Abstract
Super Enhancers (SEs) are clusters of regulatory elements associated with cell identity and disease. However, whether these elements are induced by oncogenes and can regulate gene modules cooperating for cancer cell transformation or maintenance remains elusive. To address this question, we conducted a genome-wide CRISPRi-based screening of SEs in ETO2-GLIS2+ acute megakaryoblastic leukemia. This approach revealed SEs essential for leukemic cell growth and survival that are induced by ETO2-GLIS2 expression. In particular, we identified a de novo SE specific of this leukemia subtype and regulating expression of tyrosine kinase–associated receptors KIT and PDGFRA. Combined expression of these two receptors was required for leukemic cell growth, and CRISPRi-mediated inhibition of this SE or treatment with tyrosine kinase inhibitors impaired progression of leukemia in vivo in patient-derived xenografts experiments. Our results show that fusion oncogenes, such as ETO2-GLIS2, can induce activation of SEs regulating essential gene modules synergizing for leukemia progression.
CRISPRi screening in AMKL identified specific Super Enhancers controlling expression of targetable cooperative dependencies.
INTRODUCTION
In the recent years, massively parallel sequencing approaches identified hundreds of mutated genes in cancer (1) providing an unprecedented amount of information about mechanisms of cancer cell maintenance and progression. However, while it is widely accepted that transformation processes result from oncogenic cooperation between deregulated genes and pathways, the functional characterization of candidate key players is mostly performed at the single gene level, which is generally inadequate to identify these oncogene circuitries. In addition, studies aimed at depicting oncogenic cooperation involve the generation of challenging mouse models or the deployment of tedious screening pipelines. Genome-wide mapping of epigenomic modifications on histone tails or binding of factors such as MED1 and BRD4 allowed identification of clusters of regulatory elements, also termed super-enhancers (SEs) (2). Functional annotation of these regions revealed their high relevance during normal tissue development and cancer ontogeny (3). SEs are clusters of regulatory elements characterized by high intensity of enhancer-related histone tail modifications, such as histone H3 lysine 27 acetylation or lysine 4 mono-methylation (H3K27ac and H3K4me1, respectively) and binding of enhancer-associated factors such as the mediator complex, particularly MED1, or bromodomain-containing proteins such as BRD4 (4). These clustered regulatory regions shape the transcriptional identity of specific tissues and cell types, and their landscape often shifts in disease conditions, particularly in cancer cells, where they can control oncogene expression (5, 6). Whether these regions control expression of one or several genes and what genes are directly controlled by these regions remains elusive in most cases.
An interesting paradigm of the tumorigenic function of these SE regions comes from the fusion oncogene–driven, aggressive, acute megakaryoblastic leukemia (AMKL). AMKL is a genetically heterogeneous subtype of acute myeloid leukemia (AML), often associated with Down syndrome (DS) or characterized by gene fusions, such as the recently identified CBFA2T3-GLIS2 (also termed ETO2-GLIS2) in 20 to 30% of patients (7, 8). This fusion is the result of a cryptic inversion of chromosome 16, which fuses CBFA2T3 (also called ETO2), a member of the ETO family of nuclear co-repressors, to GLIS2, a member of the GLI-similar family of transcription factors. ETO2-GLIS2 is the most frequent chimeric oncogene identified to date in non–DS-AMKL patients, confers a poor prognosis (9), and is sufficient to induce leukemia (10). In a recent study, we identified that this fusion oncogene was associated with SEs and that this association might be important for pro-oncogenic transcriptional program induced by ETO2-GLIS2 (11). However, the exact oncogenic circuitry controlled by SEs and whether these SE-controlled genes synergize for disease induction and maintenance remain elusive.
We thus hypothesized that important regulatory regions regulated by ETO2-GLIS2 could simultaneously control expression of genes cooperating in functional modules to promote leukemia development. In an effort to identify these modules, we deployed a genome-wide CRISPR interference (CRISPRi)–based screening approach and nominated SE regions that are functionally linked to leukemia maintenance. In particular, we pinpointed a novel SE region regulating the expression of both tyrosine kinases KIT and platelet-derived growth factor receptor Alpha (PDGFRA). Whereas the inhibition of each kinase alone modestly affected leukemic cell growth, combined inhibition of both receptors synergizes to impair leukemia cell growth and progression. We were also able to show that ectopic expression of ETO2-GLIS2 fusion can induce de novo SE formation. Our results demonstrate that fusion oncogenes can rewire SEs to induce coregulated genes collaborating to promote cancer and could open new avenues to the concept of combined gene inhibition upon single hit targeting.
RESULTS
CRISPRi screening reveals essential SEs for leukemia maintenance
We hypothesized that fusion oncogene–induced SEs could control and regulate expression of clusters of genes, which might collaborate in cancer progression and/or maintenance and that impeding their activity could reveal this synergistic action. We therefore designed a screening methodology to unbiasedly identify fundamental SE involved in cancer cell growth and survival (Fig. 1A). To this end, we used the CRISPRi methodology, which relies on the use of a deactivated Cas9 fused to a KRAB domain (12, 13). Properly targeted dCas9-KRAB fusion can trigger the formation of local heterochromatin and impede enhancer activity (14).
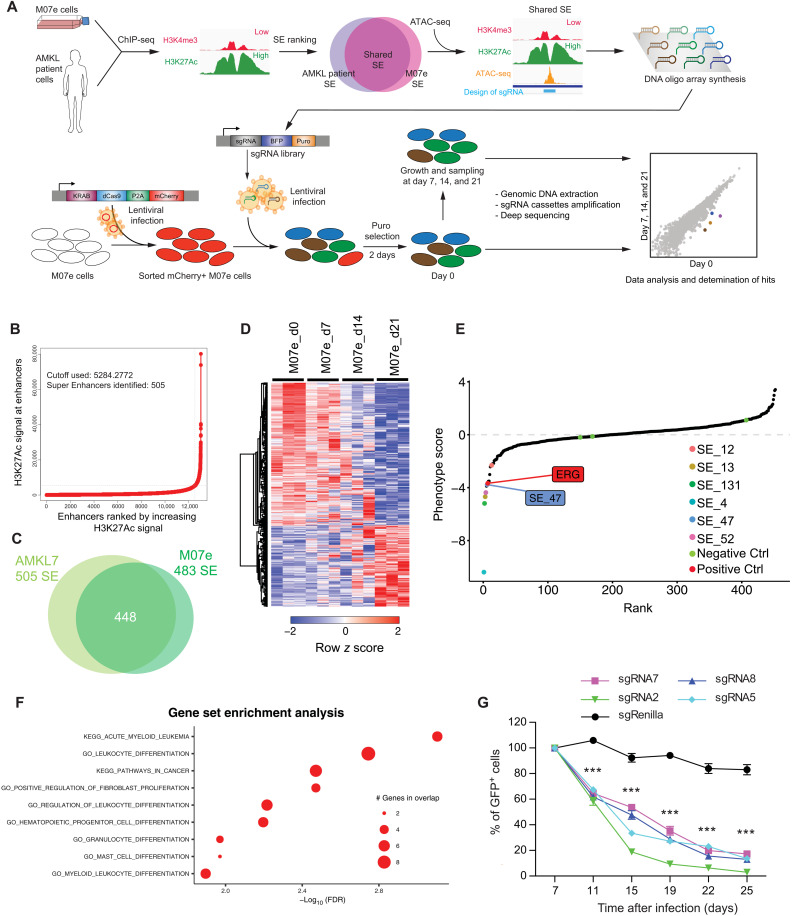
Fig. 1. Genome-wide CRISPRi screen identifies essential SEs for leukemia growth.
(A) Schematic illustration of SEs screening strategy using CRISPRi in AMKL. (B) Distribution of H3K27Ac ChIP-seq density across enhancers in AMKL7 patient cells: the 505 enhancers located above the tangent with high level of signal represent SEs. (C) Venn diagram depicting the overlap between SE in M07e and AMKL7 cells. (D) Heatmap representation of 474 significantly enriched and 278 significantly depleted sgRNAs among three replicates of SE screening experiments in M07e cells. Representation of sgRNA with a minimum of 40× coverage and P < 0.05. (E) Pooled negative selection screening depicting changes in representation of all SE ranked by the average of their depletion or enrichment score of all sgRNAs across the three replicates at day 21 compared to day 0. Significantly depleted SE are highlighted in color. Position of SE_47 and ERG-positive control are shown. Negative controls are marked in green. (F) Two dimensional dot plot representing gene set enrichment analysis (GSEA) results of top SE hits associated genes. (G) Percentage of GFP+ M07e cells following CRISPRi targeting of SE_47 with indicated sgRNAs compared to control sgRenilla and normalized to day 7 after infection. Means ± SEM, n = 3, significance is determined using Student’s t test, ***P < 0.001.
To define which SEs to target, we performed H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) in ETO2-GLIS2–expressing cell derived from an AMKL patient (AMKL7) and called SEs using ROSE algorithm (4, 5). In total, 505 SEs were identified in these patient-derived cells (Fig. 1B), of which 448 overlapped with previously defined SEs (11) in the ETO2-GLIS2–expressing patient–derived M07e cell line (Fig. 1C). Subsequently, the design of single-guide RNAs (sgRNAs) targeting open chromatin regions, as defined by ATAC (Assay for Transposase-Accessible Chromatin) sequencing (ATAC-seq) peaks within these SEs, led to a library of 7381 sgRNAs (table S1). Screening was performed in triplicate in M07e cells stably expressing the dCas9-KRAB fusion. Analysis of biological replicates showed that the representation of 474 sgRNAs targeting 265 SEs was significantly decreased over time, whereas 278 sgRNAs targeting 185 SEs were significantly overrepresented (Fig. 1D and fig. S1A). Loss of representation of sgRNAs implies that CRISPRi targeting of the associated SE impairs M07e cell growth or survival. Maximum likelihood estimation of enrichment analysis revealed that six SEs markedly and reproducibly altered M07e cell growth upon CRISPRi inhibition (Fig. 1E and fig. S1, B and C). Three sgRNAs targeting ETS-Related Gene (ERG), which is required for ETO2-GLIS2+ AMKL cell growth (11), were used as positive controls and were significantly depleted over time (fig. S1D), whereas sgRNAs targeting genes not present in the human genome (Luciferase, Renilla, and mouse Lin28) showed no significant variation (fig. S1E). Functional annotation of expressed genes [Fragments per kilobase of exon per million mapped reads (FPKM) > 3] proximal to the most significant SEs (5′ and 3′ genes within a 500-kb window) revealed their involvement in myeloid development and leukemia transformation (Fig. 1F). This functional relationship was also highlighted by evidence of multiple physical and genetic interactions and occurrence along common pathways of most of these genes (fig. S1G). Among the top hits of the screen, we decided to focus on SE_47 (Fig. 1E), not only for the robustness of the associated phenotype, with six sgRNAs targeting the two main H3K27ac peaks significantly underrepresented (fig. S1F), but also for its interesting location 5′ to the KIT oncogene on chromosome 4 (fig. S1H). To validate these results, we designed additional sgRNAs targeting the two major H3K27ac peaks of SE_47 and performed independent CRISPRi experiments in M07e cells stably expressing the dCas9-KRAB fusion. Expression of these sgRNAs reproduced the loss of representation phenotype observed in the screen, confirming that CRISPRi targeting of SE_47 impairs M07e cell growth or survival (Fig. 1G).
SE_47 regulates KIT and PDGFRA expression
To identify which genes are regulated by this SE, we performed RNA sequencing (RNA-seq) analysis after short kinetics of CRISPRi inhibition (48 hours after transduction) to limit secondary inhibitory effects. This analysis showed that short-term inhibition of this SE induces very few transcriptional changes, and mainly, KIT and PDGFRA proximal genes were significantly down-regulated (Fig. 2A and fig. S2A). The above results were confirmed by performing quantitative polymerase chain reaction (qPCR) with four independent sgRNAs that showed significant inhibition of KIT and PDGFRA expression when compared to nontargeting control (Fig. 2B), strong reduction of KIT and PDGFRA protein expression (Fig. 2C), and presentation at cell surface (fig. S2, B and C). To control for the on-target specificity of our CRISPR guides, we performed ATAC-seq and ChIP-seq for H3K27ac and H3K9me3 on M07e cells expressing each of these sgRNAs. These experiments revealed that while H3K27ac is lost at both SE_47 and promoters of proximal genes, gain of H3K9me3 and loss of chromatin accessibility are only found at sgRNA-targeted regions without affecting additional regions of the same locus or other regions of the genome (fig. S3A), suggesting that CRISPRi induces targeted significant loss of chromatin accessibility without unspecific spreading of heterochromatin. In addition, we performed CRISPRi using two independent sgRNAs in the HEL-5J20 cell line lacking the activity of this enhancer and expressing lower level of KIT. CRISPRi expression in this cell line did not induce neither inhibition of KIT expression or impaired growth of the cells, suggesting that observed phenotypes in M07e cells are on target (fig. S3, B and C). These results demonstrated that SEKIT controls the expression of KIT and PDGFRA genes. Therefore, the aforementioned SE_47 was named “SEKIT” thereafter.
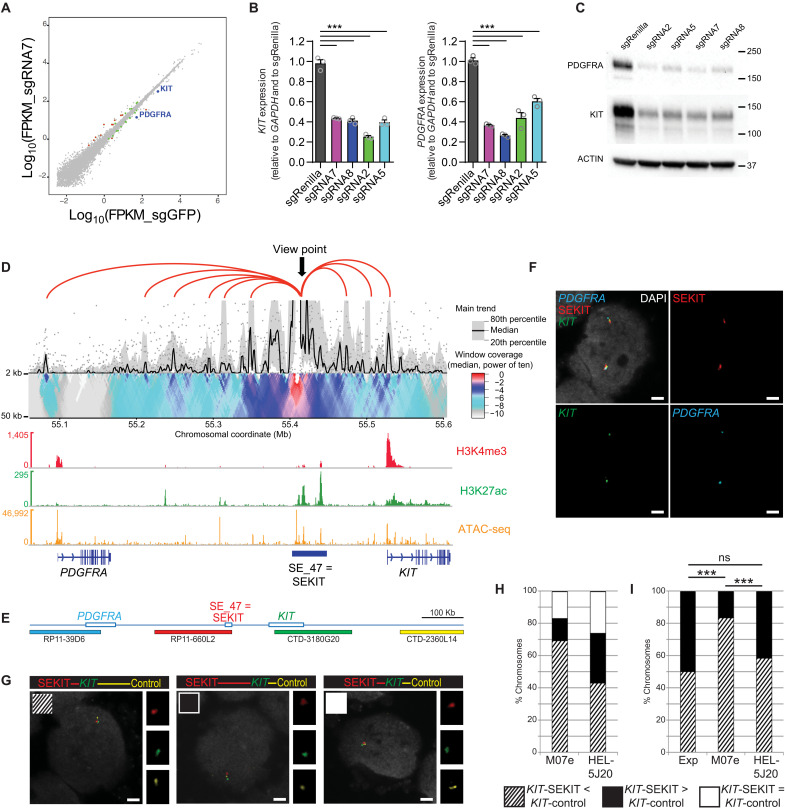
Fig. 2. SEKIT controls KIT and PDGFRA expression.
(A) Scatterplot showing gene expression variations following CRISPRi targeting with sgRNA7 compared to control sgGFP in M07e cells. Green dots P < 0.05, red dots P < 0.05, and >2-fold change. (B) qPCR showing KIT and PDGFRA expression following CRISPRi targeting of SE_47 (SEKIT) with indicated sgRNAs in M07e cells. Means ± SEM, n = 3, *** P < 0.001. (C) Western blot probing KIT, PDGFRA, and β-actin levels upon CRISPRi inhibition of SE_47 (SEKIT). (D) 4C-seq domainogram showing cis-contacts around SEKIT locus in M07e cells using 5-kb window size (top). Gene tracks with normalized read density histograms of H3K4me3 and H3K27ac ChIP-seq and ATAC-seq (bottom). (E) Scheme of the genomic region including PDGFRA (blue), SEKIT (red), KIT (green), and a control region for KIT (yellow) and corresponding BAC probes. (F) DNA FISH for PDGFRA (cyan), SEKIT (red), and KIT (green) in M07e cells. DNA was stained with DAPI (gray). Scale bars, 2 μm. (G) DNA FISH for KIT (green), SEKIT (red), and control region (yellow) in M07e or HEL-5J20 cells. DNA was stained with DAPI (gray). Scale bars, 2 μm. (H) Frequencies of chromosomes displaying the three configurations of distances in M07e and HEL-5J20 cells: KIT-SEKIT < KIT-control (striped bar), KIT-SEKIT > KIT-control (black bar), and KIT-SEKIT = KIT-control (white bar). (I) Frequencies of chromosomes displaying either the KIT-SEKIT < KIT-control (striped bar) or the KIT-SEKIT > KIT-control (black bar) configuration in M07e and HEL-5J20 cells as well as the expected distribution (Exp; 50/50%). ***P < 0.001, not significant (ns) P > 0.5.
Activity of SEKIT is linked to its enhanced physical proximity with KIT and PDGRA genes
To investigate direct regulation of proximal KIT and PDGFRA genes by SEKIT, we examined their three-dimensional (3D) physical proximity in the nuclear space. We conducted chromatin conformation capture experiments in M07e cells [4C sequencing (4C-seq) (15, 16)], using a bait located in SEKIT, and identified directly interacting proximal regions, including KIT and PDGFRA promoters (Fig. 2D). DNA fluorescence in situ hybridization (DNA FISH) experiments using bacterial artificial chromosome (BAC) probes spanning SEKIT, KIT, and PDGFRA confirmed the close proximity of these regions shown with 4C-seq (Fig. 2, E and F). To assess whether this physical proximity reflect only their proximity along the DNA fiber (KIT-SEKIT = 85 kb, PDGFRA-SEKIT = 250 kb) or whether it could be due to the presence of a bona fide functional loop between them, additional DNA FISH were performed in M07e cells, where SEKIT is activated; in control HEL-5J20 cells, where SEKIT is inactive, three BAC probes were used, spanning KIT and two regions equidistant from KIT on both sides, with SEKIT on its 5′ end and a control region on its 3′ end (Fig. 2, E and G). In M07e cells, while 17% of the chromosomes KIT-SEKIT and KIT-control distances were equal, KIT-SEKIT distance was shorter in 69.5% of the cases and longer in the remaining 13.5% (n = 200) (Fig. 2H and table S2). In HEL-5J20 cells, on the other hand, frequencies were 26, 43.3, and 30.7%, respectively (n = 150) (Fig. 2H and table S2). Thus, KIT-SEKIT distances were significantly shorter than KIT-control ones in M07e cells compared to HEL-5J20 cells (χ2 test: ***P < 0.00001; Fig. 2H and table S2). Furthermore, under the hypothesis that, in the absence of topological or functional constraint, probabilities for KIT to be closer to SEKIT or to the control should be equal, KIT appears to be closer to SEKIT than to the control region in significantly more chromosomes than expected in M07e cells, but not in HEL-5J20 cells (χ2 tests: ***P < 0.00001 in M07e cells, P = 0.71 in HEL-5J20 cells, Fig. 2I and table S2). To address whether SEKIT activation is required for this looping, we subjected M07e cells to CRISPRi inhibition using an sgRNA-targeting SEKIT (sgRNA7) or a control sgRNA-targeting Renilla (sgRenilla) and subsequently performed 4C-seq experiments using the same bait located in SEKIT. Analysis of 4C contacts showed a global reduction of cis-interactions between SEKIT and the surrounding genomic regions, particularly between SEKIT and KIT and PDGFRA promoters (fig. S4). These data strongly support that SEKIT activity is functionally required for chromatin looping, bringing it in close physical contact with KIT and PDGFRA promoters and suggest that this functional loop could then trigger their expression.
SEKIT is strongly associated with ETO2-GLIS2+ leukemic cells
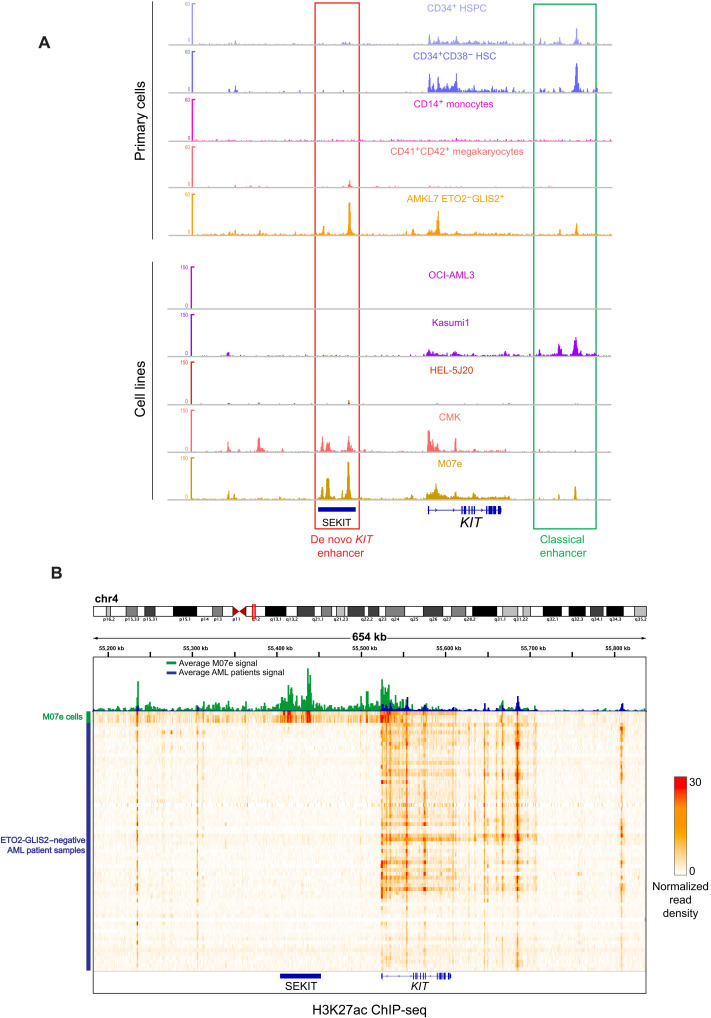
We next wondered whether this SE is normally found in other hematopoietic cells expressing the KIT gene and/or other subtypes of AML. To this end, we performed ChIP-seq analyses for H3K27ac in cells lacking ETO2-GLIS2 but expressing high level of KIT, including human whole CD34+ and CD34+CD38− cord blood hematopoietic stem and progenitor cells (HSPC), the AML1-ETO fusion expressing Kasumi-1 cell line and the DS-AMKL cell line CMK. We also used the OCI-AML3 cell line not expressing KIT and the HEL-5J20 cell line expressing low level of KIT. We then compared profiles obtained with ETO2-GLIS2+ M07e cell line and AMKL7 patient cells, as well as with KITneg CD41+CD42+ megakaryocytes and CD14+ monocytes from ENCODE dataset. This analysis showed that ETO2-GLIS2–negative primary cells do not display any H3K27ac peak located in the SEKIT region and that, instead, KIT-expressing cells show H3K27ac peaks located on the 3′ of KIT (Fig. 3A, top), in a region previously described as a KIT enhancer (17). Moreover, most AML cell lines also do not show any H3K27ac peak in SEKIT (Fig. 3A, bottom). Notably, the CMK lines derived from a DS-AMKL patient showed some low peak of H3K27ac on SEKIT, suggesting a basal activity.
Fig. 3. SEKIT is a de novo enhancer strongly associated with ETO2-GLIS2+ leukemia cells.
(A) Gene track showing normalized read density histograms of H3K27ac ChIP-seq around the KIT gene in primary hematopoietic cells (CD34+ HSPC, CD34+CD38− HSC, CD14+ monocytes, CD41+CD42+ megakaryocytes, and ETO2-GLIS2+ AMKL7; top) and AML cell lines (Kasumi-1, OCI-AML3, and HEL-5J20) and AMKL cell lines (CMK and M07e) (bottom). Classical and de novo KIT enhancers are highlighted. Read densities are shown as unique reads per million. (B) Heatmap representing normalized read density of H3K27ac ChIP-seq of 66 AML patients negative for ETO2-GLIS2 fusion and three replicates of ETO2-GLIS2+ M07e cell line at SEKIT, and KIT locus. Top panel shows average intensity signal for AML patients (blue histogram) and M07e cells (green histogram).
To investigate whether this specific SE is also active in other AML patient samples from various subtypes, we reanalyzed H3K27ac ChIP-seq data of 66 patients previously reported (18). These analyses revealed that none of the ETO2-GLIS2–negative AML samples showed any significant H3K27ac enrichment at SEKIT location (Fig. 3B) as compared to the ETO2-GLIS2–positive M07e patient–derived cell line.
These results showed that SEKIT is not active in wild-type HSPC or in any AML subtypes outside AMKL and is strongly associated with ETO2-GLIS2–positive samples, suggesting that SEKIT activity is controlled by ETO2-GLIS2 activity.
Ectopic ETO2-GLIS2 expression induces a specific enhancer program and SEKIT activity
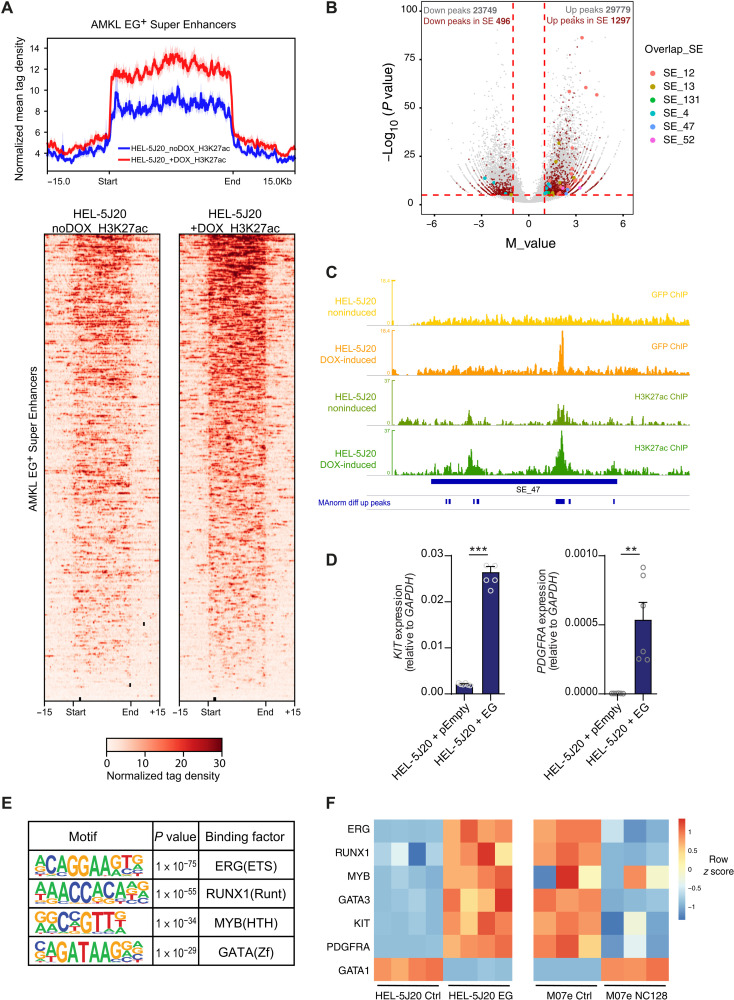
To address this question, we generated a stable HEL-5J20 cell line expressing doxycyclin-inducible green fluorescent protein (GFP)–tagged ETO2-GLIS2 fusion (fig. S5A). We performed ChIP-seq from the GFP-tag and H3K27ac upon doxycyclin induction of the fusion expression. Looking at SE regions previously identified in ETO2-GLIS2 AMKL–harboring cells, we observed a marked and significant increase of H3K27ac signal intensity after induction of the fusion expression (Fig. 4A). We further called differential H3K27ac peaks using MAnorm (19). We observed a similar overall number of peaks increased and decreased upon ETO2-GLIS2 induction (29779 and 23749, respectively), but more than 2.5-fold more increased peaks within SE regions previously identified in ETO2-GLIS2+ AMKL cells and almost all peaks overlapping top SE hits of the CRISPRi screen were increased upon the fusion expression (Fig. 4B). In particular, SEKIT was not active in noninduced HEL-5J20 cells and gained strong H3K27ac signal upon ETO2-GLIS2 expression (Fig. 4C). qPCR analyses showed that induction of ETO2-GLIS2 expression was accompanied by a strong up-regulation of KIT and PDGFRA expression (Fig. 4D). In addition, ETO2-GLIS2 inhibition in M07e cells using a nervy homology region 2 (NHR2)–interfering peptide (NC128) (11, 20) showed inhibition of KIT and PDGFRA expression (fig. S5B), further indicating that the ETO2-GLIS2 fusion activity is required to induce SEKIT activity and proximal KIT and PDGFRA gene expression. All top SE hits identified during the CRISPRi screen, including SEKIT, showed direct binding of ETO2-GLIS2 (Fig. 4C and fig. S5, C to F). To confirm the results obtained in HEL-5J20 cells, we selected OCI-AML3 AML cell line for its lack of H3K27ac at the SEKIT locus (Fig. 3A) and performed similar experiments using doxycycline (DOX)–inducible ETO2-GLIS2 expression (fig. S6A). ChIP-seq for H3K27ac upon induction of ETO2-GLIS2 expression showed a marked increase of signal at SE regions originally identified in ETO2-GLIS2+ AMKL cells (fig. S6B). Differential H3K27ac peak calling using MAnorm showed that most of the peaks located in these specific Super Enhancer regions were significantly increased following ETO2-GLIS2 expression (fig. S6C). This significant H3K27ac increase was particularly notable at the SEKIT locus and at other top SE hits identified during the CRISPRi screen (fig. S6D). Together, these data show that ectopic ETO2-GLIS2 can bind and induce activation of super enhancer regions and particularly an aberrant de novo enhancer, SEKIT, which promotes KIT and PDGFRA expression.
Fig. 4. Ectopic ETO2-GLIS2 expression induces specific Super Enhancer activation.
(A) Profile plot of normalized mean tag density (top) and heatmap of normalized tag density (bottom) of H3K27ac ChIP-seq in HEL-5J20 cells stably expressing doxycycline (DOX)–inducible ETO2-GLIS2 (EG) before and after DOX treatment at 448 SE regions defined in EG+ AMKL cells. (B) Volcano plot showing M value against −log10(P value) of H3K27ac peaks upon differential analysis. Clarets, peaks overlapping SE regions of EG+ AMKL; colors, peaks overlapping top hit SE from CRSIPRi screen. (C) Normalized read density histograms of H3K27ac ChIP-seq analysis at SEKIT locus upon ETO2-GLIS2 expression induction in HEL-5J20 cells compared to noninduced cells (khaki and green tracks, respectively) and read densities of GFP ChIP-seq in noninduced versus DOX-induced HEL-5J20 cells (yellow and orange tracks, respectively) showing ETO2-GLIS2 binding in SEKIT locus. Differentially up-regulated peaks are shown. (D) qPCR showing KIT and PDGFRA expression upon DOX-induced ETO2-GLIS2 expression in HEL-5J20 cells compared cells transduced with empty vector. Means ± SEM, representative of two independent experiments in triplicate, **P < 0.01, ***P < 0.001. (E) Motifs analysis under AMKL ATAC-seq peaks overlapping H3K27ac peaks up-regulated in HEL-5J20 cells expressing ETO2-GLIS2. (F) Heatmap showing variation of expression of selected genes in HEL-5J20 control or expressing ETO2-GLIS2 (left) or M07e cells expressing control or NC128 peptide (right).
To gain more insight into the mechanism of H3K27ac increase and SE activation induced by ETO2-GLIS2, we analyzed enriched transcription factor binding motifs present in ATAC-seq peaks found within induced H3K27ac domains in HEL-5J20 cells overexpressing ETO2-GLIS2 (from Fig. 4B). This analysis identified that binding sequences for ERG, MYB, RUNX1, and GATA3 were the most significantly enriched (Fig. 4E). Expression of these genes was up-regulated in HEL-5J20 cells overexpressing ETO2-GLIS2 for 24 hours and down-regulated in M07e cells expressing the NC128 peptide inhibiting the ETO2-GLIS2 function (Fig. 4F) (11). These results indicate that these factors, involved in enhancer regulation in other contexts, are directly up-regulated by ETO2-GLIS2.
Combined KIT and PDGFRA expression is required for ETO2-GLIS2+ cell growth
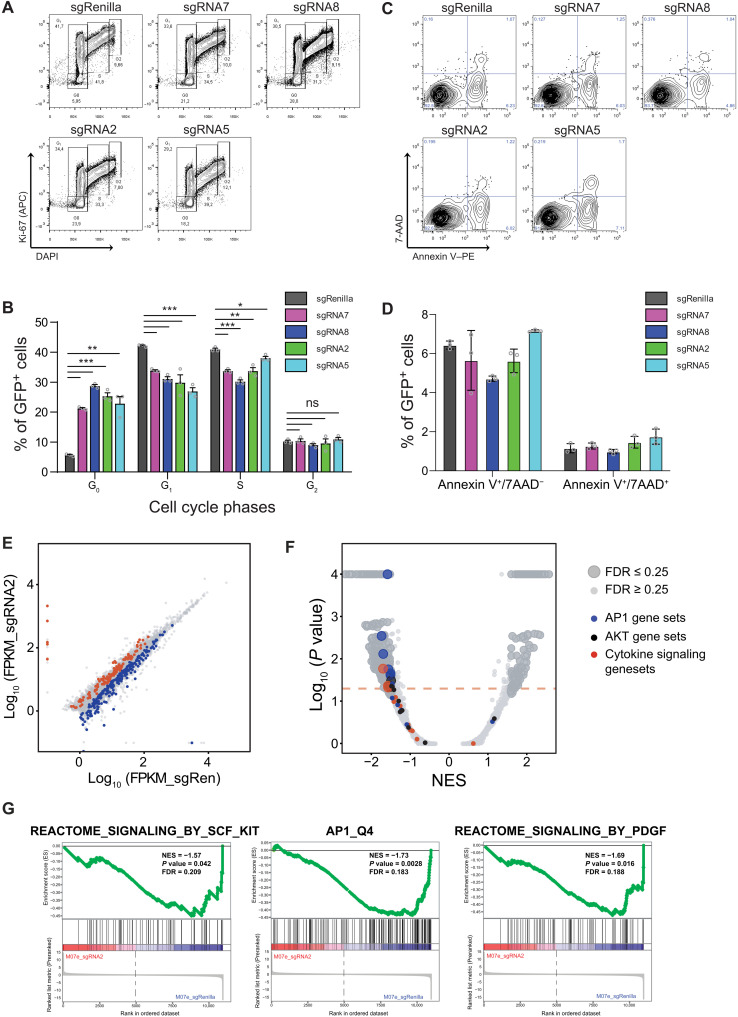
To probe mechanisms responsible for the loss of representation phenotype observed upon CRISPRi inhibition of SEKIT, we performed cell cycle analysis and observed a significant increase of cell in G0 phase and a significant decrease of cells in G1 and S phases (Fig. 5, A and B). Analysis of apoptosis using annexin V and 7-AAD (7-Aminoactinomycine D) staining showed no significant differences following SEKIT inhibition by CRISPRi (Fig. 5, C and D). To gain further insight into this growth inhibition mechanism, we performed RNA-seq experiments upon CRISPRi inhibition after longer kinetics (96 hours after infection). These transcriptomic analyses showed a higher number of genes significantly modulated upon SEKIT inhibition (Fig. 5E) than short kinetic inhibition. Gene set enrichment analysis (GSEA) showed that several gene sets related to cytokine signaling, were negatively enriched in SEKIT inhibited M07e cells, including gene sets related to stem cell factor (SCF)/KIT and PDGF pathways (Fig. 5, F and G, and table S3). In particular, gene sets related to activator protein 1 (AP-1) transcriptional complex (FOS/JUN) and AKT activity were significantly down-regulated upon SEKIT inhibition, suggesting that these pathways are important downstream mediators of KIT and PDGFRA upon SEKIT-mediated regulation (Fig. 5, F and G). Together, these data showed that SEKIT activity is required for proper growth of M07e cells through activation of KIT and PDGFRA signaling.
Fig. 5. SEKIT inhibition induces down-regulation of pro-proliferative transcriptional programs.
(A) Representative flow cytometry analysis showing Ki-67 (APC) and DAPI staining gated on transduced GFP+ M07e cells following CRISPRi targeting of SEKIT with indicated sgRNAs and compared to control sgRenilla. APC, Allophycocyanin. (B) Quantification of cell cycle phases as analyzed in (A). Means ± SEM, n = 3, significance is determined using Student’s t test, *P < 0.05, ** P < 0.01, ***P < 0.001. (C) Representative flow cytometry analysis showing annexin V–PE and 7-AAD (viable dye) staining gated on transduced GFP+ M07e cells following CRISPRi targeting of SEKIT with indicated sgRNAs and compared to control sgRenilla. (D) Quantification of apoptosis as analyzed in (C). Means ± SEM, n = 3. (E) Scatterplot showing significantly depleted (blue) or enriched (red) genes after 4 days of transduction with sgSEKIT2. Dots represent average of log10FPKM across the two replicates. (F) Volcano plot representing GSEA of expression changes in M07e cells expressing SEKIT-targeting sgRNA2 versus sgRenilla. Normalized enrichment scores (NES) are plotted against P values. Red dotted line represents threshold of P < 0.05, gene sets with false discovery rate (FDR) > 0.25 are represented as small dots and gene sets with FDR < 0.25 are represented as large dots. (G) GSEA profiles of representative gene sets negatively correlated in sgRNA2.
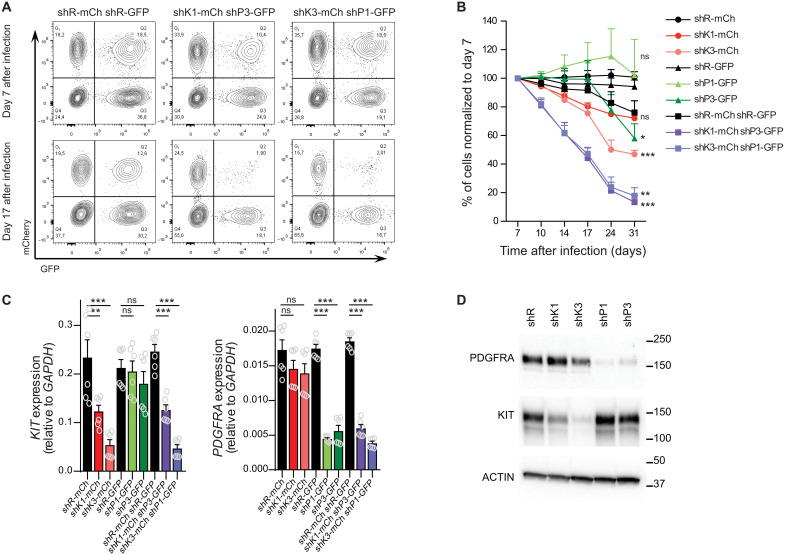
To decipher which of these receptors are essential for cell growth, shRNA against either KIT, PDGFRA, or both were transduced in M07e cells (fig. S7A). Inhibition of these genes separately only modestly affected M07e cell proliferation, but combined inhibition of both genes recapitulated growth inhibition to a similar extent as CRISPRi targeting of SEKIT (Fig. 6, A and B, and fig. S7, B and C). Targeting with shRNA was specific and efficient as shown by qPCR and Western blot analysis (Fig. 6, C and D). These results revealed a functional collaboration of these two neighboring and coregulated genes in cell growth.
Fig. 6. Combined KIT and PDGFRA expression is required for M07e cell growth.
(A to D Two independent shRNAs targeting either KIT (shK) or PDGFRA (shP) were expressed in M07e cells separately or in combination. shK are expressed with mCherry (mCh) in cells while shP are expressed with GFP. Corresponding shRNAs targeting Renilla (shR) were used as control. Cells are maintained in the presence of GM-CSF. (A) Representative flow cytometry analysis of shRNA expressing cells at days 7 and 17 after infection. (B) Percentage of shRNA expressing M07e cells normalized to day 7 after infection. Means ± SEM, n = 4, significance is determined using Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001. (C) qPCR showing KIT and PDGFRA expression at day 4 after infection. Means ± SEM, n = 3, significance is determined using Student’s t test, **P < 0.01, ***P < 0.001. (D) Western blot probing KIT and PDGFRα levels upon shRNA knockdown. Western blot against β-actin is shows as a loading control.
M07e is a cytokine-dependent cell line (21) that is generally cultured in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF). Our finding that KIT and PDGFRA expressions are required for proper M07e growth under GM-CSF stimulation raised the possibility that GM-CSF receptor activity could be dependent on KIT and PDGFRA. Canonical GM-CSF receptor is composed of a ligand-specific subunit α and a common β subunit encoded by CSF2RA and CSF2RB genes, respectively (21). Off-target effects on GM-CSF receptor subunits of shRNA targeting KIT and PDGFRA was ruled out as they did not affect CSF2RA and CSF2RB expression (fig. S7D). In addition, M07e cells can be cultured in the presence of SCF (KIT ligand) or PDGF alone, although these conditions are less efficient than combined stimulation with both ligands or with GM-CSF. Under SCF or PDGF stimulation, only shRNAs targeting KIT or PDGFRA respectively impaired cell growth, further demonstrating that shRNA silencing is on-target (fig. S7, B and C). It was previously reported that the β subunit can interact with KIT (22) and other growth factor receptors (23, 24). In addition, we found that M07e cells express lower levels of CSF2RA compared to CSF2RB (more than 40× less) using qPCR (fig. S7E). Probing absolute expression levels of these two subunits using RNA-seq confirmed that CSF2RA had a very low expression level, whereas CSF2RB is strongly expressed in M07e cells (fig. S7, F and G). Furthermore, it was shown that KIT and PDGFRα are able to interact together in gastrointestinal stromal tumors (25, 26). Together, these findings suggest that survival and proliferative signals in leukemic cells may be triggered by functional interactions between CSF2Rβ, KIT, and PDGFRα.
SEKIT activity is required for ETO2-GLIS2+ AMKL progression in vivo
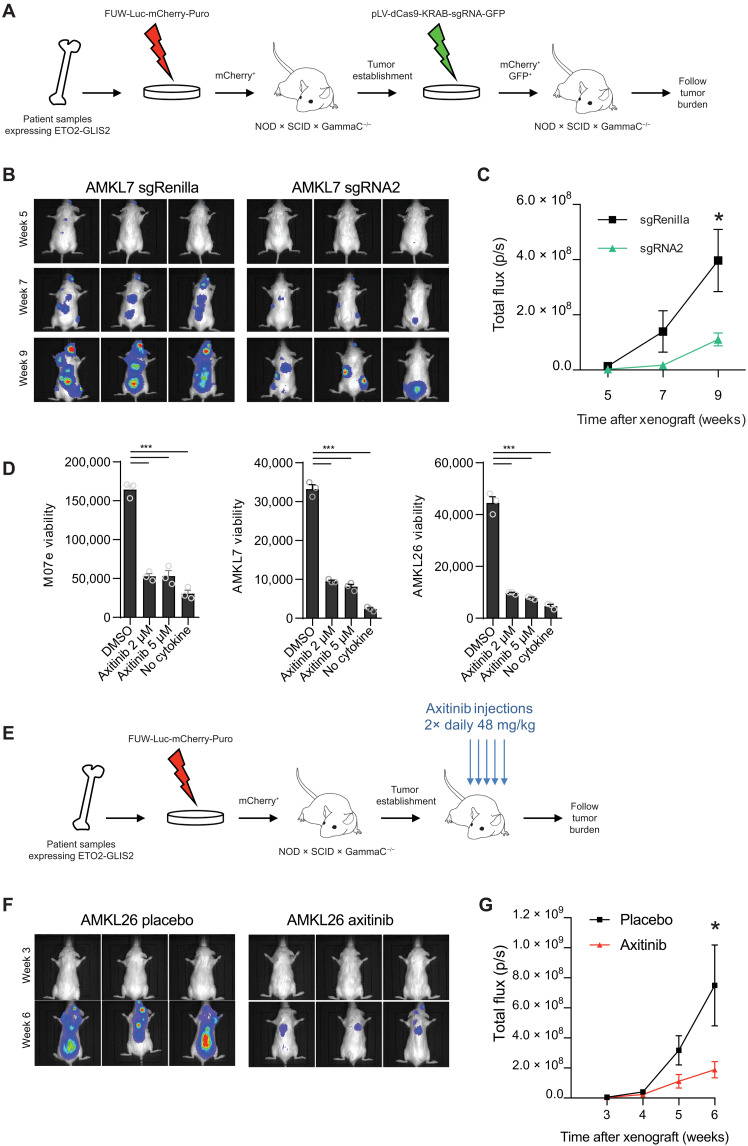
To confirm that SEKIT activity is also important for AMKL patient cell growth, we transduced AMKL7 patient–derived cells to express the SEKIT-targeting CRISPRi system. Initially, we targeted the open region of the major H3K27Ac peak in common with the M07e cell line using the sgRNA2 (fig. S8A) and performed RNA-seq and qPCR shortly after CRISPRi transduction (48 hours after infection). Similar to M07e cells, very few transcriptional changes were significant, including inhibition of KIT and PDGFRA (fig. S8, B and C). We next xenografted these transduced cells into immunocompromised NSG mice to follow leukemia progression using noninvasive bioluminescent imaging (Fig. 7A). We observed a significant delay in leukemia progression in recipients transplanted with AMKL7 cells targeted for SEKIT when compared to animals transplanted with nontargeted control (Fig. 7, B and C). Analyses of hematopoietic tissue infiltration after 12 weeks showed a reduced infiltration of leukemic cells in bone marrow, spleen, and liver (fig. S8, D and E). These results demonstrate that SEKIT inhibition can impair AMKL progression in vivo.
Fig. 7. SEKIT targeting inhibits AMKL progression in vivo.
(A) Schematic illustration describing CRISPRi targeting of SEKIT in vivo using patient-derived xenograft model. (B) Representative bioluminescent imaging of NSG recipient mice transplanted with sgRNA2- or sgRenilla-transduced AMKL7lucmCherry+ patient cells at indicated posttransplant time. (C) Quantification of bioluminescence in vivo as analyzed in (B). Means ± SEM, n = 5, statistical significance is determined using Student’s t test, *P < 0.05. (D) Viability of the M07e cell line, AMKL7, and AMKL26 patient cells treated with axitinib or vehicle (DMSO) for 96 hours. Negative control without cytokine is shown. Means ± SEM, n = 3, significance is determined using Student’s t test, ***P < 0.001. (E) Schematic illustration describing axitinib treatment strategy in vivo using patient-derived xenograft model. (F) Representative bioluminescent imaging of NSG recipient mice transplanted with AMK26lucmCherry+ patient cells treated with vehicle or axitinib at indicated posttransplant time. (G) Quantification of bioluminescence in vivo as analyzed in (F). Means ± SEM, n = 10, statistical significance is determined using Student’s t test, *P < 0.05.
Tyrosine kinase inhibitors impair ETO2-GLIS2+ AMKL growth in vivo
Together, these data indicate that ETO2-GLIS2+ leukemia growth is dependent on, at least, two tyrosine kinase receptors. These results introduced the exciting possibility that combined targeting of both receptors using dual specificity tyrosine kinase inhibitors (TKI) could be efficient for the treatment of this disease. To test this hypothesis, we treated the M07e cell line and two AMKL patient–derived samples with increasing doses of five different TKIs that can efficiently inhibit KIT and PDGFRA. Among those, avapritinib and, most particularly, axitinib potently blocked patient’s cell growth (Fig. 7D and fig. S9, A to C). Axitinib is a second-generation inhibitor of vascular endothelial growth factor receptor, KIT, and PDGFRA (27) and is currently approved for clinical usage against refractory renal cell carcinoma in the United States and Europe, raising the possibility of extension of its usage for these patients with leukemia. We tested the sensitivity of different AML cell lines expressing variable levels of KIT (fig. S9D) using increasing doses of axitinib. High axitinib sensitivity was overall correlated to high KIT expression in these cell lines, with ETO2-GLIS2+ cell line M07e being the most sensitive (fig. S9E). We also tested in vitro, sensitivity of three ETO2-GLIS2–negative AML cells from PDX (patient-derived xenograft). Once again, the highest sensitivity of these AML PDX cells to axitinib was correlated to the highest KIT expression with ETO2-GLIS2+ AMKL cells (AMKL7) being the most sensitive (fig. S9, F and G). These results indicate that within patient-derived AML cells, ETO2-GLIS2–expressing cells are highly, but not specifically, sensitive to axitinib.
To test whether axitinib treatment could affect AMKL progression in vivo, AMKL26 patient–derived cells were transplanted in NSG recipient mice, and 2 weeks after engraftment, animals were orally treated twice daily with vehicle or axitinib (48 mg/kg) 5 days a week for 4 weeks (Fig. 7E). Recipients treated with axitinib showed a significant reduction of leukemia burden when compared to vehicle-treated mice (Fig. 7, F and G). These results show that TKI treatment can impair leukemia progression in a preclinical model of ETO2-GLIS2–dependent AMKL.
DISCUSSION
In this study, we sought to design a genome-wide methodology to functionally screen SE in cancer cells to identify essential gene expression modules required for cancer cell maintenance. Using this approach, we pinpointed several enhancers whose activity is required for growth of a dismal prognosis subgroup of pediatric leukemia associated with fusion oncogenes. Functional annotation of these enhancers with neighboring expressed genes reveals a global oncogenic circuitry involving classical oncogenes related to myeloid leukemia, such as MYC, MYB, ERG, KIT, BCL2, and RUNX1. In particular, we identified a de novo SE, which we termed SEKIT, induced by ETO2-GLIS2, controlling the expression of proximal neighboring genes KIT and PDGFRA, which are required for leukemia cell growth. This control seems to be regulated through dynamic DNA looping forming interaction between SEKIT and the target promoters. Our data support a mechanism of transformation implying specific Super Enhancer activation by the ETO2-GLIS2 fusion as we were able to show that upon ectopic expression, the fusion binds and activates these regions and their associated genes. Binding motif analysis, coupled with transcriptomic data, suggests additional molecular players, including MYB and GATA3, two genes up-regulated by ETO2-GLIS2 and whose binding motifs are found together with the fusion in Super Enhancer regions. A recent report by Belver and colleagues (28) showed that GATA3 can participate in enhancer activation by doing nucleosome eviction in T-ALL (T-cell Acute Lymphoblastic Leukemia), and several reports have shown that MYB can recruit CBP/p300, the main enzymes responsible for H3K27ac (29–31), to induce enhancer activation. Investigating whether MYB and GATA3 are recruited to ETO2-GLIS2–regulated SEs to allow opening of the chromatin by nucleosome eviction should be an interesting area of future investigation. We particularly focused on SEKIT as we were able to show that this enhancer is specific of this leukemia subtype, can be induced de novo by ETO2-GLIS2 fusion and co-regulating two tyrosine kinase genes whose signaling synergizes for leukemia growth. It reveals that strong fusion oncogenes may lead, through SE-mediated regulation of functionally synergistic genes, to activation of signaling pathways essential for cancer development. In ETO2-GLIS2–driven pediatric AMKL, our findings introduce the intriguing possibility of using TKIs for the treatment of this aggressive disease. A recent study by Wagenblast and colleagues (32) identify overexpression and activation of KIT as an essential factor for induction and maintenance of DS-associated AMKL and show that TKIs could also be beneficial in this subtype. These findings, together with the basal activity on SEKIT observed here in the CMK cell line, a DS AMKL cell line, suggest converging mechanisms in yet genetically different pediatric AMKL. Emergence and progression of diseases exhibiting strong and unique driver mutations may depend on the rewiring of a modest number of SE regions that would be manageable to annotate using our screening methodology to better understand the biology involved in the transformation potential of these oncogenes and narrow down the list of gene candidates with promising therapeutic values. This approach could be particularly amenable to studying other poorly druggable oncogenes involving transcriptional and epigenetic remodeling factors, including fusion oncogenes [e.g., MLL (33) and NUP98 (34) fusions] not only in hematopoietic malignancies but also in others cancers, including ependymoma (35) and H3.3 mutants glioma (36). Together, we believe that systematic screening of essential SE can reveal coordinated regulation of genes modules involved in cancer cell transformation and cancer progression and uncover novel therapeutic strategies.
MATERIALS AND METHODS
Cells and culture
M07e, HEL-5J20, and AMKL patient cells were obtained as previously described (11). Kasumi-1 cells were obtained from American Type Culture Collection. CD34+ cord blood cells were purchased from ABCelllBio. M07e cells were cultured in minimum essential medium α (MEM α) supplemented with 20% fetal bovine serum (FBS), penicillin (100 U/ml), streptomycin (100 U/ml) (all from Gibco), and GM-CSF (5 ng/ml; PeproTech). HEL-5J20 cells were cultured in MEM α supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 U/ml). Kasumi-1 cells were cultured in RPMI 1640 (Gibco) supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 U/ml). 293T cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 U/ml). AMKL patient cells, maintained in immunodeficient mice, were cultured in StemSpan serum-free expansion medium (STEMCELL Technologies) supplemented with penicillin (100 U/ml), streptomycin (100 U/ml), and human interleukin-3 (IL3), IL6, SCF, GM-CSF, TPO (Thrombopoietin), FLT3 (Fms-like tyrosine kinase 3), and insulin-like growth factor 1 (IGF1) (10 ng/ml each; PeproTech).
The generation of DOX-inducible expression of ETO2-GLIS2 (and variants) HEL-5J20 cell lines was performed as follows. The cDNA encoding ETO2-GLIS2-GFP was cloned in a DOX-inducible LT3-GEPIR lentiviral vector (gift from J. Zuber, Austria) using Bam HI/Bgl II and Eco RI. HEL-5J20 cells were transduced with lentiviral particles produced from LT3-GFP or LT3-ETO2-GLIS2-GFP vectors, maintained in RPMI 1640 medium supplemented with 10% FBS and induced with DOX (500 ng/ml). Twenty-four hours after induction, GFP+ cells were single-cell–sorted in 96-well plates to obtain clones. Validation of the selected clones was performed by Western blotting on protein extracted from cells induced with DOX for 40 hours.
Plasmids, lentiviral, and retroviral gene transfer
pHR-SFFV-KRAB-dCas9-P2A-mCherry (Addgene #60954) was used to generate stable M07e expressing dCas9-KRAB fusion protein cell line. sgRNA library used in the screen was cloned into pU6-sgRNA-EF1Alpha-puro-T2A-BFP (Addgene #60955). The same vectors were used for screen validation or pLV-hU6-sgRNA-hUbC-dCas9-KRAB-T2a-GFP (Addgene #71237) was used. Sequence of clone sgRNA can be found in table S4. shRNAs targeting KIT, PDGFRA, or Renilla (table S5) were cloned into pLMP-Puro-IRES-GFP or pLMP-Puro-IRES-mCherry (gift from I. Aifantis, New York University).
Viral particles were produced in 293T cells by cotransfecting plasmids of interest along with a lentivirus packaging plasmid (psPAX2, Addgene #12260) and a vesicular stomatitis virus envelope expression plasmid (pMD2.G, Addgene #12259) or along with pCL-10A1 retrovirus packaging vector using calcium phosphate method. For transductions, cells were spinoculated twice with 293T supernatants harvested 24 and 48 hours after transfection and supplemented with polybrene (4 μg/ml) for 90 min at 2300 rpm and 30°C. Efficiency of knockdown was checked on homogeneous cell populations with respect to BFP, GFP, or mCherry expression after cell sorting or puromycin selection (1 μg/ml for 48 hours) by reverse transcription qPCR.
CRISPRi screen
SEs in M07e cells were identified using H3K4me3 and H3K27Ac ChIP-seq by keeping H3K27Ac peaks overlapping low enrichment or no H3K4me3 peaks and not overlapping gene transcription start sites. SEs were ranked using ROSE algorithm (4, 5) based on H3K27Ac signal. A total of 7381 sgRNAs were designed in chromatin accessible sites, as defined by ATAC-seq peaks (window centered on peak center and extended ±750bp) located in 448 SEs using CRISPR library designer (CLD) (37). Nontargeting sgRNAs were included in the pool (one shRNA targeting Renilla, one shRNA targeting Luciferase, and two shRNAs targeting mouse Lin28) as negative controls, and nine sgRNAs targeting a window of −100 bp top +500 bp around ERG TSS (Transcription Start Site) were used as positive controls. Oligos were synthesized and ordered as a pool of 73-nucleotide oligomer [GGAGAACCACCTTGTTGG(X)20GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGC, where X is the protospacer sequence] from CustomArray Inc. Double-stranded sgRNA was completed by PCR using the 73-nucleotide oligomer oligo and the following reverse oligo: CCTAGTACTCGAGAAAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC. Library was then amplified using the following primers: Lib_amp_FOR: GGAGAACCACCTTGTTGG; Lib_amp_REV: CCTAGTACTCGAGAAAAAAAGCACC. Pooled sgRNA library was cloned into pU6-sgRNA-EF1Alpha-puro-T2A-BFP using Bst XI and Xho I restriction sites. Polyclonal population of M07e dCas9-KRAB–expressing cells was sorted by flow cytometry using mCherry expression, amplified, then infected with sgRNA library in triplicate. Fifty million cells were infected with 15% transduction efficiency to achieve an effective multiplicity of infection of less than one sgRNA per cell and to ensure library coverage of 1000×. Two days after infection, cells were treated with puromycin (1 μg/ml). Two days after puromycin selection, dead cells were removed by Ficoll gradient, and 8 million BFP+ cells were sampled (day 0 of the screen). More than 16 million BFP+ cells were maintained in the presence of puromycin (0.5 μg/ml) at each passage to preserve library representation for the next samplings. Eight million BFP+ cells sampled at days 0, 7, 14, and 21 of the screen were lysed in 50 mM tris-HCl, 50 mM EDTA, 100 mM NaCl, 1% SDS supplemented with ribonuclease A (RNAse A) (0.1 mg/ml; Thermo Fisher Scientific), and proteinase K (0.2 mg/ml; Invitrogen) for 12 hours at 37°C. Genomic DNA was then isolated by phenol-chlorophorm-isoamyl alcohol extraction using PhaseLock tubes (5PRIME) followed by 100% ethanol precipitation in the presence of 0.1 M sodium acetate and glycogen (20 μg/ml). Genomic DNA was then digested with Pst I (New England Biolabs) for 12 hours at 37°C and run on 1% agarose gel to isolate 700 to 1500 bp fraction that contain the sgRNA cassettes using E.Z.N.A. Gel Extraction Kit (Omega Bio-tek). Deep sequencing libraries were generated by PCR amplification of sgRNA cassettes using sgRNA_P5_seq:
AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTTGGAGAACCACCTTGTTGG
and sgRNA_P7_barcode_seq:
CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGCCTAATGGATCCTAGTACTCGAG, where XXXXXX represents six nucleotide TruSeq indexing barcode for Illumina sequencing. We obtained at least 4 μg of 700 to 1500 bp DNA fraction from 8 million BFP+ cells that were used in total as template in multiple parallel 100 μl PCR reaction, each containing 1 μg of template, 1× Phusion HF buffer, 2 U of Phusion Hot Start II High-Fidelity DNA polymerase (Thermo Fisher Scientific), 0.2 mM deoxynucleotide triphosphate (dNTP), 3% dimethyl sulfoxide (DMSO), and 0.125 μM of each primer, which were run using the following cycling parameters: 98°C for 2 min; 29 cycles of 98°C for 30 s, 58°C for 15 s, 72°C for 15 s; and 72°C for 10 min. PCR products (286 bp) were combined for each sample, purified on 1% agarose gel using the E.Z.N.A. Gel Extraction Kit, further purified using 1.4 volume of AMPure XP beads, and sequenced on the Illumina HiSeq 4000 Sequencer (50 bp single end) at a coverage of more than 8 million reads per sample. Data were analyzed using maximum likelihood of enrichment algorithm from MAGeCK software (38). GSEA on proximal expressed genes of top SE hits was computed using online GSEA tool (https://gsea-msigdb.org/), and results were plotted in R using ggplot2 package. Network interaction analysis was performed using GENEMANIA database and visualized using Cytoscape (39).
Quantitative real-time PCR
For mRNA quantification, total RNA was isolated using the E.Z.N.A. MicroElute Total RNA Kit (Omega Bio-tekkit) and transcribed into complementary DNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR reactions were carried out using 1× GoTaq qPCR Master Mix (Promega) and 0.25 μM of forward and reverse primers (table S6) on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems).
Chromatin immunoprecipitation and sequencing
The ChIP protocol was described previously (11). Briefly, cells were fixed with 1% formaldehyde, lysed at a concentration of 20.106 cells per ml, and lastly sonicated (30-min cycle on Covaris apparatus, KBioscience). Sheared chromatin was immunoprecipitated overnight using the anti-H3K27Ac antibody (ActiveMotif, #39133). For H3K27ac ChIP in OCI-AML3 cells, cells were fixed in 1% formaldehyde, and ChIP was subsequently performed using iDeal ChIP-seq kit for histones (Diagenode, #C01010059) with anti-H3K27ac antibody from Diagenode (#C15410174) following the manufacturer’s protocol. For GFP-ChIP, GFP-Trap agarose beads (Chromotek) were used. Enriched DNA from ChIP and input DNA fragments were end-repaired, extended with an “A” base on the 3′end, ligated with indexed paired-end adaptors (NEXTflex, Bioo Scientific) using the Bravo Platform (Agilent), size-selected after 4 cycles of PCR with AMPure XP beads (Beckman Coulter), and amplified by PCR for 10 cycles more. Libraries were single-end sequenced (50 bp) using Illumina HiSeq 4000 or Illumina NextSeq 500 (Illumina, San Diego, CA).
Sequencing reads were aligned to the human hg19 version of the genome using Bowtie2 (40), duplicate reads were removed using Picard v2.25 MarkDuplicates (https://broadinstitute.github.io/picard/), and peaks were called using MACS2 (41) using default options. Normalized bigwig files for gene track representations were generated using Deeptools (42) with the --normalizeUsing RPKM option. Heatmaps and profile plots were generated using Deeptools.
Differential peak calling analysis was performed using MAnorm (19) using the following parameters: --s1 124, --s2 109, -w 500, and --summit-dis 100.
ATAC sequencing
ATAC-seq was performed as previously described (43) with some modifications. Briefly, 50,000 cells were spun at 500g for 5 min, washed with cold phosphate-buffered saline (PBS), lysed in cold lysis buffer [10 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL CA-630] and immediately spun at 500g for 10 min. The pelleted nuclei were resuspended in 50 μl of transposase reaction mix (Nextera Tn5 Transposase, Illumina) for 30 min at 37°C. Transposed DNA was purified using a DNA Clean & Concentrator-5 kit (Zymo Research) in 10 μl of nuclease-free H2O and amplified with NEBNext High-Fidelity 2× PCR Master Mix and 1.25 M of custom Nextera PCR primers as previously described (43), using the following PCR conditions: 72°C for 5 min, 98°C for 30 s, then 12 cycles of 98°C for 10 s, 63°C for 30 s, and 72°C for 1 min. Libraries were purified with AMPure XP beads (Beckman Coulter) and then subjected to high-throughput paired-end sequencing (50 bp) using the Illumina HiSeq 4000 sequencer (Illumina, San Diego, CA).
Sequencing reads were aligned to the human hg19 version of the genome using Bowtie2 (40), and accessibility peaks were called using MACS2 (41) using default options. Bedgraph files normalized for the depth of sequencing of aligned reads were generated using bedtools.
Motif enrichment analysis was performed using Homer v4.11 (44) on ATAC-seq peaks identified in AMKL7 and M07e cells located in SEs H3K27ac peaks significantly up-regulated (log2FC ≥ 0.4, P ≤ 10−5) in HEL-5J20 cells (1152 regions) using the following parameters: hg19 -size 600.
RNA sequencing
RNA from M07e and AMKL7 cells subjected to CRISPRi inactivation of KIT SE, puromycin-selected or GFP-sorted, was extracted using the RNeasy Plus Mini Kit (Qiagen). Poly(A)-selected, first-stranded Illumina libraries were prepared with a modified TruSeq protocol using deoxyuridine triphosphate (dUTP) method (45). Two biological replicates per cell type were prepared. AMPure XP size–selected libraries were amplified by PCR (maximum 16 cycles), purified with AMPure XP beads, and paired-end–sequenced (50 bp) on the Illumina HiSeq 4000 sequencer. Sequencing reads were aligned to the hg19 version of the human genome using Tophat2. Differential expression analysis was done using CuffDiff (46).
GSEA (47) was performed on a preranked list of genes after filtering out nonexpressed genes (genes with median FPKM expression below 1 in both compared conditions). Genes were ranked using their log2 fold change of expression between compared conditions. GSEA was carried out using MSigDB genesets from C2 common pathways, C3 transcription factors, C4 cancer modules, C5 molecular functions, C6, and Hallmark collections.
Cell viability assay
AML cell lines, AML, and AMKL patient cells were plated in 96-well plates (2.105 cells per well) in their respective media and treated with kinase inhibitors axitinib, amumavatinib, telatinib, avapritinib, and imatinib (all from Selleckchem). Cell viability was measured after 96 hours of treatment as adenosine triphosphate (ATP) luminescence signal readout using CellTiter-Glo Assay (Promega) according to the manufacturer’s instructions.
In vivo xenotransplantation assays
The use of human patient samples in this study was approved by the Internal Review Board of Gustave Roussy, and samples were obtained with the informed consent of the patient in accordance with good clinical practice rules and national ethics recommendations. Mice were maintained in pathogen-free conditions, and all experiments were approved by the Gustave Roussy institute animal care and use committee (Comité d’Ethique #26, projects: 2012-017 and 2017122111548235).
AMKL patient–derived xenografts have been described previously (8). For bioluminescence follow-up of human AMKL cells, human AMKL7 or AMKL26 patient cells were transduced with a FUWLuciferase-mCherry-puromycin lentiviral vector (a gift from A. L. Kung, Dana Farber Cancer Institute, Boston), sorted on mCherry expression, and then amplified in NSG recipients.
mCherry+ AMKL cells were further used for transduction with pLV-hU6-sgRNA-hUbC-dCas9-KRAB-T2a-GFP lentiviral vector for CRISPRi inactivation of KIT SE (sgRNA2) or negative control (sgRenilla). The 1.106 AMKL7lucmCherry+GFP+ blasts expressing sgRNA2 or sgRenilla were intravenously injected into 6- to 10-week-old female NSG mice (The Jackson Laboratory). For in vivo treatment with axitinib, 0.65 million of human AMKL26 patient cells (AMKL26luc) transduced with the FUW-Luc-mCherry-puro lentiviral construct are injected intravenously in 6- to-10-week-old female NSG mice (The Jackson Laboratory). Engraftment was monitored at 1 and 2 weeks after transplantation by bioluminescence. Two weeks after transplantation, drug treatment of AMKL26luc mice was performed with vehicle or axitinib (48 mg/kg) by oral gavage twice a day for 4 weeks.
Recipients were monitored weekly by bioluminescence. To this aim, mice received d-luciferin (150 mg/kg; Beetle luciferin, E1605, Promega) intraperitoneally, were anesthetized with 3% isoflurane, and imaged 1 to 5 min with an IVIS50 system (PerkinElmer). Bioluminescence intensity is expressed as photons per second (p/s).
Flow cytometry
Cells were analyzed on a BD LSR II or a BD Fortessa flow cytometer using FACS Diva software (BD Biosciences). Fluorescence-activated cell sorting data were analyzed on Flow Jo version 10 (LLC).
To assess cell proliferation after CRISPRi inactivation of KIT SE or shRNA knockdown of KIT and PDGFRA expression, infected cells were mixed with uninfected cells in equal ratio. Percentages of infected cells were followed over time using BFP, GFP, or mCherry expression, normalized to first day of follow up, and compared to negative controls targeting Renilla or GFP.
For cell cycle analysis, 1 × 106 M07e cells transduced with variants of pLV-hU6-sgRNA-hUbC-dCas9-KRAB-T2a-GFP lentiviral vector were fixed with 2% paraformaldehyde for 15 min, permeabilized with PBS, 0.5% bovine serum albumin (BSA), and 0.1% triton for 15 min, incubated in PBS and 0.5% BSA containing mouse anti-human Ki-67 (Alexa Fluor 647, clone B56; #558615, BD Pharmingen) for 30 min at 4°C, washed twice with PBS, incubated in 500 μl of PBS, 0.5% BSA containing RNAse A (5 μg/ml; Thermo Fisher Scientific), and 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/ml; #564907, BD Pharmingen) for 15 min, and then analyzed gating on GFP+ cells.
KIT and PDGFRA expression was assessed using mouse anti-human CD117 (PE-cyanine7, clone 104D2, #25-1178-42, eBioscience) and CD140a [PE (Phycoerythrin), clone αR1, #556002, BD Pharmingen], respectively. Cells were stained in PBS, 2% FBS and 2 mM EDTA for 30 min at 4°C, washed, and then analyzed. For apoptosis assay, the PE Annexin V Apoptosis Detection Kit (BD Biosciences #559763) was used following the manufacturer’s instructions.
Twelve weeks after xenotransplantation, mice were euthanized for assessment of chimerism in bone marrow, spleen, and liver. Cells from diseased mice organs were collected, subjected to red blood cell lysis, washed, resuspended in PBS, 2% FBS, and 2 mM EDTA containing DAPI (1 μg/ml), and then analyzed.
4C sequencing
The protocol was performed as previously described with minor modifications (48). Briefly, 10 to 15 × 106 cultured M07e cells were fixed with 12 ml of 2% formaldehyde (Thermo Scientific #28908) in 10% FBS for 10 min at room temperature (RT) (tumbling). Quenching of the cross-linking was performed with the addition of 1.8 ml of freshly prepared ice-cold 1 M glycine (Sigma-Aldrich #500046). Tubes were transferred directly on ice and centrifuged for 5 min 300g at 4°C. Cells were washed with 1× PBS and centrifuged for 5 min 300g at 4°C, and pellets were frozen in liquid nitrogen and stored at −80°C. Cells were then vigorously resuspended in 1 ml of fresh ice-cold lysis buffer [10 mM tris (pH 8), 10 mM NaCl, 0.2% NP-40, and 1 tablet of complete protease inhibitor (Roche #04693159001)], transferred to 9 ml of prechilled lysis buffer, and incubated for 20 min on ice. After 5 min of centrifugation at 300g at 4°C, nuclei were resuspended in 10 ml of lysis buffer, incubated for 10 min on ice, and centrifuged at 300g at 4°C. Nuclei were then resuspended in 100 μl of 0.5% SDS solution and incubated for 10 min at 62°C (under agitation 900 rpm). Twenty-five microliters of 20% Triton and 315 μl of sterile water were added to the nuclei and incubated for 15 min at 37°C (under agitation 900 rpm). After adding 50 μl of DpnII buffer, 2 μl of BSA [20 mg/ml; NEB (New England Biolabs) #B9000S], and 400 U of DpnII (NEB #R0543M), samples were incubated for 4 hours at 37°C (under agitation 900 rpm). Before incubation, 5 μl of the sample was taken as the “undigested control.” Another 400 U of DpnII was added and incubated overnight at 37°C (under agitation 900 rpm). Five microliters of the sample was taken as the “digested control.” Efficiency of chromatin digestion was verified after DNA extraction from 5 μl of undigested and digested controls and loading in a 1.5% agarose gel. After verification of chromatin digestion (smear between 0.2 and 2 kb), DpnII was deactivated by 20 min incubation at 62°C (under agitation 600 rpm). Ligation of DNA ends between the cross-linked DNA fragments was performed by addition of 992 μl of ligation master mix [150 μl of 10× ligation buffer (NEB #B0202S), 759 μl of sterile water, 75 μl of 20% Triton, 8 μl of BSA (20 mg/ml; NEB #B9000S), and 100 U of T4 ligase (NEB #M0202M)]. Samples were incubated overnight at 16°C followed by 30 min at RT. One hundred microliters of the ligated sample was tested as “ligated control,” on a 1.5% agarose gel. After an overnight reverse cross-linking at 65°C (under agitation 900 rpm), samples were divided in two safe lock tubes (approximately 500 μl each) and incubated at 45°C for 4 hours under agitation (900 rpm) in the presence of 50 μl of proteinase K (20 mg/ml; Sigma-Aldrich #P2308) and 30 μl of 20% SDS. Each tube was then sequentially incubated with 0.5 M NaCl for 1 hour at 65°C and with 15 μl of RNase A (10 mg/ml; Sigma-Aldrich #R4642) for 1 hour at 37°C (both under agitation 900 rpm). Samples were pooled back, and DNA was purified with phenol chloroform (PCI), ethanol-precipitated, resuspended in 200 μl of H20, and incubated for 1 hour at 37°C. Efficiency of extraction and purification were verified on a 1.5% agarose gel. The DpnII-ligated 3C template was diluted to a concentration of 100 ng/μl in 1× Cutsmart buffer (NEB #B7204S), and digested overnight at 37°C with 1 U of NlaIII enzyme per microgram of DNA (NEB #R0125L) (under agitation 600 rpm). NlaIII was inactivated at 65°C for 20 min, and DNA fragmentation was tested on 1.5% agarose gel. Samples were transferred to 50-ml falcon tubes, diluted up to 12.6 ml in sterile water, and DNA was ligated in the presence of 1.4 ml 10× ligation buffer and 200 U of T4 ligase overnight at 16°C. After 30 min of incubation at RT, samples were PCI-extracted, ethanol-precipitated, resuspended in 200 μl of sterile water, and purified using the Qiaquick PCR Purification Kit (Qiagen). DNA concentration of each digested sample was calculated using the Qubit brDNA HS assay kit (Invitrogen). Specific primers used to PCR-amplify the 4C DNA for peak1 VIEWPOINT: PEAK1-1-DpnII-F-2, 5′-GGGTAAGCAAAGGTTAGGAA-3′ and PEAK1-1-NlaIII-R-2, 5′-ATTAGCCCACTCTCTCACAT-3′. PCR reactions were performed using the Expand Long Template PCR system (Roche #11759060001) with the following PCR conditions: 94°C for 2 min; 30 cycles of 94°C for 15 s, 55°C for 1 min, and 68°C for 3 min, followed by a final step of 68°C for 7 min. The 4C library was sequenced on a NextSeq 500 Illumina sequencer (75 bp, single-end). Data were analyzed as previously described (16).
3D DNA FISH
DNA FISH probes
Bacterial artificial chromosomes CTD-3180G20 (KIT), RP11-660 L2 (SEKIT), CTD-2360 L14 (KIT control region), and RP11-39D6 (PDGFRA) were used as templates and directly labeled by nick translation with fluorescent Atto-488, Atto-550, or Atto-647N dUTP (Jena Bioscience), respectively, following the manufacturer’s instructions (Vysis kit, Abbott Molecular). For one hybridization, 0.25 μg of nick-translation product was precipitated with 2.5 μg of human Cot-1 DNA (Life technologies) and resuspended in 20 μl of hybridization buffer [50% formamide, 10% dextran sulphate, BSA (2 mg/ml; NEB #B9000S), and 2× SSC].
Cell preparation and hybridization
3D DNA FISH were carried out on cells adhered to poly-l-lysine–coated 22 mm by 22 mm coverslips, as previously described with some modifications (49, 50). Briefly, cells were fixed with 2% paraformaldehyde/1× PBS (pH 7 to 7.4) for 10 min at RT and permeabilized for 5 min with 0.5% Triton/1× PBS on ice before storage in 70% ethanol at −20°C. Coverslips were thawed at RT, dehydrated through an ethanol series (70, 85, 95, and 100%), and air-dried. Cells were then incubated in 0.7% Triton/0.1 M HCl for 10 min on ice and rinsed twice in 2× SSC. Cells were then denatured simultaneously with the probes at 75°C for 3 min: A superfrost glass slide was prewarmed on a hot plate at 75°C for 30 s, probes were dropped on it, and the coverslip was placed cell-side down onto the drop of probes. Slides were then placed in a dark and humid chamber at 42°C for overnight hybridization. The next day, cells were rinsed three times in 50% formamide/2× SSC (pH 7 to 7.4) and three times in 2× SSC for 5 min each at 42°C. After DAPI staining, coverslips were mounted onto slides in ProLong Gold mounting medium (Invitrogen).
Microscopy
3D image stacks were acquired using an Olympus IXplore spinning disk microscope with a Hamamatsu sCMOS ORCA-Flash 4.0 V3 fluorescent camera and a 100× objective (z step = 250 nm). Images were analyzed using Fiji Is Just ImageJ (FIJI) (51).
Western blotting
Cell were lysed in cell lysis buffer [50 mM tris (pH8), 150 mM NaCl, 2 mM EDTA (pH 8), and 1% NP-40] supplemented with cOmplete protease inhibitor cocktail (Roche). Western immunoblotting was performed using cell lysates normalized for total protein content. Lysates were boiled in Laemmli sample buffer and run on SDS–polyacrylamide gel electrophoresis (Criterion TGX Stain-Free 4 to 15%, Bio-Rad) before transfer to polyvinylidene difluoride membranes using a TransBlot Turbo (Bio-Rad) with an RTA TransBlot Turbo transfer kit (Bio-Rad) following the manufacturer’s recommendations. Immunoblotting was performed in tris-buffered saline supplemented with 0.1% Tween and 5% milk using anti-PDGFRα antibody (Cell Signaling Technologies #3164) at 1:1000 dilution or anti–c-KIT antibody (R&D #AF332) at 1:1000 dilution or anti–β-actin antibody (Sigma-Aldrich #A3854) at 1:5000 dilution.
Data availability
H3K27ac ChIP-seq from 66 AML patients was published previously (18) and is available in SRA database under accession number SRP103200. M07e H3K27Ac and H3K4me3 Chip-seq are available at ArrayExpress database (E-MTAB-4367). M07e and AMKL7 ATAC-seq and RNA-seq data, AMKL7, M07e CRISPRi H3K27ac and H3K9me3, and inducible HEL-5J20 ChIP-seq and M07e 4C-seq data are available at Gene Expression Omnibus database under accession code GSE131462. Microarray data for HEL-5J20 expressing inducible ETO2-GLIS2 and M07e cells expressing NC128 peptide were published previously (11) and are available at ArrayExpress database (E-MTAB-4332).
Statistical analyses
Results are expressed as means ± SEM. Statistical significance was calculated by two-tailed unpaired t test on two experimental conditions with P < 0.05 considered statistically significant. For DNA FISH experiments, a χ2 test was performed to compare distributions of KIT-SEKIT and KIT-control distances between M07e cells and HEL-5J20 cells, as well as to test the hypothesis (H0) that KIT is closer to SEKIT than to the control region compared to an expected distribution where probabilities of KIT to be closer to SEKIT or to the control region are equal (50/50%). Statistical significance levels are denoted as follows: ***P < 0.001; **P < 0.01; *P < 0.05. No statistical methods were used to predetermine sample size.
Acknowledgments
We would like to thank B. Jost, C. Thibault-Charpentier, M. Cerciat, and all the members of the GenomeEast sequencing facility for advice and expert sequencing. We thank D. Noordermeer and M. Dos Santos for advice with the 4C protocol. We would like to thank the members of the PFIC and PFEP of UMS AMMICA for expert sorting and animal care. We thank the IMAG’IC and GENOM’IC facilities of the Institut Cochin. We would like to thank N. Setterblad, C Doliger, S. Duchez, A. Alberdi, V. Parietti, and all members of the “Plateforme Saint Louis” for expert sorting, sequencing, and animal care.
Funding: This work was supported by the ATIP-Avenir program from French government (to C.L. and J.C.), the Fondation ARC (to C.L., J.C., and M.-C.L.), and the Fondation Gustave Roussy, PAIR-Pédiatrie/CONECT-AML (COllaborative Network for Children and Teenagers with Acute Myeloblastic Leukemia: INCa-ARC-LIGUE_11905), Association Laurette Fugain (ALF-2015/13 to T.M.), Fondation pour la Recherche Médicale (to C.K.L.), and Institut National du Cancer (PLBIO-2018-169 to T.M. and PLBIO-2020-097 to C.L.). Sequencing was performed by the GenomEast platform, a member of the ‘France Génomique’ consortium (ANR-10-INBS-0009).
Author contributions: C.L. and S.B. designed the research, performed experiments, and analyzed data. M.-C.L. helped optimize CRISPRi screening. S.L. helped with shRNA experiments. M.A. helped with CRISPRi experiments. E.S. and J.C. designed, performed, and analyzed 4C-seq and DNA FISH experiments. A.Pe. provided primary patient samples. C.K.L. and Z.A. performed in vivo experiments. B.P. and A.Pu. helped with in vivo experiments. C.T. and Z.A. performed inducible fusion experiments and ChIP-seq. Y.B., J.H., A.d.S.B., D.N.-L., and C.V. helped with revision experiments. C.L., S.B., J.C., and T.M. cowrote the paper. C.L., J.C., and T.M. supervised the research.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All high-throughput sequencing data generated for this study are available at Gene Expression Omnibus database under accession code GSE131462. The patient-derived material may be provided pending a scientific review and following the completion of a material transfer agreement [requests should be submitted to T.M. (Thomas.mercher@inserm.fr)]. All correspondence and material request should be addressed to either C.L. (Camille.lobry@inserm.fr) or T.M. (Thomas.mercher@inserm.fr).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Legend for table S1
Tables S2 to S6
Other Supplementary Material for this manuscript includes the following:
Table S1
REFERENCES AND NOTES
- 1.Meyerson M., Gabriel S., Getz G., Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11, 685–696 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Pott S., Lieb J. D., What are super-enhancers? Nat. Genet. 47, 8–12 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Sengupta S., George R. E., Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer 3, 269–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whyte W. A., Orlando D. A., Hnisz D., Abraham B. J., Lin C. Y., Kagey M. H., Rahl P. B., Lee T., Young R. A., Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovén J., Hoke H. A., Lin C. Y., Lau A., Orlando D. A., Vakoc C. R., Bradner J. E., Lee T., Young R. A., Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hnisz D., Abraham B. J., Lee T., Lau A., Saint-André V., Sigova A. A., Hoke H. A., Young R. A., Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber T. A., Gedman A. L., Zhang J., Koss C. S., Marada S., Ta H. Q., Chen S.-C., Su X., Ogden S. K., Dang J., Wu G., Gupta V., Andersson A. K., Pounds S., Shi L., Easton J., Barbato M. I., Mulder H. L., Manne J., Wang J., Rusch M., Ranade S., Ganti R., Parker M., Ma J., Radtke I., Ding L., Cazzaniga G., Biondi A., Kornblau S. M., Ravandi F., Kantarjian H., Nimer S. D., Döhner K., Döhner H., Ley T. J., Ballerini P., Shurtleff S., Tomizawa D., Adachi S., Hayashi Y., Tawa A., Shih L.-Y., Liang D.-C., Rubnitz J. E., Pui C.-H., Mardis E. R., Wilson R. K., Downing J. R., An Inv(16)(p13.3q24.3)-encoded CBFA2T3-GLIS2 fusion protein defines an aggressive subtype of pediatric acute megakaryoblastic leukemia. Cancer Cell 22, 683–697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiollier C., Lopez C. K., Gerby B., Ignacimouttou C., Poglio S., Duffourd Y., Guégan J., Rivera-Munoz P., Bluteau O., Mabialah V., Diop M., Wen Q., Petit A., Bauchet A.-L., Reinhardt D., Bornhauser B., Gautheret D., Lecluse Y., Landman-Parker J., Radford I., Vainchenker W., Dastugue N., de Botton S., Dessen P., Bourquin J.-P., Crispino J. D., Ballerini P., Bernard O. A., Pflumio F., Mercher T., Characterization of novel genomic alterations and therapeutic approaches using acute megakaryoblastic leukemia xenograft models. J. Exp. Med. 209, 2017–2031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Rooij J. D., Branstetter C., Ma J., Li Y., Walsh M. P., Cheng J., Obulkasim A., Dang J., Easton J., Verboon L. J., Mulder H. L., Zimmermann M., Koss C., Gupta P., Edmonson M., Rusch M., Lim J., Reinhardt K., Pigazzi M., Song G., Yeoh A., Shih L.-Y., Liang D.-C., Halene S., Krause D. S., Zhang J., Downing J. R., Locatelli F., Reinhardt D., van den Heuvel-Eibrink M. M., Zwaan M. C., Fornerod M., Gruber T. A., Pediatric non–Down syndrome acute megakaryoblastic leukemia is characterized by distinct genomic subsets with varying outcomes. Nat. Genet. 49, 451–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez C. K., Noguera E., Stavropoulou V., Robert E., AID Z., Ballerini P., Bilhou-Nabera C., Lapillonne H., Boudia F., Thirant C., Fagnan A., Arcangeli M.-L., Kinston S. J., Diop M., Job B., Lecluse Y., Brunet E., Babin L., Villeval J., Delabesse E., Peters A., Vainchenker W., Gaudry M., Masetti R., Locatelli F., Malinge S., Nerlov C., Droin N., Lobry C., Godin I., Bernard O. A., Gottgens B., Petit A., Pflumio F., Schwaller J., Mercher T., Ontogenic changes in hematopoietic hierarchy determine pediatric specificity and disease phenotype in fusion oncogene-driven myeloid leukemia. Cancer Discov. 9, 1736–1753 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Thirant C., Ignacimouttou C., Lopez C. K. K., Diop M., Mouël L. L., Thiollier C., Siret A., Dessen P., Aid Z., Rivière J., Rameau P., Lefebvre C., Khaled M., Leverger G., Ballerini P., Petit A., Raslova H., Carmichael C. L., Kile B. T., Soler E., Crispino J. D., Wichmann C., Pflumio F., Schwaller J., Vainchenker W., Lobry C., Droin N., Bernard O. A., Malinge S., Mercher T., ETO2-GLIS2 hijacks transcriptional complexes to drive cellular identity and self-renewal in pediatric acute megakaryoblastic leukemia. Cancer Cell 31, 452–465 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., Lim W. A., Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., Torres S. E., Stern-Ginossar N., Brandman O., Whitehead E. H., Doudna J. A., Lim W. A., Weissman J. S., Qi L. S., CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulco C. P., Munschauer M., Anyoha R., Munson G., Grossman S. R., Perez E. M., Kane M., Cleary B., Lander E. S., Engreitz J. M., Systematic mapping of functional enhancer–promoter connections with CRISPR interference. Science 354, 769–773 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W., Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture–on-chip (4C). Nat Genet 38, ng1896 (2006). [DOI] [PubMed] [Google Scholar]
- 16.van de Werken H. J., Landan G., Holwerda S. J., Hoichman M., Klous P., Chachik R., Splinter E., Valdes-Quezada C., Öz Y., Bouwman B. A., Verstegen M., de Wit E., Tanay A., de Laat W., Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods 9, 969–972 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Aranda-Orgilles B., Saldaña-Meyer R., Wang E., Trompouki E., Fassl A., Lau S., Mullenders J., Rocha P. P., Raviram R., Guillamot M., Sánchez-Díaz M., Wang K., Kayembe C., Zhang N., Amoasii L., Choudhuri A., Skok J. A., Schober M., Reinberg D., Sicinski P., Schrewe H., Tsirigos A., Zon L. I., Aifantis I., MED12 regulates HSC-specific enhancers independently of mediator kinase activity to control hematopoiesis. Cell Stem Cell 19, 784–799 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKeown M. R., Corces R. M., Eaton M. L., Fiore C., Lee E., Lopez J. T., Chen M., Smith D., Chan S. M., Koenig J. L., Austgen K., Guenther M. G., Orlando D. A., Lovén J., Fritz C. C., Majeti R., Superenhancer analysis defines novel epigenomic subtypes of non-APL AML, including an RARα dependency targetable by SY-1425, a potent and selective RARα agonist. Cancer Discov. 7, 1136–1153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao Z., Zhang Y., Yuan G.-C., Orkin S. H., Waxman D. J., MAnorm: A robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 13, R16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wichmann C., Chen L., Heinrich M., Baus D., Pfitzner E., Zörnig M., Ottmann O. G., Grez M., Targeting the oligomerization domain of ETO interferes with RUNX1/ETO oncogenic activity in t(8;21)-positive leukemic cells. Cancer Res. 67, 2280–2289 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Hercus T. R., Kan W. L. T., Broughton S. E., Tvorogov D., Ramshaw H. S., Sandow J. J., Nero T. L., Dhagat U., Thompson E. J., Shing K. S. C. T., McKenzie D. R., Wilson N. J., Owczarek C. M., Vairo G., Nash A. D., Tergaonkar V., Hughes T., Ekert P. G., Samuel M. S., Bonder C. S., Grimbaldeston M. A., Parker M. W., Lopez A. F., Role of the β common (βc) family of cytokines in health and disease. Cold Spring Harb. Perspect. Biol. 10, a028514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lennartsson J., Shivakrupa R., Linnekin D., Synergistic growth of stem cell factor and granulocyte macrophage colony-stimulating factor involves kinase-dependent and -independent contributions from c-Kit. J. Biol. Chem. 279, 44544–44553 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Dentelli P., Rosso A., Garbarino G., Calvi C., Lombard E., Stefano P., Defilippi P., Pegoraro L., Brizzi M., The interaction between KDR and interleukin-3 receptor (IL-3R) beta common modulates tumor neovascularization. Oncogene 24, 6394–6405 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Blake T. J., Jenkins B. J., D’Andrea R. J., Gonda T. J., Functional cross-talk between cytokine receptors revealed by activating mutations in the extracellular domain of the beta-subunit of the GM-CSF receptor. J. Leukoc. Biol. 72, 1246–1255 (2002). [PubMed] [Google Scholar]
- 25.Hirota S., Ohashi A., Nishida T., Isozaki K., Kinoshita K., Shinomura Y., Kitamura Y., Gain-of-function mutations of platelet-derived growth factor receptor α gene in gastrointestinal stromal tumors. Gastroenterology 125, 660–667 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Zhu M.-J., Ou W.-B., Fletcher C., Cohen P., Demetri G., Fletcher J., KIT oncoprotein interactions in gastrointestinal stromal tumors: Therapeutic relevance. Oncogene 26, 1210464 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Hu-Lowe D. D., Zou H. Y., Grazzini M. L., Hallin M. E., Wickman G. R., Amundson K., Chen J. H., Rewolinski D. A., Yamazaki S., Wu E. Y., McTigue M. A., Murray B. W., Kania R. S., O’Connor P., Shalinsky D. R., Bender S. L., Nonclinical antiangiogenesis and antitumor activities of axitinib (AG-013736), an oral, potent, and selective inhibitor of vascular endothelial growth factor receptor tyrosine kinases 1, 2, 3. Clin. Cancer Res. 14, 7272–7283 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Belver L., Yang A. Y., Albero R., Herranz D., Brundu F. G., Quinn A. S., Perez-Duran P., Alvarez S., Gianni F., Rashkovan M., Gurung D., Rocha P. P., Raviram R., Reglero C., Cortes J. R., Cooke A. J., Wendorff A. A., Cordo V., Meijerink J. P., Rabadan R., Ferrando A. A., Gata3-controlled nucleosome eviction drives MYC enhancer activity in T-cell development and leukemia. Cancer Discov. 9, 1774–1791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai P., Akimaru H., Tanaka Y., Hou D. X., Yasukawa T., Kanei-Ishii C., Takahashi T., Ishii S., CBP as a transcriptional coactivator of c-Myb. Genes Dev. 10, 528–540 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Smeenk L., Ottema S., Mulet-Lazaro R., Ebert A., Havermans M., Varea A. A., Fellner M., Pastoors D., van Herk S., Erpelinck-Verschueren C., Grob T., Hoogenboezem R. M., Kavelaars F. G., Matson D. R., Bresnick E. H., Bindels E. M., Kentsis A., Zuber J., Delwel R., Selective requirement of MYB for oncogenic hyperactivation of a translocated enhancer in leukemia. Cancer Discov. 11, 2868–2883 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pattabiraman D. R., McGirr C., Shakhbazov K., Barbier V., Krishnan K., Mukhopadhyay P., Hawthorne P., Trezise A., Ding J., Grimmond S. M., Papathanasiou P., Alexander W. S., Perkins A. C., Levesque J.-P., Winkler I. G., Gonda T. J., Interaction of c-Myb with p300 is required for the induction of acute myeloid leukemia (AML) by human AML oncogenes. Blood 123, 2682–2690 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Wagenblast E., Araújo J., Gan O. I., Cutting S. K., Murison A., Krivdova G., Azkanaz M., McLeod J. L., Smith S. A., Gratton B. A., Marhon S. A., Gabra M., Medeiros J. J. F., Manteghi S., Chen J., Chan-Seng-Yue M., Garcia-Prat L., Salmena L., Carvalho D. D. D., Abelson S., Abdelhaleem M., Chong K., Roifman M., Shannon P., Wang J. C. Y., Hitzler J. K., Chitayat D., Dick J. E., Lechman E. R., Mapping the cellular origin and early evolution of leukemia in Down syndrome. Science 373, eabf6202 (2021). [DOI] [PubMed] [Google Scholar]
- 33.Bahr C., von Paleske L., Uslu V. V., Remeseiro S., Takayama N., Ng S. W., Murison A., Langenfeld K., Petretich M., Scognamiglio R., Zeisberger P., Benk A. S., Amit I., Zandstra P. W., Lupien M., Dick J. E., Trumpp A., Spitz F., A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553, 515–520 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Rio-Machin A., Gómez-López G., Muñoz J., Garcia-Martinez F., Maiques-Diaz A., Alvarez S., Salgado R. N., Shrestha M., Torres-Ruiz R., Haferlach C., Larráyoz M. J., Calasanz M. J., Fitzgibbon J., Cigudosa J. C., The molecular pathogenesis of the NUP98-HOXA9 fusion protein in acute myeloid leukemia. Leukemia 31, 2000–2005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mack S. C., Pajtler K. W., Chavez L., Okonechnikov K., Bertrand K. C., Wang X., Erkek S., Federation A., Song A., Lee C., Wang X., McDonald L., Morrow J. J., Saiakhova A., Sin-Chan P., Wu Q., Michaelraj K., Miller T. E., Hubert C. G., Ryzhova M., Garzia L., Donovan L., Dombrowski S., Factor D. C., Luu B., Valentim C. L., Gimple R. C., Morton A., Kim L., Prager B. C., Lee J. J., Wu X., Zuccaro J., Thompson Y., Holgado B. L., Reimand J., Ke S. Q., Tropper A., Lai S., Vijayarajah S., Doan S., Mahadev V., Miñan A., Gröbner S. N., Lienhard M., Zapatka M., Huang Z., Aldape K. D., Carcaboso A. M., Houghton P. J., Keir S. T., Milde T., Witt H., Li Y., Li C.-J., Bian X.-W., Jones D. T., Scott I., Singh S. K., Huang A., Dirks P. B., Bouffet E., Bradner J. E., Ramaswamy V., Jabado N., Rutka J. T., Northcott P. A., Lupien M., Lichter P., Korshunov A., Scacheri P. C., Pfister S. M., Kool M., Taylor M. D., Rich J. N., Therapeutic targeting of ependymoma as informed by oncogenic enhancer profiling. Nature 553, 101–105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larson J. D., Kasper L. H., Paugh B. S., Jin H., Wu G., Kwon C.-H., Fan Y., Shaw T. I., Silveira A. B., Qu C., Xu R., Zhu X., Zhang J., Russell H. R., Peters J. L., Finkelstein D., Xu B., Lin T., Tinkle C. L., Patay Z., Onar-Thomas A., Pounds S. B., McKinnon P. J., Ellison D. W., Zhang J., Baker S. J., Histone H3.3 K27M accelerates spontaneous brainstem glioma and drives restricted changes in bivalent gene expression. Cancer Cell 35, 140–155.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heigwer F., Zhan T., Breinig M., Winter J., Brügemann D., Leible S., Boutros M., CRISPR library designer (CLD): Software for multispecies design of single guide RNA libraries. Genome Biol. 17, 55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W., Xu H., Xiao T., Cong L., Love M. I., Zhang F., Irizarry R. A., Liu J. S., Brown M., Liu S. X., MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montojo J., Zuberi K., Rodriguez H., Kazi F., Wright G., Donaldson S., Morris Q., Bader G., GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinform. Oxf. Engl. 26, 2927–2928 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkhomchuk D., Borodina T., Amstislavskiy V., Banaru M., Hallen L., Krobitsch S., Lehrach H., Soldatov A., Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37, e123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matelot M., Noordermeer D., Determination of high-resolution 3D chromatin organization using circular chromosome conformation capture (4C-seq). Methods Mol. Biol. 1480, 223–241 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Chaumeil J., Micsinai M., Skok J. A., Combined immunofluorescence and DNA FISH on 3D-preserved Interphase nuclei to study changes in 3D nuclear organization. J. Vis. Exp. 3, e50087 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salataj E., Stathopoulou C., Hafþórsson R. A., Nikolaou C., Spilianakis C. G., Developmental conservation of microRNA gene localization at the nuclear periphery. PLOS ONE 14, e0223759 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Legend for table S1
Tables S2 to S6
Table S1
Data Availability Statement
H3K27ac ChIP-seq from 66 AML patients was published previously (18) and is available in SRA database under accession number SRP103200. M07e H3K27Ac and H3K4me3 Chip-seq are available at ArrayExpress database (E-MTAB-4367). M07e and AMKL7 ATAC-seq and RNA-seq data, AMKL7, M07e CRISPRi H3K27ac and H3K9me3, and inducible HEL-5J20 ChIP-seq and M07e 4C-seq data are available at Gene Expression Omnibus database under accession code GSE131462. Microarray data for HEL-5J20 expressing inducible ETO2-GLIS2 and M07e cells expressing NC128 peptide were published previously (11) and are available at ArrayExpress database (E-MTAB-4332).