Summary
The Janssen (Johnson & Johnson) Ad26.COV2.S non-replicating viral vector vaccine has been widely deployed for COVID-19 vaccination programs in resource-limited settings. Here we confirm that neutralizing and binding antibody responses to Ad26.COV2.S vaccination are stable for 6 months post-vaccination, when tested against multiple SARS-CoV-2 variants. Secondly, using longitudinal samples from individuals who experienced clinically mild breakthrough infections 4 to 5 months after vaccination, we show dramatically boosted binding antibodies, Fc effector function, and neutralization. These high titer responses are of similar magnitude to humoral immune responses measured in convalescent donors who had been hospitalized with severe illness, and are cross-reactive against diverse SARS-CoV-2 variants, including the neutralization-resistant Omicron (B.1.1.529) variant that currently dominates global infections, as well as SARS-CoV-1. These data have implications for population immunity in areas where the Ad26.COV2.S vaccine has been widely deployed, but where ongoing infections continue to occur at high levels.
Keywords: SARS-CoV-2, breakthrough infection, Omicron, Variant of concern, Ad26.COV2.S, antibody-dependent cellular cytotoxicity
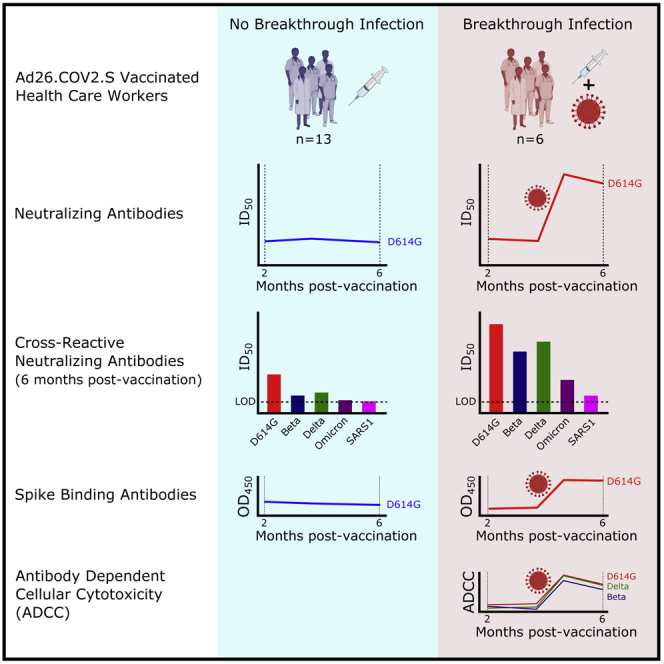
Graphical abstract
Highlights
-
•
Ad26.COV2.S neutralizing antibodies persist 6 months post-vaccination
-
•
Breakthrough infection boosts binding antibodies, ADCC, and neutralization
-
•
Boosted neutralizing antibodies cross-react with SARS-CoV-2 variants including Omicron
-
•
Vaccination and infection synergistically contribute to high levels of immunity
Kitchin, Richardson et al. show that responses to Ad26.COV2.S persist for 6 months after vaccination. Furthermore, mild breakthrough infection after Ad26.COV2.S vaccination results in dramatic boosts in binding and neutralizing antibody titers and Fc effector function. This includes high titers of neutralizing antibodies against the highly resistant Omicron variant.
Introduction
A phase 3 clinical trial of Ad26.COV2.S in eight countries demonstrated 85% protection against severe disease,1 including in South Africa, where the trial coincided with the emergence of the Beta (B.1.351) variant, which was shown to have increased resistance to neutralizing antibodies.2,3 As a result, Ad26.COV2.S was made available to South African health care workers (HCWs) in early 2021 through the Sisonke open-label, phase 3b clinical trial. Globally, this vaccine has also been used widely in several countries, including the United States and European Union member states, with 5.38, 15.68, and 16.16 million doses administered in these regions, respectively, by the beginning of November 2021.
Subsequently, South Africa has experienced a third and fourth wave of infection, driven by the Delta (B.1.617.2) and Omicron (B.1.1.529) variants, respectively, with reports of Ad26.COV2.S breakthrough infections (BTIs) occurring. Infections following mRNA vaccination result in boosted neutralizing antibody titers,4, 5, 6 but less is known about the immunological consequences of BTI after Ad26.COV2.S vaccination. With the emergence in late November 20217 of the Omicron variant, which now dominates global infections and has more spike mutations in key neutralizing epitopes than any variant to date, a key question is whether Ad26.COV2.S-vaccinated individuals who experienced breakthrough infections in the previous Delta-driven wave would have substantial neutralizing responses against this variant.
Here, we evaluated the durability and breadth of vaccine-elicited humoral responses in 19 HCWs vaccinated with Ad26.COV.2S in February-March 2021 (Figure 1A). Second, we characterized the humoral response to BTI in a subset of six individuals with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) PCR-confirmed infections 4 to 5 months (median number of months: 4.4; interquartile range [IQR]: 4.1–4.8) following vaccination. Five of these participants were followed longitudinally 2 to 6 months post-vaccination, whereas for the sixth BTI participant only 2- and 6-month post-vaccination samples were available (Table S1). These BTIs occurred between June and August 2021, during the third wave of SARS-CoV-2 infections in South Africa. This wave was driven by the more transmissible Delta variant, which accounted for between 40% and 95% of genomes sequenced in South Africa over this period.8 BTIs were thus most likely caused by the Delta variant, though sequencing data for these participants were not available. Participants, of whom 16 of 19 were female, had a median age of 34 (IQR: 30–40 years) and all presented with mild disease (Table S1). All 19 participants were SARS-CoV-2 naive prior to vaccination, as confirmed by nucleocapsid ELISA (Figure 1B).
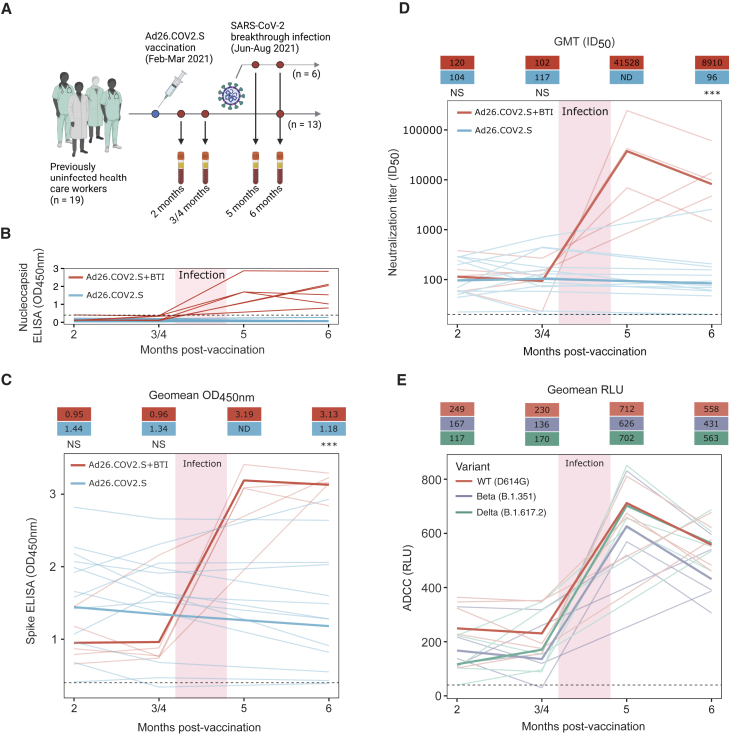
Figure 1.
Ad26.COV2.S BTI boosted plasma binding and neutralization antibody titers against the ancestral variant (D614G) and increased ADCC activity against the D614G, Beta, and Delta variants
(A) Nineteen HCWs, vaccinated with a single dose of Ad26.COV2.S, were recruited, with six having BTIs. Longitudinal blood draws occurred between 2 and 6 months post-vaccination. However, for one BTI participant, only 2- and 6-month post-vaccination plasma samples were available.
(B) Nucleocapsid ELISA binding (OD450nm), from 2 to 6 months post-vaccination, is shown for each BTI and non-BTI participant by red and blue lines, respectively.
(C) Spike binding responses to the D614G spike protein (OD450nm) by ELISA for BTI and non-BTI participants are shown in red and blue, respectively, with each line representing individual responses over time. Lines in bold show the geomean OD450nm for each group.
(D) Neutralization titers (ID50) against the D614G variant, from 2 to 6 months post-vaccination are shown for each BTI and non-BTI participant with longitudinal data by red and blue lines, respectively. Lines in bold indicate the GMTs for the BTI and non-BTI groups.
(E) Cross-reactive ADCC activity for each BTI participant with longitudinal data up to 6 months post-vaccination, shown as relative light units (RLUs). ADCC activity for each participant against the D614G, Beta, and Delta variants is shown by the red, blue, and green lines, respectively. Lines in bold show the geomean RLU for each variant. The threshold for positivity for each assay is indicated by a dashed line in each figure. All results are the mean of two independent experiments. Statistical analyses were performed using the Mann-Whitney test between groups, with ∗∗∗ denoting p < 0.001, NS for non-significant, and ND for no data.
Results
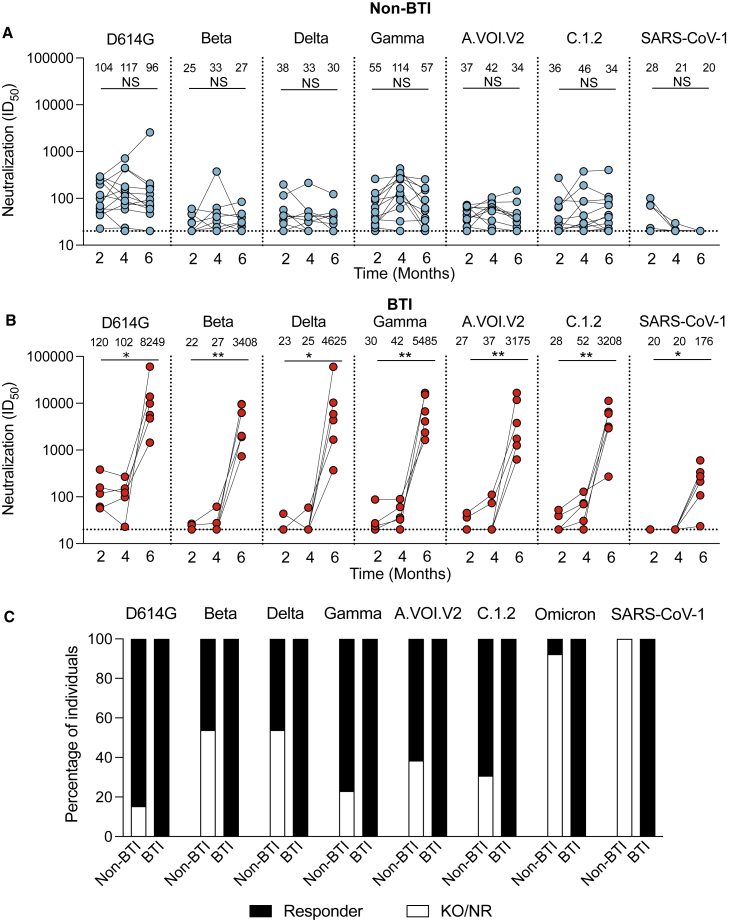
We first assessed the durability of vaccine-elicited antibody responses in individuals who were confirmed to have remained uninfected up to 6 months post-vaccination, by nucleocapsid ELISA (Figure 1B). Spike binding responses against the original D614G variant were measured at 2, 4, and 6 months post-vaccination. No significant reduction in binding was observed over this period (Figure 1C), as has been previously reported.9, 10, 11 We also used a spike-pseudotyped lentivirus neutralization assay to measure longitudinal neutralization titers against the ancestral D614G variant (which differs from the vaccine spike protein by a single D614G mutation), six SARS-CoV-2 variants with increased transmissibility and/or immune escape mutations, and SARS-CoV-1. The SARS-CoV-2 variants included Beta, Delta, Gamma (P.1), and Omicron, as well as C.1.2, and A.VOI.V2, isolated in South Africa and Angola, respectively, which share common mutations with variants of concern (VOCs) and are of local relevance to the Southern African region.12,13
For the ancestral D614G variant, geometric mean titers (GMTs) were stable up to 6 months post-vaccination (GMTs of 104, 117, and 96 at 2, 4, and 6 months post-vaccination), consistent with previous studies10,11 (Figures 1D and 2A). Where detectable, titers against the six variants were similarly stable over 6 months, showing no significant differences over time (Figure 2A), as observed in two previous studies.10,14 However, for all variants tested, titers were 1.9- to 4.2-fold lower at 2 months post-vaccination compared with the D614G variant, as reported elsewhere10,14, 15, 16, 17 (Figure 2A). For the Beta and Delta variants, in particular, half of non-BTI vaccinees showed no detectable neutralization at 6 months post-vaccination (Figure 2C). As expected, titers against SARS-CoV-1 were low, with GMTs of 28 and 21 at 2 and 4 months, respectively, and undetectable at 6 months post-vaccination (Figure 2A).
Figure 2.
Longitudinal neutralization responses over 6 months for BTI and non-BTI participants against SARS-CoV-2 variants and SARS-CoV-1
(A and B) Neutralization ID50 titers are shown for (A) Ad26.COV2.S vaccinees who did not have breakthrough infection (non-BTI) and (B) BTI Ad26.COV2.S vaccinees at 2, 3 or 4, and 6 months post-vaccination, against the D614G, Beta, Delta, Gamma, C1.2., and A.VOI.2 variants, and SARS-CoV-1. All results are the mean of two independent experiments. Significance is shown as per Friedman test, across all time points where NS denotes non-significant, ∗ denotes p < 0.05, and ∗∗ denotes p < 0.01.
(C) Percentage of individuals who are neutralization responders (Black; ID50 > 20), or are either non-responders or show knockout relative to D614G (KO/NR, ID50 < 20; white) at 6 months post-vaccination.
We next assessed the breadth and magnitude of humoral immune responses following BTI. In all participants, BTI occurred between 3 and 5 months post-vaccination. Prior to BTI, the nucleocapsid binding responses in both the BTI and non-BTI participants were negative, and only detected following BTI (Figure 1B). There were also no significant differences in D614G spike binding responses between the BTI and non-BTI participants prior to 3 to 4 months post-vaccination (Figure 1C). However, following infection, there was a 3.3-fold increase in spike responses, which peaked at approximately 2 weeks post-infection (5 months post-vaccination) and remained constant until 1 month post-infection (6 months post-vaccination) (Figure 1C).
We also assessed the impact of BTI on Fc effector functions, which have been implicated in protection from severe coronavirus disease 2019 (COVID-19) disease, and which generally retain activity against VOCs.18,19 We examined whether levels of antibody-dependent cellular cytotoxicity (ADCC), measured by ability to cross-link FcγRIIIa were boosted following BTI. Similar to binding, ADCC against D614G remained stable up to 3 to 4 months post-vaccination, with a rapid 3.1-fold increase in activity after BTI (Figure 1E). These responses peaked (geomean RLU: 712) at approximately 2 weeks post-infection (5 months post-vaccination), but declined slightly (geomean RLU: 558) by 1 month post-infection (6 months post-vaccination) (Figure 1E). Both before and after infection, ADCC was cross-reactive against D614G, Beta, and Delta variants, showing only slight decreases against VOCs relative to D614G across all time points (geomean RLUs of 712, 626, and 702 against D614G, Beta, and Delta, respectively, at 2 weeks post-infection). This illustrates the resilience of Fc effector function against VOCs in Ad26.CoV2.S BTI participants.
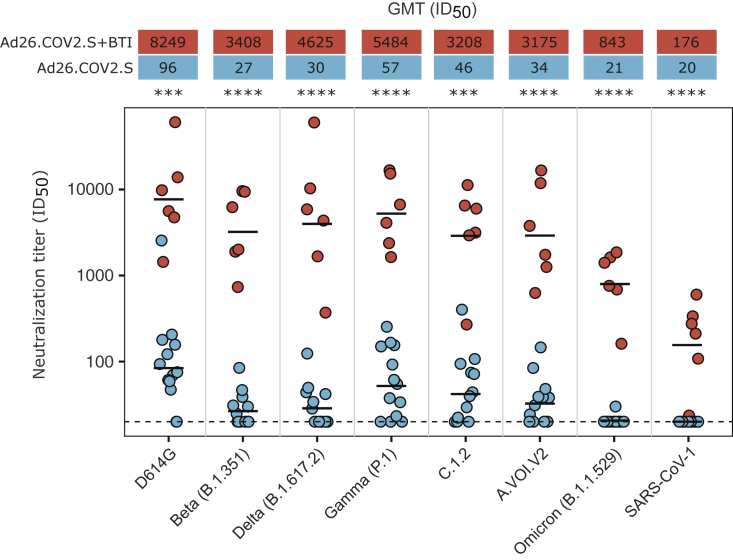
Neutralization titers against D614G closely mirrored the spike binding and ADCC response, with no significant differences in titers between the BTI and non-BTI participants prior to 3 to 4 months post-vaccination, but with a dramatic increase in titers for all participants (407-fold increase from 102 to 41,528 GMT) following infection (Figures 1D and 2B). This increase in neutralization titers is similar in magnitude to what was previously reported for a single individual with Ad26.CoV2.S BTI.10 We observed extremely high neutralization titers at approximately 2 weeks post-infection (5 months post-vaccination), which declined by approximately 4.7-fold 1 month thereafter (Figure 1D). Neutralization titers after BTI were also significantly higher against six SARS-CoV-2 variants relative to non-BTI participants (40- to 154-fold difference in GMT), and SARS-CoV-1 (9-fold difference in GMT) (Figures 2B and 3). This includes the highly neutralization-resistant Omicron variant, against which titers ranged from 161 to 1,858 (GMT of 843) for the BTI participants (Figure 3). Thus, in contrast to vaccine-elicited responses, BTI after a single dose of Ad26.COV2.S resulted in neutralization of all SARS-CoV-2 variants, with a GMT >800 against Omicron, and >3,000 against all other SARS-CoV-2 variants (Figures 2B, 2C, and 3).
Figure 3.
BTI results in increased plasma neutralization titers against all SARS-CoV-2 variants, and SARS-CoV-1, 6 months post-vaccination
The neutralization titers against the ancestral (D614G), Beta (B.1.351), Delta (B.1.617.2), Gamma (P.1), C.1.2, A.VOI.V2, and Omicron (B.1.1.529) SARS-CoV-2 variants, and SARS-CoV-1, for six BTI participants relative to 13 non-BTI participants at the 6-month post-vaccination visit (approximately 1 month post-BTI). Each dot represents the neutralization titer of a single participant, with the BTI participants and non-BTI participants shown in red and blue, respectively. The GMT for each group against each variant is shown by a black horizontal bar in the plot, with the values given in the red and blue boxes above the plot. Neutralization titers in the BTI group were significantly higher than those of the non-BTI group (40- to 154-fold higher GMT against the SARS-CoV-2 variants and 9-fold higher against SARS-CoV-1). All results are the mean of two independent experiments. Statistical analyses were performed using the Mann-Whitney test between groups, with ∗∗∗ denoting p < 0.001 and ∗∗∗∗ denoting p < 0.0001.
Discussion
Overall our data confirm durable vaccine-elicited humoral immune responses 6 months after a single dose of Ad26.COV2.S, consistent with other studies.10,14, 15, 16, 17 Moreover, despite relatively modest titers after vaccination, we observed significantly boosted binding antibodies, ADCC, and neutralization activity following BTI. This boost resulted in neutralization titers in BTI participants at 1 month post-infection (GMT 8,249) that were higher than those elicited by a two-dose Pfizer-BioNTech (BNT162b2) vaccine regimen (GMT: 1,128) 2 month post-vaccination, or those observed in convalescent donors who had previously been hospitalized with moderate (GMT: 993) or severe disease (GMT: 3,747) (Figure S1). Though we note that these comparisons differ by number of antigen exposures and timing, among other variables, this illustrates the extremely high level of boosting observed in BTIs. Similar to BTI individuals, we have previously confirmed that ADCC, binding, and neutralization are also significantly boosted following vaccination in individuals who were previously infected,20 but not to the same levels we report here for BTI (GMT of 1,372 versus 8,249 for neutralization, respectively) (Figure S1). Whether this is a result of increased affinity maturation broadening the response against variants, or rather the boosting of vaccine-elicited cross-reactive memory B cells to higher titers, remains to be determined and will be the focus of future work.
These data add to previous reports of BTI following mRNA vaccination, which results in >30-fold increased neutralization potency, suggesting broad relevance across multiple vaccine modalities.4,5,21 Affinity maturation following SARS-CoV-2 infection can lead to the development of broader and more potent neutralizing antibodies.22,23 We have previously shown that the sequence of the spike in a prior infection influenced the breadth and potency of neutralizing antibodies following vaccination with Ad26.COV2.S.20 Whether spike identity in BTIs (here, likely a heterologous exposure to the Delta variant after priming with ancestral sequence) similarly contributes to the selection of cross-reactive B cells is not yet known.24 Taken together, these findings suggest a strongly synergistic effect of vaccination and infection, which will contribute to higher levels of protective community immunity in areas with high burden of infection, and may have contributed to the low hospitalization rates seen during the Omicron-driven fourth wave in South Africa.25 As homologous and heterologous boosters are deployed, this effect may be further enhanced.26 Overall, this study provides insight into the magnitude and quality of humoral immune responses elicited by BTIs after an adenovirus-based vaccine, with implications for public health interventions in regions that have experienced high levels of SARS-CoV-2 transmission.
Limitations of study
This study is limited by the relatively small number of BTIs that were characterized. We also note that the median age of the BTI participants was higher than that of the non-BTI participants (BTI participants median age: 39 years, IQR: 32–59 years; non-BTI participants median age: 33 years, IQR: 29–36 years), which may impact comparisons between the two groups.11,27 We do not have sequencing data to determine the variant responsible for BTI, and it is possible that the infecting viral sequence may impact the quality of the response, as we have previously shown.20,28 We note that the pseudotyped virus neutralization assay used here can only assess the effectiveness of neutralizing antibodies against the spike protein in preventing viral entry into host cells, and cannot detect any effects antibodies may have on viral replication or cell-to-cell spread.29,30 Inherent differences between the pseudotyped viruses and authentic, replication-competent SARS-CoV-2 viruses, such as spike density and geometry on the virion surface, may also result in differences in sensitivity.29,30 However, neutralization titers from pseudovirus and replication-competent SARS-CoV-2 assays generally correlate well.31,32 We also focused only on humoral responses, and T cell responses following BTI will need to be defined in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CR3022 | Genscript (https://www.genscript.com) | N/A |
| BD23 | Dr Nicole Doria-Rose, VRC, USA | N/A |
| P2B-2F6 | Dr Nicole Doria-Rose, VRC, USA | N/A |
| anti-IgG APC (clone QA19A42) | Biolegend | Cat#366905 RRID:AB_2888847 |
| Palivizumab | Medimmune | Synagis; RRID: AB_2459638 |
| Bacterial and virus strains | ||
| SARS-CoV-2 pseudoviruses for ancestral (D614G), Beta, Delta, Gamma, Omicron, C.1.2, A.VOI.V2 | Wibmer et al., 2021; Richardson et al., 2021 This paper |
N/A |
| Biological samples | ||
| Convalescent hospitalized blood samples | Groote Schuur Hospital | https://www.gsh.co.za |
| Convalescent hospitalized blood samples | Steve Biko Academic Hospital | https://www.sbah.org.za |
| AD26.COV2.S vaccinee blood samples | Groote Schuur Hospital | https://www.gsh.co.za |
| AD26.COV2.S vaccinee blood samples | Steve Biko Academic Hospital | https://www.sbah.org.za |
| AD26.COV2.S vaccinee blood samples | National institute for Communicable Diseases | https://www.nicd.ac.za |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 original (D614G) spike protein | Dr Jason McKellan | N/A |
| Critical commercial assays | ||
| PEI-MAX 40,000 | Polysciences | Cat # 24765-1 |
| QUANTI-Luc luciferase | Invivogen | Cat# rep-qlc2 |
| Luciferase | Promega | Cat# PRE263B-C |
| Experimental models: Cell lines | ||
| Human Embryonic Kidney (HEK) 293F | Dr Nicole Doria-Rose, VRC, USA | N/A |
| HEK293T/ACE2.MF | Dr Michael Farzan, Scripps, USA | N/A |
| Jurkat-Lucia™ NFAT-CD16 cells | Invivogen | Cat # jktl-nfat-cd16 |
| Human Embryonic Kidney (HEK) 293T cells | Dr George Shaw, UPenn,USA | N/A |
| Recombinant DNA | ||
| Spike Hexapro plasmid | Dr Jason McKellan | N/A |
| SARS-CoV-2 ancestral variant spike (D614G) plasmid | Wibmer et al., 2021 | N/A |
| Beta spike (L18F, D80A, D215G, K417N, E484K, N501Y, D614G, A701V, 242-244 del) plasmid | Wibmer et al., 2021 | N/A |
| Delta spike (T19R, R158G L452R, T478K, D614G, P681R, D950N, 156-157 del) plasmid | Keeton et al., 2021 | N/A |
| Gamma spike (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F) | Richardson et al., 2021 | N/A |
| Omicron spike ((A67V, Δ69-70, T95I, G142D, Δ143-145, Δ211, L212I, 214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493K, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F) plasmid | This paper | N/A |
| C.1.2. spike (P9L, C136F, Δ144, R190S, D215G, Δ242-243, Y449H, E484K, N501Y, D614G, H655Y, N679K, T716I, T859N) plasmid | This paper | N/A |
| A.VOI.V2 spike (D80Y, Δ144, I210N, Δ211, D215G, R246M, Δ247-249, W258L, R346K, T478R, E484K, H655Y, P681H, Q957H) plasmid | This paper | N/A |
| SARS-CoV-1 spike plasmid | Dr Elise Landais, Scripps | N/A |
| Firefly luciferase encoding lentivirus backbone plasmid | Dr Michael Farzan, Scripps | N/A |
| Software and algorithms | ||
| Geneious software | Biomatters Ltd | https://www.geneious.com |
| FACSDiva 9 | BD Biosciences | https://www.bdbiosciences.com |
| FlowJo 10 | FlowJo, LLC | https://www.flowjo.com |
| R version 4.1.0 | The R Foundation for Statistical Computing | https://www.r-project.org |
| ggplot2 package | Tidyverse | https://ggplot2.tidyverse.org |
| Graphpad Prism 9 | Graphpad | https://www.graphpad.com |
| Biorender | Biorender | https://www.biorender.com |
Resource availability
Lead contact
Further information and reasonable requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Penny Moore (pennym@nicd.ac.za).
Materials availability
Materials will be made available by request to Penny Moore (pennym@nicd.ac.za).
Experimental model and subject details
Human subjects
HCWs vaccinated with one dose of Ad26.CoV2.S (5 × 1010 viral particles) as part of the Sisonke implementation trial were followed longitudinally and plasma sampled at 2-, 3- (in some cases 4-) and 6-months post-vaccination. An additional plasma sample was collected from BTI participants at 5-months post-vaccination, which was approximately 2-weeks post-infection. Non-BTI participants were recruited from HCWs at the National Institute for Communicable Diseases (NICD) (Johannesburg), while BTI participants were recruited from HCWs at the NICD, Steve Biko Academic Hospital (Tshwane, South Africa) and Groote Schuur Hospital (Cape Town, South Africa). Lack of prior infection in these individuals was confirmed by Nucleocapsid ELISA as described.20 Ad26.CoV2.S vaccinees with prior SARS-CoV-2 infection were recruited from a longitudinal study of healthcare workers enrolled from Groote Schuur Hospital, with plasma samples collected 2-months post-vaccination. Plasma was also collected from thirteen participants that had received two doses of the Pfizer BioNTech vaccine (BNT162b2) 2-months after they had received their last dose (Johannesburg, South Africa). Convalescent participants were recruited as part of a hospitalised cohort at the Steve Biko Academic Hospital between May and August 2020, with plasma samples collected 10-days after the initial positive PCR test. Ethics approval was obtained from the Human Research Ethics Committees of the University of the Witwatersrand (ethics reference number: M210465), University of Pretoria (ethics reference number: 247/2020) and University of Cape Town (ethics reference numbers: 190/2020 and 209/2020). Written informed consent was obtained from all participants.
Cell lines
Human embryo kidney HEK293T cells were cultured at 37°C, 5% CO2, in DMEM containing 10% heat-inactivated fetal bovine serum (Gibco BRL Life Technologies) and supplemented with 50 μg/mL gentamicin (Sigma). Cells were disrupted at confluence with 0.25% trypsin in 1 mM EDTA (Sigma) every 48–72 hours. HEK293T/ACE2.MF cells were maintained in the same way as HEK293T cells but were supplemented with 3 μg/mL puromycin for selection of stably transduced cells. HEK293F suspension cells were cultured in 293 Freestyle media (Gibco BRL Life Technologies) and cultured in a shaking incubator at 37°C, 5% CO2, 70% humidity at 125 rpm maintained between 0.2 and 0.5 million cells/ml. Jurkat-Lucia™ NFAT-CD16 cells were maintained in IMDM media with 10% heat-inactivated fetal bovine serum (Gibco, Gaithersburg, MD), 1% Penicillin Streptomycin (Gibco, Gaithersburg, MD) and 10 μg/mL of Blasticidin and 100 μg/mL of Zeocin was added to the growth medium every other passage.
Method details
SARS-CoV-2 antigens
For ELISA, SARS-CoV-2 full ancestral spike (L18F, D80A, D215G, K417N, E484K, N501Y, D614G, A701V, 242-244 del) proteins were expressed in Human Embryonic Kidney (HEK) 293F suspension cells by transfecting the cells with the respective expression plasmid. After incubating for six days at 37°C, 70% humidity and 10% CO2, proteins were first purified using a nickel resin followed by size-exclusion chromatography. Relevant fractions were collected and frozen at −80°C until use.
SARS-CoV-2 spike enzyme-linked immunosorbent assay (ELISA)
Two μg/ml of spike protein (ancestral D614G) was used to coat 96-well, high-binding plates and incubated overnight at 4°C. The plates were incubated in a blocking buffer consisting of 5% skimmed milk powder, 0.05% Tween 20, 1× PBS. Plasma samples were diluted to 1:100 starting dilution in a blocking buffer and added to the plates. IgG or IgA secondary antibody was diluted to 1:3000 or 1:1000 respectively in blocking buffer and added to the plates followed by TMB substrate (Thermofisher Scientific). Upon stopping the reaction with 1 M H2SO4, absorbance was measured at a 450 nm wavelength. In all instances, mAbs CR3022 and BD23 were used as positive controls and Palivizumab was used as a negative control.
Spike plasmid and lentiviral pseudovirus production
The SARS-CoV-2 Wuhan-1 spike, cloned into pCDNA3.1 was mutated using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies) and NEBuilder HiFi DNA Assembly Master Mix (NEB) to include D614G (ancestral) or lineage defining mutations for Beta (L18F, D80A, D215G, Δ242-244, K417N, E484K, N501Y, D614G and A701V), Delta (T19R, Δ156-157, R158G, L452R, T478K, D614G, P681R, D950N), Gamma (L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F), C1.2. (P9L, C136F, Δ144, R190S, D215G, Δ242-243, Y449H, E484K, N501Y, D614G, H655Y, N679K, T716I, T859N), A.VOI.V2 (D80Y, Δ144, I210N, Δ211, D215G, R246M, Δ247-249, W258L, R346K, T478R, E484K, H655Y, P681H, Q957H) and Omicron (A67V, Δ69-70, T95I, G142D, Δ143-145, Δ211, L212I, 214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493K, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, L981F). SARS-CoV-1 spike was also cloned into pcDNA.
Pseudotyped lentiviruses were prepared as previously described.3 Briefly, pseudoviruses were produced by co-transfecting HEK293T cell line with one of the SARS-CoV-2 variant spike plasmids or the SARS-CoV-1 spike plasmid, in conjunction with a firefly luciferase encoding lentivirus backbone plasmid (HIV-1 pNL4.luc encoding the firefly luciferase gene) with PEIMAX (Polysciences). Culture supernatants were clarified of cells by a 0.45-μM filter and stored at −70°C.
Pseudovirus neutralization assay
For the neutralization assay, plasma samples were heat-inactivated and clarified by centrifugation. Heat-inactivated plasma samples from vaccine recipients were incubated with the SARS-CoV-2 or SARS-CoV-1 pseudotyped virus for 1 hour at 37°C, 5% CO2. Subsequently, 1 × 104 HEK293T cells engineered to over-express ACE-2 (293T/ACE2.MF, kindly provided by M. Farzan at Scripps Research) were added and incubated at 37°C, 5% CO2 for 72 hours upon which the luminescence of the luciferase gene was measured. Titers were calculated as the reciprocal plasma dilution (ID50) causing 50% reduction of relative light units. CB6 and CA1 were used as positive controls, while Palivizumab was used as a negative control.
Antibody-dependent cellular cytotoxicity (ADCC) assay
The ability of plasma antibodies to cross-link and signal through FcγRIIIa (CD16) and spike expressing cells was measured as a proxy for ADCC. HEK293T cells were transfected with 5 μg of either SARS-CoV-2 original variant (D614G), Beta or Delta spike plasmids using PEI-MAX 40,000 (Polysciences) and incubated for 2 days at 37°C. Expression of spike was confirmed by differential binding of CR3022 and P2B-2F6 and their detection by anti-IgG APC staining measured by flow cytometry. Subsequently, 1 × 105 spike transfected cells per well were incubated with heat inactivated plasma (1:100 final dilution) or monoclonal antibodies (final concentration of 100 μg/mL) in RPMI 1640 media supplemented with 10% FBS 1% Pen/Strep (Gibco, Gaithersburg, MD) for 1 hour at 37°C. Jurkat-Lucia™ NFAT-CD16 cells (Invivogen) (2 × 105 cells/well) were added and incubated for 24 hours at 37°C, 5% CO2. A volume of 20 μL of supernatant was then transferred to a white 96-well plate with 50 μL of reconstituted QUANTI-Luc (Invivogen) secreted luciferase and read immediately on a Victor 3 luminometer with 1s integration time. Relative light units (RLU) of a no antibody control was subtracted as background. Palivizumab was used as a negative control, while CR3022 was used as a positive control, and P2B-2F6 to differentiate the Beta from the D614G variant. To induce the transgene 1× cell stimulation cocktail (Thermofisher Scientific, Oslo, Norway) and 2 μg/mL ionomycin in R10 was added as a positive control to confirm sufficient expression of the Fc receptor. CR3022 (for spike and RBD) or 4A8 (NTD) were used as positive controls and Palivizumab were used as negative controls. RLUs for original and Beta spikes were normalised to each other and between runs using CR3022. A cut off of 40 was determined by screening of 40 SARS-CoV-2 naive and unvaccinated individuals. All samples were run head-to-head in the same experiment as were all variants tested.
Quantification and statistical analysis
Analyses were performed in Prism (v9; GraphPad Software Inc, San Diego, CA, USA) and graphs generated using the ggplot2 package in R version 4.1.0. Where neutralization titers were below the limit of detection, these were assigned a nominal value of 20 in geometric mean titer (GMT) calculations, which is the lowest plasma dilution factor used in the neutralization assay. Non-parametric tests were used for all comparisons and all t tests were 2-sided. The Mann-Whitney test was used for unpaired comparisons between two groups, while the Kruskal-Wallis ANOVA with Dunns correction was used for multiple comparisons for unpaired groups. The Friedman test was used for multiple comparisons between paired groups. P values less than 0.05 were considered to be statistically significant.
Acknowledgments
We thank the study participants at Groote Schuur Hospital, Steve Biko Academic Hospital, and the National Institute for Communicable Diseases, who contributed samples that enabled this work. We also thank Elloise du Toit for assistance with clinical data and Nigel Garrett, Ameena Goga, and the Sisonke vaccination team. Tandile Hermanus assisted with the analysis of neutralization data. Roanne Keeton, Amkele Ngomti, Richard Baguma, and Ntombi Benede assisted with sample processing and storage. The parental soluble spike was provided by Jason McLellan (University of Texas). The parental pseudovirus plasmids were kindly provided by Elise Landais and Devin Sok (IAVI).
This work was supported by the South African Medical Research Council (grants 96825 and 96838). P.L.M. is supported by the South African Research Chairs Initiative of the Department of Science and Innovation and the National Research Foundation of South Africa (grant no. 98341). W.A.B. is supported by the EDCTP2 program of the European Union’s Horizon 2020 program (TMA2016SF-1535-CaTCH-22), the Wellcome Centre for Infectious Diseases Research in Africa (CIDRI-Africa), which is supported by core funding from the Wellcome Trust (203135/Z/16/Z), and the Poliomyelitis Research Foundation (PRF 21/65). N.A.B.N. acknowledges funding from the SAMRC, MRC UK, NRF, and the Lily and Ernst Hausmann Trust. S.I.R. is a L’Oreal/Unesco Women in Science South Africa Young Talents awardee. Related research by the authors is conducted as part of the DST-NRF Center of Excellence in HIV Prevention, which is supported by the Department of Science and Technology and the National Research Foundation.
Author contributions
D.K., S.I.R., and P.L.M. conceived the study, designed experiments, analyzed data, and wrote the paper. Z.M., F.A., B.O., and B.E.L. made molecular constructs and expressed antibodies. Z.M. and T.M.G. expressed and purified recombinant antigens. Z.M. and F.A. performed spike and nucleocapsid ELISAs. T.M., N.M., and H.K. made pseudoviruses and performed neutralization experiments. S.I.R., N.P.M., and H.S. performed ADCC assays. L.G.B. and G.G.G. conceptualized and led the Sisonke Ad26.COV2.S trial. S.I.R. established the NICD HCW cohort of vaccinees. M.A.v.d.M., M.d.P., M.T.B., T.M.R., and V.U. established the Steve Biko HCW cohort and provided samples. M.M., S.S., N.W., N.A.B.N., and W.A.B. established the Groote Schuur Hospital cohort and provided samples. All authors critically reviewed and approved the final manuscript.
Declaration of interests
P.L.M. is a member of the advisory board for Cell Reports Medicine. All other authors declare no competing interests.
Published: February 10, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100535.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
References
- 1.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 3.Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 4.Bates T.A., McBride S.K., Winders B., Schoen D., Trautmann L., Curlin M.E., Tafesse F.G. Antibody response and variant cross-neutralization after SARS-CoV-2 breakthrough infection. JAMA. 2021 doi: 10.1001/jama.2021.22898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier A.-R.Y., Brown C.M., Mcmahan K., Yu J., Liu J., Jacob-Dolan C., Chandrashekar A., Tierney D., Ansel J.L., Rowe M., et al. Immune responses in fully vaccinated individuals following breakthrough infection with the SARS-CoV-2 Delta variant in Provincetown, Massachusetts. medRxiv. 2021 doi: 10.1101/2021.10.18.21265113. https://www.medrxiv.org/content/10.1101/2021.10.18.21265113v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walls A.C., Sprouse K.R., Joshi A., Bowen J.E., Franko N., Navarro M.J., Stewart C., McCallum M., Goecker E.A., Degli-Angeli E.J., et al. Delta breakthrough infections elicit potent, broad and durable neutralizing antibody responses. bioRxiv. 2021 doi: 10.1101/2021.12.08.471707. https://www.biorxiv.org/content/10.1101/2021.12.08.471707v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., Anyaneji U.J., Bester P.A., Boni M.F., Chand M., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in Southern Africa. medRxiv. 2021 doi: 10.1101/2021.12.19.21268028. https://www.medrxiv.org/content/10.1101/2021.12.19.21268028v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tegally H., Wilkinson E., Althaus C.L., Giovanetti M., San J.E., Giandhari J., Pillay S., Naidoo Y., Ramphal U., Msomi N., et al. Rapid replacement of the beta variant by the delta variant in South Africa. medRxiv. 2021 doi: 10.1101/2021.09.23.21264018. https://www.medrxiv.org/content/10.1101/2021.09.23.21264018v1 [DOI] [Google Scholar]
- 9.Stephenson K.E., Le Gars M., Sadoff J., de Groot A.M., Heerwegh D., Truyers C., Atyeo C., Loos C., Chandrashekar A., McMahan K., et al. Immunogenicity of the Ad26.COV2.S vaccine for COVID-19. JAMA. 2021;325:1535–1544. doi: 10.1001/jama.2021.3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barouch D.H., Stephenson K.E., Sadoff J., Yu J., Chang A., Gebre M., McMahan K., Liu J., Chandrashekar A., Patel S., et al. Durable humoral and cellular immune responses 8 months after Ad26.COV2.S vaccination. N. Engl. J. Med. 2021;385:951–953. doi: 10.1056/NEJMc2108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadoff J., Le Gars M., Cardenas V., Shukarev G., Vaissiere N., Heerwegh D., Truyers C., de Groot A.M., Scheper G., Hendriks J., et al. Durability of antibody responses elicited by a single dose of Ad26.COV2.S and substantial increase following late boosting. medRxiv. 2021 doi: 10.1101/2021.08.25.21262569. https://www.medrxiv.org/content/10.1101/2021.08.25.21262569v1 [DOI] [Google Scholar]
- 12.de Oliveira T., Lutucuta S., Nkengasong J., Morais J., Paixão J.P., Neto Z., Afonso P., Miranda J., David K., Inglês L., et al. A novel variant of interest of SARS-CoV-2 with multiple spike mutations detected through travel surveillance in Africa. medRxiv. 2021 doi: 10.1101/2021.03.30.21254323. https://www.medrxiv.org/content/10.1101/2021.03.30.21254323v1 [DOI] [Google Scholar]
- 13.Scheepers C., Everatt J., Amoako D.G., Tegally H., Wibmer C.K., Mnguni A., Ismail A., Mahlangu B., Lambson B.E., Richardson S.I., et al. Emergence and phenotypic characterization of C.1.2, a globally detected lineage that rapidly accumulated mutations of concern. medRxiv. 2021 doi: 10.1101/2021.08.20.21262342. https://www.medrxiv.org/content/10.1101/2021.08.20.21262342v3 [DOI] [Google Scholar]
- 14.Jongeneelen M., Kaszas K., Veldman D., Huizingh J., van der Vlugt R., Schouten T., Zuijdgeest D., Uil T., van Roey G., Guimera N., et al. Ad26.COV2.S elicited neutralizing activity against delta and other SARS-CoV-2 variants of concern. bioRxiv. 2021 doi: 10.1101/2021.07.01.450707. [DOI] [Google Scholar]
- 15.Alter G., Yu J., Liu J., Chandrashekar A., Borducchi E.N., Tostanoski L.H., McMahan K., Jacob-Dolan C., Martinez D.R., Chang A., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021:1–5. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tada T., Zhou H., Samanovic M.I., Dcosta B.M., Cornelius A., Mulligan M.J., Landau N.R. Comparison of neutralizing antibody titers elicited by mRNA and adenoviral vector vaccine against SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.07.19.452771. [DOI] [Google Scholar]
- 17.Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Cornelius A., Herati R.S., Mulligan M.J., Landau N.R. Neutralization of Mu and C.1.2 SARS-CoV-2 variants by vaccine-elicited antibodies in individuals with and without previous history of infection. bioRxiv. 2021 doi: 10.1101/2021.10.19.463727. [DOI] [Google Scholar]
- 18.Zohar T., Loos C., Fischinger S., Atyeo C., Wang C., Slein M.D., Burke J., Yu J., Feldman J., Hauser B.M., et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell. 2020;183:1508–1519.e12. doi: 10.1016/j.cell.2020.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson S.I., Manamela N.P., Motsoeneng B.M., Kaldine H., Ayres F., Makhado Z., Mennen M., Skelem S., Williams N., Sullivan N.J., et al. A SARS-CoV-2 variant of concern triggers Fc effector function with increased cross-reactivity. medRxiv. 2021 doi: 10.1101/2021.11.05.21265853. https://www.medrxiv.org/content/10.1101/2021.11.05.21265853v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keeton R., Richardson S.I., Moyo-Gwete T., Hermanus T., Tincho M.B., Benede N., Manamela N.P., Baguma R., Makhado Z., Ngomti A., et al. Prior infection with SARS-CoV-2 boosts and broadens Ad26.COV2.S immunogenicity in a variant dependent manner. Cell Host Microbe. 2021;29:1611–1619.e5. doi: 10.1016/j.chom.2021.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacisuleyman E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., Schaefer-Babajew D.J., DaSilva J., Muecksch F., Gaebler C., et al. Vaccine breakthrough infections with SARS-CoV-2 variants. N. Engl. J. Med. 2021;384:2212–2218. doi: 10.1056/NEJMoa2105000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muecksch F., Weisblum Y., Barnes C.O., Schmidt F., Schaefer-Babajew D., Wang Z., Lorenzi J.C., Flyak A.I., DeLaitsch A.T., Huey-Tubman K.E., et al. Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity. 2021;54:1853–1868.e7. doi: 10.1016/j.immuni.2021.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., Muecksch F., Schaefer-Babajew D., Finkin S., Viant C., Gaebler C., Hoffmann H.H., Barnes C.O., Cipolla M., Ramos V., et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. 2021:1–10. doi: 10.1038/s41586-021-03696-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang J., Grubbs G., Lee Y., Huang C., Ravichandran S., Forgacs D., Golding H., Ross T.M., Khurana S. Antibody affinity maturation and cross-variant activity following SARS-CoV-2 mRNA vaccination: impact of prior exposure and sex. eBioMedicine. 2021;74:103748. doi: 10.1016/j.ebiom.2021.103748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madhi S., Kwatra G., Myers J.E., Jassat W., Dhar N., Mukendi C.K., Nana A., Blumberg L., Welch R., Ngorima-Mabhena N., Mutevedzi P.C. South African population immunity and severe covid-19 with Omicron variant. medRxiv. 2021 doi: 10.1101/2021.12.20.21268096. https://www.medrxiv.org/content/10.1101/2021.12.20.21268096v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray G.E., Collie S., Garrett N., Goga A., Champion J., Zylstra M., Reddy T., Yende N., Seocharan I., Takalani A., et al. Vaccine effectiveness against hospital admission in South African health care workers who received a homologous booster of Ad26.COV2 during an Omicron COVID19 wave: preliminary results of the Sisonke 2 study. medRxiv. 2021 doi: 10.1101/2021.12.28.21268436. https://www.medrxiv.org/content/10.1101/2021.12.28.21268436v1 [DOI] [Google Scholar]
- 27.Wei J., Stoesser N., Matthews P.C., Ayoubkhani D., Studley R., Bell I., Bell J.I., Newton J.N., Farrar J., Diamond I., et al. Antibody responses to SARS-CoV-2 vaccines in 45,965 adults from the general population of the United Kingdom. Nat. Microbiol. 2021;6:1140–1149. doi: 10.1038/s41564-021-00947-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moyo-Gwete T., Madzivhandila M., Makhado Z., Ayres F., Mhlanga D., Oosthuysen B., Lambson B.E., Kgagudi P., Tegally H., Iranzadeh A., et al. Cross-reactive neutralizing antibody responses elicited by SARS-CoV-2 501Y.V2 (B.1.351) N. Engl. J. Med. 2021;384:2161–2163. doi: 10.1056/NEJMc2104192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y., Wang J., Li Q., Hu H., Lu J., Chen Z. Advances in neutralization assays for SARS-CoV-2. Scand. J. Immunol. 2021;94:e13088. [Google Scholar]
- 30.Khoury D.S., Wheatley A.K., Ramuta M.D., Reynaldi A., Cromer D., Subbarao K., O'Connor D.H., Kent S.J., Davenport M.P. Measuring immunity to SARS-CoV-2 infection: comparing assays and animal models. Nat. Rev. Immunol. 2020;20:727–738. doi: 10.1038/s41577-020-00471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riepler L., Rössler A., Falch A., Volland A., Borena W., von Laer D., Kimpel J. Comparison of four SARS-CoV-2 neutralization assays. Vaccines. 2021;9:13. doi: 10.3390/vaccines9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.