Abstract
Penicillium species are common fungi found worldwide from diverse substrates, including soil, plant debris, food products and air. Their diversity in aquatic environments is still underexplored. With the aim to explore the fungal diversity in Spanish freshwater sediments, numerous Penicillium strains were isolated using various culture-dependent techniques. A preliminary sequence analysis of the β-tubulin (tub2) gene marker allowed us to identify several interesting species of Penicillium, which were later characterized phylogenetically with the barcodes recommended for species delimitation in the genus. Based on the multi-locus phylogeny of the internal transcribed spacer region (ITS) of the ribosomal DNA, and partial fragments of tub2, calmodulin (cmdA), and the RNA polymerase II largest subunit (rpb2) genes, in combination with phenotypic analyses, five novel species are described. These are P.ausonanum in sectionLanata-Divaricata, P.guarroi in sect.Gracilenta, P.irregulare in sect.Canescentia, P.sicoris in sect.Paradoxa and P.submersum in sect.Robsamsonia. The study of several isolates from samples collected in different locations resulted in the reinstatement of P.vaccaeorum into sectionCitrina. Finally, P.heteromorphum (sect.Exilicaulis) and P.tardochrysogenum (sect.Chrysogena) are reported, previously only known from Antarctica and China, respectively.
Keywords: 5 new species, Ascomycota, Eurotiales, fluvial sediments, phylogeny, species delimitation, taxonomy
Introduction
Fungi make up a significant component of the benthic microbial biomass in freshwater ecosystems, and play a pivotal role in decomposing organic matter in river habitats (Pascoal and Cassio 2004; Zhang et al. 2013). In general, representatives of Ascomycota account for the largest number of species recovered from water systems (Kelley et al. 2001; Liu et al. 2015; Khomich et al. 2017; Sutcliffe et al. 2018; Samson et al. 2020), their asexual forms with stauro- or scolecoconidia (aquatic hyphomycetes) being the most representative fungal group in freshwater environments (Pascoal et al. 2005; Jones and Pang 2012). Although common Ascomycota like Penicillium species are not considered aquatic fungi, they are one of the most common genera identified from freshwater (Pitt 1979; Hageskal et al. 2006; Sonjak et al. 2006; Samson et al. 2010; Heo et al. 2019; Pangging et al. 2019) and recently were also reported in marine environments (Kirichuk et al. 2017; Gonçalves et al. 2019; Grossart et al. 2019). However, in almost all of those reports, little attention has been paid to the study of river sediments as a reservoir of taxonomically interesting penicillia.
Penicillium (Eurotiomycetes, Eurotiales, Aspergillaceae) is a ubiquitous genus recovered from a wide range of substrates–including soil, plant material, indoor and outdoor air environments, a variety of food products, herbivore dung and water (Visagie et al. 2014a; Barbosa et al. 2018; Grossart et al. 2019; Guevara-Suárez et al. 2020; Rodríguez-Andrade et al. 2021). Penicillium species have a great impact on human life as agents of food spoilage, causal agents of pre- and postharvest diseases on crops (Frisvad and Samson 2004; Pitt and Hocking 2009; Samson et al. 2010), and their ability to produce toxic compounds like mycotoxins (Frisvad and Samson 2004; Perrone and Susca 2017), which can be a serious threat to human and animal health worldwide. On the contrary, positive impacts of these fungi include their use in food fermentations, the production of many bioactive compounds of medical interest (antimicrobial agents, immunosuppressants), and the production of enzymes with a variety of industrial applications (Houbraken et al. 2014; Visagie et al. 2014a; Abdelwahab et al. 2018; Park et al. 2019; Ogaki et al. 2020a, b). Other recent applications are the potential use of some species as biocontrol agents against plant pathogens or for bioremediation in polluted environments (Guijarro et al. 2017; Cecchi et al. 2019; Behera et al. 2020; Thambugala et al. 2020; Liang et al. 2021). The discovery of novel lineages in the genus Penicillium, therefore, represents an opportunity to find and characterize new fungi or molecules with a wide range of applications. As an example, more than 390 novel natural products have been isolated in recent years from marine-derived Penicillium fungi, many of which possess biological activity with potential application in new drug developments (Yang et al. 2021).
Limitations in establishing species boundaries by morphological and even molecular data are well-known in Ascomycota, mainly in those genera that comprise a huge number of species such as Aspergillus, Fusarium and Trichoderma, among many others. Therefore, the use of an integrative taxonomy that combines morphometric, phylogenetic, chemical and ecological data provides support for accurate species delimitation and formal description of novel taxa (Haelewaters et al. 2018; Aime et al. 2021; Maharachchikumbura et al 2021). Species delimitation in Penicillium is currently based on an integrative or polyphasic approach, which usually includes morphological features, extrolite profiles (when available), and multi-locus phylogenies (Visagie et al. 2014a). Phylogenetic markers used for this purpose include β-tubulin (tub2), the internal transcribed spacer region (ITS) of the ribosomal RNA gene (rDNA) and the calmodulin (cmdA) and the RNA polymerase II largest subunit (rpb2) genes. More than 480 species are accepted in the genus, distributed across 32 sections (Houbraken and Samson 2011; Visagie et al. 2014a; Houbraken et al. 2020). More recently, in a phylogenetic re-evaluation of various genera in the order Eurotiales, Houbraken et al. (2020) reinstated a series classification, and in particular for Penicillium recognized 89 series, contributing in this way to work easily with phylogenetic clades instead of large sections.
In this context, in an ongoing study of microfungal diversity from freshwater sediments collected from rivers or streams in different Spanish regions, we found several isolates of Penicillium, which could represent putative new or uncommon species of penicillia based on the sequence analysis of the recommended secondary identification marker tub2 (Visaige et al. 2014a). The present work aims to resolve the taxonomy of those isolates by using the above-mentioned polyphasic approach and following the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) criterion (Taylor et al. 2000).
Materials and methods
Sampling and fungal isolation
Sediment samples were collected between February 2018 and December 2020 from rivers and streams in natural and rural areas from various Spanish provinces (Baleares, Barcelona, Lleida, Madrid, and Tarragona) (Table 1). Sterile 100 ml plastic containers were used for collecting samples from the bunk beds or edges of the rivers selected ca 10 cm below the surface layer. Samples were transported in a refrigerated container and immediately processed in the laboratory. Samples were shaken vigorously in the same containers and, after leaving them for 1 min to settle, the water was decanted, and the sediment poured onto several layers of sterile filter paper on plastic trays to remove any water excess (Ulfig et al. 1997). To isolate a wide range of fungal species, each sample was cultured as follows: 1 g of sediment was distributed across three Petri dishes and mixed with dichloran rose-bengal-chloramphenicol agar (DRBC; 2.5 g peptone, 5 g glucose, 0.5 g KH2PO4, 0.25 g MgSO4, 12.5 mg Rose Bengal, 100 mg chloramphenicol, 1 mg dichloran, 10 g agar, 500 mL distilled water) melted at 45 °C; and in parallel, another 1 g was distributed in three other Petri dishes and mixed with melted potato dextrose agar (PDA; Pronadisa) supplemented with 2 g/L of chloramphenicol and 2 g/L of cycloheximide. Once solidified, the cultures were incubated at room temperature (22–25 °C) and examined weekly with a stereomicroscope for up to 4–5 weeks. To achieve pure cultures, conidia of Penicillium colonies were transferred with a sterile dissection needle to plates with PDA supplemented with chloramphenicol and incubated at 25 °C in darkness.
Table 1.
Strain information and GenBank/EMBL accession numbers of the Penicillium species investigated in this study.
| Species | Section | Strain no.1 | Substrate and Origin | GenBank nucleotide accession no.2: | Citation | |||
|---|---|---|---|---|---|---|---|---|
| tub2 | cmdA | ITS | rpb2 | |||||
| P.ausonanum | Lanata–Divaricata | FMR 16948T | Fluvial sediment, stream of the Guilleries National Park, Barcelona, Catalonia, Spain | LR655809 | LR655810 | LR655808 | LR655811 | This study |
| P.guarroi | Gracilenta | FMR 17747T | Fluvial sediment, Brugent River, Tarragona, Catalonia, Spain | LR814134 | LR814140 | LR814139 | LR814145 | This study |
| P.heteromorphum | Exilicaulis | CBS 226.89T | Soil, China | KJ834455 | KP016786 | KC411702 | JN406605 | Visagie et al. 2014a |
| Exilicaulis | FMR 18043 | Fluvial sediment, stream of the Cadí–Moixerò Natural Park, Lleida, Catalonia, Spain | LR861780 | LR861782 | LR861783 | LR861784 | This study | |
| P.irregulare | Canescentia | FMR 17859T | Fluvial sediment, Miraflores River, Community of Madrid, Spain | LR814144 | LR814151 | LR814181 | LR814182 | This study |
| P.sanguifluum | Citrina | CBS 127032T | Soil, Calahonda, Costa del Sol, Spain | JN606819 | JN606555 | JN617681 | – | Houbraken et al. 2011 |
| Citrina | FMR 17617 | Fluvial sediment, Mallorca Island, Spain | LR861778 | LR861781 | LR861779 | – | This study | |
| Citrina | FMR 17619 | Fluvial sediment, Mallorca Island, Spain | OU375375 | – | – | – | This study | |
| P.sicoris | Paradoxa | FMR 18076T | Fluvial sediment, Segre River, Lleida, Catalonia, Spain | LR884494 | LR884496 | LR884497 | LR884495 | This study |
| P.submersum | Robsamsonia | FMR 17140T | Fluvial sediment, stream of the Montsant Natural Park, Tarragona, Catalonia, Spain | LR814187 | LR814188 | LR814194 | LR814195 | This study |
| P.tardochrysogenum | Chrysogena | CBS 132200T | Soil, McMurdo Dry Valley, Antarctica | JX996898 | JX996239 | JX997028 | JX996634 | Houbraken et al. 2012 |
| Chrysogena | FMR 17137 | Fluvial sediment, stream of the Montsant Natural Park, Tarragona, Catalonia, Spain | HG996463 | HG996465 | HG996464 | – | This study | |
| P.vaccaeorum | Citrina | CBS 148.83T | Sandy soil under pine tree, Valladolid, Spain | JN606846 | JN606543 | MH861558 | – | Houbraken et al. 2011 |
| Citrina | FMR 17967 | Fluvial sediment, Basque Country, Spain | LR814226 | LR814227 | LR814235 | – | This study | |
| Citrina | FMR 17531 | Fluvial sediment, stream of Montseny National Park, Barcelona, Spain | LR814203 | LR814204 | LR814213 | – | This study | |
| Citrina | FMR 17534 | Fluvial sediment, stream of Montseny National Park, Barcelona, Spain | OU375168 | OU375273 | OU375272 | – | This study | |
| Citrina | FMR 17616 | Fluvial sediment, stream of Serra de Tramontana, Mallorca, Spain | LR814212 | LR814218 | LR814217 | – | This study | |
| Citrina | FMR 18100 | Fluvial sediment, Segre River, Lleida, Spain | LR814234 | LR814242 | LR814241 | – | This study | |
| Citrina | FMR 18123 | Fluvial sediment, Segre River, Lleida, Spain | LR814265 | LR814264 | LR814273 | – | This study | |
| Citrina | CBS 110.64 | Forest soil, Erzurum, Turkey | JN606829 | JN606533 | MH858377 | – | Houbraken et al. 2011 | |
| Citrina | CBS 441.88 | Sandy soil, Chile | JN606846 | JN606568 | – | – | Houbraken et al. 2011 | |
| Citrina | CBS 643.73 | Sandy soil, Manitoba, Canada | – | JN606576 | – | – | Houbraken et al. 2011 | |
| Citrina | CBS 644.73 | Sandy soil, Manitoba, Canada | LR814213 | JN606577 | – | – | Houbraken et al. 2011 | |
| Citrina | CBS 685.85 (Type of P.lacussarmientei) | Sandy soil, Torres del Paine National Park, Tierra del Fuego, Chile | JN606855 | JN606533 | JN617711 | – | Houbraken et al. 2011 | |
| Citrina | CBS 300.67 | Sandy greenhouse soil, The Netherlands | – | JN606561 | – | – | Houbraken et al. 2011 | |
| Citrina | CBS 127029 | Forest soil, Los Alerces National Park, Argentina | JN606814 | – | MH864309 | – | Houbraken et al. 2011 | |
| Citrina | CBS 118024 | Ants (Camponotus spp.), New Brunswick, Canada | JN606833 | JN606537 | – | – | Houbraken et al. 2011 | |
CBS: Culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; FMR: Facultat de Medicina i Ciències de la Salut, Reus, Spain. T Indicate ex-type strains. 1 Strain no.: strain number. 2tub2: β-tubulin; cmdA: calmodulin; ITS: Internal transcribed spacer regions of the rDNA and 5.8S region; rpb2: the DNA dependent RNA polymerase II largest subunit. Novelties and sequences generated in this study are in bold.
All isolates were preserved and deposited into the fungal collection of the Faculty of Medicine, Reus (FMR). Those isolates that were representative of novel species, ex-type cultures and holotypes (dried cultures) were prepared and deposited at the Westerdijk Fungal Biodiversity Institute (CBS, Utrecht, The Netherlands).
DNA extraction, sequencing and phylogenetic analysis
Isolates were cultured on PDA for 7–14 days at 25 °C in darkness. The DNA was extracted through the modified protocol of Werner et al. (1998). Preliminary species identifications were carried out by similarity searches of obtained tub2 DNA sequences against reference sequences on GenBank. In the case of putative new species, the ITS region and fragments of cmdA and rpb2 genes were amplified and sequenced (Visagie et al. 2014b). The primer pairs used for gene amplifications were: Bt2a/Bt2b for tub2 (Glass and Donaldson 1995), ITS5/ITS4 for ITS (White et al. 1990), CMD5/CMD6 for cmdA (Hong et al. 2006) and RPB2–5F/RPB2–7Cr for rpb2 (Liu et al. 1999). The PCR conditions were carried out using primers and methods previously described (Peterson 2008; Houbraken and Samson 2011; Visagie et al. 2014a). Amplified products were purified and sequenced at Macrogen (Madrid, Spain). Consensus sequences were obtained using SeqMan v. 7.0.0 (DNAStar Lasergene, Madison, WI, USA).
Sequences for phylogenies of Penicillium sections and series included in the study were retrieved from NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/), considering the accepted species of penicillia included in the last update of the International Commission of Penicillium and Aspergillus database (http://www.aspergilluspenicillium.org), and those more recently published (Houbraken et al. 2020; Visagie et al. 2021). Initially, a preliminary phylogenetic analysis with tub2 sequences was carried out to resolve the taxonomic position of our isolates at section level. Thereafter, single and concatenated phylogenetic analyses for each section were calculated to allocate the isolates at series level and establish phylogenetic relationships among closely related species. Individual analyses of the alternative molecular markers of the series in which the isolates under study belong are included as supplementary material.
Datasets for each locus were aligned individually in MEGA (Molecular Evolutionary Genetic Analysis) software v. 6.0. (Tamura et al. 2013) using the CLUSTALW algorithm (Thompson et al. 1994) and refined with MUSCLE (Edgar 2004) or manually adjusted, if necessary, on the same platform. Phylogenetic concordance of the four-locus datasets was evaluated individually through visual comparison of each single-locus phylogeny to assess any incongruent results among nodes with high statistical support. Once the lack of incongruence was confirmed, individual alignments were concatenated into a single data matrix with SequenceMatrix (Vaidya et al. 2011). The best substitution model for all gene matrices was estimated using MEGA software for Maximum Likelihood (ML) analysis, while for the Bayesian Inference (BI) analysis it was estimated using jModelTest v.2.1.3 according to the Akaike criterion (Guindon and Gascuel 2003; Darriba et al. 2012). The phylogenetic reconstructions were made with the combined genes using ML under RAxML-HPC2 on XSEDE v-8.2.12 (Stamatakis 2014) on the CIPRES Science gateway portal (Miller et al. 2012) and BI with MrBayes v.3.2.6 (Ronquist et al. 2012).
For ML, phylogenetic support for internal branches was assessed by 1,000 ML bootstrapped pseudoreplicates and bootstrap support (bs) ≥ 70 was considered significant (Hillis and Bull 1993). The phylogenetic reconstruction by BI was carried out using 5 million Markov chain Monte Carlo (MCMC) generations, with four runs (one cold chain and three heated chains), and samples were stored every 1,000 generations. The 50% majority-rule consensus tree and posterior probability (pp) values were calculated after discarding the first 25% of samples. A pp value of ≥ 0.95 was considered significant (Hespanhol et al. 2019). The resulting trees were plotted using FigTree v.1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
The DNA sequences and alignments generated in this study were deposited in GenBank (Table 1) and in TreeBASE under the submission number: 28954 (http://treebase.org), respectively.
Phenotypic study
Phenotypic characterization of the strains was made using standard growth conditions established previously (Visagie et al. 2014a). Strains were inoculated in three-point fashion onto Czapek Yeast Autolysate agar (CYA; Pitt 1979), 2% Malt Extract Agar (MEA; Samson et al. 2010), Yeast Extract Sucrose agar (YES; Frisvad 1981), Oatmeal agar (OA; Samson et al. 2010), Dichloran 18% Glycerol agar (DG18; Hocking and Pitt 1980) and Creatine Sucrose agar (CREA; Frisvad 1981) and incubated at 25 °C for 7d in darkness, with the exception of FMR 16948 which was incubated for 14d for sporulation to occur. Colony growth rates were measured after 7 d at 5, 15, 20, 30, 35, 37, and 40 °C on CYA. Color annotations in descriptions follow Kornerup and Wanscher (1978). Microscopic measurements and features were described from colonies grown on MEA after 7 d (14 d for FMR 16948) of incubation. Microscopic slides were mounted with Shear’s solution, after prior removal of the excess conidia with 70% ethanol. Photomicrographs were obtained using a Zeiss Axio-Imager M1 light microscope (Zeiss, Oberkochen, Germany) with a DeltaPix Infinity x digital camera. Photoplates were assembled from separate photographs using PhotoShop CS6.
Results
Phylogenetic analyses
Among the penicillium-like fungi found, we recovered 15 isolates (12 from PDA at 0.2% cycloheximide; three from DRBC), which preliminary identification, having compared their tub2 sequences through BLAST search, revealed as possibly representing putative novel or uncommon species of penicillia (Table 1). According to the tub2 phylogeny, Penicillium isolates were distributed over two major clades corresponding to the two subgenera of Penicillium (i.e. Aspergilloides and Penicillium) and all sections included in the analysis were well delineated (Suppl. material 1: Fig. S1). Five of the isolates under study, which were distributed in the sections Robsamsonia (FMR 17140), Paradoxa (FMR 18076), Canescentia (FMR 17859), Lanata-Divaricata (FMR 16948) and Gracilenta (FMR 17747), could not be assigned to any currently accepted species in the genus. Another eight isolates, which belong to the sectionCitrina (FMR 17531, FMR 17534, FMR 17616, FMR 17617, FMR 17619, FMR 17967, FMR 18100 and FMR 18123), were closely related to the ex-type strains of P.sanguifluum, P.roseopurpureum and P.vaccaeorum, the latter species currently considered synonym of P.sanguifluum (Houbraken et al. 2011). The other two isolates, FMR 17137 and FMR 18043, were placed in sections Chrysogena and Exilicaulis, respectively. However, the tub2 marker did not show enough discriminatory power to resolve the identity of both isolates concerning their counterparts, which were the ex-type strains of P.chrysogenum, P.tardochrysogenum and P.rubens for FMR 17137, and P.restrictum and P.heteromorphum for FMR 18043. Phylogenetic analyses and species delimitation were performed using ITS, tub2, cmdA, and rpb2. Dataset characteristics and substitution models for each data partition are summarized in Table 2. Since tree topologies were similar and congruent between the ML and BI analyses in all cases, we selected ML trees to represent section results. Bootstrap support values ≥70% and BI posterior probability values ≥0.95 are indicated on branches.
Table 2.
Overview and details used for phylogenetic analyses in sections of Penicillium analysed in this study.
| Dataset | Sections | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chrysogena | Robsamsonia | Paradoxa/Turbata | Canescentia | Exilicaulis | Lan.-Div. | Gracilenta | Citrina | ||
| tub2 | Length (bp) | 499 | 468 | 493 | 431 | 503 | 547 | 508 | 470 |
| tub2 | Pvar | 113 | 171 | 173 | 161 | 254 | 307 | 189 | 240 |
| tub2 | Pi | 65 | 103 | 108 | 101 | 205 | 257 | 93 | 207 |
| tub2 | Model* | K2+G | K2+G | K2+G | K2+G | HKY+G+I | HKY+G+I | HKY+G | HKY+G+I |
| tub2 | Model** | K2+G | K2+G | K2+G | K2+G | K2+G | K2+G+I | TN93+G | K2+G |
| cmdA | Length (bp) | 531 | 542 | 529 | 560 | 604 | 626 | 642 | 626 |
| cmdA | Pvar | 177 | 202 | 221 | 189 | 302 | 392 | 264 | 378 |
| cmdA | Pi | 93 | 124 | 136 | 95 | 349 | 329 | 145 | 337 |
| cmdA | Model* | K2+G | K2+I | SYM+G | K2+G | SYM+G+I | HKY+G+I | K2+I | SYM+G+I |
| cmdA | Model** | K2+G | K2+G | K2+I | K2+G | K2+G+I | K2+G+I | K2+I | K2+G+I |
| ITS | Length (bp) | 580 | 605 | 572 | 610 | 576 | 559 | 566 | 566 |
| ITS | Pvar | 25 | 60 | 39 | 45 | 102 | 180 | 44 | 119 |
| ITS | Pi | 7 | 17 | 21 | 33 | 60 | 154 | 20 | 99 |
| ITS | Model* | K2+G | K2+G | K2+I | K2 | K2+G+I | GTR+G+I | GTR+I | GTR+G+I |
| ITS | Model** | T92 | JC+G | K2+I | T92 | K2+G+I | GTR+G+I | T92+I | K2+G+I |
| rpb2 | Length (bp) | - | 936 | 929 | 915 | 950 | 837 | 978 | - |
| rpb2 | Pvar | - | 249 | 256 | 230 | 360 | 345 | 294 | - |
| rpb2 | Pi | - | 176 | 183 | 121 | 306 | 313 | 113 | - |
| rpb2 | Model* | - | SYM+G | SYM+G | K2+G | SYM+G+I | GTR+G+I | SYM+G | - |
| rpb2 | Model** | - | K2+G | K2+I | K2+G | K2+G+I | K2+G+I | TN93+G | - |
| Concatenated | Length (bp) | 1610 | 2551 | 2523 | 2516 | 2633 | 2569 | 2694 | 1662 |
| Concatenated | Pvar | 315 | 628 | 689 | 625 | 1018 | 1224 | 791 | 737 |
| Concatenated | Pi | 165 | 420 | 448 | 350 | 820 | 1053 | 371 | 643 |
| Concatenated | Model* | K2+G+I | SYM+G | SYM+G | K2+G+I | HKY+G+I | GTR+G+I | GTR+G | SYM+G+I |
| Concatenated | Model** | K2+G | K2+G | K2+G | K2+G | K2+G+I | GTR+G+I | K2+I | K2+G |
Lan.-Div. = sect.Lanata-Divaricata; Pvar = variable sites; Pi = phylogenetic informative sites; *= substitution model for Bayesian inference; **= substitution model for ML analysis; K2 = Kimura 2-parameter; HKY = Hasegawa-Kishino-Yano; SYM = Symmetrical; GTR = General Time Reversible; TN93 = Tamura-Nei; T92 = Tamura 3-parameter; JC = Jukes-Cantor; G = Gamma Distributed; I = Invariant Sites.
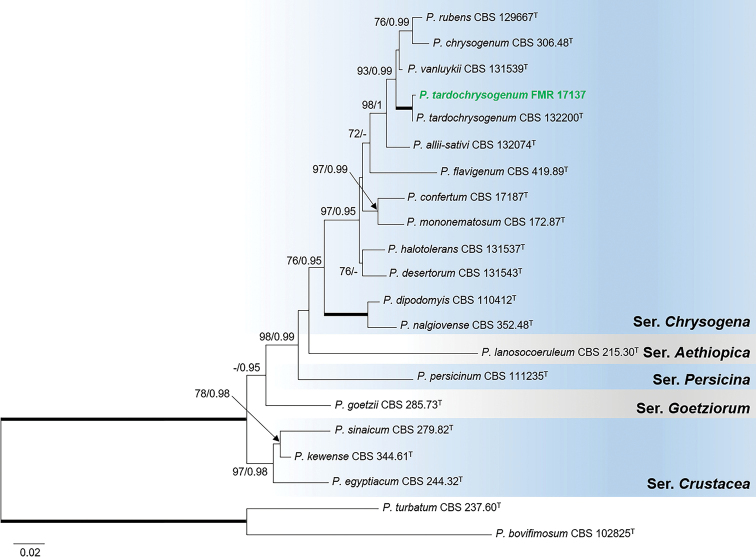
The concatenate phylogeny for the sectionChrysogena was constructed with ITS, tub2 and cmdA markers since rpb2 was not available for FMR 17137. This isolate was placed in a fully-supported clade (100 bs/1 pp) with the ex-type strain of P.tardochrysogenum (Fig. 1). Both specimens showed a high sequence similarity (99.2% for tub2, 98.9% cmdA, and 99.8% ITS). It is closely related to the type species of the section P.chrysogenum and its relatives (P.rubens, P.allii-sativa, P.rubens and P.vanluytii). Additional analyses of the series Chrysogena with tub2 and cmdA sequences, including ex-type strains and more reference strains of those mentioned species, confirmed the identity of the sediment isolate (Suppl. material 1: Fig. S2). Penicilliumtardochrysogenum has so far been isolated exclusively from soil and rock samples in Antarctica (Houbraken et al. 2012; Alves et al. 2019), and our specimen is thus the first report of the species from temperate regions.
Figure 1.
Phylogenetic tree of PenicilliumsectionChrysogena based on ML analysis obtained by RAxML inferred from the combined tub2, cmdA and ITS loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70%/0.95 are indicated on the nodes. Bold branches indicate bs/pp values 100/1. The tree is rooted to P.turbatumCBS 237.60 and P.bovifimosumCBS 102825. The name in green is the strain of P.tardochrysogenum included in this study. T= Ex-type strain.
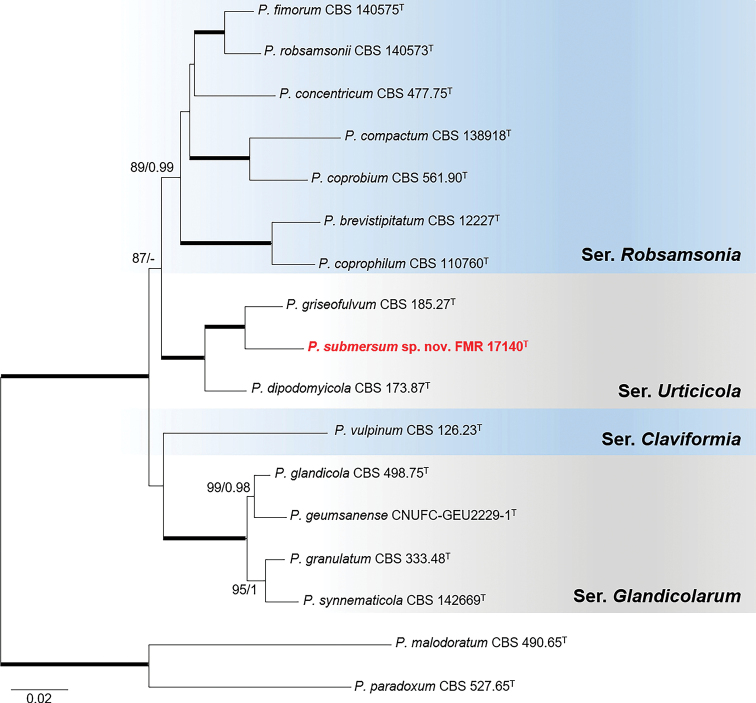
Phylogenetic reconstruction of sectionRobsamsonia with the four markers (Fig. 2) showed the unidentified isolate FMR 17140 grouped with the ex-type strain of P.griseofulvum in a fully-supported terminal clade (100 bs/1 pp), but forming an independent and single branch with enough genetic distance (97.0% tub2, 97.7% cmdA, 99.6% ITS, and 97.2% rpb2 similarity) with the ex-type of P.griseofulvulm to be considered a distinct phylogenetic species. In order to evaluate possible inter- and intraspecific variability regarding closely related species, we carried out additional analyses with tub2, cmdA and rpb2 markers of the species in series Urticicola (Suppl. material 1: Fig. S3), including more sequences of P.griseofulvum retrieved from GenBank. These analyses showed that our isolate was always placed distant to the clade representative of P.griseofulvum. Therefore, genetic and phenotypic differences, such as a faster growth rate on CYA and strong acid production compared to P.griseofulvum, allow us to propose the novel species Penicilliumsubmersum.
Figure 2.
Phylogenetic tree of PenicilliumsectionRobsamsonia based on ML analysis obtained by RAxML inferred from the combined tub2, cmdA, ITS, and rpb2 loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70%/0.95 are indicated on the nodes. Bold branches indicate bs/pp values 100/1. The tree is rooted to P.paradoxumCBS 527.65 and P.malodoratumCBS 490.65. The name in red is the new species described in this study. T= Ex-type strain.
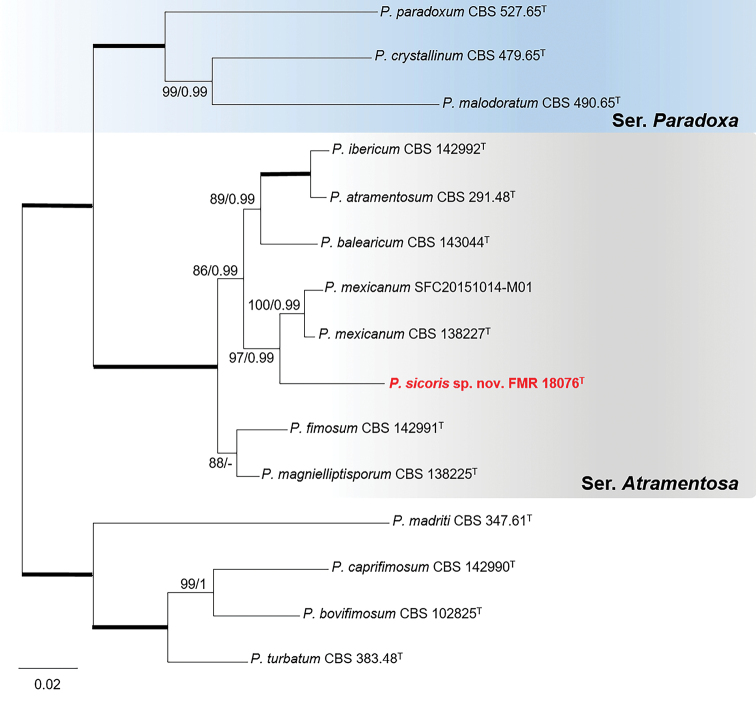
The concatenated dataset for sectionParadoxa (Fig. 3) revealed that FMR 18076 belonged to the series Atramentosa and was closely related to the lineage representative of P.mexicanum, a species recently described from house dust of which only two specimens are known (Visagie et al. 2014c; Park et al. 2019). Both lineages were also well differentiated when additional analyses of the series were carried out with the three alternative markers (tub2, cmdA and rpb2) and more representative sequences of the species in the series (Suppl. material 1: Fig. S4). The genetic distance with the ex-type strain of P.mexicanum (96.5%, 94.3% 98.2%, and 97.6% sequence similarity for tub2, cmdA, ITS, and rpb2, respectively) and their morphological differences in colony color, growth rates, and conidial shape (see Taxonomy section) allow to propose FMR 18076 as Penicilliumsicoris.
Figure 3.
Phylogenetic tree of PenicilliumsectionParadoxa based on ML analysis obtained by RAxML inferred from the combined tub2, cmdA, ITS, and rpb2 loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70%/0.95 are indicated on the nodes. Bold branches indicate bs/pp values 100/1. The tree is rooted to Penicillium species belonging to sectionTurbata (P.madritiCBS 347.61, P.caprifimosumCBS 142990, P.bovifimosumCBS 102825 and P.turbatumCBS 383.48). The name in red is the new species described in this study. T= Ex-type strain.
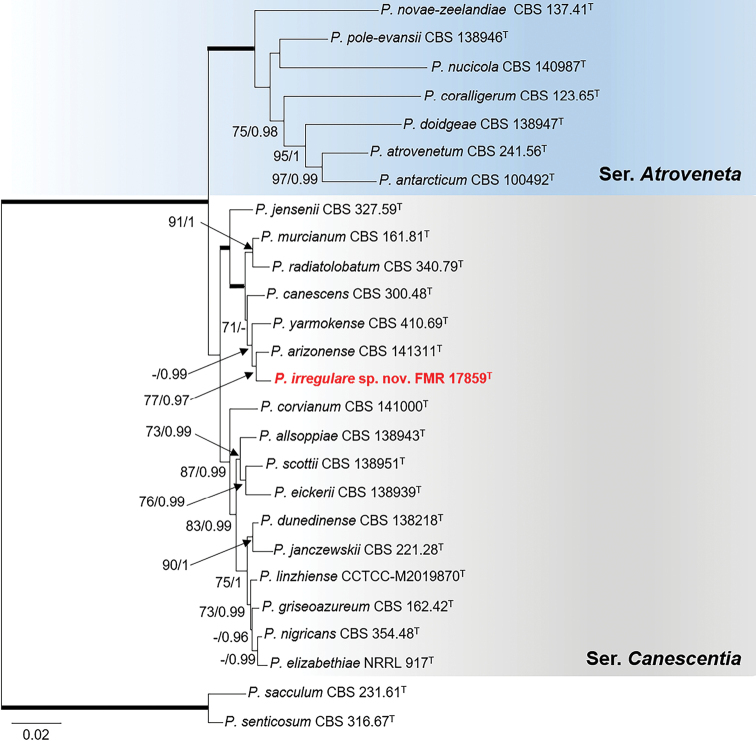
The phylogeny constructed for sectionCanescentia (Fig. 4) placed FMR 17859 in a divergent lineage closely related to the ex-type strains of P.arizonense and P.yarmokense, the three forming a terminal clade only supported with BI analysis (- bs/0.99 pp). In order to elucidate possible inter- and intraspecific variability among these close relatives, additional analyses of the series with the three alternative markers and with more sequences of reference strains of those mentioned species were carried out (Suppl. material 1: Fig. S5). All revealed that P.arizonense was the closest relative and FMR 17859 being always placed in a distant branch. Phylogentic and phenotypic differences like stipe and metulae length, its ability to grow at 37 °C and its reverse color on CYA support the proposal of the novel species Penicilliumirregulare.
Figure 4.
Phylogenetic tree of PenicilliumsectionCanescentia based on ML analysis obtained by RAxML inferred from the combined tub2, cmdA, ITS, and rpb2 loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70%/0.95 are indicated on the nodes. Bold branches indicate bs/pp values 100/1. The tree is rooted to P.sacculumCBS 231.61 and P.senticosumCBS 316.67. The name in red is the new species described in this study. T= Ex-type strain.
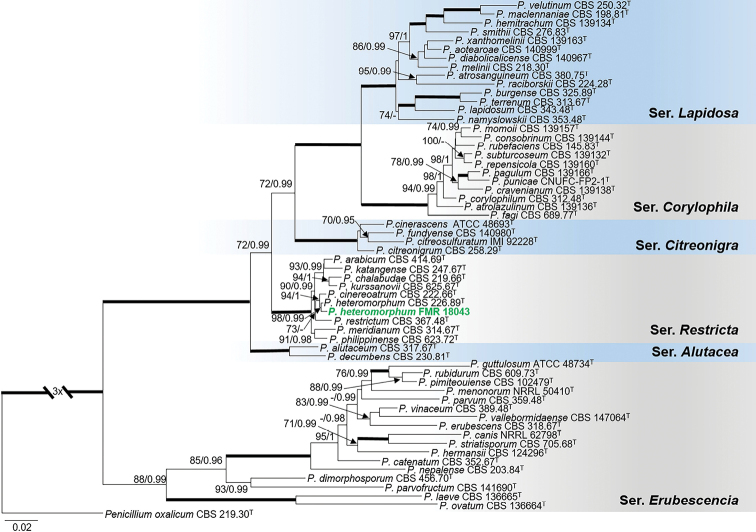
Identification of FMR 18043 as P.heteromorphum was confirmed with the multi-locus phylogeny of sectionExilicaulis (Fig. 5). This species belongs to series Restricta, which includes species with still unresolved phylogeny (Visagie et al. 2016). In the tree, our isolate grouped in a strongly supported terminal clade (98 bs/0.99 pp) with the monotypic species P.heteromorphum (Kong and Qi 1988). However, despite the high genetic similarity of both strains in tub2, cmdA, ITS, and rpb2 (99.2%, 99.8%, 99.6% and 99.4%, respectively), we observed morphological differences regarding ornamentation of conidia, length of conidiophores and its growth at 37 °C. These differences are described in the Taxonomy section. Additional analyses with more sequences of representative strains of its closely related species in series Restricta also supported the identification of our sediment isolate (Suppl. material 1: Fig. S6).
Figure 5.
Phylogenetic tree of PenicilliumsectionExilicaulis based on ML analysis obtained by RAxML inferred from the combined tub2, cmdA, ITS, and rpb2 loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70%/0.95 are indicated on the nodes. Bold branches indicate bs/pp values 100/1. The tree is rooted to P.oxalicumCBS 219.30. The name in green is the strain of P.heteromorphum included in this study. T= Ex-type strain.
Phylogenetic reconstruction of sectionLanata-Divaricata (Fig. 6) resolved FMR 16948 in series Dalearum closely related to the ex-type strains of P.amphipolaria and P.viridissimum (78 bs/0.98 pp), but the three specimens were placed in distant lineages. In addition, FMR 16948 showed a similarity of 97.1% tub2, 97.2% cmdA and 97.4% rpb2 with the ex-type strain of P.amphipolaria and of 97.8% tub2, 97.7% cmdA and 98.4% rpb2 with the ex-type of P.viridissimum. Similar results were obtained in analyses of the series with alternative markers and more sequences of representative strains of its closest relatives (Suppl. material 1: Fig. S7). Our isolate also differed from its relatives by strong acid production and by the predominance of mono- and biverticillate conidiophores. Therefore, morphological and phylogenetic differences support to propose our isolate as Penicilliumausonanum.
Figure 6.
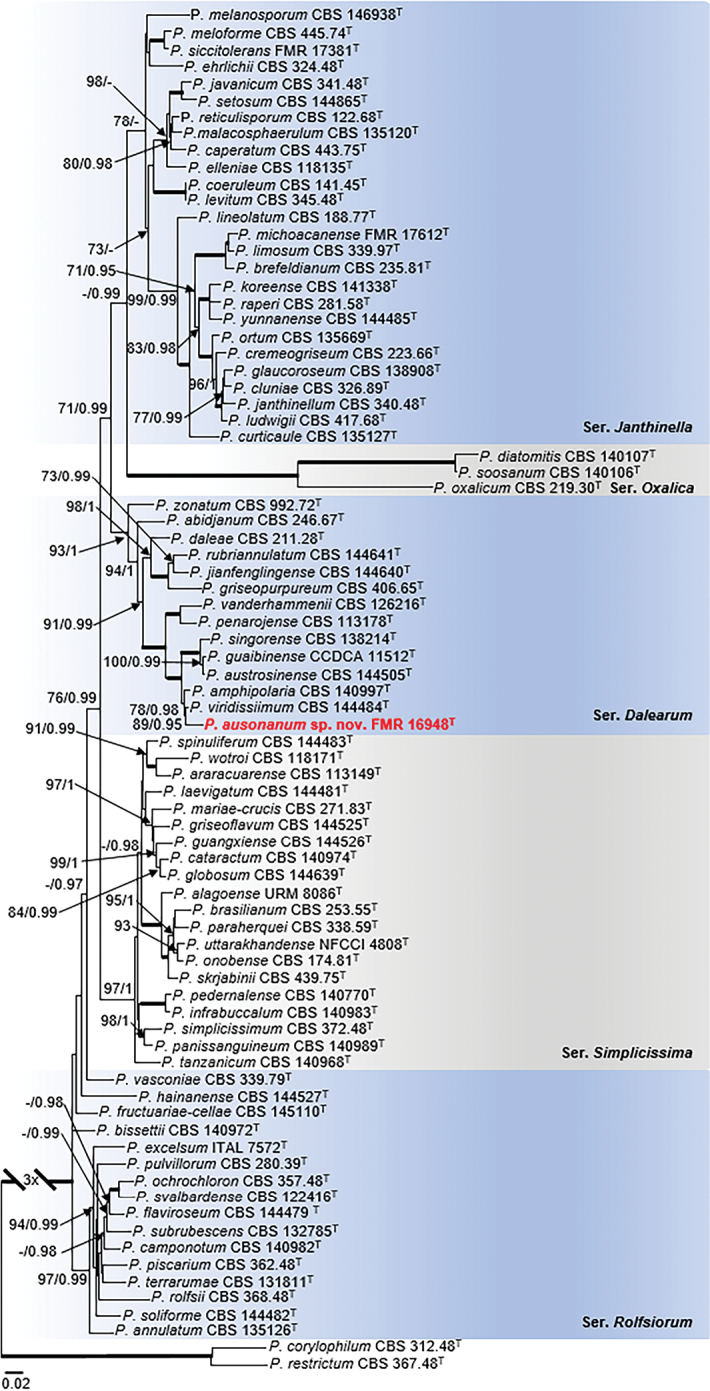
Phylogenetic tree of PenicilliumsectionLanata-Divaricata based on ML analysis obtained by RAxML inferred from the combined tub2, cmdA, ITS, and rpb2 loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70%/0.95 are indicated on the nodes. Bold branches indicate bs/pp values 100/1. The tree is rooted to P.restrictumCBS 367.48 and P.corylophilumCBS 312.48. The name in red is the new species described in this study. T= Ex-type strain.
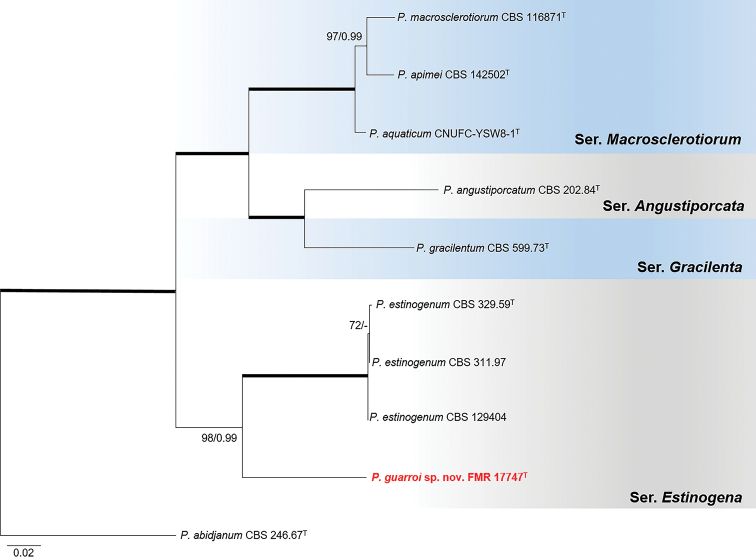
The phylogenetic analysis of sectionGracilenta (Fig. 7), comprising the six species P.aquaticum, P.angustiporcatum, P.apimei, P.estinogenum, P.gracilentum and P.macroesclerotiorum with considerable genetic distance between them, revealed that FMR 17747 did not belong to any of the lineages of these known species and formed a basal and distant branch neighboring the fully-supported clade representative of P.estinogenum. Similarity values of the sediment isolate with respect to the ex-type strain of this latter species were 86.5% for tub2, 82.5% for cmdA, and 96.8% for ITS. Rpb2 sequences of P.estinogenum were not available for comparison. Individual analyses of the alternative gene markers including sequences of all reference strains available for the species in the section are shown in Suppl. material 1: Fig. S8. This undescribed monophyletic lineage is proposed as Penicilliumguarroi, which differed morphologically from its counterpart mainly by its smooth-walled globose conidia.
Figure 7.
Phylogenetic tree of PenicilliumsectionGracilenta based on ML analysis obtained by RAxML inferred from the combined tub2, cmdA, ITS, and rpb2 loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70%/0.95 are indicated on the nodes. Bold branches indicate bs/pp values 100/1. The tree is rooted to P.abidjanumCBS 246.67. The name in red is the new species described in this study. T = Ex-type strain.
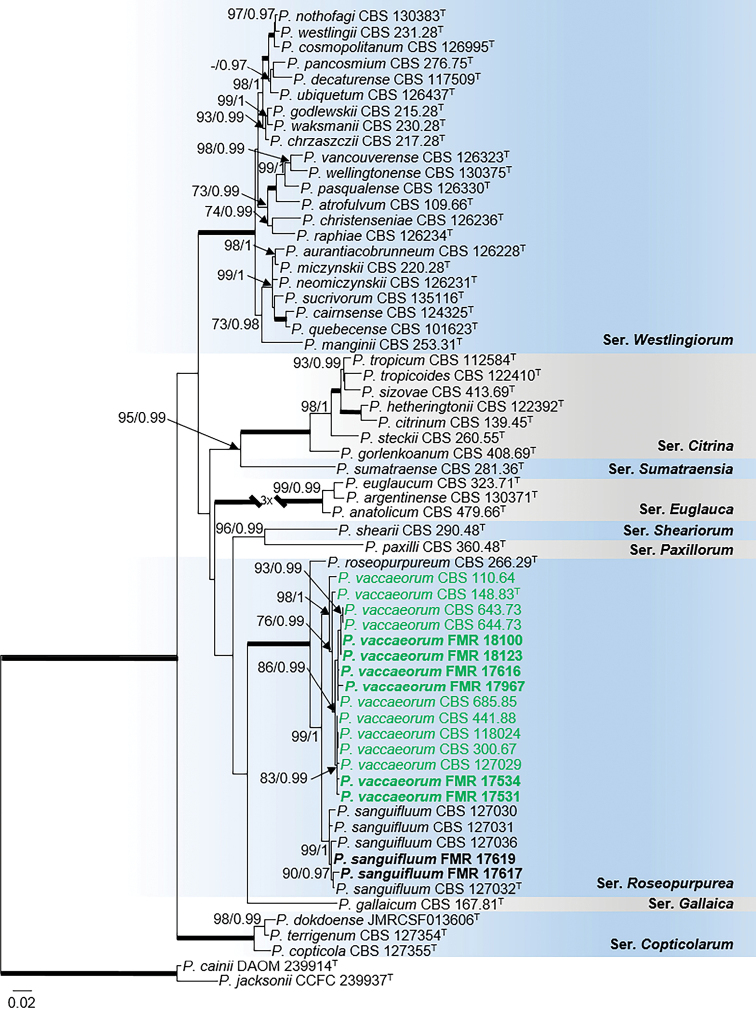
The concatenated alignment for sectionCitrina (Fig. 8) was carried out with tub2, cmdA and ITS loci, since sequences of the rpb2 marker were not available for many species in the section. This multi-locus analysis placed the sediment isolates (FMR 17531, FMR 17534, FMR 17616, FMR 17967, FMR 18100 and FMR 18123) in the series Roseopurpurea and grouped them in a well-supported clade (98 bs/1 pp) with the ex-type strains of P.vaccaeorum (CBS 148.83) and P.lacussarmientei (CBS 685.85), together with other strains (CBS 110.64, CBS 127029, CBS 441.88, CBS 300.67, CBS 118024, CBS 644.73, CBS 643.73) from soil and insects of different countries, and previously considered as P.sanguifluum by Houbraken et al. (2011). This terminal clade was sister to the also strongly supported clade with the ex-type strain of P.sanguifluum (CBS 127032) and other reference strains from different origins, including some collected in our survey from Spanish fluvial sediments (FMR 17617 and 17619). Both monophyletic lineages showed a genetic distance of 3.4%, which supports considering them distinct species. This result was corroborated with additional analyses of the alternative markers tub2 and cmdA, including more sequences of reference strains for the species in the series (Suppl. material 1: Fig. S9). In addition, features such as faster growth on YES agar, the ability to grow at 35 °C and longer stipes, allow us to distinguish the clade represented by P.vaccaeorum from that of P.sanguifluum. Since we accept P.vaccaeorum as a distinct species, a detailed description is provided below.
Figure 8.
Phylogenetic tree of PenicilliumsectionCitrina based on ML analysis obtained by RAxML inferred from the combined tub2, cmdA, and ITS loci. Branch lengths are proportional to phylogenetic distance. Bootstrap support values/Bayesian posterior probability scores above 70%/0.95 are indicated on the nodes. Bold branches indicate bs/pp values 100/1. The tree is rooted to P.cainii DAOM 239914 and P.jacksonii CCFC 239937. The name in green is the resurrected species P.vaccaeorum included in this study. T= Ex-type strain.
Taxonomy
. Penicillium ausonanum
Torres-Garcia, Gené and Dania García sp. nov.
840556
Figure 9.
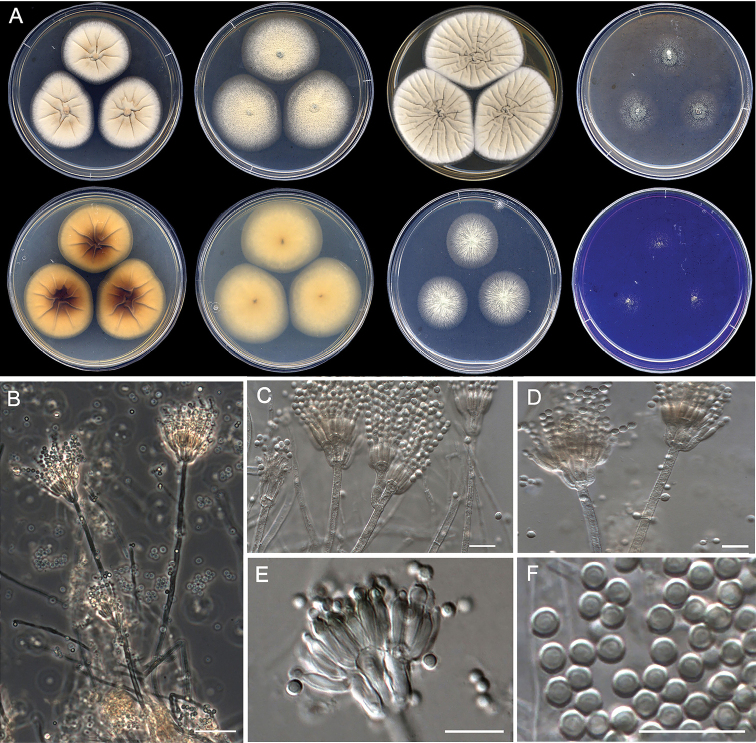
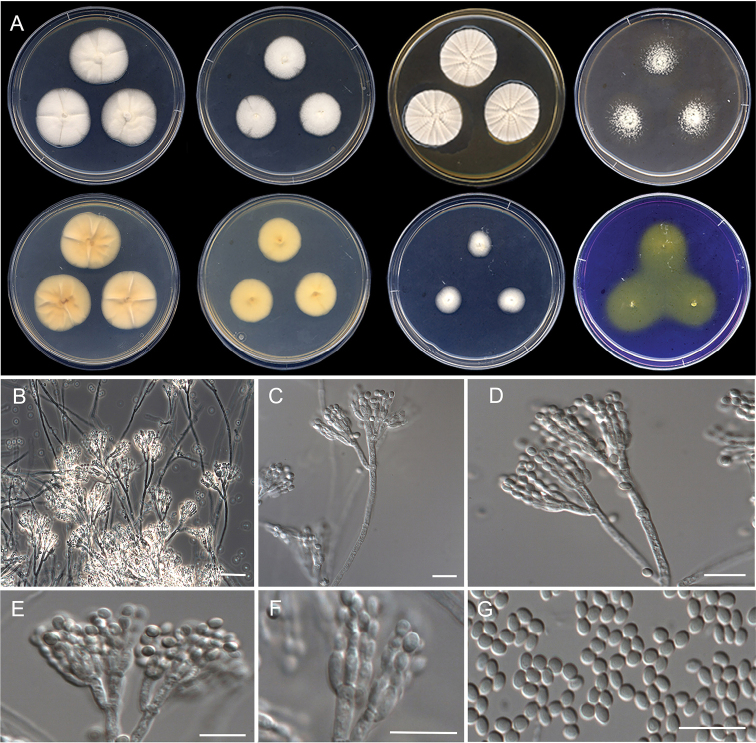
Morphological characters of Penicilliumausonanum sp. nov. (ex-type FMR 16948). A colonies from left to right (top row) CYA, MEA, YES, and OA; (bottom row) CYA reverse, MEA reverse, DG18, and CREAB colony on OA under stereoscopic magnifying glass C–H conidiophores I conidia. Scale Bars: 25 μm (C), 10 μm (D–I).
Etymology.
Referring to Ausona (Osona), the region of Catalonia where the fungus was collected.
Type.
Spain, Catalonia, Osona, Guilleries-Savassona Natural Park, Malafogassa, Major Stream, from sediments, Nov. 2018, E. Carvalho & J. Gené (holotypeCBS H-24781, cultures ex-type CBS 148237 = FMR 16948).
Subgeneric classification.
SubgenusAspergilloides, sectionLanata-Divaricata, series Dalearum.
Description.
Mycelium superficial and immersed, composed of septate, smooth-walled, hyaline hyphae, 2–3 μm wide. Conidiophores monoverticillate and divaricate, minor proportion biverticillate; stipes smooth-walled, 20–120 × 2–2.5 μm; metulae slightly appressed to divergent, mostly 2, occasionally 3 per stipe, 10–18 × 2–3 μm, occasionally a solitary phialide borne on the same level as metulae; phialides 2–5 per stipe/metula, ampulliform to cylindrical, 9–12 × 2–3 μm; conidia smooth-walled, globose to subglobose, 2–3 × 2–3 μm.
Culture characteristics (14 d at 25 °C).
Colonies on CYA, 58–59 mm diam., slightly radially and concentrically sulcate, velvety to floccose, whitish (5A1), margins regular and slightly fimbriate, sporulation absent to sparse, when present conidial masses brownish gray (6C2); reverse grayish yellow (4B4); little production of exudates hyaline, soluble pigment absent. On MEA, 61–62 mm diam., slightly raised, floccose, white (3A1), margins fimbriate, sporulation absent to sparse, conidial masses brownish gray (5E3); reverse light yellow (4A4); exudate absent, soluble pigment absent. On YES, 67–71 mm diam., slightly raised, radially sulcate, randomly furrowed as well, velvety to floccose, dull yellow (3B3) at center and white (3B1) towards periphery, margins slightly fimbriate, sporulation absent to sparse, conidial masses grayish to dull green (25C4–5C); reverse brownish yellow (5C8), exudates and soluble pigment absent. On OA, 63–65 mm diam., slightly raised, white (3A1) with gray (3E1) to olive (3F3) areas, velvety, margins slightly fimbriate, sporulation abundant, conidial masses grayish to dull green (25C5–D5); reverse grayish-yellow (3B5); exudates and soluble pigment absent. On DG18, 10–12 mm diam., randomly furrowed at the center, radially sulcate towards periphery, velvety, yellowish gray (2B2), margins entire, sporulation absent to sparse, conidial masses grayish to dull green (25C4–5C); reverse grayish yellow (2C3); exudates and soluble pigment absent. On CREA, 61–63 mm diam., slightly raised, floccose, gray (4B1) at center to yellowish gray (4B2) and white (4A1) towards periphery, margins slightly fimbriate, sporulation abundant, conidial masses brownish gray (6C2); reverse vivid yellow (3A8); exudates absent, acid production strong.
Colony diameter on CYA after 7d (mm).
5 °C 3–2, 15 °C 41–43, 20 °C 46–48, 30 °C 56–57, 35 °C 50–51, 37 °C 38–39, 40 °C no growth.
Distribution.
Spain.
Notes.
Penicilliumausonanum formed a phylogenetically supported group together with P.amphipolaria and P.viridissimum in series Dalearum (Fig. 6). These are two species recently described, the former from soil in Antarctica and Canada, and the latter from acidic and forest soil from China (Visagie et al. 2016a, Diao et al. 2019). The new species can be morphologically differentiated from them by its equal proportion of monoverticillate and divaricate conidiophores, which are mostly with a complex branching pattern in P.amphipolaria (biverticillate and divaricate) (Visagie et al. 2016a), and mono- to terverticillate in P.viridissimum (Diao et al. 2019). Both P.amphipolaria (6.5–10 µm) and P.viridissimum (6.5–10 µm) have slightly shorter phialides than P.ausonanum (9–12 µm). Also, P.amphipolaria (240–460 µm) and P.viridissimum (40–125 µm) have longer stipes (Visagie et al. 2016a, Diao et al. 2019) in comparison to those of P.ausonanum (20–120 µm). Furthermore, the three species also differed in acid production on CREA, which is strong in P.ausonanum, moderate in P.amphipolaria and absent in the neighboring species P.viridissimum.
. Penicillium guarroi
Torres-Garcia, Gené and Dania García sp. nov.
840567
Figure 10.
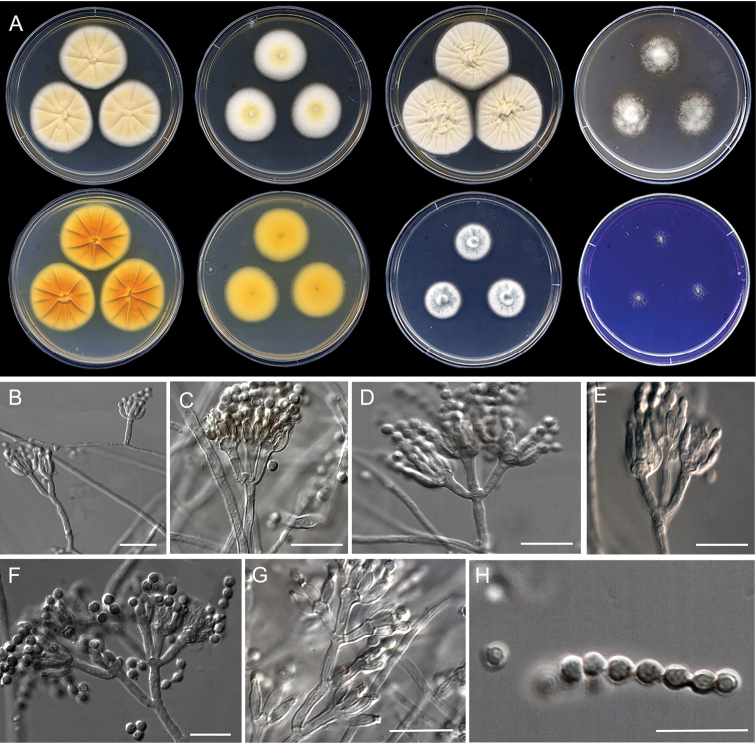
Morphological characters of Penicilliumguarroi sp. nov. (ex-type FMR 17747). A colonies from left to right (top row) CYA, MEA, YES, and OA; (bottom row) CYA reverse, MEA reverse, DG18, and CREAB–E conidiophores on MEAF conidia. Scale Bars: 25 μm (B), 10 μm (C–F).
Etymology.
Named in honor of Josep Guarro for his contributions to our knowledge of microfungi.
Type.
Spain, Catalonia, Alt Camp, Alcover, Brugent River, sediments, Mar. 2019, D. Torres & J. Gené (holotypeCBS H-24782, cultures ex-type CBS 148238 = FMR 17747).
Subgeneric classification.
Subgenus Aspergilloides, sectionGracilenta, series Estinogena.
Description.
Mycelium superficial and immersed, composed of septate, smooth-walled, hyaline hyphae, 2.5–3.5 μm wide. Conidiophores predominantly symmetrically biverticillate, occasionally with subterminal branches; stipes smooth- to rough-walled, 88–215 × 3–4 μm; metulae appressed, 2–4 per stipe, vesiculate, 5–10 × 2–4.5 μm (vesicle up to 5.5 μm wide); phialides 3–6 per metula, ampulliform, 6–9 × 1.5–3 μm; conidia smooth-walled, globose, 2–2.5 × 2–2.5 μm.
Culture characteristics (7 d at 25 °C).
Colonies on CYA, 38–40 mm diam., slightly raised at center, radially sulcate, velvety, brownish gray (6C2) and white (1A1) towards periphery, margins fimbriate, sporulation moderate, conidial masses greenish gray (28C2); reverse dark brown (6F6) and light brown (6D6) at periphery, becoming entirely brown after 14 d; soluble pigment absent. On MEA, 41–43 mm diam., slightly raised, granular, yellowish green (29B7) and white (1A1) towards periphery, margins slightly fimbriate, sporulation moderate, conidial masses grayish green (28D2); reverse yellowish brown (5E6) at center to grayish yellow at periphery; soluble pigment absent. On YES, 49–51 mm diam., raised at center, radially sulcate, velvety, brownish gray (5C2) and white (1A1) at periphery, margins entire, sporulation sparse, conidial masses greenish gray (28D2); reverse dark green (30F5) and yellowish brown (5D5) towards periphery; soluble pigment absent. On OA, 24–26 mm diam., elevated at center, velvety, white (1A1) at center and dull green (25E3) towards periphery, margins regular, sporulation moderate, conidial masses dull green (25D4); reverse brown (6E4) and yellowish gray (4B2) at periphery; soluble pigment absent. On DG18, 22–25 mm diam., flattened, granular, grayish green (30C3) at center, and dull green (29D49) towards periphery, margins fimbriate, sporulation moderate, conidial masses greenish gray (27D2); reverse grayish green (30E5) and white (1A1) at periphery, soluble pigment absent. On CREA, 22–25 mm diam., flattened, floccose, yellowish green (29B7) and white (1A1) at periphery, margins irregular, sporulation moderate, conidial masses grayish green (27B3–D3); reverse dark gray (1F1); soluble pigment absent, no acid production.
Colony diameter on CYA after 7d (mm).
5 °C no growth, 15 °C 17–19, 20 °C 26–28, 30 °C 34–36, 35 °C 4–5, 37 °C no growth, 40 °C no growth.
Distribution.
Spain.
Notes.
Penicilliumguarroi is the second species included in sectionGracilenta series Estinogena (Fig. 7). This species shows morphological attributes of the series based on its type P.estinogenum, namely that both have symmetrically appressed biverticillate conidiophores with rough-walled stipes (Houbraken et al. 2020). However, P.guarroi mainly differs from P.estinogenum by producing strictly smooth-walled globose conidia, which are ellipsoidal to ovate and with smooth to finely roughened walls in the latter (Abe 1956; Houbraken et al. 2020). In addition to their phylogenetic distance, other members of sectionGracilenta (i.e., series Gracilenta and Macrosclerotiorum) can be differentiated morphologically by the production of monoverticillate conidiophores and the lack of growth at 37 °C, with the exception of P.apimei and P.aquaticum, which are able to grow at this temperature (Barbosa et al. 2018; Houbraken et al. 2020). Penicilliumguarroi was unable to grow at 37 °C, but it shows a maximum temperature for growth at 35 °C (4–5 mm), like other members in the section (i.e., P.macrosclerotiorum, P.angustiporcatum, P.gracilentum and P.estinogenum).
. Penicillium heteromorphum
H.Z. Kong and Z.T. Qi. Mycosystema 1:107. 1988.
Figure 11.
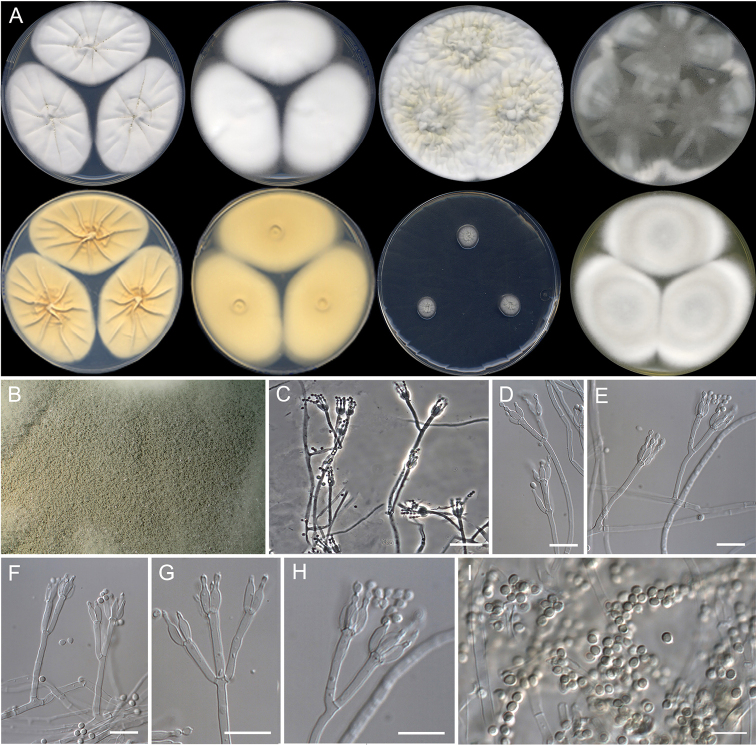
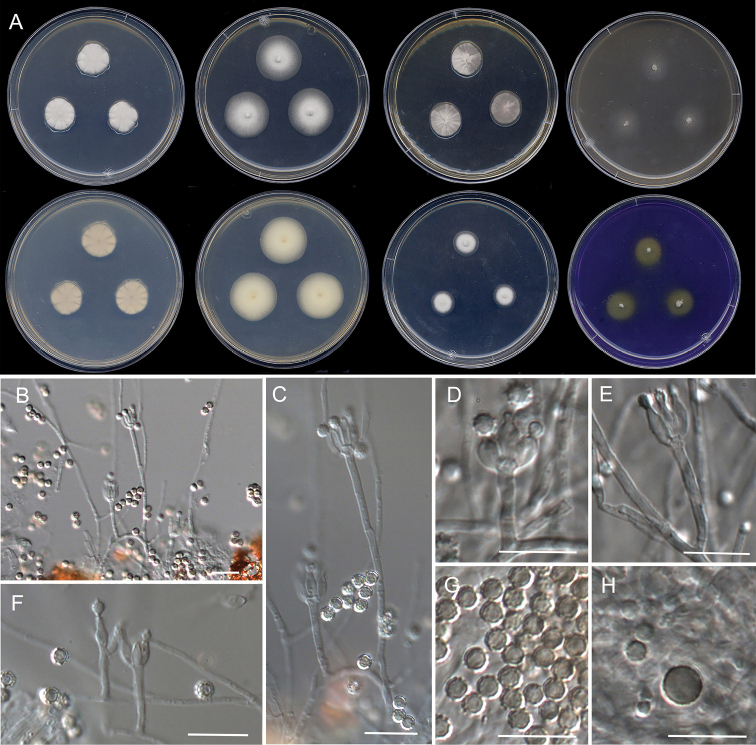
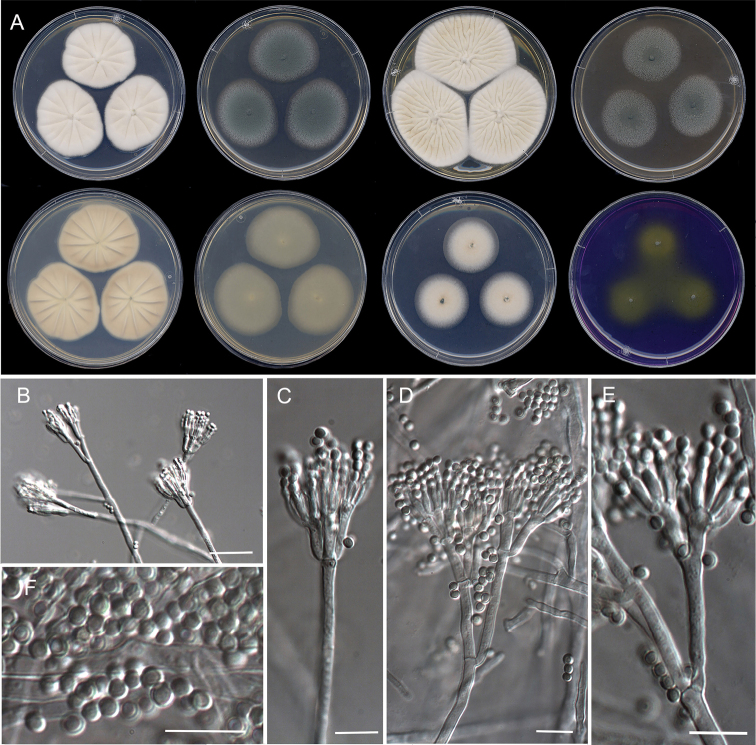
Morphological characters of Penicilliumheteromorphum (FMR 18043). A colonies from left to right (top row) CYA, MEA, YES, and OA; (bottom row) CYA reverse, MEA reverse, DG18, and CREAB, C, F conidiophores on OA. D, E conidiophores on Czapek’s Agar G–H conidia. Scale Bars: 25 μm (B), 10 μm (C–H).
Subgeneric classification.
Subgenus Aspergilloides, sectionExilicaulis. series Restricta.
Description.
Mycelium superficial and immersed, composed of septate, smooth-walled, hyaline hyphae, 1.5–3 μm wide. Conidiophores monoverticillate, occasionally irregularly branched; stipes smooth-walled, thin, 6–47.5 × 1.5–2 μm; phialides 2–3 per stipe, ampulliform, 3–7 × 1.5–2.5 μm; conidia roughened, globose to subglobose, 2.5–3 × 2.5–3 μm, occasionally conidia up to 5 μm were observed.
Culture characteristics (7 d at 25 °C).
Colonies on CYA, 19–20 mm diam., slightly raised at center, velvety, radially sulcate, yellowish gray (4B2) at center and white (1A1) at periphery, margins slightly undulate, sporulation absent; reverse pale yellow (4A3); soluble pigment absent. On MEA, 27–28 mm diam., slightly raised at center, velvety, white (1A1) at center, ash blond (3C3) towards periphery, margins entire, sporulation absent; reverse champagne colored (4A4) at center, pastel yellow (3A4) towards periphery; soluble pigment absent. On YES, 22–21 mm diam., slightly raised at center, velvety, radially sulcate, yellowish gray (4B2) and Sahara colored (6C5), margins entire, sporulation absent; reverse grayish orange (5B5) at center and champagne colored (4B4) towards periphery; soluble pigment absent. On OA, 23–24 mm diam., slightly elevated at center, floccose, greenish gray (28B2) at center and beige (4C3) towards periphery, margins fimbriate, sporulation abundant, conidial masses dull green (29E3); reverse beige (4C3); soluble pigment absent. On DG18, 14–15 mm diam., slightly raised at center, velvety, yellowish white (1A2) at center and white (1A1) towards periphery, margins regular, sporulation absent; reverse wine yellow (3B3) at center and yellowish white (3A2) towards periphery; soluble pigment absent. On CREA reaching 17–19 mm diam., slightly raised at center, floccose, white (1A1) at center and lemon yellow (3B8) towards periphery, margins fimbriate, sporulation absent; reverse lemon yellow (3B8); soluble pigment absent and acid production moderate. Colonies on Czapek’s agar reaching 13–14 mm diam., flattened, floccose, white (1A1) at center to ash gray (1B2) towards periphery, margins entire, sporulation abundant, conidial masses dull green (29D3); reverse ash gray (1B2); soluble pigment absent.
Colony diameter on CYA after 7d (mm).
5 °C no growth, 15 °C 9–11, 20 °C 12–13, 30 °C 23–24, 35 °C 16–19, 37 °C 4–7, 40 °C no growth.
Specimen examined.
Spain, Catalonia, Berguedà, Gósol, from stream sediments, Nov. 2019, J. Gené (CBS 148239, FMR 18043).
Distribution.
China and Spain.
Notes.
Penicilliumheteromorphum was first described from a soil sample collected in Shennongjia, China. FMR 18043 is thus the second isolate of this species. The protologue of P.heteromorphum, which was based on CYA and Czapek’s agar, noted that it does not grow at 37 °C, has strictly monoverticillate conidiophores with stipes up to 60 μm long, and produce conidia that are globose to subglobose, smooth or nearly, which show two well-differentiated measures on Czapek’s agar (ones of 2–2.5 (–3) μm diam, and the largest of 4–10 μm) (Kong and Qi 1988). By contrast, despite the high sequence similarity to the ex-type strain, our isolate differs phenotypically in its ability to grow at 37 °C, and in the production of shorter conidiophores and rough-walled conidia in all media studied; some larger conidia (up to 5 μm diam.) were only observed on Czapek’s agar. Nevertheless, features we observed in the sediment isolate P.heteromorphum match those of the species of series Restricta, which briefly consisted in growing restricted to moderately fast, producing generally short monoverticillate conidiophores with smooth stipes, globose to subglobose or (broadly) ellipsoidal, smooth or roughened conidia and they commonly grow at 37 °C (Houbraken et al. 2020). Based on the production of two types of conidia, Kong and Qi (1988) compared P.heteromorphum with P.dimorphosporum. However, although both species belongs in sectionExilicaulis, the current taxonomy of the genus places P.dimorphosporum in the genetically distant series Erubescentia (Visage et al. 2016b, Houbraken et al. 2020).
. Penicillium irregulare
Torres-Garcia, Gené and Dania García sp. nov.
840558
Figure 12.
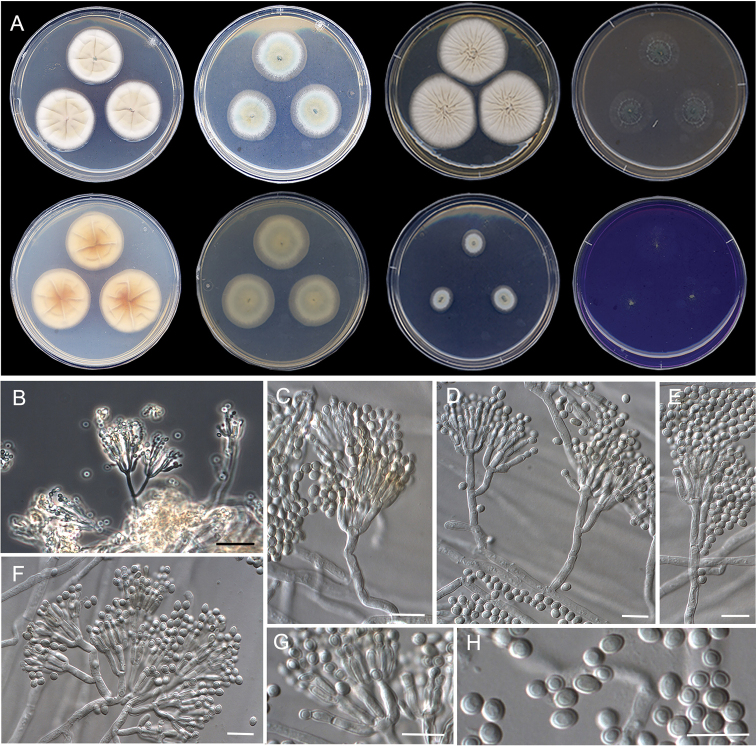
Morphological characters of Penicilliumirregulare sp. nov. (ex-type FMR 17859). A colonies from left to right (top row) CYA, MEA, YES, and OA; (bottom row) CYA reverse, MEA reverse, DG18, and CREAB–G conidiophores on MEAH conidia. Scale Bars: 10 μm (B–H).
Etymology.
Referring to the variable branching pattern of the conidiophores of the species.
Type.
Spain, Comunidad de Madrid, Miraflores de la Sierra, Miraflores River, from sediments, Jun. 2019, J.F. Cano (holotypeCBS H-24783, cultures ex-type CBS 148240 = FMR 17859).
Subgeneric classification.
Subgenus Penicillium, sectionCanescentia, series Canescentia.
Description.
Mycelium superficial and immersed, composed of septate, smooth-walled, hyaline hyphae, 2–3.5 μm wide. Conidiophores biverticillate, in minor proportion monoverticillate, terverticillate or divaricate; stipes smooth-walled, 13–152 × 1.5–2 μm; metulae divergent, 2–3 per stipe/branch, unequal in length, vesiculate, 7–10 × 1.5–2.5 μm (vesicle up to 4 μm wide), occasionally a solitary phialide borne on same level as metulae; phialides 5–8 per metula, ampulliform, 6–7.5 × 1.5–2.5 μm; conidia smooth- to finely rough-walled, globose to subglobose, somewhat ellipsoidal, 1.5–3 × 1.5–2 μm.
Culture characteristics (7 d at 25 °C).
Colonies on CYA, 36–38 mm diam, slightly elevated, radially sulcate, velvety, grayish orange (6B3) at center and white (1A1) towards periphery, margins entire, sporulation abundant, conidial masses grayish green (25B3); reverse brownish yellow (5C8) at center and vivid yellow towards periphery (3A8); exudate and soluble pigment absent. On MEA, 33–34 mm diam, elevated, floccose, light yellow (4A4) at center and white (1A1) towards periphery, margins fimbriate, sporulation abundant, conidial masses grayish green (25C3); reverse yellowish orange (4A7); exudate and soluble pigment absent. On YES, 43–44 mm diam, raised, concentrically sulcate and pale yellow (4A3) at center, radially sulcate and white (1A1) towards periphery, velvety, margins entire, sporulation absent to sparse, conidial masses grayish green (25D2); reverse brownish yellow (5C8) and white (1A1) at periphery; exudate and soluble pigment absent. On OA, 25–27 mm, slightly elevated at center, cottony and fasciculate, dull green (25E4) at center, gray (1D1) and white (1A1) towards periphery, margins entire, sporulation abundant, conidial masses dull green (30E3); reverse yellowish brown (5F4), golden brown (5D7) towards periphery; soluble pigment absent. On DG18, 14–15 mm, elevated, velvety, white (1A1) with grayish turquoise (24E4) areas, margins slightly fimbriate, sporulation abundant, conidial masses grayish turquoise (24B3–C5); reverse yellowish green (30B8) at center to pale green (30A3) and white (1A1) at periphery; soluble pigment absent. On CREA, 12–13 mm, flat, floccose, yellowish green (29B7), margins regular, sporulation sparse, conidial masses grayish green (27B3–C3); reverse dark gray (1F1); soluble pigment and acid production absent.
Colony diameter on CYA after 7d (mm).
5 °C no growth, 15 °C 19–20, 20 °C 25–27, 30 °C 31–33, 35 °C 11–12, 37 °C 5–10, 40 °C no growth.
Distribution.
Spain.
Notes.
Penicilliumirregulare is related to P.arizonense, P.yarmokense and P.canescens, all belonging to series Canescentia (Fig. 4). Species of this series are characterized by the production of biverticillate conidiophores, that occasionally produce additional branching stages (divaricate), with smooth- or rough-walled stipes (Houbraken et al. 2020; Visage et al. 2021). Penicilliumirregulare can be differentiated from its closest relative P.arizonense by the production of shorter stipes (13–152 μm vs 50–400 μm) and metulae (7–10 μm vs. 8–16 μm), and colony reverse yellowish to orange, in contrast to brown shades, even red brown to violet brown on YES agar in P.arizonense (Grijseels et al. 2016). In addition, P.irregulare was able to grow at 37°C on CYA, but restrictedly (5–10 mm diam. 7 d), while P.arizonense does not grow at this temperature.
. Penicillium sicoris
Torres-Garcia, Gené and Dania García sp. nov.
840559
Figure 13.
Morphological characters of Penicilliumsicoris sp. nov. (ex-type FMR 18076). A colonies from left to right (top row) CYA, MEA, YES, and OA; (bottom row) CYA reverse, MEA reverse, DG18, and CREAB–G conidiophores on MEAH conidia. Scale Bars: 25 μm (B), 10 μm (C–H).
Etymology.
Referring to the Segre River where the fungus was found.
Type.
Spain, Catalonia, La Noguera, Camarassa, Segre river, from sediments, Dec. 2019, D. Torres & J. Gené (holotypeCBS H-24784, cultures ex-type CBS 148241 = FMR 18076).
Subgeneric classification.
Subgenus Penicillium, sectionParadoxa, series Atramentosa.
Description.
Mycelium superficial and immersed, composed of septate, smooth-walled, hyaline hyphae, 3–5 μm wide. Conidiophores biverticillate or terverticilliate, occasionally irregularly branched with phialides growing directly from branches and divaricate; stipes smooth-walled, 25–215 × 3–4.5 μm; metulae divergent, 2–3 per branch, vesiculate, 7–20 × 2.5–4 μm (vesicle up to 5.5 μm wide); phialides 1–6 per metula, ampulliform, 4–7.5 × 2.5–4 μm; conidia smooth-walled, usually globose to subglobose, some broadly ellipsoidal, 2–4.5 × 2–3.5 μm.
Culture characteristics (7 d at 25 °C).
Colonies on CYA, 32–34 mm diam., raised at center, radially sulcate, velvety, brownish violet (11D8) at center, pale orange (5A3) and white (1A1) towards periphery, margins entire, sporulation abundant, conidial masses grayish green (28B3); reverse light orange (6A5) to orange (6B7) at center and grayish yellow (4B4) towards periphery; soluble pigment absent. On MEA, 28–30 mm diam., flat, velvety, grayish green (30D6) at center, bluish green (25C8), and white (1A1) at periphery, margins entire, sporulation abundant, conidial masses grayish turquoise (24C3–C4); reverse pea green (29D5), yellowish white (4A2); soluble pigment absent. On YES, 39–43 mm diam., raised at center, radially sulcate, velvety, orange gray (5B2) at center and white (1A1) towards periphery, margins entire, sporulation sparse, conidial masses grayish green (28C3); reverse grayish yellow (4B4) and pale yellow (4A3) at periphery; soluble pigment absent. On OA, 23–24 mm diam., slightly elevated at center, floccose, grayish green (26E6), opaline green (25C6) and brownish gray (5C2) towards periphery, margins slightly fimbriate, sporulation abundant, conidial masses dull green (27D3); reverse pea green (29D5) at center and brownish gray (5C2) towards periphery; soluble pigment absent. On DG18, 13–16 mm diam., slightly raised at center, velvety, olive (3D3) at center, grayish turquoise (24B3) and white (1A1) towards periphery, margins entire, sporulation abundant, conidial masses grayish turquoise (24B3); reverse, grayish green (1C4) and white (1A1) at periphery; soluble pigment absent. On CREA, 21–27 mm diam., slightly elevated at center, velutinous, apple green (29C7), margins regular, sporulation abundant, conidial masses grayish green (26B3–C3); reverse colorless; soluble pigment absent, acid production absent.
Colony diameter on CYA after 7d (mm).
5 °C 3–4, 15 °C 25–26, 20 °C 30–31, 30 °C 29–31, 35 °C no growth, 37 °C no growth, 40 °C no growth.
Distribution.
Spain.
Notes.
Penicilliumsicoris is closely related to P.mexicanum in series Atramentosa (Fig. 3). Phenotypically, species of this series share a moderately fast colony growth and brown reverse on CYA and YES, and good growth on CREA without acid production (Houbraken et al. 2020). However, our species differs in having an orange to grayish yellow reverse on CYA. In addition, P.sicoris also differs from its counterpart in several micromorphological features: i.e., its conidiophores are bi- or terverticillate, whereas in P.mexicanum they are ter- or quaterverticillate, stipes are shorter (25–215 vs. 65–370 μm), phialides shorter (4–7.5 vs. 7–10 μm) and metulae longer (7–20 vs. 8.5–15.5 μm) than those of P.mexicanum, and its conidia are predominantly globose to subglobose, whereas in P.mexicanum they are broadly ellipsoidal to ellipsoidal (Visagie et al. 2014c). Moreover, P.mexicanum has a more restrictive growth on CREA than P.sicoris (5–8 vs. 21–27 mm diam. after 7 d).
. Penicillium submersum
Torres-Garcia, Gené and Dania García sp. nov.
840560
Figure 14.
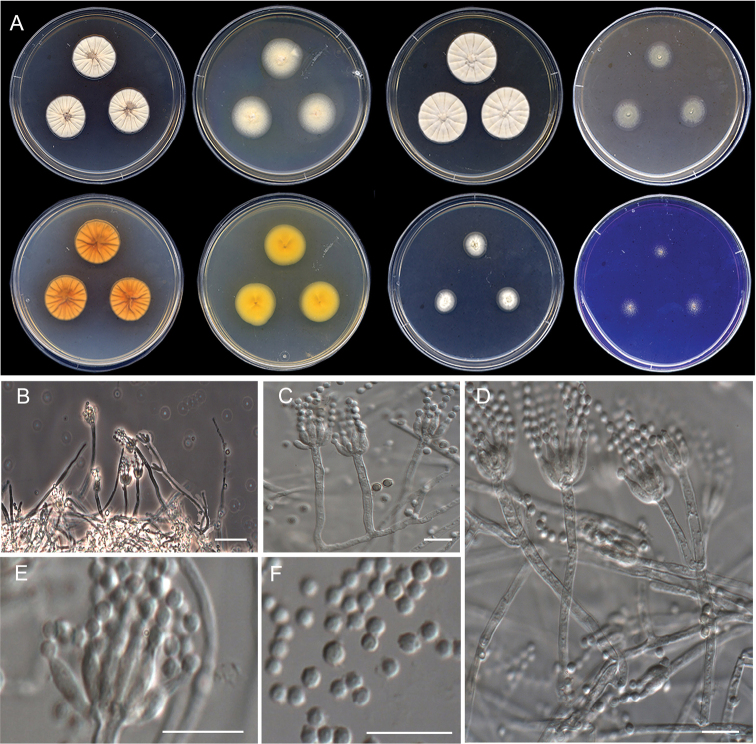
Morphological characters of Penicilliumsubmersum sp. nov. (ex-type FMR 17140). A colonies from left to right (top row) CYA, MEA, YES, and OA; (bottom row) CYA reverse, MEA reverse, DG18, and CREAB–F conidiophores on MEAG conidia. Scale Bars: 25 μm (B), 10 μm (C–G).
Etymology.
Referring to the submerged sediment sample where the fungus was isolated.
Type.
Spain, Catalonia, Montsant Natural Park, Siurana’s Swamp, from sediments, Feb. 2018, E. Carvalho & J. Gené (holotypeCBS H-24785, cultures ex-type CBS 148242 = FMR 17140).
Subgeneric classification.
Subgenus Penicillium, sectionRobsamsonia, series Urticicola.
Description.
Mycelium superficial and immersed composed of septate, smooth-walled, hyaline hyphae, 2–2.5 μm wide. Conidiophores mostly terverticillate, in minor proportion biverticillate and quarterverticillate; stipes smooth-walled, 29–142 × 1.5–2.5 μm; metulae divergent, mostly 2, occasionally 3 per stipe/branch, 5.5–7.5 × 1.5–4 μm; phialides 2–5 per metula, ampulliform, 4–5.5 × 1.5–2.5 μm; conidia smooth-walled, ellipsoidal, 3–3.5 × 2–2.5 μm.
Culture characteristics (7 d at 25 °C).
Colonies on CYA, 34–37 mm diam., elevated, with some radially furrow, floccose, light yellow (4A5) and yellowish white (4A2) towards periphery, margins entire, sporulation sparse, conidial masses grayish green (28C4); reverse golden brown (5D7) and orange (5A6) at periphery; soluble pigment absent. On MEA, 28–29 mm diam., slightly elevated, floccose, white (1A1) to light yellow (4A5) at periphery, margins entire, sporulation sparse, conidial masses grayish green (27C3); reverse light yellow (4A5); soluble pigment absent. On YES, 33–36 mm diam., slightly elevated at center, radially sulcate, velvety, light brown (6D4) and white (1A1) towards periphery, margins slightly lobate, sporulation sparse, conidial masses grayish green (28C3); reverse grayish orange (5B5); soluble pigment absent. On OA, 18–20 mm diam., elevated at center, fasciculate, yellowish white (4A2) and pale gray towards periphery, margins low and entire, sporulation abundant, conidial masses grayish green (28B3); reverse grayish yellow (4C5); soluble pigment absent. On DG18, 11–13 mm diam., elevated, floccose, white (1A1) at center, pale yellow (4A3) and grayish yellow (4C3) towards periphery, margins entire, sporulation abundant, conidial masses grayish green (27C3); reverse light yellow (4A5) and yellowish white (2A2) at periphery; soluble pigment absent. On CREA, 15–19 mm diam., flattened, floccose, white (1A1) and pale yellow (3A3), margins low and irregular, sporulation sparse, conidial masses grayish green (28B3–C3); reverse white (1A1) and pale yellow (3A3); soluble pigment absent, acid production strong.
Colony diameter on CYA after 7d (mm).
5 °C no growth, 15 °C 20–21, 20 °C 25–26, 30 °C 28–30, 35 °C 17–16, 37 °C 9–11, 40 °C no growth.
Distribution.
Spain.
Notes.
Species in sectionRobsamsonia were characterized by restricted to moderately fast growth rate on CYA at 25 °C (15–32 mm diam in 7 d) and lack or slow of growth on CYA at 30 °C (up to 19 mm diam) (Houbraken et al. 2016; Houbraken et al. 2020). However, the novel species showed faster growth rates on CYA at both temperatures (i.e., 34–37 mm and 28–30 mm diam., respectively). Penicilliumsubmersum shares morphological features with the other two species (P.griseofulvum and P.dipodomycola) of the series Urticicola where it is classified (Fig. 2), which consisted in having bi-, ter, or quarterverticillate, divergent, smooth-walled conidiophores and short phialides (up to 7 µm) (Houbraken et al. 2020). However, P.submersum shows the shortest phialides within the group (4–5.5 vs. 5–7 µm). In addition, our species has strong acid production on CREA, in contrast to the lack of acid production of P.griseofulvum and P.dipodomycola in the same medium (Houbraken et al. 2016, 2020); and colony reverse on CYA and YES in P.submersum is golden brown to orange and grayish orange, respectively, while in P.griseofulvum and P.dipodomycola it is beige brown to dark brown in both culture media (Houbraken et al. 2020). Furthermore, P.griseofulvum differs from P.submersum in its gray colony color, especially on CYA, which is in shades of yellow in our species.
. Penicillium tardochrysogenum
Frisvad, Houbraken & Samson . Persoonia 29: 93. 2012.
Figure 15.
Morphological characters of Penicilliumtardochrysogenum (FMR 17137). A colonies from left to right (top row) CYA, MEA, YES, and OA; (bottom row) CYA reverse, MEA reverse, DG18, and CREAB–E conidiophores on MEAF conidia. Scale Bars: 25 μm (B), 10 μm (C–F).
Subgeneric classification.
Subgenus Penicillium, sectionChrysogena, series Chrysogena.
Description.
Mycelium superficial and immersed composed of septate, smooth-walled, hyaline hyphae, 2.5–5.5 μm wide. Conidiophores biverticillate, terverticillate or quaterverticillate; stipes smooth-walled, 40–200 × 2.5–4 μm; metulae appressed to slightly divergent, 2–4 per branch or stipe, vesiculate, 6–12.5 × 2–4 μm (vesicle up to 4.5 μm wide); phialides 3–6 per metulae, ampulliform, 5.5–7.5 × 1.5–2.5 μm; conidia smooth-walled, globose to subglobose, 2.5–3 × 2.5–3 μm.
Culture characteristics (7 d at 25 °C).
Colonies on CYA reaching 41–43 mm diam., slightly raised at center, floccose, radially sulcate, yellowish white (1A2) at center to orange white (6A2) and white (1A1) towards periphery, margins slightly lobate, sporulation sparse, conidial masses grayish green (28B3); reverse champagne (4B4) at center to red-haired (6C4) towards periphery; soluble pigment absent. On MEA, 38–39 mm diam., flattened, velvety, grayish green (26D2-E4), white (1A1) towards periphery, margins low and slightly fimbriate, sporulation abundant, conidial masses dull green (25D4); reverse colorless; soluble pigment absent. On YES, 56–57 mm diam., slightly raised at center, floccose, radially sulcate, yellowish white (4A2) at center to champagne (4B4) towards periphery, margins entire, sporulation sparse, conidial masses grayish green (27B3); reverse amber yellow (4B6) at center to maize (yellow) (4A5) towards periphery; soluble pigment absent. On OA, 35–37 mm diam., flattened, velvety, dark green (28F8) at center to greenish gray (28D4) and white (1A1) towards periphery, margins low and entire, sporulation abundant, conidial masses dull green (25D3); reverse honey yellow (4D6) at center and sand yellow (4B3) towards periphery; soluble pigment absent. On DG18, 30–32 mm diam., flattened, floccose, dark green (27F8) at center to pale orange (5A3) and white (1A1) towards periphery, margins low and entire, sporulation abundant, conidial masses grayish green (28C3) at the center; reverse wax yellow (3B5) at center to white (1A1) towards periphery; soluble pigment absent. On CREA, 24–25 mm diam., flattened, floccose, jade green (27E5) at center to yellowish green (30B8) towards periphery, margins slightly fimbriate, sporulation sparse, conidial masses grayish green (27C3–C4); reverse yellowish white (30B8), soluble pigment absent and acid production moderately strong.
Colony diameter on CYA after 7d (mm).
5 °C 3–4, 15 °C 23–24, 20 °C 27–28, 30 °C 31–33, 35 °C 16–19, 37 °C 8–9, 40 °C no growth.
Specimen examined.
Spain, Catalonia, Montsant Natural Park, Siurana’s Swamp, from sediments, Feb 2018, E. Carvalho & J. Gené (FMR 17137).
Distribution.
Antarctica and Spain.
Notes.
Although P.tardochrysogenum was introduced based only on the type specimen collected in the Antarctica, the species was later described as endemic of that continent since it was isolated at high densities on rocks from several Islands and Continental Antarctica (Houbraken et al. 2012; Alves et al. 2019). The Spanish isolate from freshwater sediments represents the first report of this species in temperate regions. Of note, however, is that recently the species has also been reported from historical manuscripts preserved in Iraq (Jasim et al. 2019), but only the ITS barcode was used for confirming the identity of isolates, a well-known gene marker unable to distinguish between closely related penicillia (Houbraken et al. 2012; Visagie et al 2014a). Penicilliumtardochrysogenum belongs to series Chrysogena and is closely related to P.allii-sativi, P.chrysogenum, P.rubens and P.vanluykii (Fig. 1), but it was distinguished from these species and other members of the series by more restricted and floccose colonies on MEA, the lack of sporulation on YES and the production of finely roughened conidia (Houbraken et al. 2012). Despite the high sequence similarity of the markers analyzed with the ex-type strain of P.tardochrysogenum, our isolate showed some phenotypic variation regarding the protologue; i.e., faster growth rate after 7 d on MEA (38–39 vs. 18–24 mm), sporulation (sparse) on YES, smooth-walled conidia, and shorter stipes (40–200 × 2.5–4 vs. 150–400 × 2–3 µm) and metulae (6–12.5 × 2–4 vs. 10–13(–18) × 2.5–3.5 µm). These differences suggest that more specimens should be examined for a more accurate morphological characterization of this fungus.
. Penicillium vaccaeorum
Quintanilla, Mycopathol. 80: 74. 1982.
109999
Figure 16.
Morphological characters of Penicilliumvaccaeorum (FMR 17967). A colonies from left to right (top row) CYA, MEA, YES, and OA; (bottom row) CYA reverse, MEA reverse, DG18, and CREAB–E conidiophores on MEAF conidia. Scale Bars: 25 μm (B), 10 μm (C–F).
=Penicilliumlacussarmientei Ramírez, Mycopathol. 96: 29. 1986.
Type.
Spain, Valladolid, San Miguel del Arroyo, from sandy soil under pine tree; J.A. Quintanilla (holotypeCBS H-148.83, cultures ex-type CBS 148.83, DTO 9E2, CECT 2753).
Subgeneric classification.
Subgenus Aspergilloides, sectionCitrina, series Roseopurpurea
Description.
Mycelium superficial and immersed composed of septate, smooth-walled, hyaline hyphae of 1.5–2.5 μm wide. Conidiophores monoverticillate, rarely biverticillate and divaricate; stipes smooth-walled, vesiculate, 22.5–103 × 1.5–2.5 μm (vesicle up to 4.5 µm); metulae divergent 2–3, unequal in length, 7–37 × 1.5–3 µm; phialides 2–5 per stipe/metula, ampulliform, 6–8.5 × 2–2.5 μm; conidia smooth- or finely roughened, globose, 2–2.5 × 2–2.5 μm.
Culture characteristics (7 d at 25 °C).
Colonies on CYA, 20–22 mm diam., slightly raised, velvety, radially sulcate, dull red (8C3) at center to light yellow (4A5) and white (1A1) towards periphery, margins slightly undulate, sporulation sparse, conidial masses grayish green (28B3); reverse brownish orange (5C6); with reddish soluble pigment. On MEA, 24–27 mm diam., slightly elevated, velvety, light yellow (4A5) and pale orange (5A2) at periphery, margins low and entire, sporulation sparse, conidial masses grayish green (27C4); reverse golden yellow (5B7) and reddish-yellow (4A6) at periphery; soluble pigment absent. On YES, 30–32 mm diam., slightly raised at center, velvety, radially sulcate, pale yellow (3A3) to white (1A1) and brownish orange (5C3) towards periphery, margins slightly undulate, sporulation abundant, conidial masses grayish green (28B3); reverse brownish yellow (5C8); soluble pigment absent. On OA, 18–20 mm diam., flattened, velvety, dark green (28F4) to light gray (25D1) and white (1A1) towards periphery, margins low and entire, sporulation abundant, conidial masses dull green (25D3); reverse brown (6E4) and yellowish gray (4B2) at periphery; soluble pigment absent. On DG18, 12–13 mm, slightly raised at center, velvety, radially sulcate, white (1A1) and yellowish white (1A2) towards periphery, margins regular, sporulation sparse, conidial masses grayish green (27C3); reverse light yellow (4A5) and white (1A1) at periphery; soluble pigment absent. On CREA, 9–11 mm diam., flattened, floccose, yellowish green (29B7) and white (1A1) towards periphery, margins entire, sporulation sparse, conidial masses grayish green (28B3); reverse dark gray (1F1); soluble pigment and production of acid absent.
Colony diameter on CYA after 7d (mm).
On CYA: 5 °C no growth, 15 °C 13–14, 20 °C 16–17, 30 °C 18–20, 35 °C 6–11, 37 °C no growth, 40 °C no growth.
Specimens examined.
Spain, Catalonia, Fogars de Montclús, La Costa de l’Infern, from stream sediments, Oct 2018, D. Torres (FMR 17531); Fogars de Montclús, La Costa de l’Infern, from stream sediments, Oct 2018, D. Torres (FMR 17534); Aitona, Segre River, from sediments, Dec 2020, D. Torres & J. Gené (FMR 18100); La Granja d’Escarp, Segre River, from sediments, Dec 2020, D. Torres & J. Gené (FMR 18123); Balearic Islands, Mallorca, Serra de Tramontana, from stream sediments, Dec 2018, J. F. Cano (FMR 17616); Basque Country, from stream sediments, Aug 2019, J. Gené (FMR 17967).
Distribution.
Argentina, Canada, Chile, Spain, The Netherlands and Turkey.
Notes.
Penicilliumvaccaeorum and P.lacussarmientei, two species described from sandy soils in Spain and Chile (Quintanilla 1982; Ramírez 1986), respectively, were considered synonyms of P.roseopurpureum by Frisvad et al. (1990), noting that both species were fast growing variants of P.roseopurpureum. Later on, based on that criterion and the lack of morphological differences, Houbraken et al. (2011) considered the two former species synonyms of P.sanguifluum despite some sequence variation where P.vaccaeorum and P.lacussarmientei clustered together in a clade sister to that of P.sanguifluum. Our phylogeny correlates with Houbraken et al. (2011) who found the same topology. Having the opportunity to examine specimens from both monophyletic sister clades (Fig. 8), we observed consistent phenotypic features to distinguish them. For instance, isolates of P.vaccaeorum had longer stipes (up to 103 μm; up to 120 μm in the protologue of the species) (Quintanilla 1982), they were able to grow on CYA at 35 °C (6–11 mm diam. after 7 d), had good sporulation and faster growth on YES agar (30–32 mm diam. 7 d) and more restricted on DG18 (12–13 mm diam. 7 d). In contrast, isolates of the P.sanguifluum clade showed considerably shorter conidiophores (15–50 µm long), they were unable to grow above 30 °C, and the colonies on YES and DG18 showed sparsely or absent sporulation and attained 18–28 mm and 16–22 mm diam., respectively (Houbraken et al. 2011). Hence, genetic and phenotypic differences support the reinstatement of P.vaccaeorum as an accepted species, with P.lacussarmientei considered synonym. This species together with P.sanguifluum and P.roseopurpureum are classified in series Roseopurpurea, which differs from almost all series of the sectionCitrina by species’ monoverticillate conidiophores. The only other series in the section with monoverticillate conidiophores is Gallaica, represented exclusively by P.gallaicum, which differs from the former series mainly by the production of sclerotia (Houbraken et al. 2020).
According to the revised data, P.vaccaeorum occurs worldwide, and is commonly isolated from sandy soils of beaches and forests, and even associated with ants (Table 1).
Discussion
Due to nomenclatural revisions of Penicillium, and efforts to release extensive reference sequences for both ex-types and other reference strains (Houbraken and Samson 2011; Visagie et al. 2014a; Houbraken et al. 2020), the number of Penicillium species newly described in the last decade has significantly increased, particularly with the examination of fungi recovered from poorly studied substrates or unexplored areas (sensu Hawksworth and Lücking 2017; Wijayawardene et al. 2020). Although Penicillium species are commonly isolated from aquatic environments (Niewolak 1975; Tóthová 1999; Amaral-Zettler et al. 2002), studies exploring their diversity are scarce. For instance, Heo et al. (2019) recovered P.brasilianum, P.crustosum, P.expansum, P.oxalicum and P.piscarium from freshwater environments in Korea. Only recently, Penicillium diversity has begun to be investigated in marine sediments (Gonçalves et al. 2013; Kirichuk et al. 2017; Ogaki et al. 2020a), and some novel species have even been described from this substrate, such as P.piltunense, P.ochotense and P.attenuatum (Kirichuk et al. 2017), although they are currently considered conspecific with P.antarcticum (Visagie et al. 2021).
Several studies reveal that the fungal biomass and, subsequently, ascomycetes inhabiting freshwater sediments are fundamental to the decomposition of deposited organically bound C and N, using carbohydrates, phenolic compounds and carboxylic acids as carbon sources (Huang et al. 2010; Zhang et al. 2013; Zhang et al. 2014). The penicillia might also contribute greatly to this activity, although further studies are needed to define the spectrum of species and their role in this particular substrate.
In our contribution to the diversity of Penicillium from freshwater sediments, we identified several interesting species, such as P.heteromorphum and P.tardochrysogenum only so far known from China and Antarctica, respectively (Kong and Qi 1988; Alves et al. 2019). Our finding reveals a wider distribution of these taxa, being first reports in freshwater sediment samples, as in the case of the resurrected species P.vaccaeorum and its counterpart P.sanguifluum. These two latter species (classified in sectionCitrina) seem to be common penicillia inhabiting soil (Houbraken et al. 2011). Furthermore, we recovered several isolates representative of new species described above as P.ausonanum, P.guarroi, P.irregulare, P.sicoris and P.submersum. Of note is that of the fifteen Penicillium isolates investigated (Table 1), twelve were obtained from PDA supplemented with 0.2% cycloheximide, a culture technique previously used to recover keratinophylic or extremotolerant fungi such as black yeasts from various substrates (Ulfig et al. 1997; Salgado et al. 2004; Madrid et al. 2016). Isolates obtained using this culture medium, and therefore resistant to a relatively high concentration of cycloheximide, corresponded to P.ausonanum, P.irregulare, P.submersum, P.tardochrysogenum, as well as all those identified as P.vaccaeorum and P.sanguifluum. Cycloheximide tolerance in Penicillium has been previously studied by Seifert and Giuseppin (2000), even recently two novel species, P.krskae and P.silybi, have been described as resistant to cycloheximide (Labuda et al. 2021). Other fungi with the ability to grow at high doses of that antimicrobial agent can cause human infections, such as numerous dermatophytes, black yeasts, or members in the Ophiostomatales (de Hoog et al. 2020). However, the meaning of the cycloheximide resistance in penicillia would deserve further studies, for instance at genomic level. On the other hand, the isolates of P.guarroi, P.heteromorphum and P.sicoris were obtained from DRBC. It is well-known that this culture medium restricts fast-growing moulds due mainly to the Rose Bengal effect, allowing them to recover fungi with slower growth rates, a feature shown by the mentioned species in comparison with other Penicillium species that have been previously recovered from aquatic environments (Gonçalves et al. 2019; Heo et al. 2019). The efficacy of the DRBC for the detection of filamentous fungi and yeasts in aquatic environments was proven in a study investigating the yield across different media, in which it was found to be the best in quantity and diversity of fungi detection in different water samples tested (Pereira et al. 2010). According to our experience, it is important to use different media in order to investigate and recover the greatest diversity of species as possible from environmental samples.
Most of the new species described here were mainly recovered from samples collected in rivers and streams of the north-western Mediterranean region, an area recognized as one of the most species-rich regions in Southern Europe (Máiz-Tomé et al. 2017; Rundel et al. 2018). This fact was particularly highlighted in the study of Guevara-Suárez et al. 2020 on species diversity of coprophilous penicillium-like fungi, from which the highest number of species identified and novel taxa described were recovered from samples collected in Mediterranean localities. Therefore, regarding fluvial sediments as an accumulative substrate in biodiversity of the environment, it is not surprising that freshwater sediments from the Mediterranean region comprise a great reservoir of novel fungal lineages in general.
Supplementary Material
Acknowledgements
This study was supported by the Spanish Ministerio de Economía y Competitividad, grant CGL2017–88094-P.
Citation
Torres-Garcia D, Gené J, García D (2022) New and interesting species of Penicillium (Eurotiomycetes, Aspergillaceae) in freshwater sediments from Spain. MycoKeys 86: 103–145. https://doi.org/10.3897/mycokeys.86.73861
Supplementary materials
Figures S1–S9
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
D Torres-Garcia, J Gené, D García
Data type
phylogenetic trees
Explanation note
Additional phylogenetic trees of the Penicillium series corresponding to the individual analyses of the different molecular markers with additional reference strains representing the species closely related to penicillia described here.
References
- Abdelwahab MF, Fouad MA, Kamel MS, Özkaya FC, Kalscheuer R, Müller WEG, Lin W, Liu Z, Ebrahim W, Daletos G, Proksch P. (2018) Tanzawaic acid derivatives from freshwater sediment-derived fungus Penicillium sp. Fitoterapia 128: 258–264. 10.1016/j.fitote.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Abe S. (1956) Studies on the classification of the Penicillia. The Journal of General and Applied Microbiology 2: 1–194. 10.2323/jgam.2.1 [DOI] [Google Scholar]
- Aime MC, Miller AN, Aoki T, Bensch K, Cai L, Crous PW, Hawksworth DL, Hyde KD, Kirk PM, Lücking R, May TW, Malosso E, Redhead SA, Rossman AY, Stadler M, Thines M, Yurkov AM, Zhang N, Schoch CL. (2021) How to publish a new fungal species, or name, version 3.0. IMA Fungus, 12: e11. 10.1186/s43008-021-00063-1 [DOI] [PMC free article] [PubMed]
- Alves IM, Gonçalves VN, Oliveira FS, Schaefer CE, Rosa CA, Rosa LH. (2019) The diversity, distribution, and pathogenic potential of cultivable fungi present in rocks from the South Shetlands archipelago, Maritime Antarctica. Extremophiles 23: 327–336. 10.1007/s00792-019-01086-8 [DOI] [PubMed] [Google Scholar]
- Amaral-Zettler LA, Gómez F, Zettler E, Keenan BG, Amils R, Sogin ML. (2002) Microbiology: eukaryotic diversity in Spain’s River of Fire. Nature 417: 137–137. 10.1038/417137a [DOI] [PubMed] [Google Scholar]
- Barbosa RN, Bezerra JD, Souza-Motta CM, Frisvad JC, Samson RA, Oliveira NT, Houbraken J. (2018) New Penicillium and Talaromyces species from honey, pollen and nests of stingless bees. Antonie van Leeuwenhoek 111: 1883–1912. 10.1007/s10482-018-1081-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera BK, Chakraborty HJ, Patra B, Rout AK, Dehury B, Das BK, Sarkar DJ, Parida PK, Raman RK, Rao AR, Rai A, Mohapatra T. (2020) Metagenomic analysis reveals bacterial and fungal diversity and their bioremediation potential from sediments of river Ganga and Yamuna in India. Frontiers in Microbiology 11: e556136. 10.3389/fmicb.2020.556136 [DOI] [PMC free article] [PubMed]
- Cecchi G, Vagge G, Cutroneo L, Greco G, Piazza SD, Faga M, Zotti M, Capello M. (2019) fungi as potential tool for polluted port sediment remediation. Environmental Science and Pollution Research 26: 35602–35609. 10.1007/s11356-019-04844-5 [DOI] [PubMed] [Google Scholar]
- Chen AJ, Sun B D, Houbraken J, Frisvad JC, Yilmaz N, Zhou YG. (2016) New Talaromyces species from indoor environments in China. Studies in Mycology 84: 119–144. 10.1016/j.simyco.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoog GS, Guarro J, Gené J, Ahmed S, Al-Hatmi AMS, Figueras MJ, Vitale RG. (2020) Atlas of Clinical fungi, 4th edn. Foundation Atlas of Clinical fungi: Hilversum, The Netherlands, 1599 pp. [Google Scholar]
- Diao YZ, Chen Q, Jiang XZ, Houbraken J, Barbosa RN, Cai L, Wu WP. (2019) PenicilliumsectionLanata-Divaricata from acidic soil. Cladistics 35: 514–549. 10.1111/cla.12365 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS, Källersjö M, Kluge AG, Bult C. (1994) Testing significance of incongruence. Cladistics 10: 315–319. 10.1111/j.1096-0031.1994.tb00181.x [DOI] [Google Scholar]
- Frisvad JC. (1981) Physiological criteria and mycotoxin production as aids in identification of common asymmetric Penicillia. Applied and Environmental Microbiology 41: 568–579. 10.1128/aem.41.3.568579.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad JC, Samson RA, Stolk AC. (1990) Disposition of recently described species of Penicillium. Persoonia 14: 209–232. [Google Scholar]
- Frisvad JC, Samson RA. (2004) Polyphasic taxonomy of PenicilliumsubgenusPenicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Studies in Mycology 49: 1–174. [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. 10.1128/aem.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves MFM, Santos L, Silva BMV, Abreu AC, Vicente TF, Esteves AC, Alves A. (2019) Biodiversity of Penicillium species from marine environments in Portugal and description of Penicilliumlusitanum sp. nov., a novel species isolated from sea water. International Journal of Systematic and Evolutionary Microbiology 69: 3014–3021. 10.1099/ijsem. 0.003535 [DOI] [PubMed] [Google Scholar]
- Gonçalves VN, Campos LS, Melo IS, Pellizari VH, Rosa CA, Rosa LH. (2013) Penicilliumsolitum: a mesophilic, psychrotolerant fungus present in marine sediments from Antarctica. Polar Biology 36: 1823–1831. 10.1007/s00300-013-1403-8 [DOI] [Google Scholar]
- Grijseels S, Nielsen JC, Randelovic M, Nielsen J, Nielsen KF, Workman M, Frisvad JC. (2016) Penicilliumarizonense, a new, genome sequenced fungal species, reveals a high chemical diversity in secreted metabolites. Scientific Reports – Nature 6: 1–13. 10.1038/srep35112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossart HP, Van den Wyngaert S, Kagami M, Wurzbacher C, Cunliffe M, Rojas-Jimenez K. (2019) fungi in aquatic ecosystems. Nature Reviews Microbiology 17: 339–354. 10.1038/s41579-019-0175-8 [DOI] [PubMed] [Google Scholar]
- Guevara-Suárez M, García D, Cano-Lira JF, Guarro J, Gené J. (2020) Species diversity in Penicillium and Talaromyces from herbivore dung, and the proposal of two new genera of penicillium-like fungi in Aspergillaceae. Fungal Systematics and Evolution 5: 39–75. 10.3114/fuse.2020.05.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijarro B, Larena I, Melgarejo P, De Cal A. (2017) Adaptive conditions and safety of the application of Penicilliumfrequentans as a biocontrol agent on stone fruit. International Journal of Food Microbiology 254: 25–35. 10.1016/j.ijfoodmicro.2017.05.004 [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. System 52: 696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Haelewaters D, De Kesel A, Pfister DH. (2018) Integrative taxonomy reveals hidden species within a common fungal parasite of ladybirds. Scientific Reports 8(1): 1–16. 10.1038/s41598-018-34319-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageskal G, Knutsen AK, Gaustad P, de Hoog GS, Skaar I. (2006) Diversity and significance of mold species in Norwegian drinking water. Applied Environmental Microbiology 72: 7586–7593. 10.1128/AEM.01628-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL, Lücking R. (2017) Fungal diversity revisited: 2.2 to 3.8 million species. In: Heitman J, Howlett B, Crous P, Stukenbrock E, James T, Gow N. (Eds) The Fungal Kingdom.ASM Press, Washington, 79–95. 10.1128/microbiolspec.FUNK-0052-2016 [DOI] [PubMed]
- Heo I, Hong K, Yang H, Lee HB, Choi YJ, Hong SB. (2019) Diversity of Aspergillus, Penicillium, and Talaromyces species isolated from freshwater environments in Korea. Mycobiology 47: 12–19. 10.1080/12298093.2019.1572262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hespanhol L, Vallio CS, Costa LM, Saragiotto BT. (2019) Understanding and interpreting confidence and credible intervals around effect estimates. Brazilian Journal of Physical Therapy 23: 290–301. 10.1016/j.bjpt.2018.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Hocking AD, Pitt JI. (1980) Dichloran-glycerol medium for enumeration of xerophilic fungi from low-moisture foods. Applied Environmental Microbiology 39: 488–492. 10.1128/aem.39.3.488-492.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SB, Cho HS, Shin HD, Frisvad JC, Samson RA. (2006) Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology 56: 477–486. 10.1099/ijs.0.63980-0 [DOI] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Samson RA. (2011) Taxonomy of PenicilliumsectionCitrina. Studies in Mycology 70: 53–138. 10.3114/sim.2011.70.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Samson RA. (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology 70: 1–51. 10.3114/sim.2011.70.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Frisvad JC, Seifert KA, Overy DP, Tuthill DM, Valdez JG, Samson RA. (2012) New penicillin-producing Penicillium species and an overview of sectionChrysogena. Persoonia 29: 78–100. 10.3767/003158512X660571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, de Vries RP, Samson RA. (2014) Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Applied and Environmental Microbiology 86: 199–249. 10.1016/B978-0-12-800262-9.00004-4 [DOI] [PubMed] [Google Scholar]
- Houbraken J, Wang L, Lee HB, Frisvad JC. (2016) New sections in Penicillium containing novel species producing patulin, pyripyropens or other bioactive compounds. Persoonia 36: 299–314. 10.3767/003158516X692040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J, Kocsubé S, Visagie CM, Yilmaz N, Wang XC, Meijer M, Kraak B, Hubka V, Bensch K, Samson RA, Frisvad JC. (2020) Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families, genera, subgenera, sections, series and species. Studies in Mycology 95: 5–169. 10.1016/j.simyco.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TL, Chai BB, Qiu ES, Zhou WH. (2010) Microbial effects on phosphorus release from sediments on the multi-phase interface of water-sediment-biofacies. Journal of Basic Science and Engineering 18: 61–70. [Google Scholar]
- Jasim AA, Mohammed BT, Lahuf AA. (2019) Molecular and enzymatic properties of fungi isolated from historical manuscripts preserved at the Al-Hussein Holy Shrine. Biochemical and Cellular Archives 19: 4295–4305. [Google Scholar]
- Jones EBG, Pang KL. (2012) Tropical aquatic fungi. Biodiversity and Conservation 21: 2403–2423. 10.1007/s10531-011-0198-6 [DOI] [Google Scholar]
- Kelley J, Kinsey GC, Paterson RRM, Pitchers R. (2001) Identification and control of fungi in distribution systems. AWWA Research Foundation. Denver, USA, 150 pp. [Google Scholar]
- Khomich M, Davey ML, Kauserud H, Rasconi S, Andersen T. (2017) Fungal communities in Scandinavian lakes along a longitudinal gradient. Fungal Ecology 27: 36–46. 10.1016/j.funeco.2017.01.008 [DOI] [Google Scholar]
- Kirichuk NN, Pivkin MV, Matveeva TV. (2017) Three new Penicillium species from marine subaqueous soils. Mycological Progress 16: 15–26. 10.1007/s11557-016-1247-z [DOI] [Google Scholar]
- Kong HZ, Qi ZT. (1988) Three new species of Penicillium. Mycosystema 1: 107 114.
- Kornerup A, Wanscher JH. (1978) Methuen Handbook of Colour, 3rd edn. ; Methuen: London, UK, 256 pp. [Google Scholar]
- Labuda R, Bacher M, Rosenau T, Gasparotto E, Gratzl H, Doppler M, Sulyok M, Kubátova A, Berger H, Cank K, Raja HA, Oberlies NH, Schüller C, Strauss J. (2021) Polyphasic approach utilized for the identification of two new toxigenic members of PenicilliumsectionExilicaulis, P.krskae and P.silybi spp. nov. Journal of fungi, 7: e557. 10.3390/jof7070557 [DOI] [PMC free article] [PubMed]
- Liang LJ, Jeewon R, Dhandevi P, Durairajan SSK, Li H, Lin FC, Wang HK. (2021) A novel species of Penicillium with inhibitory effects against Pyriculariaoryzae and fungal pathogens inducing Citrus diseases. Frontiers in Cellular and Infection Microbiology 10: e604504. 10.3389/fcimb.2020.604504 [DOI] [PMC free article] [PubMed]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Liu J, Wang J, Gao G, Bartlam MG, Wang Y. (2015) Distribution and diversity of fungi in freshwater sediments on a river catchment scale. Frontiers in Microbiology. 6: e329. 10.3389/fmicb.2015.00329 [DOI] [PMC free article] [PubMed]
- Madrid H, Hernández-Restrepo M, Gené J, Cano J, Guarro J, Silva V. (2016) New and interesting Chaetothyrialean fungi from Spain. Mycological Progress 15: 1179–1201. 10.1007/s11557-016-1239-z [DOI] [Google Scholar]
- Maharachchikumbura SSN, Chen Y, Ariyawansa HA, Hyde KD, Haelewaters D, Perera RH, Samarakoon MC, Wanasinghe DN, Bustamante DE, Liu J‑K, Lawrence DP, Cheewangkoon R, Stadler M. (2021) Integrative approaches for species delimitation in Ascomycota. Fungal Diversity, 109: 155–179. 10.1007/s13225-021-00486-6 [DOI] [Google Scholar]
- Máiz-Tomé L, Darwall W, Numa C, Barrios V, Smith KG. (2017) Freshwater key biodiversity areas in the north-western Mediterranean sub-region. IUCN, Gland, Switzerland, Cambridge, UK and Malaga, Spain 64: 1–48. 10.2305/IUCN.CH.2017.SSC-OP.64.en [DOI] [Google Scholar]
- Miller MA, Pfeifferm W, Schwartz T. (2012) The CIPRES science gateway: Enabling high-impact science for phylogenetics researchers with limited resources. In Proceedings of the 1st Conference of the extreme science and engineering discovery environment: Bridging from the extreme to the campus and beyond, Chicago, IL, USA, 16 July 2012; Association for Computing Machinery, New York, USA, 39: 1–8. 10.1145/2335755.2335836 [DOI]
- Niewolak S. (1975) The occurrence of microorganisms in the water of some lakes in the district of Wegorzewo. Acta Hydrobiologica 17: 371–390. [Google Scholar]
- Ogaki MB, Coelho LC, Vieira R, Neto AA. (2020a) Cultivable fungi present in deep-sea sediments of Antarctica: taxonomy, diversity, and bioprospecting of bioactive compounds. Extremophiles 24: 227–238. 10.1007/s00792-019-01148-x [DOI] [PubMed] [Google Scholar]
- Ogaki MB, Vieira R, Muniz MC, Zani CL. (2020b) Diversity, ecology, and bioprospecting of culturable fungi in lakes impacted by anthropogenic activities in Maritime Antarctica. Extremophiles 24: 637–655. 10.1007/s00792-020-01183-z [DOI] [PubMed] [Google Scholar]
- Park MS, Oh SY, Fong JJ, Houbraken J, Lim YW. (2019) The diversity and ecological roles of Penicillium in intertidal zones. Scientific Reports 9: 1–11. 10.1038/s41598-019-49966-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangging M, Nguyen TT, Lee HB. (2019) New records of four species belonging to Eurotiales from soil and freshwater in Korea. Mycobiology 47: 154–164. 10.1080/12298093.2018.1554777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoal C, Cássio F. (2004) Contribution of fungi and bacteria to leaf litter decomposition in a polluted river. Applied Environmental. Microbiology 70: 5266–5273. 10.1128/AEM.70.9.5266-5273.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoal C, Marvanová L, Cássio F. (2005) Aquatic hyphomycete diversity in streams of Northwest Portugal. Fungal Diversity 19: 109–128. [Google Scholar]
- Pereira VJ, Fernandes D, Carvalho G, Benoliel MJ. (2010) Assessment of the presence and dynamics of fungi in drinking water sources using cultural and molecular methods. Water Research 44: 4850–4859. 10.1016/j.watres.2010.07.018 [DOI] [PubMed] [Google Scholar]
- Perrone G, Susca A. (2017) Penicillium species and their associated mycotoxins. In: Mycotoxigenic fungi. Methods in Molecular Biology Moretti A., Susca A. (Eds). Humana Press: New York, NY, USA, Volume 1542, 107–119. 10.1007/978-1-4939-6707-0_5 [DOI] [PubMed]
- Peterson SW. (2008) Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100: 205–226. 10.3852/mycologia.100.2.205 [DOI] [PubMed] [Google Scholar]
- Pitt JI. (1979) The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. Academic Press Inc. Ltd. : London, UK, 634 pp. [Google Scholar]
- Pitt JI, Hocking AD. (2009) fungi and food spoilage. 3rd edn. ; Springer US: New York, NY, USA, 388 pp. 10.1007/978-0-387-92207-2_2 [DOI] [Google Scholar]
- Quintanilla JA. (1982) Three new species of Penicillium isolated from soil. Mycopathologia 80: 73–82. 10.1007/BF00641181 [DOI] [PubMed] [Google Scholar]
- Ramírez C. (1986) A new species of Penicillium from the Chilean Tierra del Fuego. Mycopathologia 96: 29–32. 10.1007/BF00467682 [DOI] [Google Scholar]
- Rodríguez-Andrade E, Stchigel AM, Cano-Lira JF. (2021) New xerophilic species of Penicillium from Soil. Journal of fungi 7: e126. 10.3390/jof7020126 [DOI] [PMC free article] [PubMed]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. System 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundel PW, Arroyo MTK, Cowling RM, Keeley JE, Lamont BB, Pausas JG, Vargas P. (2018) Fire and plant diversification in Mediterranean-climate regions. Frontiers in Plant Science 9: e851. 10.3389/fpls.2018.00851 [DOI] [PMC free article] [PubMed]
- Salgado CG, da Silva JP, Picanço-Diniz JA, da Silva MB, da Costa PF, Teixeira C, Salgado UI. (2004) Isolation of Fonsecaeapedrosoi from thorns of Mimosapudica, a probable natural source of chromoblastomycosis. Revista do Instituto de Medicina Tropical de São Paulo 46: 33–36. 10.1590/S0036-46652004000100006 [DOI] [PubMed] [Google Scholar]
- Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. (2010) CBS Laboratory Manual Series 2: Food and indoor fungi. The Netherlands: CBS-KNAW Fungal Biodiversity Centre Utrecht, 269 pp.
- Samson R, Rajput V, Shah M, Yadav R, Sarode P, Dastager SG, Dharne MS, Khairnar K. (2020) Deciphering taxonomic and functional diversity of fungi as potential bioindicators within confluence stretch of Ganges and Yamuna rivers, impacted by anthropogenic activities. Chemosphere 252: e126507. 10.1016/j.chemosphere.2020.126507 [DOI] [PubMed]
- Seifert KA, Giuseppin S. (2000) . Cycloheximide tolerance as a taxonomic character in Penicillium. In: Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Samson RA, Pitt JI (Eds), Harword Publishers; Amsterdam, The Netherlands, 259–263
- Sonjak S, Frisvad JC, Gunde-Cimerman N. (2006) Penicillium mycobiota in Arctic subglacial ice. Microbial Ecology 52: 207–216. 10.1007/s00248-006-9086-0 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe B, Chariton AA, Harford AJ, Hose GC, Greenfield P, Midgley DJ, Paulsen IT. (2018) Diverse fungal lineages in subtropical ponds are altered by sediment-bound copper. Fungal Ecology 34: 28–42. 10.1016/j.funeco.2018.03.003 [DOI] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbeti DS, Fisher MC. (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32. 10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- Thambugala KM, Daranagama DA, Phillips AJL, Kannangara SD, Promputtha I. (2020) fungi vs. fungi in biocontrol: an overview of fungal antagonists applied against fungal plant pathogens. Frontiers in Cellular and Infection Microbiology 10: e604923. 10.3389/fcimb.2020.604923 [DOI] [PMC free article] [PubMed]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóthová L. (1999) Occurrence of mitosporic fungi in the Slovak section of the Danube river. Biologia 54: 379–385. [Google Scholar]
- Ulfig K, Guarro J, Cano J, Gené J, Vidal P, Figueras MJ. (1997) General assessment of the occurrence of keratinolytic fungi in river and marine beach sediments of Catalonian waters (Spain). Water Air Soil Pollution 94: 275–287. 10.1007/BF02406063 [DOI] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Visagie CM, Houbraken J, Frisvad JC, Hong SB, Klaassen CHW, Perrone G, Seifert KA, Varga J, Yaguchi T, Samson RA. (2014a) Identification and nomenclature of the genus Penicillium. Studies in Mycology 78: 343–371. 10.1016/j.simyco.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Seifert KA, Houbraken J, Samson RA, Jacobs K. (2014b) Diversity of PenicilliumsectionCitrina within the fynbos biome of South Africa, including a new species from a Protearepens infructescence. Mycologia 106: 537–552. 10.3852/13-256 [DOI] [PubMed] [Google Scholar]
- Visagie CM, Hirooka Y, Tanney JB, Whitfield E, Mwange K, Meijer M, Asmend AS, Seifert KA, Samson RA. (2014c) Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology 78: 63–139. 10.1016/j.simyco.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Renaud JB, Burgess KMN, Malloch DW, Clark D, Ketch L, Urb M, Louis-Seize G, Assabgui R, Sumarah MW, Seifert KA. (2016a) Fifteen new species of Penicillium. Persoonia 36: 247–280. 10.3767/003158516X691627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Seifert KA, Houbraken J, Samson RA, Jacobs K. (2016b) A phylogenetic revision of Penicilliumsect.Exilicaulis, including nine new species from fynbos in South Africa. IMA fungus 7: 75–117. 10.5598/imafungus.2016.07.01.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie CM, Frisvad JC, Houbraken J, Visagie A, Samson RA, Jacobs K. (2021) A re-evaluation of PenicilliumsectionCanescentia, including the description of five new species. Persoonia 46: 163–187. 10.3767/persoonia.2021.46.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner KE, Kasai M, Francesconi A, Mu FC, Chanock SJ, Walsh TJ. (1998) Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. Journal of Clinical Microbiology 36: 1625–1629. 10.1128/JCM.36.6.1625-1629.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR Protocols: A guide to methods and applications.Academic Press, San Diego, CA, USA, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Kukwa M, Issi IV, Madrid H, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Bensch K, Kirk PM, Kolaříková K, Raja HA, Radek R, Papp V, Dima B, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska J, Humber RA, Kodsueb R, Sánchez-Castro I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipol S, Lateef AA, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Xu J, Wang Y, Tian F, Alvarado P, Li DW, Kušan I, Matočec N, Maharachchikumbura SSN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VP, Lawrey JD, Santiago ALCMA, Bezerra JDP, Souza-Motta CM, Firmino AL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AJ, Monteiro JS, Grossart HP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Vázquez V, Mungai P, Damm U, Li QR, Zhang H, Boonmee S, Lu YZ, Becerra AG, Kendrick B, Brearley FQ, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DSA, Tang LZ, He MQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MP, McKenzie EHC, Stadler M, Bhat DJ, Liu JK, Raza M, Jeewon R, Nassonova ES, Prieto M, Jayalal RGU, Erdoğdu M, Yurkov A, Schnittler M, Shchepin ON, Novozhilov YK, Silva-Filho AGS, Liu P, Cavender JC, Kang Y, Mohammad S, Zhang LF, Xu RF, Li YM, Dayarathne MC, Ekanayaka AH, Wen TC, Deng CY, Pereira OL, Navathe S, Hawksworth DL, Fan XL, Dissanayake LS, Kuhnert E, Grossart HP, Thines M. (2020) Outline of fungi and fungus-like taxa. Mycosphere 11: 1060–1456. 10.5943/mycosphere/11/1/8 [DOI] [Google Scholar]
- Yang X, Liu J, Mei J, Jiang R, Tu S, Deng H, Liu J, Yang S, Li J. (2021) Origins, structures, and bioactivities of secondary metabolites from marine-derived Penicillium fungi. Mini-Reviews in Medicinal Chemistry 21(15): 1–20. 10.2174/1389557521666210217093517 [DOI] [PubMed] [Google Scholar]
- Zhang H, Huang T, Chen S, Guo L, Liu T, Yang X. (2014) Microbial community functional diversity and enzymatic activity in the sediments of drinking water reservoirs, Northwest China. Desalination and Water Treatment 52: 1608–1614. 10.1080/19443994.2013.827383 [DOI] [Google Scholar]
- Zhang H, Huang T, Liu T. (2013) Sediment enzyme activities and microbial community diversity in an oligotrophic drinking water reservoir, eastern China. PLoS One 8: e78571. 10.1371/journal.pone.0078571 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S9
This dataset is made available under the Open Database License (http://opendatacommons.org/licenses/odbl/1.0/). The Open Database License (ODbL) is a license agreement intended to allow users to freely share, modify, and use this Dataset while maintaining this same freedom for others, provided that the original source and author(s) are credited.
D Torres-Garcia, J Gené, D García
Data type
phylogenetic trees
Explanation note
Additional phylogenetic trees of the Penicillium series corresponding to the individual analyses of the different molecular markers with additional reference strains representing the species closely related to penicillia described here.