Abstract
The xanthophyll cycle in the antenna of photosynthetic organisms under light stress is one of the most well-known processes in photosynthesis, but its role is not well understood. In the xanthophyll cycle, violaxanthin (Vio) is reversibly transformed to zeaxanthin (Zea) that occupies Vio binding sites of light-harvesting antenna proteins. Higher monomer/trimer ratios of the most abundant light-harvesting protein, the light-harvesting complex II (LHCII), usually occur in Zea accumulating membranes and have been observed in plants after prolonged illumination and during high-light acclimation. We present a combined NMR and coarse-grained simulation study on monomeric LHCII from the npq2 mutant that constitutively binds Zea in the Vio binding pocket. LHCII was isolated from 13C-enriched npq2 Chlamydomonas reinhardtii (Cr) cells and reconstituted in thylakoid lipid membranes. NMR results reveal selective changes in the fold and dynamics of npq2 LHCII compared with the trimeric, wild-type and show that npq2 LHCII contains multiple mono- or digalactosyl diacylglycerol lipids (MGDG and DGDG) that are strongly protein bound. Coarse-grained simulations on npq2 LHCII embedded in a thylakoid lipid membrane agree with these observations. The simulations show that LHCII monomers have more extensive lipid contacts than LHCII trimers and that protein-lipid contacts are influenced by Zea. We propose that both monomerization and Zea binding could have a functional role in modulating membrane fluidity and influence the aggregation and conformational dynamics of LHCII with a likely impact on photoprotection ability.
Significance
As a response to high light, photosynthetic light-harvesting proteins can reorganize themselves in the photosynthetic membrane and reversibly bind zeaxanthin. The most abundant light-harvesting complex is the trimeric light-harvesting complex II (LHCII), for which also monomerization under high-light conditions has been reported. The roles of zeaxanthin binding and monomerization under high-light conditions are not well understood. We report that monomerization of LHCII and binding of zeaxanthin have a significant effect on the structural dynamics and protein-lipid interactions of membrane-embedded LHCII. These effects are potentially relevant for understanding light regulation in plants and algae.
Introduction
Photosynthetic organisms have the challenging task to perform the conversion of light into chemical energy under fluctuating sunlight conditions. To cope with this challenge, they evolved several strategies by which they adjust their photosynthetic efficiency to the environmental conditions and efficiently exploit sunlight in low light while protecting photosystem II (PSII) from photodamage in high light (1). Light-harvesting antennas are pigment binding proteins that form flexible structures that adapt their conformation, size, and self-assembled architectures to the light environment. In high-light conditions, several processes are activated in the photosynthetic thylakoid membrane to prevent over excitation, including nonphotochemical quenching (NPQ), which causes dissipation of singlet chlorophyll (Chl) excited-state energies in the PSII antenna as heat (2). Two processes contribute to NPQ on a second to minute timescale, both controlled by the thylakoid lumen pH: rapid (1–3 min) induced quenching termed as qE through activation of the proteins photosystem II subunit S and light-harvesting complex stress-related protein (LHCSR), and zeaxanthin (Zea)-dependent quenching, known as qZ that is activated by the xanthophyll cycle, which reversibly converts the carotenoid violaxanthin (Vio) into Zea via antheraxanthin (3, 4, 5). Molecular sites involved in qE and qZ are integrated in the light-harvesting antenna complexes of PSII, of which the most abundant ones are the Chl a/b binding protein complexes (light-harvesting complex II [LHCII], see Fig. 1).
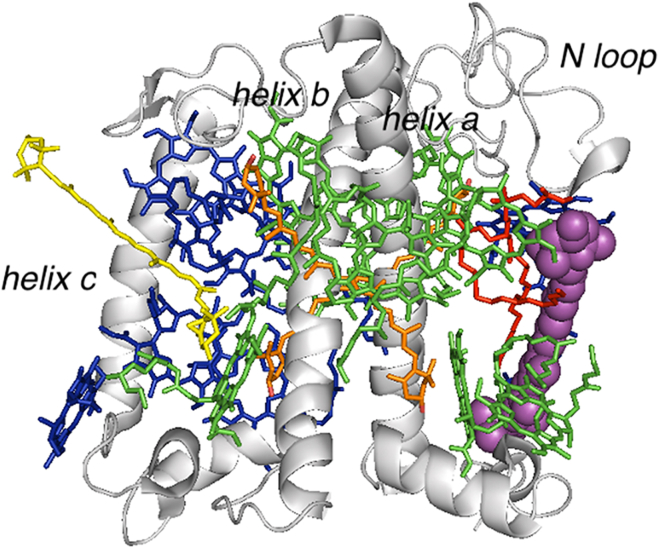
Figure 1.
Monomeric structure of the pigment-protein complex LHCII (PDB: 2bhw (24)). The V1 carotenoid (violaxanthin [Vio]) is highlighted as purple spheres. The other protein-bound pigments are shown as sticks (green, Chl a; blue, Chl b; yellow, neoxanthin; orange, lutein). A phosphatidyl glycerol (PG) structural lipid is shown as red stick.
The effect of Zea replacement in the Vio binding pocket of LHCII, called V1, is still unclear. Zea might interact directly with Chls inside the antenna proteins, preventing over excitation in excess light conditions by quenching excitations via Chl-to-Zea excitation energy transfer or charge-transfer states (6,7). The xanthophyll could also act between the thylakoid membrane complexes to create a variety of quenching sites (8). Furthermore, Zea has been proposed to act as an allosteric regulator of NPQ induction (qE) (9). In this latter role as allosteric regulator of qE, Zea binding to light-harvesting complexes might promote a protein conformational change due to its more hydrophobic nature (10). Besides, the presence of Zea appears to have a rigidifying effect on membrane fluidity (11), and it has been suggested that Zea binding rigidifies LHCII and stabilizes its photoprotective, quenched state (12).
Many studies on the effects of Zea make use of carotenoid-less mutants, of which the most well-studied ones are those of the npq2 strains (4,13,14). The npq2 mutants, which contain a defect in the enzyme zeaxanthin deepoxidase and bind constitutively Zea in the V1 binding pocket, lack the carotenoid neoxanthin (Neo). Whereas native LHCII complexes form trimers in thylakoid membranes (15, 16, 17), npq2 membranes show a reduced content of trimeric LHCII compared with the parental strains in optimal light conditions. Higher monomer/trimer ratios of LHCII usually occur in Zea-accumulating membranes (18,19), as has been observed in wild-type (wt) plants after prolonged illumination (20,21). The ratio increases even further upon high-light acclimation (22). Janik et al. also report on light-induced dimerization of LHCII (20), forming quenched dimers. Trimerization of LHCII increases its protein stability and has been proposed to optimize light-harvesting, by controlling the environment of the lutein 2 domain in LHCII that faces the trimer interior (23). Light-induced monomerization or dimerization has been inferred to facilitate excitation quenching and the NPQ state or be important for regulating proteolytic degradation (21).
Structural studies on LHCII have been performed on crystals of trimeric LHCII from plants (15,25) and structural data have been obtained through single-particle cryoelectron microscopy (cryo-EM) analyses of LHCII-PSII supercomplexes of plants and algae (17,26). The LHCII pigment-protein structures always contain a structural lipid, phosphatidyl glycerol (PG), per monomer, which is known to mediate trimerization (27). While crystallography and cryo-EM studies are not suited to capture LHCII protein and pigment dynamics or dynamic protein-protein or protein-lipid interactions, the structural dynamics, and lipid and protein association of monomeric and trimeric LHCII, has been assessed in molecular dynamics (MD) and coarse-grained (CG) MD simulations (28, 29, 30, 31, 32). We recently demonstrated that magic angle spinning (MAS) NMR spectroscopy can be applied to evaluate the conformational dynamics of membrane-embedded LHCII trimers (33). MAS NMR spectroscopy is a sensitive probe for detecting atomistic changes in protein and pigment structural dynamics. In addition, with this technique, intrinsically bound lipids can be detected that are copurified together with the target protein from the original thylakoid membranes.
In this work, we present a structural analysis of monomeric, Zea-containing LHCII complexes that were reconstituted in thylakoid lipid membranes. 13C-15N isotope-labeled LHCII complexes were isolated from the npq2 strain of Chlamydomonas reinhardtii (Cr). Sucrose gradient analysis shows that LHCII from the npq2 Cr strain, further denoted as npq2 LHCII, has a reduced trimeric content with respect to the cw15 strain, further denoted as the wt, as reported in the literature (18). The npq2 LHCII fractions were thus collected as monomers. Using solid-state NMR spectroscopy supported by CG MD simulations, we analyzed the conformational dynamics and lipid association of npq2 LHCII. We show that both LHCII monomerization and Zea binding influence the protein conformation and dynamics and have a pronounced effect on its lipid-binding properties. This is of particular importance for our structural understanding of light regulation in plants and algae.
Materials and methods
LHCII extraction
Cr strains of npq2 were grown under conditions as previously described for the wt cells (33). After thylakoid isolation, Cr npq2 thylakoids were resuspended in buffer (50 mM HEPES-KOH [pH 7.5], 5 mM MgCl2 with 50% glycerol). For isolation of the LHCII fractions, thylakoid membranes corresponding to 3 mg/ml of total chlorophylls (based on the optical density at 680 nm) were washed with 50 mM ethylenediaminetetraacetic acid and solubilized for 20 min on ice in 3 ml of 1.2% α-DM in 10 mM HEPES buffer (pH 7.5), after vortexing for 1 min. The solubilized samples were centrifuged at 15,000 × g for 30 min to eliminate any insolubilized material and the supernatant containing the photosynthetic complexes was then fractionated by ultracentrifugation in a 0–1 M sucrose gradient containing 0.06% α-DM and 10 mM HEPES (pH 7.5), at 141,000 × g for 40 h at 4°C. The green fraction corresponding to monomeric npq2 LHCII proteins (Fig. S1 A) was harvested with a syringe and the Chl concentration was adjusted to 2 mg/ml with buffer (50 mM HEPES, 5 mM MgCl2 [pH 7.5]).
Preparation of liposomes
Npq2 LHCII proteins solubilized in α-DM were reconstituted in lipid membranes, the lipid composition of which mimics the average native thylakoid membrane. The proteoliposomes contained 47% monogalactosyl diacylglycerol (MGDG), 12% sulfoquinovosyl diacylglycerol (SQDG), 14% PG, and 27% digalactosyl diacylglycerol (DGDG) at a molar protein/lipid ratio of 1:55 (34). The chosen protein/lipid ratio is in the range of native protein packing densities in thylakoid membranes, where 70–80% of the membrane surface area is occupied by proteins (35). The lipids were dissolved in chloroform and dried into a thin film using a rotary evaporator at 40°C. The lipid film was hydrated by reconstitution buffer (50 mM HEPES, 5 mM MgCl2 [pH 7.5], and 0.03% β-DM) and was exposed to 10 freeze-thaw cycles. After that, npq2 LHCII was inserted into liposomes and detergent was removed by 72 h of dialysis against detergent-free buffer. During the dialysis bio beads (SM-2, Bio-Rad) were added to the buffer to speed up the process.
Pigment analysis
The content of individual carotenoids of npq2 LHCII was determined using high-performance liquid chromatography (Beckman System Gold, Indianapolis, Indiana, USA) according to Farber and Jahns (36). The peaks of each sample were identified through the retention time and absorption spectrum.
Time-resolved fluorescence spectroscopy
Time-resolved fluorescence measurements were performed using a FluoTime 300 (PicoQuant) time-correlated photon counter spectrometer. Samples were held in a 1 × 1 cm quartz cuvette that was thermostated at 20°C and excited at 440 nm using a diode laser (PicoQuant). Fluorescence decay traces convoluted with the instrument response function were fitted with multi-exponentials using a χ2 least-square fitting procedure (Fig. S2 A and Table S1).
UV-visible spectroscopy
Absorption spectra of npq2 LHCII in α-DM and in proteoliposomes were recorded on a Cary 60 UV-visible spectrophotometer (Agilent Technologies, Santa Clara, California, USA) with the wavelength range set from 350 to 750 nm (Fig. S2 B).
NMR sample preparation
For the NMR samples, 18 ml of npq2 LHCII proteoliposomes, containing approximately 6 mg LHCII and 1.5 mg Chl (as determined by the optical density at 680 nm of the Chls), was pelleted by ultracentrifugation (223,000 × g, 4°C, 90 min) and transferred to a thin-walled 3.2 mm solid-state NMR MAS rotor through centrifugation.
Solid-state NMR experiments
Solid-state NMR spectra were recorded on an ultrahigh field 950-MHz 1H Larmor frequency spectrometer (Bruker, BioSpin, Billerica, MA) equipped with a triple-channel 1H, 13C, 15N 3.2 mm MAS probe. 13C-13C PARIS and 13C-13C INEPT on 13C-15N npq2 LHCII in lipid bilayers were recorded under the same MAS rate, temperature, and parameters as 13C-15N wt LHCII in lipid bilayers (33). Typical π/2 pulses were 3 μs for 1H, 5 μs for 13C, and 8 μs for 15N. The 1H/15N and 1H/13C cross-polarization (CP) (37) contact times were 800 μs and 1 ms, respectively, with a constant radio frequency (rf) field of 35 and 50 kHz on nitrogen and carbon, respectively, while the proton lock field was ramped linearly around the n = 1 Hartmann/Hahn condition (38). The 15N/13Ca SPECIFIC-CP transfer (39) was implemented with an optimized contact time of 4.2 ms with a constant lock field of 2.5 × νr applied on 15N, while the 13C field was ramped linearly (10% ramp) around 1.5 × νr. 1H decoupling during direct and indirect acquisition was performed using SPINAL64 (40) with ∼83 kHz irradiation. The presented 2D 13C-13C PARIS (41) spectra were collected with a mixing time of 30 ms at 17 kHz MAS at a set temperature of −18°C. The J-coupling-based 2D 13C-13C INEPT-TOBSY (42,43) experiments were recorded at −3°C with TOBSY mixing of 6 ms at 14 kHz MAS. Spectra were processed using a Bruker TopSpin 3.2 (Bruker, Germany) with LPfr linear prediction, and fqc modes for Fourier transformation spectra were analyzed using Sparky version 3.114 (44) and MestReNova 11.0 (Mestrelab Research SL, Santiago de Compostela, Spain).
CG modeling
We performed two sets of simulations to study the lipid-binding capacity and MD of wt and npq2 LHCII. In the first set, we simulated single proteins, namely wt LHCII trimers, wt LHCII monomers, LHCII trimers containing Zea instead of Vio, and npq2 LHCII monomers (containing Zea instead of Vio and lacking Neo), embedded in thylakoid lipid membrane patches. In the second set, we simulated multiple protein copies, namely 4 wt LHCII trimers and 12 npq2 LHCII monomers, respectively, embedded in thylakoid lipid membrane patches. In the first set, we focused on the composition of the annular lipid shell and the protein-lipid interactions. In the second set, we analyzed the lipids being trapped at the protein-protein interfaces during protein aggregation and the protein flexibility in the aggregate.
Simulation details
All simulations were performed using the CG force field Martini 2.2 (45) and the program package Gromacs (versions 2016.1 and 2018.1) (46). The protein was described using a Gō-like model in combination with Martini (32,47,48). We used the crystal structure of spinach LHCII (15) to generate the CG model as well as the trimer starting structures. The missing residues were modeled with the I-TASSER server (49). The detailed setup of the LHCII model and its validation are described in (32); the cofactor parametrization is based on the cofactors of the PSII (32). To mimic the LHCII complexes extracted experimentally from the npq2 mutant, we used monomeric LHCII, replaced Vio by Zea, and removed Neo. To investigate intermediate states between wt trimer and npq2 mutant, we simulated LHCII trimers in which Vio was replaced by Zea as well as monomeric wt LHCII with Vio and Neo in their original crystal structure position. For all simulations, the proteins were embedded in a plant-like thylakoid membrane model (50), which contained four major lipid species in the following ratio: 40% MGDG, 30% DGDG, 15% PG, and 15% SQDG. Due to different lipid tail combinations, the detailed composition was 18:3(9,12,15)-16:0 MGDG, di18:3(9,12,15) MGDG, 18:3(9,12,15)-16:0 DGDG, di18:3(9,12,15) DGDG, 16:1(3t)-16:0 PG, 16:1(3t)-18:3(9,12,15) PG, and 18:3(9,12,15)-16:0 SQDG in the ratio of 14/100/14/72/14/28/42.
Two different setups were simulated: First, a single copy of the respective protein was embedded in thylakoid membrane (for monomers: patch size 11 × 11 nm2 containing ∼330 lipids; for trimers: patch size 20 × 20 nm2 containing ∼1200 lipids), neutralized, and solvated in 0.15 M NaCl solution. Second, four wt LHCII trimers (corresponding to 12 monomers, patch size 30 × 30 nm2 containing ∼2400 lipids) or 12 npq2 monomers were embedded in thylakoid membrane (patch size 32 × 24 nm2 containing ∼1900 lipids), neutralized, and solvated in 0.15 M NaCl solution. The simulation parameters were identical to those given in (32). For the Verlet cutoff scheme a buffer tolerance of 0.005 kJ/mol was used. The reaction-field method was applied for the Coulomb interactions with a cutoff of 1.1 nm and a dielectric constant of εr = 15 for water (51). For the van der Waals interactions a cutoff of 1.1 nm was used. The reference temperature for the velocity rescaling thermostat was set to T = 310 K (τT = 1 ps) (52). During the equilibrations, the pressure was controlled with a Berendsen barostat (p = 1 bar, τp = 3 ps) (53), and with a Parrinello-Rahman barostat (p = 1 bar, τp = 12 ps) (54) during the production runs.
After minimization, three equilibration stages with 1) a time step of 1 fs and 100,000 steps, 2) 2 fs and 250,000 steps, and 3) 10 fs and 106 steps were performed. In the production runs, the systems with one protein copy were simulated for 15 μs each (two replicas for wt trimer and trimer with Zea; four replicas for wt and npq2 monomer). The systems containing multiple protein copies were simulated for 20 μs each (four replicas for wt trimers; two replicas for npq2 monomers). For all production runs, a time step of 10 fs was used.
Analysis
To identify the annular lipid shell of the respective proteins—their lipid fingerprint (55)—the lipid composition around the proteins with a distance ≤0.7 nm within the last 13 μs of each single-copy system was analyzed. In addition, we analyzed the number of contacts between the lipid beads and the protein backbone of the middle part of helices a and b with a distance ≤0.7 nm again within the last 13 μs. In the case of the wt trimer and the trimer with Zea, the three PG lipids at the monomer-monomer interfaces were excluded for the calculation of the number of contacts.
To evaluate the flexibility of the protein in a crowded thylakoid membrane after protein aggregation, we calculated the root mean-square fluctuations (RMSF) of the backbone beads within the last 900 ns of the simulations containing multiple proteins. Before calculating the RMSF, the three transmembrane helices of each LHCII monomer were aligned separately. The RMSF were calculated for time intervals of 30 ns and averaged subsequently. To quantify the number and type of lipids being present at a protein-protein interface, we analyzed the lipids which are simultaneously in contact with two different protein copies during the last 5 μs of simulation using the Gromacs tool gmx select. A contact was taken into account if the glycerol linker of the lipid had a distance ≤0.7 nm to two proteins. Existing interfaces between the monomers within an LHCII trimer were excluded.
Results
The Zea-containing LHCII complexes were purified as monomers from the sucrose gradient, and the incorporation of Zea was confirmed by high-performance liquid chromatography analysis (Fig. S1). The purified pigment-protein complexes were reconstituted in lipid membranes composed of 47% MGDG, 27% DGDG, 12% SQDG, and 14% PG, mimicking the lipid composition of native thylakoid membranes. The liposomes for protein insertion had a protein/lipid ratio of 1:55 (mol:mol) to mirror native protein packing densities (35).
Fluorescence spectroscopy
The intrinsic Chl fluorescence of LHCII is a reporter for its functional condition that can alternate between a light-harvesting state and a photoprotective, quenched state (2). In the latter state, Chl excitations are rapidly quenched, resulting in reduced fluorescence lifetimes. Isolated LHCII in detergent micelles has long-lived Chl fluorescence lifetimes in the range of 3–4 ns, while those are shortened to ∼2 ns in native thylakoid membranes (2). A fluorescence lifetime analysis on our npq2 LHCII proteoliposomes shows that the average fluorescence lifetime is shortened to 1.1 ns compared with a lifetime of 3.3 ns of npq2 LHCII in α-DM detergent (Fig. S2 A and Table S1). Reduced fluorescence lifetimes of LHCII in proteoliposomes have been reported in various studies and are attributed to protein self-clustering inside lipid bilayers, which can also occur in native thylakoid membranes and which is known to induce quencher states (34,56, 57, 58). We conclude that the npq2 LHCII proteoliposomes are moderately fluorescence quenched, while isolated npq2 LHCII form fully light-harvesting states in detergent micelles. This indicates that binding of Zea by itself does not change the LHCII photophysical state, in line with the observations of Son et al. (59), but that quencher states are generated via protein and lipid interactions in the membrane.
MAS NMR spectroscopy
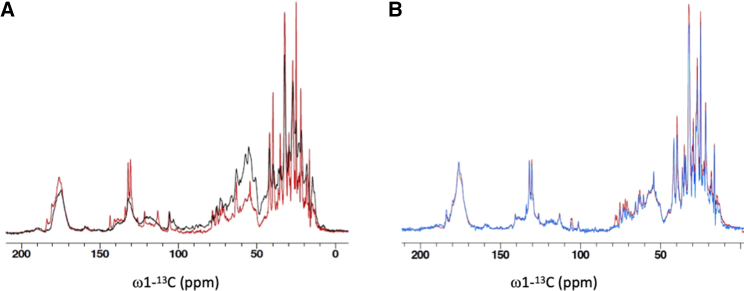
MAS NMR spectroscopy is a unique tool for investigation of structure and dynamic disorder of noncrystalline membrane protein complexes embedded in lipid bilayers (60, 61, 62). Here, we performed MAS NMR spectroscopy to address the structural dynamics of Zea-containing npq2 LHCII. The Zea-LHCII proteoliposomes were spun in a 4 mm MAS NMR rotor as described in the materials and methods and subjected to solid-state NMR spectroscopy. Fig. 2 A shows the 1D 13C NMR spectrum of the npq2 LHCII proteoliposome sample. Two spectra are presented that were obtained by 1H-13C CP and by 13C direct polarization (DP), respectively. In the CP experiment, magnetization is transferred from protons to carbons, which gives a fourfold signal enhancement in the case of a pure solid owing to the larger gyromagnetic ratio of 1H compared with 13C. For flexible molecules, the CP efficiency, which relies on dipolar couplings that are orientation dependent, will be reduced and for highly dynamic molecules the orientations of the dipolar couplings will be averaged to zero. Therefore, the CP NMR spectrum only contains the contributions from rigid molecules. DP of 13C carbons on the other hand produces a spectrum that contains the signals of all 13-carbon atoms. Fig. 2 A presents the CP and DP spectrum collected at −3°C readout temperature. The CP intensities are enhanced in the aliphatic region, confirming good CP efficiency. The DP spectrum shows additional peaks from flexible protein sites and dynamic pigment molecules. In addition, lipid peaks can be seen as sharp peaks in the regions 0–50 ppm (CH2 carbons), 60–80 ppm (carbohydrate carbons), and 100–150 ppm (CH carbons) that are typical of galactolipids (63). This indicates that thylakoid lipids were copurified together with the LHCII proteins because the lipids used for membrane reconstitution were not enriched in 13C atoms.
Figure 2.
CP and DP NMR spectral intensities of npq2 LHCII and comparison of NMR spectra of wt and npq2 LHCII, showing extensive lipid binding to npq2 LHCII. (A) 13C DP (red) and CP (black) spectrum of npq2 LHCIa. (B) 13C DP spectrum of npq2 LHCII (red) and of wt LHCII (blue), normalized at the carbonyl peak at 175 ppm. Sharp peaks in the DP spectra are attributed to (galacto)lipid signals and are more intense for npq2 LHCII.
Fig. 2B shows a comparison of the 13C DP NMR spectrum of monomeric, npq2 LHCII in liposomes (red) and the 13C DP NMR spectrum of trimeric, wt LHCII in liposomes (blue) prepared according to the same methods and using the same lipid composition and protein/lipid ratio. The wt LHCII trimers were isolated from the cw15 strain that was also used for the npq2 mutant. The two spectra in Fig. 2 B are normalized to the protein carbonyl peak at 175 ppm. Comparing npq2 LHCII to the wt, the relative lipid content is higher in the spectrum of npq2 LHCII (red spectrum in Fig. 2 B), indicating that more thylakoid lipids were copurified together with npq2 LHCII than with wt LHCII.
We continued with 2D NMR experiments, which enable better separation of protein and lipid signals and to investigate the structural dynamics of npq2 LHCII. To identify dynamic protein sites, we collected 13C-13C spectra using two types of complementary NMR pulse experiments, which rely on the use of dipolar-based versus J-coupling-based polarization transfer efficiencies. In dipolar-based CP experiments, as explained above, the transfer efficiency relies on dipolar couplings that are orientation dependent and will average to zero for rapidly tumbling molecules. J-Coupling-based NMR experiments rely on hetero-nuclear J-coupling between molecules and effectively work for molecules that display (sub)nanosecond large-amplitude motions. As a result, in the J-based NMR spectra only highly dynamic molecules will be detected. The two types of experiments selectively probe rigid molecules (CP, rotational correlation times exceeding 0.1 ms and/or anisotropic molecules with high-order parameters) or highly dynamic molecules (J-based, rotational correlation times in the nano- to picosecond range and/or anisotropic molecules with low order parameters).
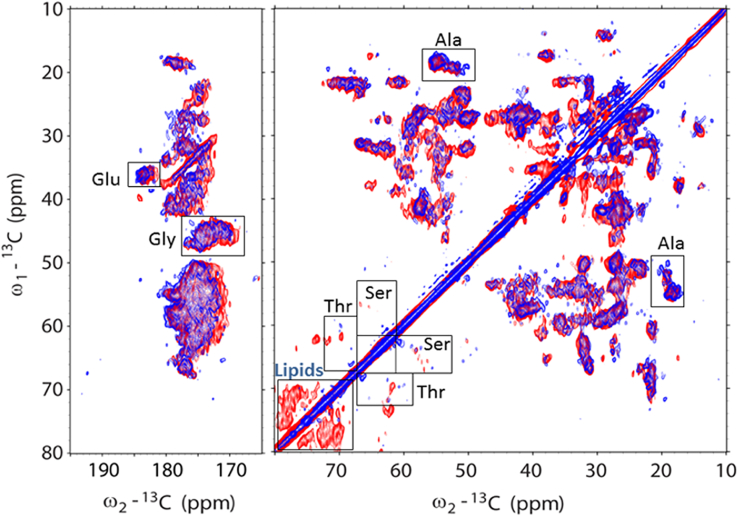
Fig. 3 shows the CP-based 13C-13C (CP-PARIS) NMR spectrum of Zea-LHCII in the aliphatic (right) and carbonyl (left) region. For the 2D PARIS NMR experiments, the readout temperature was set to −18°C to compare the spectrum with that of wt LHCII proteoliposomes under identical conditions. The spectrum of npq2 LHCII in red is overlaid with a spectrum of wt LHCII proteoliposomes (blue spectrum) that was recorded and processed using identical parameters. Data results on the wt LHCII proteoliposomes have been presented in previous work (33). The spectrum of npq2 LHCII (red) has a strong overlap with the spectrum of wt LHCII (blue), indicating that the overall protein conformation is preserved. However, several differences can be observed, as follows.
Figure 3.
13C-13C CP-PARIS spectrum of npq2 LHCII proteoliposomes (red) with the spectrum of wt LHCII proteoliposomes (blue), showing conformational changes for Thr and Ser residues and strong presence of bound lipids in npq2 LHCII. Spectra were collected at −18°C with a 17 kHz MAS frequency. Selective regions with Ala, Thr, Ser, Gly, and Glu chemical shifts are indicated with gray boxes. Signals of galactolipids are also indicated. The blue spectrum of the wt LHCII proteoliposomes has been presented in previous work (33).
The peaks in the CP spectrum of npq2 LHCII are broadened compared with the spectrum of the wt LHCII (Fig. S3 C). Apparent line broadening is an indication of conformational disorder (64) and suggests that the npq2 LHCII complexes overall have more conformational disorder than the wt complexes, which could be a result of the monomerization.
We can identify cross-correlation signals of alanine (Ala), threonine (Thr), serine (Ser), and glycine (Gly) in the crowded spectrum owing to spectral dispersion by which they fall in distinct spectral regions and are separated from the signals of other amino acid residues (indicated with boxes in Fig. 3). In Fig. S3, the Ala, Thr, Ser, and Gly regions of the spectra are presented in close-up for a detailed comparison of the wt and npq2 LHCII NMR chemical shifts. Dependent on the spectral region, they fall into amino acid signals can be attributed to helix, coil, or strand conformation, as indicated in Fig. S3. Figs. 3 and S3 show that three Ala peaks appearing in the wt spectrum are lacking in the npq2 spectrum. In addition, a different pattern is observed for the Thr and Ser coil amino acid signals. In the npq2 LHCII spectrum, notably, strong Ser correlation signals are observed in a region far away from the diagonal, where signals from amino acids in strand conformation typically accumulate (65). This indicates that npq2 LHCII contains a Ser-containing strand segment. Fig. S4 shows the spectral region where Chl and carotenoid signals accumulate. In the Chl and carotenoid region, also different patterns are observed for Chl and carotenoid cross-correlations in npq2 LHCII. For the carotenoid signals, those changes are attributed to the difference in xanthophyll composition.
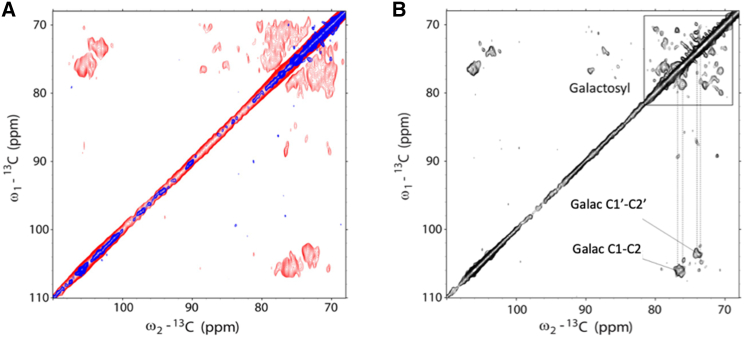
A further prominent difference between the spectra of wt and npq2 LHCII are the signals accumulating between 70 and 80 ppm that are more pronounced in the npq2 spectrum and of which the chemical-shift correlations are representative of the galactosyl heads of galactolipids. The appearance of lipid signals in the 2D NMR spectrum gives further evidence that the detected galactolipids are not liposome lipids, as the probability of 13C-13C polarization transfer that would give rise to crosspeaks under natural abundance 13C conditions (i.e. without 13C enrichment) is very low. The lipids therefore must originate from the original, 13C-labeled thylakoid membranes and have been copurified together with the protein. Fig. 4 A shows the spectral region where strong lipid signals occur, correlating between 73–76 and 104–106 ppm, are only seen in the npq2 spectrum. NMR 13C chemical shifts between 100 and 110 ppm are typical of enantiomeric carbon atoms, as can be found in glucose ring structures. Considering the constituents of our sample, such structures are attributable to the headgroups of galactolipids. Both the headgroups of MGDG and DGDG contain enantiomeric carbons with NMR chemical shifts in the range 103–106 ppm (66). DGDG molecules should give additional signals from the second ring, with the enantiomeric carbons resonating around 100 ppm (67). Because no correlating peaks are observed in this region, we tentatively attribute the strong lipid galactosyl signals to MGDG molecules. We noted that the lipid headgroup signals are better NMR resolved at higher temperature. Fig. 4 B shows a spectrum that was collected at a higher readout temperature (−3°C instead of −18°C) for better visualization of the galactolipid signals. Fig. S5 shows an enlarged part of the region where galactosyl resonance signals occur. A connecting walk can be made starting from the enantiomeric C1 carbon correlations to C2, C3, C4, and C5 of the galactosyl headgroup.
Figure 4.
Identification of galactolipid signals in the CP-PARIS spectrum of npq2 LHCII. (A) 13C-13C CP-PARIS spectrum of npq2 LHCII proteoliposomes (red) with the spectrum of wt LHCII proteoliposomes (blue) overlaid. Spectra were collected at −18°C with a 17 kHz MAS frequency. (B) 13C-13C CP-PARIS spectrum of npq2 LHCII proteoliposomes collected at −3°C. Galactosyl (Galac) resonances are indicated in the figure. The blue spectrum of the wt LHCII proteoliposomes in (A) has been presented in previous work (33).
Next, we collected a J-based 13C-13C spectrum of npq2 LHCII to identify highly mobile sites. Amino acids that appear in a J-based NMR spectrum are referred to as “J amino acids,” which are characterized by high mobility. The J-based INEPT-TOBSY spectrum of npq2 LHCII is shown in Fig. 5 in red, with the spectrum of the wt LHCII overlaid in blue. While most of the correlation signals overlap, the npq2 spectrum lacks the signals from one Phe and two Thr residues and several amino acid signals are shifted as indicated by the encircled areas in Fig. 5 (right panel). NMR assignments of overlapping protein, pigment, and lipid signals can be found in previous work (68). Only the wt spectrum contains two sets of correlation signals of two highly mobile Chl phytol tails (Fig. 5, left panel). Those signals in the wt spectrum were previously attributed to Chl605 and Chl606 (68), of which the phytol chains are not resolved in the LHCII crystal structure, emphasizing their dynamics (15). The J-based spectra of wt and npq2 LHCII both contain signals of mobile galactolipids that can be seen in the right panel in Fig. 5 in the region 60–80 ppm. We assume that those lipids are copurified lipids that formed an annular shell around the protein in the original membranes and may have exchanged with the liposome lipids upon membrane reconstitution of LHCII. Signals of unsaturated lipid double-bond carbons are indicated in the blue box in the left panel in Fig. 5. It is remarkable that several of those signals, which arise from unsaturated bonds of mobile lipid tails, are not seen in the npq2 spectrum. This indicates that the unsaturated lipids are limited in their tail movements inside the membranes.
Figure 5.
13C-13C INEPT-TOBSY spectrum of npq2 LHCII proteoliposomes (red) with the spectrum of wt LHCII proteoliposomes (blue), highlighting changes of dynamic protein and pigment sites. Spectra were collected at −3°C and with 14 kHz MAS frequency. Amino acid and Chl signals that are shifted or invisible for npq2 LHCII are indicated by the encircled areas. The blue square highlights double-bond signals of unsaturated lipids, highlighting signals that are invisible for npq2 LHCII. The spectrum of wt LHCII has been presented in (33).
Summarizing, compared with wt trimeric LHCII, monomeric npq2 LHCII has more overall conformational disorder and adopts a somewhat different conformation containing one or more strand segments. Second, npq2 LHCII more strongly binds thylakoid MGDG or DGDG lipids. Third, while overall NMR line broadening suggests that npq2 LHCII has more conformational disorder, selective highly dynamic protein, lipid, and Chl sites in wt LHCII are motion limited in npq2 LHCII proteoliposomes.
MD simulations
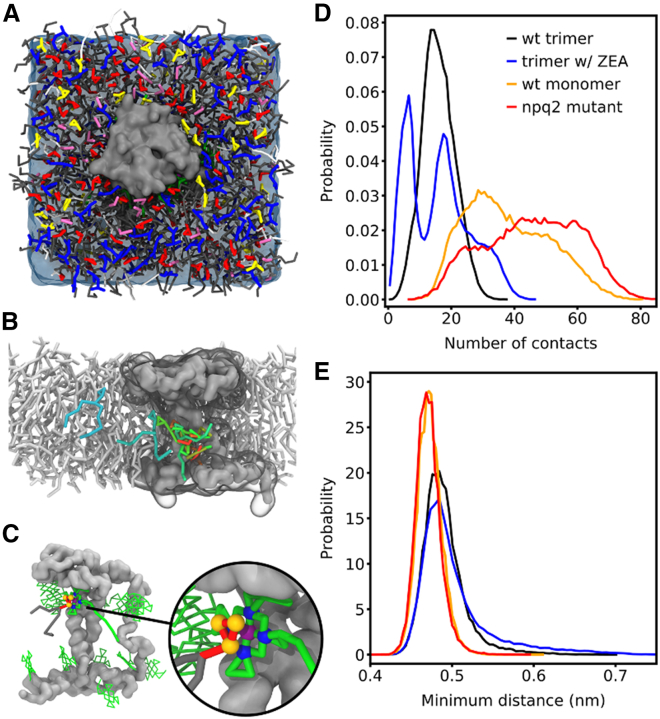
The most striking difference between the wt and npq2 LHCII proteoliposome NMR spectra are the strong galactolipid signals of npq2 LHCII that are seen in the CP-based spectrum, which are from associated lipids that do not exchange on an NMR timescale (∼0.1 ms). We hypothesized that the lipids may form tight associates with npq2 LHCII due to their monomerization. Compared with wt trimers, LHCII monomers will have more membrane-exposed hydrophobic sites that form potential lipid-binding sites. Another possibility is that Zea increases the affinity of LHCII for lipids, owing to its more hydrophobic nature compared with Vio. To understand the structural basis for strong lipid binding to npq2 LHCII, CG MD simulations were performed on Vio-(wt) and Zea-LHCII trimers as well as on npq2 LHCII and wt LHCII monomers, which are well suited to investigate dynamical processes in photosynthesis on the ns to (sub)ms regime (30). Fig. 6 A depicts the CG simulation setup of one npq2 LHCII monomer embedded in a plant thylakoid membrane patch. Based on simulations of single proteins in the plant thylakoid membrane, we analyzed the annular lipid shell. Table 1 summarizes the annular lipid shell at a distance of ≤0.7 nm. While the lipids around wt LHCII trimers and Zea-LHCII trimers are very similar within the accuracy of the employed CG force field, the values for npq2 LHCII and wt LHCII monomer differ from their trimeric counterparts, in particular for PG and MGDG lipids. The difference in PG lipids can be attributed to the three PG lipids that are located at the monomer-monomer interfaces in the LHCII trimers. They are coordinating the Mg2+ ion of one chlorophyll (Chl611) and do not exchange during the whole simulation time of the trimer. They have a substantial role in stabilizing the interface and are stably bound. In contrast, these PG lipids can easily exchange with other lipids in the case of monomeric LHCIIs. The population of the negatively charged SQDG lipids is slightly reduced as well enhancing further the MGDG preference.
Figure 6.
Lipids approaching the central crossing helices of trimeric and monomeric LHCII containing different carotenoids, showing more extensive lipid-protein contacts for monomeric wt and npq2 LHCII. (A) CG simulation setup of an npq2 LHCII monomer (gray surface) embedded in plant thylakoid membrane. The colors of the lipid headgroups are: PG, orchid; MGDG, red; DGDG, blue; SQDG, yellow. Chls are colored green, carotenoids orange, and water below the membrane is shown as transparent blue surface. (B) Selected snapshots of an MGDG lipid approaching and binding to helices a and b in the bilayer middle. (C) Selected snapshot highlighting the interaction between Chl602 and the headgroup of an MGDG lipid, which dived into the bilayer middle. (D) Number of contacts between the protein backbone of the middle part of transmembrane helices a and b and lipid beads with a distance ≤0.7 nm. (E) Minimum distance between the protein backbone and the lipids for these contacts.
Table 1.
Composition of the annular lipid shell (all values are given in %); errors are below 0.1%
| Sample | PG | MGDG | DGDG | SQDG |
|---|---|---|---|---|
| wt LHCII trimer | 10.9 | 51.7 | 19.3 | 18.0 |
| wt LHCII monomer | 4.6 | 61.8 | 19.2 | 14.5 |
| Zea-LHCII trimer | 10.1 | 49.4 | 20.4 | 20.0 |
| npq2 LHCII | 4.2 | 62.3 | 18.0 | 15.5 |
Visual inspection of the simulations of monomeric LHCII revealed that lipids are able to enter the protein-cofactor ensemble toward the crossing helices a and b. Fig. 6 D shows the number of contacts between the lipid beads and the protein backbone of the central part of helices a and b with a distance ≤0.7 nm. Both monomeric LHCIIs (wt, orange; npq2, red) show significantly more lipid contacts compared with the wt LHCII trimer (black) and the Zea-LHCII trimer (blue), indicative of stronger binding. Moreover, the minimum distance between the lipids and the protein backbone is shorter in the case of the monomeric proteins when these contacts are present (Fig. 6 E). This is particularly pronounced for npq2 LHCII. Fig. 6 B shows a series of snapshots of an MGDG lipid that approached the two helices a and b of npq2 LHCII and stayed in contact with them until the end of the simulation (contact time >10 μs). The colors of the lipid show the time evolution from blue to red. It can be easily recognized that the headgroup of the lipid dives into the bilayer middle, where it can also interact with the cofactors of the npq2 LHCII. Fig. 6 C shows an exemplary snapshot highlighting the interaction between Chl602 and the headgroup of another MGDG lipid which dived into the bilayer middle. In the trimer, these cofactors are better stabilized and shielded from the lipids due to protein-protein contacts. Taken together, our simulations of isolated proteins support the observation from NMR experiments that monomeric npq2 LHCII exhibits a stronger lipid binding than wt LHCII trimers. Moreover, the comparison with simulations of wt LHCII monomers indicates that a monomer formation rather than a carotenoid exchange is responsible for this observation. A preference for MGDG was also observed in CG simulations of another photosynthetic complex, PSII embedded in thylakoid membrane, where the annular shell around PSII was enriched of MGDG and SQDG lipids that formed distinct binding sites (69).
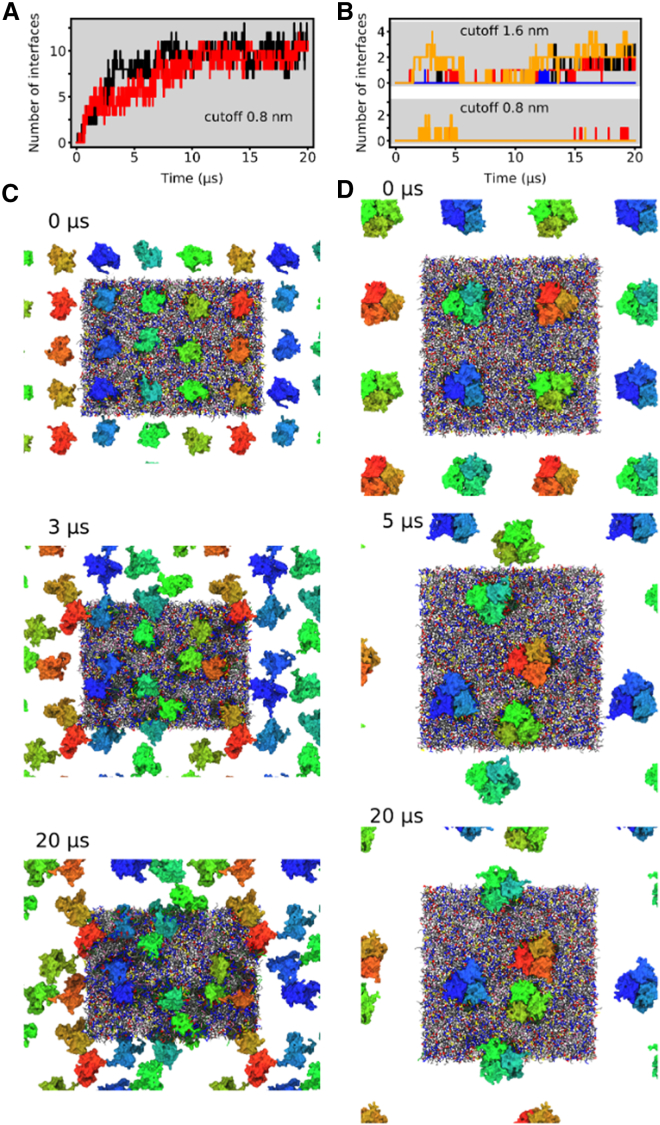
To compare the formation of aggregates of monomeric npq2 LHCII and of wt LHCII trimers, we performed additional CG MD simulations of 12 npq2 LHCII monomers and 4 wt LHCII trimers (corresponding to 12 monomers) in the thylakoid membrane, respectively. Npq2 LHCII formed persistent protein-protein interfaces within about 5 μs in both replicas, which were stable up to the total simulation time of 20 μs (Fig. 7). Representative snapshots from one replica depicted in Fig. 7 C show the formation of different oligomers, including dimers, trimers, and higher-order oligomers. However, trimeric arrangements corresponding to the arrangement in LHCII crystal structures were not observed. In contrast, the wt LHCII trimers were much less prone to form protein-protein interfaces within the total simulation time of 20 μs (Fig. 7 B). Only very few direct protein-protein contacts (cutoff distance 0.8 nm) of temporary nature were formed in the four replicas. Note that the larger cutoff distance of 1.6 nm for a protein-protein interface shown in Fig. 7 B also includes proteins that are separated by a thin lipid solvation shell. The snapshots depicted in Fig. 7 D show that the proteins approach each other but they remain separated by at least one layer of lipids.
Figure 7.
CG simulations of the aggregation of monomeric npq2 LHCII and trimeric wt LHCII in thylakoid membrane, showing more extensive aggregation for monomeric npq2 LHCII. (A and B) Time evolution of the protein-protein interfaces of 12 npq2 LHCII (A) (cutoff distance 0.8 nm) and 4 wt LHCII (B) (cutoff distance 0.8 and 1.6 nm), respectively. (C and D) Representative snapshots showing the aggregation of npq2 LHCII (C) and wt LHCII (D), respectively.
We further analyzed if aggregation of npq2 LHCII monomers results in thylakoid lipids being trapped at protein-protein interfaces during the last 5 μs of the CG simulation. Because we are particularly interested in lipids that are tightly bound at the interface and thus would remain in the protein fraction during protein purification, we applied a cutoff distance of 0.7 nm between the lipid linker beads (GL1 and GL2) and the backbone and side chain beads (BB and SCx, x ∈ [1,5]) of two protein copies. Only lipids that are simultaneously in close contact with two npq2 LHCII are taken into account in Table 2. On average, 0.94 lipids are tightly bound per protein-protein interface. The composition of the lipids being bound is in good agreement with the composition of the annular lipid shell of npq2 LHCII (see Table 1). MGDG is the most prominent lipid at the interfaces (59%), followed by SQDG (20%) and DGDG (17%). For wt LHCII trimers, we could only identify one protein-protein interface that trapped lipids temporarily according to the chosen cutoff criteria. Nevertheless, this interface is not stably formed (Fig. 7 B). Overall, our CG simulations of multiple protein copies in thylakoid membrane reveal that monomeric npq2 LHCII is more strongly liable to form aggregates than wt LHCII trimers. Note that the strong npq2 LHCII aggregation might in parts be due to overestimated interactions and in particular protein-protein interactions in Martini 2 (70, 71, 72). Nevertheless, the comparison between monomeric npq2 LHCII and wt LHCII trimers strongly indicates a significant difference between the two states of LHCII. Moreover, thylakoid lipids are tightly bound at the interfaces between npq2 LHCII monomers.
Table 2.
Absolute and relative number of lipids being present at the protein-protein interfaces of npq2 LHCII during the last 5 μs of CG simulation (relative values are given in %)
| No. of Lipids/Interface | No. of Interfaces | PG | MGDG | DGDG | SQDG | Sum | |
|---|---|---|---|---|---|---|---|
| npq2 LHCII | absolute | 27 | 0.04 | 0.56 | 0.16 | 0.18 | 0.94 |
| relative | 4 | 59 | 17 | 20 | 100 |
The CP NMR spectrum of npq2 LHCII indicates the formation of a strand segment. Secondary structure changes are challenging to be captured by CG simulations but the strand formation results in a reduced flexibility of the corresponding residues. To see if our CG simulations indicate changes in protein flexibility, we analyzed the RMSF of the protein backbone in the aggregation simulations of multiple copies of wt and npq2 LHCII. Fig. S6 compares these RMSF of the protein backbone. Overall, trimer formation clearly reduces the protein flexibility; in particular in the regions with higher RMSF, which are loops located at the membrane-water interface (32). This is in line with the NMR-observed overall increase of conformational disorder for monomeric, npq2 LHCII compared with wt LHCII trimers. Only the first few residues at the N-terminus of npq2 LHCII exhibit fewer fluctuations compared with the wt LHCII trimer (see inlay of Fig. S6). Atomistic simulations of single-monomeric wt LHCII in nonthylakoid membrane revealed high flexibility of the N-terminus (31). Moreover, local conformational changes of the N-terminus observed in the simulations correlate with altered interactions at two putative quenching sites (Chl 611 and 612). It is noted, however, that the simulations were performed on plant LHCII, whereas the NMR experiments were run on Cr LHCII of which the protein amino acid sequences are not identical.
Discussion
Protein and pigment dynamics in npq2 LHCII
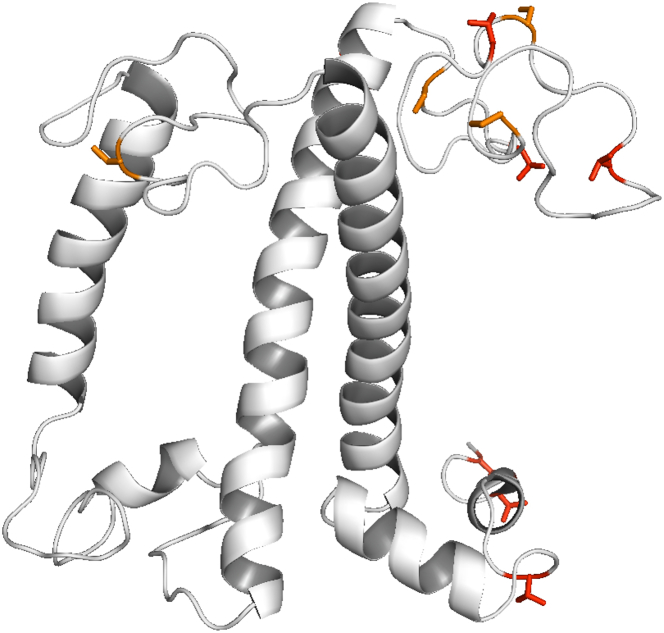
The NMR results report that the conformation of npq2 LHCII is not the same as that of wt LHCII trimers and contains one or more strand segments. Furthermore, fast motions of two Thr residues and one Phe residue are suppressed and several J residues have shifted correlation signals, indicating local changes of structured, dynamic protein sites. For wt LHCII, it was concluded that signals of the flexible N-tail that have not been resolved by crystallography are visible using J-based NMR (33). Because many of the J signals are also visible for npq2 LHCII, including those of three Ser amino acids, we conclude that npq2 LHCII also has a dynamic N-tail. In addition to two Ser residues in the N-tail, the amino acid sequence of the most abundant polypeptide of Cr LHCII, LHCBM1, contains four Ser residues outside the helical regions: one Ser residue in the stromal ac loop and three Ser residues in the stromal N-loop connected to the N-terminus. We consider those as probable sites for local folding into a strand. The N stromal loop, together with an amphipathic 310 helix near the C-terminus on the luminal site, stabilizes Vio in the V1 pocket and may refold upon xanthophyll exchange.
Fig. 8 presents the native LHCII structure with Thr and Ser residues highlighted to indicate their locations. The structure is based on the amino acid sequence of LHCBM1. Vio is also shown in the structure. The N-loop and C-terminus contain several Thr amino acids, of which selective ones may rigidify upon a conformational change. In addition to Zea binding, monomerization of LHCII could induce a local conformational change as was observed in a combined EPR spin label and modeling study for the luminal loop domain connecting helices b and c (73). The bc loop domain contains two Phe amino acids, and a conformational change of this domain could alternatively explain the observed rigidification of a Phe residue. Taken together, we speculate that the binding of Zea in the V1 pocket induces a conformational change in the stromal N-loop, producing a strand segment, and that Zea binding and monomerization affect the LHCII structure of the luminal sites.
Figure 8.
LHCII structure with Thr (red) and Ser (orange) residues outside the helical regions highlighted. The V1 xanthophyll (shown in purple sphere presentation) is stabilized via the N-loop and the C-terminus. The NMR observation of Ser strand residues in the stromal N-loop suggests that this loop forms a strand segment in npq2 LHCII.
wt LHCII contains two highly flexible Chl tails according to their visibility in the J-based NMR spectrum (68). The dynamic Chl tails were attributed to Chl 605 and Chl 606 that are located at the exterior of the LHCII complex. In the J-based spectrum of npq2 LHCII, those two Chl tails are not visible and thus appear to be rigidified. The Chl tails were also not visible in J spectra of LHCII in native thylakoid membranes, suggesting that their dynamics are restrained by specific protein-lipid or protein-protein contacts in native membranes (68). The simulations, however, show increased RMSF for most of the Chl tails in membrane-embedded, aggregated npq2 LHCII monomers with respect to aggregated wt LHCII trimers. In the simulation, the size of the aggregate clusters is limited by the number of proteins in one simulation box (12 in our case) and, thus, the effects of interprotein contacts on the pigment and protein dynamics may not be fully reproduced.
Lipid association and dynamics
The relative content of copurified lipids, distinguished from the liposome lipids by their 13C labels, is larger for npq2 LHCII monomers than for wt LHCII trimers. Only for npq2 LHCII, we observe MGDG and/or DGDG lipids that do not exchange on an NMR timescale and thus seem to be strongly bound to the proteins or entrapped between the proteins. In the CG simulations, the composition of the annular lipid shell does not change dramatically between the wt trimers and npq2 monomers. The annular lipid shell of wt LHCII trimers also compares well with that reported previously (48). The similarity of the annular lipid shell of wt and npq2 LHCII is in agreement with the observed overlap of lipid signals in their J-based NMR spectra, confirming the presence of the same types of mobile, exchangeable lipids. The decisive difference between the wt trimers and npq2 monomers in the simulation is the number of lipid-protein contacts and their distance, with more frequent lipid-protein contacts between the lipids and npq2 LHCII and closer distance of the lipids to the a and b transmembrane helices. Furthermore, simulations of multiple LHCII complexes show that npq2 LHCII monomers form aggregates that have lipids entrapped between the protein complexes. In contrast, this was not observed for wt LHCII trimers within the simulation time of 20 μs. At the interfaces of the monomers, predominantly glycolipids are present, with a strong preference for MGDG. Both effects—the increased interaction with transmembrane helices a and b and the increased number of trapped lipids at the monomer interfaces—agree with the NMR observation that substantially more lipids are copurified with npq2 LHCII, and that those are immobilized in the npq2 LHCII proteoliposomes. We suspect that, in particular, those lipids trapped at the monomer interfaces might be immobilized for a long time so that they become visible in the CP-based NMR spectrum. The lipids bound to the transmembrane helices are potentially an intermediate state that could eventually cause the increased number of trapped lipids upon monomer aggregation.
Implications for LHCII function
According to the CG simulations, monomeric LHCII has more frequent lipid-protein contacts in the central region of transmembrane helices a and b than trimeric LHCII and, for both cases, the protein-lipid interactions are influenced by Zea. It is not clear to which extent LHCII complexes exist as monomers in the original npq2 thylakoid membranes, or whether lower stability of npq2 LHCII trimers causes their monomerization upon purification. The native lipids that are copurified with npq2 LHCII must have been tightly associated with LHCII in the original membranes, supporting the idea that the LHCII complexes form monomers or clusters of monomers in the original npq2 membranes. For wt LHCII, light-induced trimer to monomer transitions have been observed (21), and it has been reported that high light induces monomerization of LHCII trimers in plant leaves, which could lead to more quenching and less efficient transfer of excitations to the reaction center (22). The monomeric state of LHCII or clusters of monomers thus represents a state that may also be present under stress conditions in vivo. In cryo-EM structures of PSII-LHCII supercomplexes, well-defined lipid molecules are resolved, which contributes to interactions between the LHCII and PSII core (27). Furthermore, a simulation study showed that the binding of DGDG lipids to trimeric LHCII at low pH reduces protein mobility (28). LHCII interprotein interactions and dynamics thus are influenced by lipid binding, which is controlled by monomerization, Zea binding, and pH conditions. Elevated levels of Zea associated with LHCII oligomers also enhance resistance to photooxidative stress by a lipid-protective mechanism (74). Increased affinity of lipids for Zea-containing LHCII therefore may serve as a way to protect thylakoid lipids under light stress.
In earlier work, we detected strong signals of immobilized lipids in the CP-based NMR spectrum of npq2 thylakoid membranes (63) and rigidification of thylakoid membranes has been observed for npq2 strains of A. thaliana (11,75). Our results here explain these observations, as we note that lipids can be trapped between (monomeric) npq2 LHCII complexes in galactolipid membranes. In thylakoid membranes, lipid densities are low and proteins occupy 70–80% of the total membrane surface area (76,77). In recent work, we estimated that, of the nonstructural lipids in Cr thylakoid membranes, ∼40% occupy the lipid annular shells around the proteins and ∼60% are bulk lipids with high mobility (78). Extensive binding of lipids to LHCIIs or entrapment of substantial number of lipids inside clusters of LHCIIs may significantly reduce the pool of mobile lipids and affect thylakoid membrane fluidity.
Conclusions
We report that lipid association is enhanced in Zea binding, monomeric LHCII and show that local conformation and dynamics of the protein matrix differ from that of membrane-embedded Vio-containing LHCII trimer. CG simulations of LHCII-lipid interactions are in agreement with the NMR data and show extensive lipid binding to LHCII monomers.
Zea binding by itself does not induce the photoprotective, quenched state (59), but promotes qE quenching by stabilizing the NPQ state (79). Considering the significant increase in lipid binding to npq2 LHCII, reversible Zea accumulation and monomerization of LHCII could play an important role in the control of protein-lipid interactions and conformational dynamics.
Author contributions
A.P. and F.A.-C. designed the research, with input from T.M., S.T., and M.B. F.A.-C. and M.E.W. performed the NMR experiments. S.T. designed and analyzed the CG simulations. S.T and S.J.M. discussed the simulation results. G.P. prepared the U-13C-npq2 LHCII complexes and F.A.-C. performed the protein membrane reconstitution. F.A.-C., S.T., and A.P. wrote the manuscript with input from all authors.
Acknowledgments
A.P. and F.A.-C. were financially supported by a CW-VIDI grant of the Netherlands Organization of Scientific Research (NWO) under grant no. 723.012.103. We thank Emanuela Crisafi for assistance with the liposome preparations and Diana Simionato for help with growing of the cells and preparation of the U-13C-npq2 LHCII complexes. M.E.W. is a recipient of a Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship. This work was supported in part by uNMR-NL, an NWO-funded National Roadmap Large-Scale Facility of the Netherlands (grant no. 184.032.207). S.T. thanks the European Commission for financial support via a Marie Skłodowska-Curie Actions Individual Fellowship (MicroMod-PSII, grant agreement 748895). S.T. and S.J.M. thank the Center for Information Technology of the University of Groningen for providing access to the Peregrine high-performance computing cluster and the NWO for providing access to the Dutch national supercomputer Cartesius.
Editor: Alemayehu Gorfe.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.12.039.
Supporting material
References
- 1.Ruban A.V., Johnson M.P., Duffy C.D.P. The photoprotective molecular switch in the photosystem II antenna. Biochim. Biophys. Acta Bioenerg. 2012;1817:167–181. doi: 10.1016/j.bbabio.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Ruban A.V. Nonphotochemical chlorophyll fluorescence quenching: mechanism and effectiveness in protecting plants from photodamage. Plant Physiol. 2016;170:1903–1916. doi: 10.1104/pp.15.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müller P., Li X.-P., Niyogi K.K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001;125:1558. doi: 10.1104/pp.125.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niyogi K.K., Grossman A.R., Björkman O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell. 1998;10:1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szabó I., Bergantino E., Giacometti G.M. Light and oxygenic photosynthesis: energy dissipation as a protection mechanism against photo-oxidation. EMBO Rep. 2005;6:629–634. doi: 10.1038/sj.embor.7400460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holt N.E., Zigmantas D., et al. Fleming G.R. Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science. 2005;307:433–436. doi: 10.1126/science.1105833. [DOI] [PubMed] [Google Scholar]
- 7.Park S., Fischer A.L., et al. Fleming G.R. Snapshot transient absorption spectroscopy of carotenoid radical cations in high-light-acclimating thylakoid membranes. J. Phys. Chem. Lett. 2017;8:5548–5554. doi: 10.1021/acs.jpclett.7b02486. [DOI] [PubMed] [Google Scholar]
- 8.Xu P., Tian L., et al. Croce R. Molecular insights into zeaxanthin-dependent quenching in higher plants. Sci. Rep. 2015;5:13679. doi: 10.1038/srep13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kress E., Jahns P. The dynamics of energy dissipation and xanthophyll conversion in arabidopsis indicate an indirect photoprotective role of zeaxanthin in slowly inducible and relaxing components of non-photochemical quenching of excitation energy. Front. Plant Sci. 2017;8:2094. doi: 10.3389/fpls.2017.02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruban A.V. Light harvesting control in plants. FEBS Lett. 2018;592:3030–3039. doi: 10.1002/1873-3468.13111. [DOI] [PubMed] [Google Scholar]
- 11.Tardy F., Havaux M. Thylakoid membrane fluidity and thermostability during the operation of the xanthophyll cycle in higher-plant chloroplasts. Biochim. Biophys. Acta. 1997;1330:179–193. doi: 10.1016/s0005-2736(97)00168-5. [DOI] [PubMed] [Google Scholar]
- 12.Krüger T.P., Ilioaia C., et al. van Grondelle R. Controlled disorder in plant light-harvesting complex II explains its photoprotective role. Biophys. J. 2012;102:2669–2676. doi: 10.1016/j.bpj.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niyogi K.K., Bjorkman O., Grossman A.R. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell. 1997;9:1369–1380. doi: 10.1105/tpc.9.8.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govindjee, Seufferheld M.J. Non-photochemical quenching of chlorophyll a fluorescence: early history and characterization of two xanthophyll-cycle mutants of Chlamydomonas reinhardtii. Funct. Plant Biol. 2002;29:1141–1155. doi: 10.1071/FP02061. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z., Yan H., et al. Chang W. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 16.Barros T., Kühlbrandt W. Crystallisation, structure and function of plant light-harvesting complex II. Biochim. Biophys. Acta. 2009;1787:753–772. doi: 10.1016/j.bbabio.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Wei X., Su X., et al. Liu Z. Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature. 2016;534:69–74. doi: 10.1038/nature18020. [DOI] [PubMed] [Google Scholar]
- 18.Tardy F., Havaux M. Photosynthesis, chlorophyll fluorescence, light-harvesting system and photoinhibition resistance of a zeaxanthin-accumulating mutant of Arabidopsis thaliana. J. Photochem. Photobiol. B. 1996;34:87–94. doi: 10.1016/1011-1344(95)07272-1. [DOI] [PubMed] [Google Scholar]
- 19.Lokstein H., Tian L., et al. DellaPenna D. Xanthophyll biosynthetic mutants of Arabidopsis thaliana: altered nonphotochemical quenching of chlorophyll fluorescence is due to changes in photosystem II antenna size and stability. Biochim. Biophys. Acta. 2002;1553:309–319. doi: 10.1016/s0005-2728(02)00184-6. [DOI] [PubMed] [Google Scholar]
- 20.Janik E., Bednarska J., et al. Gruszecki W.I. Light-induced formation of dimeric LHCII. Photosynth. Res. 2017;132:265–276. doi: 10.1007/s11120-017-0387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garab G., Cseh Z., et al. Horton P. Light-induced trimer to monomer transition in the main light-harvesting antenna complex of plants: thermo-optic mechanism. Biochemistry. 2002;41:15121–15129. doi: 10.1021/bi026157g. [DOI] [PubMed] [Google Scholar]
- 22.Bielczynski L.W., Schansker G., Croce R. Effect of light acclimation on the organization of photosystem II super- and sub-complexes in Arabidopsis thaliana. Front. Plant Sci. 2016;7:105. doi: 10.3389/fpls.2016.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wentworth M., Ruban A.V., Horton P. The functional significance of the monomeric and trimeric states of the photosystem II light harvesting complexes. Biochemistry. 2004;43:501–509. doi: 10.1021/bi034975i. [DOI] [PubMed] [Google Scholar]
- 24.Barros T., Royant A., et al. Kühlbrandt W. Crystal structure of plant light-harvesting complex shows the active, energy-transmitting state. EMBO J. 2009;28:298–306. doi: 10.1038/emboj.2008.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Standfuss J., Terwisscha van Scheltinga A.C., et al. Kühlbrandt W. Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 A resolution. EMBO J. 2005;24:919–928. doi: 10.1038/sj.emboj.7600585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen L., Huang Z., et al. Zhang X. Structure of a C2S2M2N2-type PSII–LHCII supercomplex from the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U S A. 2019;116:21246. doi: 10.1073/pnas.1912462116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng X., Liu X., et al. Liu Z. Structural roles of lipid molecules in the assembly of plant PSII−LHCII supercomplex. Biophys. Rep. 2018;4:189–203. doi: 10.1007/s41048-018-0068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daskalakis V., Papadatos S., Kleinekathöfer U. Fine tuning of the photosystem II major antenna mobility within the thylakoid membrane of higher plants. Biochim. Biophys. Acta Biomembr. 2019;1861:183059. doi: 10.1016/j.bbamem.2019.183059. [DOI] [PubMed] [Google Scholar]
- 29.Daskalakis V., Papadatos S., Stergiannakos T. The conformational phase space of the photoprotective switch in the major light harvesting complex II. Chem. Commun. 2020;56:11215–11218. doi: 10.1039/d0cc04486e. [DOI] [PubMed] [Google Scholar]
- 30.Liguori N., Croce R., et al. Thallmair S. Molecular dynamics simulations in photosynthesis. Photosynth. Res. 2020;144:273–295. doi: 10.1007/s11120-020-00741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liguori N., Periole X., et al. Croce R. From light-harvesting to photoprotection: structural basis of the dynamic switch of the major antenna complex of plants (LHCII) Sci. Rep. 2015;5:15661. doi: 10.1038/srep15661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thallmair S., Vainikka P.A., Marrink S.J. Lipid fingerprints and cofactor dynamics of light-harvesting complex II in different membranes. Biophys. J. 2019;116:1446–1455. doi: 10.1016/j.bpj.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azadi-Chegeni F., Ward M.E., et al. Pandit A. Conformational dynamics of light-harvesting complex II in a native membrane environment. Biophys. J. 2021;120:270–283. doi: 10.1016/j.bpj.2020.11.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crisafi E., Pandit A. Disentangling protein and lipid interactions that control a molecular switch in photosynthetic light harvesting. Biochim. Biophys. Acta. 2017;1859:40–47. doi: 10.1016/j.bbamem.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Kirchhoff H., Mukherjee U., Galla H.J. Molecular architecture of the thylakoid membrane: lipid diffusion space for plastoquinone. Biochemistry. 2002;41:4872–4882. doi: 10.1021/bi011650y. [DOI] [PubMed] [Google Scholar]
- 36.Färber A., Jahns P. The xanthophyll cycle of higher plants: influence of antenna size and membrane organization. Biochim. Biophys. Acta. 1998;1363:47–58. doi: 10.1016/s0005-2728(97)00093-5. [DOI] [PubMed] [Google Scholar]
- 37.Pines A., Gibby M.G., Waugh J.S. Proton-enhanced NMR of dilute spins in solids. J. Chem. Phys. 1973;59:569–590. [Google Scholar]
- 38.Hartmann S.R., Hahn E.L. Nuclear double resonance in the rotating frame. Phys. Rev. 1962;128:2042–2053. [Google Scholar]
- 39.Baldus M., Petkova A.T., et al. Griffin R.G. Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 1998;95:1197–1207. [Google Scholar]
- 40.Fung B.M., Khitrin A.K., Ermolaev K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 2000;142:97–101. doi: 10.1006/jmre.1999.1896. [DOI] [PubMed] [Google Scholar]
- 41.Weingarth M., Demco D.E., et al. Tekely P. Improved magnetization transfer in solid-state NMR with fast magic angle spinning. Chem. Phys. Lett. 2009;469:342–348. [Google Scholar]
- 42.Baldus M., Meier B.H. Total correlation spectroscopy in the solid state. The use of scalar couplings to determine the through-bond connectivity. J. Magn. Reson. Ser. A. 1996;121:65–69. [Google Scholar]
- 43.Morris G.A., Freeman R. Enhancement of nuclear magnetic resonance signals by polarization transfer. J. Am. Chem. Soc. 1979;101:760–762. [Google Scholar]
- 44.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Jong D.H., Singh G., et al. Marrink S.J. Improved parameters for the Martini coarse-grained protein force field. J. Chem. Theory Comput. 2013;9:687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- 46.Abraham M.J., Murtola T., et al. Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 47.Poma A.B., Cieplak M., Theodorakis P.E. Combining the MARTINI and structure-based coarse-grained approaches for the molecular dynamics studies of conformational transitions in proteins. J. Chem. Theory Comput. 2017;13:1366–1374. doi: 10.1021/acs.jctc.6b00986. [DOI] [PubMed] [Google Scholar]
- 48.Souza P.C.T., Thallmair S., et al. Mera-Adasme R. An allosteric pathway in copper, zinc superoxide dismutase unravels the molecular mechanism of the G93A amyotrophic lateral sclerosis-linked mutation. J. Phys. Chem. Lett. 2019;10:7740–7744. doi: 10.1021/acs.jpclett.9b02868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J., Yan R., et al. Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Eerden F.J., de Jong D.H., et al. Marrink S.J. Characterization of thylakoid lipid membranes from cyanobacteria and higher plants by molecular dynamics simulations. Biochim. Biophys. Acta. 2015;1848:1319–1330. doi: 10.1016/j.bbamem.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 51.de Jong D.H., Baoukina S., et al. Marrink S.J. Martini straight: boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 2016;199:1–7. [Google Scholar]
- 52.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 53.Berendsen H.J.C., Postma J.P.M., et al. Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 54.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 55.Corradi V., Mendez-Villuendas E., et al. Tieleman D.P. Lipid–protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 2018;4:709–717. doi: 10.1021/acscentsci.8b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Oort B., van Hoek A., et al. van Amerongen H. Aggregation of light-harvesting complex II leads to formation of efficient excitation energy traps in monomeric and trimeric complexes. FEBS Lett. 2007;581:3528–3532. doi: 10.1016/j.febslet.2007.06.070. [DOI] [PubMed] [Google Scholar]
- 57.Moya I., Silvestri M., et al. Bassi R. Time-resolved fluorescence analysis of the photosystem II antenna proteins in detergent micelles and liposomes. Biochemistry. 2001;40:12552–12561. doi: 10.1021/bi010342x. [DOI] [PubMed] [Google Scholar]
- 58.Natali A., Gruber J.M., et al. Croce R. Light-harvesting complexes (LHCs) cluster spontaneously in membrane environment leading to shortening of their excited state lifetimes. J. Biol. Chem. 2016;291:16730–16739. doi: 10.1074/jbc.M116.730101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Son M., Pinnola A., Schlau-Cohen G.S. Zeaxanthin independence of photophysics in light-harvesting complex II in a membrane environment. Biochim. Biophys. Acta Bioenerg. 2020;1861:148115. doi: 10.1016/j.bbabio.2019.148115. [DOI] [PubMed] [Google Scholar]
- 60.Bonaccorsi M., Le Marchand T., Pintacuda G. Protein structural dynamics by magic-angle spinning NMR. Curr. Opin. Struct. Biol. 2021;70:34–43. doi: 10.1016/j.sbi.2021.02.008. [DOI] [PubMed] [Google Scholar]
- 61.Shi C., Öster C., et al. Lange A. Structure and dynamics of the rhomboid protease GlpG in liposomes studied by solid-state NMR. J. Am. Chem. Soc. 2019;141:17314–17321. doi: 10.1021/jacs.9b08952. [DOI] [PubMed] [Google Scholar]
- 62.Pinto C., Mance D., et al. Baldus M. Studying assembly of the BAM complex in native membranes by cellular solid-state NMR spectroscopy. J. Struct. Biol. 2019;206:1–11. doi: 10.1016/j.jsb.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 63.Azadi Chegeni F., Perin G., et al. Pandit A. Protein and lipid dynamics in photosynthetic thylakoid membranes investigated by in-situ solid-state NMR. Biochim. Biophys. Acta. 2016;1857:1849–1859. doi: 10.1016/j.bbabio.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 64.Su Y., Hong M. Conformational disorder of membrane peptides investigated from solid-state NMR line widths and line shapes. J. Phys. Chem. B. 2011;115:10758–10767. doi: 10.1021/jp205002n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wishart D.S., Nip A.M. Protein chemical shift analysis: a practical guide. Biochem. Cell Biol. 1998;76:153–163. doi: 10.1139/bcb-76-2-3-153. [DOI] [PubMed] [Google Scholar]
- 66.de Souza L.M., Iacomini M., et al. Sassaki G.L. Glyco- and sphingophosphonolipids from the medusa Phyllorhiza punctata: NMR and ESI-MS/MS fingerprints. Chem. Phys. Lipids. 2007;145:85–96. doi: 10.1016/j.chemphyslip.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 67.Marcolongo G., De Appolonia F., et al. Ceschi Berrini C. Diacylglycerolipids isolated from a thermophile cyanobacterium from the Euganean hot springs. Nat. Product Res. 2006;20:766–774. doi: 10.1080/14786410500176393. [DOI] [PubMed] [Google Scholar]
- 68.Azadi-Chegeni F., Ward M.E., Pandit A., et al. Conformational dynamics of a photosynthetic light-harvesting complex in native thylakoid membranes. Biophys. J. 2021;120:270–283. doi: 10.1016/j.bpj.2020.11.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van Eerden F.J., Melo M.N., et al. Marrink S.J. Prediction of thylakoid lipid binding sites on photosystem II. Biophys. J. 2017;113:2669–2681. doi: 10.1016/j.bpj.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Javanainen M., Martinez-Seara H., Vattulainen I. Excessive aggregation of membrane proteins in the Martini model. PLoS One. 2017;12:e0187936. doi: 10.1371/journal.pone.0187936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alessandri R., Souza P.C.T., et al. Marrink S.J. Pitfalls of the Martini model. J. Chem. Theory Comput. 2019;15:5448–5460. doi: 10.1021/acs.jctc.9b00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Souza P.C.T., Alessandri R., et al. Marrink S.J. Martini 3: a general purpose force field for coarse-grained molecular dynamics. Nat. Methods. 2021;18:382–388. doi: 10.1038/s41592-021-01098-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fehr N., Dietz C., et al. Paulsen H. Modeling of the N-terminal section and the lumenal loop of trimeric light harvesting complex II (LHCII) by using EPR. J. Biol. Chem. 2015;290:26007–26020. doi: 10.1074/jbc.M115.669804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson M.P., Havaux M., et al. Horton P. Elevated zeaxanthin bound to oligomeric LHCII enhances the resistance of Arabidopsis to photooxidative stress by a lipid-protective, antioxidant mechanism. J. Biol. Chem. 2007;282:22605–22618. doi: 10.1074/jbc.M702831200. [DOI] [PubMed] [Google Scholar]
- 75.Havaux M. Carotenoids as membrane stabilizers in chloroplasts. Trends Plant Sci. 1998;3:147–151. [Google Scholar]
- 76.Kirchhoff H. Molecular crowding and order in photosynthetic membranes. Trends Plant Sci. 2008;13:201–207. doi: 10.1016/j.tplants.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 77.Kirchhoff H. Structural changes of the thylakoid membrane network induced by high light stress in plant chloroplasts. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130225. doi: 10.1098/rstb.2013.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nami F., Tian L., et al. Pandit A. Lipid and protein dynamics of stacked and cation-depletion induced unstacked thylakoid membranes. BBA Adv. 2021;1:100015. doi: 10.1016/j.bbadva.2021.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jahns P., Holzwarth A.R. The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim. Biophys. Acta. 2012;1817:182–193. doi: 10.1016/j.bbabio.2011.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.