Abstract
With the development of proteomics and epigenetics, a large number of RNA‐binding proteins (RBPs) have been discovered in recent years, and the interaction between long non‐coding RNAs (lncRNAs) and RBPs has also received increasing attention. It is extremely important to conduct in‐depth research on the lncRNA‐RBP interaction network, especially in the context of its role in the occurrence and development of cancer. Increasing evidence has demonstrated that lncRNA‐RBP interactions play a vital role in cancer progression; therefore, targeting these interactions could provide new insights for cancer drug discovery. In this review, we discussed how lncRNAs can interact with RBPs to regulate their localization, modification, stability, and activity and discussed the effects of RBPs on the stability, transport, transcription, and localization of lncRNAs. Moreover, we explored the regulation and influence of these interactions on lncRNAs, RBPs, and downstream pathways that are related to cancer development, such as N6‐methyladenosine (m6A) modification of lncRNAs. In addition, we discussed how the lncRNA‐RBP interaction network regulates cancer cell phenotypes, such as proliferation, apoptosis, metastasis, drug resistance, immunity, tumor environment, and metabolism. Furthermore, we summarized the therapeutic strategies that target the lncRNA‐RBP interaction network. Although these treatments are still in the experimental stage and various theories and processes are still being studied, we believe that these strategies may provide new ideas for cancer treatment.
Keywords: cancer epigenetics, cancer, interaction network, lncRNA‐RBP, long non‐coding RNA, RNA‐binding protein, treatment strategy
List of abbreviations
- miRNA
microRNA
- siRNA
small interfering RNA
- lncRNA
long non‐coding RNA
- XIST
X‐inactive specific transcript
- RBP
RNA‐binding protein
- SFPQ
Splicing factor proline‐ and glutamine‐rich
- NEAT1
nuclear enriched abundant transcript 1
- IL
interleukin
- WDR5
WD repeat domain 5
- GADD45A
growth arrest and DNA damage inducible alpha
- SATB2‐AS1
special AT‐rich binding protein 2 antisense transcript 1
- SATB2
special AT‐rich binding protein 2
- KAT2A
lysine acetyltransferase 2A
- SOD2
superoxide dismutase 2
- OIP5‐AS1
opa interacting protein 5‐antisense transcript 1
- RMST
rhabdomyosarcoma 2‐associated transcript
- AR
androgen receptor
- MDM2
murine double minute 2
- PLK1
polo like kinase 1
- MSC
mesenchymal stem cell
- MUF
mesenchymal stem cell upregulated factor
- GSK‐3β
glycogen synthase kinase‐3β
- ANXA2
annexin A2
- HCC
hepatocellular carcinoma
- PTM
post‐translational modification
- MALAT1
metastasis‐associated in lung adenocarcinoma transcript 1
- NSCLC
non‐small cell lung cancer
- SIRT1
sirtuin 1
- DBC1
deleted in breast cancer 1
- EZH2
zeste homolog 2
- PRC2
polycomb repressive complex 2
- STAT3
signal transducer and activator of transcription 3
- NF‐κB
nuclear factor‐kappa B
- IκB
inhibitor of κB
- ANCR
association of the nordic cancer registries
- CDK1
cyclin‐dependent kinase 1
- GAS5
growth arrest specific 5
- YAP
yes‐associated protein
- CRC
colorectal cancer
- hnRNP
heterogeneous nuclear ribonucleoprotein
- TBK1‐IRF3
TANK‐binding kinase 1‐interferon regulatory factor 3
- IFN
inflammatory factors interferon
- cGAS
cyclic guanosine‐adenosine synthetase
- STING
stimulator of interferon gene
- IFI16
interferon gamma inducible protein 16
- PKM2
pyruvate kinase M2
- FEZF1‐AS1
FEZ family zinc finger 1‐antisense transcript 1
- SNHG1
small nucleolar RNA host gene 1
- HNRNPL
heterogeneous nuclear ribonucleoprotein L
- EMT
epithelial‐mesenchymal transition
- Pca
prostate cancer
- eIF 1‐6
eukaryotic initiation factor 1‐6
- IF 1‐3
initiation factor 1‐3
- PHD2
prolyl hydroxylase domain 2
- HIF‐1α
hypoxia inducible factor 1 alpha
- METTL3
methyltransferase‐like 3
- FTO
fat mass and obesity‐associated protein
- ALKBH5
α‐ketoglutarate‐dependent dioxygenase homolog
- YTHDC1
YTH domain containing 1
- IGF2BP2
insulin like growth factor 2 mRNA binding protein 2
- CBP
CREB binding protein
- CASC9
cancer susceptibility candidate 9
- LAMC2
laminin subunit gamma 2
- PI3K
phosphoinositide 3‐kinase
- ESCC
esophageal squamous cell carcinoma
- PTBP1
Polypyrimidine tract binding protein 1
- MACC1‐AS1
metastasis‐associated in colon cancer 1 antisense transcript 1
- SChLAP1
second chromosome locus associated with prostate‐1
- ACTN4
α actinin 4
- CTNNB1
catenin beta‐1
- YTHDF2
YTH domain family 2
- HNRNPK
heterogeneous nuclear ribonucleoprotein K
- SINE
short scattered nuclear element
- TAR
transactive responsive
- KHDRBS1
KH RNA binding domain containing, signal transduction associated 1
- TRA2A
transformer‐2 protein homolog alpha
- RBAT1
retinoblastoma associated transcript‐1
- LSD1
lysine‐specific demethylase 1
- PRSS8
repression of protease serine 8
- NCL
Nucleolin
- CYTOR
cytoskeleton regulator RNA
- KLF2
krüppel‐like factor 2
- SP1
specificity protein 1
- APOC1P1‐3
apolipoprotein C1 pseudogene 1‐3
- HMGB1
high mobility group box 1
- P53RRA
p53 related lncRNA
- G3BP1
GTPase‐activating protein‐binding protein 1
- SFPQ
splicing factor proline/glutamine‐rich
- AGR2
anterior gradient 2
- AXL
anexelekto
- DRAIC
downregulated RNA in cancer
- UCHL5
ubiquitin C‐Terminal hydrolase L5
- NFRKB
nuclear factor related to kappa‐B
- NF90
nuclear factor 90
- VEGFA
vascular endothelial growth factor A
- SRSF6
serine and arginine‐rich splicing factor 6
- TMZ
Temozolomide
- PTBP1
polypyrimidine tract binding protein 1
- MIR155HG
miR155 host gene
- ER
estrogen receptor
- AFAP1‐AS1
actin filament‐associated protein 1‐antisense RNA1
- AUF1
AU‐binding factor 1
- ERBB2
erb‐b2 receptor tyrosine kinase 2
- HOXD‐AS1
Homebox D antisense RNA 1
- AGAP2‐AS1
ankyrin repeat and PH domain 2‐antisense transcript 1
- RIG‐1
retinoic acid‐inducible gene I
- lnc‐Lsm3b
lncRNA‐sm‐like 3b
- Th1
T helper type 1
- MRPL23‐AS1
mitochondrial ribosomal protein L23‐antisense RNA 1
- SACC
salivary adenoid cystic carcinoma
- LNMAT2
Lymph node metastasis‐associated transcript 2
- PROX1
prospero homeobox 1
- SMAD4
small mother against decapentaplegic family member 4
- PDK1
pyruvate dehydrogenase kinase 1
- FTX
five prime to XIST
- LDH
lactate dehydrogenase
- PFKL
phosphofructokinase, liver type
- WFDC21p
WAP four‐disulfide core domain 21, pseudogene
- FILNC1
FoxO‐induced long non‐coding RNA 1
- FoxO
forkhead box O
- AUF1
ARE/poly(U)‐binding/degradation factor 1
- PTENP1
phosphatase and tensin homolog pseudogene 1
- ceRNA
competing endogenous RNA
- HER2
human epidermal growth factor receptor 2
- ASO
anti‐sense oligonucleotide
- LNA
locked nucleic acid
- PVT1
plasmacytoma variant translocation 1
- PCAT1
prostate cancer associated transcript 1
- PRNCR1
prostate cancer associated non‐coding RNA 1
- PCGEM1
prostate cancer gene expression marker 1
- PlncRNA1
prostate cancer‐up‐regulated long noncoding RNA 1
- RNAi
RNA interference
1. BACKGROUND
It is now widely recognized that less than 2% of the human genome is transcribed as coding RNA, while more than 98% is transcribed as non‐coding RNA [1], indicating that there are a large number of non‐coding RNA genes stored in the human genome. Non‐coding RNAs can be divided into microRNAs (miRNAs), small interfering RNAs (siRNAs), and long non‐coding RNAs (lncRNAs) according to size. lncRNAs are non‐coding RNAs of more than 200 nt in length. They are transcribed by RNA polymerase II and have a conserved secondary structure. However, they encode little or no protein due to the absence of effective open reading frames [2, 3, 4, 5, 6]. lncRNAs can be produced in several ways, including from gene editing regions, non‐coding regions, exons, introns and plus, and antisense chains. The intensive studies on lncRNAs have confirmed that lncRNAs play an important role in various biological regulation processes. With the advancement of sequencing technology and the discovery of functional characterization genomic elements, an increasing number of lncRNAs, such as X‐inactive specific transcript (XIST) [7] and H19 [8], have been identified.
lncRNAs can bind to protein, RNA, and DNA and thereby control gene expression levels in epigenetics, transcriptional regulation, transcriptional regulation, and other mechanisms through genetic imprinting, chromatin remodeling, cell cycle regulation, splicing regulation, mRNA degradation, and translation regulation [9, 10]. Some lncRNAs can cis‐regulate transcriptional activation and expression regulation of adjacent protein‐coding genes. lncRNA binds to the chromatin gap of the undissolved strand or to the DNA strand of the undissolved strand according to the principle of sequence complementarity. Transcription of genes near the lncRNA site is affected by lncRNAs on the transcript or spliceosome, and the transcription initiation elements of genes appear at the transcription sites of adjacent lncRNAs. In addition, some lncRNAs can regulate the expression of distal genes. LncRNAs can act as scaffolders and histone complexes and perform corresponding functions as subcellular structures, regulating the activity of binding proteins or RNA in the nucleus and cytoplasm in a dose‐dependent manner. The exact physical relationship between lncRNAs and various biological macromolecules remains to be clarified. Since the functions of a large number of lncRNAs are still unknown, they have obvious potential in the extensive regulation of gene expression.
Based on the ubiquity of protein‐RNA interactions, many studies have emphasized how their perturbations are related to pathology, including autoimmune diseases, neurological diseases, and cancer. Cancer is a complex disease caused by genetic changes or epigenetic abnormalities. In the field of RNA research, the search for the role of RNAs and RNA‐binding proteins (RBPs) in cancer has been a focus of research. RBPs are proteins that can bind to specific RNAs, and their interaction with RNA is an important checkpoint for regulating gene expression at the RNA level [11]. It has been reported that lncRNAs specifically combined with RBPs and influenced the related functions of RBPs [12]. Conversely, certain specific RBPs can combine with lncRNAs to influence the function of lncRNAs to regulate downstream gene expression. Moreover, RBPs can regulate the expression of lncRNAs at the transcriptional level [13]. However, research on the impact of RBPs on the fate of lncRNAs is still limited, and further exploration of the associated regulatory mechanism is needed.
The lack of information on lncRNA‐RBP interactions, even from the interaction of a single protein with RNA to the interaction of multiple proteins with RNA, hinders a better understanding of the mechanism of the lncRNA‐RBP interaction network in the occurrence and development of cancer. As research progresses, more RNA‐binding domains in RBPs have been discovered, which provides a theoretical basis for the development of more drugs to target RNA‐binding domains in proteins. In this review, we described how lncRNAs and RBPs influence each other and clarified the mechanism of their interaction network abnormalities in cancer development. Understanding the possible commonalities of the underlying mechanisms may help develop drugs targeting the binding sites of lncRNAs and RBPs and provide guiding recommendations.
2. FUNCTIONS OF LNCRNAS IN REGULATING RBPS
The conventional view is that RBPs regulate RNA, but it has been proven that RNA can also regulate the function of RBPs [12]. Emerging studies have revealed that lncRNAs can target many types of proteins through direct interactions with components, such as transcription factors, ribonucleoproteins, and chromatin modification complexes [14, 15, 16]. lncRNAs act as decoys, scaffolds, or guides for RBPs in the process of epigenetic regulation and affect the modification, stability, localization, and activity of their binding proteins, which regulate genes at the transcriptional and translational levels [17] (Figure 1).
FIGURE 1.
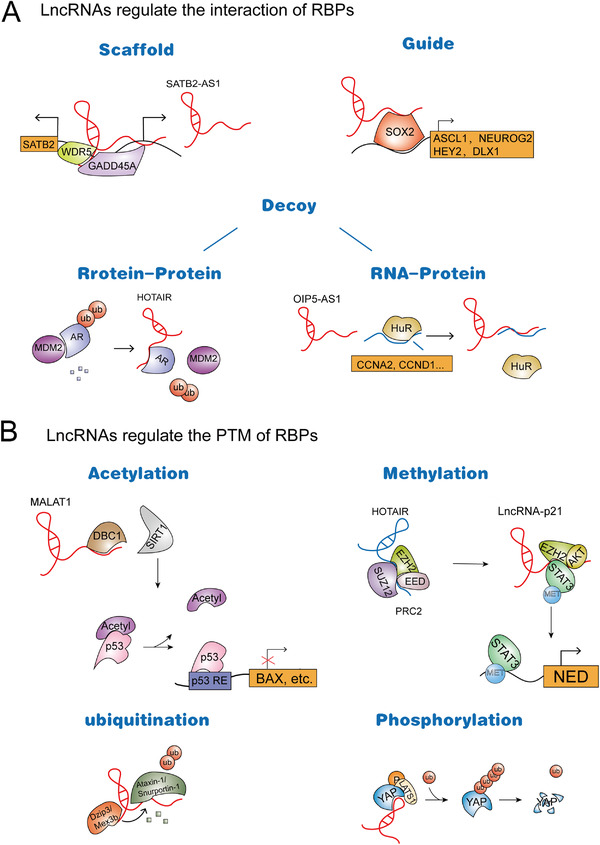
Functions of lncRNAs in regulating RBPs. (A) lncRNAs regulate the interaction of RBPs. Type I: as a scaffold, lncRNA SATB2‐AS1 can bind to WDR5 and GADD45A and recruit them to the promoter region of SATB2, thus cis‐activating the expression of SATB2. Type II: as a guide, RMST binds to the promoter regions of some SOX2 target genes and regulates the transcription of many common downstream target genes. Type III: as a decoy, HOTAIR blocks the interaction between AR and the E3 ubiquitin ligase MDM2, which prevents AR ubiquitination and protein degradation, thereby stabilizing the AR protein. OIP5‐AS1 can function as an endogenous competing RNA for HuR, which sequesters it away from target mRNAs. (B) lncRNAs regulate the PTM of RBPs. I: Acetylation and MALAT1 reduce the acetylation of p53 by competing for the interaction with SIRT1 and DBC1. II: Methylation: lncRNA‐p21 not only disrupts the PRC2 complex, causing the release of EZH2, but also enhances the interaction of EZH2 and STAT3 to trigger STAT3 methylation. III: Ubiquitination and HOTAIR can interact with E3 ubiquitin ligases and their substrates, which enhances the ubiquitination and degradation of Ataxin‐1 and Snurportin‐1. IV: Phosphorylation: GAS5 interacts with YAP and enhances YAP phosphorylation, which facilitates its ubiquitination and degradation, thereby inhibiting YAP signaling. Abbreviations: lncRNA, Long non‐coding RNA; RBP, RNA‐binding proteins; SATB2‐AS1, Special AT‐rich binding protein 2 antisense transcript 1; WDR5, WD repeat domain 5; GADD45A, Growth arrest and DNA damage inducible alpha; SOX2, SRY‐box transcription factor 2; ASCL1, Achaete‐scute family BHLH transcription Factor 1; NEUROG2, Neurogenin 2; HEY2, Hes related family BHLH transcription factor with YRPW motif 2; DLX1, Distal‐Less homeobox 1; MDM2, Murine double minute2; Ub, Ubiquitination; AR, Androgen receptor; CCNA2, Cyclin A2; CCND1, Cyclin D1; MALAT1, Metastasis associated lung adenocarcinoma transcript 1; DBC1,Deleted in breast cancer‐1; SIRT1, Sirtuin 1; Acetyl, Acetyl‐CoA acetyltransferase 1; EZH2, Enhancer of zeste 2 polycomb repressive complex 2 subunit; SUZ12, Suppressor of zeste 12 homolog; EED, Embryonic ectoderm development; PRC2, Polycomb repressive complex 2; NED, Neuroendocrine differentiation; MET, Methyltransferases; YAP, Yes‐associated protein
2.1. lncRNAs regulate the interaction of RBPs
lncRNAs can act as scaffolds to promote the assembly of protein complexes. In addition, lncRNAs can also be used as decoys to weaken the interaction between proteins and biological macromolecules (DNA, RNA, and proteins). Studies have shown that a large number of lncRNAs are involved in the regulation of a variety of biological processes, especially playing an important regulatory role in tumors. Although we have just begun to understand the function of lncRNAs, increasing evidence shows that lncRNAs bind to RBPs and thereby regulate gene transcription and translation processes in cis or trans.
At the transcriptional and/or post‐transcriptional level, lncRNAs can recruit interacting proteins to specific sites in the genome to regulate cis or trans gene expression [18, 19, 20]. Paraspeckles are subnuclear structures of the nucleus that are usually dynamic, and their abnormal formation is closely related to cancer [21, 22, 23]. Splicing factor proline‐ and glutamine‐rich (SFPQ), a nuclear enriched abundant transcript 1 (NEAT1)‐binding paraspeckle protein, serves as a repressor of interleukin (IL)‐8 transcription. Importantly, NEAT1 can induce the relocation of SFPQ from the IL‐8 promoter to paraspeckles, which activates the transcription of IL‐8 [24]. lncRNAs not only bind to transcription factors but also recruit them to promoters. At the transcriptional level, a new pattern has been widely observed, in which lncRNAs bind to histone‐modified complexes and recruit histone‐modified complexes to specific locations [19, 25]. This provides an explanation for how histone complexes recognize specific binding regions in their chromatin. It has been reported that many antisense lncRNAs can exert their function by regulating the expression of their neighboring genes. WD repeat domain 5 (WDR5) and growth arrest and DNA damage inducible alpha (GADD45A) are important chromatin regulators. lncRNA special AT‐rich binding protein 2 antisense transcript 1 (SATB2‐AS1) can bind to them and recruit them to the promoter region of special AT‐rich binding protein 2 (SATB2), thus cis‐activating the expression of SATB2 . GClnc1 can also act as a molecular link between WDR5 and the lysine acetyltransferase 2A (KAT2A) complex, thus upregulating the transcription level of target genes such as superoxide dismutase 2 (SOD2). GClnc1 alters the histone modification pattern, coordinates WDR5/KAT2A complex localization, and recruits WDR5 and KAT2A to the promoter of the SOD2 gene [26]. lncRNAs can sponge not only miRNAs but also RBPs and act as endogenous competitive RNAs; that is, lncRNAs act as “decoys” to prevent proteins from binding to target RNAs; these lncRNAs can bind to RBPs and regulate the post‐transcriptional fate of the target mRNAs. For example, opa interacting protein 5‐antisense transcript 1 (OIP5‐AS1) can function as an endogenous competing RNA for HuR, which sequesters it away from target mRNAs [27]. In addition to the decoy model, lncRNAs can act as “recruiters” to affect the interaction between histones or chromatin modifiers and DNA, thus playing an important role in the transcriptional regulation of genes. For example, rhabdomyosarcoma 2‐associated transcript (RMST) can act as a transcriptional coactivator of SOX2 by specifically binding to SOX2. In addition, RMST is required for SOX2 to bind to some genomic sites. It binds to the promoter regions of some SOX2 target genes and regulates the transcription of many common downstream target genes [28].
At the translational level, as one of the first known lncRNAs related to cancer, homeobox transcript antisense RNA (HOTAIR) is abnormally expressed in many tumors, including breast cancer [29] and gastric cancer [30], and is closely related to the occurrence and development of tumors. Zhang et al. [31] showed that HOTAIR blocks the interaction between androgen receptor (AR) and the E3 ubiquitin ligase murine double minute 2 (MDM2), which prevents AR ubiquitination and protein degradation, thereby stabilizing the AR protein. Likewise, APAL, termed polo like kinase 1 (PLK1)‐associated lncRNA, promotes the phosphorylation of PLK1 mediated by Aurora A by increasing the interaction between PLK1 and Aurora A [32]. Additionally, lncRNA mesenchymal stem cell (MSC) upregulated factor (MUF) interacts specifically with glycogen synthase kinase‐3β (GSK‐3β) and annexin A2 (ANXA2) through distinct but partially overlapping sites, which modulates hepatocellular carcinoma (HCC) progression. In addition, increased expression of lncRNA‐MUF promotes the interactions between ANXA2 and GSK‐3β. Conversely, depleting lncRNA‐MUF inhibits the binding effect [33].
2.2. lncRNAs regulate the post‐translational modification (PTM) of RBPs
The function of proteins is usually regulated by introducing chemical modifications after translation. Different types of PTMs will affect many functions of proteins, which will affect cell function and eventually lead to the occurrence and development of cancer [34, 35]. Different types of PTMs will affect many functions of proteins, thus affecting cell function. Recently, the regulation of PTM has become the focus of attention. To date, it has been found that some lncRNAs can regulate the PTM of their binding proteins via mechanisms, such as phosphorylation, ubiquitination, and acetylation, to regulate the degradation or production of proteins and then affect the expression level and activity of proteins. The p53 can determine the fate of cells by regulating the transcriptional ability of many target genes, and the effect of its PTM on its activity is very important. However, how the PTM of p53 is dysregulated is still a question worth exploring. It has been reported that metastasis‐associated in lung adenocarcinoma transcript 1 (MALAT1) can participate in the PTM of p53 and affect the occurrence and development of tumors. MALAT1 was reported to exist widely in a variety of cancers, including non‐small cell lung cancer (NSCLC) and HCC [36, 37, 38] and is related to the proliferation and metastasis of cancer cells. MALAT1 reduces the acetylation of p53 by competing for the interaction with sirtuin 1 (SIRT1) and deleted in breast cancer 1 (DBC1) to release SIRT1, which regulates SIRT1 activity [33].
Early studies have found that lncRNAs may interact with zeste homolog 2 (EZH2) to regulate EZH2 polycomb repressive complex 2 (PRC2)‐dependent activity, while Luo et al. [39] found that lncRNA‐p21 could release EZH2 from the PRC2 complex. By competing with HOTAIR to bind EZH2, lncRNA‐p21 not only disrupts the PRC2 complex to release EZH2 but also enhances the interaction of EZH2 and signal transducer and activator of transcription 3 (STAT3) to trigger STAT3 methylation. Furthermore, it has been suggested that nuclear factor‐kappa B (NF‐κB)‐interacting long non‐coding RNA (NKILA) regulates NF‐κB activation and post‐translationally modifies NF‐κB signaling proteins by preventing inhibitor of κB (IκB) phosphorylation via physical interaction with the NF‐κB/IκB complex, which suppresses breast cancer metastasis [40]. In addition, association of the nordic cancer registries (ANCR) mediates the ubiquitylation degradation of EZH2 by interacting with EZH2 and promoting the interaction between EZH2 and cyclin‐dependent kinase 1 (CDK1) and then exerts an anticancer effect in breast cancer [41]. It is worth noting that growth arrest specific 5 (GAS5) interacts with yes‐associated protein (YAP) and enhances YAP phosphorylation, which facilitates its ubiquitination and degradation, thereby inhibiting YAP signaling. Therefore, lncRNA GAS5 suppresses colorectal cancer (CRC) progression by dysregulating YAP in vivo and in vitro [39]. It has been reported that lncRNAs can be used as regulators of ubiquitin‐mediated proteolysis to promote ubiquitin‐binding proteins and increase their degradation. HOTAIR can interact with E3 ubiquitin ligases and their substrates, which enhances the ubiquitination and degradation of Ataxin‐1 and Snurportin‐1 [42].
3. ROLES OF RBPS IN REGULATING LNCRNAS
Most currently known lncRNAs bind to proteins to form RNA‐protein complexes. RBPs can change the fate and function of lncRNAs by regulating their stability, transport and transcription. m6A is the most common modification of lncRNAs [43, 44], and m6A readers can act as RBPs to recognize and target m6A‐modified lncRNAs and regulate lncRNA degradation and transcription. Revealing the interactions between lncRNAs and proteins and studying the regulation of RBPs on lncRNAs are key to exploring the function of lncRNAs [43, 45] (Figure 2).
FIGURE 2.
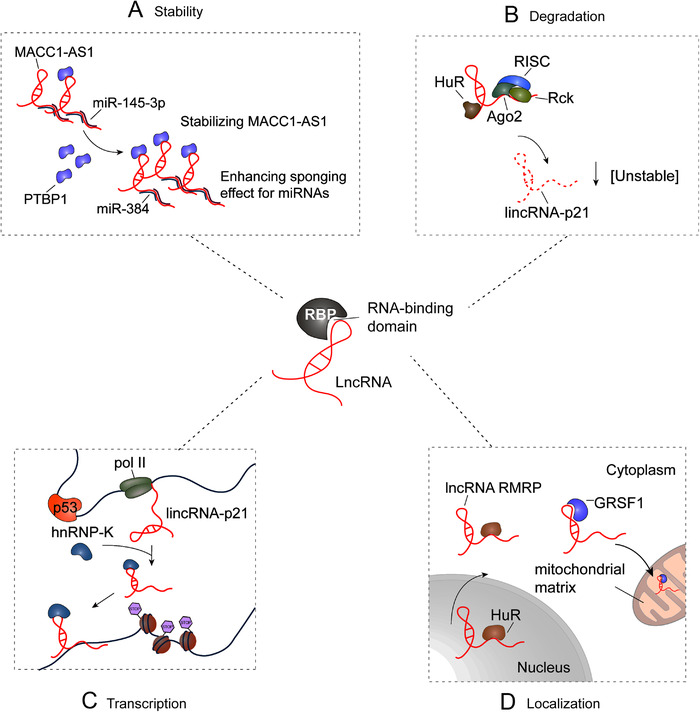
Roles of RBPs in regulating lncRNAs. (A) PTBP1 can act as a MACC1‐AS1‐binding partner to stabilize MACC1‐AS1 and enhance the sponging effect for miRNAs via a conserved pyrimidine‐rich motif within this lncRNA. (B) HuR and let‐7/Ago2 can bind to lincRNA‐p21, which cooperatively enhances lincRNA‐p21 decay. (C) p53 directly interacts with the lincRNA‐p21 promoter, inducing transcriptional activation in response to DNA damage. (D) RMRP can be recruited into the cytosol and the mitochondrial matrix by interacting with HuR and GRSF1, respectively. Abbreviations: lncRNA, Long non‐coding RNA; MACC1‐AS1, Metastasis‐associated in colon cancer 1 antisense transcript 1; PTBP1, Polypyrimidine tract binding protein 1; HuR, Human antigen R; RISC, RNA miRNA‐induced silencing complex; RMRP, RNA component of mitochondrial RNA processing endoribonuclease; GRSF1, Guanine‐rich RNA sequence binding factor 1
3.1. Quantity and classification of RBP
Based on existing research, scientists believe that there are approximately 1,000‐2,000 RBPs in mammals. Dr Stefanie Gerstberger of Columbia University further mapped RBPs in human cells based on mass spectrometry data regions from RNA‐protein cross‐linking experiments. According to Gerstberger's estimates, there are 1,542 RBPs in human cells, and perhaps many more, which may be individual proteins or families of proteins that bind to RNA in different ways (specific or non‐specific) [46].
RBPs can be classified into at least two groups: classical RBPs and non‐classical RBPs. Classical RBPs have a broad range of substrates, such as the heterogeneous nuclear ribonucleoprotein (hnRNP) family. Within this group, some may only interact with RNA molecules. For example, hnRNP K positively regulates the transcriptional activity of lncRNA‐OG by promoting the acetylation of the 27th lysine (H3K27) of histone H3 of the lncRNA‐OG (osteogenesis‐related lncRNA) promoter, thereby regulating the bone morphogenetic protein (activation of bone morphogenetic protein signaling pathway) [47]. In addition, hnRNP can also interact with lncRNAs to inhibit gene expression. For example, in macrophages, lnc13 forms a complex with the P42 subtype of hnRNP D, which can promote HDAC1 to bind the lnc13‐regulated promoters and ultimately to inhibit the expression of inflammatory factors, indicating that hnRNP D and lnc13 play an important role in recruiting HDAC1 to combine with the promoter [48].
However, other RBPs may interact with both RNA and DNA molecules. For example, Wang et al. [49] discovered that hnRNPA2B1 in the hnRNP family can recognize foreign viral DNA. Then, the TANK‐binding kinase 1‐interferon regulatory factor 3 (TBK1‐IRF3) signaling pathway is activated to promote the production of the inflammatory factors interferon (IFN)‐α/β. In addition, hnRNPA2B1 can regulate the methylation modification of cyclic guanosine‐adenosine synthetase (cGAS), stimulator of interferon genes (STING) and interferon gamma inducible protein 16 (IFI16) mRNA levels, as well as nucleoplasmic transport, to directly regulate STING‐dependent antiviral innate immunity. In this example, hnRNP simultaneously interacts with DNA and RNA to further regulate gene expression. From this point of view, RBPs can not only interact with RNA but also regulate the process of life by interacting with DNA.
The non‐classical RBPs usually have a narrow range of substrates or may even interact with only one or two substrates, such as pyruvate kinase M2 (PKM2). PKM2 is an isoenzyme of glycolytic pyruvate kinase. Studies have demonstrated that lncRNA FEZ family zinc finger 1‐antisense transcript 1 (FEZF1‐AS1) can bind to and increase the stability of PKM2 protein, leading to increased levels of cytoplasmic and nuclear PKM2. Increased cytoplasmic PKM2 promotes pyruvate kinase activity and lactate production (aerobic glycolysis), while the upregulation of nuclear PKM2 induced by FEZF1‐AS1 further activates STAT3 signaling [50]. This type of RBP only needs to bind to specific lncRNAs to initiate the corresponding biological function. In addition, small nucleolar RNA host gene 1 (SNHG1) interactively competes with heterogeneous nuclear ribonucleoprotein L (HNRNPL) and reduces the translation of the E‐cadherin protein, thereby activating the effect of SNHG1 on the epithelial‐mesenchymal transition (EMT) pathway and ultimately promoting the transfer of prostate cancer (PCa) [51]. The above examples all show that when this type of RBP interacts with a specific lncRNA, it will always cause some changes in the configuration of lncRNA, which will affect its activity and ultimately affect the signal transduction of the tumor.
However, in our opinion, there is another way to classify RBPs. One is based on the RNA bound to the RBP; it classifies RBPs as mRNA‐, tRNA‐, snoRNA‐, snRNA‐, pre‐rRNA‐ and ncRNA‐binding proteins; they are involved in the regulation of different signaling pathways. Some RBPs bind to mRNA, tRNA, or ribosomal proteins eukaryotic initiation factor 1‐6 (eIF 1‐6), translation initiation factor 1‐3 (IF 1‐3) to regulate gene transcription and translation (transcription factors, translation factors). Other RBPs are related to rRNA biosynthesis (Trm112, 18S rRNA methyltransferase) [52]. In addition, it has been shown that RBPs can indirectly regulate the process of transcription by interacting with transcription factors, some of which are involved in snRNA formation (snRNP; snRNA‐protein complex). For ncRNAs, lncRNAs have been a hot topic of research in recent years, and accordingly, proteins that bind to lncRNAs have also been the subject of recent researches. For example, spen family transcriptional repressor (SPEN) promotes gene silencing by interacting with the A‐repeat domain in XIST [53], DBC1 binds to MALAT1 to decrease p53 acetylation [33], and prolyl hydroxylase domain 2 (PHD2) in tumor‐associated macrophages binds to hypoxia inducible factor 1 alpha (HIF‐1α) stable long non‐coding RNA (HISLA) and blocks the interaction of PHD2 and HIF‐1α to inhibit the hydroxylation and degradation of HIF‐1α, ultimately leading to increased HIF‐1α protein levels and elevated aerobic glycolysis in tumor cells [54]. In tumor cells, there are many other lncRNA‐binding proteins that bind directly or indirectly to lncRNAs, causing several downstream biological events (including protein synthesis, RNA maturation and transport, and, as recently discovered, transcribed gene silencing through the regulation of chromatin structure) and further regulating the expression of downstream genes, ultimately influencing tumor progression [17].
However, if RBPs are classified according to the gene expression process in which they are involved, they can be divided into RBPs involved in transcription, RBPs involved in post‐transcriptional modification, and RBPs involved in translation. The study of the post‐transcriptional modification of RBPs has been a hot topic in recent years. By studying the mechanism by which RBPs interact with RNA at the transcriptional or post‐transcriptional level, we can further understand how RBPs regulate gene expression and participate in disease processes. Post‐transcriptional modifications of RNA include N1‐methyladenosine (m1A), m6A, and N4‐acetylcytidine (ac4C) [55]. In recent years, there has been significant interest in the study of RBPs involved in the m6A modification of lncRNAs. There are three types of RBPs involved in the m6A modification of lncRNAs: methyltransferases (writers), demethylases (erasers), and readers [43]. The methyltransferases involved in the m6A modification of lncRNAs include methyltransferase‐like 3 (METTL3) and METTL16 [56]; the demethylases include fat mass and obesity‐associated protein (FTO) and α‐ketoglutarate‐dependent dioxygenase homolog (ALKBH5) [57]; and the reader includes YTH domain containing 1 (YTHDC1) [58] and insulin like growth factor 2 mRNA binding protein 2 (IGF2BP2) [59]. These RBPs play an important regulatory role in the process of lncRNA m6A modification. In addition to methylation modifications, LncRBP is also involved in acetylcytidine modifications of lncRNAs, which is another popular research focus. For example, CREB binding protein (CBP) can bind to the lncRNA cancer susceptibility candidate 9 (CASC9) to recruit CBP and promote the transcriptional expression of laminin subunit gamma 2 (LAMC2), ultimately activating the downstream FAK‐phosphoinositide 3‐kinase (PI3K)/Akt signaling pathway to promote esophageal squamous cell carcinoma (ESCC) metastasis [60]. In summary, regardless of the mechanism, m6A modification or acetylation modification of lncRNAs further modulates their stability or localization through the interactions between RBPs and lncRNAs. However, we still need to explore the underlying mechanisms by which RBPs are involved in the regulation of the m6A modification or acetylation of lncRNAs and the components required in this process.
There are still a large number of RBPs waiting to be discovered. However, as Gerstberger argues, the discovery of new RBPs and potential RBPs is of little significance, and the more important discovery lies in the regulatory and physiological functions of RBPs. Moreover, examination of the role played by the lncRNA‐RBP interaction network in the regulation of gene expression can further reveal therapeutic targets for related diseases, such as cancer, and provide a theoretical basis for cancer treatment [46].
3.2. RBPs regulate the stability of lncRNAs
At the post‐transcriptional level, proteins modulate the stability, degradation, and function of lncRNAs by interacting with them to form a ribonucleoprotein complex [61]. Polypyrimidine tract binding protein 1 (PTBP1) can act as a metastasis‐associated in colon cancer 1 antisense transcript 1 (MACC1‐AS1)‐binding partner to stabilize MACC1‐AS1 and enhance the sponging effect for miRNAs via a conserved pyrimidine‐rich motif within this lncRNA (Figure 2A) [62]. Likewise, HNRNPL can serve as a protein partner of second chromosome locus associated with prostate‐1 (SChLAP1), which enhances the stabilization of lncRNA, leading to the promotion of the interaction with α actinin 4 (ACTN4). Additionally, the lncRNA SChLAP1 facilitates the formation of the HNRNPL/ACTN4 complex and plays a potential role in the progression of human glioblastoma [57]. The SChLAP1‐HNRNPL complex stabilizes ACTN4 by inhibiting proteasome degradation, leading to increased nuclear localization of the p65 subunit of NF‐κB and activation of NF‐κB signaling. In addition, HuR and let‐7/Ago2 can bind to lincRNA‐p21, which cooperatively enhances lincRNA‐p21 decay (Figure 2B) [63]. The association between the RBP HuR and lincRNA‐p21 facilitates the recruitment of let‐7/Ago2 to lincRNA‐p21, thereby promoting the decay of lincRNA‐p21. Under reduced HuR levels, lincRNA‐p21 is increased in HeLa human cervical cancer cells, promoting its association with junB proto‐oncogene (JUNB) and catenin beta‐1 (CTNNB1) mRNA and selectively inhibiting their translation. Similarly, IGF2BP1 serves as an adaptor protein to negatively regulate the expression of lncRNA highly up‐regulated in liver cancer by binding to HULC. Mechanistically, the carbon catabolite repressor protein 4 (CCR4)‑negative on TATA (NOT) complex (CNOT1) protein acts as a novel interaction partner of IGF2BP1. The consumption of CNOT1 increases the half‐life and expression of HULC. Therefore, IGF2BP1, as an adaptor protein, initiates the decay of lncRNA HULC by recruiting the carbon catabolite‐repression 4‐negative on TATA‐less (CCR4‐NOT) complex [61]. m6A readers have been shown to play a key role in the stability of lncRNAs.
YTH domain family 2 (YTHDF2), the first identified m6A reader, has been the most extensively studied. Within the cytoplasm, YTHDF2 was reported to mediate m6A‐dependent mRNA degradation under normal and stress conditions [64, 65]. Furthermore, YTHDF3 can directly bind to and recognize m6A‐modified GAS5 and promote the degradation of GAS5 in a methylation‐dependent manner [39]. GAS5 promotes the phosphorylation of YAP and subsequent ubiquitin‐mediated degradation and promotes the translocation of endogenous YAP from the nucleus to the cytoplasm by directly interacting with the WW domain of YAP, thereby inhibiting the progression of CRC in vitro and in vivo. In addition, recent research has shown that inhibiting the expression of METTL14 can eliminate the m6A level of XIST and enhance the expression of XIST. The interaction of YTHDF2 with XIST facilitates the degradation of XIST, the expression of which negatively correlates with that of METTL14 and YTHDF2 [66].
3.3. RBPs regulate the transcription and localization of lncRNAs
m6A has been suggested to mediate key aspects of XIST function since lncRNA XIST has more mapped m6A residues than any other RNAs. m6A modification of XIST is essential for recruiting the nuclear m6A reader YTHDC1, which may serve to stabilize the assembly of silencing components on XIST. Additionally, YTHDC1 has been shown to mediate transcriptional repression through its binding to XIST in a METTL3/RBM15/15B‐dependent manner [67]. Numerous key transcription factors are involved in the transcription of protein‐coding genes and non‐coding RNA genes. To date, numerous lncRNAs regulated by p53 have been reported, and they can affect various cellular processes [13]. P53 directly interacts with the lincRNA‐p21 promoter, inducing transcriptional activation in response to DNA damage. It has been suggested that lincRNA‐p21 is a new p53 target gene and plays a critical role in the p53 pathway (Figure 2C) [68].
Notably, p53, a well‐known transcription factor, could regulate the transcription of lncRNA loc285194 through its interaction with the p53 response element in the upstream region of loc285194, inhibiting the development of cancer [69]. Increasing evidence has demonstrated that RBPs could affect the localization of lncRNAs. For example, it has been indicated that the RBP HuR could influence the stability and export of its target RNAs. The lncRNA ribonuclease mitochondrial RNA processing (RMRP), transcribed from nuclear DNA, is related to the RBPs HuR and guanine‐rich RNA sequence binding factor 1 (GRSF1). Furthermore, RMRP could be recruited to the cytosol and mitochondrial matrix by interacting with HuR and GRSF1, respectively (Figure 2D) [70]. Additionally, heterogeneous nuclear ribonucleoprotein K (HNRNPK) has been identified as a driver of RNA nuclear retention, and it has been reported that it could drive RNA nuclear retention. MALAT1 lacks short scattered nuclear elements (SINEs), easily translocates to the cytoplasm, binds to TDP‐43 more strongly, and may even be guided to the cytoplasm by cytoplasmic MALAT1, resulting in a decrease in transactive responsive (TAR) DNA‐binding protein 43 (TDP‐43). Interestingly, the SINE of MALTA1 could modulate the interaction between HNRNPK and MALAT1, which increases the nuclear retention of MALAT1 [71]. This may occur through the direct interaction of SINE with KH RNA binding domain containing, signal transduction associated 1 (KHDRBS1) and transformer‐2 protein homolog alpha (TRA2A), which binds to HNRNPK [71].
4. THE LNCRNA‐RBP INTERACTION NETWORK IN CANCER
Although lncRNAs function as major gene regulators, they do not function alone [20]. With further research, an increasing number of lncRNAs may be found to drive many important cancer phenotypes by interacting with other cellular macromolecules, including DNA, RNA, and protein [72, 73]. Accumulating evidence indicates that cellular processes are regulated by lncRNA‐RBP interactions, including cell proliferation and apoptosis, as well as cancer metastasis. LncRNAs carry out cellular functions by forming macromolecular complexes with proteins [74]. There is a causal relationship between the disturbance of the lncRNA‐RBP interaction network and the occurrence and development of cancer. Deciphering the interaction network between cancer‐related lncRNAs and their binding proteins will reveal new cancer treatment targets (Figure 3).
FIGURE 3.
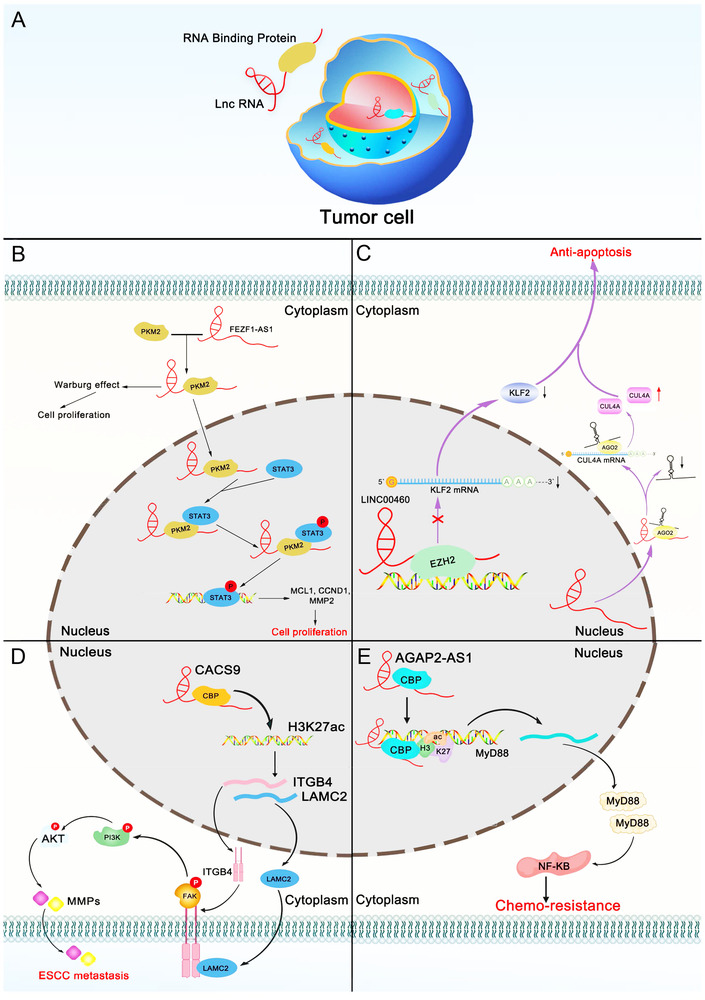
The lncRNA‐RBP interaction network in cancer phenotypes. (A) lncRNAs interact with RBPs to influence tumor cell development and progression. (B) FEZF1‐AS1 increases the stability of PKM2 by binding to PKM2, promoting the production of PKM2 in the nucleus, activating STAT3 signaling, and promoting aerobic glycolysis (Warburg effect) and, ultimately, cancer cell proliferation. (C) LINC00460 binds to EZH2, which inhibits the transcription of KLF2. LINC00460 also sponges miR‐149‐5p and antagonizes its translational inhibition of the cullin 4A (CUL4A) protein. Through these two pathways, apoptosis of tumor cells is ultimately inhibited. (D) In the nucleus, lncRNA CACS9 recruits CBP to regulate the level of H3K27ac, which in turn promotes the transcription of ITGB4 and LAMC2. LAMC2 exits in the nucleus and moves to the extracellular matrix, where it binds to ITGB4 in the cell membrane, further activating the FAK‐PI3K/Akt signaling pathway and ultimately promoting esophageal cancer metastasis. (E) The binding of AGAP2‐AS1 to CBP (a transcriptional coactivator) promotes H3K27ac enrichment in the promoter region of MyD88, leading to the upregulation of MyD88 expression. MyD88 further activates the downstream NF‐κB pathway, leading to an increase in drug resistance in tumor cells. Abbreviations: lncRNA, Long non‐coding RNA; RBP, RNA‐binding protein; PKM2, Pyruvate kinase M2; FEZF1‐AS1, FEZ family zinc finger 1‐antisense RNA 1; STAT3, Signal transducer and activator of transcription 3; MCL1, Myeloid cell leukemia sequence 1; CCND1, Cyclin D1; MMP2, Matrix metallopeptidase 2; CUL4A, Cullin 4A; EZH2, Enhancer of zeste homolog 2; AGO2, Argonaute 2; CACS9, Cancer susceptibility candidate 9; CBP, CREB‐binding protein; ITGB4, Integrin subunit beta 4; LAMC2, Laminin subunit gamma 2; ESCC, Esophageal squamous cell carcinoma; CBP, CREB binding protein; AGAP2‐AS1, Arf GAP with GTP‐binding protein‐like domain, Ankyrin repeat and PH domain 2‐antisense transcript 1
4.1. The lncRNA‐RBP interaction network in cancer proliferation
Unlimited proliferation is a characteristic of cancer cells. Uncontrolled cell proliferation leads to uncontrolled expansion or even metastasis [75]. As key regulatory factors, lncRNAs can be positively or negatively correlated with tumor cell proliferation [76]. The lncRNA‐RBP network is involved in a wide range of biological processes via diverse mechanisms. It has been reported that an imbalance in the non‐coding RNA‐RBP interaction network plays a major role in tumor proliferation. Colon cancer 1 (OCC‐1) was found to affect the stability of the RBP HuR and CRC cell growth through the regulation of mRNAs at the post‐transcriptional level. OCC‐1 can reduce the level of HuR and its target mRNA by promoting the binding of ubiquitin E3 ligase β‐TrCP1 to HuR, including mRNA related to tumor cell growth. This shows that lncRNA OCC‐1 can regulate the expression level of a large number of mRNAs at the post‐transcriptional level by regulating the stability of RBP HuR [77]. As a new non‐coding transcript, retinoblastoma associated transcript‐1 (RBAT1) recruits HNRNPL protein to the E2F3 promoter to activate E2F3 transcription. This mechanism plays a key role in cell proliferation in vitro and in vivo [78]. High expression of CASC9 is significantly related to the overall survival rate of HCC patients. The results from RNA affinity purification and native RNA immunoprecipitation confirmed the interaction between HNRNPL and CASC9, which together act as a clinically relevant lncRNA/protein complex to regulate AKT signaling in HCC [79].
It was observed that lncRNA homeobox A11 antisense RNA (HOXA11‐AS) is overexpressed in gastric cancer cells. HOXA11‐AS silencing suppressed gastric cancer progression. Furthermore, HOXA11‐AS serves as a scaffold to facilitate the formation of the EZH2 and lysine‐specific demethylase 1 (LSD1) protein complex, inducing the repression of protease serine 8 (PRSS8) expression [80]. Scientists also found that the upregulation of FEZF1‐AS1 in CRC is closely related to poor survival. FEZF1‐AS1 was found to bind to PKM2 and activate STAT3 and aerobic glycolysis, ultimately promoting the proliferation and metastasis of CRC cells [81]. Nucleolin (NCL) and src‐associated in mitosis of 68 kDa (Sam68) bind to cytoskeleton regulator RNA (CYTOR) Exon 1 by recognizing their specific motifs. Exon 1 is a key site that mediates the interaction of CYTOR with NCL and Sam68. NCL and Sam68 can promote the progression of CRC. lncRNA CYTOR can bind to NCL and Sam68 and activate the NF‐κB pathway to promote cancer invasion [82]. Another study showed that overexpression of antisense non‐coding RNA in the INK4 locus (ANRIL) is associated with the poor prognosis of patients with NSCLC. Mechanistic studies have revealed that ANRIL binds to EZH2, which can directly bind to the krüppel‐like factor 2 (KLF2) and p21 promoter regions and mediate H3K27me3 modification, which decreases the transcript levels of KLF2 and p21, thus affecting the proliferation and apoptosis of NSCLC cells [83]. Based on the above‐discussed literature, the key function of lncRNA and RBP interactions in the proliferation of a variety of tumor cells is clarified. Pharmacological targeting of the binding target of lncRNAs and RBPs can provide a theoretical basis for the diagnosis and treatment of various cancers.
4.2. The lncRNA‐RBP interaction network in cancer apoptosis
Apoptosis is a mechanistically distinct pathway of programmed cell death. Moreover, the poor regulation of apoptotic cell death is involved in the development of cancer, and the imbalance of lncRNAs involved in apoptosis may provide a potential mechanism for the development and treatment of cancer. A deeper understanding of the relationship between the lncRNA‐RBP network and cell apoptosis may reveal an effective treatment for cancer [84, 85, 86, 87, 88]. Several studies have shown that lncRNA‐RBP interactions are related to the regulation of apoptosis. For example, the lncRNA terminal differentiation‐induced ncRNA (TINCR) is overexpressed in gastric cancer tissues and cell lines and is regulated by specificity protein 1 (SP1). TINCR can directly bind to the Staufen 1 (STAU1) protein to affect the stability and expression of KLF2 mRNA and to regulate cell cycle progression and apoptosis [89]. Researchers have used microarrays to identify the abnormally expressed long intergenic non‐coding RNA apolipoprotein C1 pseudogene 1‐3 (APOC1P1‐3) in breast cancer. Moreover, in breast cancer, the promoter region of APOC1P1‐3 is hypomethylated, which contributes to the transcriptional activation and overexpression of APOC1P1‐3. In addition, APOC1P1‐3 can directly bind to α‐tubulin and promote cell proliferation by inhibiting apoptosis by changing its acetylation level [90]. It has also been demonstrated that MALAT1 can bind to high mobility group box 1 (HMGB1) and promote the expression of HMGB1 at the post‐translational level, thereby increasing autophagy and suppressing apoptosis. Additionally, MALAT1 knockdown facilitates the ubiquitination and degradation of HMGB1 [91].
LINC00460 suppresses KLF2 expression and participates in the pathophysiological processes in the progression of CRC by interacting with EZH2, suggesting its potential role in CRC. When LINC00460 expression was downregulated, proliferation was inhibited, but apoptosis occurred [65]. lncRNA p53 related lncRNA (P53RRA) is downregulated in cancers and functions as a tumor suppressor by binding Ras‐GTPase‐activating protein‐binding protein 1 (G3BP1), which interacts with p53 to displace p53 from a G3BP1 complex, resulting in greater p53 retention in the nucleus, which leads to cell cycle arrest, apoptosis, and ferroptosis [92]. Furthermore, lncRNA NEAT1 can serve as an important mediator in imatinib mesylate‐induced apoptosis. In addition, NEAT1 is a direct transcriptional target of c‐Myc, and c‐Myc can directly bind to the promoter of NEAT1 and inhibit the expression of NEAT1. It has been reported that splicing factor proline/glutamine‐rich (SFPQ) is the protein component of speckles [93]. NEAT1 can sequester SFPQ within paraspeckles by binding to SFPQ, which restricts the pro‐apoptotic effect of SFPQ [94]. In short, targeting lncRNAs and their proteins to regulate the apoptosis of cancer cells is a new idea for tumor therapy (Table 1).
TABLE 1.
The list of lncRNA‐RBP interaction network in cancer proliferation and apoptosis
| lncRNA | RBP | Cancer type | Function* | Mechanism | Reference |
|---|---|---|---|---|---|
| OCC‐1 | HuR | Colorectal cancer | Proliferation ↓ | Enhances the ubiquitination and degradation of HuR to regulate mRNAs related to the growth of cancer cells | [77] |
| CASC9 | HNRNPL | Hepatocellular carcinoma | Proliferation ↑, apoptosis ↓ | Regulates a joint set of target genes associated with PI3K/AKT signaling and DNA damage signaling | [79] |
| HOXA11‐AS | EZH2, LSD1 | Gastric cancer | Proliferation, migration and invasion ↑ | Serves as a scaffold to facilitate the formation of EZH2 and LSD1 protein complex and inhibit the expression of PRSS8 | [80] |
| FEZF1‐AS1 | PKM2 | Colorectal cancer | Proliferation and metastasis ↑ | Enhances the STAT3 signaling and aerobic glycolysis (Warburg effect) | [81] |
| CYTOR | NCL, Sam68 | Colorectal cancer | Proliferation and metastasis ↑ | Activates the NF‐κB signaling pathway and EMT | [82] |
| ANRIL | EZH2 | Non‐small cell lung cancer | Proliferation ↑, apoptosis ↓ | Silences the transcriptional level of KLF2 and p21 | [83] |
| TINCR | STAU1 | Gastric cancer | Proliferation ↑, apoptosis ↓ | Affects the stability and expression of KLF2 mRNA | [89] |
| APOC1P1‐3 | α‐tubulin | Breast cancer | Apoptosis ↓ | Increases the acetylation level of α‐tubulin | [90] |
| MALAT‐1 | HMGB1 | Multiple myeloma | Autophagy ↑, apoptosis ↓ | Promotes the expression level of HMGB1 at the post‐translational level | [91] |
| LINC00460 | EZH2 | Colorectal cancer | Proliferation ↓, apoptosis ↑ | Suppresses KLF1 expression | [65] |
| P53RRA | G3BP1 | Lung cancer | Apoptosis ↑ | Activates the p53 pathway | [92] |
| NEAT1 | SFPQ | Chronic myeloid leukemia | Apoptosis ↑ | Limits the effectiveness of SFPQ | [94] |
↑ and ↓ indicate increase and decrease, respectively.
Abbreviations: lncRNA, Long non‐coding RNA; RBP, RNA‐binding proteins; HNRNPL, Heterogeneous nuclear ribonucleoprotein L; EZH2, Enhancer of zeste homolog 2; LSD1, Lysine demethylase 1A; PKM2, Pyruvate kinase M2;NCL, Nucleolin; EZH2, Enhancer of zeste homolog 2; STAU1,Staufen1; HMGB1, High mobility group box 1; EZH2, Enhancer of zeste homolog 2; G3BP1, GTPase‐activating protein‐binding protein 1; SFPQ, Splicing factor proline/glutamine‐ric; PRSS8, Recombinant protease, serine 8; STAT3, Signal transducer and activator of transcription 3; EMT, Epithelial‐mesenchymal transition; NF‐κB, Nuclear factor‐kappa B; KLF2, Kruppel like factor 2.
Although this evidence shows that the interaction between lncRNA and RBP plays an important role in tumor cell apoptosis, there are many mechanisms of action that are still unknown. Therefore, the current challenge is not only to discover new lncRNAs and RBPs that are involved in cell apoptosis but also to determine the method of interaction and design corresponding drugs for its target of action for treatment. The idea of targeting lncRNAs and RBPs to interact as targets to regulate cancer cell apoptosis has now opened up a challenging field for drug discovery.
4.3. The lncRNA‐RBP interaction network in cancer metastasis
Metastasis is the cause of most cancer‐related deaths. Tumor cells penetrate blood vessels or lymphatic channels, spread from the primary site, and form new tumors (metastases) in distant organs. The metastasis process includes tumor cell infiltration, distant organ infiltration, angiogenesis, and unrestricted growth, and clarification of the molecular participants that regulate this process may provide valuable insights into the treatment of tumor metastasis [66, 95‐97]. Currently, an increasing number of studies have illustrated that some lncRNAs are significantly involved in the regulation of cancer metastasis. Liang et al. [60] found that CASC9 and LAMC2 were significantly upregulated in ESCC tissues and were associated with ESCC prognosis and metastasis. Knockdown of CASC9 can inhibit the migration and invasion of ESCC. The underlying mechanism is that CASC9 specifically binds to CBP and regulates histone acetylation, therefore increasing LAMC2 expression to promote ESCC metastasis. Similarly, one study identified a novel lncRNA, LINC02273, that was stabilized by the protein hnRNPL, the expression of which was elevated in metastatic lesions. Further study revealed that hnRNPL can form a complex with LINC02273 to increase anterior gradient 2 (AGR2) expression at the transcriptional level, leading to enhanced metastasis of breast cancer cells [98]. Because anexelekto (AXL) is overexpressed in a variety of cancer types, AXL has also become a drug target. It has also been reported that context‐based, adaptive, lossless image codec (CALIC) was upregulated in the metastatic subpopulation of HCT116 cells, and the CALIC/hnRNPL complex can significantly affect the metastasis of CRC cells through the regulation of AXL [99]. Current research has revealed that SNHG7 is a potential oncogene whose overexpression could promote the migration and invasion of ovarian cancer cells. Mechanistically, it could inhibit the expression of KLF2 by binding EZH2 [100]. In addition, downregulated RNA in cancer (DRAIC) can combine with ubiquitin C‐Terminal hydrolase L5 (UCHL5), interfere with the ubiquitination of nuclear factor related to kappa‐B (NFRKB), promote the degradation of NFRKB, and suppress the proliferation and metastasis of gastric cancer cells [73].
Furthermore, it was found that lncRNA tumor protein translationally controlled 1 antisense RNA 1 (TPT1‐AS1) binds to nuclear factor 90 (NF90) proteins and enhances the association between NF90 and vascular endothelial growth factor A (VEGFA) mRNA, which upregulates the stability of VEGFA mRNA. Therefore, TPT1‐AS1 enhances CRC angiogenesis and metastasis via the TPT1‐AS1/NF90/VEGFA axis [101]. In NSCLC, the lncRNA HOXA transcript induced by transforming growth factor β (TGFβ) (HIT) acts as a prometastatic oncogene that binds to ZEB1, regulates the stability of ZEB1, and promotes NSCLC cell invasion and migration. HIT acts as a metastatic oncogene by regulating the expression of ZEB1. Additionally, their interaction may provide potential treatment strategies for NSCLC [102]. Another example is LINC01133, which serves as a tumor suppressor. It was reported that LINC01133 could bind to serine and arginine‑rich splicing factor 6 (SRSF6) to inhibit EMT and suppress CRC cell migration and invasion. In addition, the expression of LINC01133 was positively correlated with E‐cadherin and negatively correlated with vimentin. Therefore, LINC01133 could be a novel prognostic biomarker and clinical target for antimetastatic therapies for CRC [103]. The dysregulation of lncRNAs is related to the occurrence of tumors and the progression of cancer, but the mechanism of action of lncRNAs in the occurrence of cancer has not yet been elucidated. Cancer metastasis is the leading cause of death and recurrence of cancer patients. lncRNAs participate in cancer metastasis mainly through interaction with RBPs, epigenetic gene regulation, and participation in the EMT process. Finally, we note that the lncRNA‐RBP interaction may be a very reliable entry point for cancer metastasis. Therefore, more studies are needed to better clarify the function and key mechanism of lncRNAs and RBPs in the progression of cancer metastasis. In the future, the lncRNA‐RBP interaction network will provide a theoretical basis for efficient, specific, and rapid diagnosis and treatment.
4.4. The lncRNA‐RBP interaction network in cancer chemoresistance
Chemotherapy is currently a major cancer therapy. Unfortunately, it is the main reason for treatment failure and indirect promotion of tumor progression. Although the mechanism of tumor drug resistance has been widely explored, it has not been fully characterized. As a hot spot in the field of tumor drug resistance, the lncRNA‐RBP interaction network is closely related to resistance to many anticancer drugs. Therefore, it is very important to link the molecular alterations of cancer with chemical resistance and decipher the potential molecular mechanism to overcome this obstacle [19, 102, 104‐111]. Temozolomide (TMZ) is one of the commonly used first‐line drugs that are widely used clinically in the treatment of gliomas. However, the development of TMZ resistance limits its application. In addition, polypyrimidine tract binding protein 1 (PTBP1) has been reported to modulate TMZ resistance by binding to the lncRNA miR155 host gene (MIR155HG). MIR155H is highly expressed in gliomas and is related to a shorter survival time. In addition, MIR155H binds to PTBP1, affects the expression of PTBP1, and then activates the Wnt/β‐Catenin pathway to endow gliomas with chemical resistance to TMZ [112]. In a study of breast cancer, the overexpression of HOTAIR was associated with the proliferation of breast cancer cells, and its knockdown seriously decreased the survival of breast cancer cells. Furthermore, HOTAIR can enhance the transcriptional activity of estrogen receptor (ER) through direct interaction with estrogen receptors (ERs) and further promote the progression of tamoxifen‐resistant breast cancer [29]. In another study of breast cancer, actin filament‐associated protein 1‐antisense RNA1 (AFAP1‐AS1) was highly expressed in trastuzumab‐resistant cells. In addition, AFAP1‐AS1 could be packaged into exosomes and transmitted to trastuzumab‐sensitive cells through exosomes. Mechanistic studies have shown that AFAP1‐AS1 can bind to the AU‐binding factor 1 (AUF1) protein and enhance the translational activity of erb‐b2 receptor tyrosine kinase 2 (ERBB2) [111].
Sorafenib is the first‐line standard treatment for patients with advanced HCC. However, there is an important problem in the treatment process, namely, resistance to targeted drugs. Therefore, it is important to clarify the underlying mechanism of sorafenib resistance. In addition, sorafenib resistance‐associated lncRNA (SRLR) is overexpressed in sorafenib‐resistant renal cell carcinoma. SRLR could promote sorafenib resistance by directly binding NF‐κB and activating the STAT3 pathway, promoting IL‐6 transcription, which leads to the activation of STAT3 and the occurrence of sorafenib tolerance. This shows that SRLR can be used as a biomarker of inherent sorafenib resistance [113]. Homebox D antisense RNA 1 (HOXD‐AS1) was identified as an lncRNA related to castration‐resistant prostate cancer. HOXD‐AS1 binds WDR5 and activates gene expression by mediating H3K4me3 in the promoter region of the target gene, which inhibits the tumorigenicity and chemoresistance of prostate cancer in vivo. The knockdown of WDR5 eliminates the effect of HOXD‐AS1, indicating that HOXD‐AS1 exerts its regulatory role in a WDR5‐dependent manner [114]. Myeloid differentiation factor 88 (MyD88) is a key linker molecule in the Toll‐like receptor signaling pathway. It is defined as an oncoprotein and plays an important role in information transmission and the growth and metastasis of various tumors, making it a prognostic and therapeutic target [115]. lncRNA arf GAP with GTP‐binding protein‐like domain, ankyrin repeat and PH domain 2‐antisense transcript 1 (AGAP2‐AS1) has been reported to play a carcinogenic role in breast cancer. AGAP2‐AS1 could promote the growth of breast cancer and the resistance to trastuzumab by upregulating the expression of MyD88 and activating the NF‐κB signaling pathway [116]. In summary, these findings reveal potential biomarkers for predicting the response of tumor patients to chemicals and reveal novel tumor therapeutic targets (Table 2). With the progress of drug therapy, the emergence and development of drug resistance seriously affect the prognosis of patients. Increasing evidence shows that lncRNAs play a key role in the progression of various cancers and the resistance to multiple drugs. Because lncRNAs have specific biological functions, especially in terms of regulating gene expression at the epigenetic, transcriptional, and post‐transcriptional levels, the relationship between lncRNAs and tumor cell drug sensitivity has received extensive attention. Understanding the lncRNAs related to tumor drug resistance will promote the development of an understanding of multiple factors that affect drug resistance, which will help break the barriers of tumor drug resistance.
TABLE 2.
The list of lncRNA‐RBP interaction network in cancer metastasis and drug resistance
| lncRNA | RBP | Cancer type | Function* | Mechanism | Reference |
|---|---|---|---|---|---|
| CASC9 | CBP | Esophageal cancer | Metastasis ↑ | Upregulates LAMC2 expression by binding with CBP and modifying histone acetylation | [60] |
| CALIC | hnRNP‐L | Colon cancer | Metastasis ↑ | Induces the expression of AXL by binding hnRNP‐L | [99] |
| SNHG7 | EZH2 | Ovarian cancer | Migration and invasion ↑ | Inhibits the expression of KLF2 by binding EZH2 | [100] |
| DRAIC | UCHL5 | Gastric cancer | Proliferation and metastasis ↓ | Promotes the degradation of NFRKB by attenuating binding of UCHL5 and NFRKB | [73] |
| TPT1‐AS1 | NF90 | Colorectal cancer | Metastasis ↑ | Enhances the association between NF90 and VEGFA mRNA | [101] |
| lncRNA‐HIT | ZEB1 | Non‐small cell lung cancer | Invasion and migration ↑ | Binds to ZEB1 and regulates the stability of ZEB1 | [102] |
| LINC01133 | SRSF6 | Colorectal cancer | Metastasis ↓ | Binds to SRSF6 to inhibit EMT | [103] |
| MIR155H | PTBP1 | Glioma | Endows gliomas with chemical resistance to TMZ | Activates the Wnt/β‐Catenin pathway | [112] |
| HOTAIR | ER | Breast cancer | Contributes to tamoxifen resistance | Promotes ligand‐independent ER activities | [29] |
| AFAP1‐AS1 | AUF1 | Breast cancer | Induce trastuzumab resistance | Enhances the translation activity of ERBB2 | [111] |
| SRLR | NF‐κB | Renal cell carcinoma | Elicits intrinsic sorafenib resistance | Promotes IL‐6 transcription and activates the STAT3 pathway | [113] |
| HOXD‐AS1 | WDR5 | Prostate cancer | Promotes chemo‐resistance | Regulates the expression of target genes by mediating H3K4me3 | [114] |
| AGAP2‐AS1 | CBP | Breast cancer | Promotes the drug resistance of trastuzumab | Up‐regulates the expression of MyD88 and activating NF‐κB signal pathway | [116] |
↑ and ↓ indicate increase and decrease, respectively.
Abbreviations: CBP, CREB binding protein; EZH2, Enhancer of zeste homolog 2; UCHL5, Ubiquitin C‐Terminal hydrolase L5; NF90, Nuclear factor 90; ZEB1, Zinc finger E‐Box binding homeobox 1; SRSF6, Serine and arginine rich splicing factor 6; PTBP1, Polypyrimidine tract binding protein 1; ER, Estrogen receptor; AUF1, AU‐binding factor 1; NF‐κB, Nuclear factor kappa‐B; WDR5, WD repeat domain 5; CBP, CREB binding protein.
4.5. The lncRNA‐RBP interaction network in cancer immunity
The imbalance of immunity in cancer plays an important role in the occurrence and progression of cancer. Besides, cancer cells will create an immunosuppressive environment to increase survival rate [117]. The association of the lncRNA‐RBP network with cancer immunity has been confirmed. For example, lncRNAs have been shown to affect cancer immunity by helping tumors evade immune surveillance. These findings may provide novel insights into cancer immunotherapy. LncRNA‐CCAAT enhancer binding protein beta (Lnc‐C/EBPβ) is a lncRNA that can negatively regulate cell immunosuppression. It has been reported that lnc‐C/ EBPβ binds to the c/ EBPβ protein to affect the activity of c/EBPβ, inhibiting the immunosuppression of myeloid‐derived suppressor cells. Lnc‐C/EBPβ upregulation increases IFN‐γ production by infiltrating T cells, which inhibits immunoregulatory enzyme secretion [118]. Another finding demonstrated that NKILA, an NF‐κB‐binding lncRNA, can interact specifically with NF‐κB and regulate the sensitivity of T cells to tumor‐mediated activation‐induced cell death (AICD) by inhibiting NF‐κB activity in the breast and lung cancer microenvironments. However, the elimination of this interaction will in turn inhibit breast cancer cell death. Thus, inhibiting NKILA may improve cancer immunotherapy effects by providing a novel strategy with which to protect transferred T cells from tumor‐mediated AICD [119].
Furthermore, a study revealed that lnc‐Tim3 (ENST00000443947.1) plays a vital role in CD8+ T cell exhaustion. Mechanistically, lnc‐Tim3 can specifically bind to the intracellular tail of Tim‐3 and then inhibit the downstream pathway, leading to T cell exhaustion, which is associated with compromised antitumor immunity [120]. The RNA sensor retinoic acid‐inducible gene I (RIG‐1) has been reported to recognize “nonself” viral RNAs and induce an immune response by producing IFNs [121]. A study showed that lncRNA‐sm‐like 3b (lnc‐Lsm3b) could bind to RIG‐1, inhibiting the conformational shift of RIG‐1 and suppressing the downstream signaling pathway, therefore inhibiting the production of type I IFNs and maintaining immune homeostasis [120]. lncRNA SATB2‐AS1 is reported to control T helper type 1 (Th1)‐type chemokine production and immune cell density in CRC. Mechanistically, SATB2‐AS1 exerts its function by directly binding to WDR5 and GADD45A, cis‐activating SATB2 [122]. In conclusion, lncRNAs act as important regulators in cancer and may play considerable roles in cancer immune regulation.
4.6. The lncRNA‐RBP interaction network in the tumor microenvironment
The tumor microenvironment is the local living environment around tumor cells and is composed of immune cells, endothelial cells, fibroblasts, extracellular matrix, and signaling molecules surrounding tumor cells. The tumor microenvironment is a protective layer of tumor cells that can help cancer cells evade the immune system and can promote tumor growth and the angiogenesis and metastasis of tumor cells [123, 124, 125, 126]. The lncRNA‐RBP interaction network can mediate cell‐to‐cell communication in the tumor microenvironment and play an important role in tumor growth and progression [127]. Studies have shown that tumor cells can secrete lncRNAs in exosomes, and these exogenous lncRNAs can be taken up by nearby or distant cells through endocytosis and regulate their functions [128]. Numerous studies have reported the abnormal expression of exogenous lncRNAs in cancers. Additionally, this may be related to a favorable tumor microenvironment. These findings indicate that lncRNAs have potential in the development of biomarkers and therapeutic targets [119, 129, 130].
lncRNA mitochondrial ribosomal protein L23‐antisense RNA 1 (MRPL23‐AS1) is highly expressed in exosomes secreted by adenoid cystic carcinoma cells. Furthermore, MRPL23‐AS1 forms a complex with the EZH2 protein, and the nucleic acid‐protein complex increases the H3K27me3 promoter region of E‐cadherin, which results in the initiation of EMT. In addition, exogenous MRPL23‐AS1 can promote the microvascular permeability of pulmonary microvascular endothelial cells. In conclusion, salivary adenoid cystic carcinoma (SACC) patients are more prone to lung metastasis and reduced overall survival under the action of exosomal MRPL23‐AS1 [130].
Lymph node metastasis‐associated transcript 2 (LNMAT2) is a lncRNA that interacts with hnRNPA2B1 in bladder cancer cells. Under the action of hnRNPA2B1, LNMAT2 can be specifically packaged into exosomes and delivered to human lymphatic endothelial cells (HLECs). In HLECs, LNMAT2 interacts with the promoter of prospero homeobox 1 (PROX1) and integrates to form a tricomplex. Prox1 transcription is enhanced by H3K4me3 epigenetic modification mediated by HRNPA2B1 [131]. Therefore, LNMAT2 mediates lymphangiogenesis and lymph node metastasis in bladder cancer and plays an important role in the tumor microenvironment. Wu et al. [132] found that LINC00941 can interact with small mother against decapentaplegic family member 4 (SMAD4) protein to enhance the stability of SMAD4 protein by prolonging its degradation half‐life, activating the TGF‐β/SMAD2/3 signaling pathway, and promoting CRC metastasis. However, the removal of this interaction will also reduce the transferability of CRC. In summary, the lncRNA‐RBP interaction network is a crucial component of the tumor microenvironment.
4.7. The lncRNA‐RBP interaction network in cancer metabolism
An increasing number of studies have reported that the lncRNA‐RBP network plays an important role in a variety of tumors. Although the roles of the lncRNA‐RBP network in regulating gene expression has been widely studied, the role of this network in metabolism remains unclear.
Peroxisome proliferator‐activated receptor γ (PPARγ), a member of the PPAR family, plays an important regulatory role in tumor metabolism [133]. Studies have shown that ectopic expression of PPARγ inhibits the Wnt/β‐catenin pathway, which in turn downregulates pyruvate dehydrogenase kinase 1 (PDK1), thereby inhibiting the glycolysis process in tumor cells [134]. lncRNA five prime to XIST (FTX) has also been shown to promote glucose consumption through a glucose transporter GLUT and inhibit the expression of tumor necrosis factor α (TNFα) and leptin in HCC cells by targeting PPARγ [135]. In addition, lncRNA FTX promoted glycolysis in HCC cells by directly targeting PPARγ, thereby increasing the activity and expression of glycolytic enzymes, including lactate dehydrogenase (LDH) and phosphofructokinase, liver type (PFKL). Subsequently, the molecules associated with the tricarboxylic acid cycle (TNFα, leptin, and PDK1). In contrast, studies have shown that if the interaction network between lncRNA FTX and PPARγ is inhibited, glycolysis in HCC cells will be inhibited.
Meanwhile, studies have shown that lncRNA WAP four‐disulfide core domain 21, pseudogene (WFDC21p) is also involved in glucose metabolism in tumor cells. lncRNA WFDC21p reduces glycolysis by decreasing the expression and activity of human platelet‐type phosphofructokinase and PKM2 [136]. FoxO‐induced long non‐coding RNA 1 (FILNC1) is a forkhead box O (FoxO)‐induced lncRNA that has been found in renal cancer cells. It has been reported that a lack of FILNC1 can upregulate c‐myc, leading to increased glucose uptake and lactic acid production. In the absence of energy, FILNC1 interacts with ARE/poly(U)‐binding/degradation factor 1 (AUF1), a c‐myc mRNA‐binding protein, separating AUF1 from c‐myc mRNA‐binding, resulting in the downregulation of c‐myc protein [137]. This downregulation can be restored with the destruction of the FILNC1‐AUF1 interaction network, indicating that FILNC1 is a negative regulator of renal cancer, which has value in research applications, and demonstrating a regulatory mechanism of lncRNAs controlling energy metabolism. Similarly, LINC01554 could accelerate the degradation of PKM2 while reducing the rate of glycolysis [138]. The lncRNA FTX participates in glycolysis in HCC by enhancing the activity and expression of PFKL [135].
5. CANCER THERAPIES TARGETING THE LNCRNA‐RBP INTERACTION NETWORK
Many studies have shown that the signaling pathways in cancer activities are strictly regulated by lncRNAs, so lncRNAs are important targets for cancer treatment. In recent years, some studies have shown that the “sponge” molecule of lncRNAs has some anticancer activity in both in vivo and in vitro experiments. Experimental methods were used on clear cell renal carcinoma cell lines with high miR‐21 expression. It was found that overexpression of phosphatase and tensin homolog pseudogene 1 (PTENP1) reduces the effective concentration of miR‐21 and thereby inhibits the proliferation and invasion of tumor cells [139]. Similarly, Zhang et al. [140] found that lncRNA‐GAS5 Exon 4 can interact with miR‐21 to inhibit the appearance and development of breast cancer cells [141]. In addition, papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) acts as a tumor suppressor gene in thyroid cancer cell lines, and with the interaction improving, seine expression is significantly reduced. Under the experimental conditions, when PTCSC3 was overexpressed, the expression of the tumor‐promoting miRNA miR‐574‐5p was significantly reduced in thyroid cancer cell lines. The above experimental results and bioinformatics analyses show that PTCSC3 interacts with miR‐574‐5p and that other specific cancer‐promoting miRNAs have a tumor suppressor effect. In addition, competing endogenous RNA (ceRNA) acts as a tumor suppressor or clinical treatment by changing the expression of the corresponding ceRNA. For example, human epidermal growth factor receptor 2 (HER2) is an important prognostic factor for breast cancer [142]. There are currently clinically targeted therapeutics for HER2‐positive (overexpression or amplification) patients, such as trastuzumab [30]. Recent studies have shown that HOTAIR can promote HER2 expression through competitive binding of miR‐331‐3p. Therefore, the expression of HOTAIR can theoretically be achieved through interaction with HER2. Silencing can be reduced by the mode of action of ceRNA to power the gene, creating a new platform for potential RNA‐based tumor therapy applications, reducing the binding between lncRNAs and RBPs and reducing the interaction between them. Currently, a variety of targeted treatments for lncRNAs have been developed, such as anti‐sense oligonucleotides (ASOs), liposome/nanoparticle‐delivered siRNAs, and small‐molecule inhibitors of lncRNAs [143, 144].
5.1. ASOs
ASOs are DNA molecule analogs that can specifically bind to lncRNA transcripts through base complementation, thereby inhibiting gene expression and achieving regulation at the gene level. ASOs include ASO gapmers [145], double‐stranded DNA, and locked nucleic acids (LNAs) [146]. An LNA is an oligonucleotide derivative that is formed from the 2'‐O,4'‐C position of β‐D‐ribofuranose via a dehydration condensation reaction. It also has six bases containing A, C, G, T, U, and mC, which are often used to modify RNA nucleotides [147, 148]. In cell line and animal model experiments, scientists have discovered that mixed LNA‐DNA‐LNA gapmers can specifically bind to RNA and achieve RNA silencing [149]. This feature provides a theoretical basis for potential lncRNA‐targeted therapy; specifically, ASOs can be used to knock down lncRNAs related to tumor development in vivo, thereby inhibiting tumor growth. Some similar experiments have been performed in various cancer models. Leucci et al. [150] confirmed the successful silencing of survival associated mitochondrial melanoma specific oncogenic non‐coding RNA (SAMMSON), a melanoma‐specific lncRNA that has been replicated or amplified in approximately 10% of cases) by using LNA‐modified ASOs in vitro and in vivo. It triggers RNase‐H‐mediated degradation of the target, thereby inhibiting the growth of melanoma cells. According to the study by Iden et al. [151], knocking down plasmacytoma variant translocation 1 (PVT1) with LNA can significantly increase the sensitivity of ovarian cancer cells (SiHa) to cisplatin. In addition, Qu et al. [152] found that lncRNA regulator of AKT signaling associated with HCC and RCC (lncARSR) is a therapeutic target that can overcome the resistance of renal cancer to sunitinib. Specifically, lncARSR can promote the resistance of renal cancer patients to sunitinib, and using LNA to target lncARSR will re‐establish the sensitivity of the tumor to sunitinib. Arun et al. [153] indicated that using ASO to knock out MALAT1 in breast cancer mice can lead tumor growth to be slowed and accompanied by a reduction in the differentiation and metastasis of tumor cells. Correspondingly, antagonists (ASOs that target antisense lncRNAs) are used to upregulate specific mRNAs/proteins by silencing the corresponding antisense lncRNAs. Inhibition/perturbation of endogenous NATs by modified oligonucleotides (AntagoNATs) are modified not only at their 5' and 3' ends but also in their backbone to make them more stable and enhance their cellular uptake. Therefore, ASOs have advantages over siRNAs that are generally unstable and have difficulty targeting tumor cells in vivo. It is worth noting that in the mouse xenograft model, despite the promise of this system, cellular uptake and cytotoxicity are still concerns related to ASOs. Currently, there are many other clinical trials on ASOs targeting lncRNAs in cancer therapy, which are still being evaluated and show good prospects.
5.2. Small‐molecule inhibitors
lncRNAs are complex tertiary structures, and there are some conserved regions between the secondary and tertiary structures that constitute lncRNAs. It is speculated that these conserved regions are related to lncRNA‐binding proteins (as an example, the stem loop for protein binding); however, currently, the study of these regions is limited [154]. Research on the molecular structure of lncRNAs related to cancer can promote the discovery of small‐molecule compounds that can be targeted to inhibit lncRNAs. Currently, scientists have tried to identify small‐molecule compounds that can inhibit RNA with small‐molecule microarray screening and dynamic combinatorial screening [155]. For instance, small‐molecule compounds can inhibit the lncRNA‐RBP interactions by targeting action regions. Blocking certain sequences of HOTAIR binding to PRC2 or LSD1 with small‐molecule inhibitors and the interaction between PRC2 and LSD1 can inhibit breast cancer cell metastasis [156, 157]. Alternatively, inhibitors that can bind to lncRNAs can change the structure of lncRNAs and prevent them from binding to target molecules. These strategies will be verified in further clinical trials.
5.3. Nanoparticle‐delivered siRNAs
siRNAs are novel tools for cancer treatment. They can bind to target genes and subsequently lead to gene silencing. Since siRNAs are easily degraded and not easily absorbed by cells, natural nanoparticles composed of lipids, polymers, and metals are required for siRNA therapy. Delivery of ASOs and siRNAs to tumors is still challenging because they are unstable in plasma and have negative charges and high molecular weights [158]. Therefore, scientists have developed many different types of nanoparticles to better facilitate the delivery of small‐molecule inhibitors. For example, research on the application of new oligonucleotide aptamers has become a hot topic [159]. The current research has proven that using aptamers to mediate siRNA delivery into the cell leads to silencing of the corresponding gene to achieve a therapeutic effect. This carrier can accumulate near tumor cells under the attraction of tumor surface antigens. The aptamer carrying siRNA or ASO can specifically recognize and bind to the protein on the target cell membrane, enter tumor cells through chimerization or endocytosis, and further release small‐molecule drugs, siRNAs, and ASOs [160, 161]. The entry of specific siRNAs into tumor cells will inhibit the expression of the target mRNA [162]. siRNAs targeting lncRNAs can also be encapsulated in nanoparticles to improve the tissue distribution and efficacy of anticancer drugs. Rupaimoole et al. [163] used dipalmitoylphosphatidylcholine nanoparticles containing siRNA targeting lncRNA ceruloplasmin (NRCP) in ovarian cancer‐bearing mice. They found that this strategy can indeed lead to the silencing of the target lncRNA and can further reduce the growth of ovarian cancer cells and increase the sensitivity of cancer cells to cisplatin. lncRNA HOTAIR is upregulated and can be used as a biomarker for the diagnosis and prognosis of breast, liver, stomach, pancreatic, lung, prostate, cervical, and colon cancers. In breast, hepatocellular, and pancreatic cancers, siRNA downregulation of HOTAIR expression is associated with reduced tumor cell viability, reduced invasiveness, and increased cell apoptosis. In addition, disabling HOTAIR may increase the sensitivity of tumor cells to a TNFα‐based immune response and may increase the sensitivity of chemotherapeutic agents such as cisplatin and doxorubicin [164]. The expression of PCA3 in prostate cancer is highly upregulated and is an effective biomarker that can be detected in urine [165]. siRNA‐mediated downregulation of PCA3 can significantly suppress the growth and proliferation of prostate cancer cells and reduce the expression of AR target genes, indicating a potential targeted therapy. lncRNAs prostate cancer associated transcript 1 (PCAT1), prostate cancer associated non‐coding RNA 1 (PRNCR1), prostate cancer gene expression marker 1 (PCGEM1), prostate cancer‐up‐regulated long noncoding RNA 1 (PlncRNA1) [166], and PCAT18 are all upregulated in advanced prostate cancer and are considered specific biomarkers and therapeutic targets. In prostate cancer cell lines, silencing siRNA/shRNA‐based lncRNAs can inhibit cell proliferation and induce cell apoptosis by reducing AR expression [165]. lncRNAs H19, HULC, hepatocellular carcinoma up‐regulated EZH2‐associated long non‐coding RNA (HEIH), and microvascular invasion in hepatocellular carcinoma (MVIH) are strongly upregulated in HCC and are biomarkers of the same value. Silencing of these transcripts mediated by siRNA/shRNA results in altered expression of some genes and reduced tumor growth in xenografts, indicating that they are potential therapeutic targets.
Additionally, to reduce the risks of treatment, some scientists have made full use of the high specificity of certain lncRNAs in tumor cells. For example, plasmid BC‐819 (DTA‐19) exploits the characteristic that lncRNA H19 is expressed only in tumors. This plasmid was cloned into the diphtheria toxin A subunit gene under the H19 promoter. After intratumoral injection of the plasmid, its expression product shrunk the tumor without affecting other normal tissues [167].
5.4. Plasmid‐based therapy
In this new treatment method, scientists have developed the plasmid BC‐819/DTAH19 system, which carries the diphtheria toxin subunit regulated by the H19 promoter. When the plasmid is injected into a tumor, it produces high levels of diphtheria toxin in human bladder cancer research, causing the tumor to shrink [167]. This method is usually used to shrink tumors rather than to target specific lncRNAs. It is now showing promising results in the treatment of other tumors, such as lung, colon, pancreatic, and ovarian cancers.
6. CONCLUSIONS
In the past few decades, numerous insights into the lncRNA‐RBP interaction network have revealed complex relationships and mechanisms, including the functions of lncRNAs in regulating RBPs, the roles of RBPs in regulating lncRNAs, and the roles of the lncRNA‐RBP interaction network in cancer, increasing the understanding of the occurrence and development of different types of human cancers. Exploring the mechanism of cancer development and finding effective methods for treating cancer have been the focus of modern tumor molecular biology research. Currently, the exploration of the lncRNA‐RBP interactions has become a new hotspot in tumor biology. In particular, the lncRNA‐RBP interaction network provides a new theoretical framework for researching the proliferation, metastasis, migration, and apoptosis mechanisms of cancer cells and a new strategy for researching corresponding anticancer drugs. Currently, the most important task is to find new lncRNA‐binding proteins and to study the interaction mechanisms between lncRNAs and RBPs at the levels of structure and function as well as RNA modification and protein PTM (a newly discovered role of RBPs). More importantly, the role of the lncRNA‐RBP interaction network in the development of cancer and its role in regulating gene expression in cancer cells have been explored by various means, such as RNA immunoprecipitation and crosslinking immunoprecipitation.
lncRNA and RBP therapy has been the focus of research in the past ten years; however, no lncRNA‐targeted therapy has entered clinical development. As a biomarker, researchers are actively looking for the universal correlation of lncRNAs and RBPs with disease. In the future, lncRNAs are expected to broaden the number of RNA interference (RNAi) and CRISPR targets. Some unique lncRNA therapies, such as circular RNA and natural antisense transcripts, represent cutting‐edge methods of RNA therapy.
All processes of RNA‐based therapy entering the clinic are hampered by issues related to specificity, administration methods, and immunogenicity. Specific problems include off‐target effects caused by uptake of drugs by cells other than the target cells, off‐target effects caused by similar sequences or excessively high levels of endogenous protein expression. The transport problem is related to three main issues: the structural instability of RNA without chemical modification, the low intracellular transport efficiency of RNA using the endosomal escape mechanism, and the lack of organs and cell types suitable for targeted concentration. For the reasons mentioned above, clinical trials are often discontinued due to a lack of effectiveness.
In addition, tolerance problems (immunogenic problems) are caused by pathogen‐associated molecular pattern receptors that recognize RNA structures, such as Toll‐like receptors, which can cause immune reactions. We believe that this approach maybe give helps to further explore cancer treatment strategies.
DECLARATIONS
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
COMPETING INTERESTS
The authors declare that they have no competing interests.
FUNDING
This work was supported by the National Key Research and Development Program of China (2021YFC2501000 and 2017YFA0505100) and the National Natural Science Foundation of China (31961160727, 81973339, and 81773085).
AUTHORS' CONTRIBUTIONS
Z.T.Y., Y.M.Y., M.M.S., Y.H., and L.L. wrote the manuscript. B.L. and K.S.C. designed and revised the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
Not applicable.
Yao Z‐T, Yang Y‐M, Sun M‐M, He Y, Liao L, Chen K‐S, et al. New insights into the interplay between long non‐coding RNAs and RNA‐binding proteins in cancer. Cancer Commun. 2022;42:117–140. 10.1002/cac2.12254
Contributor Information
Kui‐Sheng Chen, Email: chenksh2002@163.com.
Bin Li, Email: lib2128@163.com.
REFERENCES
- 1. Slack FJ. Regulatory RNAs and the demise of ‘junk’ DNA. Genome Biol. 2006;7(9):328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, et al. Ab initio reconstruction of cell type‐specific transcriptomes in mouse reveals the conserved multi‐exonic structure of lincRNAs. Nat Biotechnol. 2010;28(5):503‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon‐Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Slack FJ, Chinnaiyan AM. The Role of Non‐coding RNAs in Oncology. Cell. 2019;179(5):1033‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145‐66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, et al. The human XIST gene: analysis of a 17 kb inactive X‐specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71(3):527‐42. [DOI] [PubMed] [Google Scholar]
- 8. Feil R, Walter J, Allen ND, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994;120(10):2933‐43. [DOI] [PubMed] [Google Scholar]
- 9. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097‐109. [DOI] [PubMed] [Google Scholar]
- 10. Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA‐mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). 2021;41(2):109‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Köster T, Marondedze C, Meyer K, Staiger D. RNA‐Binding Proteins Revisited ‐ The Emerging Arabidopsis mRNA Interactome. Trends Plant Sci. 2017;22(6):512‐26. [DOI] [PubMed] [Google Scholar]
- 12. Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA‐binding proteins. Nat Rev Mol Cell Biol. 2018;19(5):327‐41. [DOI] [PubMed] [Google Scholar]
- 13. Peng WX, Koirala P, Mo YY. LncRNA‐mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hudson WH, Ortlund EA. The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Biol. 2014;15(11):749‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cech TR, Steitz JA. The noncoding RNA revolution‐trashing old rules to forge new ones. Cell. 2014;157(1):77‐94. [DOI] [PubMed] [Google Scholar]
- 16. Ferrè F, Colantoni A, Helmer‐Citterich M. Revealing protein‐lncRNA interaction. Brief Bioinform. 2016;17(1):106‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43(6):904‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell‐cycle promoters. Nat Genet. 2011;43(7):621‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29(4):452‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non‐coding RNA in cancer. Cancer Sci. 2018;109(7):2093‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106(8):2525‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35(4):467‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717‐26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, et al. Long noncoding RNA NEAT1‐dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol Cell. 2014;53(3):393‐406. [DOI] [PubMed] [Google Scholar]
- 25. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin‐modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6(7):784‐801. [DOI] [PubMed] [Google Scholar]
- 27. Kim J, Abdelmohsen K, Yang X, De S, Grammatikakis I, Noh JH, et al. LncRNA OIP5‐AS1/cyrano sponges RNA‐binding protein HuR. Nucleic Acids Res. 2016;44(5):2378‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ng SY, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51(3):349‐59. [DOI] [PubMed] [Google Scholar]
- 29. Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35(21):2746‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR‐331‐3p in gastric cancer. Mol Cancer. 2014;13:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang A, Zhao JC, Kim J, Fong KW, Yang YA, Chakravarti D, et al. LncRNA HOTAIR Enhances the Androgen‐Receptor‐Mediated Transcriptional Program and Drives Castration‐Resistant Prostate Cancer. Cell Rep. 2015;13(1):209‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luo ML, Li J, Shen L, Chu J, Guo Q, Liang G, et al. The Role of APAL/ST8SIA6‐AS1 lncRNA in PLK1 Activation and Mitotic Catastrophe of Tumor Cells. J Natl Cancer Inst. 2020;112(4):356‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y, et al. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA‐MUF Interaction with ANXA2 and miR‐34a. Cancer Res. 2017;77(23):6704‐16. [DOI] [PubMed] [Google Scholar]
- 34. Duan G, Walther D. The roles of post‐translational modifications in the context of protein interaction networks. PLoS Comput Biol. 2015;11(2):e1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ji J, Xu R, Ding K, Bao G, Zhang X, Huang B, et al. Long Noncoding RNA SChLAP1 Forms a Growth‐Promoting Complex with HNRNPL in Human Glioblastoma through Stabilization of ACTN4 and Activation of NF‐κB Signaling. Clin Cancer Res. 2019;25(22):6868‐81. [DOI] [PubMed] [Google Scholar]
- 36. Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, et al. MALAT‐1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene. 2003;22(39):8031‐41. [DOI] [PubMed] [Google Scholar]
- 37. Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non‐coding RNA in cancer. Biochim Biophys Acta. 2016;1859(1):192‐9. [DOI] [PubMed] [Google Scholar]
- 38. Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26(6):851‐8. [DOI] [PubMed] [Google Scholar]
- 39. Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y, et al. LncRNA‐p21 alters the antiandrogen enzalutamide‐induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun. 2019;10(1):2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A cytoplasmic NF‐κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370‐81. [DOI] [PubMed] [Google Scholar]
- 41. Li Z, Hou P, Fan D, Dong M, Ma M, Li H, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017;24(1):59‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga‐Yamanaka K, et al. Scaffold function of long non‐coding RNA HOTAIR in protein ubiquitination. Nat Commun. 2013;4:2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang H, Weng H, Chen J. m6A Modification in Coding and Non‐coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37(3):270‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patil DP, Pickering BF, Jaffrey SR. Reading m6A in the Transcriptome: m6A‐Binding Proteins. Trends Cell Biol. 2018;28(2):113‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lan Q, Liu PY, Haase J, Bell JL, Hüttelmaier S, Liu T. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res. 2019;79(7):1285‐92. [DOI] [PubMed] [Google Scholar]
- 46. Gerstberger S, Hafner M, Tuschl T. A census of human RNA‐binding proteins. Nat Rev Genet. 2014;15(12):829‐45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang S, Xie Z, Wang P, Li J, Wang S, Liu W, et al. LncRNA‐OG Promotes the Osteogenic Differentiation of Bone Marrow‐Derived Mesenchymal Stem Cells Under the Regulation of hnRNPK. Stem Cells. 2019;37(2):270‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Castellanos‐Rubio A, Fernandez‐Jimenez N, Kratchmarov R, Luo X, Bhagat G, Green PH, et al. A long noncoding RNA associated with susceptibility to celiac disease. Science. 2016;352(6281):91‐5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang L, Wen M, Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science. 2019;365(6454). [DOI] [PubMed] [Google Scholar]
- 50. Quandt D, Fiedler E, Boettcher D, Marsch W, Seliger B. B7‐h4 expression in human melanoma: its association with patients' survival and antitumor immune response. Clin Cancer Res. 2011;17(10):3100‐11. [DOI] [PubMed] [Google Scholar]
- 51. Tan X, Chen WB, Lv DJ, Yang TW, Wu KH, Zou LB, et al. LncRNA SNHG1 and RNA binding protein hnRNPL form a complex and coregulate CDH1 to boost the growth and metastasis of prostate cancer. Cell Death Dis. 2021;12(2):138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tafforeau L, Zorbas C, Langhendries J‐L, Mullineux S‐T, Stamatopoulou V, Mullier R, et al. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre‐rRNA processing factors. Molecular cell. 2013;51(4):539‐51. [DOI] [PubMed] [Google Scholar]
- 53. Dossin F, Pinheiro I, Żylicz JJ, Roensch J, Collombet S, Le Saux A, et al. SPEN integrates transcriptional and epigenetic control of X‐inactivation. Nature. 2020;578(7795):455‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen F, Chen J, Yang L, Liu J, Zhang X, Zhang Y, et al. Extracellular vesicle‐packaged HIF‐1α‐stabilizing lncRNA from tumour‐associated macrophages regulates aerobic glycolysis of breast cancer cells. Nature cell biology. 2019;21(4):498‐510. [DOI] [PubMed] [Google Scholar]
- 55. Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175(7):1872‐86. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zuo X, Chen Z, Gao W, Zhang Y, Wang J, Wang J, et al. M6A‐mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Journal of hematology & oncology. 2020;13(1):1‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F, et al. m6A‐induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Molecular cancer. 2019;18(1):1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Patil D, Chen C, Pickering B, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST‐mediated transcriptional repression. Nature. 2016;537(7620):369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hu X, Peng W‐X, Zhou H, Jiang J, Zhou X, Huang D, et al. IGF2BP2 regulates DANCR by serving as an N6‐methyladenosine reader. Cell Death & Differentiation. 2020;27(6):1782‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang K, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB‐binding protein. Cell Death Differ. 2018;25(11):1980‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hämmerle M, Gutschner T, Uckelmann H, Ozgur S, Fiskin E, Gross M, et al. Posttranscriptional destabilization of the liver‐specific long noncoding RNA HULC by the IGF2 mRNA‐binding protein 1 (IGF2BP1). Hepatology. 2013;58(5):1703‐12. [DOI] [PubMed] [Google Scholar]
- 62. Zhang X, Zhou Y, Chen S, Li W, Chen W, Gu W. LncRNA MACC1‐AS1 sponges multiple miRNAs and RNA‐binding protein PTBP1. Oncogenesis. 2019;8(12):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA‐p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6‐methyladenosine‐dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lian Y, Yan C, Xu H, Yang J, Yu Y, Zhou J, et al. A Novel lncRNA, LINC00460, Affects Cell Proliferation and Apoptosis by Regulating KLF2 and CUL4A Expression in Colorectal Cancer. Mol Ther Nucleic Acids. 2018;12:684‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu WW, Huang ZH, Liao L, Zhang QH, Li JQ, Zheng CC, et al. Direct Targeting of CREB1 with Imperatorin Inhibits TGFβ2‐ERK Signaling to Suppress Esophageal Cancer Metastasis. Adv Sci (Weinh). 2020;7(16):2000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST‐mediated transcriptional repression. Nature. 2016;537(7620):369‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann‐Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, et al. LncRNA loc285194 is a p53‐regulated tumor suppressor. Nucleic Acids Res. 2013;41(9):4976‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016;30(10):1224‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nguyen TM, Kabotyanski EB, Reineke LC, Shao J, Xiong F, Lee JH, et al. The SINEB1 element in the long non‐coding RNA Malat1 is necessary for TDP‐43 proteostasis. Nucleic Acids Res. 2020;48(5):2621‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang X, Lai Q, He J, Li Q, Ding J, Lan Z, et al. LncRNA SNHG6 promotes proliferation, invasion and migration in colorectal cancer cells by activating TGF‐β/Smad signaling pathway via targeting UPF1 and inducing EMT via regulation of ZEB1. Int J Med Sci. 2019;16(1):51‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhang Z, Hu X, Kuang J, Liao J, Yuan Q. LncRNA DRAIC inhibits proliferation and metastasis of gastric cancer cells through interfering with NFRKB deubiquitination mediated by UCHL5. Cell Mol Biol Lett. 2020;25:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Engreitz JM, Ollikainen N, Guttman M. Long non‐coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17(12):756‐70. [DOI] [PubMed] [Google Scholar]
- 75. Hu HF, Xu WW, Li YJ, He Y, Zhang WX, Liao L, et al. Anti‐allergic drug azelastine suppresses colon tumorigenesis by directly targeting ARF1 to inhibit IQGAP1‐ERK‐Drp1‐mediated mitochondrial fission. Theranostics. 2021;11(4):1828‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Evan GI, Vousden KH. Proliferation, cell cycle and apoptosis in cancer. Nature. 2001;411(6835):342‐8. [DOI] [PubMed] [Google Scholar]
- 77. Lan Y, Xiao X, He Z, Luo Y, Wu C, Li L, et al. Long noncoding RNA OCC‐1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46(11):5809‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. He X, Chai P, Li F, Zhang L, Zhou C, Yuan X, et al. A novel LncRNA transcript, RBAT1, accelerates tumorigenesis through interacting with HNRNPL and cis‐activating E2F3. Mol Cancer. 2020;19(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Klingenberg M, Groß M, Goyal A, Polycarpou‐Schwarz M, Miersch T, Ernst AS, et al. The Long Noncoding RNA Cancer Susceptibility 9 and RNA Binding Protein Heterogeneous Nuclear Ribonucleoprotein L Form a Complex and Coregulate Genes Linked to AKT Signaling. Hepatology. 2018;68(5):1817‐32. [DOI] [PubMed] [Google Scholar]
- 80. Sun M, Nie F, Wang Y, Zhang Z, Hou J, He D, et al. LncRNA HOXA11‐AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76(21):6299‐310. [DOI] [PubMed] [Google Scholar]
- 81. Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, et al. LncRNA‐FEZF1‐AS1 Promotes Tumor Proliferation and Metastasis in Colorectal Cancer by Regulating PKM2 Signaling. Clin Cancer Res. 2018;24(19):4808‐19. [DOI] [PubMed] [Google Scholar]
- 82. Wang X, Yu H, Sun W, Kong J, Zhang L, Tang J, et al. The long non‐coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68. Mol Cancer. 2018;17(1):110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, et al. Long noncoding RNA ANRIL promotes non‐small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14(1):268‐77. [DOI] [PubMed] [Google Scholar]
- 84. Wong RS. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hong P, Liu QW, Xie Y, Zhang QH, Liao L, He QY, et al. Echinatin suppresses esophageal cancer tumor growth and invasion through inducing AKT/mTOR‐dependent autophagy and apoptosis. Cell Death Dis. 2020;11(7):524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Elmore S. Apoptosis: a review of programmed cell death. Toxicologic pathology. 2007;35(4):495‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. John C, Reed J. Mechanisms of apoptosis. Am J Pathol. 2000;157:1415‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Carson DA. Ribeiro JM. Apoptosis and disease. Lancet. 1993;341(8855):1251‐4. [DOI] [PubMed] [Google Scholar]
- 89. Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM, et al. SP1‐induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34(45):5648‐61. [DOI] [PubMed] [Google Scholar]
- 90. Liao XH, Wang JG, Li LY, Zhou DM, Ren KH, Jin YT, et al. Long intergenic non‐coding RNA APOC1P1‐3 inhibits apoptosis by decreasing α‐tubulin acetylation in breast cancer. Cell Death Dis. 2016;7(5):e2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gao D, Lv AE, Li HP, Han DH, Zhang YP. LncRNA MALAT‐1 Elevates HMGB1 to Promote Autophagy Resulting in Inhibition of Tumor Cell Apoptosis in Multiple Myeloma. J Cell Biochem. 2017;118(10):3341‐8. [DOI] [PubMed] [Google Scholar]
- 92. Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, et al. A G3BP1‐Interacting lncRNA Promotes Ferroptosis and Apoptosis in Cancer via Nuclear Sequestration of p53. Cancer Res. 2018;78(13):3484‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25(1):169‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zeng C, Liu S, Lu S, Yu X, Lai J, Wu Y, et al. The c‐Myc‐regulated lncRNA NEAT1 and paraspeckles modulate imatinib‐induced apoptosis in CML cells. Mol Cancer. 2018;17(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2(8):563‐72. [DOI] [PubMed] [Google Scholar]
- 96. Kim JJ, Diamond DM. The stressed hippocampus, synaptic plasticity and lost memories. Nature Reviews Neuroscience. 2002;3(6):453. [DOI] [PubMed] [Google Scholar]
- 97. Hu HF, Xu WW, Zhang WX, Yan X, Li YJ, Li B, et al. Identification of miR‐515‐3p and its targets, vimentin and MMP3, as a key regulatory mechanism in esophageal cancer metastasis: functional and clinical significance. Signal Transduct Target Ther. 2020;5(1):271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xiu B, Chi Y, Liu L, Chi W, Zhang Q, Chen J, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18(1):187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kawasaki Y, Miyamoto M, Oda T, Matsumura K, Negishi L, Nakato R, et al. The novel lncRNA CALIC upregulates AXL to promote colon cancer metastasis. EMBO Rep. 2019;20(8):e47052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bai Z, Wu Y, Bai S, Yan Y, Kang H, Ma W, et al. Long non‐coding RNA SNGH7 Is activated by SP1 and exerts oncogenic properties by interacting with EZH2 in ovarian cancer. J Cell Mol Med. 2020;24(13):7479‐89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang Y, Sun J, Qi Y, Wang Y, Ding Y, Wang K, et al. Long non‐coding RNA TPT1‐AS1 promotes angiogenesis and metastasis of colorectal cancer through TPT1‐AS1/NF90/VEGFA signaling pathway. Aging (Albany NY). 2020;12(7):6191‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jia X, Wang Z, Qiu L, Yang Y, Wang Y, Chen Z, et al. Upregulation of LncRNA‐HIT promotes migration and invasion of non‐small cell lung cancer cells by association with ZEB1. Cancer Med. 2016;5(12):3555‐63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kong J, Sun W, Li C, Wan L, Wang S, Wu Y, et al. Long non‐coding RNA LINC01133 inhibits epithelial‐mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016;380(2):476‐84. [DOI] [PubMed] [Google Scholar]
- 104. Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, et al. An Integrated Genome‐wide CRISPRa Approach to Functionalize lncRNAs in Drug Resistance. Cell. 2018;173(3):649‐64. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Corrà F, Agnoletto C, Minotti L, Baldassari F, Volinia S. The Network of Non‐coding RNAs in Cancer Drug Resistance. Front Oncol. 2018;8:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Deng H, Zhang J, Shi J, Guo Z, He C, Ding L, et al. Role of long non‐coding RNA in tumor drug resistance. Tumour Biol. 2016;37(9):11623‐31. [DOI] [PubMed] [Google Scholar]
- 107. Jiang W, Xia J, Xie S, Zou R, Pan S, Wang ZW, et al. Long non‐coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist Updat. 2020;50:100683. [DOI] [PubMed] [Google Scholar]
- 108. Yu WD, Wang H, He QF, Xu Y, Wang XC. Long noncoding RNAs in cancer‐immunity cycle. J Cell Physiol. 2018;233(9):6518‐23. [DOI] [PubMed] [Google Scholar]
- 109. Li B, Hong P, Zheng CC, Dai W, Chen WY, Yang QS, et al. Identification of miR‐29c and its Target FBXO31 as a Key Regulatory Mechanism in Esophageal Cancer Chemoresistance: Functional Validation and Clinical Significance. Theranostics. 2019;9(6):1599‐613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Li B, Xu WW, Guan XY, Qin YR, Law S, Lee NP, et al. Competitive Binding Between Id1 and E2F1 to Cdc20 Regulates E2F1 Degradation and Thymidylate Synthase Expression to Promote Esophageal Cancer Chemoresistance. Clin Cancer Res. 2016;22(5):1243‐55. [DOI] [PubMed] [Google Scholar]
- 111. Han M, Gu Y, Lu P, Li J, Cao H, Li X, et al. Exosome‐mediated lncRNA AFAP1‐AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Mol Cancer. 2020;19(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 112. He X, Sheng J, Yu W, Wang K, Zhu S, Liu Q. LncRNA MIR155HG Promotes Temozolomide Resistance by Activating the Wnt/β‐Catenin Pathway Via Binding to PTBP1 in Glioma. Cell Mol Neurobiol. 2021.41 (6):1271–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Xu Z, Yang F, Wei D, Liu B, Chen C, Bao Y, et al. Long noncoding RNA‐SRLR elicits intrinsic sorafenib resistance via evoking IL‐6/STAT3 axis in renal cell carcinoma. Oncogene. 2017;36(14):1965‐77. [DOI] [PubMed] [Google Scholar]
- 114. Gu P, Chen X, Xie R, Han J, Xie W, Wang B, et al. lncRNA HOXD‐AS1 Regulates Proliferation and Chemo‐Resistance of Castration‐Resistant Prostate Cancer via Recruiting WDR5. Mol Ther. 2017;25(8):1959‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wang JQ, Jeelall YS, Ferguson LL, Horikawa K. Toll‐Like Receptors and Cancer: MYD88 Mutation and Inflammation. Front Immunol. 2014;5:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Dong H, Wang W, Mo S, Chen R, Zou K, Han J, et al. SP1‐induced lncRNA AGAP2‐AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37(1):202. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 117. Lu C, Rong D, Zhang B, Zheng W, Wang X, Chen Z, et al. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: challenges and opportunities. Mol Cancer. 2019;18(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gao Y, Sun W, Shang W, Li Y, Zhang D, Wang T, et al. Lnc‐C/EBPβ Negatively Regulates the Suppressive Function of Myeloid‐Derived Suppressor Cells. Cancer Immunol Res. 2018;6(11):1352‐63. [DOI] [PubMed] [Google Scholar]
- 119. Huang D, Chen J, Yang L, Ouyang Q, Li J, Lao L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation‐induced cell death. Nat Immunol. 2018;19(10):1112‐25. [DOI] [PubMed] [Google Scholar]
- 120. Jiang M, Zhang S, Yang Z, Lin H, Zhu J, Liu L, et al. Self‐Recognition of an Inducible Host lncRNA by RIG‐I Feedback Restricts Innate Immune Response. Cell. 2018;173(4):906‐19.e13. [DOI] [PubMed] [Google Scholar]
- 121. Chen W, Han C, Xie B, Hu X, Yu Q, Shi L, et al. Induction of Siglec‐G by RNA viruses inhibits the innate immune response by promoting RIG‐I degradation. Cell. 2013;152(3):467‐78. [DOI] [PubMed] [Google Scholar]
- 122. Xu M, Xu X, Pan B, Chen X, Lin K, Zeng K, et al. LncRNA SATB2‐AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer. 2019;18(1):135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ferrone S, Whiteside TL. Tumor microenvironment and immune escape. Surg Oncol Clin N Am. 2007;16(4):755‐74, viii. [DOI] [PubMed] [Google Scholar]
- 124. Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27(45):5904‐12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Watnick RS. The role of the tumor microenvironment in regulating angiogenesis. Cold Spring Harb Perspect Med. 2012;2(12):a006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Chen D, Lu T, Tan J, Li H, Wang Q, Wei L. Long Non‐coding RNAs as Communicators and Mediators Between the Tumor Microenvironment and Cancer Cells. Front Oncol. 2019;9:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Ni C, Fang QQ, Chen WZ, Jiang JX, Jiang Z, Ye J, et al. Breast cancer‐derived exosomes transmit lncRNA SNHG16 to induce CD73+γδ1 Treg cells. Signal Transduct Target Ther. 2020;5(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Işın M, Uysaler E, Özgür E, Köseoğlu H, Şanlı Ö, Yücel Ö B, et al. Exosomal lncRNA‐p21 levels may help to distinguish prostate cancer from benign disease. Front Genet. 2015;6:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L, et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR‐1303/PTBP3 axis. Mol Cancer. 2020;19(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 131. Chen CW, Fu M, Du ZH, Zhao F, Yang WW, Xu LH, et al. Long Noncoding RNA MRPL23‐AS1 Promotes Adenoid Cystic Carcinoma Lung Metastasis. Cancer Res. 2020;80(11):2273‐85. [DOI] [PubMed] [Google Scholar]
- 132. Wu N, Jiang M, Liu H, Chu Y, Wang D, Cao J, et al. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF‐β/SMAD2/3 signaling pathway. Cell Death Differ. 2021;28(1):219‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Patitucci C, Couchy G, Bagattin A, Cañeque T, de Reyniès A, Scoazec JY, et al. Hepatocyte nuclear factor 1α suppresses steatosis‐associated liver cancer by inhibiting PPARγ transcription. J Clin Invest. 2017;127(5):1873‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zuo Q, He J, Zhang S, Wang H, Jin G, Jin H, et al. PPARγ Coactivator‐1α Suppresses Metastasis of Hepatocellular Carcinoma by Inhibiting Warburg Effect by PPARγ‐Dependent WNT/β‐Catenin/Pyruvate Dehydrogenase Kinase Isozyme 1 Axis. Hepatology. 2021;73(2):644‐60. [DOI] [PubMed] [Google Scholar]
- 135. Li X, Lei Y, Wu M, Li N. Regulation of Macrophage Activation and Polarization by HCC‐Derived Exosomal lncRNA TUC339. Int J Mol Sci. 2018;19(10):2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Guan YF, Huang QL, Ai YL, Chen QT, Zhao WX, Wang XM, et al. Nur77‐activated lncRNA WFDC21P attenuates hepatocarcinogenesis via modulating glycolysis. Oncogene. 2020;39(11):2408‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Xiao ZD, Han L, Lee H, Zhuang L, Zhang Y, Baddour J, et al. Energy stress‐induced lncRNA FILNC1 represses c‐Myc‐mediated energy metabolism and inhibits renal tumor development. Nat Commun. 2017;8(1):783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zheng YL, Li L, Jia YX, Zhang BZ, Li JC, Zhu YH, et al. LINC01554‐Mediated Glucose Metabolism Reprogramming Suppresses Tumorigenicity in Hepatocellular Carcinoma via Downregulating PKM2 Expression and Inhibiting Akt/mTOR Signaling Pathway. Theranostics. 2019;9(3):796‐810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Yu G, Yao W, Gumireddy K, Li A, Wang J, Xiao W, et al. Pseudogene PTENP1 functions as a competing endogenous RNA to suppress clear‐cell renal cell carcinoma progression. Mol Cancer Ther. 2014;13(12):3086‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, et al. Negative regulation of lncRNA GAS5 by miR‐21. Cell Death Differ. 2013;20(11):1558‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Fan M, Li X, Jiang W, Huang Y, Li J, Wang Z. A long non‐coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. 2013;5(4):1143‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ow TJ, Sandulache VC, Skinner HD, Myers JN. Integration of cancer genomics with treatment selection: from the genome to predictive biomarkers. Cancer. 2013;119(22):3914‐28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Qiu JJ, Lin XJ, Tang XY, Zheng TT, Lin YY, Hua KQ. Exosomal Metastasis‑Associated Lung Adenocarcinoma Transcript 1 Promotes Angiogenesis and Predicts Poor Prognosis in Epithelial Ovarian Cancer. Int J Biol Sci. 2018;14(14):1960‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yang YM, Hong P, Xu WW, He QY, Li B. Advances in targeted therapy for esophageal cancer. Signal Transduct Target Ther. 2020;5(1):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Fatehi Hassanabad A, Chehade R, Breadner D, Raphael J. Esophageal carcinoma: Towards targeted therapies. Cell Oncol (Dordr). 2020;43(2):195‐209. [DOI] [PubMed] [Google Scholar]
- 146. Vester B, Wengel J. LNA (locked nucleic acid): high‐affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43(42):13233‐41. [DOI] [PubMed] [Google Scholar]
- 147. Tereshko V, Teplova M, Brunzelle J, Watterson DM, Egli M. Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression. Nat Struct Biol. 2001;8(10):899‐907. [DOI] [PubMed] [Google Scholar]
- 148. Kumar R, Singh SK, Koshkin AA, Rajwanshi VK, Meldgaard M, Wengel J. The first analogues of LNA (locked nucleic acids): phosphorothioate‐LNA and 2′‐thio‐LNA. Bioorganic & Medicinal Chemistry Letters. 1998;8(16):2219‐22. [DOI] [PubMed] [Google Scholar]
- 149. Sekiguchi M, Obika S, Somjing R, Imanishi T. Synthesis and properties of a novel bridged nucleic acid analogue bearing a P3'→N5' phosphoroamidate linkage, 5'‐amino‐3', 5'‐BNA. Nucleic Acids Symp Ser (Oxf). 2004(48):7‐8. [DOI] [PubMed] [Google Scholar]
- 150. Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, et al. Melanoma addiction to the long non‐coding RNA SAMMSON. Nature. 2016;531(7595):518‐22. [DOI] [PubMed] [Google Scholar]
- 151. Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 Contributes to the Cervical Cancer Phenotype and Associates with Poor Patient Prognosis. PLoS One. 2016;11(5):e0156274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome‐Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29(5):653‐68. [DOI] [PubMed] [Google Scholar]
- 153. Arun G, Diermeier S, Akerman M, Chang K‐C, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes & development. 2016;30(1):34‐51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Rivas E, Clements J, Eddy SR. A statistical test for conserved RNA structure shows lack of evidence for structure in lncRNAs. Nature methods. 2017;14(1):45‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Connelly CM, Moon MH, Schneekloth JS, Jr. The Emerging Role of RNA as a Therapeutic Target for Small Molecules. Cell Chem Biol. 2016;23(9):1077‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Cai B, Song X, Cai J, Zhang S. HOTAIR: a cancer‐related long non‐coding RNA. Neoplasma. 2014;61(4):379‐91. [DOI] [PubMed] [Google Scholar]
- 158. Zhou J, Rossi JJ. Bivalent aptamers deliver the punch. Chem Biol. 2008;15(7):644‐5. [DOI] [PubMed] [Google Scholar]
- 159. Zhu G, Chen X. Aptamer‐based targeted therapy. Adv Drug Deliv Rev. 2018;134:65‐78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Park KS. Nucleic acid aptamer‐based methods for diagnosis of infections. Biosens Bioelectron. 2018;102:179‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, et al. Systemic administration of optimized aptamer‐siRNA chimeras promotes regression of PSMA‐expressing tumors. Nat Biotechnol. 2009;27(9):839‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Ozpolat B, Sood AK, Lopez‐Berestein G. Liposomal siRNA nanocarriers for cancer therapy. Adv Drug Deliv Rev. 2014;66:110‐6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Rupaimoole R, Lee J, Haemmerle M, Ling H, Previs RA, Pradeep S, et al. Long Noncoding RNA Ceruloplasmin Promotes Cancer Growth by Altering Glycolysis. Cell Rep. 2015;13(11):2395‐402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Cui Y, Cao W, Li Q, Shen H, Liu C, Deng J, et al. Evaluation of prostate cancer antigen 3 for detecting prostate cancer: a systematic review and meta‐analysis. Sci Rep. 2016;6:25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ferreira LB, Palumbo A, de Mello KD, Sternberg C, Caetano MS, de Oliveira FL, et al. PCA3 noncoding RNA is involved in the control of prostate‐cancer cell survival and modulates androgen receptor signaling. BMC Cancer. 2012;12:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Dong L, Ni J, Hu W, Yu C, Li H. Upregulation of Long Non‐Coding RNA PlncRNA‐1 Promotes Metastasis and Induces Epithelial‐Mesenchymal Transition in Hepatocellular Carcinoma. Cell Physiol Biochem. 2016;38(2):836‐46. [DOI] [PubMed] [Google Scholar]
- 167. Smaldone MC, Davies BJ. BC‐819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12(5):607‐16. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.


