Abstract
Radiomics analysis quantifies the interpolation of multiple and invisible molecular features present in diagnostic and therapeutic images. Implementation of 18-fluorine-fluorodeoxyglucose positron emission tomography/computed tomography (18F-FDG PET/CT) radiomics captures various disorders in non-invasive and high-throughput manner. 18F-FDG PET/CT accurately identifies the metabolic and anatomical changes during cancer progression. Therefore, the application of 18F-FDG PET/CT in the field of oncology is well established. Clinical application of 18F-FDG PET/CT radiomics in lung infection and inflammation is also an emerging field. Combination of bioinformatics approaches or textual analysis allows radiomics to extract additional information to predict cell biology at the micro-level. However, radiomics texture analysis is affected by several factors associated with image acquisition and processing. At present, researchers are working on mitigating these interrupters and developing standardised workflow for texture biomarker establishment. This review article focuses on the application of 18F-FDG PET/CT in detecting lung diseases specifically on cancer, infection and inflammation. An overview of different approaches and challenges encountered on standardisation of 18F-FDG PET/CT technique has also been highlighted. The review article provides insights about radiomics standardisation and application of 18F-FDG PET/CT in lung disease management.
Keywords: 18F-FDG PET/CT, Biomarker, Lung diseases, Radiomics feature, Standardisation
Key points
The micro-level changes in lung disease cannot be detected by CT alone.
Employment of 18F-FDG PET/CT radiomics moves toward patient-specific of lung diseases management.
However, numerous features and techniques of feature extraction have raised tremendous complexity.
This complexity can only be removed by standardisation of radiomics analysis.
The artificial intelligence and machine learning with radiomics analysis improve diagnosis and treatment.
Background
Molecular haracterisation of physiological abnormality is conventionally performed by biopsy or random sample collection of the suspected site for diagnosis and identification. However, biopsy is an invasive process, and it has risk of complication such as infection [1, 2]. Lung cancer is one of the leading causes of respiratory morbidity and mortality among adults [3]. Thus, to avoid this risk, medical imaging techniques, for instance magnetic resonance imaging (MRI), computed tomography (CT) and positron emission tomography (PET), and hybrid imaging modalities such as PET/CT and PET/MRI are widely used to determine the tumour heterogeneity and morphological abnormalities in a non-invasive manner [4, 5]. 18F-Fluorodeoxyglucose (18F-FDG) PET/CT is one of the prevailing diagnostic tools, especially in the field of oncology and other clinical disorders owing to its higher accuracy in diagnosis, prognosis and therapeutic response assessment [6–11]. Lung cancer is one of the primary causes of death around the world [12]. The application of 18F-FDG PET/CT is recognised to reduce the morbidity rate [13]. PET imaging has also gathered attention in the field of neuroimaging and cardiac imaging and vascular abnormality detection [14–16].
Medical images have the ability to capture cellular- and molecular-level tumour characteristics reflected in phenotype [17–20]. A study on CT image of anaplastic lymphoma kinase mutations in lung tumours captured substantial pulmonary fluid and no pulmonary tails [21]. In another study, CT images of complementary contrast demonstrated that the metamorphosis of von Hippel–Lindau located in renal cell carcinoma is considerably correlated with the total intra-tumoral vasculariSation, sharp edges of the tumour and nodular advancement of the tumour [22]. Conventionally, CT scan is performed on different stages of cancer treatment to understand the drug efficacy [23]. Before surgery, the severity of lung cancer is assessed performing therapeutic techniques and invasive diagnosis. These are termed as conventional workup. Integration of 18F-FDG PET with therapeutic techniques and invasive diagnosis leads to 51% consequential decline of impractical thoracotomy such as eliminating one in five critical surgeries comprehensively when compared with conventional workup exclusively, in the PET in lung cancer staging multicentre randomised trial [24].
Rise of glucose metabolism is a well-recognised hallmark of cancer, and molecular PET imaging mainly captures these increased metabolism for diagnostic purpose [25]. Cell sites exposed to abnormal proliferation, infection and inflammation can be determined by identifying the high uptake and accumulation of the glucose analogue, 18F-FDG [13, 26]. 18F-FDG combining with the glucose transporters present in cell and phosphorylated by hexokinase results in 18F-FDG-6-phosphate misses 2-hydroxyl group needed for glycolysis [13, 27, 28]. This complex chemical component remains metabolically trapped in the cell and can be detected through PET imaging [13, 14, 29]. 18F-FDG uptake in PET imaging depends on the number of active cancer cells, histopathology of tumour, and biological processes responsible for continuous oncogenesis [30–33]. Therefore, studies have shown that heterogeneity of tumour may be correlated with the sparse distribution 18F-FDG distribution [17, 34, 35]. The 18F-FDG PET has been proven to have greater mediastinal staging capability than CT in according to a meta-analysis study [36]. Furthermore, 18F-FDG PET/CT produces more precise image quantification information compared to either imaging modality individually [37].
Radiomics analysis is an emerging field in the medical imaging sector, and it is recognised as a promising classification tool that holds the innate potential of revolutionising disease diagnosis specially cancer [38–42]. Radiomics has been introduced in imaging field to strengthen the conventional and manual image comprehension by recognising features and patterns, which largely remains undetected to the human eye [43, 44]. Radiomics enables extraction, collection and evaluation of higher order and statistical datasets through radiographic information conversion into large-scale and mineable entities [44–47]. Generally, radiomics analysis process is impartial to the disease under investigation, and it is performed in the order of data acquisition, data pre-filtration, region of interest (ROI) selection, feature derivation, post-filtration following data investigation [37]. The field of radiomics mainly targets improving patient management such as disease-type prediction, survival rate and efficacy of therapy [45, 48–55]. Detailed investigation of single nodule alongside more nodules within the region of interest in nearly real-time result production is some of the many technical advantages of radiomics [56]. Previous studies focused on the development and validation of machine-learning-based clinical models to predict the patient outcome to ensure that it becomes feasible and practical [54]. However, these multicentric models consist of enormous amount of higher-order and diverse functionality-based image features; as a result, interpreting and understanding these features become overwhelming even for the experts in the field [57, 58]. At present, researchers are working on upgrading the readability of quantitative information of the radiomics model so that radiologists and physicians can comprehend the data effortlessly [59]. To replace the current diagnostic assessment of imaging technique (human eye interpretation), the features must be accurate, robust and reproducible. Dedicated research works are performed to attain this goal, and some of the published works have very inspiring results [53, 59, 60].
Radiomics can serve as biomarkers, incorporated with artificial intelligence (AI), and it can be used to develop prediction models that may enable a far more detailed, precise and micro-level assessment, well beyond the predominantly methodological techniques used in medical image evaluation at present [42, 61]. The 18F-FDG PET as a potential biomarker for therapeutic response evaluation was acknowledged way back in 1999 [62]. The 18F-FDG PET/CT biomarkers facilitate immunotherapy response prediction in advanced stages of non-small cell lung cancer (NSCLC) [63]. However, ubiquitous establishment and recognition of 18F-FDG PET/CT as a computable biomarker are lacking due to the absence of standardised imaging and data exploration techniques [64]. On a brighter side, literature review confirms that researches are working on overcoming these challenges; thus, the 18F-FDG PET/CT can be established as a quantitative biomarker in near future [56, 61].
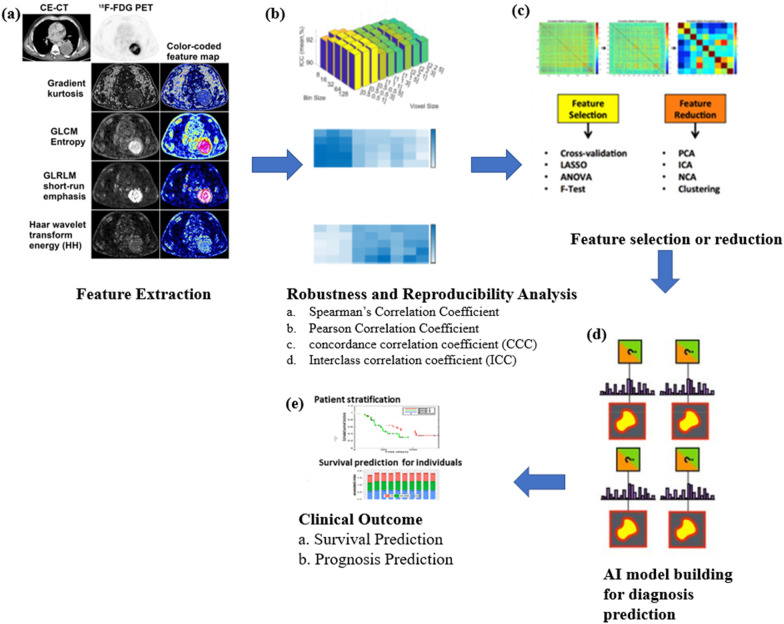
Transition of radiomics finding into therapeutic practice is the ultimate goal of the field of radiomics and texture analysis. Figure 1 shows the overview of optimisation of radiomics feature for clinical practice translation. The process begins with feature extraction from the region of interest through a computerised method (Fig. 1a) [65]. Next, robustness and the reproducibility of the features are determined by evaluating Spearman’s correlation coefficient, Pearson correlation coefficient, concordance correlation coefficient or interclass correlation coefficient (Fig. 1b). Based on the robustness and reproducibility findings, the optimum features are selected and redundant features are removed (Fig. 1c) [66]. For automisation of the clinical practice, artificial intelligence-based model is developed for lung disease prediction, prognosis and diagnosis (Fig. 1d) [65]. Finally, clinical outcomes such as survival prediction and prognosis prediction can be achieved by the application of radiomics (Fig. 1e) [67].
Fig. 1.
The overview of optimisation of radiomics feature for its translation in clinical practice
In this review paper, we discuss the significant aspects of 18F-FDG PET/CT radiomics in proper management of lung diseases (cancer, infection and inflammation) and standardisation initiatives, progress and challenges. This review article is written appreciating the importance of 18F-FDG PET/CT so that it can be recognised as quantitative biomarker. The article has been divided into two parts: The first part addresses the application of radiomics in assessing and diagnosing lung diseases and the factors affection radiomics analysis. The second part of this paper addresses the challenges face in standardisation of radiomics features and approaches involving standardisation of morphological, textural and statistical radiomics features.
Application of 18F-FDG PET/CT radiomics
Lung cancer detection and assessment
Cancer is the uncontrolled cell proliferation that follows death without proper detection, assessment and treatment during the earlier stages [68]. Application of PET in the field of oncology has been well recognised as PET imaging has the ability to extract phenotypic and functional tumour heterogeneity information [61]. During cancer treatment, the metabolic transformation usually takes place before anatomical changes, and this can be spotted through 18F-FDG PET [69]. Commonly used 18F-FDG PET/CT parameters such as standardised uptake value derivatives, metabolically active tumour volume and total lesion glycolysis have widespread application in oncological medicine [70, 71]. Tumours tend to have intricate topological arrangement termed as intra-tumour heterogeneity, and tumour phenotype assessment by intra-tumour heterogeneity quantification is beyond the scope of these matrices [71]. Prediction models have been established to distinguish benign, malignant and inflammatory pulmonary condition emphasising computable imaging features [42, 72–75]. Texture analysis of radiotracer uptake value significantly increased specificity compared to maximum standardised uptake value alone [7, 76]. Texture analysis has been adopted for primary and metastatic lesions distinction on a considerably large patient cohort (n = 545) in a retrospective study [77].
In the field of oncology, researchers have acknowledged the exceptional tumour-to-background ratio and greater tumour examination capacity of 18F-FDG PET/CT compared to CT imaging [16]. Investigation on medical application of 18F-FDG PET/CT in oncological patients showed that high concentration of 18F-FDG in pulmonary nodules might represent malignancy of the investigated lesion [78]. Among lung cancer variations, NSCLC holds major concern [79]. Adenocarcinoma, squamous cell carcinoma and large cell carcinoma are its primary subtypes [37]. Healthcare professionals, radiologists and scientists are working hand in hand to overcome its danger and trying to manage cancer risk by extracting and interpreting information from these associating histological subtypes, micro-level tumour characteristics and understanding the tumour stage using CT and PET radiomic signatures [37, 71, 80, 81]. 18F-FDG PET facilitates detailed apprehension and evaluation of carcinoma traits such as its metabolism and receptor recognition on the microscopic level and so its application in NSCLC patient management is clinically momentous [37]. 18F-FDG PET/CT has received appreciation from the National Comprehensive Cancer Network for having NSCLC patient evaluation capability [82]. American College of Radiology Appropriateness Criteria and American College of Chest Physicians guidelines have also recommended 18F-FDG-PET/CT for NSCLC staging due to its well-recognised effectiveness [83, 84]. Minuscule 18F-FDG uptake by subcentimetre pulmonary nodules was reported to be benign in 98% cases in a study performed on large cohort of population [71, 85]. Abatement of tumour size detection might be extremely slow or remain hidden in the targeted drugs; for example, thymidine kinase inhibitors’ treatment period and evaluation of the competence these drugs can be done earlier by 18F-FDG PET/CT [86]. Computable 18F-FDG PET/CT investigation studies might promote observer-independent appraisal of tracer uptake, thus expanding its capacity of turning into image biomarker [87, 88]. Moreover, combination of CT and 18F-FDG PET enhances the NSCLC patient management by combining anatomic and biologic information [89].
Texture analysis is a specialised branch of radiomics concentrating on quantitative analysis and regional topology variation discretisation of the image voxel densities [47, 90, 91]. Initially, researchers became enthusiastic about texture when they realised that phenotypic characteristics present in diagnostic images can be distinguished though higher-order statistical aspects remain unidentified by visual perception alone [91, 92].
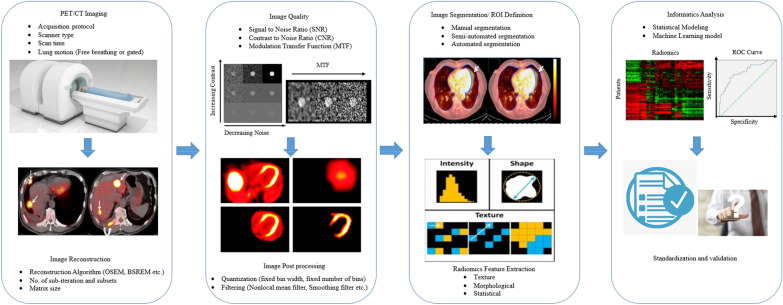
Figure 2 shows the workflow of radiomics texture analysis. The workflow of texture analysis begins with image acquisition. Afterwards, the acquired image is reconstructed using different software platforms. During image reconstruction, filtering process includes sharpening and smoothing. Next, delineation of region of interest (ROI) also known as segmentation is performed where the location of tumour is defined. Textural features are extracted from the ROI and finally statistical model, or machine learning algorithm is developed. The 18F-FDG PET/CT-based texture traits correlated with regional reappearance and cause-specific survival of patients undergoing radiotherapy and forecasted disease-free survival in NSCLC patients after invasive surgery [6, 93, 94].
Fig. 2.
Workflow of radiomics texture analysis
Lung infection and inflammation detection and diagnosis
There are a number of studies emphasised on the oncological role of 18F-FDG PET/CT [44]. However, function of 18F-FDG PET/CT in analysing other medical conditions is also worthy of attention [26, 27, 95]. Differentiation of tuberculosis from lung cancer could be perplexing as both diseases share some similar phenotypic traits and consists of solid nodule [96, 97]. Critically malignant tumour such as lung cancer results in severe clinical condition than solid active pulmonary tuberculosis without early diagnosis [98]. In situation even worse, diagnostic error due to unsuccessful detection of tuberculosis from lung cancer could result in inconvenient, expensive let alone unnecessary invasive medical procedure. Contrarily, diagnosis delay of lung cancer would oversight required treatment leading towards uncontrollable tumour progression and fatal consequences [99]. SUVmax, a metabolic PET parameter, might barely differentiate these two conditions as to vast aggregation of activated macrophages and lymphocytes promotes high 18F-FDG avidity in both conditions [100, 101]. Addressing these issue, a research team demonstrated that personalised and distinctive diagnosis of solid active pulmonary TB and solid LC might be performed using the radiomics nomogram [102].
Histoplasmosis a fungal infection frequently appears as pleural lesion on radiographic imaging as it resembles various lung cancer characteristics such as ring-shaped calcification layers on CT and higher avidity on 18F-FDG PET [103–105]. According to literature review, there are some limitation on identification of histoplasmosis nodule [105]. Identification of histoplasmosis in apprehensive lung effusion might be possible using the documented radiological features [103, 106, 107]. Previous studies suggested that radiomics might perform a complex work of identifying histoplasmosis selectively from lung cancer [103]. The work was based on the hypothesis that the nodule and surrounding perinodular parenchyma-based radiomic features might be able to differentiate suspicious histoplasmosis lung nodules and NSCLC precisely [103]. CT and MRI imaging were found to be incompetent for distinguishing post-surgical tumours from inflammatory sites [108–110]. In addition, glucose metabolism escalation due to soft tissue inflammation increases difficulty in distinction between these health conditions [111]. A limited number of investigation have addressed the potential of feature designation and framework design combination on radiomics analysis-assisted medical assessment [54, 72, 112]. Furthermore, harmonised machine learning would require systematic evaluation as contrasting features highly influence learning algorithms application.
Recently, COVID-19 has drove researchers towards the assessment of the potential capacity of radiomics in differentiating corona virus infection (COVID) from non-COVID [113, 114]. Radiomics feature extracted from CT image of COVID-19 patients was found to bear noticeable similarities with pneumonia-identifying radiomic features [113]. Focusing on this issue, a preliminary investigation showed there is a distinction between COVID-19 pneumonia and other pneumonias such as flu, bacterial and mycoplasma-dependent pneumonias that might be achieved by radiomic feature-based models [113]. In another study, COVID-19 and non-COVID-19 patients were classified using radiomics feature extracted from CT scan images of lung. They employed a dual machine learning technique to distinguish COVID-19 from non-COVID-19 patients [115].
Evaluation of variables that affect radiomics features
Radiomics is the computable expression of the clinical imaging including PET, CT, PET/CT. Comprehensively, establishing quantitative feature-based classification and/or regression model is the purpose of radiomics. Sophisticated and subtle traces of diseases remain unnoticed when examining images in the conventional manner. Selection of reproducible and robust features is an arduous and prudent work, and radiomics has the potential to mine and detect those feature so that imaging modalities can be utilised thoroughly [37, 44]. In general, there are several factors that affect radiomics feature analysis of lung diseases as shown in Fig. 3. 18F-FDG PET/CT imaging protocol, post-processing techniques, image reconstruction, image quality, segmentation technique and the information analysis affect the radiomic feature analysis. The primary factor that affects radiomics is the 18F-FDG PET/CT imaging, i.e. the image acquisition protocol, scanner type, scan time, presence of lung motion [13].
Fig. 3.
List of factors that affect radiomics feature analysis of lung diseases
Variation of protocol, scanner type and difference in scan time generate different outcomes even for the same subject. Lung motion influences image data collected from PET scan resulting artifact in texture features analysis as PET acquisitions take place typically for a few minutes in every bed position [61, 116–118]. The presence of lung motion induces noise (blur image) as a result the radiomics analysis becomes challenging. Respiratory gating is introduced to minimise the effect of lung motion. Accuracy of imaging increases by respiratory gating as blurring caused by breathing motion becomes negligible, but the application of respiratory gating in medical settings is not well established yet [119–121]. The 4D PET imaging might remove the effect of respiratory motion causing textural feature distortion present on 3D PET image [122].
However, the assumption of advanced textures might be interpreted from 4D-PET that requires future assessment and investigation [122]. Factors affecting CT acquisition such as metal artefacts [123], X-ray tube peak voltage and current [124], matrix size, and attenuation correction factor also impact PET radiomic feature quantification. Regardless of the apparent influence of these factors, robust analytical indications of the features could be perceived [122, 123, 125–127]. Reconstruction is one of the key factors that impacts radiomics analysis of the 18F-FDG PET/CT image [128]. There are two iterative reconstruction algorithms commonly used in clinical settings, and these are Ordered Subset Expectation Maximization (OSEM) and Block Sequential Regularised Expectation Maximization (BSREM). The BSREM has been proven to be more sensitive towards reconstruction [71, 192]. A study recently examined the role of deep learning for lung nodule detection in PET/CT as in almost 80% of cases higher 18F-FDG uptake by pulmonary nodule turns to become malignant. They suggested that deep learning may pose potential in this field and artificial intelligence performed better on BSREM than OSEM in detecting lesion, thus resulting in greater sensitivity [71]. Studies conducted by Yan et al. demonstrated that PET image reconstruction configuration variation does not alter some features such as normalised grey-level co-occurrence matrix, run-length matrix and size zone matrix [129, 130]. However, further elaboration is needed to determine the cause behind sensitivity variation of radiomic features [129, 130]. PET images have larger voxels than MRI and CT.
Thus, quantification of heterogeneity becomes biased [119, 131, 132] and it results in coarser tumour sampling. A large number of radiological features are sensitive to voxel dimension and so steady and equal voxel spacing is important for reproducing distinct image feature [45]. A study with the view of omitting the bias of voxel size in PET imaging concluded that the lower limit of volume for unbiased tumour sampling is 45 cm3 with the compromise of sensitivity of second-order entropy [132]. Contrast, resolution and noise are integral parts of any digital imaging technique. Image matrices such as signal-to-noise ratio (SNR) and contrast-to-noise ratio (CNR) are used to understand image quality. Radiomics analysis is affected by the low SNR and CNR of PET scan. In the case of PET imaging, CNR could be optimised by selecting the best possible segmentation method. In the case of SNR, PET images inherent high noise; therefore, achieving high SNR is always challenging. However, optimisation of scanner sensitivity, administered dose and scan time may lead towards high SNR. Application of time of flight (TOF) and point spread function (PSF) modelling play a vital role in optimising SNR and contrast. However, a limited number of studies have incorporated these two techniques as shown in Table 1.
Table 1.
A summary of previous findings on potential feature exploration based on 18F-FDG PET/CT image of lung diseases
| No. | Dose (MBq) | Scanner type | Scan time (min) | Respiratory gating | Reconstruction algorithm | Matrix size | PSF Modelling | Time of flight (TOF) | Image processing filter | Type of segmentation | Type of quantisation | Tumour size (cm3) | REF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3.70–4.81 | Discovery ST4, GE Healthcare | 16–12 | No | OSEM | CT: 512 × 512 | No | No | Exponential filter, square filter, square root filter, logarithm filter and wavelet decomposition | Manual | – | – | [42] |
| 2 | VD: 370 FD: 400 | Validation DS: VCT-XT-Discovery, GE-Healthcare Feasibility DS: GE Discovery ST PETCT system (GE Healthcare, Waukesha, WI, USA) | – | No | OSEM | – | No | No | None | Automated | – | – | [50] |
| 3 | 210–620 | Discovery ST, GE Healthcare, Waukesha, WI) | – | No | OSEM | – | No | No | – | Manual | Equal-probability and Lloyd–Max quantisation | – | [55] |
| 4 | 220 – 690 | Philips GE PET/CT scanner (Philips Medical Systems, USA) | – | No | RAMLA | – | No | No | 5-mm full-width at half-maximum Gaussian | Semi-automated | 64 grey-level quantisation | – | [57] |
| 5 | 350–550 | Siemens Biograph 6 LSO (Siemens, Erlangen Germany) or a General Electric Discovery 690 (General Electric Healthcare, Waukesha, WI, USA) | – | No | Iterative, TOF, sharp IR | PET: 128 × 128, CT: 512 × 512 | No | Yes | None | Semi-automated | PET: 64 bins from 0 to 25,CT: 400 bins from − 1000 to 3000 | 1.64 ± 0.78 | [77] |
| 6 | 340–450 | 24–28 | No | OSEM | CT: 512 × 512 128 × 128 | No | No | None | Manual | 256 bins quantisation | < 3 | [80] | |
| 7 | 370 | ECAT EXACT 47 scanner (CTI/ Siemens, Munich, Germany) | – | No | OSEM | – | No | No | – | – | – | – | [86] |
| 8 | 350–550 | Siemens Biograph 6 LSO (Siemens, Erlangen, Germany), General Electric Discovery 690 (General Electric Healthcare, Waukesha, WI, USA) | – | No | Iterative, TOF, sharp IR | PET: 128 × 128, 256 × 256 CT: 512 × 512 | No | Yes | – | Semi-automatic | Tonal discretisation (64 bins) | – | [94] |
| 9 | – | GE I PET/CT scanner (Philips), | – | No | RAMLA | – | No | No | 5-mm full-width-at-half-maximum Gaussian | Automatic | – | – | [117] |
| 10 | 370–555 | General Electric Medical Systems, Waukesha, WI | 35 | Yes | OSEM | 128 × 128 | No | No | – | Automatic | – | – | [119] |
| 11 | 618–814 | Siemens Biograph PET/CT scanner (Siemens AG, Erlangen, Germany) | Non-gated: 18–35 Gated: 20–30 | Yes | 3D PET: OSEM, 4D PET: OSEM | No-gated: 168 × 168 Gated: 256 × 256 | No | No | Non-gated: 7 mm full-width half- maximum Gaussian Gated: 5 mm full-width half-maximum Gaussian | – | 32 discrete values quantisation | – | [122] |
| 12 | 223–690 | GE I PET/CT scanner (Philips) | – | No | RAMLA | – | No | No | 5 mm in full width at half maximum Gaussian | Automatic | – | – | [138] |
| 13 | 550 | Philips GE I PET/CT, Philips Health Care, Cleveland, Ohio, USA | – | No | RAMLA | PET: 144 × 144, CT: 512 × 512 | No | No | – | Manual | – | 0.7–5.8 | [6] |
| 14 | 150–310 | Discovery MI, GE Healthcare | 15–20 | No | OSEM BSREM | 256 × 256 | Yes | Yes | 6.4-mm Gaussian filter with time-of-flight and PSF, and BSREM with a beta-value of 450 | Manual | – | ≤ 2 | [71] |
| 15 | 555 | GE discovery LS 4 PET/CT scanner | – | No | OSEM | 128 × 128 | No | No | 8 mm full-width at half-maximum Gaussian | Manual | 256-bin discretisation | – | [76] |
| 16 | 350 – 450 | Biograph 16 Siemens Medical Solutions | 20–25 | No | OSEM | PET: 128 × 128, CT: 512 × 512 | No | No | 5 mm full-width at half-maximum Gaussian | Manual | – | 1.7–6.8 | [81] |
| 17 | – | Biograph 16 PET/CT scanner | 18–21 | No | OSEM | 164 × 164 | No | No | – | Manual | 64 bins quantisation | 1.7 – 6.8 | [93] |
| 18 | 270–410 | Biograph mCT scanner (Siemens, Germany) | – | No | OSEM | – | Yes | Yes | 4-mm full-width-at-half-maximum Gaussian | Manual | – | > 3 | [102] |
| 19 | 440 ± 2.0 | GE Discovery STE PET/CT Scanner, GE Discovery 600 PET/CT Scanner | – | Yes | OSEM | – | No | No | 4.29 mm, 7 mm, or 10 mm full-width-at-half-maximum Gaussian filter | Automated | – | – | [123] |
| 20 | 370 | GE Discovery VCT scanner (Waukesha, WI) | – | No | 2D PET: OSEM 3D PET: Iterative- Vue Point algorithm | 2D: 128 × 128 3D: 256 × 256 | No | No | 3 mm, 5 mm, 6 mm post-filtration width | Semi-automated | – | – | [126] |
| 21 | 229.4 ± 22.2 | Biograph 64 mCT scanner (Siemens) | – | No | FBP and OSEM | 256 × 256,128 × 128 | Yes | Yes | 2.5 mm, 3.5 mm, 4.5 mm, 5.5 mm full-width at half-maximum Gaussian | Semi-automated | – | < 5 | [127] |
| 22 | 740 | Siemens Biograph PET/CT scanner | 21–40 | Yes | OSEM | 3D: 168 × 168 4D: 256 × 256 | No | No | 3D scan: 7 mm full-width at half-maximum Gaussian 4D scan: 5 mm full-width at half-maximum Gaussian | Automated | – | < 3 | [147] |
Higher SNR can be attained by TOF resulting in heterogeneity generated by noise and improving image quality. On the other hand, higher resolution is obtained by PSF modelling as it models the matrix physical processes, producing detailed structures within lesion. The post-processing techniques involve smoothing by averaging the pixels, application of Gaussian filters to improve the scan image quality, image noise regulation and image enhancement by the virtue of histogram equalisation, deblurring and resampling [55, 133]. Quantisation performed for image-acquired noise-suppression is an important step of tomographic modification for traceable calculation of texture features, which also impacts radiomics feature analysis [45].
Conventionally, fixed number of bins and fixed bin width are the two approaches of quantisation, and both techniques come with their specific characteristics accommodating their use per requirement [45, 134]. The effect of segmentation is well recognised as the selection of segmentation method determines the balance between accuracy and reproducibility. In the case of manual segmentation, inter- and intra-observer variability is always present without question [135–139]. Feature nomenclature and feature extraction guideline are yet to be established. Therefore, variation is present in extracted features causing variation of radiomics analysis. Three types of features are mainly extracted from the ROI. These are texture, morphological features and statistical features. Texture provides information in the spatial arrangement of intensities in an image. Texture feature computation involves dataset comparison and rotationally even voxel spacing distribution [45].
Common set of textural features are those derived from the grey-level co-occurrence matrix (GLCM) and grey-level run-length matrix (GLRLM). Morphological features describe the shape of the delineated ROI and properties including its volume, maximum diameter, maximum surface, tumour compactness and sphericity. Statistical features include mean, median, skewness, kurtosis, uniformity and entropy. Another key factor that influences radiomics study is statistical calculation [13]. Correction for multiple testing is one of the important steps of accuracy of feature while working with large dataset [119, 140]. Surprisingly, a retrospective systemic review by Alic et al. showed that a significant number of radiomics trait become statistically inconsequential when the correction factor is applied [141].
Validation and standardisation of radiomics features pose significant challenge [142–144]. Some studies comprise validation-level limitations such as inadequate statistical study such as asynchronous p-value for multiple tests, insufficient independent validation dataset resulting in biased discovery rates [7, 145]. Validation of radiomics approaches requires ample multicentre datasets [7]. Overstatement of positive results against negative ones is also another crucial factor [146, 147]. Quantification of radiomic feature with identical names might have different implementation due to the lack of standard definition in radiomic studies. For example, calculation of GLCM could be done by averaging matrix values of 13 distinct directions or a single matrix encompassing tumour co-occurrence values in all 13 directions [131]. Indistinct feature terminology and feature definition variation caused by different operating systems (MaZda1, CGITA2, IBEX3, LIFEx4, MITK Phenotyping5, RaCaT6, CERR radiomic extension7 and Pyradiomics8) also affect the radiomics analysis [148]. These issues have been addressed by the IBSI initiative [148].
Importance of 18F-FDG PET/CT image biomarker standardisation
Radiomics has received much attention and interest in the field of medical science. Nonetheless, reproducibility and validation of the published work are still a big challenge [44, 149–152]. The absence of unanimously recognised reference values and definitions has hampered clinical use of 18F-FDG PET/CT image biomarker. Furthermore, there is lack of uniformity of the image processing platforms required to analyse features [153–155]. Manipulation and assessment of a single image set in two different software platforms result in dissimilar feature values [156]. Variation of imaging procedure, 18F-FDG activity, image reconstruction, data comprehension and uptake time are significant [128, 157, 158]. Reproducibly has been challenged frequently as there is a lack of detailed report of the reproducibility of the experiments. There are various factors including the absence of open-source data and standardised protocol that limit the reproducibility studies of the radiomics features [159–161]. The situation can be solved by standardisation of the radiomics features definition with supportable references and coherent execution of image assessment strategies for feature quantification [103, 154, 156, 162]. The mainstream quantification of 18F-FDG PET/CT is accomplished using the quantitative index of tracer uptake called SUV.
Quantification by SUV is well recognised despite having variance of factor [64]. In the interest of strengthening, the application of 18F-FDG PET/CT as imaging biomarkers guidelines on tumour imaging using 18F-FDG PET/CT has been published and revised [163, 164]. Currently, it is well understood that harmonisation of imaging modalities is vital alongside standardising imaging performance for standardising computation of 18F-FDG PET/CT as biomarker [64]. Reproducibility and validation of radiomic features are hard to achieve without standardising the software platforms used across different research facilities. A study on the level of agreement between IBSI guideline and the Image Biomarker Explorer (IBEX) that is an open-source radiomic software was performed alongside development and validation of S-IBEX [165, 166]. The software platform achieved validation by employing the five different pre-processing configurations proposed by IBSI [143, 166].
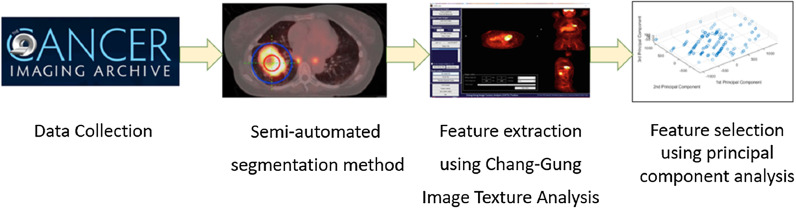
From the literature, it is well understood that application of radiomics in lung disease management has been able to attract great interests for the past few years. For example, a study was performed on feature selection to identify adenocarcinoma histologic subtype present in non-small cell lung cancer (NSCLC) [167]. Figure 4 illustrates the detail procedure of feature selection. In this study, the initial PET/CT data were collected from The Cancer Imaging Achieve [168]. Semi-automated segmentation was applied on the images to delineate the region of interest. Chang-Gung image texture analysis (CGITA), an open-source platform, was then used for extraction of textural features form the segmented ROI. Principal component analysis (PCA) was performed in MATLAB before feature selection, to minimise the feature space and maximise the relevant information. The feature selection criteria were set to select features having a coefficient with one principal component with the major variance and the normalised value retaining 99% of the variability.
Fig. 4.
Adenocarcinoma in non-small cell lung cancer (NSCLC) detection using radiomics [167]
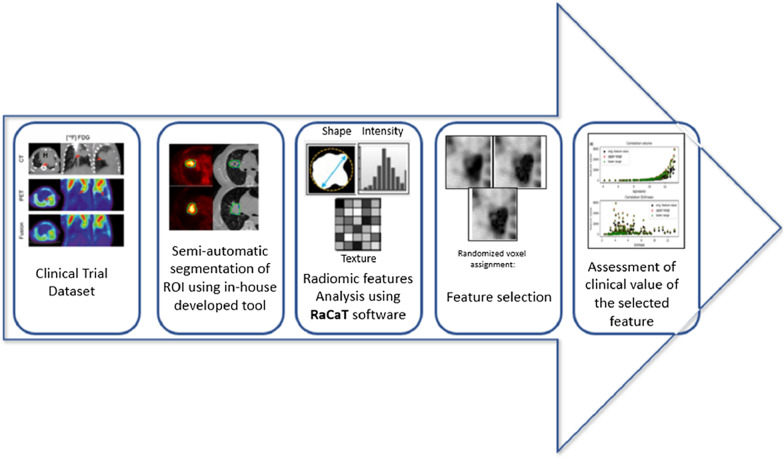
In another study, a prediction outcome of locally advanced NSCLC was investigated [169]. The detail workflow is illustrated in Fig. 5. This study highlighted the importance of PET acquisition standardisation. They emphasised the role of preselection in the case of determining robust radiomic features. The method started by extracting 1404 radiomic features. The dataset included either pre-treatment 18F-FDG PET scans of stage IIIA/N2 or IIIB NSCLC patients. In this prospective study, robustness was determined against tumour motion, delineation variation and attenuation correction. Finally, the training of regression models was performed using standardised imaging. Validation was done in two ways. It includes separate single-centre dataset and fivefold cross-validation. The performance of the model was denoted by area under the receiver operating characteristic curve (AUC).
Fig. 5.
Workflow PET radiomics model for prediction of event-free survival in locally advanced NSCLC using multicentre datasets [169]
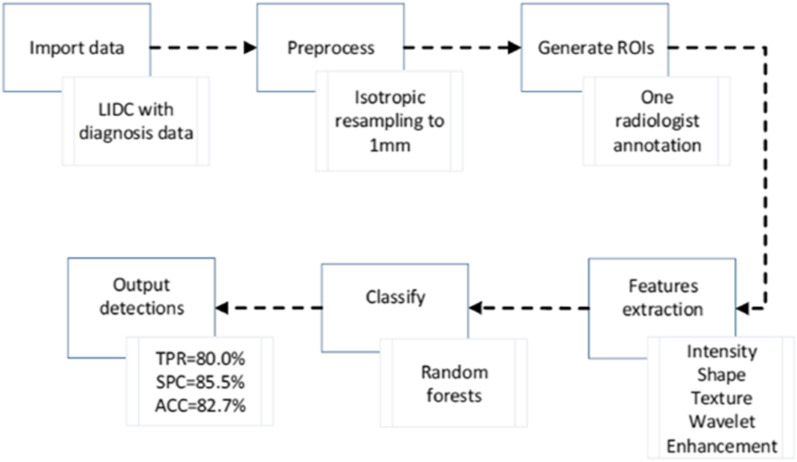
Another study proposed a computer-aided diagnostic (CAD) method for identifying the benign and malignant lung cancer utilising radiomics from CT images [170]. The method attained 82.7% accuracy in distinguishing between benign and malignant primary lung nodules. The intensity, heterogeneity information and shape of the suspected nodules were quantified using 583 features, at multi-frequencies. Random forest method was then applied to identify benign or malignant nodules by analysing all these features. The step-by-step flow chart is shown in Fig. 6.
Fig. 6.
Workflow of automatic lung nodule classification with radiomics approach [170]
In addition, Fig. 7 illustrates another robust feature selection method for NSCLC diagnosis [171]. The method is free of false-positive findings and overfitting. In this method, a semi-automated segmentation method was applied before extracting radiomics features. The features were then analysed using an open-source platform, RaCaT which follows the IBSI [166]. Finally, data analysis was performed using Python.
Fig. 7.
Workflow of feature selection procedure for reproducible textural feature identification describing relevant texture and independent of conventional PET metrics [191]
Challenges to overcome in 18F-FDG PET/CT image biomarker standardisation
Standardisation is the prime solution of any challenges faced in enhancing quality and safety of clinical care. In this era of technological advancement, when new findings meet reality and new information gets stacked with every passing second, standardisation of 18F-FDG PET/CT is far from being easy. Optimisation of feature calculation is a significant step towards reproducible radiomics. In radiomics analysis, image acquisition, reconstruction and segmentation present considerable influences for heterogeneity [16]. Characteristics that can be replicated using optimised radiomics tools from the same image can still lack reproducibility in multicentric or multi-scanner configuration unless the parameters associated with image acquisition, reconstruction and segmentation attain standardisation and harmonisation [159, 172]. Key factors impacting 18F-FDG PET/CT feature standardisation are illustrated in Fig. 8.
Fig. 8.
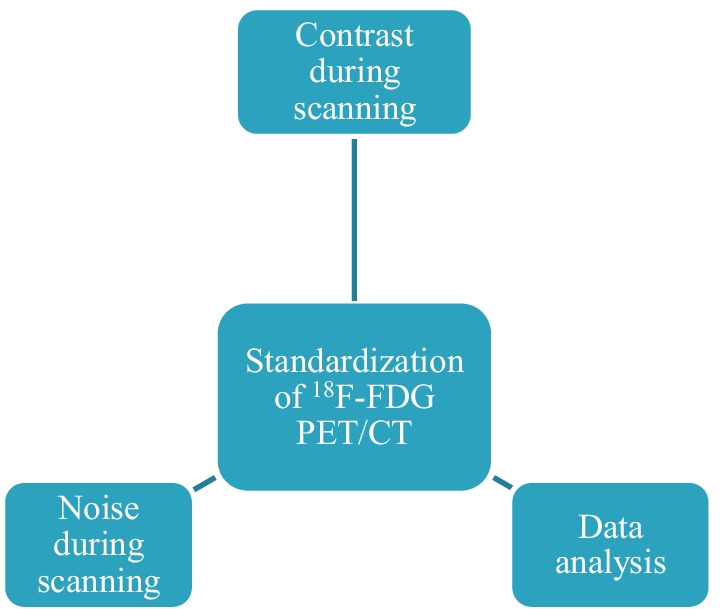
Factors impacting 18F-FDG PET/CT feature standardisation
As the figure shows, standardisation of features can be achieved using well-defined combination of image characteristics (resolution, noise) and data analysis method. Change of any of these factors results in change of feature calculation. During the initial stage of lung disease, the change in the molecular level takes place before anatomical changes happen. This change at molecular level is responsible for the heterogeneity. When 18F-FDG PET/CT scan is performed for detecting the biological heterogeneity, mechanical heterogeneity is introduced by the scanner. Image resolution, noise and the data analysis technique induce the mechanical heterogeneity. By optimising resolution and noise during 18F-FDG PET/CT imaging and harmonising, the data analysis technique will eventually result in standardisation of 18F-FDG PET/CT.
Accuracy, feasibility and precision are the must meet criteria for establishing biomarker and moving towards standardisation. In 2001, the biomarkers definition working group defined the biomarker as objectively measured and evaluated characteristic as an indicator of homeostatic biologic, pathogenic processes, or drug responses duding treatment [173]. Thus, biomarker is the assertive diagnostic and treatment standard which characterises biological and functional activities [64]. It shows that image feature can be acknowledged as biomarker when it is standardised. Accuracy is defined by the correctness of a measurement and determined by comparing the measurement against the true or accepted value. From the view point of medical science, accuracy is the ratio of true positive and true negative in all calculated cases under a specific investigation [174].
Accuracy in the case of radiomics can be assured by comparison between computed values and mathematically or theoretically correct value, which requires consistent and focused effort. Feasibility denotes that the image feature contains logical, analytical and extrapolative value. Radiomics being a computational analysis, overwhelming number of features are obtained by image processing and so selecting feasible features from data large cohort of data is of highly challenging. Precision in achieved by meeting the criteria of acceptable fidelity, repeatability and reproducibility. The systemic review conducted by Traverso et al. confirmed that the repeatability and reproducibility of radiomic features depend on acquisition, reconstruction algorithm, preprocessing and software platform employed for computing the features [159]. Reproducibility is one of the big challenges in 18F-FDG PET/CT radiomics studies. Factors affecting the reproducibility of biomarkers have been unanimously agreed [154, 159, 163, 172, 175]. In multicentre clinical trials, incorporation of 18F-FDG PET biomarkers and treatment response would be impossible without calibration and optimisation of the quantitative 18F-FDG PET parameters. A study conducted with the aim of 18F-FDG PET/CT uptake test–retest reproducibility in cancer patients based on multicentre qualification processing found that 18F-FDG PET/CT scanner quality and settings may result in significantly reproducible test–retest tumour SUV measurements [176]. However, kinetic behaviour of the tracer uptake is not considered in SUVs [177].
Standardisation of radiomics analysis depends on the optimisation of the PET image quality parameters such as contrast-to-noise ratio (CNR) and signal-to-noise ratio (SNR) [178]. However, the PET scan inherently has low SNR and CNR compared to other diagnostic imaging modalities [179]. Reconstruction algorithm types and parameters also impact the radiomics features. Likelihood expectation maximization (MLEM) or ordered subset expectation maximization (OSEM)-type algorithm are highly affected by the minute changes of initial data. Consequently, the resulted outputs become noisy as the iteration converges. To minimise this effect, the iteration is typically stopped before it reaches full convergence, which may introduce bias in the reconstructed images [180]. Block sequential regularised expectation maximization (BSREM), which includes an edge-preserving penalty term, can be used to obviate all these problems. BSREM algorithm achieves optimal SNR by using the penalty term as it employs low smoothing in higher activity areas (such as tumour) as well as in neighbourhood with the high-intensity edges and high smoothing in lower activity regions (such as background) [180].
In addition, a study was performed by Gabriel Reynés-Llompart et al., to prove the promising role of a radiomics approach to assess image quality of abdominal PET imaging by using new reconstruction algorithms with BSRM methods and testing the utility of a radiomics approach. This study found for the OSEM + PSF and especially for the BSRM reconstructions; the image quality parameters presented only at best moderated correlations with the subjective image quality. None of the studied parameters presented a good predictive power for image quality, while a simple radiomics model increased the performance of the image quality prediction [192].
Statistical method application could narrow down the influence of cohort size on radiomics features [181], and artificial intelligence would improve conversion between reconstruction kernels in CT imaging [182]. Incorporating AI into the image analysis field comes with its challenges. Computer-aided detection system always generates false-positive results, which increases the workload of false finding elimination for radiologists and physician. The task is also time-consuming though it does not affect the patient care to a great extent. Medical imaging has still to collect ample amount of data so that AI algorithm can be trained rigorously. This challenge can be dealt by introducing neural network-based transfer learning. Successful implementation of neural network relies on unbiased and large training set and assimilation of different settings [183, 184].
Overview of standardisation methods of PET image biomarkers
The advancement of 18F-FDG PET/CT standardisation in oncologic imaging is noteworthy [10, 148, 149, 185, 186]. Research works dedicated towards developing novel imaging biomarkers have been proposed [4, 187, 188]. Standardisation methods have been employed during the scan time (image reconstruction modification, scan framework reformation), patient level (blood glucose level regulation and amelioration, supervising tolerable delay time of radiotracer dose and uptake) and image post-processing level (z-score, transformation method) [10, 186]. Alleviation from the undesired nonetheless unavoidable image acquisition interrupters, namely body weight, radiotracer uptake interval, partial volume [4], is the goal of the studies focusing diagnostic and prognostic image standardisation.
In recent times, specific guidance for addressing the limitations faced in application of radiomics analysis has also been published [154]. Study on steadiness of nearly 100 radiomic features and inter-observer variability in lung tumour identification [123] showed that the PET-based stable features were also robust to interobserver variability. From their observation, they suggested that poorly reproduced features might also be sensitive to other factors as well [122]. Scientists have also confirmed the invariability of some features, regardless of the reconstruction configuration applied [61, 121]. Standardisation is required for image acquisition, reconstruction, segmentation and feature calculation. In this present work, we focused on standardisation initiative by the international collaboration such as Image Biomarker Standardisation Initiative (IBSI) alongside post-acquisition standardisation method and histology standardisation techniques.
Figure 9 shows a flowchart of standardisation process applied on the collaborative investigation by the researchers of Tokushima university and the university of Tokyo hospital jointly [186]. They were able to successfully upgrade the accuracy of histology extrapolation. The study was performed on CT image sets (training set and test set) of patient having confirmed adenocarcinoma, squamous cell carcinoma and NSCLC [186]. They considered the inter-observer variation by considering four segmentations of a tumour. In the first step, CT images were transformed into three-dimensional wavelets. Then, 476 features were generated from the raw and the transformed images. Next, using univariate assessment of a fixed threshold, feature selection was performed. To eliminate the effect of imaging condition the team performed normalisation of features. Random forest model was used to build the histology prediction model, and its performance was verified by test cohort specificity, sensitivity, accuracy and receiver observed characteristic curve. However, the standardisation technique built, should be performed on large cohort of patients for further validation.
Fig. 9.
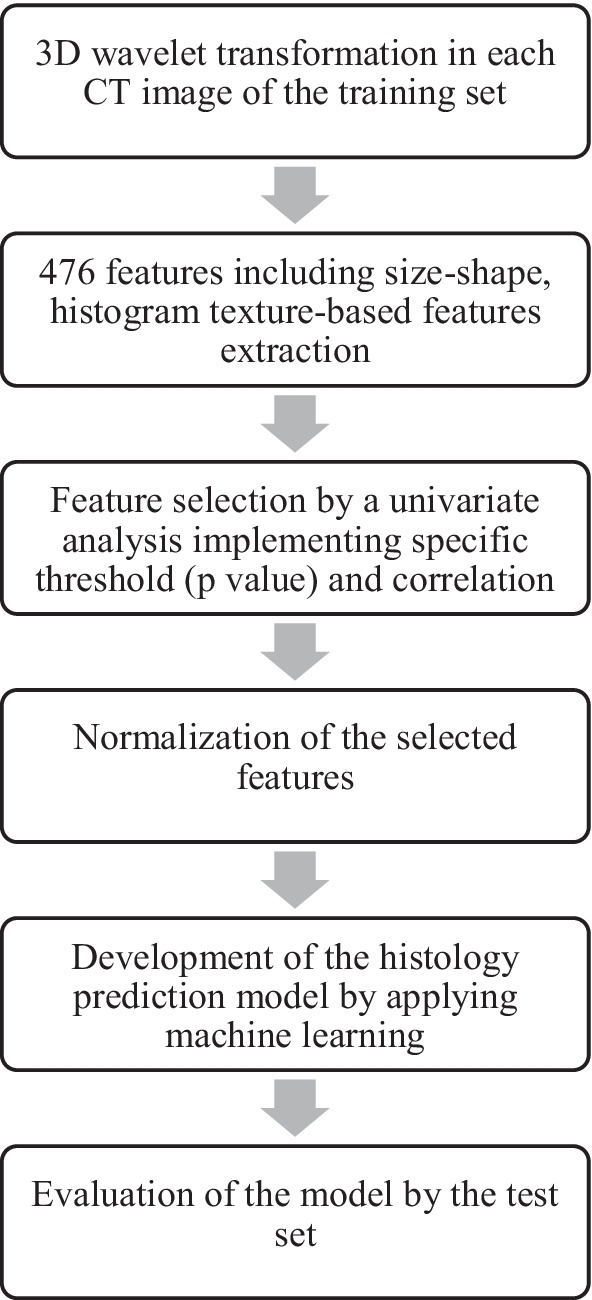
Flowchart of feature extraction study based on CT images performed by Haga, Akihiro et al. [173]
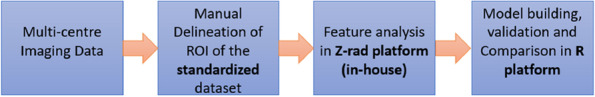
In 2007, Radiological Society of Northern America organised the Quantitative Imaging Biomarkers Alliance for systematic validation and standardisation of a number of radiological biomarkers [4]. The achievement of standardising 169 radiomics feature and standardising image processing framework by this researcher alliance is definitely a breakthrough towards automation of disease diagnosis. They accomplished the enormous work of defining 174 radiomic features and efficaciously reproducing thus standardising 169 features. Figure 10 demonstrates the workflow of the study.
Fig. 10.
Flowchart of validation study by Zwanenburg, Alex et al., overview [145]
The study was performed in three stages. Digital phantom with specific ROI mask (74 voxel) was used as the dataset during the first phase. In the next phase, the dataset was taken from open-source CT images where the ROI was defined by the segmented gross tumour volume. Research groups calculated feature values from the ROI directly and applied predefined image processing pathway as per requirement. Standardisation of the computed values was achieved by comparing the degree of concurrence and stability of the concurrence. In the first two stages, the feature reference values were achieved through iterative filtering level of concurrence. The features calculated in these two phases was recognised as standard only when the degree of concurrence was high enough. In the third stage, dataset of patients diagnosed with soft-tissue sarcoma was selected from Cancer imaging Archive consisting of multimodality imaging (T1-weighted MRI and 18F-FDG PET/CT). Similar to the previous phase, the images were accompanied by segmentation of the gross tumour volume. In this stage, the standardised features (achieved from the first two phases) were validated by reproducing the features using predefined image processing configuration on the dataset. Finally, IBSI achieved the standardisation of 169 features out of 174 features examined in this research study. However, the study excluded uncommon features and traits such as fractals and image filters for feasibility purpose.
A research team recently presented a post-acquisition standardisation workflow. Their proposed principle is based on the modified MRI standardisation method recommended by Nyul et al. [189, 190]. The workflow followed during this study is illustrated in Fig. 11. Two sets of 18F-FDG PET/CT scan data were utilised in this study as the training set and validation set. First, a standardised intensity scale was defined for the image set. This was achieved by initially computing the low percentile, 50th percentile and high percentile intensities and mean values of these intensities from the training image dataset. Then, the intensity of images from test dataset was mapped nonlinearly in the mean value interval obtained from the training dataset.
Fig. 11.
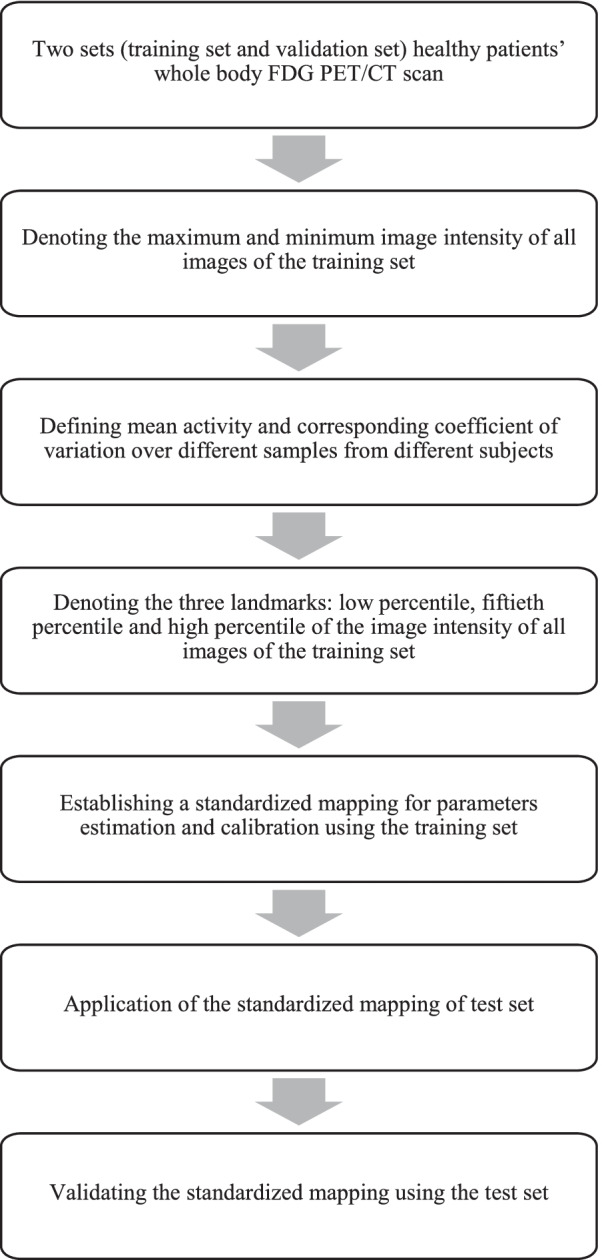
Flowchart of establishment of standardised mapping for whole-body FDG PET/CT scan study by Mortazi et al. [10]
Performance of the standardisation process was determined by implementing the coefficient of variation of mean metabolic activity and coefficient of variation of mean activity computed from the training dataset. One of the utilities of this technique is that it is free from the effect of patient and image acquisition parameters. The study result showed significant decrease of the mean metabolic activity coefficient variation was achieved by standardised PET (sPET). Additionally, sPET was proven to be superior to conventional standardisation methods such as SUV and z-score normalisation [10]. However, the validation data set adopted in the study was healthy cohort of patient. The developed sPET scheme should also be applied on diseased patients in clinical trial to further investigate its feasibility.
Conclusion
The micro-level changes in lung disease such as tumour, infection or inflammation progresses cannot be detected by CT scan alone. 18F-FDG PET/CT diagnosis carefully examines detailed and diverse cell anomalies in the field of biology. Nevertheless, low-grade resolution and irregular noise of PET images upholds added methodological boundary. In the field of tumour treatment response assessment, the use of 18F-FDG PET/CT is not as common as CT. This is because the globally approved PET equivalent of CT-Based Response Evaluation Criteria in Solid Tumours guideline is yet to be established. Application of radiomics might enhance the diagnostic capability of the imaging techniques as it extracts a large number of quantitative features from the images which otherwise remains unattended. Biologic changes at the molecular lever can be traced back by intelligent assessment of the computed features. Employment of 18F-FDG PET/CT radiomics, especially texture analysis in lung abnormalities management, directs its gradual steps towards a patient-specific approach of lung diseases management.
Recently, application of 18F-FDG PET/CT radiomics is also being applied for lung infection and inflammation diagnosis. However, numerous numbers of features and various techniques of feature extraction have raised tremendous complexity. This complexity can only be removed by standardisation of radiomics analysis. Hence, introduction of radiomics in the medical practice is impossible without standardisation and harmonisation through sufficient and convincing scientific evidence. Importance of standardisation, reproducibility, and validation of radiomics in clinical trials cannot be overlooked.
To achieve the ultimate goal to employ radiomics analysis as an integral part of the routine medical diagnosis and prognosis, validation of its robustness across reconstruction algorithm and parameters is crucial. However, the absence of appropriate cross-validation of the radiomics studies till date raises the concern of false-positive results. Radiomics textural features are a set of numeric and their interpretation by human, which are often difficult. Elucidation of these feature statistics is not beyond mistakes, e.g. assumption of correlation implies the causation, misinterpretation of correlation, over generalisation [161]. Acceptance of appropriate radiomic features will only be achieved once these challenges are properly addressed. For the time being, comparison of the findings across different studies is unattainable due to different protocols and practices. Only the establishment of a uniform prognostic and predictive factors of feature analysis can promote the transition of radiomics into the field of clinical practice. Basically, the available studies on radiomics are mainly retrospective and hence demonstrate the perception of radiomics. Adaptation of prospective research is essential to establish radiomics into the medical support system. Acceptance of radiomics can only be achieved upon proper addressing of these challenges.
The standardisation and quantification of 18F-FDG PET/CT radiomics will increase its potential field of application even more. The combination of artificial intelligence and machine-learning techniques with radiomics research will hold the ground of disease diagnosis and treatment evaluation robustly and speed up medical translation.
Acknowledgements
The authors would like to thank Universiti Sains Malaysia and Deputyship for Research & Innovation, Ministry of Education Kingdom of Saudi Arabia for the financial assistance through International Collaboration Fund for this study.
Abbreviations
- 18F-FDG PET/CT
18-Fluorine-fluorodeoxyglucose positron emission tomography/computed tomography
- CT
Computed tomography
- GLCM
Grey-level co-occurrence matrix
- GLRLM
Grey-level run-length matrix
- IBEX
Image Biomarker Explorer
- MRI
Magnetic resonance imaging
- NSCLC
Non-small cell lung cancer
- PET
Positron emission tomography
- PET/CT
Positron emission tomography/computed tomography
- ROI
Region of interest
- sPET
Standardised PET
- SUVmax
Maximum standard uptake value
Authors' contributions
NA prepared original draft writing; RZ contributed to the methodology, supervision, reviewing and editing; MT providing the funding acquisition, supervision and reviewing.
Funding
The authors received research funding from Deputyship for Research & Innovation, Ministry of Education Kingdom of Saudi Arabia (Grant No. 304.CIPPT.6501089.S158).
Availability of data and materials
All data and materials concerning this work are available in the submitted manuscript.
Declarations
Ethics approval and consent to participate
Ethics approval and consent to participate approval are not required for this manuscript.
Consent for publication
Consent for publication is given.
Competing interests
There are no competing interests among authors related to the submitted manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/28/2022
A Correction to this paper has been published: 10.1186/s13244-022-01186-8
References
- 1.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson E, Leahy O, Cheng AC, Grummet J. Risk factors for infection following prostate biopsy-a case control study. BMC Infect Dis. 2015;15(1):1–6. doi: 10.1186/s12879-015-1328-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Troeger C, Blacker B, Khalil IA, et al. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18(11):1191–1210. doi: 10.1016/S1473-3099(18)30310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC, Group A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology. 2011;258(3):906–914. doi: 10.1148/radiol.10100799. [DOI] [PubMed] [Google Scholar]
- 5.Kurland BF, Gerstner ER, Mountz JM, et al. Promise and pitfalls of quantitative imaging in oncology clinical trials. Magn Reson Imaging. 2012;30(9):1301–1312. doi: 10.1016/j.mri.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oikonomou A, Khalvati F, Tyrrell PN, et al. Radiomics analysis at PET/CT contributes to prognosis of recurrence and survival in lung cancer treated with stereotactic body radiotherapy. Sci Rep. 2018;8(1):1–11. doi: 10.1038/s41598-018-22357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianconi F, Palumbo I, Spanu A, Nuvoli S, Fravolini ML, Palumbo B. PET/CT radiomics in lung cancer: an overview. Appl Sci. 2020;10(5):1718. [Google Scholar]
- 8.Torigian DA, Zaidi H, Kwee TC, et al. PET/MR imaging: technical aspects and potential clinical applications. Radiology. 2013;267(1):26–44. doi: 10.1148/radiol.13121038. [DOI] [PubMed] [Google Scholar]
- 9.Kwee TC, Basu S, Saboury B, Alavi A, Torigian DA. Functional oncoimaging techniques with potential clinical applications. Front Biosci (Elite Ed) 2012;4:1081–1096. doi: 10.2741/443. [DOI] [PubMed] [Google Scholar]
- 10.Mortazi A, Udupa JK, Tong Y, Torigian DA (2020) A post-acquisition standardization method for positron emission tomography images. In: Medical imaging 2020: computer-aided diagnosis. International Society for Optics and Photonics
- 11.Kelloff GJ, Hoffman JM, Johnson B, et al. Progress and promise of FDG-PET imaging for cancer patient management and oncologic drug development. Clin Cancer Res. 2005;11(8):2785–2808. doi: 10.1158/1078-0432.CCR-04-2626. [DOI] [PubMed] [Google Scholar]
- 12.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2020;70(4):313. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 13.Voigt W. Advanced PET imaging in oncology: status and developments with current and future relevance to lung cancer care. Curr Opin Oncol. 2018;30(2):77–83. doi: 10.1097/CCO.0000000000000430. [DOI] [PubMed] [Google Scholar]
- 14.Pucar D, Liu C. Standardization and quantification is a key to the future of atherosclerosis FDG PET/CT imaging. Berlin: Springer; 2019. [DOI] [PubMed] [Google Scholar]
- 15.Doris MK, Otaki Y, Krishnan SK, et al. Optimization of reconstruction and quantification of motion-corrected coronary PET-CT. J Nucl Cardiol. 2020;27(2):494–504. doi: 10.1007/s12350-018-1317-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morbelli S, Arbizu J, Booij J, et al. The need of standardization and of large clinical studies in an emerging indication of [18 F] FDG PET: the autoimmune encephalitis. Berlin: Springer; 2017. [DOI] [PubMed] [Google Scholar]
- 17.Henriksson EV, Kjellen E, Wahlberg P, Ohlsson T, Wennerberg J, Brun EV. 2-Deoxy-2-[18F] fluoro-D-glucose uptake and correlation to intratumoral heterogeneity. Anticancer Res. 2007;27(4B):2155–2159. [PubMed] [Google Scholar]
- 18.Yang X, Knopp MV. Quantifying tumor vascular heterogeneity with dynamic contrast-enhanced magnetic resonance imaging: a review. BioMed Res Int. 2011 doi: 10.1155/2011/732848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu S, Kwee TC, Gatenby R, Saboury B, Torigian DA, Alavi A. Evolving role of molecular imaging with PET in detecting and characterizing heterogeneity of cancer tissue at the primary and metastatic sites, a plausible explanation for failed attempts to cure malignant disorders. Berlin: Springer; 2011. [DOI] [PubMed] [Google Scholar]
- 20.Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A. 2008;105(13):5213–5218. doi: 10.1073/pnas.0801279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto S, Korn RL, Oklu R, et al. ALK molecular phenotype in non-small cell lung cancer: CT radiogenomic characterization. Radiology. 2014;272(2):568–576. doi: 10.1148/radiol.14140789. [DOI] [PubMed] [Google Scholar]
- 22.Karlo CA, Di Paolo PL, Chaim J, et al. Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology. 2014;270(2):464–471. doi: 10.1148/radiol.13130663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber WA, Figlin R. Monitoring cancer treatment with PET/CT: does it make a difference? J Nucl Med. 2007;48(1 suppl):36S–44S. [PubMed] [Google Scholar]
- 24.Van Tinteren H, Hoekstra OS, Smit EF, et al. Effectiveness of positron emission tomography in the preoperative assessment of patients with suspected non-small-cell lung cancer: the PLUS multicentre randomised trial. Lancet. 2002;359(9315):1388–1392. doi: 10.1016/s0140-6736(02)08352-6. [DOI] [PubMed] [Google Scholar]
- 25.Fouad YA, Aanei C. Revisiting the hallmarks of cancer. Am J Cancer Res. 2017;7(5):1016. [PMC free article] [PubMed] [Google Scholar]
- 26.Vaidyanathan S, Patel CN, Scarsbrook AF, Chowdhury FU. FDG PET/CT in infection and inflammation—current and emerging clinical applications. Clin Radiol. 2015;70(7):787–800. doi: 10.1016/j.crad.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Signore A, Glaudemans AW. The molecular imaging approach to image infections and inflammation by nuclear medicine techniques. Ann Nucl Med. 2011;25(10):681–700. doi: 10.1007/s12149-011-0521-z. [DOI] [PubMed] [Google Scholar]
- 28.Glaudemans AW, de Vries EF, Galli F, Dierckx RA, Slart RH, Signore A. The use of F-FDG-PET/CT for diagnosis and treatment monitoring of inflammatory and infectious diseases. Clin Dev Immunol. 2013 doi: 10.1155/2013/623036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figueroa AL, Subramanian SS, Cury RC, et al. Distribution of inflammation within carotid atherosclerotic plaques with high-risk morphological features: a comparison between positron emission tomography activity, plaque morphology, and histopathology. Circ Cardiovasc Imaging. 2012;5(1):69–77. doi: 10.1161/CIRCIMAGING.110.959478. [DOI] [PubMed] [Google Scholar]
- 30.Higashi K, Clavo AC, Wahl RL. Does FDG uptake measure proliferative activity of human cancer cells? In vitro comparison with DNA flow cytometry and tritiated thymidine uptake. J Nucl Med. 1993;34(3):414–419. [PubMed] [Google Scholar]
- 31.Haberkorn U, Ziegler SI, Oberdorfer F, et al. FDG uptake, tumor proliferation and expression of glycolysis associated genes in animal tumor models. Nucl Med Biol. 1994;21(6):827–834. doi: 10.1016/0969-8051(94)90162-7. [DOI] [PubMed] [Google Scholar]
- 32.Rajendran JG, Mankoff DA, O’Sullivan F, et al. Hypoxia and glucose metabolism in malignant tumors: evaluation by [18F] fluoromisonidazole and [18F] fluorodeoxyglucose positron emission tomography imaging. Clin Cancer Res. 2004;10(7):2245–2252. doi: 10.1158/1078-0432.ccr-0688-3. [DOI] [PubMed] [Google Scholar]
- 33.Fanchon LM, Dogan S, Moreira AL, et al. Feasibility of in situ, high-resolution correlation of tracer uptake with histopathology by quantitative autoradiography of biopsy specimens obtained under 18F-FDG PET/CT guidance. J Nucl Med. 2015;56(4):538–544. doi: 10.2967/jnumed.114.148668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tixier F, Le Rest CC, Hatt M, et al. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med. 2011;52(3):369–378. doi: 10.2967/jnumed.110.082404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, Cook GJ. Quantifying tumour heterogeneity in 18 F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging. 2013;40(1):133–140. doi: 10.1007/s00259-012-2247-0. [DOI] [PubMed] [Google Scholar]
- 36.Gould MK, Kuschner WG, Rydzak CE, et al. Test performance of positron emission tomography and computed tomography for mediastinal staging in patients with non-small-cell lung cancer: a meta-analysis. Ann Intern Med. 2003;139(11):879–892. doi: 10.7326/0003-4819-139-11-200311180-00013. [DOI] [PubMed] [Google Scholar]
- 37.Kaseda K. Recent and current advances in FDG-PET imaging within the field of clinical oncology in NSCLC: a review of the literature. Diagnostics. 2020;10(8):561. doi: 10.3390/diagnostics10080561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sollini M, Cozzi L, Antunovic L, Chiti A, Kirienko M. PET Radiomics in NSCLC: state of the art and a proposal for harmonization of methodology. Sci Rep. 2017;7(1):1–15. doi: 10.1038/s41598-017-00426-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bailly C, Bodet-Milin C, Bourgeois M, et al. Exploring tumor heterogeneity using PET imaging: the big picture. Cancers. 2019;11(9):1282. doi: 10.3390/cancers11091282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee G, Lee HY, Park H, et al. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: state of the art. Eur J Radiol. 2017;86:297–307. doi: 10.1016/j.ejrad.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Chen B, Zhang R, Gan Y, Yang L, Li W. Development and clinical application of radiomics in lung cancer. Radiat Oncol. 2017;12(1):154. doi: 10.1186/s13014-017-0885-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R, Zhu L, Cai Z, et al. Potential feature exploration and model development based on 18F-FDG PET/CT images for differentiating benign and malignant lung lesions. Eur J Radiol. 2019;121:108735. doi: 10.1016/j.ejrad.2019.108735. [DOI] [PubMed] [Google Scholar]
- 43.Keek SA, Leijenaar RT, Jochems A, Woodruff HC. A review on radiomics and the future of theranostics for patient selection in precision medicine. Br J Radiol. 2018;91(1091):20170926. doi: 10.1259/bjr.20170926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forghani R, Savadjiev P, Chatterjee A, Muthukrishnan N, Reinhold C, Forghani B. Radiomics and artificial intelligence for biomarker and prediction model development in oncology. Comput Struct Biotechnol J. 2019;17:995. doi: 10.1016/j.csbj.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forghani R, Savadjiev P, Chatterjee A, Muthukrishnan N, Reinhold C, Forghani B. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 47.Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ. CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics. 2017;37(5):1483–1503. doi: 10.1148/rg.2017170056. [DOI] [PubMed] [Google Scholar]
- 48.Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2(1):1–8. doi: 10.1186/s41747-018-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aiello M, Cavaliere C, D’Albore A, Salvatore M. The challenges of diagnostic imaging in the era of big data. J Clin Med. 2019;8(3):316. doi: 10.3390/jcm8030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Win T, Miles KA, Janes SM, et al. Tumor heterogeneity and permeability as measured on the CT component of PET/CT predict survival in patients with non-small cell lung cancer. Clin Cancer Res. 2013;19(13):3591–3599. doi: 10.1158/1078-0432.CCR-12-1307. [DOI] [PubMed] [Google Scholar]
- 51.Forghani R, Chatterjee A, Reinhold C, et al. Head and neck squamous cell carcinoma: prediction of cervical lymph node metastasis by dual-energy CT texture analysis with machine learning. Eur Radiol. 2019;29(11):6172–6181. doi: 10.1007/s00330-019-06159-y. [DOI] [PubMed] [Google Scholar]
- 52.Al Ajmi E, Forghani B, Reinhold C, Bayat M, Forghani R. Spectral multi-energy CT texture analysis with machine learning for tissue classification: an investigation using classification of benign parotid tumours as a testing paradigm. Eur Radiol. 2018;28(6):2604–2611. doi: 10.1007/s00330-017-5214-0. [DOI] [PubMed] [Google Scholar]
- 53.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5(1):1–9. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJ. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5:13087. doi: 10.1038/srep13087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60(14):5471. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
- 56.Castiglioni I, Gilardi MC. Radiomics: is it time to compose the puzzle? Clin Transl Imaging. 2018;6(5):411–413. doi: 10.1007/s40336-018-0302-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Desseroit MC, Visvikis D, Tixier F, et al. Development of a nomogram combining clinical staging with 18 F-FDG PET/CT image features in non-small-cell lung cancer stage I-III. Eur J Nucl Med Mol Imaging. 2016;43(8):1477–1485. doi: 10.1007/s00259-016-3325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Upadhaya T, Morvan Y, Stindel E, Le Reste PJ, Hatt M. A framework for multimodal imaging-based prognostic model building: preliminary study on multimodal MRI in glioblastoma multiforme. Irbm. 2015;36(6):345–350. [Google Scholar]
- 59.Hatt M, Tixier F, Visvikis D, Le Rest CC. Radiomics in PET/CT: more than meets the eye? J Nucl Med. 2017;58(3):365–366. doi: 10.2967/jnumed.116.184655. [DOI] [PubMed] [Google Scholar]
- 60.Desseroit MC, Tixier F, Visvikis D, Le Rest CC, Hatt M. Nomogram for NSCLC exploiting clinical staging, tumor volume and PET/CT heterogeneity features: development using support vector machines in a retrospective cohort and first validation results in prospectively recruited patients. J Nucl Med. 2016;57(supplement 2):437–437. [Google Scholar]
- 61.Ha S, Choi H, Paeng JC, Cheon GJ. Radiomics in oncological PET/CT: a methodological overview. Nucl Med Mol Imaging. 2019;53(1):14–29. doi: 10.1007/s13139-019-00571-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young H, Baum R, Cremerius U, et al. Measurement of clinical and subclinical tumour response using [18F]-fluorodeoxyglucose and positron emission tomography: review and 1999 EORTC recommendations. Eur J Cancer. 1999;35(13):1773–1782. doi: 10.1016/s0959-8049(99)00229-4. [DOI] [PubMed] [Google Scholar]
- 63.Mu W, Qi J, Lu H, Schabath M, Balagurunathan Y, Tunali I, Gillies RJ (2018) Radiomic biomarkers from PET/CT multi-modality fusion images for the prediction of immunotherapy response in advanced non-small cell lung cancer patients. In: Medical imaging 2018: computer-aided diagnosis. International Society for Optics and Photonics
- 64.Boellaard R (2017) Standardization of imaging biomarkers: the FDG PET/CT example. In: Quality in nuclear medicine. Springer, pp 227–240
- 65.Mayerhoefer ME, Materka A, Langs G, et al. Introduction to radiomics. J Nucl Med. 2020;61(4):488–495. doi: 10.2967/jnumed.118.222893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cutaia G, La Tona G, Comelli A, et al. Radiomics and prostate MRI: current role and future applications. J Imaging. 2021;7(2):34. doi: 10.3390/jimaging7020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Galperin-Aizenberg M, Pryma D, Simone CB, II, Fan Y. Unsupervised machine learning of radiomic features for predicting treatment response and overall survival of early stage non-small cell lung cancer patients treated with stereotactic body radiation therapy. Radiother Oncol. 2018;129(2):218–226. doi: 10.1016/j.radonc.2018.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21(2):137–148. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 69.Weber W. PET for response assessment in oncology: radiotherapy and chemotherapy. Br J Radiol. 2005;1:42–49. [Google Scholar]
- 70.Schwyzer M, Ferraro DA, Muehlematter UJ, et al. Automated detection of lung cancer at ultralow dose PET/CT by deep neural networks–initial results. Lung Cancer. 2018;126:170–173. doi: 10.1016/j.lungcan.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 71.Schwyzer M, Martini K, Benz DC, et al. Artificial intelligence for detecting small FDG-positive lung nodules in digital PET/CT: impact of image reconstructions on diagnostic performance. Eur Radiol. 2020;30(4):2031–2040. doi: 10.1007/s00330-019-06498-w. [DOI] [PubMed] [Google Scholar]
- 72.Hawkins S, Wang H, Liu Y, et al. Predicting malignant nodules from screening CT scans. J Thorac Oncol. 2016;11(12):2120–2128. doi: 10.1016/j.jtho.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu W, Hu H, Gong J, Li X, Huang G, Nie S. Malignant-benign classification of pulmonary nodules based on random forest aided by clustering analysis. Phys Med Biol. 2019;64(3):035017. doi: 10.1088/1361-6560/aafab0. [DOI] [PubMed] [Google Scholar]
- 74.Suo S, Cheng J, Cao M, et al. Assessment of heterogeneity difference between edge and core by using texture analysis: differentiation of malignant from inflammatory pulmonary nodules and masses. Acad Radiol. 2016;23(9):1115–1122. doi: 10.1016/j.acra.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 75.Balagurunathan Y, Schabath MB, Wang H, Liu Y, Gillies RJ. Quantitative imaging features improve discrimination of malignancy in pulmonary nodules. Sci Rep. 2019;9(1):8528. doi: 10.1038/s41598-019-44562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen S, Harmon S, Perk T, Li X, Chen M, Li Y, Jeraj R. Diagnostic classification of solitary pulmonary nodules using dual time 18 F-FDG PET/CT image texture features in granuloma-endemic regions. Sci Rep. 2017;7(1):1–8. doi: 10.1038/s41598-017-08764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kirienko M, Cozzi L, Rossi A, et al. Ability of FDG PET and CT radiomics features to differentiate between primary and metastatic lung lesions. Eur J Nucl Med Mol Imaging. 2018;45(10):1649–1660. doi: 10.1007/s00259-018-3987-2. [DOI] [PubMed] [Google Scholar]
- 78.Taralli S, Scolozzi V, Triumbari EK, et al. Is 18F-fluorodeoxyglucose positron emission tomography/computed tomography useful to discriminate metachronous lung cancer from metastasis in patients with oncological history? Q J Nucl Med Mol. 2019;64(3):291–298. doi: 10.23736/S1824-4785.19.03140-6. [DOI] [PubMed] [Google Scholar]
- 79.Fischbach F, Knollmann F, Griesshaber V, et al. Detection of pulmonary nodules by multislice computed tomography: improved detection rate with reduced slice thickness. Eur Radiol. 2003;13(10):2378–2383. doi: 10.1007/s00330-003-1915-7. [DOI] [PubMed] [Google Scholar]
- 80.Bianconi F, Palumbo I, Fravolini ML, et al. Texture analysis on [18 F] FDG PET/CT in non-small-cell lung cancer: correlations between PET features, CT features, and histological types. Mol Imaging Biol. 2019;21(6):1200–1209. doi: 10.1007/s11307-019-01336-3. [DOI] [PubMed] [Google Scholar]
- 81.Brunese L, Greco B, Setola FR, et al. Non-small cell lung cancer evaluated with quantitative contrast-enhanced CT and PET-CT: net enhancement and standardized uptake values are related to tumour size and histology. Med Sci Monit. 2013;19:95. doi: 10.12659/MSM.883759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ettinger DS, Wood DE, Aisner DL, et al. Non-small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Cancer Netw. 2017;15(4):504–535. doi: 10.6004/jnccn.2017.0050. [DOI] [PubMed] [Google Scholar]
- 83.Silvestri GA, Gonzalez AV, Jantz MA, et al. Methods for staging non-small cell lung cancer: diagnosis and management of lung cancer: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5):e211S–e250S. doi: 10.1378/chest.12-2355. [DOI] [PubMed] [Google Scholar]
- 84.Ravenel JG, Rosenzweig KE, Kirsch J, et al. ACR Appropriateness Criteria non-invasive clinical staging of bronchogenic carcinoma. J Am Coll Radiol. 2014;11(9):849–856. doi: 10.1016/j.jacr.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 85.Chang ST, Nguyen DC, Raptis C, et al. Natural history of preoperative subcentimeter pulmonary nodules in patients with resectable pancreatic adenocarcinoma: a retrospective cohort study. Ann Surg. 2015;261(5):970–975. doi: 10.1097/SLA.0000000000000719. [DOI] [PubMed] [Google Scholar]
- 86.Zander T, Scheffler M, Nogova L, et al. Early prediction of nonprogression in advanced non-small-cell lung cancer treated with erlotinib by using [(18) F] fluorodeoxyglucose and [(18) F] fluorothymidine positron emission tomography. J Clin Oncol. 2011;29(13):1701–1708. doi: 10.1200/JCO.2010.32.4939. [DOI] [PubMed] [Google Scholar]
- 87.Itti E, Meignan M, Berriolo-Riedinger A, et al. An international confirmatory study of the prognostic value of early PET/CT in diffuse large B-cell lymphoma: comparison between Deauville criteria and ΔSUVmax. Eur J Nucl Med Mol Imaging. 2013;40(9):1312–1320. doi: 10.1007/s00259-013-2435-6. [DOI] [PubMed] [Google Scholar]
- 88.Hoekstra CJ, Paglianiti I, Hoekstra OS, et al. Monitoring response to therapy in cancer using [18 F]-2-fluoro-2-deoxy-D-glucose and positron emission tomography: an overview of different analytical methods. Eur J Nucl Med. 2000;27(6):731–743. doi: 10.1007/s002590050570. [DOI] [PubMed] [Google Scholar]
- 89.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat Rev Cancer. 2002;2(9):683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 90.Ganeshan B, Miles KA. Quantifying tumour heterogeneity with CT. Cancer Imaging. 2013;13(1):140. doi: 10.1102/1470-7330.2013.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tourassi GD. Journey toward computer-aided diagnosis: role of image texture analysis. Radiology. 1999;213(2):317–320. doi: 10.1148/radiology.213.2.r99nv49317. [DOI] [PubMed] [Google Scholar]
- 92.Julesz B, Gilbert EN, Shepp LA, Frisch HL, et al. Inability of humans to discriminate between visual textures that agree in second-order statistics—revisited. Perception. 1973;2(4):391–405. doi: 10.1068/p020391. [DOI] [PubMed] [Google Scholar]
- 93.Pyka T, Bundschuh RA, Andratschke N, et al. Textural features in pre-treatment [F18]-FDG-PET/CT are correlated with risk of local recurrence and disease-specific survival in early stage NSCLC patients receiving primary stereotactic radiation therapy. Radiat Oncol. 2015;10(1):100. doi: 10.1186/s13014-015-0407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kirienko M, Cozzi L, Antunovic L, et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging. 2018;45(2):207–217. doi: 10.1007/s00259-017-3837-7. [DOI] [PubMed] [Google Scholar]
- 95.Kumar R, Basu S, Torigian D, Anand V, Zhuang H, Alavi A. Role of modern imaging techniques for diagnosis of infection in the era of 18F-fluorodeoxyglucose positron emission tomography. Clin Microbiol Rev. 2008;21(1):209–224. doi: 10.1128/CMR.00025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Parker CS, Siracuse CG, Litle VR. Identifying lung cancer in patients with active pulmonary tuberculosis. J Thorac Dis. 2018;10(Suppl 28):S3392. doi: 10.21037/jtd.2018.07.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bhatt M, Kant S, Bhaskar R. Pulmonary tuberculosis as differential diagnosis of lung cancer. South Asian J Cancer. 2012;1(1):36. doi: 10.4103/2278-330X.96507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA (2008) Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. In: Mayo clinic proceedings. Elsevier [DOI] [PMC free article] [PubMed]
- 99.Akgul AG, Liman ST, Topcu S, Yuksel M. False positive PET scan deserves attention. J Buon. 2014;19(3):836e41. [PubMed] [Google Scholar]
- 100.Boyaci H, Basyigit I, Baris SA. Positron emission tomography/computed tomography in cases with tuberculosis mimicking lung cancer. Braz J Infect Dis. 2013;17(2):267–269. doi: 10.1016/j.bjid.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Du D, Gu J, Chen X, et al. Integration of PET/CT radiomics and semantic features for differentiation between active pulmonary tuberculosis and lung cancer. Mol Imaging Biol. 2020;23:287–298. doi: 10.1007/s11307-020-01550-4. [DOI] [PubMed] [Google Scholar]
- 102.Uthoff J, Nagpal P, Sanchez R, Gross TJ, Lee C, Sieren JC, et al. Differentiation of non-small cell lung cancer and histoplasmosis pulmonary nodules: insights from radiomics model performance compared with clinician observers. Transl Lung Cancer Res. 2019;8(6):979. doi: 10.21037/tlcr.2019.12.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dall Bello AG, Severo CB, Guazzelli LS, Oliveira FM, Hochhegger B, Severo LC. Histoplasmosis mimicking primary lung cancer or pulmonary metastases. J Bras Pneumol. 2013;39(1):63–68. doi: 10.1590/S1806-37132013000100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gazzoni FF, Severo LC, Marchiori E. Fungal diseases mimicking primary lung cancer: radiologic–pathologic correlation. Mycoses. 2014;57(4):197–208. doi: 10.1111/myc.12150. [DOI] [PubMed] [Google Scholar]
- 105.Khoo T, Clarke G, Psevdos G. Lung cancer screening reveals a nonspiculated nodule: histoplasmosis. J Glob Infect Dis. 2018;10(4):226–227. doi: 10.4103/jgid.jgid_92_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rolston KV, Rodriguez S, Dholakia N, Whimbey E, Raad I. Pulmonary infections mimicking cancer: a retrospective, three-year review. Support Care Cancer. 1997;5(2):90–93. doi: 10.1007/BF01262563. [DOI] [PubMed] [Google Scholar]
- 107.Yen RF, Hung RL, Pan MH, et al. 18-Fluoro-2-deoxyglucose positron emission tomography in detecting residual/recurrent nasopharyngeal carcinomas and comparison with magnetic resonance imaging. Cancer. 2003;98(2):283–287. doi: 10.1002/cncr.11519. [DOI] [PubMed] [Google Scholar]
- 108.Zhou H, Shen G, Zhang W, Cai H, Zhou Y, Li L. 18F-FDG PET/CT for the diagnosis of residual or recurrent nasopharyngeal carcinoma after radiotherapy: a metaanalysis. J Nucl Med. 2016;57(3):342–347. doi: 10.2967/jnumed.115.165407. [DOI] [PubMed] [Google Scholar]
- 109.Du D, Feng H, Lv W, et al. Machine learning methods for optimal radiomics-based differentiation between recurrence and inflammation: application to nasopharyngeal carcinoma post-therapy PET/CT images. Mol Imaging Biol. 2020;22(3):730–738. doi: 10.1007/s11307-019-01411-9. [DOI] [PubMed] [Google Scholar]
- 110.Lee N, Yoo IR, Park SY, Yoon H, Lee Y, Oh JK. Significance of incidental nasopharyngeal uptake on 18 F-FDG PET/CT: patterns of benign/physiologic uptake and differentiation from malignancy. Nucl Med Mol Imaging. 2015;49(1):11–18. doi: 10.1007/s13139-014-0299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJ. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front Oncol. 2015;5:272. doi: 10.3389/fonc.2015.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fang M, He B, Li L, et al. CT radiomics can help screen the coronavirus disease 2019 (COVID-19): a preliminary study. Sci China Inf Sci. 2020;63(7):1–8. [Google Scholar]
- 113.Yang N, Liu F, Li C, et al. Diagnostic classification of coronavirus disease 2019 (COVID-19) and other pneumonias using radiomics features in CT chest images. Sci Rep. 2020;11:1–7. doi: 10.1038/s41598-021-97497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cardobi N, Benetti G, Cardano G et al (2021) CT radiomic models to distinguish COVID-19 pneumonia from other interstitial pneumonias. Radiol Med 1–7 [DOI] [PMC free article] [PubMed]
- 115.Nehmeh SA, Erdi YE, Ling CC, et al. Effect of respiratory gating on quantifying PET images of lung cancer. J Nucl Med. 2002;43(7):876–881. [PubMed] [Google Scholar]
- 116.Huang T-C, Wang Y-C. Deformation effect on SUV max changes in thoracic tumors using 4-D PET/CT scan. PLoS One. 2013;8(3):e58886. doi: 10.1371/journal.pone.0058886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aristophanous M, Yong Y, Yap JT, et al. Evaluating FDG uptake changes between pre and post therapy respiratory gated PET scans. Radiother Oncol. 2012;102(3):377–382. doi: 10.1016/j.radonc.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 118.Cook GJ, Azad G, Owczarczyk K, Siddique M, Goh V. Challenges and promises of PET radiomics. Int J Radiat Oncol Biol Phys. 2018;102(4):1083–1089. doi: 10.1016/j.ijrobp.2017.12.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yip S, McCall K, Aristophanous M, Chen AB, Aerts HJ, Berbeco R. Comparison of texture features derived from static and respiratory-gated PET images in non-small cell lung cancer. PLoS One. 2014;9(12):e115510. doi: 10.1371/journal.pone.0115510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oliver JA, Budzevich M, Zhang GG, Dilling TJ, Latifi K, Moros EG. Variability of image features computed from conventional and respiratory-gated PET/CT images of lung cancer. Transl Oncol. 2015;8(6):524–534. doi: 10.1016/j.tranon.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Leijenaar RT, Carvalho S, Hoebers FJ, et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol. 2015;54(9):1423–1429. doi: 10.3109/0284186X.2015.1061214. [DOI] [PubMed] [Google Scholar]
- 123.Fave X, Cook M, Frederick A, et al. Preliminary investigation into sources of uncertainty in quantitative imaging features. Comput Med Imaging Graph. 2015;44:54–61. doi: 10.1016/j.compmedimag.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 124.Cheng NM, Fang YH, Chang JT, et al. Textural features of pretreatment 18F-FDG PET/CT images: prognostic significance in patients with advanced T-stage oropharyngeal squamous cell carcinoma. J Nucl Med. 2013;54(10):1703–1709. doi: 10.2967/jnumed.112.119289. [DOI] [PubMed] [Google Scholar]
- 125.Cheng NM, Fang YH, Lee LY, et al. Zone-size nonuniformity of 18 F-FDG PET regional textural features predicts survival in patients with oropharyngeal cancer. Eur J Nucl Med Mol Imaging. 2015;42(3):419–428. doi: 10.1007/s00259-014-2933-1. [DOI] [PubMed] [Google Scholar]
- 126.Parmar C, Leijenaar RT, Grossmann P, et al. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci Rep. 2015;5:11044. doi: 10.1038/srep11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Messerli M, Kotasidis F, Burger IA, et al. Impact of different image reconstructions on PET quantification in non-small cell lung cancer: a comparison of adenocarcinoma and squamous cell carcinoma. Br J Radiol. 2019;92(1096):20180792. doi: 10.1259/bjr.20180792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. 2010;49(7):1012–1016. doi: 10.3109/0284186X.2010.498437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yan J, Chu-Shern JL, Loi HY, et al. Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med. 2015;56(11):1667–1673. doi: 10.2967/jnumed.115.156927. [DOI] [PubMed] [Google Scholar]
- 130.Hatt M, Majdoub M, Vallières M, et al. 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi–cancer site patient cohort. J Nucl Med. 2015;56(1):38–44. doi: 10.2967/jnumed.114.144055. [DOI] [PubMed] [Google Scholar]
- 131.Brooks FJ, Grigsby PW. The effect of small tumor volumes on studies of intratumoral heterogeneity of tracer uptake. J Nucl Med. 2014;55(1):37–42. doi: 10.2967/jnumed.112.116715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang L, Fried DV, Fave XJ, et al. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys. 2015;42(3):1341–1353. doi: 10.1118/1.4908210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bagher-Ebadian H, Chetty IJ. ROdiomiX: a validated software for radiomics analysis of medical images in radiation oncology. Med Phys. 2020;48:354–365. doi: 10.1002/mp.14590. [DOI] [PubMed] [Google Scholar]
- 134.Davnall F, Yip CS, Ljungqvist G, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012;3(6):573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.De Jong J, Van Diest P, Baak J. Heterogeneity and reproducibility of microvessel counts in breast cancer. Lab Invest. 1995;73(6):922–926. [PubMed] [Google Scholar]
- 136.Mussurakis S, Buckley DL, Coady AM, Turnbull LW, Horsman A. Observer variability in the interpretation of contrast enhanced MRI of the breast. Br J Radiol. 1996;69(827):1009–1016. doi: 10.1259/0007-1285-69-827-1009. [DOI] [PubMed] [Google Scholar]
- 137.Tixier F, Hatt M, Valla C, et al. Visual versus quantitative assessment of intratumor 18F-FDG PET uptake heterogeneity: prognostic value in non-small cell lung cancer. J Nucl Med. 2014;55(8):1235–1241. doi: 10.2967/jnumed.113.133389. [DOI] [PubMed] [Google Scholar]
- 138.Wetzel SG, Cha S, Johnson G, et al. Relative cerebral blood volume measurements in intracranial mass lesions: interobserver and intraobserver reproducibility study. Radiology. 2002;224(3):797–803. doi: 10.1148/radiol.2243011014. [DOI] [PubMed] [Google Scholar]
- 139.Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol) 1995;57(1):289–300. [Google Scholar]
- 140.Alic L, van Vliet M, Van Dijke CF, Eggermont AM, Veenland JF, Niessen WJ. Heterogeneity in DCE-MRI parametric maps: a biomarker for treatment response? Phys Med Biol. 2011;56(6):1601. doi: 10.1088/0031-9155/56/6/006. [DOI] [PubMed] [Google Scholar]
- 141.Leger S, Zwanenburg A, Pilz K, et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep. 2017;7(1):1–11. doi: 10.1038/s41598-017-13448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bettinelli A, Branchini M, De Monte F, Scaggion A, Paiusco M. An IBEX adaption toward image biomarker standardization. Med Phys. 2020;47(3):1167–1173. doi: 10.1002/mp.13956. [DOI] [PubMed] [Google Scholar]
- 143.Götz M, Nolden M, Maier-Hein K. MITK Phenotyping: an open-source toolchain for image-based personalized medicine with radiomics. Radiother Oncol. 2019;131:108–111. doi: 10.1016/j.radonc.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 144.Chalkidou A, O’Doherty MJ, Marsden PK. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS One. 2015;10(5):e0124165. doi: 10.1371/journal.pone.0124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Joober R, Schmitz N, Annable L, Boksa P. Publication bias: what are the challenges and can they be overcome? J Psychiatry Neurosci. 2012;37(3):149. doi: 10.1503/jpn.120065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Buvat I, Orlhac F. The dark side of radiomics: on the paramount importance of publishing negative results. J Nucl Med. 2019;60(11):1543–1544. doi: 10.2967/jnumed.119.235325. [DOI] [PubMed] [Google Scholar]
- 147.Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Boellaard R, Delgado-Bolton R, Oyen WJ, et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: version 2.0. Eur J Nucl Med Mol Imaging. 2015;42(2):328–354. doi: 10.1007/s00259-014-2961-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Berenguer R, Pastor-Juan MD, Canales-Vázquez J, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288(2):407–415. doi: 10.1148/radiol.2018172361. [DOI] [PubMed] [Google Scholar]
- 150.Welch ML, McIntosh C, Haibe-Kains B, et al. Vulnerabilities of radiomic signature development: the need for safeguards. Radiother Oncol. 2019;130:2–9. doi: 10.1016/j.radonc.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 151.Meyer M, Ronald J, Vernuccio F, et al. Reproducibility of CT radiomic features within the same patient: influence of radiation dose and CT reconstruction settings. Radiology. 2019;293(3):583–591. doi: 10.1148/radiol.2019190928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vallieres M, Visvikis D, Hatt M. Dependency of a validated radiomics signature on tumor volume and potential corrections. J Nucl Med. 2018;59(supplement 1):640–640. [Google Scholar]
- 153.Hatt M, Tixier F, Pierce L, et al. Characterization of PET/CT images using texture analysis: the past, the present… any future? Eur J Nucl Med Mol Imaging. 2017;44(1):151–165. doi: 10.1007/s00259-016-3427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bousabarah K, Temming S, Hoevels M, et al. Radiomic analysis of planning computed tomograms for predicting radiation-induced lung injury and outcome in lung cancer patients treated with robotic stereotactic body radiation therapy. Strahlenther Onkol. 2019;195(9):830–842. doi: 10.1007/s00066-019-01452-7. [DOI] [PubMed] [Google Scholar]
- 155.Foy JJ, Robinson KR, Li H, Giger ML, Al-Hallaq H, Armato SG. Variation in algorithm implementation across radiomics software. J Med Imaging. 2018;5(4):044505. doi: 10.1117/1.JMI.5.4.044505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Beyer T, Czernin J, Freudenberg LS. Variations in clinical PET/CT operations: results of an international survey of active PET/CT users. J Nucl Med. 2011;52(2):303–310. doi: 10.2967/jnumed.110.079624. [DOI] [PubMed] [Google Scholar]
- 157.Graham MM, Badawi RD, Wahl RL. Variations in PET/CT methodology for oncologic imaging at US academic medical centers: an imaging response assessment team survey. J Nucl Med. 2011;52(2):311–317. doi: 10.2967/jnumed.109.074104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys. 2018;102(4):1143–1158. doi: 10.1016/j.ijrobp.2018.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reynés-Llompart G, Sabaté-Llobera A, Llinares-Tello E, Martí-Climent JM, Gámez-Cenzano C. Image quality evaluation in a modern PET system: impact of new reconstructions methods and a radiomics approach. Sci Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-46937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging—“How-to” guide and critical reflection. Insights Imaging. 2020;11(1):1–16. doi: 10.1186/s13244-020-00887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bogowicz M, Leijenaar RT, Tanadini-Lang S, et al. Post-radiochemotherapy PET radiomics in head and neck cancer–the influence of radiomics implementation on the reproducibility of local control tumor models. Radiother Oncol. 2017;125(3):385–391. doi: 10.1016/j.radonc.2017.10.023. [DOI] [PubMed] [Google Scholar]
- 162.Boellaard R. Standards for PET image acquisition and quantitative data analysis. J Nucl Med. 2009;50(Suppl 1):11S–20S. doi: 10.2967/jnumed.108.057182. [DOI] [PubMed] [Google Scholar]
- 163.Schelbert HR, Hoh CK, Royal HD, et al. Procedure guideline for tumor imaging using fluorine-18-FDG. J Nucl Med. 1998;39(7):1302–1305. [PubMed] [Google Scholar]
- 164.Partarrieu I, Morris D, Jackson A, Matthews J. Technical validation of heterogeneity features for PET imaging using phantom measurements. J Nucl Med. 2017;58(supplement 1):504–504. [Google Scholar]
- 165.Hatt M, Vallieres M, Visvikis D, Zwanenburg A. IBSI: an international community radiomics standardization initiative. J Nucl Med. 2018;59(supplement 1):287–287. [Google Scholar]
- 166.Lima MA, Motta CF, de Sá AM, Ichinose RM (2019) Radiomic features selection from PET/CT images for the adenocarcinoma histologic subtype identification in non-small cell lung cancer. In: XXVI brazilian congress on biomedical engineering. Springer
- 167.Clark K, Vendt B, Smith K, et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(6):1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Oliveira C, Amstutz F, Vuong D, et al. Preselection of robust radiomic features does not improve outcome modelling in non-small cell lung cancer based on clinical routine FDG-PET imaging. EJNMMI Res. 2021;11(1):1–12. doi: 10.1186/s13550-021-00809-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ma J, Wang Q, Ren Y, Hu H, Zhao J (2016) Automatic lung nodule classification with radiomics approach. In: Medical imaging 2016: PACS and imaging informatics: next generation and innovations. International Society for Optics and Photonics
- 170.Pfaehler E, Zwanenburg A, de Jong JR, Boellaard R. RaCaT: an open source and easy to use radiomics calculator tool. PLoS One. 2019;14(2):e0212223. doi: 10.1371/journal.pone.0212223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zwanenburg A. Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging. 2019;46(13):2638–2655. doi: 10.1007/s00259-019-04391-8. [DOI] [PubMed] [Google Scholar]
- 172.Downing G. Biomarkers definitions working group. Biomarkers and surrogate endpoints. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 173.Gunjan VK, Zurada JM, Raman B, Gangadharan GR. Modern approaches in machine learning and cognitive science: a walkthrough. Berlin: Springer; 2020. [Google Scholar]
- 174.Lovinfosse P, Visvikis D, Hustinx R, Hatt M. FDG PET radiomics: a review of the methodological aspects. Clin Transl Imaging. 2018;6(5):379–391. [Google Scholar]
- 175.Kurland BF, Peterson LM, Shields AT, et al. Test–retest reproducibility of 18F-FDG PET/CT uptake in cancer patients within a qualified and calibrated local network. J Nucl Med. 2019;60(5):608–614. doi: 10.2967/jnumed.118.209544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vriens D, Visser EP, de Geus-Oei LF, Oyen WJ. Methodological considerations in quantification of oncological FDG PET studies. Eur J Nucl Med Mol Imaging. 2010;37(7):1408–1425. doi: 10.1007/s00259-009-1306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gao X, Tham IW, Yan J. Quantitative accuracy of radiomic features of low-dose 18F-FDG PET imaging. Transl Cancer Res. 2020;9(8):4646–4655. doi: 10.21037/tcr-20-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.An L, Zhang P, Adeli E, et al. Multi-level canonical correlation analysis for standard-dose PET image estimation. IEEE Trans Image Process. 2016;25(7):3303–3315. doi: 10.1109/TIP.2016.2567072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Reynés-Llompart G, Gámez-Cenzano C, Vercher-Conejero JL, Sabaté-Llobera A, Calvo-Malvar N, Martí-Climent JM. Phantom, clinical, and texture indices evaluation and optimization of a penalized-likelihood image reconstruction method (Q. Clear) on a BGO PET/CT scanner. Med Phys. 2018;45(7):3214–3222. doi: 10.1002/mp.12986. [DOI] [PubMed] [Google Scholar]
- 180.Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I. Validation of a method to compensate multicenter effects affecting CT radiomics. Radiology. 2019;291(1):53–59. doi: 10.1148/radiol.2019182023. [DOI] [PubMed] [Google Scholar]
- 181.Choe J, Lee SM, Do KH, et al. Deep learning–based image conversion of CT reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology. 2019;292(2):365–373. doi: 10.1148/radiol.2019181960. [DOI] [PubMed] [Google Scholar]
- 182.Chen D-R, Chang R-F, Huang Y-L. Computer-aided diagnosis applied to US of solid breast nodules by using neural networks. Radiology. 1999;213(2):407–412. doi: 10.1148/radiology.213.2.r99nv13407. [DOI] [PubMed] [Google Scholar]
- 183.Antropova N, Huynh B, Giger M. SU-D-207B-06: predicting breast cancer malignancy on DCE-MRI data using pre-trained convolutional neural networks. Med Phys. 2016;43(6Part4):3349–3350. [Google Scholar]
- 184.Shankar LK, Hoffman JM, Bacharach S, et al. Consensus recommendations for the use of 18F-FDG PET as an indicator of therapeutic response in patients in National Cancer Institute Trials. J Nucl Med. 2006;47(6):1059–1066. [PubMed] [Google Scholar]
- 185.Haga A, Takahashi W, Aoki S, et al. Standardization of imaging features for radiomics analysis. J Med Investig. 2019;66(1.2):35–37. doi: 10.2152/jmi.66.35. [DOI] [PubMed] [Google Scholar]
- 186.Clarke LP, Nordstrom RJ, Zhang H, et al. The quantitative imaging network: NCI's historical perspective and planned goals. Transl Oncol. 2014;7(1):1–4. doi: 10.1593/tlo.13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.O'Connor JP, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017;14(3):169–186. doi: 10.1038/nrclinonc.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nyúl LG, Udupa JK. On standardizing the MR image intensity scale. Magn Reson Med. 1999;42(6):1072–1081. doi: 10.1002/(sici)1522-2594(199912)42:6<1072::aid-mrm11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 189.Nyúl LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Med Imaging. 2000;19(2):143–150. doi: 10.1109/42.836373. [DOI] [PubMed] [Google Scholar]
- 190.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 191.Pfaehler E, Mesotten L, Zhovannik I, et al. Plausibility and redundancy analysis to select FDG-PET textural features in non-small cell lung cancer. Med Phys. 2021;48(3):1226–1238. doi: 10.1002/mp.14684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Reynés-Llompart G, Sabaté-Llobera A, Llinares-Tello E, Martí-Climent JM, Gámez-Cenzano C. Image quality evaluation in a modern PET system: impact of new reconstructions methods and a radiomics approach. Sci Rep. 2019;9:10640. doi: 10.1038/s41598-019-46937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials concerning this work are available in the submitted manuscript.