Keywords: Crohn’s disease, gut microbiome, inflammatory bowel disease, machine learning, ulcerative colitis
Abstract
Despite the availability of various diagnostic tests for inflammatory bowel diseases (IBD), misdiagnosis of IBD occurs frequently, and thus, there is a clinical need to further improve the diagnosis of IBD. As gut dysbiosis is reported in patients with IBD, we hypothesized that supervised machine learning (ML) could be used to analyze gut microbiome data for predictive diagnostics of IBD. To test our hypothesis, fecal 16S metagenomic data of 729 subjects with IBD and 700 subjects without IBD from the American Gut Project were analyzed using five different ML algorithms. Fifty differential bacterial taxa were identified [linear discriminant analysis effect size (LEfSe): linear discriminant analysis (LDA) score > 3] between the IBD and non-IBD groups, and ML classifications trained with these taxonomic features using random forest (RF) achieved a testing area under the receiver operating characteristic curves (AUC) of ∼0.80. Next, we tested if operational taxonomic units (OTUs), instead of bacterial taxa, could be used as ML features for diagnostic classification of IBD. Top 500 high-variance OTUs were used for ML training, and an improved testing AUC of ∼0.82 (RF) was achieved. Lastly, we tested if supervised ML could be used for differentiating Crohn’s disease (CD) and ulcerative colitis (UC). Using 331 CD and 141 UC samples, 117 differential bacterial taxa (LEfSe: LDA score > 3) were identified, and the RF model trained with differential taxonomic features or high-variance OTU features achieved a testing AUC > 0.90. In summary, our study demonstrates the promising potential of artificial intelligence via supervised ML modeling for predictive diagnostics of IBD using gut microbiome data.
NEW & NOTEWORTHY Our study demonstrates the promising potential of artificial intelligence via supervised machine learning modeling for predictive diagnostics of different types of inflammatory bowel diseases using fecal gut microbiome data.
INTRODUCTION
Inflammatory bowel disease (IBD), characterized by chronic gastrointestinal inflammation, has two major clinical presentations, Crohn’s disease (CD) and ulcerative colitis (UC). Although exact pathogenesis of IBD is still unknown, one of the potential causes of IBD has been proposed as altered immune response to symbiotic microbiota due to host genome susceptibility (1) and abnormal gut microbiota composition (2–4). Since both CD and UC could lead to mild to serious complications such as Clostridium difficile infection (5), extraintestinal fibrosis (6), and intestinal fibrosis (7), early diagnosis is important for ensuring appropriate treatment. Current clinical diagnosis of IBD involves various procedures such as blood tests, colonoscopy, and MRI (8), but misdiagnosis of IBD, as one of the idiopathic diseases known for nonspecific symptoms (9), is common in clinical practice. For example, IBD can be misdiagnosed with irritable bowel syndrome (10) and diverticular disease (11). Furthermore, timely diagnostic classification of IBD into CD and UC still remains difficult (12–14). Therefore, exploration and development of novel diagnostic approaches for IBD and its subtypes are urgently needed.
Previous studies have shown strong associations of IBD with dysregulated gut microbiota (15, 16). Some studies showed that alteration of gut microbiota involves a reduction of Firmicutes and an enrichment of Proteobacteria in patients with IBD (17–20). Similarly, another study reported reduced abundance of butyrate-producing bacteria, Roseburia hominis and Faecalibacterium prausnitzii, in patients with UC (21). Further, the American Gut Project (22) has cataloged microbiome data from a large cohort of humans including those affected with IBD, which therefore serves as a valuable platform for assessing the overall population relationships between microbiota and IBD. In the current study, we sought to not only identify microbial signatures indicative of IBD and its clinical presentations as CD or UC but also to apply gut microbiome features to train machine learning (ML) models. ML, which is a major branch of artificial intelligence (AI), has been used in gastroenterology to detect polyps, lesions, and cancer (23, 24), but to date, has not been applied to detect IBD in clinics. Notably, ML has been used to analyze large-scale metagenomics data (25). In this study, we hypothesized that supervised machine learning (ML) models could be trained with gut microbiome data for diagnostic classifications of IBDs including CD and UC. To test our hypothesis, we obtained stool 16S rRNA sequencing data collected from human subjects diagnosed with IBD through the American Gut Project and trained the large-scale data of bacterial taxa or operational taxonomic units (OTUs) with five different supervised ML models for diagnostic classifications of IBD versus non-IBD and CD versus UC. Our study further demonstrates the promising potential of training supervised ML models using large-scale fecal microbiome data for a convenient diagnostic screening of IBD and its subtypes.
MATERIALS AND METHODS
Data Collection and Processing
The workflow of data collection, processing, and analysis is summarized in Fig. 1. The 16S rRNA metagenomics data were collected from the American Gut Project (22) using Redbiom (26). Out of a total of 19,978 stool samples (as of February 5, 2020, Qiita study ID: 10317), 934 samples were collected from the participants diagnosed with IBD [ibd = “Diagnosed by a medical professional (doctor, physician assistant)”] and 19,044 samples were collected from the participants with no IBD (ibd = “I do not have this condition”). Out of 19,044 non-IBD samples, 941 samples were randomly selected through random shuffling of all the non-IBD samples, to match the final sample size of the IBD group after quality controlling. For subtype classification experiments, 406 and 179 samples from the participants diagnosed with CD and UC were found, respectively (as of May 28, 2020). Our study excluded the IBD and subtype samples that were marked as either self-diagnosed or not provided. Metadata and BIOM files were obtained using the “redbiom fetch” function with the context “Deblur-Illumina-16S-V4-150nt-780653.” The BIOM file was further filtered using the QIIME 2 (27) (v. 2019.10) for quality controlling to remove the samples with a total frequency fewer than 10,000. The OTU table was generated using the filtered BIOM file with the BIOM format tool (28). We obtained a total of 71,199 OTUs for IBD versus non-IBD and a total of 18,627 OTUs for CD versus UC. The 16S microbiome data of stool samples collected from 729 IBD, 700 non-IBD, 331 CD, and 141 UC subjects were eventually used for subsequent analyses. Additional two distinct non-IBD subsets of the randomly selected 713 and 740 non-IBD samples were used for the validation purpose.
Figure 1.
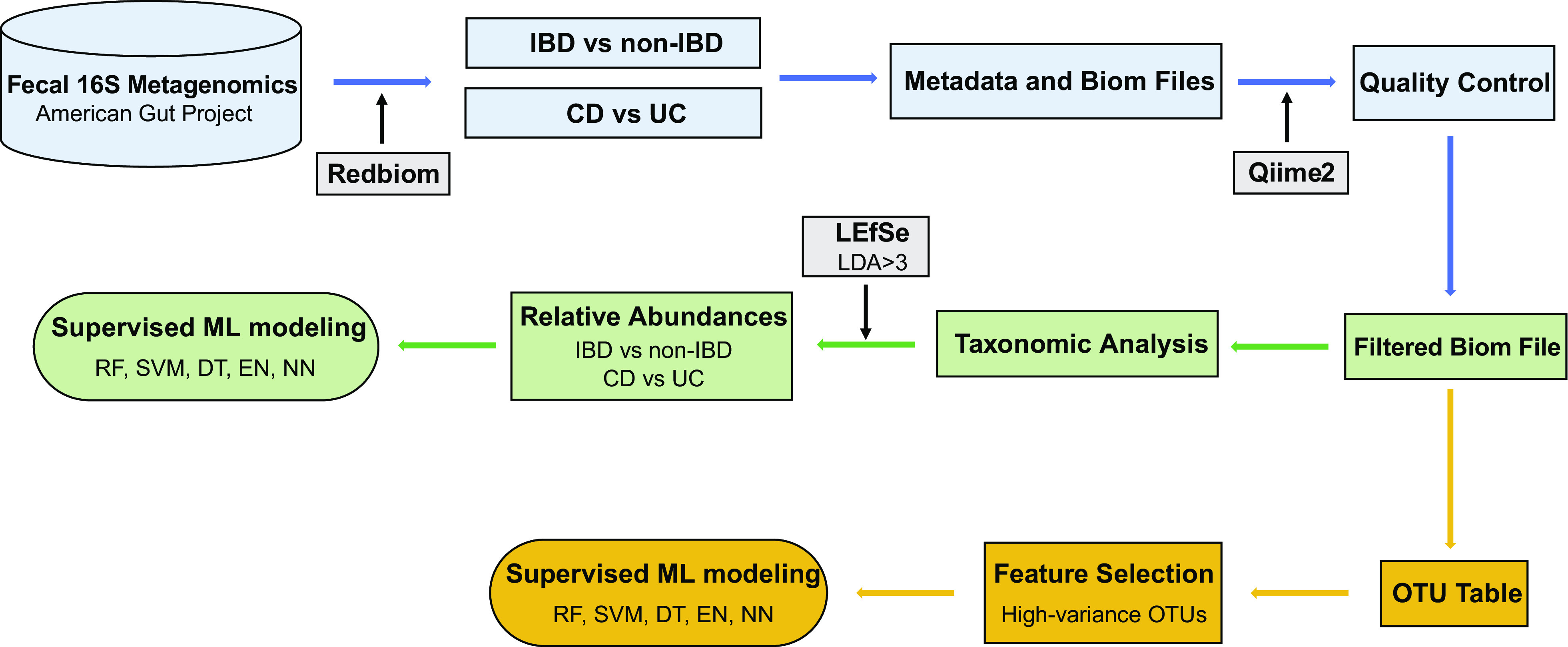
The study workflow of data collection and supervised machine learning experiments. CD, Crohn’s disease; DT, decision tree; EN, elastic net; UC, ulcerative colitis; IBD, inflammatory bowel disease; LEfSe, linear discriminant analysis effect size; NN, neural networks; OTU, operational taxonomic units; RF, random forest; SVM, support vector machine with radial kernel.
Taxonomic Analysis
The taxonomic assignment was implemented using QIIME 2 with a pretrained Naive Bayes classifier on the Greengenes (v. 13.8) at a 99% similarity threshold for OTU clustering (29). Linear discriminant analysis effect size (LEfSe: https://huttenhower.sph.harvard.edu/galaxy/) was used to identify differentially abundant taxonomic features (30). The LEfSe bar graph and cladogram of taxonomical features with linear discriminant analysis (LDA) score threshold greater than 3.0 were generated.
Supervised Machine Learning
In the classification of IBD versus non-IBD, five different supervised ML algorithms, which are random forest (RF), decision tree (DT), elastic net (EN), support vector machine with radial kernel (SVM), and neural networks (NN), were trained with the features of bacterial taxa or OTUs using the caret R package (31). For the classification of CD versus UC, RF model was trained with the features of bacterial taxa or OTUs. Kernlab (32), randomForest (33), glmnet (34), and rpart (35) were deployed as the assistant R packages. Data splitting into the training (70%) and testing (30%) samples was performed after data shuffling. To reduce the dimensionality of the feature space and computational complexity, OTU-wise variance was computed for each OTU, and the top 500 high-variance OTUs across all the samples were selected for training the ML models. In the training stage, a 10-time repetition of 10-fold cross-validation was applied to assess performances of the ML models using only the training samples. Hyperparameter tuning was performed automatically using caret testing 10 different values for each hyperparameter. In the testing stage, trained ML models were evaluated on the testing samples for the commonly used ML performance parameters: area under the receiver operating characteristic curves (AUC), accuracy, sensitivity, specificity, precision, and F1. The whole procedures, including data shuffling, data splitting, training, and testing, were performed for 50 independent iterations. The average values and standard deviations were computed for all the testing performance parameters collected from the 50 iterations. The box-plot depictions of the values of AUC and accuracy were generated using the ggplot2 package (36) in R.
RESULTS
Differential Taxonomic Composition between the IBD and Non-IBD Groups
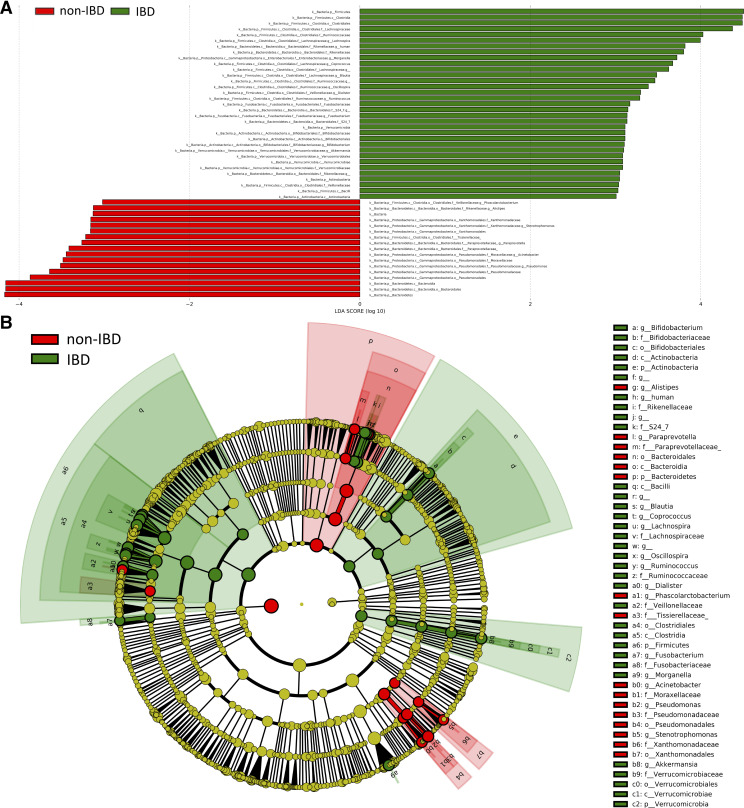
Significant differences in gut microbiota were observed between the subjects with IBD and those without IBD (Fig. 2). Fifty taxonomic features (LDA > 3.0) were identified to be enriched in either IBD or non-IBD groups [Fig. 2A, Supplemental Table S1 (all Supplemental material is available at https://doi.org/10.5281/zenodo.4420108)]. For example, at the bacterial phylum level, Firmicutes, Verrucomicrobia, and Actinobacteria were more abundant in the IBD group, whereas Bacteroidetes was more abundant in the non-IBD group (Fig. 2A). Increased levels of bacterial genus, including Lachnospira, Morganella, Coprococcus, Blautia, Oscillospira, Dialister, Ruminococcus, Fusobacterium, Bifidobacterium, and Akkermansia, were observed in the IBD group, whereas Pseudomonas, Acinetobacter, Paraprevotella, Stenotrophomonas, Alistipes, and Phascolarctobacterium were more enriched in the non-IBD group (Fig. 2A). The cladogram in Fig. 2B presents the significantly differential taxonomic signatures and their phylogenetic relationships.
Figure 2.
Differential bacterial taxa between the inflammatory bowel disease (IBD) and non-IBD groups. A: linear discriminant analysis effect size (LEfSe) bar plot (LDA > 3.0) showing enriched taxa in different groups. B: cladogram (LDA > 3.0) showing differential taxa with their phylogenetic relationships.
Supervised ML Models Trained with Taxonomic Features for Classifying IBD and Non-IBD
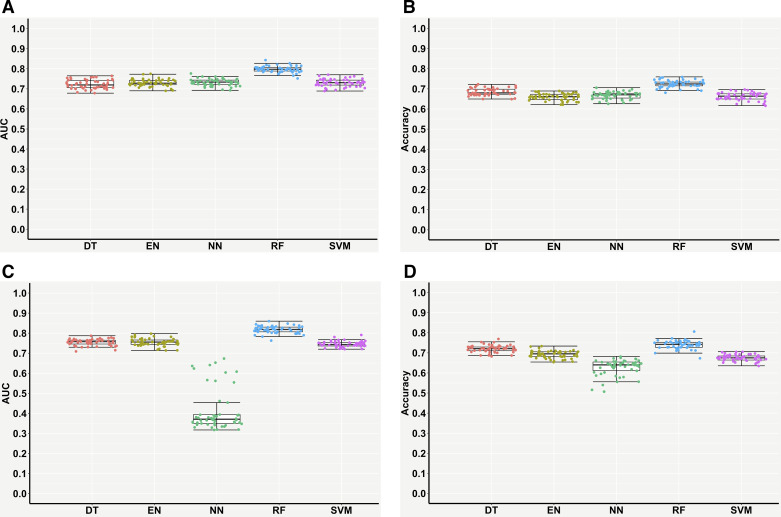
Supervised ML models were trained with the 50 differential taxonomic features (Fig. 2A, Supplemental Table S1) for classifying the IBD and non-IBD samples. Table 1 and Fig. 3, A and B, present the performances measures of the five ML models evaluated on the testing samples. RF performed best and achieved a testing AUC of ∼0.80, followed by EN (∼0.73), NN (∼0.73), SVM (∼0.73), and DT (∼0.72) ( Table 1, Fig. 3A). In terms of testing accuracy, RF achieved ∼72% accuracy in classifying the IBD and non-IBD subjects, followed by DT (∼68%), NN (∼67%), EN (∼66%), and SVM (∼66%) (Table 1, Fig. 3B). Overall, RF outperformed other ML models for predictive classifications of IBD versus non-IBD.
Table 1.
Performance measures of supervised ML models for classifying the subjects with IBD and those without IBD using differential taxonomic features
| Models | AUC | Accuracy | Sensitivity | Specificity | Precision | F1 |
|---|---|---|---|---|---|---|
| DT | 0.72 ± 0.02 | 0.68 ± 0.02 | 0.80 ± 0.06 | 0.57 ± 0.06 | 0.64 ± 0.02 | 0.71 ± 0.02 |
| EN | 0.73 ± 0.02 | 0.66 ± 0.02 | 0.82 ± 0.05 | 0.50 ± 0.06 | 0.62 ± 0.02 | 0.70 ± 0.02 |
| NN | 0.73 ± 0.02 | 0.67 ± 0.02 | 0.75 ± 0.06 | 0.59 ± 0.06 | 0.64 ± 0.02 | 0.69 ± 0.02 |
| RF | 0.80 ± 0.01 | 0.72 ± 0.02 | 0.80 ± 0.03 | 0.64 ± 0.03 | 0.69 ± 0.02 | 0.74 ± 0.02 |
| SVM | 0.73 ± 0.02 | 0.66 ± 0.02 | 0.74 ± 0.08 | 0.59 ± 0.08 | 0.74 ± 0.02 | 0.68 ± 0.03 |
Values are presented as means ± SD calculated from 50 independent iterations. AUC, area under the receiver operating characteristic curve; DT, decision tree; EN, elastic net; IBD, inflammatory bowel disease; ML, machine learning; NN, neural networks; RF, random forest; SVM, support vector machine with radial kernel.
Figure 3.
Performance measures of supervised machine learning models for classifying subjects with inflammatory bowel disease (IBD) and those without IBD (non-IBD) using gut microbiome features. Differential taxonomic features: area under the receiver operating characteristic curve (AUC) (A) and accuracy (B); high-variance OTU features: AUC (C) and accuracy (D). Each point in the box plot represents each performance measure in one iteration (total 50 iterations). OTU, operational taxonomic units.
Supervised ML Models Trained with High-Variance OTU Features for Classifying IBD and Non-IBD
Next, we examined if OTUs, instead of bacterial taxa, could be used for diagnostic classification of IBD. The five ML models above were trained with the top 500 high-variance OTU features across all the samples. Slight testing performance improvements were observed in DT, EN, RF, and SVM (Table 2, Fig. 3, C and D). Surprisingly, the performance of NN significantly decreased to ∼0.41 AUC and ∼63% accuracy (Table 2, Fig. 3, C and D). Overall, by any performance measure, RF still performed best, with the highest outcomes represented by ∼0.82 AUC and ∼74% accuracy (Table 2). Using another two distinct subsets of 713 (subset 1) and 740 (subset 2) non-IBD samples with 729 IBD samples for RF modeling, similar testing performances of ∼0.84 AUC and ∼76% accuracy confirmed the above results (Supplemental Fig. S1 and Supplemental Table S2).
Table 2.
Performance measures of supervised ML models for classifying the subjects with IBD and those without IBD using the top 500 high-variance OTU features
| Models | AUC | Accuracy | Sensitivity | Specificity | Precision | F1 |
|---|---|---|---|---|---|---|
| DT | 0.75 ± 0.02 | 0.72 ± 0.02 | 0.81 ± 0.04 | 0.63 ± 0.04 | 0.68 ± 0.02 | 0.74 ± 0.02 |
| EN | 0.75 ± 0.02 | 0.69 ± 0.02 | 0.77 ± 0.05 | 0.62 ± 0.06 | 0.66 ± 0.02 | 0.70 ± 0.02 |
| NN | 0.41 ± 0.10 | 0.63 ± 0.04 | 0.80 ± 0.22 | 0.46 ± 0.18 | 0.60 ± 0.03 | 0.66 ± 0.11 |
| RF | 0.82 ± 0.02 | 0.74 ± 0.02 | 0.84 ± 0.03 | 0.64 ± 0.04 | 0.70 ± 0.02 | 0.76 ± 0.02 |
| SVM | 0.74 ± 0.01 | 0.67 ± 0.02 | 0.77 ± 0.06 | 0.58 ± 0.06 | 0.64 ± 0.02 | 0.70 ± 0.02 |
Values are presented as means ± SD calculated from 50 independent iterations. AUC, area under the receiver operating characteristic curve; DT, decision tree; EN, elastic net; IBD, inflammatory bowel disease; ML, machine learning; NN, neural networks; OTU, operational taxonomic units; RF, random forest; SVM, support vector machine with radial kernel.
Taxonomic Analysis and Supervised ML Modeling for Classifying CD and UC
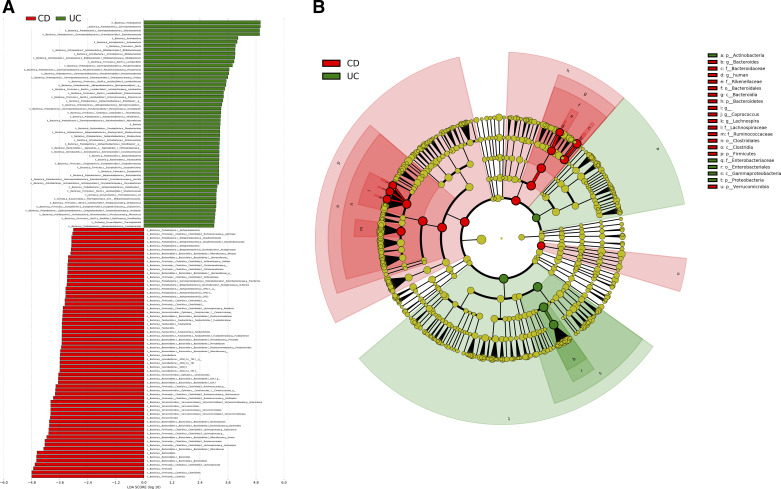
We next examined if the two IBD subtypes, CD and UC, could be differentially classified using the above approaches. A total of 117 bacterial taxa (LDA > 3.0) were identified to be significantly differential between CD and UC (Fig. 4A, Supplemental Table S3). For example, multiple bacterial genera, such as Bifidobacterium, Pseudomonas, Proteus, Lactobacillus, Enterococcus, Acinetobacter, Serratia, Corynebacterium, Streptococcus, Eubacterium, and Arcobacter, were significantly enriched in the UC group, whereas Lachnospira, Coprococcus, Bacteriodes, Akkermansia, Oscillospira, Ruminococcus, Parabacteroides, Prevotella, Fusobacterium, Roseburia, Sutterella, Providencia, Dialister, Alistipes, and Gemmiger were more abundant in the CD group (Fig. 4A). The cladogram (LDA > 4.0) shown in Fig. 4B distinctly presented the overall differential taxa and their phylogenetic relationships.
Figure 4.
Differential bacterial taxa between the Crohn’s disease (CD) and ulcerative colitis (UC) groups. A: linear discriminant analysis effect size (LEfSe) bar plot (LDA > 3.0) showing enriched taxa in different groups. B: cladogram (LDA > 4.0) showing differential taxa with their phylogenetic relationships.
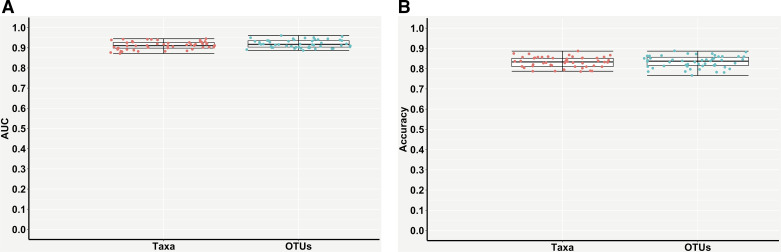
The RF model was reimplemented using either the differential taxonomic features or the top 500 high-variance OTU features for classifying the subjects with CD and UC. Table 3 and Fig. 5 present the performance measures of the trained RF model evaluated on the testing CD and UC samples. Impressively, the RF models trained with either taxonomic features or OTU features achieved ∼0.91 AUC and ∼0.92 AUC, respectively (Table 3, Fig. 5A). In terms of testing accuracy, both models achieved ∼83% accuracy of differentiating CD and UC (Table 3, Fig. 5B). Due to the imbalanced ratio of the numbers of the UC and CD samples, 331 CD samples were randomly divided into two subsets of 165 (subset 1) and 166 (subset 2) CD samples for RF modeling with 141 UC samples; similar testing performances of ∼0.91 AUC and ∼83% accuracy confirmed the above results (Supplemental Fig. S2 and Supplemental Table S4).
Table 3.
Performance measures of the RF models for classifying CD and UC using differential taxonomic features or the top 500 high-variance OTU features
| Features | AUC | Accuracy | Sensitivity | Specificity | Precision | F1 |
|---|---|---|---|---|---|---|
| Taxa | 0.91 ± 0.02 | 0.83 ± 0.03 | 0.85 ± 0.03 | 0.79 ± 0.06 | 0.90 ± 0.02 | 0.88 ± 0.02 |
| OTUs | 0.92 ± 0.02 | 0.83 ± 0.03 | 0.85 ± 0.04 | 0.80 ± 0.06 | 0.90 ± 0.03 | 0.88 ± 0.02 |
Values are presented as means ± SD calculated from 50 independent iterations. AUC, area under the receiver operating characteristic curve; CD, Crohn’s disease; OTU, operational taxonomic units; RF, random forest; UC, ulcerative colitis.
Figure 5.
Performance measures of the random forest (RF) models for classifying Crohn’s disease (CD) and ulcerative colitis (UC) groups using differential taxonomic features or the top 500 high-variance OTU features. A: area under the receiver operating characteristic curve (AUC). B: accuracy. Each point in the box plot represents each performance measure in one iteration (total 50 iterations). OTU, operational taxonomic units.
DISCUSSION
Unlike routine invasive procedures such as colonoscopies and endoscopies, our study provides a noninvasive approach of gut microbiome-based machine learning classifications for a convenient and efficient diagnostic screening of IBD and its subtypes. To our knowledge, our study demonstrates the promising potential of utilizing fecal gut microbiome data to distinguish between subjects with IBD and those without IBD as well as between CD and UC. Despite that the gut microbiome composition and diversity can vary greatly among individuals of different characteristics such as sex, age, diet habit, residential area, and health condition (37–39), the ability to distinguish between gut physiology and pathophysiology in the host solely by assessing gut microbiota compositions as presented in the current work is to be viewed as a significant advancement. The ML models developed here represent a robust and generalized diagnostic screening tool, which is applicable across a wide range of human populations.
Significant gut microbiota alterations were observed between the subjects with IBD and those without IBD (Fig. 2). Main gut dysbiosis features, as reported previously (17, 40, 41), were represented by increased levels of Firmicutes and decreased levels of Bacteroidetes in subjects with IBD (Fig. 2A, Supplemental Table S1). We also observed enriched phyla Verrucomicrobia and Actinobacteria in the IBD group (Fig. 2A, Supplemental Table S1), which is consistent with previous reports (17, 42, 43). Moreover, at the bacterial family level, more abundant Fusobacteriaceae and Veillonellaceae were previously reported to be associated with IBD (40, 44) and also found in our study (Fig. 2A, Supplemental Table S1). Lachnospiraceae was previously reported to be less abundant in patients with IBD (17, 45), whereas our studies showed its increased abundance in the IBD group (Fig. 2A, Supplemental Table S1). As different subtypes of IBD can have similar symptoms, but they require different diagnostic standards, we further investigated and compared gut microbiota signatures between two major subtypes of IBD, CD and UC. Surprisingly, the comparison of CD versus UC showed significantly more taxonomic differences than the comparison between IBD and non-IBD (Fig. 2 and Fig. 4). Several bacterial phyla, such as Firmicutes, Bacteroidetes, Verrucomicrobia, Cyanobactetria, and Fusobacteria, were more abundant in CD than UC (Fig. 4A, Supplemental Table S3). Enriched Actinobacteria and Bifidobacterium were reported in patients with UC (46–48), and we observed the same associations in our UC group (Fig. 4A, Supplemental Table S3). Increased Deltaproteobacteria, a bacterial class of sulfate-reducing bacteria, was previously reported in patients with UC (49, 50), whereas we observed more abundant Deltaproteobacteria in the CD group (Fig. 4A, Supplemental Table S3). Therefore, although not all of the taxa are consistent with previous reports, our study takes into account the overall composition and has detected overall gut microbiome signatures, which are associated with IBD and its subtypes. These are more meaningful when one considers that bacteria have quorum-sensing properties whereby they are interdependent on each other for adjusting their individual compositions based on their immediate environmental milieu in the gut. Moreover, as gut microbiota is a highly variable and dynamic feature, individual or a few statistical disease-associated bacterial taxa may not be reliable biomarkers for diagnostic applications. For example, an altered Firmicutes/Bacteroidetes ratio was reported to be associated with several diseases (51–53), but the ratio can greatly change with aging (54) and diets (55), and the altered ratio can relate to several coexisting disease conditions in a single individual, and thus a targeted diagnostic indication of a specific disease cannot simply rely on alterations in only a few taxa of gut microbiota. Therefore, we examined gut microbiome-based supervised ML modeling for evaluating its diagnostic potential of IBD and its subtypes over a significant amount of diverse population with distinct characteristics, such as sex and age. We tested the capacity of supervised ML models for differentiating subjects with IBD and those without IBD based on their gut microbiota features, and our results indicate that ML modeling could achieve ∼0.82 AUC for diagnostic classifications of IBD and non-IBD (Table 2).
In prior reports, data of histological and endoscopic findings (56), genomic database (57), and gut microbiota (58–60) have been used to train ML models for IBD diagnosis. For example, ML models were trained with gut microbiota data from pediatric stool samples of 67 subjects with IBD and 24 subjects without IBD and achieved an AUC of 0.83 (61). However, most of these studies had limited sample size to rigorously train and validate the reliability of ML predictive diagnostics, especially over a diverse population distribution. In contrast, our study trained and tested ML models with a significant number of individual samples (729 subjects with IBD and 700 subjects without IBD) to develop a robust ML diagnostic screening tool. Another recent study applied unsupervised ML approaches for identifying and quantifying taxon co-occurrence patterns for diagnosing subjects with IBD (62). It should be noted that our study used nonnormalized bacterial taxonomic or OTU data for ML modeling, as we aimed to test the capacity and adaptability of ML models trained with raw microbiome data to classify and predict new unknown samples without requiring repeated processing of the previous samples with the new samples in future.
Currently, there is still limited evidence regarding the performance and reliability of ML classification and diagnosis of IBD subtypes. A previous study, which trained supervised ML models with bacterial genus and OTU data collected from only 20 patients with CD and 19 patients with UC, achieved 0.79 AUC and 0.72 AUC, respectively (63). In our study, we performed the RF training and testing on a significant number of human samples (331 subjects with CD and 141 subjects with UC) and achieved significantly better prediction performances of >0.90 AUC (Table 3). It should be noted that we only used the fecal metagenomics data of the patients with IBD who were indicated to be diagnosed by a medical profession (doctor, physician assistant) in the database of the American Gut Project, but we could not rule out the possibility of misdiagnosed IBD cases. Even so, our study still demonstrates the promising potential of applying gut microbiome-based supervised ML approaches for diagnostic differentiation of clinical IBDs. Interestingly, we found that the RF model performed best not only for the IBD versus non-IBD classification but also for the CD versus UC classification, which is consistent with pervious observations that RF performed well in metagenome-based classification (64–66).
In summary, using newly identified distinct gut microbiota signatures in IBD and its subtypes, CD and UC, our study demonstrates the promising potential of using large-scale gut microbiota data, as a noninvasive approach, to train supervised ML models for efficient diagnostic screening of different types of clinical IBD.
SUPPLEMENTAL DATA
Supplemental Figures and Supplemental Tables S1–S4: https://doi.org/10.5281/zenodo.4420108.
GRANTS
The work was supported by the Dean’s Postdoctoral to Faculty Fellowship from University of Toledo College of Medicine and Life Sciences to X. Cheng. X. Cheng also acknowledges funding support from the P30 Core Center Pilot Grant from National Institute on Drug Abuse (NIDA) Center of Excellence in Omics, Systems Genetics, and the Addictome. B. Joe acknowledges grant support from the National Heart, Lung, and Blood Institute (HL143082). P. B. Munroe acknowledges support from the National Institute of Health Research Cardiovascular Biomedical Research Centre at Barts and Queen Mary University of London.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
X.C. conceived and designed research; I.M. and X.C. performed experiments; I.M. and X.C. analyzed data; I.M. and X.C. interpreted results of experiments; I.M. prepared figures; I.M. drafted manuscript; I.M., A.A., S.A., P.B.M., B.J., and X.C. edited and revised manuscript; I.M., A.A., S.A., P.B.M., B.J., and X.C. approved final version of manuscript.
REFERENCES
- 1.Kaplan GG, Ng SC. Understanding and preventing the global increase of inflammatory bowel disease. Gastroenterology 152: 313–321, 2017. [Erratum in Gastroenterology 152: 2084, 2017]. doi: 10.1053/j.gastro.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Hold GL, Smith M, Grange C, Watt ER, El-Omar EM, Mukhopadhya I. Role of the gut microbiota in inflammatory bowel disease pathogenesis: what have we learnt in the past 10 years? World J Gastroenterol 20: 1192–1210, 2014. doi: 10.3748/wjg.v20.i5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. In: Seminars in immunopathology, edited by Ohno H. Springer, 2015, p. 47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seksik P. Gut microbiota and IBD. Gastroenterol Clin Biol 34: S44–S51, 2010. doi: 10.1016/S0399-8320(10)70020-8. [DOI] [PubMed] [Google Scholar]
- 5.Khanna S, Pardi DS. IBD: poor outcomes after Clostridium difficile infection in IBD. Nat Rev Gastroenterol Hepatol 9: 307–308, 2012. doi: 10.1038/nrgastro.2012.87. [DOI] [PubMed] [Google Scholar]
- 6.Ott C, Schölmerich J. Extraintestinal manifestations and complications in IBD. Nat Rev Gastroenterol Hepatol 10: 585–595, 2013. doi: 10.1038/nrgastro.2013.117. [DOI] [PubMed] [Google Scholar]
- 7.Rieder F, Fiocchi C. Intestinal fibrosis in IBD—a dynamic, multifactorial process. Nat Rev Gastroenterol Hepatol 6: 228–235, 2009. doi: 10.1038/nrgastro.2009.31. [DOI] [PubMed] [Google Scholar]
- 8.Inflammatory bowel disease (IBD). (Online). https://www.mayoclinic.org/diseases-conditions/inflammatory-bowel-disease/diagnosis-treatment/drc-20353320 [19 May 2020].
- 9.Waljee AK, Lipson R, Wiitala WL, Zhang Y, Liu B, Zhu J, Wallace B, Govani SM, Stidham RW, Hayward R, Higgins PDR. Predicting hospitalization and outpatient corticosteroid use in inflammatory bowel disease patients using machine learning. Inflamm Bowel Dis 24: 45–53, 2017. doi: 10.1093/ibd/izx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Card TR, Siffledeen J, Fleming KM. Are IBD patients more likely to have a prior diagnosis of irritable bowel syndrome? Report of a case-control study in the General Practice Research Database. United European Gastroenterol J 2: 505–512, 2014. doi: 10.1177/2050640614554217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shivashankar R, Lichtenstein GR. Mimics of inflammatory bowel disease. Inflamm Bowel Dis 24: 2315–2321, 2018. doi: 10.1093/ibd/izy168. [DOI] [PubMed] [Google Scholar]
- 12.Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet 380: 1590–1605, 2012. [Erratum in Lancet 381: 204, 2013]. doi: 10.1016/S0140-6736(12)60026-9. [DOI] [PubMed] [Google Scholar]
- 13.Bousvaros A, Antonioli DA, Colletti RB, Dubinsky MC, Glickman JN, Gold BD. Differentiating ulcerative colitis from Crohn’s disease in children and young adults: report of the working group of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the Crohn’s and Colitis Foundation of America. J Pediatr Gastroenterol Nutr 44: 653–674, 2007. doi: 10.1097/mpg.0b013e31805563f3. [DOI] [PubMed] [Google Scholar]
- 14.Kasperczuk A, Daniluk J, Dardzinska A. Smart model to distinguish Crohn’s disease from ulcerative colitis. Appl Sci 9: 1650, 2019. doi: 10.3390/app9081650. [DOI] [Google Scholar]
- 15.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 9: 599–608, 2012. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 16.Matijašić M, Meštrović T, Perić M, Čipčić Paljetak H, Panek M, Vranešić Bender D, Ljubas Kelečić D, Krznarić Ž, Verbanac D. Modulating composition and metabolic activity of the gut microbiota in IBD patients. Int J Mol Sci 17: 578, 2016. doi: 10.3390/ijms17040578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785, 2007. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, van Zanten SJOV. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J Clin Microbiol 44: 4136–4141, 2006. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55: 205–211, 2006. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vester-Andersen MK, Mirsepasi-Lauridsen HC, Prosberg MV, Mortensen CO, Träger C, Skovsen K, Thorkilgaard T, Nøjgaard C, Vind I, Krogfelt KA, Sørensen N, Bendtsen F, Petersen AM. Increased abundance of proteobacteria in aggressive Crohn’s disease seven years after diagnosis. Sci Rep 9: 13473, 2019. doi: 10.1038/s41598-019-49833-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, Ferrante M, Verhaegen J, Rutgeerts P, Vermeire S. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63: 1275–1283, 2014. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 22.McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, , et al. American gut: an open platform for citizen science microbiome research. mSystems 3: e00031-18, 2018. doi: 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Berre C, Sandborn WJ, Aridhi S, Devignes M-D, Fournier L, Smail-Tabbone M, Danese S, Peyrin-Biroulet L. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology 158: 76–94.e2, 2020. doi: 10.1053/j.gastro.2019.08.058. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Qian J-M. Artificial intelligence in inflammatory bowel disease: current status and opportunities. Chin Med J (Engl) 133: 757–759, 2020. doi: 10.1097/CM9.0000000000000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasolli E, Truong DT, Malik F, Waldron L, Segata N. Machine learning meta-analysis of large metagenomic datasets: tools and biological insights. PLoS Comput Biol 12: e1004977, 2016. doi: 10.1371/journal.pcbi.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald D, Kaehler B, Gonzalez A, DeReus J, Ackermann G, Marotz C, Huttley G, Knight R. Redbiom: a rapid sample discovery and feature characterization system. mSystems 4: e00215-19, 2019. doi: 10.1128/mSystems.00215-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, , et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37: 852–857, 2019. [Erratum in Nat Biotechnol 37: 1091, 2019]. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D, Wilke A, Huse S, Hufnagle J, Meyer F, Knight R, Caporaso JG. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience 1: 7, 2012. doi: 10.1186/2047-217X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618, 2012. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol 12: R60, 2011. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhn M. Predictive modeling with R and the caret Package. 2013. https://www.r-project.org/conferences/useR-2013/Tutorials/kuhn/user_caret_2up.pdf.
- 32.Karatzoglou A, Smola A, Hornik K, Zeileis A. kernlab-an S4 package for kernel methods in R. J Stat Softw 11: 1–20, 2004. doi: 10.18637/jss.v011.i09. [DOI] [Google Scholar]
- 33.Liaw A, Wiener M. Classification and regression by randomForest. R news 2: 18–22, 2002. https://cogns.northwestern.edu/cbmg/LiawAndWiener2002.pdf [Google Scholar]
- 34.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 33: 1–22, 2010. [PMC free article] [PubMed] [Google Scholar]
- 35.Therneau T, Atkinson B, Ripley B, Ripley MB. Recursive Partitioning and Regression Trees. Package ‘rpart.’ 2015. https://cran.pau.edu.tr/web/packages/rpart/rpart.pdf.
- 36.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer, 2016. [Google Scholar]
- 37.Gupta VK, Paul S, Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol 8: 1162, 2017. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y-Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108, 2011. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature 486: 222–227, 2012. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alam MT, Amos GCA, Murphy ARJ, Murch S, Wellington EMH, Arasaradnam RP. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog 12: 1, 2020. doi: 10.1186/s13099-019-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sha S, Xu B, Wang X, Zhang Y, Wang H, Kong X, Zhu H, Wu K. The biodiversity and composition of the dominant fecal microbiota in patients with inflammatory bowel disease. Diagn Microbiol Infect Dis 75: 245–251, 2013. doi: 10.1016/j.diagmicrobio.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 42.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13: R79, 2012. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santoru ML, Piras C, Murgia A, Palmas V, Camboni T, Liggi S, Ibba I, Lai MA, Orrù S, Blois S, Loizedda AL, Griffin JL, Usai P, Caboni P, Atzori L, Manzin A. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci Rep 7: 1–14, 2017. doi: 10.1038/s41598-017-10034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, Gonzalez A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15: 382–392, 2014. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Angelberger S, Reinisch W, Makristathis A, Lichtenberger C, Dejaco C, Papay P, Novacek G, Trauner M, Loy A, Berry D. Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol 108: 1620–1630, 2013. doi: 10.1038/ajg.2013.257. [DOI] [PubMed] [Google Scholar]
- 46.Lepage P, Häsler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, Ott S, Kupcinskas L, Doré J, Raedler A, Schreiber S. Twin study indicates loss of interaction between microbiota and mucosa of patients with ulcerative colitis. Gastroenterology 141: 227–236, 2011. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 47.Tong M, Li X, Parfrey LW, Roth B, Ippoliti A, Wei B, Borneman J, McGovern DPB, Frank DN, Li E, Horvath S, Knight R, Braun J. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS One 8: e80702, 2013. doi: 10.1371/journal.pone.0080702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, Wang G, Xia B, Forbes BA. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J Clin Microbiol 52: 398–406, 2014. doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibson GR, Cummings JH, Macfarlane GT. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Lett 86: 103–111, 1991. doi: 10.1111/j.1574-6968.1991.tb04799.x. [DOI] [Google Scholar]
- 50.Roediger WE, Moore J, Babidge W. Colonic sulfide in pathogenesis and treatment of ulcerative colitis. Dig Dis Sci 42: 1571–1579, 1997. doi: 10.1023/A:1018851723920. [DOI] [PubMed] [Google Scholar]
- 51.Kasselman LJ, Vernice NA, DeLeon J, Reiss AB. The gut microbiome and elevated cardiovascular risk in obesity and autoimmunity. Atherosclerosis 271: 203–213, 2018. doi: 10.1016/j.atherosclerosis.2018.02.036. [DOI] [PubMed] [Google Scholar]
- 52.López-Cepero AA, Palacios C. Association of the intestinal microbiota and obesity. P R Health Sci J 34: 60–64, 2015. [PubMed] [Google Scholar]
- 53.Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, Zadeh M, Gong M, Qi Y, Zubcevic J, Sahay B, Pepine CJ, Raizada MK, Mohamadzadeh M. Gut dysbiosis is linked to hypertension. Hypertension 65: 1331–1340, 2015. doi: 10.1161/HYPERTENSIONAHA.115.05315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mariat D, Firmesse O, Levenez F, Guimarăes VD, Sokol H, Doré J, Corthier G, Furet JP. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9: 123, 2009. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr 94: 58–65, 2011. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mossotto E, Ashton JJ, Coelho T, Beattie RM, MacArthur BD, Ennis S. Classification of paediatric inflammatory bowel disease using machine learning. Sci Rep 7: 1, 2017. doi: 10.1038/s41598-017-02606-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wei Z, Wang W, Bradfield J, Li J, Cardinale C, Frackelton E, Kim C, Mentch F, Van Steen K, Visscher PM, Baldassano RN, Hakonarson H; International IBD Genetics Consortium. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Hum Genet 92: 1008–1012, 2013. doi: 10.1016/j.ajhg.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douglas GM, Hansen R, Jones CMA, Dunn KA, Comeau AM, Bielawski JP, Tayler R, El-Omar EM, Russell RK, Hold GL, Langille MGI, Van Limbergen J. Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn’s disease. Microbiome 6: 13, 2018. doi: 10.1186/s40168-018-0398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hacılar H, Nalbantoğlu OU, Bakir-Güngör B. Machine Learning Analysis of Inflammatory Bowel Disease-Associated Metagenomics Dataset. In: 2018 3rd International Conference on Computer Science and Engineering (UBMK). IEEE, 2018, p. 434–438. [Google Scholar]
- 60.Oh M, Zhang L. DeepMicro: deep representation learning for disease prediction based on microbiome data. Sci Rep 10: 1–9, 2020. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papa E, Docktor M, Smillie C, Weber S, Preheim SP, Gevers D, Giannoukos G, Ciulla D, Tabbaa D, Ingram J, Schauer DB, Ward DV, Korzenik JR, Xavier RJ, Bousvaros A, Alm EJ. Non-invasive mapping of the gastrointestinal microbiota identifies children with inflammatory bowel disease. PLoS One 7: e39242, 2012. doi: 10.1371/journal.pone.0039242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tataru CA, David MM. Decoding the language of microbiomes using word-embedding techniques, and applications in inflammatory bowel disease. PLOS Comput Biol 16: e1007859, 2020. [Erratum in PLoS Comput Biol 16: e1008423, 2020]. doi: 10.1371/journal.pcbi.1007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forbes JD, Chen C-Y, Knox NC, Marrie R-A, El-Gabalawy H, de Kievit T, Alfa M, Bernstein CN, Van Domselaar G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases—does a common dysbiosis exist? Microbiome 6: 221, 2018. doi: 10.1186/s40168-018-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev 35: 343–359, 2011. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 65.Soueidan H, Nikolski M. Machine learning for metagenomics: methods and tools. 2015. https://arxiv.org/abs/1510.06621
- 66.Statnikov A, Wang L, Aliferis CF. A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification. BMC Bioinformatics 9: 319, 2008. doi: 10.1186/1471-2105-9-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figures and Supplemental Tables S1–S4: https://doi.org/10.5281/zenodo.4420108.