Summary
Tetraspanins are four-pass transmembrane proteins that function by regulating trafficking of partner proteins and organizing signaling complexes in the membrane. Tspan15, one of a six-member TspanC8 subfamily, forms a complex that regulates the trafficking, maturation and substrate selectivity of the transmembrane protease ADAM10, an essential enzyme in mammalian physiology that cleaves a wide variety of membrane-anchored substrates including Notch receptors, amyloid precursor protein, cadherins and growth factors. We present here crystal structures of the Tspan15 large extracellular loop (LEL) required for functional association with ADAM10 both in isolation and in complex with the Fab fragment of an anti-Tspan15 antibody. Comparison of the Tspan15 LEL with other tetraspanin LEL structures shows that a core helical framework buttresses a variable region that structurally diverges among LELs. Using co-immunoprecipitation and a cellular N-cadherin cleavage assay, we identify a site on Tspan15 required for both ADAM10 binding and promoting substrate cleavage.
Graphical Abstract
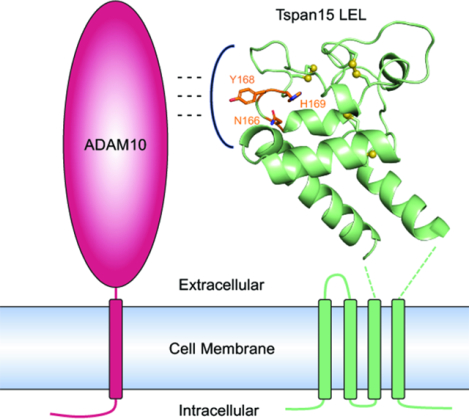
eTOC blurb
Tspan15 is a member of the TspanC8 subfamily of tetraspanins that regulate the essential transmembrane protease ADAM10. Lipper, et al. report crystal structures of the extracellular domain of Tspan15 both in isolation and in complex with a Fab fragment. They use cell-based assays to identify a conserved ADAM10 binding site.
Introduction
The tetraspanins are a family of membrane proteins found in all metazoans, with 33 members in humans (Charrin et al., 2014). Tetraspanins play critical roles in diverse physiological and pathophysiological processes, including cell morphology (Yamada et al., 2008), immune regulation (van Spriel, 2011), adhesion (Reyes et al., 2018; Yáñez-Mó et al., 2001), signal transduction (Termini and Gillette, 2017), viral infection (Martin et al., 2005) and cancer progression (Hemler, 2014). Tetraspanins form specific interactions with partner proteins and regulate their trafficking and function (Levy et al., 2012; Matthews et al., 2017; te Molder et al., 2019). They organize signaling complexes in the membrane through secondary interactions with other tetraspanins, forming what has been referred to as tetraspanin enriched microdomains or the tetraspanin web (Hemler, 2005; Levy and Shoham, 2005).
Tetraspanins are defined by a structural organization consisting of four membrane-spanning helices and two extracellular domains, with a small extracellular loop (SEL/EC1) connecting TM1 and TM2 and a large extracellular loop (LEL/EC2) connecting TM3 and TM4 (Hemler, 2005). The LEL domain has a conserved helical motif and a variable region that forms specific contacts with binding partners (Susa et al., 2020, 2021; Yang et al., 2020). Tetraspanins can be classified as C4, C6 or C8, based on the number of disulfide bond forming cysteines in their LEL domain (DeSalle et al., 2010). To date structures of three C4 tetraspanins, CD9 (Umeda et al., 2020), CD53 (Yang et al., 2020) and CD81 (Zimmerman et al., 2016) have been reported, each adopting a similar cone-shaped conformation with a central intramembrane lipid-binding cavity in their apo, uncomplexed form. Molecular dynamics simulations of CD81 suggested that these proteins can adopt “open” and “closed” conformations, with the LEL domain shifting up away from the transmembrane region in the open form, possibly acting as a molecular switch (Zimmerman et al., 2016). Our group recently reported the cryo-EM structure of CD81 bound to its partner CD19, showing directly that CD81 can adopt such an open form when bound (Susa et al., 2021). Low resolution structures of CD9 in complex with EWI-2 (Oosterheert et al., 2020; Umeda et al., 2020) and EWI-F (Oosterheert et al., 2020) are also consistent with the conclusion that it adopts a more open conformation when bound to its partners.
An important role for the six TspanC8 subfamily members (Tspan5, Tspan10, Tspan14, Tspan15, Tspan17 and Tspan33), named for the presence of eight cysteine residues in the LEL, is to regulate the function of the single-pass transmembrane protease ADAM10 (A Disintegrin and Metalloproteinase 10). ADAM10 is produced as a zymogen, and when processed into its mature, active form, it cleaves the extracellular domains of membrane-anchored proteins in a process termed ectodomain shedding (Seegar and Blacklow, 2019), processing a number of substrates involved in diverse biological processes (Kuhn et al., 2016; Pruessmeyer and Ludwig, 2009). It functions in physiological Notch signaling by cleaving the Notch receptor upon ligand engagement (Sprinzak and Blacklow, 2021), and ADAM10 knockout is embryonically lethal in mice with phenotypes attributed to loss of Notch signaling activity (Hartmann et al., 2002). Dysregulation of ADAM10 has implications in Alzheimer’s disease because it acts as the constitutive α-secretase in non-amyloidogenic proteolysis of the amyloid precursor protein (APP), preventing β-secretase cleavage and toxic β-amyloid peptide production (Kuhn et al., 2010). ADAM10 also functions in the regulation of cell adhesion through the shedding of cadherins (Kohutek et al., 2009; Maretzky et al., 2005). Studies suggest that dysregulation of ADAM10-dependent cadherin cleavage promotes cancer metastasis due to increased cell migration (Kohutek et al., 2009; Yuan et al., 2020).
Current evidence suggests that each TspanC8 protein binds to ADAM10 in the endoplasmic reticulum (ER), and promotes export of ADAM10 to the Golgi where it is processed into the mature form by pro-protein convertases (Saint-Pol et al., 2017a). ADAM10 then appears to be directed to different compartments depending on the cell type and the identity of the bound TspanC8 (Dornier et al., 2012). It has also been suggested that TspanC8s differentially regulate substrate selectivity of ADAM10 (Jouannet et al., 2016; Matthews et al., 2017; Noy et al., 2016). Whether this selectivity derives from differences in subcellular localization or by differences in the site of ADAM10 binding, however, remains unclear.
This work focuses on Tspan15 (also known as NET-7, TM4SF15) as a representative member of the TspanC8 subfamily. It forms a complex with ADAM10 that is active as a protease in the plasma membrane (Koo et al., 2020) and selectively promotes N-cadherin shedding by ADAM10 both in cell lines and in vivo (Jouannet et al., 2016; Noy et al., 2016; Prox et al., 2012; Seipold et al., 2018). The Tspan15-dependence of N-cadherin cleavage may have particular relevance to cancer progression, as high Tspan15 expression has recently been reported to promote cancer cell metastasis in oral and esophageal squamous cell carcinomas (Hiroshima et al., 2019; Zhang et al., 2018), and knockdown of Tspan15 exhibited reduced N-cadherin shedding along with reduced cell invasion and migration in oral squamous cell carcinomas cell lines (Hiroshima et al., 2019).
We present here the crystal structure of the Tspan15 LEL domain both in isolation and complexed with the Fab fragment of the anti-Tspan15 antibody 1C12 (Koo et al., 2020). The core helical framework of the LEL supports a variable region that diverges substantially in structure from that of other known tetraspanin LEL domains. Structure-guided analysis of Tspan15 mutants in two cell-based assays identifies a conserved site required for functional association of the two proteins.
Results
Crystal structure of the Tspan15 LEL
The LEL domain of Tspan15 (Figure 1A, colored residues) was expressed in Expi293 cells, purified to homogeneity and used for crystallization. The amino acid sequence of the purified protein has two modifications from the native sequence: the glycan at N189 was removed with PNGase F, producing an Asp residue, and N118 was mutated to Gln to prevent non-native glycosylation at that site. Although there are three predicted glycosylation sites in the sequence, Tspan15 only has a single glycan at N189 (Figure S1), perhaps due to the close proximity of N118 and N230 to the membrane (Nilsson and von Heijne, 1993).
Figure 1. Crystal structure of the Tspan15 large extracellular loop (LEL).
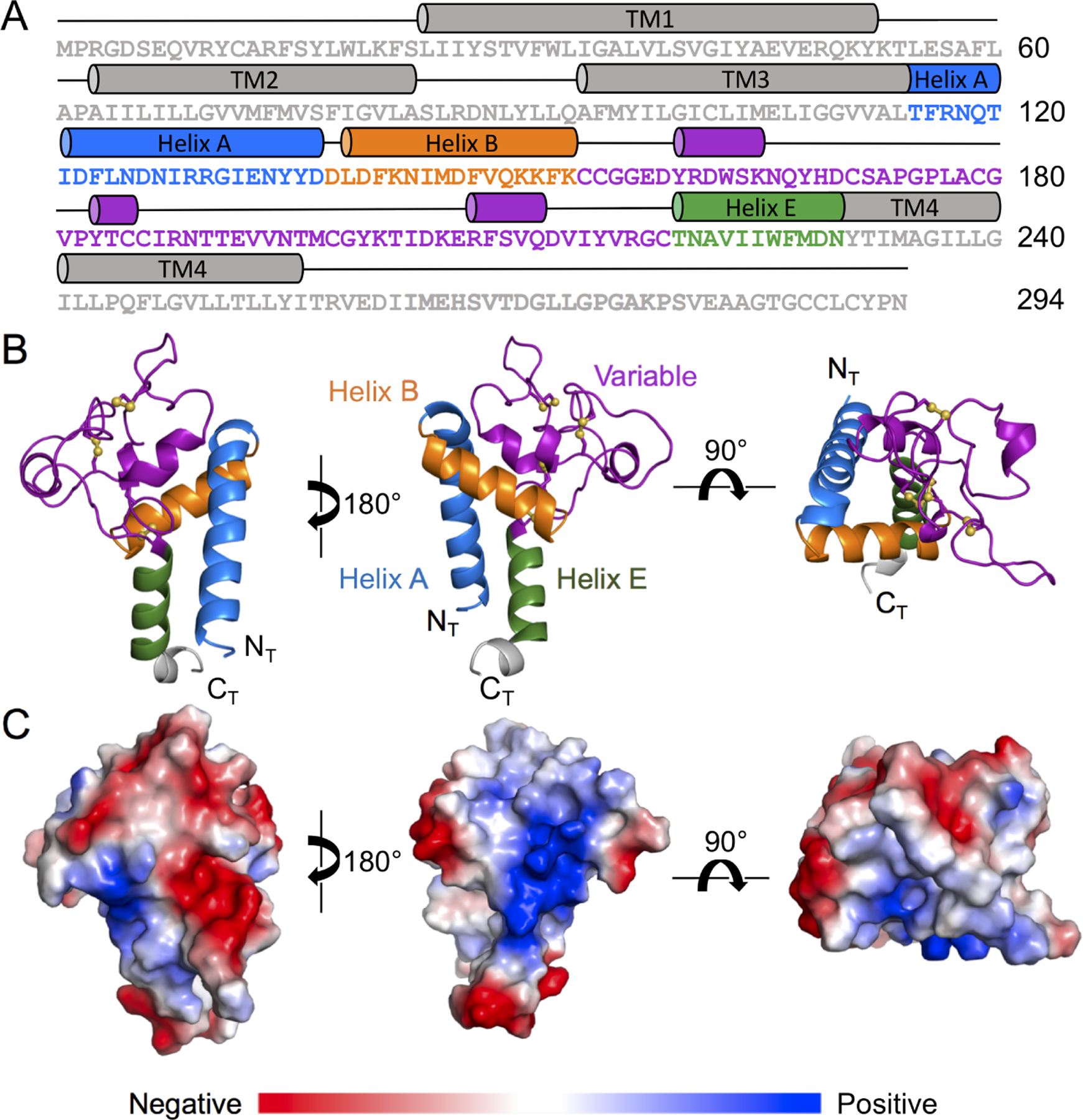
(A) Tspan15 amino acid sequence and domain organization highlighting the structural elements of the LEL domain. (B) Cartoon representation of the Tspan15 LEL domain crystal structure, colored according to the domain organization schematic in (A). The sulfur atoms of the Tspan15 cysteine residues engaged in disulfide bonds are shown as yellow spheres. (C) Surface representation of the Tspan15 LEL colored by electrostatic potential on a scale from red (acidic) to blue (basic).
The structure of the LEL domain was determined to 2.52 Å by x-ray crystallography in space group P1 with 24 copies in the asymmetric unit (Table 1). The 24 copies superimpose with an average backbone RMSD of 0.317Å and maximum RMSD of 0.395 (Figure S2); we use copy B as representative for all figures and structure descriptions (for representative electron density maps, see Figure S3). The overall architecture of the LEL contains a framework of 3 conserved helices, here termed helix A, helix B, and helix E by analogy to other tetraspanins (Figure 1B). The variable region, which connects helices B and E, consists mainly of loops with some short helical segments.
Table 1.
Data Collection and Refinement Statistics
| Tspan15 LEL | Tspan15 LEL – 1C12 Fab complex | |
|---|---|---|
| Wavelength (Å) | 0.979 | 0.979 |
| Resolution range (Å) | 48.67 – 2.52 (2.61 – 2.52) | 44.71 – 3.6 (3.729 – 3.6) |
| Space group | P 1 | C 1 2 1 |
| Unit cell (Å, °) | 83.57, 91.95, 112.38, 87.552, 86.519, 86.241 | 171.3, 124.64, 116.12, 90, 98.171, 90 |
| Total reflections | 641293 (51951) | 68508 (6908) |
| Unique reflections | 111846 (11044) | 27150 (2715) |
| Multiplicity | 5.7 (4.7) | 2.5 (2.5) |
| Completeness (%) | 99.38 (97.51) | 95.77 (97.17) |
| Mean I/sigma(I) | 6.85 (1.34) | 4.63 (0.86) |
| Wilson B-factor | 76.66 | 121.58 |
| R-merge | 0.1461 (1.29) | 0.1577 (1.08) |
| R-meas | 0.1601 (1.451) | 0.1991 (1.364) |
| R-pim | 0.06505 (0.6453) | 0.1196 (0.8215) |
| CC1/2 | 0.996 (0.441) | 0.995 (0.52) |
| CC* | 0.999 (0.782) | 0.999 (0.827) |
| Reflections used in refinement | 111803 (11044) | 26966 (2715) |
| Reflections used for R-free | 1971 (202) | 1980 (200) |
| R-work | 0.2350 (0.3650) | 0.2701 (0.3910) |
| R-free | 0.2736 (0.4137) | 0.3088 (0.4077) |
| CC(work) | 0.952 (0.538) | 0.920 (0.596) |
| CC(free) | 0.938 (0.491) | 0.905 (0.571) |
| Number of non-hydrogen atoms | 22771 | 8279 |
| macromolecules | 22730 | 8279 |
| ligands | 0 | 0 |
| solvent | 41 | 0 |
| Protein residues | 2919 | 1136 |
| RMS(bonds) | 0.012 | 0.003 |
| RMS(angles) | 1.41 | 0.74 |
| Ramachandran favored (%) | 93.97 | 94.84 |
| Ramachandran allowed (%) | 6.03 | 4.80 |
| Ramachandran outliers (%) | 0.00 | 0.36 |
| Rotamer outliers (%) | 1.08 | 0.12 |
| Clashscore | 12.57 | 8.98 |
| Average B-factor | 95.75 | 126.93 |
| macromolecules | 95.80 | 126.93 |
| solvent | 69.06 | N/A |
| Number of TLS groups | 129 | 29 |
Highest shell statistics are reported in parentheses.
The defining feature of the TspanC8 subfamily is the presence of eight cysteines in the LEL domain that form four disulfide bonds. The first two disulfides are conserved among tetraspanins, the first connecting the C-terminal end of Helix B to the N-terminal end of Helix E (C154 to C219), and the second connecting C155, just after Helix A, to C185 in a small helical segment in the domain core. The two disulfide bonds specific to TspanC8 proteins form two loops between pairs of adjacent cysteines, C171-C179 and C186-C198. A surface representation of the LEL colored by surface potential highlights distinct acidic and basic surfaces on opposing sides of the domain (Figure 1C). There is also a hydrophobic, solvent-exposed region within the variable domain adjacent to the TspanC8 specific disulfide bonds.
The variable region of the Tspan15 LEL differs substantially from reported tetraspanin structures
Structures of three other tetraspanin proteins - CD9, CD81 and CD53 - have been solved (Umeda et al., 2020; Yang et al., 2020; Zimmerman et al., 2016). All of these tetraspanins have four extracellular cysteines and are in the C4 subgroup. Structural alignment of our Tspan15 structure with the LEL domains of the other tetraspanins shows that the A, B, and E helices form a shared structural framework, whereas the variable region of Tspan15 is strikingly divergent from the others (Figure 2). Tspan15 has a longer variable region with additional loops formed between the TspanC8 specific disulfide bonds, which may impart additional capacity to bind to diverse partner proteins.
Figure 2. Comparison of the Tspan15 LEL with other Tspan LEL structures.
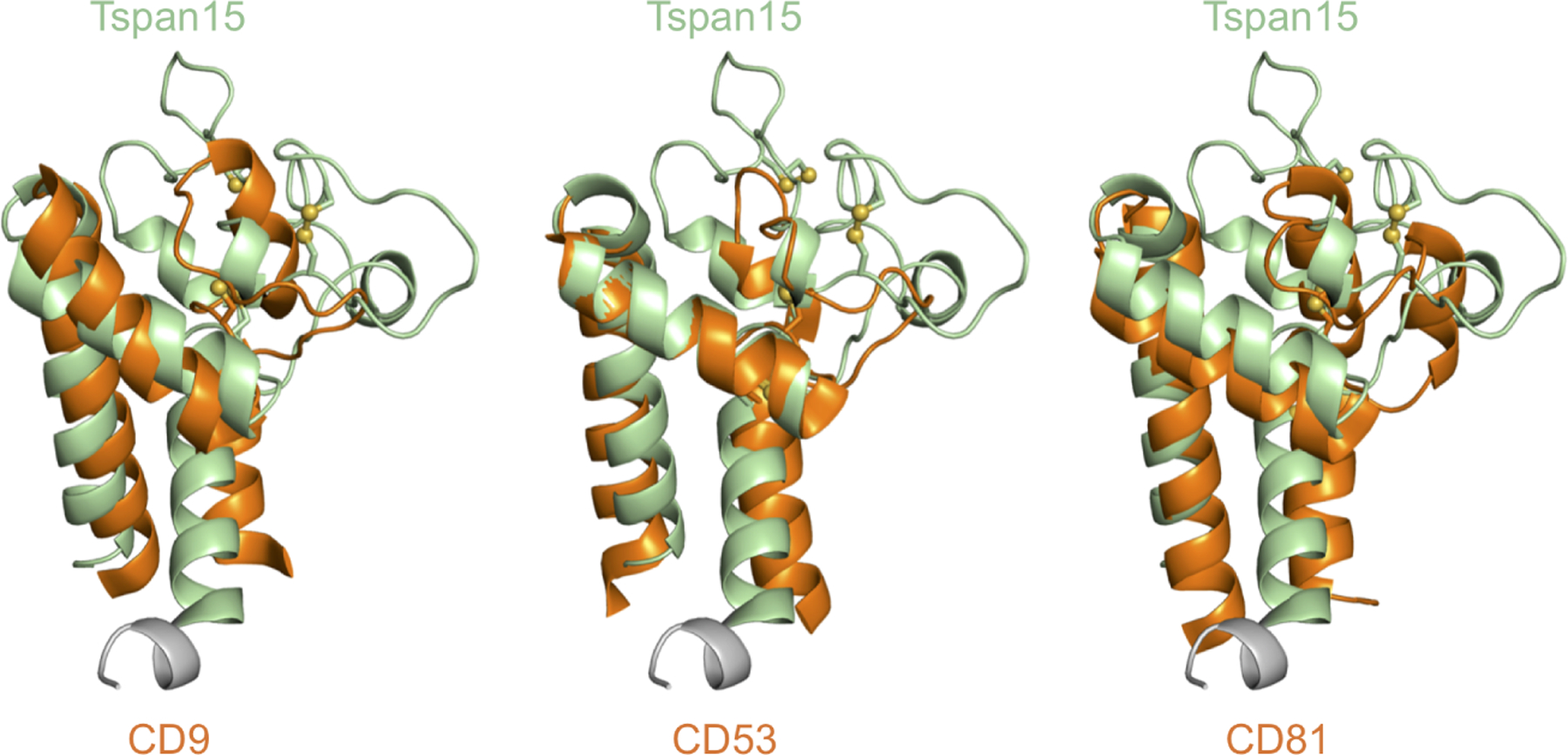
The crystal structure of the Tspan15 LEL domain is aligned with the LEL structures of CD81 (PDB: 5TCX), CD53 (PDB: 6WVG) and CD9 (PDB: 6K4J). The Tspan15 LEL is shown in pale green with each of the other tetraspanins shown in orange.
Crystal structure of the Tspan15 LEL bound to the 1C12 Fab
We also determined the crystal structure of the Tspan15 LEL domain in complex with the Fab fragment of the α-Tspan15 1C12 antibody to 3.6 Å resolution (Figure 3A; Table 1). 1C12 was previously reported to partially inhibit VE-cadherin cleavage by ADAM10 in a cell-based cleavage assay (Koo et al., 2020). The antibody contact site lies primarily in a loop formed by the C198-C219 disulfide bond, which also encompasses a small helical segment (Figure 3B). Additional residues in contact with the Fab are also adjacent to this loop. Overlay of the Fab complex with the free LEL domain structure shows that the binding site conformation is unaffected by complexation with 1C12. The major difference between the Fab bound and free structures is a 5.6 Å shift in the loop between I187 and N195, and there is unidentified electron density in the loop that may be due to bound co-solvent or other small molecule. Due to the modest resolution of the structure, however, we cannot definitively assign the identity of this molecule.
Figure 3. Crystal structure of Tspan15 LEL bound to the 1C12 Fab fragment.
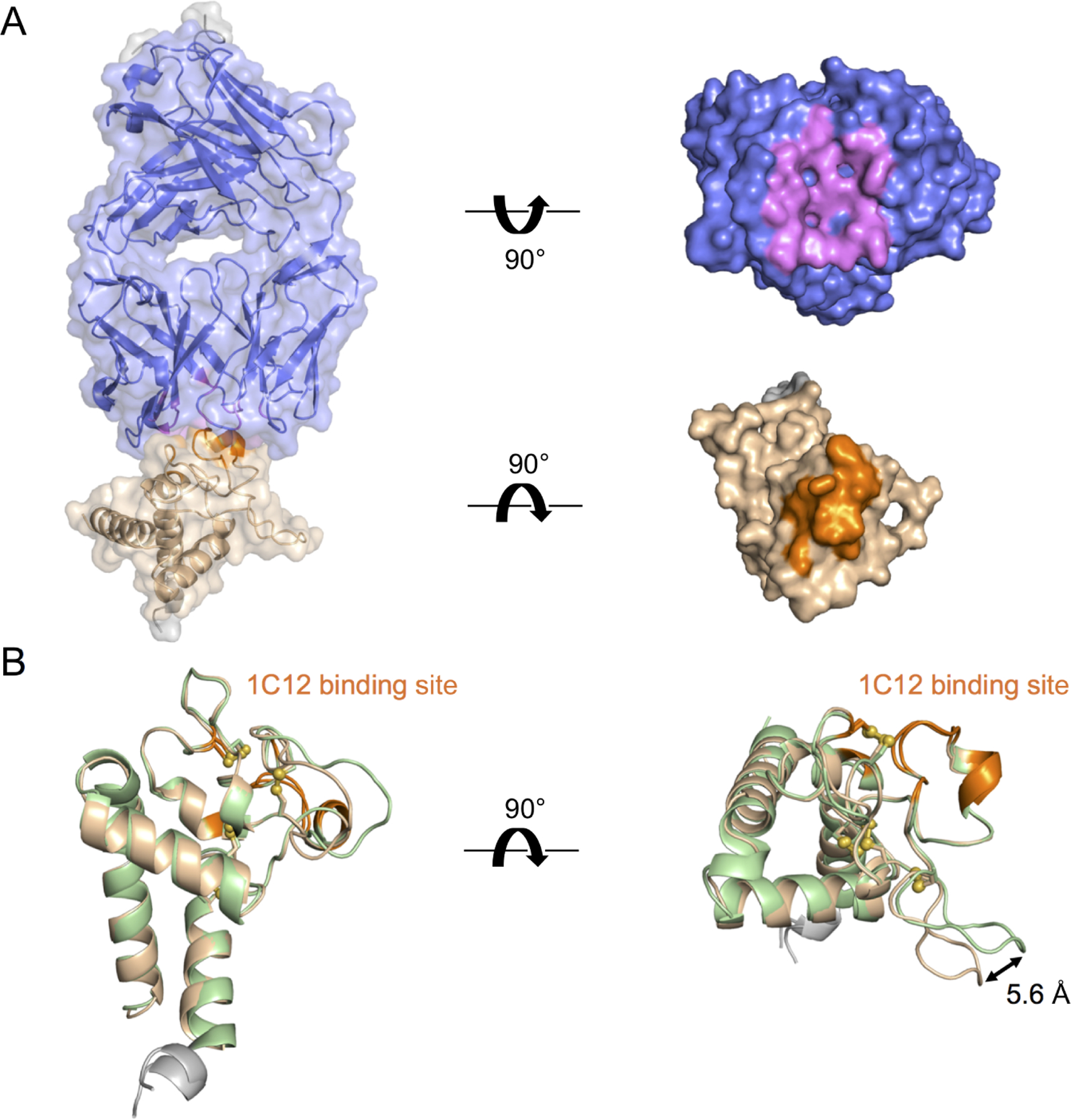
(A) The crystal structure of the Tspan15 LEL bound to the 1C12 Fab is shown as a transparent surface over a cartoon representation (left), and as a solid surface with the two proteins displayed in an “open book” view to highlight the binding interface (right). The Tspan15 LEL is colored light brown with contacting residues in orange. 1C12 is colored slate blue with contacting residues in violet. Contact residues are defined as those approaching within 4 Å. (B) The structure of the Tspan15 LEL bound to 1C12 (light brown) is superimposed on that of the isolated LEL domain (pale green). The 1C12 binding site is colored in orange on both structures. The major difference between the two structures is a 5.6 Å shift in the loop between I187 and N195.
Mutation at conserved site impairs binding to ADAM10
To identify conserved, surface-exposed regions of Tspan15 that might comprise an ADAM10 binding site, we used the program Consurf (Ashkenazy et al., 2016; Figure 4A). Based on the structural and conservation data, we designed triple alanine mutations at two highly conserved sites in the variable region. The first mutant has the N166A, Y168A and H169A substitutions (166/168/169), and the second has the V193A, N195A and T196A (193/195/196) substitutions. We designed an additional triple alanine mutant, D204A, K205A and E206A (204/205/206), at a non-conserved patch overlapping the 1C12 antibody binding site.
Figure 4. Mapping of an ADAM10 interaction site on the Tspan15 LEL.
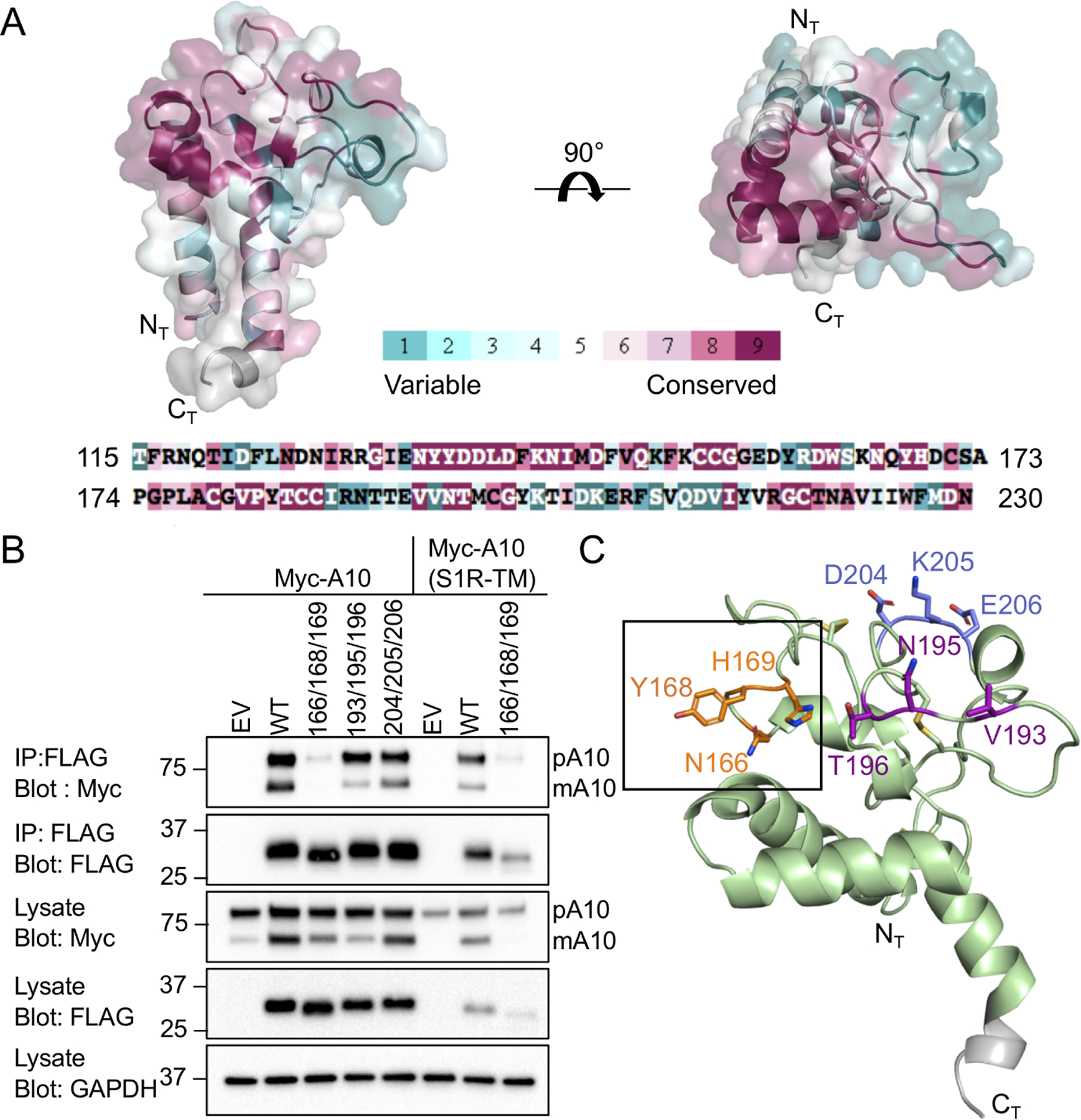
(A) Conservation analysis of the Tspan15 LEL domain, performed using the Consurf server. 150 sequences that sample the list of homologs with between 95 and 35 percent sequence identity were used. The crystal structure and amino acid sequence of the Tspan15 LEL are colored by the Consurf conservation score on a scale from teal (not conserved) to maroon (most conserved). (B) Co-Immunoprecipitation of ADAM10 with wild-type and mutant forms of FLAG-tagged Tspan15. HEK293T cells were co-transfected with FLAG-Tspan15 or the indicated triple alanine mutants with either myc-tagged ADAM10 or ADAM10(S1R-TM). Cleared lysates were subjected to immunoprecipitation with anti-FLAG resin. Lysates and immunoprecipitates were western blotted and probed with the indicated antibodies. Empty pcDNA3.1/Hygro(+) plasmid vector (EV) was used as a negative control. Bands for pro-ADAM10 (pA10) and mature ADAM10 (mA10) are labeled. (C) The three mutated sites are highlighted in different colors on the Tspan15 LEL crystal structure. The site of the mutation that does not co-immunoprecipitate ADAM10 is boxed.
To test for the effect of the mutations on the interaction of Tspan15 with ADAM10, plasmids encoding DNA constructs for expression of these proteins with a FLAG tag were co-transfected with a plasmid encoding Myc-tagged ADAM10 in HEK-293T cells, and anti-FLAG immunoprecipitates were probed for the amount of recovered Tspan15 and ADAM10 by Western blot with anti-FLAG and anti-Myc antibodies, respectively (Figure 4B). Immature and mature ADAM10 have previously been shown to run at ~90 kDa and ~65 kDa respectively (Noy et al., 2016). Wild-type Tspan15 increased ADAM10 maturation and immunoprecipitated both mature and immature ADAM10. When the 166/168/169 mutant and ADAM10 were co-expressed, co-immunoprecipitation of ADAM10 with the mutant was nearly undetectable. Maturation of ADAM10 was diminished by comparison to that seen with co-transfected WT Tspan15, but increased relative to empty vector, suggesting that binding was greatly reduced but not completely abolished. The 193/195/196 mutant co-immunoprecipitated ADAM10, although maturation of ADAM10 was reduced relative to wild-type. As anticipated, the 204/205/206 mutation at the 1C12 binding site did not have an effect on ADAM10 co-immunoprecipitation or maturation. These three sites are highlighted on the crystal structure (Figure 4C). The steady state protein levels of both WT Tspan15 and the 166/168/169 mutant were similarly increased with overexpression of ADAM10 (Figure S4). This effect on 166/168/169 protein amount together with increased ADAM10 maturation (relative to empty vector) is also consistent with the conclusion that the mutant is properly folded and forms a weak interaction with ADAM10 that is not stable to immunoprecipitation. The increased mobility of the 166/168/169 band seen in Figure 4B is likely due to different glycoforms in the free and ADAM10 bound states. This interpretation is consistent with a reported mobility difference for Tspan5 that was immunoprecipitated with an antibody that only recognized the fraction not associated with ADAM10 (Eschenbrenner et al., 2020).
To assess whether the transmembrane region (TM) of ADAM10 contributes to its association with Tspan15, we expressed an ADAM10 chimera in which the TM of ADAM10 was replaced with the TM of the unrelated sigma 1 receptor (S1R). When the 166/168/169 mutant and the ADAM10-S1R chimera were co-expressed, co-immunoprecipitation of the ADAM10-S1R chimera with the mutant was virtually undetectable and the ADAM10-S1R chimera failed to mature. The overall abundance of the Tspan15 mutant was also greatly reduced when co-expressed with the ADAM10-S1R chimera, when compared to its abundance when co-expressed with wild-type ADAM10 (Figure 4B). These results suggest that there may be additional contacts in the transmembrane region that are lost in the ADAM10-S1R-TM chimera, with the loss of these contacts leading to decreased steady-state Tspan15 protein abundance and loss of ADAM10 maturation. These findings are consistent with the lower steady-state abundance of Tspan15 when ADAM10 was not co-expressed (Figure S4), and was also consistent with other work investigating the Tspan15-ADAM10 interaction, in which the steady-state amounts of the two proteins depended on their association (Koo et al., 2020).
N-cadherin shedding is selectively promoted by Tspan15 expression
Multiple studies have shown that Tspan15 promotes shedding of N-cadherin by ADAM10 (Jouannet et al., 2016; Noy et al., 2016; Prox et al., 2012; Seipold et al., 2018). Using the known ADAM10 activating compound N-ethylmaleimide (NEM), we observe near complete processing of N-cadherin in U251 glioblastoma cells (measured by appearance of a ~40 kDa C-terminal fragment), whereas cleavage is severely reduced in Tspan15 knockout U251 cells (Figure 5). When each of the six TspanC8 proteins was individually added back to the knockout cells, Tspan15 restored cleavage most effectively (Figure 5A). Tspan10, Tspan17 and Tspan33 were detected at similar or higher abundance when compared to Tspan15, but did not increase N-cadherin cleavage above the basal amount seen in the empty vector control. There was no detectable expression of Tspan5 or Tspan14, however (Figure 5A). The incomplete cleavage upon re-addition of Tspan15 is most likely due to transfection efficiency in these cells that is less than 100%.
Figure 5. A Tspan15 requirement for N-cadherin cleavage by ADAM10.
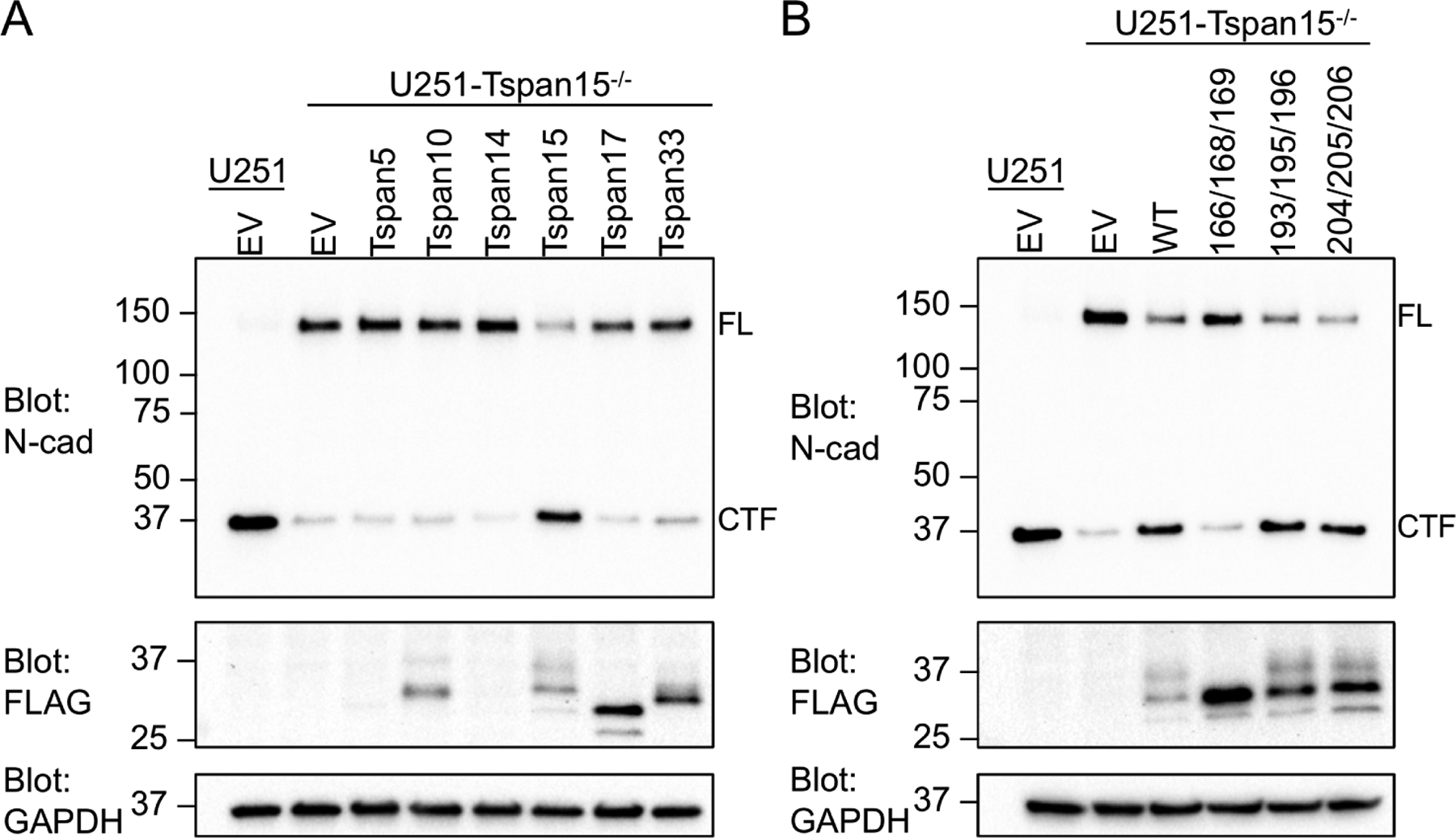
(A, B) N-cadherin cleavage assay. U251 cells and U251-Tspan15−/− cells were transfected with the indicated FLAG-tagged tetraspanin construct or empty vector. Cells were pre-treated with the gamma-secretase inhibitor Compound E for one hour prior to addition of 2 mM NEM. Cells were incubated for 40 minutes, subjected to western blotting, and probed with an antibody against the cytoplasmic region of N-cadherin (N-cad). Assay results are shown following transfection with (A) human TspanC8 proteins and (B) Tspan15 alanine mutants. Assay samples were also probed with anti-FLAG (middle panel) and GAPDH (lower panel) antibodies. Bands for full-length (FL) and C-terminal fragment (CTF) of N-cadherin are labeled on the blots.
Tspan15 binding mutant is unable to rescue N-cadherin cleavage
To examine the functional effect of the Tspan15 LEL mutants, we compared their effect on N-cadherin cleavage in Tspan15 knockout cells with that of wild-type Tspan15 or empty vector. Whereas the 193/195/196 and 204/205/206 mutants, which both co-immunoprecipitated ADAM10 comparably to wild-type Tspan15, rescued N-cadherin cleavage like wild-type protein, the 166/168/169 mutant, which was highly deficient in ADAM10 immunoprecipitation (Figure 4B), also failed to rescue N-cadherin cleavage, indicating that it does not form a functional complex (Figure 5B).
Discussion
Tspan15 is a member of the tetraspanin-C8 subfamily of proteins, which have eight extracellular cysteine residues and function by directing ADAM10 trafficking, maturation, and substrate selectivity (Matthews et al., 2017). Tspan15 binds to ADAM10 and forms a functional, plasma-membrane complex that carries out ectodomain shedding of membrane-bound substrates. This work reports X-ray crystal structures of the LEL domain of Tspan15 both in isolation and in complex with the 1C12 antibody Fab fragment, identifies a conserved ADAM10 binding site in the variable region of the LEL, and confirms the functional importance of this site in an ectodomain shedding assay for N-cadherin.
The structure of the Tspan15 LEL is distinct from that of known structures of other tetraspanins, which are all from the C4 group with two disulfide bonds (Umeda et al., 2020; Yang et al., 2020; Zimmerman et al., 2016). Like all tetraspanins, Tspan15 has the same core three-helix motif formed by the A, B and E-helices, but differs from the C4 tetraspanins by having a substantially larger variable region with two additional loops formed by the disulfide bonds unique to the TspanC8 proteins. Interestingly, the loop formed between C171 and C179 is present in Tspan15 and Tpsan10, where the loop is hydrophobic, and diverges in Tspan5, Tspan14, Tspan17 and Tspan33, where the sequence is less hydrophobic and has a two-residue insertion (Figure S5).
Our structure of the Tspan15 LEL-1C12 Fab complex reveals that the antibody binding site is located away from the experimentally determined ADAM10 interaction site. Furthermore, mutation at the 1C12 binding site had no effect on ADAM10 binding or N-cadherin cleavage. These findings are consistent with the observation that 1C12 can bind to the ADAM10-Tspan15 complex at the cell surface (Koo et al., 2020). 1C12 was reported to have a modest inhibitory effect on VE-cadherin cleavage by ADAM10 (Koo et al., 2020), but the mechanism of inhibition is not clear from our structure of the LEL-Fab complex.
The results of our co-immunoprecipitation and cleavage assays show that functional association between ADAM10 and Tspan15 is primarily mediated by the LEL domain, with additional contacts potentially present between the transmembrane regions. Mutation of the conserved 166/168/169 site impairs, but does not completely eliminate binding to ADAM10, as judged by the partial maturation of ADAM10 and the increased amount of Tspan15 present when co-expressed with ADAM10. The equivalent site in Tspan5 has also been reported to impair ADAM10 binding (Saint-Pol et al., 2017b), fully consistent with the results reported here. This three-residue site is highly conserved across the TspanC8 subfamily and likely constitutes an important part of the ADAM10 binding interface for all TspanC8s.
Our results show that ADAM10 specifically requires Tspan15 in order to cleave N-cadherin in U251 cells. This observation raises the question of how TspanC8 family members promote ADAM10 processing of specific substrates. One possibility is that the bound TspanC8 directs ADAM10 to a membrane microdomain containing its substrate, possibly through interactions with additional proteins. One study provides an example for how the different TspanC8s can affect ADAM10 membrane compartmentalization: Tspan33 clusters ADAM10 in cell-cell junctions through interaction of its C-terminal cytoplasmic tail with the PLEKHA7-PDZD11 complex in mouse CCD cells (Shah et al., 2018). An alternative possibility is that different TspanC8 proteins can engage ADAM10 in different ways, potentially altering its conformation. Co-immunoprecipitation experiments with a series of ADAM10 ectodomain truncations show differences in ability to bind TspanC8s, with Tspan15 as the only one that immunoprecipitated with ADAM10 truncated to the membrane-proximal stalk domain (Noy et al., 2016). It is also plausible that localization differences and variability in the ADAM10 contact interface both contribute to substrate selectivity.
Structures have been reported of full-length tetraspanins both in isolation and in complex with binding partners (Oosterheert et al., 2020; Susa et al., 2021; Umeda et al., 2020; Yang et al., 2020; Zimmerman et al., 2016), with the structure of one such complex at sufficiently high resolution to build an atomic model (Susa et al., 2021). The isolated tetraspanin proteins adopt closed conformations in which the LEL covers a cavity formed by the transmembrane helices, whereas the bound forms of the tetraspanins adopt an open conformation in which the central cavity either becomes exposed or occluded by closer approximation of the transmembrane helices. The crystal structure of CD81 contained a bound cholesterol in the intramembrane cavity, but when bound to CD19, the cavity collapsed and there was no pocket available for cholesterol binding (Susa et al., 2021; Zimmerman et al., 2016). Mutations to the cholesterol binding site increased the ability to traffic CD19 to the cell surface, suggesting that binding to cholesterol could regulate CD19 binding by CD81 (Zimmerman et al., 2016). Whether Tspan15 undergoes a similar conformational opening in response to ADAM10 binding remains unresolved, and structures of full-length Tspan15 in its free and ADAM10-bound states will ultimately resolve this question.
STAR Methods
Resource availability
Lead contact
Further information and requests for either resources or reagents should be directed to and will be fulfilled by the lead contact Stephen C. Blacklow (stephen_blacklow@hms.harvard.edu).
Materials availability
Requests for materials generated in this study should be directed to and will be fulfilled by the lead contact Stephen C. Blacklow (stephen_blacklow@hms.harvard.edu).
Data and code availability
Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (PDB) under the accession numbers 7RDB (Tspan15 LEL) and 7RD5 (Tspan15 LEL-Fab complex).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Protein for crystallography was isolated from conditioned media of Expi293F cells. Expi293 cells were maintained in suspension at 37 °C in Expi293 expression medium (ThermoFisher). Co-immunoprecipitation experiments were performed in HEK293T cells. N-cadherin cleavage assays were performed in U251 cells. HEK293T and U251 cells were maintained at 37 °C in DMEM (VWR) supplemented with 10% fetal calf serum and pen/strep.
Method details
Plasmid construction
The LEL domain of Tspan15 (residues 115–230) was cloned into the pFUSE-hIgG1-Fc2 vector containing a C-terminal human IgG Fc tag after a 3C protease site. The mutation N118Q was introduced using site directed mutagenesis. A single DNA insert containing the 1C12 light chain followed by a P2A ribosomal skipping sequence and then the heavy chain Fab sequence was chemically synthesized (Integrated DNA Technologies) and inserted into the pFUSE-hIgG1-Fc2 plasmid containing the human IgG Fc. The sequence contains a 3C protease site between the light chain and the P2A sequence, and another 3C site between the heavy chain and the Fc. The light chain uses the IL2 signal sequence in the vector and the heavy chain uses the native signal sequence. 1C12 contains the N5Q mutation in the light chain to remove a glycosylation site. N-terminal Flag-tagged Tspan5, Tspan10, Tspan14, Tspan15, Tspan17 and Tspan33 were cloned into pcDNA3.1/Hygro(+) using synthetic genes (gBlocks) from Integrated DNA Technologies. Tspan15 mutations were made using site-directed mutagenesis. ADAM10-myc in the pRK5m plasmid was obtained from Addgene.
Protein expression
Tspan15 LEL and 1C12 antibody were expressed in Expi293F cells. For each protein, Expi293F cells were grown in Expi293 growth medium to a density of 3.0×106 cells/mL and then transfected with 1 mg DNA / L of culture and PEI MAX 40K reagent at a 1:3 DNA/PEI ratio. 24 hours following transfection the cells were fed with 6 mL of 45% D-(+)-Glucose solution (Sigma-Aldrich) and 3 mM valproic acid sodium salt (Sigma-Aldrich). Cells were incubated for 5–6 days following transfection, then centrifuged at 4000×g for 15 minutes to harvest the medium.
Tspan15 LEL purification
The protein was harvested from the culture medium on a column containing protein A agarose resin. After the protein was loaded onto the column, the column was washed with buffer containing 20 mM HEPES, 150 mM NaCl pH 7.4. Protein was then eluted with buffer containing 20 mM glycine, 150 mM NaCl pH 3.0, and the eluate was rapidly neutralized with 1M HEPES buffer, pH 7.4 to a final buffer concentration of 100 mM. After the Tspan15 LEL was concentrated to 1–2 mg/ml, 3C protease and PNGaseF were added at 1:5 and 1:2 ratios (w/w) respectively to remove the Fc tag and the glycan. The Tspan15 LEL enzyme mixture was incubated at room temperature overnight on a rotator. The his-tagged enzymes and Fc were removed by flowing over HisPur Ni-NTA resin and Protein A resin. Tspan15 LEL was then purified using a Mono-Q 10/100 GL anion exchange column with 20 mM HEPES buffer using a gradient of NaCl. The protein was further purified by size exclusion chromatography (SEC) on a Superdex-75 10/300 column equilibrated with 20 mM HEPES, 150 mM NaCl pH 7.4. Peak fractions containing purified protein were concentrated using a 3 kDa cutoff centrifugal concentrator.
Tspan15 LEL 1C12 Fab complex purification
1C12 was purified using Protein-A resin with the same protocol as for the Tspan15 LEL. The Fc was removed by cleavage with 3C protease at a ratio of 40:1 (w/w) at room temperature overnight. The Fc tag and enzyme were removed by passage over Ni-NTA resin and Protein A resin. 1C12-Fab was mixed 1:1 with Tspan15 LEL and incubated at 4 C for 1 hour. The complex was then purified by size exclusion chromatography using a Superdex-200 10/300 column equilibrated with 20 mM HEPES, 150 mM NaCl pH 7.4. The protein complex was concentrated using a 10 kDa cutoff centrifugal concentrator.
Protein crystallization and data collection
24 well hanging drop trays with 10 mg/ml Tspan15 LEL were set at room temperature with 0.5 μl protein and 0.5 μl reservoir solution. P1 space group crystals were grown in 0.1 M MES pH 6.9, 34–35 % polypropylene glycol p-400. P321 crystals were grown in 0.1 M HEPES pH 7.4, 37 % polypropylene glycol p-400. Crystals appeared overnight and started to degrade beyond 24 hours. Tspan15 LEL crystals were cryoprotected in reservoir solution and flash frozen in liquid nitrogen. Tspan15 LEL-1C12 complex crystals were grown in 24 well hanging drop trays with 2.5 mg/ml protein. Drops were set with 0.5 μl protein and 0.5 μl reservoir solution. Crystals grew in 0.1 M sodium acetate pH 4.6, 3.4 M sodium nitrate. Tspan15 LEL-1C12 crystals were cryoprotected in reservoir solution with the addition of 20 % glycerol and flash frozen in liquid nitrogen. X-ray data was collected at Advanced Photon Source NE-CAT beamlines 24 ID-C and ID-E.
Structure determination
X-ray diffraction images were processed and scaled using XDS (Kabsch, 2010). The Phenix software suite (Afonine et al., 2012) was used for phasing (Phaser) and refinement for all data sets. Coot (Emsley and Cowtan, 2004) was used for manual building. All crystallographic data processing, refinement, and analysis software was compiled and supported by the SBGrid Consortium (Morin et al., 2013). The coordinates of the anti-ADAM10 11G2 Fab (PDB: 6BDZ; Seegar et al., 2017) were used to make a homology model (SWISS-MODEL; Waterhouse et al., 2018) of the anti-Tspan15 11C2 Fab. Tspan15 LEL-1C12 complex data was phased using this modeled Fab as a search model for molecular replacement. Initially we built a partial model of the Tspan15 LEL using Coot, but it was incomplete in the variable region due to low resolution data. The partial model of the Tspan15 LEL was used as a search model to phase a low resolution P321 data set of the Tspan15 LEL alone. Two copies (of four in the asymmetric unit) from the P321 partial model were then used as a search model to solve a data set that diffracted to higher resolution in space group P1 with 24 copies in the asymmetric unit. Data from two crystals were combined for the P1 space group data set. The complete model was built with iterative cycles of manual building in Coot and refinement in Phenix Refine. Refinement used optimization of real space and reciprocal space xyz coordinates, individual B-factor refinement, TLS and NCS restraints. The final cycles of refinement were done without NCS restraints. The complete high-resolution structure of Tspan15 LEL was then used to fit the LEL for the Fab complex data set. The Fab complex structure was refined with optimization of real space and reciprocal space xyz coordinates, individual B-factor refinement, TLS, NCS and secondary structure restraints. Quality of the models was evaluated using composite omit density maps. Final models were validated using MolProbity in the Phenix suite and the PDB validation server. Structures were visualized using Pymol (Schrödinger, 2010).
Sequence conservation analysis
The Consurf server using the Clean Uniprot database was used for conservation analysis. 150 sequences that sample the list of homologs with between 95 and 35 percent sequence identity were used. The sequence conservation output was mapped onto the Tspan15 LEL crystal structure using Consurf default parameters.
Immunoprecipitation and western blotting
HEK-293T cells were plated in 6-well tissue culture dishes and cultured in DMEM with 10% BSA. Cells were transfected with 2 μg total DNA and PEI MAX 40K transfection reagent at a 1:3 ratio. After 48 hours cells were washed in ice cold PBS and lysed in buffer containing 20 mM HEPES pH 7.4, 150 mM NaCl, 1% DDM and 10 μM BB94 and centrifuged at 20000 × g for 10 minutes. Lysates were added to 20 μl of M2 anti-FLAG resin and incubated for one hour at 4 °C. The resin was washed three times with buffer containing 20 mM HEPES pH 7.4, 150 mM NaCl, 0.1% DDM and 1 μM BB94. 2X SDS loading dye was added directly to the washed resin. Samples were run on an SDS-PAGE gel, and transferred to a nitrocellulose membrane. Membranes were blocked in tris-buffered saline containing 0.1% tween 20 (TBST) containing 5% non-fat milk for one hour, and then incubated in primary antibody overnight at 4 °C. Primary antibodies were anti-FLAG (1:2000 dilution), anti-Myc (1:2000 dilution), and anti-GAPDH (1:2000 dilution). Membranes were washed three times with TBST and incubated with anti-rabbit-HRP secondary antibody at 1:10000 dilution for one hour. Membranes were again washed three times with TBST. Western blots were developed using Western Lightning Plus-ECL detection Kit. Experiments were performed in duplicate.
Development of U251 Tspan15 knockout cell line
The guide RNA sequence GCGCGCGCUUCUCCUACCUC was cloned into the pSpCas9(BB)-2A-GFP vector. The resulting plasmid, coding for the guide RNA, spCas9 and GFP was used to transfect U251 cells. GFP expressing cells were single-cell sorted into 96 well plates and clonal populations were expanded for 3 weeks. Genomic DNA was extracted from the cells and the targeted locus was amplified by PCR and Topo-cloned (Topo TA cloning kit) for sequencing. A minimum of ten clones were sequenced to confirm the targeted mutation.
N-cadherin cleavage assay
U251 cells were plated in 6-well tissue culture dishes in DMEM with 10% BSA. The cells were transfected with 2 μg DNA using 3.2 μl U251 Avalanche transfection reagent. 48 hours after transfection, fresh media containing the gamma-secretase inhibitor compound E (500 nM) was added to the cells. After one hour, 2 mM N-ethylmaleimide (NEM) was added to the cells. 40 minutes after adding NEM, the cells were washed in PBS and lysed directly in 500 μl SDS-loading dye containing 5% β-mercaptoethanol and 2 mM EDTA. Samples were sonicated and run on an SDS-PAGE gel. Proteins were then transferred to a nitrocellulose membrane. Membranes were blocked in TBST containing 5% non-fat milk for one hour, then incubated in anti-N-cadherin antibody (mouse, 1:1000 dilution), anti-FLAG (rabbit, 1:2000 dilution) or GAPDH (rabbit, 1:5000) overnight at 4 °C. Membranes were washed three times with TBST and incubated with anti-mouse-HRP or anti-rabbit-HRP secondary antibody at 1:10000 dilution for one hour. Membranes were again washed three times with TBST. Western blots were developed using Western Lightning Plus-ECL detection Kit. Experiments were performed in duplicate.
Quantification and Statistical Analysis
Each Western blot is representative of at least n=2 biological replicates.
Additional Resources
None.
Supplementary Material
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-FLAG | Cell Signaling Technology | Cat # 214793 RRID:AB_2572291 |
| Rabbit anti-myc-tag (clone 71D10) | Cell Signaling Technology | Cat # 2278 RRID:AB_490778 |
| Rabbit anti-GAPDH (clone 14C10) | Cell Signaling Technology | Cat # 2118s RRID:AB_561053 |
| Mouse monoclonal anti-N-Cadherin (Clone 32) | BD Biosciences | Cat # 610920 RRID:AB_398236 |
| Goat anti-rabbit-HRP | Abcam | Cat # ab6721 |
| Goat anti-mouse-HRP | ThermoFisher | Cat # 62-6520 |
| ANTI-FLAG M2 Affinity Gel | Sigma-Aldrich | Cat # A2220 RRID: AB_10063035 |
| Chemicals, peptides, and recombinant proteins | ||
| Batimastat (BB94) | Sigma-Aldrich | Cat # 19440 |
| Compound E | EMD Millipore | Cat # 565790 |
| U251 Avalanche transfection reagent | EZ Biosystems | Cat # EZT-U251-1 |
| PEI Max 40000 | VWR | Cat # 75800-188 |
| n-Dodecyl-b-D-Maltoside (DDM) | Anatrace | Cat # D310 |
| polypropylene glycol p-400 | Hampton Research | Cat # HR2-771 |
| sodium nitrate | Hampton Research | Cat # HR2-661 |
| sodium acetate trihydrate pH 4.6 | Hampton Research | Cat # HR2-731 |
| StockOptions MES pH 6.9 | Hampton Research | Cat # HR2-243 |
| StockOptions HEPES pH 7.4 | Hampton Research | Cat # HR2-102 |
| N-ethylmaleimide (NEM) | Sigma-Aldrich | Cat # E3876 |
| Expi293 expression medium | ThermoFisher | Cat # A1435103 |
| HisPur Ni-NTA resin | ThermoFisher | Cat # 88222 |
| Protein A agarose resin | EMD Millipore | Cat # 16-125 |
| 3C protease recombinant protein | Produced in-house | N/A |
| PNGaseF recombinant protein | Produced in-house | N/A |
| Critical commercial assays | ||
| Western Lightning Plus-ECL detection Kit | PerkinElmer | Cat # NEL103001EA |
| TOPO TA Cloning Kit | ThermoFisher | Cat # 450030 |
| Deposited data | ||
| Human Tspan15 LEL structure coordinates | This paper | PDB: 7RDB |
| Human Tspan15 LEL-1C12Fab structure coordinates | This Paper | PDB: 7RD5 |
| CD9 structure coordinates | (Umeda et al., 2020) | PDB: 6K4J |
| CD53 structure coordinates | (Yang et al., 2020) | PDB: 6WVG |
| CD81 structure coordinates | (Zimmerman et al., 2016) | PDB: 5TCX |
| ADAM10 ectodomain-11G2Fab complex structure coordinates | (Seegar et al., 2017) | PDB: 6BDZ |
| Experimental models: Cell lines | ||
| HEK293T cells | ATCC | Cat # CRL-3216 |
| Expi293F cells | ThermoFisher | Cat # A14527 |
| U251 cells | Gift from Thomas M. Roberts at the Dana-Farber Cancer Institute | N/A |
| U251-Ts15−/− cells | This paper | N/A |
| Recombinant DNA | ||
| pcDNA3.1/Hygro(+) | ThermoFischer | Cat # V87020 |
| pRK5M-ADAM10-myc | (Liu et al., 2009) | Addgene Cat # 31717 |
| pFUSE-hIgG1-Fc2 | Invivogen | Cat # pfuse-hg1fc2 |
| pSpCas9(BB)-2A-GFP (PX458) | (Ran et al., 2013) | Addgene Cat # 48138 |
| gBlocks | Integrated DNA Technologies | N/A |
| Software and algorithms | ||
| Phenix | (Afonine et al., 2012) | https://sbgrid.org/software/ |
| Coot | (Emsley and Cowtan, 2004) | https://sbgrid.org/software/ |
| XDS | (Kabsch, 2010) | https://sbgrid.org/software/ |
| Pymol | (Schrödinger, 2010) | https://sbgrid.org/software/ |
| SWISS-MODEL | (Waterhouse et al., 2018) | https://swissmodel.expasy.org/ |
| SBGrid Consortium | (Morin et al., 2013) | https://sbgrid.org/software/ |
| PDB validation server | World Wide Protein Data Bank | https://www.wwpdb.org/ |
Highlights.
Crystal structures of Tspan15 LEL domain both in isolation and in complex with 1C12 Fab
Structures reveal a variable region supported by a conserved three-helix motif
A conserved site in the variable region of Tspan15 LEL participates in ADAM10 binding
This site is required for rescue of N-cadherin cleavage in Tspan15 knockout cells
Acknowledgments
We thank members of the Blacklow laboratory for helpful discussions. Financial support for this work was provided by NIH grants R35 CA220340 (S.C.B.), a gift from Edward B. Goodnow to S.C.B. and BBSRC grant BB/P00783X/1 to M.G.T. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the NIH (P41 GM103403). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science user facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Declaration of Interests
S.C.B. receives funding for an unrelated project from Novartis, is on the scientific advisory board for Erasca, Inc., is an advisor to MPM Capital, and is a consultant for IFM, Scorpion Therapeutics, Odyssey Therapeutics, and Ayala Pharmaceuticals for unrelated projects.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, and Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, and Ben-Tal N (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44, W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S, Jouannet S, Boucheix C, and Rubinstein E (2014). Tetraspanins at a glance. J. Cell Sci 127, 3641–3648. [DOI] [PubMed] [Google Scholar]
- DeSalle R, Mares R, and Garcia-España A (2010). Evolution of cysteine patterns in the large extracellular loop of tetraspanins from animals, fungi, plants and single-celled eukaryotes. Mol. Phylogenet. Evol 56, 486–491. [DOI] [PubMed] [Google Scholar]
- Dornier E, Coumailleau F, Ottavi J-F, Moretti J, Boucheix C, Mauduit P, Schweisguth F, and Rubinstein E (2012). TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell Biol 199, 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Eschenbrenner E, Jouannet S, Clay D, Chaker J, Boucheix C, Brou C, Tomlinson MG, Charrin S, and Rubinstein E (2020). TspanC8 tetraspanins differentially regulate ADAM10 endocytosis and half-life. Life Sci. Alliance 3, e201900444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lübke T, Lena Illert A, von Figura K, et al. (2002). The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for α-secretase activity in fibroblasts. Hum. Mol. Genet 11, 2615–2624. [DOI] [PubMed] [Google Scholar]
- Hemler ME (2005). Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol 6, 801–811. [DOI] [PubMed] [Google Scholar]
- Hemler ME (2014). Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer 14, 49–60. [DOI] [PubMed] [Google Scholar]
- Hiroshima K, Shiiba M, Oka N, Hayashi F, Ishida S, Fukushima R, Koike K, Iyoda M, Nakashima D, Tanzawa H, et al. (2019). Tspan15 plays a crucial role in metastasis in oral squamous cell carcinoma. Exp. Cell Res 384, 111622. [DOI] [PubMed] [Google Scholar]
- Jouannet S, Saint-Pol J, Fernandez L, Nguyen V, Charrin S, Boucheix C, Brou C, Milhiet P-E, and Rubinstein E (2016). TspanC8 tetraspanins differentially regulate the cleavage of ADAM10 substrates, Notch activation and ADAM10 membrane compartmentalization. Cell. Mol. Life Sci 73, 1895–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohutek ZA, diPierro CG, Redpath GT, and Hussaini IM (2009). ADAM-10-Mediated N-Cadherin Cleavage Is Protein Kinase C- Dependent and Promotes Glioblastoma Cell Migration. J. Neurosci 29, 4605–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo CZ, Harrison N, Noy PJ, Szyroka J, Matthews AL, Hsia H-E, Müller SA, Tüshaus J, Goulding J, Willis K, et al. (2020). The tetraspanin Tspan15 is an essential subunit of an ADAM10 scissor complex. J. Biol. Chem 295, 12822–12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P-H, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Roßner S, and Lichtenthaler SF (2010). ADAM10 is the physiologically relevant, constitutive α-secretase of the amyloid precursor protein in primary neurons. EMBO J 29, 3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn P-H, Colombo AV, Schusser B, Dreymueller D, Wetzel S, Schepers U, Herber J, Ludwig A, Kremmer E, Montag D, et al. (2016). Systematic substrate identification indicates a central role for the metalloprotease ADAM10 in axon targeting and synapse function. ELife 5, e12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, and Shoham T (2005). The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol 5, 136–148. [DOI] [PubMed] [Google Scholar]
- Levy S, Kuo C-C, Sagi Y, Chen H, Kela-Madar N, van Zelm M, and van Dongen JJM (2012). CD81-Dependent Trafficking of CD19: Restoration of CD19 Surface Expression in Human B Cells Harboring A CD81 Mutation. Blood 120, 1049–1049. [Google Scholar]
- Liu C, Xu P, Lamouille S, Xu J, and Derynck R (2009). TACE-mediated ectodomain shedding of the type I TGF-beta receptor downregulates TGF-beta signaling. Mol. Cell 35, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, Strooper B. de, Hartmann D, and Saftig P (2005). ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and β-catenin translocation. Proc. Natl. Acad. Sci 102, 9182–9187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Roth DM, Jans DA, Pouton CW, Partridge LJ, Monk PN, and Moseley GW (2005). Tetraspanins in Viral Infections: a Fundamental Role in Viral Biology? J. Virol 79, 10839–10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AL, Szyroka J, Collier R, Noy PJ, and Tomlinson MG (2017). Scissor sisters: regulation of ADAM10 by the TspanC8 tetraspanins. Biochem. Soc. Trans 45, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Molder L, Juksar J, Harkes R, Wang W, Kreft M, and Sonnenberg A (2019). Tetraspanin CD151 and integrin α3β1 contribute to the stabilization of integrin α6β4-containing cell-matrix adhesions. J. Cell Sci 132. [DOI] [PubMed] [Google Scholar]
- Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, and Sliz P (2013). Collaboration gets the most out of software. ELife 2, e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson IM, and von Heijne G (1993). Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem 268, 5798–5801. [PubMed] [Google Scholar]
- Noy PJ, Yang J, Reyat JS, Matthews AL, Charlton AE, Furmston J, Rogers DA, Rainger GE, and Tomlinson MG (2016). TspanC8 Tetraspanins and A Disintegrin and Metalloprotease 10 (ADAM10) Interact via Their Extracellular Regions. J. Biol. Chem 291, 3145–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterheert W, Xenaki KT, Neviani V, Pos W, Doulkeridou S, Manshande J, Pearce NM, Kroon-Batenburg LM, Lutz M, Henegouwen van B. en PM, et al. (2020). Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F. Life Sci. Alliance 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prox J, Willenbrock M, Weber S, Lehmann T, Schmidt-Arras D, Schwanbeck R, Saftig P, and Schwake M (2012). Tetraspanin15 regulates cellular trafficking and activity of the ectodomain sheddase ADAM10. Cell. Mol. Life Sci 69, 2919–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessmeyer J, and Ludwig A (2009). The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin. Cell Dev. Biol 20, 164–174. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes R, Cardeñes B, Machado-Pineda Y, and Cabañas C (2018). Tetraspanin CD9: A Key Regulator of Cell Adhesion in the Immune System. Front. Immunol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Pol J, Eschenbrenner E, Dornier E, Boucheix C, Charrin S, and Rubinstein E (2017a). Regulation of the trafficking and the function of the metalloprotease ADAM10 by tetraspanins. Biochem. Soc. Trans 45, 937–944. [DOI] [PubMed] [Google Scholar]
- Saint-Pol J, Billard M, Dornier E, Eschenbrenner E, Danglot L, Boucheix C, Charrin S, and Rubinstein E (2017b). New insights into the tetraspanin Tspan5 using novel monoclonal antibodies. J. Biol. Chem 292, 9551–9566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L (2010). The PyMOL Molecular Graphics System, Version 2.5.1
- Seegar TC, and Blacklow SC (2019). Domain integration of ADAM family proteins: Emerging themes from structural studies. Exp. Biol. Med. Maywood NJ 244, 1510–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegar TCM, Killingsworth LB, Saha N, Meyer PA, Patra D, Zimmerman B, Janes PW, Rubinstein E, Nikolov DB, Skiniotis G, et al. (2017). Structural Basis for Regulated Proteolysis by the α-Secretase ADAM10. Cell 171, 1638–1648.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipold L, Altmeppen H, Koudelka T, Tholey A, Kasparek P, Sedlacek R, Schweizer M, Bär J, Mikhaylova M, Glatzel M, et al. (2018). In vivo regulation of the A disintegrin and metalloproteinase 10 (ADAM10) by the tetraspanin 15. Cell. Mol. Life Sci 75, 3251–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Rouaud F, Guerrera D, Vasileva E, Popov LM, Kelley WL, Rubinstein E, Carette JE, Amieva MR, and Citi S (2018). A Dock-and-Lock Mechanism Clusters ADAM10 at Cell-Cell Junctions to Promote α-Toxin Cytotoxicity. Cell Rep 25, 2132–2147.e7. [DOI] [PubMed] [Google Scholar]
- van Spriel AB (2011). Tetraspanins in the humoral immune response. Biochem. Soc. Trans 39, 512–517. [DOI] [PubMed] [Google Scholar]
- Sprinzak D, and Blacklow SC (2021). Biophysics of Notch Signaling. Annu. Rev. Biophys [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susa KJ, Seegar TC, Blacklow SC, and Kruse AC (2020). A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking. ELife 9, e52337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susa KJ, Rawson S, Kruse AC, and Blacklow SC (2021). Cryo-EM structure of the B cell co-receptor CD19 bound to the tetraspanin CD81. Science 371, 300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termini CM, and Gillette JM (2017). Tetraspanins Function as Regulators of Cellular Signaling. Front. Cell Dev. Biol 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda R, Satouh Y, Takemoto M, Nakada-Nakura Y, Liu K, Yokoyama T, Shirouzu M, Iwata S, Nomura N, Sato K, et al. (2020). Structural insights into tetraspanin CD9 function. Nat. Commun 11, 1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Sumida Y, Fujibayashi A, Fukaguchi K, Sanzen N, Nishiuchi R, and Sekiguchi K (2008). The tetraspanin CD151 regulates cell morphology and intracellular signaling on laminin-511. FEBS J 275, 3335–3351. [DOI] [PubMed] [Google Scholar]
- Yáñez-Mó M, Tejedor R, Rousselle P, and Sánchez -Madrid F (2001). Tetraspanins in intercellular adhesion of polarized epithelial cells: spatial and functional relationship to integrins and cadherins. J. Cell Sci 114, 577–587. [DOI] [PubMed] [Google Scholar]
- Yang Y, Liu XR, Greenberg ZJ, Zhou F, He P, Fan L, Liu S, Shen G, Egawa T, Gross ML, et al. (2020). Open conformation of tetraspanins shapes interaction partner networks on cell membranes. EMBO J 39, e105246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q, Yu H, Chen J, Song X, and Sun L (2020). ADAM10 promotes cell growth, migration, and invasion in osteosarcoma via regulating E-cadherin/β-catenin signaling pathway and is regulated by miR-122–5p. Cancer Cell Int 20, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhang Z, Li L, Qin Y-R, Liu H, Jiang C, Zeng T-T, Li M-Q, Xie D, Li Y, et al. (2018). TSPAN15 interacts with BTRC to promote oesophageal squamous cell carcinoma metastasis via activating NF-κB signaling. Nat. Commun 9, 1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman B, Kelly B, McMillan BJ, Seegar TCM, Dror RO, Kruse AC, and Blacklow SC (2016). Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket. Cell 167, 1041–1051.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the RCSB Protein Data Bank (PDB) under the accession numbers 7RDB (Tspan15 LEL) and 7RD5 (Tspan15 LEL-Fab complex).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


