ABSTRACT
Bacteriocins are ribosomally synthesized bacterial antimicrobial peptides that have a narrow spectrum of antibacterial activity against species closely related to the producers. Pediocin-like (or class IIa) bacteriocins (PLBs) exhibit antibacterial activity against several Gram-positive bacterial strains by forming pores in the cytoplasmic membrane of target cells with a specific receptor, the mannose phosphotransferase system (man-PTS). In this study, we report the cryo-electron microscopy structures of man-PTS from Listeria monocytogenes alone and its complex with pediocin PA-1, the first and most extensively studied representative PLB, at resolutions of 3.12 and 2.45 Å, respectively. The structures revealed that the binding of pediocin PA-1 opens the Core domain of man-PTS away from its Vmotif domain, creating a pore through the cytoplasmic membranes of target cells. During this process, the N-terminal β-sheet region of pediocin PA-1 can specifically attach to the extracellular surface of the man-PTS Core domain, whereas the C-terminal half penetrates the membrane and cracks the man-PTS like a wedge. Thus, our findings shed light on a design of novel PLBs that can kill the target pathogenic bacteria.
IMPORTANCE Listeria monocytogenes is a ubiquitous microorganism responsible for listeriosis, a rare but severe disease in humans, who become infected by ingesting contaminated food products (i.e., dairy, meat, fish, and vegetables): the disease has a fatality rate of 33%. Pediocin PA-1 is an important commercial additive used in food production to inhibit Listeria species. The mannose phosphotransferase system (man-PTS) is responsible for the sensitivity of Listeria monocytogenes to pediocin PA-1. In this study, we report the cryo-EM structures of man-PTS from Listeria monocytogenes alone and its complex with pediocin PA-1 at resolutions of 3.12 and 2.45 Å, respectively. Our results facilitate the understanding of the mode of action of class IIa bacteriocins as an alternative to antibiotics.
KEYWORDS: antimicrobial peptides, bacteriocins, pediocin PA-1, pediocin-like/class IIa bacteriocins, antibiotic resistance, mannose phosphotransferase, man-PTS
INTRODUCTION
Bacteriocins are ribosomally synthesized peptides produced by bacteria of all genera, which have antimicrobial activity on related bacterial species (1–3). Bacteriocins from Gram-positive bacteria are typically classified into peptides that undergo lanthionine-containing posttranslational modifications (class I) and unmodified peptides (class II) (4). Class II bacteriocins are further divided into four subclasses: IIa, IIb, IIc, and IId (4). Class IIa bacteriocins, linear peptides with 25 to 58 residues containing one or two disulfide bridges and a conserved Y-G-N-G-V/L region (“pediocin box” motif) (5), have been termed the “pediocin family” after the first and most extensively studied representative of this class, pediocin PA-1 (6). The peptide chain of pediocin-like bacteriocins (PLBs) can be divided into two regions: (i) a hydrophilic, cationic, and highly conserved N-terminal region that contains the “pediocin box” motif and (ii) a less-well-conserved hydrophobic/amphiphilic C-terminal region (7). The conserved N-terminal region forms a three-stranded anti-parallel β-sheet-like structure that is stabilized by a conserved disulfide bridge. The more hydrophobic C-terminal region is a major specificity determinant in PLBs, which forms a hairpin-like structure in most cases and consists of an amphiphilic α-helix followed by an extended C-terminal tail that folds back onto the central α-helix (8).
PLBs are grouped into four subgroups according to sequence similarities and differences in the less-well-conserved C-terminal region (8). They attract attention due to their high activity exerted at the nanomolar range and low toxicity in humans (9). High bacterial sensitivity at nanomolar concentrations was related to a high affinity to specific receptors or docking molecules. It was found that the receptor was a mannose phosphotransferase system (man-PTS) that phosphorylates and transports carbohydrates and other related substances (10, 11). Together, the membrane components of man-PTS, ManY/IIC and ManZ/IID, form a membrane-located complex (12). This complex serves as a receptor for several different bacteriocins, including pediocin-like class IIa bacteriocins and class IId bacteriocin lactococcin-A (13), as well as some class IIb microcins (14) (e.g., MccE492 [15] or MccN/24 [16]). A previous study demonstrated that class IIa bacteriocins act on the cytoplasmic membrane of Gram-positive cells, which dissipates the transmembrane electrical potential and results in intracellular ATP depletion (17). Moreover, bacteriocin-producing cells are protected from the lethal effect of their own bacteriocin by the expression of an immunity protein (18).
It remains unknown how PLBs specifically recognize the man-PTS receptor and the mechanism by which the pore is formed on the cytoplasmic membrane of target cells. To elucidate the specific interaction between PLBs and man-PTS, as well as the conformational changes made to the receptor accompanied by PLB binding, we sought to determine both structures of man-PTS from Listeria monocytogenes alone and its complex with pediocin PA-1. Bacteriocin pediocin PA-1/AcH (also termed PedA; UniProtKB no. P29430) is a 4.6-kDa cationic antimicrobial peptide with 44 amino acids produced by the Pediococcus acidilactici PAC-1.0 strain (19). It shows particularly strong activity against Listeria monocytogenes, which is a foodborne pathogen of special concern within the food industry (20).
RESULTS AND DISCUSSION
Cryo-EM structures of Listeria man-PTS and its complex with pediocin PA-1.
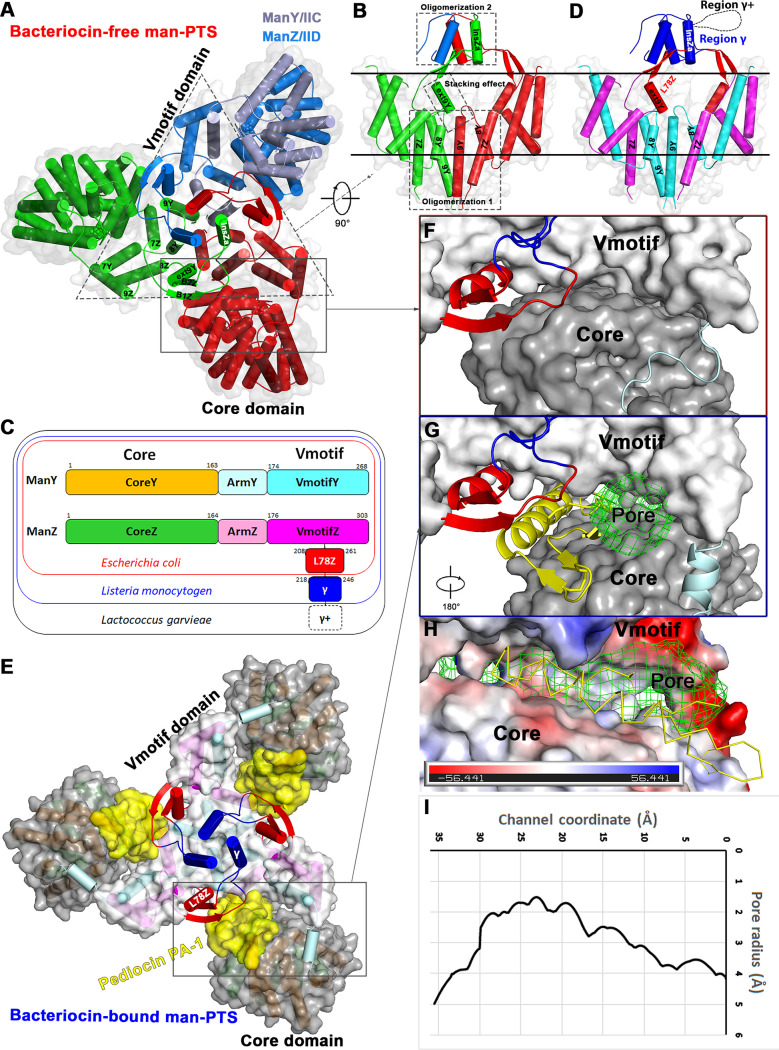
Multiple sequence alignments revealed that an extracellular loop (region γ) was unique in pediocin-like bacteriocin receptors compared with the man-PTS from Escherichia coli (here referred to as ecManYZ). In addition, a novel loop (region γ+) of ManZ from Lactococcus garvieae was identified to be important for sensitivity to the bacteriocin GarQ (21) (see Fig. S1 in the supplemental material). Although the cryo-EM structures of ecManYZ have been reported (12, 22), the role of region γ is difficult to predict solely based on an E. coli homologue. Therefore, we first sought to identify the structure of the bacteriocin-free Listeria man-PTS (lmManYZ). We coexpressed lmManY and N-terminal 6×His-tagged lmManZ and determined the structure of lmManYZ to a 3.12-Å resolution (Fig. 1A and Table 1; see Fig. S2 in the supplemental material). Due to the flexibility and limited local resolution (Fig. S2D), the C terminus of lmManY (residues 248 to 268) and the N terminus of lmManZ (residues 1 to 5) were not observed in the cryo-EM map. Since lmManYZ possessed the same topology as ecManYZ, lmManYZ was also spatially organized into the Vmotif and Core domains (Fig. 1A). It was proposed that sugar transportation across membranes uses an elevator-type mechanism, in which the Core domain moves vertically against a stationary Vmotif domain (11). The extrainsertional region γ is composed of an α-helix (InsZa) that extrudes outside the membrane from a two-stranded β-sheet of B1Z and B2Z in loop L78Z (Fig. 1A and B). InsZa helices in the lmManYZ trimer form a helical bundle (Fig. 1B), which provides additional stabilization to the extracellular end of the L78Z loop and lmManYZ’s oligomerization. The opposite end of the B2Z strand was linked to the short helix of extTM9Y, which was arranged in a head-to-tail orientation to the neighboring protomer transmembrane helix, TM9Y (Fig. 1B). Without the immobilization effect of InsZa, loop L78Z in E. coli swings away while binding to MccE492 (22). TM8Y and TM9Y from lmManY contributed to the cytoplasmic oligomerization within the trimer (Fig. 1B). From a structural perspective, lmManY and lmManZ could be further classified as the CoreY (orange), ArmY (pale cyan), and VmotifY (cyan) domains, as well as the CoreZ (lime), ArmZ (pink), VmotifZ (magenta) domains, loop L78Z, and region γ, respectively (Fig. 1C) (12). CoreY/CoreZ and VmotifY/VmotifZ share a similar fold (see Fig. S3 in the supplemental material). VmotifY and VmotifZ (including L78Z and region γ) interlock to form a Vmotif domain, whereas CoreY and CoreZ clamp the substrate to form the Core domain (11). Region γ is unique in Listeria monocytogenes as it provides additional stability to the configuration of loop L78Z (Fig. 1D).
FIG 1.
Cryo-EM structures of man-PTS from Listeria monocytogenes and its complex with antilisterial bacteriocin pediocin PA-1. (A) The structure of the bacteriocin-free man-PTS trimer is represented as viewed from the extracellular side of the membrane, with each protomer differently colored. man-PTS is spatially organized into the Vmotif and Core domains. (B) The Vmotif domain is viewed within the plane of the membrane, with parts of the structural components on one protomer removed for clarity. Three stabilizing interactions between protomers are identifiable. (C) The color-coded domain architecture of ManY and ManZ. ManY and ManZ from E. coli have a similar fold, except for loop L78Z. Regions γ and γ+, which were successively inserted into L78Z, resulted in ManZs from Listeria monocytogenes and Lactococcus garvieae. (D) The Vmotif domain is color-coded based on the domain architecture as in panel C. Region γ+ from Lactococcus garvieae is indicated by a dashed loop. Loop L78Z is stabilized through one end by stacking the effect of extTM9Y onto the neighboring TM9Y and the other end by helical bundles of region γ. (E) Top view of a man-PTS complex of Listeria monocytogenes with pediocin PA-1. Pediocin PA-1s are shown as surface representations colored in yellow. Man-PTS is color-coded in accordance with panel C. (F) Surface representation of the interface between the Vmotif and Core domains in the bacteriocin-free man-PTS. (G) Surface representation of the interface between the Vmotif domain and Core domain in the bacteriocin-bound man-PTS. The transmembrane pore is shown in green mesh as generated with HOLE software (51). (H) The electrostatic potential surface representation of the interface between the Vmotif domain and Core domain. (I) The pore radii along the potential transport path are shown. The minimal radius of the pore is approximately 1.5 Å.
TABLE 1.
Cryo-EM data collection, refinement, and validation
| Parameter | Result for man-PTS: |
|
|---|---|---|
| Bacteriocin free | Bacteriocin bound | |
| Data collection and processing | ||
| Voltage (kV) | 300 | 300 |
| Electron exposure (e−/Å2) | 50 | 50 |
| Defocus range (μm) | −1.0 to ∼−2.0 | −1.0 to ∼−2.0 |
| Pixel size (Å) | 1.0742 | 0.8433 |
| Symmetry imposed | C3 | C3 |
| Final data set (no. of particles) | 136,729 | 659,531 |
| Map resolution (Å), FSC01.43 | 3.12 | 2.45 |
| Refinement | ||
| Initial model used | EMD-9906 | EMD-9906 |
| Map-sharpening B-factor (Å2) | −116.0 | −99.0 |
| Model composition (no.) | ||
| Non-hydrogen atoms | 12,303 | 13,602 |
| Protein residues | 1,635 | 1,773 |
| Ligands (no.) | 3 Man | 3 Man, 3 pediocin PA-1, 10 H2O |
| RMSDa | ||
| Bond lengths (Å) | 0.004 | 0.002 |
| Bond angles (°) | 0.731 | 0.584 |
| Validation | ||
| MolProbity score | 2.08 | 1.65 |
| Clash score | 11.65 | 5.81 |
| Poor rotamers (%) | 0.0 | 0.0 |
| Ramachandran plot (%) | ||
| Favored | 91.31 | 95.38 |
| Allowed | 8.5 | 4.62 |
| Outliers | 0.18 | 0.88 |
| Model to map fit CC | 0.8113 | 0.8197 |
| EMRinger scoreb | 2.64 | 3.30 |
| PDB code | 7VLX | 7VLY |
| EMDB code | EMD-32030 | EMD-32031 |
RMSD, root mean square deviation.
As described in reference 50.
To form a complex with pediocin PA-1, maltose-binding protein (MBP) fuses to the N terminus of functional mature pediocin PA-1 (23) and is coexpressed with lmManYZ. The complex structure was determined at a 2.45-Å resolution (Fig. 1E and Table 1; see Fig. S4 in the supplemental material). Vmotifs from the bacteriocin-free and bacteriocin-bound structures overlap each other with a root square mean deviation (RMSD) of 0.67 Å (Cα atoms) (see Fig. S5A in the supplemental material). The major difference between these two structures can be described as a rigid-body rotation of approximately 33° of the Core relative to the Vmotif (inset in Fig. S5A). The substrates are accessible from the cytoplasmic side of the membrane and localized at approximately the same distance from the cytoplasmic membrane (Fig. S5A). Therefore, both structures were identified as having an inward-facing conformation. Pediocin PA-1 dips into the membrane and is embedded between the Core and Vmotif domains (Fig. 1E), both of which are tightly held together in the bacteriocin-free man-PTS (Fig. 1F). A boomerang-shaped hydrophilic pore was surrounded by pediocin PA-1, Core, and Vmotif (Fig. 1G and H), with a minimum diameter of approximately 3 Å (Fig. 1I), and led to membrane permeabilization and cell death.
Pediocin-like bacteriocin structure.
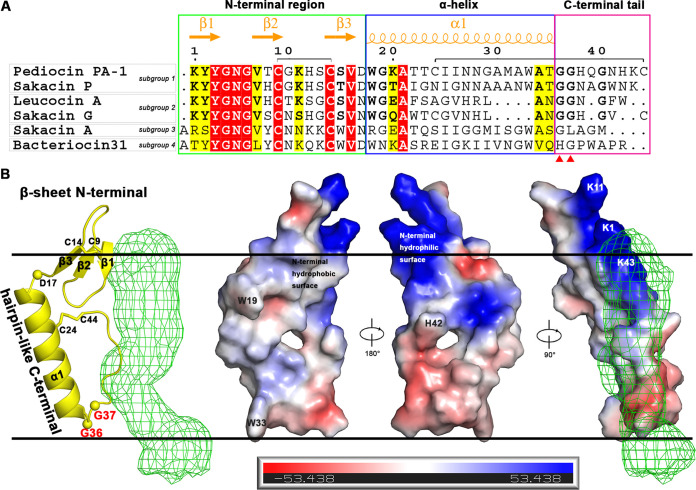
Members of the pediocin-like bacteriocins exhibit very strong antilisterial activity and have 40 to 60% sequence similarity. In particular, the N-terminal region is particularly well conserved and contains a conserved Y-G-N-G-V/L “pediocin box” motif (24) (Fig. 2A). The conserved N-terminal region of pediocin PA-1 (up to around residue 17) forms a three-stranded anti-parallel β-sheet-like structure that is stabilized by a conserved disulfide bridge (Cys9-Cys14) (Fig. 2B). This N-terminal anti-parallel β-sheet contains hydrophobic residues on a single side and hydrophilic ones on the other side. A flexible hinge (Asp/Asn-17) exists between the β-sheet N-terminal and C-terminal regions. Therefore, the two domains may move in relation to each other, which is important for the functions of the N-terminal recognition of the target cell and C-terminal penetration of the membrane. Starting from residue 18 in the C-terminal portion of pediocin PA-1, a central α-helix with 18 residues is repeated for five helical cycles (Fig. 2B). Subsequently, two consecutive Gly-Gly motifs made a sharp turn and facilitated the formation of the hairpin-like structure of the C-terminal tail (Fig. 2B). The C-terminal tail was fixed by a disulfide bridge between a cysteine in the middle of the α-helix and a cysteine at the C terminus, which together with the central α-helix formed a triangular prism or wedge (Fig. 2B). Sakacin G from subgroup 2 (8) is four residues shorter (more than one α-helical turn) in its α-helix region than pediocin PA-1, which likely results in a correspondingly shorter C-terminal tail as for its final cysteine to reach the helical middle cysteine (Cys24) to form a disulfide bridge. Both sakacin P from subgroup 1 and leucocin A from subgroup 2 lack the C-terminal hairpin-stabilizing cysteine. Therefore, the hairpin structure must be stabilized by another mechanism, which was possibly aided by a membrane receptor. It was previously shown that an insertion of a C-terminal cysteine into sakacin P rendered it more thermostable (25). The removal of at least one C-terminal residue (Trp) of mesentericin Y105 (from subgroup 2) was found to elevate the MIC by 4 orders of magnitude (26). Sakacin A (termed curvacin A) from subgroup 3 and bacteriocin 31 from subgroup 4 lack the Gly-Gly sharp turn motif, and it is possible that their C-terminal tails could not fold back to form the hairpin structure like other PLBs. The C-terminal domain (from about residue 18) is less conserved and represents the major determinant of target cell specificity and the region that is specifically recognized by immunity proteins (7, 27).
FIG 2.
Structure and orientation in membrane of pediocin PA-1. (A) Sequence alignment of pediocin-like bacteriocins from the four representative subgroups (8). The pediocin PA-1 peptide chain is divided into three regions: a cationic, hydrophilic, and highly conserved N-terminal region, a less-conserved more hydrophobic C-terminal α-helical region, and a hairpin-like C-terminal tail. (B) An illustration and electrostatic potential surface depiction of the structure and orientation of pediocin PA-1 in membranes. The N-terminal β-sheet has two distinct hydrophobic/hydrophilic surfaces.
Interaction between the N-terminal β-sheet-like domain with man-PTS.
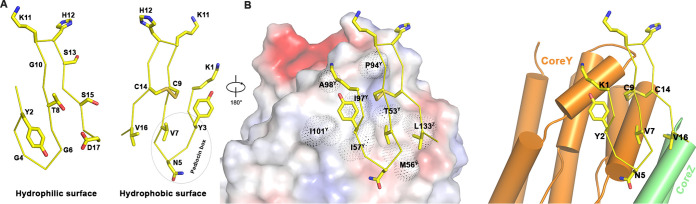
One side of the N-terminal β-sheet contains conserved hydrophobic residues, including two cysteines, whereas the opposite surface was comprised of less-conserved hydrophilic residues (Fig. 3A). The conserved pediocin box corresponds to the first β-turn, whereas the second β-turn contains two positively charged residues (Lys11 and His12). These residues, together with K1, contributed to the positive charge of the periplasmic side of the pore (Fig. 2B). In the complex structure, only the hydrophobic side of the N-terminal β-sheet interacted with the hydrophobic surface of the Core domain of man-PTS, whereas the less-conserved opposite side made no contact with the receptor (Fig. 1G). The conserved residues of pediocin PA-1 (Val7, Cys9, Cys14, and Tyr3) were inserted into the hydrophobic pockets on the CoreY subdomain of man-PTS, and Val16 was located between Met56Y and Leu133Z (Fig. 3B). A carbocyclic leucocin A analog contained a covalent carbon-carbon bond that was stronger than a disulfide bond and differed from the original peptide by an approximately 10-fold-higher MIC (28). Asn5 was located in the “pediocin box” motif and exhibited hydrogen bonding with the carboxyl oxygens of the helical C-terminal end and was close to the binding pocket of the substrate.
FIG 3.
The recognition of the N-terminal region of pediocin PA-1 by man-PTS. (A) The highly conserved residues are hydrophobic, including the disulfide bond from the hydrophobic surface, whereas the other surface is hydrophilic, without any observable interactions with the receptor, man-PTS. (B) The N-terminal part of pediocin PA-1 is attached to the Core domain of man-PTS, primarily through hydrophobic interactions.
Once the lmManYZ receptor possessed an inward-facing conformation, as shown in Fig. 1A, the N-terminal β-sheet domain of pediocin PA-1 was unable to directly bind to the surface of the target cell through hydrophobic interactions in Fig. 3B. This effect was due to the binding site that was buried deep between the Core and Vmotif domains in the current configuration (see Fig. S6 in the supplemental material). Therefore, for the recognition surface on the Core domain to be exposed, pediocin PA-1 must wait for the elevator movement of the Core domain outward or in an occluded conformation for binding (12). Once the target surface of the Core domain is exposed, the N terminus of PLBs attaches to the conserved hydrophobic side.
Interaction between the membrane-penetrating C-terminal domain and man-PTS.
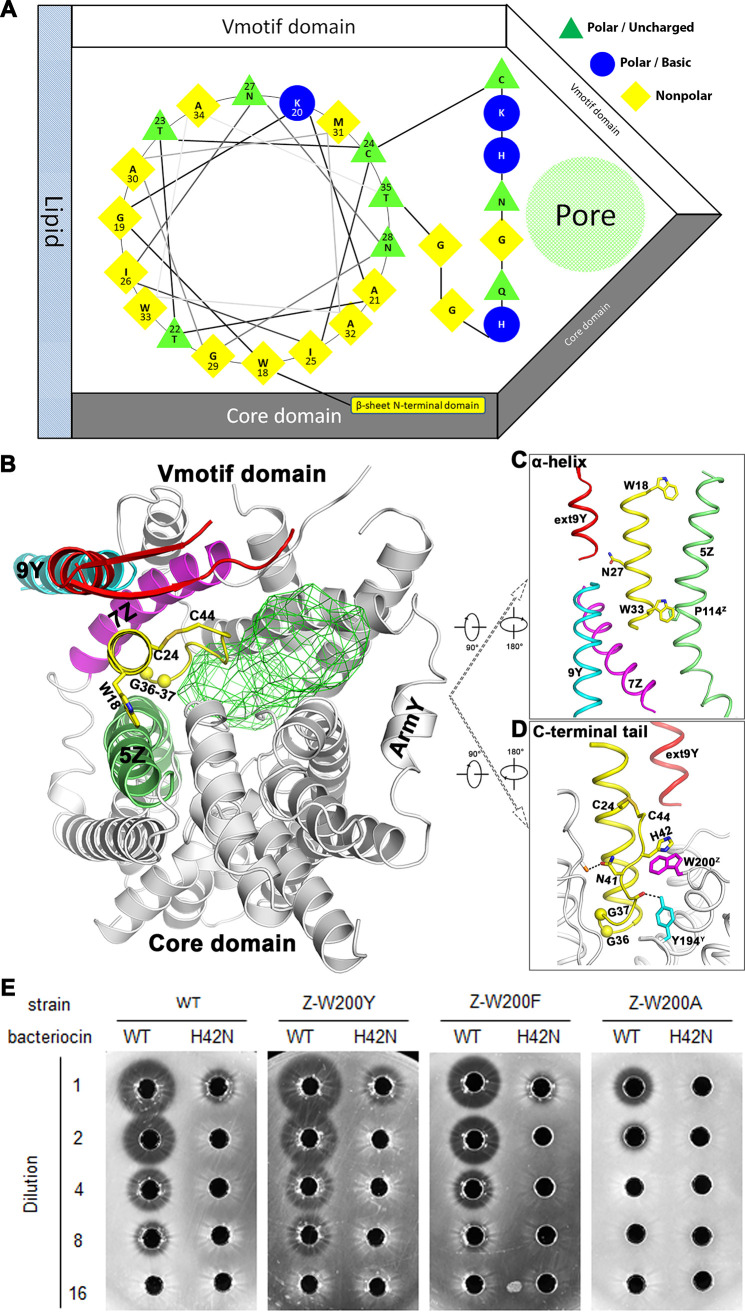
The membrane-penetrating C-terminal domain is an important determinant of target cell specificity (7). It was previously shown that the hybrid bacteriocin exhibited an inhibition spectrum similar to that of the bacteriocin from which its C-terminal module was derived and differed from the bacteriocin that provided the N-terminal section (29). The C-terminal domain of pediocin PA-1 forms a wedge-like hairpin embedded between the Core and Vmotif domains (Fig. 4A and B). The central α-helix subdomain begins at residue W18 and ends at residue T35. It was shown that fragments 20 to 34 could inhibit the activity of full-sized pediocin PA-1, with a MIC value that increased by 2- to 20-fold (30). The Trp18 side chain faces into the crevice on TM5Z of the receptor (Fig. 4C). Any mutation in Trp18 reduced the activity of sakacin P (31). While the C-terminal domain penetrates the membrane, Trp18 may act as an anchor point that correctly positions pediocin PA-1. Interestingly, Asn27 pointed toward the junction between TM9Y and extTM9Y (Fig. 4C). The number of helical cycles extensible within the α-helix subdomain was limited by the N-terminal end of TM7Z in lmManZ (Fig. 4C). Molecular modeling suggests that at most, one additional cycle would additionally be allowed within the α-helix at the current width between the Core and Vmotif domains (see Fig. S7A in the supplemental material).
FIG 4.
Interactions between the C-terminal region of pediocin PA-1 and man-PTS. (A) The interaction of the C-terminal portion of pediocin PA-1 and man-PTS is shown schematically. The central α-helix region is shown in the helical wheel. (B) The specific interactions between man-PTS and the central α-helix, as well as the C-terminal tail of pediocin PA-1. The transmembrane pore is shown in green mesh. (C) Enlarged view of the interactions between the central α-helix of pediocin PA-1 and man-PTS. (D) Interactions between the C-terminal tail of pediocin PA-1 and man-PTS. (E) Bactericidal activity of the wild type (WT) and various lmManZ mutants and the C-terminal tail of pediocin PA-1. Serial dilutions of overnight cultures were spotted (5 μL) on LB plates.
The C-terminal tail introduces a sharp turn and folds back onto the α-helix to create a wedge-like hairpin structure embedded between the Core and Vmotif domains (Fig. 4A). The conformation of the C-terminal tail is stabilized by the disulfide bridge between Cys24 and Cys44 (Fig. 4B). While this disulfide bond is not an obligatory structural component, the presence of this bond extends its spectrum of activity (32) and elevates the upper limit of temperature-dependent activity (25), since the breaking of this disulfide bridge reduced the activity by approximately 200-fold (31). However, some pediocin-like bacteriocins (e.g., sakacin P and leucocin A) (Fig. 2A) lack this last cysteine and contain a C-terminal tryptophan residue instead. In the bacteriocin-bound lmManYZ complex, the His42 of pediocin PA-1 and Trp200Z of lmManZ interact through a π-stacking interaction (Fig. 4D), thereby contributing to the additional stabilization of the hairpin structure. Breaking this π stacking by mutations from either the receptor (Z-W200A) or bacteriocin (H42N) increases the MIC by 4- to 8-fold, whereas aromatic mutations (Z-W200Y or Z-W200F) retain bactericidal activity (Fig. 4E and Table 2). Therefore, sakacin P and leucocin A might also employ π-stacking interactions to stabilize its C-terminal tail and subsequently maintain a hairpin structure. Since sakacin G is shorter by one turn in the α-helix subdomain than pediocin PA-1 (Fig. S7B), to maintain a similar disulfide bridge, its C-terminal tail must be correspondingly shortened, as its sequence suggested (Fig. 2A).
TABLE 2.
Activity of bacteriocin variants toward various lmManYZ mutant strains
| Bacteriocin | MIC (BU mL−1) for straina: |
|||
|---|---|---|---|---|
| WT | lmManYZ mutant |
|||
| Z-W200Y | Z-W200F | Z-W200A | ||
| WT | 10 | 10 | 20 | 40 |
| H42N mutant | 80 | 80 | NI | NI |
aBacteriocin activity was measured as described in Materials and Methods. The numbers indicate the minimum concentrations required to produce zones of growth inhibition. The MIC values were determined at least twice with virtually the same results. BU, bacteriocin units; NI, no inhibition was observed at the highest bacteriocin concentration tested (80 BU mL−1).
Mechanism of PLB action.
Class IIa bacteriocins (PLBs) are membranotropic agents that cause pore formation, the dissipation of transmembrane potential, and ATP depot exhaustion, as well as the loss of inorganic ions, amino acids, and other low-molecular-weight substances (6, 11). A hinge region joining the conserved hydrophilic N-terminal and variable C-terminal regions allows the angle between these domains to change (29). In this study, the antimicrobial effect of PLBs through the formation of transmembrane pores may be that the N terminus of PLBs waits for the exposure of the extracellular surface on the Core domain of the receptor while it switches to an outward or occluded state via an elevator mechanism (Fig. 5A). Once the recognition surface is exposed, the conserved hydrophobic side of the N terminus of PLBs is attached to it (the first state in Fig. 5B). Then the C-terminal half of PLBs penetrates the membrane. As a result, the Core domain is pried open away from the Vmotif domain, which results in the formation of a pore (the second state in Fig. 5B). Concomitantly, the Core domain experienced transmembrane movement in order to return to an inward-facing state. To protect the producer strain, a corresponding immunity protein may be synthesized to block the pore (the third state in Fig. 5B) (29). In particular, several polar or charged residues in the extreme C terminus of immunity proteins are crucial for the specific recognition of bacteriocin and its receptor (33). Bacteriocin blocks the elevator, thereby inhibiting the normal man-PTS transporter function (10).
FIG 5.
The proposed mechanism of pediocin PA-1 antibacterial activity. (A) An elevator-type mechanism of sugar transportation across membranes. The Core domain moves vertically against a stationary Vmotif domain. (B) Once the pediocin PA-1 binding site on the Core domain of the receptor is exposed, the N-terminal region of pediocin PA-1 specifically recognizes the Core domain for binding. Subsequently, the C-terminal wedge-like region is embedded between the Vmotif and Core domains to form a pore. The immunity protein potentially blocks the channel from the cytoplasmic side to protect the producing strain.
Conclusions.
There is an urgent need for solutions to the increasing number of infections caused by antibiotic-resistant bacteria. Bacteriocins are antimicrobial peptides produced by certain bacteria and may warrant serious consideration as an alternative to traditional antibiotics (34). Investigation of PLBs offers a novel and promising means of creating narrow-spectrum antibiotics. Pediocin PA-1 is a heat-stable lactic acid bacterial bacteriocin that exhibits a particularly strong activity against Listeria monocytogenes, a foodborne pathogen of particular concern within the food industry. Membrane-located proteins of the mannose phosphotransferase system (man-PTS) serve as target receptors for pediocin PA-1. The cationic hydrophilic N terminus of the pediocin-like bacteriocin specifically recognizes man-PTS. The C-terminal half of PA-1 is embedded between the Core and Vmotif domains of man-PTS, which forms a pore in the cytoplasmic membranes of target cells. To protect the producer of bacteriocin from its own bacteriocin, a cognate immunity protein was synthesized to block the hole from the cytoplasmic side.
MATERIALS AND METHODS
Expression and purification of man-PTS from Listeria monocytogenes alone and in a complex with pediocin PA-1.
ManYZ from Listeria monocytogenes was induced into E. coli strain BL21(DE3) with 0.4 mM isopropyl-β-d-thiogalactoside (IPTG) and 1 mM mannose at an optical density at 600 nm (OD600) of 1.2. After overnight growth at 18°C, the E. coli cells were harvested and homogenized in a buffer containing 25 mM Tris-HCl (pH 8.0) and 150 mM NaCl and disrupted by sonication. Cell debris was removed by low-speed centrifugation for 10 min. The supernatant was ultracentrifuged at 150,000 × g for 1 h. The membrane pellets were collected and homogenized in buffer A (25 mM Tris [pH 8.0], 150 mM NaCl, and 2 mM mannose) and solubilized with 2% (wt/vol) n-dodecyl-β-d-maltoside (DDM) (Anatrace) at 4°C for 2 h. The insoluble fraction was precipitated by ultracentrifugation (150,000 × g) for 30 min at 4°C. The supernatant was collected and loaded onto Ni-nitrilotriacetic acid (NTA) affinity resin (Qiagen) three times at 4°C. The protein was washed with buffer A plus 20 mM imidazole and eluted with washing buffer plus 250 mM imidazole. The protein was further concentrated to 1.5 mL and subsequently applied to Superdex 200 10/300 GL (SD200; GE Healthcare) preequilibrated with buffer containing 25 mM Tris (pH 8.0), 150 mM NaCl, 2 mM mannose, and 0.07% digitonin. The peak fractions were collected.
To obtain the complex with pediocin PA-1, maltose-binding protein (MBP) was fused to the N terminus of the functional mature domain of pediocin PA-1 and coexpressed with lmManY and lmManZ. The purification method of the lmManYZ and pediocin PA-1 complex was similar to that of lmManYZ, except that the protein was loaded onto amylose affinity resin (Qiagen) following the Ni-NTA affinity resin to remove redundant bacteriocin-free lmManYZ.
Cryo-EM sample preparation and data acquisition.
For negative staining, aliquots of 4-μL samples containing 0.01 mg/mL purified bacteriocin-free lmManYZ or bacteriocin-bound lmManYZ were applied on a carbon film grid (Beijing XXBR Technology Co., Ltd.) after plasma glow discharge. The grids were stained in uranyl acetate (2% [wt/vol]) and stored at room temperature. Negatively stained sample was imaged on a Tecnai Spirit Bio TWIN microscope (Thermo Fisher) operating at 120 kV to verify the sample quality.
Aliquots (4 μL) of concentrated protein complex (bacteriocin-free or bacteriocin-bound lmManYZ) were loaded onto holey carbon grids (Quantifoil Cu R1.2/1.3, 300 mesh) overlaid with graphene oxide (GO grid) (35) or holey carbon grids with 2-nm continuous carbon grids (Quantifoil Cu R1.2/1.3, 300 mesh plus 2-nm carbon) (carbon grid). The samples were blotted for 5 to 10 s and plunge-frozen in liquid ethane cooled by liquid nitrogen using Vitrobot Mark IV (Thermo Fisher) at 8°C and 100% humidity. For bacteriocin-bound lmManYZ cryo-sample preparation, a similar condition was used, and another extra graphene grid (36, 37) (kindly provided by Hailin Peng from Peking University) was also applied. The sample quality of the grids was verified with a Tecnai Arctica 200-kV electron microscope equipped with a Falcon III camera (Thermo Fisher). The verified grids with optimal ice thickness and particle density were transferred to a Titan Krios electron microscope (Thermo Fisher) operating at 300 kV and equipped with a Gatan GIf Quantum energy filter (slit width, 20 eV).
Micrographs were recorded using a K3 Summit counting camera (Gatan Company) in superresolution mode with a nominal magnification of 105,000×, resulting in a calibrated pixel size of 0.5371 Å. Each stack contains 32 frames and has a total accumulated dose of 50 e−/Å2. AutoEMation (38) was used for fully automated data collection. All 32 frames in each stack were aligned and totaled using the whole-image motion correction program MotionCorr2 (39) and binned to a pixel size of 1.0742 Å, and dose weighting was performed (40). The defocus values were set from −1.0 to −2.0 μm and estimated by Gctf (41). Motion correction, contrast transfer function (CTF) estimation, and micrograph inspection were automatically executed by the TsingTitan.py program, which was developed by Fan Yang from the Tsinghua cryo-EM facility.
Cryo-EM image processing.
All particles of the bacteriocin-free complex were automatically picked using Gautomatch (developed by Kai Zhang, https://www2.mrc-lmb.cam.ac.uk/download/gautomatch-056/) and REION-3.0 (42) from 2,962 micrographs, including 2,068 micrographs from the GO grid and 894 micrographs from the carbon grid. All subsequent 2D and 3D classifications and refinements were performed using RELION-3.0 and EMAutoMask (43). Multiple rounds of reference-free two-dimensional (2D) classification were performed to remove ice spots, contaminants, and aggregates, which yielded a total of 2,477,182 good particles. The particles were reextracted with a bin2 box size of 128 and processed with a 3D Auto-Refine procedure, with a previous ecManYZ (EMDB no. EMD-9906) (12) low-pass filtered to 60 Å as the initial model. The resulting data set was subsequently subjected to a local search 3D classification with 5 to 10 classes, with an angular sampling step of 3.7° and searching range of 15°. A total of 1,560,116 good particles were combined and reextracted with a bin1 box size of 256, which yielded a reconstruction at 3.87 Å with C3 symmetry. To reduce the data set and improve the resolution for final reconstruction, three additional rounds of 3D classification resulted in 136,729 particles and yielded the reconstruction at an improved resolution of 3.12 Å. Further CTF refinement and Bayesian polishing provided a marginal improvement for a final resolution.
The data set of the bacteriocin-bound complex was processed in a similar manner. A total of 5,561 micrographs were used, including 1,390 micrographs from the GO grid, 2,227 micrographs from the carbon grid, and 1,944 micrographs from the graphene grid. A total of 4,250,899 particles were selected from multiple rounds of reference-free 2D classification, and 1,877,843 particles were combined from multiple rounds of 3D classification and resulted in a resolution of 2.665 Å. Further 3D classifications reduced the particle number to 659,531 and improved the resolution to 2.45 Å.
Reported resolutions were calculated based on the FSC0.143 criterion, with a high-resolution noise substitution method. Prior to visualization, all density maps were corrected for the modulation transfer function (MTF) of the detector and sharpened by applying a negative B-factor estimated using automated procedures (44). The local resolution variations were estimated using ResMap (45).
Model building and structure refinement.
The reconstruction maps were used to build a de novo model in EMBuilder (46) and manually adjusted in COOT (47). Structure refinements were carried out using PHENIX in real space with a secondary structure and geometry restraints to prevent structure overfitting (48). The statistics for 3D reconstruction and model refinement are listed in Table 1. All structural figures were prepared using PyMol (49).
Bacteriocin assay.
Bacteriocin activity was assayed using a plate diffusion bioassay, and different lmManYZ constructs were cloned into a pNZ8148 vector and transformed into L. lactis NZ9000 by electroporation as the indicator organisms. Cultures of L. lactis NZ9000-expressing strains were grown to saturation overnight in liquid medium plus the required antibiotics. On the following day, 1 ng/mL nisin was added to induce the expression of lmManYZ. The cells were diluted and added to molten 0.5% M17 soft agar. The mixtures were poured into plates, and wells were formed and filled with bacteriocin solutions at 2-fold dilutions. The plates were incubated overnight at 30°C and examined for zones of growth inhibition. One bacteriocin unit (BU) was defined as the amount of bacteriocin required to produce zones of growth inhibition in plates containing the wild-type (WT) indicator strain. The MIC was defined as the minimum concentration of bacteriocin that inhibited the growth of indicator organisms to form a zone of growth inhibition.
Data availability.
The cryo-EM maps and the structures have been deposited in the Electron Microscopy Data Bank (EMD-32030 and EMD-32031) and the Protein Data Bank (PDB no. 7VLX and 7VLY), respectively.
ACKNOWLEDGMENTS
We thank Xiaochun Li (UT Southwestern Medical Center) for comments on the manuscript. We also thank Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for kindly providing the cryo-EM facility support and the computational facility support on the cluster of Bio-Computing Platform. We appreciate J. Lei, X. Li, F. Yang, and J. Wen for technical support.
This work was supported by grants from the National Natural Science Foundation of China (grant no. 32171190) and the Ministry of Science and Technology of China (grant no. 2016YFA0501103).
L.Z., J.Z., and J.W. conceived the project. L.Z., C.W., and J.Z. designed and performed experiments. J.W. wrote the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Jiawei Wang, Email: jwwang@tsinghua.edu.cn.
Nicole R. Buan, University of Nebraska—Lincoln
REFERENCES
- 1.O'Connor PM, Kuniyoshi TM, Oliveira RP, Hill C, Ross RP, Cotter PD. 2020. Antimicrobials for food and feed; a bacteriocin perspective. Curr Opin Biotechnol 61:160–167. 10.1016/j.copbio.2019.12.023. [DOI] [PubMed] [Google Scholar]
- 2.Cao L, Do T, Link AJ. 2021. Mechanisms of action of ribosomally synthesized and posttranslationally modified peptides (RiPPs). J Ind Microbiol Biotechnol 48:kuab005. 10.1093/jimb/kuab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rebuffat S. 2021. Ribosomally synthesized peptides, foreground players in microbial interactions: recent developments and unanswered questions. Nat Prod Rep 10.1039/d1np00052g. [DOI] [PubMed] [Google Scholar]
- 4.Ríos Colombo NS, Chalón MC, Navarro SA, Bellomio A. 2018. Pediocin-like bacteriocins: new perspectives on mechanism of action and immunity. Curr Genet 64:345–351. 10.1007/s00294-017-0757-9. [DOI] [PubMed] [Google Scholar]
- 5.Fimland G, Johnsen L, Dalhus B, Nissen-Meyer J. 2005. Pediocin-like antimicrobial peptides (class IIa bacteriocins) and their immunity proteins: biosynthesis, structure, and mode of action. J Pept Sci 11:688–696. 10.1002/psc.699. [DOI] [PubMed] [Google Scholar]
- 6.Balandin SV, Sheremeteva EV, Ovchinnikova TV. 2019. Pediocin-like antimicrobial peptides of bacteria. Biochemistry (Mosc) 84:464–478. 10.1134/S000629791905002X. [DOI] [PubMed] [Google Scholar]
- 7.Fimland G, Blingsmo OR, Sletten K, Jung G, Nes IF, Nissen-Meyer J. 1996. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol 62:3313–3318. 10.1128/aem.62.9.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissen-Meyer J, Rogne P, Oppegård C, Haugen HS, Kristiansen PE. 2009. Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by Gram-positive bacteria. Curr Pharm Biotechnol 10:19–37. 10.2174/138920109787048661. [DOI] [PubMed] [Google Scholar]
- 9.Cleveland J, Montville TJ, Nes IF, Chikindas ML. 2001. Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol 71:1–20. 10.1016/s0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- 10.Diep DB, Skaugen M, Salehian Z, Holo H, Nes IF. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc Natl Acad Sci USA 104:2384–2389. 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeckelmann J-M, Erni B. 2020. The mannose phosphotransferase system (Man-PTS)—mannose transporter and receptor for bacteriocins and bacteriophages. Biochim Biophys Acta Biomembr 1862:183412. 10.1016/j.bbamem.2020.183412. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Zeng J, Huang K, Wang J. 2019. Structure of the mannose transporter of the bacterial phosphotransferase system. Cell Res 29:680–682. 10.1038/s41422-019-0194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kjos M, Nes IF, Diep DB. 2011. Mechanisms of resistance to bacteriocins targeting the mannose phosphotransferase system. Appl Environ Microbiol 77:3335–3342. 10.1128/AEM.02602-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Telhig S, Ben Said L, Zirah S, Fliss I, Rebuffat S. 2020. Bacteriocins to thwart bacterial resistance in Gram negative bacteria. Front Microbiol 11:586433. 10.3389/fmicb.2020.586433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagos R, Tello M, Mercado G, García V, Monasterio O. 2009. Antibacterial and antitumorigenic properties of microcin E492, a pore-forming bacteriocin. Curr Pharm Biotechnol 10:74–85. 10.2174/138920109787048643. [DOI] [PubMed] [Google Scholar]
- 16.O'Brien GJ, Mahanty HK. 1994. Colicin 24, a new plasmid-borne colicin from a uropathogenic strain of Escherichia coli. Plasmid 31:288–296. 10.1006/plas.1994.1030. [DOI] [PubMed] [Google Scholar]
- 17.Baindara P, Korpole S, Grover V. 2018. Bacteriocins: perspective for the development of novel anticancer drugs. Appl Microbiol Biotechnol 102:10393–10408. 10.1007/s00253-018-9420-8. [DOI] [PubMed] [Google Scholar]
- 18.Fimland G, Eijsink VGH, Nissen-Meyer J. 2002. Comparative studies of immunity proteins of pediocin-like bacteriocins. Microbiology (Reading) 148:3661–3670. 10.1099/00221287-148-11-3661. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez CF, Kunka BS. 1987. Plasmid-associated bacteriocin production and sucrose fermentation in Pediococcus acidilactici. Appl Environ Microbiol 53:2534–2538. 10.1128/aem.53.10.2534-2538.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodríguez JM, Martínez MI, Kok J. 2002. Pediocin PA-1, a wide-spectrum bacteriocin from lactic acid bacteria. Crit Rev Food Sci Nutr 42:91–121. 10.1080/10408690290825475. [DOI] [PubMed] [Google Scholar]
- 21.Tymoszewska A, Diep DB, Aleksandrzak-Piekarczyk T. 2018. The extracellular loop of Man-PTS subunit IID is responsible for the sensitivity of Lactococcus garvieae to garvicins A, B and C. Sci Rep 8:15790. 10.1038/s41598-018-34087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang K, Zeng J, Liu X, Jiang T, Wang J. 2021. Structure of the mannose phosphotransferase system (man-PTS) complexed with microcin E492, a pore-forming bacteriocin. Cell Discov 7:20. 10.1038/s41421-021-00253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller KW, Schamber R, Chen Y, Ray B. 1998. Production of active chimeric pediocin AcH in Escherichia coli in the absence of processing and secretion genes from the Pediococcus pap operon. Appl Environ Microbiol 64:14–20. 10.1128/AEM.64.1.14-20.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nissen-Meyer J, Nes IF. 1997. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch Microbiol 167:67–77. 10.1007/s002030050418. [DOI] [PubMed] [Google Scholar]
- 25.Fimland G, Johnsen L, Axelsson L, Brurberg MB, Nes IF, Eijsink VG, Nissen-Meyer J. 2000. A C-terminal disulfide bridge in pediocin-like bacteriocins renders bacteriocin activity less temperature dependent and is a major determinant of the antimicrobial spectrum. J Bacteriol 182:2643–2648. 10.1128/JB.182.9.2643-2648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleury Y, Dayem MA, Montagne JJ, Chaboisseau E, Le Caer JP, Nicolas P, Delfour A. 1996. Covalent structure, synthesis, and structure-function studies of mesentericin Y 105(37), a defensive peptide from Gram-positive bacteria Leuconostoc mesenteroides. J Biol Chem 271:14421–14429. 10.1074/jbc.271.24.14421. [DOI] [PubMed] [Google Scholar]
- 27.Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H. 2006. The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev 70:564–582. 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derksen DJ, Stymiest JL, Vederas JC. 2006. Antimicrobial leucocin analogues with a disulfide bridge replaced by a carbocycle or by noncovalent interactions of allyl glycine residues. J Am Chem Soc 128:14252–14253. 10.1021/ja066203q. [DOI] [PubMed] [Google Scholar]
- 29.Johnsen L, Fimland G, Nissen-Meyer J. 2005. The C-terminal domain of pediocin-like antimicrobial peptides (class IIa bacteriocins) is involved in specific recognition of the C-terminal part of cognate immunity proteins and in determining the antimicrobial spectrum. J Biol Chem 280:9243–9250. 10.1074/jbc.M412712200. [DOI] [PubMed] [Google Scholar]
- 30.Fimland G, Jack R, Jung G, Nes IF, Nissen-Meyer J. 1998. The bactericidal activity of pediocin PA-1 is specifically inhibited by a 15-mer fragment that spans the bacteriocin from the center toward the C terminus. Appl Environ Microbiol 64:5057–5060. 10.1128/AEM.64.12.5057-5060.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fimland G, Eijsink VG, Nissen-Meyer J. 2002. Mutational analysis of the role of tryptophan residues in an antimicrobial peptide. Biochemistry 41:9508–9515. 10.1021/bi025856q. [DOI] [PubMed] [Google Scholar]
- 32.Richard C, Cañon R, Naghmouchi K, Bertrand D, Prévost H, Drider D. 2006. Evidence on correlation between number of disulfide bridge and toxicity of class IIa bacteriocins. Food Microbiol 23:175–183. 10.1016/j.fm.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Kim IK, Kim MK, Kim JH, Yim HS, Cha SS, Kang SO. 2007. High resolution crystal structure of PedB: a structural basis for the classification of pediocin-like immunity proteins. BMC Struct Biol 7:35. 10.1186/1472-6807-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 35.Bokori-Brown M, Martin TG, Naylor CE, Basak AK, Titball RW, Savva CG. 2016. Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein. Nat Commun 7:11293. 10.1038/ncomms11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Lin L, Sun L, Huang Y, Koh AL, Dang W, Yin J, Wang M, Tan C, Li T, Tan Z, Liu Z, Peng H. 2017. Clean transfer of large graphene single crystals for high-intactness suspended membranes and liquid cells. Adv Mater 10.1002/adma.201700639. [DOI] [PubMed] [Google Scholar]
- 37.Zheng L, Chen Y, Li N, Zhang J, Liu N, Liu J, Dang W, Deng B, Li Y, Gao X, Tan C, Yang Z, Xu S, Wang M, Yang H, Sun L, Cui Y, Wei X, Gao P, Wang HW, Peng H. 2020. Robust ultraclean atomically thin membranes for atomic-resolution electron microscopy. Nat Commun 11:541. 10.1038/s41467-020-14359-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei J, Frank J. 2005. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J Struct Biol 150:69–80. 10.1016/j.jsb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 39.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14:331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grant T, Grigorieff N. 2015. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife 4:e06980. 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang K. 2016. Gctf: real-time CTF determination and correction. J Struct Biol 193:1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, Scheres SH. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7:e42166. 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su R, Zhang SY, Wang JW. 2019. Automatically generating mask for mapping out the density map in cryoEM single-particle reconstruction. Prog Biochem Biophys 46:1020–1030. [Google Scholar]
- 44.Rosenthal PB, Henderson R. 2003. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333:721–745. 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 45.Kucukelbir A, Sigworth FJ, Tagare HD. 2014. Quantifying the local resolution of cryo-EM density maps. Nat Methods 11:63–65. 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou N, Wang H, Wang J. 2017. EMBuilder: a template matching-based automatic model-building program for high-resolution cryo-electron microscopy maps. Sci Rep 7:2664. 10.1038/s41598-017-02725-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD. 2018. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74:531–544. 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLano WL. 2002. The PyMOL molecular graphics system. https://www.pymol.org.
- 50.Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, Fraser JS. 2015. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods 12:943–946. 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. 1996. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14:354–360, 376. 10.1016/S0263-7855(97)00009-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S7. Download AEM.01992-21-s0001.pdf, PDF file, 3.1 MB (3.1MB, pdf)
Data Availability Statement
The cryo-EM maps and the structures have been deposited in the Electron Microscopy Data Bank (EMD-32030 and EMD-32031) and the Protein Data Bank (PDB no. 7VLX and 7VLY), respectively.