Abstract
Abstract
Species of the genera Bacteroides and Phocaeicola play an important role in the human colon. The organisms contribute to the degradation of complex heteropolysaccharides to small chain fatty acids, which are in part utilized by the human body. Furthermore, these organisms are involved in the synthesis of vitamins and other bioactive compounds. Of special interest is Phocaeicola vulgatus, originally classified as a Bacteroides species, due to its abundance in the human intestinal tract and its ability to degrade many plant-derived heteropolysaccharides. We analyzed different tools for the genetic modification of this microorganism, with respect to homologous gene expression of the ldh gene encoding a D-lactate dehydrogenase (LDH). Therefore, the ldh gene was cloned into the integration vector pMM656 and the shuttle vector pG106 for homologous gene expression in P. vulgatus. We determined the ldh copy number, transcript abundance, and the enzyme activity of the wild type and the mutants. The strain containing the shuttle vector showed an approx. 1500-fold increase in the ldh transcript concentration and an enhanced LDH activity that was about 200-fold higher compared to the parental strain. Overall, the proportion of lactate in the general catabolic carbon flow increased from 2.9% (wild type) to 28.5% in the LDH-overproducing mutant. This approach is a proof of concept, verifying the genetic accessibility of P. vulgatus and could form the basis for targeted genetic optimization.
Key points
• A lactate dehydrogenase was overexpressed in Phocaeicola (Bacteroides) vulgatus.
• The ldh transcript abundance and the LDH activity increased sharply in the mutant.
• The proportion of lactate in the catabolic carbon flow increased to about 30%.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00253-022-11777-6.
Keywords: Microbiota, Human health, Bacteroidaceae, Short-chain fatty acids, Carbon flux
Introduction
The human colon contains a versatile and complex microbial flora, and the majority of these organisms proliferate only under anaerobic conditions (Sender et al. 2016; Lu and Imlay 2021). Species of the genera Bacteroides and Phocaeicola are ubiquitous commensals, comprising about thirty percent of the human gut microbiota (Salyers 1984). Hence, these microorganisms play an essential role in the colonic ecosystem (Wexler and Goodman 2017). Phocaeicola vulgatus (P. vulgatus), originally classified as a Bacteroides species (García-López et al. 2019), is one of the most numerous organisms within the family Bacteroidaceae in the colon (up to 1010 per g stool) (Salyers 1984).
To date, members of the phylum Bacteriodetes have not been used for biotechnological processes. However, species belonging to the family Bacteroidaceae, including P. vulgatus, are known to be highly effective succinate and propionate producers (Flint et al. 2015; Koh et al. 2016; Ríos-Covián et al. 2016; de Vadder and Mithieux 2018) and possess enzyme systems for the degradation of complex polysaccharides (Chassard et al. 2007; Flint et al. 2008; Makki et al. 2018). There is also strong evidence that these organisms are involved in the synthesis of prebiotic and bioactive compounds (Smith and Macfarlane 1996; Russell et al. 2013), which are often associated with human and animal health benefits. Therefore, species of the family Bacteroidaceae could be used as platform organisms for the efficient and sustainable conversion of renewable resources, such as xylan and other hemicelluloses (Dodd et al. 2011; Robert et al. 2007), into bioactive compounds and important bulk chemicals. For the production of such substances at biotechnological scales, the accessibility of these organisms for targeted genetic manipulation is desirable. However, few genetic tools are available. Replicative plasmids and integrative transposons have been constructed for increased gene expression (Smith et al. 1992; Wang et al. 2000; Gupta et al. 2003). However, in heterologous gene expression studies, strong Escherichia coli (E. coli) promoters were shown not to function in B. fragilis (Smith et al. 1992). Unlike most other prokaryotes, such as E. coli, the unique major sigma factor in Bacteroidaceae species binds to a different consensus sequence, separated by a spacer of variable length (generally 19–21 nucleotides) (Bayley et al. 2000; Mastropaolo et al. 2009). Similarly, the ribosome binding site (RBS) differs in comparison to the RBS of E. coli. Accordingly, the RBS from species of the family Bacteroidaceae has a lower GC content, which results in a lower tendency to form secondary structures (Accetto and Avguštin 2011). These unique promoters and RBS structures make it difficult to transfer genetic systems evolved in bacteria of other phyla (Smith et al. 1992). Furthermore, in genetic and phenotypic studies with intestinal Bacteroidaceae, predominantly only two species have been analyzed: B. fragilis and B. thetaiotaomicron (Salyers et al. 2000; Wexler and Goodman 2017). However, members of the genera Bacteroides and Phocaeicola exhibit high intra-specific genomic and metabolic diversity (Pasolli et al. 2019). The heterogeneity highlights the need for genetic studies on different species belonging to the family Bacteroidaceae.
In this study, we analyzed different tools for the genetic modification of P. vulgatus, to enable homologous gene expression and protein production. Besides succinate and propionate, the organism also produces acetate, formate, and lactate. In the central metabolism of P. vulgatus, lactate is formed from the reduction of pyruvate. Since lactate is a metabolic end product and is not metabolized further, overproduction of this compound should not have a negative effect on the organism and its metabolic properties. Therefore, homologous expression of the ldh gene encoding a D-lactate dehydrogenase was chosen as a proof of concept in order to accomplish the genetic accessibility of P. vulgatus. In addition, the overproduction of lactate also provides a link to industrial application as it can be converted into biodegradable polyesters, making it a potential material for environmentally friendly plastics (Chang et al. 1999). Here, we demonstrate that different plasmids can be used as a platform for gene expression in P. vulgatus, allowing selective manipulation of the metabolism and thus shifting the ratio of metabolic end products towards the synthesis of lactate.
Materials and methods
Materials
All reagents, chemicals, and substrates used in this study were purchased from Carl Roth GmbH (Karlsruhe, Germany) and Sigma-Aldrich (St. Louis, USA). Q5 and One Taq DNA polymerase, restriction endonucleases, T4 ligase, and PCR reagents were bought from New England Biolabs (Ipswich, USA). Oligonucleotides were synthesized by Eurofins Scientific (Ebersberg, Germany).
Bacterial strains and standard culture conditions
E. coli strains were cultivated in lysogeny broth (LB) medium (Miller 1972) at 37 °C, 180 rpm with 300 μg ml−1 erythromycin or 100 μg ml−1 ampicillin, respectively, for plasmid maintenance. P. vulgatus DSM 1447 was obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ; Brunswick, Germany). Cultures were grown anaerobically either in complex brain heart infusion (BHI) medium or in modified Defined Minimal Medium with Glucose (DMMG) in serum flasks sealed with butyl rubber stoppers under an N2/CO2 atmosphere (80%/20%) (Varel and Bryant 1974). Prior to inoculation, glucose (30 mM), L-cysteine (0.5 g l−1) as reducing agent, vitamin K1 (0.1% (v/v)), and hemin (5 mg l−1) were added. DMMG was additionally supplemented with 1 ml l−1 vitamin solution (Wolin et al. 1963) and potassium butyrate (2 mM). Cells were grown at 37 °C, and growth was quantified by measuring the optical density at 600 nm (OD600). Plasmid-containing P. vulgatus strains were grown in broth with the addition of erythromycin (100 μg ml−1) and gentamycin (200 μg ml−1) during selection after conjugation with E. coli.
Molecular cloning
Plasmids used in this study are listed in Table 1. The shuttle vector pG106 was kindly provided by Kevin R. Jones (The Philips Institute for Oral Health Research, Virginia Commonwealth University, Richmond, VA, USA). BLASTp analysis (https://www.ncbi.nlm.nih.gov/) using the biochemically characterized D-LDH from E. coli (b1380) was performed resulting in the identification of protein BVU_2499 as a potential D-LDH in P. vulgatus. The corresponding gene (ldh) was amplified including 250 bp of the upstream region by PCR using Q5 High-fidelity DNA polymerase and primers ldh_nativP_SalI_for and ldh_SphI_rev (Table 1) with genomic DNA of P. vulgatus as a template. For cloning of the fragment in pG106, the PCR products contained the endonuclease restriction sites SalI and SphI at the 5′ and 3′ end, respectively. The fragments were ligated into the corresponding restriction sites of pG106, resulting in the expression vector pG106_ldhnP. For insertion of ldh in pMM656, the DNA fragment was cloned via the NEBuilder® HiFi DNA Assembly Cloning Kit into the vector following the manufacturer’s protocol. The following primers were used: (1) bb_pMM656_ldhnP_fwd and bb_pMM656_ldhnP_rev for the amplification of the vector backbone. (2) as_ldhnP_pMM656_fwd and as_ldhnP_pMM656_rev to amplify the ldh gene. The PRhaKIPAO promoter and the downstream encoded gene nanoluc of pMM656 were exchanged for the ldh gene including 250 bp of the upstream region of the gene. The corresponding plasmid was referred to as pMM656_ldhnP. For overproduction of ldh, pASK5_ldh was constructed. The amplification of ldh was performed using primers ldh_pASK5_for and ldh_pASK5_rev (Table 1), and the fragment was cloned in-frame to the N-terminal Strep-tag into pASK5 vector by restriction with BsaI. Ligated DNA was transformed via heat shock or electroporation in E. coli DH5α (pG106) or E. coli λpir S17-1 (pMM656). The plasmids were purified from E. coli host strains using a plasmid miniprep Kit from New England Biolabs (Ipswich, USA) and were used for transformation in P. vulgatus.
Table 1.
Strains, plasmids, and primers used in this study
| Strains | Description | Source |
|---|---|---|
| P. vulgatus DSM 1447 | Wild type | DSMZ1 |
| P. vulgatus pMM656_ ldhnP | ΔtRNASer derivate of P. vulgatus with chromosomally integrated plasmid pMM656_ldhnP | This study |
| P. vulgatus pG106_ldhnP | P. vulgatus containing plasmid pG106_ldhnP | This study |
| E. coli NEB®5-alpha (referred to as E. coli) | fhuA2 Δ(argF-lacZ) U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | New England Biolabs (Ipswich, USA) |
| E. coli pG106_ldhnP | E. coli NEB®5-alpha containing plasmid pG106_ldhnP | This study |
| E. coli λpir S17-1 | sup E44, ΔlacU169 (ΦlacZΔM15), recA1, endA1, hsdR17, thi-1, gyrA96, relA1, λpir phage lysogen | DSMZ1) |
| E. coli λpir S17-1 pMM656_ldhnP | E. coli λpir S17-1 containing plasmid pMM656_ldhnP | This study |
| Plasmids | Description | Source |
| pASK-IBA.5 (pASK5) | Vector with inducible tetracycline promoter/operator, ampicillin resistance cassette, f1 origin, MCS, and Strep-Tag for N-terminal fusion to a recombinant protein | IBA Göttingen, Germany |
| pASK5_ldh | pASK-IBA5 derivative containing BVU_2499 from P. vulgatus DSM 1447 | This study |
| pG106 | Vector with erythromycin resistance cassette (ermF and ermAM), f1 origin, MCS, repB-mob region for Bacteroides replication | Jones et al. 2020 |
| pG106_ldhnP | pG106 derivative containing BVU_2499 including 250 bp upstream sequence from P. vulgatus DSM 1447 | This study |
| pMM656 | Vector with erythromycin and ampicillin resistance cassette, NBU2 integrase, luciferase (NanoLuc), R6KOrigin | Mimee et al. 2015 |
| pMM656_ldhnP | pMM656 derivative containing BVU_2499 including 250 bp upstream sequence from P. vulgatus DSM 1447 | This study |
| Primer | Sequence (5’ ➔ 3’) | Restriction site |
| bb_pMM656_ldhnP_fwd | TGAACTGCACTTGCTTTG | - |
| bb_pMM656_ldhnP_rev | TTTTATGCAAAAAAAGCATGATTTATG | - |
| as_ldhnP_pMM656_fwd | CATGCTTTTTTTGCATAAAATAACGTTTGCAGAATTGTCCG | - |
| as_ldhnP_pMM656_rev | ATCAAAGCAAGTGCAGTTCATTAGACTCTCCCCACCGC | - |
| ldh_nativP_SalI_for | ATTAGTCGACTAACGTTTGCAGAATTGTCC | SalI |
| ldh_SphI_rev | AATAGCATGCTTAGACTCTCCCCACCGCTA | SphI |
| ldh_pASK5_for | ATGGTAGGTCTCAGCGCCGCTTATAAAATAGCTTTTTACGACAC | BsaI |
| ldh_pASK5_rev | ATGGTAGGTCTCATATCAGACTCTCCCCACCGCTACTTC | BsaI |
| qPCR_LDH_for | TTACGGTGGCGTTGATGTTA | - |
| qPCR_LDH_rev | TGTTCACGGGCAAAATTGTA | - |
| qPCR_L23_for | TCGATTCGGCTTTATTGTACG | - |
| qPCR_L23_rev | CGCCTTCTTTCAATGTTACGA | - |
| attB2451_for | TGTTCCAGCCATTCATTAATA | - |
| attB2094_for | ATCTCCTTTATTTAAATAG | - |
| attB_inErm_rev | ATGGTTTTGCTAAAATGTTA | - |
1DSMZ, German Collection of Microorganisms, Brunswick, Germany
Generation of the ldh overexpression mutant P. vulgatus pG106_ldhnP
Transformation of pG106 via electroporation in P. vulgates was performed as described (Smith 1995) in an anaerobic chamber (Coy Laboratory Products, Grass Lakewood MI, USA) under a 49% N2/49% CO2/2% H2 atmosphere. An overnight culture (50 ml) was harvested by centrifugation at 8000 × g for 15 min at 4 °C. The pellet was washed twice in 4 ml of cold electroporation buffer (10% glycerol, 1 mM MgCl2) and resuspended in 0.5 ml of the same buffer. Fifty microliters of cell suspension and 5 μl plasmid DNA were added to a pre-chilled cuvette (0.2 cm) and incubated on ice for 5 min. The cuvette was placed in an electroporation chamber and pulsed for 6 ms using settings of 2.5 kV and 400 Ω on a Biorad Gene Pulser II (Biorad, Feldkirchen, Germany). Immediately after electroporation, 500 μl of prewarmed BHI medium was added to the cuvette and transferred into 2 ml of prewarmed BHI medium. Regeneration occurred overnight at 37 °C. The next day, 50 μl of each sample was plated on BHI agar plates with erythromycin and incubated anaerobically for 72 h at 37 °C. Transformants were screened by PCR. A positive clone was verified by sequencing and was referred to as P. vulgatus pG106_ldhnP.
Genome integration of pMM656_ldhnP
pMM656 constructs were conjugated into P. vulgatus using E. coli S17 λpir, which contains the conjugative machinery of the plasmid RP4 integrated into the chromosome. For mating, an overnight culture of E. coli S17 λpir was added to P. vulgatus at a cell number ratio of 1:5 (v/v). The mating mixture was pelleted, resuspended in a 200 μl BHI medium, spotted onto a filter on a BHI agar plate, and incubated overnight at 37 °C under aerobic conditions. After overnight incubation, cells on the filter were collected in a 3 ml BHI medium, plated on BHI agar plates with gentamycin and erythromycin, and incubated for 48 h at 37 °C under anaerobic conditions. Colony screening by PCR resulted in the identification of clones containing pMM656 or pMM656_ldhnP inserted in the correct chromosomal location at the tRNASer gene (BVU_2094). The mutants were referred to as P. vulgatus pMM656 and P. vulgatus pMM656_ldhnP and were verified by PCR and sequencing (Fig. S1). For this purpose, the primer pair attB2451_for or attB2094_for and attB_inErm_rev was used, which annealed at the plasmid backbone as well as at BVU_2451 or BVU_2094 located on the chromosome.
Overexpression and purification of ldh
For protein production, overnight cultures of E. coli DH5α (5 ml) harbouring plasmid pASK5_ldh were used to inoculate 500 ml LB medium and were incubated at 37 °C and 180 rpm in shaker flasks. Cells were grown to an OD600 of 0.4 and induced by adding 0.2 μg ml−1 anhydrotetracycline to start production of D-LDH (16 h, 16 °C, 180 rpm). Cells were harvested by centrifugation at 8000 × g and 4 °C for 20 min and resuspended in 5 ml buffer W, containing 100 mM Tris-HCl, 150 mM NaCl, and pH 8. Cells were lysed by sonification and cell debris were separated by centrifugation at 13,000 × g and 4 °C for 10 min. For purification of D-LDH, the culture supernatant was applied to a gravity flow Strep-Tactin Superflow affinity column (IBA GmbH, Göttingen, Germany). Buffer W supplemented with 2.5 mM D-desthiobiotin was used as eluent. The concentration of purified D-LDH was determined by a Bradford assay (Bradford 1976). For protein visualization, polyacrylamide gel electrophoresis was performed (Laemmli 1970) and protein bands were detected via silver staining as described (Blum et al. 1987). Immunoblotting was done according to Towbin et al. (1979) and the protein was detected colorimetrically by the fused Strep-Tag using the Strep-Tactin® HRP Conjugate (IBA GmbH, Göttingen, Germany).
Luciferase assay
Two hundred microliters of cultures of P. vulgatus pMM656 were incubated in 96-well plates with different concentrations of rhamnose for induction of the PRhaKIPOA promoter overnight at 37 °C in an Infinite 200 PRO NanoQuant Microplate Reader (Tecan, Männedorf, Switzerland) in an anaerobic chamber filled with N2/CO2/H2 (49%/49%/2%). The cultures were lysed by adding 20 μl of PopCulture reagent (EMD Millipore, Darmstadt, Germany) supplemented with lysozyme solution (160 U). The NanoLuc® luciferase assay (Promega, Walldorf, Germany) was performed according to the manufacturer’s instructions using 10 μl P. vulgatus cell lysate and 10 μl NanoLuc reaction buffer including the substrate furimazine. NanoLuc oxidizes furimazine and produces bioluminescence (Emax = 460 nm) with a signal half-life of 2 h. Luciferase activity was measured with an integration time of 1 s in an Infinite 200 PRO NanoQuant Microplate Reader (Tecan, Männedorf, Switzerland) under aerobic conditions. The corresponding relative light units (RLU) were normalized to OD1.
Determination of the ldh copy number
The relative number of ldh genes in P. vulgatus pG106_ldhnP, P. vulgatus pMM656_ldhnP, and the wild type was determined by qPCR. The gene l23 (BVU_0803), encoding the ribosomal protein L23, is present once on the chromosome of P. vulgatus and was used as a reference. Gene-specific primers (qPCR_LDH_for, qPCR_LDH_rev, qPCR_L23_for, and qPCR_L23_rev; Table 1) for qPCR experiments were designed using the primer3 software (https://bioinfo.ut.ee/primer3/) (Koressaar and Remm 2007; Untergasser et al. 2012; Koressaar et al. 2018). All primers had an almost identical annealing temperature, and their amplicons had a similar size (180–200 bp), a similar melting temperature (58–60°C), and a similar GC % content. Strains were grown overnight in the BHI medium. After 24 h, each culture was diluted to OD600 = 0.5, and 1 ml was centrifuged and washed in a phosphate-buffered saline solution (8.0 g NaCl, 0.2 g KCl, 1.4 g Na2HPO4, 0.27 g KH2PO4 per 1 l H2O). Cell pellets were resuspended in 1 ml nuclease-free water and boiled for 10 min to lyse the cells. The cell lysates were diluted and supplemented with the Luna® Universal qPCR Master Mix (New England Biolabs, Ipswich, USA). qPCR was performed on a CFX Connect Real-Timer PCR Detection System (BioRad, Munich, Germany). For calculations of ΔCt values, the quantification cycle (Ct) of the gene of interest was subtracted from the Ct value of the reference gene (l23). Subsequently, ΔΔCt values and the corresponding fold change were calculated in relation to the wild type. The fold change was calculated using the formula 2−ΔΔCt.
RT-qPCR analysis of gene expression
The analysis of transcript abundance of the ldh gene of P. vulgatus strains was performed by RT-qPCR experiments. Total RNA from P. vulgatus was isolated from 50 ml cultures grown in the BHI medium to mid-exponential phase. The total RNA Miniprep Kit (New England Biolabs, Ipswich, USA) was used to purify RNA from cells following the manufacturer’s protocol. RNA samples were treated with DNase I to remove residual DNA, and RNA concentrations were measured spectrophotometrically using a BioSpectrometer® (Eppendorf, Hamburg, Germany). Additionally, control PCR experiments were performed to confirm that RNA samples did not contain DNA contaminations. Gene-specific primers used for RT-qPCR experiments were the same as for qPCR experiments. The gene l23, encoding the ribosomal protein L23, was chosen as a reference. RT-qPCR reactions were performed with a Luna® Universal One-Step RT-qPCR Kit (New England Biolabs, Ipswich, USA). Each PCR reaction contained 200 ng of purified RNA. For temperature cycling and fluorescence measurement, a cycler CFX Connect™ and a suitable software (BioRad, Munich, Germany) were used. Melting curve analysis resulted in single peaks for the respective PCR fragments, confirming specific products from PCR reactions. To determine transcript abundance, the ΔΔCt values and the fold change were evaluated as described above.
Preparation of cell-free extract and enzyme assay
Cell-free extract was prepared from P. vulgatus cells grown in a 50 ml BHI medium. Cells were harvested in the exponential growth phase by centrifugation (10,000 × g, 15 min, 4°C) and resuspended in 10 ml buffer W, containing 100 mM Tris-HCl, 150 mM NaCl, and pH 8. Cells were lysed by sonification and cellular debris were removed by centrifugation for 15 min at 8000 × g and 4 °C. Protein concentrations of the cell-free extracts were determined as described (Bradford 1976). The 1 ml assay contained 100 mM potassium phosphate buffer pH 7, 2.5 μM NADH, 5 mM pyruvate, and different concentrations of cell-free extract. The activity was measured photometrically at 340 nm. The temperature optimum of purified D-LDH was determined at pH 7 and temperatures ranging from 30 to 80 °C. The optimum pH of D-LDH was determined using a combined buffer system containing sodium acetate, Tris-HCl, potassium dihydrogen phosphate, and dipotassium hydrogen phosphate (50 mM each) in the range from pH 4 to 9 at 37 °C. Enzyme kinetics were measured by varying substrate concentration (0–50 mM) at physiological pH (7) and temperature (37 °C).
HPLC analysis of culture supernatants
The analysis of substrate and product concentrations was performed by HPLC (Knauer Smartline HPLC system, Knauer GmbH, Berlin, Germany) with an Aminex HPX-87H column (BioRad, Munich, Germany, 300 mm × 7.8 mm) using 5 mM H2SO4 as mobile phase. One milliliter of culture was harvested by centrifugation (10,000 × g, 2 min and 10 °C) at different OD600 in the exponential growth phase. Metabolic end products and the substrate glucose were separated at a column temperature of 65 °C with a flow rate of 0.6 ml min−1 and detected by a refractive index detector. Concentrations were calculated by comparison to corresponding calibration curves.
Results
Genetic tools for homologous gene expression in P. vulgatus
For homologous expression of genes, the shuttle vector pG106 (Jones et al. 2020) and the genome-integrative vector pMM656 (Mimee et al. 2015) were used. Plasmid pG106 comprises a 5 kb Mob-Rep region, which contains an oriV for the initiation of plasmid replication in P. vulgatus. For the selection of E. coli and P. vulgatus, the vector encodes an ermF-ermAM cassette. Stable transformation of pG106 into P. vulgatus was achieved by electroporation.
The pMM656 plasmid encodes an intN2 tyrosine integrase, which mediates sequence-specific recombination between the attN2 site of pMM656 and one of two attPV sites located in the 3′ ends of the two tRNASer genes, BVU_2451 and BVU_2094, on the P. vulgatus chromosome (Mimee et al. 2015). Insertion of the pMM656 plasmid inactivates one tRNASer gene, so simultaneous insertion in BVU_2451 and BVU_2094 is unlikely due to the essentiality of tRNASer. The gene encoding the luciferase NanoLuc functions as a reporter gene. Transformation of pMM656 into P. vulgatus was performed by biparental conjugation.
To evaluate the functionality of pMM656 in P. vulgatus, the luciferase reporter system encoded on the integrated vector was used. Expression of the nanoluc gene was mediated by the upstream promoter-RBS region (PRhaKIPAO) of the rhamnulose kinase gene (BT_3763) of B. thetaiotaomicron (Mimee et al. 2015). Gene expression was dependent on the concentration of rhamnose (0–4 mM) and demonstrated a response curve with an output dynamic range of 525-fold. Without induction by rhamnose, transcription of nanoluc could not be detected (Fig. 1). Above a rhamnose concentration of 4 mM maximal expression was observed, which did not influence the growth of P. vulgatus (data not shown). In this context, it is to mention that the functionality of the nanoluc gene was already shown for several Bacteroides strains and P. vulgatus by using anhydrotetracycline-inducible tet promoter constructs (Lim et al. 2017).
Fig. 1.
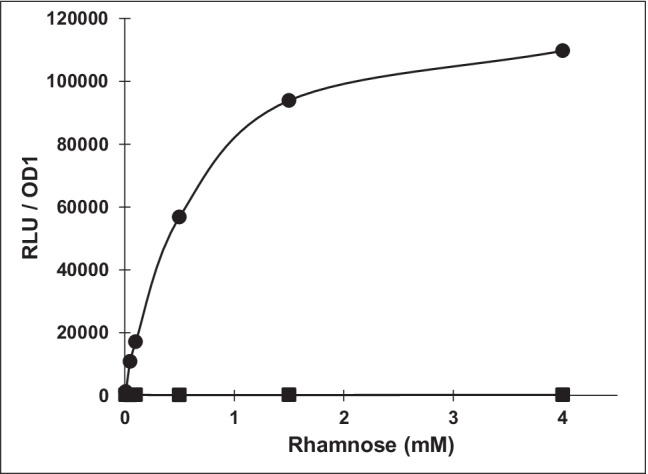
NanoLuc activity under the control of the rhamnose inducible promoter. P. vulgatus pMM656 cells were incubated for 24 h at 37 °C without rhamnose or serial dilutions of rhamnose starting at 0.05 mM. After lysis, the cell lysate was treated as described in “Materials and methods”. Relative light unit (RLU) values were normalized to OD 1. The experiment was conducted four times and the respective standard deviations are indicated by error bars
Application of expression vector pG106 and integration vector pMM656 for the redirection of the carbon flow in P. vulgatus
The metabolic versatile P. vulgatus produces acetate, formate, and lactate by fermentation and succinate by fumarate respiration. Furthermore, succinate can be decarboxylated to propionate. Substrates are complex heteropolysaccharides, which are degraded to pentoses or hexoses. When growing on a DMMG medium with glucose as substrate, the lactate production is very low compared to the other fermentative end products.
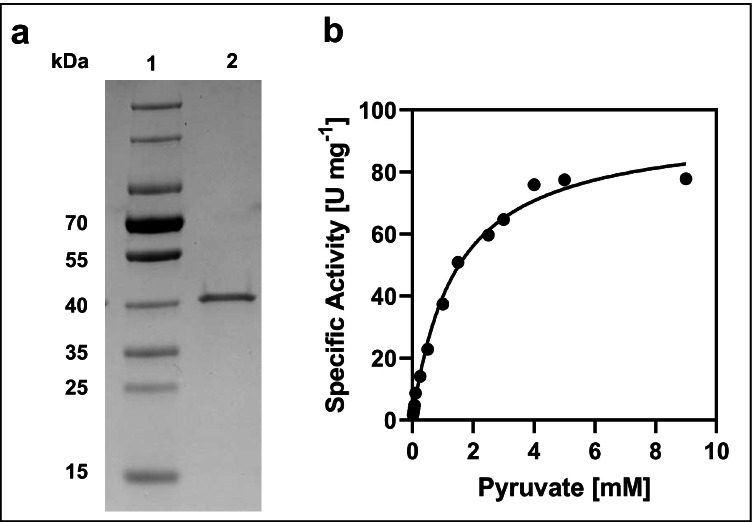
The question arose whether it is possible to increase the concentration of lactate by overexpressing a lactate dehydrogenase and thus modifying the central metabolism of P. vulgatus. To answer this question, the genome of P. vulgatus was screened for the presence of a gene encoding a lactate dehydrogenase. Based on bioinformatic analyses, we identified a ldh gene (BVU_2499) that potentially codes for a D-lactate dehydrogenase (D-LDH, EC 1.1.1.28). In general, NAD-dependent D-LDH catalyse the reduction of pyruvate to D-lactate coupled to the oxidation of NADH to NAD+ (Holbrook et al 1975). Compared to L-LDHs (EC 1.1.1.27), which are widely distributed in vertebrates and higher plants (Holbrook et al. 1975; Garvie 1980), D-LDHs are found only in some invertebrates and mainly in microorganisms (Garvie 1980). The corresponding motifs that characterize D-LDHs (Furukawa et al. 2018) can be found in the D-LDH amino acid sequence of P. vulgatus (Fig. S2). To verify the function of BVU_2499, the corresponding gene was amplified, ligated into the expression vector pASK-IBA5, and overproduced in E. coli. The N-terminal fused Strep-tag sequence allowed purification via Strep-Tactin affinity chromatography, leading to a total yield of 27 mg recombinant protein per l culture. The predicted molecular mass of the recombinant tagged protein was 39.1 kDa. Separation by polyacrylamide gel electrophoresis and subsequent silver staining revealed a single band within the elution fraction that corresponded to the expected size (Fig. 2a).
Fig. 2.
Characterization of purified D-LDH from P. vulgatus, overproduced in E. coli. a SDS/PAGE analysis of recombinant D-LDH using silver staining. Lane 1: PageRulerTM Prestained Protein Ladder (ThermoFisher Scientific). Lane 2: purified D-LDH (1.5 μg protein). b Michaelis–Menten kinetics. For the detection of NADH oxidation, purified D-LDH and pyruvate in different concentrations were added and the change in absorbance at 340 nm was analyzed at 37 °C. The enzyme assay (1 ml) contained 100 mM K-phosphate buffer, pH 7, and 250 μM NADH. The pyruvate concentration was varied between 0.02 and 9 mM. Nonlinear regression of the Michaelis–Menten data was used to calculate the kinetic constants Vmax and Km with the program GraphPad Prism
To analyze the activity of the enzyme, the oxidation of NADH to NAD+ during the conversion of pyruvate to D-lactate was measured at a wavelength of 340 nm. Purified D-LDH showed the highest specific activity at 60 °C and pH 7. However, the half-life of the enzyme was less than 2 min at this temperature, indicating that the enzyme was not thermostable. The optimal growth temperature of P. vulgatus is 37 °C, which correlates with the physiological temperature of the human gut, the natural habitat of P. vulgatus. At this temperature, the enzyme was stable over several hours and showed 86.6% of the activity at 60 °C (data not shown). Kinetic parameters were determined from the nonlinear regression of the Michaelis–Menten data (Fig. 2b). Using pyruvate as a substrate, the KM and Vmax values were 1.4 ± 0.1 mM and 95.6 ± 3.0 U/ mg protein at a temperature of 37 °C, respectively. These results correspond to values given in the literature (Stoll et al. 1998; Furukawa et al. 2014; Kochhar et al. 1992).
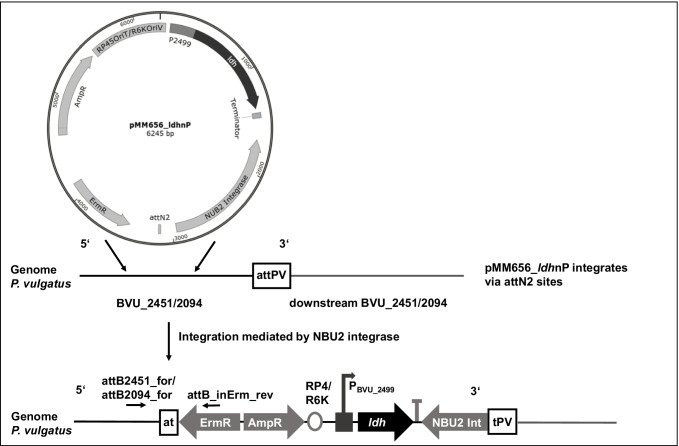
For homologous expression of ldh from P. vulgatus, the gene was cloned into the multiple cloning site of the shuttle vector pG106, resulting in the plasmid pG106_ldhnP. Since little is known about promoter structures in P. vulgatus, 250 bp of the upstream region of the gene was also cloned for initiation of transcription. For homologous expression by the genome-integrative vector pMM656, the B. thetaiotaomicron-derived promoter PRhaKIPAO and the coding gene for NanoLuc from pMM656 were substituted against ldh (BVU_2499) including the 250 bp upstream region with the presumed promoter region (Fig. 3). The corresponding plasmid was referred to as pMM656_ldhnP. After conjugation, the DNA sequence of the PCR fragment generated by the primer pair attB2094_for and attB_inErm_rev demonstrated that the vector was integrated into BVU_2094 on the genome of P. vulgatus, mediated via the attN2 site of pMM656 (Fig. S1).
Fig. 3.
Schematic representation of the integration of pMM656_ldhnP into the genome of P. vulgatus. The promoter PRhaKIPAO and the coding gene of NanoLuc of pMM656 were replaced with the ldh gene (BVU_2499) of P. vulgatus including 250 bp of the upstream region. The tyrosine integrase NBU2 Int catalyzed the stable integration of pMM656_ldhnP into one of two attN2 sites in the P. vulgatus genome. The two attN2 sites are located at the 3′ ends of tRNASer genes (BVU_2451 at nt 3,152,564 and BVU_2094 at nt 2,710,348, respectively). AmpR, ampicillin resistance cassette; ErmR, erythromycin resistance cassette; RP4, origin of transfer; R6K, origin of replication. Black horizontal arrows indicate the position of the primer pair attB2451_for/attB2094_for and attB_inErm_rev. The primers were used to generate a DNA fragment for sequencing and to determine the position of integration
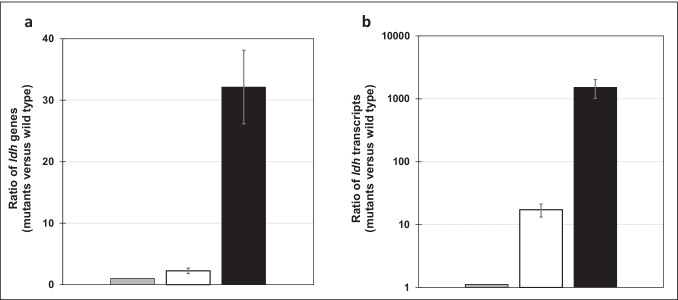
To analyze the conditions for homologous overproduction of D-LDH in P. vulgatus, the copy number of the ldh gene in each construct was determined by qPCR. The quantification cycle (Ct) of the gene encoding the ribosomal protein L23 (BVU_0803) was used as a reference. It should be noted that the ldh gene and the l23 gene are encoded only once in the genome of the wild type. As expected, the highest amount of amplicons of the ldh gene was detected in P. vulgatus pG106_ldhnP containing the plasmid-encoded version of the ldh gene (Fig. 4a). The number of PCR fragments from the ldh gene was 32 ± 6.0-fold higher in P. vulgatus pG106_ldhnP and 2.2 ± 0.4-fold higher in P. vulgatus pMM656_ldhnP, respectively compared to the wild type containing a single ldh copy. Hence, it is evident that besides the chromosomal version of the ldh gene, a second copy was present due to the integration of the vector pMM656_ldhnP into the genome of P. vulgatus pMM656_ldhnP via one of the two attN2 sites. It can also be concluded that 32 ± 6.0 copies of the ldh gene were present in the plasmid-containing strain, indicating that the copy number of the shuttle vector pG106_ldhnP is in the range of 30.
Fig. 4.
ldh copy number (a) and ldh transcripts abundance (b) in the wild type and genetically modified mutants of P. vulgatus. a To estimate the copy number, qPCR was performed to compare chromosomal and plasmid-encoded ldh of the mutant strains with the single-copy chromosomal ldh of the wild type strain. b The relative amount of ldh transcripts was analyzed by RT-qPCR. Experiments shown in a and b were conducted in duplicate using DNA or RNA preparation from three different cultures harvested in the mid-exponential growth phase. ΔCt values were determined by subtracting the average Ct values of the ldh gene (BVU_2499) from the reference gene encoding the ribosomal protein L23 (BVU_0803). Ratios were calculated from ΔΔCt values using the function 2–ΔΔCt. Standard deviations are indicated by error bars. Gray bars, wild type; white bars, P. vulgatus pMM656_ldhnP; black bars, P. vulgatus pG106_ldhnP
To determine the transcript abundance, RT-qPCR was performed to compare the expression level of chromosomal and plasmid-encoded ldh of the mutant strains with the expression level of the single-copy chromosomal ldh gene of the wild-type strain. Total RNA was extracted from each P. vulgatus mutant and the wild type grown in BHI and harvested in the mid-log phase. mRNA concentrations were quantified using the same gene-specific primers used in qPCR experiments (Table 1) to determine the copy number of the ldh gene. Compared to the wild type, a 1521 ± 506-fold higher transcript abundance was detected in P. vulgatus carrying the shuttle vector pG106_ldhnP. In P. vulgatus harbouring the genome-integrative vector pMM656_ldhnP, the transcript level of the ldh gene was 17 ± 4-fold higher in comparison to the wild-type strain (Fig. 4b). These data indicate that P. vulgatus expressed ldh in considerably higher amounts in the shuttle vector containing strain, and is a suitable system for the overproduction of LDH as a model for the modulation of the intracellular carbon flow.
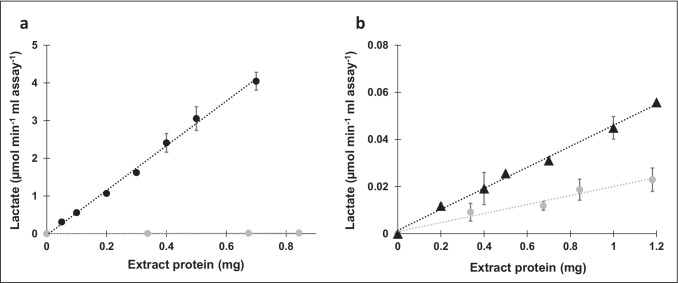
Enzyme activity of LDH in cell-free extract of P. vulgatus
To evaluate the effect of the ldh transcription levels on the corresponding enzyme yields, D-LDH activity assays were performed using cytoplasmic fractions of the investigated P. vulgatus strains. The change of absorbance at a wavelength of 340 nm was measured, corresponding to the oxidation of NADH to NAD+ during the conversion of pyruvate to D-lactate. A much higher D-LDH activity was detected in the cytoplasm of P. vulgatus pG106_ldhnP (5.9 ± 0.4 U/mg protein) compared to P. vulgatus pMM656_ldhnP (0.046 ± 0.002 U/mg protein) and the wild-type strain of P. vulgatus (0.025 ± 0.002 U/mg protein) (Fig. 5). All assays revealed a stringent correlation between the amount of D-LDH and the lactate formation rate. Considering the ratios of enzyme activities of the D-LDH normalized to that of the wild type, the activity of the D-LDH overexpressing mutant P. vulgatus pG106_ldhnP and the genome-integrative mutant P. vulgatus pMM656_ldhnP was 236 and 1.8 times higher, respectively.
Fig. 5.
Photometric analysis of enzyme activity of D-LDH in cell-free extract. Determination of the activity of D-LDH from P. vulgatus pG106_ldhnP (a) and P. vulgatus pMM656_ldhnP (b) compared to wild type. Enzyme activities were assayed using cell-free extract by measuring the change in absorbance at 340 nm corresponding to the oxidation of NADH to NAD+ during the conversion of pyruvate to D-lactate. The experiment was performed in duplicate using the cell-free extract from two different cultures and the respective standard deviations are indicated by error bars. Black circles, P. vulgatus pG106_ldhnP; black triangles, P. vulgatus pMM656_ldhnP; gray circles, wild type
Influence of LDH activity on the ratio of metabolic end products of P. vulgatus
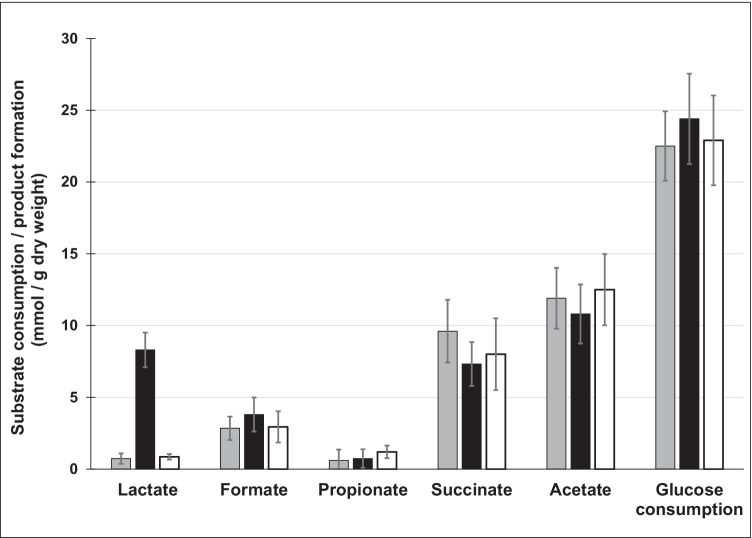
To test the effect of the increased abundance of ldh transcripts and the elevated activity of D-LDH on the central metabolism of P. vulgatus pG106_ldhnP and P. vulgatus pMM656_ldhnP in comparison to the wild type, the end products of glucose conversion were determined by HPLC analysis. Several cultures of the respective strains were grown in a minimal medium with glucose as the substrate. Cells were harvested at different optical densities in the exponential growth phase, and the supernatant was used for product analysis. For the exact determination of the growth behaviour, product and substrate concentrations were correlated with the dry weight (DW) of the respective culture.
As expected, succinate, acetate, lactate, formate, and propionate were found as products of glucose fermentation. However, a major difference was observed with respect to the formation of lactate (Fig. 6). The wild-type strain of P. vulgatus produced 0.73 ± 0.36 mmol lactate/g DW. With 0.85 ± 0.2 mmol/g DW, P. vulgatus pMM656_ldhnP formed a little more lactate than the wild-type strain. In the shuttle vector carrying mutant P. vulgatus pG106_ldhnP, a level of 8.3 ± 1.2 mmol/g DW lactate was detected. Compared to the wild type, this corresponded to a ratio of 1:11.4, and compared to P. vulgatus pMM656_ldhnP, 9.8 times as much lactate was produced (Fig. 6). Substrate consumption of the individual strains was also different (Fig. 7). The P. vulgatus wild type and the mutant containing a second chromosomal copy of the ldh gene consumed 22.5 ± 2.4 and 22.9 ± 3.13 mmol glucose/g DW, respectively, whereas the plasmid-harbouring strain metabolized significantly (p ≤ 0.05) more glucose (24.4 ± 3.15 mmol glucose/g DW) compared to the wild type (Figs. 7, S3). A closer look at the products indicated that the formation of succinate and acetate was slightly lower in P. vulgatus pG106_ldhnP compared to the wild type and P. vulgatus pMM656_ldhnP. The concentrations of formate and propionate were in the same range of 1–2 mmol/g DW.
Fig. 6.
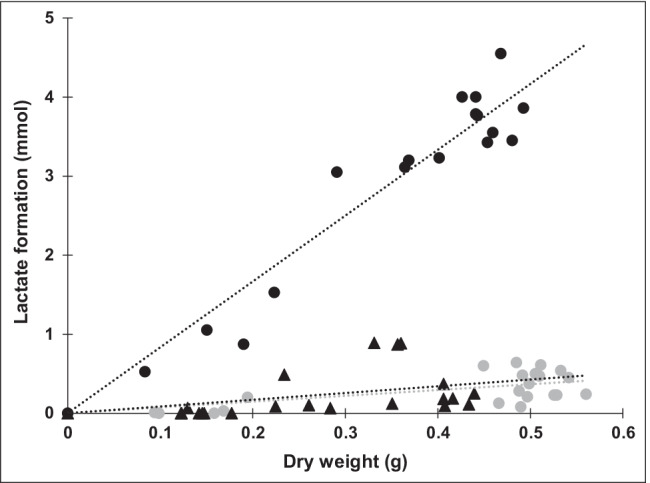
Lactate formation by the wild type and the mutants. P. vulgatus pG106_ldhnP (black circles), P. vulgatus pMM656_ldhnP (black triangles), and the wild type (gray circles) were grown in minimal medium with glucose as substrate. At least 16 cultures of each strain were harvested in the exponential growth phase and the supernatants were analyzed by HPLC. The lactate concentration was correlated to the dry weight of the corresponding culture. For a culture with an optical density of 1.0, the DW was 360 mg per liter culture. Lactate yields per g DW were calculated from the slope of the regression lines
Fig. 7.
Substrate consumption and product formation. P. vulgatus pG106_ldhnP (black), P. vulgatus pMM656_ldhnP (white), and the wild type (gray) were grown in DMMG medium. Cultures were harvested and the supernatants analyzed by HPLC. The concentrations of the metabolic end products were correlated to the dry weight of the corresponding cultures. The values represent the average of at least 16 different cultures for each strain (cultures from Fig. 6). Standard deviations are indicated by error bars
The additional carbon required for the increased amount of lactate was reflected in a slightly increased glucose consumption and a decreased amount of succinate and acetate produced by P. vulgatus pG106_ldhnP. In summary, the data clearly indicate that the plasmid-based overexpression of the ldh gene resulted in a highly increased number of transcripts and enhanced activity of the D-LDH within the cells of P. vulgatus pG106_ldhnP. This in turn leads to a shift in the product spectrum towards the increased production of lactate.
Discussion
The human gastrointestinal tract represents one of the densest microbial communities known in nature. This habitat is dominated mainly by the two bacterial phyla Firmicutes and Bacteroidetes (Faith et al. 2013; Human Microbiome Project Consortium 2012). The latter phylum comprises genera such as Bacteroides, Alistipes, Parabacteroides, Phocaeicola, and Prevotella (Wexler and Goodman 2017). Recently, some members of the genus Bacteroides were taxonomically separated and assigned to the genus Phocaeicola. Among these species is P. vulgatus as one of the most prominent gut bacteria (García-López et al. 2019).
Members of the mentioned genera possess enzyme systems for the degradation of complex polysaccharides (Hill 1995; Ley et al. 2008; Flint et al. 2012), which cannot be digested by the human host. Fermentation of these dietary fibres ultimately leads to the formation of short-chain fatty acids (mainly butyrate, propionate, and acetate), which are released into the intestinal lumen and taken up by host colonocytes (Fischbach and Sonnenburg 2011; Tremaroli and Bäckhed 2012; den Besten et al. 2013). This production of organic acids from biomass conversion is gaining economic attention (McKinlay et al. 2007; Baumann and Westermann 2016). Since Bacteroides spp. and Phocaeicola spp. are known to be highly effective producers of succinate, acetate, and propionate (Scheifinger and Wolin 1973; Salonen et al. 2014), the production of these organic acids by the corresponding organisms could be of great interest in future biotechnological applications.
In general, the production of these platform chemicals on a biotechnological scale by members of these genera requires genetic manipulation of the metabolism towards the improvement of the corresponding biochemical pathways. There is already a variety of genetic tools available for members of the family Bacteriodaceae, which includes the genera Bacteroides and Phocaeicola. However, most of the genetic tools were designed for B. thetaiotaomicron, B. fragilis, and B. ovatus, all classified as risk group 2 organisms and therefore not applicable for biotechnological applications. Moreover, it was shown that Bacteroides species possess unique promoter structures (Bayley et al. 2000; Vingadassalom et al. 2005) and ribosome binding sites (Smith et al. 1992; Accetto and Avguštin 2011; Wegmann et al. 2013). Hence, genetic systems for targeted gene manipulation developed for members of the Proteobacteria, especially E. coli, cannot be used in members of the family Bacteriodaceae. Many Bacteroides spp. possess small cryptic plasmids associated with transmissible antibiotic resistance and genes for conjugal transfer (Smith 1995), which can function as a basis for the construction of conjugative shuttle vectors. Some shuttle vectors, with constitutive or inducible promoters, have already been employed for regulated gene expression in B. thetaiotaomicron, B. fragilis, and B. ovatus (Smith 1995; Hamady et al. 2008; Parker and Smith 2012; Horn et al. 2016; Lim et al. 2017; Jones et al. 2020), but most are not commercially available. In addition, a group of plasmids was identified that enable transposon mutagenesis based on a marine transposon (Goodman et al. 2009). Deletion mutants in Bacteroides spp. were generated by standard allelic replacement techniques using conjugal suicide vectors in combination with erythromycin selection (Chatzidaki-Livanis et al. 2009; Nakayama-Imaohji et al. 2012) or antibacterial counterselection markers (Bencivenga-Barry et al. 2020).
Here, we used the commercially available integration vector pMM656 (Mimee et al. 2015) and the shuttle vector pG106 (Jones et al. 2020) for homologous gene expression in P. vulgatus. The function of pMM656 is based on the integration of the vectors into the so-called “attN sites”, which are located at the 3’ end of the genes of the tRNAs for serine. Bioinformatic analysis revealed that these attN sites are also present in P. vulgatus. Our results showed that the B. thetaiotaomicron–specific and rhamnose-induced promoter upstream of the NanoLuc reporter gene worked as expected, indicated by the rhamnose-dependent expression of the luciferase. Furthermore, the exchange of this promoter/gene sequence against the ldh gene led to an increase of the corresponding mRNA by 17-fold. Hence, pMM656 is a suitable tool for applications of molecular cloning including complementation and gene expression studies in P. vulgatus.
The second plasmid pG106 applied in this study is used as a shuttle vector for genetic manipulation in B. thetaiotaomicron or Porphyromonas spp. (Jones et al. 2020). A copy number of about six was determined for B. thetaiotaomicron whereas we detected about 30 copies indicating that the plasmid has a medium copy number in P. vulgatus. To test the genetic accessibility of P. vulgatus, homologous overexpression of the ldh gene was chosen as a proof of concept in this study. Therefore, the ldh gene and its upstream region (250 bp) were cloned into pG106 replacing the NanoLuc reporter gene and its promoter. The plasmid was referred to as pG106_ldhnP. In relation to the copy number of the ldh gene, an approx. 1500-fold increase in the ldh transcript concentration was observed in P. vulgatus pG106_ldhnP in comparison to the parental strain. Hence, the results also indicate that the 5’ UTR of the ldh gene encodes a strong promoter. Representatives of the Bacteroidetes were shown to typically have promoter structures with a TAnn(T)TTG and (T)TTG region located 7 base pairs and 33 base pairs upstream of the transcription start of the corresponding gene, respectively (Chen et al. 2007; Lim et al. 2017; Bayley et al. 2000). Corresponding motifs are highly conserved in the 5’ UTR region of the ldh gene of P. vulgatus. In addition to the enhanced transcript concentration, the enzyme activity of D-LDH increased by a factor of more than 200 in the overexpression strain, compared to the wild type. This result suggests that this tool is a suitable system for the overproduction of proteins in P. vulgatus.
The increased amount of D-LDH in the cells had a severe impact on the end product composition. Under the growth condition employed, P. vulgatus produced only minor amounts of lactate (< 1 mmol/g DW). The enhanced activity of D-LDH in P. vulgatus pG106_ldhnP led to an 11.4 times higher production rate of lactate compared to the parental strain. The final concentration was in the range of 8.3 mmol/g DW, which was comparable to the concentration of the major end products succinate and acetate. Considering the amount of glucose used for the catabolic metabolism (14.4 mmol glucose/g DW), the proportion of lactate in the overall catabolic carbon flow increased from 2.9% (wild type) to 28.8% in the LDH-overproducing mutant. The increased amount of lactate formed by P. vulgatus pG106_ldhnP compared to the wild-type strain was probably compensated by the higher consumption of glucose (plus 1.9 mmol/g DW) and the lower amount of the metabolic end products succinate (minus 2.3 mmol/g DW) and acetate (minus 1.1 mmol/g DW).
To date, strains of the family Bacteroidaceae have not been used for biotechnological processes. However, it is known that members of this taxonomical group are highly effective succinate and propionate producers and possess sophisticated enzyme systems for the degradation of complex polysaccharides. In the future, these species could function as platform organisms for the efficient and sustainable conversion of renewable resources into bioactive compounds and important bulk chemicals. However, a genetic system would be desirable to allow a defined modelling of the intracellular carbon flux. Here, we demonstrated that P. vulgatus was genetically accessible and that appropriate expression systems were applicable. Thus, this approach formed the basis for targeted genetic optimization of P. vulgatus, namely the homologous expression of the ldh gene as a proof of concept for the manipulation of the central carbon flow. Furthermore, improvement of lactate production could be possible by the deletion of genes encoding enzymes, which are involved in the synthesis of other metabolic end products. Examples are the phosphoenolpyruvate carboxykinase or fumarate reductase to prevent the formation of succinate and propionate as well as the pyruvate: formate lyase to stop the synthesis of formate (Franke and Deppenmeier 2018). In the long run, the results could also strengthen the research on the gut microbiota in relation to medically relevant Bacteroides and Phocaeicola spp..
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Natalie Thum-Schmitz (Institute of Microbiology and Biotechnology, University of Bonn, Germany) for technical assistance. We also thank Kevin R. Jones (The Philips Institute for Oral Health Research, Virginia Commonwealth University, Richmond, VA, USA) for providing the shuttle vector pG106.
Author contribution
RL planned experiments, performed experiments, analyzed data, and wrote the paper. UD designed the study, planned experiments, analyzed data, wrote the paper, and supervised the study.
Funding
Open Access funding enabled and organized by Projekt DEAL. The study was supported by fundings of the BMBF project BaPro (031B0846A). The funding agency was not involved in the research.
Availability of data and material
Data are available upon request from the authors.
Code availability
Not applicable.
Declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Accetto T, Avguštin G. Inability of Prevotella bryantii to form a functional Shine-Dalgarno interaction reflects unique evolution of ribosome binding sites in Bacteroidetes. PLoS ONE. 2011;6:e22914. doi: 10.1371/journal.pone.0022914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley DP, Rocha ER, Smith CJ. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol Lett. 2000;193:149–154. doi: 10.1111/j.1574-6968.2000.tb09417.x. [DOI] [PubMed] [Google Scholar]
- Baumann I, Westermann P. Microbial production of short chain fatty acids from lignocellulosic biomass: current processes and market. Biomed Res Int. 2016;2016:8469357. doi: 10.1155/2016/8469357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga-Barry NA, Lim B, Herrera CM, Trent MS, Goodman AL. Genetic manipulation of wild human gut bacteroides. J Bacteriol. 2020;202:e00544–19. doi: 10.1128/JB.00544-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. doi: 10.1002/elps.1150080203. [DOI] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chang DE, Jung HC, Rhee JS, Pan JG. Homofermentative production of D- or L-lactate in metabolically engineered Escherichia coli RR1. Appl Environ Microbiol. 1999;65:1384–1389. doi: 10.1128/AEM.65.4.1384-1389.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C, Goumy V, Leclerc M, Del'homme C, Bernalier-Donadille A. Characterization of the xylan-degrading microbial community from human faeces. FEMS Microbiol Ecol. 2007;61:121–131. doi: 10.1111/j.1574-6941.2007.00314.x. [DOI] [PubMed] [Google Scholar]
- Chatzidaki-Livanis M, Coyne MJ, Comstock LE. A family of transcriptional antitermination factors necessary for synthesis of the capsular polysaccharides of Bacteroides fragilis. J Bacteriol. 2009;191:7288–7295. doi: 10.1128/JB.00500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bagdasarian M, Kaufman MG, Walker ED. Characterization of strong promoters from an environmental Flavobacterium hibernum strain by using a green fluorescent protein-based reporter system. Appl Environ Microbiol. 2007;73:1089–1100. doi: 10.1128/JB.00401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vadder F, Mithieux G. Gut-brain signaling in energy homeostasis: the unexpected role of microbiota-derived succinate. J Endocrinol. 2018;236:R105–R108. doi: 10.1530/JOE-17-0542. [DOI] [PubMed] [Google Scholar]
- Dodd D, Mackie RI, Cann IK. Xylan degradation, a metabolic property shared by rumen and human colonic Bacteroidetes. Mol Microbiol. 2011;79:292–304. doi: 10.1111/j.1365-2958.2010.07473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. 2015;74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- Franke T, Deppenmeier U. Physiology and central carbon metabolism of the gut bacterium Prevotella copri. Mol Microbiol. 2018;109(4):528–540. doi: 10.1111/mmi.14058. [DOI] [PubMed] [Google Scholar]
- Furukawa N, Miyanaga A, Togawa M, Nakajima M, Taguchi H. Diverse allosteric and catalytic functions of tetrameric d-lactate dehydrogenases from three Gram-negative bacteria. AMB Express. 2014;4:76. doi: 10.1186/s13568-014-0076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa N, Miyanaga A, Nakajima M, Taguchi H. Structural basis of sequential allosteric transitions in tetrameric d-lactate dehydrogenases from three gram-negative bacteria. Biochemistry. 2018;57:5388–5406. doi: 10.1021/acs.biochem.8b00557. [DOI] [PubMed] [Google Scholar]
- García-López M, Meier-Kolthoff JP, Tindall BJ, Gronow S, Woyke T, Kyrpides NC, Hahnke RL, Göker M. Analysis of 1,000 type-strain genomes improves taxonomic classification of Bacteroidetes. Front Microbiol. 2019;10:2083. doi: 10.3389/fmicb.2019.02083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvie EI. Bacterial lactate dehydrogenases. Microbiol Rev. 1980;44:106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Vlamakis H, Shoemaker N, Salyers AA. A new Bacteroides conjugative transposon that carries an ermB gene. Appl Environ Microbiol. 2003;69:6455–6463. doi: 10.1128/AEM.69.11.6455-6463.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady ZZR, Farrar MD, Whitehead TR, Holland KT, Lodge JPA, Carding SR. Identification and use of the putative Bacteroides ovatus xylanase promoter for the inducible production of recombinant human proteins. Microbiology (Reading) 2008;154:3165–3174. doi: 10.1099/mic.0.2008/019109-0. [DOI] [PubMed] [Google Scholar]
- Hill MJ. Bacterial fermentation of complex carbohydrate in the human colon. Eur J Cancer Prev. 1995;4:353–358. doi: 10.1097/00008469-199510000-00004. [DOI] [PubMed] [Google Scholar]
- Holbrook JJ, Liljas A, Steindel SJ, Rossmann MG (1975) Lactate dehydrogenase. In: The Enzymes (Boyer PD, ed.) 3rd ed., Vol. 11, pp 191−292, Academic Press, New York
- Horn N, Carvalho AL, Overweg K, Wegmann U, Carding SR, Stentz R. A novel tightly regulated gene expression system for the human intestinal symbiont Bacteroides thetaiotaomicron. Front Microbiol. 2016;7:1080. doi: 10.3389/fmicb.2016.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KR, Belvin BR, Macrina FL, Lewis JP. Sequence and characterization of shuttle vectors for molecular cloning in Porphyromonas, Bacteroides and related bacteria. Mol Oral Microbiol. 2020;35:181–191. doi: 10.1111/omi.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochhar S, Chuard N, Hottinger H. Cloning and overexpression of the Lactobacillus bulgaricus NAD(+)-dependent D-lactate dehydrogenase gene in Escherichia coli: purification and characterization of the recombinant enzyme. Biochem Biophys Res Commun. 1992;185:705–12. doi: 10.1016/0006-291x(92)91683-h. [DOI] [PubMed] [Google Scholar]
- Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;15(23):1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Koressaar T, Lepamets M, Kaplinski L, Raime K, Andreson R, Remm M. Primer3_masker: integrating masking of template sequence with primer design software. Bioinformatics. 2018;34:1937–1938. doi: 10.1093/bioinformatics/bty036. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Zimmermann M, Barry NA, Goodman AL. Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell. 2017;169:547–558. doi: 10.1016/j.cell.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Imlay JA (2021) When anaerobes encounter oxygen: mechanisms of oxygen toxicity, tolerance and defence. Nat Rev Microbiol 10.1038/s41579-021-00583-y [DOI] [PMC free article] [PubMed]
- Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23:705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- Mastropaolo MD, Thorson ML, Stevens AM. Comparison of Bacteroides thetaiotaomicron and Escherichia coli 16S rRNA gene expression signals. Microbiology (Reading) 2009;155:2683–2693. doi: 10.1099/mic.0.027748-0. [DOI] [PubMed] [Google Scholar]
- McKinlay JB, Vieille C, Zeikus JG. Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol. 2007;76:727–740. doi: 10.1007/s00253-007-1057-y. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mimee M, Tucker AC, Voigt CA, Lu TK. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 2015;1:62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama-Imaohji H, Ichimura M, Iwasa T, Okada N, Ohnishi Y, Kuwahara T. Characterization of a gene cluster for sialoglycoconjugate utilization in Bacteroides fragilis. J Med Invest. 2012;59:79–94. doi: 10.2152/jmi.59.79. [DOI] [PubMed] [Google Scholar]
- Parker AC, Smith JC. Development of an IPTG inducible expression vector adapted for Bacteroides fragilis. Plasmid. 2012;68:86–92. doi: 10.1016/j.plasmid.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasolli E, Asnicar F, Manara S, Zolfo M, Karcher N, Armanini F, Beghini F, Manghi P, Tett A, Ghensi P, Collado MC, Rice BL, DuLong C, Morgan XC, Golden CD, Quince C, Huttenhower C, Segata N. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176:649–662. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Chassard C, Lawson PA, Bernalier-Donadille A (2007) Bacteroides cellulosilyticus sp. nov., a cellulolytic bacterium from the human gut microbial community. Int J Syst Evol Microbiol. 57(Pt 7):1516-1520 10.1099/ijs.0.64998-0 [DOI] [PubMed]
- Russell WR, Duncan SH, Scobbie L, Duncan G, Cantlay L, Calder AG, Anderson SE, Flint HJ. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol Nutr Food Res. 2013;57:523–535. doi: 10.1002/mnfr.201200594. [DOI] [PubMed] [Google Scholar]
- Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, Date P, Farquharson F, Johnstone AM, Lobley GE, Louis P, Flint HJ, de Vos WM. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA. Bacteroides of the human lower intestinal tract. Annu Rev Microbiol. 1984;38:293–313. doi: 10.1146/annurev.mi.38.100184.001453. [DOI] [PubMed] [Google Scholar]
- Salyers AA, Bonheyo G, Shoemaker NB. Starting a new genetic system: lessons from Bacteroides. Methods. 2000;20:35–46. doi: 10.1006/meth.1999.0903. [DOI] [PubMed] [Google Scholar]
- Scheifinger CC, Wolin MJ. Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl Environ Microbiol. 1973;26:789–795. doi: 10.1128/am.26.5.789-795.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, Rogers MB, McKee ML. Heterologous gene expression in Bacteroides fragilis. Plasmid. 1992;27:141–154. doi: 10.1016/0147-619x(92)90014-2. [DOI] [PubMed] [Google Scholar]
- Smith CJ. Genetic transformation of Bacteroides spp. using electroporation. Methods Mol Biol. 1995;47:161–169. doi: 10.1385/0-89603-310-4:161. [DOI] [PubMed] [Google Scholar]
- Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996;81:288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- Stoll VS, Manohar AV, Gillon W, MacFarlane EL, Hynes RC, Pai EF. A thioredoxin fusion protein of VanH, a D-lactate dehydrogenase from Enterococcus faecium: cloning, expression, purification, kinetic analysis, and crystallization. Protein Sci. 1998;7(5):1147–55. doi: 10.1002/pro.5560070508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;7:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Untergasser S, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Research 40(15, 1) e115 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed]
- Valguarnera E, Wardenburg JB. Good gone bad: one toxin away from disease for Bacteroides fragilis. J Mol Biol. 2020;432:765–785. doi: 10.1016/j.jmb.2019.12.003. [DOI] [PubMed] [Google Scholar]
- Varel VH, Bryant MP. Nutritional features of Bacteroides fragilis subsp. fragilis. Appl Microbiol. 1974;28:251–257. doi: 10.1128/am.28.2.251-257.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vingadassalom D, Kolb A, Mayer C, Rybkine T, Collatz E, Podglajen I. An unusual primary sigma factor in the Bacteroidetes phylum. Mol Microbiol. 2005;56:888–902. doi: 10.1111/j.1365-2958.2005.04590.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Shoemaker NB, Wang GR, Salyers AA. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J Bacteriol. 2000;182:3559–3571. doi: 10.1128/JB.182.12.3559-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann U, Horn N, Carding SR. Defining the Bacteroides ribosomal binding site. Appl Environ Microbiol. 2013;79:1980–1989. doi: 10.1128/AEM.03086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler A, Goodman A. An insider’s perspective: Bacteroides as a window into the microbiome. Nat Microbiol. 2017;2:17026. doi: 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin EA, Wolin MJ, Wolfe RS. Formation of methane by bacterial extracts. J Biol Chem. 1963;238:2882–2886. doi: 10.1016/S0021-9258(18)67912-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request from the authors.
Not applicable.