Abstract
Longitudinal bone growth is achieved by a tightly controlled process termed endochondral bone formation. C-type natriuretic peptide (CNP) stimulates endochondral bone formation through binding to its specific receptor, guanylyl cyclase (GC)-B. However, CNP/GC-B signaling dynamics in different stages of endochondral bone formation have not been fully clarified, especially in terms of the interaction between the cyclic guanine monophosphate (cGMP) and cyclic adenosine monophosphate (cAMP) pathways. Here, we demonstrated that CNP activates the cAMP/protein kinase A (PKA) pathway and that this activation contributed to the elongation of the hypertrophic zone in the growth plate. Cells of the chondrogenic line ATDC5 were transfected with Förster resonance energy transfer (FRET)–based cGMP and PKA biosensors. Dual-FRET imaging revealed that CNP increased intracellular cGMP levels and PKA activities in chondrocytes. Further, CNP-induced PKA activation was enhanced following differentiation of ATDC5 cells. Live imaging of the fetal growth plate of transgenic mice, expressing a FRET biosensor for PKA, PKAchu mice, showed that CNP predominantly activates the PKA in the hypertrophic chondrocytes. Additionally, histological analysis of the growth plate of PKAchu mice demonstrated that CNP increased the length of the growth plate, but coadministration of a PKA inhibitor, H89, inhibited the growth-promoting effect of CNP only in the hypertrophic zone. In summary, we revealed that CNP-induced cGMP elevation activated the cAMP/PKA pathway, and clarified that this PKA activation contributed to the bone growth–promoting effect of CNP in hypertrophic chondrocytes. These results provide insights regarding the cross-talk between cGMP and cAMP signaling in endochondral bone formation and in the physiological role of the CNP/GC-B system.
Keywords: C-type natriuretic peptide, endochondral bone formation, protein kinase A, live imaging, FRET biosensor
In vertebrates, long bones are formed through a process of endochondral bone formation. In this process, chondrocytes in the growth plate undergo dynamic changes in their biological and morphometric properties. Within the growth plate, resting chondrocytes, which are spherical and distally located immature chondrocytes, initially differentiate into proliferative chondrocytes, which represent flattened cell shapes and a columnar arrangement parallel to the longitudinal axis (1). The proliferative chondrocytes then actively proliferate and express chondrogenic markers, namely Sox9 and type II collagen (Col2a1). Subsequently, these chondrocytes exit the cell cycle and differentiate into the enlarged prehypertrophic and hypertrophic chondrocytes, followed by an increase in the expression of hypertrophic markers, such as Runx2 and type X collagen (Col10a1). After coordinated and sequential differentiation, mature chondrocytes undergo apoptotic cell death and are replaced by osseous tissue (2).
C-type natriuretic peptide (CNP) is a member of the natriuretic peptide family and has a prominent bone growth effect during the process of endochondral bone formation (3, 4). CNP rescues the bone growth defect known as achondroplasia, which is 1 of the most common forms of dwarfism (5). Recently, a biological analog of CNP was shown to have therapeutic efficacy in patients with achondroplasia in phase III trials (6). CNP and its receptor guanylyl cyclase (GC)-B are predominantly expressed in proliferative and prehypertrophic chondrocytes in the growth plate. Consistent with the gene expression pattern, the CNP/GC-B system enhances longitudinal bone growth by increasing chondrocyte proliferation, hypertrophy, and cartilage matrix production (4, 7). This biological action of CNP is considered to be mediated by the production of intracellular cyclic guanine monophosphate (cGMP) through the activation of GC-B. However, the precise molecular mechanisms and the dynamics of CNP/GC-B signaling in different stages of endochondral bone formation remain to elucidated.
Several reports have revealed that CNP induces signaling cross-talk between the cGMP and cyclic adenosine monophosphate (cAMP) pathways in gastric smooth muscle cells and heart muscle cells (8, 9). The role played by cAMP in the bone growth–promoting effect of CNP has not been fully clarified, especially in the context of endochondral bone formation. To address this issue, we employed Förster resonance energy transfer (FRET) biosensors, which are versatile tools for monitoring intracellular signal dynamics in living cells and tissues (10). Among the various cGMP biosensors, protein kinase G (PKG) #7 is 1 of the most highly sensitive (11). PKA is a major protein kinase activated by cAMP. Booster-PKA is a red FRET biosensor of PKA activity, which is compatible with cyan fluorescent protein (CFP)/yellow fluorescent protein (YFP)-based FRET biosensors (12). ATDC5 cells recapitulate the differentiation process of growth plate chondrocytes and thus provide a useful model for studying the growth plate chondrocytes in each differentiation stage (13, 14). In addition, by employing PKAchu mice, a transgenic mouse line systemically expressing a FRET biosensor for PKA (15), we were able to examine PKA activity at each differentiation stage of chondrocytes in living growth plates.
The results of dual-FRET imaging with ATDC5 cells expressing PKG #7 and Booster-PKA revealed that CNP-induced cGMP elevation activated the cAMP/PKA pathway. In addition, live imaging of the fetal growth plate of PKAchu mice demonstrated that CNP-induced PKA activation contributed to the bone growth–promoting effect of CNP in hypertrophic chondrocytes.
Materials and Methods
Cell Lines and Culture Conditions
ATDC5 cells were kindly provided by Professor Chisa Shukunami (Hiroshima University, Hiroshima, Japan). The cells were maintained in Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 (DMEM/F-12) (Gibco, New York, NY) containing 5% fetal bovine serum (Sigma, St. Louis, MO) (13, 14). The cells were plated at a density of 5.0 × 104 cells/cm2 in a 6-well plate for RNA-Seq and quantitative real-time reverse transcription polymerase chain reaction (RT-PCR), a 10-cm dish for western blotting, and a 24-well glass bottom plate (Iwaki, Tokyo, Japan) for imaging analysis. For induction of chondrogenesis, the cells were cultured in a differentiation medium which was composed of DMEM/F-12 containing 5% fetal bovine serum, 10 μg/mL insulin solution from bovine pancreas (Sigma-Aldrich, St. Louis, MO), 10 μg/mL human apo-transferrin (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan), and 3 × 10–8 M sodium selenite (FUJIFILM Wako Pure Chemical Corporation) from the cell seeding.
Plasmids and Establishment of Cell Lines
The cDNAs of PKA biosensor Booster-PKA (12), cGMP biosensor PKG #7 (11), and calcium biosensor RGECO1.0 (17) were inserted into the pCSIIpuro, pCSIIbsr, and pCSIIhyg lentiviral vectors (46), respectively. The packaging vector psPAX2 was purchased from Addgene (#12260, RRID:Addgene_12260) (47). pCSII and the envelope plasmid pCMV-VSV-G-RSV-Rev were gifted by Dr. Miyoshi (RIKEN BioResource Center). For lentiviral production, Lenti-X 293T cells were cotransfected with the pCSII plasmids, psPAX2, and pCMV-VSV-G-RSV-Rev with polyethylenimine (Polyscience Inc., Warrington, PA). ATDC5 cells were infected with lentiviruses encoding Booster and PKG #7, followed by drug selection with 4 μg/mL puromycin (InvivoGen, San Diego, CA) and 10 μg/mL blasticidin (FUJIFILM Wako Pure Chemical Corporation). ATDC5 cells were further infected with a lentivirus encoding RGECO1.0 and selected with 200 μg/mL hygromycin (FUJIFILM Wako Pure Chemical Corporation).
RNA-Seq and Quantitative RT-PCR
RNA was extracted from ATDC5 cells using an RNeasy Mini Kit (Qiagen, Hilden, Germany) and from radial, ulnar, and tibial growth plates of fetal mice using an RNeasy Micro Kit (Qiagen), and libraries for RNA-Seq were prepared using a NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (New England Biolabs, MA, USA) with the NEBNext Poly(A) mRNA Magnetic Isolation Module (New England Biolabs) and sequenced on the Illumina Nextseq500 as 75 bp single-end reads. RNA-Seq data were processed using the TrimGalore toolkit, which employs Cutadapt to trim low-quality bases and Illumina sequencing adapters from the 3′ end of the reads. The reads were mapped to a reference using HISAT2 software, and the resulting sam files were converted to bam files and sorted, then indexed using SAMtools. Relative abundances of genes were measured in FPKM using StringTie. Nucleotide sequence data reported are available in the DNA Data Bank of Japan Sequenced Read Archive under accession numbers DRR322305 to DRR322328 (48). For quantitative real-time RT-PCR, the extracted total RNA was reverse transcribed into cDNA using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). Quantitative real-time RT-PCR was performed using an Applied Biosystems StepOne Real-Time PCR System and Power SYBR Green PCR Master Mix (Thermo Fisher Scientific). The expression data were normalized to those of Hprt. The primers used in this analysis are as follows: Sox9 forward: 5′-ATCGGTGAACTGAGCAGCGAC-3′, Sox9 reverse: 5′-GCCTGCTGCTTCGACATCCA-3′, Col2a1 forward: 5′-TTGAGACAGCACGACGTGGAG-3′, Col2a1 reverse: 5′- AGCCAGGTTGCCATCGCCATA-3′, Runx2 forward: 5′- TCCACAAGGACAGAGTCAGATTAC-3′, Runx2 reverse: 5′- TGGCTCAGATAGGAGGGGTA-3′, Col10a1 forward: 5′- GCATCTCCCAGCACCAGA-3′, Col10a1 reverse: 5′- CCATGAACCAGGGTCAAGAA-3′, Npr2 forward: 5′- GAGGCCCTGCTGTACCAAATTCTA-3′, Npr2 reverse: 5′- GAAGCCCACGATGTCACTGAAG-3′, Pthrp forward: 5′- TTCAGCAGTGGAGTGTCCTG-3′, Pthrp reverse: 5′-AGCTCTGATTTCGGCTGTGT-3′, Pth1r forward: 5′-GCACTGCACGCGCAACTACA-3′, Pth1r reverse: 5′- ACCTGCGCGATGATATGCAAC-3′, Ihh forward: 5′-TGCATTGCTCTGTCAAGTCTG-3′, Ihh reverse: 5′- GCTCCCCGTTCTCTAGGC-3′, Pde3a forward: 5′- CGACTCCGATTCTGACAGTG-3′, Pde3a reverse: 5′- ATATTCCCAGACAGGCATCC-3′, Pde3b forward: 5′- CCTCAGGCAGTTTTATACAATG-3′, Pde3b reverse: 5′- TGCTTCTTCATCTCCCTGCTC-3′, Hprt forward: 5′- GGACCTCTCGAAGTGTTGGATAC-3′, Hprt reverse: 5′- GCTCATCTTAGGCTTTGTATTTGGCT-3′.
Western Blotting
Total membrane protein was extracted from ATDC5 cells using the MemPER Plus membrane protein extraction kit (Thermo Fisher Scientific). Lysates were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to Immobilon-FL PVDF membranes (Millipore, Bedford, MA). After blocking with Intercept TBS blocking buffer (LI-COR Biosciences, Lincoln, NE), the membranes were immunoblotted with the following primary antibodies: rabbit anti-NPR-B (1:1000, Abcam, Cat# ab139188, RRID:AB_2895557) (49); rabbit anti-β-Actin (1:1000, Cell Signaling Technology, Cat# 4970, RRID:AB_2223172) (50). Fluorescence was detected with IRDye 800CW goat antirabbit IgG secondary antibody (1:10 000; LI-COR Biosciences, Cat# 926-32211, RRID:AB_621843) (51). Images were obtained using an Odyssey infrared image system (LI-COR Biosciences). Western blot quantification was performed using ImageJ software.
Confocal and Wide-field Fluorescence Imaging of Cultured Cells
The ATDC5 cells expressing biosensors were plated on a 24-well glass bottom plate coated with collagen type I at a density of 5.0 × 104 cells/cm2. The cells were starved for 1 hour with phenol red–free DMEM/F12 medium containing 0.5% bovine serum albumin. Starved cells were treated with each concentration of CNP (LKT Laboratories, St. Paul, MN), following the addition of 32 μM 1,2-Bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester) (BAPTA/AM) (Enzo Life Sciences, Farmingdale, NY) or 1 μM cilostamide (Abcam), if necessary. Confocal fluorescence images were obtained with an IX81 inverted microscope (Olympus, Tokyo, Japan) equipped with an FV1000 confocal imaging system (Olympus) and a 40×/0.90 NA dry objective lens (UPLSAPO 40X; Olympus) under the following conditions. For multiplexed FRET imaging, a 405-nm excitation laser was used for CFP and YFP and a 559-nm excitation laser was used for mKOκ and mKate2, and a DM 405/488/559 dichroic mirror, and 460 to 490, 520 to 550, 570 to 625, and 655 to 755 nm were applied for CFP, YFP, mKOκ, and mKate2, respectively. Wide-field fluorescence images were acquired with an IX83 inverted microscope (Olympus) equipped with a 20×/0.75 NA dry objective lens (UPLSAPO 20X; Olympus) under the following conditions. For RGECO1.0 imaging, a 575/25 excitation filter (incorporated in the Spectra-X light engine), a glass dichromatic mirror (Olympus), and FF01-624/40-25 (Semrock, Rochester, NY) emission filters were used. The imaging data are available in the Systems Science of Biological Dynamics: repository (52).
Organ Culture and Live Imaging of Radial Growth Plates
PKAchu mice were crossed with B6N-Tyrc-Brd/BrdCrCrl mice (Japan SLC, Shizuoka, Japan). On day 17 of pregnancy, mice were sacrificed by cervical dislocation. Bilateral radial bones were aseptically dissected from mouse fetuses. Organ cultures of fetal mouse radial bones were performed using the static culture technique in BGJb Medium, Fitton–Jackson Modified without Phenol Red (United States Biological, Swampscott, MA) containing 6 mg/mL bovine serum albumin (Wako Pure Chemical Industries), 150 μg/mL ascorbic acid (Wako Pure Chemical Industries), 100 units/mL penicillin, and 100 μg/mL streptomycin (Nacalai Tesque, Kyoto, Japan). Radial explants were incubated in CELLview, 4-compartment 35 × 10 mm glass bottom dishes (Greiner Bio-One, Frickenhausen, Germany). The metaphysis of the specimen was fixed by agarose gel to avoid artificial movement. Proximal growth plates were observed using an incubator-integrated multiphoton fluorescence microscope system (LCV-MPE; Olympus) with a 25×/1.05 NA water immersion lens (XLPLN25XWMP2; Olympus). The excitation wavelength was set to 840 nm (InSight DeepSee, Spectra-Physics) and the filter sets used were as follows: IR cut filter, RDM690 (Olympus); dichroic mirrors, DM505 and DM570 (Olympus); and emission filters: BA460-500 for CFP and BA520-560 for FRET (Olympus). Radial explants were treated with vehicle or CNP (100 nM), following the addition of vehicle, H89 (20 nM), or 100 nM parathyroid hormone–related protein (PTHrP) (hypercalcemia of malignancy factor fragment 1-34 amide human, Sigma-Aldrich). The imaging data are available in the SSBD:repository (52). All experimental procedures using mice were performed in accordance with the ethical guidelines of Kyoto University, and this study was approved by the Animal Research Committee, Graduate School of Medicine, Kyoto University (Med Kyo19090, 20081, and 21043).
Histology and Definition of Resting, Proliferative, Prehypertrophic, and Hypertrophic Zones
Fetal mouse radial bones were fixed in 4% formaldehyde neutral buffer solution (Wako Pure Chemical Industries) for 24 hours, then embedded in O.C.T Compound (Sakura Finetek, Torrance, CA) and frozen at –20°C. Sections (10 μm thick) were cut from OCT-embedded specimens and stained using immunofluorescence. The concentration of primary antibodies for the immunofluorescence are rabbit anticollagen X antibody (1:100, Abcam, Cat# ab58632, RRID:AB_879742) (53) and mouse anti-NPR-B (1:50, Santa Cruz Biotechnology, Cat# sc-293451, RRID:AB_2847874) (54). Fluorescence was detected with Alexa Fluor 568 goat antirabbit (1:1000; Molecular Probes, Cat# A-11036, RRID:AB_10563566) (55) and Alexa Fluor 555 goat antimouse (1:1000; Molecular Probes, Cat# A-21424, RRID:AB_141780) (56). Each segmented area of the growth plate was defined by a comparison between the histology acquired from static and live imaging analysis. Areas composed of enlarged cytoplasm were defined as hypertrophic zones (HZs). Areas adjoining an HZ and without enlargement of cytoplasm or columnar array structures were defined as prehypertrophic zones. Areas adjacent to a prehypertrophic zone and composed of flattened and columnar arrayed cells were defined as proliferative zones. Areas adjacent to a proliferative zone and composed of randomly arranged spherical cells were defined as resting zones (57). In this study, prehypertrophic zones were included in HZs, and proliferative and resting zones were included in nonhypertrophic zones (non-HZs), following the definition in previous studies (58). The widths of the HZs and non-HZs were analyzed using MetaMorph software (Molecular Devices, Sunnyvale, CA). The fluorescence intensity of type X collagen was measured using line scan analysis. Intensity data were smoothed with a 35 pixel moving average and differentiated. We defined the proximal peak of the intensity derivative as the end of the HZs and the proximal dip as a border between the HZs and non-HZs. Consistent with the distal peak of the intensity moving average, the point of the intensity derivative whose value is 0 was defined as the end of the epiphysis. The width of the HZ was set as the difference between the end of the HZ and the border between the hypertrophic and non-HZs, and the width of the proliferative zone was set as the difference between the border between the hypertrophic and non-HZs and the end of the epiphysis.
Statistical Analysis
All statistical analyses were performed using Excel, and an R. Shirley–Williams test was used to evaluate statistically significant differences in the results of quantitative RT-PCR in Fig. 2. Student’s t-test was used for the analyses in Figs. 3C-3F, and 4G, 4L, and 4M. One-way factorial analysis of variance followed by the Turkey–Kramer test were used for the other experiments. P < 0.05 was considered to indicate statistical significance.
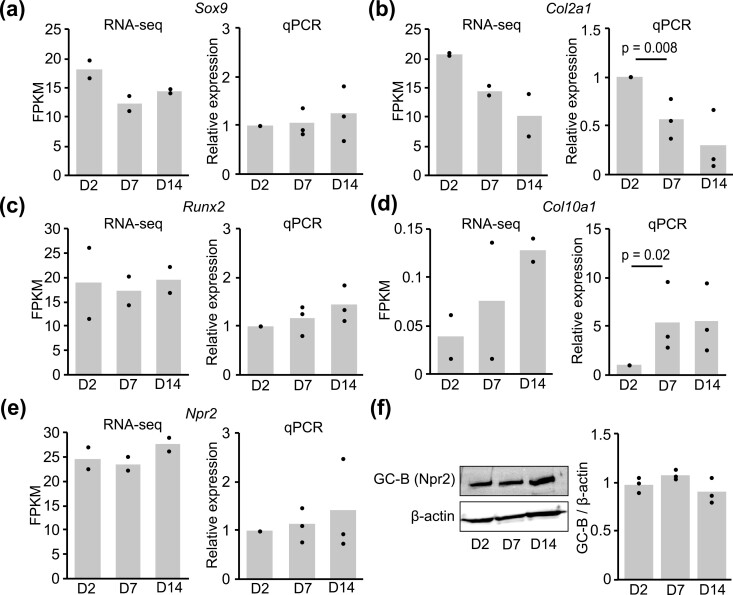
Figure 2.
Expression of chondrogenic differentiation marker genes. FPKM values and relative expression levels of Sox9 (A), Col2a1 (B), Runx2 (C), Col10a1 (D), and Npr2 (E) on days 2, 7, and 14 after induction of differentiation were obtained by RNA-Seq and quantitative real-time RT-PCR. In the results of RNA-Seq, each column represents the mean of 2 independent experiments, and plots represent each sample. In the results of RT-PCR, each column represents the mean of 3 independent experiments, and plots represent the mean of duplicate samples. Data are normalized against Hprt. (F) Total membrane proteins were extracted from ATDC5 cells on days 2, 7, and 14 after induction of differentiation and subjected to western blotting. The protein levels of GC-B (Npr2) were obtained and densitometry was carried out to evaluate the ratio of the intensity of the signals corresponding to GC-B to that of β-actin in the cells. Each column represents the mean of 3 independent experiments, and plots represent each sample.
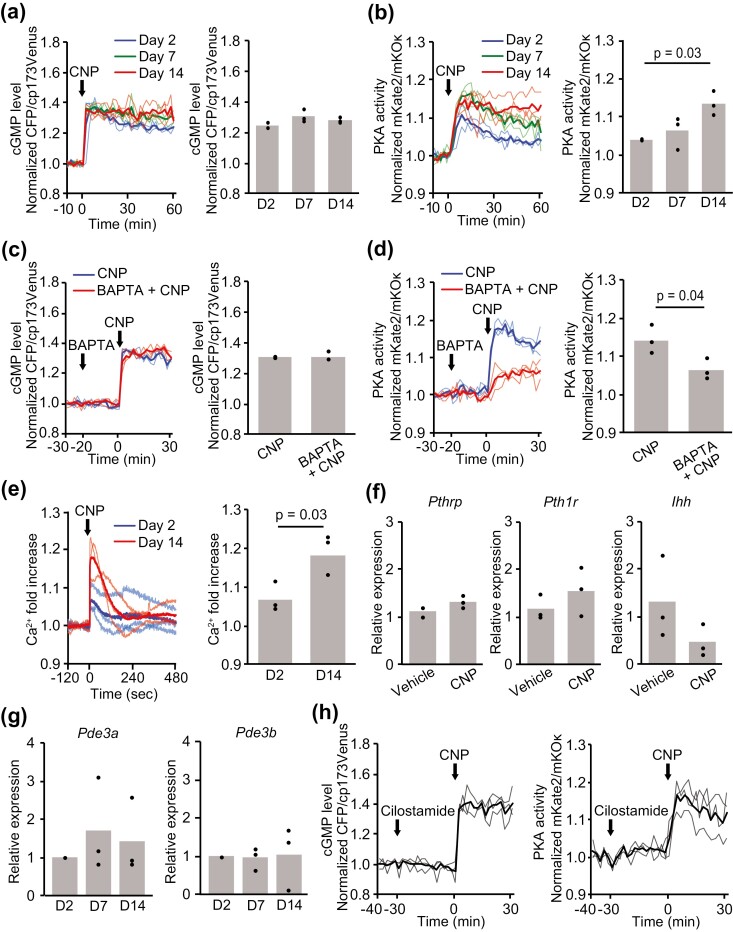
Figure 3.
Cell responses to CNP in each chondrogenic differentiation stage. (A, B) ATDC5 cells coexpressing cGMP and PKA biosensors were stimulated with CNP (100 nM) on days 2, 7, and 14 after the induction of differentiation. (A) Time course of the intracellular cGMP level represented as CFP/cp173Venus ratios and values at 60 minutes after CNP stimulation. (B) Time course of PKA activity represented as mKate2/mKOκ ratios and values at 60 minutes after CNP stimulation. CFP/cp173Venus and mKate2/mKOκ ratios are normalized to the average before CNP stimulation. (C, D) On day 14, ATDC5 cells coexpressing cGMP and PKA biosensors were stimulated with CNP (100 nM), following the addition of BAPTA-AM (32 μM). (C) Time course of the intracellular cGMP level represented as the CFP/cp173Venus ratio and (D) time course of PKA activity represented as the mKate2/mKOκ ratio normalized to the average before CNP stimulation. (E) ATDC5 cells expressing the Ca2+ biosensor were stimulated with CNP (100 nM) on days 2 and 14. The time course of the normalized fold increase of Ca2+ and the peak values after CNP stimulation are shown. Values were normalized to the average before CNP stimulation. (F) The relative expression levels of Pthrp, Pth1r, and Ihh in ATDC5 cells after 24 hours of treatment with CNP (100 nM) were obtained on day 14 by quantitative real-time RT-PCR. (G) The relative expression levels of Pde3a and Pde3b on days 2, 7, and 14 after induction of differentiation were obtained by quantitative real-time RT-PCR. (H) On day 14, ATDC5 cells coexpressing cGMP and PKA biosensors were stimulated with CNP (100 nM), following the addition of cilostamide (1 μM). Time course of the intracellular cGMP level represented as the CFP/cp173Venus ratio and time course of PKA activity represented as the mKate2/mKOκ ratio normalized to the average before cilostamide stimulation. In (A-G), each column represents the mean of 3 independent experiments, and plots represent the value of each sample.
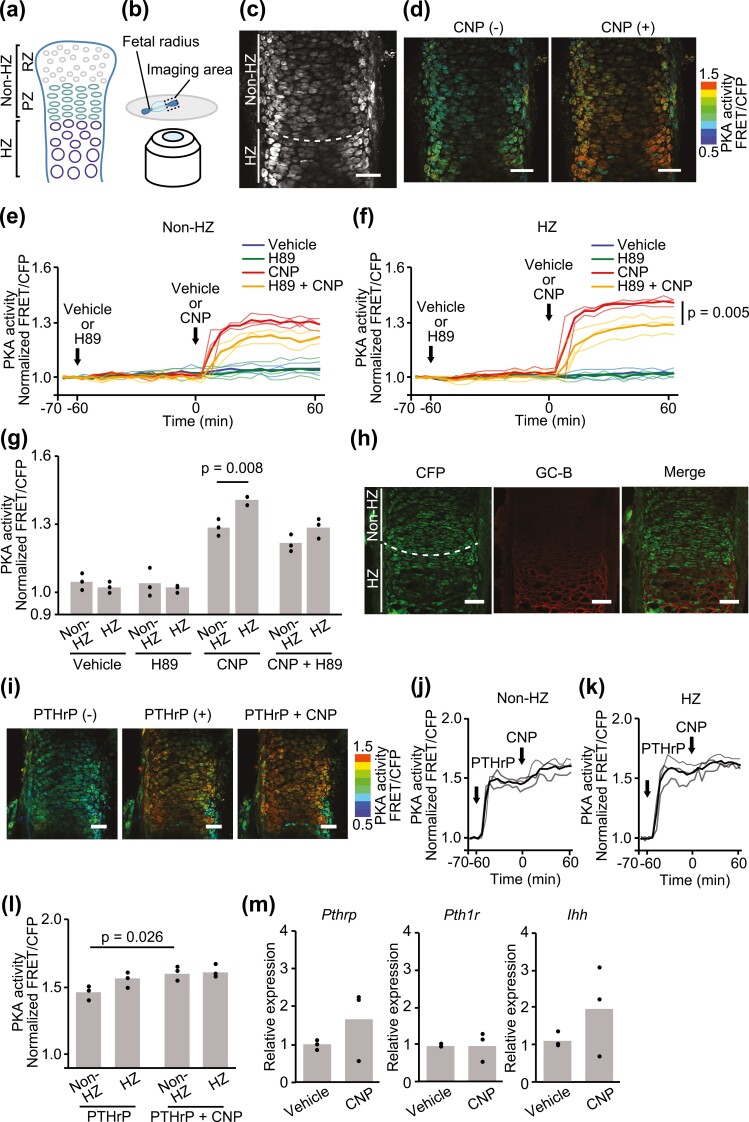
Figure 4.
PKA response to CNP in the PKAchu growth plate. (A) Structure of the growth plate. The growth plate was composed of resting (RZ), proliferative (PZ), prehypertrophic, and hypertrophic (HZ) zones. In this study, RZ and PZ were included in the nonhypertrophic zone (non-HZ) and the prehypertrophic zones were included in the HZ. (B) A schematic diagram of the imaging system. Radial explants of PKAchu mice were fixed on the glass bottom dish using agarose. Proximal growth plates were observed using a multiphoton fluorescence microscope under ex vivo culture conditions. (C) CFP image of the fetal proximal radial growth plate of PKAchu mice. A white dashed line divides the growth plate into the non-HZ and HZ as indicated. (D) The radial growth plates of PKAchu mice were stimulated with CNP (100 nM). FRET/CFP ratio images are shown in the IMD mode. Bar, 50 μm. (E-G) The radial growth plates of PKAchu mice were stimulated with CNP (100 nM), following the addition of vehicle or H89 (20 nM). (E, F) Time course of PKA activity represented as the normalized FRET/CFP ratio in the (E) non-HZ and (f) HZ. (G) Normalized FRET/CFP ratios at 60 min. (h) GC-B staining of the proximal radial growth plates at E17.5. Bar, 50 μm. (I-L) The radial growth plates of PKAchu mice were stimulated with CNP (100 nM), following the addition of PTHrP (100 nM). (I) FRET/CFP ratio images are shown in the IMD mode. Bar, 50 μm. (J, K) Time course of PKA activity represented as the normalized FRET/CFP ratio in the (J) non-HZ and (K) HZ. (G) Normalized FRET/CFP ratios at 0 minutes (PTHrP) and 60 minutes (PTHrP + CNP). (M) The relative expression levels of Pthrp, Pth1r, and Ihh in growth plate chondrocytes after 24 hours of treatment with CNP (100 nM) under organ culture conditions were obtained by quantitative real-time RT-PCR. Each column represents the mean of 3 independent experiments, and plots represent the value of each sample. FRET/CFP ratios are normalized to the average value before drug treatment.
Results
CNP Activated cGMP and cAMP/PKA Signaling in Chondrocytes
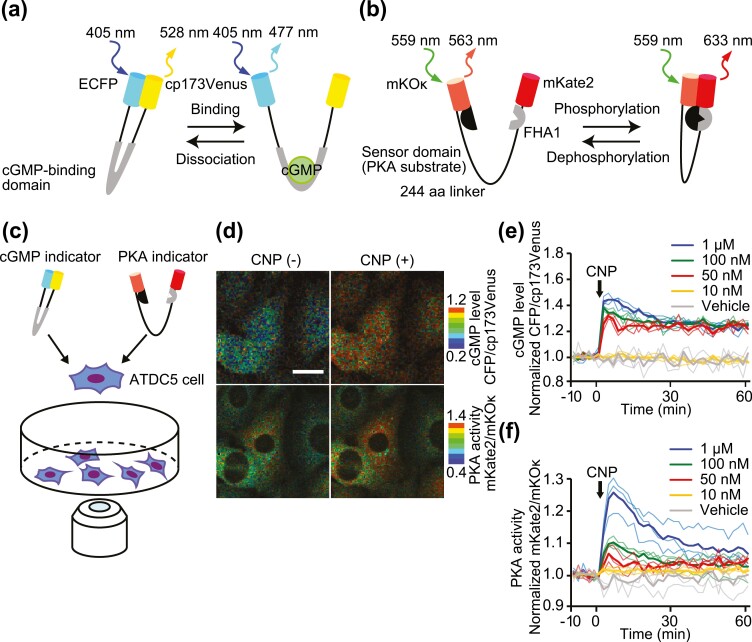
To verify the interaction between CNP-induced cGMP and cAMP/PKA dynamics in chondrocytes at the single cell level, the FRET-based cGMP biosensor PKG #7 (11) and PKA sensor Booster-PKA (12) were coexpressed in the mouse chondrogenic cell line ATDC5. PKG #7 is composed of cGMP-binding domain B from cGMP-dependent protein kinase IB, which binds specifically to intracellular cGMP, and of the ECFP and cp variant of Venus, cp173Venus, as the FRET donor and acceptor, respectively (Fig. 1A). Booster-PKA comprises the FRET acceptor mKate2, an FHA1 phosphothreonine–binding domain, a 244 amino acid flexible linker, the FRET donor mKOκ, and a PKA substrate peptide. PKA activation induces conformational change of the PKA substrate peptide and binding to the FHA1 domain (Fig. 1B). Biosensor-expressing ATDC5 cells were observed under a confocal microscope to obtain the fluorescent intensity ratio of acceptor vs donor and thereby monitor intracellular cGMP concentration and PKA activity (Fig. 1C). Upon CNP stimulation, rapid increases in cGMP and PKA activity were represented by the ratio of CFP to cp173Venus and that of mKate2 to mKOκ, respectively (Fig. 1D). Next, ATDC5 cells were stimulated with increasing concentrations of CNP and observed for 60 minutes. Interestingly, cGMP exhibited an almost all-or-none response with a threshold between 10 nM to 50 nM. Meanwhile, PKA activity increased in a dose-dependent manner from 50 nM to 1 µM (Fig. 1E and 1F). These results imply that CNP may transduce a signal to the PKA pathway downstream of cGMP.
Figure 1.
Dual-FRET imaging of cGMP concentration and PKA activity in ATDC5 cells. (A, B) Mechanism of action of the FRET-based cGMP (A) or PKA (B) biosensors. (A) cGMP binding to the cGMP biosensor induces conformational change and decreases in FRET. (B) Phosphorylation of the PKA biosensor by PKA induces increases in FRET. (C) Schematic diagram of the live cell imaging of ATDC5 cells co-expressing cGMP and PKA biosensors using the confocal microscope. (D) ATDC5 cells coexpressing cGMP and PKA biosensors were stimulated with CNP (100 nM). Images of the CFP/cp173Venus (upper panels) and mKate2/mKOκ (lower panels) ratios are shown in the IMD mode. Bar, 20 μm. (E, F) Time course of the CFP/cp173Venus (E) and mKate2/mKOκ (F) ratios normalized to the average before CNP stimulation. The cells were stimulated by each concentration of CNP. Solid lines represent the means; translucent lines represent the value of each experiment (n = 3).
Hypertrophic Differentiation of ATDC5 Cells as Examined by the Marker Genes
ATDC5 cells recapitulate the differentiation process of growth plate chondrocytes, providing a useful model for studying the growth plate chondrocytes in each differentiation stage. To monitor the differentiation stage of ATDC5 cells, RNA-Seq and quantitative real-time RT-PCR analyses were conducted (Fig. 2A-2E). The results were represented as FPKM (fragments per kilobases per million reads) and relative expression levels normalized to Hprt. Among the known chondrogenic markers, namely Sox9, Col2a1, Runx2, and Col10a1, the level of Col2a1, which is expressed in immature chondrocytes, was decreased (Fig. 2B) and the level of Col10a1, which is expressed in hypertrophic chondrocytes, was increased (Fig. 2D) at day 7. Meanwhile, expression and protein levels of the CNP receptor GC-B (Npr2) were not significantly altered during differentiation (Fig. 2E and 2F). Taken together, these results indicated that ATDC5 cells differentiated into hypertrophic chondrocytes during the 14-day differentiation.
Differential Responses to CNP in each Chondrogenic Differentiation Stage
To clarify whether CNP-induced cGMP elevation or PKA activation is affected by the differentiation stage, ATDC5 cells were treated with 100 nM CNP and observed for 60 minutes at 2, 7, and 14 days after differentiation induction. The cGMP level was rapidly increased in all differentiation stages after CNP stimulation (Fig. 3A). There was no significant difference in cGMP level among the differentiation stages. Meanwhile, CNP-induced PKA activation was significantly enhanced during differentiation (Fig. 3B). This all-or-none response of cGMP and gradual increase in PKA activity resembled the dose response at day 2 (Fig. 1E and 1F).
Intracellular Ca2+ elevation has been shown to stimulate cAMP production (16). Therefore, we hypothesized that CNP would activate PKA through the elevation of cytoplasmic Ca2+ levels. Thus, ATDC5 cells at day 14 were treated with the calcium chelator BAPTA/AM and stimulated with CNP. Although cells treated with BAPTA/AM exhibited a faint cGMP response (Fig. 3C), their PKA activation was almost canceled (Fig. 3D). This observation suggests a relationship between the calcium response of cells and their differentiation. To examine this possibility, we used the calcium biosensor R-GECO1.0 (17). In accordance with CNP-induced PKA activation during the differentiation process, CNP significantly increased intracellular Ca2+ levels after the induction of differentiation (Fig. 3E).
It is well established that PTHrP and its receptor parathyroid hormone 1 receptor (PTH1R) regulate endochondral bone formation via the cAMP/PKA pathway (1, 18). Nevertheless, CNP did not affect the expression levels of Pthrp, Pth1r, and Ihh in ATDC5 cells on day 14 (Fig. 3F). CNP increases intracellular cAMP via inhibition of phosphodiesterase (PDE) 3, which is a dual-specificity phosphodiesterase (8, 9). However, the expression levels of Pde3a and Pde3b were not altered during differentiation (Fig. 3G). Further, PDE 3 inhibitor cilostamide did not apparently change the CNP-induced cGMP elevation and PKA activation (Fig. 3H). Collectively, these results indicate that CNP-induced elevation of cytoplasmic Ca2+ activated the cAMP/PKA pathway, and the calcium response was enhanced upon differentiation of chondrocytes.
PKA Response to CNP in the Growth Plate
We next examined whether CNP activates PKA in the mouse growth plate using PKAchu mice, a transgenic mouse line expressing the PKA biosensor AKAR3EV (15). The growth plate consists of resting zones, proliferative zones, prehypertrophic zones, and HZs. In this study, resting and proliferative zones were analyzed collectively as the non-HZ, and the prehypertrophic and HZs were analyzed collectively as the HZ (Fig. 4A). Fetal radial explants of PKAchu mice were observed under a 2-photon excitation microscope to monitor PKA activity (Fig. 4B). The non-HZ and HZ were distinguished by cell size, shape, and alignment in fluorescence images (Fig. 4C). Radial explants pretreated with the PKA inhibitor H89 or vehicle alone were stimulated with CNP (Fig. 4D). In the non-HZ, PKA activity was increased after CNP addition, and H89 partially attenuated its activation. The values of PKA activity at 60 minutes were significantly increased in the CNP-treated group compared with the vehicle-treated group. H89 pretreatment attenuated PKA activation of the CNP-treated group, though the difference in PKA activation between the H89 + CNP–treated group and CNP-treated group did not reach the level of statistical significance (Fig. 4E). Similarly, but unlike in the non-HZ, the attenuation of CNP-induced PKA activation by H89 pretreatment reached the level of statistical significance in the HZ (Fig. 4F). Furthermore, the peak value of PKA activity was higher in the HZ than in the non-HZ in the CNP-treated groups (Fig. 4G). The distribution of GC-B was identified by immunofluorescence staining. Consistent with CNP-induced PKA activation, GC-B was mainly detected in the HZ (Fig. 4H).
The effect of PTHrP on CNP-induced PKA activation is of great interest and was demonstrated by live imaging of the fetal radial explants of PKAchu mice. Radial explants pretreated with PTHrP were stimulated with CNP (Fig. 4I). PKA activities were predominantly increased in the HZ after PTHrP addition, though the difference between the non-HZ and HZ was not statistically significant. Subsequently, CNP slightly increased its activation only in the non-HZ (Fig. 4J-L). In agreement with the result of the ATDC5 cells, CNP did not alter the expression levels of Pthrp, Pth1r, and Ihh in cultured explants of fetal growth plates (Fig. 4M).
Collectively, these results clarified that, even under an ex vivo conditions, CNP-induced PKA activation was increased following differentiation of growth plate chondrocytes and significantly attenuated by H89 in hypertrophic chondrocytes. Further, PTHrP-induced PKA activation was revealed to be potent, and additional PKA activation by CNP was found to be small.
Effect of PKA Inhibitor on the Growth Plate
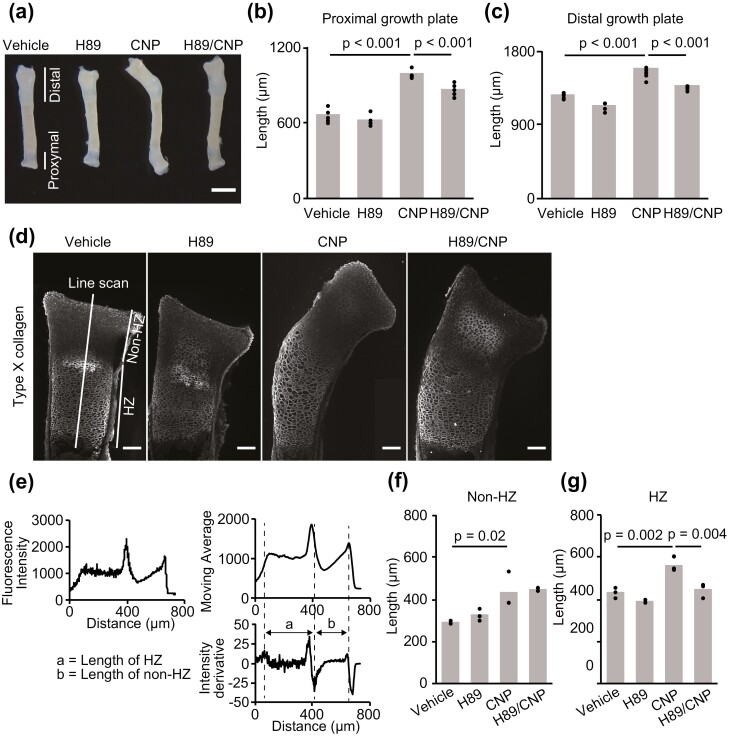
To examine the physiological role of CNP-induced PKA activation on bone growth, the radial explants were incubated in the presence or absence of CNP and/or H89 for 3 days. CNP induced marked elongation of both the proximal and distal growth plates (Fig. 5A-5C). This CNP-induced elongation could be attenuated by H89, indicating that CNP promotes elongation of the growth plate at least partially through PKA activation. To further delineate the region responsible for CNP-induced growth plate elongation, we identified hypertrophic chondrocytes by immunostaining with anti-Col10a1 antibody (Fig. 5D) and measured the lengths of the non-HZ and HZ as described in the “Materials and Methods” (Fig. 5E). Consequently, we found that the lengths of both the non-HZ and HZ were increased in a CNP-dependent manner (Fig. 5F and 5G). However, only the HZ was sensitive to the PKA inhibitor H89, indicating that CNP-induced PKA activation partially contributed to the growth-promoting effect of CNP in the HZ.
Figure 5.
Histological analysis of the growth plate of PKAchu mice. (A) Gross appearance of radial explants incubated with vehicle, H89, CNP, or both H89 and CNP (H89/CNP) for 3 days. Bar, 1 mm. (B, C) Length of the (B) proximal and (C) distal growth plates of the vehicle, H89, CNP, or H89/CNP-treated groups at the end of the organ culture period. Each column represents the mean of 5 or 6 independent experiments, and plots represent the value of each sample. (D) Type X collagen staining of the proximal growth plates of each group at the end of the organ culture period. Fluorescence intensity along the long axis of the proximal growth plate was acquired using the line scan function in MetaMorph. Bar, 50 μm. (E) Scheme of the measurement of histological sections. The value of fluorescence intensity is shown in the left upper panel. Right upper and lower panels: moving average of fluorescence intensity and its derivative. Dashed lines represent the end of the HZ, the border between the HZ and non-HZ, and the end of the epiphysis, respectively. The length of the HZ is shown with the letter a and the length of the non-HZ is shown with the letter b in the right lower panel. (F, G) Length of the (F) non-HZ and (G) HZ of the vehicle, H89, CNP, and H89/CNP-treated groups. Each column represents the mean of 3 independent experiments, and plots represent the value of each sample.
Discussion
In this study, we revealed that CNP-induced PKA activation was enhanced following the chondrogenic differentiation and this activation contributed to the elongation of the HZ of the growth plate. ATDC5 cells coexpressing biosensors for intracellular cGMP concentration and PKA activity visualized the dynamics of intracellular cGMP levels and PKA activity in CNP-stimulated chondrocytes at a single cell level (Figs. 1D-F). CNP binds to its specific receptor GC-B and exerts physiological effects by increasing intracellular cGMP levels and activating type II cGMP dependent protein kinase (PKG II) (4, 19). The bone growth–promoting effect of CNP is generally thought to be mainly mediated by an inhibition of the ERK pathway by PKG II at the level of Raf-1, although smaller contributions from the p38 and Akt/GSK3β pathways have also been reported (5, 20-23). Further, an interaction between cGMP and cAMP signaling has been demonstrated in CNP-stimulated gastric tract and CNP-stimulated heart. CNP-induced cGMP elevation inhibits PDE 3, and increases intracellular cAMP, resulting in attenuation of gastric contraction and enhancement of β-adrenoceptor–mediated lusitropic and inotropic responses in the heart (8, 9). The interaction between cGMP and cAMP signaling in the bone growth–promoting effect of CNP has not been fully elucidated. However, we herein demonstrated that CNP activated the cAMP/PKA pathway concomitant with the elevation of cGMP in chondrocytes.
In the present study, CNP-induced cGMP elevations were not significantly different among the differentiation stages (Fig. 3A), whereas the PKA response to CNP was increased in hypertrophic chondrocytes (Fig. 3B). Moreover, expression and protein levels of the GC-B were not altered during differentiation (Fig. 2E and 2F). These results suggest that CNP-induced cAMP/PKA activation is enhanced downstream of cGMP following the differentiation of chondrocytes. The production of cAMP is regulated by intracellular calcium concentrations. Adenylate cyclase (AC) synthesizes cAMP from ATP, and its activity is regulated by G protein subunits, serine/threonine and tyrosine protein kinases, and Ca2+ (16). AC has 9 membrane-bound and 1 soluble isoform, and only AC1, AC3, and AC8 are stimulated by calmodulin in a Ca2+-dependent manner (24-26). Therefore, we hypothesized that CNP increases intracellular Ca2+, resulting in cAMP/PKA activation. Intriguingly, CNP-induced cGMP elevation was not affected, whereas PKA activation was attenuated by the chelation of intracellular Ca2+ by BAPTA-AM in ATDC5 cells (Figs. 3C, 3D). Additionally, CNP-induced Ca2+ elevation was enhanced by hypertrophic differentiation in ATDC5 cells expressing RGECO1.0 (Fig. 3E). CNP-induced Ca2+ elevation has been reported in renal, pituitary, and cardiac cells (27-30). In cardiomyocytes, CNP-induced cGMP elevation activates sarcoplasmic reticulum Ca2+ ATPase2 (SERCA2) via phospholamban Ser16 phosphorylation and increases the amplitude of the Ca2+ transient in response to electrical stimulation (29, 30). Similarly, mobilization of Ca2+ from intracellular stores is presumed to contribute to the CNP-induced Ca2+ elevation in renal and pituitary cells (27, 28). These results suggest that CNP mobilizes Ca2+ from intracellular stores of chondrocytes. CNP-induced SERCA2 activation is mediated by the cGMP/type I cGMP–dependent protein kinase (PKGI) pathway (30). In the growth plate, PKGII is broadly distributed but predominantly expressed in the border between proliferative and HZs. Meanwhile, PKGI expression is confined to the HZ (31) and coincides with enhancement of Ca2+ elevation in the HZ. This might account for enhancement of the effect of CNP on Ca2+ and PKA signaling. Mice with knockout of either PKGI (32) or PKGII (31) would provide a better understanding of this finding. On the other hand, CNP did not affect the expression levels of Pthrp, Pth1r, and Ihh in ATDC5 cells and cultured explants of fetal growth plates (Figs. 3F and 4M). Expression levels of Pde3a and Pde3b were also not altered during differentiation (Fig. 3G), and cilostamide did not affect basal PKA activity and CNP-induced PKA activation in ATDC5 cells (Fig. 3H). These findings indicated that PTHrP/PTH1R signaling and PDE 3 inhibition were not involved in the mechanism of CNP-induced PKA activation during chondrogenic differentiation. Collectively, these results led us to conclude that CNP-induced Ca2+ elevation mediates cAMP production downstream of cGMP, and this effect is enhanced by hypertrophic differentiation.
Live imaging of fetal radial growth plates of PKAchu mice revealed that CNP activated the cAMP/PKA pathway predominantly in the HZ (Fig. 4E-4G). In a previous study, we established a live-imaging system of growth plate cartilage in cultured murine long bones using 2-photon excitation microscopy and demonstrated that CNP mainly affects the HZ and promotes growth plate elongation (33). Furthermore, we generated transgenic mice expressing FRET biosensors for PKA, PKAchu mice, which allowed us to monitor PKA activity in vivo (15). By adapting PKAchu mice to the live-imaging system of growth plate cartilage, the PKA response to CNP was successfully measured in different differentiation stages of the growth plate (Fig. 4E and 4F). This approach revealed that CNP activated PKA predominantly in hypertrophic chondrocytes, which agreed with the results obtained from ATDC5 cells (Fig. 4G). To our knowledge, this is the first demonstration of PKA activation mediated by CNP in the growth plate. Recently, Yamamoto et al revealed that cyclic AMP–responsive element binding protein (CREB), which is activated by PKA-mediated phosphorylation following cAMP accumulation, is phosphorylated by GC-B activation, resulting in an increase in cyclin D1 expression and persistence of proliferation in matured chondrocytes. They also demonstrated that a cell permeable cGMP analog, 8-Br-cGMP, induced phosphorylation of CREB (34). This is in agreement with the pattern of PKA activation observed in this study. However, cAMP production was not affected by CNP treatment in an enzyme immunoassay using primary chondrocytes isolated from ribcages in their study. This difference between their study and ours might be attributable to the different cell types or the different detection limits of the methods employed. As for CNP/GC-B signaling in the growth plate, Shuhaibar et al reported that cGMP production via the CNP/GC-B system is decreased by fibroblast growth factor receptor 3–mediated dephosphorylation of GC-B, based on live imaging of a tibia isolated from transgenic mice expressing the cGMP biosensor, cGi500 (35, 36). In their report, cGMP production was collectively measured in proliferative and prehypertrophic zones that express both GC-B and fibroblast growth factor receptor 3, and CNP-induced cGMP elevation was robustly observed in this region. In our study, GC-B was mainly detected in the HZ of the growth plate (Fig. 4H), even though protein levels of the GC-B and CNP-induced cGMP elevation did not change at each differentiation stage in ATDC5 cells (Figs. 2F and 3A). Although the results obtained from ATDC5 cells suggested that CNP-induced PKA activation was enhanced downstream of cGMP, the use of transgenic mice expressing the cGMP biosensor, as demonstrated in their study, would allow the analysis of the discrete differences in cGMP dynamics between the non-HZ and HZ in the growth plate, and could uncover additional insights into the interaction between cGMP and cAMP/PKA signaling in endochondral bone elongation through the CNP/GC-B system.
Finally, histological analysis revealed that PKA activation contributes to CNP-induced elongation of the growth plate in the HZ (Fig. 5D and 5G). Regarding the physiological role of CNP in bone growth, systemic and cartilage-specific CNP or GC-B knockout mice both exhibit a prominent short stature phenotype due to impaired skeletal growth, whereas transgenic mice with cartilage-specific overexpression of CNP or increased circulating levels of CNP exhibit a skeletal overgrowth phenotype (7, 37). In humans, homozygous loss-of-function mutations in the GC-B gene cause acromesomelic dysplasia–type Maroteaux, a form of skeletal dysplasia characterized by severely impaired endochondral bone growth, as well as heterozygous mutations in GC-B, which have been associated with idiopathic short stature (38-40). Conversely, both gain-of-function mutations in GC-B and expression of CNP by balanced chromosomal translocations have been reported to cause skeletal overgrowth in humans (41, 42). Collectively, these results underscore that CNP is a crucial enhancer of endochondral bone growth in rodents and humans.
A previous report revealed that CNP increases the length of the proliferative zone and HZ by promoting the proliferation and hypertrophic differentiation of the growth plate chondrocytes. This growth-promoting effect is predominantly observed in the HZ and mimicked by 8-bromo-cGMP (4). Therefore, cGMP production via activation of the CNP/GC-B system is considered to be a potent regulator of endochondral bone formation, especially hypertrophic differentiation. Regarding this matter, Miyazawa et al reported that PKGII knockout mice show a disorganized growth plate which is characterized by irregular and broadened HZs with mixed nonhypertrophic and hypertrophic chondrocytes (19). This abnormal structure is caused by impairment of the function of PKGII which promotes differentiation from proliferative to hypertrophic chondrocytes (22). PKGII knockout mice show high levels of cGMP (19). These results suggested that compensatory elevated cGMP stimulates hypertrophy of the chondrocytes in the disorganized HZ via unimpeded PKGI and downstream PKA activation, and supported the growth-promoting effect of CNP-induced cAMP/PKA activation in the HZ.
Further, we demonstrated that PTHrP increased PKA activity predominantly in the HZ, though the difference between the non-HZ and HZ was not statistically significant (Fig. 4J-4L). This result was consistent with the distribution of PTH1R, which is mainly localized in the HZ (43). Intriguingly, CNP additionally increased PKA activity only in the non-HZ. As shown by the range of increase in PKA activity, PTHrP potently activated PKA compared with CNP. However, PKA activation in the non-HZ was weaker than in the HZ, suggesting that there was room for additional PKA activation by CNP. In the process of endochondral bone formation, the role of the cAMP/PKA pathway has been studied from the perspective of the PTHrP and its receptor PTH1R in detail (18, 44, 45). PTHrP-induced cAMP/PKA activation increases chondrocyte proliferation and delays hypertrophic differentiation (18). In addition, PTHrP decreases apoptosis of the hypertrophic chondrocytes by an increase in expression of Bcl-2 (44) under the control of Zfp521 (45). CNP increases proliferation and decreases apoptosis of growth plate chondrocytes (4, 34). Collectively, this evidence and our present results lead us to propose that CNP-induced cAMP/PKA activation promotes elongation of the HZ by increasing proliferation and survival of hypertrophic chondrocytes.
In summary, we have shown that PKA activation contributes to the growth-promoting effect of CNP in the HZ. This observation provides evidence of cross-talk between cGMP and cAMP signaling in endochondral bone formation and sheds light on the physiological role of CNP.
Acknowledgments
We thank Takefumi Kondo and Yukari Sando (NGS Core Facility of the Graduate Schools of Biostudies, Kyoto University) for supporting the RNA-seq analysis.
Glossary
Abbreviations
- AC
adenylate cyclase
- cAMP
cyclic adenosine monophosphate
- CFP
cyan fluorescent protein
- cGMP
cyclic guanine monophosphate
- CNP
C-type natriuretic peptide
- Col10a1
type X collagen
- Col2a1
type II collagen
- DMEM
Dulbecco’s modified Eagle’s medium
- FRET
Förster resonance energy transfer
- GC
guanylyl cyclase
- HZ
hypertrophic zone
- PDE
phosphodiesterase
- PKA
protein kinase A
- PKG
protein kinase G
- PTH1R
parathyroid hormone 1 receptor
- PTHrP
parathyroid hormone–related protein
- RT-PCR
reverse transcription polymerase chain reaction
- YFP
yellow fluorescent protein
Financial Support
This work was supported by the Kyoto University Live Imaging Center. Financial support was provided in the form of JSPS KAKENHI grants (nos. 20K17532 to K.H., 20H05898 to M.M., and 19H00993 to M.M.), a JST CREST grant (no. JPMJCR1654), and a Moonshot R&D grant (no. JPMJPS2022 to M.M.).
Disclosure Summary
The authors have nothing to disclose.
Data Availability
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in the References.
References
- 1. Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332-336. [DOI] [PubMed] [Google Scholar]
- 2. Kozhemyakina E, Lassar AB, Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142(5):817-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakao K, Ogawa Y, Suga S, Imura H. Molecular biology and biochemistry of the natriuretic peptide system. II: natriuretic peptide receptors. J Hypertens. 1992;10(10):1111-1114. [DOI] [PubMed] [Google Scholar]
- 4. Yasoda A, Ogawa Y, Suda M, et al. Natriuretic peptide regulation of endochondral ossification. Evidence for possible roles of the C-type natriuretic peptide/guanylyl cyclase-B pathway. J Biol Chem. 1998;273(19):11695-11700. [DOI] [PubMed] [Google Scholar]
- 5. Yasoda A, Komatsu Y, Chusho H, et al. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat Med. 2004;10(1):80-86. [DOI] [PubMed] [Google Scholar]
- 6. Savarirayan R, Tofts L, Irving M, et al. Once-daily, subcutaneous vosoritide therapy in children with achondroplasia: a randomised, double-blind, phase 3, placebo-controlled, multicentre trial. Lancet. 2020;396(10252):684-692. [DOI] [PubMed] [Google Scholar]
- 7. Chusho H, Tamura N, Ogawa Y, et al. Dwarfism and early death in mice lacking C-type natriuretic peptide. Proc Natl Acad Sci USA. 2001;98(7):4016-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai YL, Sun Q, Huang X, et al. cGMP-PDE3-cAMP signal pathway involved in the inhibitory effect of CNP on gastric motility in rat. Regul Pept. 2013;180:43-49. [DOI] [PubMed] [Google Scholar]
- 9. Meier S, Andressen KW, Aronsen JM, et al. PDE3 inhibition by C-type natriuretic peptide-induced cGMP enhances cAMP-mediated signaling in both non-failing and failing hearts. Eur J Pharmacol. 2017;812:174-183. [DOI] [PubMed] [Google Scholar]
- 10. Terai K, Imanishi A, Li C, Matsuda M. Two decades of genetically encoded biosensors based on Förster resonance energy transfer. Cell Struct Funct. 2019;44(2):153-169. [DOI] [PubMed] [Google Scholar]
- 11. Ohta Y, Kamagata T, Mukai A, Takada S, Nagai T, Horikawa K. Nontrivial effect of the color-exchange of a donor/acceptor pair in the engineering of Förster resonance energy transfer (FRET)-based indicators. ACS Chem Biol. 2016;11(7):1816-1822. [DOI] [PubMed] [Google Scholar]
- 12. Watabe T, Terai K, Sumiyama K, Matsuda M. Booster, a red-shifted genetically encoded Förster resonance energy transfer (FRET) biosensor compatible with cyan fluorescent protein/yellow fluorescent protein-based FRET biosensors and blue light-responsive optogenetic tools. ACS Sens. 2020;5(3):719-730. [DOI] [PubMed] [Google Scholar]
- 13. Shukunami C, Shigeno C, Atsumi T, Ishizeki K, Suzuki F, Hiraki Y. Chondrogenic differentiation of clonal mouse embryonic cell line ATDC5 in vitro: differentiation-dependent gene expression of parathyroid hormone (PTH)/PTH-related peptide receptor. J Cell Biol. 1996;133(2):457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shukunami C, Ishizeki K, Atsumi T, Ohta Y, Suzuki F, Hiraki Y. Cellular hypertrophy and calcification of embryonal carcinoma-derived chondrogenic cell line ATDC5 in vitro. J Bone Miner Res. 1997;12(8):1174-1188. [DOI] [PubMed] [Google Scholar]
- 15. Kamioka Y, Sumiyama K, Mizuno R, et al. Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell Struct Funct. 2012;37(1):65-73. [DOI] [PubMed] [Google Scholar]
- 16. Willoughby D, Cooper DM. Organization and Ca2+ regulation of adenylyl cyclases in cAMP microdomains. Physiol Rev. 2007;87(3):965-1010. [DOI] [PubMed] [Google Scholar]
- 17. Zhao Y, Araki S, Wu J, et al. An expanded palette of genetically encoded Ca² + indicators. Science. 2011;333(6051):1888-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo J, Chung UI, Kondo H, Bringhurst FR, Kronenberg HM. The PTH/PTHrP receptor can delay chondrocyte hypertrophy in vivo without activating phospholipase C. Dev Cell. 2002;3(2):183-194 [DOI] [PubMed] [Google Scholar]
- 19. Miyazawa T, Ogawa Y, Chusho H, et al. Cyclic GMP-dependent protein kinase II plays a critical role in C-type natriuretic peptide-mediated endochondral ossification. Endocrinology. 2002;143(9):3604-3610. [DOI] [PubMed] [Google Scholar]
- 20. Krejci P, Masri B, Fontaine V, et al. Interaction of fibroblast growth factor and C-natriuretic peptide signaling in regulation of chondrocyte proliferation and extracellular matrix homeostasis. J Cell Sci. 2005;118(Pt 21):5089-5100. [DOI] [PubMed] [Google Scholar]
- 21. Agoston H, Khan S, James CG, et al. C-type natriuretic peptide regulates endochondral bone growth through p38 MAP kinase-dependent and -independent pathways. BMC Dev Biol. 2007;7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chikuda H, Kugimiya F, Hoshi K, et al. Cyclic GMP-dependent protein kinase II is a molecular switch from proliferation to hypertrophic differentiation of chondrocytes. Genes Dev. 2004;18(19):2418-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kawasaki Y, Kugimiya F, Chikuda H, et al. Phosphorylation of GSK-3beta by cGMP-dependent protein kinase II promotes hypertrophic differentiation of murine chondrocytes. J Clin Invest. 2008;118(7):2506-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang WJ, Krupinski J, Gilman AG. Expression and characterization of calmodulin-activated (type I) adenylylcyclase. J Biol Chem. 1991;266(13):8595-8603. [PubMed] [Google Scholar]
- 25. Choi EJ, Xia Z, Storm DR. Stimulation of the type III olfactory adenylyl cyclase by calcium and calmodulin. Biochemistry. 1992;31(28):6492-6498. [DOI] [PubMed] [Google Scholar]
- 26. Cali JJ, Zwaagstra JC, Mons N, Cooper DM, Krupinski J. Type VIII adenylyl cyclase. A Ca2+/calmodulin-stimulated enzyme expressed in discrete regions of rat brain. J Biol Chem. 1994;269(16):12190-12195. [PubMed] [Google Scholar]
- 27. Dai LJ, Quamme GA. Atrial natriuretic peptide initiates Ca2+ transients in isolated renal cortical thick ascending limb cells. Am J Physiol. 1993;265(4 Pt 2):F592-F597. [DOI] [PubMed] [Google Scholar]
- 28. Fowkes RC, Forrest-Owen W, Williams B, McArdle CA. C-type natriuretic peptide (CNP) effects on intracellular calcium [Ca2+]i in mouse gonadotrope-derived alphaT3-1 cell line. Regul Pept. 1999;84(1-3):43-49. [DOI] [PubMed] [Google Scholar]
- 29. Wollert KC, Yurukova S, Kilic A, et al. Increased effects of C-type natriuretic peptide on contractility and calcium regulation in murine hearts overexpressing cyclic GMP-dependent protein kinase I. Br J Pharmacol. 2003;140(7):1227-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moltzau LR, Aronsen JM, Meier S, et al. SERCA2 activity is involved in the CNP-mediated functional responses in failing rat myocardium. Br J Pharmacol. 2013;170(2):366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfeifer A, Aszódi A, Seidler U, Ruth P, Hofmann F, Fässler R. Intestinal secretory defects and dwarfism in mice lacking cGMP-dependent protein kinase II. Science. 1996;274(5295):2082-2086. [DOI] [PubMed] [Google Scholar]
- 32. Pfeifer A, Klatt P, Massberg S, et al. Defective smooth muscle regulation in cGMP kinase I-deficient mice. EMBO J. 1998;17(11):3045-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hirota K, Yasoda A, Kanai Y, et al. Live imaging analysis of the growth plate in a murine long bone explanted culture system. Sci Rep. 2018;8(1):10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yamamoto K, Kawai M, Yamazaki M, et al. CREB activation in hypertrophic chondrocytes is involved in the skeletal overgrowth in epiphyseal chondrodysplasia Miura type caused by activating mutations of natriuretic peptide receptor B. Hum Mol Genet. 2019;28(7):1183-1198. [DOI] [PubMed] [Google Scholar]
- 35. Thunemann M, Wen L, Hillenbrand M, et al. Transgenic mice for cGMP imaging. Circ Res. 2013;113(4):365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shuhaibar LC, Robinson JW, Vigone G, et al. Dephosphorylation of the NPR2 guanylyl cyclase contributes to inhibition of bone growth by fibroblast growth factor. Elife. 2017;6:e31343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nakao K, Osawa K, Yasoda A, et al. The Local CNP/GC-B system in growth plate is responsible for physiological endochondral bone growth. Sci Rep. 2015;5:10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bartels CF, Bükülmez H, Padayatti P, et al. Mutations in the transmembrane natriuretic peptide receptor NPR-B impair skeletal growth and cause acromesomelic dysplasia, type Maroteaux. Am J Hum Genet. 2004;75(1):27-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vasques GA, Amano N, Docko AJ, et al. Heterozygous mutations in natriuretic peptide receptor-B (NPR2) gene as a cause of short stature in patients initially classified as idiopathic short stature. J Clin Endocrinol Metab. 2013;98(10): E1636-E1644. [DOI] [PubMed] [Google Scholar]
- 40. Amano N, Mukai T, Ito Y, et al. Identification and functional characterization of two novel NPR2 mutations in Japanese patients with short stature. J Clin Endocrinol Metab. 2014;99(4): E713-E718. [DOI] [PubMed] [Google Scholar]
- 41. Miura K, Namba N, Fujiwara M, et al. An overgrowth disorder associated with excessive production of cGMP due to a gain-of-function mutation of the natriuretic peptide receptor 2 gene. PLoS One. 2012;7(8):e42180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bocciardi R, Giorda R, Buttgereit J, et al. Overexpression of the C-type natriuretic peptide (CNP) is associated with overgrowth and bone anomalies in an individual with balanced t(2;7) translocation. Hum Mutat. 2007;28(7):724-731. [DOI] [PubMed] [Google Scholar]
- 43. Lee K, Lanske B, Karaplis AC, et al. Parathyroid hormone-related peptide delays terminal differentiation of chondrocytes during endochondral bone development. Endocrinology. 1996;137(11):5109-5118. [DOI] [PubMed] [Google Scholar]
- 44. Amling M, Neff L, Tanaka S, et al. Bcl-2 lies downstream of parathyroid hormone-related peptide in a signaling pathway that regulates chondrocyte maturation during skeletal development. J Cell Biol. 1997;136(1):205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Correa D, Hesse E, Seriwatanachai D, et al. Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev Cell. 2010;19(4):533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miyoshi H, Blömer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72(10):8150-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. RRID:Addgene_12260.
- 48. DDBJ Sequenced Read Archive. https://www.ddbj.nig.ac.jp/ Uploaded October 7, 2021.
- 49. RRID:AB_2895557.
- 50. RRID:AB_2223172.
- 51. RRID:AB_621843.
- 52. SSBD:repository. Deposited December 20, 2021. 10.24631/ssbd.repos.2021.12.221 [DOI]
- 53. RRID:AB_879742.
- 54. RRID:AB_2847874.
- 55. RRID:AB_10563566.
- 56. RRID:AB_141780.
- 57. Buckwalter JA, Mower D, Schafer J, Ungar R, Ginsberg B, Moore K. Growth-plate-chondrocyte profiles and their orientation. J Bone Joint Surg Am. 1985;67(6):942-955. [PubMed] [Google Scholar]
- 58. Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2(4):389-406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all data generated or analyzed during this study are included in this published article or in the data repositories listed in the References.