.3389/fimmu.2019.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schett G. Resolution of inflammation in arthritis. Seminars in Immunopathology. 2019;41:675–679. doi: 10.1007/s00281-019-00768-x. [DOI] [PubMed] [Google Scholar]
- Scumpia PO, Botten GA, Norman JS, Kelly-Scumpia KM, Spreafico R, Ruccia AR, Purbey PK, Thomas BJ, Modlin RL, Smale ST. Opposing roles of Toll-like receptor and cytosolic DNA-STING signaling pathways for Staphylococcus aureus cutaneous host defense. PLOS Pathogens. 2017;13:e1006496. doi: 10.1371/journal.ppat.1006496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalés J, Perdiguero E, Serrano AL, Sousa-Victor P, Ortet L, Jardí M, Budanov AV, Garcia-Prat L, Sandri M, Thomson DM, Karin M, Hee Lee J, Muñoz-Cánoves P. Sestrin prevents atrophy of disused and aging muscles by integrating anabolic and catabolic signals. Nature Communications. 2020;11:189. doi: 10.1038/s41467-019-13832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Yamasaki S, Sakurai M, Yumoto N, Ikeda M, Mishima-Tsumagari C, Kukimoto-Niino M, Watanabe T, Kawamura M, Shirouzu M, Fujii S-I. Granzyme A Stimulates pDCs to Promote Adaptive Immunity via Induction of Type I IFN. Frontiers in Immunology. 2019;10:1450. doi: 10.3389/fimmu.2019.01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shresta S, Goda P, Wesselschmidt R, Ley TJ. Residual cytotoxicity and granzyme K expression in granzyme A-deficient cytotoxic lymphocytes. The Journal of Biological Chemistry. 1997;272:20236–20244. doi: 10.1074/jbc.272.32.20236. [DOI] [PubMed] [Google Scholar]
- Shresta S, Graubert TA, Thomas DA, Raptis SZ, Ley TJ. Granzyme A initiates an alternative pathway for granule-mediated apoptosis. Immunity. 1999;10:595–605. doi: 10.1016/s1074-7613(00)80059-x. [DOI] [PubMed] [Google Scholar]
- Silver LM. In Mouse Genetics Concepts and Applications. The Jackson Laboratory. 2008 http://www.informatics.jax.org/silver/index.shtml
- Simon MS. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biology. 2013;14:R82. doi: 10.1186/gb-2013-14-7-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons A. FastQC: A quality control tool for high throughput sequence data. FastQC 2010
- Smyth MJ, Street SEA, Trapani JA. Cutting Edge: Granzymes A and B Are Not Essential for Perforin-Mediated Tumor Rejection. The Journal of Immunology. 2003;171:515–518. doi: 10.4049/jimmunol.171.2.515. [DOI] [PubMed] [Google Scholar]
- Souza-Fonseca-Guimaraes F, Young A, Mittal D, Martinet L, Bruedigam C, Takeda K, Andoniou CE, Degli-Esposti MA, Hill GR, Smyth MJ. NK cells require IL-28R for optimal in vivo activity. PNAS. 2015;112:E2376–E2384. doi: 10.1073/pnas.1424241112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sower LE, Froelich CJ, Allegretto N, Rose PM, Hanna WD, Klimpel GR. Extracellular activities of human granzyme A. Monocyte activation by granzyme A versus alpha-thrombin. Journal of Immunology. 1996;156:2585–2590. [PubMed] [Google Scholar]
- Spaeny-Dekking EH, Hanna WL, Wolbink AM, Wever PC, Kummer JA, Kummer AJ, Swaak AJ, Middeldorp JM, Huisman HG, Froelich CJ, Hack CE. Extracellular granzymes A and B in humans: detection of native species during CTL responses in vitro and in vivo. Journal of Immunology. 1998;160:3610–3616. [PubMed] [Google Scholar]
- Spaeny-Dekking EH, Kamp AM, Froelich CJ, Hack CE. Extracellular granzyme A, complexed to proteoglycans, is protected against inactivation by protease inhibitors. Blood. 2000;95:1465–1472. [PubMed] [Google Scholar]
- Spencer CT, Abate G, Sakala IG, Xia M, Truscott SM, Eickhoff CS, Linn R, Blazevic A, Metkar SS, Peng G, Froelich CJ, Hoft DF. Granzyme A produced by γ(9)δ(2) T cells induces human macrophages to inhibit growth of an intracellular pathogen. PLOS Pathogens. 2013;9:e1003119. doi: 10.1371/journal.ppat.1003119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhubl SR. Why have antioxidants failed in clinical trials? The American Journal of Cardiology. 2008;101:14D–19D. doi: 10.1016/j.amjcard.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Stewart SE, D’Angelo ME, Bird PI. Intercellular communication via the endo-lysosomal system: translocation of granzymes through membrane barriers. Biochimica et Biophysica Acta. 2012;1824:59–67. doi: 10.1016/j.bbapap.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Strik MCM, Wolbink A, Wouters D, Bladergroen BA, Verlaan AR, van Houdt IS, Hijlkema S, Hack CE, Kummer JA. Intracellular serpin SERPINB6 (PI6) is abundantly expressed by human mast cells and forms complexes with beta-tryptase monomers. Blood. 2004;103:2710–2717. doi: 10.1182/blood-2003-08-2981. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhrbier A, Fernan A, Burrows SR, Saul A, Moss DJ. BLT esterase activity as an alternative to chromium release in cytotoxic T cell assays. Journal of Immunological Methods. 1991;145:43–53. doi: 10.1016/0022-1759(91)90309-4. [DOI] [PubMed] [Google Scholar]
- Suhrbier A, Jaffar-Bandjee M-C, Gasque P. Arthritogenic alphaviruses--an overview. Nature Reviews. Rheumatology. 2012;8:420–429. doi: 10.1038/nrrheum.2012.64. [DOI] [PubMed] [Google Scholar]
- Suhrbier A. Rheumatic manifestations of chikungunya: emerging concepts and interventions. Nature Reviews. Rheumatology. 2019;15:597–611. doi: 10.1038/s41584-019-0276-9. [DOI] [PubMed] [Google Scholar]
- Suidan HS, Bouvier J, Schaerer E, Stone SR, Monard D, Tschopp J. Granzyme A released upon stimulation of cytotoxic T lymphocytes activates the thrombin receptor on neuronal cells and astrocytes. PNAS. 1994;91:8112–8116. doi: 10.1073/pnas.91.17.8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suidan HS, Clemetson KJ, Brown-Luedi M, Niclou SP, Clemetson JM, Tschopp J, Monard D. The serine protease granzyme A does not induce platelet aggregation but inhibits responses triggered by thrombin. The Biochemical Journal. 1996;315 (Pt 3):939–945. doi: 10.1042/bj3150939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang X, Saredy J, Yuan Z, Yang X, Wang H. Innate-adaptive immunity interplay and redox regulation in immune response. Redox Biology. 2020;37:101759. doi: 10.1016/j.redox.2020.101759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susanto O, Stewart SE, Voskoboinik I, Brasacchio D, Hagn M, Ellis S, Asquith S, Sedelies KA, Bird PI, Waterhouse NJ, Trapani JA. Mouse granzyme A induces a novel death with writhing morphology that is mechanistically distinct from granzyme B-induced apoptosis. Cell Death and Differentiation. 2013;20:1183–1193. doi: 10.1038/cdd.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann JB, Hayakawa Y, Zerafa N, Sheehan KCF, Scott B, Schreiber RD, Hertzog P, Smyth MJ. Type I IFN contributes to NK cell homeostasis, activation, and antitumor function. Journal of Immunology. 2007;178:7540–7549. doi: 10.4049/jimmunol.178.12.7540. [DOI] [PubMed] [Google Scholar]
- Szabo R, Lantsman T, Peters DE, Bugge TH. Delineation of proteolytic and non-proteolytic functions of the membrane-anchored serine protease prostasin. Development. 2016;143:2818–2828. doi: 10.1242/dev.137968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering C von. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh SSY, Vieusseux J, Prakash M, Berkowicz S, Luu J, Bird CH, Law RHP, Rosado C, Price JT, Whisstock JC, Bird PI. Maspin is not required for embryonic development or tumour suppression. Nature Communications. 2014;5:3164. doi: 10.1038/ncomms4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To EE, Erlich JR, Liong F, Luong R, Liong S, Esaq F, Oseghale O, Anthony D, McQualter J, Bozinovski S, Vlahos R, O’Leary JJ, Brooks DA, Selemidis S. Mitochondrial Reactive Oxygen Species Contribute to Pathological Inflammation During Influenza A Virus Infection in Mice. Antioxidants & Redox Signaling. 2020;32:929–942. doi: 10.1089/ars.2019.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toye AA, Lippiat JD, Proks P, Shimomura K, Bentley L, Hugill A, Mijat V, Goldsworthy M, Moir L, Haynes A, Quarterman J, Freeman HC, Ashcroft FM, Cox RD. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia. 2005;48:675–686. doi: 10.1007/s00125-005-1680-z. [DOI] [PubMed] [Google Scholar]
- Tumiati M, Hemmes A, Uusivirta S, Koopal S, Kankainen M, Lehtonen E, Kuznetsov SG. Loss of Rad51c accelerates tumourigenesis in sebaceous glands of Trp53-mutant mice. The Journal of Pathology. 2015;235:136–146. doi: 10.1002/path.4455. [DOI] [PubMed] [Google Scholar]
- Ubaida-Mohien C, Lyashkov A, Gonzalez-Freire M, Tharakan R, Shardell M, Moaddel R, Semba RD, Chia CW, Gorospe M, Sen R, Ferrucci L. Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria. eLife. 2019;8:e49874. doi: 10.7554/eLife.49874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uranga S, Marinova D, Martin C, Pardo J, Aguilo N. Granzyme A Is Expressed in Mouse Lungs during Mycobacterium tuberculosis Infection but Does Not Contribute to Protection In Vivo. PLOS ONE. 2016;11:e0153028. doi: 10.1371/journal.pone.0153028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daalen KR, Reijneveld JF, Bovenschen N. Modulation of Inflammation by Extracellular Granzyme A. Frontiers in Immunology. 2020;11:931. doi: 10.3389/fimmu.2020.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boogaard FE, van Gisbergen K, Vernooy JH, Medema JP, Roelofs J, van Zoelen MAD, Endeman H, Biesma DH, Boon L, Van’t Veer C, de Vos AF, van der Poll T. Granzyme A impairs host defense during Streptococcus pneumoniae pneumonia. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2016;311:L507–L516. doi: 10.1152/ajplung.00116.2016. [DOI] [PubMed] [Google Scholar]
- van Eck JA, Shan L, Meeldijk J, Hack CE, Bovenschen N. A novel proinflammatory role for granzyme A. Cell Death & Disease. 2017;8:e2630. doi: 10.1038/cddis.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet H, Brillet-Guéguen L, Coppée J-Y, Dillies M-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLOS ONE. 2016;11:e0157022. doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent G, Novak EA, Siow VS, Cunningham KE, Griffith BD, Comerford TE, Mentrup HL, Stolz DB, Loughran P, Ranganathan S, Mollen KP. Nix-Mediated Mitophagy Modulates Mitochondrial Damage During Intestinal Inflammation. Antioxidants & Redox Signaling. 2020;33:1–19. doi: 10.1089/ars.2018.7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vozenilek AE, Vetkoetter M, Green JM, Shen X, Traylor JG, Klein RL, Orr AW, Woolard MD, Krzywanski DM. Absence of Nicotinamide Nucleotide Transhydrogenase in C57BL/6J Mice Exacerbates Experimental Atherosclerosis. Journal of Vascular Research. 2018;55:98–110. doi: 10.1159/000486337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward NP, Kang YP, Falzone A, Boyle TA, DeNicola GM. Nicotinamide nucleotide transhydrogenase regulates mitochondrial metabolism in NSCLC through maintenance of Fe-S protein function. The Journal of Experimental Medicine. 2020;217:e20191689. doi: 10.1084/jem.20191689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb LV, Barbarulo A, Huysentruyt J, Vanden Berghe T, Takahashi N, Ley S, Vandenabeele P, Seddon B. Survival of Single Positive Thymocytes Depends upon Developmental Control of RIPK1 Kinase Signaling by the IKK Complex Independent of NF-κB. Immunity. 2019;50:348–361. doi: 10.1016/j.immuni.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wensink AC, Hack CE, Bovenschen N. Granzymes regulate proinflammatory cytokine responses. Journal of Immunology. 2015;194:491–497. doi: 10.4049/jimmunol.1401214. [DOI] [PubMed] [Google Scholar]
- Williams JL, Hall CL, Meimaridou E, Metherell LA. Loss of Nnt Increases Expression of Oxidative Phosphorylation Complexes in C57BL/6J Hearts. International Journal of Molecular Sciences. 2021;22:6101. doi: 10.3390/ijms22116101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JAC, Prow NA, Schroder WA, Ellis JJ, Cumming HE, Gearing LJ, Poo YS, Taylor A, Hertzog PJ, Di Giallonardo F, Hueston L, Le Grand R, Tang B, Le TT, Gardner J, Mahalingam S, Roques P, Bird PI, Suhrbier A. RNA-Seq analysis of chikungunya virus infection and identification of granzyme A as a major promoter of arthritic inflammation. PLOS Pathogens. 2017;13:e1006155. doi: 10.1371/journal.ppat.1006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Hainz N, Beckmann A, Maack C, Menger MD, Tschernig T, Meier C. Brain damage resulting from postnatal hypoxic-ischemic brain injury is reduced in C57BL/6J mice as compared to C57BL/6N mice. Brain Research. 2016;1650:224–231. doi: 10.1016/j.brainres.2016.09.013. [DOI] [PubMed] [Google Scholar]
- Wu CH, Li J, Li L, Sun J, Fabbri M, Wayne AS, Seeger RC, Jong AY. Extracellular vesicles derived from natural killer cells use multiple cytotoxic proteins and killing mechanisms to target cancer cells. Journal of Extracellular Vesicles. 2019;8:1588538. doi: 10.1080/20013078.2019.1588538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zheng L, Mo J, Yao X, Fan C, Bao Y. Protective Effects of MitoTEMPO on Nonalcoholic Fatty Liver Disease via Regulating Myeloid-Derived Suppressor Cells and Inflammation in Mice. BioMed Research International. 2020;2020:9329427. doi: 10.1155/2020/9329427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Vet LJ, Tang B, Hobson-Peters J, Rawle DJ, Le TT, Larcher T, Hall RA, Suhrbier A. A Yellow Fever Virus 17D Infection and Disease Mouse Model Used to Evaluate a Chimeric Binjari-Yellow Fever Virus Vaccine. Vaccines. 2020;8:E368. doi: 10.3390/vaccines8030368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yodoi J, Matsuo Y, Tian H, Masutani H, Inamoto T. Anti-Inflammatory Thioredoxin Family Proteins for Medicare, Healthcare and Aging Care. Nutrients. 2017;9:E1081. doi: 10.3390/nu9101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaid A, Burt FJ, Liu X, Poo YS, Zandi K, Suhrbier A, Weaver SC, Texeira MM, Mahalingam S. Arthritogenic alphaviruses: epidemiological and clinical perspective on emerging arboviruses. The Lancet. Infectious Diseases. 2021;21:e123–e133. doi: 10.1016/S1473-3099(20)30491-6. [DOI] [PubMed] [Google Scholar]
- Zhou Z, He H, Wang K, Shi X, Wang Y, Su Y, Wang Y, Li D, Liu W, Zhang Y, Shen L, Han W, Shen L, Ding J, Shao F. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368:eaaz7548. doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]