ABSTRACT
Biological organisms carry a rich potential for removing toxins from our environment, but identifying suitable candidates and improving them remain challenging. We explore the use of computational tools to discover strains and enzymes that detoxify harmful compounds. In particular, we focus on mycotoxins—fungus-produced toxins that contaminate food and feed—and biological enzymes that are capable of rendering them less harmful. We discuss the use of established and novel computational tools to complement existing empirical data in three directions: discovering the prospect of detoxification among underexplored organisms, finding important cellular processes that contribute to detoxification, and improving the performance of detoxifying enzymes. We hope to create a synergistic conversation between researchers in computational biology and those in the bioremediation field. We showcase open bioremediation questions where computational researchers can contribute and highlight relevant existing and emerging computational tools that could benefit bioremediation researchers.
KEYWORDS: biodegradation, bioinformatics, bioremediation, computational biology, enzymes, exoenzymes, mycotoxins, toxins
INTRODUCTION
BACKGROUND AND MOTIVATION
Context: detoxifying contaminated food and feed.
Fungi that grow on foodstuffs are one of the major sources of contamination in food and feed; these fungus-produced toxins are called mycotoxins. Currently, an estimated 25% of world crops is thought to get contaminated with mycotoxins each year (1, 2), putting a major burden on agriculture and public health. Preventing contamination or detoxifying mycotoxins is a major safety priority (3). In what follows, we briefly describe the threat of mycotoxins and the potentials of biological organisms to address this threat via detoxifying enzymes. We then explore the use of computational approaches to discover and improve such potentials. We primarily discuss three aspects: (i) the use of bioinformatics tools to search genomic databases for candidate species and enzymes, (ii) the use of genetics and genomics data to investigate how the detoxification performance can be improved, and (iii) the use of computational tools to improve the detoxifying enzymes. While we discuss established computational methods used in identifying mycotoxin-degrading enzymes, we also consider the use of novel, field-adjacent methods that have potential in mycotoxin detoxification.
Mycotoxins are prevalent and harmful.
Mycotoxins are secondary metabolites produced by a variety of filamentous fungi that contaminate common food crops and cause negative health effects in animals and humans. More than 300 types of mycotoxins have been identified so far, all of which would be candidates for detoxification (1). Among these, six major types are of particular interest and the focus of this review because of their detrimental health impact and because they routinely contaminate foods and animal feed (4, 5): aflatoxin (AF), ochratoxin (OT), zearalenone (ZEA), fumonisin, deoxynivalenol (DON), and patulin.
Aflatoxins, produced by Aspergillus species, are among the most carcinogenic naturally occurring substances and active inducers of mutations, liver cancer, congenital malformations, hormone disorders, and immunodepression (6, 7). Ochratoxin is also produced by Aspergillus species, as well as certain Penicillium species, and is a nephrotoxin, immunosuppressant, potent teratogen, and renal carcinogen (6, 8, 9). Zearalenone and fumonisins are produced by Fusarium species. ZEA acts through estrogen mimicry to dysregulate the hormone receptor and antagonize the estrogen pathway, leading to reproductive disorders, hormone imbalance, and breast cancer (6, 10). Fumonisins have been linked to esophageal cancer in humans as well as a variety of health complications in animals such as pulmonary edema and hepatotoxicity (6, 11). Produced by Fusarium graminearum, DON is a vomitoxin, causing emetic and nauseous effects after ingestion (12). Finally, patulin is produced by ascomycetes such as Penicillium, Aspergillus, and Byssochlamys species and is commonly found in fruit and vegetable products, especially rotten apples and apple juice (13). Patulin ingestion is linked to a number of health complications, namely, immune suppression, ulcers, gastrointestinal inflammation, and embryotoxicity (13). There are a variety of food crops that these mycotoxins contaminate, including cereal crops such as wheat, barley, corn, and oats (6, 11). Due to the serious health implications of mycotoxin contamination, economic losses arise from reduction of crop and livestock yields as well as the cost of decontamination efforts. Annually, the United States faces an estimated $932 million in economic losses from AFs, fumonisins, and DON alone (14). This sizable economic burden is faced across agriculture and livestock producers globally and requires efficient and cost-effective measures as a solution.
Mycotoxin buildup on foodstuff necessitates methods of decontamination in order to supply safe foods for consumption. Currently, decontamination is limited to physical and chemical methods. Physical methods, including sorting and cleaning, have been shown to be effective in some, but not all, cases of mycotoxin contamination. Chemical methods, which use chemical agents to reduce or convert mycotoxins into less toxic by-products, include ozonation and ammoniation. While these physical and chemical methods have been used to reduce mycotoxin contamination, they suffer from high operational costs and limited reliability and may decrease the quality or nutritional value of the food (3, 14–16). These limitations expose the need to look for better solutions.
Toxin removal by biological processes is a promising solution.
Bioremediation, or the use of biological entities to detoxify or remove toxins in the environment, is a promising alternative to current decontamination methods. Bioremediation offers lower costs, fewer undesired environmental side effects, and potentially higher efficiency and reliability (17–19). The use of microbes is a particularly attractive choice in bioremediation, offering faster activity and the feasibility of strain evolution and engineering for improved performance (20). There are six key factors that make a good bioremediator: (i) it is fast and efficient at degradation, (ii) it yields safe degradation products, (iii) it is nonpathogenic to plants, animals, and humans, (iv) it is not detrimental to the quality of the food/feed, (v) it is applicable outside lab settings, and (vi) it is applicable to multiple pollutants (17). Among identified mycotoxin degraders, none effectively has all of these factors, with speed and efficiency often being subpar. Additionally, the mechanisms of degradation by these identified microorganisms are often unknown or understudied, limiting the ability to improve upon the native degradation performance. Therefore, identifying new species that possess mycotoxin degradation ability and elucidating the mechanisms of degradation are beneficial in making this capability effective and commercially viable.
Modes of biological detoxification.
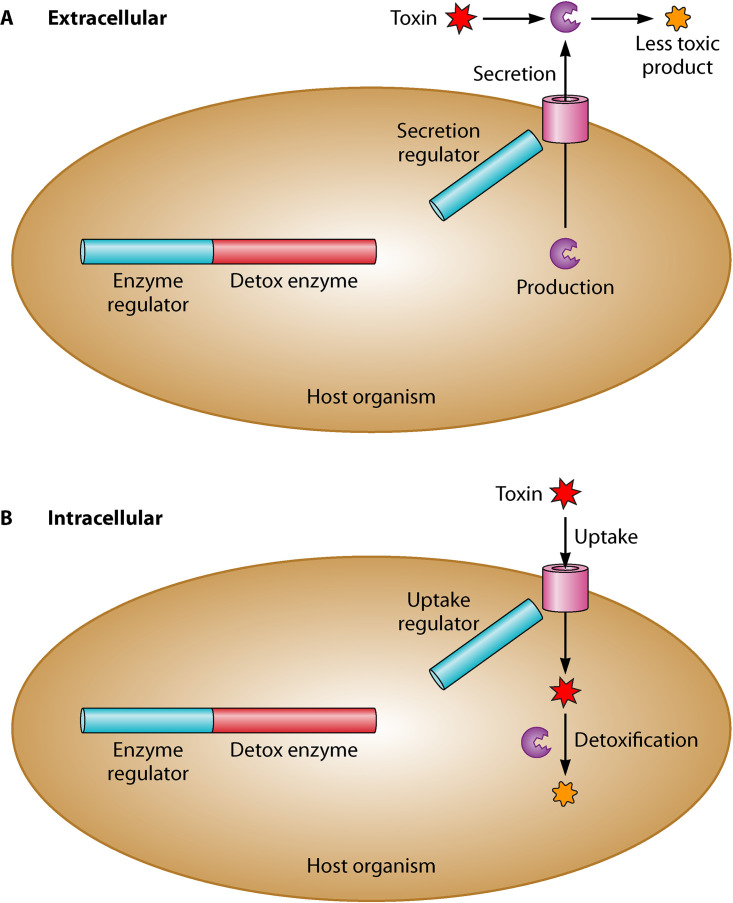
In the context of microbial interventions for removing mycotoxins, the two main modes of detoxification are adsorption and biotransformation. In adsorption, mycotoxins are physically bound to polysaccharides and proteins in the outer cell structures (21, 22). Biotransformation utilizes microbes and their enzymes to convert mycotoxins into nontoxic compounds (23, 24). In this paper we are solely concerned with methods to identify and improve biotransformation processes. Biotransformation can be further broken into two categories (schematically shown in Fig. 1): secretion of enzymes (extracellular degradation) and uptake of the toxin into the cell (intracellular degradation). Intracellular degradation of toxins more closely follows normal metabolic processing of molecules by microbes inside the cell. Microbes that mitigate mycotoxins through extracellular degradation are more likely to produce stable enzymes that can be isolated and used in practice; this has been the strategy for several existing commercial products (25–27). Table 1 shows some of the bacterial and fungal enzymes that have been found to degrade major mycotoxins.
FIG 1.
Simplified representation of the cellular machinery involved in extracellular (A) versus intracellular (B) detoxification.
TABLE 1.
Representative examples of identified bacterial and fungal enzymes with the capability to degrade major mycotoxinsa
| Mycotoxin category | Enzyme family | Organism(s) | Reference |
|---|---|---|---|
| Aflatoxin | Reductase | Mycobacterium smegmatis | 28 |
| Manganese peroxidase | Phanerochaete sordida | 29 | |
| Laccase (oxidase) | Trametes versicolor | 30 | |
| Manganese peroxidase | Irpex lacteus | 31 | |
| Pseudomonas AFB1-degrading enzyme | Pseudomonas aeruginosa | 32 | |
| Deoxynivalenol | Peroxidase | Aspergillus oryzae, Rhizopus oryzae | 33 |
| Fumonisin | Fumonisin esterase | Sphingopyxis sp. | 34 |
| Manganese peroxidase | Irpex lacteus | 31 | |
| Ochratoxin | Carboxypeptidase* | Aspergillus spp. | 35 |
| Peptidase* | Pediococcus parvulus | 36 | |
| Carboxypeptidases | Yarrowia lipolytica | 37 | |
| Patulin | Orotate phosphoribosyltransferase | Rhodotorula mucilaginosa | 38 |
| Zearalenone | Zearalenone detoxification gene(s) | Pseudomonas putida | 39 |
| Zearalenone lactonohydrolase | Clonostachys rosea | 40 | |
| Esterase* | Lactobacillus plantarum | 41 | |
| Manganese peroxidase | Irpex lacteus | 31 | |
| Zearalenone lactonohydrolase | Pichia pastoris | 42 |
Enzymes hypothesized but not yet confirmed are marked by an asterisk.
Enzymatic degradation has been suggested in a number of studies; however, identification of the degrading enzymes has proven difficult. Sangare et al. describe a Pseudomonas species capable of degrading aflatoxin B1 (AFB1) from cell-free culture supernatant, suggesting that an extracellular enzyme is responsible for the degradation (43). Screening for the effect of common functional cofactors may potentially help identify the enzyme class. Similar extracellular degradation has been reported for Rhodococcus spp., Stenotrophomonas spp., and Myxococcus spp. (44–46). DON has been observed to be assimilated as a carbon source in some, but not all, strains (47). Other extracellular enzymes with mycotoxin-degrading abilities include oxidoreductase, dehydrogenase, aldo-keto reductases, and peroxidases (48–50). While there has been less focus on intracellular mechanisms, intracellular enzymatic degradation has been shown by Zhu et al. (51).
Bacteria and fungi carry a rich repertoire of enzymes capable of removing mycotoxins.
Biotransformation of mycotoxins into nontoxic products by bacterial and fungal enzymes has already been demonstrated (19, 48, 52, 53). The detoxification performance can be improved by identifying and characterizing the enzymes with degradation/detoxification capability. On one side, uncovering the cellular machinery of degradation (schematically shown in Fig. 1 and explored in “Regulation: even when the detoxification capability exists in an organism, its availability may be under regulation” below) allows us to select conditions to express the enzyme (when searching for candidates) or engineer strains to improve their performance. On the other hand, the enzyme itself can be modified and improved. Structural modeling and design-of-experiments (DOE) techniques can shed light on the identification of key structural components that contribute to degradation (52).
In the remainder of this work, we limit the scope to extracellular bacterial and fungal detoxifying enzymes. We make this choice to offer a more focused view on recent developments in computational tools for biological enzymes, but also because deploying enzymes (versus live organisms) in food/feed applications is a more practical approach (23, 52). The use of enzymes for reducing the threat of mycotoxins has reached industrial applications, even if only in a few cases. The Mycofix line of products (27) combines different modalities, including biotransformation and adsorption, to remove several mycotoxins from feed. FUMzyme is a commercially available fumonisin esterase produced in a genetically modified strain of Komagataella pastoris (54) that has shown success in removing contamination from feed (26). However, more research is still needed to improve the performance of mycotoxin removal.
Several previous reports have cataloged specific enzymes that act on mycotoxins (17, 48, 55), and Table 1 lists representative examples for the major mycotoxins explored in this review. Here, instead, we focus on current challenges and questions in the field of mycotoxin detoxification that can be addressed by computational tools. In this context, we survey some of the existing tools that have already been applied in this field and then propose emerging tools that have the potential to lead to transformative progress.
CURRENT CHALLENGES AND COMPUTATIONAL SOLUTIONS
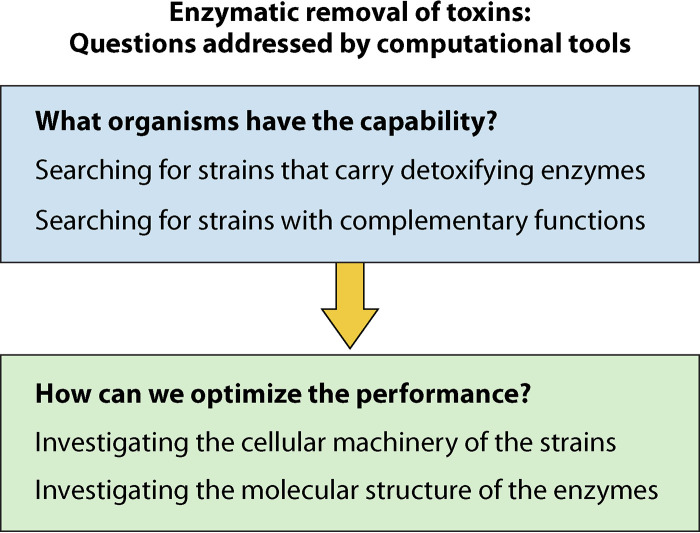
How can we effectively remove mycotoxins using biological organisms? Conceptually, we break down this search into two steps: (i) finding organisms that have this capability and (ii) optimizing the performance by modifying the environmental conditions, the detoxifying strain, or the target enzymes. We survey existing computational tools that can facilitate this process (Fig. 2). We focus our discussions on genomic and structural biology tools. We acknowledge that there are other useful tools—including proteomics—that can offer additional insights but are beyond the scope of this minireview.
FIG 2.
Conceptual breakdown of major questions of interest where computational tools can facilitate more efficient removal of toxins.
Finding candidate organisms: who can do the job?
Discovering organisms that can degrade mycotoxins poses a number of challenges that can be met through both experimental and computational approaches. In terms of enzymatic degradation, there are three challenges to be addressed. First, organisms must have the genes necessary to produce enzymes and possibly cofactors involved in degradation. Second, the organisms must have favorable regulatory mechanisms for these enzymes. Third, the method of obtaining and isolating the enzymes must be favorable to the end use case. One can describe the search space as being largely defined by these characteristics that may be specific to the use cases but are still conceptually similar among different cases.
From the experimental front, high-throughput screening may be used both to identify candidate organisms and to explore mutations for optimizing degradation potential. Environmental isolates are a traditional source for identifying mycotoxin degraders. Isolates can be cultivated and tested for degradation, especially when high-throughput screening is possible. As an example, Ciegler et al. screened ∼1,000 organisms, both prokaryotes and eukaryotes, for their aflatoxin degradation capability (56). Screening can also be used for optimizing the environmental conditions or the enzyme itself. However, unless feasible high-throughput assays are available, this process is resource and time expensive. Therefore, looking to computational methods to screen for new organisms will be beneficial.
As an example, there is a known, highly specific two-step enzymatic process in the detoxification of fumonisin, which involves a carboxylesterase and an aminotransferase (34). This becomes a useful bottleneck in the search space, as candidate organisms must contain both enzyme-encoding genes to be viable degraders. Toward this end, tools such as BLASTp (57) can be utilized in cases where genome sequences are available. Simply put, the presence of these two genes largely dictates whether or not an organism is a fumonisin degrader. On the other hand, in the example of AF detoxification, many species can possess hydrolases or oxidases related to those that are known to degrade AF (24, 48, 58). The search space is instead constrained on a separate manifold involving the specificity and affinity of the hydrolase for AFs. That is, the presence of the same hydrolase gene may not be sufficient to identify degradation potential, since it may be optimized for a different substrate. The sequence-to-function relationship then becomes critical, which is not guaranteed to be captured by sequence similarity à la BLASTp. This shortcoming can be thought of as a signal-to-noise ratio, where key amino acids involved in the active-site mechanism are sparse signals, and the rest of the sequence functions primarily to provide the correct structural shape and may be noisy in this regard. This is witnessed in the work by Dellafiora and colleagues (59), in which two related, AF-degrading oxidases shared only 72% sequence similarity, despite using the same mechanism for degradation. In a more extreme example, a recently identified carboxylesterase that degrades fumonisin showed only around 34% sequence similarity to previously reported fumonisin-degrading carboxylesterases (60).
Similarity in sequence does not necessarily overlap similarity in function. Sequence similarity may be used to imply functional similarity; however, such a predicate does not include enzymes that share functional similarity without sequence similarity. High sequence similarity among closely related species might not fully overlap functional similarity either. Therefore, searches should be conducted on a sequence-to-function relationship model. While this method loses the high-throughput optimizations of BLAST-based sequence similarity, it may be modeled via a reductive filter pipeline to maintain reasonable complexity. It also loses the generalizability of sequence similarity, and instead, pipelines must be custom designed for each case. Dellafiora et al. have combined an in silico screen with an enzymatic assay to address this challenge in search of hydrolyzing enzymes that can degrade ochratoxin A (61). In the example of AFs, initial work has been performed to design a structure-to-function reductive filter model using a number of filters. Furthermore, this model does not necessarily require a labeled, positive enzyme to seed the search; rather, it only requires characteristics to build the filters. Prior research by Risa and colleagues (62) has revealed that excreted enzymes can be responsible for degrading AFs. SignalP is able to predict protein excretion in bacteria and can be used as an initial filter to narrow down proteomes. These sequences can be passed through both size- and sequence-based enzyme classification filters based on facile experimental determinations to further reduce the candidate pool. From here, three-dimensional (3D) structures may be built, the binding pockets predicted, and AF docked to identify high-affinity interactions that then may be confirmed experimentally. These computational processes are expanded below. The reductive filter model uses low-complexity tools at its head, increasing in complexity toward the tail to ensure efficiency. Similarly, its modular nature allows for easy insertion or upgrading of components as advances occur in each domain.
Community-level detoxification: when the task needs to be divided.
Mycotoxin degradation may require multiple reactions to reach by-products with significantly decreased toxicity. There are several examples in which a single enzyme is insufficient for complete degradation and two or more enzymatic steps are required for the detoxification process. In such cases, we need to better understand how multiple enzymes from the same, or even different, species are required for degradation of a single mycotoxin. While this increases the difficulty and cost of searching for degrading enzymes that can work together, the outcome of complete degradation and reduced toxicity is desirable for application in agriculture, in which mycotoxin levels must fall under set regulatory limits. For degradation of fumonisin B1 by Sphingopyxis sp. strain MTA144, Heinl et al. found that two enzymes were involved (34). A carboxylesterase facilitated the initial deesterification step to form a hydrolyzed fumonisin B1, which is less active in its known ceramide synthase inhibitory pathway but still possesses a significant toxic effect (34, 63). A second enzyme, an aminotransferase, deaminated the hydrolyzed by-product of the first reaction resulting in complete degradation and loss of toxic effects (34). Similarly, Carere et al. elucidated a two-component enzymatic pathway involved in the epimerization of DON by Devosia mutans 17-2-E-8 (64, 65). The enzymes, designated DepA and DepB, first oxidize DON into 3-keto-DON (DepA) (64) and subsequently reduce 3-keto-DON into 3-epi-DON (DepB) (65), significantly reducing toxicity. These examples highlight the need to understand all the enzymes playing a role in complete degradation.
In some instances, mycotoxin biotransformation does not lead to complete detoxification (52); DON degradation, above as an example, leads to end products that are less toxic than the starting substrate but still retain some toxicity. In biotransformation of ZEA, there are cases in which microbial breakdown results in by-products, α-zearalenol and β-zearalenol, that are even more toxic than the original compound (39, 66, 67). In such cases, we need to identify additional species or enzymes that can take the by-products and convert them into nontoxic compounds in a multistep process.
Multistep degradation underscores the possible need to look beyond single microorganisms and employ microbial consortia to complete the job; as an example, Wang et al. discovered a microbial consortium that utilizes multiple species across various taxa working in unison to transform ZEA to nontoxic by-products (68). Bioinformatic searches for identifying multiple enzymes necessary for a particular case would be an extension of the single-enzyme searches discussed in the previous section, using similar tools. Of note could be searching for individual organisms that carry two or more necessary enzymes that have previously been identified in multiple species/strains.
Regulation: even when the detoxification capability exists in an organism, its availability may be under regulation.
Even after organisms have been identified that are capable of detoxifying target pollutants, the availability of the relevant enzymes depends on whether the environmental context induces the relevant genes of enzyme production and secretion effectively. These considerations point to the need to explore the internal regulation of the production and secretion of detoxification enzymes. Microorganisms respond to cellular and environmental changes through regulatory decisions that could impact the availability of degradation machinery for target pollutants (69). Production of enzymes is regulated through different mechanisms, such as transcription factors binding in and around promoter regions. These mechanisms are likely influenced by nutrient availability and overall conditions of the cell (i.e., growth phase) (70). Secreted enzymes have an added layer of regulation due to the high energy cost of secretion. While these enzymes have beneficial effects, often being employed to break down macromolecules in the environment for cellular uptake, they also incur an energy/biomass cost (71). Therefore, certain enzymes targeted for secretion are up- or downregulated by the presence of nutrients in the environment that, respectively, do or do not require extracellular breakdown.
Here, we primarily emphasize the existing native potential as the starting point, even though ultimately the deployment likely happens in a safe and tractable host organism. Our discussion on regulation and the detoxification machinery in the native context has two purposes. (i) It reveals the preferred conditions for the expression of the detoxification machinery to enable more effective screening for functions of interest. (ii) It allows us to better understand the diversity of possibilities and the ideal machinery to be transferred to a host organism. Understanding the influence of regulation on production and secretion of the enzyme is also necessary for strain optimization to factor in the cost-benefit balance of increased enzyme production and secretion.
Several existing bioinformatic tools can help us uncover aspects of bacterial gene regulation, such as promoter and DNA binding sites, operon regions, and secretion signals, which are touched on in the following sections. The usefulness of these tools in the context of bioremediation is that they allow researchers to uncover possible mechanisms of regulation that control the detoxification process. Insight from regulation, for example, similarity to a known catabolic pathway, can also be used to choose suitable environmental conditions or infer the mechanism of degradation.
(i) Promoter prediction.
Identifying promoter regions and DNA binding sites is important in that transcription initiation is the most frequently regulated step in gene expression. Promoters contain an intrinsic strength that governs the amount of transcription a gene undergoes and when transcription occurs according to environmental factors such as nutrient availability (70). It is important to properly regulate gene expression to ensure that the degrading enzyme is sufficiently expressed, but only when the particular substrate is present to limit wasteful production of enzymes that are disadvantageous to the cell without the substrate (72). By uncovering promoters associated with genes/enzymes of interest in bioremediation, we can understand how the cell naturally regulates its expression and better manipulate it toward improved expression for application in agriculture. There are several existing tools for predicting and cataloging promoter regions in different organisms, such as phiSITE (73, 74), SAPPHIRE (75), PRODORIC2 (76), BacPP (77), and Promoter Prediction Convolutional Neural Network (PPCNN) (78). We expand here on the last three.
PRODORIC2 is a transcription factor binding site (TFBS) database that possesses one of the largest collections of DNA binding sites in prokaryotic organisms (76). In 2018, its most recent update, PRODORIC2 expanded its database to host the genomic information of 2,274 bacterial strains and their 5,191 replicons (76). This database is curated to include only experimentally validated binding sites, limiting the expanse of bacterial species it contains but ensuring accuracy in its TFBS inventory. De Avila e Silva et al. created a bioinformatic tool, BacPP, to predict promoter sequences in Escherichia coli strains through neural network simulations (77). BacPP is able to recognize and predict promoter sites with various levels of accuracy (all above 83%) across the different sigma factors crucial for prokaryotic transcription initiation (77). Additionally, BacPP has 76% prediction accuracy among other enterobacterial species (77). The advantage of this method is its ability to classify promoter sequences by its sigma factor, an important distinguishing feature that was a shortcoming of previous tools. However, BacPP is currently limited to E. coli and, to a lower degree of accuracy, enterobacteria. Another promoter prediction tool is PPCNN, developed for both eukaryotic and prokaryotic prediction and implemented into the CNNProm program. This approach uses deep learning neural networks for its prediction models (78). For prokaryotes, PPCNN was trained on E. coli and Bacillus subtilis, offering insight into both Gram-positive and Gram-negative species. A highlight of this method is its applicability to other sequenced species because it predicts promoters without prior knowledge of specific promoter features (78).
(ii) Operon prediction.
Metabolically or functionally related genes within prokaryotic genomes are often arranged in contiguous segments called operons and are cotranscribed along the same mRNA (79). This organization imparts an added layer of regulation on the genes within the operon. Specifically, in the context of bioremediation, if an enzyme of interest is encoded within an operon, it opens up new genes that could help play a role in degradation, either functionally or through regulation. As an example, Heinl et al. identified two fumonisin degrading enzymes that were held within a gene cluster organized in two operons and subsequently determined that other genes in the operon held importance to transcriptional regulation and transport of the degrading enzymes, as well as additional enzymes that might play a role in further breakdown of the degradation by-products (34). Additionally, downstream utilization of the enzyme-encoding gene(s) can be affected by its placement within an operon. For example, Altahli and El-Deeb transferred ZEA degradation capability in Pseudomonas putida into E. coli via a plasmid encoding detoxification genes (39). Multiple genes were shown to be expressed for detoxification; however, the authors were unable to separate these genes due to their organization in operons. Therefore, understanding the genomic organization of these genes within operons can aid in their use for degradation. Determining operons computationally has been a field of interest for a number of years, leading to tools such as Operon DataBase (80, 81), OperomeDB (82, 83), Operon Hunter (84), and Operon-mapper (85, 86), with recent advances in de novo prediction of operons from genomic data, which is expanded on below.
Operon-mapper, a web-based server for operon prediction, was developed in 2018 and is the first publicly available tool for operon prediction that requires only genome sequences as the input (85, 86). Operon-mapper uses a five-step procedure: (i) open reading frame (ORF) prediction using Prokka software (87, 88), (ii) homology gene determination using the hmmsearch program based on hidden Markov models (85, 88), (iii) intergenic-distance evaluation using a custom program (85), (iv) operon prediction using an artificial neural network with intergenic distance and a score defining functional relatedness of protein products as the input arguments (85, 89, 90), and (v) gene function assignment using the DIAMOND algorithm (91). The accuracy of this method in predicting operons was ∼90% across eight tested genomes with various sizes and GC contents, and this method outperformed other algorithms in a recent evaluation of correlation to experimentally validated operons (92). Operon-mapper also has the advantage of providing ORF identification and functional annotation of protein (85).
(iii) Secreted-protein prediction.
A signal peptide (SP) is a sequence of amino acids in a newly synthesized protein that targets the protein into or across the membranes in the cell (93). Determining whether and how an enzyme is secreted outside the cell enables better utilization of the degradation machinery (schematically represented in Fig. 1A). To predict secreted proteins, several algorithms to identify SPs within a proteome have been developed: SignalP (94), Psort (95), Pred-Tat (96), and TatP (97).
Of note, SignalP is able to determine these secretion signals and distinguish between the type of secretion pathway. The current version, SignalP 5.0, uses deep neural networks in combination with conditional random field classification and optimized transfer learning to determine SPs in prokaryotes, eukaryotes, and archaea (94). This update builds upon previous versions based on artificial neural networks (98), with added improvements of hidden Markov models (99), enhanced cleavage site predictions (100), and discrimination of signal peptides and transmembrane helices (101). For prokaryotes, there are two main secretion pathways, Sec and Tat, with three enzymes, signal peptidases I to III (SPase I to SPase III), needed to cleave proteins for secretion. SignalP 5.0 is able to distinguish between three types of SPs: (i) Sec substrates cleaved by SPase I, (ii) Sec substrates cleaved by SPase II, and (iii) Tat substrates cleaved by SPase I (94). Unfortunately, due to limited training data sets, SignalP 5.0 is unable to predict Sec substrates processed by SPase III or Tat substrates processed by SPase II. However, the current ability to determine between the three secretion pathways is important in understanding how the protein will be secreted and the regulation of the secretion process. SignalP 5.0 is available either through its web server or as a standalone package, making it an accessible tool for secreted protein prediction. SignalP has already been used in the context of determining mycotoxin degrading enzymes: Carere et al. utilized this predictive power in conjunction with an experimental approach to narrow down gene candidates for the identification of DepA in the DON degradation pathway by D. mutans (64). This example highlights the application this tool has in aiding mycotoxin degradation research.
Suboptimal enzymes: naturally evolved enzymes may not be the best match.
Enzymes found to be capable of degrading mycotoxins may not be naturally optimized for targeting the mycotoxin of interest. Importantly, some of the detoxifying enzymes belong to common categories such as oxidases and hydrolases; however, it is not well understood what features of the particular enzymes separate efficient detoxifiers from nonefficient ones. Thus, there is a need to better understand what aspects determine the efficacy of the enzymes and how they can be improved. Enzyme optimization often involves adaptation of a wild-type isolate to a new substrate or reaction environment. New reaction environments often involve changes of temperature, pH, and solvent conditions, all of which nontrivially affect the structure and activity of the enzyme. One technique that is agnostic to fundamental understanding of these effects is directed evolution (102–104). In directed evolution, genetic diversity is introduced via random mutations and the resultant mutant proteins are screened/selected for improved performance. There is some evidence that restricting directed evolution to residues close to the active site leads to a higher probability of displaying meaningful contributions to its activity (105). However, it remains unclear how such a process is achieved through traditionally structure-agnostic in vitro mutagenesis. Often, directed evolution is applied iteratively to further improve strongly performing mutants (106). Though directed evolution conveniently creates a black-box optimization method, it does so at the cost of efficiency, where screening for fitness can become a major bottleneck in the process (107). As an alternative, a variety of computational tools have been developed for targeted enzyme engineering (e.g., those reviewed in references 108 and 109).
Protein sequence activity relationship (ProSAR) models can assist the search algorithm by creating a statistical model that links the protein sequence to its activity (i.e., fitness) (110, 111). ProSAR relies on a mutant library generated from mutagenesis with a constraint of constant protein sequence length, along with the corresponding activities of interest (catalytic constant, thermostability, etc.). A statistical model is built that links the presence or absence of individual mutations to a contribution to the activity, from which some subset of the highest contributing mutations can be fixed for the next round of mutagenesis. Unlike the close mutations described earlier by Morley and Kazlauskas (105), this method is able to link individual mutations to activity contributions without explicit knowledge of the 3D structure. The traditional statistical methods for ProSAR involved partial least square regression and genetic algorithm, while more recently traditional statistical methods could be replaced with recurrent neural network architectures (112).
Focused evolution, in which targeted mutations are introduced based on rational mutation hypotheses, can increase the efficiency of optimization by narrowing the search space; however, current robust methods require 3D structures of the enzyme. When optimizing for known properties such as thermostability and where reasonable 3D models are available, such as homology models, a small subset of rational mutations can feasibly be explored through computational methods and the final mutations evaluated experimentally. Rational mutation methods rely on heuristic evaluation methods like FoldX (113) to predict changes in Gibbs free energy from mutations or predictive methods like DbD2 (114), which predicts mutations to introduce disulfide bonds that potentially have stabilizing effects on the protein for given conditions. Potential mutations identified via heuristic methods are then commonly evaluated as a narrow combinatorial library. Although not strictly necessary, to reduce cost and labor for the in vitro experiments, the mutated proteins are often computationally evaluated for stability to further narrow down viable mutations. Because of their heuristic nature, it is always necessary to be able to introduce the mutations in vitro and evaluate them experimentally under the target conditions to confirm that the mutated protein is improved.
FUTURE OUTLOOK
Computational biology tools we have discussed above—although not comprehensive—represent a range of traditional applications for better understanding the mechanisms and ultimately improving the performance of toxin biodegradation. Some of these tools have already been used in this context, whereas others have the potential to yield helpful insights. Table 2 captures the current landscape, using representative examples from the literature. Next, we explore ongoing and future advancements in computational methods that would further facilitate answering pertinent questions in the field of mycotoxin bioremediation.
TABLE 2.
Representative examples of applications of computational biology tools for usages outlined in the previous sectiona
| Category | Function | Computational tool | Reference(s) describing examples of use |
|---|---|---|---|
| Functional-gene level/community level | Protein sequence homology search | BLASTp* | 50, 64, 115, 116 |
| Regulation | Promoter prediction | BacPP | 117, 118 |
| PRODORIC2 | 119, 120 | ||
| PPCNN | 121 | ||
| Operon prediction | Operon-mapper | 122, 123 | |
| Secretion prediction | SignalP* | 64, 124 | |
| Suboptimal enzymes | Optimizing existing enzymes | Response-surface-methodology* | 125, 126 |
| MD/QM studies | 127 – 129 | ||
| Discovery of novel enzymes | Biopanning | 130 | |
| ML generative models | 131 | ||
| Directed evolution* | 132 | ||
| ProSAR | 133 | ||
| TD-MS/shotgun MS | 134 |
Tools used for bioremediation are marked by an asterisk. TD-MS, thermal desorption-mass spectrometry.
Taking the next step: combining machine learning with high-throughput experimentation.
Both the use of machine learning and automated, high-throughput laboratory experiments are becoming increasingly prevalent for enzyme optimization. Enzyme engineering may become a useful tool for the optimization of known degrading enzymes, especially when only sequences, rather than solved crystallographic structures, are known (135). Models for directed evolution can be experimentally realized in parallel and incrementally updated, moving toward an optimal sequence. Like directed evolution, biopanning assays, also known as phage display assays, are a technique often used to determine novel antibodies with high affinity to known antigens (136, 137). Biopanning involves washing a random peptide library over a target ligand immobilized on some substrate. The nonbinding peptides may be washed away, after which the peptides with high affinity remain bound to the ligand and can be separately identified. Like a genetic algorithm, these peptides form the seed for the next round of mutation and panning. While this technique does not offer per-sequence performance metrics, we obtain partitioned sequence data sets resulting from the pannings. Such partitioned data sets have been used in unsupervised, autoregressive sequence models for nanobodies to generate novel sequences that overlap the high-affinity partition without a need to perform additional physical experiments (138, 139). While further evaluation is needed to obtain specific performance estimates for these novel sequences, the method aims to narrow the search space needed in optimization. Biopanning has been previously shown to optimize TEM-1 beta-lactamase and biotin ligase, indicating that it may be feasible to use in optimizing mycotoxin degrading enzymes (140–143).
Complemented by high-throughput assays, machine learning approaches are gradually taking charge to bring out patterns, similarities, and dependencies—for example, in sequence-function relation of an enzyme family—that may otherwise be too cryptic. The use of machine learning is in particular expanding in situations when an a priori model does not exist.
Computational chemistry can further advance our understanding of enzymatic processes.
Towards the understanding of enzymatic mechanisms, advancements in quantum mechanics (QM) and molecular mechanics (MM, atomistic) studies will be vital for characterizing reaction mechanisms and exploring the chemical space available via mutations (144). Additionally, crystallographic structures can be slow and expensive to solve; therefore, recent advances in protein 3D structure prediction will be instrumental to develop high-throughput pipelines.
Molecular mechanics provides a view of a system at the atomic level. It is often used for molecular dynamics (MD) simulations, in which a system (e.g., a protein-substrate interaction) is studied using Newtonian physics, often at nanosecond to microsecond timescales. For some protein systems, this timescale is sufficient to study the relevant mechanisms, such as in the case of using steered MD simulations to characterize an aflatoxin oxidase enzyme isolated from Armillariella tabescens as a member of the dipeptidyl peptidase III family of enzymes (145). However, for larger proteins, or proteins involving large conformational shifts, extensive computation may be needed. For these systems, a coarse-grained approach is taken in which moieties in the system are combined to reduce the total atom count, reducing the computational cost (146, 147). Some examples are coarse-grained water models, as well as proteins in which the side chains are often reduced to a single pseudoatom. Coarse-grained models face issues in faithfully reproducing the system, and current research is focused on this area (147).
Atomistic models allow some insight into the interaction between the protein and the toxin. Such models are often sufficient to determine if the toxin will sterically fit in the binding pocket and may also help to determine pose, electrostatic favorability of the binding, and conformational changes of the protein-ligand complex (148, 149). Unlike the more common use for MM in evaluating noncovalent inhibitors, some difficulty emerges in the inherent covalent nature of detoxification, which cannot be captured by an atomistic view (150). This issue may preclude some energetic effects brought about by the changes in electronic structure, raising concerns about how realistic such a model is. This concern may be partially solved by using QM/MM methods, in which part of the system is partitioned into a QM region and the rest remains in MM views (151). The QM region then can model electronic changes, and the rest can remain in lower-cost MM regions. However, the QM region cannot be too large, which precludes cases that require large, complex QM regions (e.g., in metalloenzymes like laccases). Additionally, the QM region adds computational cost and cannot be well integrated into microsecond timescale calculations.
At a relatively high computational cost, QM calculations provide a detailed and comprehensive view of the electronic state of the system. They can provide information about covalent and electronic changes, often necessary for detoxification studies. An example of this is calculating a Fukui function of a molecule, which describes the change in a frontier orbital as the molecule undergoes a redox reaction. Fukui functions have been used to identify the location of redox in an AF-laccase system (152). QM may also be used to study electron transfer in the protein. As a tool for microbiologists, however, QM remains prohibitively expensive both in computational cost and in learning curve, and it is often used for fine-grained mechanistic studies in collaboration with a QM expert.
ACKNOWLEDGMENTS
This work was supported by Boston College through a startup fund and an Ignite grant. This material is partially based upon work supported by the National Science Foundation (NSF-CBET) under grant no. 2103545. N.S. is supported by a NIFA-AFRI predoctoral fellowship from the USDA (award no. 2021-67034-35108).
Contributor Information
Babak Momeni, Email: momeni@bc.edu.
Ning-Yi Zhou, Shanghai Jiao Tong University.
REFERENCES
- 1.Alshannaq A, Yu J. 2017. Occurrence, toxicity, and analysis of major mycotoxins in food. Int J Environ Res Public Health 14:632. 10.3390/ijerph14060632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marin S, Ramos AJ, Cano-Sancho G, Sanchis V. 2013. Mycotoxins: occurrence, toxicology, and exposure assessment. Food Chem Toxicol 60:218–237. 10.1016/j.fct.2013.07.047. [DOI] [PubMed] [Google Scholar]
- 3.Čolović R, Puvača N, Cheli F, Avantaggiato G, Greco D, Đuragić O, Kos J, Pinotti L. 2019. Decontamination of mycotoxin-contaminated feedstuffs and compound feed. Toxins (Basel) 11:617. 10.3390/toxins11110617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kebede H, Liu X, Jin J, Xing F. 2020. Current status of major mycotoxins contamination in food and feed in Africa. Food Control 110:106975. 10.1016/j.foodcont.2019.106975. [DOI] [Google Scholar]
- 5.Edite Bezerra da Rocha M, da Chagas Oliveira Freire F, Erlan Feitosa Maia F, Izabel Florindo Guedes M, Rondina D. 2014. Mycotoxins and their effects on human and animal health. Food Control 36:159–165. 10.1016/j.foodcont.2013.08.021. [DOI] [Google Scholar]
- 6.Bennett JWW, Klich M. 2003. Mycotoxins. Clin Microbiol Rev 16:497–516. 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahato DK, Lee KE, Kamle M, Devi S, Dewangan KN, Kumar P, Kang SG. 2019. Aflatoxins in food and feed: an overview on prevalence, detection and control strategies. Front Microbiol 10:2266. 10.3389/fmicb.2019.02266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liew WPP, Mohd-Redzwan S. 2018. Mycotoxin: its impact on gut health and microbiota. Front Cell Infect Microbiol 8:60. 10.3389/fcimb.2018.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bui-Klimke TR, Wu F. 2015. Ochratoxin A and human health risk: a review of the evidence. Crit Rev Food Sci Nutr 55:1860–1869. 10.1080/10408398.2012.724480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hueza IM, Raspantini PCF, Raspantini LER, Latorre AO, Górniak SL. 2014. Zearalenone, an estrogenic mycotoxin, is an immunotoxic compound. Toxins (Basel) 6:1080–1095. 10.3390/toxins6031080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sydenham EW, Shephard GS, Thiel PG, Marasas WFO, Stockenström S. 1991. Fumonisin contamination of commercial corn-based human foodstuffs. J Agric Food Chem 39:2014–2018. 10.1021/jf00011a028. [DOI] [Google Scholar]
- 12.Sobrova P, Adam V, Vasatkova A, Beklova M, Zeman L, Kizek R. 2010. Deoxynivalenol and its toxicity. Interdiscip Toxicol 3:94–99. 10.2478/v10102-010-0019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pal S, Singh N, Ansari KM. 2017. Toxicological effects of patulin mycotoxin on the mammalian system: an overview. Toxicol Res (Camb) 6:764–771. 10.1039/c7tx00138j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhatnagar D, Payne G, Cleveland TE, Robens J. 2003. Mycotoxins: current issues in U.S.A. https://www.ars.usda.gov/research/publications/publication/?seqNo115=151980.
- 15.Karlovsky P, Suman M, Berthiller F, De Meester J, Eisenbrand G, Perrin I, Oswald IP, Speijers G, Chiodini A, Recker T, Dussort P. 2016. Impact of food processing and detoxification treatments on mycotoxin contamination. Mycotoxin Res 32:179–205. 10.1007/s12550-016-0257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pankaj SK, Shi H, Keener KM. 2018. A review of novel physical and chemical decontamination technologies for aflatoxin in food. Trends Food Sci Technol 71:73–83. 10.1016/j.tifs.2017.11.007. [DOI] [Google Scholar]
- 17.Wu Q, Jezkova A, Yuan Z, Pavlikova L, Dohnal V, Kuca K. 2009. Biological degradation of aflatoxins. Drug Metab Rev 41:1–7. 10.1080/03602530802563850. [DOI] [PubMed] [Google Scholar]
- 18.Singh R, Singh P, Sharma R. 2014. Microorganism as a tool of bioremediation technology for cleaning environment: a review. Proc Int Acad Ecol Environ Sci 4:1–6. [Google Scholar]
- 19.Wang J, Xie Y. 2020. Review on microbial degradation of zearalenone and aflatoxins. Grain Oil Sci Technol 3:117–125. 10.1016/j.gaost.2020.05.002. [DOI] [Google Scholar]
- 20.Grenier B, Loureiro-Bracarense A-P, Leslie JF, Oswald IP. 2014. Physical and chemical methods for mycotoxin decontamination in maize, p 116–129. In Leslie JF, Logrieco AF (ed), Mycotoxin reduction in grain chains. John Wiley & Sons, Ltd, Chichester, UK. [Google Scholar]
- 21.Sadiq FA, Yan B, Tian F, Zhao J, Zhang H, Chen W. 2019. Lactic acid bacteria as antifungal and anti‐mycotoxigenic agents: a comprehensive review. Compr Rev Food Sci Food Saf 18:1403–1436. 10.1111/1541-4337.12481. [DOI] [PubMed] [Google Scholar]
- 22.de Melo Nazareth T, Luz C, Torrijos R, Quiles JM, Luciano FB, Mañes J, Meca G. 2019. Potential application of lactic acid bacteria to reduce aflatoxin B1 and fumonisin B1 occurrence on corn kernels and corn ears. Toxins (Basel) 12:21. 10.3390/toxins12010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vega MF, Dieguez SN, Riccio B, Aranguren S, Giordano A, Denzoin L, Soraci AL, Tapia MO, Ross R, Apás A, González SN. 2017. Zearalenone adsorption capacity of lactic acid bacteria isolated from pigs. Braz J Microbiol 48:715–723. 10.1016/j.bjm.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alberts JFF, Gelderblom W, Botha A, van Zyl WHH. 2009. Degradation of aflatoxin B1 by fungal laccase enzymes. Int J Food Microbiol 135:47–52. 10.1016/j.ijfoodmicro.2009.07.022. [DOI] [PubMed] [Google Scholar]
- 25.Moll D. 2019. Enzyme technology for detoxification of mycotoxins in animal feed, p 219–254. In Vogel A, May O (ed), Industrial enzyme applications. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 26.Alberts J, Schatzmayr G, Moll W-D, Davids I, Rheeder J, Burger H-M, Shephard G, Gelderblom W. 2019. Detoxification of the fumonisin mycotoxins in maize: an enzymatic approach. Toxins (Basel) 11:523. 10.3390/toxins11090523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biomin. Mycofix. https://www.biomin.net/solutions/mycotoxin-risk-management/mycotoxin-deactivation/mycofix/.
- 28.Taylor MC, Jackson CJ, Tattersall DB, French N, Peat TS, Newman J, Briggs LJ, Lapalikar GV, Campbell PM, Scott C, Russell RJ, Oakeshott JG. 2010. Identification and characterization of two families of F420H2-dependent reductases from mycobacteria that catalyse aflatoxin degradation. Mol Microbiol 78:561–575. 10.1111/j.1365-2958.2010.07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Ogata M, Hirai H, Kawagishi H. 2011. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol Lett 314:164–169. 10.1111/j.1574-6968.2010.02158.x. [DOI] [PubMed] [Google Scholar]
- 30.Scarpari M, Bello C, Pietricola C, Zaccaria M, Bertocchi L, Angelucci A, Ricciardi MR, Scala V, Parroni A, Fabbri AA, Reverberi M, Zjalic S, Fanelli C. 2014. Aflatoxin control in maize by Trametes versicolor. Toxins (Basel) 6:3426–3437. 10.3390/toxins6123426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Qin X, Hao Z, Luo H, Yao B, Su X. 2019. Degradation of four major mycotoxins by eight manganese peroxidases in presence of a dicarboxylic acid. Toxins 11:566. 10.3390/toxins11100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song J, Zhang S, Xie Y, Li Q. 2019. Purification and characteristics of an aflatoxin B1 degradation enzyme isolated from Pseudomonas aeruginosa. FEMS Microbiol Lett 366:fnz034. 10.1093/femsle/fnz034. [DOI] [PubMed] [Google Scholar]
- 33.Garda-Buffon J, Kupski L, Badiale-Furlong E. 2011. Deoxynivalenol (DON) degradation and peroxidase enzyme activity in submerged fermentation. Ciênc Tecnol Aliment 31:198–203. 10.1590/S0101-20612011000100030. [DOI] [Google Scholar]
- 34.Heinl S, Hartinger D, Thamhesl M, Vekiru E, Krska R, Schatzmayr G, Moll WD, Grabherr R. 2010. Degradation of fumonisin B1 by the consecutive action of two bacterial enzymes. J Biotechnol 145:120–129. 10.1016/j.jbiotec.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Abrunhosa L, Serra R, Venâncio A. 2002. Biodegradation of ochratoxin A by fungi isolated from grapes. J Agric Food Chem 50:7493–7496. 10.1021/jf025747i. [DOI] [PubMed] [Google Scholar]
- 36.Abrunhosa L, Inês A, Rodrigues AI, Guimarães A, Pereira VL, Parpot P, Mendes-Faia A, Venâncio A. 2014. Biodegradation of ochratoxin A by Pediococcus parvulus isolated from Douro wines. Int J Food Microbiol 188:45–52. 10.1016/j.ijfoodmicro.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Yang H, Apaliya MT, Zhao L, Gu X, Zheng X, Hu W, Zhang H. 2018. The mechanisms involved in ochratoxin A elimination by Yarrowia lipolytica Y-2. Ann Appl Biol 173:164–174. 10.1111/aab.12452. [DOI] [Google Scholar]
- 38.Tang H, Li X, Zhang F, Meng X, Liu B. 2019. Biodegradation of the mycotoxin patulin in apple juice by orotate phosphoribosyltransferase from Rhodotorula mucilaginosa. Food Control 100:158–164. 10.1016/j.foodcont.2019.01.020. [DOI] [Google Scholar]
- 39.Altalhi AD, El-Deeb B. 2009. Localization of zearalenone detoxification gene(s) in pZEA-1 plasmid of Pseudomonas putida ZEA-1 and expressed in Escherichia coli. J Hazard Mater 161:1166–1172. 10.1016/j.jhazmat.2008.04.068. [DOI] [PubMed] [Google Scholar]
- 40.Kosawang C, Karlsson M, Vélëz H, Rasmussen PH, Collinge DB, Jensen B, Jensen DF. 2014. Zearalenone detoxification by zearalenone hydrolase is important for the antagonistic ability of Clonostachys rosea against mycotoxigenic Fusarium graminearum. Fungal Biol 118:364–373. 10.1016/j.funbio.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 41.Chen S-W, Hsu J-T, Chou Y-A, Wang H-T. 2018. The application of digestive tract lactic acid bacteria with high esterase activity for zearalenone detoxification. J Sci Food Agric 98:3870–3879. 10.1002/jsfa.8904. [DOI] [PubMed] [Google Scholar]
- 42.Chang X, Liu H, Sun J, Wang J, Zhao C, Zhang W, Zhang J, Sun C. 2020. Zearalenone removal from corn oil by an enzymatic strategy. Toxins 12:117. 10.3390/toxins12020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sangare L, Zhao Y, Folly YM, Inni E, Chang J, Li J, Ima Selvaraj JN, Xing F, Zhou L, Wang Y, Liu Y. 2014. Aflatoxin B1 degradation by a Pseudomonas strain. Toxins (Basel) 6:3028–3040. 10.3390/toxins6103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alberts J, Engelbrecht Y, Steyn P, Holzapfel W, Van Zyl W. 2006. Biological degradation of aflatoxin B1 by Rhodococcus erythropolis cultures. Int J Food Microbiol 109:121–126. 10.1016/j.ijfoodmicro.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 45.Guan S, Ji C, Zhou T, Li J, Ma Q, Niu T. 2008. Aflatoxin B1 degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int J Mol Sci 9:1489–1503. 10.3390/ijms9081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guan S, Zhao L, Ma Q, Zhou T, Wang N, Hu X, Ji C. 2010. In vitro efficacy of Myxococcus fulvus ANSM068 to biotransform aflatoxin B1. Int J Mol Sci 11:4063–4079. 10.3390/ijms11104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato I, Ito M, Ishizaka M, Ikunaga Y, Sato Y, Yoshida S, Koitabashi M, Tsushima S. 2012. Thirteen novel deoxynivalenol-degrading bacteria are classified within two genera with distinct degradation mechanisms. FEMS Microbiol Lett 327:110–117. 10.1111/j.1574-6968.2011.02461.x. [DOI] [PubMed] [Google Scholar]
- 48.Adebo OA, Njobeh PB, Gbashi S, Nwinyi OC, Mavumengwana V. 2017. Review on microbial degradation of aflatoxins. Crit Rev Food Sci Nutr 57:3208–3217. 10.1080/10408398.2015.1106440. [DOI] [PubMed] [Google Scholar]
- 49.Ji C, Fan Y, Zhao L. 2016. Review on biological degradation of mycotoxins. Anim Nutr 2:127–133. 10.1016/j.aninu.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyagin I, Efremenko E. 2019. Enzymes for detoxification of various mycotoxins: origins and mechanisms of catalytic action. Molecules 24:2362. 10.3390/molecules24132362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu R, Feussner K, Wu T, Yan F, Karlovsky P, Zheng X. 2015. Detoxification of mycotoxin patulin by the yeast Rhodosporidium paludigenum. Food Chem 179:1–5. 10.1016/j.foodchem.2015.01.066. [DOI] [PubMed] [Google Scholar]
- 52.Loi M, Fanelli F, Liuzzi VC, Logrieco AF, Mulè G. 2017. Mycotoxin biotransformation by native and commercial enzymes: present and future perspectives. Toxins (Basel) 9:111. 10.3390/toxins9040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murugesan GR, Ledoux DR, Naehrer K, Berthiller F, Applegate TJ, Grenier B, Phillips TD, Schatzmayr G. 2015. Prevalence and effects of mycotoxins on poultry health and performance, and recent development in mycotoxin counteracting strategies. Poultry Sci 94:1298–1315. 10.3382/ps/pev075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rychen G, Aquilina G, Azimonti G, Bampidis V, de Lourdes Bastos M, Bories G, Chesson A, Cocconcelli PS, Flachowsky G, Gropp J, Kolar B, Kouba M, López-Alonso M, Mantovani A, Mayo B, Ramos F, Saarela M, Villa RE, Wallace RJ, Wester P, Martelli G, Renshaw D, López Puente S. 2016. Safety and efficacy of fumonisin esterase (FUMzyme®) as a technological feed additive for all avian species. EFSA J 14:e04617. [Google Scholar]
- 55.Agriopoulou S, Stamatelopoulou E, Varzakas T. 2020. Advances in occurrence, importance, and mycotoxin control strategies: prevention and detoxification in foods. Foods 9:137. 10.3390/foods9020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciegler A, Lillehoj EB, Peterson RE, Hall HH. 1966. Microbial detoxification of aflatoxin. Appl Microbiol 14:934–939. 10.1128/am.14.6.934-939.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Protein BLAST. BLAST® >> blastp suite. https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins.
- 58.Afsharmanesh H, Perez-Garcia A, Zeriouh H, Ahmadzadeh M, Romero D. 2018. Aflatoxin degradation by Bacillus subtilis UTB1 is based on production of an oxidoreductase involved in bacilysin biosynthesis. Food Control 94:48–55. 10.1016/j.foodcont.2018.03.002. [DOI] [Google Scholar]
- 59.Dellafiora L, Galaverna G, Reverberi M, Dall’Asta C. 2017. Degradation of aflatoxins by means of laccases from Trametes versicolor: an in silico insight. Toxins (Basel) 9:17. 10.3390/toxins9010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z, Wang Y, Liu Z, Jin S, Pan K, Liu H, Liu T, Li X, Zhang C, Luo X, Song Y, Zhao J, Zhang T. 2021. Biological detoxification of fumonisin by a novel carboxylesterase from Sphingomonadales bacterium and its biochemical characterization. Int J Biol Macromol 169:18–27. 10.1016/j.ijbiomac.2020.12.033. [DOI] [PubMed] [Google Scholar]
- 61.Dellafiora L, Gonaus C, Streit B, Galaverna G, Moll WD, Vogtentanz G, Schatzmayr G, Dall’Asta C, Prasad S. 2020. An in silico target fishing approach to identify novel ochratoxin A hydrolyzing enzyme. Toxins 12:258. 10.3390/toxins12040258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Risa A, Divinyi DM, Baka E, Krifaton C. 2017. Aflatoxin B1 detoxification by cell-free extracts of Rhodococcus strains. Acta Microbiol Immunol Hung 64:423–438. 10.1556/030.64.2017.023. [DOI] [PubMed] [Google Scholar]
- 63.Humpf HU, Schmelz EM, Meredith FI, Vesper H, Vales TR, Wang E, Menaldino DS, Liotta DC, Merrill AH. 1998. Acylation of naturally occurring and synthetic 1-deoxysphinganines by ceramide synthase: formation of N-palmitoyl-aminopentol produces a toxic metabolite of hydrolyzed fumonisin, AP1, and a new category of ceramide synthase inhibitor. J Biol Chem 273:19060–19064. 10.1074/jbc.273.30.19060. [DOI] [PubMed] [Google Scholar]
- 64.Carere J, Hassan YI, Lepp D, Zhou T. 2018. The enzymatic detoxification of the mycotoxin deoxynivalenol: identification of DepA from the DON epimerization pathway. Microb Biotechnol 11:1106–1111. 10.1111/1751-7915.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carere J, Hassan YI, Lepp D, Zhou T. 2018. The identification of DepB: an enzyme responsible for the final detoxification step in the deoxynivalenol epimerization pathway in Devosia mutans 17-2-E-8. Front Microbiol 9:1573. 10.3389/fmicb.2018.01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hahn I, Kunz-Vekiru E, Twarużek M, Grajewski J, Krska R, Berthiller F. 2015. Aerobic and anaerobic in vitro testing of feed additives claiming to detoxify deoxynivalenol and zearalenone. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 32:922–933. 10.1080/19440049.2015.1023741. [DOI] [PubMed] [Google Scholar]
- 67.Catteuw A, Broekaert N, De Baere S, Lauwers M, Gasthuys E, Huybrechts B, Callebaut A, Ivanova L, Uhlig S, De Boevre M, De Saeger S, Gehring R, Devreese M, Croubels S. 2019. Insights into in vivo absolute oral bioavailability, biotransformation, and toxicokinetics of zearalenone, α-zearalenol, β-zearalenol, zearalenone-14-glucoside, and zearalenone-14-sulfate in pigs. J Agric Food Chem 67:3448–3458. 10.1021/acs.jafc.8b05838. [DOI] [PubMed] [Google Scholar]
- 68.Wang Y, Zhao C, Zhang D, Zhao M, Peng M, Guo P, Cui Z. 2020. Microbial degradation of zearalenone by a novel microbial consortium, NZDC-6, and its application on contaminated corncob by semisolid fermentation. J Agric Food Chem 68:1634–1644. 10.1021/acs.jafc.9b05343. [DOI] [PubMed] [Google Scholar]
- 69.Bervoets I, Charlier D. 2019. Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol Rev 43:304–339. 10.1093/femsre/fuz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haugen SP, Ross W, Gourse RL. 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6:507–519. 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maffei B, Francetic O, Subtil A. 2017. Tracking proteins secreted by bacteria: what’s in the toolbox? Front Cell Infect Microbiol 7:221. 10.3389/fcimb.2017.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Santero E, Díaz E. 2020. Special Issue: Genetics of biodegradation and bioremediation. Genes 11:441. 10.3390/genes11040441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Klucar L, Stano M, Hajduk M. 2010. PhiSITE: database of gene regulation in bacteriophages. Nucleic Acids Res 38:D366–D370. 10.1093/nar/gkp911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stano M, Klucar L. 2011. PhiGENOME: an integrative navigation throughout bacteriophage genomes. Genomics 98:376–380. 10.1016/j.ygeno.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 75.Coppens L, Lavigne R. 2020. SAPPHIRE: a neural network based classifier for σ70 promoter prediction in Pseudomonas. BMC Bioinformatics 21:415. 10.1186/s12859-020-03730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eckweiler D, Dudek CA, Hartlich J, Brötje D, Jahn D. 2018. PRODORIC2: the bacterial gene regulation database in 2018. Nucleic Acids Res 46:D320–D326. 10.1093/nar/gkx1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Avila e Silva S, Echeverrigaray S, Gerhardt GJL. 2011. BacPP: bacterial promoter prediction—a tool for accurate sigma-factor specific assignment in enterobacteria. J Theor Biol 287:92–99. 10.1016/j.jtbi.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 78.Umarov RK, Solovyev VV. 2017. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS One 12:e0171410. 10.1371/journal.pone.0171410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Osbourn AE, Field B. 2009. Operons. Cell Mol Life Sci 66:3755–3775. 10.1007/s00018-009-0114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okuda S, Yoshizawa AC. 2011. ODB: a database for operon organizations, 2011 update. Nucleic Acids Res 39:D552–D555. 10.1093/nar/gkq1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shujiro Okuda Laboratory. OBD4. Operon Database Version 4. https://operondb.jp/.
- 82.Chetal K, Janga SC. 2015. OperomeDB: a database of condition-specific transcription units in prokaryotic genomes. Biomed Res Int 2015:318217. 10.1155/2015/318217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janga SC, Chetal K. Operon Prediction for Bacterial Genomes Database (OperomeDB). https://sysbio.informatics.iupui.edu/operomeDB/#/.
- 84.Assaf R, Xia F, Stevens R. 2021. Detecting operons in bacterial genomes via visual representation learning. Sci Rep 11:2124. 10.1038/s41598-021-81169-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taboada B, Estrada K, Ciria R, Merino E. 2018. Operon-mapper: a web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 34:4118–4120. 10.1093/bioinformatics/bty496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Computational Genomic Group, Instituto de Biotecnología, Universidad Nacional Autónoma de México. Operon Mapper. https://biocomputo.ibt.unam.mx/operon_mapper/.
- 87.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 88.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37:D412–D416. 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taboada B, Verde C, Merino E. 2010. High accuracy operon prediction method based on STRING database scores. Nucleic Acids Res 38:e130. 10.1093/nar/gkq254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 92.Zaidi SSA, Zhang X. 2017. Computational operon prediction in whole-genomes and metagenomes. Brief Funct Genomics 16:181–193. 10.1093/bfgp/elw034. [DOI] [PubMed] [Google Scholar]
- 93.Owji H, Nezafat N, Negahdaripour M, Hajiebrahimi A, Ghasemi Y. 2018. A comprehensive review of signal peptides: structure, roles, and applications. Eur J Cell Biol 97:422–441. 10.1016/j.ejcb.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 94.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 95.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Cenk Sahinalp S, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bagos PG, Nikolaou EP, Liakopoulos TD, Tsirigos KD. 2010. Combined prediction of Tat and Sec signal peptides with hidden Markov models. Bioinformatics 26:2811–2817. 10.1093/bioinformatics/btq530. [DOI] [PubMed] [Google Scholar]
- 97.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. 2005. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6:167. 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nielsen H, Engelbrecht J, Brunak S, Von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6. 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 99.Krogh AS, Nielsen H. 1998. Prediction of signal peptides and signal anchors by a hidden Markov model, p 122–130. In Proc Int Conf Intell Syst Mol Biol. AAAI Press, Palo Alto, CA. [PubMed] [Google Scholar]
- 100.Bendtsen JD, Nielsen H, Von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795. 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 101.Petersen TN, Brunak S, Von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 102.Farinas ET, Bulter T, Arnold FH. 2001. Directed enzyme evolution. Curr Opin Biotechnol 12:545–551. 10.1016/s0958-1669(01)00261-0. [DOI] [PubMed] [Google Scholar]
- 103.Lutz S, Patrick WM. 2004. Novel methods for directed evolution of enzymes: quality, not quantity. Curr Opin Biotechnol 15:291–297. 10.1016/j.copbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 104.Goldsmith M, Tawfik DS. 2012. Directed enzyme evolution: beyond the low-hanging fruit. Curr Opin Struct Biol 22:406–412. 10.1016/j.sbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 105.Morley KL, Kazlauskas RJ. 2005. Improving enzyme properties: when are closer mutations better? Trends Biotechnol 23:231–237. 10.1016/j.tibtech.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 106.Chica RA, Doucet N, Pelletier JN. 2005. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr Opin Biotechnol 16:378–384. 10.1016/j.copbio.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 107.Siedhoff NE, Schwaneberg U, Davari MD. 2020. Machine learning-assisted enzyme engineering. Methods Enzymol 643:281–315. 10.1016/bs.mie.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 108.Damborsky J, Brezovsky J. 2014. Computational tools for designing and engineering enzymes. Curr Opin Chem Biol 19:8–16. 10.1016/j.cbpa.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 109.Welborn VV, Head-Gordon T. 2019. Computational design of synthetic enzymes. Chem Rev 119:6613–6630. 10.1021/acs.chemrev.8b00399. [DOI] [PubMed] [Google Scholar]
- 110.Berland M, Offmann B, André I, Remaud-Siméon M, Charton P, Arnold F. 2014. A web-based tool for rational screening of mutants libraries using ProSAR. Protein Eng Des Sel 27:375–381. 10.1093/protein/gzu035. [DOI] [PubMed] [Google Scholar]
- 111.Fox RJ, Davis SC, Mundorff EC, Newman LM, Gavrilovic V, Ma SK, Chung LM, Ching C, Tam S, Muley S, Grate J, Gruber J, Whitman JC, Sheldon RA, Huisman GW. 2007. Improving catalytic function by ProSAR-driven enzyme evolution. Nat Biotechnol 25:338–344. 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- 112.Alley EC, Khimulya G, Biswas S, AlQuraishi M, Church GM. 2019. Unified rational protein engineering with sequence-based deep representation learning. Nat Methods 16:1315–1322. 10.1038/s41592-019-0598-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. 2005. The FoldX web server: an online force field. Nucleic Acids Res 33:W382–W388. 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Craig DB, Dombkowski AA. 2013. Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinformatics 14:346. 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.He W-J, Zhang L, Yi S-Y, Tang X-L, Yuan Q-S, Guo M-W, Wu A-B, Qu B, Li H-P, Liao Y-C. 2017. An aldo-keto reductase is responsible for Fusarium toxin-degrading activity in a soil Sphingomonas strain. Sci Rep 7:9549. 10.1038/s41598-017-08799-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun J, Xia Y, Ming D. 2020. Whole-genome sequencing and bioinformatics analysis of Apiotrichum mycotoxinivorans: predicting putative zearalenone-degradation enzymes. Front Microbiol 10:1866. 10.3389/fmicb.2020.01866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Millacura FA, Cárdenas F, Mendez V, Seeger M, Rojas LA. 2017. Degradation of benzene by the heavy-metal resistant bacterium Cupriavidus metallidurans CH34 reveals its catabolic potential for aromatic compounds. bioRxiv 2017:164517. [Google Scholar]
- 118.Kernan T, West AC, Banta S. 2017. Characterization of endogenous promoters for control of recombinant gene expression in Acidithiobacillus ferrooxidans. Biotechnol Appl Biochem 64:793–802. 10.1002/bab.1546. [DOI] [PubMed] [Google Scholar]
- 119.Ibraim IC, Parise MTD, Parise D, Sfeir MZT, de Paula Castro TL, Wattam AR, Ghosh P, Barh D, Souza EM, Góes-Neto A, Gomide ACP, Azevedo V. 2019. Transcriptome profile of Corynebacterium pseudotuberculosis in response to iron limitation. BMC Genomics 20:663. 10.1186/s12864-019-6018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee M, Ryu M, Joo M, Seo Y-J, Lee J, Kim H-M, Shin E, Yeom J-H, Kim Y-H, Bae J, Lee K. 2021. Endoribonuclease-mediated control of hns mRNA stability constitutes a key regulatory pathway for Salmonella Typhimurium pathogenicity island 1 expression. PLoS Pathog 17:e1009263. 10.1371/journal.ppat.1009263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wuisan ZG, Kresna IDM, Böhringer N, Lewis K, Schäberle TF. 2021. Optimization of heterologous Darobactin A expression and identification of the minimal biosynthetic gene cluster. Metab Eng 66:123–136. 10.1016/j.ymben.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 122.Martinez-Amador P, Castañeda N, Loza A, Soto L, Merino E, Gutierrez-Rios RM. 2019. Prediction of protein architectures involved in the signaling-pathway initiating sporulation in Firmicutes. BMC Res Notes 12:686. 10.1186/s13104-019-4712-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grünberger F, Reichelt R, Bunk B, Spröer C, Overmann J, Rachel R, Grohmann D, Hausner W. 2019. Next generation DNA-Seq and differential RNA-Seq allow re-annotation of the Pyrococcus furiosus DSM 3638 genome and provide insights into archaeal antisense transcription. Front Microbiol 10:1603. 10.3389/fmicb.2019.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Otero IVR, Ferro M, Bacci M, Ferreira H, Sette LD. 2017. De novo transcriptome assembly: a new laccase multigene family from the marine-derived basidiomycete Peniophora sp. CBMAI 1063. AMB Expr 7:222. 10.1186/s13568-017-0526-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Khatoon H, Rai JPN. 2020. Optimization studies on biodegradation of atrazine by Bacillus badius ABP6 strain using response surface methodology. Biotechnol Rep 26:e00459. 10.1016/j.btre.2020.e00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zaveri P, Iyer AR, Patel R, Munshi NS. 2021. Uncovering competitive and restorative effects of macro- and micronutrients on sodium benzoate biodegradation. Front Microbiol 12:634753. 10.3389/fmicb.2021.634753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lonsdale R, Harvey JN, Mulholland AJ. 2012. A practical guide to modelling enzyme-catalysed reactions. Chem Soc Rev 41:3025–3038. 10.1039/c2cs15297e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang Z, Mehmood R, Wang M, Qi HW, Steeves AH, Kulik HJ. 2019. Revealing quantum mechanical effects in enzyme catalysis with large-scale electronic structure simulation. React Chem Eng 4:298–315. 10.1039/C8RE00213D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang X, Li R, Cui W, Li Q, Yao J. 2018. QM/MM free energy simulations of an efficient gluten hydrolase (Kuma030) implicate for a reactant-state based protein-design strategy for general acid/base catalysis. Sci Rep 8:7042. 10.1038/s41598-018-25471-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Frietze KM, Roden RBS, Lee J-H, Shi Y, Peabody DS, Chackerian B. 2016. Identification of anti-CA125 antibody responses in ovarian cancer patients by a novel deep sequence–coupled biopanning platform. Cancer Immunol Res 4:157–164. 10.1158/2326-6066.CIR-15-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nian R, Kim DS, Nguyen T, Tan L, Kim CW, Yoo IK, Choe WS. 2010. Chromatographic biopanning for the selection of peptides with high specificity to Pb2+ from phage displayed peptide library. J Chromatogr A 1217:5940–5949. 10.1016/j.chroma.2010.07.048. [DOI] [PubMed] [Google Scholar]
- 132.Ang EL, Obbard JP, Zhao H. 2009. Directed evolution of aniline dioxygenase for enhanced bioremediation of aromatic amines. Appl Microbiol Biotechnol 81:1063–1070. 10.1007/s00253-008-1710-0. [DOI] [PubMed] [Google Scholar]
- 133.Yang Y, Zhang S, Howe K, Wilson DB, Moser F, Irwin D, Thannhauser TW. 2007. A comparison of nLC-ESI-MS/MS and nLC-MALDI-MS/MS for GeLC-based protein identification and iTRAQ-based shotgun quantitative proteomics. J Biomol Tech 18:226–237. [PMC free article] [PubMed] [Google Scholar]
- 134.Fornelli L, Parra J, Hartmer R, Stoermer C, Lubeck M, Tsybin YO. 2013. Top-down analysis of 30–80 kDa proteins by electron transfer dissociation time-of-flight mass spectrometry. Anal Bioanal Chem 405:8505–8514. 10.1007/s00216-013-7267-5. [DOI] [PubMed] [Google Scholar]
- 135.Chowdhury R, Maranas CD. 2020. From directed evolution to computational enzyme engineering—a review. AIChE J 66:e16847. 10.1002/aic.16847. [DOI] [Google Scholar]
- 136.Hu D, Hu S, Wan W, Xu M, Du R, Zhao W, Gao X, Liu J, Liu H, Hong J. 2015. Effective optimization of antibody affinity by phage display integrated with high-throughput DNA synthesis and sequencing technologies. PLoS One 10:e0129125. 10.1371/journal.pone.0129125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lim CC, Woo PCY, Lim TS. 2019. Development of a phage display panning strategy utilizing crude antigens: isolation of MERS-CoV nucleoprotein human antibodies. Sci Rep 9:6088. 10.1038/s41598-019-42628-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shin J-E, Riesselman AJ, Kollasch AW, McMahon C, Simon E, Sander C, Manglik A, Kruse AC, Marks DS. 2021. Protein design and variant prediction using autoregressive generative models. Nat Commun 12:2403. 10.1038/s41467-021-22732-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saka K, Kakuzaki T, Metsugi S, Kashiwagi D, Yoshida K, Wada M, Tsunoda H, Teramoto R. 2021. Antibody design using LSTM based deep generative model from phage display library for affinity maturation. Sci Rep 11:5852. 10.1038/s41598-021-85274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Richman SA, Healan SJ, Weber KS, Donermeyer DL, Dossett ML, Greenberg PD, Allen PM, Kranz DM. 2006. Development of a novel strategy for engineering high-affinity proteins by yeast display. Protein Eng Des Sel 19:255–264. 10.1093/protein/gzl008. [DOI] [PubMed] [Google Scholar]
- 141.Legendre D, Soumillion P, Fastrez J. 1999. Engineering a regulatable enzyme for homogeneous immunoassays. Nat Biotechnol 17:67–72. 10.1038/5243. [DOI] [PubMed] [Google Scholar]
- 142.Mathieu V, Fastrez J, Soumillion P. 2010. Engineering allosteric regulation into the hinge region of a circularly permuted TEM-1 β-lactamase. Protein Eng Des Sel 23:699–709. 10.1093/protein/gzq041. [DOI] [PubMed] [Google Scholar]
- 143.Huang W, Beharry Z, Zhang Z, Palzkill T. 2003. A broad‐spectrum peptide inhibitor of β‐lactamase identified using phage display and peptide arrays. Protein Eng 16:853–860. 10.1093/protein/gzg108. [DOI] [PubMed] [Google Scholar]
- 144.Zhou J, Zhu L, Chen J, Wang W, Zhang R, Li Y, Zhang Q, Wang W. 2020. Degradation mechanism for zearalenone ring-cleavage by zearalenone hydrolase RmZHD: a QM/MM study. Sci Total Environ 709:135897. 10.1016/j.scitotenv.2019.135897. [DOI] [PubMed] [Google Scholar]
- 145.Tomin M, Tomić S. 2019. Oxidase or peptidase? A computational insight into a putative aflatoxin oxidase from Armillariella tabescens. Proteins 87:390–400. 10.1002/prot.25661. [DOI] [PubMed] [Google Scholar]
- 146.Souza PCT, Alessandri R, Barnoud J, Thallmair S, Faustino I, Grünewald F, Patmanidis I, Abdizadeh H, Bruininks BMH, Wassenaar TA, Kroon PC, Melcr J, Nieto V, Corradi V, Khan HM, Domański J, Javanainen M, Martinez-Seara H, Reuter N, Best RB, Vattulainen I, Monticelli L, Periole X, Tieleman DP, de Vries AH, Marrink SJ. 2021. Martini 3: a general purpose force field for coarse-grained molecular dynamics. Nat Methods 18:382–388. 10.1038/s41592-021-01098-3. [DOI] [PubMed] [Google Scholar]
- 147.Wang J, Chmiela S, Müller KR, Noé F, Clementi C. 2020. Ensemble learning of coarse-grained molecular dynamics force fields with a kernel approach. J Chem Phys 152:194106. 10.1063/5.0007276. [DOI] [PubMed] [Google Scholar]
- 148.Guterres H, Im W. 2020. Improving protein-ligand docking results with high-throughput molecular dynamics simulations. J Chem Inf Model 60:2189–2198. 10.1021/acs.jcim.0c00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Peterson L. 2020. In silico molecular dynamics docking of drugs to the inhibitory active site of SARS-CoV-2 protease and their predicted toxicology and ADME. SSRN 10.2139/ssrn.3580951. [DOI]
- 150.Aljoundi A, Bjij I, El Rashedy A, Soliman MES. 2020. Covalent versus non-covalent enzyme inhibition: which route should we take? A justification of the good and bad from molecular modelling perspective. Protein J 39:97–105. 10.1007/s10930-020-09884-2. [DOI] [PubMed] [Google Scholar]
- 151.Yu T, Guo H. 2020. Understanding enzyme catalysis mechanism using QM/MM simulation methods. ACS Symp Ser Am Chem Soc 1357:121–137. [Google Scholar]
- 152.Zaccaria M, Dawson W, Kish DR, Reverberi M, Bonaccorsi Di Patti MC, Domin M, Cristiglio V, Dellafiora L, Gabel F, Nakajima T, Genovese L, Momeni B. 2021. Mechanistic insight from full quantum mechanical modeling: laccase as a detoxifier of aflatoxins. bioRxiv 2021. 10.1101/2020.12.31.424992. [DOI] [Google Scholar]