Abstract
Motivation
InterARTIC is an interactive web application for the analysis of viral whole-genome sequencing (WGS) data generated on Oxford Nanopore Technologies (ONT) devices. A graphical interface enables users with no bioinformatics expertise to analyze WGS experiments and reconstruct consensus genome sequences from individual isolates of viruses, such as SARS-CoV-2. InterARTIC is intended to facilitate widespread adoption and standardization of ONT sequencing for viral surveillance and molecular epidemiology.
Results
We demonstrate the use of InterARTIC for the analysis of ONT viral WGS data from SARS-CoV-2 and Ebola virus, using a laptop computer or the internal computer on an ONT GridION sequencing device. We showcase the intuitive graphical interface, workflow customization capabilities and job-scheduling system that facilitate execution of small- and large-scale WGS projects on any common virus.
Availability and implementation
InterARTIC is a free, open-source web application implemented in Python that executes best-practice command line workflows from the ARTIC network. The application can be downloaded as a set of pre-compiled binaries that are compatible with all common Linux distributions, Windows with Linux subsystems, MacOSX and ARM systems. All code can be found on GitHub at https://github.com/Psy-Fer/interARTIC/ and documentation can be found at https://github.com/Psy-Fer/interARTIC/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Viral whole-genome sequencing (WGS) has become a critical tool used to guide local, national and international public health responses to the ongoing COVID-19 pandemic, as well as Ebola, Zika, Dengue and other viral epidemics (du Plessis et al., 2021; Eden et al., 2020; Fauver et al., 2020; Gonzalez-Reiche et al., 2020; Gudbjartsson et al., 2020; Lu et al., 2020; Meredith et al., 2020; Msomi et al., 2020; Quick et al., 2016, 2017; Rockett et al., 2020; Stubbs et al., 2020). Viral WGS is used to define the phylogenetic structure of disease outbreaks in order to better understand geographical spread, resolve transmission networks and infer the origin of unknown cases (Eden et al., 2020; Fauver et al., 2020; Gonzalez-Reiche et al., 2020; Rockett et al., 2020). Viral WGS can also identify novel strains, and monitor virus evolution over time or in response to public health interventions, such as vaccines (du Plessis et al., 2021; Fiorentini et al., 2021; Tegally et al., 2021; Williams and Burgers, 2021).
Oxford Nanopore Technologies (ONT) is an emerging sequencing platform that can be used to perform viral WGS. ONT devices are small, cheap, require minimal supporting laboratory infrastructure or technical expertise for sample preparation, and can be used to perform rapid viral WGS with high consensus accuracy (Bull et al., 2020).
The ARTIC network for viral surveillance and molecular epidemiology has been instrumental in driving the adoption of ONT sequencing for viral WGS. ARTIC is an international consortium that has developed standardized protocols for WGS analysis of multiple common viruses, including SARS-CoV-2. ARTIC researchers have also developed best-practice, open-source bioinformatics workflows for the reconstruction of consensus viral genome sequences from ONT sequencing data. While these command line-driven workflows are a critical resource for the community, they require specialist bioinformatics expertise that is a barrier for some users, particularly in under-resourced areas. Moreover, the dependency of ARTIC pipelines on multiple third-party software packages often creates difficulties during installation.
To help address these issues, we have developed InterARTIC, an interactive local web application for the analysis of viral WGS data generated on ONT devices. InterARTIC provides an intuitive graphical interface for configuration and execution of command line workflows from the ARTIC network, enabling novice users to reconstruct consensus genome sequences from viral isolates, including SARS-CoV-2. The workflows are fully customizable, ensuring compatibility with different viruses and/or upstream laboratory preparations and an intelligent job-scheduling system facilitates efficient handling of large projects.
2 Results
2.1 Software implementation
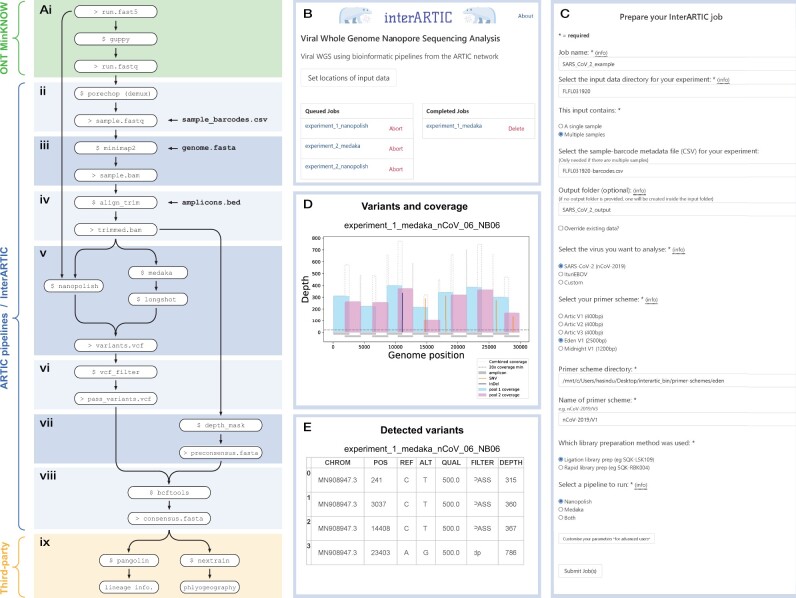
InterARTIC is a local (offline) web application for viral genomics analysis with established ARTIC pipelines. The central component is a simple and intuitive graphical user interface developed using Python, HTML, CSS and JavaScript that encapsulates current best-practice command-line bioinformatics pipelines from ARTIC (Fig. 1A). InterARTIC uses Flask as the web framework and features an asynchronous task queue implemented using Celery with Redis as the database backend. InterARTIC is provided as a standalone package that bundles the aforementioned interface, the ARTIC pipeline (fieldbioinformatics toolkit) and all dependencies.
Fig. 1.
Overview of the InterARTIC workflow and graphical interface. (A) Schematic summary of a typical workflow for the analysis of ONT viral genome-sequencing data. Workflow steps (ii–viii) (blue) are performed within ARTIC command line tools or carried out by InterARTIC. (B) Example screenshot of the InterARTIC interface hompage, with queued and completed jobs displayed. (C) Example of the project setup page within the interface, where the user may customize their workflow. Note: full parameter selection options for advanced users are hidden. (D) Example of a coverage profile plot for SARS-CoV-2 that is generated and displayed by InterARTIC. (E) Example of the variant detection table that is generated and displayed
InterARTIC requires no installation of third-party software and itself can be downloaded as a set of pre-compiled binaries for the two common CPU architectures, x86_64 and arm64 (aarch64) and for all major operating systems—all Linux distributions (Ubuntu, Debian, Fedora, CentOS, etc.), Windows (requires Windows Subsystem for Linux) and macOS. Developers can alternatively build InterARTIC from source.
The application can execute all analyses on a standard laptop or desktop PC, after receiving sequencing data from an ONT device, such as a MinION. Alternatively, InterARTIC can run the analysis on the internal computer of a GridION/PromethION benchtop sequencer, generating all outputs without the need for data transfer. Importantly, InterARTIC was implemented with a novel software compilation/containment strategy that prevents potential conflicts with software on a user’s PC or ONT device (Supplementary Note S1).
2.2 Launching the application
To use InterARTIC, the binaries should be downloaded, extracted and launched by running the following simple commands in a bash terminal:
$ wget\
https://github.com/Psy−Fer/interARTIC/releases/download/v0 .4/interartic−v0.4−linux−x86 − 64−binaries.tar.gz\
−O interartic_bin.tar.gz
$ tar xf interartic_bin.tar.gz
$ cd interartic_bin
$ ./run.sh
This launches an interactive session that can be accessed through a standard web browser, via a link that is printed to the standard output (by default: http://127.0.0.1:5000), as follows:
$ ./run.sh
InterARTIC is now running on your machine:)
To launch InterARTIC web interface visit http://127.0.0.1:5000 on your browser
To keep your InterARTIC active this terminal must remain open.
To terminate InterARTIC type CTRL−C or close the terminal.
After navigating to the URL, the user can interact with the graphical interface in order to set up and launch their analysis (Fig. 1B and C). Note that, while the user interacts with InterARTIC through their web browser (via localhost), all analysis is performed securely on the local hardware, where the application is hosted, and no external Internet connection is required.
A simple video tutorial is available to assist new users in setting up InterARTIC, configuring and running their workflow, and viewing the outputs: https://youtu.be/RCArn-xOkHg
2.3 Workflow summary
InterARTIC analyses amplicon-based viral WGS data generated on ONT devices using best-practice analysis pipelines developed by ARTIC (Fig. 1A). It is compatible with any virus genome, and any standard or custom amplicon primer scheme. InterARTIC inputs base-called sequencing reads (FASTQ format), which are typically generated during a sequencing run, using the Guppy base-caller within ONT’s MinKNOW sequencing manager software [Fig. 1A(i)]. If multiple samples were sequenced on a single flow cell, InterARTIC uses Porechop to demultiplex the library into individual sample barcodes, then runs each sample separately through the analytic workflow [Fig. 1A(ii)]. Alternatively, if the user’s data have already been demultiplexed with Guppy, this step will be skipped.
InterARTIC first uses Minimap2 to align sequencing reads to a viral reference genome [e.g. MN908947 for SARS-CoV-2; Fig. 1A(iii)]. Primer sequences are then trimmed, and alignments are down-sampled to a maximum coverage-depth threshold that can be specified by the user [Fig. 1A(iv)]. The genome (FASTA format) and primer site (BED format) reference files used during these steps can be selected from a range of popular options or supplied as custom files, according to the user’s needs. Genetic variants (SNVs and indels) are then identified relative to the reference genome. InterARTIC currently supports two alternative workflow choices for variant detection that utilize the popular tools Nanopolish or Medaka/Longshot, with the user selecting either or both workflows [Fig. 1A(v)]. Low-quality variant candidates are then excluded [Fig. 1A(vi)] and regions of low coverage are masked from the reference genome (Fig. 1A(vii)). Finally, variant candidates are incorporated into the masked reference genome using Bcftools to generate a consensus genome sequence for the viral isolate under analysis [Fig. 1A(viii)]. This consensus genome sequence is suitable for downstream applications including lineage classification and phylogeographic analysis [Fig. 1A(ix)], and ready for upload to public repositories (e.g. GISAID).
In addition to the detected variants (VCF format) and consensus genome (FASTA format) output files from the workflow, InterARTIC also generates and displays a data plot and summary table to facilitate quick inspection of the data and longitudinal quality control (Fig. 1D and E).
InterARTIC employs an asynchronous job queue system with Celery to allow multiple jobs of the same, or differing virus and/or amplicon scheme, to be launched to allow for efficient use of compute resources (Fig. 1B). This is especially important if running analyses on a GridION/PromethION benchtop device, alongside other sequencing jobs.
2.4 Example data for testing InterARTIC
To demonstrate the use of InterARTIC, we analyzed publicly available nanopore WGS datasets from SARS-CoV-2 (10× isolates, sequenced on an ONT GridION; see (Bull et al., 2020) and Ebola [2× isolates, sequenced on an ONT MinION; see (Quick et al., 2016, 2017)]. Example screenshots of the workflow configuration process for each experiment are provided in Supplementary Figures S1 and S2, and example output files generated by InterARTIC are also available for download.
Both projects were analyzed using InterARTIC on a standard laptop PC, and separately on a GridION sequencing device (Supplementary Table S1). Indicative run-time statistics on both devices are reported in Supplementary Table S2. We encourage users to download these example datasets to test InterARTIC on their own machine.
3 Discussion
With new SARS-CoV-2 strains (e.g. Delta variant) emerging with increasing regularity, and showing evidence in some cases of immune escape (Garcia-Beltran et al., 2021), there is a critically important ongoing role for viral genomics in the international response to COVID19. Nanopore sequencing enables rapid, cost-effective and decentralized viral WGS (Bull et al., 2020) and has become a useful tool for genomic surveillance initiatives across the globe.
InterARTIC is intended to help facilitate this process by solving two key issues. First, the application enables users with no bioinformatics experience to analyze their own nanopore sequencing data to assemble complete viral genomes from individual patient isolates. This removes a major barrier for teams without dedicated bioinformatic scientists on their staff. Second, InterARTIC requires no installation of third-party software. Installation of the multiple third-party tools (and their underlying dependencies) currently required for WGS analysis can be a major challenge, even for experienced users. During this process, dependency conflicts are frequently encountered between the various software environments on a user’s PC. InterARTIC employs a novel containment approach, described in Supplementary Note S1, which removes any risk of dependency conflicts. Therefore, the application can be easily and safely run on any PC, including the on-board computer of an ONT GridION/PromethION device, removing a resource barrier for teams lacking access to high-performance computing infrastructure.
While InterARTIC was originally designed for SARS-CoV-2 genomics, it is equally suitable for the analysis of any other virus. Users can supply custom inputs specific to their own virus or amplicon scheme and, to date, InterARTIC has been tested with SARS-CoV-2, Ebola, Dengue, Hepatitis C and Ross River viruses. We provide InterARTIC as a free, open-source tool to facilitate further adoption and standardization of ONT sequencing for viral surveillance during the ongoing COVID-19 pandemic and future disease outbreaks.
Supplementary Material
Acknowledgements
The authors thank Charles Foster, Ellen de Vries, Han N. Phan, Meutia Kumaheri, Igor Stevanovski and Jillian Hammond for providing feedback on InterARTIC.
Funding
The authors acknowledge the following funding support: MRFF Investigator [MRF1173594 to I.W.D.] and philanthropic support from The Kinghorn Foundation.
Conflict of Interest: J.M.F. and H.G. have previously received travel and accommodation expenses to speak at Oxford Nanopore Technologies conferences. The authors declare no other competing financial or non-financial interests.
Contributor Information
James M Ferguson, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia.
Hasindu Gamaarachchi, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia; School of Computer Science and Engineering, University of New South Wales, Sydney, NSW, Australia.
Thanh Nguyen, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia; School of Computer Science and Engineering, University of New South Wales, Sydney, NSW, Australia.
Alyne Gollon, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia; School of Computer Science and Engineering, University of New South Wales, Sydney, NSW, Australia.
Stephanie Tong, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia; School of Computer Science and Engineering, University of New South Wales, Sydney, NSW, Australia.
Chiara Aquilina-Reid, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia; School of Computer Science and Engineering, University of New South Wales, Sydney, NSW, Australia.
Rachel Bowen-James, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia; School of Computer Science and Engineering, University of New South Wales, Sydney, NSW, Australia.
Ira W Deveson, Kinghorn Centre for Clinical Genomics, Garvan Institute of Medical Research, Sydney, NSW, Australia; St Vincent’s Clinical School, Faculty of Medicine, University of New South Wales, Sydney, NSW, Australia.
References
- Bull R.A. et al. (2020) Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat. Commun., 11, 6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden J.-S. et al. ; 2019-nCoV Study Group. (2020) An emergent clade of SARS-CoV-2 linked to returned travellers from Iran. Virus Evol., 6, veaa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauver J.R. et al. (2020) Coast-to-Coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell, 181, 990–996. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorentini S. et al. (2021) First detection of SARS-CoV-2 spike protein N501 mutation in Italy in August, 2020. Lancet Infect. Dis., 21, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F. et al. (2021) Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell, 184, 2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reiche A.S. et al. (2020) Introductions and early spread of SARS-CoV-2 in the New York City area. Science, 369, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson D.F. et al. (2020) Spread of SARS-CoV-2 in the Icelandic Population. N. Engl. J. Med., 382, 2302–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. et al. (2020) Genomic epidemiology of SARS-CoV-2 in Guangdong province, China. Cell, 181, 997–1003. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith L.W. et al. (2020) Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect. Dis., 20, 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Msomi N. et al. ; Network for Genomic Surveillance in South Africa Writing Group. (2020) A genomics network established to respond rapidly to public health threats in South Africa. Lancet Microbe, 1, e229–e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Plessis L. et al. ; COVID-19 Genomics UK (COG-UK) Consortium. (2021) Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science, 371, 708–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J. et al. (2016) Real-time, portable genome sequencing for Ebola surveillance. Nature, 530, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J. et al. (2017) Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc., 12, 1261–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockett R.J. et al. (2020) Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat. Med., 26, 1398–1404. [DOI] [PubMed] [Google Scholar]
- Stubbs S.C.B. et al. (2020) Assessment of a multiplex PCR and Nanopore-based method for dengue virus sequencing in Indonesia. Virol. J., 17, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H. et al. (2021) Detection of a SARS-CoV-2 variant of concern in South Africa. Nature, 592, 438–443. [DOI] [PubMed] [Google Scholar]
- Williams T.C., Burgers W.A. (2021) SARS-CoV-2 evolution and vaccines: cause for concern? Lancet Respir. Med., 9, 333–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.