Summary
Background
Intestinal barrier impairment plays an essential role in the pathogenesis of Crohn's disease (CD), and claudins (CLDNs) dysfunction contributes to intestinal mucosa injury. SOX9, an important transcription factor, is upregulated in the disease-affected colon of patients with CD; however, its precise role in CD remains largely unknown. Our aim was to explore the interaction between SOX9 and CLDNs, and further elucidate the underlying mechanisms in CD.
Methods
SOX9 expression in patients with CD was evaluated using quantitative polymerase chain reaction, immunoblotting, and immunohistochemistry. The regulatory relationship between SOX9 and CLDNs was analyzed via a dual-luciferase reporter assay, chromatin immunoprecipitation, overexpression, and RNA interference methods. MicroRNAs (miRNAs) involved in the SOX9-CLDN pathway were predicted with bioinformatics analysis, and the upstream molecular mechanism was interpreted using MassARRAY methylation detection.
Findings
Upregulated expression of SOX9 in the disease-affected intestine mucosa was identified in both patients with CD and mice challenged with trinitrobenzene sulfonic acid (TNBS). SOX9 negatively regulated the expression of CLDN8, accompanying reduced intestinal permeability. MiR-145-5p downregulation was found in patients with CD and TNBS-induced colitis mice owing to an aberrant miR-145 promoter hypermethylation, which subsequently interfered the SOX9-CLDN8 pathway. MiR-145-5p agomir treatment alleviated TNBS-induced colitis in wild-type mice by inhibiting Sox9 expression and restoring Cldn8 expression, whereas similar findings were not apparent in the Cldn8−/− mice.
Interpretation
SOX9 mediates the crosstalk between upstream miR-145-5p and downstream CLDN8, and further impairs intestinal mucosal barrier homeostasis in CD. Targeting the miR-145-5p/SOX9/CLDN8 pathway represents a promising therapeutic strategy for CD.
Funding
The National Natural Science Foundation of China (#81870374, #81670498, #81630018, #82070538, #8210031148), the Guangdong Science and Technology (#2017A030306021, #2020A1515111087), the Guangzhou Science and Technology Department (#202002030041), and the Fundamental Research Funds for the Central Universities (#19ykzd11).
Keywords: Crohn's disease, Intestinal mucosal barrier, SOX9, CLDN8, miR-145-5p
Research in context.
Evidence before this study
The impaired intestinal mucosal barrier is an essential molecular event in the pathogenesis of inflammatory bowel disease (IBD), and previous studies have demonstrated that an aberrant alteration of certain tight junction (TJ) proteins existed in patients with IBD. The claudin family includes more than 20 members and claudins are an integral part of TJs. The structural and functional defects in the claudin family of proteins could result in dysregulated intercellular permeability of the intestinal epithelium. Our microarray chip analysis data showed that the expression of Claudin 8 is extremely downregulated in the disease-affected intestinal mucosa of patients with IBD, especially those with active disease. Moreover, the effect of IL23/miR223/Claudin 8 and IL-9/miR21/Claudin 8 pathways on regulating intestinal mucosal barrier function in CD have been proven in our previous research.
Added value of this study
In the current study, we mainly attempted to explore the specific role of miR-145/SOX9/Claudin 8 pathway in CD via series of experiments in-vitro and in-vivo. Our findings illustrated that an aberrant promoter hypermethylation of miR-145-encoding gene-mediated activation of SOX9 expression negatively regulate Claudin 8, which ultimately impairs the intestinal epithelial TJ barrier and then aggravates intestinal inflammation.
Implications of all available evidence
Our findings give new ideas for investigating the influence of Claudin 8 on the occurrence and development of CD. Taken together, the expression of Claudin 8 in patients with CD is regulated by several signalling pathways, and a complete view of Claudin 8 may provide treatment guidance in intestinal mucosal barrier repairment for CD.
Alt-text: Unlabelled box
Introduction
Inflammatory bowel disease (IBD) as a chronic and relapsing-remitting inflammatory intestinal disorder, mainly comprises two distinct categories: Crohn's disease (CD) and ulcerative colitis (UC).1,2 Although the aetiology and pathogenesis of this disease remain largely unidentified, the most accepted consensus is that complex environmental factor disturbance can induce a disordered immune response in genetically susceptible individuals, eventually contributing to a deficiency in intestinal barrier function.3, 4, 5, 6 An impaired intestinal mucosal barrier could further trigger abnormally persistent immune activation and then aggravate intestinal inflammation.7 Achieving mucosal healing or deep remission has been highlighted as a more ideal treatment goal for IBD.8 Hence, elucidating the molecular mechanisms involved in intestinal epithelial barrier regulation and developing novel therapeutic approaches for IBD are urgently needed.
Barrier dysfunction in the intestinal mucosa plays a major role in the pathogenesis of IBD, and structural and functional defects in certain tight junctions (TJs) have been highly emphasised according to several studies.9, 10, 11 TJs are essential ingredients of the intestinal epithelial barrier, and the claudin (CLDN) family accounts for the major proportion.12, 13, 14 Claudins have been categorised into barrier-forming and channel-/por-forming ones, generating a complex network to control intestinal epithelium homeostasis.15 A dysregulated expression of several CLDNs has been identified in patients with IBD and experimental colitis models, which are known to induce persistent mucosal barrier damage and unachievable mucosal healing.16, 17, 18 Our previous studies and data from Clark et al. both confirmed that, among the CLDN family proteins, CLDN8 is significantly downregulated in IBD through multiple relevant signalling pathways activation.19,20 In the colon, CLDN8 could inhibit back leakage of absorbed Na+ into the lumen by sealing lateral paracellular space and forming a barrier. However, potential factors regulating CLDN8 expression and the precise biological role of this gene in IBD remain largely unknown.
SOX9, belonging to the sex-determining region of the Y chromosome-related high-mobility group (HMG) box family, is a superfamily of transcription factors with an HMG domain.21,22 SOX9 is expressed in multiple organs and involved in diverse physiological and pathological processes of organ development.23, 24, 25 A large body of evidence indicates that SOX9 is selectively located at the intestinal crypts and contributes to the homeostatic states of intestinal stem cells.26, 27, 28 Therefore, abnormally altered SOX9 could reduce the growth capacity of intestine epithelium cell, which can limit its repairing effect on the intestine.29 In addition, SOX9 can directly or indirectly regulate corresponding downstream target genes via binding to a consensus sequence of the promoter region, or inducing DNA bending.30 The SOX family weakens the function of TJs, and previous studies showed that SOX may regulate CLDN expression directly or indirectly as a transcription factor.31, 32, 33 To date, the biological role of SOX9 has rarely been reported in patients with IBD presenting controversial results, and the precise regulatory mechanism of SOX9 remains unclear in IBD.34,35
This current study aimed to explore the interaction between SOX9 and CLDNs and further elucidate the underlying molecular mechanisms, identify SOX9 as a molecule that mediate upstream miR-145-5p and downstream CLDN8 in CD, and provide new insights into a novel mechanism of the miR-145-5p/SOX9/CLDN8 pathway-mediated the intestinal mucosal barrier regulation in CD.
Methods
Intestinal mucosal tissues
Intestinal mucosal specimens were clamped from patients with CD and healthy controls (HCs) undergoing endoscopy at our hospital (The First Affiliated Hospital of Sun Yat-sen University, Guangdong, China), from October 2017 to October 2018. Biopsy samples were collected from the affected lesion sites in CD, including erosions and ulcerations. HCs underwent colonoscopy for physical examination purposes, and their intestinal mucosa were sampled at similar locations as those in patients with CD. All included patients received physical, biochemical, radiographic and histopathological examinations before they were diagnosed with CD by our multi-disciplinary treatment group. For CD patients with inflammatory lesions in the terminal ileum only, biopsy samples were obtained from the ileum; those from other patients were collected from the colon. In addition, lesion tissues obtained from the surgical removal of the intestinal segment were also collected in patients with CD during surgery for further research. The demographic and clinical characteristics of patients with CD and HCs are compared in Supplementary Table 1.
Cell lines and cell culture
Human colon adenocarcinoma cell line (Caco-2, RRID: CVCL_0025) and human colonic-cancer cell line (SW480, RRID: CVCL_0546) were acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA), normal human colon epithelial cell line (NCM460, RRID: CVCL_0460) were purchased from the INCELL Corporation LLC (San Antonio, TX, USA), and human embryonic kidney cell line (293T, RRID: CVCL_0063) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cell line identification was performed before the series of experiments in this study (Supplementary materials). Caco-2 and SW480 cell lines were maintained in Dulbecco's Modified Eagle's Medium, NCM460 cells were cultured in M3: BaseF medium (INCELL, San Antonio, TX, USA), and 293T cells were incubated in 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA). Each medium was supplemented with 10% foetal bovine serum (FBS; Thermo Fisher Scientific, Waltham, MA, USA), respectively. In addition, 100 U/mL penicillin G and 100 µg/mL streptomycin sulphate (Invitrogen, Carlsbad, CA, USA) were added to cells to avoid potential contamination, when necessary. All cell lines were cultured in a 5% CO2 humidified incubator at 37°C.
Cell transfection
Intestinal epithelial cell lines at the exponential growth phase were first digested and inoculated in a 6-well plate at a density of 1×105 cells/well, and then were incubated for subsequent transfections. The pcDNA3-Flag-SOX9 plasmid (OBiO, Shanghai, China), Si-SOX9, miR-145-5p mimic, miR-145-5p inhibitor, or corresponding negative controls (RiboBio, Guangzhou, China) were applied to cell transfection in accordance with different purposes of the respective experiments. All transfection processes were conducted using Lipofectamine™ 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to relevant protocol. The transfected cells were incubated for 48 h or 72 h for the subsequent analyses. In addition, the transfection efficiency was evaluated via determining the distribution and localisation of the FLAG-llabeled plasmid using immunofluorescence.
Real-time quantitative polymerase chain reaction analysis (RT-qPCR)
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's protocol. Next, the extracted RNA was reverse transcribed to cDNA via Transcriptor cDNA Synthesis Kit (Roche, Basel, BS, Switzerland). RT-qPCR was then carried out for the quantitative study of gene expression with the Fast Start Universal SYBR Green Master kit (Roche, Basel, BS, Switzerland) and the LightCycler 480 fluorescence quantitative PCR software (Roche, Basel, BS, Switzerland). The expression of the target mRNAs and miRNAs was assessed, and β-actin and U6 were selected as internal references, respectively. The relative expressions of target and reference genes were calculated via the 2−△△CT method. Primer sequences used in this study are provided in Supplementary Table 2.
Western blotting assay
Total protein was extracted using RIPA Lysis Buffer (Millipore, Billerica, MA, USA) containing a protease/phosphatase inhibitor cocktail (Cell Signalling Technology, Boston, MA, USA) from the intestinal mucosal samples on ice. The protein concentration was quantified using a BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Subsequently, 30 μg protein was boiled for 5 min at 95°C and then was separated by 10% sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE, Bio-Rad, Hercules, CA, USA) at the appropriate voltage. Next, the resolved proteins were transferred to a PVDF membrane (Millipore, Billerica, MA, USA), and blocked with 5% skim milk (Bio-Rad, Hercules, CA, USA). Subsequently, individual membranes were incubated with primary antibodies against SOX9 (1:1000 dilution, Abcam, Cat# ab185966, RRID: AB_2728660), GAPDH (1:1000 dilution, Cell Signalling Technology, Cat# 5174, RRID: AB_10622025), and CLDN8 (1:1000 dilution, Gene Tex, Cat# GTX77832, RRID: AB_424780). After extensive washing with TBST three times, all membranes were further incubated with HRP-conjugated anti-rabbit IgG antibody (1:2000 dilution, Cell Signalling Technology, Cat# 7074, RRID: AB_2099233). Finally, specific bands were visualised using an ECL detection system (Tanon, Shanghai, China). Protein expression was normalised to that of the reference. Each assay was repeated thrice.
Immunohistochemistry (IHC) and immunofluorescence (IF)
The paraffin sections were formalin- or 4% paraformaldehyde-fixed, and IHC staining for SOX9 was conducted according to standard protocols. Briefly, paraffin sections were deparaffinised and rehydrated via incubation in decreasing ethanol concentrations. Antigen retrieval, endogenous peroxidase activity quench, immunostaining, counterstaining, and image capture were performed according to the methodology used in our previous studies.19, 20 Subsequently, paraffin sections were incubated with the IgG antibody (rabbit monoclonal SOX9 IgG antibody, 1:300 dilution, Abcam, Cat# ab185966, RRID: AB_2728660) overnight at 4 ℃, and then with an anti-species secondary antibody (ZSGB-BIO, Beijing, China). Positive reactions in IHC staining were defined as the cells showing brown signals. The scoring system of intensity were as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. Each section was independently assessed by two blinded pathologists.
In addition, dual-label immunofluorescence staining was performed to observe SOX9-CLDN8 interaction in patients with CD and HCs. After overnight incubation with primary antibodies (SOX9, 1:300 dilution, Abcam, Cat# ab185966, RRID: AB_2728660; CLDN8, 1:300 dilution, Gene Tex, Cat# GTX77832, RRID: AB_424780), specific fluorescence-conjugated secondary antibodies were applied against the primary antibodies (SOX9 was labelled with Cy3 dyes and CLDN8 was labelled with Cy5 dyes). All sections were concurrently stained with DAPI (Vector Laboratories, Cat# H-1200, RRID: AB_2336790).
Fluorescence in-situ hybridisation (FISH)
FISH for miR-145-5p and SOX9 was conducted on paraffin sections of biopsy samples as described in our previous studies.19, 20 Briefly, hybridisation was performed using digoxigenin-labelled locked nucleic acid probes (Servicebio, Wuhan, China) targeting miR-145-5p at 55°C overnight, and simultaneous immunostaining for SOX9 was conducted after deparaffinisation. Subsequently, cy3 and cy5-conjugated anti-rabbit secondary antibodies were used for SOX9 and CLDN8, respectively. Next, slides were counterstained with DAPI for 1 min at room temperature. Finally, images were captured via a fluorescence image system (Zeiss LSM710, Oberkochen, Baden-Wuerttemberg, Germany). The sequence of the probe targeting miR-145-5p was 5′-AGGGATTCCTGGGAAAACTGGAC-3′.
Dual-luciferase reporter assay
Bioinformatic analysis showed that the 3′-UTR of SOX9 gene possessed a binding site for miR-145-5p using TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do). Wild-type (WT) or mutant (MUT) 3′-UTR containing binding sites of miR-145-5p was cloned into a pmiR-RB-Report vector (RiboBio, Guangzhou, China). Next, 3’-UTR-WT and 3′-UTR-MUT plasmids were transfected into 293T cells when they reached 50-70% confluence, and then treated with the mimic of miR-145-5p and its negative control, respectively. After 48 h of incubation, the luciferase activities of firefly and renilla were detected in harvested 293T cells using a Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions. All experiments were repeated thrice independently.
SOX9 binding site prediction to the CLDN8 promoter was performed using the JASPAR database shown in Supplementary Table 3. The 2.0-kb CLDN8 promoter fragments were amplified, and the products were then cloned into the vector. Functional assays between SOX9 and the CLDN8 promoter were performed via transient transfection of 293T cells. Transfections included the pGL3-CLDN8/pcDNA-SOX9/pRL-TK group, the pGL3-CLDN8/pcDNA-control vector/pRL-TK group, the pGL3-basic vector/pcDNA-SOX9/pRL-TK group, and the pGL3-SV40/pRL-TK group. Forty-eight hours later, colonic epithelial cells were harvested to quantify luciferase activity as described above.
Chromatin immunoprecipitation (ChIP) assay
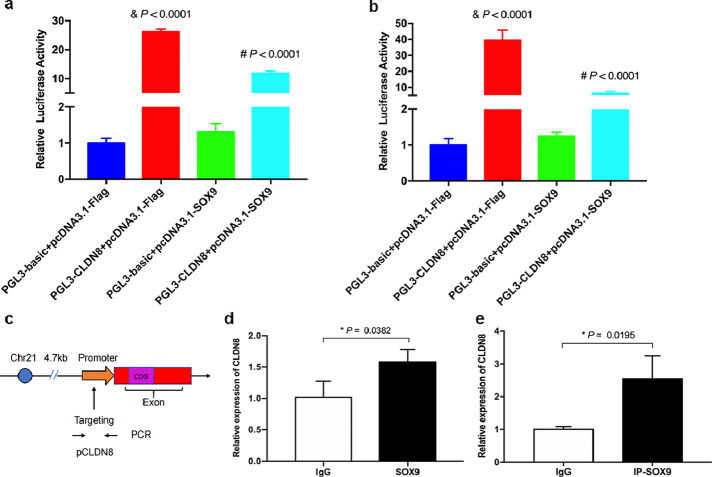
A ChIP assay was performed with the Pierce Magnetic ChIP Kit (Thermo Fisher Scientific, Waltham, MA, USA) with an anti-SOX9 antibody (ChIP Grade; Abcam, Cat# ab3697, RRID: AB_304012) according to the manufacturer's instructions. ChIP-qPCR assay was carried out using the Brilliant SYBR Green QPCR Master Mix with Stratagene Mx300P (Agilent Technologies Inc., Palo Alto, CA, USA). The cycle threshold amplification values and the percentages of sample input were calculated. The putative SOX9-binding sites located within the CLDN8 promoter region targeted by the ChIP primers was represented in Figure 3c, and the PCR primer sequences were 5′- AGCCGATCCCTTGTAATGTG-3′ and 3′-CCCCTACTCTCGTTTCACTTTG-5′.
Figure 3.
SOX9 negatively regulates CLDN8 in colonic epithelial cells by binding CLDN8 promoter. a and b, Luciferase reporter assays in Caco-2 and NCM460 cells. & P < 0.0001 compared with pGL3-basic + pcDNA3.1-Flag group; # P < 0.0001 compared with pGL3-CLDN8 + pcDNA3.1-Flag group. c, The putative SOX9-binding sites located within the CLDN8 promoter region targeted by the ChIP primers. d and e, ChIP-qPCR analysis in Caco-2 and NCM460 cells. ChIP-qPCR shows SOX9 occupancy on the CLDN8 promoter region compared with immunoglobulin-G (IgG). All Data are shown as means ± SEM in (a), (b), (d) and (e). Student's t-test and one-way ANOVA were used for comparison, respectively.
MassARRAY methylation analysis
To quantitatively evaluate the DNA methylation levels of the promoter of miR-145, genomic DNA from colonic tissues was extracted using TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) based on the manufacturer's protocols. Sequence-specific primers were designed via Agena EpiDesigner (http://www.epidesigner.org/) and Methprimer (http://www.urogene.org/methprimer2/tester-invitation.html). PCR experiments were conducted, and the products were subsequently incubated with Shrimp Alkaline Phosphatase (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer's protocol. Promoter methylation of the miR-145 gene was measured using quantitative methylation analysis (MassARRAY Analyser; Agena Bioscience, San Diego, CA, USA), and the methylation ratios were calculated using the MassARRAY EpiTYPER software (Agena Bioscience, San Diego, CA, USA).
Transepithelial electrical resistance (TEER) experiment
TEER was performed to assess the paracellular permeability and the tightness of TJs in the intestinal epithelium cells, as previously mentioned.19,20 Digested Caco-2 cells were counted and inoculated on the apex side of polycarbonate membrane filters purchased from Corning (Pickerington, NY, USA) at a density of 2×105 cells/cm2. After 21 days of cell growth, the Caco-2 cell monolayers model was structured successfully with a TEER of ≥ 200 Ω×cm2.36 Subsequently, cell monolayers were exposed to various treatments for different purposes, and then, the corresponding TEER measurement was performed using Millicell ERS-2 (Millipore, Billerica, MA, USA).
FITC-labelled dextran permeability assay
Fluorescein isothiocyanate (FITC)-labelled dextran (FD4000; Sigma, St. Louis, MO, USA) effusion detection was used to observe the paracellular permeability of Caco-2 cell monolayers after different treatments. First, 1 mL FD4000-OptiMEM mixed medium (2 mg/mL) was added to the apical compartment, while another 2 mL of pure Opti-MEM medium was supplemented into the lower compartment. After incubation for 4 h, 200 μL of the medium was sucked from the lower compartment to detect the fluorescence intensity of FD4000 at 490/530 nm using Spectra Max M5 (Molecular Devices Corporation, San Jose, CA, USA).
A flux assay with FITC-labelled dextran, molecular weight 40 kD (FD40000, Sigma, St. Louis, MO, USA), was used to measure the intestinal barrier function of colitis mice.37 In brief, FITC-labelled dextran FD40000 at a dose of 0.6 mg per gram of body weight was administered to TNBS-induced colitis mice before sacrifice by intragastric gavage. Next, blood was collected after 4 h for fluorescence intensity measurement to estimate intestinal mucosal barrier permeability alterations in mice. A standard curve was simultaneously established by serially diluting the serum concentrations of FD40000.
Trinitrobenzene sulfonic acid (TNBS)-induced colitis model
The TNBS-induced colitis model was conducted in male BALB/c mice (week age: 6-8 w; weight: 20-22 g, Nanjing Biomedical Research Institute of Nanjing University, Nanjing, China), and raised under specific pathogen-free conditions in the Animal Experimental Center of our hospital. This colitis mouse model was established following a previously published protocol.19,20 Experimental groups were pre-sensitised with 1% (w/v) TNBS acid (Sigma, St. Louis, MO, USA) for 7 days, and then challenged with 100 μL 2.5% TNBS mixture (one volume 5% TNBS in one volume of absolute ethanol) on day 8 by enema, whereas the control group were administered with same volume of 50% ethanol solution. After another 7 days, all mice were sacrificed for further research. Furthermore, the TNBS-induced colitis model was also established in Cldn8−/− mice to demonstrate the role of the miR-145/SOX9/CLDN8 pathway in colitis initiation.
MiRNA agomir (RiboBio, Shanghai, China) was administered to TNBS-induced colitis mice to observe the effect of miR-145-5p on regulating the expression of Sox9 in vivo. MiR-145-5p agomir and its corresponding negative control at a dose of 10 mg/kg were administered to TNBS-induced colitis mice via intraperitoneal injection on days 8, 9, and 10. In addition, the sequences of miR-145-5p agomir could be required from RiboBio (Shanghai, China).
Evaluation of colitis in mice
Haematoxylin and eosin (H&E) staining was conducted to analyse the histological scores for colonic tissue of TNBS-induced colitis mice. In addition, colonic inflammation was evaluated using a standardised scoring system (Supplementary Table 4) and the Myeloperoxidase (MPO) activity assay as previously published.19,20 The absorbance of MPO was calculated using the SpectraMax M5 (Molecular Devices Corporation, San Jose, CA, USA) at 460 nm.
Statistical analyses
The statistical analyses were performed using Graph Prism version 7.0 (GraphPad Software, La Jolla, CA, USA) and SPSS version 23.0 (IBM Corporation, Armonk, NY, USA). Qualitative variables were reported as numbers or percentages, and quantitative data were expressed as mean ± standard deviation (SD) or median (interquartile ratio [IQR]). Correlation analysis was conducted via Pearson's or Spearman's correlation coefficient. Statistical differences were two sided, and they were established using Student's t-test or one-way analysis of variance, where appropriate. P < 0.05 was considered statistically significant.
Ethics statement
The protocol of this study complied with the ethical guidelines of the 1975 Declaration of Helsinki principles and was approved by the Ethics Committee of our hospital (approved protocol number: No. 2017[062]). All human subjects included in this study completed written informed consent before enrolment. All experimental procedures and protocols were carried out following the ethical standards of our hospital at which the studies were conducted.
Role of funding sources
Funders had no contribution to study design, experiment operation, data analyses, manuscript preparation, and publish decisions of this manuscript.
Results
SOX9 expression is upregulated in the disease-affected mucosa of CD
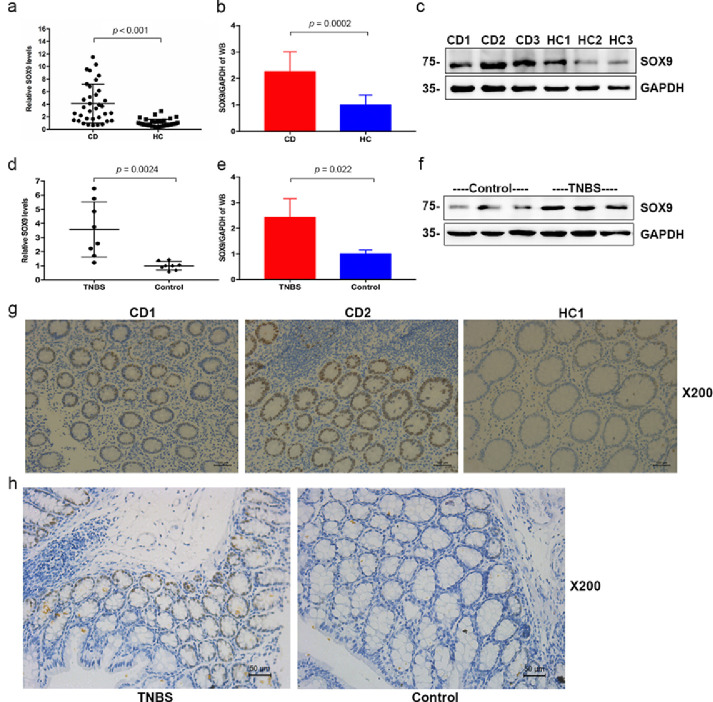
First, a gene expression analysis was performed to investigate the expression profile of SOX9 in patients with CD using the Gene Expression Omnibus (GEO) database. As shown in Fig. S1a, the expression profiling from one study showed that SOX9 expression in patients with CD appeared to be higher than that in HCs (Dataset Accession number: GSE2461), even though this array only included two patients with CD and two HCs. Moreover, expression profiling of another 36 samples (including active and inactive UC and CD, patients with infectious colitis and HCs) revealed that higher SOX9 expression was found in the disease-affected colon than in the unaffected colon (Dataset Accession number: GSE6731; P < 0.0001, Student's t-test, Fig. S1b). To further investigate the expression of SOX9 in the CD intestinal mucosa, as well as in the normal mucosa of HCs, we conducted RT-qPCR (P < 0.001, Student's t-test, Figure 1a), immunoblotting (P = 0.0002, Student's t-test, Figure 1b-c), and IHC (Figure 1g) to compare the expression of SOX9 in 35 CD patients and 35 HCs at both mRNA and protein levels. In the clinically collected mucosal biopsy samples, we identified that SOX9 was significantly upregulated (approximately four times) in patients with CD compared to HCs at the mRNA level. However, we found no significant association between the expression of SOX9 in the colon and the terminal ileum (Fig. S2a). To determine whether highly expressed SOX9 is exclusive to CD, we also determined the expression of SOX9 in patients with UC. Notably, a decreased expression of SOX9 was not observed in patients with UC (P = 0.542, Student's t-test), even if they experienced mucosal barrier damage with downregulated levels of CLDN8 (Fig. S2b-S2c).
Figure 1.
SOX9 upregulation in Crohn's disease and TNBS-induced colitis mice. a, SOX9 expression in CD and HC by RT-qPCR (CD, n = 35; HC, n = 35); b-c, SOX9 expression in CD and HC by immunoblotting (CD, n = 10; HC, n = 10); d-f, SOX9 expression in TNBS-induced colitis mice and normal controls by RT-qPCR and immunoblotting (TNBS, n = 8; Controls, n = 8); g-h, IHC staining for SOX9 expression (CD, n = 10; HC, n = 10; TNBS, n = 8; Controls, n = 8). Scale bars, 100 μm in (g) and 50 μm in (h). All data are reported as means ± SEM in (a), (b), (d), (e) and Student's t-test were used for comparison.
Subsequently, a series of IHC examinations was performed using paraffin sections from the intestinal tissue of patients with CD and HCs to accurately determine the spatial pattern of SOX9. In CD, upregulated SOX9 expression extended from the base of crypt to the surface, covering both the cell nucleus and cytoplasm, and was associated with active intestinal inflammation (Figure 1g). Furthermore, correlation analysis was performed to investigate the relationship between SOX9 expression and the degree of clinical and endoscopic disease activity. As shown in Figure S3, we identified that the level of SOX9 was positively associated with clinical disease activity of CD (Crohn's Disease Activity Index; CDAI) with an r value of 0.65, endoscopic activity (Crohn's Disease Endoscopic Index of Severity, CDEIS) with an r value of 0.61. In addition, other important inflammatory mediators including calprotectin, C-reactive protein, erythrocyte sedimentation rate and platelet counts also correlate positively with SOX9 expression. Consistent with these findings, an upregulation of Sox9 expression was also observed in the intestinal mucosa of the TNBS-induced colitis mouse model, at the mRNA and protein levels (P = 0.0024 and P = 0.022, Student's t-test, Figure 1d-f and h). Collectively, upregulated SOX9 expression was identified in patients with CD and the expression of this gene was positively correlated with clinical and endoscopic activity.
SOX9 induces junction tightness damage in colonic cells
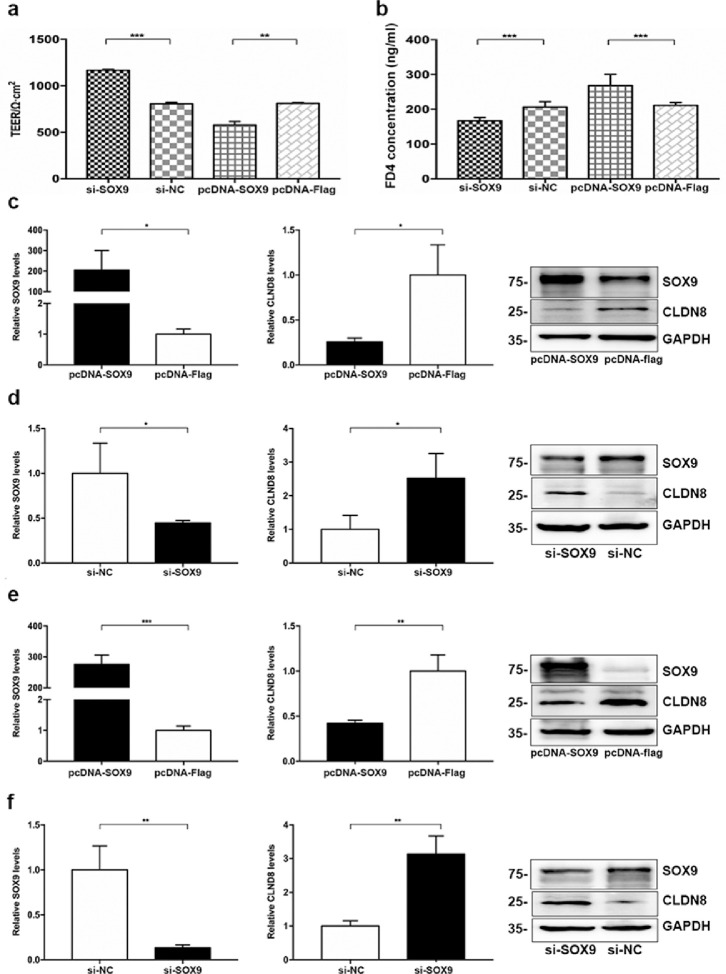
TEER was detected, and the paracellular permeability was measured using FITC-labelled dextran (FD4000) to evaluate the dysregulation of intestinal mucosal barrier in a Caco-2 cell monolayer model. We investigated the role of SOX9 in regulating TJs by overexpressing or knocking down SOX9 in Caco-2 cells. Using the TEER assay, we observed that SOX9 overexpression (pcDNA-SOX9) significantly reduced TEER when compared to the control group, whereas SOX9 knockdown (si-SOX9) significantly enhanced the TJs of epithelial cells (Figure 2a). In addition, we measured FD4 permeability to investigate the effect of SOX9 on intestinal barrier function. Compared with the control group, SOX9 knockdown significantly decreased FD4 permeability, whereas overexpression of SOX9 led to increased FD4 permeability (Figure 2b). Thus, our data indicated that SOX9 might have an adverse effect on the intestinal epithelial barrier function by regulating potential TJs. To confirm our hypothesis and observe the regulatory effect of SOX9 on TJ structure (mainly the Claudin family) in intestinal epithelial cells (Caco-2 cell), SOX9 overexpression experiments were conducted. Our results showed that the expression of CLDN8 was significantly decreased in Caco-2 cells (P = 0.0213, Student's t-test), and a significantly higher expression of CLDN4 was also detected after transfection with pcDNA-SOX9 (P = 0.0034, Student's t-test), whereas other members were not significantly altered (Fig. S4). Altogether, although SOX9 regulated other members of the claudin family, downregulation of CLDN8 was more prevalently observed.
Figure 2.
SOX9 impairs the intestinal mucosal barrier integrity by targeting CLDN8. a-b, TEER and FD4 permeability alterations in the Caco-2 cell monolayer. c-d, Overexpression and knockdown of SOX9 in Caco-2 cells. SOX9 mRNA (left panel) and CLDN8 mRNA (middle panel), and relevant proteins (right panel). e-f, Overexpression and knockdown of SOX9 in NCM460 cells. All Data are shown as means ± SEM in (a), (b), (c), (d), (e) and (f). Student's t-test were used for comparison between two groups. ns: no significant difference, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as compared with the control.
SOX9 expression is negatively correlated with CLDN8 in the CD mucosa
Sequencing analyses of the CLDN8 promoter region using bioinformatics tools revealed several putative binding sites for SOX9. Additionally, a downregulated expression of CLDN8 was observed in patients with CD and TNBS-induced colitis (Fig. S5). Therefore, we paid close attention to the SOX9-CLDN8 pathway. To explore the interaction between upregulated SOX9 and downregulated CLDN8 in CD, we analysed the correlation between SOX9 and CLDN8 expressions at the mRNA level, and observed the expression patterns of SOX9 and CLDN8 using immunofluorescence. As shown in Fig. S6a, SOX9 and CLDN8 expression levels were negatively correlated shown in the intestinal mucosa tissues of patients with CD. Compared to the normal intestinal mucosa, SOX9 expression was higher and CLDN8 expression was lower in the CD mucosa, and similar results were also observed in TNBS-induced colitis (Fig. S6b). These results indicated that abnormal activation of the SOX9-CLDN8 pathway affected the intestinal barrier function in CD.
To further observe the effect of SOX9 on regulating CLDN8 expression, we knocked down or overexpressed SOX9 in Caco-2 and NCM460 cells. As shown in Figure S7, a transfection efficiency of up to 75% was achieved for plasmid transfection in intestinal epithelial cells (Caco-2 cells). Significantly decreased CLDN8 were detected in Caco-2 and NCM460 cells after being transfected with pcDNA-SOX9 at mRNA and protein levels, in contrast to the those transfected with pcDNA-NC (Figure 2c and e). On the contrary, the mRNA and protein expressions of CLDN8 were elevated when SOX9 was knocked down in intestinal epithelial cells transfected with si-SOX9 (Figure 2d and f). Because CLDN8 was detected in intestinal epithelial cells that express SOX9, we determined whether SOX9 directly regulates the transcription of CLDN8. To elucidate the potential mechanism underlying SOX9-mediated CLDN8 downregulation, we performed dual-luciferase reporter assays and ChIP-qPCR experiments to investigate the regulatory role of SOX9 in CLDN8 promoter activity as a transcription factor. A significantly diminished luciferase activity was obtained using a plasmid containing a 2.0-kb section of the CLDN8 promoter in 293T cells co-transfected with SOX9 overexpression plasmids (all P < 0.0001, Student's t-test, Figure 3a and b). Furthermore, we used a SOX9 antibody and primers designed for the putative SOX-binding and SOX-coding regions for ChIP-qPCR (Figure 3c). As shown in Figure 3d and e, we found that SOX9 associated with the chromatin at the CLDN8 promoter region but not at a more distant coding region that does not contain the putative SOX9 binding motif (P = 0.0382 and P = 0.0195, Student's t-test). This suggested that SOX9 might directly influence CLDN8 expression.
miR-145-5p targets SOX9-CLDN8 pathway in colonic epithelial cells
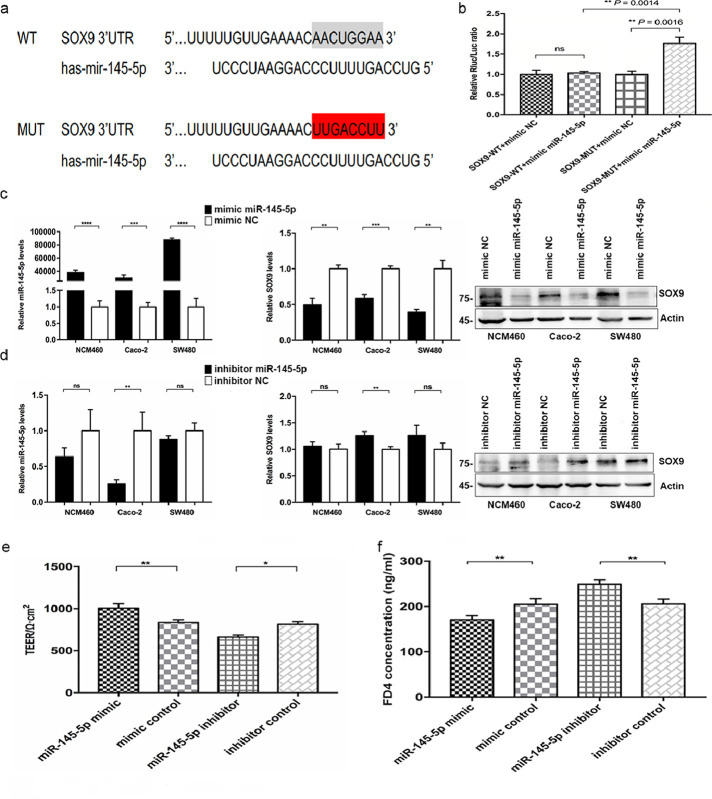
To further delineate the upstream regulatory mechanism targeting the SOX9-CLDN8 pathway in regulating the intestinal epithelial barrier of patients with CD, we focused on the role of miRNAs. By searching microRNA prediction algorithms, miR-145-5p was identified as a potential upstream molecular that targets SOX9 at the 3’UTR region (Figure 4a). In addition, we compared the expression of miR-145-5p in the intestinal mucosa between patients with CD and HCs using RT-qPCR. As shown in Figure 7a, miR-145-5p expression was dramatically decreased in patients with CD compared to that in the normal intestinal mucosa. Similarly, in the intestinal mucosa of TNBS-induced colitis mice, the expression of miR-145-5p was 2.2-fold lower than that in normal controls (P = 0.0109, Student's t-test, Fig. S8).
Figure 4.
Identification of miR-145-5p in targeting SOX9 to regulate the intestinal mucosal barrier function. a, Target binding sequences of miR-145-5p in the wild-type (WT) and mutant (MUT) 3’-UTR of human SOX9. b, Luciferase activity in 293T cells co-transfected with miR-145-5p mimic and luciferase reporters containing SOX9. c, miR-145-5p mimic downregulates SOX9 expression in NCM460, Caco-2 and SW480 cells. d, miR-145-5p inhibitor upregulates SOX9 expression in Caco-2, NCM460, and SW480 cells. e-f, TEER and FD4 Permeability of the Caco-2 cell monolayer. All Data are shown as means ± SEM in (a), (b), (c) and (d). Student's t-test and one-way ANOVA were used for comparison, respectively. ns: no significant difference, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as compared with the control.
Figure 7.
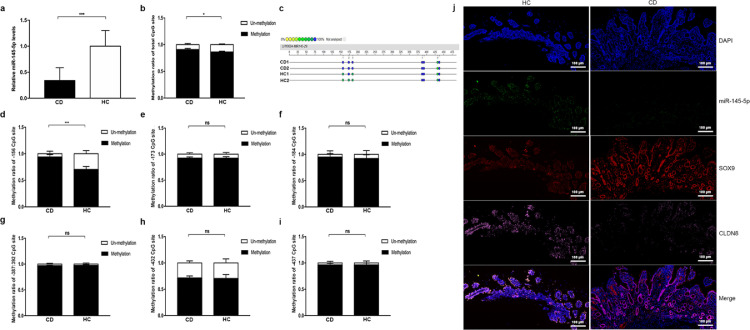
Hypermethylation of miR-145 promoter suppresses miR-145-5p expression. a, miR-145-5p expression in CD and HC (CD, n = 10; HC, n = 10). b-i, Methylation analysis of total methylation ratio and relative methylation ratio of each CpG site. j, FISH of miR-145-5p (green stain), and IF of SOX9 (red stain) and CLDN8 (pink stain) in the colonic mucosa of patients with CD and HCs. Scale bars, 100 μm. All Data are shown as means ± SEM in (a), (b), (d), (e), (f), (g), (h) and (i). Student's t-test were used for com-parison between two groups. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as compared with the control.
Subsequently, we observed the effect of miR-145-5p on regulating the 3’UTR of SOX9 by performing luciferase assays. Then, a plasmid encoding the firefly luciferase transcript with either WT-3’UTR or MUT-3’UTR of SOX9 was constructed. Decreased luciferase activity was found in the WT-3’UTR-transfected 293T cells when co-transfected with the mimic of miR-145-5p (P = 0.0014, Student's t-test), whereas this effect could not be observed in the cells expressing mutant SOX9 3’UTR (Figure 4b). Therefore, the above data indicated that miR-145-5p specifically targeted the SOX9 3’UTR to restrain the corresponding mRNA expression.
Next, we explored the regulatory role of miR-145-5p in SOX9 by transfecting a mimic and inhibitor or corresponding negative controls into Caco-2, SW480, and NCM460 cells. We found that miR-145-5p overexpression inhibited the expression of SOX9 at both mRNA and protein levels in all intestinal epithelial cells as compared to those transfected with negative control (Figure 4c). However, an elevated level of SOX9 expression was observed only in Caco-2 cells when these cell lines were transfected with the inhibitor of miR-145-5p (Figure 4d). In addition, SW480 and NCM460 cells showed an enhanced expression of SOX9 at the protein level rather than mRNA level. These results proved the view that the expression of SOX9 in intestinal epithelial cells was negatively regulated by upstream miR-145-5p.
Subsequently, both TEER and transepithelial FITC-FD4 permeability assays were applied to investigate the influence of miR-145-5p on TJs among Caco-2 cells via transfection with mimic and inhibitor (Figure 4e-f). Significantly increased the TEER value and decreased the FD4 permeability were detected in the Caco-2 cell monolayer model after transfection with the mimic; on the contrary, a decrease in TEER and an increase in FD4 permeability occurred in the monolayer cells when being transfected with the inhibitor. Moreover, we repeated overexpression experiments to observe whether miR-145-5p also affects the expression of other members of the TJ protein family in Caco-2 cells, and found that the expressions of CLDN8 (P = 0.002, Student's t-test) and CLDN15 (P = 0.02, Student's t-test) were significantly increased in Caco-2 cells after transfection with mimic of miR-145-5p (Fig. S9). Overall, we found that miR-145-5p was required for maintaining a normal intestinal epithelial barrier function and that it might play an essential role in the SOX9-CLDN8 pathway.
miR-145-5p agomir attenuates TNBS-induced colitis in WT mice
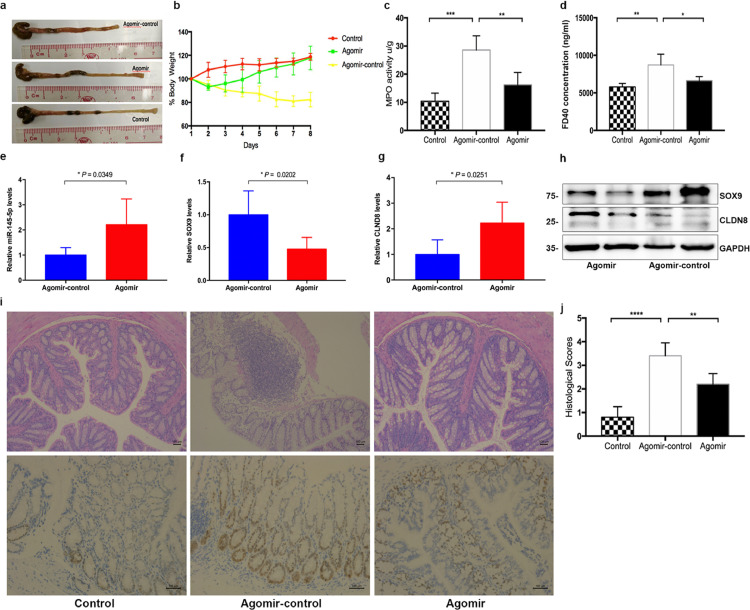
To elucidate the protective role of miR-145-5p in CD and investigate the therapeutic efficacy of its inhibitor, TNBS-induced colitis mice were intraperitoneally injected with miR-145-5p agomir. As shown in Figure 5, treatment with miRNA agomir reduced inflammation in the colons of mice and significantly recovered the weight loss, whereas the control group failed to show the therapeutic effect (Figure 5a and b). In addition, alleviated intestinal inflammation and colonic myeloperoxidase (MPO) activity was observed in the treatment group after being treated with miR-145-5p agomir (Figure 5c), and descending histological scores were obtained (Figure 5i-j). Moreover, a significantly greater decrease in FD40 permeability was detected in the TNBS-induced mice challenged with miR-145-5p agomir (Figure 5d). Importantly, miRNA agomir supplementation decreased Sox9 expression and restored Cldn8 expression via enriching the level of miR-145-5p in TNBS-induced colitis mice, detected using RT-qPCR, immunoblotting, and FISH (Figure 5e-h and Fig. S10). TNBS-induced colitis mice showed weight loss recovery, histological appearance improvement, MPO activity reduction, and reconstruction of intestinal mucosal barrier homeostasis upon miR-145-5p supplementation to regulate the Sox9-Cldn8 pathway.
Figure 5.
Therapeutic treatment of miR-145-5p agomirs in TNBS-induced colitis mice. a. Colon lengths. b, Recovery of body weight. c, MPO activity of the colon. d, Serum FITC-dextran FD40. e-h, miR-145-5p agomirs upregulates CLDN8 by inhibiting the expression of SOX9. i, H&E and IHC staining for SOX9 in the colon tissues of colitic mice. j, Histological scores were assessed by H&E staining of colon tissues (Agomir group, n = 5; Agomir-control group, n = 5; Controls group, n = 5). Scale bars, 100 μm. All Data are shown as means ± SEM in (c), (d), (e), (f), (g) and (j). Student's t-test and one-way ANOVA were used for comparison, respectively. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as compared with the control.
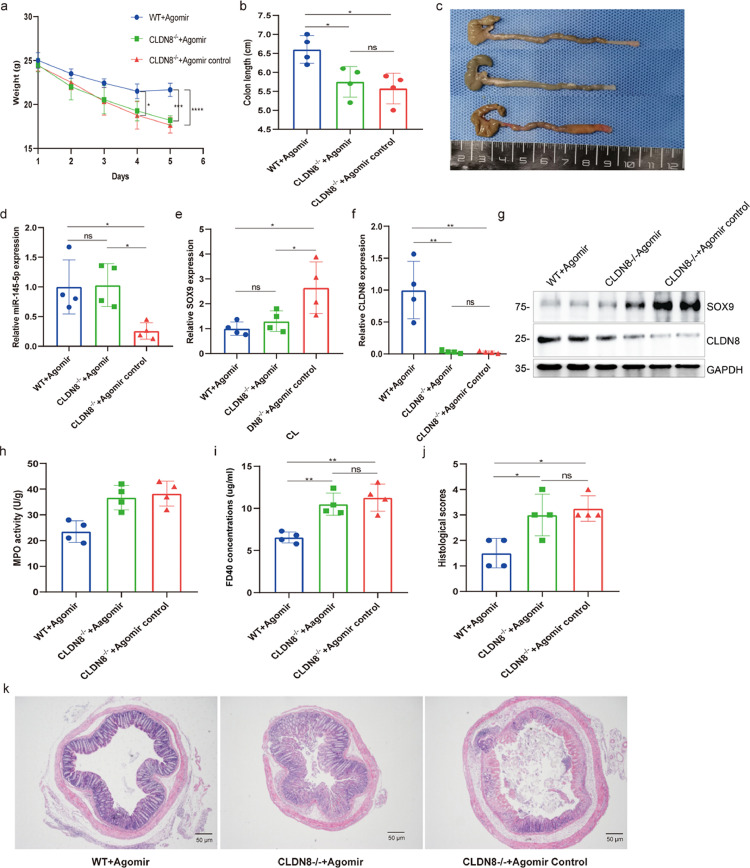
To demonstrate the role of the miR-145/SOX9/CLDN8 pathway in colitis initiation, we further conducted a series of experiments in Cldn8-knockout mice to confirm its role in colitis initiation. Our results showed that miR-145-5p agomir treatment could effectively alleviate TNBS-induced colitis in WT mice by inhibiting Sox9 expression and restoring Cldn8 expression, whereas similar improvements were not found in a TNBS-induced colitis model of Cldn8−/− mice, although miR-145-5p agomir treatment down-regulated Sox9 expression. As shown in Figure 6, Cldn8−/- mice could not reduce intestinal inflammation and improve various cardinal signs of colitis after being treated with miR-145-5p agomir. On the contrary, weight loss, MPO activity, and histological scores in Cldn8−/- mice were not significantly recovered when administered with miR-145-5p agomir, accompanied by an increase of FD40 permeability. Notably, exogenous miR-145-5p supplement inhibited Sox9 expression in the colon tissues of the animal model. Taken together, the results from animal experiments in this study suggested that miR-145-5p agomir treatment improved the inflammation of TNBS-induced colitis in vivo by downregulating the Sox9 target gene under the premise that the expression of Cldn8 was not affected.
Figure 6.
Therapeutic treatment of miR-145-5p agomirs in Cldn8-knockout mice. a. Body weight in Cldn8−/− mice. b-c, Colon lengths in Cldn8−/− mice. d-g, miR-145-5p, SOX9 and CLDN8 expressions at mRNA and protein levels. h, MPO activity of the colon. i, Serum FITC-dextran FD40. j-k, Histological scores were assessed by H&E staining of colon tissues (WT+Agomir group, n = 4; Cldn8−/−+Agomir group, n = 4; Cldn8−/−+Agomir-control group, n = 4). Scale bars, 50 μm. All Data are shown as means ± SEM in (a), (b), (d), (e), (f), (h), (i) and (j). Student's t-test and one-way ANOVA were used for comparison, respectively. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as compared with the control.
Crosstalk between miR-145-5p, SOX9 and CLDN8 in CD
Given that miR-145-5p plays a major role in regulating the SOX9 expression, we then assessed its expression in individuals with CD. To precisely depict the locations of miR-145-5p, SOX9 and CLDN8 in the intestinal mucosa, we determined the spatial pattern via FISH. We found that miR-145-5p and CLDN8 were downregulated in patients with CD, whereas SOX9 was abundantly expressed (Figure 7j). All FISH data suggested that miR-145-5p held an important role in maintaining intestinal mucosal barrier homeostasis by regulating SOX9-CLDN8 pathway.
Promoter hypermethylation of miR-145 results in its downregulation in CD
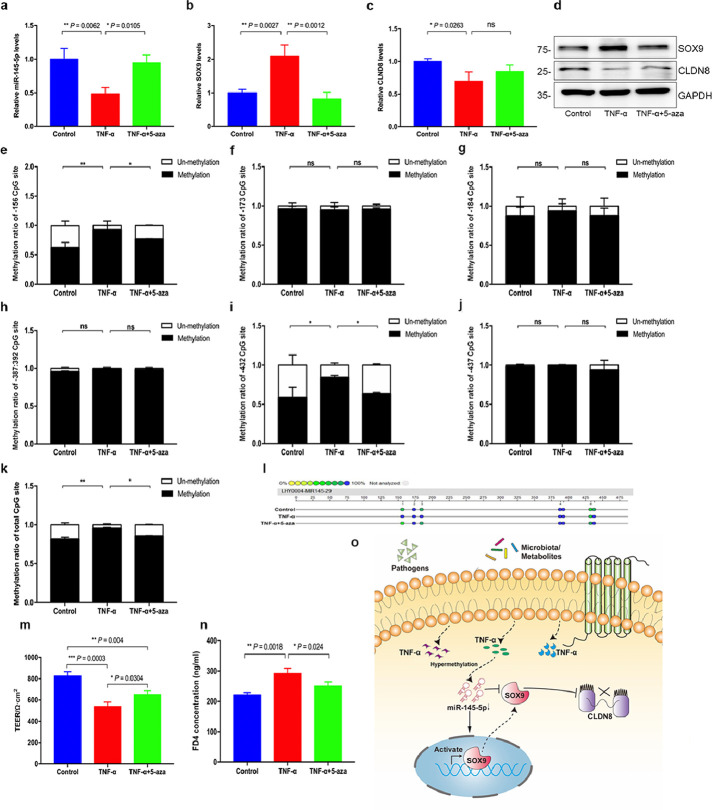
Finally, we sought to explore the potential mechanism associated with downregulated miR-145-5p in patients with CD. By searching the National Centre for Biotechnology Information and MethPrimer databases, no CpG island was identified in the promoter of miR-145, but six CpG sites with potential methylation properties were detected (Fig. S11). To determine the presence of methylation, we quantified the methylation ratio of the promoter in mucosal biopsy samples via pyrosequencing. We found that the total DNA methylation ratio was significantly elevated in patients with CD compared to that in the normal mucosae from healthy individuals, using MassARRAY methylation analysis (Figure 7b-c). Moreover, the -156 CpG site was more methylated than other CpG sites (Figure 7d-i). Subsequently, promoter methylation levels of the miR-145-encoding gene under inflammatory condition were analysed in vitro. We used different concentrations of TNF-α and 5-Aza-deoxycytidine (5-aza) to stimulate NCM460 cells for 48 h and found that 50 ng/mL of TNF-α and 5 μmol/L of 5-aza showed a significantly effect on miR-145-5p expression; accordingly, these concentrations were used for subsequent experiments (Fig. S12).
Furthermore, we investigated whether decreased miR-145-5p expression induced by 50 ng/mL of TNF-α was mediated via aberrant methylation, and 5 μmol/L of 5-aza was used to reverse hypermethylation. Interestingly, the expression of miR-145-5p (P = 0.002, one-way ANOVA test) was rescued in NCM460 cells challenged with TNF-α when 5-aza was added, and expression of SOX9 and CLDN8 was also reversed at mRNA and protein levels (Figure 8a-d). We also performed MassARRAY methylation analysis to define the methylation ratio of miR-145 promoter in NCM460 cells stimulated by TNF-α with or without 5-aza treatment. Consistent with human studies, TNF-α stimulation resulted in an aberrant hypermethylation at the promoter region of miR-145, and NCM460 cells treated with TNF-α combined with 5-aza prevented hypermethylation onset. However, unlike patients with CD, the -156 and -432 CpG sites were more likely to be methylated than other sites (Figure 8e-l). Together, these findings suggested that miR-145 promoter inactivation occurred after hypermethylation, which could underpin the decreased expression of miR-145-5p in CD.
Figure 8.
Hypermethylation of miR-145 promoter-mediated SOX9-CLDN8 pathway. a-d, The expression of miR-145-5p, SOX9 and CLDN8 in NCM460 cells treated with TNF-a (50 ng/ml) or 5-aza (5 μmol/L). e-l, Methylation analysis of total methylation ratio and relative methylation ratio of each CpG site of miR-145 promoter in NCM460 cells treated with TNF-a (50 ng/ml) or 5-aza (5 μmol/L). m-n, TEER and FD4 permeability of the Caco-2 cell monolayer. o, The proposed model of the miR-145-5p/SOX9/CLDN8 pathway. Aberrant hypermethylation of miR-145-5p promoter mediated activation of SOX9 expression to negatively regulate CLDN8, which aggravates the intestinal epithelial TJ barrier dysfunction. All Data are shown as means ± SEM in (a), (b), (c) and (d). Student's t-test and one-way ANOVA were used for comparison, respectively. ns: no significant difference, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 as compared with the control.
TEER and transepithelial FD4 permeability assays were conducted in Caco-2 cell monolayers to further investigate the effect of promoter hypermethylation of miR-145 on TJs between Caco-2 cells (Figure 8m-n). A decrease in TEER and an increase in FD4 permeability were observed in Caco-2 cells after being treated with TNF-α. However, co-treatment with 5-aza significantly attenuated this effect to the intestinal barrier, showing increased TEER and decreased FD4 permeability. Furthermore, we repeated TNF-α treatment experiments in intestinal epithelial cells to observe its effect on other TJ proteins. Expression levels of several TJ proteins in NCM460 cells were indeed altered upon treatment with TNF-α, including CLDN1, CLDN2, CLDN4, CLDN5, CLDN7, CLDN8, CLDN15, CLDN18 (Fig. S13), whereas altered expressions of CLDN1, CLDN2, CLDN3, CLDN5, CLDN8, CLDN12, CLDN15, and CLDN18 were found in Caco-2 cells (Fig. S14). These results further confirmed the hypothesis that hypermethylation occurred in the miR-145 promoter mediated the SOX9-CLDN8 pathway to regulate the intestinal mucosal barrier in CD (Figure 7o).
Discussion
In this study, we identified miR-145-5p and SOX9 as two important upstream regulatory drivers of CLDN8 in CD and the TNBS-induced colitis model. We finally draw a conclusion that the miR-145-5p/SOX9/CLDN8 pathway might play a significant role in the intestinal mucosal barrier homeostasis of CD via a series of experiments conducted in vivo and in vitro.
In our previous research, we found that a downregulated CLDN8 expression was regulated by IL-23/miR-223 or IL-9/miR-21 pathways, which ultimately results in inflammation-induced intestinal mucosal damage.19,20 These findings suggested that the link between intestinal inflammation and epithelial barrier function damage is not only through a single single-pathway, but a network with multiple pathways. In this context, more potential upstream regulators associated with CLDN8 expression are urgently needed, to determine their role in intestinal barrier function regulation.
SOX9, as a key member of the HMG transcription factor family, exhibits a key role in IEC differentiation.38,39 Previous expression profiling in the GEO database showed that SOX9 expression in patients with CD is upregulated when compared with HC, and SOX9 expression in the affected colon is higher than that in the unaffected colon. In addition, SOX9 is indispensable in repairing the damaged mucosa caused by high-dose irradiation, indicating it might promote intestinal mucosal repair upon other types of damage.40 However, SOX9 is considered to play a dual role in the pathogenesis of diverse diseases in previous studies.34 For example, adverse effects of SOX9 were reported in several studies on switching adult stem cell modulation to cancer stem cell activation.41, 42, 43 In this study, we observed upregulated expression of SOX9 in patients with CD and its mRNA expression was positively associated with the clinical and endoscopic disease activity and multiple inflammatory markers, which was not entirely consistent with a previous study reporting that SOX9 expression at the protein level rather than mRNA level increases in the CD mucosa.34 In addition, an increased expression of Sox9 was observed in TNBS-induced colitis mice, suggesting that dysregulated Sox9 played an important role in the intestinal inflammation of the experimental colitis model.
Previous work from our laboratory has demonstrated that CLDN8, an essential member of the claudin family, is significantly downregulated in patients with CD and TNBS-induced colitis mice. The crucial finding of our study was that SOX9 negatively regulated the expression of CLDN8, resulting in dysfunction of intestinal mucosal barrier. Bioinformatics analysis demonstrated that SOX9 also functions as an important transcriptional regulator of claudin expression.44 Previous studies showed that SOX9 is an important mediator of the Tcf4/CLDN7 pathway in colorectal cancer cells.31 However, we did not observe an altered expression of CLDN7 in intestinal epithelial cells upon transfection with the plasmid overexpressing SOX9. In addition, a significant increase in CLDN4 was found in Caco-2 cells after transfection with pcDNA-SOX9. Altogether, although SOX9 regulated the expression of other claudin members, downregulated CLDN8 was more prevalently observed. Furthermore, the interaction between SOX18 and CLDN15 has been reported by Fontijn et al.32 The Sox transcription factor family regulates claudins via a complex interplay of different regulatory effects.
The regulatory mechanisms between SOX9 and CLDN8 in CD remain undefined. Knocking down SOX9 using siRNA increased TEER and reduced FD4 permeability in Caco-2 cell monolayers combined with an elevated CLDN8 expression in our study. In contrast, an ectopic expression of SOX9 weakened the TJs of colonic epithelial cells owing to diminished CLDN8 expression, presenting significantly decreased TEER and increased FD4 permeability. TEER and FD4 measurements assess the barrier function in different aspects and they complement each other. TEER is used to measure the cell monolayer integrity and give information about permeability for ions and other charged solutes; it reflects the tightness and leakiness of the epithelium. FD4 is used to assess the paracellular permeability of the epithelium, but it also provides information about the integrity by acting as a paracellular flux marker.45 Furthermore, a negative correlation between SOX9 and CLDN8 in the inflamed mucosa was confirmed. Recent ChIP-seq analyses have shown that SOX9 may possess many tissue-specific transcriptional targets; however, few studies have identified SOX9 targets in the intestinal epithelium. Using luciferase reporter gene and ChIP-qPCR assays, we proved that the promoter of CLDN8 harboured a potential binding site for SOX9, and SOX9 had a negative regulatory effect on CLDN8 by directly binding to its promoter region. Our results showed that SOX9 was an important mediator to downregulate the expression of CLDN8, which is consistent with the findings reported by Qi et al.46
To further elucidate the underlying mechanism of the specific SOX9-CLDN8 pathway, epigenetic modifications have provided innovative insights. MicroRNAs were emphasised in our study because they provided new strategies for exploring the pathogenesis of CD. Additionally, an altered expression of SOX9 in a variety of human inflammatory diseases regulated by a complex miRNA network has been reported in recent studies.47,48 Among these miRNAs, the potential candidate miR-145-5p is significantly decreased in UC as a putative regulator of inflammation and protooncogenes.49, 50, 51 In this study, we characterised miR-145-5p for the first time as an important player that targeted the SOX9-CLDN8 pathway to maintain the mucosal epithelial barrier integrity. Using loss-of-function and gain-of-function analysis, miR-145-5p was identified to downregulate the SOX9 expression, leading to CLDN8 recovery. Subsequently, we verified that SOX9 as the downstream target gene of miR-145-5p via a dual-luciferase reporter assay. Importantly, the expression trend and pattern of miR-145-5p, SOX9, and CLDN8 in the intestinal mucosa were shown using FISH. In the TNBS-induced colitis mouse model, miR-145-5p mimic treatment reduced weight loss, improved intestinal inflammation, and restored intestinal epithelial barrier integrity, whereas similar improvements were not observed in the Cldn8−/− mice model, although miR-145-5p agomir treatment downregulated Sox9 expression.
Finally, we explored the possible mechanism involved in miR-145-5p downregulation of CD. DNA hypermethylation in CpG-rich promoters has been recognised as a gene silencing mechanism, and promoter hypermethylation of the miR-145-encoding gene is reportedly responsible for the downregulated expression.52, 53, 54, 55 Our study still supported that aberrant promoter hypermethylation of miR-145-5p promoter occurred in patients with CD compared with normal tissues. Additionally, it is well known that an inflammatory environment could lead to a lower expression of miR-145-5p as a momentous initiator and driver, and its expression decreases significantly under TNF-α-induced inflammatory conditions in vitro.56 We also found a differentially downregulated miR-145-5p in NCM460 cells challenged with TNF-α. In addition, 5-aza treatment elevated miR-145-5p expression by demethylating the promoter of miR-145, which ultimately resulted in lower repression of SOX9 and reduced the damage to the intestinal epithelial barrier by reversing the expression of CLDN8. These findings indicate that the promoter region of the miR-145 gene in inflammatory conditions could be methylated, and the 156 CpG site remains the most common candidate. Therapeutic measures targeting the 156 CpG site of the miR-145 promoter region might prevent hypermethylation, providing therapeutic evidence for blocking the dysregulated miR-145/SOX9/CLDN8 pathway.
In this study, we identified aberrantly downregulated miR-145-5p in intestinal mucosal tissues and its expression was regulated by the hypermethylation of its promoter. The decreased expression of miR-145-5p resulted in SOX9 upregulation, which induced a negative regulatory effect on the upstream promoter region of CLDN8 via the direct binding site. Our findings provide new insights linking the miR-145-5p/SOX9/CLDN8 pathway to intestinal mucosal barrier homeostasis and will lead to the development of new therapies for patients with CD. We highlight potential strategies to inhibit the aberrant promoter methylation of miR-145 and maintain miR-145-5p-SOX9-CLDN8 pathway homeostasis as a potential therapeutic avenue for regulating the intestinal mucosal barrier.
Contributors
Guarantor of the article: Shenghong Zhang.
SHZ designed and oversaw the study. XJZ performed most of the experiments and wrote the manuscript. SSH, JH, GSZ and SX helped perform experiments and data analysis. BLC contributed methodology and experimental resources. MHC, ZRZ, and SHZ have verified the underlying data. All authors have read and approved the final version of the manuscript.
Declaration of interests
The authors have no conflicts of interest to disclose.
Acknowledgments
Acknowledgements
We want to thank the Digestion Laboratory of the First Affiliated Hospital of Sun Yat-sen University (Guangzhou, Guangdong, China) for providing guidance and technical assistance in the study. We would like to acknowledge the funding of the National Natural Science Foundation of China (#81870374, #81670498, #81630018, #82070538, #8210031148), Guangdong Science and Technology (#2017A030306021, #2020A1515111087), Guangzhou Science and Technology Department (#202002030041), and the Fundamental Research Funds for the Central Universities (#19ykzd11).
Data sharing statement
Data, materials, and software information are included within the article and its supplementary materials.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103846.
Contributor Information
Zhirong Zeng, Email: zengzhirong@mail.sysu.edu.cn.
Shenghong Zhang, Email: shenghongzhang@163.com, zhshh3@mail.sysu.edu.cn.
Appendix. Supplementary materials
References
- 1.Torres J, Mehandru S, Colombel JF, et al. Crohn's disease. Lancet. 2017;389(10080):1741–1755. doi: 10.1016/S0140-6736(16)31711-1. [DOI] [PubMed] [Google Scholar]
- 2.Ungaro R, Mehandru S, Allen PB, et al. Ulcerative colitis. Lancet. 2017;389(10080):1756–1770. doi: 10.1016/S0140-6736(16)32126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature. 2020;578(7796):527–539. doi: 10.1038/s41586-020-2025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12(4):205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 5.Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martini E, Krug SM, Siegmund B, et al. Mend your fences: the epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4(1):33–46. doi: 10.1016/j.jcmgh.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang J, Leong RW, Wasinger VC, et al. Impaired intestinal permeability contributes to ongoing bowel symptoms in patients with inflammatory bowel disease and mucosal healing. Gastroenterology. 2017;153(3):723–731. doi: 10.1053/j.gastro.2017.05.056. [DOI] [PubMed] [Google Scholar]
- 8.Kotla NG, Rana S, Sivaraman G, et al. Bioresponsive drug delivery systems in intestinal inflammation: State-of-the-art and future perspectives. Adv Drug Deliv Rev. 2019;146:248–266. doi: 10.1016/j.addr.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 9.Michielan A, D'Inca R. Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediators Inflamm. 2015;2015 doi: 10.1155/2015/628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capaldo CT, Powell DN, Kalman D. Layered defense: how mucus and tight junctions seal the intestinal barrier. J Mol Med (Berl) 2017;95(9):927–934. doi: 10.1007/s00109-017-1557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landy J, Ronde E, English N, et al. Tight junctions in inflammatory bowel diseases and inflammatory bowel disease associated colorectal cancer. World J Gastroenterol. 2016;22(11):3117–3126. doi: 10.3748/wjg.v22.i11.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Hernandez V, Quiros M, Nusrat A. Intestinal epithelial claudins: expression and regulation in homeostasis and inflammation. Ann N Y Acad Sci. 2017;1397(1):66–79. doi: 10.1111/nyas.13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slifer ZM, Blikslager AT. The integral role of tight junction proteins in the repair of injured intestinal epithelium. Int J Mol Sci. 2020;21(3):972. doi: 10.3390/ijms21030972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Itallie CM, Anderson JM. Claudin interactions in and out of the tight junction. Tissue Barriers. 2013;1(3):e25247. doi: 10.4161/tisb.25247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka H, Tamura A, Suzuki K, et al. Site-specific distribution of claudin-based paracellular channels with roles in biological fluid flow and metabolism. Ann N Y Acad Sci. 2017;1405(1):44–52. doi: 10.1111/nyas.13438. [DOI] [PubMed] [Google Scholar]
- 16.Barmeyer C, Fromm M, Schulzke JD. Active and passive involvement of claudins in the pathophysiology of intestinal inflammatory diseases. Pflugers Arch. 2017;469(1):15–26. doi: 10.1007/s00424-016-1914-6. [DOI] [PubMed] [Google Scholar]
- 17.Zhu LG, Han J, Li L, et al. Claudin family participates in the pathogenesis of inflammatory bowel diseases and colitis-associated colorectal cancer. Front Immunol. 2019;10:1441. doi: 10.3389/fimmu.2019.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim TI. The role of barrier dysfunction and change of claudin expression in inflammatory bowel disease. Gut Liver. 2015;9(6):699–700. doi: 10.5009/gnl15430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HL, Chao K, Ng SC, et al. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. 2016;17:58. doi: 10.1186/s13059-016-0901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Huang SS, Wang HL, et al. Cytokine IL9 triggers the pathogenesis of inflammatory bowel disease through the miR21-CLDN8 pathway. Inflamm Bowel Dis. 2018;24(10):2211–2223. doi: 10.1093/ibd/izy187. [DOI] [PubMed] [Google Scholar]
- 21.Sinclair AH, Berta P, Palmer MS, et al. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. 1990; 346(6281):240-244. [DOI] [PubMed]
- 22.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227(2):239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 23.Pritchett J, Athwal V, Roberts N, et al. Understanding the role of SOX9 in acquired diseases: lessons from development. Trends Mol Med. 2011;17(3):166–174. doi: 10.1016/j.molmed.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Lefebvre V, Angelozzi M, Haseeb A. SOX9 in cartilage development and disease. Curr Opin Cell Biol. 2019;61:39–47. doi: 10.1016/j.ceb.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richtig G, Aigelsreiter A, Schwarzenbacher D, et al. SOX9 is a proliferation and stem cell factor in hepatocellular carcinoma and possess widespread prognostic significance in different cancer types. 2017; 12(11):e0187814. [DOI] [PMC free article] [PubMed]
- 26.Blache P, van de Wetering M, Duluc I, et al. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J Cell Biol. 2004;166(1):37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bastide P, Darido C, Pannequin J, et al. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol. 2007;178(4):635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bankaitis ED, Ha A, Kuo CJ, et al. Reserve stem cells in intestinal homeostasis and injury. Gastroenterology. 2018;155(5):1348–1361. doi: 10.1053/j.gastro.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prevostel C, Blache P. The dose-dependent effect of SOX9 and its incidence in colorectal cancer. Eur J Cancer. 2017;86:150–157. doi: 10.1016/j.ejca.2017.08.037. [DOI] [PubMed] [Google Scholar]
- 30.Haseeb A, Lefebvre V. The SOXE transcription factors-SOX8, SOX9 and SOX10-share a bi-partite transactivation mechanism. Nucleic Acids Res. 2019;47(13):6917–6931. doi: 10.1093/nar/gkz523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Darido C, Buchert M, Pannequin J, et al. Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res. 2008;68(11):4258–4268. doi: 10.1158/0008-5472.CAN-07-5805. [DOI] [PubMed] [Google Scholar]
- 32.Fontijn RD, Volger OL, Fledderus JO. SOX-18 controls endothelial-specific claudin-5 gene expression and barrier function. Am J Physiol Heart Circ Physiol. 2008;294(2):891–900. doi: 10.1152/ajpheart.01248.2007. [DOI] [PubMed] [Google Scholar]
- 33.Singh AP, Cummings CA, Mishina Y, et al. SOX8 regulates permeability of the blood-testes barrier that affects adult male fertility in the mouse. Biol Reprod. 2013;88(5):133. doi: 10.1095/biolreprod.112.107284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roda G, Dahan S, Mezzanotte L, et al. The defect in CEACAM family member expression in Crohn's disease IECs is regulated by the transcription factor SOX9. Inflamm Bowel Dis. 2009;15(12):1775–1783. doi: 10.1002/ibd.21023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanoli A, Di Sabatino A, Martino M, et al. Small bowel carcinomas in celiac or Crohn's disease: distinctive histophenotypic, molecular and histogenetic patterns. Mod Pathol. 2017;30(10):1453–1466. doi: 10.1038/modpathol.2017.40. [DOI] [PubMed] [Google Scholar]
- 36.Iftikhar M, Iftikhar A, Zhang HJ, et al. Transport, metabolism and remedial potential of functional food extracts (FFEs) in Caco-2 cells monolayer: A review. Food Res Int. 2020;136 doi: 10.1016/j.foodres.2020.109240. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Chen SW, Zhu J, et al. Intestinal alkaline phosphatase inhibits the translocation of bacteria of gut-origin in mice with peritonitis: mechanism of action. PLoS One. 2015;10(5):e0124835. doi: 10.1371/journal.pone.0124835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang CZ, Xu JH, Zhong W, et al. Sox9 transcriptionally regulates Wnt signaling in intestinal epithelial stem cells in hypomethylated crypts in the diabetic state. Stem Cell Res Ther. 2017;8(1):60. doi: 10.1186/s13287-017-0507-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Formeister EJ, Sionas AL, Lorance DK, et al. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296(5):1108–1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roche KC, Gracz AD, Liu XF, et al. SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroenterology. 2015;149(6):1553–1563. doi: 10.1053/j.gastro.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He CL, Zhao C, Liu S, et al. Down-regulation of MiR-138-5p protects chondrocytes ATDC5 and CHON-001 from IL-1 β-induced inflammation Via Up-regulating SOX9. Curr Pharm Des. 2020;25(43):4613–4621. doi: 10.2174/1381612825666190905163046. [DOI] [PubMed] [Google Scholar]
- 42.Zhou H, Qin Y, Ji S, et al. SOX9 activity is induced by oncogenic Kras to affect MDC1 and MCMs expression in pancreatic cancer. Oncogene. 2018;37(7):912–923. doi: 10.1038/onc.2017.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santos JC, Carrasco-Garcia E, Garcia-Puga M, et al. SOX9 elevation acts with canonical WNT signaling to drive gastric cancer progression. Cancer Res. 2016;76(22):6735–6746. doi: 10.1158/0008-5472.CAN-16-1120. [DOI] [PubMed] [Google Scholar]
- 44.Mathelier A, Zhao XB, Zhang AW, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:142–147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spalinger MR, Sayoc-Becerra A, Santos AN, et al. PTPN2 regulates interactions between macrophages and intestinal epithelial cells to promote intestinal barrier function. Gastroenterology. 2020;159(5):1763–1777. doi: 10.1053/j.gastro.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi J, Yang YQ, Hao P, Xu JQ. Transcription factor SOX9 promotes osteosarcoma cell growth by repressing Claudin-8 expression. Tohoku J Exp Med. 2017;241(1):55–63. doi: 10.1620/tjem.241.55. [DOI] [PubMed] [Google Scholar]
- 47.He CL, Zhao C, Liu S, et al. Down-regulation of MiR-138-5p Protects Chondrocytes ATDC5 and CHON-001 from IL-1 β-induced Inflammation Via Up-regulating SOX9. Curr Pharm Des. 2020;25(43):4613–4621. doi: 10.2174/1381612825666190905163046. [DOI] [PubMed] [Google Scholar]
- 48.Zhang WK, Cheng P, Hu WH, et al. Inhibition of microRNA-384-5p alleviates osteoarthritis through its effects on inhibiting apoptosis of cartilage cells via the NF-κB signaling pathway by targeting SOX9. Cancer Gene Ther. 2020;27(10-11):836–837. doi: 10.1038/s41417-020-0202-y. [DOI] [PubMed] [Google Scholar]
- 49.Pekow JR, Dougherty U, Mustafi R, et al. miR-143 and miR-145 are downregulated in ulcerative colitis: putative regulators of inflammation and protooncogenes. Inflamm Bowel Dis. 2012;18(1):94–100. doi: 10.1002/ibd.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan YG, Zhang YF, Guo CJ, et al. Screening of differentially expressed microRNA in ulcerative colitis related colorectal cancer. Asian Pac J Trop Med. 2013;6(12):972–976. doi: 10.1016/S1995-7645(13)60174-1. [DOI] [PubMed] [Google Scholar]
- 51.Verbus EA, Kenyon JD, Sergeeva O, et al. Expression of miR-145-5p during chondrogenesis of mesenchymal stem cells. J Stem Cell Res. 2017;1(3):1–10. [PMC free article] [PubMed] [Google Scholar]
- 52.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 53.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4(12):988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 54.Gao W, Zhang CM, Li WQ, et al. Promoter methylation-regulated miR-145-5p inhibits laryngeal squamous cell carcinoma progression by targeting FSCN1. Mol Ther. 2019;27(2):365–379. doi: 10.1016/j.ymthe.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong W, Li B, Xu Y, et al. Hypermethylation of the Micro-RNA 145 promoter is the key regulator for NLRP3 inflammasome-induced activation and plaque formation. JACC Basic Transl Sci. 2018;3(5):604–624. doi: 10.1016/j.jacbts.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu JB, He Y, Luo YN, et al. MiR-145-5p inhibits proliferation and inflammatory responses of RMC through regulating AKT/GSK pathway by targeting CXCL16. J Cell Physiol. 2018;233(4):3648–3659. doi: 10.1002/jcp.26228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.