Abstract
In Arabidopsis, de novo organogenesis of lateral roots is patterned by an oscillatory mechanism called the root clock, which is dependent on unidentified metabolites. To determine if retinoids regulate the root clock, we used a chemical reporter for retinaldehyde (retinal) binding proteins. We found that retinal binding precedes the root clock and predicts sites of lateral root organogenesis. Application of retinal increased root clock oscillations and promoted lateral root formation. Expression of an Arabidopsis protein with homology to vertebrate retinoid binding proteins, TEMPERATURE INDUCED LIPOCALIN (TIL) oscillates in the region of retinal binding to the reporter, confers retinal binding activity in a heterologous system, and when mutated, decreases retinal sensitivity. These results demonstrate a role for retinal and its binding partner in lateral root organogenesis.
Plants continuously develop post-embryonic lateral roots in order to forage for water and nutrients. The root clock, a temporal series of oscillating changes in gene expression, pre-patterns sites for initiation of lateral root primordia (1, 2). Blocking carotenoid metabolism genetically or with inhibitors is sufficient to dampen root clock oscillations and prevent lateral root initiation (3, 4). Known apocarotenoid regulators of root development, including abscisic acid, strigolactone, anchorene, and β-cyclocitral, do not rescue lateral root initiation in plants with reduced carotenoid metabolism (3, 5–8). In vertebrate development, β-carotene-derived retinoic acid has important functions in the oscillatory somitogenesis clock (9), neurogenesis, and vasculature development (10–12). Therefore, we hypothesized that retinoids may have similar developmental functions across different domains of life, and that retinal signaling may play a role in the root clock.
To examine the potential for retinal signaling in the root, we deployed merocyanine aldehyde (MCA), a chemical reporter designed to measure vertebrate retinoid binding protein activity (13). MCA has a Cyanine-5 (Cy5) head group fused to a retinal-like moiety that fluoresces when bound to a lysine residue in retinoid binding proteins (Fig. 1A). To test for the presence of proteins that interact with retinal in the root, we treated Arabidopsis plants with 10 μM MCA. The majority of the mature root remained non-fluorescent with MCA treatment. However, MCA-induced fluorescence exhibited spatial and temporal patterns in developing regions of the root (movie S1 and Fig. 1B). Root meristems, regions of actively dividing cells, consistently displayed high levels of MCA fluorescence (Fig. 1B and fig. S1, A to C). In the early differentiation zone, pulses of MCA fluorescence were observed (Fig. 1B and fig. S1, A and B), which were reminiscent of the root clock. To determine if MCA fluorescence pulses overlapped spatially or temporally with the root clock, we simultaneously tracked both processes using MCA treatment in roots expressing pDR5:LUC, an auxin-responsive reporter. The pDR5:LUC reporter line tracks the root clock and marks the pre-branch site, the region of primary root tissue where lateral root organogenesis is initiated (1). MCA fluorescence was measured in the region of the root that oscillates (exhibits maximal luminescence of DR5 expression) and forms the pre-branch site. We found that MCA signal preceded the root clock and marked a presumptive pre-branch site by an average of 5 hours before the DR5 pre-branch site (Fig. 1, C to E). Furthermore, like DR5 oscillations, MCA presumptive pre-branch sites accurately predict the sites of lateral root primordia (fig. S2, A and B). These results indicate that Arabidopsis proteins capable of binding retinal are active in the specific region of the root that undergoes de novo organogenesis and that this activity precedes the peak of expression of auxin signaling during the root clock.
Fig. 1. A reporter for retinal binding proteins predicts sites of lateral root organogenesis in Arabidopsis.
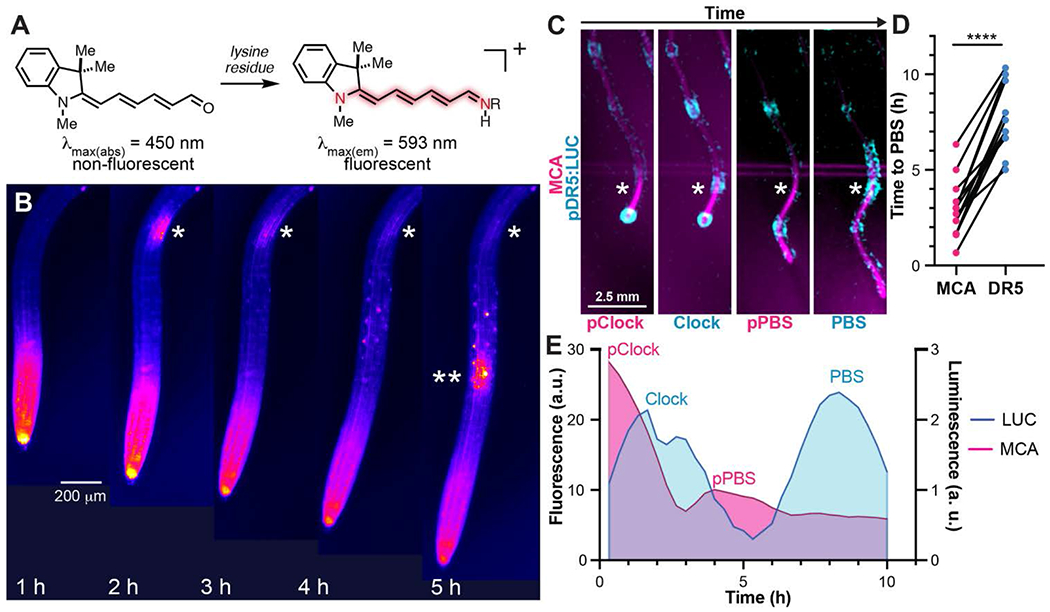
(A) The chemical structure and fluorescence properties of merocyanine aldehyde (MCA) in the absence and presence of a lysine residue from a retinal binding protein. (B) Time course of MCA fluorescence dynamics in Arabidopsis roots. * and ** indicate the site of fluorescent oscillations in the early differentiation zone of the root. (C) Representative images showing the spatial and temporal dynamics of MCA oscillations and the root clock (pDR5:LUC) in a growing root. (D) The pulse of MCA fluorescence indicates the presumptive pre-branch site (PBS) before pDR5:LUC (time is measured from the start of the pDR5:LUC root clock oscillation). (E) A representative example of the temporal signal from MCA and pDR5:LUC in the region of the root that becomes the PBS (this graph was derived from the root shown in panel C).
To further test the connection between putative retinal binding protein dynamics and lateral root primordia development, we characterized MCA dynamics in roots in which DR5 oscillations are inhibited. D15 is a chemical inhibitor of carotenoid metabolism that reduces DR5 clock oscillations, blocks lateral root initiation, and decreases cell elongation (3, 4). D15 had two effects on MCA spatial and temporal patterning. First, it decreased MCA fluorescence in the meristem (fig. S3, A and B). Second, it reduced the frequency of MCA pulses in the differentiation zone (fig. S3C). These results suggest that carotenoid metabolites are necessary for retinal binding in the root meristem and at sites of lateral root specification. D15 did not affect the amplitude of rare MCA pulses (fig. S3D), suggesting that the effect of D15 on MCA pulses is binary – pulses either occur at full amplitude or they do not occur at all. These findings are consistent with a model in which retinal biosynthesis is critical for retinal-binding protein activity. Previous work demonstrates that apocarotenoid synthesis is necessary for initiating DR5 oscillations (3, 4), further suggesting that retinal signaling precedes the root clock. These data are consistent with the hypothesis that retinal binding proteins are involved in lateral root formation.
The correlation between MCA dynamics and lateral root development suggests a role for retinal in lateral root organogenesis (Fig. 2A and fig. S4A). To determine if retinoids naturally occur in Arabidopsis and are affected by D15 treatment, we analyzed extracts from plants treated with D15 or a mock control using HPLC-MS. We found four compounds present in Arabidopsis that are decreased in D15-treated plants: retinal (apo16), 14’-apo-β-carotenal (apo14), 12’-apo-β-carotenal (apo12), and 10’-apo-β-carotenal (apo10) (fig. S4, A and B). To determine if retinal, apo14, apo12, or apo10 could rescue D15 inhibition of the lateral root clock, we treated D15-inhibited roots with these compounds and quantified resulting changes in DR5 oscillations. We found that retinal, apo12, and apo14 significantly increased the amplitude of DR5 oscillations in D15-inhibited roots (fig. S4C). 1 μM of retinal added to D15-treated roots was sufficient to fully rescue the amplitude of root clock oscillations within 24 hours (Fig. 2, B and C, and movie S2). At this concentration, retinal also rescued cell elongation in D15 inhibited roots (fig. S4D). We next monitored the ability of retinal, apo12, and apo14 to rescue D15 inhibition of lateral root development. Only retinal and apo14 fully rescued D15 inhibition of primary root growth and lateral root capacity, which is defined as the ability of plants to form lateral roots after the tip of the primary root has been excised (Fig. 2, D and E). Retinal and apo14 significantly increased the number of lateral root primordia in untreated plants (Fig. 2F), indicating that exogenous application of either retinal or apo14 is sufficient to induce ectopic organogenesis. Exogenous retinal treatment increased lateral root density (fig. S4E) as well as the total number of root primordia. These findings reveal that retinal and apo14 are sufficient to induce lateral root organogenesis. Enzymatic or non-enzymatic oxidation of the terminal double bond in apo14 would generate retinal. Therefore, it is possible that retinal is the active signaling molecule during apo14 treatments.
Fig. 2. Retinal rescues D15 inhibition of the root clock and lateral root organogenesis.
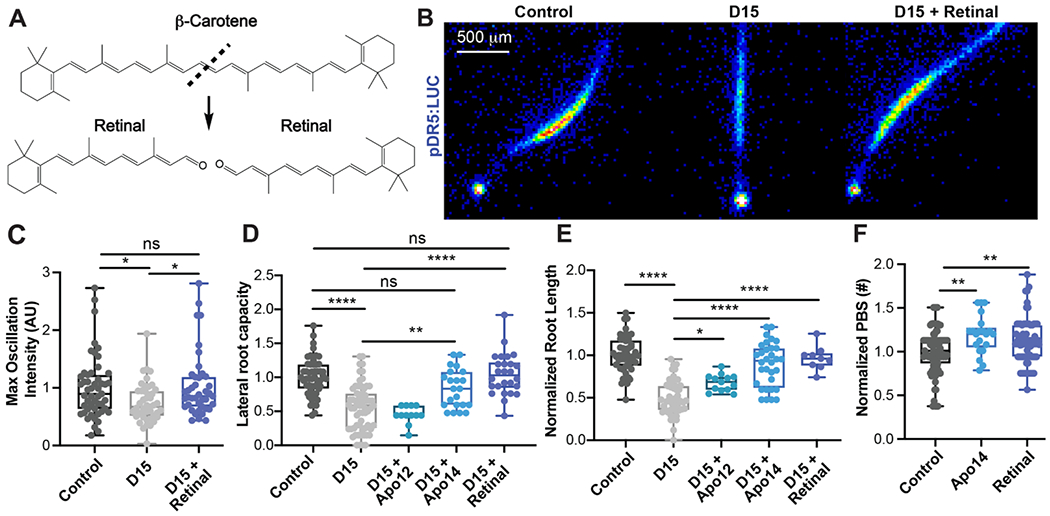
(A) Chemical structures of retinal and its precursor, β-carotene. (B) Luminescent images of the maximum root clock oscillation in pDR5:LUC roots with the following treatments: vehicle control, D15, and D15 in combination with retinal. (C) Quantification of the maximum root clock oscillation intensity in pDR5:LUC roots. (D) Quantification of the capacity of plants to form lateral roots in D15 inhibited roots (normalized to the vehicle control). (E) Root length in plants exposed to D15 (normalized to the vehicle control). (F) The number of lateral root pre-branch sites in pDR5:LUC roots treated with apo14 or retinal (normalized to the vehicle control).
To test whether retinal rescues D15 inhibition of MCA signal intensity, we examined MCA fluorescence in roots treated with D15 and retinal. Retinal rescued the MCA signal intensity in the meristem (fig. S5A), One interpretation is that retinal regulates activity of retinoid binding proteins. An alternative hypothesis is that retinal induces the expression of a retinal binding protein. To determine whether retinal biosynthesis occurs locally in the root, lateral root capacity was measured in roots where the entire plant was grown on D15 media except for the root tip (fig. S5B). Isolating the root tip from D15 treatment was sufficient to restore lateral root branching (fig. S5C), indicating that retinal synthesis in immature root tissue regulates organogenesis. These data are consistent with the hypothesis that retinal biosynthesis, protein binding, and function in lateral root development are localized to the tip of the root.
To further understand the role of retinal in lateral root formation, we searched for Arabidopsis proteins with sequence homology to retinal binding proteins in algae and vertebrates. We did not find significant homology between algae opsins and Arabidopsis proteins. However, we did identify homology between vertebrate and plant lipocalins, cross-kingdom protein transporters of small, hydrophobic molecules (14, 15). TIL (AT5G58070), the primary lipocalin expressed in the Arabidopsis root, has sequence homology (E-value = 9e-04, % identity = 25%) and predicted structural homology (TM-score = 0.72) to RETINOL BINDING PROTEIN 4 (RBP4), a vertebrate lipocalin (Fig. 3A and fig. S6). In plants, TIL has been implicated in preventing lipid peroxidation, particularly during light and heat stress (16). Analysis of an RNA-seq expression map of the root during DR5 oscillations revealed that there is spatiotemporal overlap in TIL expression and the root clock (17) (fig. S7). To investigate the TIL expression pattern dynamics more precisely, we generated pTIL:LUC lines and monitored expression in the root. We found that the TIL promoter confers expression that overlaps with the MCA fluorescence signal intensity (Fig. 3B). TIL expression is strongest in the meristem and oscillates in the early differentiation zone (Fig. 3, C and D, and movie S4). Together, these data suggest that TIL is a good candidate for a plant retinal binding protein.
Fig. 3. TIL is a plant lipocalin that interacts with retinal.
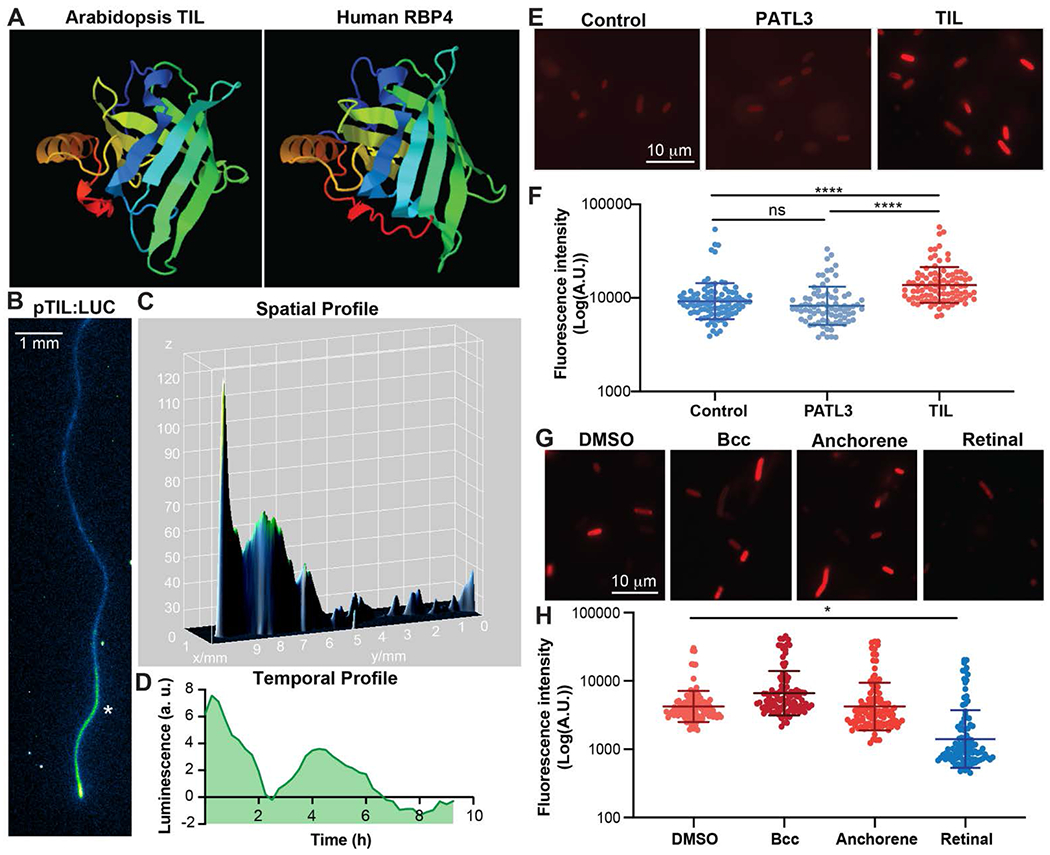
(A) Predicted protein structure of Arabidopsis TIL and Human RBP4 using the protein homology/analogy recognition engine V 2.0 (PHYRE2) (19). (B) The TIL expression pattern in a representative pTIL:LUC root. The asterisk indicates the spatial region analyzed in panel D. (C) A 3D surface plot showing the spatial profile of the pTIL:LUC root shown in panel B. (D) The luminescence intensity over time in the region of root highlighted by the asterisk in panel B. (E) Representative images of E. Coli cells transformed with Arabidopsis genes. (F) Fluorescence intensity of MCA-treated E. Coli transformed with genes from Arabidopsis. The control cells were not transformed. (G) Representative images of E. Coli cells expressing TIL that were pre-treated with DMSO, β-cyclocitral (Bcc), anchorene, or retinal. (H) Fluorescence intensity of MCA treated E. Coli transformed with TIL and pre-treated with DMSO, β-cyclocitral (Bcc), anchorene, or retinal.
To test if TIL binds retinal, we heterologously expressed TIL in E. coli and measured the effect on MCA fluorescence. As a control, we expressed PATL3, a plant lipid binding protein with a cellular retinaldehyde binding protein (CRAL)-TRIO domain. PATL3 expression did not change MCA fluorescence, indicating that the ability to bind lipids and homology to retinal binding protein domains are not sufficient to induce MCA fluorescence (Fig. 3, E and F). In contrast, TIL significantly increased MCA fluorescence, indicating that it interacts with MCA (Fig. 3, E and F). Next, we hypothesized that if TIL binds to retinal, application of retinal should disrupt TIL-induced MCA fluorescence. Application of retinal to TIL-expressing E. coli significantly reduced MCA fluorescence, indicating that retinal and MCA compete for TIL binding (Fig. 3, G and H). In contrast, cells treated with anchorene or β-cyclocitral, carotenoid-derivatives with chemical similarities to retinal (fig. S8), did not diminish MCA fluorescence. These results suggest that TIL interacts with retinal but not to structurally-similar apocarotenoids.
To determine the role of TIL in organogenesis, we examined root development in previously characterized TIL mutant alleles (16). Because both til-1 and til-2 mutant alleles had similar significant reductions in lateral root branching (Fig. 4, A and B), we focused further experimentation on the til-1 allele. To determine if TIL was important for establishing the root clock or for later stages of primordia development, we examined the expression of DR5 in til-1. We found that root clock amplitude is significantly inhibited in the til-1 mutant, suggesting that TIL regulates the root clock (Fig. 4, C and D). In til-1 mutants treated with MCA, fluorescence was significantly decreased (Fig. 4, E and F), indicating that meristematic and differentiation zone retinal-binding is decreased in til-1 roots. Furthermore, the til-1 mutant has a defect in primary root growth due to a reduction in cell elongation, which phenocopies inhibition of carotenoid biosynthesis with D15 (Fig. 4G). Additionally, til-1 mutants are slower to initiate lateral root primordia after the peak of the DR5 oscillation (Fig. 4H, fig. S9, and movie S3). This indicates that TIL contributes to the MCA signal in plants. These results support a model in which TIL regulates retinal signaling, root clock oscillations, and pre-branch site formation.
Fig. 4. TIL is essential for proper lateral root development.
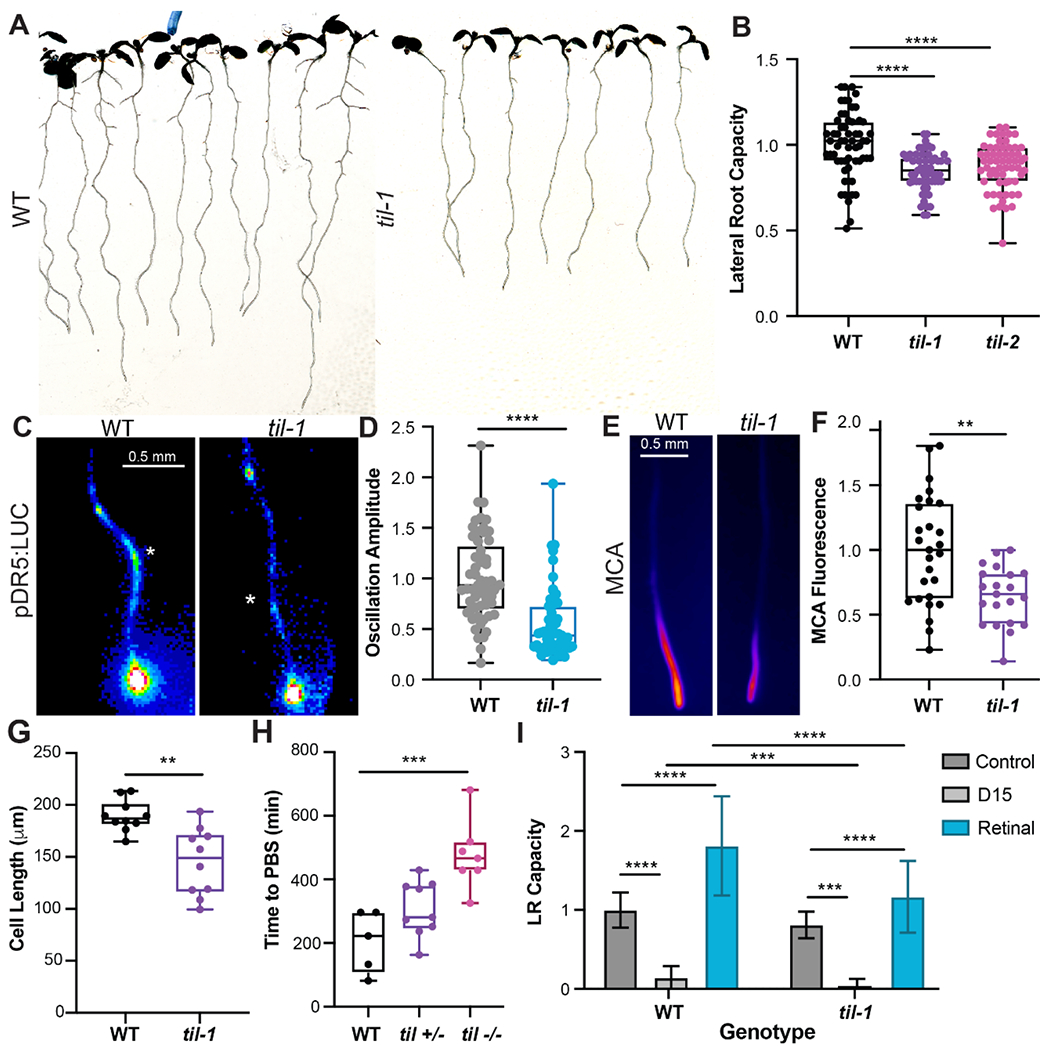
(A) Images of WT and til-1 mutant seedlings. (B) Lateral root capacity (LRC) in WT and til mutants (normalized to WT roots). (C) Images showing the root clock oscillation maxima in WT and til-1 mutant roots. (D) Quantification of the root clock oscillation amplitude in WT and til-1 mutant roots. (E) Fluorescence images showing MCA signal intensity in WT and til-1 mutant roots. (F) Quantification of the MCA fluorescence in the root tip (1 mm region) in WT and til-1. (G) Average length of mature cortex cells in WT and til-1 plants. (H) The time between the root clock maxima and the formation of a lateral root pre-branch site, measured in pDR5:LUC roots. (I) Quantification of lateral root (LR) capacity in response to retinal and D15. Values are normalized with respect to the untreated roots for each genotype. Genotype by treatment interactions were significant for both treatments (p ≤ 0.0001). For each sample, n > 64.
Previous work identified TIL as being involved in heat stress tolerance (15). To test whether lateral root organogenesis is temperature dependent, we quantified the root clock oscillation before and after heat shock. The amplitude of the root clock significantly decreased after heat shock stress, indicating that organogenesis is a temperature-dependent process. To determine if TIL has a role in this temperature dependency, root clock amplitude was measured in til-1 pDR5:LUC plants. Temperature sensitivity of the root clock is suppressed in the til mutant (Two-way ANOVA genotype by environment interaction p value = 0.008), suggesting that TIL is important for proper response to heat stress during lateral root organogenesis (fig. S10).
To further explore the relationship between the plant lipocalins and the retinal response, we treated til-1 roots with both retinal and D15 and quantified the effects on lateral root development. We found that til-1 roots had significantly decreased sensitivity to retinal (Two-way ANOVA genotype by treatment interaction p value < 0.0001), indicating that TIL is a key player in retinal-induced root organogenesis (Fig. 4I). In contrast, D15 treatment caused an increase in inhibition of lateral root capacity in til-1 mutants (Fig. 4I). Mutants were significantly more sensitive to D15 than WT plants (Two-way ANOVA genotype by treatment interaction p < 0.0001), suggesting that til-1 is more sensitive to D15-induced retinal reduction compared to WT. This suggests that additional retinal perception pathways are still functional in til-1 mutants. Overall, the changes in sensitivity in til-1 provide genetic evidence that TIL is involved in retinal-mediated effects on lateral root development.
We identified retinal as an endogenous metabolite that is sufficient to induce post-embryonic root organogenesis. Reduction of retinal biosynthesis leads to inhibition of the root clock, the first known stage of lateral root organogenesis. Oscillations of activity of a chemical retinal binding protein reporter predicted sites of lateral root organogenesis. We identified TIL, a plant lipocalin able to bind retinal and discovered that it is essential for retinal perception and modulation of root clock oscillations. To date, at least three pathways have been identified that play a role in regulating the root clock. In addition to apocarotenals, these are pectin modification (17) and auxin (18). Whether these pathways act independently or in interlocking processes remains unknown. Our results show the convergent use of retinal related compounds in the regulation of developmental clocks across the plant and animal kingdoms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dominique Bergmann, Miguel Moreno-Risueno, Loredana Quadro and Isaiah Taylor for critical reading of the manuscript. We thank Elizabeth Sattely for use of her lab equipment and Medhavinee Mijar for technical support.
Funding:
This work was supported by the Howard Hughes Medical Institute and the US National Institutes of Health under grant MIRA 1R35GM131725 (to PNB) and by an Arnold and Mabel Beckman Postdoctoral Fellowship (to AJD). The research of JRD was supported in part by a Faculty Scholar grant from the Howard Hughes Medical Institute and the Simons Foundation. ML and MS are supported by the Intramural Research Program of the National Institutes of Health (NIH), the National Cancer Institute, and the Center for Cancer Research.
Footnotes
Competing interests: Authors declare no competing interests. PNB is the co-founder and Chair of the Scientific Advisory Board of Hi Fidelity Genetics, Inc, a company that works on crop root growth. Alexandra J. Dickinson and Philip N. Benfey are inventors on a patent application (63110088) submitted by Duke University that covers the use of retinal and related compounds in root growth.
Data and materials availability:
All data are available in the main text or the supplementary materials.
REFERENCES AND NOTES
- 1.Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN, Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329, 1306–1311 (2010). 10.1126/science.1191937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xuan W, De Gernier H, Beeckman T, The dynamic nature and regulation of the root clock. Development 147, dev181446 (2020). 10.1242/dev.181446 [DOI] [PubMed] [Google Scholar]
- 3.Dickinson AJ, Lehner K, Mi J, Jia K-P, Mijar M, Dinneny J, Al-Babili S, Benfey PN, β-Cyclocitral is a conserved root growth regulator. Proc. Natl. Acad. Sci. U.S.A 116, 10563–10567 (2019). 10.1073/pnas.1821445116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Norman JM, Zhang J, Cazzonelli CI, Pogson BJ, Harrison PJ, Bugg TDH, Chan KX, Thompson AJ, Benfey PN, Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proc. Natl. Acad. Sci. U.S.A 111, E1300–E1309 (2014). 10.1073/pnas.1403016111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia KP, Dickinson AJ, Mi J, Cui G, Xiao TT, Kharbatia NM, Guo X, Sugiono E, Aranda M, Blilou I, Rueping M, Benfey PN, Al-Babili S, Anchorene is a carotenoid-derived regulatory metabolite required for anchor root formation in Arabidopsis. Sci. Adv 5, eaaw6787 (2019). 10.1126/sciadv.aaw6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruyter-Spira C, Kohlen W, Charnikhova T, van Zeijl A, van Bezouwen L, de Ruijter N, Cardoso C, Lopez-Raez JA, Matusova R, Bours R, Verstappen F, Bouwmeester H, Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 155, 721–734 (2011). 10.1104/pp.110.166645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agusti J, Herold S, Schwarz M, Sanchez P, Ljung K, Dun EA, Brewer PB, Beveridge CA, Sieberer T, Sehr EM, Greb T, Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. U.S.A 108, 20242–20247 (2011). 10.1073/pnas.1111902108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Smet I, Signora L, Beeckman T, Inzé D, Foyer CH, Zhang H, An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33, 543–555 (2003). 10.1046/j.1365-313X.2003.01652.x [DOI] [PubMed] [Google Scholar]
- 9.Wahl MB, Deng C, Lewandoski M, Pourquié O, FGF signaling acts upstream of the NOTCH and WNT signaling pathways to control segmentation clock oscillations in mouse somitogenesis. Development 134, 4033–4041 (2007). 10.1242/dev.009167 [DOI] [PubMed] [Google Scholar]
- 10.Vermot J, Pourquié O, Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature 435, 215–220 (2005). 10.1038/nature03488 [DOI] [PubMed] [Google Scholar]
- 11.Maden M, Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat. Rev. Neurosci 8, 755–765 (2007). 10.1038/nrn2212 [DOI] [PubMed] [Google Scholar]
- 12.Lin SC, Dollé P, Ryckebüsch L, Noseda M, Zaffran S, Schneider MD, Niederreither K, Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc. Natl. Acad. Sci. U.S.A 107, 9234–9239 (2010). 10.1073/pnas.0910430107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yapici I, Lee KSS, Berbasova T, Nosrati M, Jia X, Vasileiou C, Wang W, Santos EM, Geiger JH, Borhan B, “Turn-on” protein fluorescence: In situ formation of cyanine dyes. J. Am. Chem. Soc 137, 1073–1080 (2015). 10.1021/ja506376j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charron J-BF, Ouellet F, Pelletier M, Danyluk J, Chauve C, Sarhan F, Identification, expression, and evolutionary analyses of plant lipocalins. Plant Physiol. 139, 2017–2028 (2005). 10.1104/pp.105.070466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi WT, Fung RWM, Liu HC, Hsu CC, Charng YY, Temperature-induced lipocalin is required for basal and acquired thermotolerance in Arabidopsis. Plant Cell Environ. 32, 917–927 (2009). 10.1111/j.1365-3040.2009.01972.x [DOI] [PubMed] [Google Scholar]
- 16.Boca S, Koestler F, Ksas B, Chevalier A, Leymarie J, Fekete A, Mueller MJ, Havaux M, Arabidopsis lipocalins AtCHL and AtTIL have distinct but overlapping functions essential for lipid protection and seed longevity. Plant Cell Environ. 37, 368–381 (2014). 10.1111/pce.12159 [DOI] [PubMed] [Google Scholar]
- 17.Wachsman G, Zhang J, Moreno-Risueno MA, Anderson CT, Benfey PN, Cell wall remodeling and vesicle trafficking mediate the root clock in Arabidopsis. Science 370, 819–823 (2020). 10.1126/science.abb7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E, Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. U.S.A 105, 8790–8794 (2008). 10.1073/pnas.0712307105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ, The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 10, 845–858 (2015). 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berezin MY, Guo K, Teng B, Edwards WB, Anderson CJ, Vasalatiy O, Gandjbakhche A, Griffiths GL, Achilefu S, Radioactivity-synchronized fluorescence enhancement using a radionuclide fluorescence-quenched dye. J. Am. Chem. Soc 131, 9198–9200 (2009). 10.1021/ja903685b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herwig L, Rice AJ, Bedbrook CN, Zhang RK, Lignell A, Cahn JKB, Renata H, Dodani SC, Cho I, Cai L, Gradinaru V, Arnold FH, Directed evolution of a bright near-infrared fluorescent rhodopsin using a synthetic chromophore. Cell Chem. Biol 24, 415–425 (2017). 10.1016/j.chembiol.2017.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, Möller B, Wilson M, Holman T, Van Isterdael G, Brunoud G, Vuylsteke M, Vernoux T, De Veylder L, Inzé D, Weijers D, Bennett MJ, Beeckman T, A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol 20, 1697–1706 (2010). 10.1016/j.cub.2010.09.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials.


