Abstract
Recent taxonomic developments, based on 16s and 23s rRNA gene sequences, have divided the family Chlamydiaceae into two genera and nine species, of which five have been found to infect humans. Few simple methods are available to detect and identify all species sensitively and specifically. In this study the suitability of the omp2 gene as a target for molecular identification of Chlamydiaceae is demonstrated. Phylogenetic analysis of partial omp2 gene sequences from all nine species agrees with the recently published taxonomic changes based on the ribosomal genes. The use of a family-specific PCR primer pair, which is able to amplify the 5′ end of the omp2 gene from all Chlamydiaceae except some Chlamydophila pecorum strains, is described. Identification of all nine species was achieved using restriction fragment length polymorphism analysis with a single enzyme, AluI, confirmed by DNA sequencing. A PCR enzyme-linked oligonucleotide assay was developed which can detect a single chlamydial genome and may be applied to DNA extracts from any specimen or culture for the detection of single or mixed human chlamydial infection.
The family Chlamydiaceae has recently been reclassified into two genera and nine species (4). Current human infection by Chlamydiaceae is frequently diagnosed as Chlamydia trachomatis, but less frequently as Chlamydophila pneumoniae (formerly Chlamydia pneumoniae), and rarely as Chlamydophila psittaci (formerly Chlamydia psittaci avian group), Chlamydophila abortus (formerly Chlamydia psittaci abortion group), or Chlamydophila felis (formerly Chlamydia psittaci feline pneumonitis agent). No observation has been made of human infection with Chlamydophila caviae (formerly Chlamydia psittaci guinea pig inclusion conjunctivitis agent), Chlamydophila pecorum (formerly Chlamydia pecorum), Chlamydia suis (formerly porcine Chlamydia trachomatis), or Chlamydophila muridarum (formerly Chlamydia trachomatis of mice).
Other than for C. trachomatis, laboratory diagnostic methods are poorly developed and not standardized. Serology is commonly used for diagnosis (6, 15), but the immune response to chlamydial infection is often delayed and variable. Microimmunofluorescence, the reference serology method, which was designed for the serotyping of C. trachomatis, is difficult to perform and not fully standardized and does not always correlate with cell culture or nucleic acid detection (8, 12, 15, 16, 20). Commercial reagents available for the detection of Chlamydiaceae by direct immunofluorescence (DIF) include the family-specific antibody against the lipopolysaccharide (e.g., Imagen Chlamydia; Dako Diagnostica GmbH, Hamburg, Germany) and a C. trachomatis-specific antibody directed against the major outer membrane protein (MOMP) (e.g., Syva Microtrak; Bearing Diagnostics Inc., Cupertino, Calif.). Use of these antibodies in DIF for the detection of elementary bodies (EBs) from clinical material requires skilled staff and is subjective. Enzyme immunoassays for detection of chlamydial antigens have been shown to be generally of modest sensitivity (13).
Commercially available sensitive and specific nucleic acid amplification tests are primarily designed for C. trachomatis (13). Specific in-house PCR assays have been developed for other species (1, 10), but multiple or multiplex PCRs are needed to screen specimens collected from sites that are associated with infection by more than one species, such as conjunctivae and the respiratory tract. In addition, most PCR-based assays for the former C. psittaci group do not distinguish the recently described species (10).
C. trachomatis may be isolated when cell culture is available, but C. pneumoniae is difficult to culture, with only two reported isolates from patients in the United Kingdom (2). Other Chlamydophila species are more easily isolated, when attempted, but require level 3 (P3) containment facilities for their propagation. When a Chlamydiaceae species is isolated by culture, there are few phenotypic methods available for determining the species (17). Production of a glycogen vacuole and sensitivity to sulfonamides have been used to identify Chlamydia spp. Species-specific antisera have been produced but are not freely available.
Recent methods for typing chlamydiae have been based on DNA sequencing. The new taxonomic description of the Chlamydiaceae uses the 16s and 23s rRNA gene sequences, and a typing system based on PCR and sequencing of the rRNA genes has been proposed, although other genes may be suitable for identification of species (4, 5, 18).
The omp2 gene of the Chlamydiaceae has two conserved regions at the 5′ end bordering a variable segment, and primers designed to be complementary to these conserved regions have been used to amplify the omp2 gene from many Chlamydiaceae (18). The sequence variation has been used to design species identification tests using the omp2 PCR product, e.g., PCR-restriction fragment length polymorphism (PCR-RFLP) analysis and heminested PCR (14, 18) and agarose gel electrophoresis with bisbenzimide-polyethylene glycol (PEG) (3). Although no publications were found distinguishing all the current species, PCR-RFLP patterns after digestion with AluI do produce characteristic patterns for six species (3, 14), but no published patterns have been observed for C. felis, C. muridarum, or C. suis.
The purpose of this work was to determine whether a sensitive and species-specific PCR–enzyme-linked oligonucleotide assay (PCR-ELONA) and/or PCR-RFLP assay could be designed, based on the omp2 gene, which would be able to detect and identify any Chlamydiaceae sp. To confirm the suitability of the omp2 gene as a target, partial omp2 gene sequences were generated for the two species not previously sequenced (C. felis and C. suis), and the taxonomic relationship of all species was investigated by cluster analysis.
MATERIALS AND METHODS
Bacterial cultures and DNA extraction.
C. pneumonia (IOL207, TW183, and V1355), C. trachomatis (SA2F), C. muridarum (MoPn), and C. psittaci (Z10*) were grown in McCoy cell culture, and supernatants were clarified by low-speed centrifugation (400 × g) to produce suspensions of EBs. DNA was extracted by boiling. C. felis (FePn*) and C. pecorum (P787*, 11/88*, VR628*, and R69*) were grown in McCoy cell culture and stored in cell culture growth medium (minimal essential medium [MEM]). DNA was extracted by the QIAamp blood kit protocol (Qiagen Ltd., Crawley, England) from these isolates and from uninfected McCoy cells in MEM. C. suis S45* was obtained as an egg yolk sac preparation of EBs. (Isolates originally provided by Garath Jones, Yaba Ltd., are marked with an asterisk.) C. abortus (AB7) and C. caviae (GPIC) cell culture supernatants were provided by P. Bavoil, London School of Hygiene and Tropical Medicine. DNA was extracted by boiling for 15 min and then diluted in water. C. caviae and C. suis boilates were further purified on a Qiagen spin column (Qiagen Ltd.). Fourteen serotypes of C. trachomatis, A (SA1), B (TW5), C (UW1), D (ICCa18), E (DK20), F (MRC301), G (IOL238), H (UW4), I (UW12), J (UW36), K (UW31), L1 (440L), L2 (434B), and L3 (404L), were obtained as egg-grown formaldehyde-treated EB preparations (originally from the Institute of Ophthalmology, London). Aliquots (5 μl) were diluted in 200 μl of water and boiled for 15 min.
PCR specificity was confirmed by cross-testing against previously characterized DNA extracts from Mycoplasma pneumoniae, Ureaplasma urealyticum, Staphylococcus epidermidis, Streptococcus pneumoniae, Streptococcus agalactiae, Streptococcus pyogenes, Streptococcus mitis, Stenotrophomonas maltophilia, Burkholderia cepacia, Escherichia coli, and Neisseria meningitidis, provided by Kathryn Harris, Great Ormond Street Hospital, London. Additionally, DNA was extracted from boiled suspensions of pure cultures of Pseudomonas aeruginosa, Listeria monocytogenes, Klebsiella pneumoniae, Staphylococcus aureus, Haemophilus influenzae, Moraxella catarrhalis, Shigella sonnei, Shigella flexneri, Vibrio cholerae, Bacteroides fragilis, Salmonella sp., Campylobacter jejeni, Clostridium perfringens, Peptostreptococcus sp., Enterococcus faecalis, Neisseria gonorrhoeae, and Candida albicans.
Uncharacterized chlamydial isolates.
Two chlamydiae isolated in the Chlamydia Laboratory, University College London Hospital, were provided for analysis as cell culture supernatants. Culture confirmation had been positive with the family-specific antilipopolysaceharide antibody but negative with the C. trachomatis anti-MOMP antibody, confirming the presence of Chlamydiaceae other than C. trachomatis. DNA was extracted by boiling.
Production of enumerated suspensions of EBs of C. trachomatis and C. pneumoniae.
A suspension of EBs (which are 0.2 to 0.4 μm in diameter) was prepared by a differential filtration method. C. trachomatis SA2F (L2) and C. pneumoniae TW187 were propagated in McCoy cells. An aliquot of fresh cell culture supernatant was passed through a 0.45-μm filter (Minisart; Sartorius), and the filtrate was retained. The filtrate was then passed through a 0.2-μm filter (Minisart; Sartorius), followed by 10 ml of phosphate-buffered saline (PBS), and the filter was back eluted with 5 ml of PBS to give a purified EB suspension. The stock suspension was diluted in PBS, counted, and stored in aliquots at −70°C. The concentration of EBs was determined by counting of organisms stained with fluorescein isothiocyanate-labeled anti-LPS antibody (Imagen Chlamydia) and by endpoint dilution PCR. Quantification by endpoint dilution PCR was performed by amplifying multiple replicates of each dilution of a boiled suspension of EBs using the PCR and ELONA conditions described below.
PCR primer and probe design.
Primer and probe design was based on alignment of published omp2 gene sequences (sequence data for C. felis, C. muridarum, C. suis, and the primer-binding sites of C. pecorum was not available initially). Primers were selected to allow the amplification of all target species in a single reaction by identifying conserved sequences in the omp2 gene. The primer sequences selected, designated Ch1 and Ch2, were modified from ones described previously (D1 and D2 in reference 18), although D2 was also used. Family-specific and species-specific probes were designed for use in a stringent ELONA. We have subsequently sequenced the corresponding omp2 regions of C. felis (FePn) and C. suis (S45), while C. muridarum (MoPn) and the complete 3′ end of C. pecorum strain W73 have since been submitted to GenBank.
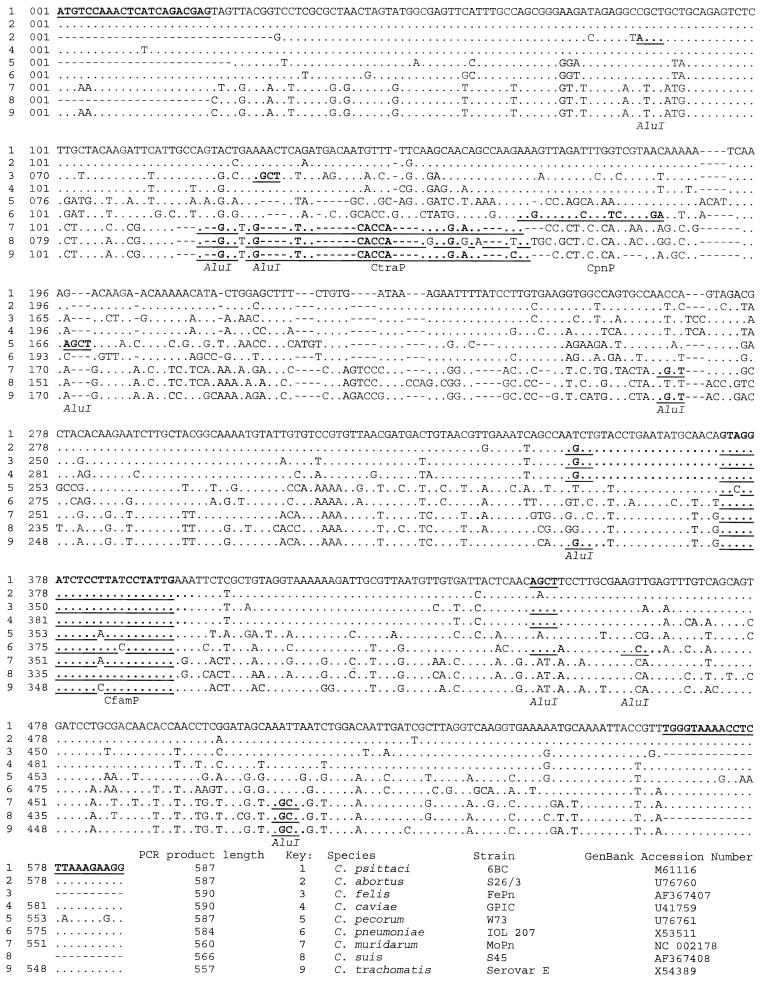
Alignment of sequences from all nine species is shown in Fig. 1. Primer and probe sequences selected are highlighted in Fig. 1 and shown in Table 1.
FIG. 1.
Alignment of partial omp2 nucleotide sequences from all nine species of Chlamydiaceae. Primer and probe sequences and AluI restriction sites (motif AGCT) are highlighted. Dashes represent areas of predicted DNA deletions. (Initial alignment produced by submission to the ClustalW programme on the European Bioinformatics Institute website.) Additional sequences available: C. trachomatis, eight sequences (GenBank loci CTOMP2, CHTOMPE, CHTOMPA, CHTCRPA, CTSL3CRP, CTSCCRP, CHTOMP2A, and AE001317); C. pneumoniae, 10 sequences (8 in reference 7, and genomes of AR39 and CW1029 in Genbank); C. psittaci, 2 sequences (Genbank CPSCROMP and reference 14); C. abortus, 1 sequence (14); C. pecorum, 4 partial sequences (U56927, AF111199, and reference 14). All additional sequences show 98.5 to 100% homology with the sequence shown above, with no alterations in restriction sites; however, the 3′ primer-binding site of C. pecorum strain VR268 (accession no. U56927) is altered at the first 3 bases (TGG to GCC).
TABLE 1.
omp2 primer and probe sequencesa
| Designation | Specificity | Sequence (5′ → 3′) |
|---|---|---|
| Ch1 | Family-specific PCR primer (sense) | ATG TCC AAA CTC ATC AGA CGA G |
| Ch2 | Family-specific PCR primer (antisense) | Bio-CCT TCT TTA AGA GGT TTT ACC CA |
| D2 | Family-specific PCR primer (antisense) | CCT TCT TTA AGA GGT TTT ACC |
| CfamP | Family-specific probeb | GTA GGA TCT CCT TAT CCT ATT G |
| CtraP | C. trachomatis probeb; also detects animal isolates C. muridarum and C. suis | TGA CAC CAA AGC GAA AGA CAA |
| CpnP | C. pneumoniae probeb | AAG GTT AGA CTT GTC CGT AGA |
| Tail sequenceb | GCACAACTCATTATGAA | |
| AP-COM | Common alkaline phosphatase probe | AP-TTCATAATGAGTTGTGC |
All oligonucleotides were purchased from Oswel DNA Service, Southampton, England.
Each probe was extended at the 3′ end with the same tail sequence.
PCR conditions.
All reactions were carried out in a Perkin-Elmer type 480 thermal cycler in 50μl reaction volumes. Reactions contained 1 to 10 μl of processed sample, 200 μM each of a mix of deoxynucleoside triphosphates (Amersham-Pharmacia), 5 pmol of each primer, 5 μl of 10× buffer with 15 mM MgCl2 and 1.25 U of Taq or Taq Gold (used during specificity tests with nonchlamydia isolates) polymerase (Roche Biochemicals). Standard amplification conditions for primers Ch1 and Ch2 were 94°C for 4 min (or 95°C for 10 min for Taq Gold) for 1 cycle; 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min for 40 cycles; and a final extension step of 72°C for 7 min for 1 cycle. Additional amplifications were performed with Ch1/Ch2 and with primer pair Ch1/D2, Taq Gold polymerase, and an annealing temperature of 45°C.
Chemiluminescent ELONA.
The ELONA method was that described by Whitby and Garson (19). Briefly, 5 μl of the biotinylated PCR product was captured on a streptavidin-coated microtiter well. After capture and washing, the double-stranded DNA was denatured by the addition of 0.15 M NaOH, and the second strand was washed away. A specific enzyme-labeled oligonucleotide probe was then hybridized to the captured antisense strand. Unbound label was removed by washing, and bound label was detected with a chemiluminescent substrate (Lumiphos; Lumigen Inc.). The final result is given as a numerical value in counts per second (cps) produced by a Cunberra Packard Top Count scintillation counter.
In a modified protocol designed to reduce the expense of synthesizing multiple enzyme-labeled probes, unlabeled probes were synthesized with a common 3′ tail sequence to which a complementary enzyme-labeled oligonucleotide could be annealed. Sequence details are given in Table 1. After washing away the unbound probe, the alkaline phosphatase-labeled oligonucleotide AP-COM, diluted in the same buffer as the probe (see Whitby and Garson 1995 for details [19]), was added to the wells and incubated at 37°C for 30 min before washing and addition of substrate as with a directly labeled probe.
RFLP analysis.
A discriminatory restriction enzyme was sought by analysis of cutting sites from available sequence data. Restriction with AluI was predicted to give species-specific band lengths, shown in Table 2. Digestion was performed by incubating a 10-μl aliquot of PCR product with 1 U of enzyme (Promega), 2 μl of 10× buffer, and 7 μl of water for 1 h at 37°C. The products were analyzed by electrophoresis on a 4% Metaphor gel (FMC Bioproducts, Rockland, Maine), stained with ethidium bromide, and compared to the predicted analysis.
TABLE 2.
Predicted fragment lengths for chlamydial omp2 gene PCR products following digestion with AluIa
| Species | Fragment lengths (bp) |
|---|---|
| Chlamydia trachomatis | 158, 119, 114, 84, 77 |
| Chlamydia muridarum | 216, 117, 97, 77, 26, 22, 5 |
| Chlamydia suis | 339, 119, 77, 36, 5 |
| Chlamydophila abortus | 352, 235 |
| Chlamydophila psittaci | 227, 220, 140 |
| Chlamydophila caviae | 355, 140, 105 |
| Chlamydophila felis | 224, 142, 93, 85, 47, 46 |
| Chlamydophila pneumoniae | 444, 127 |
| Chlamydophila pecorum | 397, 193 |
Restriction sites were detected by submission of sequence data to Webcutter, found at www.medkem.gu.se/cutter/.
Sequencing.
Sequencing of C. felis (FePn), C. suis (S45), C. psittaci (Z10), and unknown isolate A was performed using primers Ch1 and Ch2 on PCR products amplified from the omp2 gene and analyzed on an ABI 377 automated sequencer (Big Dye). Isolate B was sequenced using Texas red-labeled Ch1 and Ch2 primers and analyzed on a Vistra 725 automated sequencer (Amersham). Sequences have been submitted to GenBank. C. felis and C. suis are shown within the sequence alignment (Fig. 1).
Phylogenetic analysis.
The corresponding partial omp2 gene sequences available for all species (Fig. 1) were cluster analyzed by using the ClustalW algorithm in the Megalign program.
RESULTS
PCR specificity.
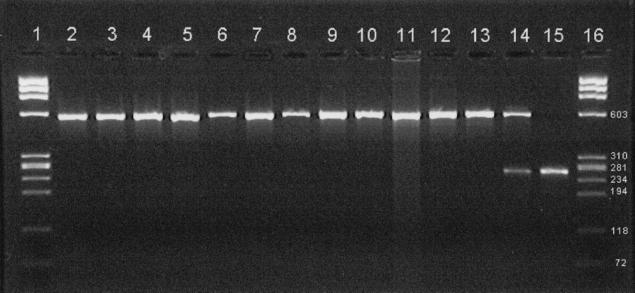
PCR with primers Ch1 and Ch2 was performed on DNA extracts of Chlamydiaceae species and analyzed on a 2% agarose gel stained with ethidium bromide. With an annealing temperature of 55°C, the primers amplified a single product band from all species except C. pecorum; no visible bands were seen from the four C. pecorum strains. At the lower annealing temperature of 45°C, all chlamydial strains except C. pecorum VR628 were amplified, producing a band of the expected length; however, an additional smaller band was produced with all four extracts of C. pecorum and the uninfected McCoy cells in FCM. Results are shown in Fig. 2. Amplification with the primer pair Ch1/D2 at an annealing temperature of 45°C yielded a single product of the expected length from all species except C. pecorum VR628 (gel not shown), which produced no visible product. CH1/D2 produced no product from any C. pecorum at 55°C, although C. trachomatis L2 gave the expected omp2 product (gel not shown).
FIG. 2.
Gel electrophoresis of partial omp2 PCR product from Chlamydiaceae. Lanes 1 and 16, size markers (φX174 HaeIII digest), with sizes (in base pairs) of weight markers indicated at the right); lane 2, C. trachomatis serotype D; lane 3, C. trachomatis serotype L2; lane 4, C. suis S45; lane 5, C. muridarum MoPn; lane 6, C. pneumoniae TW183; lane 7, C. pneumoniae V1355; lane 8, unidentified isolate A; lane 9, C. psittaci Z10; lane 10, C. abortus AB7; lane 11, C. caviae GPIC; lane 12, C. felis FePn; lane 13, unidentified isolate B; lane 14, C. pecorum P787 (C. pecorum 11/88 and R69 also gave this pattern); lane 15, C. pecorum VR628 (uninfected McCoy cells also gave this pattern). All amplifications were performed with primers Ch1 and Ch2 with an annealing temperature of 55°C except for the C. pecorum strains and uninfected McCoy cells, for which the annealing temperature was 45°C.
Amplification of DNA extracts from other bacterial isolates listed, using primers Ch1 and Ch2, Taq Gold, and an annealing temperature of 55°C, yielded no detectable products by gel electrophoresis or ELONA. All extracts were shown not to be inhibitory by spiking with C. abortus DNA (gel not shown). DNA from V. cholerae, S. sonnei, S. flexneri, Bacteroides sp., and Salmonella sp. gave nonspecific bands when amplified with Taq (gel not shown).
PCR-ELONA specificity.
All DNA extracts were amplified with Ch1/Ch2 and an annealing temperature of 55°C and then probed in parallel with three probes. The C. pecorum PCR products from the lower annealing temperature reaction were also probed. ELONA results are shown in Table 3. PCR products from all Chlamydiaceae species, including C. pecorum when amplified well, gave a strong signal (100× background) with the family-specific probe (CfamP). The species-specific probe for C. pneumoniae (CpnP) specifically detected only this species. The C. trachomatis (CtraP) probe detected C. trachomatis, C. muridarum, and, weakly, C. suis. The signal strength for the family-specific probe used with C. trachomatis was lower than with the other species. Comparison with the C. trachomatis species-specific probe indicates that the signal reduction occurred during the ELONA, possibly due to the single-base mismatch (Fig. 1). The low CtraP signal for C. suis (formerly C. trachomatis of pigs) compared to the family-specific probe was explained by a four-base difference in the gene sequence complementary to the probe.
TABLE 3.
ELONA top count results after assay of omp2 PCR products with three hybridization probesa
| Annealing temp | Species | Isolate | Counts per s with probe:
|
||
|---|---|---|---|---|---|
| CfamP | CtraP | CpnP | |||
| 55°C | C. trachomatis serotype A | SA1 P36 31.10.97 | 252,380 | 2,772,491 | 647 |
| C. trachomatis serotype B | TW5 27.10.97 P24 | 280,450 | 2,739,010 | 632 | |
| C. trachomatis serotype C | UW1 P25 24.6.78 | 236,082 | 2,645,840 | 705 | |
| C. trachomatis serotype D | ICCa18 | 349,967 | 3,035,122 | 684 | |
| C. trachomatis serotype E | DK20 | 126,135 | 999,432 | 1,263 | |
| C. trachomatis serotype F | MRC 301 | 418,897 | 3,011,100 | 777 | |
| C. trachomatis serotype G | IOL238 | 309,902 | 3,064,016 | 716 | |
| C. trachomatis serotype H | UW4 | 105,042 | 953,883 | 1,247 | |
| C. trachomatis serotype I | UW12 | 311,720 | 3,103,978 | 724 | |
| C. trachomatis serotype J | UW36 P18 5.12.97 | 513,429 | 3,045,417 | 768 | |
| C. trachomatis serotype K | UW31 | 339,222 | 3,116,967 | 713 | |
| C. trachomatis serotype L1 | 440L L1 | 141,605 | 2,660,000 | 743 | |
| C. trachomatis serotype L2 | 434B L2 | 177,258 | 2,899,205 | 797 | |
| C. trachomatis serotype L2 | SA2F | 183,431 | 3,199,281 | 1,388 | |
| C. trachomatis serotype L1 | 404L L3 P20 | 115,142 | 2,690,211 | 687 | |
| C. suis | S45 | 721,944 | 24,374 | 1,824 | |
| C. muridarum | MoPn | 1,762,822 | 5,160,565 | 2,096 | |
| C. pneumoniae | IOL207 | 321,050 | 921 | 243,502 | |
| C. pneumoniae | TW183 | 1,470,481 | 983 | 1,256,855 | |
| C. pneumoniae | V1355 | 2,851,556 | 1,633 | 1,833,771 | |
| C. psittaci | Z10 | 1,525,444 | 1,220 | 1,164 | |
| C. abortus | AB7 | 879,980 | 1,055 | 1,139 | |
| C. caviae | GPIC | 1,036,393 | 1,283 | 1,129 | |
| C. felis | FePn | 2,263,371 | 1,260 | 1,129 | |
| C. pecorumb | P787 | 10,392 | 1,332 | 1,943 | |
| C. pecorumb | 11/88 | 2,841 | 1,261 | 1,499 | |
| C. pecorumb | R69 | 1,817 | 1,051 | 1,685 | |
| C. pecorumb | VR628 | 1,707 | 1,253 | 1,601 | |
| McCoy cells in MEMb | 1,748 | 1,379 | 1,760 | ||
| Isolate A | 1,241,830 | 939 | 766,181 | ||
| Isolate B | 1,214,531 | 851 | 1,102 | ||
| 45°C | C. pecorum | P787 | 410,807 | 921 | 1,144 |
| C. pecorum | 11/88 | 47,354 | 755 | 1,097 | |
| C. pecorum | R69 | 75,572 | 672 | 1,111 | |
| C. pecorumc | VR628 | 11,286 | 648 | 1,220 | |
| McCoy cells in MEMc | 1,431 | 1,573 | 2,321 | ||
DNA extracts (of various amounts) were PCR amplified with primers Ch1 and Ch2 using an annealing temperature of 55°C. C. pecorum strains and McCoy cells in MEM were additionally amplified at 45°C. Hybridization probes: CfamP, Chlamydiaceae family probe; CtraP, C. trachomatis probe; CpnP, C. pneumoniae probe. Negative ELONA controls gave values of 600 to 2000 cps.
No products visible on 2% agarose gel.
No specific omp2 product visible but low-molecular-weight band present.
Test of PCR sensitivity.
Absolute sensitivity was tested for C. pneumoniae and C. trachomatis using enumerated suspensions of EBs amplified with Ch1/Ch2 and an annealing temperature of 55°C. The previously enumerated stock EB suspensions were extracted by boiling for 15 min, and limiting-dilution PCR was performed using a half-log dilution series in water. Each dilution was amplified by PCR in up to four reactions, and amplification was detected by ELONA. The results of the assay were also used to determine a test/control ratio which enabled a cutoff ratio to be assigned. An example of the ELONA results is shown in Table 4, showing the titration of a counted suspension of C. pneumoniae EBs. ELONA titrations of C. pneumoniae and C. trachomatis showed that the method was able to detect a single input DNA target molecule and show a positive/negative ratio for a single molecule of between 5 and 15.
TABLE 4.
PCR-ELONA limiting dilution titration for C. pneumoniae EB suspensiona
| EB input into PCR | ELONA cps
|
No. positive/4 tested | |||
|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | Replicate 4 | ||
| 30 | 197,435 | 91,228 | 91,351 | 169,439 | 4 |
| 10 | 65,743 | 15,039 | 101,287 | 87,291 | 4 |
| 3 | 8,027 | 28,837 | 34,542 | 14,435 | 4 |
| 1 | 549 | 553 | 620 | 14,430 | 1 |
| 0.3 | 577 | 616 | 567 | 553 | 0 |
| 0.1 | 609 | 607 | 616 | 666 | 0 |
PCR was performed with primers Ch1 and Ch2 and an annealing temperature of 55°C.
Identification by omp2 PCR-RFLP.
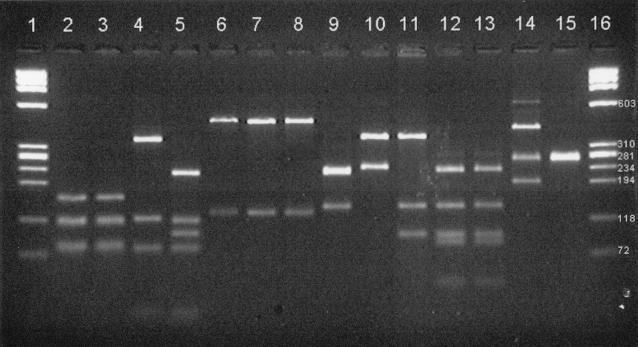
The products from the omp2 PCRs (C. pecorum was amplified at 45°C) were restriction digested with AluI as described in Materials and Methods. The products of the digestion are shown in Fig. 3, demonstrating that the bands generated correspond to the predicted sizes shown in Table 2 and are characteristic for all nine species.
FIG. 3.
Gel electrophoresis of omp2 PCR products of Chlamydiaceae after restriction digestion with AluI. Lanes 1 and 16, size markers (as in Fig. 2); lane 2, C. trachomatis serotype D; lane 3, C. trachomatis serotype L2; lane 4, C. suis S45; lane 5, C. muridarum MoPn; lane 6, C. pneumoniae TW183; lane 7, C. pneumoniae V1355; lane 8, unidentified isolate A; lane 9, C. psittaci Z10; lane 10, C. abortus AB7; lane 11, C. caviae GPIC; lane 12, C. felis FePn; lane 13, unidentified isolate B; lane 14, C. pecorum P787 (C. pecorum 11/88 and R69 also gave this pattern); lane 15, C. pecorum VR628 (uninfected McCoy cells also gave this pattern). All amplifications were performed with primers Ch1 and Ch2 with an annealing temperature of 55°C except for the C. pecorum strains and uninfected McCoy cells, for which the annealing temperature was 45°C. The predicted sizes (in base pairs) of the digest fragments are listed in Table 2.
Sequence of omp2 gene products of C. felis (FePn), C. suis (S45), and C. psittaci (Z10); sequence cluster analysis, taxonomic classification, and species identification.
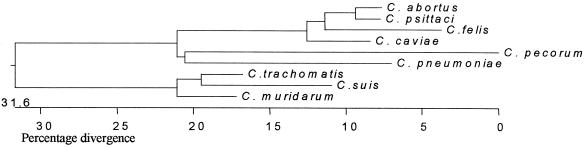
The newly generated sequences of C. felis and C. suis (GenBank accession nos. AF367407 and AF367408, respectively) are shown aligned with the other Chlamydiaceae in Fig. 1. C. psittaci Z10 sequencing produced a 536-bp sequence identical to bases 29 to 564 of the C. psittaci sequence shown. The phylogenetic relationship of all species based on the partial omp2 sequence clustering is shown in Fig. 4. The dendrogram produced demonstrates the divergence between the designated species and mirrors that derived by analysis of the rRNA sequences (4). The similarity of the Z10 sequence to other C. psittaci sequences demonstrates the conservation of the partial omp2 sequence within this species.
FIG. 4.
Unrooted tree of the omp2 sequences from Chlamydiaceae species. Dendrogram produced using the ClustalW algorithm in the Megalign program.
Uncharacterized chlamydial isolates.
The two nontrachomatis isolates were analyzed by the PCR-ELONA and PCR-RFLP methods. Results are shown in Table 3 and Fig. 3. In the ELONA, both isolates gave strong signals with the family-specific probe (CgenP). Isolate A was positive with the C. pneumoniae probe but not with the C. trachomatis probe (CtraP), while isolate B was negative with both of the specific probes used. The RFLP band pattern of isolate A corresponded to that predicted for C. pneumoniae, and isolate B corresponded to C. felis. Isolate A sequencing produced a 536-bp sequence identical to bases 29 to 564 of C. pneumoniae; the isolate B sequence (GenBank AF367406) was 471 bp long and identical to the corresponding C. felis sequence (bases 38 to 508 in Fig. 1). DNA sequence analysis confirmed that isolate A is C. pneumoniae and isolate B is C. felis.
DISCUSSION
Infection with chlamydiae other than C. trachomatis is often difficult to diagnose. Culture requires special facilities, while the serological response is variable and the tests are poorly standardized. Nucleic acid amplification tests are being used more frequently (1, 10, 13, 15). However, the majority of diagnostic tests are designed for C. trachomatis, and methods for the detection and identification to species level of all nontrachomatis isolates are not readily available. Recent taxonomic changes to the family were based on the 16s and 23s rRNA genes; however, it may be possible to distinguish all species by the detection of specific sequences within other genes. Outer membrane protein 2 is an integral component of the cell wall of all chlamydiae, containing a variable N-terminal fragment which is species specific but does not induce a species-specific humoral response (11). It had been shown that the 5′ end of the gene for the Omp2 protein contains two short, highly conserved sequences suitable for use as primer-binding sites for the amplification of many chlamydiae (3, 14, 18). We designed primers (Ch1 and Ch2) to target the same conserved region. The product amplified from a single optimized PCR could then be detected and identified by ELONA or RFLP designed around the differences in the variable section, as long as these differences were sufficient and conserved within a species.
Submitted sequence data (shown in Fig. 1) confirmed the highly conserved nature of the primer-binding sequences for all species tested except C. pecorum, for which variation was revealed at the 3′ end. C. pecorum W73 shows five base changes within the 3′ primer-binding site, while C. pecorum VR268 has a complete mismatch at the 5′ end of the 3′ primer-binding site. As a result, primers CH1 and CH2, at an annealing temperature of 55°C, efficiently amplified all Chlamydiaceae except C. pecorum. At 45°C, some strains of C. pecorum were also amplified.
With the sequencing of the 5′ end of C. felis and C. suis, data are now available for the variable region in all nine chlamydial species (Fig. 1). The nucleotide sequence between the primer sites has been determined a number of times for some species. It is identical for all C. pneumoniae isolates (7) and almost identical for C. trachomatis (eight GenBank accessions). Smaller numbers of the other Chlamydophila spp. have been analyzed (listed in Fig. 1), and they also show a high degree of conservation of the partial omp2 gene sequence within a species, as we have demonstrated with C. felis and C. psittaci. Conservation may be maintained, as Omp2 does not induce a species-specific humoral response (11). In contrast to the conserved intraspecies sequence of omp2, alignment of the sequences from all species demonstrates a high level of interspecies divergence, with the same phylogenetic relationship to that produced by alignment of the rRNA gene sequences (4). The combination of interspecies variation with intraspecies conservation makes this locus a suitable candidate gene for the identification of all nine species, including C. pecorum strains, when amplified.
The present study has used a PCR based on the family-specific primers and combined the reaction with family- or species-specific oligonucleotide probes or with restriction digestion with a single enzyme, AluI, for the detection and identification of Chlamydiaceae. The omp2 gene was originally amplified with shorter primers, D1 and D2 (18), and an annealing temperature of 45°C. We modified these primers to provide better balance in a planned quantitative assay and to allow use of a higher annealing temperature. Restriction digestion of the partial omp2 gene PCR product with the enzyme AluI was shown to produce the predicted species-specific patterns for all nine species. The PCR-RFLP is therefore able to determine the species of isolates in agreement with the new taxonomic classification. This was demonstrated with two unknown laboratory isolates, which were shown to be strains of C. felis and C. pneumoniae.
For use in clinical diagnosis, single-round reactions and analysis by agarose gel electrophoresis may lack sensitivity and specificity. Therefore, our PCR was combined with an ELONA using a chemiluminescent substrate to provide family- or species-specific detection at high sensitivity. The method can also be used for quantification (19). Using suspensions of enumerated EBs of C. pneumoniae and C. trachomatis, the sensitivity of the species-specific assay approached one genome copy input into the PCR. The two species-specific hybridization probes designed for C. pneumoniae and C. trachomatis were both specific and sensitive for chlamydiae infecting humans. The C. trachomatis probe also detected mouse and, less sensitively, pig chlamydial infection. The PCR-ELONA as designed classifies samples as containing chlamydial DNA using the family-specific probe and as containing C. pneumoniae, C. trachomatis, or some other species using the species-specific probes. Probes may be combined for ease in a screening assay (data not shown), and additional species-specific probes could easily be incorporated into the method.
The format of a PCR-ELONA using a family-specific primer pair and identification during the sensitive detection stage has other advantages in addition to sensitivity and specificity. C. pneumoniae DNA has been shown to be difficult to recover from specimens, with up to 99% loss demonstrated during spiking experiments with various commercial and in-house procedures (9). The addition of a known number of C. trachomatis EBs to the specimen before extraction can be used as an internal control for all stages of the assay, since the control and target can be separately detected and quantified in the ELONA detection stage. Furthermore, the chemiluminescent assay can be used to quantify the DNA input either against an external calibration curve or through the competitive quantitative PCR that occurs with the internal control (unpublished data). In laboratories where a microtiter luminometer is unavailable, colorimetric substrates may be substituted for the chemiluminescent substrate used in the described method.
In conclusion, we have demonstrated that the omp2 gene of Chlamydiaceae is a suitable locus to which molecular detection and identification methods may be targeted. The alignment of previously available and newly generated genome sequences confirmed the taxonomic relationship between the newly described species within the Chlamydiaceae family. The standard PCR-RFLP and PCR-ELONA (annealing temperature of 55°C) were able to detect and identify all previous and newly described species except C. pecorum. Reduction of the annealing temperature permitted detection and identification of most C. pecorum strains, but within-species variation in the 3′ primer-binding site prevents amplification of all strains. The combination of the family-specific PCR with a chemiluminescent ELONA detection system gives a highly sensitive and specific assay. The single optimized method avoids the need for multiplex PCR, with its reduced sensitivity, or for multiple PCRs, with their additional resource demand. This assay has the potential to provide a simple and reliable means for the detection and identification of Chlamydiaceae from clinical specimens and cultures for the investigation of human chlamydial disease.
ACKNOWLEDGMENT
We thank K. Harris, Great Ormond Street Hospital, for performing the cluster analysis.
REFERENCES
- 1.Boman J, Gaydos C A, Quinn T C. Molecular diagnosis of Chlamydia pneumonia infection. J Clin Microbiol. 1999;37:3791–3799. doi: 10.1128/jcm.37.12.3791-3799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter M W, Harrison T G, Shuja Shafi M, George R C. Typing strains of Chlamydia pneumoniae by AFLP. Clin Microbiol Infect. 1998;4:663–664. doi: 10.1111/j.1469-0691.1998.tb00352.x. [DOI] [PubMed] [Google Scholar]
- 3.Demkin V V, Edelstein M V, Zimin A L, Edelstein I A, Suvorov M M. Detection of sequence variation in PCR-amplified fragments of omp2 gene from three species of the family Chlamydiaceae using agarose gel electrophoresis containing bisbenzimide-PEG. FEMS Microbiol Lett. 2000;184:215–218. doi: 10.1111/j.1574-6968.2000.tb09016.x. [DOI] [PubMed] [Google Scholar]
- 4.Everett K D A, Bush R M, Anderson A A. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49:415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 5.Everett K D A, Hornung L J, Anderson A A. Rapid detection of the Chlamydiaceae and other families in order Chlamydiales: three PCR tests. J Clin Microbiol. 1999;37:575–580. doi: 10.1128/jcm.37.3.575-580.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freidank H N, Vögele H, Eckert K. Evaluation of a new commercial microimmunofluorescence test for detection of antibodies to Chlamydia pneumoniae, Chlamydia trachomatis, and Chlamydia psittaci. Eur J Clin Microbiol Infect Dis. 1997;16:685–688. doi: 10.1007/BF01708561. [DOI] [PubMed] [Google Scholar]
- 7.Jantos C A, Heck S, Roggendorf R, Sen-Gupta M, Hegemann H. Antigenic and molecular analyses of different Chlamydia pneumoniae strains. J Clin Microbiol. 1997;35:620–623. doi: 10.1128/jcm.35.3.620-623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutlin A, Roblin P M, Hammerschlag M R. Antibody response to Chlamydia pneumoniae in children with respiratory illness. J Infect Dis. 1998;177:720–724. doi: 10.1086/514223. [DOI] [PubMed] [Google Scholar]
- 9.Lindholt J S, Ostergard L, Henneberg E W, Fasting H, Anderson P. Failure to demonstrate Chlamydia pneumoniae in symptomatic abdominal aortic aneurysms by a nested polymerase chain reaction (PCR) Eur J Vasc Endovasc Surg. 1998;15:161–164. doi: 10.1016/s1078-5884(98)80138-x. [DOI] [PubMed] [Google Scholar]
- 10.Messmer T O, Skelton S K, Moroney J F, Daugharty H, Fields B S. Application of a nested, multiplex PCR to psittacosis outbreaks. J Clin Microbiol. 1997;35:2043–2046. doi: 10.1128/jcm.35.8.2043-2046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mygind P, Christiansen G, Persson K, Birkelund S. Analysis of the humoral immune response to Chlamydia outer membrane protein 2. Clin Diagn Lab Immunol. 1998;5:313–318. doi: 10.1128/cdli.5.3.313-318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Persson K, Haidl S. Evaluation of a commercial test for antibodies to the chlamydial lipopolysaccharide (Medac) for serodiagnosis of acute infections by Chlamydia pneumoniae (TWAR) and Chlamydia psittaci. APMIS. 2000;108:131–138. doi: 10.1034/j.1600-0463.2000.d01-36.x. [DOI] [PubMed] [Google Scholar]
- 13.Schachter J. Which test is best for chlamydia? Curr Opin Infect Dis. 1999;12:41–45. doi: 10.1097/00001432-199902000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Sheehy N, Markey B, Gleeson M, Quinn P J. Differentiation of Chlamydia psittaci and C. pecorum strains by species-specific PCR. J Clin Microbiol. 1996;34:3175–3179. doi: 10.1128/jcm.34.12.3175-3179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verkooyen R P, Willemse D, Hiepvan Casteren S C A M, Mousave Joulandan S A, Snijder R J, van den Bosch J M M, et al. Evaluation of PCR, culture, and serology for diagnosis of Chlamydia pneumoniae respiratory infections. J Clin Microbiol. 1998;36:2301–2307. doi: 10.1128/jcm.36.8.2301-2307.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S P. The microimmunofluorescence test for Chlamydia pneumoniae infection: technique and interpretion. J Infect Dis. 2000;181(Suppl 3):S421–S425. doi: 10.1086/315622. [DOI] [PubMed] [Google Scholar]
- 17.Ward M E, Ridgway G. Chlamydia. In: Collier L, Balows A, Sussman M, editors. Topley and Wilson's microbiology and microbial infections. 9th ed. New York, N.Y: Arnold; 1998. pp. 1331–1346. [Google Scholar]
- 18.Watson M W, Lambden P R, Clarke I N. Genetic diversity and identification of human infection by amplification of chlamydial 60-kilodalton cysteine-rich outer membrane protein gene. J Clin Microbiol. 1991;29:1188–1193. doi: 10.1128/jcm.29.6.1188-1193.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitby K, Garson J A. Optimisation and evaluation of a quantitative chemiluminescent polymerase chain reaction assay for hepatitis C virus RNA. J Virol Methods. 1995;51:75–88. doi: 10.1016/0166-0934(94)00144-6. [DOI] [PubMed] [Google Scholar]
- 20.Wong Y K, Sueur J M, Fall C H, Orfila J, Ward M E. The species specificity of the microimmunofluorescence antibody test and comparisons with a time resolved fluoroscopic immunoassay for measuring IgG antibodies against Chlamydia pneumoniae. J Clin Pathol. 1999;52:99–102. doi: 10.1136/jcp.52.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]