Abstract
Gene-editing by CRISPR/Cas systems has revolutionized plant biology by serving as a functional genomics tool. It has tremendously advanced plant breeding and crop improvement by accelerating the development of improved cultivars, creating genetic variability, and aiding in domestication of wild and orphan crops. Gene-editing is a rapidly evolving field. Several advancements include development of different Cas effectors with increased target range, efficacy, and enhanced capacity for precise DNA modifications with base editing and prime editing. The existing toolbox of various CRISPR reagents facilitate gene knockouts, targeted gene insertions, precise base substitutions, and multiplexing. However, the major challenge in plant genome-editing remains the efficient delivery of these reagents into plant cells. Plants have larger and more complex genome structures compared to other living systems due to the common occurrence of polyploidy and other genome re-arrangements. Further, rigid cell walls surrounding plant cells deter the entry of any foreign biomolecules. Unfortunately, genetic transformation to deliver gene-editing reagents has been established only in a limited number of plant species. Recently, there has been significant progress in CRISPR reagents delivery in plants. This review focuses on exploring these delivery mechanisms categorized into Agrobacterium-mediated delivery and breakthroughs, particle bombardment-based delivery of biomolecules and recent improvements, and protoplasts, a versatile system for gene-editing and regeneration in plants. The ultimate goal in plant gene-editing is to establish highly efficient and genotype-independent reagent delivery mechanisms for editing multiple targets simultaneously and achieve DNA-free gene-edited plants at scale.
Keywords: gene-editing, CRISPR-Cas9, gene targeting, agrobacterium-mediated transformation, biolistics, protoplasts, nanoparticles
Introduction
CRISPR/Cas9 derived from Streptococcus pyogenes (SpCas9) is the most used gene-editing reagent in plants. Unlike its predecessors, zinc finger nucleases (Gao et al., 2010; Osakabe et al., 2010; Zhang et al., 2010) and Transcription Activator-like Effector Nucleases (TALENs) (Cermak et al., 2011; Li et al., 2012), which rely on protein-based DNA recognition mechanisms, CRISPR/Cas systems are RNA-guided endonucleases. The resulting versatility, simplicity, and cost-effectiveness brought about by CRISPR led to significant advances in plant genome engineering. In the CRISPR/Cas9 system, a chimeric single guide RNA (sgRNA), formed by fusion of CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA), directs the SpCas9 nuclease to generate blunt double-strand breaks (DSBs) at the genomic DNA target site three bases upstream of Protospacer Adjacent Motif (PAM) sequence of ‘NGG’ (Jinek et al., 2012). The DSBs are repaired either by error-prone non-homologous end joining (NHEJ) resulting in insertion-deletion mutations (InDels) leading to gene knock-out or by precise, albeit inefficient, homology-directed repair (HDR) through which DNA insertions are achieved by providing an external donor repair template (DRT) (Atkins and Voytas, 2020; Dong and Ronald, 2021). In addition to Cas9, multiple other Cas variants with alternative PAM requirements have been identified and successfully utilized in plants expanding the range of DNA recognition (Kaya et al., 2016; Jia et al., 2017; Steinert et al., 2017; Zhang Y. et al., 2019; Veillet et al., 2020). Furthermore, base editors (BEs), including cytosine, adenine, and glycosylase BEs can precisely convert one target DNA base to another without a DSB. BEs rely on base excision repair, facilitating both transition and transversion mutations, and are increasingly being used in plant systems (Shimatani et al., 2017; Zong et al., 2017; Shan and Voytas, 2018; Zhang R. et al., 2019; Li et al., 2020; Zhao et al., 2020). In addition, prime editing (PE), a versatile “search-and-replace” strategy, was also developed (Anzalone et al., 2019) and optimized in plants (Butt et al., 2020; Lin et al., 2020; Tang et al., 2020; Xu et al., 2020). PEs copy desired edits incorporated into the PE gRNA (PegRNA) directly into the genomic DNA by target primed reverse transcription. With this existing toolbox of various CRISPR reagents, the biggest challenge in plant genome-editing remains to be the efficient delivery of these reagents into plant cells.
Several plant species have larger and more complex genome structures compared to other living systems. Polyploidy and genomic rearrangements are common in plants, and rigid cell walls surrounding the plant cells deter the entry of any foreign biomolecules. Furthermore, genetic transformation to deliver transgenes has only been established in a limited number of plant species and genotypes within each species. This is currently considered the biggest bottleneck in plant genome engineering. Gene-editing reagents are delivered into plants, most commonly as plasmid DNA constructs and predominantly by Agrobacterium-mediated transformation or particle bombardment are summarized in tables recently (Sandhya et al., 2020; Ghogare et al., 2021). In both methods, the plasmid DNA with CRISPR/Cas expression cassette is likely to get integrated into a random genomic site(s), leading to continued expression in host genomes. With the revision of the regulatory landscape of gene-edited lines in the US (USDA press release 1 ) and across the world (Nadakuduti et al., 2018; Lassoued et al., 2021), developing gene-edited lines without integrating foreign genomic DNA into the host plant is gaining prominence. DNA-free delivery of in vitro transcripts (IVTs), pre-assembled ribonucleoprotein complexes (RNPs), or transient expression of plasmid DNA constructs delivered into protoplasts, and subsequent regeneration of gene-edited plants have been successful in several plant species (Liang et al., 2017; Andersson et al., 2018; González et al., 2020, 2021; Sidorov et al., 2021; Zhang et al., 2021). This review will focus on various advances in CRISPR delivery mechanisms in plants categorized into Agrobacterium-mediated delivery and breakthroughs for efficient and heritable mutagenesis and gene targeting (GT) in plants; particle bombardment mediated delivery of DNA, RNA, and protein biomolecules for plant gene-editing, and protoplast transfection and regeneration of transgene-free gene-edited plants. The ultimate goal in plant gene-editing is to establish highly efficient and species non-specific reagent delivery mechanisms for editing multiple targets simultaneously and achieve DNA-free gene-edited plants at scale.
Breakthroughs in Agrobacterium-Mediated Delivery of CRISPR Reagents for Efficient and Heritable Mutagenesis and Gene Targeting
Agrobacterium-mediated genetic transformation remains the principal means of delivering gene-editing reagents including CRISPR/Cas variants, base editing and prime editing reagents, into plants (Lin et al., 2020). This method typically involves inoculating the explants with Agrobacterium expressing gene-editing cassettes integrated into its T-DNA (Figure 1A). Upon infection of plant cells, the T-DNA containing the CRISPR cassette likely gets integrated into the host plant genome leading to stable genetic transformation. Transgene-free gene-editing has been achieved by transient expression of CRISPR reagents by regenerating events without employing selection (Chen et al., 2018). This is important for generating edited plants with no foreign DNA to avoid regulatory oversight and for vegetatively propagated plants, where segregating out the integrated transgene by making crosses is not feasible. Agrobacterium has a limited host range, and several plant species are recalcitrant to Agrobacterium-mediated transformation. Furthermore, the regeneration process involving tissue culture leads to undesirable somaclonal variations in edited lines. Floral dip method of transformation, only amenable to Arabidopsis thaliana and some related species (Clough and Bent, 1998; Lu and Kang, 2008) can generate transformed seeds, bypassing the need for regeneration. Other means of avoiding regeneration process include, the use of A. rhizogenes, which can drastically reduce time between reagent delivery and mutation evaluation, as well as widening the range of species transformed (Yoshida et al., 2015; Triozzi et al., 2021).
FIGURE 1.
Agrobacterium mediated delivery of CRISPR gene-editing reagents in plants. (A) Conventional Agrobacterium-mediated transformation consisting of T-DNA carrying expression cassette for Streptococcus pyogenes Cas9 and kanamycin resistance gene NptII, both driven by cauliflower mosaic virus 35S promoter (CaMV 35S), and a single guide RNA (sgRNA) driven by the U6 promoter targeting the phytoene desaturase (PDS) gene. Explants are infected and co-cultivated with agrobacterium cultures, then placed on selective media for callus induction and regeneration. The resulting gene-edited lines are transgenic and have photobleaching phenotype. (B) A. tumefaciens T-DNA harboring sgRNA targeting PDS along with plant developmental regulators (DRs) Wuschel2 (Wus2) driven by nopaline synthase (nos) promoter, and isopentenyl synthase (ipt) driven by 35S promoter are injected in Cas9 expressing soil grown plants after meristem removal. DRs induce new meristems at the wounded site and pds phenotype is visible in edited meristems. Offspring from seeds produced on de novo meristems show segregation for photobleaching phenotype. Maher et al. (2020) found that de novo meristems with bi-allelic mutations did not set viable seeds, and edited offspring are only recovered from meristems exhibiting mosaicism. (C) Tobacco rattle virus (TRV) is a bipartite RNA virus: TRV1 encodes replicases RNA-dependent RNA polymerase (RDRP), a movement protein (MP), a 16 KDa cysteine rich protein, and a ribozyme (RZ) and can independently replicate itself and move within the plant during infection. TRV2, encodes a coat protein (CP) and, a sgRNA targeting PDS fused to Flowering locus T (FT) driven by a pea early browning virus promoter (PeBv). FT is a mobile RNA which increases infection spread by reaching the shoot apical meristem (SAM). TRV1 and TRV2 are introduced into T-DNA regions of agrobacterium and infiltrated into 35S:Cas9 transgenic plants. Systemic infection of the plant leads to editing of somatic and germline cells thereby increasing heritability. Infected plants exhibit photobleaching and pds phenotype segregates in progeny. (D) Sonchus yellow net rhabdovirus (SYNV) is a negative-strand RNA virus encoding the core structural proteins nucleoprotein (N), phosphoprotein (P), and the large RNA polymerase (L), and Sc4 protein, matrix protein (M), glycoprotein (G) which are involved in cell-to-cell movement. The viral cassette is manipulated to express a Cas9 nuclease and a tRNA-gRNA-tRNA (tgtRNA) which is processed to release the sgRNA targeting the PDS gene by tRNA processing enzymes. Soil grown plants are infiltrated with agrobacterium harboring the SYNV plasmid. Explants from systemically infected leaves are prepared and placed on non-selective regeneration medium. Regenerants are then transferred to soil. Since Cas9 is delivered virally and SYNV does not integrate into the host genome nor have a DNA-phase, the resulting plants are non-transgenic.
Co-delivery of Developmental Regulators with CRISPR Reagents via Agrobacterium to Expedite and Improve Gene-editing Efficiency in Plants
Developmental regulators (DRs) are genes involved in dictating meristem identity in plants. Ectopic expression of DRs in plants has resulted in somatic embryogenesis, formation of embryos from somatic tissues (Lowe et al., 2016). Overexpression of DRs such as Baby Boom (Bbm) and Wuschel2 (Wus2) enhanced regeneration and transformation frequency in both dicot and monocot plants (Srinivasan et al., 2007; Deng et al., 2009; Lowe et al., 2016). This phenomenon was leveraged to induce de novo meristems in somatic tissues by injecting Agrobacterium cultures co-delivering DRs and gene-editing cassettes directly into soil-grown plants (Figure 1B). Wus2 and Isopentenyl transferase (Ipt), when co-delivered with gene-editing reagents by Agrobacterium injections into dicot plants generated meristems in somatic tissues with edits, enabling tissue culture free gene-editing (Maher et al., 2020). This can potentially be a high throughput and less tedious approach when Cas9 expressing plants are generated. Alternatively, Growth Regulating Factor (GRF) and GRF-interacting Factor (GIF) cofactor when expressed as GRF4-GIF chimera increased the speed and efficiency of regeneration (Debernardi et al., 2020). Co-delivery of GRF4-GIF chimera and CRISPR-Cas9 on the same T-DNA increased the regeneration efficiency in both monocots and dicots and produced fertile edited plants (Debernardi et al., 2020). The expression of DRs is extremely beneficial in plant species that are recalcitrant to regeneration or ones with long regeneration periods to reduce the time and cost of plant gene-editing.
Viral Vectors and Mobile RNAs for Systemic Delivery of CRISPR Reagents for Heritable Gene-Editing
Recently, viral vectors showed promise for efficient delivery of CRISPR reagents into germline cells to achieve heritable and DNA-free gene-editing (Ali Z. et al., 2015; Ellison et al., 2020; Ma et al., 2020; Kujur et al., 2021; Li et al., 2021). Traditionally, heritable modifications are accomplished by stable expression of the CRISPR cassettes and generating transgenic lines through regeneration. Autonomously replicating viral vectors delivered into plants via Agrobacterium offer an alternative for heritable gene-editing in plants. RNA viruses don’t integrate into the plant genome but have lower cargo capacity impeding their use for Cas9 delivery. Tobacco rattle virus (TRV), a bipartite positive-strand RNA virus is widely used in plants. TRV mediated sgRNA delivery into Cas9 overexpressing lines by agroinfiltration has been optimized in dicots, albeit with low heritability of edits (Ali et al., 2015; Cody et al., 2017). To improve heritability, the endogenous mobile RNA Flowering Locus T (FT) has been fused to sgRNA to enhance mobility and facilitate systemic distribution within plant to reach germline cells (Figure 1C) (Ellison et al., 2020). Barley stripe mosaic virus (BMSV) has been engineered to deliver sgRNAs into wheat to achieve heritable genome editing. Furthermore, by co-infiltration of a pool of BMSV vectors harboring different sgRNAs resulted in multiplexed mutagenesis in the progeny (Li et al., 2021). Sonchus yellow net rhabdovirus (SYNV), a negative-strand RNA virus with higher cargo capacity, has been engineered to carry both Cas9 and sgRNA for DNA-free in planta editing (Figure 1D) (Ma et al., 2020).
Enhancing Gene Targeting by Agrobacterium-Mediated Delivery of CRISPR Reagents
GT includes precise DNA modifications based on HDR using a DRT with homology to the host target DNA on both ends. DSBs generated by CRISPR/Cas reagents initiate the cell repair process. However, NHEJ is the predominant repair mechanism in plants cells to repair these DSBs as HDR is not active throughout the cell cycle. This, in combination with inefficient delivery of DRT to facilitate HDR, make GT very inefficient in plants. To increase GT frequencies, viral replicons including Bean Yellow Dwarf Virus (BeYDV) (Baltes et al., 2014; Butler et al., 2015; Čermák et al., 2015; Cermak et al., 2017; Wang et al., 2017; Vu et al., 2020) or wheat Dwarf virus (WDV) (Gil-Humanes et al., 2017) have been successfully used in several dicot and monocot plants. These viral replicons carrying the CRISPR expression cassette and DRT undergo rolling-circle replication in the host cells thereby increasing the abundance of nuclease and availability of DRT for HDR (Baltes et al., 2014). The GT event is not heritable if it doesn’t occur in the germline cells. To increase the heritability of GT, germline-specific promoters including the egg-cell, early embryo-specific promoter and pollen-specific promoters or promoters active in the shoot apical meristems (SAM) have been employed to drive Cas9 expression (Wang et al., 2015; Yan et al., 2015; Mao et al., 2016). Furthermore, to improve the efficiency of heritable in-frame gene insertions and amino acid substitutions by HDR, plants expressing Cas9 from germline-specific promoters are used for sequential transformation with HDR constructs containing DRT and sgRNA targeting the gene of interest. This led to an increase in GT efficiency of up to 9% (Miki et al., 2018). Since GT is a rare phenomenon, even with all the advances to improve efficiency, selection must still be employed to detect positive GT events. A piggyBac transposition system from T-DNA has been used to eliminate the GT selection marker from host plant genome. In this method, a transposon integrates into the host genome at TTAA element and excises without a footprint (Nishizawa-Yokoi et al., 2015). Recently, a novel marker elimination system was developed wherein the excision is based on I-SceI recognition site. By overlapping this recognition site on 5′ and 3′ homology arms of the DRT, seamless marker elimination and precise GT have been achieved (Endo et al., 2021). To this end, the same research group also developed a piggyBac-mediated transgenesis system to temporarily express CRISPR and selection marker cassettes from T-DNA with subsequent excision of piggyBac via transposase after successful editing and selection had occurred (Nishizawa-Yokoi and Toki, 2021).
Biolistics for Delivery of CRISPR Reagents Into Plants as DNA, RNA, or Proteins
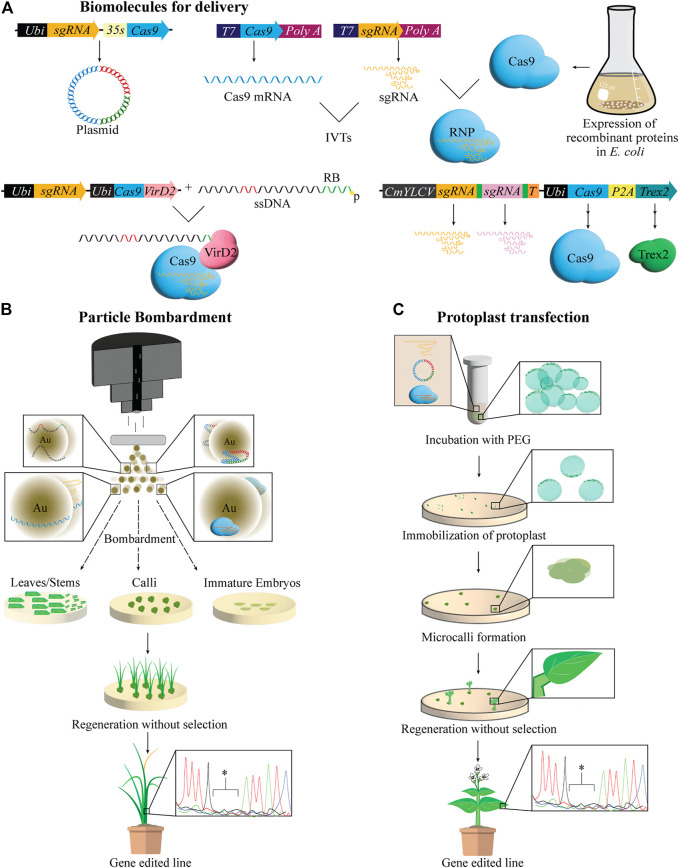
Biolistics or particle-bombardment, is a common alternative for transforming plants recalcitrant to Agrobacterium-infection. It relies on physically breaching the plant cell wall and membrane with gold or tungsten microprojectiles coated with biomolecules accelerated to very high velocities. Biolistics offers the possibility of delivering a variety of cargo including plasmid DNA, ssDNA, RNA, or ribonucleic proteins (RNPs) assembled from IVTs and recombinant proteins (Figure 2A). Major drawbacks of biolistic delivery include random integration of cargo at multiple genomic sites when delivered as DNA and labor-intensive preparation of explants such as calli or immature embryos with the capability to regenerate.
FIGURE 2.
Biomolecules delivered via biolistics and protoplast transfections for regenerating gene-edited plants. (A) Biomolecules used for gene-editing are delivered into plants cells in a variety of forms including plasmid DNA, ssDNA, mRNA or ssRNA, prepared via in vitro transcription (IVT), and preassembled ribonucleic proteins (RNPs) using IVTs and recombinant proteins. Targeted mutagenesis and gene targeting (GT) can be enhanced by various mechanisms. For example, fusion of Cas9 to VirD2, one component of the agrobacterium relaxosome complex integral to the cleavage of T-DNA from the Ti plasmid, as well as its localization and integration in the plant genome, has been shown to increase homology-directed repair (HDR) mediated GT using a donor repair template (DRT). DRT in this case is a single stranded DNA (ssDNA) harboring the desired edits (red) and the canonical 25 bp right border (RB) sequence (green), and is delivered to the plant cell along with the Cas-VirD2 fusion protein. VirD2 will covalently bind the template, thus bringing it in close proximity to the DSB induced by Cas9. Delivering Trex2 exonuclease has also been shown to increase HDR as well as the efficiency of multiplex editing when sgRNA are co-delivered and processed by t-RNA system, illustrated by green boxes between sgRNA. p = phosphorylation. (B) Particle bombardment or biolistics, rely on the physical disruption of plant cell walls by metal particles, often gold, coated with ssDNA or dsDNA, IVTs or RNPs, which are introduced to the cell. Bombarded explants can be regenerated in tissue culture with or without selection to recover gene edited plants. Au = gold particles. (C) Protoplast transfection and regeneration is shown. polyethylene glycol (PEG) mediated transfection is the most common way to deliver biomolecules for gene-editing to protoplasts. Post transfection, protoplasts are immobilized on culture media where protoplasts undergo cell divisions to form microcalli, followed by shoot and root formation and finally resulting in regeneration of entire gene-edited plants. Editing at the target site is confirmed by sequencing represented in the chromatogram * = deletions.
Advances in Biolistic Delivery for DNA-free Gene-Editing and Chromosome Engineering
Instead of plasmid DNA, bombarding RNPs was successfully demonstrated to produce transgene-free gene-edited lines in cereal crops (Svitashev et al., 2016; Liang et al., 2017; Banakar et al., 2019, 2020; Zhang et al., 2021) (Figure 2B). In addition, when a single base pair mismatch was present in the protospacer of sgRNA targeting homeologs, a dramatic decrease in off-target editing was observed with RNPs as compared to plasmid DNA delivery indicating high specificity of RNPs (Liang et al., 2017). Furthermore, RNPs also facilitated large heritable inversion of 75.5 Mb in maize chromosome 2, when assembled with guide RNAs flanking the junctions of the desired inversion (Schwartz et al., 2020). Such precise chromosomal engineering in invaluable to crop breeding. To avoid labor-intensive preparation of explants, in planta biolistic delivery using SAM as a target tissue (Hamada et al., 2017) for germline transmission was employed as an alternative (Hamada et al., 2018; Imai et al., 2020). Embryonic SAM exposed mature wheat seeds were bombarded with plasmid DNA expressing CRISPR cassettes to generate gene-edited lines (Hamada et al., 2018; Imai et al., 2020). Alternatively, when pollen was used as a target tissue to bombard plasmid DNA for gene-editing in Nicotiana benthamiana, the bombarded pollen retained fertility and delivered the cargo into the ovules (Nagahara et al., 2021). Furthermore, technical improvements have also been made to overcome variability between bombardments. A double-barreled gene gun in combination with cell counting software was developed to scale bombardment experiments with an internal standard, thereby reducing standard deviation between bombardments by half (Miller et al., 2021).
Gene Insertion or Replacement by Intron Targeting and Determining Genomic Safe Harbors
To leverage the relatively more efficient NHEJ compared to HDR for targeted insertions, DNA fragments were inserted in selected introns such that any mutations by NHEJ would not affect protein-coding sequences of either endogenous or inserted genes. By bombarding calli with plasmids expressing pairs of sgRNA targeting adjacent introns of target genes and DRT harboring desired mutations flanked by the same intronic sgRNA sites, replacement of endogenous gene has been achieved at 2% frequency. Additionally, the gene replacement events were heritable (Li et al., 2016). Enhancers and promoters up to 2 Kb were introduced into the target site using these modified DRTs (Lu et al., 2020). Another strategy for targeted insertion by NHEJ is to determine the genomic safe harbors (GSH) in the host plant genome, within which integrations of transgenes do not cause any genic disruptions or adverse morphological effects. A 5.2 Kb carotenoid biosynthesis cassette was inserted at targeted GSH to generate marker-free rice with high carotenoid containing seeds and no-off target mutations observed (Dong et al., 2020).
Enhancing HDR by Delivery of Transcript-Donor Templates or by VirD2 Relaxase-Cas9 Fusion
Recent advances in HDR by particle bombardment include delivery of ssDNA, including a canonical 25 bp right border (RB) sequence of T-DNA, as DRT co-delivered with a plasmid expressing Cas9-VirD2 fusion protein (Ali et al., 2020) (Figure 2A). Achieving a 20.8% HDR efficiency, this method relies on the ability of the VirD2 protein, an Agrobacterium virulence factor, to covalently bind the RB of DRT, thus bringing it in close proximity to the DSB induced by Cas9 (Figure 2A) (Ali et al., 2020). Other attempts to improve HDR include the delivery of DRT as transcripts. RNA-DRT was shown to result in higher HDR efficiency than DNA-DRT when delivered to rice calli, possibly due to the high stability of RNA:DNA complexes, resulting in edited rice with two desired point mutations in the ALS gene conferring herbicide tolerance (Li et al., 2019). This transcript-templated HDR (TT-HDR), approach improves not only HDR efficiency but also creates a DNA-free path to HDR-mediated gene-editing, which may avoid regulatory hurdles.
Protoplasts Provide a Versatile System for DNA-free Genome Editing in Plants
Protoplasts are plant cells devoid of cell walls, which offer a versatile platform for DNA-free GE and a good transient system to evaluate the activity of gene-editing reagents before moving into a more-labor intensive transformation pipeline (Nadakuduti et al., 2019; Lin et al., 2020). Polyethylene glycol (PEG)-mediated transfection and electro-transfection are two common methods to deliver plasmid DNA, IVTs, or RNPs into protoplasts for transient expression of CRISPR cassettes. Subsequently, edited plants can be regenerated from transfected protoplasts by tissue culture procedures (Figure 2C). Plasmid DNA may integrate into the host genome randomly as filler DNA during protoplast transfection (Gorbunova and Levy, 1997; Kim and Kim, 2016). However, IVTs or RNPs offer DNA-free gene-editing by immediately editing the target site, bypassing transcription and translational machinery respectively in the cell and rapidly degrade (Liang et al., 2017, 2018; Andersson et al., 2018; González et al., 2020, 2020, 2020; Lee et al., 2020; Sidorov et al., 2021; Zhang et al., 2021). However, plant regeneration from protoplast remains unestablished in many plant species. In addition, somaclonal variations and genome instability is reported in regenerated lines (Fossi et al., 2019). Once efficient protoplast isolation, transfection, and regeneration have been established in a plant species, it could be a high throughput platform by combining with flow cytometry and omic analyses for optimizing gene-editing. Furthermore, multiplexing, editing multiple genes at a time has been achieved using protoplasts (Klimek-Chodacka et al., 2021; Nicolia et al., 2021; Yu et al., 2021; Zhang et al., 2021). By co-delivering Three Prime Repair exonuclease 2 (TREX2) and CRISPR/Cas9 into protoplasts, targeted mutagenesis using a multiplexing strategy was further improved (Weiss et al., 2020) (Figure 2A).
Nanocarrier-Mediated Delivery of CRISPR/Cas Reagents in Plants
Nanotechnology has evolved in the past decade in the field of plant genetic engineering. Nanomaterials including carbon nanotubes (CNTs), carbondots, mesosporous silicon nanoparticles (MSNs) etc have been used to deliver biomolecules such as DNA, RNA, RNPs and proteins etc., discussed in recent reviews (Kumari and Singh, 2021; Mujtaba et al., 2021). Nanoparticle-mediated delivery of DNA and proteins into both nuclear and chloroplast genomes has been achieved in plants (Demirer et al., 2019, 2020; Kwak et al., 2019). Furthermore, Cre protein was previously delivered via MSNs for maize GE via loxP site demonstrating the feasibility of gene-editing (Martin-Ortigosa et al., 2014). Gene-editing using RNPs delivered by nanoparticles has been achieved in human cells (Wang et al., 2016; Lee et al., 2017; Mout et al., 2017). However, it has yet to be achieved in plants mainly due to high delivery efficiencies required for GE.
Future Aspects of Delivering Plant-Gene Editing Reagents
Relying on tissue culture-based plant genetic transformation methods and inefficient reagent delivery mechanisms are the major bottle necks to overcome before we realize the full potential of gene-editing in plants. Current advancements in delivery mechanisms, including de novo meristem induction or use of viral vectors to circumvent tissue culture, rely on Agrobacterium for delivery and have been demonstrated only in dicots and need to be expanded to monocots. Delivering repair templates for HDR through these innovative methods is also a future possibility. Furthermore, smaller sized Cas9 alternatives would overcome the cargo capacity of some of these viral vectors. Agrobacterium, however, has a narrow host range for infection and several species are recalcitrant to Agrobacterium transformation. Particle bombardment has been shown to be better equipped for co-delivery of cargo for simultaneous editing than Agrobacterium and is universally applicable to all plant species and cell types (Kuang et al., 2020). Chromosomal inversions achieved via bombardment could revolutionize breeding by unlocking regions for chromosomal cross overs, creating novel linkage groups and facilitating targeted recombination to maximize genetic gain in crops. However, complex segregation patterns of DNA integrated in bombarded plant genomes might complicate downstream uses of transformed plants. Agrobacterium and biolistic transformation of pollen also bypasses regeneration but often results in pollen with lower viability (Wang et al., 2008; Zhao et al., 2017). In addition, pollen-tube transformations may result in chimerism (Ali A. et al., 2015). While pollen magnetofection has improved on these drawbacks (Zhao et al., 2017), its application remains constrained to dicots (Vejlupkova et al., 2020). The prospects of nanoparticles as delivery engines for plant genome editing are also encouraging (Demirer et al., 2021) and further advances are essential to facilitate plant gene-editing.
Acknowledgments
We thank Dr. Felix Enciso-Rodriguez from the Colombian Corporation for Agricultural Research for reviewing the manuscript and editorial suggestions.
Footnotes
Author Contributions
SN conceived the idea; SN and LL wrote the manuscript. All authors read and approved the manuscript.
Funding
Funding for SN was provided by the Biotechnology Risk Assessment Grant Program competitive grant no. 2018-33522-28736 from the USDA National Institute of Food and Agriculture and start-up funds from the Environmental Horticulture Department, University of Florida.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Ali A., Bang S. W., Chung S.-M., Staub J. E. (2015). Plant Transformation via Pollen Tube-Mediated Gene Transfer. Plant Mol. Biol. Rep. 33, 742–747. 10.1007/S11105-014-0839-5/TABLES/2 [DOI] [Google Scholar]
- Ali Z., Abul-Faraj A., Li L., Ghosh N., Piatek M., Mahjoub A., et al. (2015). Efficient Virus-Mediated Genome Editing in Plants Using the CRISPR/Cas9 System. Mol. Plant 8, 1288–1291. 10.1016/j.molp.2015.02.011 [DOI] [PubMed] [Google Scholar]
- Ali Z., Shami A., Sedeek K., Kamel R., Alhabsi A., Tehseen M., et al. (2020). Fusion of the Cas9 Endonuclease and the VirD2 Relaxase Facilitates Homology-Directed Repair for Precise Genome Engineering in rice. Commun. Biol. 3, 1–13. 10.1038/s42003-020-0768-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M., Turesson H., Olsson N., Fält A.-S., Ohlsson P., Gonzalez M. N., et al. (2018). Genome Editing in Potato via CRISPR-Cas9 Ribonucleoprotein Delivery. Physiol. Plantarum 164, 378–384. 10.1111/ppl.12731 [DOI] [PubMed] [Google Scholar]
- Anzalone A. V., Randolph P. B., Davis J. R., Sousa A. A., Koblan L. W., Levy J. M., et al. (2019). Search-and-replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 576, 149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins P. A., Voytas D. F. (2020). Overcoming Bottlenecks in Plant Gene Editing. Curr. Opin. Plant Biol. 54, 79–84. 10.1016/j.pbi.2020.01.002 [DOI] [PubMed] [Google Scholar]
- Baltes N. J., Gil-Humanes J., Cermak T., Atkins P. A., Voytas D. F. (2014). DNA Replicons for Plant Genome Engineering. Plant Cell 26, 151–163. 10.1105/tpc.113.119792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banakar R., Eggenberger A. L., Lee K., Wright D. A., Murugan K., Zarecor S., et al. (2019). High-frequency Random DNA Insertions upon Co-delivery of CRISPR-Cas9 Ribonucleoprotein and Selectable Marker Plasmid in rice. Sci. Rep. 9, 1–13. 10.1038/s41598-019-55681-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banakar R., Schubert M., Collingwood M., Vakulskas C., Eggenberger A. L., Wang K. (2020). Comparison of CRISPR-Cas9/Cas12a Ribonucleoprotein Complexes for Genome Editing Efficiency in the Rice Phytoene Desaturase (OsPDS) Gene. Rice 13, 1–7. 10.1186/s12284-019-0365-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler N. M., Atkins P. A., Voytas D. F., Douches D. S. (2015). Generation and Inheritance of Targeted Mutations in Potato (Solanum tuberosum L.) Using the CRISPR/Cas System. PLoS One 10, e0144591–12. 10.1371/journal.pone.0144591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H., Rao G. S., Sedeek K., Aman R., Kamel R., Mahfouz M. (2020). Engineering Herbicide Resistance via Prime Editing in rice. Plant Biotechnol. J. 18, 2370–2372. 10.1111/pbi.13399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T., Baltes N. J., Čegan R., Zhang Y., Voytas D. F. (2015). High-frequency, Precise Modification of the Tomato Genome. Genome Biol. 16, 1–14. 10.1186/s13059-015-0796-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T., Curtin S. J., Gil-Humanes J., Čegan R., Kono T. J. Y., Konečná E., et al. (2017). A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 29, 1196–1217. 10.1105/tpc.16.00922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E. L., Christian M., Wang L., Zhang Y., Schmidt C., et al. (2011). Efficient Design and Assembly of Custom TALEN and Other TAL Effector-Based Constructs for DNA Targeting. Nucleic Acids Res. 39, e82. 10.1093/nar/gkr73910.1093/nar/gkr218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Li W., Katin-Grazzini L., Ding J., Gu X., Li Y., et al. (2018). A Method for the Production and Expedient Screening of CRISPR/Cas9-mediated Non-transgenic Mutant Plants. Hortic. Res. 5, 1–12. 10.1038/s41438-018-0023-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral Dip: a Simplified Method forAgrobacterium-Mediated Transformation ofArabidopsis Thaliana. Plant J. 16, 735–743. 10.1046/J.1365-313X.1998.00343.X [DOI] [PubMed] [Google Scholar]
- Cody W. B., Scholthof H. B., Mirkov T. E. (2017). Multiplexed Gene Editing and Protein Overexpression Using a Tobacco Mosaic Virus Viral Vector. Plant Physiol. 175, 23–35. 10.1104/pp.17.00411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi J. M., Tricoli D. M., Ercoli M. F., Hayta S., Ronald P., Palatnik J. F., et al. (2020). A GRF-GIF Chimeric Protein Improves the Regeneration Efficiency of Transgenic Plants. Nat. Biotechnol. 38, 1274–1279. 10.1038/s41587-020-0703-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer G. S., Silva T. N., Jackson C. T., Thomas J. B., Ehrhardt D. W., Mortimer J. C., et al. (2021). Nanotechnology to advance CRISPR-Cas Genetic Engineering of Plants. Nat. Nanotechnol. 16, 243–250. 10.1038/s41565-021-00854-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer G. S., Zhang H., Goh N. S., Pinals R. L., Chang R., Landry M. P. (2020). Carbon Nanocarriers Deliver siRNA to Intact Plant Cells for Efficient Gene Knockdown. Sci. Adv. 6, eaaz0495. 10.1126/sciadv.aaz0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer G. S., Zhang H., Matos J. L., Goh N. S., Cunningham F. J., Sung Y., et al. (2019). High Aspect Ratio Nanomaterials Enable Delivery of Functional Genetic Material without DNA Integration in Mature Plants. Nat. Nanotechnol. 14, 456–464. 10.1038/s41565-019-0382-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Luo K., Li Z., Yang Y. (2009). A Novel Method for Induction of Plant Regeneration via Somatic Embryogenesis. Plant Sci. 177, 43–48. 10.1016/j.plantsci.2009.03.009 [DOI] [Google Scholar]
- Dong O. X., Ronald P. C. (2021). Targeted DNA Insertion in Plants. Proc. Natl. Acad. Sci. USA 118, e2004834117–9. 10.1073/pnas.2004834117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong O. X., Yu S., Jain R., Zhang N., Duong P. Q., Butler C., et al. (2020). Marker-free Carotenoid-Enriched rice Generated through Targeted Gene Insertion Using CRISPR-Cas9. Nat. Commun. 11, 1–10. 10.1038/s41467-020-14981-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison E. E., Nagalakshmi U., Gamo M. E., Huang P.-j., Dinesh-Kumar S., Voytas D. F. (2020). Multiplexed Heritable Gene Editing Using RNA Viruses and mobile Single Guide RNAs. Nat. Plants 6, 620–624. 10.1038/s41477-020-0670-y [DOI] [PubMed] [Google Scholar]
- Endo M., Iwakami S., Toki S. (2021). Precision Genome Editing in Plants via Gene Targeting and Subsequent Break‐induced Single‐strand Annealing. Plant Biotechnol. J. 19, 563–574. 10.1111/pbi.13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossi M., Amundson K., Kuppu S., Britt A., Comai L. (2019). Regeneration of Solanum Tuberosum Plants from Protoplasts Induces Widespread Genome Instability. Plant Physiol. 180, 78–86. 10.1104/pp.18.00906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Smith J., Yang M., Jones S., Djukanovic V., Nicholson M. G., et al. (2010). Heritable Targeted Mutagenesis in maize Using a Designed Endonuclease. Plant J. 61, 176–187. 10.1111/j.1365-313X.2009.04041.x [DOI] [PubMed] [Google Scholar]
- Ghogare R., Ludwig Y., Bueno G. M., Slamet-Loedin I. H., Dhingra A. (2021). Genome Editing Reagent Delivery in Plants. Transgenic Res. 30, 321–335. 10.1007/S11248-021-00239-W [DOI] [PubMed] [Google Scholar]
- Gil‐Humanes J., Wang Y., Liang Z., Shan Q., Ozuna C. V., Sánchez‐León S., et al. (2017). High‐efficiency Gene Targeting in Hexaploid Wheat Using DNA Replicons and CRISPR/Cas9. Plant J. 89, 1251–1262. 10.1111/tpj.13446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González M. N., Massa G. A., Andersson M., Décima Oneto C. A., Turesson H., Storani L., et al. (2021). Comparative Potato Genome Editing: Agrobacterium Tumefaciens-Mediated Transformation and Protoplasts Transfection Delivery of CRISPR/Cas9 Components Directed to StPPO2 Gene. Plant Cel Tiss Organ. Cult 145, 291–305. 10.1007/s11240-020-02008-9 [DOI] [Google Scholar]
- González M. N., Massa G. A., Andersson M., Turesson H., Olsson N., Fält A.-S., et al. (2020). Reduced Enzymatic Browning in Potato Tubers by Specific Editing of a Polyphenol Oxidase Gene via Ribonucleoprotein Complexes Delivery of the CRISPR/Cas9 System. Front. Plant Sci. 10, 1649. 10.3389/fpls.2019.01649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V., Levy A. A. (1997). Non-homologous DNA End Joining in Plant Cells Is Associated with Deletions and Filler DNA Insertions. Nucleic Acids Res. 25, 4650–4657. 10.1093/nar/25.22.4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Linghu Q., Nagira Y., Miki R., Taoka N., Imai R. (2017). An in Planta Biolistic Method for Stable Wheat Transformation. Sci. Rep. 7, 1–8. 10.1038/s41598-017-11936-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Liu Y., Nagira Y., Miki R., Taoka N., Imai R. (2018). Biolistic-delivery-based Transient CRISPR/Cas9 Expression Enables in Planta Genome Editing in Wheat. Sci. Rep. 8, 1–7. 10.1038/s41598-018-32714-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai R., Hamada H., Liu Y., Linghu Q., Kumagai Y., Nagira Y., et al. (2020). In Planta Particle Bombardment (iPB): A New Method for Plant Transformation and Genome Editing. Plant Biotechnol. 37, 171–176. 10.5511/PLANTBIOTECHNOLOGY.20.0206A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Xu J., Orbović V., Zhang Y., Wang N. (2017). Editing Citrus Genome via SaCas9/sgRNA System. Front. Plant Sci. 8, 2135. 10.3389/fpls.2017.02135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337, 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya H., Mikami M., Endo A., Endo M., Toki S. (2016). Highly Specific Targeted Mutagenesis in Plants Using Staphylococcus aureus Cas9. Sci. Rep. 6, 1–9. 10.1038/srep26871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kim J.-S. (2016). Bypassing GMO Regulations with CRISPR Gene Editing. Nat. Biotechnol. 34, 1014–1015. 10.1038/nbt.3680 [DOI] [PubMed] [Google Scholar]
- Klimek-Chodacka M., Gieniec M., Baranski R. (2021). Multiplex Site-Directed Gene Editing Using Polyethylene Glycol-Mediated Delivery of CRISPR gRNA:Cas9 Ribonucleoprotein (RNP) Complexes to Carrot Protoplasts. Ijms 22, 10740. 10.3390/ijms221910740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Y., Li S., Ren B., Yan F., Spetz C., Li X., et al. (2020). Base-Editing-Mediated Artificial Evolution of OsALS1 in Planta to Develop Novel Herbicide-Tolerant Rice Germplasms. Mol. Plant 13, 565–572. 10.1016/J.MOLP.2020.01.010 [DOI] [PubMed] [Google Scholar]
- Kujur S., Senthil-Kumar M., Kumar R. (2021). Plant Viral Vectors: Expanding the Possibilities of Precise Gene Editing in Plant Genomes. Plant Cel Rep 40, 931–934. 10.1007/s00299-021-02697-2 [DOI] [PubMed] [Google Scholar]
- Kumari R., Singh D. P. (2021). Nano-Biotechnological Approach of Plant Genetic Engineering,” in Policy Issues in Genetically Modified Crops (Cambridge, MA: Academic Press; ), 481–494. 10.1016/B978-0-12-820780-2.00021-2 [DOI] [Google Scholar]
- Kwak S.-Y., Lew T. T. S., Sweeney C. J., Koman V. B., Wong M. H., Bohmert-Tatarev K., et al. (2019). Chloroplast-selective Gene Delivery and Expression in Planta Using Chitosan-Complexed Single-Walled Carbon Nanotube Carriers. Nat. Nanotechnol. 14, 447–455. 10.1038/s41565-019-0375-4 [DOI] [PubMed] [Google Scholar]
- Lassoued R., Phillips P. W. B., Macall D. M., Hesseln H., Smyth S. J. (2021). Expert Opinions on the Regulation of Plant Genome Editing. Plant Biotechnol. J. 19, 1104–1109. 10.1111/pbi.13597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Conboy M., Park H. M., Jiang F., Kim H. J., Dewitt M. A., et al. (2017). Nanoparticle Delivery of Cas9 Ribonucleoprotein and Donor DNA In Vivo Induces Homology-Directed DNA Repair. Nat. Biomed. Eng. 1, 889–901. 10.1038/s41551-017-0137-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Lee J., Choi S. A., Kim Y.-S., Koo O., Choi S. H., et al. (2020). Efficient Genome Editing Using CRISPR-Cas9 RNP Delivery into Cabbage Protoplasts via Electro-Transfection. Plant Biotechnol. Rep. 14, 695–702. 10.1007/s11816-020-00645-2 [DOI] [Google Scholar]
- Li C., Zhang R., Meng X., Chen S., Zong Y., Lu C., et al. (2020). Targeted, Random Mutagenesis of Plant Genes with Dual Cytosine and Adenine Base Editors. Nat. Biotechnol. 38, 875–882. 10.1038/s41587-019-0393-7 [DOI] [PubMed] [Google Scholar]
- Li J., Meng X., Zong Y., Chen K., Zhang H., Liu J., et al. (2016). Gene Replacements and Insertions in rice by Intron Targeting Using CRISPR-Cas9. Nat. Plants 2, 139. 10.1038/nplants.2016.139 [DOI] [PubMed] [Google Scholar]
- Li S., Li J., He Y., Xu M., Zhang J., Du W., et al. (2019). Precise Gene Replacement in rice by RNA Transcript-Templated Homologous Recombination. Nat. Biotechnol. 37, 445–450. 10.1038/s41587-019-0065-7 [DOI] [PubMed] [Google Scholar]
- Li T., Hu J., Sun Y., Li B., Zhang D., Li W., et al. (2021). Highly Efficient Heritable Genome Editing in Wheat Using an RNA Virus and Bypassing Tissue Culture. Mol. Plant. 14, 1787–1798. 10.1016/J.MOLP.2021.07.010 [DOI] [PubMed] [Google Scholar]
- Li T., Liu B., Spalding M. H., Weeks D. P., Yang B. (2012). High-efficiency TALEN-Based Gene Editing Produces Disease-Resistant rice. Nat. Biotechnol. 30, 390–392. 10.1038/nbt.2199 [DOI] [PubMed] [Google Scholar]
- Liang Z., Chen K., Li T., Zhang Y., Wang Y., Zhao Q., et al. (2017). Efficient DNA-free Genome Editing of Bread Wheat Using CRISPR/Cas9 Ribonucleoprotein Complexes. Nat. Commun. 8, 1–5. 10.1038/ncomms14261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Chen K., Zhang Y., Liu J., Yin K., Qiu J.-L., et al. (2018). Genome Editing of Bread Wheat Using Biolistic Delivery of CRISPR/Cas9 In Vitro Transcripts or Ribonucleoproteins. Nat. Protoc. 13, 413–430. 10.1038/nprot.2017.145 [DOI] [PubMed] [Google Scholar]
- Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., et al. (2020). Prime Genome Editing in rice and Wheat. Nat. Biotechnol. 38, 582–585. 10.1038/s41587-020-0455-x [DOI] [PubMed] [Google Scholar]
- Lowe K., Wu E., Wang N., Hoerster G., Hastings C., Cho M.-J., et al. (2016). Morphogenic Regulators Baby Boom and Wuschel Improve Monocot Transformation. Plant Cell 28, 1998–2015. 10.1105/tpc.16.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Kang J. (2008). Generation of Transgenic Plants of a Potential Oilseed Crop Camelina Sativa by Agrobacterium-Mediated Transformation. Plant Cel Rep 27, 273–278. 10.1007/S00299-007-0454-0/TABLES/2 [DOI] [PubMed] [Google Scholar]
- Lu Y., Tian Y., Shen R., Yao Q., Wang M., Chen M., et al. (2020). Targeted, Efficient Sequence Insertion and Replacement in rice. Nat. Biotechnol. 38, 1402–1407. 10.1038/s41587-020-0581-5 [DOI] [PubMed] [Google Scholar]
- Ma X., Zhang X., Liu H., Li Z. (2020). Highly Efficient DNA-free Plant Genome Editing Using Virally Delivered CRISPR-Cas9. Nat. Plants 6, 773–779. 10.1038/s41477-020-0704-5 [DOI] [PubMed] [Google Scholar]
- Maher M. F., Nasti R. A., Vollbrecht M., Starker C. G., Clark M. D., Voytas D. F. (2020). Plant Gene Editing through De Novo Induction of Meristems. Nat. Biotechnol. 38, 84–89. 10.1038/s41587-019-0337-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Zhang Z., Feng Z., Wei P., Zhang H., Botella J. R., et al. (2016). Development of Germ-line-specific CRISPR-Cas9 Systems to Improve the Production of Heritable Gene Modifications inArabidopsis. Plant Biotechnol. J. 14, 519–532. 10.1111/pbi.12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Ortigosa S., Peterson D. J., Valenstein J. S., Lin V. S.-Y., Trewyn B. G., Lyznik L. A., et al. (2014). Mesoporous Silica Nanoparticle-Mediated Intracellular Cre Protein Delivery for Maize Genome Editing via loxP Site Excision,. Plant Physiol. 164, 537–547. 10.1104/pp.113.233650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D., Zhang W., Zeng W., Feng Z., Zhu J.-K. (2018). CRISPR/Cas9-mediated Gene Targeting in Arabidopsis Using Sequential Transformation. Nat. Commun. 9, 1967. 10.1038/s41467-018-04416-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K., Eggenberger A. L., Lee K., Liu F., Kang M., Drent M., et al. (2021). An Improved Biolistic Delivery and Analysis Method for Evaluation of DNA and CRISPR-Cas Delivery Efficacy in Plant Tissue. Sci. Rep. 11, 1–11. 10.1038/s41598-021-86549-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mout R., Ray M., Yesilbag Tonga G., Lee Y.-W., Tay T., Sasaki K., et al. (2017). Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano 11, 2452–2458. 10.1021/acsnano.6b07600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujtaba M., Wang D., Carvalho L. B., Oliveira J. L., Espirito Santo Pereira A. d., Sharif R., et al. (2021). Nanocarrier-Mediated Delivery of miRNA, RNAi, and CRISPR-Cas for Plant Protection: Current Trends and Future Directions. ACS Agric. Sci. Technol. 1, 417–435. 10.1021/ACSAGSCITECH.1C00146 [DOI] [Google Scholar]
- Nadakuduti S. S., Buell C. R., Voytas D. F., Starker C. G., Douches D. S. (2018). Genome Editing for Crop Improvement - Applications in Clonally Propagated Polyploids with a Focus on Potato (Solanum tuberosum L.). Front. Plant Sci. 9, 1607. 10.3389/fpls.2018.01607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti S. S., Starker C. G., Ko D. K., Jayakody T. B., Buell C. R., Voytas D. F., et al. (2019). Evaluation of Methods to Assess In Vivo Activity of Engineered Genome-Editing Nucleases in Protoplasts. Front. Plant Sci. 10, 110. 10.3389/fpls.2019.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahara S., Higashiyama T., Mizuta Y. (2021). Detection of a Biolistic Delivery of Fluorescent Markers and CRISPR/Cas9 to the Pollen Tube. Plant Reprod. 34, 191–205. 10.1007/S00497-021-00418-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolia A., Andersson M., Hofvander P., Festa G., Cardi T. (2021). Tomato Protoplasts as Cell Target for Ribonucleoprotein (RNP)-mediated Multiplexed Genome Editing. Plant Cel Tiss Organ. Cult 144, 463–467. 10.1007/s11240-020-01954-8 [DOI] [Google Scholar]
- Nishizawa‐Yokoi A., Endo M., Ohtsuki N., Saika H., Toki S. (2015). Precision Genome Editing in Plants via Gene Targeting and Piggy B Ac ‐mediated Marker Excision. Plant J. 81, 160–168. 10.1111/tpj.12693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa‐Yokoi A., Toki S. (2021). A piggyBac ‐mediated Transgenesis System for the Temporary Expression of CRISPR/Cas9 in rice. Plant Biotechnol. J. 19, 1386–1395. 10.1111/pbi.13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K., Osakabe Y., Toki S. (2010). Site-directed Mutagenesis in Arabidopsis Using Custom-Designed Zinc finger Nucleases. Proc. Natl. Acad. Sci. 107, 12034–12039. 10.1073/pnas.1000234107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhya D., Jogam P., Allini V. R., Abbagani S., Alok A. (2020). The Present and Potential Future Methods for Delivering CRISPR/Cas9 Components in Plants. J. Genet. Eng. Biotechnol. 18, 25. 10.1186/S43141-020-00036-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C., Lenderts B., Feigenbutz L., Barone P., Llaca V., Fengler K., et al. (2020). CRISPR-Cas9-mediated 75.5-Mb Inversion in maize. Nat. Plants 6, 1427–1431. 10.1038/s41477-020-00817-6 [DOI] [PubMed] [Google Scholar]
- Shan Q., Voytas D. F. (2018). Editing Plant Genes One Base at a Time. Nat. Plants 4, 412–413. 10.1038/s41477-018-0177-y [DOI] [PubMed] [Google Scholar]
- Shimatani Z., Kashojiya S., Takayama M., Terada R., Arazoe T., Ishii H., et al. (2017). Targeted Base Editing in rice and Tomato Using a CRISPR-Cas9 Cytidine Deaminase Fusion. Nat. Biotechnol. 35, 441–443. 10.1038/nbt.3833 [DOI] [PubMed] [Google Scholar]
- Sidorov V., Wang D., Nagy E. D., Armstrong C., Beach S., Zhang Y., et al. (2021). Heritable DNA-free Genome Editing of Canola (Brassica Napus L.) Using PEG-Mediated Transfection of Isolated Protoplasts. In Vitro Cell.Dev.Biol.-Plant, 1–10. 10.1007/s11627-021-10236-7 [DOI] [Google Scholar]
- Srinivasan C., Liu Z., Heidmann I., Supena E. D. J., Fukuoka H., Joosen R., et al. (2006). Heterologous Expression of the BABY BOOM AP2/ERF Transcription Factor Enhances the Regeneration Capacity of Tobacco (Nicotiana Tabacum L.). Planta 225, 341–351. 10.1007/s00425-006-0358-1 [DOI] [PubMed] [Google Scholar]
- Steinert J., Schmidt C., Puchta H. (2017). “Use of the Cas9 Orthologs from Streptococcus Thermophilus and Staphylococcus aureus for Non-homologous End-Joining Mediated Site-specific Mutagenesis in Arabidopsis thaliana ,” in Methods Mol. Biol. Editor Schmidt A. (New York, NY: Springer New York; ), 365–376. 10.1007/978-1-4939-7286-9_27 [DOI] [PubMed] [Google Scholar]
- Svitashev S., Schwartz C., Lenderts B., Young J. K., Mark Cigan A. (2016). Genome Editing in maize Directed by CRISPR-Cas9 Ribonucleoprotein Complexes. Nat. Commun. 7, 1–7. 10.1038/ncomms13274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Sretenovic S., Ren Q., Jia X., Li M., Fan T., et al. (2020). Plant Prime Editors Enable Precise Gene Editing in Rice Cells. Mol. Plant 13, 667–670. 10.1016/j.molp.2020.03.010 [DOI] [PubMed] [Google Scholar]
- Triozzi P. M., Schmidt H. W., Dervinis C., Kirst M., Conde D. (2021). Simple, Efficient and Open-Source CRISPR/Cas9 Strategy for Multi-Site Genome Editing in Populus Tremula × alba. Tree Physiol. 41, 2216–2227. 10.1093/TREEPHYS/TPAB066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillet F., Kermarrec M.-P., Chauvin L., Chauvin J.-E., Nogué F. (2020). CRISPR-induced Indels and Base Editing Using the Staphylococcus aureus Cas9 in Potato. PLoS One 15, e0235942. 10.1371/journal.pone.0235942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejlupkova Z., Warman C., Sharma R., Scheller H. V., Mortimer J. C., Fowler J. E. (2020). No Evidence for Transient Transformation via Pollen Magnetofection in Several Monocot Species. Nat. Plants 6 (6), 1323–1324. 10.1038/s41477-020-00798-6 [DOI] [PubMed] [Google Scholar]
- Vu T. V., Sivankalyani V., Kim E. J., Doan D. T. H., Tran M. T., Kim J., et al. (2020). Highly Efficient Homology‐directed Repair Using CRISPR/Cpf1‐geminiviral Replicon in Tomato. Plant Biotechnol. J. 18, 2133–2143. 10.1111/pbi.13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Li Y., Liang C. (2008). Recovery of Transgenic Plants by Pollen-Mediated Transformation in Brassica Juncea. Transgenic Res. 17, 417–424. 10.1007/S11248-007-9115-X/TABLES/1 [DOI] [PubMed] [Google Scholar]
- Wang M., Lu Y., Botella J. R., Mao Y., Hua K., Zhu J.-k. (2017). Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant 10, 1007–1010. 10.1016/j.molp.2017.03.002 [DOI] [PubMed] [Google Scholar]
- Wang M., Zuris J. A., Meng F., Rees H., Sun S., Deng P., et al. (2016). Efficient Delivery of Genome-Editing Proteins Using Bioreducible Lipid Nanoparticles. Proc. Natl. Acad. Sci. USA 113, 2868–2873. 10.1073/pnas.1520244113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-P., Xing H.-L., Dong L., Zhang H.-Y., Han C.-Y., Wang X.-C., et al. (2015). Egg Cell-specific Promoter-Controlled CRISPR/Cas9 Efficiently Generates Homozygous Mutants for Multiple Target Genes in Arabidopsis in a Single Generation. Genome Biol. 16, 1–12. 10.1186/s13059-015-0715-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss T., Wang C., Kang X., Zhao H., Elena Gamo M., Starker C. G., et al. (2020). Optimization of Multiplexed CRISPR/Cas9 System for Highly Efficient Genome Editing in Setaria Viridis. Plant J. 104, 828–838. 10.1111/tpj.14949 [DOI] [PubMed] [Google Scholar]
- Xu R., Li J., Liu X., Shan T., Qin R., Wei P. (2020). Development of Plant Prime-Editing Systems for Precise Genome Editing. Plant Commun. 1, 100043. 10.1016/j.xplc.2020.100043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Wei S., Wu Y., Hu R., Li H., Yang W., et al. (2015). High-Efficiency Genome Editing in Arabidopsis Using YAO Promoter-Driven CRISPR/Cas9 System. Mol. Plant 8, 1820–1823. 10.1016/j.molp.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Yoshida K., Ma D., Constabel C. P. (2015). The MYB182 Protein Down-Regulates Proanthocyanidin and Anthocyanin Biosynthesis in Poplar by Repressing Both Structural and Regulatory Flavonoid Genes. Plant Physiol. 167, 693–710. 10.1104/PP.114.253674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Tu L., Subburaj S., Bae S., Lee G.-J. (2021). Simultaneous Targeting of Duplicated Genes in Petunia Protoplasts for Flower Color Modification via CRISPR-Cas9 Ribonucleoproteins. Plant Cel Rep 40, 1037–1045. 10.1007/s00299-020-02593-1 [DOI] [PubMed] [Google Scholar]
- Zhang F., Maeder M. L., Unger-Wallace E., Hoshaw J. P., Reyon D., Christian M., et al. (2010). High Frequency Targeted Mutagenesis in Arabidopsis thaliana Using Zinc finger Nucleases. Proc. Natl. Acad. Sci. 107, 12028–12033. 10.1073/pnas.0914991107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Liu J., Chai Z., Chen S., Bai Y., Zong Y., et al. (2019a). Generation of Herbicide Tolerance Traits and a New Selectable Marker in Wheat Using Base Editing. Nat. Plants 5, 480–485. 10.1038/s41477-019-0405-0 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Iaffaldano B., Qi Y. (2021). CRISPR Ribonucleoprotein-Mediated Genetic Engineering in Plants. Plant Commun. 2, 100168. 10.1016/j.xplc.2021.100168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Malzahn A. A., Sretenovic S., Qi Y. (2019b). The Emerging and Uncultivated Potential of CRISPR Technology in Plant Science. Nat. Plants 5, 778–794. 10.1038/s41477-019-0461-5 [DOI] [PubMed] [Google Scholar]
- Zhao D., Li J., Li S., Xin X., Hu M., Price M. A., et al. (2020). Glycosylase Base Editors Enable C-To-A and C-To-G Base Changes. Nat. Biotechnolnat. Biotechnol. 39, 35–40. 10.1038/s41587-020-0592-210.1038/s41587-020-0648-3 [DOI] [PubMed] [Google Scholar]
- Zhao X., Meng Z., Wang Y., Chen W., Sun C., Cui B., et al. (2017). Pollen Magnetofection for Genetic Modification with Magnetic Nanoparticles as Gene Carriers. Nat. Plants 3 (3), 956–964. 10.1038/s41477-017-0063-z [DOI] [PubMed] [Google Scholar]
- Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., et al. (2017). Precise Base Editing in rice, Wheat and maize with a Cas9-Cytidine Deaminase Fusion. Nat. Biotechnol. 35, 438–440. 10.1038/nbt.3811 [DOI] [PubMed] [Google Scholar]