Abstract
The use of molecular data for living groups is vital for interpreting fossils, especially when morphology-only analyses retrieve problematic phylogenies for living forms. These topological discrepancies impact on the inferred phylogenetic position of many fossil taxa. In Crocodylia, morphology-based phylogenetic inferences differ fundamentally in placing Gavialis basal to all other living forms, whereas molecular data consistently unite it with crocodylids. The Cenomanian Portugalosuchus azenhae was recently described as the oldest crown crocodilian, with affinities to Gavialis, based on morphology-only analyses, thus representing a potentially important new molecular clock calibration. Here, we performed analyses incorporating DNA data into these morphological datasets, using scaffold and supermatrix (total evidence) approaches, in order to evaluate the position of basal crocodylians, including Portugalosuchus. Our analyses incorporating DNA data robustly recovered Portugalosuchus outside Crocodylia (as well as thoracosaurs, planocraniids and Borealosuchus spp.), questioning the status of Portugalosuchus as crown crocodilian and any future use as a node calibration in molecular clock studies. Finally, we discuss the impact of ambiguous fossil calibration and how, with the increasing size of phylogenomic datasets, the molecular scaffold might be an efficient (though imperfect) approximation of more rigorous but demanding supermatrix analyses.
Keywords: Crocodylia, molecular data, divergence-age, calibration, Portugalosuchus
1. Introduction
In phylogenetic analyses, DNA data for living groups are often vital for interpreting the position of fossil taxa, as adaptive convergence in morphological characters can strongly mislead phylogenetic analyses [1–7]. Although molecular analysis in Crocodylia using both mitochondrial and nuclear DNA has consistently favoured a common topology [1,8–12], analyses focused on fossil crocodylians often continue to use morphology-only datasets, which do not retrieve the molecular tree for living crocodilians (e.g. [13–17]). Phylogenetic inference based on morphology alone places Gavialis gangeticus sister to all other living crocodilians (i.e. alligators and crocodiles) [18–21], while molecular data unite Gavialis with Tomistoma as sister to Crocodylidae alone [1,8–10,22–25]. Recent morphological studies, however, have presented strong evidence that numerous apparently plesiomorphic character states in Gavialis are instead atavistic, consistent with the molecular tree [16,21,26]. Fossil taxa close to the root of Crocodylia, and/or with a Gavialis-like morphology, are particularly susceptible to considerable changes in phylogenetic position with the addition of molecular data as the polarization and optimization of key morphological characters are likely to shift. This in turn can affect their utility as age calibrations for molecular divergence age estimations.
Portugalosuchus azenhae, recently described from an incomplete skull from the upper Cenomanian (ca 95 Ma) of Portugal, represents a notable example of this phenomenon. Based on morphology alone, Portugalosuchus was potentially the oldest member of Crocodylia (i.e. the crown group; the least-inclusive clade that contains all living crocodilians) [27]. This would pre-date the previous oldest crown crocodilian fossils (e.g. Brachychampsa sealeyi, [28]) and imply substantial ghost lineages. Furthermore, it would influence the age of Crocodylia if used as a node calibration for molecular divergence dating (e.g. [29]). Considering the topological conflict between morphological and molecular data in Crocodylia, relying on morphology alone to interpret the putative position of Portugalosuchus as the oldest crocodylian is potentially problematic, and indeed the original description acknowledged this conclusion had some uncertainty [27].
Here, we use DNA-informed analyses to investigate whether molecular data substantially alters the phylogenetic interpretation of Portugalosuchus. We added molecular data into the original, and updated, morphological datasets of [27] using different phylogenetic analytical approaches, including parsimony, undated Bayesian and tip-dating Bayesian analyses. All analyses robustly exclude Portugalosuchus from Crocodylia, instead placing it as a non-crocodylian eusuchian. We highlight the importance of DNA-informed phylogenetic inference for basal crocodylian relationships and divergence age estimates together with the use of well-justified fossil calibrations.
2. Material and methods
(a) . Morphological, molecular and stratigraphic data
The morphological datasets analysed here include (i) [30] as modified by [27] (abbreviated as NM), and (ii) [31] as modified by [27] (TM). In addition, we analysed a modified version of NM (mNM) by changing 16 characters scorings of Portugalosuchus to ‘unknown' that cannot be confirmed using published information (see electronic supplementary material, file S1 for a list of modified characters scorings).
For total evidence analyses of the NM and mNM datasets, we added molecular data for all 16 living species: a total of 9284 base pairs of mtDNA and nucDNA compiled from published data, especially [1,10]; details of sources and alignment are in a previous study [11].
For tip-dated analyses, stratigraphic data for the taxa were collected from the literature. A table with ages of taxa and references is provided in the electronic supplementary material, file S2.
(b) . Analyses
We performed eleven additional phylogenetic analyses (table 1) employing the NM and mNM morphological datasets using different approaches: maximum-parsimony and Bayesian analyses, both with and without molecular information, either as a molecular scaffold [32] using the topology of [10] or added as a DNA alignment in a supermatrix [33]. Tip-dating Bayesian analysis was also performed on the morphology + molecular supermatrices. We also re-analysed the TM matrix under parsimony and undated Bayesian approaches (but did not add DNA information as this dataset only included three living species). Furthermore, the taxon and character sampling for the TM dataset were not aimed at resolving crown crocodylian relationships, so we focused on the NM and mNM datasets.
Table 1.
Phylogenetic position of Portugalosuchus azenhae relative to Crocodylia according to different data types and analytical approaches of the present study.
| data type | analytic method | position of Portugalosuchus | referred phylogenies |
|---|---|---|---|
| morphology, original (NM) | parsimony | crown | Fig.11 in Mateus et al. [27] |
| morphology, modified (mNM) | parsimony | unresolved | electronic supplementary material, figure S1 |
| morphology, original (NM) | Bayesian undated | crown | electronic supplementary material, figure S2 |
| morphology, modified (mNM) | Bayesian undated | crown | electronic supplementary material, figure S3 |
| scaffold + morphology, original (NM + DNA) | parsimony | stem | electronic supplementary material, figure S4 |
| scaffold + morphology, modified (mNM + DNA) | parsimony | stem | electronic supplementary material, figure S5 |
| DNA + morphology, original (NM + DNA) | parsimony | stem | electronic supplementary material, figure S6 |
| DNA + morphology, modified (mNM + DNA) | parsimony | stem | electronic supplementary material, figure S7 |
| DNA + morphology, original (NM + DNA) | Bayesian undated | stem | electronic supplementary material, figure S8 |
| DNA + morphology, modified (mNM + DNA) | Bayesian undated | stem | electronic supplementary material, figure S9 |
| DNA + morphology, original (NM + DNA) | Bayesian tip-dated | stem | electronic supplementary material, figure S10 and figure 1 |
| DNA + morphology, modified (mNM + DNA) | Bayesian tip-dated | stem | electronic supplementary material, figure S11 |
All parsimony analyses were conducted in TNT v. 1.5 [34] following the same search settings as [27]; undated and tip-dated Bayesian analyses were performed, respectively, in MrBayes [35] and BEAST 2.5 [36]. The optimal partitioning scheme and substitution models for the molecular data were obtained by PartitionFinder [37]. The tip-dating analysis co-estimated topologies, branch lengths, divergence dates and evolutionary rates. The divergence age estimations for the nodes incorporate the phenotypic and stratigraphic information contained in the fossil taxa (tips). A full description of all analyses is provided in the electronic supplementary material (electronic supplementary material, file S3).
3. Results
Parsimony analysis of the original morphological dataset (NM) reproduces Mateus et al. [27] by placing Portugalosuchus within Crocodylia, but the modified dataset with some character state-codings re-scored to unknown (mNM) finds Portugalosuchus forming a polytomy with Crocodylia (electronic supplementary material, figure S1). The (undated) Bayesian analyses of morphology alone (NM and mNM) also place Portugalosuchus within Crocodylia, along the Gavialis lineage (electronic supplementary material, figures S2 and S3).
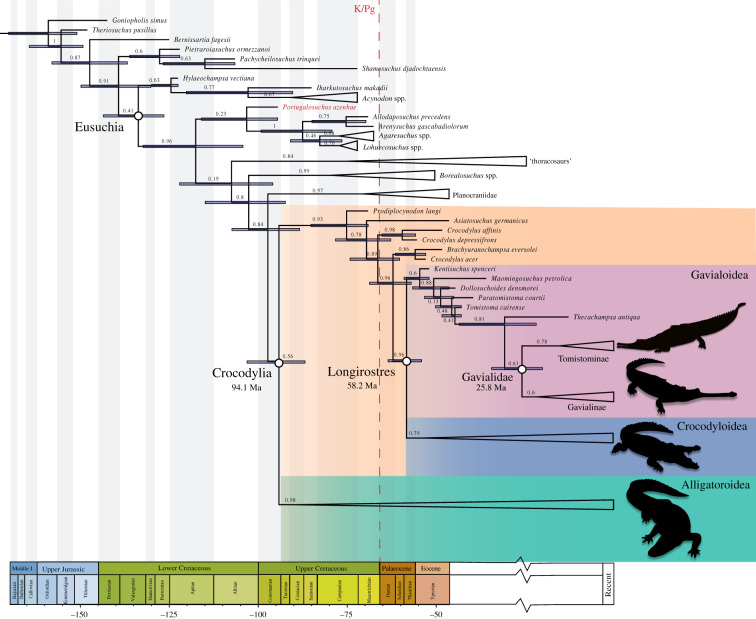
However, these two morphological datasets (NM, mNM) consistently place Portugalosuchus outside Crocodylia when molecular data are considered, either as a scaffold or incorporated in a supermatrix in a total evidence framework. These results hold under scaffold + parsimony (electronic supplementary material, figures S4 and S5), total evidence parsimony (electronic supplementary material, figures S6 and S7), total evidence undated Bayesian (electronic supplementary material, figures S8 and S9) and total evidence tip-dated Bayesian (figure 1; electronic supplementary material, figures S10 and S11). In all analyses employing molecular data (scaffold or total evidence), Planocraniidae and Borealosuchus spp. are likewise recovered outside Crocodylia. Finally, our tip-dated analysis estimate ca 95 Ma (Cenomanian, early Late Cretaceous) as the age for Crocodylia.
Figure 1.
Simplified phylogeny of crocodylians based on total evidence tip-dated Bayesian analysis using the original morphology dataset (NM) of [27]. Numbers indicate posterior probability support values for the clades. Horizontal blue–grey bars indicate 95% highest posterior density interval (HPD) for age estimate. Full phylogeny is available in
electronic supplementary material, figure S10.
For the TM dataset, parsimony analysis found Portugalosuchus as part of a large polytomy that included living crocodylians (again in agreement with [27]); however, we note that in several most parsimonious trees, Portugalosuchus was outside of crown Crocodylia (electronic supplementary material, figure S14). All supplementary figures can be found in the electronic supplementary material, file S4.
4. Discussion
(a) . The impact of DNA data on the phylogenetic position of Portugalosuchus
Previous morphological phylogenies inferred the Cenomanian P. azenhae to represent the oldest known crown crocodilian, either as sister to all other non-gavialoid crocodylians [27] or in a clade with gavialoids [16,17,26]. In each case, ghost lineages are inferred extending into the mid to latest Early Cretaceous. The addition of molecular data abruptly moves Gavialis from a basal to a nested position with respect to other living crocodilans and is thus expected to most affect taxa around the ‘gavialoid' region of the morphological tree. For instance, putative synapomorphies placing Portugalosuchus closer to Alligatoridae + Crocodylidae (to the exclusion of gavialoids) are likely to reoptimize as symplesiomorphies for all Crocodylia under the molecular topology (e.g. [1]). Indeed, in our analyses (both dated and undated), Portugalosuchus together with Borealosuchidae and Planocraniidae are consistently recovered outside Crocodylia when we add molecular information to the morphological dataset of [27], either under total evidence or under a scaffold (figure 1; electronic supplementary material, figures S4–S13; cf. also electronic supplementary material, figure S15 with [38]). In the total evidence tip-dated tree (NM), Portugalosuchus and its sister group, Allodaposuchidae, are excluded from the clade formed by Borealosuchidae, Planocraniidae and Crocodylia owing to the absence of an enlarged external mandibular fenestra (63 : 0–1). The slit-like condition score for P. azenhae instead [27] was acquired four times independently according to this topology but never within Crococodylia, except for Deinosuchus riograndensis. Additionally, the presence of a postorbital process divided into two spines (134 : 0) and postorbital bar flush with lateral jugal surface (135 : 0) (both reversed in Gavialinae, electronic supplementary material, file S5) contributes in placing P. azenhae outside the crown and more inclusive clades.
Phylogenies finding Portugalosuchus inside Crocodylia place it near ‘thoracosaurs’ (in either a clade or a grade) and are questionable given that tip-dating Bayesian analyses ([11]; this study) suggest that most if not all ‘thoracosaurs’ are not crown crocodylians related to gavialids, but are outside crown Crocodylia. As pointed out by [26], Portugalosuchus does exclusively share character states with ‘thoracosaurs’ that distinguish these taxa from gavialids. Portugalosuchus therefore likely represents a non-crocodylian eusuchian and should be avoided as a fossil age constraint for Crocodylia in calibration databases (e.g. [29]).
Supermatrix (‘combined’, ‘total evidence') approaches are the most rigorous way to integrate multiple sources of phylogenetic data, such as morphology and DNA (e.g. [33]). However, the increasing size of phylogenomic datasets is making these approaches harder to implement, owing to computational demands as well as additional bioinformatic expertise required. However, this trend also means that phylogenetic relationships between living taxa are increasingly dictated by DNA (e.g. [39]); this asymmetry means that interactions between morphological and molecular datasets that manifest themselves in a supermatrix framework (e.g. [40]) might be less important. If so, molecular scaffolds might be an efficient way forward. In this study, the molecular scaffold analysis generated topologies identical to those retrieved by combined morphological and molecular data with respect to the placement of both fossil and extant taxa (electronic supplementary material, figures S4–S7); in accordance with expectation, mapping morphology and fossils onto a molecular scaffold will be highly comparable to the results of a simultaneous analysis where the DNA data essentially constrain the relationships among living taxa (e.g. [7,39,41]). As phylogenomic data become commonplace, molecular scaffolds therefore comprise an increasingly pragmatic way of integrating molecular information into fossil phylogenies.
(b) . The age of Crocodylia and the impact of ambiguous fossil calibrations
In our tip-dated analyses (using both original and modified datasets), the age for Crocodylia is approximately 94 Ma (Cenomanian, Late Cretaceous), with a 95% highest posterior density interval (HPD) of approx. 87–104 Ma. If Portugalosuchus were assumed to be a crown crocodilian, tip- or node-dated molecular divergence dating would estimate a substantially older age for the clade; the age of Portugalosuchus (95 Ma) would form the hard minimum age for the clade. The present estimate is consistent with recent tip-dating estimates of approximately 100 Ma [11] despite substantial differences in taxon and character sampling. Our estimate is also broadly consistent with molecular studies implicitly or explicitly following best practices [42] for fossil calibrations (table 2) as well as fossil divergence age estimates (e.g. [13,43]). All these converge upon an interval between 90 and 100 Ma for the age of Crocodylia. On the other hand, controversial choices for fossil calibrations of some published molecular divergence studies have led to much earlier inferred ages for Crocodylia. Brachychampsa sealeyi, a taxon conventionally regarded as a basal alligatoroid (e.g. [28,44–49]), has been used as a constraint for a much smaller clade, Caimaninae, following some weakly supported phylogenies (see [49] for a review), resulting in inflated age estimations for Alligatoridae (split between Caimaninae and Alligatorinae, 71.61–129.7 Ma; [50,51]). Similarly, the same studies overlooked that some crocodylian clades have stem-based definitions and therefore include stem fossils [45,52] and thus calibrated crown-Alligatorinae (Alligator mississippiensis–A. sinensis split) with the stem-alligatorine Navajosuchus mooki [13,45,53]; this leads to overestimating crown-alligatorine divergence ages.
Table 2.
Compilation of divergence age estimations for Crocodylia. Except for Roos et al. [9], all the age intervals represent the 95% highest posterior densities (HPD).
| reference | Crocodylia divergence age estimates (Ma) | time-calibrated technique | clock model | tree model |
|---|---|---|---|---|
| Roos et al. [9] | 101 ± 3.0 | molecular clock (node calibration) | r8s non-parametric rate smoothing (NPRS) | |
| Oaks [10] | 81.08–90.00 | molecular clock (node calibration) | relaxed uncorrelated lognormal | Yule or birth–death process |
| Turner et al. [43] | 81.02–114.25 | tip-dating (morphology only) | relaxed uncorrelated lognormal | birth–death process |
| Lee & Yates [11] | 90.0–110.0 | tip-dating (total evidence) | relaxed uncorrelated lognormal | birth–death process |
| Pan et al. [12] | 83.6–90.02 | molecular clock (node calibration) | relaxed uncorrelated lognormal | Yule process |
| this work | 86.77–103.09 | tip-dating (total evidence) | relaxed uncorrelated lognormal | birth–death process |
Our analyses demonstrate how the integration of molecular and morphological data/topologies plays an important role in interpreting the phylogenetic position of basal crocodylians and suggest avoiding inferences based exclusively on morphological data, especially when morphology-based relationships among living taxa are robustly contradicted by genomic data. While it is intriguing that a recent morphological phylogeny resolved the Tomistoma–Gavialis conflict [26], its use of quantitative characters in turn resulted in unconventional placement of some extant and fossil taxa (e.g. polyphyletic Jacarea as opposed to all previous morphological and molecular phylogenies). Integrating DNA and morphological data/topologies therefore remains a vital approach for phylogenetic inference as well as reconstruction of character evolution and divergence ages, and molecular scaffolds can be an efficient way to approximate more rigorous supermatrix approaches.
Acknowledgements
We thank G. Ferreira, S. Onary, P. Godoy, C. Brochu, W. G. Joyce and J. Sterli for insightful discussions, and the Willi Henning Society for supporting free availability of the TNT software. We also thank the handling editor and three anonymous referees for the feedback and comments on the manuscript.
Contributor Information
Gustavo Darlim, Email: gustavo.darlim@gmail.com.
Márton Rabi, Email: marton.rabi@uni-tuebingen.de.
Data accessibility
Full details of the methods and results, along with datasets and analyses executables, can be found in the eletronic supplementary material and in the Dryad Digital Repository (http://doi.org/10.5061/dryad.q2bvq83mf) [54].
Authors' contributions
G.D.: conceptualization, data curation, formal analysis, investigation, project administration, writing—original draft; M.S.Y.L.: conceptualization, data curation, formal analysis, methodology, software, writing—review and editing; J.W.: conceptualization, data curation, formal analysis, writing—review and editing; M.R.: conceptualization, funding acquisition, investigation, project administration, supervision, visualization, writing—original draft.
All authors gave final approval for publication and agreed to be held accountable for the work described herein.
Competing interests
We declare we have no competing interests.
Funding
This study was funded by the Deutsche Forschungsgemeinschaft (grant no. 417629144) and the Volkswagen Stiftung (grant no. Az. 90 978), both awarded to M.R.
References
- 1.Gatesy J, Amato G, Norell M, DeSalle R, Hayashi C. 2003. Combined support for wholesale taxic atavism in gavialine crocodylians. Syst. Biol. 52, 403-422. ( 10.1080/10635150390197037) [DOI] [PubMed] [Google Scholar]
- 2.Asher RJ, Hofreiter M. 2006. Tenrec phylogeny and the noninvasive extraction of nuclear DNA. Syst. Biol. 55, 181-194. ( 10.1080/10635150500433649) [DOI] [PubMed] [Google Scholar]
- 3.Koepfli KP, Gompper ME, Eizirik E, Ho CC, Linden L, Maldonado JE, Wayne RK. 2007. Phylogeny of the Procyonidae (Mammalia: Carnivora): molecules, morphology and the great American interchange. Mol. Phylogenet. Evol. 43, 1076-1095. ( 10.1016/j.ympev.2006.10.003) [DOI] [PubMed] [Google Scholar]
- 4.Havermans C, Nagy ZT, Sonet G, De Broyer C, Martin P. 2010. Incongruence between molecular phylogeny and morphological classification in amphipod crustaceans: a case study of Antarctic lysianassoids. Mol. Phylogenet. Evol. 55, 202-209. ( 10.1016/j.ympev.2009.10.025) [DOI] [PubMed] [Google Scholar]
- 5.Dávalos LM, Velazco PM, Warsi OM, Smits PD, Simmons NB. 2014. Integrating incomplete fossils by isolating conflicting signal in saturated and non-independent morphological characters. Syst. Biol. 63, 582-600. ( 10.1093/sysbio/syu022) [DOI] [PubMed] [Google Scholar]
- 6.Zou Z, Zhang J. 2016. Morphological and molecular convergences in mammalian phylogenetics. Nat. Commun. 7, 12758. ( 10.1038/ncomms12758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou CF, Rabi M. 2015. A sinemydid turtle from the Jehol Biota provides insights into the basal divergence of crown turtles. Scient. Rep. 5, 16299. ( 10.1038/srep16299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harshman J, Huddleston CJ, Bollback JP, Parsons TJ, Braun MJ. 2003. True and false gharials: a nuclear gene phylogeny of Crocodylia. Syst. Biol. 52, 386-402. ( 10.1080/10635150390197028) [DOI] [PubMed] [Google Scholar]
- 9.Roos J, Aggarwal RK, Janke A. 2007. Extended mitogenomic phylogenetic analyses yield new insight into crocodylian evolution and their survival of the Cretaceous–Tertiary boundary. Mol. Phyl. Evol. 45, 663-673. ( 10.1016/j.ympev.2007.06.018) [DOI] [PubMed] [Google Scholar]
- 10.Oaks JR. 2011. A time-calibrated species tree of Crocodylia reveals a recent radiation of the true crocodiles. Evolution 65, 3285-3297. ( 10.1111/j.1558-5646.2011.01373.x) [DOI] [PubMed] [Google Scholar]
- 11.Lee MSY, Yates AM. 2018. Tip dating and homoplasy: reconciling the shallow molecular divergences of modern gharials with their long fossil record. Proc. R. Soc. B. 285, 20181071. ( 10.1098/rspb.2018.1071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan T, et al. 2020. Near-complete phylogeny of extant Crocodylia (Reptilia) using mitogenome-based data. Zool. J. Linn. Soc. 191, 1075-1089. ( 10.1093/zoolinnean/zlaa074) [DOI] [Google Scholar]
- 13.Bona P, Ezcurra MD, Barrios F, Fernandez Blanco MV. 2018. A new Palaeocene crocodylian from southern Argentina sheds light on the early history of caimanines. Proc. R. Soc. B 285, 20180843. ( 10.1098/rspb.2018.0843) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cossette AP, Brochu CA. 2020. A systematic review of the giant alligatoroid Deinosuchus from the Campanian of North America and its implications for the relationships at the root of Crocodylia. J. Vertebr. Paleontol. 40, e1767638. ( 10.1080/02724634.2020.1767638) [DOI] [Google Scholar]
- 15.Godoy PL, Cidade GM, Montefeltro FC, Langer MC, Norell MA. 2021. Redescription and phylogenetic affinities of the caimanine Eocaiman cavernensis (Crocodylia, Alligatoroidea) from the Eocene of Argentina. Pap. Palaeontol. 7, 1205-1231. ( 10.1002/spp2.1339)) [DOI] [Google Scholar]
- 16.Ristevski J, Yates AM, Price GJ, Molnar RE, Weisbecker V, Salisbury SW. 2020. Australia's prehistoric ‘swamp king’: revision of the Plio-Pleistocene crocodylian genus Pallimnarchus de Vis, 1886. PeerJ. 8, e10466. ( 10.7717/peerj.10466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ristevski J, Price GJ, Weisbecker V, Salisbury SW. 2021. First record of a tomistomine crocodylian from Australia. Scient. Rep. 11, 12158. ( 10.1038/s41598-021-91717-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norell MA. 1989. The higher level relationships of the extant Crocodylia. J. Herpetol. 23, 325-335. ( 10.2307/1564042) [DOI] [Google Scholar]
- 19.Brochu CA. 1997. Morphology, fossils, divergence timing, and the phylogenetic relationships of Gavialis. Syst. Biol. 46, 479-522. ( 10.1093/sysbio/46.3.479) [DOI] [PubMed] [Google Scholar]
- 20.Piras P, Colangelo P, Adams DC, Buscalioni A, Cubo J, Kotsakis T, Meloro C, Raia P. 2010. The Gavialis–Tomistoma debate: the contribution of skull ontogenetic allometry and growth trajectories to the study of crocodylian relationships. Evol. Dev. 12, 568-579. ( 10.1111/j.1525-142X.2010.00442.x) [DOI] [PubMed] [Google Scholar]
- 21.Iijima M, Kobayashi Y. 2019. Mosaic nature in the skeleton of East Asian crocodylians fills the morphological gap between ‘Tomistominae’ and Gavialinae. Cladistics 35, 623-632. ( 10.1111/cla.12372) [DOI] [PubMed] [Google Scholar]
- 22.Gatesy J, Amato GD. 1992. Sequence similarity of 12S ribosomal segment of mitochondrial DNAs of gharial and false gharial. Copeia 1992, 241-243. ( 10.2307/1446560) [DOI] [Google Scholar]
- 23.Janke A, Gullberg A, Hughes S, Aggarwal RK, Arnason U. 2005. Mitogenomic analyses place the gharial (Gavialis gangeticus) on the crocodile tree and provide pre-K/T divergence times for most crocodilians. J. Mol. Evol. 61, 620-626. ( 10.1007/s00239-004-0336-9) [DOI] [PubMed] [Google Scholar]
- 24.Willis RE, McAliley LR, Neeley ED, Densmore LD III. 2007. Evidence for placing the false gharial (Tomistoma schlegelii) into the family Gavialidae: inferences from nuclear gene sequences. Mol. Phylogenet. Evol. 43, 787-794. ( 10.1016/j.ympev.2007.02.005) [DOI] [PubMed] [Google Scholar]
- 25.Willis RE. 2009. Transthyretin gene (TTR) intron 1 elucidates crocodylian phylogenetic relationships. Mol. Phylogenet. Evol. 53, 1049-1054. (doi:10.1016%2Fj.ympev.2009.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rio JP, Mannion PD. 2021. Phylogenetic analysis of a new morphological dataset elucidates the evolutionary history of Crocodylia and resolves the long-standing gharial problem. PeerJ. 9, e12094. ( 10.7717/peerj.12094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateus O, Puértolas-Pascual E, Callapez PM. 2019. A new eusuchian crocodylomorph from the Cenomanian (Late Cretaceous) of Portugal reveals novel implications on the origin of Crocodylia. Zool. J. Linn. Soc. 186, 501-528. ( 10.1093/zoolinnean/zly064) [DOI] [Google Scholar]
- 28.Williamson TE. 1996. ?Brachychampsa sealeyi, sp nov., (Crocodylia, Alligatoroidea) from the Upper Cretaceous (lower Campanian) Menefee Formation, northwestern New Mexico. J. Vertebr. Paleontol. 16, 421-431. ( 10.1080/02724634.1996.10011331) [DOI] [Google Scholar]
- 29.Benton MJ, Donoghue PC, Asher RJ, Friedman M, Near TJ, Vinther J. 2015. Constraints on the timescale of animal evolutionary history. Palaeontol. Electron. 18, 1-106. ( 10.26879/424) [DOI] [Google Scholar]
- 30.Narváez I, Brochu CA, Escaso F, Pérez-García A, Ortega F. 2016. New Spanish Late Cretaceous eusuchian reveals the synchronic and sympatric presence of two allodaposuchids. Cretac. Res. 65, 112-125. ( 10.1016/j.cretres.2016.04.018) [DOI] [Google Scholar]
- 31.Turner AH. 2015. A review of Shamosuchus and Paralligator (Crocodyliformes, Neosuchia) from the Cretaceous of Asia. PLoS ONE 10, e0118116. ( 10.1371/journal.pone.0118116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Springer MS, Teeling EC, Madsen O, Stanhope MJ, de Jong WW. 2001. Integrated fossil and molecular data reconstruct bat echolocation. Proc. Natl Acad. Sci. USA 98, 6241-6246. ( 10.1073/pnas.111551998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Queiroz A, Gatesy J. 2007. The supermatrix approach to systematics. Trends Ecol. Evol. 22, 34-41. ( 10.1016/j.tree.2006.10.002) [DOI] [PubMed] [Google Scholar]
- 34.Goloboff PA, Catalano SA. 2016. TNT version 1.5, including a full implementation of phylogenetic morphometrics. Cladistics 32, 221-238. ( 10.1111/cla.12160) [DOI] [PubMed] [Google Scholar]
- 35.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539-542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouckaert R, et al. 2019. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15, e1006650. ( 10.1371/journal.pcbi.1006650) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 6, 1695-1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 38.Blanco A. 2021. Importance of the postcranial skeleton in eusuchian phylogeny: reassessing the systematics of allodaposuchid crocodylians. PLoS ONE 16, e0251900. ( 10.1371/journal.pone.0251900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee MSY, Palci A. 2015. Morphological phylogenetics in the genomic age. Curr. Biol. 25, R922-R929. ( 10.1016/j.cub.2015.07.009) [DOI] [PubMed] [Google Scholar]
- 40.Gatesy J, O'Grady P, Baker RH. 1999. Corroboration among data sets in simultaneous analysis: hidden support for phylogenetic relationships among higher level artiodactyl taxa. Cladistics 15, 271-313. ( 10.1111/j.1096-0031.1999.tb00268.x) [DOI] [PubMed] [Google Scholar]
- 41.Crawford NG, Parham JF, Sellas AB, Faircloth BC, Glenn TC, Papenfuss TJ, Henderson JB, Hansen MH, Simison WB. 2015. A phylogenomic analysis of turtles. Mol. Phyl. Evol. 83, 250-257. ( 10.1016/j.ympev.2014.10.021) [DOI] [PubMed] [Google Scholar]
- 42.Parham JF, et al. 2012. Best practices for justifying fossil calibrations. Syst. Biol. 61, 346-359. ( 10.1093/sysbio/syr107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner AH, Pritchard AC, Matzke NJ. 2017. Empirical and Bayesian approaches to fossil-only divergence times: a study across three reptile clades. PLoS ONE 12, e0169885. ( 10.1371/journal.pone.0169885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Norell M, Clark JM, Hutchison JH. 1994. The Late Cretaceous alligatoroid Brachychampsa montana (Crocodylia): new material and putative relationships. Am. Mus. Novit. 3116, 1-26. [Google Scholar]
- 45.Brochu CA. 1999. Phylogenetics, taxonomy, and historical biogeography of Alligatoroidea. J. Vertebr. Paleontol. 19, 9-100. ( 10.1080/02724634.1999.10011201) [DOI] [Google Scholar]
- 46.Brochu CA. 2010. A new alligatorid from the lower Eocene Green River Formation of Wyoming and the origin of caimans. J. Vertebr. Paleontol. 30, 1109-1126. ( 10.1080/02724634.2010.483569) [DOI] [Google Scholar]
- 47.Brochu CA. 2011. Phylogenetic relationships of Necrosuchus ionensis Simpson, 1937 and the early history of caimanines. Zool. J. Linn. Soc. 163, S228-S256. ( 10.1111/j.1096-3642.2011.00716.x) [DOI] [Google Scholar]
- 48.Massonne T, Vasilyan D, Rabi M, Böhme M. 2019. A new alligatoroid from the Eocene of Vietnam highlights an extinct Asian clade independent from extant Alligator sinensis. PeerJ. 7, e7562. ( 10.7717/peerj.7562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walter J, Darlim G, Massonne T, Aese A, Frey E, Rabi M. In press.. On the origin of Caimaninae: insights from new fossils of Tsoabichi greenriverensis and a review of the evidence. Hist. Biol. ( 10.1080/08912963.2021.1938563) [DOI] [Google Scholar]
- 50.Bittencourt PS, Campos Z, Muniz FL, Marioni B, Souza BC, Da Silveira R, De Thoisy B, Hrbek B, Farias IP. 2019. Evidence of cryptic lineages within a small South American crocodilian: the Schneider's dwarf caiman Paleosuchus trigonatus (Alligatoridae: Caimaninae). PeerJ. 7, e6580. ( 10.7717/peerj.6580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberto IJ, et al. 2020. Unexpected but unsurprising lineage diversity within the most widespread Neotropical crocodilian genus Caiman (Crocodylia, Alligatoridae). Syst. Biodivers. 18, 377-395. ( 10.1080/14772000.2020.1769222) [DOI] [Google Scholar]
- 52.deQueiroz KD, Gauthier J. 1992. Phylogenetic taxonomy. Annu. Rev. Ecol. Syst. 23, 449-480. ( 10.1146/annurev.es.23.110192.002313) [DOI] [Google Scholar]
- 53.Brochu CA. 2004. Alligatorine phylogeny and the status of Allognathosuchus Mook, 1921. J. Vertebr. Paleontol. 24, 857-873. ( 10.1671/0272-4634(2004)024[0857:APATSO]2.0.CO;2) [DOI] [Google Scholar]
- 54.Darlim G, Lee MSY, Walter J, Rabi M. 2022. Data from: The impact of molecular data on the phylogenetic position of the putative oldest crown crocodilian and the age of the clade. Dryad Digital Repository. ( 10.5061/dryad.q2bvq83mf) [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Full details of the methods and results, along with datasets and analyses executables, can be found in the eletronic supplementary material and in the Dryad Digital Repository (http://doi.org/10.5061/dryad.q2bvq83mf) [54].